Abstract
Microorganisms play a dominant role in the biogeochemical cycling of nutrients. They are rightly praised for their facility at fixing both carbon and nitrogen into organic matter, and microbial driven processes have tangibly altered the chemical composition of the biosphere and its surrounding atmosphere. Despite their prodigious capacity for molecular transformations, microorganisms are powerless in the face of the immutability of the elements. Limitations for specific elements, either fleeting or persisting over eons, have left an indelible trace on microbial genomes, physiology, and their very atomic composition. We here review the impact of elemental limitation on microbes, with a focus on selected genetic model systems and representative microbes from the ocean ecosystem. Evolutionary adaptations that enhance growth in the face of persistent or recurrent elemental limitations are evident from genome and proteome analyses. These range from the extreme (such as dispensing with a requirement for a hard to obtain element) to the extremely subtle (changes in protein amino acid sequences that slightly, but significantly, reduce cellular carbon, nitrogen, or sulfur demand). One near universal adaptation is the development of sophisticated acclimation programs by which cells adjust their chemical composition in response to a changing environment. When specific elements become limiting, acclimation typically begins with an increased commitment to acquisition and a concomitant mobilization of stored resources. If elemental limitation persists, the cell implements austerity measures including elemental-sparing and elemental-recycling. Insights into these fundamental cellular properties have emerged from studies at many different levels; including ecology, biological oceanography, biogeochemistry, molecular genetics, genomics, and microbial physiology. Here, we present a synthesis of these diverse studies and attempt to discern some overarching themes.
Keywords: metal homeostasis, sparing, phosphorus, sulfur, iron, zinc, copper, cyanobacteria, diatom, Chlamydomonas, Bacillus
I. Overview: Mendeleev meets Darwin
One of the great achievements of 20th century science was the melding of two largely distinct disciplines, chemistry and biology, leading to a renaissance in the molecular life sciences. Chemistry underlies the myriad processes that power and enable cell growth, and an understanding of chemical principles is imperative for the modern biologist. One of the most fundamental principles of chemistry is that elements are immutable, at least under the conditions conducive to life processes. Transformation of one element into another occurs either rarely and stochastically, in the case of radioactive decay and nuclear fission, or under extremes of temperature and pressure, in the case of fusion-based nucleosynthesis.
It is ironic that the very foundations of Chemistry are rooted in the notion of elemental transmutation and the countless years of effort devoted to alchemical pursuits including, most famously, efforts to turn base metals into gold. Isaac Newton was one of the more ardent practitioneers; it is estimated that he devoted far more attention to alchemy than to mathematics and physics combined (Dobbs, 1983). Ultimately, of course, the atomic theory as espoused by John Dalton (1766–1844), with the notion of atoms as indestructible and indivisible, was hailed for its explanatory power and provided a foundation for modern chemistry.
The atomic theory set the stage for an increased understanding of the properties of the elements, which displayed a periodic pattern when arranged, in early versions, relative to their masses. While many scientists contributed to the development of the periodic table, Dmitri Mendeleev (1834–1907) is generally credited with this synthesis (ca. 1869). In biology, a contemporary of Mendeleev, Charles Darwin (1809–1882) published his landmark "Origin of Species" in 1859 with its description of evolution by natural selection. Here, we consider the conceptual intersection of these two great ideas <Figure 1>. Our theme will be the wide range of remarkable adaptations found in the microbial world that have resulted from limitations for elemental nutrients.
Figure 1.
Mendeleev meets Darwin. Dmitri Mendeleev (1834–1907) (left) developed the periodic table of the elements (ca. 1869). Charles Darwin (1809–1882) (right) developed the theory of evolution by natural selection.
A. The Elemental Composition of Life
Living cells rely on only a small, and somewhat variable, subset of the periodic table <Figure 2>. The elements of life can be divided into the macronutrients (C,H,N,O,P,S), major cations (K,Mg,Ca), and the so-called micronutrients (including many metal ions) (Frieden, 1985, Frausto da Silva and Williams, 2001). Many familiar elements are dispensable to living cells and in many cases can be deleterious if present.
Figure 2.
A cellular perspective on the periodic table. Essential macronutrients are in white against a black background and universally essential cations (Zn, Mg) in white against a grey background. Elements that have important biological roles in many but likely not all cells are shown in boldface against a dark grey background. These include the key transition metals (Mn, Fe, Co, Cu, Mo) and cations (K). Elements that are used for specialized purposes in some microbes are shown against a light grey background. These include a requirement for boron (B) in plants for cell wall structure and in some bacteria for quorum sensing. Elements that may have specialized uses, but are not known to contribute to growth are in large font against a white background (Cr,Cl, I).
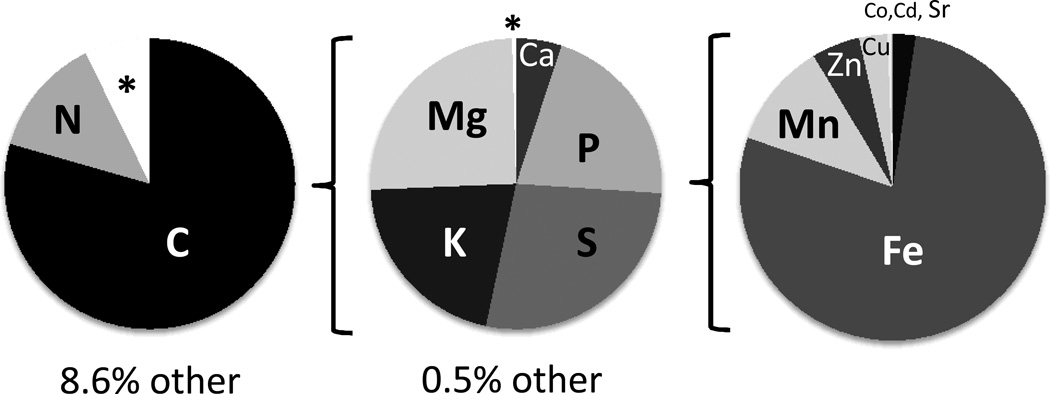
It is unexpectedly difficult to define the minimum set of elements absolutely essential for life, although there is little doubt that all cells require the so-called macronutrients of CHNOPS (note that all elements will be referred to by their atomic symbols with ionization state indicated only where relevant). The requirements for metal ions and other elements is more idiosyncratic. It is likely that all cells require Zn and Mg and nearly all also require Fe. The requirements for Mn, Cu, Co, Ni, Ca, K, Na, Mo, Se, and other elements are likely to be variable and are often unestablished. Some elements have very specialized but beneficial or even essential functions in a very select subset of organisms. Examples include the use of Si in plant and diatom cell walls, B in plants, and Cd in certain marine organisms <Table 1>. The atomic inventory has been measured for numerous organisms and is relatively constant for the most abundant macronutrients, but remarkably flexible for the much less abundant micronutrients. As an example, the elemental composition of a representative marine cyanobacterium, Synechococcus sp. CCMP835, is shown in <Figure 3> (Quigg et al., 2011).
Table 1.
The elements of microbial cells
| Element (symbol) |
Major functions and uses in microbial cells |
|---|---|
| Required for All Cells | |
| C | basis of all organic molecules |
| H | H2O, organic molecules |
| N | organic molecules, esp. proteins and nucleic acids |
| O | H2O, organic molecules |
| P | nucleic acids, NTPs, metabolites, phospholipids |
| S | proteins, glutathione and LMW thiols, biotin, lipoic acid, thiamin |
| Mg | major cation; cofactor for phosphotransferase reactions |
| Zn | enzyme cofactor, protein folding |
| Required for Most Cells | |
| K | major cation, common in cells |
| Ca | major cation, required by many eukaryotes |
| Mn | enzyme cofactor, ribonucleotide reductase, SOD, PS II |
| Fe | heme, iron-sulfur cluster, non-heme enzymes |
| Co | enzyme cofactor, B12-dependent enzymes |
| Cu | enzyme cofactor, electron carrier, respiration, SOD |
| Mo | FeMoCo cofactor (nitrogenase), Mo cofactor enzymes |
| Required for Specialized Functions in Some Cells | |
| Se | selenocysteine in proteins |
| B | plant cell wall, quorum sensing (some AI-2) |
| Na | used for ion potential in halophiles |
| Si | some plant cell walls, diatom walls (frustules) |
| Cl,Br,Fl,I | some bacterial secondary metabolites |
| V | nitrogenase, haloperoxidases |
| Ni | urease, SOD (SodN), glyoxalase |
| Cd | cofactor for carbonic anhydrase (CA) in some marine microbes |
| W | tungstoenzymes (aldehyde oxidoreductase, formate dehydrogenase, acetyl hydratase) |
Figure 3.
Atomic composition of Synechococcus sp., a representative of the bacterial phytoplankton. For each pie chart, the portion indicated by the asterisk (other) is expanded in the pie chart to the right. On the far right, Sr is indicated by the black slice and the thin white slice between Cu and Sr includes Co and Cd. Plotted with data from (Quigg et al. 2011).
The requirements for macronutrient elements are well established and easily understood. Life is fundamentally based on aqueous chemistry and this requirement alone provides an absolute requirement for H and O, although of course not all organisms require molecular oxygen (O2). The chemistry of proteins, nucleic acids, carbohydrates, and lipids accounts for the major requirements for C, N, P and S. In no case can cells be assembled in the absence of these crucial elements although, as we will see, there are sophisticated mechanisms for minimizing (to the extent possible) the requirements for these macronutrients when they might otherwise be limiting.
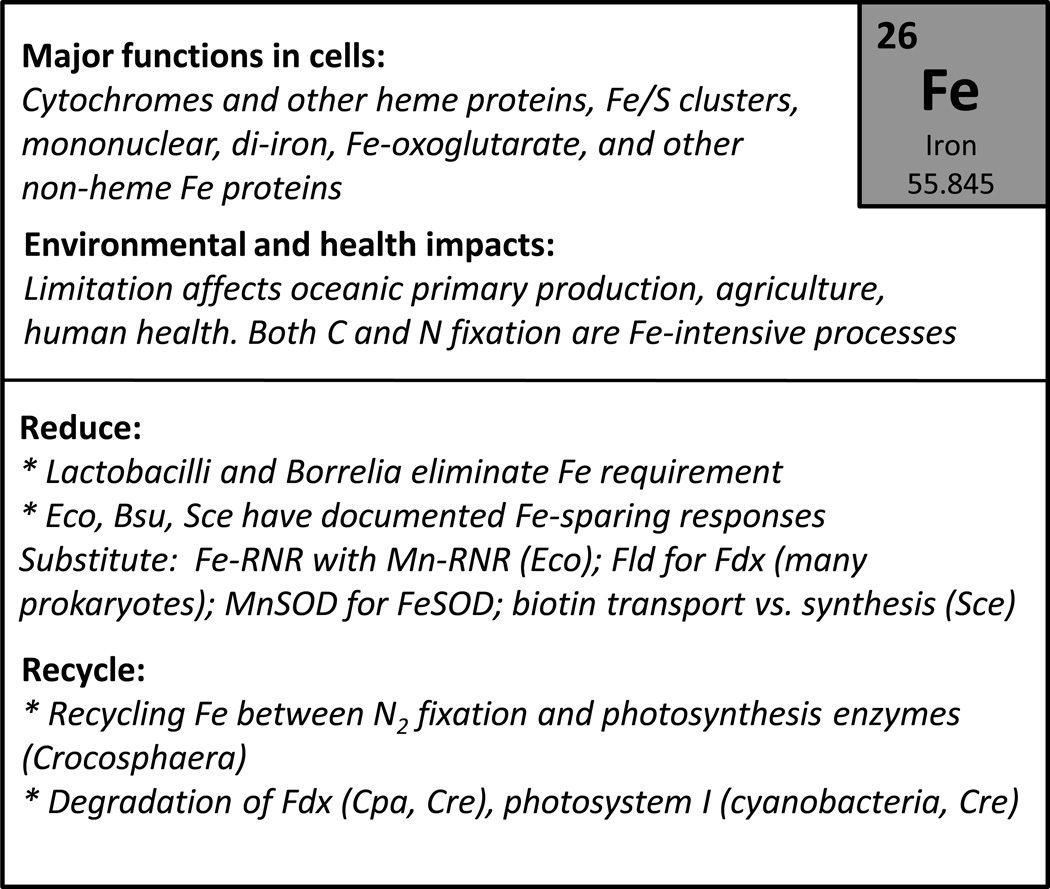
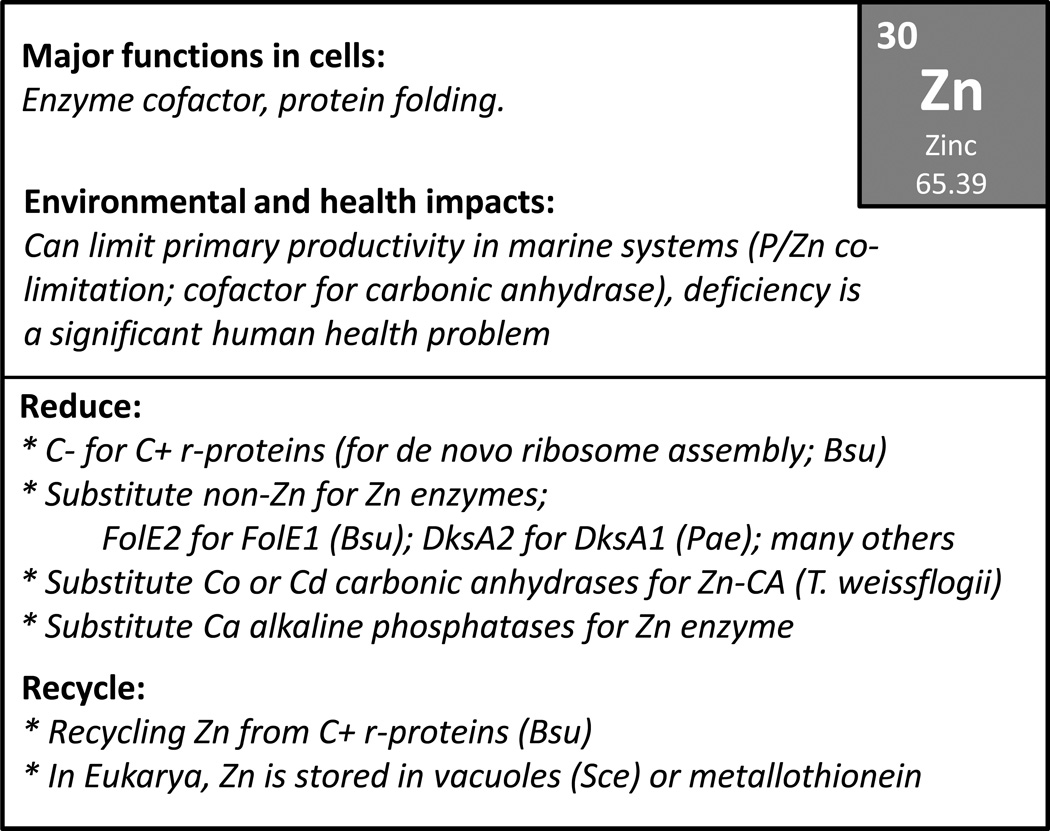
Requirements for the remaining elements are less firmly established and few, if any, are likely to be universal requirements (Table 1). Zn is widely used as a cofactor for protein folding and as a Lewis acid cofactor for several families of enzymes (Andreini et al., 2009). Mg(II) is the major biological cation in many microbial systems. We suggest that both of these are likely essential for all life, at least until a counterexample is identified. Fe is essential for growth of nearly all microbes and is present in heme-cofactored proteins, iron-sulfur cluster (Fe/S) proteins, and di-iron and mononuclear enzymes, amongst others. However, there are organisms, including the lactobacilli and Borrelia burdorferi, that have dispensed with an Fe requirement (Posey and Gherardini, 2000). Other cations (e.g. Ca, Cu, Co, Mn, K) are often needed or at are least stimulatory for growth, but a requirement is not necessarily universal. Note that in most cells the major anions are likely organic compounds (acetate, glutamate), so a requirement for Cl− or other inorganic anions is not universal.
Microorganisms are the consummate experimentalists; over the course of several billion years of evolution single-celled life forms learned to exploit a wide variety of niches that often differed in the availability of the "elements of life." As organisms adapted to new environments where previously accessible elements were now limited in abundance or absent, they had to evolve alternative solutions for life's chemistry. It is the results of these evolutionary experiments, as embodied in the physiology and genetics of contemporary microbes, that form the basis for this review.
B. Reference Systems for Understanding Microbial Responses to Elemental Limitation
We will here explore the molecular mechanisms by which microbial cells adapt and acclimate to elemental limitation. Note that throughout this review, the term elemental refers to the atomic elements. We will use adaptation to refer specifically to changes in the DNA that occur in response in selection pressures, potentially acting over very long time scales, and acclimation to refer to those changes in cellular physiology that allow individual cells to better utilize available nutrients. Adaptations may include, for example, changes in protein sequence or the evolution of isozymes with alternative cofactor dependencies. The conditional expression of alternative enzymes or pathways, in response to changes in elemental availability, is a major feature of the acclimation response for many elemental limitations.
Access to the elements of life plays a large role in defining the microbial composition of the diverse habitats within the biosphere. For some environments, such as the surface waters of the open ocean, elemental composition (and hence limitations) may be relatively constant, and are therefore a driving force for adaptation. Perhaps two-thirds of the ocean's surface waters have biologically limiting levels of P, and for much of the rest, it is the availability of Fe that limits primary productivity. The most successful organisms in these environments have evolved adaptations that allow them to maximize growth by reducing demand for limiting nutrients. These organisms are specialists, and their genomes and their metabolism reflect an evolutionary optimization for their environment. For other cosmopolitan organisms the environment is ever-changing and a more generalist approach is advantageous. Where elemental availability is variable, cells have evolved genetic mechanisms to acclimate their physiology to the changing local environment. In many cases, these types of acclimation involve changes in gene expression.
1. Genetic model systems for investigating mechanisms of elemental economy
Not surprisingly, many of the best understood mechanisms of adaptation and acclimation have been revealed in well-studied reference or “model” organisms <Table 2>. Here, we will focus on the Gram-negative bacterium Escherichia coli and the Gram-positive bacterium Bacillus subtilis. Eukaryotic microbes will be represented by the eukaryote Saccharomyces cerevisiae and the photosynthetic eukaryote, Chlamydomonas reinhardtii, a soil-resident alga. Each of these four organisms displays sophisticated mechanisms for acclimation to changing nutrient availability (Table 2a). Work in the Archaea is still not well developed in this area, although this diverse group of organisms clearly has sophisticated systems for the optimization of nutrient utilization and, in particular, for metal homeostasis (Bini, 2010). Emerging Archaeal model systems include the halophiles Halobacterium salinarum and Haloferax volcanii and the methanogens Methanococcus maripaludis and Methanosarcina acetivorans (reviewed in Leigh et al., 2011). Of note, systems level studies of Fe homeostasis have been initiated in H. salinarum (Schmid et al., 2011) and methanogenesis is an intensively metal-utilizing process.
Table 3.
Microbial Reference Systems for Investigations of Elemental Economy
| Domain:Phylum | Organism (abbreviation) | Environment | Nutrition | Notable mechanisms for elemental economy |
|---|---|---|---|---|
| A. Model systems for molecular genetic studies | ||||
| B: Proteobacteria | Escherichia coli (Eco) | mammalian intestine | facultative anaerobe | C/N - elemental optimization; recycling by ribophagy S - elemental optimization Fe - Fur regulon; Fe-sparing (RyhB sRNA); isozyme substitution, MnSOD vs. FeSOD; Mn RNR vs. Fe RNR |
| B: Firmicutes | Bacillus subtilis (Bsu) | soil, rhizosphere | facultative anaerobe | C/N - recycling by cannibalism P - P-sparing and recycling by cell wall remodeling Fe - Fur regulon; Fe sparing (FsrA sRNA); flavodoxin substitution for ferredoxin Zn - Zur regulon; ribosomal protein mobilization, FolE2 |
| A: Euryarchaeota | Halobacterium salinarum (Hsa) | halophilic archaeon | Fe - Fe acclimation response defined | |
| A: Euryarchaeota | Methanococcus maripaludis (Mma); Methanosarcina acetivorans (Mac) | methanogenesis | Ni - Ni sparing response for methanogenesis enzymes Mo/W-isozymes of formylmethanofuran dehydrogenases |
|
| E: Ascomycota | Saccharomyces cerevisiae (Sce) | yeast, model eukaryote | respiration, fermentation | C/N - elemental optimization; recycling S - S-sparing, substitution with S-depleted isozymes Fe - Fe-sparing (Aft1/Aft2 and Cth1/Cth2 regulons); isozyme substitution Zn - Zn-sparing by Zap1 repression of Adh1 and Adh3 |
| E: Chlorophyta | Chlamydomonas reinhardtii (Cre) | soil | photosynthetic and heterotrophic | N - ribophagy during gametogenesis S - S-sparing, substitution with isozymes; recycling of S from sulfolipids P - substitute P-lipids with S-lipids; recycle P from chloroplast DNA by copy number reduction Fe – down-regulation of PS I and Fd, prioritizing respiration over photosynthesis under heterotrophic conditions, chloroplast MnSOD induced Cu – Cyt c6 for plastocyanin, flavin amine oxidase instead of Cu amine oxidase |
| B. Reference systems for environmental and genomics-based studies in marine ecosystems | ||||
| B: Cyanobacteria | Prochlorococcus marinus (Pma) | oligotrophic open ocean | smallest known phototroph, | P - substitute P-lipids with S-lipids Fe - PS I remodeling |
| B: Cyanobacteria | Trichodesmium (Tri) | oligotrophic open ocean | phototroph, diazotroph | Fe- Fe-sparing by down-regulation of nitrogen fixation; Fe mobilization from Dps miniferritin |
| B: Cyanobacteria | Synechococcus (Syn) | mesophilic ocean and freshwater species | phototroph | N - recycling by phycobilisome degradation Fe - Fe sparing; flavodoxin substitution for ferredoxin Ni - recycling of Ni from urease to SodN (proposed) |
| B. Cyanobacteria | Synechocystis (Syc) | freshwater lake | phototroph | Cu - Cyt c6 for plastocyanin |
| B: Cyanobacteria | Crocosphaera watsonii (Cwa) | oligotrophic open ocean | phototroph, diazotroph | Fe - diurnal cycling of iron between photosynthetic and nitrogen fixation complexes (reduces Fe quota by 40%) |
2. Phytoplankton and the marine ecosystem: the global impacts of elemental limitation
Life is thought to have evolved in shallow seas and perhaps other surface waters and the atomic composition of cells reflects this archaic environment (Dupont et al., 2010). As life diversified and colonized a wider variety of environments, the ability to adapt to changing elemental availability likely assumed greater importance (Armbrust, 2009, Sohm et al., 2011). The abundance of life enabled by the evolution of mechanisms to access the most abundant forms of C (photosynthetic fixation of atmospheric CO2) and N (fixation of atmospheric N2) inevitably led to growth restriction by other nutrients (Morel and Price, 2003, Konhauser et al., 2009). The result has been the evolution of mechanisms to either bypass these restrictions or, minimally, to increase the efficiency of utilization of limiting nutrients. Moreover, elemental availability has changed dramatically on geological timescales (Quigg et al., 2003, Konhauser et al., 2009). One major driving force was the great oxidation event (~2.4 billion years ago) and the subsequent depletion of soluble ferrous Fe from ocean surface waters and other aerobic environments. It is estimated that ~30% of the open ocean is Fe-limited and this left a major imprint on the genetics and physiology of the constituent plankton (Armbrust, 2009, Behrenfeld et al., 2009, Sohm et al., 2011).
We will discuss the marine microbial community in some detail, including cyanobacteria and other phytoplankton, since these organisms provide particularly informative examples of adaptations to limiting P, Fe, and Zn (Table 2b). Because of the severity of nutrient limitations in this environment, and the long timescales over which these limitations have persisted, the resident organisms have evolved unique adaptations to bypass elemental restrictions. These adaptations are, in many cases, complemented by acclimation responses.
The dominant microorganisms in the open ocean vary depending on location, light fluxes, and availability of both macro- and micronutrients. In many areas of the open ocean, the numerically dominant microorganisms include members of the cyanobacteria (in particular, Synechococcus spp. and Prochlorococcus spp.) (Zehr et al., 2007) and diatoms (Bowler et al., 2010, Partensky and Garczarek, 2010). Diatoms are eukaryotic phytoplankton notable for their silica-containing cell walls (frustules) and are responsible for perhaps 20% of total primary productivity globally (reviewed in Armbrust, 2009). Representative organisms which have helped illuminate mechanisms of elemental economy, and where genomics-enabled approaches are now coming to the fore, are summarized in Table 2b.
C. General Strategies for Dealing with Elemental Limitation
Insights into how microbes adapt to elemental limitations emerged in laboratory studies of microbial metabolism on the one hand (e.g. Hutber et al., 1977, Wood, 1978, Schönheit et al., 1979, Bishop et al., 1980, Ragsdale and Ljungdahl, 1984) and the study of the microbial ecology, and in particular the ocean ecosystem, on the other (Morel and Price, 2003). Increasingly, genomic and molecular genetic investigations of model organisms (Table 2a) are revealing mechanisms of adaptation and programs for acclimation to elemental limitation. These same mechanisms likely occur in globally important but less tractable organisms such as the marine phytoplankton. Studies of representative marine microbes, many of which are now amenable to genomics-based investigations (Table 2b), reminds us of the impact of elemental limitations on global ecology and primary productivity. Comparing these two bodies of largely distinct literature reveals that similar strategies for elemental economy have evolved independently in many systems.
With their ability to double their population in as little as 20 minutes or less (under optimal growth conditions), responses to resource limitation are deeply ingrained in the genetics and physiology of microorganisms. The impact of nutrient limitation is apparent both in cellular adaptations and in the processes of acclimation. At the level of adaptation, macronutrient limitation has led microbes to alter the elemental composition of selected constituents through a process of elemental optimization. As one example, signatures of macronutrient limitation are recorded in the protein sequences of the elemental acquisition machinery of phylogenetically diverse organisms (Baudouin-Cornu et al., 2001). At the level of acclimation, nutrient limitation leads to several predictable responses that may be activated either simultaneously or sequentially, depending on the system. These responses can be classified as elemental acquisition, mobilization, sparing, and recycling.
