Abstract
Orofacial clefts are among the commonest birth defects. Among many genetic contributors to orofacial clefting, Interferon Regulatory Factor 6 (IRF6) is unique since mutations in this gene cause Van der Woude (VWS), the most common clefting syndrome. Furthermore, variants in IRF6 contribute to increased risk for non-syndromic cleft lip and/or palate (NSCL/P). Our previous work shows that individuals with either VWS or NSCL/P may have cerebral anomalies (larger anterior, smaller posterior regions), and a smaller cerebellum. The objective of this study was to test the hypothesis that disrupting Irf6 in the mouse will result in quantitative brain changes similar to those reported for humans with VWS and NSCL/P. Male mice heterozygous for Irf6 (Irf6gt1/+; n = 9) and wild type (Irf6+/+; n = 6) mice at comparable age underwent a 4.7T MRI scan to obtain quantitative measures of cortical and subcortical brain structures. There was no difference in total brain volume between groups. However, the frontal cortex was enlarged in the Irf6gt1/+ mice compared to that of wild types (p = 0.028) while the posterior cortex did not differ. In addition, the volume of the cerebellum of Irf6gt1/+ mice was decreased (p = 0.004). Mice that were heterozygous for Irf6 showed a similar pattern of brain anomalies previously reported in humans with VWS and NSCL/P. These structural differences were present in the absence of overt oral clefts. These results support a role for IRF6 in brain morphometry and provide evidence for a potential genetic link to abnormal brain development in orofacial clefting.
Keywords: IRF6, oral clefts, brain development, magnetic resonance imaging, mouse model
INTRODUCTION
Orofacial clefts are a disturbance of craniofacial development. This malformation is thought to result from a combination of genetic and environmental factors affecting cell migration, proliferation, adhesion, differentiation, and/or apoptosis [Burdi, 2006; Knight et al., 2006]. Collectively, orofacial clefting affects 1 in 700 live births making it one of the most common birth defects worldwide [Canfield et al., 2006]. Clefts can occur as part of a defined syndrome or in an isolated manner. Isolated clefts are commonly termed non-syndromic clefts of the lip and/or palate (NSCL/P) and comprise roughly 70% of all oral clefts [Jones, 1988].
In addition to the facial cleft, many individuals affected with NSCL/P manifest functional problems, including speech abnormalities [Timmons et al., 2001], behavioral abnormalities [Hunt et al., 2005], and cognitive deficits [Jocelyn et al., 1996]. The cognitive deficits are specifically language-based and often manifest in language-based learning difficulties. In fact, the rate of learning disabilities in this population is nearly ten times greater than the general population [Richman et al., 1988].
The cause of speech, behavioral, and cognitive deficits in NSCL/P has been debated. The traditional view is that these deficits are secondary issues, due to other factors related to the cleft such as chronic otitis media, exposure to anesthesia during operations, or even the psychological stress of living with a facial cleft [Jocelyn et al., 1996; Millard and Richman, 2001; Riski, 2006]. But more recently, it has been suggested that deficits in speech, cognition, and behavior are a direct result of abnormal brain development and are therefore primary neurodevelopmental issues that present in parallel with the cleft. Initial support for this theory was provided by the observation that the development of the face and the brain are intimately linked [Kjaer, 1995]. Our laboratory has focused on brain structure and function in children and adults with NSCL/P hypothesizing that the factors responsible for disruption in the development of craniofacial structures may also adversely affect the development of the brain [Rutter et al., 2004]. In a study evaluating brain structure using Magnetic Resonance Imaging (MRI), men with NSCL/P were compared to matched controls, and a unique pattern of brain morphology was found. The two groups did not differ in overall brain size, however there was a shift in tissue distribution in which the anterior cerebrum (in particular, the gray matter) was enlarged while the posterior cerebrum was abnormally small (in particular, the white matter). In addition, there was a significant deficit in cerebellar volume [Nopoulos et al., 2000; 2002].