1. Elemental optimization
Organisms adapt to elemental limitation by altering the atomic composition of their constituents; a trend noted in protein primary sequences. For example, proteins necessary for the acquisition of S are selectively depleted of Cys and Met in their amino acid sequences (Baudouin-Cornu et al., 2001). Clearly, this enhances the ability of the cell to express these proteins under S limiting conditions where translation of proteins requiring high levels of Cys and Met might be impaired. Parenthetically, we note that an analogous selection pressure also operates at higher levels of organization: the amino acids themselves. In the "cognate bias hypothesis" it has been noted that amino acid biosynthetic enzymes are under selection pressure to minimize use of the cognate amino acid in their coding sequences (Alves and Savageau, 2005). Other notable examples of elemental optimization include the elimination of an Fe requirement in organisms that grow in severely Fe-limited environments (Posey and Gherardini, 2000), and the substantial reduction in phospholipid content in cells that have adapted to persistent P limitation (Van Mooy et al., 2009). As defined here, elemental optimization is a fixed adaptation and is therefore most characteristic of specialist organisms that inhabit environments of relatively constant elemental composition. However, analogous changes in elemental composition also contribute to the conditionally expressed programs by which organisms acclimate to nutrient limitation.
2. Acclimation: acquisition and mobilization
Generally, the first major cellular response during acclimation to nutrient limitation is an increased expression of acquisition pathways. Acquisition pathways are dominated by transporters for the limiting element or compounds that are rich in the limiting element. Associated functions include enzymes to help mobilize nutrients, such as proteases, nucleases, and various hydrolases. Acquisition pathways also include, for many organisms, induction of pathways to access resources that are chemically recalcitrant or more energetically prohibitive and therefore only used as a last resort. One well characterized example is the repression of nitrogen fixation by ammonium (Dixon and Kahn, 2004). A second example is the derepression of enzymes to cleave direct C-P bonds (phosphonates) when P (as phosphate) is limiting (Jiang et al., 1995, Baek and Lee, 2007), an adaptation that appears widespread in marine bacteria (Martinez et al., 2010). Phosphonates may comprise up to 25% of the high molecular weight organic P pool in the ocean and uptake and utilization of phosphonates is widespread in the important marine cyanobacterium Prochlorococcus as well as in Synechococcus spp. in microbial mats in hot springs (Adams et al., 2008, Feingersch et al., 2011).
Concomitant with the expression of acquisition pathways, organisms will mobilize stored resources. Examples of this pathway include release of Fe from ferritin, degradation of polyphosphates to release inorganic P, and mobilization of excess C stored, for example, in lipid bodies. In each of these examples, mobilization is from a source whose primary, if not exclusive, function is elemental (or energy) storage. Once dedicated stores are depleted, cells may resort to more drastic measures that involve the selective consumption of their own organelles (autophagy) which we classify as elemental recycling (see below).
Acquisition and mobilization from stores are both typically activated when nutrients first start to become limiting for growth, a state that can be defined as elemental deficiency. If the acquisition and mobilization strategies are ineffective, and the deficiency is not relieved, the lack of the specific required element will ultimately lead to a cessation of growth or of key metabolic activities, a state that can be defined as elemental limitation (La Fontaine et al., 2002). While interesting in their own right, processes of nutrient acquisition and mobilization have been well reviewed elsewhere and we focus our attention on the elemental sparing and recycling responses.
3. Acclimation by elemental sparing
When nutrients are not accessible in the environment, and intracellular stores are depleted, austerity measures are implemented following the precepts of reduce, reuse, and recycle. Elemental sparing refers to responses that serve to reduce cellular demand for limiting nutrients by selectively repressing synthesis of non-essential proteins and macromolecules. This is, in essence, a prioritization mechanism by which the cell distinguishes between high priority and often essential functions and those that are of lower priority for survival. Elemental sparing responses are common for both macronutrient (P,S) and micronutrient limitations.
To be effective, and therefore a target of evolutionary selection, elemental sparing frequently targets the most abundant macromolecules in the cell. As a result, limitations for some of the key macronutrients (C,N,S) typically lead to changes in protein composition (the proteome) and targets highly abundant proteins. Limitation for P often leads to changes in the membrane and cell envelope, which can contain abundant phospholipids and other P-containing macromolecules. P-limited cells may also recycle P from nucleic acids (either DNA in the case of polyploid plastids or RNA in ribosomes). Limitation for specific metal ions often leads to a shift to enzymes that use alternative metal cofactors. Alternatively, some cells may simply dispense with a requirement for what would otherwise be an essential and hard to access nutrient.
While it is often possible for microorganisms to reduce their dependency on particular elements by the elimination of specific proteins or other macromolecules, in many cases these changes must be compensated for by the expression of alternative pathways. As a result, a common feature of many elemental sparing responses is functional substitution. For example, in response to metal ion limitation, substitute pathways may take advantage of an alternative, non-metal-based chemistry, or may rely instead on a more abundant metal in place of a scarce one. In response to P limitation, simply repressing the synthesis of phospholipid membrane synthesis would impede cell growth. Expression of alternative biosynthetic pathways for lipids lacking P enables continued cell growth, while sparing P for its more indispensable functions.
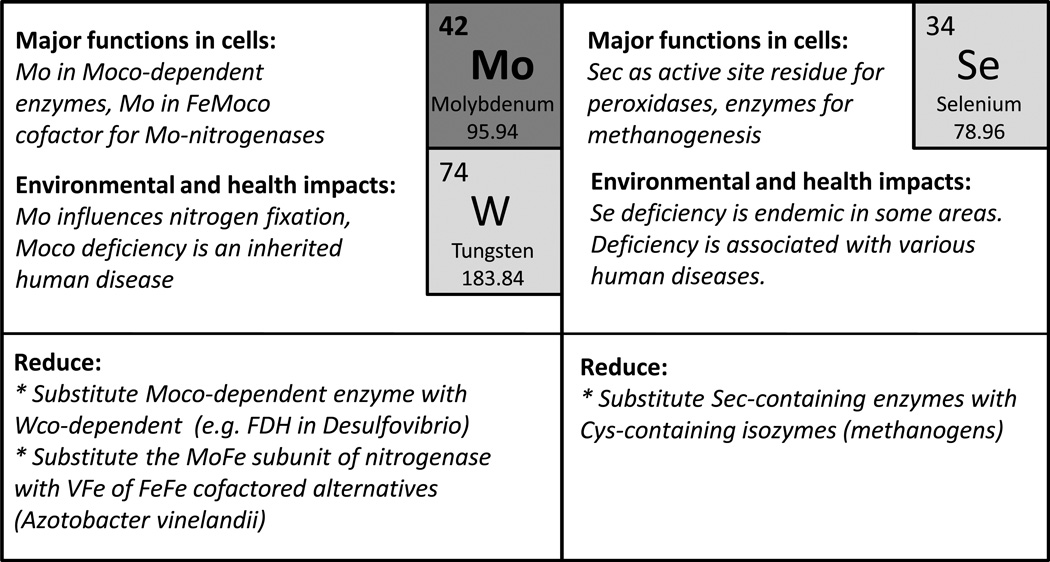
Not all elemental substitution processes in cells contribute to elemental sparing. In many organisms, S is substituted with Se, usually at a single active site residue. Since most organisms contain between one and three selenoproteins (Zhang and Gladyshev, 2011), this is not sufficient to affect the overall S budget. In the case of P, there has been a report of its replacement by As with an apparent reduction in cellular P demand (Wolfe-Simon et al., 2011), but this has been widely challenged (see below). Indeed, there is not a single documented example of As replacing P in a biological molecule that retains its normal function in the cell.
4. Acclimation by elemental recycling
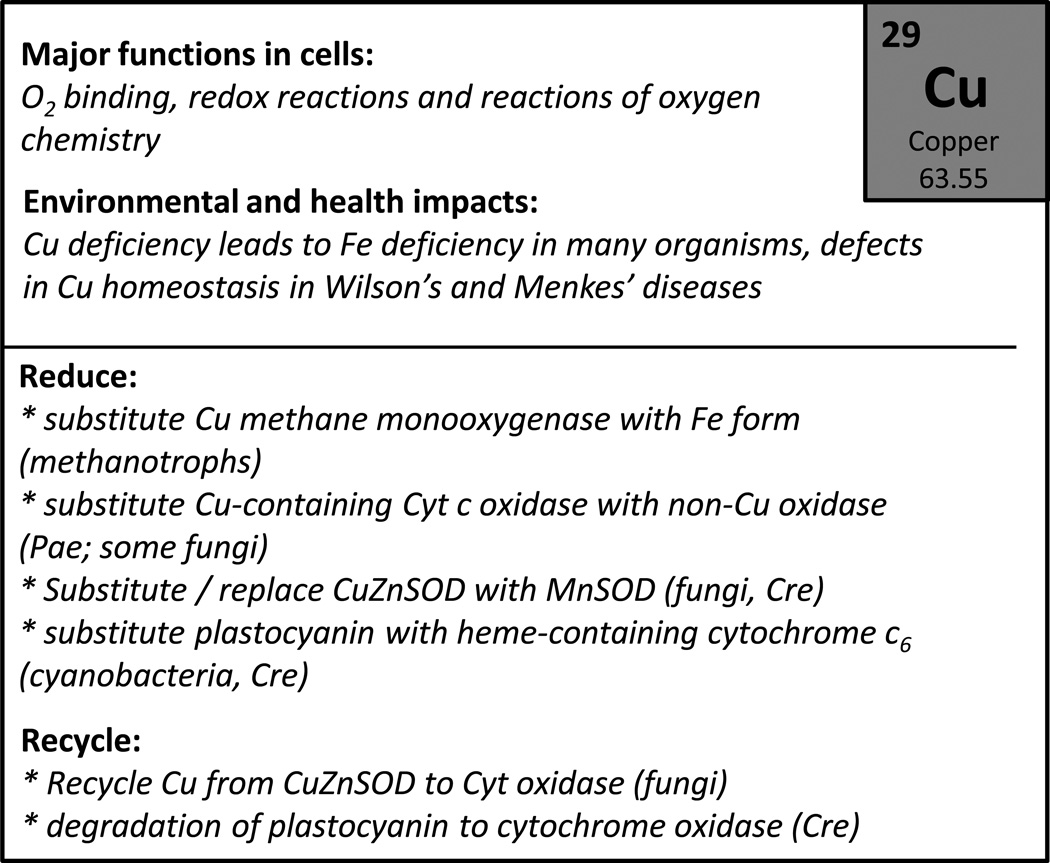
In those systems where it is difficult to dispense with an elemental requirement, and the gains to be obtained by elemental sparing are limited, cells may additionally implement an elemental recycling program. Such strategies may include the degradation of key cellular components (proteins, the ribosome, photosynthetic complexes, and even chromosomal DNA in polyploid plastids) for the sole apparent purpose of recovering the nutrients stored therein to supply new biosynthetic processes <Figures 4 and 5>. One notable example is the degradation of ribosomes (ribophagy) upon starvation, which frees up significant stores of C, N, P, and possibly Mg (Martin et al., 1976, Kraft et al., 2008). Similarly, in C. reinhardtii and in a marine cyanobacterium, abundant metalloenzymes may be proteolytically degraded simply to release and recycle the valuable copper and iron cofactors (Merchant and Bogorad, 1986a, Saito et al., 2011).
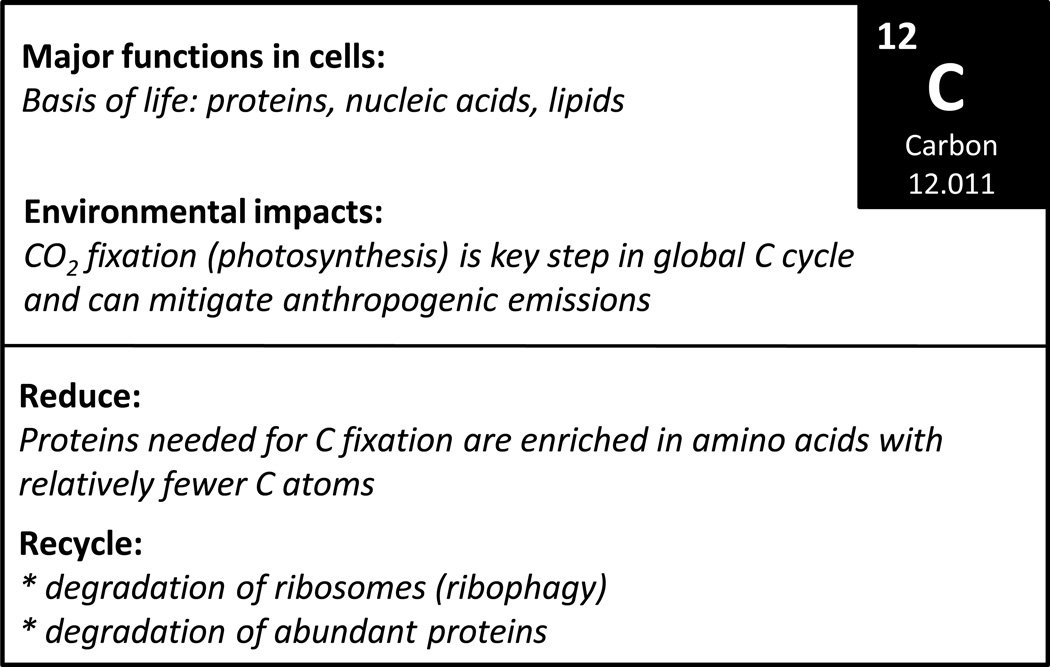
Figure 4.
Overview of the biological roles of C and known sparing and recycling mechanisms (see text for details.)
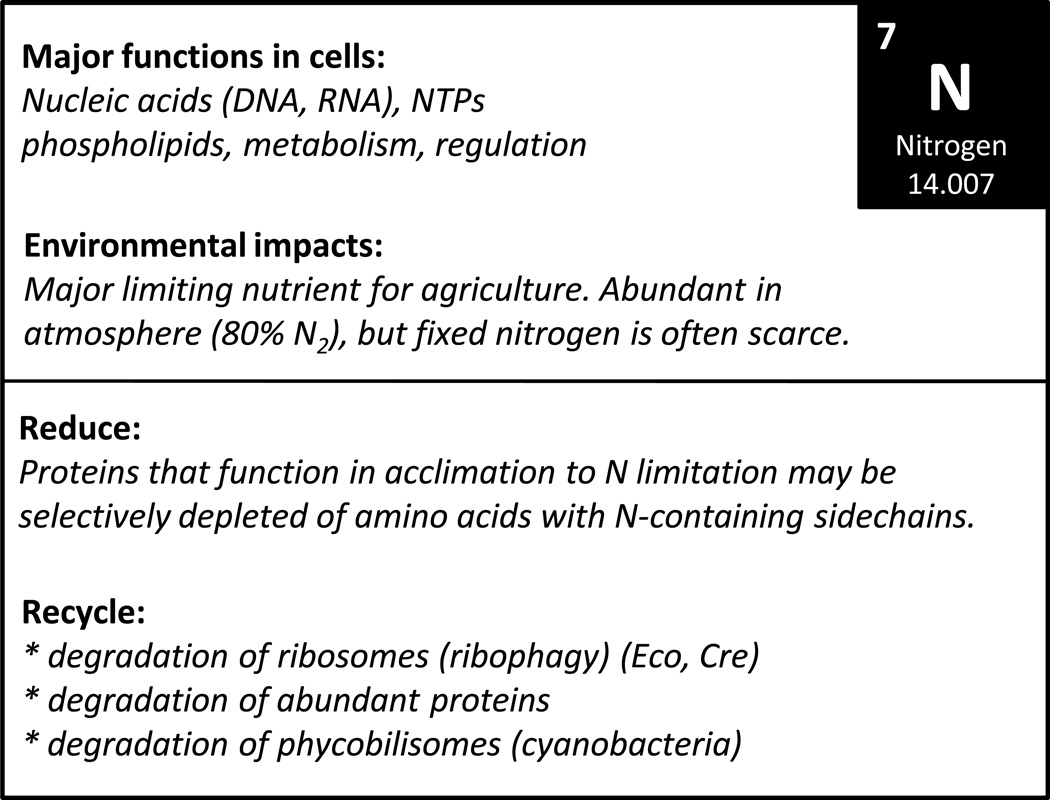
Figure 5.
Overview of the biological roles of N and known sparing and recycling mechanisms.
In the following sections, we will highlight numerous examples, in both model organisms and representative environmentally significant microbes, of these major strategies for dealing with elemental limitation. We will begin with some of the best understood processes that enable efficient macronutrient utilization and conclude with studies of metal homeostasis and the evolution of pathways to bypass metal limitations on growth.
II. Microbial Adaptations to Macronutrient Limitation
By weight, cells are mostly water and this accounts for, and provides a source of, both H and O. For all of the remaining macronutrients (C,N,P,S) there are well-documented processes of both adaptation and acclimation in response to limitation. The effects of macronutrient limitation on cell growth have long been appreciated in ecology and were perhaps first formalized in terms of crop production by Professor Carl Sprengel of Göttingen (1839) in what became known as the "law of the minimum" as popularized by Justus von Leibig. The impact of elemental limitations was highlighted by the influential work of the oceanographer Alfred Redfield who noted that the averaged ratio of macronutrients in biomass (largely phytoplankton) from the oceans (C:N:P=106:16:1) was very similar to the elemental composition of the ocean surface waters. This so-called Redfield ratio is a founding concept in the development of "ecological stoichiometry": a discipline which traces the impact of elemental composition and limitations on ecosystem dynamics (Jeyasingh and Weider, 2007).
Despite a long and influential history, ecological stoichiometry has remained a largely descriptive approach. However, insights into the underlying mechanisms by which microorganisms adapt and acclimate to elemental limitation are emerging due to both molecular biological studies and, increasingly, bioinformatic analyses. Genome sequence information, especially when coupled with expression (transcriptome and proteome) data, allows insights into the protein composition of cells from different environments and under different growth regimens. Remarkably, the evolutionary impact of both macro- and micronutrient limitation is often discernable by a careful analysis of genomes: an emerging science for which the name "stoichiogenomics" has been proposed (Elser et al., 2011).
A. Carbon, Nitrogen, and Sulfur Limitation: adaptation and acclimation mechanisms
The bulk of cellular C, N, and S demand is in support of protein synthesis. While C and N are constituents of all amino acids, these elements are not equally distributed due to variations in the length and composition of the side-chains. Two amino acids, Cys and Met, contain S and together these account for the bulk of cellular S requirements. The ability of changes in protein sequence to spare limiting macronutrients was first hinted at from protein sequence analyses of individual proteins, but has more recently been inferred on a genome-wide basis.
1. Adaptation and acclimation to S limitation
a. Elemental optimization: selective reduction of Cys and Met content in proteins
For any given protein, a small subset of amino acids is typically essential for catalytic function, and many others may contribute to optimal expression, protein folding, or stability. However, at many positions, identified as variable in alignments of homologous proteins, substitutions may occur more or less freely (e.g. Wen et al., 1996). The identity of the amino acids at these variable positions can change and become established in response to a variety of selection pressures, including elemental availability.
One of the first noted examples of such an effect emerged from the protein composition analysis of the abundant sulfate binding protein induced in E. coli upon S starvation (Pardee, 1966). Amino acid analysis suggested that this polypeptide, which can constitute 1% of cellular protein when cells are grown under S limitation, was lacking Cys and Met. The lack of these amino acids (in the mature protein) ensures that its synthesis in response to S limitation is not impaired by restrictions on Cys and Met availability. Numerous other examples are now apparent where proteins expressed under S limitation either lack or have a reduced content of Cys and Met <Figure 6>. Indeed, at a proteomic level, proteins that are involved in S assimilation may be selectively depleted in Cys and Met relative to other proteins. For example, analysis of proteins annotated as having likely roles in S assimilation in both E. coli (23 proteins) and S. cerevisiae (20 proteins) revealed a substantial decrease (up to 2-fold) in the fraction of amino acids that contain S relative to the rest of the proteome (Baudouin-Cornu et al., 2001). No such difference was seen in the orthologous enzymes (where present) from human, which was interpreted as evidence that S limitation has not been a selective pressure in mammals. The selective reduction of S content in S assimilation proteins is just one of many ways in which macronutrient limitations and energy constraints can influence protein sequences. A survey of ~150 different species (mostly Bacteria) suggests that there are also substantial differences (approaching 2-fold) in the computed fractional S content of proteomes amongst species. One notable trend is a slightly higher S content, on average, amongst anaerobes, although the significance of this observation is not yet clear (Bragg et al., 2006).
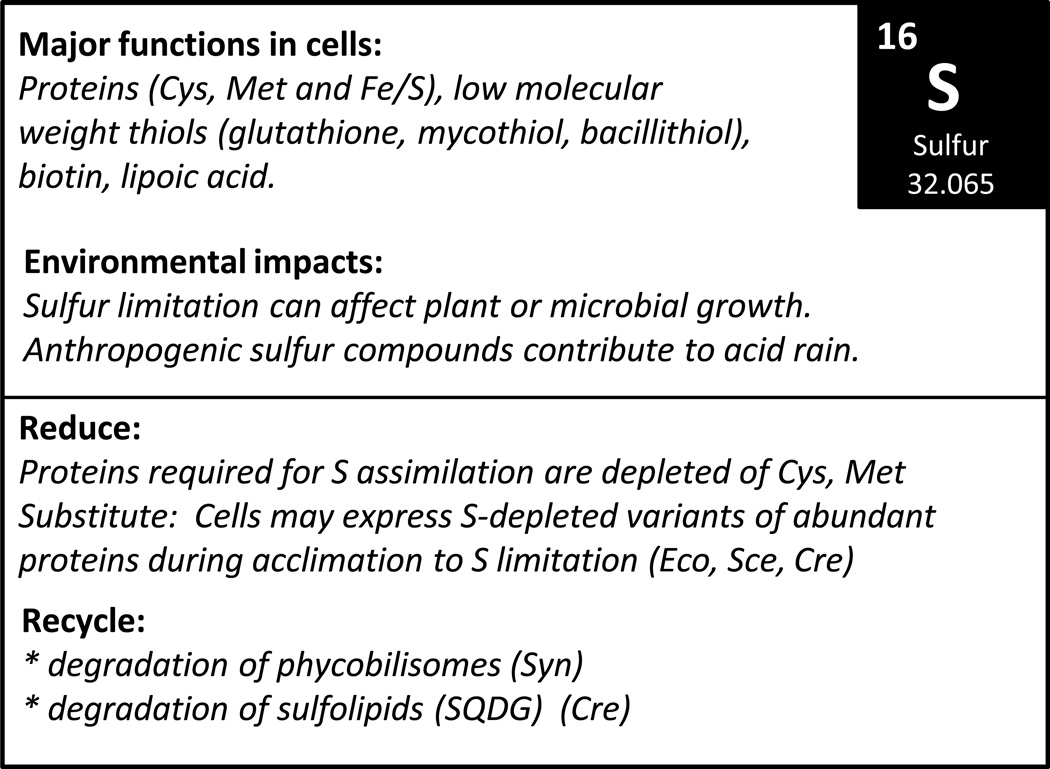
Figure 6.
Overview of the biological roles of S and known sparing and recycling mechanisms.
b. Remodeling of the proteome as a S sparing mechanism
Expression of S depleted protein variants or isozymes is also an important mechanism of acclimation to S limiting growth conditions. For example, in cyanobacteria, the light-harvesting phycobilisome proteins can account for ~50–60% of soluble protein. In the cyanobacterium Calothrix sp. PCC7601, the phycocyanins, a key phycobilisome constituent, are encoded by three differentially expressed operons (Mazel and Marlière, 1989). The cpc3 operon is specifically induced under conditions of S limitation. The encoded phycocyanins have been selectively depleted of S-containing amino acids relative to the phycocyanins encoded by the cpc1 and cpc2 operons. In over 1000 amino acids of five proteins encoded by cpc3, there are only five Met initiation codons and three indispensable Cys used as attachment sites via thioether linkage for phycobilin (Mazel and Marlière, 1989). A similar response has been documented in a freshwater cyanobacterium, Fremyella diplosiphon (Gutu et al., 2011). This organism also remodels its abundant phycobilisome complex in response to S depletion to replace proteins relatively rich in Cys and Met with a paralog that is depleted of these S-containing amino acids. The replaced phycobilisome proteins are likely degraded which thereby provides the cell with a source of S (and potentially N) under conditions of nutrient deprivation (elemental recycling).
A comparable acclimation response has been noted in the yeast S. cerevisiae. In this organism, intracellular S demand was modified by exposure to Cd, which induces synthesis of the Cys-containing tripeptide glutathione and hence creates a draw on the S metabolite pool. Cd is toxic to cells because of its affinity for intracellular thiols (e.g. at the active site of enzymes) and is detoxified by tight binding to glutathione. The strength of the binding (essentially irreversible), combined with the transport of the glutathione-Cd complexes into vacuoles, results in sequestration of Cd and depletion of intracellular S. Analysis of changes in the proteome of Cd-treated cells revealed the induction of isoforms of three central metabolic enzymes that are selectively depleted in S-containing amino acids (Fauchon et al., 2002). These include an alternative pyruvate decarboxylase, an enolase, and an aldehyde dehydrogenase. These induced isozymes contain only 22 S atoms as compared to 42 for their counterparts. The usual isoforms are among the most abundant proteins in yeast, constituting >6% of the soluble proteome. For pyruvate decarboxylase, down-regulation of the usual isoform was also documented. In addition, several other mRNAs that were induced in Cd-exposed cells were noted to be significantly depleted in Cys- and Met-encoding capacity. These responses are dependent on Met4p, a transcriptional regulator of S assimilation. Interestingly, of the 66 transcripts induced by Cd and dependent on Met4p, just 13 (encoding the most abundant proteins) were significantly depleted of sulfur amino acids. Analysis of the fate of sulfate nutrient in Cd-treated cells indicated a 70% distribution of S in favour of GSH biosynthesis as compared to a 79% distribution towards protein in untreated cells. These S-sparing changes are estimated to lead to an ~30% reduction in cellular S allocation towards protein, which increases S availability for Cd-detoxification pathways (Fauchon et al., 2002).