Of the many known genetic factors involved in cleft lip and palate (CLP), Interferon Regulatory Factor 6 (IRF6) is unique because DNA variants in this gene contribute to syndromic and non-syndromic forms of orofacial clefting [Kondo et al., 2002; Zucchero et al., 2004]. Loss-of-function mutations in IRF6 cause Van der Woude syndrome, the most common syndromic form of CLP, comprising lower lip fistulas in addition to the cleft [Kondo et al., 2002]. Furthermore, a common single nucleotide polymorphism in MCS9.7, an enhancer element near IRF6, contributes to the risk of NSCL/P [Rahimov et al., 2008]. The pattern of brain structure changes in men with VWS was very similar to the pattern seen in the men with NSCL/P; compared to matched controls, the VWS subjects had significant enlargement of the anterior regions of the cerebrum (in particular gray matter), and a volumetric reduction of the cerebellum [Nopoulos et al., 2007]. This pattern of structural brain changes was observed in a subset of patients with VWS who had no cleft, but lip pits only. This suggests that the abnormal brain morphology seen in VWS is a pleiotropic manifestation and maybe present even in the absence of a cleft.
In an effort to better understand the relationship between Irf6 and the brain changes associated with orofacial clefting, we assessed quantitatively the brain phenotype in Irf6 mutant mice. Mice homozygous for the null allele die perinatally with limb, craniofacial and epidermal anomalies [Ingraham et al., 2006; Richardson et al., 2006]. However, mice heterozygous for Irf6 survive to adulthood, superficially appear normal, and lack an orofacial cleft and lip pits [Ingraham et al., 2006]. Therefore, the heterozygous mouse constitutes a unique approach to assess potential alteration in brain structure in the context of genetic alteration in Irf6 without a clefting phenotype. Furthermore, the mice present with the advantage of being free of the environmental influences (chronic otitis media, exposure to anesthesia, or psychological stress) encountered in humans with clefts.
We evaluated total brain and regional brain structures of the Irf6 heterozygous mouse compared to wild type controls using volumetric MRI in order to test the hypothesis that disrupting Irf6 in the mouse will result in quantitative brain changes similar to those reported for humans with VWS and NSCL/P.
MATERIALS AND METHODS
A total of nine male mice heterozygous for the Irf6gt1 allele were compared to six wild type mice (Irf6+/+). All mice were cared for according to the Animal Care and Use Review Form (ACURF) at the University of Iowa, Iowa City, IA. Mice were housed in an environment under light-, temperature-, and humidity-controlled conditions. Food and water were available ad libidum. In order to avoid variability in brain structure due to age or sex, a sample of age-matched male mice was used. All mice were 25 weeks old, which corresponds to fully mature animals. The Irf6gt1 allele was previously described and is maintained in a C57BL/6J background [Ingraham et al., 2006].
Magnetic resonance imaging (MRI) was performed on a 4.7 Tesla Varian small-bore scanner. All acquisitions utilized a 25 mm diameter transmit/receive coil for high-resolution imaging. Mice were anesthetized with isoflurane (3% induction, 1.5% maintenance) and transferred to the scanner for imaging. After a series of three localizer scans (each about five seconds long), a set of T2-weighted fast spin-echo images was acquired in the axial plane. The protocol parameters were TR/TE = 2100/60 ms, echo train length of eight, 0.5 mm thick contiguous slices with in-plane resolution of 0.16 mm × 0.16 mm over a 256 × 256 matrix using 12 signal averages. The total time for the entire protocol was about 40 minutes.
All MRI data were processed using BRAINS software developed locally at the University of Iowa [Magnotta et al., 2002]. The mouse brain atlas used for segmentation purposes was the mouse Biomedical Informatics Research Network (mBIRN) atlas, which was constructed using T2-weighted magnetic resonance microscopy (MRM) from 11 WT C57BL/6J mice at the University of California, Los Angeles [MacKenzie-Graham et al., 2006].