In the alga C. reinhardtii, S-starvation induces massive changes in the proteome and transcriptome (González-Ballester et al., 2010). As expected, many of the changes relate to S acquisition, such as mobilization of S from esterified organic sulfates, utilization of less preferred S sources, and sulfate transport (reviewed in Irihimovitch and Yehudai-Resheff, 2008). There is also evidence for S recycling (González-Ballester et al., 2010). For instance, enzymes involved in S mobilization (including CDO1, TAUD1 and TAUD2 encoding cysteine and taurine/α-ketoglutarate dioxygenases, respectively) are strongly up-regulated. These responses are dependent on a plant-specific SNF-related kinase, which is central to the S starvation signaling pathway, indicating the direct relevance of these responses to S metabolism.
Down-regulation of transcripts encoding biosynthesis of S-containing vitamins, thiamin and biotin, and the cofactor S-adenosyl methionine, is likely to be part of an S sparing response in which the limited pool of intracellular S metabolites is directed towards critical processes. Several extracellular proteins (Ecp56, Ecp61, Ecp76 and Ecp88) and a particular isoform of chlorophyll binding light harvesting protein are induced by S deficiency (Takahashi et al., 2001, Nguyen et al., 2008, González-Ballester et al., 2010). In fact, the mRNA for the light-harvesting chlorophyll-binding protein Lhcb9 increases as much as 103-fold to become the 2nd most abundant transcript in the cell. In each case, the induced proteins are depleted for Cys and Met. The induced Ecps have at most one S-containing amino acid out of 500–600 (0 to 0.2%) compared to 4.1 to 7.5% for other cell wall proteins. In the case of Lhcb9, out of five S-containing amino acids that are invariant in the other 8 major Lhcb proteins, this isoform retains only two: Met157, Met213 and Cys101 are replaced by Leu, Ser and Ile, respectively. Interestingly, the depletion is noted only in the mature part of the protein which is the form that accumulates and is hence subject to selective pressure, but not in the signal sequence for export or the transit peptide for thylakoid membrane targeting, both of which are cleaved, with the constituent amino acids recycled. The authors estimate that this form of S sparing reduces from 4.2% to 2.3% the S amino acid content of the C. reinhardtii proteome (González-Ballester et al., 2010), reminiscent of the yeast study discussed above.
S sparing has been documented in multicellular organisms as well (Petrucco et al., 1996, Kim et al., 1999). In the soybean seed, the S content of storage proteins in seed is determined by the level of S nutrition: S-amino acid-rich glycinin is the major protein if S is available, but S-amino acid depleted β-conglycinin accumulates in the S-poor situation. The abundance of a key metabolite, O-acetyl-Ser, signals the change in gene expression (Kim et al., 1999).
2. Elemental optimization: signatures of C and N limitation in proteomes
Just as selection in the face of S limitation has led to a reduction or even eradication of S-containing amino acids from proteins, both C and N limitation can affect protein composition (Figs. 4 and 5). It has been noted, for example, that a single amino acid change in a protein can add up to three N atoms (Gly to Arg) or nine C atoms (Gly to Trp). When considered in the context of protein abundance, mathemetical modeling suggests that a single Arg to Gly change, for example, would have a sufficient effect on N balance to be targeted for selection in the most abundant ~8% of yeast proteins and a single Trp to Gly change would provide sufficient C savings to be visible to selection in ~4% of proteins. The effects of depleting proteins for S (by elimination of one or more Cys or Met residues) is even more dramatic and is potentially subject to selection in more than half of all yeast proteins, although this value depends on the precise parameters used (Bragg and Wagner, 2009). Such changes are particularly apparent when one considers only the subset of proteins expressed in response to a particular elemental limitation, as noted for S assimilation proteins above (Baudouin-Cornu et al., 2001). In both S. cerevisiae and E. coli, those enzymes specifically involved in processes of C assimilation have, on average, slightly shorter (less C-rich) side chains than the bulk proteome (or S assimilation enzymes). Conversely, enzymes for N assimilation are built, on average, using amino acids with fewer N atoms (Baudouin-Cornu et al., 2001). These effects are relatively small (5–10% reduction), but the fact that this signature of selection is visible at the proteomic level is rather remarkable. Elemental composition is not the only factor that affects amino acid selection at otherwise neutral positions in protein sequences. It has been shown, for example, that amino acid selection is also driven by the energetic demands of amino acid biosynthesis (Akashi and Gojobori, 2002) or even periodic fluctuations in the predominant metabolic pathways at different metabolic phases of the yeast cell cycle (de Bivort et al., 2009).
3. Recycling: macromolecular turnover as a way of redistributing C, N, and S
Elemental sparing responses can significantly reduce cellular demand for S, but are relatively ineffective in the face of the ubiquitous use of C and N in cell constituents. While elemental optimization of protein sequences provides some additional efficiency for utilization for all three of these macronutrients (C,N,S), the gains are incremental. Ultimately, when no external sources can be scavenged, and cell growth becomes severely limited for macronutrients, processes of recycling assume great importance.
Recycling of nutrients within and between cells typically involves the degradation of abundant cellular components to release their molecular (e.g. amino acid and nucleotide) and elemental constituents. In multicellular populations, entire cells may serve as nutrients in this way. Even if we restrict ourselves to the microbial world, this type of process is likely widespread as noted, for example, in the cannibalism response of B. subtilis. When growing populations of B. subtilis become nutrient limited, they synthesize toxins that kill and lyse non-starved cells of the population, which thereby delays entry into sporulation (Gonzalez-Pastor, 2011). Similar processes of fratricide occur in other, genetically homogeneous populations of cells (Claverys and Havarstein, 2007) and it has been suggested that toxin:antitoxin modules may contribute to programmed cell death in a sort of altruistic suicide triggered, at least under some conditions, by nutritional stress (Engelberg-Kulka et al., 2006).
Individual cells recycle nutrients by the selective degradation of surplus or replaceable constituents. This general process is termed autophagy (eating of self), but can be further classified as ribophagy, mitophagy, and so forth, depending on the structure or organelle that is targeted for destruction. The physiological role of autophagy is not always clear, although recycling of nutrients is one obvious benefit. Autophagy is also related to quality control and serves to selectively degrade non-functioning or damaged organelles. Studies in animal models indicate that autophagy helps prevent numerous degenerative diseases and may be a key mediator of the beneficial effects of exercise (He et al., 2012).
a. Recycling of ribosomes (ribophagy)
Ribosomes represent a major fraction of cell mass in rapidly growing bacteria and the cell’s requirement for ribosomes scales linearly with growth rate. During rapid growth, ribosomes may comprise close to 1/3 of the dry mass of a rapidly growing bacterium, but far smaller numbers are needed for the slow growth during elemental deprivation or for maintanence of the cell during non-growing (stationary) phases that are imposed by elemental limitations. Early studies in E. coli revealed that ribosomes are degraded in cells presented with C or N limitation (Kaplan and Apirion, 1975, Zundel et al., 2009). Degradation appears to occur during the transition to stationary phase, and is correlated with the formation of free ribosomal subunits (Zundel et al., 2009, Piir et al., 2011).
Recent studies have begun to reveal the pathways of rRNA degradation and their regulation (Deutscher, 2009). Degradation of ribosomal subunits in response to nutrient starvation is initiated by a pathway distinct from the ones operating for quality control during growth (Basturea et al., 2011). Turnover of ribosome subunits begins, in the case of the 30S subunit, with the trimming of the 16S rRNA 3'-end by RNase PH. Since this region contains the anti-Shine-Dalgarno sequence, this functionally inactivates the subunit. Another early step in ribosome degradation is endonucleolytic cleavage which, in both subunits, targets the RNA-rich subunit interface. Although the identity of the relevant endonuclease is not yet clear, this may be part of the mechanism by which un-associated subunits are selectively targeted for destruction. Once cleaved, the rRNA molecules are degraded by processive exonucleases including RNase II, RNase R and polynucleotide phosphorylase. These RNases may work in concert with proteins related to the Ro autoantigen (Wurtmann and Wolin, 2010) and may themselves be regulated. For example, RNase R is regulated by protein acetylation which leads to instability during exponential growth and stabilization during stationary phase (Liang et al., 2011).
It is not clear whether degradation of ribosomes serves primarily to liberate nutrients associated with the abundant rRNA (which is ~50% of the ribosome mass), the ribosomal proteins, or both. In most cases, it is rRNA that is monitored as this is technically easier. However, it has also been noted that macronutrient limitation leads to a starvation-specific proteolysis of ~20–40% of total cell protein in E. coli (Nath and Koch, 1971), which would be consistent with degradation of released ribosomal proteins (r-proteins).
Ribophagy is also increasingly appreciated for its role in nutrient cycling in eukaryotic cells. In C. reinhardtii, nitrogen starvation triggers a conversion of vegetatively growing cells into gametes and this differentiation process involves extensive remodeling of the ribosome pool (Martin et al., 1976). Indeed, it has been suggested that mobilization of precursors (nucleotides) from rRNA degradation is needed for DNA replication under these conditions. However, this remodeling of the ribosome pool may also have other roles in the cell such as modulation of translational accuracy (Bulté and Bennoun, 1990).
In S. cerevisiae, the induction of ribophagy upon nutrient limitation involves de-ubiquitination of ribosomal proteins by the Ubp3p/Bre5p ubiquitin protease that then triggers engulfment of the large ribosomal subunit into a vacuole for degradation (Kraft et al., 2008, Lafontaine, 2010). This process, for which the term ribophagy was first coined, involves the targeted delivery of both ribosome subunits to the vacuole, the presumed degradation of their protein and nucleic acid components, and the recycling of macronutrients. Genetic studies indicate that the process of ribophagy contributes to survival during prolonged periods of starvation (Kraft et al., 2008).
b. Recycling of other abundant protein components
In addition to ribosomes, other abundant proteins may be targeted for proteolytic destruction upon nutrient limitation. Indeed, in mammalian cells, protein degradation mediated by the proteasome provides an important source of amino acids for ongoing protein synthesis when amino acid availability becomes limiting (Vabulas and Hartl, 2005). Photosynthetic organisms may also take advantage of protein degradation to release macronutrients when they find themselves N limited. In Synechococcus strain DC2 the light-harvesting phycobiliproteins may constitute 50% of total protein. Phycobilisome complexes in Synechococcus can be targeted for degradation in response to either N or S limitation (Collier and Grossman, 1992) and that degradation appears to be a highly regulated and ordered process (Grossman et al., 1993). It is likely that similar targeted recycling processes will be present in many cell types that devote a large fraction of their biosynthetic resources to one or a few specific proteins.
c. Recycling of sulfolipids as a S source
Many oxygenic photosynthetic organism produce S-containing lipids as part of their chloroplast membrane. For example, sulfoquinovosyl diacylglycerol (SQDG) is a major component of the thylakoid membrane in cyanobacteria and in C. reinhardtii. In the latter organism, SQDG accounts for ~13% of total cell S, and up to 85% of this lipid is degraded within 6–12 hours in response to S limitation. This S recycling mechanism, which is regulated as part of the S acclimation response (Sugimoto et al., 2010), provides a large fraction of the S needed for ongoing protein synthesis. It is formally possible that degradation of an abundant protein (such as Rubisco) could provide a comparable amount of S (and in the more convenient form of Cys and Met) (Sugimoto et al., 2007). Indeed, S starvation does lead to a decrease in Rubisco levels, but this response appears to occur on a much longer timescale than the mobilization of S from SQDG, suggesting that this may be a secondary mechanism (Sugimoto et al., 2007).
B. Phosphorous (P): An indispensable element for information and energy transfer
Phosphorous is required for life and exists in cells primarily as the phosphate anion (PO43−) in various states of protonation and esterification. At neutral pH, phosphate is predominantly in the HPO42− and H2PO4− states; phosphate esters also carry a net negative charge in cells. Phosphate forms the linkage unit of nucleic acids and is therefore essential for the storage, transmission, and expression of genetic information <Figure 7>. Nucleoside triphosphates (NTPs) not only serve as precursors for the synthesis of DNA and RNA, but also function as the universal energy currency in the cell with most biosynthetic processes fueled directly or indirectly by NTP hydrolysis. In E. coli, the total P content is ~3% of dry weight, making P one of the major macronutrients for cell growth (Neidhardt et al., 1990). The ribonucleotides are present at millimolar concentrations and their γ-phosphoryl groups turnover rapidly during growth. Most ATP is synthesized in respiring cells by ATP synthase using the energy of the proton-motive force. The free energy of hydrolysis of the phosphoanhydride linkages drives anabolic metabolism, while energy yielding catabolic metabolism can be coupled to ATP or GTP synthesis. GTP hydrolysis fuels the process of translation, one of the single most energy intensive processes in growing cells.
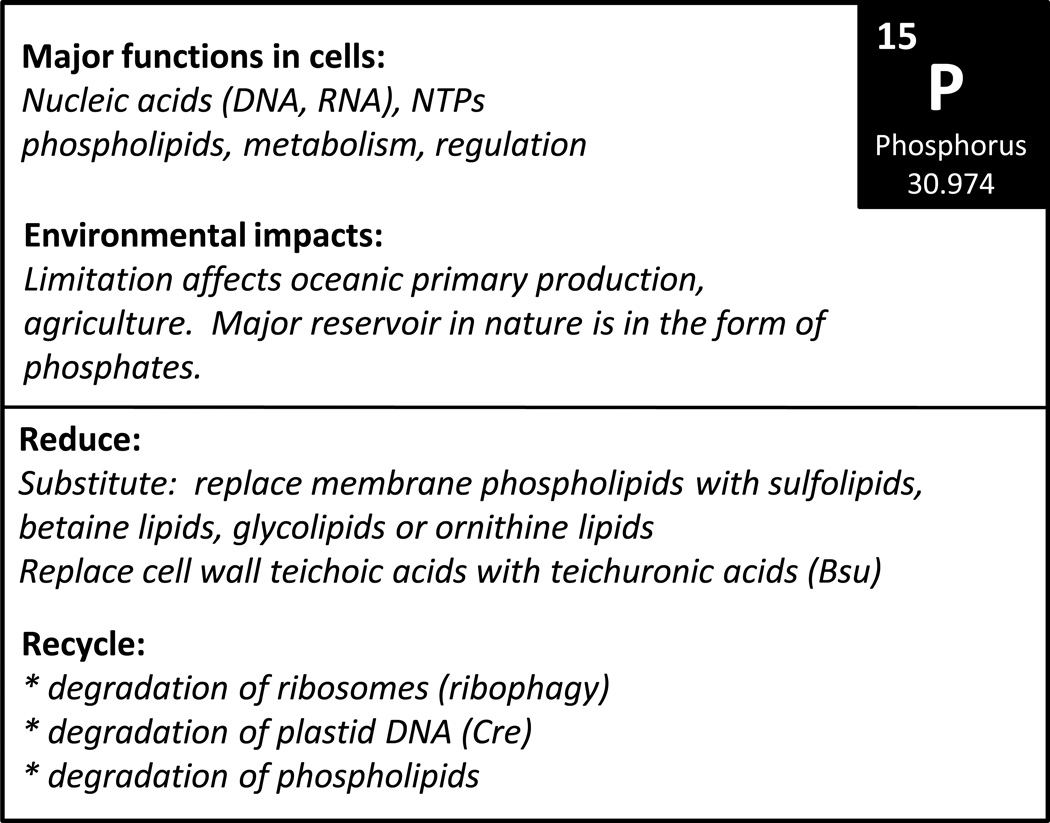
Figure 7.
Overview of the biological roles of P and known sparing and recycling mechanisms.
Phosphorous can also be used for a variety of other functions including most commonly the synthesis of the phospholipids of the membrane lipid bilayer. Phosphorus containing polymers are also abundant in the cell walls of Gram positive bacteria (in teichoic acids). When in excess, phosphate can be polymerized into polyphosphate, which is a potential storage form of P. Although each of these P-containing molecules can be a significant fraction of total cellular P, they are not universally present and may, in some cases, be expressed conditional on P availability (Fig. 7).
While it is generally accepted that P is required for life, it has been speculated that alternative types of biochemistry might be feasible in which P is substituted with As (Wolfe-Simon et al., 2009). This idea grew out of an exercise in trying to imagine what types of alternative biochemical processes might be able to evolve in extreme habitats (or on planets) where the availability of elements is substantially different (Davies et al., 2009). The authors postulate that there might exist on Earth a "shadow biosphere" of alternative chemistries and analysis of these "weird" organisms could shed light on possible lifeforms on other planets. This concept rose to international prominence recently with the publication of an article (Wolfe-Simon et al., 2011) with the unfortunately misleading title of "A Bacterium That Can Grow by Using Arsenic Instead of Phosphorus." In this specific case, a bacterium tolerant to growth in very high concentrations of As was suggested to use As in place of P in some of its macromolecules, and a specific claim was made for incorporation (albeit at a very low level) into DNA. Nevertheless, the evidence presented did not meet biochemical criteria for establishing chemical linkages (see Comments published in response to the article). One of the key concerns is that replacement of a significant amount of P by As is chemically implausible. The phosphoanhydride linkage is special because its hydrolysis has a high activation energy (requiring catalysis in vivo) but a negative free energy change, which drives group transfer reactions. Interested readers will find further discussions of these results in numerous online blogs, news commentaries, and several published articles (Danchin, 2010, Rosen et al., 2011, Silver and Phung le, 2011). As described below, there are in fact numerous examples of elegant solutions to the problem of limited P availability, and these can reduce the P requirements for growth significantly, but not eliminate this requirement completely. These responses often target P-rich components of the cell wall or membrane rather than NTPs or nucleic acids.
1. Acclimation to phosphorous limitation: cell wall remodeling in Bacillus subtilis
Growth of B. subtilis in phosphate limiting conditions activates a complex acclimation process regulated by the PhoPR two-component system (Hulett, 1996). A central feature of this acclimation mechanism is a remodeling of the cell wall as a mechanism to optimize growth. In media containing sufficient phosphate (~2 mM or more), B. subtilis cell walls contain an abundant anionic polymer known as teichoic acid that accounts for nearly 50% of cell wall weight (Bhavsar and Brown, 2006). Teichoic acids are alternating copolymers of glycerol and phosphate and can be linked to a glycolipid carrier (in lipoteichoic acid) or to the peptidoglycan cell wall (in wall teichoic acid). Cells grown in P limited medium repress the expression of the wall teichoic acid biosynthetic pathway and activate the expression of an alternative anionic polymer, teichuronic acid (Qi and Hulett, 1998, Lahooti and Harwood, 1999). In teichuronic acid, the carboxylates provide the negative charge that the phosphates provide in the teichoic acids. This is not necessarily a complete replacement, since the synthesis of teichoic acids may continue at a low maintenence level even under P limitation (Botella et al., 2011). Nevertheless, this remodeling of the cell wall is found to significantly reduce cellular P demand. Cells grown with limiting P contain only 32–47% as much P as cells grown in P-replete medium and the amount of P in the cell wall fraction was reduced by 25-fold (Lang et al., 1982). The ability of cells to reduce their P demand by a factor of 2 or more, simply by altering the composition of the cell wall (while maintaining a comparable level of anionic polymers in the wall), is an efficient mechanism for optimizing P usage. It is not yet clear to what extent other P-containing macromolecules (including lipoteichoic acid) may be functionally replaced, nor is it clear whether pre-existing teichoic acid is scavenged as a P source, although this seems reasonable.
2. Adaptation and acclimation by membrane phospholipid remodeling in Bacteria
Membrane phospholipids are responsible for a large fraction of the phosphorus content of both prokaryotic and eukaryotic cells. Heterotrophic and phototrophic microorganisms face P deficient growth conditions in unfertilized soils as well as in aquatic environments (Bieleski, 1973). P deficiency has been described for many regions of the open ocean, yet it does not appear to limit productivity because the inhabiting Prochlorococcus species have adapted to the low P content of that niche by reducing their P quota to 1:500, which is substantially lower than the Redfield ratio of 1:106 (Bertilsson et al., 2003). This is accomplished by replacement of membrane anionic phospholipids by anionic sulfolipids, specifically sulfoquinovosyl diglyceride (SQDG): less than 1% of the assimilated P is incorporated into membrane lipids, which spares P for nucleic acid and nucleotides, where its function is irreplaceable (Van Mooy et al., 2006). In comparing the fate of P assimilate in the Sargasso sea where P levels are <10 nM to the South Pacific subtropical gyre with 10-fold higher P concentrations, the authors noted 1.3% allocation to phospholipid in the former vs. 17% in the latter (Van Mooy et al., 2009). In parallel, analysis of membrane lipids from these locations showed that S- and N-containing lipids were more abundant in the former vs. the latter, and it was suggested that this P sparing adaptation is important for the success of prokaryotic Prochlorococcus and cyanobacterial species as well as eukaryotic phytoplankton in these environments. Measurements of various lipid types in laboratory experiments with P-replete vs. –deplete conditions indicate that the ability of the prokaryotes to synthesize a sulfolipid spares up to 43% of the P quota.
The replacement of phospholipids is a common acclimation response in bacteria faced with P deficiency. It has been documented in rhizobia, where SQDG, ornithine-containing lipids and diacylglyceryl trimethylhomoserine are used as substitutes, in Pseudomonas, where acidic glycolipids replace phospholipids, in gram positive Marinococcus species, where sulfolipid replaces phosphatidylglycerol, and anoxygenic photosynthetic bacteria where each of these replaces phospholipids in response to P deficiency (e.g. Minnikin et al., 1974, Benning et al., 1995, Geiger et al., 1999, Sprott et al., 2006). Cyanobacterial mutants blocked in sulfolipid synthesis or rhizobium mutants that are unable to synthesize ornithine or betaine lipids are growth compromised only in P-deficiency, indicating the importance of lipid substitution as a P-sparing mechanism (Güler et al., 1996, López-Lara et al., 2005).
Lipid substitution is an effective P-sparing response and degradation of pre-existing phospholipids also provides a P-recycling mechanism. Molecular genetic analysis in Rhizobium meliloti indicates that P-recycling is part of the phosphate-deficiency program mediated by the response regulator PhoB. A specific intracellular phospholipase C is induced so that the phospholipids (whose function can be covered by non-P-containing molecules) can be used as a pool of mobilizable P, which is recycled for molecules in which P is essential (like nucleotides) (Geiger et al., 1999, Zavaleta-Pastor et al., 2010).
3. P-sparing and recycling in Chlamydomonas
Because of the importance of P in agriculture (it is one of the major constituents of fertilizer), there is an excellent understanding of P metabolism and its regulation in reference organisms like C. reinhardtii (reviewed in Irihimovitch and Yehudai-Resheff, 2008, Moseley and Grossman, 2009). Three phospholipids (phosphatidylglycerol, -ethanolamine and –inositol) are prevalent in membranes of P-replete cells. In P-deficiency, phosphatidylglycerol is reduced by as much as 50%, concomitant with a greater than 2-fold increase in sulfolipids. The importance of this substitution is evident from the phenotype of an sqd1 sulfolipid biosynthesis mutant, which grows poorly under P starvation (Riekhof et al., 2003). Nevertheless, because of specific binding sites for lipids in membrane protein complexes (e.g. phosphatidylglycerol in photosystem II; PS II), sulfolipid cannot completely replace phospholipid in the thylakoid membrane (Yu et al., 2002).
P sparing via membrane lipid re-modelling is conserved throughout the plant lineage, and has been documented in moss, Arabidopsis and perennial rye grass, species that are separated from Chlamydomonas by a billion years of evolution (e.g. Yu et al., 2002, Wang et al., 2008, Byrne et al., 2011). In Arabidopsis, SQD1 mRNA and protein are dramatically increased in P-deficient plants and promoter-reporter fusions implicate transcriptional regulation (Essigmann et al., 1998, Hammond et al., 2003). In bacterial, algal, as well as Arabidopsis sqd mutants, the level of glycerolipids is maintained (or even increased), recycling of P is precluded, and mutant cells grow poorly in P-deficient conditions. The eukaryotic phytoplankton, such as the diatoms Thalassiosira pseudonana and Chaetoceros affinis and the coccolithophorid Emiliana huxleyi, use both sulfolipids and betaine lipids as substitutes for phospholipids, which spares about 10–30% of the P quota (Van Mooy et al., 2009, Martin et al., 2011).
Nucleic acids represent the other major reservoir of P. In C. reinhardtii, where the plastid genome is polyploid with up to 80 copies per cell, there is evidence for copy number reduction, which could release P for recycling to other processes. Interestingly, plastid mRNA abundance increases under P limitation because of down-regulation of a polynucleotide phosphorylase, a phosphorylytic enzyme responsible for cpRNA degradation which requires phosphate as a substrate. The regulation is dependent on Psr1, a Myb-domain transcriptional activator, which also turns on extracellular phosphatases and assimilatory transporters in P-deficient C. reinhardtii (Wykoff et al., 1999, Yehudai-Resheff et al., 2007). The up-regulation of nucleases in P-deficient plants has been noted in moss and Arabidopsis, but specific intracellular targets have not yet been identified and therefore whether this nucleic-acid based acclimation response extends beyond C. reinhardtii is not known.
III. Microbial Adaptation and Acclimation to Metal Ion Limitation
Some metal ions are essential or beneficial for life, but others are neutral or even harmful, and some of the beneficial ones can become harmful when they are present in excess. The importance of metals in biology is reflected in the maturation of bioinorganic chemistry as a distinct discipline with its own meetings, societies, and journals including, for example, Bioinorganic Chemistry (Elsevier; initiated 1970), Biometals (Springer; 1997) and, more recently, Metallomics (RSC publishing, 2009).
It is estimated that 30% or more of proteins contain at least one metal cofactor in most organisms (Waldron et al., 2009, Seravalli and Ragsdale, 2010). The Zn metalloproteome is typically 5–6% of proteins in Bacteria to near 10% in Eukaryotes, with Fe-containing proteins having an inverse trend (Andreini et al., 2009). The key chemical properties of metal ions that contribute to their essential roles are their ability to serve as electron carriers and to function as electrophilic centers in catalysis. The midpoint potentials of many biological Fe and Cu centers are well suited for their electron carrier roles, and Fe, Zn, Mn, Co, and Mg are well suited for roles as electrophilic catalysts. Metal ions can also serve as organizing centers for the folding of small protein domains as exemplified by Zn finger proteins or Ca-binding domains, which are abundantly represented in eukaryotes.
Microbial responses to metal ions typically follow a gradient including states that can be defined as metal-limited, metal-deficient, metal-replete, or metal-excess. Limitation, also known as starvation, refers to a lack of an essential metal that leads typically to a cessation in the ability of the cell to grow and may lead to a loss of viability. Deficiency refers to a sub-optimal level of metal availability that impacts cell physiology and leads to measurable alterations in metabolism. Metal-replete conditions are those that provide enough metal to support growth and the accumulation of the full quota of proteins that require that metal, whereas metal excess refers to conditions where high concentrations of metals begin to negatively impact growth or may lead to cell death. As cells transition from replete to deficient to limited conditions they engage a variety of acclimation responses which include, as for the macronutrients, elemental acquisition, mobilization, sparing, and recycling. Conversely, as cells transition from replete to excess, they engage mechanisms to store excess metals for future use or may efflux the ions from the cell. Although detailed metal requirements vary significantly between species, similar acclimation strategies have evolved in multiple organisms across phylogenetic boundaries.
A. Metal homeostasis across three domains of life
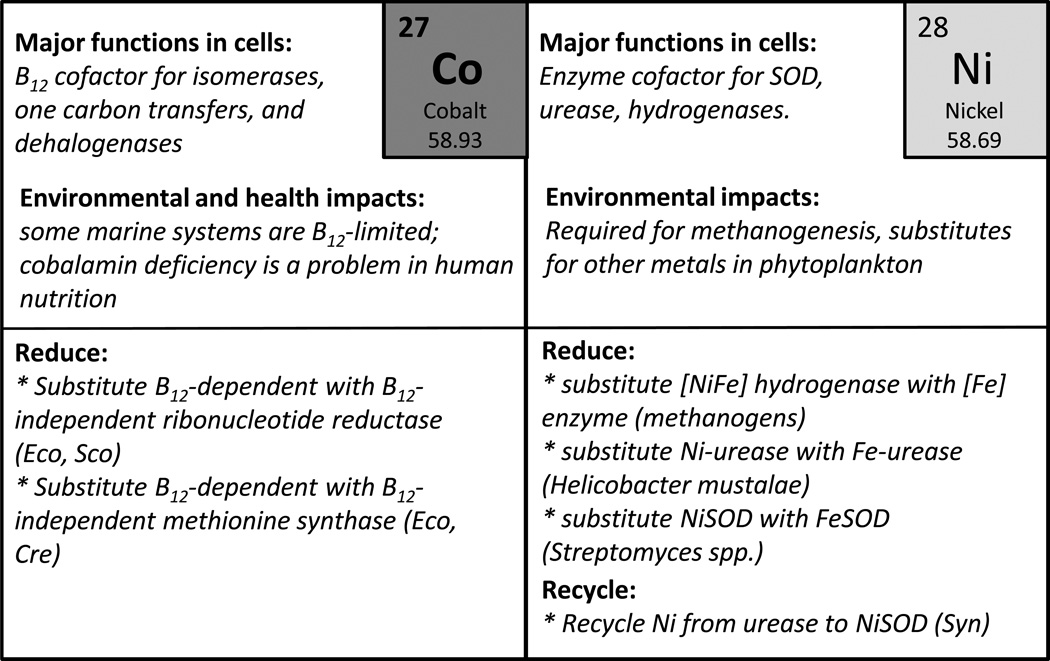
The use of metals within biology differs substantially between the Bacteria and Archaea and the Eukarya. In some cases, these differences reflect adaptations to specific niches. For example, the Archaeon Ferroplasma acidiphilum grows in highly acidic environments rich in Fe(II), the more bioavailable oxidation state, and routinely uses Fe in place of Zn to organize metalloprotein domains (Ferrer et al., 2007). More generally, the prevalence of different metal ions in various organisms reflects their evolutionary history. Life evolved in the ancient oceans and, in many ways, cells reflect the chemical composition of this environment (Dupont et al., 2010). The ancient (Archaen) ocean in which life evolved (beginning ca. ~4.5 billion years ago; GYA) was anoxic and chemically reducing with relatively high concentrations of Fe, Mn, Ni, and Co. The evolution of oxygenic photosynthesis contributed to an increase in atmospheric oxygen (~2.4 GYA) leading to an eventual transition to an ocean with oxygen-rich surface waters. As the ocean waters became more oxidizing (by ~0.8 to 0.5 GYA), Cu, Zn, and Mo became abundant whereas Fe, Mn, Ni, and Co were greatly reduced in abundance. Thus, Fe-containing proteins like cytochromes and ferredoxins and many Ni-containing enzymes are ancient, while Cu proteins are a more recent innovation (Crichton and Pierre, 2001).
With the availability of thousands of microbial genomes, and an ever improving ability to discern metal specificity from protein sequence, it has been possible to distinguish several trends in microbial metal ion utilization (Dupont et al., 2010). Bacteria and Archaea contain a core group of Zn proteins, often involved in central metabolism (transcription, translation), and comparatively more Fe, Mn, and Mo proteins. Assigning metal specificity to protein domains is not trivial, but it is notable that many of the earliest evolving protein domains appear to be cambialistic (able to use or function with multiple metal cofactors). Later evolving Eukaryotes make much greater use of Ca, Cu, and Zn, with the latter represented by the proliferation of Zn finger proteins. Whereas many non-heme Fe enzymes are conserved in all Domains of life, consistent with their early appearance in evolution, many Cu enzymes are specific to the eukaryotes and likely evolved after the great oxidation event when the bioavailability of Cu as cupric ion increased (Crichton and Pierre, 2001, Andreini et al., 2009). Conversely, the use of Ni as an enzyme cofactor appears to have declined over time (Zhang et al., 2009).
Metal ion deficiencies are also widely appreciated in both medicine and agriculture. For instance, Fe deficiency anemia alone is estimated to affect nearly 1.7 billion people (25% of the population) (Benoist et al., 2008). Similarly, Fe-deficiency chlorosis is a common problem in agriculture, especially in alkaline soils. The consequences of excess exposure can also be severe, even for those metals required for life. Sophisticated metal ion homeostasis systems are therefore operative in cells to prevent the adverse effects of either deficiency or excess. Defects in metal ion homeostasis pathways, often involving proteins first defined in model systems, are increasingly recognized as a source of genetic disorders (Bleackley and Macgillivray, 2011).
Competition for limiting metal ions can be a determining factor for the outcome of host-pathogen interactions. The Fe-withholding response is an important feature of the innate immune system in mammals (Ganz, 2009). Bacterial pathogens have responded to the low levels of available Fe in the human host by elaboration of very high affinity siderophores such as enterobactin made by E. coli and its relatives (Fischbach et al., 2006). Humans have responded by the synthesis of a high affinity siderophore binding protein known as lipocalin or siderocalin. As this evolutionary arms race has continued, Salmonella has evolved the ability to decorate enterobactin with glycosyl groups (generating a family of compounds known as Salmochelins), thereby rendering lipocalin ineffective (Fischbach et al., 2006, Muller et al., 2009). The success of macrophages in killing engulfed bacteria can also depend on metal ion competition: the NRAMP family of divalent metal ion transporters were named for their role as "natural resistance associated macrophage proteins." Subsequent to engulfment, NRAMP proteins deplete the phagocytic vacuole of Mn (and possibly Fe) thereby limiting bacterial growth (Cellier et al., 2007). Many bacteria, in turn, also have NRAMP family transporters working to import these very same cations. Neutrophils also play a role in the competition for metals by secreting a protein, calprotectin, that sequesters Mn and Zn (Corbin et al., 2008, Kehl-Fie and Skaar, 2010, Kehl-Fie et al., 2011). Metal toxicity is also employed as part of the macrophage killing arsenal by the delivery of redox active Cu to the phagosomal compartment (White et al., 2009, Wakeman and Skaar, 2011). Genetic studies indicate that, individually and collectively, these mechanisms can have a large impact on the outcome of infection (White et al., 2009, Haley and Skaar, 2011, Hammer and Skaar, 2011).
B. Challenges in defining the roles of metals in biology
The concept of the metallome, as first coined by RJP Williams (Williams, 2001), refers to the quantitative description of the metal contents of cells. Ideally, metallomics seeks to describe both the amounts of each metal required for life and their distribution within the cell and its sub-cellular compartments. Clearly, the metallome differs between organisms and depends on the precise growth conditions studied. For many metals, the major fraction of the metallome is bound to proteins (the metalloproteome). However, efforts to define the nature of the metalloproteome are still in their early stages and there are many surprises still in store (Cvetkovic et al., 2010, Seravalli and Ragsdale, 2010).
The application of high sensitivity techniques for elemental analysis, such as inductively-coupled plasma mass spectroscopy (ICP-MS), allows the quantitation of elements down to at least the 1 part-per-billion (ppb) range. It is thereby possible to determine the elemental composition of cells with very high sensitivity and over many orders of magnitude of concentration. The amount of metal ion per cell is sometimes referred to as the metal quota. The very sensitivity of this technique, however, leads to additional challenges because of the ease with which samples can be contaminated. Analyses of trace elements in environmental samples, and in particular ocean waters (Sohrin and Bruland, 2011), have required the development of sophisticated sampling methods and the corollary laboratory studies typically require extensive efforts to prevent contamination including the use of clean-rooms (e.g. Shiller and Boyle, 1985, Trefry et al., 1985, Tortell and Price, 1996).
While elemental analyses of cells can define metal ion quotas, these measurements do not distinguish metal ions bound to proteins or other macromolecules relative to those that are hydrated or bound to low molecular weight ligands. Nor do these measurements, by themselves, distinguish between cell compartment and organelles or between metal that is required for metabolism or stored for future use. When grown under replete conditions, much of the metal may be in storage compartments rather than in active use. Conversely, the metal remaining in metal-limited cells is likely to define a lower limit capable of supporting growth under the tested conditions. The metal quota of a replete cell was used recently in an approach to devise a trace element mix for C. reinhardtii (Kropat et al., 2011). This resulted in removal of non-beneficial elements (Co, B) and reduction of others (Zn, Mn) to provide only 3-fold the quota of a healthy cell, which allows room for accommodating an increased quota in situations of altered physiology.
The distribution of metals between their various possible coordination environments (referred to as speciation) varies enormously between metals, but is often quite similar between cells, governed as it is by the fundamental properties of the ions themselves (Waldron and Robinson, 2009). For example, Mg(II) is very soluble as a hydrated ion and is often present at high concentrations in cells where it is frequently complexed with phosphoryl groups in nucleic acids and other metabolites. Conversely, metals like Zn and Cu are tightly bound either to proteins or other chelating groups within the cell (Colvin et al., 2010, Robinson and Winge, 2010). As a result, the equilibrium concentration of free ions is sub-picomolar, although there is nevertheless a substantial pool of ions within the cell that is kinetically accessible for incorporation into nascent metalloproteins (Finney and O'Halloran, 2003). For some metals, specific protein "chaperones" ferry metals from their sites of uptake to their target proteins (O'Halloran and Culotta, 2000). Such metallochaperones play important roles in the delivery of Cu to specific proteins (Robinson and Winge, 2010), in the assembly of Fe/S clusters (Subramanian et al., 2011), in the insertion of Fe into protoporphyrin to generate heme, and the insertion of Ni into urease (Carter et al., 2009).
The speciation of metals in cells is governed by both kinetics and thermodynamics. The thermodynamics is described by the Irving-Williams series (Mn(II) < Fe(II) < Co(II) < Ni(II) < Cu(II) > Zn(II)). The ability of Cu to bind more tightly to ligands than other ions leads to toxicity, in part owing to the disruption of Fe/S centers in enzymes (Macomber and Imlay, 2009, Chillappagari et al., 2010). Kinetics is also important, since the ease with which metal-ligand bonds can be exchanged can determine the availability of the metal, both environmentally and within the cell. Intracellular movement of Cu is controlled by protein-protein interactions involving metal binding domains on target proteins and metallochaperones (Tottey et al., 2005, Boal and Rosenzweig, 2009). Proteins that require Cu for function are therefore metallated only by a specific Cu chaperone or after export from the cytosol (Tottey et al., 2008).
Defining which metals are absolutely essential for growth is experimentally challenging even in the best understood model systems. Since many microorganisms can be grown in chemically defined media, it would appear to be easy to define the minimal elemental requirements for growth. Indeed, in the simplest cases the requirements are sufficiently high that they can be determined by monitoring growth as a function of added metal ion. However, in other cases the low level requirements, contamination of reagents and growth chambers, and the presence of high affinity acquisition systems make this approach problematic. Indeed, the impressive ability of cells to scavenge miniscule quantities of essential metals from their environment is a recurring challenge in metal ion nutrition studies. High affinity transport systems are likely present for all essential metals and may involve the secretion of high affinity chelators such as siderophores for Fe. Researchers sometimes rely on chelators to impose metal limitation, but chelation reduces but does not eliminate the bioavailability of the metal and has the added complication that chelators are generally not specific for a single element. The use of high purity chemicals and acid-washed glassware is important for reproducibility in laboratory experiments (e.g. Cox, 1994, Quinn and Merchant, 1998). However, metal ions tend to slowly leach even from acid-washed glassware and for some studies more stringent conditions are needed.
In one notable example, experiments to study the effects of Zn depletion in E. coli required the pretreatment of all non-metal media components with a solid-phase metal chelator (Chelex-100) and the use of a chemostat made from non-metal components (Graham et al., 2009). Cells were pregrown in Zn free chemically defined medium and yet, when the cells were recovered they contained more Zn than was added to the entire volume of growth medium used for their culture. This anomaly can be accounted for by the ability of cells to leach Zn from within the glass walls of the culture flask, despite the prior removal of bound metals by acid-washing. Indeed, the extent of Zn limitation increased with subsequent uses of the same flask, suggesting that E. coli is more efficient at removal of excess Zn from the culture flask than chemical chelation and acid-washing (Graham et al., 2009). Similar observations were made in laboratory experiments to generate Mn-deficiency in C. reinhardtii (Allen et al., 2007b).
Ultimately, the goal of metallome studies is to define both the metals needed for cellular functions, and to identify the enzymes that require metals for their activity. While the study of metalloenzymes in vitro has a long history, there are now several examples where the cofactor required for in vivo function was initially misassigned (Jain et al., 2005, Chai et al., 2008). This reflects the fact that metalloenzymes are often assayed under conditions that may not mimic the levels of metal ion availability in the cell. Moreover, many Fe-containing enzymes must be assayed anaerobically and, when assays are done aerobically, the strongest activation will often be provided by a different ion (Tripp et al., 2004).
An additional complexity in defining the metals essential for life is that, for some functions, metal ions play redundant roles. For example, two enzymes may each use a different metal ion to catalyze the same reaction and thereby be functionally redundant. This is the case, for example, with the multiple SOD isozymes in many organisms. In other cases, a single protein may be able to function with more than one ion. Such cambialistic enzymes include a subset of SODs (Priya et al., 2007), some carbonic anhydrases (Lane and Morel, 2000b), and a lipid A biosynthetic enzyme (Gattis et al., 2010). More generally, it has been suggested that many E. coli enzymes that may normally function with a non-heme Fe as cofactor may, under conditions of oxidative stress, use Mn instead (Anjem et al., 2009). Although there are relatively few well defined examples to date, these studies suggest that functional redundancy is likely to be more widespread than generally appreciated. One result is that the required levels of each metal may be interdependent, especially for abundant metalloenzymes. For example, for some phytoplankton limitation for Zn can be partially alleviated by Co or Cd (Morel, 2008). In marine systems, low Zn levels are also correlated with low P availability, which is problematic since alkaline phosphatase, a key P acquisition enzyme, is itself a Zn enzyme. To circumvent this Zn-P colimitation, many Prochlorococcus species contain a distinct Ca-dependent phosphatase, PhoX (Kathuria and Martiny, 2011). These complex interactions between elements emphasize the challenges inherent in defining a minimal or representative metallome for any organism.
Recent years have seen the development of powerful new techniques for monitoring metal ion speciation in cells, including both genetically encoded and chemical sensors to monitor metal availability in living cells (reviewed in Cook et al., 2008, Domaille et al., 2008, McRae et al., 2009, Vinkenborg et al., 2010, Palmer et al., 2011), nanometer-scale imaging Secondary Ion Mass Spectrometry (nanoSIMS) and related techniques to monitor spatial distributions of metals on a sub-micron scale (e.g. Orphan and House, 2009, Byrne et al., 2010), and fractionation techniques for monitoring metalloproteomes (Cvetkovic et al., 2010). These analytical techniques, particularly when applied to genetically amenable model organisms, will likely enable new insights into the diverse roles of metal ions in cells and, as a corollary, increase our understanding of the corresponding acclimation mechanism when metals become limiting.
C. Iron (Fe): A near universal transition metal and redox center
Iron is required for the growth of nearly all cells. In cells, Fe is found in heme proteins such as catalase and cytochromes, iron-sulfur cluster (Fe/S)-containing electron carriers and enzymes, and non-heme Fe enzymes. In most cell types, Fe has multiple essential roles in the cell. For instance, both heme and Fe/S clusters are commonly essential for cell growth <Figure 8>.
Figure 8.
Overview of the biological roles of Fe and known sparing and recycling mechanisms.
The only known exceptions to this general requirement are certain Bacteria that grow in severely Fe-limited environments. For example, the Lactobacilli and the spirochete Borrelia burgdorferi (causative agent of Lyme disease) are thought not to require Fe (Weinberg, 1997, Posey and Gherardini, 2000). In the case of B. burgdorferi, there are at most a handful of Fe atoms present in the cell and no known or demonstrable requirement for Fe for growth (Posey and Gherardini, 2000). In addition to dispensing with heme and Fe/S-containing proteins, enzymes that in other organisms often require Fe for catalytic activity use a different metal in these systems. For example, in both L. plantarum and B. burgdorferi peptide deformylase contains a catalytically essential Zn in place of what would normally be Fe (Nguyen et al., 2007). This is an illustration of the general principle that one way in which cells adapt or acclimate to metal limitation is to replace one metal cofactor with another.
1. Overview of molecular mechanisms of adaptation and acclimation to Fe-limitation
Most bacteria, and all known eukaryotes, do not have the luxury of simply dispensing with a requirement for Fe. For these organisms, Fe is essential for growth and the response to Fe limitation is to reduce, to the maximal extent possible, the burden placed on the cell by the need to obtain Fe. In many different microbial systems, acclimation to Fe limitation involves Fe-sparing and Fe-recycling mechanisms analogous to those discussed above for macronutrients. The Fe-sparing response reduces the requirement for Fe-containing proteins by synthesizing alternative enzymes using either organic cofactors or other metal ions in place of Fe, and concurrently shutting off the synthesis of low-priority Fe proteins. In this context, low priority refers to those functions that are not absolutely essential for growth or which can be functionally replaced by other proteins and pathways. In some cases, as we will see, cells may also recycle Fe: they actively degrade pre-existing Fe-containing protein complexes simply to recover the valuable sequestered Fe.
Acclimation to Fe limitation requires, first, that cells have mechanisms to monitor their Fe status and alter gene expression appropriately. In Bacteria, iron homeostasis is regulated by specific Fe-sensing metalloregulatory proteins, such as the ferric uptake repressor (Fur) and the diphtheria toxin repressor (DtxR), which serve to sense the cytosolic availability of Fe(II) (Hantke, 2001, Andrews et al., 2003). In S. cerevisiae and other fungi, Fe-acclimation responses are coordinated by functionally equivalent metalloregulatory proteins such as Aft1 and Aft2 (Philpott and Protchenko, 2008), whereas C. reinhardtii may use orthologs of the regulatory proteins found in Arabidopsis (Long et al., 2010).
Fur is representative of a large group of metalloregulators that sense metal ions such as Fe, Zn, Mn, and Ni to regulate metal homeostasis (Lee and Helmann, 2007). Typically, Fur family regulators act as metal-dependent transcriptional repressors, although there are exceptional organisms where Fur proteins can also act as direct activators by binding upstream of target genes and enhancing the recruitment or activity of RNA polymerase (Delany et al., 2004, Danielli et al., 2006). Under Fe replete conditions, Fur binds Fe(II) and represses the expression of uptake function and activates the expression of Fe storage proteins. Conversely, when Fe levels drop, iron uptake functions are derepressed and pathways contributing to acclimation are induced.
In response to Fe deficiency, induction of the Fur regulon leads to the expression of iron acquisition systems. Fur directly regulates the synthesis of high affinity uptake systems and chelators (siderophores) for import of Fe. This can potentially relieve the shortage, but only if accessible Fe is present in the environment. Fe may also be mobilized from intracellular stores including ferritins, bacterioferritins, and mini-ferritin/Dps family proteins. Ferritins can sequester up to ~5000 atoms of Fe while the smaller miniferritins can sequester 500 atoms within their spherical protein cores (Bevers and Theil, 2011). Although the pathways of Fe mineralization into these storage proteins are well studied, the processes by which this Fe is mobilized upon starvation are less clear (Smith, 2004). Fe mobilization likely involves the gating of specific pores (Liu et al., 2003, Bevers and Theil, 2011), and presumably also requires either a specific or non-specific reduction of the oxo-Fe core to release soluble Fe(II). In mammals, autophagy by degradation in the lysosome also plays a role in Fe recovery (De Domenico et al., 2009). As cells transition from iron deficient to iron limited conditions, they additionally engage programs of Fe-sparing and, in some systems, Fe-recycling.
Insights into mechanisms of adaptation and acclimation to Fe limitation emerged early in ecological studies of Fe-limited systems and, in particular, studies of the marine ecosystem and its constituent microorganisms. These studies are complemented by detailed analyses of molecular mechanisms in reference organisms (including the Bacteria E. coli and B. subtilis and the Eukarya S. cerevisiae and C. reinhardtii) that have provided insights into how cells acclimate to changes in Fe availability.
2. Fe-sparing and Fe-recycling: insights from marine picoplankton
Primary productivity of about 30% of the world’s ocean is Fe-limited, due in part to the high Fe-demand for photosynthesis (Raven et al., 1999, Behrenfeld et al., 2009). In addition, Fe-containing proteins are widely used for various redox reactions in cellular metabolism. Ocean surface waters often contain very low levels of Fe, with measurements indicating between 0.01 and 2 nM (Sandy and Butler, 2009). Marine bacteria, like many other Fe-limited organisms, synthesize and secrete siderophores (Sandy and Butler, 2009), express high affinity transport systems, and implement complex acclimation strategies to maximize growth in the face of elemental limitations. Here, we focus specifically on adaptations that have emerged in this environment and on documented examples of acclimation by Fe-sparing and Fe-recycling.
a. Fe-sparing by substitution of ferredoxins with flavodoxins
Perhaps the most widespread acclimation strategy for Fe-limitation is replacement of the electron carrier protein ferredoxin with an Fe-free alternative, flavodoxin. Ferredoxins are Fe/S proteins with negative midpoint potentials and they serve as electron donors in many biosynthetic reactions and, accordingly, are often abundant. In 1966, a protein with ferredoxin activity was isolated from Clostridium pasteuranium cultured in iron-poor medium (Knight et al., 1966). This protein had a bound flavin and was named flavodoxin. Subsequent studies revealed that the substitution of ferredoxin with flavodoxin allows the former to be actively degraded in order to release and recycle Fe for maintenance of pyruvate synthase (another Fe/S protein) (Schönheit et al., 1979).
The loss of ferredoxins appears to have emerged as a genome adaptation for a clade of Prochlorococcus that is associated with very low Fe (estimated at <0.5 nM) in high nutrient, low chlorophyll (HNLC) regions of the ocean (Rusch et al., 2010). Metagenomic analyses suggest that Prochlorococcus strains from this environment have lost ~10% of the estimated 60 or so predicted Fe-containing proteins found in similar strains from more Fe-rich environments. The missing genes encode two ferredoxins, a plastoquinol terminal oxidase, and a cytochrome. Thus, one mechanism for adapting to a chronic limitation for Fe is to reduce the number of Fe requiring proteins encoded in the genome (Rusch et al., 2010).
A more common evolutionary strategy is to retain the genes for both ferredoxins and flavodoxins, but only express the former when Fe is relatively abundant. The Fe-regulated substitution of ferredoxin with flavodoxin as a major electron carrier has been noted in numerous organisms including a chlorophyte alga, several cyanobacterial species, cryptomonads, and diatoms (e.g. Zumft and Spiller, 1971, Hutber et al., 1977, Sandmann and Malkin, 1983, Ragsdale and Ljungdahl, 1984, McKay et al., 1997, Li et al., 2004). The reciprocal pattern of expression is consistent with an Fe-sparing mechanism, since the substitution will decrease the Fe quota of the cell. The phenomenon is so widespread in nature that flavodoxin abundance is now the accepted standard biomarker for assessing Fe-status in the marine environment (La Roche et al., 1996). Down-regulation of ferredoxin in Fe-deficiency occurs even in organisms that do not have genetic information for flavodoxin. In these situations ferredoxin is essential and Fe-deficient cells reduce the abundance of ferredoxin, but do not eliminate it entirely (e.g. Pardo et al., 1990, Terauchi et al., 2010).
In Synechococcus sp. PCC7942, the regulation of ferredoxin occurs by Fe-dependent stabilization of the petF transcript. Induction of flavodoxin, encoded by the iron-starvation inducible gene isiB, occurs at the level of transcription (Leonhardt and Straus, 1992, Bovy et al., 1993). In many cyanobacteria, isiB is part of an Fe-deficiency stress operon that also encodes a modified antenna for the Fe-rich photosystem I (PS I) (discussed below).