For our pipeline process we employed a directed acyclic pipeline architecture using Nipype [Gorgolewski et al., 2011], a Python-based wrapping library for neuroimaging applications. The mBRIN atlas was registered to the input T1 file using b-spline warping within BRAINSFit [Johnson et al., 2007], a mutual information driven application developed under the ITK framework. The atlas was then resampled using BRAINSResample to match the voxel lattice of the T1 image, thereby allowing one-to-one correspondence between atlas and image. Finally, we computed the volume measurements for each desired region of our atlas as the sum of voxels within a given label times the volume of a voxel (Fig. 1).
Figure 1. Volumetric Labels of mBIRN Atlas use for Segmentation of Mouse Brain.
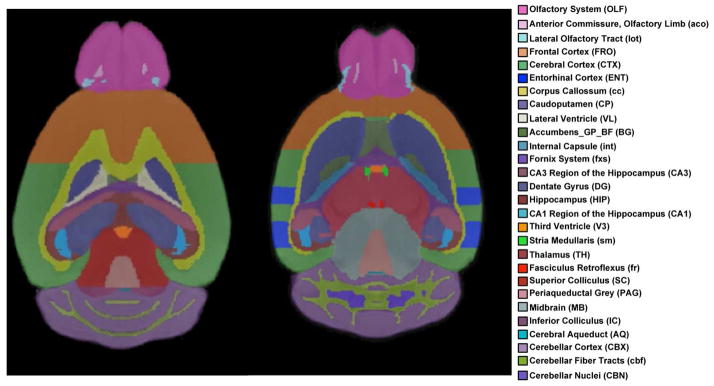
Horizontal sections of mouse brains showing the volumetric labels of the mouse Biomedical Informatics Research Network (mBIRN) Atlas. The image on the left shows a more dorsal section passing through the hippocampus; the image on the right shows a more ventral section passing through the thalamus.
The atlas defined 43 regions of interest. The regions were then grouped into the following areas: amygdala, hypothalamus, pituitary, thalamus, total brain volume, basal ganglia, brainstem, cerebrospinal fluid (CSF), cerebellum, hippocampus, white matter tracts, anterior cortex, and posterior cortex. The anterior cortex was further subdivided into the Olfactory and Frontal Cortices. The posterior cortex was similarly subdivided into the posterior, entorhinal, and perirhinal cortices. Volumes were reported in mm3.
All analyses were performed using Statistical Package for Social Science (SPSS), version 19.0 for Windows (SPSS Inc., Chicago Ill). Due to the small sample size, non-parametric analysis using the rank of all measures was utilized in order to minimize any effects of outliers. Analysis of variance (ANOVA) was used to evaluate total brain volume across groups. The remainder of the brain regions was compared across the two groups using Analysis of Covariance (ANCOVA), controlling for total brain volume.
RESULTS
Table I shows the results of the analysis comparing Irf6gt1/+ and wild type mice on general brain measures. Both groups had a similar total brain and white matter volume. However, the Irf6gt1/+ mice showed a statistically significant increase in cortical volume, (F = 9.80, p = 0.008). In contrast to the cortical enlargement, the cerebellum in the Irf6gt1/+ mice displayed markedly smaller volumes compared to wild type mice, (F = 12.43, p = 0.004). Significant differences were also found in the amygdala indicating it was enlarged in the Irf6gt1/+ mice (F = 6.04, p = 0.030).
Table I.
Results of general brain measures and regional cortex measures, between group comparison.
| Irf6+/+ (Wild type, n = 6) | Irf6gt1/+ (n = 9) | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Volume (mm3) | S.D. | Volume (mm3) | S.D. | F* | P* | |
| Total Brain | 457.89 | 17.18 | 465.47 | 9.81 | 0.12# | 0.737# |
| Cortex | 171.58 | 8.87 | 178.97 | 4.44 | 9.80 | 0.008 |
| Anterior | 69.22 | 7.33 | 77.23 | 5.65 | 6.21 | 0.028 |
| Olfactory | 27.27 | 4.84 | 32.43 | 3.65 | 4.44 | 0.056 |
| Frontal | 41.96 | 2.90 | 44.80 | 2.38 | 9.13 | 0.010 |
| Posterior | 102.35 | 4.00 | 101.74 | 3.23 | 0.31 | 0.585 |
| Cortex | 93.90 | 3.61 | 93.68 | 3.04 | 0.14 | 0.712 |
| Entorhinal | 5.05 | 0.60 | 4.83 | 0.37 | 0.25 | 0.626 |
| Perirhinal | 3.40 | 0.29 | 3.23 | 0.24 | 2.08 | 0.174 |
| White Matter | 124.59 | 6.98 | 126.63 | 4.15 | 0.02 | 0.888 |
| CSF | 3.07 | 0.35 | 3.08 | 0.31 | 0.12 | 0.731 |
| Cerebellum | 60.52 | 5.06 | 56.17 | 3.34 | 12.43 | 0.004 |
| Amygdala | 1.03 | 0.08 | 1.12 | 0.05 | 6.04 | 0.030 |
| Basal Ganglia | 35.07 | 1.75 | 35.88 | 1.33 | 1.97 | 0.186 |
| Hippocampus | 26.35 | 1.32 | 26.39 | 2.05 | 0.27 | 0.618 |
| Hypothalamus | 12.50 | 1.29 | 13.68 | 0.79 | 4.41 | 0.057 |
| Pituitary | 2.06 | 0.38 | 2.23 | 0.37 | 0.39 | 0.542 |
| Thalamus | 21.12 | 0.79 | 21.32 | 1.62 | 0.12 | 0.739 |
ANOVA based on ranked variables
ANCOVA based on ranked variables, controlling for Total Brain volume
Regional analysis of the cortex in the Irf6gt1/+ is also found in Table I. When the cortex was further divided into an anterior and posterior portion, the overall cortical enlargement was accounted for by greater volumes of the anterior cortex (F = 6.21, p = 0.028). Posterior cortex volumes were not significantly different from those of wild type mice. When the anterior cortex was further subdivided, the frontal region (F = 9.13, p = 0.010) was significantly enlarged. The olfactory cortex was also enlarged, but this finding was not statistically significant.
DISCUSSION
These findings largely mirror those from human studies on the brains of adult males with NSCL/P and VWS. Although the Irf6gt1/+ mice are without orofacial clefts or lip pits (the clinical hallmarks of VWS), both the mice and humans with NSCL/P and VWS showed an almost identical pattern of brain structure abnormalities, including a subset of adult patients with VWS and lip pits only (no cleft). Specifically, an increase in volume in the anterior portions of the cerebrum (cortical gray matter in particular) and a reduction in overall cerebellum volume were common to humans and mice (amygdala volume was not measured in the adult humans). These results support a potential link between Irf6 and the abnormal brain phenotype observed in human orofacial clefting.
Although preliminary, these results lend support to our working hypothesis that the structural brain changes observed in NSCL/P are not simply the secondary consequences of hearing loss, psychosocial factors, or anesthesia. Rather, they suggest that at least some of the genetic factors that underlie NSCL/P risk may also result in the characteristic pattern of brain morphology that is typically observed in this population. Structural factors during early embryogenesis may link this observed pattern of brain dysmorphology, characterized by disproportionate increases in forebrain regions, to the failure of fusion between adjacent facial prominences. Several studies have suggested that the size and position of the developing frontonasal mass exerts an influence on the position and orientation of immediately adjacent tissue components of the nascent face responsible for forming the upper lip and palate, components required to approximate and contact one another in a precise spatiotemporal sequence to ensure the structural integrity of the midface [Diewert et al., 1993; Jiang et al., 2006 Weinberg et al., 2009; Parsons et al., 2011]. An improved characterization of the complex network of molecular signals coordinating the forebrain and face during development will be essential to understanding how such structural integration is achieved [Creuzet et al., 2004; Marcucio et al., 2005].