In a cyanobacterial strain, Nostoc sp., Fe deficiency had a different impact on two ferredoxins of different midpoint potentials, leading to the suggestion that flavodoxin may not substitute for all the reactions of ferredoxin. Accordingly, the ferredoxin whose activity could be replaced by flavodoxin was more rapidly lost upon transition to Fe deficiency. Flavodoxins can replace ferredoxin in photosynthesis, where the proteins are acceptors from PS I and donors to NADP+ via a ferredoxin NADP+ oxidoreductase, and as substrates for a number of enzymes like ribonucleotide reductase, nitrate reductase, pyruvate formate oxidoreductase, hydrogenase, nitrogenase and the fatty acid desaturation reactions (e.g. Sandmann et al., 1990, Gangeswaran and Eady, 1996, Cotruvo and Stubbe, 2008, Chazarreta-Cifre et al., 2011). Biochemical comparison of ferredoxin and flavodoxin from Synechocystis sp. 6803 indicates very similar physical properties of the two proteins (with respect to isoelectric point and midpoint potentials) compatible with co-evolution in response to common reaction partners (Bottin and Lagoutte, 1992).
Curiously, although this Fe-sparing mechanism is widespread in microbes, it appears to have been lost in land plants, even though Fe can be a limiting nutrient in the soil environment (Yi and Guerinot, 1996). Transgenic expression of a chloroplast-targeted flavodoxin complements a ferredoxin mutant, indicating that the protein can function in the plant chloroplast, so its absence in plant genomes remains a mystery (Blanco et al., 2011). Most plants, including algae, express multiple ferredoxin isoforms with specificity for a subset of the many ferredoxin-dependent reactions in the chloroplast (e.g. Hanke et al., 2004, Terauchi et al., 2009). The slightly reduced specificity of flavodoxin interaction with reaction partners compared to that of ferredoxin may have led to its loss in the land plant lineage in the absence of selective pressure for Fe sparing (Zurbriggen et al., 2007, Goñi et al., 2008).
b. Fe-sparing by substitution of FeSOD with MnSOD and NiSOD
The replacement of FeSOD by MnSOD is another widespread Fe-sparing mechanism. It has been noted in bacteria, algae, as well as diatoms (Privalle and Fridovich, 1993, Wolfe-Simon et al., 2006, Allen et al., 2007b). In organisms adapted to the low Fe content of the open ocean, the use of MnSOD may contribute to their higher Mn quota (Peers and Price, 2004). The two SODs evolved from a common ancestor and are structurally highly similar, with specificity for Fe vs. Mn determined by the second shell ligands (Wintjens et al., 2004). Accordingly, in bacteria with an Fe-sparing genetic program, there are separate Fur-regulated genes, sodA and sodB, for each form (see below). Interestingly, it was reported that Cu can replace Fe or Mn, but the resulting enzyme is less active (Meier et al., 1994). This type of substitution may be adventitious rather than programmed.
An adaptation that has emerged in some organisms faced with chronic Fe limitation is the evolution of NiSOD (SodN). SodN is a structurally distinct protein that can functionally replace FeSOD. It has been noted that genes encoding SodN are widespread in the marine cyanobacteria and are correlated with the presence of urease, another (typically) Ni-requiring enzyme. Together, these two enzymes contribute to a Ni-requirement for bacteria in the ocean (Dupont et al., 2008b) while concomittantly reducing the cellular Fe quota. Streptomyces griseus and S. coelicolor also use a NiSOD to replace an Fe/ZnSOD and an FeSOD, respectively, but its use is controlled by Ni availability rather than Fe unavailability (see below).
c. Fe-sparing and Fe-recycling by remodeling of PS I
In aerobic photosynthetic organisms, about half the Fe in the photosynthetic apparatus is found in PS I, which has three Fe4S4 centers. Accordingly, down-regulation of PS I is a common Fe-sparing acclimation response to Fe-deficiency in cyanobacteria, diatoms, and algae (Moseley et al., 2002, Strzepek and Harrison, 2004). In cyanobacteria, the down-regulation of PS I is accompanied by a complete re-modelling of the PS I-associated peripheral antenna to change the supply of excitation energy to that photosystem (Park et al., 1999, Ivanov et al., 2000, Havaux et al., 2005). The new antenna protein is encoded by the isiA (for iron stress induced) gene and is often co-transcribed with the previously-mentioned isiB gene encoding flavodoxin (Laudenbach et al., 1988, and see above). Indeed, isiA sequences are retrieved with the highest frequencies in metatranscriptome surveys of plankton communities in the oligotrophic open ocean. It ranked in the top 4 during a bloom of C. watsonii in the SW Pacific (Hewson et al., 2009). In Synechocystis 6803, the transcription of isiA is repressed by Fur in the Fe replete situation. When Fe is limited, the isiA operon is derepressed, although expression is limited, at least initially, by an antisense RNA (isrR) encoded on the opposite strand (Dühring et al., 2006). Diatoms adapted to low Fe content in the ocean appear to have lowered the ratio of PS I to PS II and also reduced the abundance of cytochrome-containing complexes to reduce the cellular Fe quota (Strzepek and Harrison, 2004).
A fascinating example of Fe-sparing and recycling to reduce the cellular Fe quota was recently described in a marine cyanobacterium (Saito et al., 2011). The constitutive expression of flavodoxin in this organism suggests that it is adapted to persistent Fe-limitation. In many photosynthetic diazotrophs, photosynthesis and N2-fixation are separated either in space or time because of the incompatibility of oxygenic photosynthesis with the O2-sensitive metalloclusters in dinitrogenase (e.g. Schneegurt et al., 1994, Steunou et al., 2008). Temporal separation in Crocosphaera watsonii offers an opportunity for reducing the Fe quota. Proteomic analysis showed that nitrogenase metalloproteins changed from being the most abundant in the dark phase to being undetectable in the light phase while the Psa proteins of the Fe/S containing PS I complex and cytochromes showed the opposite pattern. The authors calculate that the “sharing” of intracellular Fe by daily degradation of the iron proteins and recycling of the released Fe reduces the Fe requirement by 40%, which would clearly be advantageous in a low Fe environment.
When an organism is never in a state of Fe luxury owing to persistent (over evolutionary time scales) poor Fe supply in its niche, it may simply dispense with the gene for an Fe-containing protein, as suggested by the constitutive use of Cu-containing plastocyanin in place of cytochrome (Cyt) c6 in the diatom T. oceanica (Peers and Price, 2006). The trade-off is that this organism is now more sensitive to Cu deficiency due to an increased requirement for Cu for photosynthesis. However, since Cu levels in the open ocean (0.4–1 nM) are often higher than Fe levels, this is favorable adaptation (Peers and Price, 2006). A converse example is offered by a subset of the chlorophyte (green) algae, which appear to have dispensed with the genes encoding CuZnSODs in favour of Cu allocation to plastocyanin and Cyt oxidase, owing perhaps to persistent Cu-deficiency in their environments (see below) (Asada et al., 1977). In these organisms, SOD function is provided by Fe- and Mn-containing enzymes.
3. Fe homeostasis in E. coli
Bacterial iron homeostasis is exceptionally well understood in E. coli, which is the reference organism for many different aspects of bacterial metabolism (Andrews et al., 2003). E. coli normally accumulates Fe to a level corresponding to ~1 mM averaged over the cell volume, with the majority of this Fe bound to proteins. The acclimation of E. coli to Fe limitation is regulated by Fur, which senses the cytosolic availability of Fe(II) (Hantke, 2001).
E. coli mutants unable to maintain Fe homeostasis, due to inactivation of the fur gene, have a number of striking phenotypes. They display an elevated sensitivity to reactive oxygen species, such as hydrogen peroxide (Touati et al., 1995), are unable to grow on succinate, and are relatively resistant to elevated levels of Mn which presumably acts as a Fur agonist and inappropriately represses Fe uptake (Hantke, 1987). Although essential for the function of numerous enzymes, Fe(II) in excess is toxic due to its ability to catalyze (via Fenton chemistry) the formation of destructive hydroxyl radicals (Imlay and Linn, 1988). This problem is exacerbacted in E. coli, since even low levels of endogenously produced oxidants have the potential to inactivate the Fur:Fe(II) complex leading to further elevation of intracellular Fe levels (Varghese et al., 2007).
The peroxide-sensitivity of fur mutant cells provided early evidence that cytosolic Fe levels are elevated (Touati et al., 1995), as might be expected from the constitutive expression of Fe import functions. Evidence for elevated cytosolic Fe was obtained using a whole cell electron paramagnetic resonance (EPR) approach in which desferrioxamine is added to stabilize chelatable (bioavailable) Fe as Fe(III) (Keyer and Imlay, 1996). This analysis led to an initial estimate of the level of Fe in the E. coli cytosol as ~10 µM with this increasing to ~70 µM in the fur mutant strain (Keyer and Imlay, 1996), although subsequent studies have measured values as much as 10-times higher (Jacques et al., 2006). This chelatable Fe pool represents an approximation of the labile Fe pool (presumably Fe in transit from uptake systems to protein targets) and may also include some Fe stripped by the chelator from enzymes with a relatively weak Fe affinity. The nature of the labile Fe pool is poorly understood, and indeed the whole issue of metal speciation in the cell is largely unexplored. The development of whole cell spectroscopic approaches, which is particularly amenable to genetically-tractable microorganisms, is promising in this regard (reviewed by Lindahl and Holmes-Hampton, 2011).
Although the labile Fe pool is elevated in fur mutants, S. Andrews and coworkers have noted that fur mutant cells are actually Fe-deficient with total Fe levels reduced 2.5-fold relative to wild-type (WT) (Abdul-Tehrani et al., 1999). These observations can be reconciled, since the labile Fe pool measured by EPR is a small subset (routinely less than 10%) of total cellular Fe: although fur mutants have an elevated chelatable Fe pool, their total Fe content is significantly reduced due to repression (mediated by RyhB, see below) of numerous Fe-containing enzymes. The same type of remodeling of the proteome, to efficiently utilize Fe, occurs during the acclimation of E. coli to Fe limitation. Indeed, sudden imposition of Fe starvation leads to large changes in gene expression which can be modeled as resulting from transient changes in a labile Fe pool that represents ~1% of the total Fe needed by the cell and that directly controls Fur activity (Amir et al., 2010).
Fur contributes to Fe homeostasis in numerous ways. First, Fur directly represses, under replete conditions, Fe acquisition pathways. Second, Fur indirectly activates synthesis of the Fe-storge protein ferritin under Fe replete conditions. Fur binds to the region upstream of the ftnA gene and acts to prevent gene repression by the H-NS nucleoid protein (Nandal et al., 2010). Third, Fur regulates RyhB, a small, non-coding RNA (sRNA) with wide-ranging effects that serves to remodel the proteome as part of a global Fe-sparing response (see below). Fourth, Fur regulates the expression of enzymes that can substitute for what would otherwise be Fe-dependent pathways. The synthesis of deoxyribonucleotides (from their ribonucleotide precursors) is essential for DNA replication, and most bacterial ribonucleotide reductases contain Fe as an essential catalytic cofactor. An elegant example of a Fur-regulated substitution pathway is the utilization in Fe-deficient cells of an alternative, Mn-utilizing ribonucleotide reductase (RNR) encoded by the nrdEF operon (Andrews, 2011, Cotruvo and Stubbe, 2011, Martin and Imlay, 2011). The ability of the NrdEF enzymes to be derepressed under Fe limitation provides a mechanism to sustain growth. Consistent with its expression under Fe replete conditions, the class Ia Fe-dependent RNR requires an Fe2S2 ferredoxin for maintenence and likely assembly of the diferric-tyrosyl radical at the heart of the enzyme (Wu et al., 2007). In contrast, assembly of the class Ib Mn-dependent enzymes (which contains a dimanganese(III)-tyrosyl radical) requires a flavodoxin (NrdI) as oxidant (Cotruvo and Stubbe, 2008, Boal et al., 2010). Iron limitation alone is rather inefficient at supporting active NrdEF, since E. coli normally does not actively accumulate significant amounts of Mn. Uptake of Mn, through the MntH proton-coupled importer, is required for efficient activation of NrdEF. Activation of the Mn-dependent RNR is more efficient under mild oxidative stress conditions which inactivate Fur and thereby induce mntH transcription (Martin and Imlay, 2011).
An additional, and earlier, example of substitution of an Fe-containing enzyme by its Mn counterpart is the inverse regulation of the sodB and sodA genes encoding, respectively, an Fe-superoxide dismutase (FeSOD) and MnSOD (e.g. Niederhoffer et al., 1990, Privalle and Fridovich, 1993). The sodA gene is repressed by Fur in response to Fe and this gene is preferentially expressed when Fe is limited. Conversely, the sodB gene is expressed when Fe is available and is positively regulated by Fur. Although sodA can be expressed in response to Fe limitation, up to 95% of the enzyme is inactive under standard growth conditions since, as noted above, E. coli does not normally import sufficient Mn for SodA metallation unless oxidative stress is imposed (Anjem et al., 2009).
The mechanism of Fe-dependent activation of sodB expression was initially quite puzzling. Early experiments to test the hypothesis that Fur might bind to the sodB promoter region to activate transcription were inconclusive (Fee, 1991, Dubrac and Touati, 2000) and subsequent studies suggested that much of the regulation was likely post-transcriptional (Dubrac and Touati, 2000, Dubrac and Touati, 2002). This work set the stage for the description of RyhB, the sRNA that serves as the key mediator of the Fe sparing response in E. coli (Masse and Gottesman, 2002).
a. Fe-sparing by RyhB-dependent proteome remodeling
The RyhB sRNA was originally discovered in a survey of E. coli for small, non-coding RNAs (sRNA). Fur links the synthesis of the RyhB sRNA to Fe availability: when cells are Fe-limited the Fur repressor is inactive and RyhB accumulates (Masse and Gottesman, 2002). RyhB, in turn, down-regulates the synthesis of several abundant Fe-containing proteins. The initial characterization of RyhB noted that this sRNA has extended complementarity to a region near the end of the first gene of the sdhCDAB operon encoding succinate dehydrogenase (SDH). As predicted for an antisense RNA-based mechanism, the effect of RyhB on expression of SDH is post-transcriptional and relies on Hfq, an RNA chaperone that facilitates the annealing of sRNAs with their mRNA targets, often leading to degradation (Masse and Gottesman, 2002). The destruction of the sdhCDAB mRNA under Fe-limiting conditions can therefore account for the inability of fur mutants to grow on succinate as a carbon source. Repression of SDH synthesis reduces Fe demand since this enzyme complex contains ten Fe atoms including one heme and a complex electron transfer chain ([Fe2S2], [Fe4S4], and [Fe3S4]) within the SdhB subunit (Cecchini, 2003).
In addition to sdh, several other target operons share a similar pattern of positive regulation by Fur in the presence of Fe (Hantke, 2001, Masse and Gottesman, 2002). These have in common the fact that they encode abundant proteins that bind Fe as cofactor (aconitase, fumarase A, FeSOD) or store Fe (bacterioferritin and ferritin). It is interesting to note that in some cases (aconitase, fumarase, and SOD), the Fe-regulation targets only one of mutiple paralogs. In most cases, this apparent positive regulation is attributed to RyhB.
The regulation of gene expression by RyhB is one of the best understood sRNA regulons and serves as a paradigm for thinking about Fe-sparing mechanisms in Bacteria (Masse et al., 2007). Homologs of RyhB are found in several related bacterial species and the overall outlines of the response are largely similar, although there are unique aspects to each system. Altogether, RyhB targets at least 18 distinct mRNA targets leading to the down-regulation of >50 proteins (Masse et al., 2005). In addition, RyhB targets several other sites leading to more nuanced effects on gene expression (Salvail and Masse, 2012). RyhB is complementary to an upstream open reading frame that is translationally coupled to fur and thereby down-regulates Fur expression. RyhB targets an intergenic region in the iscRSUA mRNA (encoding proteins for iron sulfur cluster assembly and regulation) leading to degradation of the iscSUA portion of the mRNA and stabilization of the iscR portion which encodes an operon-specific repressor (Desnoyers et al., 2009). Finally, RyhB can activate gene expression in the case of targets required for the optimal synthesis of enterobactin, the major E. coli siderophore (Prevost et al., 2007, Salvail et al., 2010).
The physiological impact of the RyhB-mediated Fe sparing response on E. coli is quantitatively significant and amounts to a greater than 2-fold reduction in Fe demand. This process allows E. coli to make the most efficient use of a scarce resource and direct the available Fe to the most high priority enzymes. These are presumably those essential functions that can not be replaced by alternative enzymes or pathways, although the enzymes that constitute these essential functions will likely vary depending on the precise growth conditions. In other cases, such as for RNR, E. coli has evolved a backup Fe-independent pathway that allows growth even when the Fe-cofactored enzyme is not able to be metallated. The analysis of the E. coli Fe sparing response also illustrates another recurring theme: discovery of the transcription factors that respond to elemental limitations, and thereby the regulons that they control, often points directly to key players in the acclimation process.
b. Fe-sparing in other Gram-negative bacterial systems
Iron-sparing responses are widespread in the Bacteria (Salvail and Masse, 2012), although the molecular details are not as well defined as for E. coli. For those organisms more closely related to E. coli, RyhB orthologs are common and a similar overall pathway is likely present. For example, RyhB orthologs have been described for Shigella (Africa et al., 2011) and Vibrio spp. (Davis et al., 2005, Mey et al., 2005). A more divergent system is found in Pseudomonas aeruginosa that requires two, tandemly encoded sRNAs designated PrrF1 and PrrF2. These two sRNAs (116 and 114 nt) are nearly identical in sequence and, as far as is known, completely redundant in function (Oglesby-Sherrouse and Vasil, 2010). Functionally, they are analogs of RyhB and target the mRNAs for some of the same types of Fe-utilizing proteins including SDH, SodB, and a bacterioferritin (Wilderman et al., 2004). Curiously, in response to combined heme and iron limitation a longer RNA (prrH) is produced, perhaps by a regulated antitermination mechanism, that spans the prrF1 and prrF2 genes, and includes what would otherwise be the intergenic region. This additional sequence information may allow this longer sRNA to additionally target heme biosynthesis functions (Oglesby-Sherrouse and Vasil, 2010). PrrF-like loci are also involved in controlling Fe-sparing in other bacteria including ArrF in Azotobacter vinelandii (Jung and Kwon, 2008) and NrrF in Neisseria meningitidis (Mellin et al., 2007). The regulons for each of these sRNAs are likely distinct, although they also target abundant, Fe-using proteins such as SOD or SDH.
4. Fe homeostasis in B. subtilis
B. subtilis also responds to Fe limitation with derepression of a large and complex regulon of genes controlled by the Fe-sensing repressor, Fur (Baichoo et al., 2002). The induced operons lead to the synthesis of siderophore (bacillibactin) and expression of transport systems for ferric-bacillibactin and other siderophores (Ollinger et al., 2006). Clearly, the major function of these operons is Fe acquisition. In addition, other operons function in Fe-sparing by encoding proteins that can substitute for functions that might be disabled under Fe limitation.
One function likely to be compromised in Fe-limited cells is electron transfer, and B. subtilis, like the marine phytoplankton discussed previously, substitutes ferredoxin with flavodoxins. Recently, a ferredoxin has been described as the single most abundant Fe-containing protein in the soluble fraction of B. anthracis cells (Tu et al., 2011), which makes this protein a logical target for an Fe-sparing response. When B. subtilis is Fe-limited, the ykuNOP operon is strongly derepressed, which leads to the synthesis of two flavodoxins (YkuN and YkuP) that are proposed to functionally substitute for at least some functions of ferredoxin. These flavodoxins have been biochemically characterized and can function with BioI, a P450 enzyme (Lawson et al., 2004), nitric oxide synthase (Wang et al., 2007), and the Fe-dependent Δ-5-acyl lipid desaturase for remodeling membrane lipids (Chazarreta-Cifre et al., 2011). In the latter case, both the ferredoxin and flavodoxin proteins are catalytically active, and it can be imagined that when Fe is limiting, the flavin-containing protein may functionally replace the Fe-containing protein for at least this and possibly other functions. The fate of previously synthesized ferredoxin is not known in B. subtilis, but in Clostridium pasteurianum it has been found to be degraded in response to Fe limitation to serve as an endogenous source of Fe (Schönheit et al., 1979).
a. The FsrA-dependent Fe-sparing response
B. subtilis has a robust Fe-sparing response that is activated when Fe is limiting for growth (Gaballa et al., 2008). In this organism, Fur represses the synthesis of an sRNA designated FsrA which is distinct in sequence, but similar in function, to RyhB. The FsrA sRNA is highly complementary to the leader region of the sdhCAB operon and, as for E. coli, fur mutants are unable to grow on succinate. This growth defect is due to FsrA since a fur fsrA double mutant regains the ability to grow on succinate (Gaballa et al., 2008). A number of other FsrA targets have been identified using proteomic and transcriptomic approaches and in at least one case the predicted RNA:RNA pairing interactions have been confirmed by genetic analysis. Some of the notable FsrA-regulated targets include SDH, and several other Fe/S-containing enzymes including aconitase, a lactate oxidase complex, glutamate synthase, and dehydratases involved in branched chain amino acid biosynthesis (Gaballa et al., 2008). As a result of the wide range of metabolic activities targeted by FsrA, a fur mutant strain is significantly impaired in growth even when Fe is sufficient. This is due to the inappropriate (under these conditions) repression of metabolic enzymes. One notable consequence of this is a significant reduction in the activity of the TCA cycle as monitored using metabolomics (Fischer and Sauer, 2005).
The B. subtilis Fe-sparing response differs from that of the enteric bacteria in that it also involves three small, basic (positively charged) proteins named FbpA, B, and C (for Fur-regulated basic proteins). The ability of FsrA to target mRNAs for regulation is independent of a presumed Hfq homolog in B. subtilis, but is at least partially dependent on the FbpABC proteins, which are postulated to function as Fur-regulated RNA chaperones (Gaballa et al., 2008). However, direct evidence for chaperone activity (the ability to increase the rate of RNA:RNA annealing) is still lacking. Transcriptome and proteome studies of mutant strains lacking Fur, FsrA, and/or one or more Fbp proteins indicate that these regulators overlap in their specificity, and that several FsrA targets are co-regulated by one or more Fbp. Mutations have been introduced into the start codons of the fbpA and fbpB genes that prevent protein expression while having negligible effect on RNA sequence stability. Using these constructs, and complementation studies, it has been demonstrated that the requirement for the fbpAB operon for repression of lactate utilization (lutABC) genes during Fe limitation requires the FbpB protein, but not FbpA (Smaldone et al., 2012). In contrast, for some targets (e.g. sdh) there is no apparent requirement for Fbp proteins (even a triple fbpABC mutant still regulates SDH normally).
The B. subtilis Fe sparing response has drastic effects on cell physiology due to extensive remodeling of the proteome as visualized using gel-based proteomics (Gaballa et al., 2008). Comparable to E. coli, the Fe content of the B. subtilis cell corresponds to ~1 mM averaged over the cell volume and it is likely that only a very small fraction of this corresponds to the labile Fe pool sensed by Fur. Ultimately, a complete metallomics analysis of B. subtilis is needed to quantify the distribution of Fe between heme-, Fe/S- and non-heme-Fe-containing proteins and to better understand how this distribution is remodeled by the Fe-sparing response. Fur itself is quite abundant (~10,000 molecules per cell) and elevation of this level by even two-fold grossly perturbs Fe homeostasis (Faulkner et al., 2011). The fur gene is repressed by a paralogous metalloregulatory protein, PerR, and in the absence of perR, Fur levels are elevated. As a result, perR mutants are severely Fe starved. This is due, in part, to an abundant catalase (KatA) which is highly expressed in perR mutants and likely accounts for ~10% of the total Fe quota in these cells (Faulkner et al., 2011).
5. A protein-mediated Fe-sparing response in Corynebacterium glutamicum
Iron sparing responses are also likely to have evolved in other Gram positive bacteria although these are, in general, not well defined. Of note, Corynebacterium glutamicum has a protein rather than an sRNA-based Fe-sparing response (Wennerhold et al., 2005). In this organism, the DtxR protein functions as a global regulator of Fe homeostasis (analogous to Fur) and upon Fe limitation an AraC-type repressor named RipA (regulator of iron proteins A) is derepressed (Wennerhold and Bott, 2006). RipA, in turn, binds directly to the promoter regions of numerous Fe-utilizing enzymes to block their expression. The RipA targets include aconitase, succinate dehydrogenase (sdhCAB), nitrate/nitrite transporter and nitrate reductase (narKGHJI), isopropylmalate dehydratase (leuCD), catechol 1,2-dioxygenase (catA), and phosphotransacetylase (pta). The DtxR-RipA regulatory circuit is thereby analogous in function to the Fur-RyhB (E. coli) or Fur-FsrA (B. subtilis) systems (Wennerhold et al., 2005). Repression of aconitase and SDH in response to Fe limitation has also been noted in one of two studied strains of Mycobacterium avium (Janagama et al., 2010), although the mechanism remains to be defined.
6. Fe homeostasis in S. cerevisiae
S. cerevisiae is an exceptionally well understood model system for defining processes of Fe homeostasis in a model Eukaryote (reviewed in Kaplan et al., 2006, Bird, 2008, Philpott and Protchenko, 2008). Under Fe replete conditions, most of the Fe that enters cells is transported by low affinity pathways including the Fet4 and Smf1 proteins. Under these conditions, aerobic respiration pathways are active, the cellular Fe quota is relatively high, and the Ccc1 protein imports Fe into vacuoles (Li et al., 2001).