Based on our prior work, the pattern of brain abnormalities is not identical across all cleft types [van der Plaas et al., 2010; DeVolder et al., 2013]. In particular, individuals with cleft palate only show a different pattern in which the cerebellum is not reduced in volume but instead shows a different distribution of cerebellar tissue [DeVolder et al., 2013]. This is especially interesting when considering that variation in IRF6 is associated with an increased risk for cleft lip only or combined cleft lip and palate and not with an increased risk of cleft palate only [Rahimov et al., 2008]. Thus our finding of reduced cerebellar volume in the heterozygous mouse more closely resembles the pattern found in the cleft types most strongly associated with alterations in IRF6 (cleft lip only and combined cleft lip/palate).
We chose to work on the Irf6gt1/+ mouse because it is the best model for the evaluation of the role of IRF6 in human disease. We recognize that this mutant murine strain does not have a cleft phenotype, but a number of practical limitations required us to work with this model. As mentioned earlier, although mice homozygous for the null allele have clefts, they die perinatally [Ingraham et al., 2006; Richardson et al., 2006]. Embryological studies of these mice are not possible due to technical limitations. In addition, many mice that are heterozygous for mutations in other cleft-related genes (Tp63, Tfap2a, Tbx1, etc.) similarly lack an overt orofacial cleft [Lindsay et al., 2001; Mills et al., 1999; Zhang et al., 1996]. Even in human studies, the subset of individuals with VWS and no cleft (only lip pits) exhibited the same abnormal brain morphology as those with NSCL/P [Nopoulos et al., 2007]. The fact that our mouse model (which lacked a clefting phenotype) showed a similar pattern of brain structure abnormalities provides even further evidence that mutations in IRF6 alone (even in the absence of an orofacial cleft) are sufficient to cause aberrant neurodevelopment.
The biological mechanism by which disruptions in Irf6 translate into altered brain phenotypes is unknown. However, in a recent study, a mouse Irf6 enhancer (MCS9.7) was fused to the B-galactosidase gene and showed strong expression in the hindbrain and some expression in the anterior pole of the forebrain early in development [Fakhouri et al., 2012]. Additional work is needed to connect the observed phenotypic changes to altered gene expression and to pinpoint the timing and sequencing of events leading to abnormal brain morphogenesis. A recent paper found no significant change in mandibular morphometry in Irf6 adult het mice using micro-CT [Boell et al., 2013], although mandibular hypoplasia was observed in knockout embryos [Ingraham et al., 2006]. This suggests that development of the brain is more sensitive to Irf6 dosage than development of the mandible. Similarly, landmark-based morphometric analyses would be an excellent follow-up study and provide additional information on how the brain is developing in response to alterations in Irf6. However, this type of analysis would require a larger sample size than the current study. Hopefully the findings presented here can provide a basis for landmark selection in future morphometric studies in this mouse model.
The current finding of abnormal brain morphology in Irf6gt1/+ mice parallels the abnormal brain morphology of adult human NSCL/P and VWS subjects. Taken together, our data provide strong evidence that abnormalities in IRF6 are directly related to changes in brain structure in NSCL/P and VWS subjects. Though the underlying functional mechanism is still unclear, it appears that IRF6 may provide a genetic link between abnormal craniofacial and abnormal neurodevelopment common to both VWS and NSCL/P.
Acknowledgments
This study was supported by the Carver College of Medicine Collaborative Grant awarded to Dr. Nopoulos and Dr. Schutte. Partial support was provided by NIH AR-055313 (M.D.).
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no conflicts of interest to declare.