The primary response to Fe deprivation is the activation of the Aft1 and Aft2 transcription factors, which coordinate a multifaceted acclimation response that can reduce the cellular Fe quota by two-fold or more. Most genes activated by Fe-deprivation are under the transcriptional control of Aft1, which is translocated to the nucleus in response to declining Fe status. Conversely, in the presence of Fe, Aft1 is oxidized to form mixed disulfides with a monothiol glutaredoxin and interacts with an exporter that mediates Aft1 translocation to the cytosol (Kaplan et al., 2006, Philpott and Protchenko, 2008). Aft2, an Aft1 paralog, may activate many of the same genes as Aft1 does, but appears to play a secondary role most apparent in strains lacking Aft1 (Philpott and Protchenko, 2008). Note that homologues of Aft1/2 are found only in the fungi, and therefore other Fe-sensing regulators function in animals and plants (Long et al., 2010, Wang and Pantopoulos, 2011).
Aft1 coordinates the activation of a complex and multilayered Fe-sparing response. First, Aft1 activates the transcription of genes for Fe acquisition and mobilization (Philpott and Protchenko, 2008). Second, Aft1 activates transcription of two RNA-binding proteins, Cth1 and Cth2, that target numerous mRNAs for functional inactivation (Puig et al., 2005, Puig et al., 2008). Third, the metabolic changes resulting from both the direct and indirect effects of Aft1 alter the activity of other regulons in the cell that may also impact Fe homeostasis (Ihrig et al., 2010).
Genes for Fe acquisition and mobilization functions, including many encoding high affinity transporters for Fe-chelates and elemental Fe, are under the direct transcriptional control of Aft1. In yeast, elemental Fe uptake is mediated by a reductive system that depends on a copper-containing oxidase (Fet3p) and a specific permease (Ftr1) (Kosman, 2003). Aft1 also activates the expression of an Fe transporter (Fet5/Fth1) homologous to the Fet3/Ftr1 system that localizes to the vacuolar member for Fe mobilization from this site of storage. In addition, Aft2 activates expression of an NRAMP family divalent cation transporter, Smf3, that also functions in Fe mobilization from the vacuole (Singh et al., 2007).
The mitochondria are the sites for both heme and Fe/S biosynthesis and this can be a significant drain on cellular Fe pools. Aft1 activates the expression of the heme oxygenase Hmx1 that degrades heme. This can serve dual roles in the cell. First, heme degradation can release Fe, and thereby allow its recycling for other purposes. Second, heme normally serves as a signal molecule to activate the expression of respiratory cytochromes. Thus, degradation of heme also serves to down-regulate the expression of a major Fe-consuming pathway in the cell (Kaplan et al., 2006). As an additional Fe-sparing mechanism, Aft1 up-regulates a biotin importer which compensates for the transcriptional repression of biotin biosynthetic functions (including the Fe/S containing Bio2 protein) under Fe deprivation.
Aft1 also activates synthesis of Cth2 that coordinates a large-scale Fe-sparing and prioritization response in which numerous Fe-consuming proteins are post-transcriptionally down-regulated (Vergara and Thiele, 2008). Conceptually, this is analogous to the RyhB-mediated Fe-sparing response of E. coli and the similar FsrA-mediated response of B. subtilis. However, rather than relying on a small RNA, the yeast Cth2 RNA-binding protein recognizes AU-rich elements in the 3'-untranslated regions of target mRNAs, recruits the Dhh1 helicase, and activates an mRNA decay pathway (Pedro-Segura et al., 2008). Yeast also contain a Cth2 paralog, Cth1, that preferentially down-regulates mitochondrial functions (Puig et al., 2008). Both Cth1 and Cth2 contain two tandem Zn-finger domains involved in nucleic acid binding.
Cth2 is transcriptionally induced ~200-fold in response to Fe deficiency. Studies with deletion mutants lacking Cth2 revealed an increase in the level of mRNAs for numerous Fe-consuming processes including TCA cycle enzymes (e.g. SDH), mitochondrial respiration, fatty acid synthesis, heme biosynthesis, and Fe/S proteins (Puig et al., 2005). The down-regulation of these many Fe-consuming processes presumably functions to spare Fe for more essential processes, likely including the synthesis of Fe/S cluster enzymes and an Fe-dependent RNR. Cth2, together with Cth1, ensures that limited Fe is directed towards RNR by targeting the nuclear tethering protein Wtm1 for degradation (Sanvisens et al., 2011, Seguin et al., 2011). Wtm1 binds to the R2 subunit of RNR thereby limiting its translocation to the cytosol where it is active. Thus, Fe deprivation leads to Cth2-mediated down-regulation of numerous Fe-consuming reactions while simultaneously stimulating the translocation and thereby activity of an essential, Fe-requiring enzyme (Sanvisens et al., 2011, Seguin et al., 2011).
Global studies have revealed that Fe deprivation, and in particular the Aft1-mediated pathways noted, lead to many pleiotropic effects on transcription. In addition to the direct effects of Aft1 on gene expression, and the indirect effects of Cth2-mediated RNA destabilization, there are additional effects that result from changes in metabolites that themselves have regulatory roles. As noted above, Aft1 induces heme oxygenase, which leads to a decrease in heme levels. Heme is sensed by the Hap1 transcription factor and decreasing heme availability leads to down-regulation of cytochrome c (Ihrig et al., 2010). Similarly, decreased activity of Fe/S containing biosynthetic enzymes can lead to changes in metabolite pools that alter gene expression (Ihrig et al., 2010).
7. Fe homeostasis in Chlamydomonas
Fe homeostasis in C. reinhardtii is regulated at multiple levels. The iron assimilation pathway consists of a cell surface ferrireductase, a high affinity Fe-selective transporter consisting of a multicopper oxidase (Fox1) and a trivalent cation specific permease Ftr1 analogous to the S. cerevisiae Fet3/Ftr1, as well as Irt1/Irt2, members of the ZIP family of divalent cation transporters, and a periplasmic presumed Fe binding protein (Allen et al., 2007a, Blaby-Haas and Merchant, 2012). Each of the corresponding genes is under transcriptional control, although the Fe sensor and transcription factor remain to be discovered. Iron status in photosynthetic cells is readily visualized as “chlorosis” or loss of chlorophyll (Chl) pigment. Merchant and co-workers used chlorosis and growth rate to define “stages” of poor Fe nutrition in C. reinhardtii: the replete situation where all Fe-containing proteins are satisfied, Fe-deficiency where chlorosis is not evident and the growth rate is not impacted but where the genes for assimilation are fully turned on, and Fe-limitation where cells are chlorotic and growth inhibited because there is insufficient Fe for maintaining activities of all essential enzymes (La Fontaine et al., 2002). In the absence of known transcription factors, these operational definitions help to distinguish primary (and perhaps direct) responses to Fe-deficiency from secondary responses resulting from physiological stress caused by Fe-limitation.
Fe is required for both photosynthesis and respiration in C. reinhardtii, with PS I contributing to 50% of the Fe quota in the photosynthetic apparatus and NADH-dehydrogenase/complex I contributing to most of the Fe demand of the respiratory electron transfer chain. In a situation of respiratory growth (acetate as a carbon source), a program of Fe sparing in the chloroplast is activated (Moseley et al., 2002). This involves loss of PS I (with its 3 Fe4S4 centers) and cytochrome complexes (containing heme and an Fe2S2 Rieske center) so that their abundance is reduced to <1% of that noted in Fe-replete cultures. In contrast, the abundance of ferredoxin is decreased but not as drastically. The different impact on PS I and the Cyt b6f complex as compared to ferredoxin is consistent with the fact that on acetate-containing medium, photosynthesis is non-essential. On the other hand, ferredoxin is used also as a reaction partner for O2-utilizing oxidations in various biosynthetic reactions, including fatty acid desaturation, N-assimilation and dNTP synthesis. Since there is no evidence for a flavodoxin encoded in the C. reinhardtii genome, ferredoxin is an essential (high priority) function.
Interestingly, the activity and abundance of FeSOD is also maintained under Fe deficiency, further pointing to prioritization of Fe utilization in the chloroplast (Page et al., 2012). Five MnSOD isoforms are encoded in the C. reinhardtii genome by MSD1 through MSD5. The individual gene products are differently localized with one isoform assigned to the plastid. The corresponding gene, MSD3, is dramatically up-regulated (102–103-fold) in Fe-starved cells at the transcriptional level, with the response initiated already in a situation of deficiency rather than limitation, suggestive of a direct response to Fe status. An increased demand for SOD is expected in photosynthetic cells with compromised PS I Fe4S4 clusters, since the midpoint potentials of the PS I acceptors are sufficiently negative to reduce O2 to superoxide (Herbert et al., 1992, Asada, 2006). The use of a Mn-isoform for this protective response would spare Fe for use in other pathways. Another photoprotective mechanism that operates in this situation is the disconnection of the light harvesting Chl-binding proteins from the PS I complex (Moseley et al., 2002). This process is a selective program that responds to the activity of a di-iron enzyme in the Chl biosynthetic pathway.
When cells are shifted from phototrophic to heterotrophic growth, Fe is recycled by degradation of the photosynthetic apparatus. Released Fe is buffered by ferritin in the chloroplast stroma and can presumably be mobilized as needed for maintenance of respiratory complexes in the mitochondria (Naumann et al., 2007, Busch et al., 2008). In contrast, when cells are grown phototrophically (on CO2 and light), the photosynthetic apparatus is essential and is accordingly maintained (Terauchi et al., 2010). Other Fe-sparing and recycling mechanisms are also presumed to operate, but these are not yet well-investigated.
D. Zinc (Zn): an essential metal for life
Zinc is an essential element for life and, unlike Fe, there are no known examples of organisms that have completely dispensed with a Zn requirement. Zn is not redox active under biological conditions, and serves as an electrophilic catalyst (Lewis acid) in numerous enzymes and as a scaffold for organizing protein domains. It is this latter function that has expanded tremendously in eukaryotes leading to the proliferation of Zn-finger proteins encoded in many plants and animal genomes. Zn deficiency is considered the most common mineral deficiency of plants (Assuncao et al., 2010) and is one of the major health threats in the developing world where it contributes to the death of 800,000 children annually (Black, 2003, King, 2011).
Although Zn is required for all cells, distinguishing between its essential and its dispensable roles is not easy. E. coli is estimated to contain perhaps 100 distinct Zn-containing enzymes. A survey of the M. tuberculosis genome for likely essential Zn functions highlights the involvement of this metal in the synthesis of DNA, leucine, inositol derivatives, and mycothiol and in key metabolic reactions such as aldolase in glycolysis and methionine aminopeptidase (Riccardi et al., 2008). Zn is also found as a component in RNAP, some ribosomal proteins, and some aminoacyl tRNA synthetases that together may account for its essential nature. Because Zn has so many different roles, cells have evolved complex responses to acclimate to Zn deprivation. The major common mechanisms involve increased uptake of Zn, the synthesis of alternative, Zn-independent enzymes and proteins (where possible), and the mobilization of intracellular Zn <Figure 9>.
Figure 9.
Overview of the biological roles of Zn and known sparing and recycling mechanisms.
1. Zn homeostasis in E. coli
E. coli contains a total of ~0.2 mM Zn when averaged over the cell volume. However, only a tiny fraction of this total is in a labile Zn pool. The labile Zn pool has been estimated at ~20 pM using a genetically-encoded fluorescent biosensor based on carbonic anydrase (Wang et al., 2011a). This small, labile Zn pool is bound to small molecules in the cytosol and there is no "free" Zn in the cell at equilibrium (Outten and O'Halloran, 2001). Zn homeostasis in E. coli is governed by the Fur-like metalloregulatory protein, Zur, which binds to DNA target sites when Zn levels in the cell are sufficient. Since Zur binds Zn with an affinity in the femtomolar range, it can be concluded that Zur and other molecular ligands in the cell maintain the small labile Zn pool in a largely bound, but kinetically labile state. When Zn levels rise, the ZntR metalloregulator senses this excess and activates efflux.
Insights into the processes of acclimation to Zn deficiency can be obtained from the analysis of the Zur regulon (Graham et al., 2009). In E. coli, the primary function of the Zur regulon appears to be increased Zn uptake. Zur also regulates at least one alternative ribosomal protein, which presumably functions to replace a normally Zn-containing one (Hensley et al., 2011), as described in detail for B. subtilis (see below). Unlike the RyhB Fe-sparing response, there is no evidence for a global Zn-sparing response in E. coli, which may simply mean that Zn limitation is not a stress commonly encountered in the natural habitats of this organism.
2. Zn homeostasis in B. subtilis
Zn homeostasis in B. subtilis is also regulated by Zur which was discovered as a Fur-like regulator that represses the synthesis of an ABC transporter for Zn uptake (Gaballa and Helmann, 1998). B. subtilis Zur, like that from E. coli, binds Zn with sub-picomolar affinity (Ma et al., 2011) and monitors a small, labile pool of Zn to coordinate responses to Zn limitation. In response to sufficient cellular Zn, Zur represses the synthesis of a high affinity Zn uptake ABC transporter (YcdHI-YceA) and a putative metallochaperone (YciC) (Gaballa et al., 2002), a GTP cyclohydrolase (FolE2) (Sankaran et al., 2009), and three alternative ribosomal proteins (Gabriel and Helmann, 2009).
a. Expression of Zn-independent isozymes
The Zur-regulated yciABC genes form a complex operon that is transcribed from Zur-regulated promoters upstream of both the yciA and yciC genes (Gabriel et al., 2008). YciC is a representative of a widely distributed family of putative metallochaperones that belong to the COG0523 (conserved orthologous group) proteins found in Bacteria, Archaea, and Eukarya (Haas et al., 2009). YciC itself is a highly abundant protein and was originally identified by SDS-PAGE analysis of zur mutant cells (Gaballa and Helmann, 1998). Based on homology, it is proposed that YciC is a GTP-dependent metal insertase, although neither the relevant metal nor the relevant client proteins are known. Since YciC is expressed under Zn limitation, one hypothesis is that this protein serves to direct Zn to the most essential enzymes in the cell. However, it remains possible that YciC may function with a different metal(s) to activate enzymes that replace Zn-dependent functions (Haas et al., 2009). Indeed, some cells have more than one COG0523 family member, raising the possibility that they may have different metal selectivities.
The YciA protein was assigned as an alternative GTP cyclohydrolase based on the original observation that some cells contain a complete folate biosynthetic pathway except for the apparent absence of a folE gene encoding GTP cyclohydrolase (GCYH-IA), the first step in folate biosynthesis. Phylogenomic comparisons revealed that cells lacking folE contained instead a member of the COG1469 family, which was therefore suggested to be a non-orthologous replacement designated GCYH-IB (El Yacoubi et al., 2006). Most Bacteria (65%) encode a GCYH-IA protein, a subset (14%) encode only GCYH-IB, and others (12%) encode both (the remainder are symbionts or other organisms that rely on uptake of folate from their environment) (Sankaran et al., 2009). For those organisms that encode both types of GTP cyclohydrolase (such as B. subtilis), the GCYH-IB isozyme is typically regulated by Zur (or its functional equivalent) and is induced by Zn limitation. Biochemical analyses have revealed that GCYH-IA proteins are Zn-dependent enzymes whereas GCYH-IB can function with various other divalent metal ions, including Mg (Sankaran et al., 2009). Experiments with B. subtilis confirmed that the derepression of YciA in a zur mutant strain can compensate for the absence of the Zn-dependent FolE enzyme (Sankaran et al., 2009). Collectively, these results support a model where Zn activation of GTP cyclohydrolase is sensitive to cellular Zn depletion and the resulting inability to synthesize folate, which is essential for synthesis of dTMP and other key metabolites, imposes a growth restriction. One evolutionary solution to this elemental limitation has been the emergence of an alternative isozyme that relies instead on a different metal ion. Other candidates for this type of response in B. subtilis include peptide deformylase and methionine aminopeptidase encoded by paralogous genes. Although both enzymes are known to be metal-dependent, in this case there is no evidence for metal-dependent regulation that might provide hints as to the adaptive value of the gene duplication.
b. Expression of alternative ribosomal proteins and acclimation to Zn limitation
Bacteria have a remarkable but under-appreciated mechanism for acclimating to Zn limitation. Several ribosomomal proteins (r-proteins) contain a bound Zn ion and these proteins can be replaced by paralogs that lack Zn, thereby freeing Zn for use in other proteins (Panina et al., 2003). Ribosomes are highly abundant in growing cells and it is estimated that they contain 50% or more of the cellular Zn quota (Gabriel and Helmann, 2009, Hensley et al., 2011). This represents a physiologically significant form of stored Zn that can be re-partitioned, as needed, to allow assembly of Zn-cofactored enzymes.
The role of the ribosome in Zn homeostasis emerged from two independent lines of investigation. In the first, Panina and coworkers performed a comparative genomics analysis of Zn-regulated genes and noted that, in several organisms, there were r-proteins that were predicted to be up-regulated under conditions of Zn limitation (Panina et al., 2003). In each case, the corresponding r-protein was encoded by a duplicated gene with one copy under Zn-regulation and the other apparently constitutively expressed. r-proteins are also known to exist in variants that either have (C+) or lack (C−) potential Zn-binding motifs (CxxC motifs) (Makarova et al., 2001). This led Panina to propose that, under conditions of Zn limitation, Zn-containing (C+) r-proteins could be replaced by their non-Zn-containing (C−) counterparts (Panina et al., 2003).
The second set of experiments focused on the proteomic composition of the B. subtilis ribosome as a function of growth conditions (Nanamiya et al., 2004). When B. subtilis cells entered stationary phase the large subunit r-protein L31 disappeared and was replaced with a highly similar (C−) variant, encoded by the ytiA gene. The molecular basis of this protein switch was traced to derepression of the ytiA gene, which was shown to be a member of the Zur regulon (Nanamiya et al., 2004). In retrospect, the medium used for these growth studies was noted to lack added Zn which led, serendipitously, to the discovery of r-protein displacement. Since L31 is surface exposed on B. subtilis ribosomes, this r-protein can be displaced by newly synthesized YtiA protein and the released L31 thereby provides a potential source of Zn (Akanuma et al., 2006). A second large subunit protein, L33, is likely to be regulated similarly although the C− paralog (rpmGc) has a frameshift mutation in laboratory strains of B. subtilis (Gabriel and Helmann, 2009). Interestingly, neither L31 nor L33 is required for growth since mutants lacking all genes for either protein are still viable (Gabriel and Helmann, 2009).
A related substitution mechanism also occurs for r-protein S14 in B. subtilis (Natori et al., 2007). Unlike L31, the S14 protein assembles early into the ribosome and can not be replaced once the ribosome is synthesized. Under conditions of Zn limitation, the inability to metallate new S14 protein would thereby block the synthesis of new ribosomes. The cell has evolved a "fail-safe" mechanism in which derepression of a Zur-regulated paralog (YhzA) can functionally substitute for S14 and allow new ribosomes to be synthesized. Both the L31:YtiA and S14:YhzA protein pairs are nearly identical in sequence, but differ in the selective removal of Zn-binding Cys ligands from the C− partner.
Regulation of the YtiA(L31) and YhzA(S14) r-protein paralogs by Zur facilitates growth under Zn limitation by two distinct mechanisms. The displacement of surface-exposed large subunit r-proteins (L31 and potentially L33) releases substantial Zn for recycling into other target proteins (Nanamiya and Kawamura, 2010). The derepression of YhzA enables the continued synthesis of new ribosomes. The physiological impact of these acclimation mechanisms has been confirmed using strains lacking high affinity Zn uptake, which can thereby more readily be rendered Zn-limited (Gabriel and Helmann, 2009).
Acclimation to Zn limitation by derepression of C- family r-proteins is widespread, although the proteins induced, and the potential number of Zn atoms mobilized per ribosome, varies (Panina et al., 2003, Chen et al., 2009). Paralogous pairs of C+ and C− r-proteins are found for S4, S14, and S18 in the small subunit and L28, L31, L32, L36 in the large subunit (Chen et al., 2009). Note that the large subunit proteins were originally numbered in order of decreasing molecular mass and those that have paralogs are amongst the smallest subunits. Indeed, these r-proteins are little more than Zn-finger peptides and they are, at least in some cases, dispensable for growth (Gabriel and Helmann, 2009). One view is that these are simply small, positively charged Zn-finger peptides with a primary function in Zn storage and mobilization, and that binding to the surface of the ribosome is expedient for stabilizing the peptides against degradation. Alternatively, these may be dual function proteins with legitimate roles in translation or even in co-translational metal-loading into nascent proteins.
Paralogous pairs of r-proteins likely involved in Zn homeostasis have been documented in E. coli (Hensley et al., 2011), S. coelicolor (Owen et al., 2007, Shin et al., 2007), and M. tuberculosis (Maciag et al., 2007). The record for the most pairs of C+/C− r-proteins in a single organism is S. coelicolor with seven. Most of these are regulated by Zn mediated by Zur, but in at least one case the induction may be regulated instead by the σR oxidative stress response (Owen et al., 2007). Although an analogous Zn mobilization mechanism has yet to be reported in eukaryotes, the presence of duplicated genes for selected r-proteins suggests that such a mechanism may be present in some species.
3. Zn homeostasis in S. cerevisiae
S. cerevisiae is an outstanding model for the study of eukaryotic metal ion metabolism (Bird, 2008, Bleackley and Macgillivray, 2011), and Zn homeostasis in particular (Eide, 2009). When Zn is relatively abundant, Zn uptake is mediated by the Fet4 transporter (which also imports Fe) and Zrt2, a representative of the ZIP family of transporters that are conserved throughout all three domains of life. Under Zn replete conditions, import of excess Zn into the vacuole (up to 109 ions) provides a store for future use (Simm et al., 2007).
When cells encounter Zn deficiency, high affinity uptake systems are induced as is Zn mobilization from the vacuole (Bird, 2008, Eide, 2009). The key regulator of Zn homeostasis in yeast is the metalloregulatory protein Zap1 which contains seven carboxyl-terminal Zn finger motifs implicated in both DNA-binding and Zn sensing (Bird et al., 2000). Zap1 is thought to sense Zn directly by binding to a centrally located activation domain and a second activation domain that includes at least the first two Zn fingers, but the detailed mechanism has yet to be resolved (Frey et al., 2011).
Under Zn deficient conditions, Zap1 activates a regulon of ~80 genes (De Nicola et al., 2007, Eide, 2009). To facilitate Zn acquisition, Zap1 activates the expression of a high affinity importer, Zrt1. To enable mobilization of stored Zn, Zap1 activates expression of another ZIP transporter, Zrt3, that exports Zn stored in the vacuole. Paradoxically, Zap1 also activates expression of Zrc1 which can import Zn into the vacuole. This is proposed to protect yeast expressing high affinity uptake systems from Zn overload should Zn suddenly become available (Eide, 2009). Presumably, the affinity of the vacuolar Zn importer is poised such that import is only active when cytosolic Zn is relatively abundant.
Zap1 also coordinates a Zn-sparing response by differential regulation of isozymes of alcohol dehydrogenase. Under Zn replete conditions, yeast expresses the Adh1 and Adh3 isozymes which can bind a total of 1.5 × 106 atoms of Zn (Eide, 2009). Under conditions of Zn limitation, Zap1 induces the synthesis of RNA that traverses the promoter regions of the ADH1 and ADH3 genes and displaces factors needed for promoter activity (Bird et al., 2006). In addition to repression of Adh1 and Adh3, Zap1 induces synthesis of Adh4 which is suspected, based on sequence similarity, of being an Fe-containing alcohol dehydrogenase. Thus, this is likely a Zn-sparing mechanism (Bird, 2008, Eide, 2009). The transcription factors involved in Zn-sensing in other (non-fungal) microbial eukaryotes are not known. They are likely to be distinct, since orthologs of Zap1 have not been found in algae and diatoms, where regulatory responses to Zn-limitation have been described (see below).
4. Zn sparing by substitution of carbonic anhydrase with non-Zn alternatives
Aquatic photosynthetic organisms have carbon concentrating mechanisms for providing CO2 to the active site of Rubisco, the enzyme that initiates the Calvin cycle (reviewed in Badger and Price, 2003, Giordano et al., 2005, Roberts et al., 2007, Wang et al., 2011b). The importance of the carbon concentrating mechanism is evident from the phenotype of cyanobacterial and algal ccm mutants which are unable to grow at air levels (0.04%) of CO2. There are various types of independently-evolved carbon concentrating mechanisms. One type, common in aquatic microbes, relies on a series of bicarbonate transporters that move CO2 from the environment into Rubisco-containing compartments.
Carbonic anhydrases, which catalyze the inter-conversion of CO2 and bicarbonate, are critical enzymes in this mode of carbon acquisition, and accordingly, important for productivity of aquatic phototrophs (Cannon et al., 2010). Zn ion is the usual catalyst in carbonic anhydrases. In a Zn-deficient environment, the usual enzyme in the diatom Thalassiosira weissflogii (TWCA1) is replaced by a Cd-containing isoform, CDCA1 (Lane and Morel, 2000a). This novel enzyme, a product of convergent evolution, is found in other diatoms, based on sequence data, and its expression is stimulated by Cd availability (Lane et al., 2005, Park et al., 2007). This enzyme represents the first identification of a beneficial role for Cd in biology.
In laboratory experiments, Co was found to stimulate the growth of Zn-deficient T. weissflogii cells and this was determined to result from replacement of Zn with Co in the TWCA1 isoform (Yee and Morel, 1996, Lane and Morel, 2000c). In the coccolithophore Emiliania huxleyi, Co may be even more effective than Zn as a cofactor in carbonic anhydrase (Xu et al., 2007). Thus, Co may also support photosynthesis in a Zn-deficient natural environment. Because of limited molecular analyses in the diatoms, it is not known yet whether alternate carbonic anhydrases represent a Zn-sparing acclimation mechanism (i.e. both isoforms exist but their individual expression is reciprocally controlled by Zn nutrition) or whether, in some cases, the alternate forms are evolutionary adaptations to long term Zn limitation in their niche.