References
- Boell L, Pallares LF, Brodski C, Chen Y, Christian JL, Kousa YA, Kuss P, Nelsen S, Novikov O, Schutte BC, Wang Y, Tautz D. Exploring the effects of gene dosage on mandible shape in mice as a model for studying the genetic basis of natural variation. Dev Genes Evol. 2013;223(5):279–287. doi: 10.1007/s00427-013-0443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdi AR. Developmental biology and morphogenesis of the face, lip, and palate. In: Berkowitz S, editor. Cleft Lip and Palate. New York: Springer; 2006. pp. 3–12. [Google Scholar]
- Canfield MA, Honein MA, Yuskiv N, Xing J, Mai CT, Collins JS, Devine O, Petrini J, Ramadhani TA, Hobbs CA, Kirby RS. National estimates and race/ethnic-specific variation of selected birth defects in the United States, 1999–2001. Birth Defects Res A Clin Mol Teratol. 2006;76:747–756. doi: 10.1002/bdra.20294. [DOI] [PubMed] [Google Scholar]
- Creuzet S, Schuler B, Couly G, Le Douarin NM. Reciprocal relationships between Fgf8 and neural crest cells in facial and forebrain development. Proc Natl Acad Sci U S A. 2004;101(14):4843–4847. doi: 10.1073/pnas.0400869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVolder I, Richman L, Conrad AL, Magnotta V, Nopoulos P. Abnormal cerebellar structure is dependent on phenotype of isolated cleft of the lip and/or palate. Cerebellum. 2013;12(2):236–244. doi: 10.1007/s12311-012-0418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diewert VM, Lozanoff S, Choy V. Computer reconstructions of human embryonic craniofacial morphology showing changes in relations between the face and brain during primary palate formation. J Craniofac Genet Dev Biol. 1993;13:193–201. [PubMed] [Google Scholar]
- Fakhouri WD, Rhea L, Du T, Sweezer E, Morrison H, Fitzpatrick D, Yang B, Dunnwald M, Schutte BC. MCS9.7 enhancer activity is highly, but not completely, associated with expression of Irf6 and p63. Dev Dyn. 2012;241:340–349. doi: 10.1002/dvdy.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski K, Burns CD, Madison C, Clark D, Halchenko YO, Waskom ML, Ghosh SS. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front Neuroinform. 2011;5:13. doi: 10.3389/fninf.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt O, Burden D, Hepper P, Johnston C. The psychosocial effects of cleft lip and palate: a systematic review. Eur J Orthod. 2005;27:274–285. doi: 10.1093/ejo/cji004. [DOI] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Dev Dyn. 2006;235:1152–1166. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jocelyn LJ, Penko MA, Rode HL. Cognition, communication, and hearing in young children with cleft lip and palate and in control children: a longitudinal study. Pediatrics. 1996;97:529–534. [PubMed] [Google Scholar]
- Johnson H, Harris G, Williams K. BRAINSFIT: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit. Insight Journal. 2007;(10):1–10. [Google Scholar]
- Jones MC. Etiology of facial clefts: prospective evaluation of 428 patients. Cleft Palate J. 1988;25:16–20. [PubMed] [Google Scholar]
- Kjaer I. Human prenatal craniofacial development related to brain development under normal and pathologic conditions. Acta Odontol Scand. 1995;53:135–143. doi: 10.3109/00016359509005963. [DOI] [PubMed] [Google Scholar]
- Knight AS, Schutte BC, Jiang R, Dixon MJ. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev Dyn. 2006;235:1441–1447. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C, Houdayer C, Bahuau M, Moretti-Ferreira D, Richieri-Costa A, Dixon MJ, Murray JC. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410(6824):97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- MacKenzie-Graham A, Tinsley MR, Shah KP, Aguilar C, Strickland LV, Boline J, Martin M, Morales L, Shattuck DW, Jacobs RE, Voskuhl RR, Toga AW. Cerebellar cortical atrophy in experimental autoimmune encephalomyelitis. Neuroimage. 2006;32:1016–1023. doi: 10.1016/j.neuroimage.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Magnotta VA, Harris G, Andreasen NC, O’Leary DS, Yuh WT, Heckel D. Structural MR image processing using the BRAINS2 toolbox. Comput Med Imaging Graph. 2002;26:251–264. doi: 10.1016/s0895-6111(02)00011-3. [DOI] [PubMed] [Google Scholar]
- Marcucio RS, Cordero DR, Hu D, Helms JA. Molecular interactions coordinating the development of the forebrain and face. Dev Biol. 2005;284(1):48–61. doi: 10.1016/j.ydbio.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Millard T, Richman LC. Different cleft conditions, facial appearance, and speech: relationship to psychological variables. Cleft Palate Craniofac J. 2001;38:68–75. doi: 10.1597/1545-1569_2001_038_0068_dccfaa_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Nopoulos P, Berg S, Canady J, Richman L, Van Demark D, Andreasen NC. Abnormal brain morphology in patients with isolated cleft lip, cleft palate, or both: a preliminary analysis. The Cleft palate-craniofacial journal: official publication of the American Cleft Palate-Craniofacial Association. 2000;37:441–446. doi: 10.1597/1545-1569_2000_037_0441_abmipw_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- Nopoulos P, Berg S, Canady J, Richman L, Van Demark D, Andreasen NC. Structural brain abnormalities in adult males with clefts of the lip and/or palate. Genet Med. 2002;4(1):1–9. doi: 10.1097/00125817-200201000-00001. [DOI] [PubMed] [Google Scholar]
- Nopoulos P, Richman L, Andreasen NC, Murray JC, Schutte B. Abnormal brain structure in adults with Van der Woude syndrome. Clin Genet. 2007;71:511–517. doi: 10.1111/j.1399-0004.2007.00799.x. [DOI] [PubMed] [Google Scholar]
- Parsons TE, Schmidt EJ, Boughner JC, Jamniczky HA, Marcucio RS, Hallgrímsson B. Epigenetic integration of the developing brain and face. Dev Dyn. 2011;240(10):2233–2244. doi: 10.1002/dvdy.22729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, Melbye M, Jugessur A, Lie RT, Wilcox AJ, Fitzpatrick DR, Green ED, Mossey PA, Little J, Steegers-Theunissen RP, Pennacchio LA, Schutte BC, Murray JC. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, Shore P, Whitmarsh A, Dixon MJ. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 2006;38:1329–1334. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- Richman LC, Eliason MJ, Lindgren SD. Reading disability in children with clefts. Cleft Palate J. 1988;25:21–25. [PubMed] [Google Scholar]
- Riski JE. Speech, language, and velopharyngeal dysfunction: management throughout the life of an individual with cleft palate. In: Berkowitz S, editor. Cleft Lip and Palate. New York: Springer; 2006. pp. 705–718. [Google Scholar]
- Rutter M, Caspi A, Fergusson D, Horwood LJ, Goodman R, Maughan B, Moffitt TE, Meltzer H, Carroll J. Sex differences in developmental reading disability: new findings from 4 epidemiological studies. JAMA. 2004;291:2007–2012. doi: 10.1001/jama.291.16.2007. [DOI] [PubMed] [Google Scholar]
- Timmons MJ, Wyatt RA, Murphy T. Speech after repair of isolated cleft palate and cleft lip and palate. Br J Plast Surg. 2001;54:377–384. doi: 10.1054/bjps.2000.3599. [DOI] [PubMed] [Google Scholar]
- van der Plas E, Conrad A, Canady J, Richman L, Nopoulos P. Effects of unilateral clefts on brain structure. Arch Pediatr Adolesc Med. 2010;164(8):763–768. doi: 10.1001/archpediatrics.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SM, Andreasen NC, Nopoulos P. Three-dimensional analysis of brain shape in nonsyndromic orofacial clefting. J Anat. 2009;214(6):926–936. doi: 10.1111/j.1469-7580.2009.01084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, Flavell RA, Williams T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381(6579):238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, Natsume N, Yoshiura K, Vieira AR, Orioli IM, Castilla EE, Moreno L, Arcos-Burgos M, Lidral AC, Field LL, Liu YE, Ray A, Goldstein TH, Schultz RE, Shi M, Johnson MK, Kondo S, Schutte BC, Marazita ML, Murray JC. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]