The carbon concentrating mechanism is well-studied in C. reinhardtii (reviewed by Wang et al., 2011). There are as many as 12 carbonic anhydrases encoded in the genome with isoforms distributed in multiple compartments, including the periplasm, mitochondria, cytosol, chloroplast, stroma, and thylakoid lumen. The functions of some of these are documented by genetic studies in which individual cah mutants were shown to be growth compromised for phototrophic growth at air levels of CO2. The abundance of several carbonic anhydrases is decreased in Zn-deficient C. reinhardtii cells and the cells are accordingly growth compromised in phototrophic conditions but not heterotrophic conditions (growth on acetate) where the carbonic anhydrases are less relevant (Malasarn and Merchant, unpublished). As in yeast, there is a strong transcriptional response in C. reinhardtii to Zn-deficiency, involving up-regulation of ZIP-family transporters encoded by the ZRT genes (Hanikenne et al., 2009). In parallel, the expression of two genes encoding proteins with a COG0523 domain (see above) is also dramatically increased (Haas et al., 2009). Whether these proteins are involved in Zn-sparing/recycling pathways remains to be tested.
5. Synthesis of Zn-independent isozymes as an adaptation to Zn limitation
The development of Zn-independent alternatives is also likely prevalent in those environments where Zn is present at low levels and deficiency is common. As noted above, much of the open ocean is P-limited and many of these areas also have low Zn availability, which exacerbates P-limitation, since alkaline phosphatases (active in liberating phosphate from dissolved organic matter) are often Zn enzymes. In such environments, many bacteria encode alternative Ca-dependent alkaline phosphatases, PhoX and/or PhoD, instead of the more familiar Zn-dependent enzyme, PhoA (Luo et al., 2009, White, 2009). PhoX is widely distributed in marine bacteria (Sebastian and Ammerman, 2009). Prochlorococcus species from P-limited regions commonly substitute PhoX for PhoA (Kathuria and Martiny, 2011) as do many other marine and freshwater organisms, but it is rare for an organism to encode both types (Zaheer et al., 2009). Somewhat more than half of all phoX genes are predicted to encode secreted or periplasmically localized proteins (Luo et al., 2009). In Campylobacter jejuni, PhoX activity depends on the twin-arginine transport (TAT) export system, suggesting that metallation of this enzyme likely occurs in the cytosol prior to transport (Drozd et al., 2011).
Both metagenomic and metatranscriptomic studies indicate that induction of PhoX in response to P-depletion is a widespread acclimation mechanism in the marine system (Sebastian and Ammerman, 2009). Unexpectedly, a bioinformatics approach revealed that up to 40% of alkaline phosphatases in the marine system may be cytoplasmically located and that the import of organic phosphates, followed by hydrolysis within the cytosol, may be a widespread strategy for P acquisition (Luo et al., 2009). Many marine organisms appear to contain both a PhoX and a PhoD type alkaline phosphatase. Unlike PhoX, PhoD is thought to be predominantly cytosolic, consistent with the presence of organic phosphate uptake systems in this environment (Luo et al., 2009). Note that in these systems, the induction of the PhoD and PhoX enzymes is an acclimation strategy for P-limitation, whereas the use of these Ca-dependent enzymes as substitutes for the corresponding Zn-dependent enzymes appears to be a Zn-sparing adaptation.
Another example of adaptation to Zn limitation is offered by the occurrence of a novel membrane-associated copper containing SOD in Mycobacterium tuberculosis (Spagnolo et al., 2004). In this organism, the structure of a prototypical CuZnSOD is modified so that the protein is stable without zinc. The authors speculate that this may be an adaptation to the metal competitive environment of the host macrophage phagosome where these pathogenic bacteria reside.
6. Synthesis of Zn-independent isozymes during acclimation to Zn limitation
As noted above, B. subtilis can acclimate to Zn limiting growth conditions by derepression of alternative proteins that lack Zn, but can functionally replace Zn-dependent homologs. These include a Zn-independent GTP cyclohydrolase and ribosomal proteins. The presence of two or more differentially expressed isozymes with different metal ion requirements may in fact be a widespread mechanism for acclimation to Zn limitation.
Hints to the roles of alternative forms of a protein can often be obtained from their regulation. Analysis of predicted Zur regulons in various proteobacterial species has identified nine Zn-dependent enzymes with paralogs likely to be regulated by Zur (Haas et al., 2009). These include HisI, PyrC, HemB, CysRS, ThrRS, QueD, carbonic anhydrase, an N-acetylmuramoyl-L-alanine amidase, and DksA. While the adaptive role of these various paralogs can be envisioned, the model has only been tested (to date) for DksA. P. aeruginosa encodes two genes encoding DksA, an RNA polymerase-binding transcription factor involved in the stringent response. One, DksA1, contains an essential Zn ion bound to four Cys residues. Under conditions of Zn limitation, induction of a Zur-regulated isozyme (DksA2) lacking associated Zn enables the cell to replace DksA function with a non-Zn-dependent protein (Blaby-Haas et al., 2011).
Further evidence for the prevalence of this substitution strategy has emerged from a bioinformatic analysis of Zn-binding proteins in the Protein Data Bank to identify apparent orthologs that lack the known or presumed Zn-binding ligands (Zhang and Gladyshev, 2011). This analysis identified C+/C− pairs of isozymes for eight different r-proteins, four subunits of DNA polymerase III holoenzyme, peptide deformylase, three different aminoacyl tRNA-synthetases (Met, Ile, Leu), methionine-R-sulfoxide reductase, DnaJ, 5-aminolevulanic acid dehydratase, and adenylate kinase (Zhang and Gladyshev, 2011). In many cases, organisms have either the presumed Zn-dependent or the Zn-independent version. However, where both forms co-occur in a single organism, their regulated expression may serve as a mechanism of acclimation analogous to those noted above for B. subtilis (Sankaran et al., 2009) and P. aeruginosa (Blaby-Haas et al., 2011). A related example is the heme biosynthesis enzyme porphobilinogen synthase, which exists in both Zn-dependent and Zn-independent forms (Jaffe, 2003, Frere et al., 2005). In this case there is no evidence of regulated expression as an acclimation mechanism, so this is perhaps better considered as an adaptation strategy.
E. Copper (Cu): a versatile redox cofactor
Cu is found as a cofactor in all kingdoms of life where it is particularly useful as a catalyst of redox reactions, such as in electron transfer proteins azurin and plastocyanin, and reactions involving O2 chemistry, such as in hemocyanin for binding of O2 for transport (Crichton and Pierre, 2001). A recent (2008) survey of 450 sequenced bacterial genomes showed that most (72%) have at least one Cu enzyme (Ridge et al., 2008). Two oxidation states are relevant in biology, Cu(I) and Cu(II), with different active site geometries and ligands stabilizing one or the other, and hence allowing a range of redox potentials in cuproproteins. Most Cu proteins are classified into one of three groups – the type I proteins include structurally well-characterized plastocyanin and azurin as prototypes with a tightly bound mononuclear Cu (Redinbo et al., 1994, Malmström and Leckner, 1998, Gray et al., 2000, Choi and Davidson, 2011). The sites are often referred to as “blue” copper because of the intense absorption around 600 nm from the thiolate ligand to Cu(II) charge transfer transition. The type 2 proteins, also mononuclear, are exemplified by CuZnSOD, galactose oxidase and amine oxidase, and these lack intense absorption bands from the metal center. Type 3 proteins are binuclear and are represented by hemocyanin and tyrosinase. Some enzymes like ascorbate oxidase and ceruloplasmin, which are in a group referred to as “multicopper oxidases,” have multiple Cu binding sites (Messerschmidt and Huber, 1990).
The catalytic potential of Cu is similar to that of Fe, except that more positive midpoint potentials are possible with Cu, compatible with reactions involving molecular O2. The evolution of cuproenzymes is believed to have occurred more recently than Fe-containing enzymes. As mentioned above, Fe was readily available at the origin of life both because of its abundance and its solubility in water in the ferrous form. By contrast Cu(I) was less bioavailable, especially in the presence of sulfide. With the build-up of molecular O2, Cu became more readily accessible to life as the more soluble Cu(II) ion. Consistent with the notion of Cu proteins being newer to the protein landscape is the observation that Firmicutes, which are considered to be relatively ancient organisms, tend not to use Cu enzymes, and among the 35 archaeal genomes analyzed, 69% showed no evidence of Cu enzymes (Ridge et al., 2008). The most commonly occurring Cu proteins are cytochrome oxidase and CuZnSOD, both of which are important only in an aerobic world. Cytochrome oxidase enables energy generation through use of O2 as an electron acceptor, and SOD is essential for detoxifying superoxide, one of the harmful side products of O2 chemistry. Among the bacteria, Cu non-users tend to be anaerobic (73% of those surveyed) while Cu users are aerobic (94%), and in the Archaea, the correlation is complete, with all nonusers being anaerobic and all users being aerobic (Ridge et al., 2008). Accordingly, the Cu-utilizing and sparing regulatory pathways are focused on enzymes involved in bioenergetics (Fig. 10).
Figure 10.
Overview of the biological role of Cu and known sparing and recycling mechanisms.
1. Cu homeostasis in methanotrophs
Methane is the most inert hydrocarbon but there are bacteria, referred to as methanotrophs, that can use methane as their sole source of carbon and energy. These bacteria are a subset of a group known as methylotrophs, which are aerobic bacteria that use one carbon compounds that are more reduced than formate as sources of carbon and energy, and they assimilate formaldehyde as a major source of cellular carbon (Hanson and Hanson, 1996). The first step in the aerobic methane oxidation pathway in methanotrophs, the generation of methanol, is catalyzed by methane monooxygenase. This enzyme comes in two flavors – a Cu containing form associated with membranes and an Fe-containing form that is soluble. The multi-subunit Cu form has mononuclear and dinuclear Cu sites in the extramembrane region of the complex (analogous to subunit II of cytochrome oxidase, see below) (Nguyen et al., 1998, Lieberman and Rosenzweig, 2005). The soluble Fe form is a carboxylate-bridged di-iron enzyme (Elango et al., 1997, Merkx et al., 2001). The Cu form is the more prevalent enzyme found in methanotrophs in both the α and γ proteobacteria clades, but a few methanotrophs (including marine and freshwater species) have both forms (Murrell et al., 2000b, Nakamura et al., 2007). The expression of one or the other is dependent on the Cu nutrition status (Murrell et al., 2000a, Hakemian and Rosenzweig, 2007). The di-iron enzyme is expressed when there is low Cu in the growth medium while the Cu-enzyme is expressed when there is an adequate supply of Cu. An NADP+-linked formaldehyde dehydrogenase is co-expressed with the soluble di-iron enzyme while a dye-linked formaldehyde dehydrogenase is co-expressed with the membrane-associated Cu form, indicating that the choice of metal cofactor for the first step dictates the subsequent route of electron flow for the entire pathway (Zahn et al., 2001).
This regulated enzyme substitution has been documented in many different species, including Methylococcus capsulatus (Bath) and Methylosinus trichosporium OB3b, representing both the γ and α proteobacterial clades, respectively. The Cu enzyme is presumed to be the preferred form because of the higher redox potential of Cu, with the Fe enzyme serving as a backup for this critical step in methane metabolism. The methanotrophs that express the backup Fe form have lower Cu requirements for growth (Graham et al., 1993, Hanson and Hanson, 1996). The Cu enzyme appears to offer an advantage based on the higher growth yields and higher affinity for methane of cells expressing this type. The importance of Cu to these organisms is underscored by the occurrence of two independent Cu acquisition pathways, one involving synthesis of a peptide based chelator, methanobactin, followed by energy-dependent uptake of the Cu-methanobactin complex, and a second route of passive transport of unchelated Cu (Kim et al., 2004, Balasubramanian et al., 2011). The former route may facilitate Cu mobilization from mineralized sources (Knapp et al., 2007).
In M. capsulatus and M. trichosporium, the expression of the gene cluster encoding the di-iron enzyme is negatively regulated by Cu ions while the expression of the pmo genes encoding the Cu enzyme is concomitantly increased, implicating a common regulator in the Cu-responsive switch between one or the other (Nielsen et al., 1997). In both organisms, the MmoR and MmoG proteins have DNA binding activity and are involved in regulation of the mmo genes (encoding the soluble Fe enzyme), but the identity of the Cu sensor is not yet known (Csáki et al., 2003, Scanlan et al., 2009).
2. Cu homeostasis and bacterial respiratory pathways
Although Cu is stimulatory or even required for growth for numerous other bacterial species, the responses to Cu deprivation have been studied in only a few systems. Just as Mn is key for oxygenic photosynthesis, Cu is a key element for aerobic respiration. In P. aeruginosa it has been noted that aerobic respiration requires one of four different terminal oxidases (Frangipani et al., 2008). Three operons encode paralogous cytochrome c oxidases of the heme-Cu superfamily. A fourth cytochrome bd-type cyanide-insensitive oxidase lacks Cu in its active site. When P. aeruginosa is grown in the absence of Cu (through repeated sub-cultures) or in the presence of strong Cu chelators, growth is only possible if the Cu-free oxidase is present. Moreover, synthesis of the Cu-free oxidase is strongly induced by Cu depletion, although this seems to be an indirect effect mediated by the failure of the Cu-dependent enzymes to function (Frangipani et al., 2008). Growth of P. aeruginosa therefore seems not to absolutely require Cu, and this organism does not scavenge Cu using chelators analogous to methanobactin. It does, however, synthesize a periplasmic protein of the ScoC family postulated to function in Cu acquisition and trafficking (Frangipani and Haas, 2009). The effects of Cu deprivation have also been reported for Synechocystis sp. where it prevents respiration while allowing continued photoautotrophic growth (Duran et al., 2004). In the marine bacterium Pseudomonas stutzeri, Cu deprivation leads to a blockage in denitrification (Matsubara et al., 1982). This is due to the presence, in nitrous oxide reductase, of both a binuclear Cu center and a novel [Cu4S2] cluster (Pomowski et al., 2011).
3. Cu homeostasis in fungi
Copper homeostasis has been studied in various fungi including S. cerevisiae, Schizosaccharomyces pombe, Dactylium dendroides, Cryptococcus neoformans, Candida spp., and Podospora anserina (Shatzman and Kosman, 1978, Bird, 2008). Copper sensors that respond to both low and high Cu were discovered by genetic approaches in S. cerevisiae and then identified by homology in other fungi. In S. cerevisiae, Mac1 controls transcription of copper assimilators in the deficiency situation, and a related protein Ace1 controls the copper toxicity response (Winge, 1998). In S. pombe the regulator Cuf1 is a hybrid between Mac1 and Ace1, in P. anserina a Mac1 orthologue is called Grisea, and in C. neoformans it is called Oxy2 (Borghouts and Osiewacz, 1998, Labbé et al., 1999, Nyhus and Jacobson, 2004). Each of these organisms has a mitochondrial Cyt oxidase, a cytosolic CuZnSOD and multicopper oxidases (ferroxidases) required for high affinity Fe transport (Gralla and Kosman, 1992, Askwith and Kaplan, 1997, Carr and Winge, 2003). In addition, there are other Cu enzymes like galactose oxidases, tyrosinase, amine oxidases and laccases, whose functions are not essential or only required for a particular physiological program (such as the laccase requirement for virulence of C. neoformans, Zhu et al., 2003).
In a pioneering early study with D. dendroides, researchers noted the phenomenon of Cu-sparing and the prioritized use of Cu (when its supply was limited), and they proposed the concept of recycling (Shatzman and Kosman, 1978). They noted that galactose oxidase was synthesized and excreted but not metallated in Cu-deficient (<10 nM Cu) cells and it accumulated as the apoform. Clearly this enzyme does not compete effectively for intracellular Cu. CuZnSOD synthesis on the other hand was reduced, and its activity was replaced by induced synthesis of a non-mitochondrial MnSOD (Shatzman and Kosman, 1979). Comparison of cyanide-sensitive (i.e. Cu-dependent) SOD activity in deficient (10–30 nM) vs. replete (5–10 µM) cells indicated that 83% of total SOD activity is attributed to the CuZn enzyme in replete cells vs. 17% in the deficient cells. This Cu-sparing mechanism allowed maintenance of the Cyt oxidase levels independent of Cu nutrition status. As the deficient cells divided and further depleted the Cu pools, only Cyt oxidase is maintained, and the authors proposed that this might occur by degradation of CuZnSOD and recycling of the constituent Cu. At the time, the genes and regulators were not known, but a similar Cu sparing response in P. anserina, where apoforms of laccase and tyrosinase accumulate and MnSOD replaces a CuZnSOD in Cu-deficiency, is controlled by Grisea (Osiewacz and Nuber, 1996, Borghouts et al., 2001).
S. cerevisiae does not replace CuZnSOD with a Mn-enzyme, which may explain the loss of Cyt oxidase in Cu-deficiency resulting in poor growth (Giorgio et al., 1963). Nevertheless, gene expression and phenotype analysis of S. cerevisiae in response to variation in Cu nutrition suggested that Cu-independent Fe assimilation pathways (involving Fe-siderophore uptake) were up-regulated, presumably to compensate for loss of the Fet3 multicopper oxidase route for Fe uptake (van Bakel et al., 2005). In S. pombe, Cuf1 down-regulates components of the Cu-dependent Fe uptake pathway, perhaps as a Cu-sparing strategy (Labbé et al., 1999). In S. cerevisiae, under conditions of fermentative growth (where respiration is no longer essential) Cyt oxidase and the respiratory chain is down-regulated (van Bakel et al., 2005). The loss of Cyt oxidase in Cu-limited cultures of bacteria and fungi is well-documented (Hubbard et al., 1989, Gabel et al., 1994).
In some fungi, Cu limitation activates the expression of an alternative oxidase, which is a Cu-independent di-iron enzyme (Downie and Garland, 1973, Scheckhuber et al., 2009). This pathway bypasses two of the three sites of proton pumping in the respiratory chain and is therefore less effective than the Cu-dependent pathway, which explains the prioritization of Cyt oxidase in organisms that rely on respiration. The replacement of Cyt oxidase with an alternative oxidase is not viewed as a Cu-sparing mechanism, since it is not directly regulated by Cu nutrition and the Cu sensor, but rather by the redox state of the respiratory chain. It is likely that the alternative oxidase functions as an electron valve to reduce the generation of reactive oxygen species resulting from loss of Cyt oxidase function.
4. Cu homeostasis in algae and cyanobacteria
Plastocyanin is a blue copper protein that was isolated from plant leaves because of its abundance and spectroscopic properties (Katoh and Takamiya, 1961). Its function in the Z-scheme of photosynthesis, where it catalyzes the transfer of electrons from the Cyt b6f complex to PS I, was established in 1963 by analysis of a plastocyanin-deficient mutant of C. reinhardtii (Gorman and Levine, 1965). Plastocyanin was biochemically characterized from many land plants, and it was the first Cu protein to have its structure solved (Boulter et al., 1977, Guss and Freeman, 1983). When researchers analyzed the algae and cyanobacteria on the other hand, they were unable to isolate plastocyanin from some of them (Wildner and Hauska, 1974, Kunert and Böger, 1975), which led to the finding that a soluble c-type cytochrome could cover its function in many green algae and cyanobacteria (Bohner et al., 1980a, Bohner et al., 1980b, Sandmann and Böger, 1980, Sandmann et al., 1983, Briggs et al., 1990, Zhang et al., 1992). This cytochrome, originally named for its α-absorption maximum (hence c-552 or c-553), was classified on the basis of its function and eventually named Cyt c6 (Pettigrew and Moore, 1987).
The synthesis of plastocyanin (Cu) and Cyt c6 (Fe) was found to be reciprocally dependent on Cu nutrition in both cyanobacteria (Prochlorothrix hollandica, Anabaena, and Synechocystis spp.) and green algae (C. reinhardtii, various Scenedesmus spp., and Pediastrium boryanum) (Merchant and Bogorad, 1986b, Bovy et al., 1992, Li and Merchant, 1992, Nakamura et al., 1992, Ghassemian et al., 1994, Arudchandran and Bullerjahn, 1996, Miramar et al., 2003). The phenomenon has been observed also in the natural environment. When soluble photosynthetic electron transfer catalysts were isolated from a cyanobacterial bloom in the Potomac, Cyt c6 was abundant, but when the organisms were cultured in the laboratory (in medium amended with Cu), plastocyanin could be isolated (D. Krogmann, pers. comm.). Functional equivalence, suggested from biochemistry, was firmly established in the plastocyanin-less mutant pcy1-ac208 of C. reinhardtii (Wood, 1978, Merchant and Bogorad, 1987a). In brief, the acetate-requiring phenotype of pcy1-ac208 could be suppressed by growth on medium lacking Cu, which induced the expression of the heme-containing cytochrome. Biochemical characterization of the two proteins purified from several cyanobacteria indicated that their pIs (which ranged from 4 to 9) co-varied suggesting that they were co-evolving in response to changes in common reaction partners (Ho and Krogmann, 1984).
The signal transduction pathway responsible for the switch between plastocyanin and Cyt c6 was shown in C. reinhardtii to respond directly to Cu rather than to feedback from the redox state of the chloroplast electron transfer chain (Merchant and Bogorad, 1987b). Since then, C. reinhardtii has become a premier reference organism for understanding mechanisms of Cu sensing and sparing in photosynthetic systems (Merchant, 1998). C. reinhardtii has 3 abundant Cu proteins, plastocyanin, Cyt oxidase and a multicopper oxidase involved in Fe assimilation, plus a number of other less abundant ones (Li et al., 1996, Chen et al., 2008, Remacle et al., 2010, Castruita et al., 2011). In an Fe-replete situation (when ferroxidase expression is repressed), the Cu quota is determined largely by plastocyanin and Cyt oxidase. The Cu-requirements of pcy1 mutants gives an indication of the large contribution of plastocyanin to the cellular Cu quota (Hill and Merchant, 1992). Regulation of plastocyanin in C. reinhardtii occurs post-translationally by regulated degradation of the protein (Merchant and Bogorad, 1986a, Li and Merchant, 1995) while the CYC6 gene encoding Cyt c6 is transcriptionally regulated by a Cu-sensing transcription factor (named CRR1 for copper response regulator) that binds Cu response elements (Quinn and Merchant, 1995, Quinn et al., 2000, Kropat et al., 2005, Sommer et al., 2010). In C. reinhardtii, CRR1 controls all known responses to Cu-deficiency, including plastocyanin degradation (Eriksson et al., 2004). The replacement of plastocyanin by Cyt c6 serves to spare Cu. Degradation of plastocyanin is important for Cu recycling as evidenced by the growth phenotype of crr1 mutants, which cannot maintain Cyt oxidase because they cannot recycle Cu from plastocyanin (J. Kropat and SM, unpublished).
The amount of Cu required to repress the CYC6 gene is precisely dependent on the abundance of intracellular Cu proteins (Merchant et al., 1991). For instance, if Cu in the medium is sufficient to support the synthesis of 50% of the cellular plastocyanin quota (~8 × 106 molecules per cell), the CYC6 gene is expressed to 50% of its maximal level. When the cells divide and hence reduce available Cu so that the plastocyanin quota is now reduced to 25% of its quota, CYC6 gene expression is increased to 75% of its maximum level, presumably to allow synthesis of just enough more Cyt c6 to compensate for the deficit.
A recent transcriptome study identified an additional example of Cu-sparing in C. reinhardtii (Castruita et al., 2011). A gene encoding a flavin-utilizing amine oxidase is a CRR1 target and is up-regulated in Cu-deficiency. It was suggested that the flavoenzyme may serve as a backup to copper amine oxidases, which function in the marine environment to mobilize N from primary amines (Palenik and Morel, 1991). Indeed, in C. reinhardtii, genes encoding amine oxidases are up-regulated in N-deficiency (Boyle and Merchant, unpublished). There are a number of other metabolic modifications in Cu-deficient C. reinhardtii, including a change in the level of desaturation of thylakoid membrane galactolipids and increase in the expression of genes encoding O2-dependent enzymes (Castruita et al., 2011). These responses are dependent on CRR1 and confirmed at the level of the proteome in every case where it was queried. The purpose of these modifications is not understood yet, but one view is that they may be required to accommodate the change from use of plastocyanin in photosynthesis to use of a structurally distinct Cyt c6. The cold sensitivity and reduced photosynthetic electron transport of a cyanobacterial plastocyanin mutant that compensates by increased expression of petJ encoding Cyt c6 is not inconsistent with this view (Clarke and Campbell, 1996). The electron transfer reactions in which they participate rely on complex diffusion steps in a spatially restricted intracellular compartment (the thylakoid lumen) and this may require membrane reorganization (Hervás et al., 1998).
Besides the sparing and recycling responses described above, C. reinhardtii also changes expression of two O2-dependent enzymes in the tetrapyrrole biosynthetic pathway in Cu-depleted conditions. The changes are dramatic and physiologically relevant, since mutants in one of the target genes display Cu-conditional chlorosis, but the underlying rationale remains a puzzle (Hill and Merchant, 1995, Moseley et al., 2000, Quinn et al., 2002). Evidently, there is a connection to anaerobiosis because CRR1 and its target genes are required also in situations of low O2 tension in C. reinhardtii (Quinn et al., 2002, Eriksson et al., 2004). Perhaps the connection persists from ancestral mechanisms where Cu(II) availability was dependent on O2 levels.
Cyt c6 has been lost in the land plant lineage, clearly pointing to an evolutionary advantage of plastocyanin. One thought is that commitment to plastocyanin, as noted in a diatom, is an adaptation to economize on Fe (Peers and Price, 2006). This may be true in the marine environment but other more prevalent and more effective Fe-sparing mechanisms (discussed above), like the replacement of ferredoxin by flavodoxin, have not been found yet in the Viridiplantae lineage, suggesting that Fe limitation is not likely to be the driving force for a commitment to plastocyanin in land plants. The evolution of plastocyanin therefore facilitated photosynthesis but increased the dependence on Cu. Although Cyt c6 is absent, CRR1 homologs have been identified (Yamasaki et al., 2009, Bernal et al., 2012). The Arabidopsis orthologue of C. reinhardtii CRR1, SPL7, functions to spare Cu for plastocyanin by replacing CuZnSOD with an FeSOD (Pilon et al., 2009). Interestingly, the green algae that express Cyt c6 as a plastocyanin replacement have also lost the otherwise ubiquitous gene(s) encoding CuZnSOD (Asada et al., 1977). This is likely a permanent adaptation to long term Cu-deficiency experienced by these organisms. C. reinhardtii uses an FeSOD in the chloroplast, which can be supplemented with a MnSOD in Fe-deficient cells, plus MnSODs in the mitochondria and cytosol instead of the Cu enzyme (Allen et al., 2007b). CuZnSOD has also been abandoned in favor of MnSOD in crustaceans where Cu is in demand as the O2-carrier in hemocyanin (Brouwer et al., 2003).
Cu sparing by replacement of plastocyanin with Cyt c6 is widespread in the cyanobacteria (cited above). Since the b6f complex in these organisms transfers electrons to both PS I and Cyt oxidase, the proteins serve both respiration and photosynthesis (Sandmann and Malkin, 1984). The regulatory pathways (.i.e. Cu sensor, cis-acting sequences and DNA binding proteins) are not yet elucidated, but the petJ gene (encoding the Cyt) responds transcriptionally to Cu nutrition status while the petE gene (encoding plastocyanin) responds transcriptionally in some cases and post-transcriptionally in others (Briggs et al., 1990, Bovy et al., 1992). In the alga P. boryanum, a 5’ truncated form of the plastocyanin-encoding mRNA (which lacks the initiation codon) is generated in Cu-depleted cells, which prevents synthesis of the polypeptide (Nakamura et al., 2000). This would serve a Cu-sparing function but whether pre-existing plastocyanin is also degraded for Cu recycling is not known.
F. Elemental substitution: a widespread adaptation for bypassing limitation
Although the regulatory pathways and mechanisms may not always be well understood, the strategy of developing alternative enzymes or pathways that can functionally substitute for a limiting metal extends beyond the Fe, Zn, and Cu homeostasis systems reviewed above. Here, we provide a brief survey of those systems where selection has led to the presence of substitutions that help cells maintain function in response to limitation for Mn, Ni, Co, Mo, V, W, or Se. Remarkably, for nearly all elements for which a biological function as been ascertained, those cells that face limitation have either adapted, in some instance by dispensing with a requirement completely, or developed substitute pathways that are either constitutively expressed or expressed conditional on elemental depletion of the preferred nutrient. The existence of these mechanisms allows life to exploit more diverse ecological niches, which is a powerful driving force.
1. Manganese (Mn): a key to oxygenic photosynthesis and cofactor for SOD
Mn is widely used in biology and is often critical for growth, but the processes that require Mn are often not well understood. The single most critical role for Mn, considered globally, is for its role in the water-splitting complex of PS II in oxygenic photosynthesis. As a result, the Mn quota for the cyanobacterium, Synechocystis sp. strain PCC 6803, is >100-fold higher that of the purple bacterium Rhodobacter capsulatus (Keren et al., 2002). Synechocystis cells grow well at levels of Mn as low as 100 nM; at lower levels they remodel their photosynthetic complexes to prevent photooxidative damage (Salomon and Keren, 2011). Mn appears to be concentrated from the environment and stored associated with the cell envelope (Keren et al., 2002), but the details of this storage and mobilization process are not well understood. However, there is little evidence for an obvious Mn-sparing response, perhaps because no other single component of the cell has such a high demand for Mn.
Phototrophic growth has a similarly high Mn requirement in the Eukaryote, C. reinhardtii (Allen et al., 2007b). In this organism, Mn deficiency is elicited by growth with <0.5 µM Mn; lower levels lead to defects in photosynthesis. The first response to declining Mn availability is the expression of acquisition pathways including a member of the NRAMP family (NRAMP1). Mn deficiency also led to induction of two PHO84 family transporters thought to import phosphate:Mn complexes. Possibly as a consequence, Mn deficient cells are secondarily P deficient. Further reduction in Mn availability leads to a loss of MnSOD activity which, in general, precedes the eventual loss of photosytem II activity. This suggests a prioritization of Mn usage, although the corresponding mechanisms are not well understood (Allen et al., 2007b).
Mn is also required for growth of many non-photosynthetic bacteria, although a universal requirement is by no means established. B. subtilis requires ~20 nM Mn for growth and sporulation requires higher levels still. In Lactobacillus plantarum, mutants defective in Mn uptake were only able to grow when the medium was supplemented with >10 mM Mn (Hao et al., 1999), orders of magnitude higher than for wild-type cells. This is consistent with the high intracellular levels of Mn present in this genus, which is notable for its lack of an apparent Fe requirement (Weinberg, 1997). The highly radioresistant bacterium Deinococcus radiodurans also accumulates high intracellular levels of Mn, which in this and other organisms is correlated with greater resistance to damage by reactive oxygen species (Daly, 2009).
Although the Mn-deprivation responses have been characterized in many different organisms, and the corresponding regulatory proteins have often been defined, there is surprising little evidence for a Mn-sparing response. The B. subtilis MntR regulator senses Mn directly and represses acquisition systems, but little else (Que and Helmann, 2000). Similarly, S. aureus up-regulates Mn import and this helps resist the inhibitory effects of the divalent metal sequestration protein calprotectin (Hammer and Skaar, 2011). Despite the fact that cells fail to grow in the absence of Mn, the essential processes requiring this metal have not been identified. One possibility is suggested by the presence of a single, Mn-dependent RNR in B. subtilis and related Firmicutes (Zhang and Stubbe, 2011).
Mn also plays a critical role in S. cerevisiae, where it serves as cofactor for a mitochondrial SOD (Sod2), but there is no evidence to suggest the presence of a specific metallochaperone (Reddi et al., 2009). Since Sod2 also can bind Fe, which leads to an inactive enzyme, mechanisms that ensure proper metallation are thought to be present in cells but are, as yet, poorly understood (Aguirre and Culotta, 2012). For these and related systems there is a relatively large Mn requirement and conditions of deprivation are easily achieved in the laboratory. We can therefore anticipate that ongoing studies will resolve the key Mn-dependent pathways required to support growth and may reveal sparing and recycling mechanisms analogous to those noted for other elements.
2. Nickel (Ni): a key cofactor for both C and N cycling
Ni is an essential trace nutrient for many Bacteria and Archaea and for many plants, but its roles are generally limited to, at most, a handful of enzymes (Ragsdale, 2009, Zhang and Gladyshev, 2010). The single most widely distributed Ni-dependent enzyme, and the only known Ni enzyme in eukaryotes (Zhang et al., 2009), is urease, which plays a key role in N-cycling <Figure 11>. Interestingly, C. reinhardtii, which can use urea as the sole N source, uses a Ni-independent enzyme for urea metabolism, although other organisms in the plant lineage use the Ni enzyme. Other Ni-dependent proteins include [NiFe] hydrogenase, a Ni form of SOD, and CO dehydrogenase (Kaluarachchi et al., 2010). Ni was relatively abundant in the ancient oceans in which life evolved and anaerobic bacteria and Archaea tend to have the most Ni-dependent enzymes (Zhang et al., 2009).
Figure 11.
Overview of the biological roles of Co (B12) and Ni and known sparing mechanisms.
Many bacteria appear to have only one or two Ni-dependent proteins, but nevertheless have dedicated Ni transport and regulation systems. For example, E. coli has a [NiFe] hydrogenase and a Ni urease, and encodes the NikR-regulated NikABCDE transport system (reviewed in Li and Zamble, 2009). In contrast, B. subtilis has only a single predicted Ni enzyme (urease) and no obvious Ni uptake system. Indeed, efforts to demonstrate a dependence on Ni for urease activity, or a requirement for Ni for growth on urea, have been unsuccessful, raising the possibility that this enzyme is, in fact, not Ni dependent (SE Gabriel and JDH, unpublished studies). This suggests that B. subtilis may have dispensed with a requirement for Ni.
a. Ni, methanogenesis, and the great oxidation event
In the oceans of the Archaean era (before 2.5 GYA), Ni was relatively abundant with estimated concentrations as high as 400 nM. Although cause and effect are uncertain, a large decline in oceanic Ni accompanied the great oxidation event (Konhauser et al., 2009) leading, eventually, to Ni concentrations in the surface waters of the present day ocean of ~1–2 nM (Saito, 2009). Despite this low level, laboratory studies combined with genomic analyses suggest that Ni is a required nutrient for many marine Synechococcus and all Prochlorococcus strains (Dupont et al., 2008a). While quite low, it should be remembered that Fe, which is used by many more organisms, may be even less available (<0.05 nM) (Saito, 2009).
Early evolving lineages, including the Archaea, have comparatively more Ni-dependent enzymes whereas some Bacteria, and many Eukaryotes (including vertebrates, insects and yeasts), may have dispensed with a Ni requirement as they lack obvious Ni transporters and urease (Zhang et al., 2009). It has been reported that mammals have a Ni requirement, but no Ni-dependent enzymes have been identified. It is notable, however, that human blood contains ~0.5 nM Ni, similar to the ocean in which our (distant) vertebrate ancestors evolved (Ragsdale, 2009).
i. Substitutions for F420-reducing [NiFe]-hydrogenase in Methanobacteria
The methanobacteria are noteworthy for having a relatively large number of Ni-dependent enzymes including some of the key enzymes in methanogenesis itself. The Ni-dependent methyl-CoM reductase is responsible for the production of all biogenic methane and thereby has implications for the global climate both in the Archaean era and as a contributor to global warming today (Singh et al., 2010).
Methanogens use H2 as a reductant to convert CO2 to methane (CH4) in a complex reaction involving several Ni enzymes. As a result, methanogens grow optimally with >1 µM Ni and become limited at lower levels (Thauer et al., 2010). Methanogenesis uses a flavin-based cofactor (F420) that is reduced by a hydrogenase. Under Ni replete conditions, reduction of F420 is catalyzed by a multi-subunit flavoprotein containing both Ni and several Fe/S centers (the [NiFe] hydrogenase). Under Ni limiting conditions, the specific activity of this enzyme declines at least 100-fold and it is functionally replaced by two other enzymes that do not require Ni and use a tetrahydromethanopterin cofactor. These two enzymes (an [Fe] hydrogenase and an F420-dependent methylenetetrahydromethanopterin dehydrogenase) are up-regulated several fold in response to Ni deficiency (Afting et al., 2000, Thauer et al., 2010). The [Fe] hydrogenase can functionally replace the [NiFe] enzyme for H2 oxidation, but it is catalytically inferior with a Km for H2 of 0.2 mM vs. 0.01 mM for the [NiFe] enzyme. As a result, Ni-limited cells must synthesize relatively more [Fe] hydrogenase to maintain the same catalytic efficiency as for the [NiFe] enzyme (Thauer et al., 2010). C. reinhardtii, which also has an active hydrogenase, uses only the [Fe] type of enzyme (Happe et al., 1994), and given its Ni-independent urease has thereby dispensed with any requirement for Ni as a nutrient.
b. Substitution of Ni-urease with an Fe-variant in Helicobacter mustelae
An interesting Ni-sparing response has been identified in an organism that is highly reliant on urease for survival, Helicobacter mustalae. H. mustalae is a gastric pathogen of ferrets and, like its human counterpart H. pylori, relies on NH3 released by urease to function as a base and locally neutralize the highly acidic pH of the stomach (Stoof et al., 2008). Urease can constitute up to 10% of soluble protein in H. pylori, which creates a significant Ni demand. Since ferrets are carnivores, and meat is much lower than vegetable matter in Ni content, H. mustalae may commonly face Ni limitation. The result is the presence of two, differentially regulated operons encoding urease. When Ni is limiting, and Fe is abundant, expression of the ureA2B2 operon is induced and this Fe-containing urease functionally replaces the Ni-dependent isozyme (Stoof et al., 2008, Carter et al., 2011). The structure of the Fe urease reveals a dinuclear metal center. Although this enzyme is not as active as the Ni isozyme, it is sufficient to provide acid resistance (Carter et al., 2011).
c. Recycling of Ni from urease for SodN in marine Synechococcus
Ni is present at low levels (ca. 2 nM) in the open ocean, but in comparison to other metals, many of which are present in even lower concentration and/or in greater demand, Ni is a comparatively available resource. Synechococcus spp. actively transport Ni from their environment and use this metal as a cofactor for both urease and NiSOD. In laboratory studies, the former is required for growth on urea as a nitrogen source, but for one studied ocean isolate (Synechococcus WH8102) Ni is required even when NH4+ is provided as N source (Dupont et al., 2008b). This Ni requirement was ascribed to the fact that this organism encodes only a single SOD, which is a Ni enzyme and essential for phototrophic growth. When transferred from urea to NH4+ as nitrogen source, Ni-limited cells were able to grow for several doublings before becoming Ni limited, suggesting that Ni was likely reallocated from urease (which was no longer needed) to NiSOD. Phylogenomic comparisons indicate that NiSOD and FeSOD are often mutually exclusive which leads to the hypothesis that all marine Prochlorococcus strains have an obligate Ni requirement (they only encode a NiSOD), whereas most Synechococcus, Trichodesmium, and Crocosphaera also encode a NiSOD and are therefore at least partially dependent upon Ni for growth (Dupont et al., 2008b).
d. Substitutions for Ni-dependent enzymes
Despite the relatively low number of Ni-dependent enzymes in most organisms (Zhang et al., 2009), the presence of alternative, non-Ni-dependent alternatives appears to be common. For example, Streptomyces griseus encodes both a NiSOD and an FeZnSOD. These two isozymes are reported to be under reciprocal regulation by Ni: elevated levels of Ni stimulate the production of NiSOD and repress the FeZnSOD. The latter is due to Ni-dependent metalloregulation that involves a complex of an ArsR-family repressor (SrnR) and a Ni-binding sensor protein (SrnQ) (Kim et al., 2003). A similar reciprocal regulation is observed between NiSOD and FeSOD in S. coelicolor, but this regulation is mediated by a Ni-sensing Fur family protein designated as Nur. Nur, when bound to Ni, directly represses the transcription of the FeSOD-encoding gene while indirectly activating transcription of the gene for NiSOD (Ahn et al., 2006). A final example of alternatives to Ni-dependent enzymes is provided by the glyoxalases, widely distributed enzymes that detoxify methylglyoxal. Most eukaryotic glyoxalases use Zn as cofacter, whereas many bacterial enzymes use Ni or Co. While most organisms have one or the other, in Pseudomonas aeruginosa both types are present (Sukdeo and Honek, 2007). This redundancy presumably allows efficient methylglyoxal detoxification over a wider range of growth conditions relative to organisms with only a single isozyme.
3. Cobalt (Co): vitamin B12 dependent and independent enzymes
Co, most commonly in the form of the corrinoid-based cofactor known as vitamin B12, is required by most prokaryotes (Bacteria and Archaea) and many Eukaryotes (Figure 11). The major exceptions appear to be plants and fungi (Rodionov et al., 2003). Many organisms (including humans) that do not synthesize B12 nevertheless have a dietary requirement for it. In humans, B12 is needed for methionine synthesis and as a cofactor for propionate metabolism. Organisms that require B12 but lack the potential for synthesis have B12-transport systems for acquiring this key nutrient. The expression of B12 uptake and synthesis functions is often translationally repressed by cytosolic B1, which can be sensed by an RNA-based (riboswitch) mechanism (Nahvi et al., 2004).
A common adaptation to B12 limitation is the substitution of B12-dependent enzymes with B12-independent isozymes. For example, E. coli encodes two forms of methionine synthase (Drummond and Matthews, 1993). In the absence of exogenous B12 (which E. coli is unable to synthesize de novo), the Zn-dependent MetE protein is required for methionine synthesis (Hondorp and Matthews, 2009). Conversely, in the presence of B12, synthesis of MetE is reduced (Wu et al., 1992). Reciprocal regulation of the two methionine synthase isozymes is also noted in M. tuberculosis. In this organism, a B12-sensing riboswitch selectively represses the transcription of the B12-independent isozyme (Warner et al., 2007). We previously noted the functional redundancy between the E. coli Fe and Mn forms of ribonucleotide reductase, an enzyme required for DNA synthesis. In S. coelicolor, two RNR enzymes are used. The class 1a enzyme is oxygen dependent while the class II enzyme requires B12. When B12 is available, the class 1a enzyme is repressed (Borovok et al., 2006).
Functional redundancy of methionine synthases is also widespread in the algae. Amongst the algae, B12 is obtained from bacterial symbionts. When present, B12-represses the synthesis of the B12-independent (METE) isozyme and functionally activates the B12-dependent enzyme (METH). Phylogenetic comparisons combined with growth assays reveal that many algal species require B12 for growth due to loss of METE, which appears to have occurred multiple times in algal evolution (Helliwell et al., 2011). Algae are estimated to be responsible for fixation of 50% of C worldwide (Field et al., 1998). Since many algae must acquire B12 from their environment, B12 limitation can influence primary productivity (Bertrand et al., 2011a, Bertrand et al., 2011b, King et al., 2011). B12 production by bacteria, in turn, can be limited by the availability of Co.
4. Metal and cofactor substitutions involving Molybdenum (Mo), Vanadium (V) and Tungsten (W)
Mo is a required element for many organisms throughout all three domains of life <Figure 12>. The high solubility of molybdate salts and their abundance in the oceans likely contributed to the adoption of Mo early in evolution as a redox cofactor. Mo often functions as part of a metal-binding pterin (MPT)-based cofactor known as molybdopterin (Moco), which is a required redox cofactor in as many as 50 different enzymes, mostly in bacteria, from four different families (Zhang and Gladyshev, 2008). In some enzymes, MPT functions instead with tungsten (W) to generate Wco (or Tuco), which is analogous to Moco.
Figure 12.
Overview of the biological roles of Mo (W) and Se and known sparing mechanisms.
Mo is very broadly required for life (Zhang and Gladyshev, 2008).The major Moco-dependent enzyme families are sulfite oxidase, xanthine oxidase, dimethylsulfoxide reductase, and aldehyde:ferredoxin oxidoreductase (Schwarz et al., 2009). Genomic surveys reveal that ~72% of bacteria encode Moco enzymes with the major exceptions being members of the Firmicutes and Chlamydia. Moco enzymes are found in ~95% of Archaeal and ~63% of Eukarya genomes. A requirement for Moco appears to have been lost in many parasites, yeast, and ciliates (Zhang and Gladyshev, 2008). The biosynthetic pathway for Moco is also conserved and most organisms that synthesize Moco have dedicated Mo uptake systems, which have been shown in some systems to be inducible by Mo deficiency.
Unlike Mo, tungsten (W) and vanadium (V) have very restricted uses in biology and those enzymes that use these metals often do so in place of Mo (Hille, 2002). The chemical properties of W and Mo are, in many ways, quite similar (W is immediately below Mo in the periodic table; Figure 2) and they can catalyze many of the same reactions. It has been suggested that ancient life may have evolved to use W, which would have been more available in the anaerobic, sulfidic conditions of the Archean era. Subsequent to the great oxidation event, increased solubility of Mo oxides may have favored a displacement of W with Mo. Today, tungstoenzymes are restricted to obligate, usually thermophilic, anaerobes. V also has restricted uses in biology and can functionally substitute, in some bacteria, for Mo by serving as cofactor for an alternative nitrogenase. Here, we focus specifically on those examples where enzyme substitutions appear to play a possible role in metal-sparing responses.
The sulfate reducing bacterium Desulfovibrio alaskensis encodes two formate dehydrogenases (members of the dimethylsulfate reductase family of enzymes). One isozyme (W-FDH) requires W, whereas the other can function with either W or Mo (Mo/W-FDH) (Mota et al., 2011). Genome analysis suggested the presence of uptake and cofactor assembly pathways for both Moco and Wco, consistent with the ability of this organism to synthesize both cofactors. Growth in the presence of Mo leads to the up-regulation of the Mo/W isozyme whereas addition of 10 µM W to the medium led to the exclusive synthesis of the W-FDH isozyme. These results suggest that this organism modulates its synthesis of these two enzymes in response to metal availability for cofactor synthesis (Mota et al., 2011). Other organisms may also have functionally redundant pathways that can utilize either Mo or W cofactored enzymes. This has been suggested, for example, in the Archaean Methanosarcina acetivorans where genome analysis suggests the presence of paralogous gene clusters encoding Mo-dependent and W-dependent forms of formylmethanofuran dehydrogenases (Rohlin and Gunsalus, 2010). It is presently unknown whether this apparent redundancy is an elemental sparing response and confers an advantage when either Mo or W is limiting in the growth medium.
Insights into acclimation mechanisms can often be obtained by noting the genes that are induced in response to elemental limitation. As noted above for both Fe and Zn, defining the regulons controlled by metalloregulatory proteins led to the identification of numerous sparing responses. Similarly, it can be anticipated that defining the regulatory consequences of Mo or W deficiency may lead to new insights. One powerful tool is provided by the observation that riboswitch elements are associated with the regulation of MPT synthesis genes in many different Bacteria (Regulski et al., 2008). To date, however, this regulatory element seems to exclusively regulate uptake and synthesis functions (acquisition), rather than controlling alternative isozymes (e.g. Mo-sparing).
a. Alternative nitrogenases and acclimation to Mo limitation
Mo is also required in nitrogenase, the key enzyme of nitrogen fixation (Hernandez et al., 2009, Schwarz et al., 2009). In the absence of fixed forms of N, diazotrophic (N2-fixing) organisms synthesize nitrogenase at levels up to 10% of total cell protein, which places a high demand for the corresponding metal ion cofactors (Dingler et al., 1988). The soil bacterium Azotobacter vinelandii has emerged as one of the premier model systems for investigating nitrogenase enzymology and regulation. This organism secretes a variety of catechols (metallophores) originally identified as siderophores but which are thought to also help mobilize Mo and other metals to support nitrogenase synthesis (Kraepiel et al., 2009). When Mo is available, and there is no fixed nitrogen, A. vinelandii induces the synthesis of a Mo-nitrogenase and, conversely, if Mo is unavailable alternative nitrogenases are synthesized instead.
In the Mo-nitrogenase, Mo does not bind as a complex with MPT, but instead forms a complex polynuclear cluster with Fe. The resulting cofactor (FeMoco, with formula MoFe7S9X-homocitrate; where X=C,O, or N), together with the [Fe8S7] P cluster, is part of the MoFe subunit of nitrogenase. The MoFe subunit functions together with an Fe protein (a homodimeric protein with a bridging [Fe4S4] cluster) to catalyze what is ultimately the eight electron reduction of N2 to 2 NH3. Two alternative nitrogenases are expressed under conditions of Mo limitation (Joerger and Bishop, 1988, Dos Santos and Dean, 2011). Nitrogenase-2 contains an Fe protein very similar to the Fe protein of the Mo-nitrogenase and a vanadium-containing VFe subunit that functionally replaces the MoFe subunit. When both Mo and V are limiting, nitrogenase 3 is expressed which is sometimes referred to as an Fe-only enzyme. Similarly, Rhodobacter capsulatus expresses an alternative Fe-only nitrogenase (containing a cofactor designated FeFe-co) in response to Mo limitation. In general, these alternative nitrogenases are not as efficient as is the MoFe enzyme (Hernandez et al., 2009).
The expression of alternative nitrogenases is regulated by metal ion availability (Bellenger et al., 2011). In A. vinelandii grown in the presence of both Mo (7 nM) and V (6.5 nM), the cells first import Mo until this metal is exhausted and then they import V such that the total (Mo + V) maintains a near constant quota in the growing cells. Since both Mo and V can be complexed by metallophores, it appears that the metallophore:Mo complex is selectively recognized and transported prior to expression of a presumed transporter specific for the metallophore:V complex (Bellenger et al., 2011). The Mo nitrogenase is the most active of the three nitrogenase forms and cells hyperaccumulate Mo in excess of their immediate needs to maintain synthesis of this preferred enzyme. Cytosolic Mo activates expression of the Mo-nitrogenase while repressing expression of activator proteins needed for expression of both the V-nitrogenase and Fe-nitrogenase (Masepohl and Hallenbeck, 2010). Although not defined for A. vinelandii, in R. capsulatus Mo is sensed by two molybdate-binding metalloregulatory proteins (MopA and MopB) that are related to the E. coli ModE protein (Masepohl et al., 2002). The V-nitrogenase is only slightly less active, but V does not appear to be stored within the cells and may be toxic in excess (Bellenger et al., 2011).
Studies in these model organisms, together with metagenomic surveys of diverse environments (Zehr et al., 2003, Gaby and Buckley, 2011), indicate that many organisms contain alternative nitrogenase systems to maintain diazotrophy even when Mo is not readily available. The major N2 fixing species in the open ocean are likely cyanobacteria such as Trichodesmium, but a newly identified small, unicellular cyanobacterium group (UCYN-A) (Moisander et al., 2010, Zehr, 2011), as well as proteobacteria (Farnelid et al., 2011) may also be a major contributors. The impact of metal availability on N2 fixation is not well understood, and is further complicated by the fact that some N2-fixing ocean organisms likely exist in still poorly defined symbiotic relationships (Tripp et al., 2010, Zehr and Kudela, 2011).
5. Selenium (Se): substitutions for acclimation to Se deprivation
Selenium is an essential trace element for many organisms where it is cotranslationally inserted into proteins as the 21st amino acid, selenocysteine (Sec) (Figure 12). Insertion of Sec occurs at UGA stop codons within the specific context of mRNAs with a selenocysteine insertion sequence (SECIS). Most selenoproteins have a single Sec residue, which typically serves a catalytic role as in, for example, Se-containing glutathione peroxidases. However, once an mRNA is targeted for Sec insertion, it is possible for other Sec-encoding UGA codons to be tolerated and additional, non-catalytic Sec residues have also been identified (Lee et al., 2011), although these are rare.
Selenoproteins are found throughout all three domains of life although there are exceptions including, notably, yeast and higher plants (Kryukov and Gladyshev, 2004, Lobanov et al., 2009). Sec insertion can be readily inferred from the occurrence of UGA codons within reading frames, typically in place of a Cys codon in orthologs, which (in combination with SECIS elements) is diagnostic of Sec. Bioinformatic analyses (ca. 2003) identified Sec-containing proteins in ~20% of completed Bacterial and ~14% of Archaeal genomes (Kryukov and Gladyshev, 2004). For those organisms that lack Sec, the corresponding selenoproteins may still be present but typically have a Cys residue in place of Sec. Substitution of Sec for Cys has occurred many times in evolution, as has the converse. At least two rationales, not mutually exclusive, have been advanced for the substitution of Cys with Sec. First, selenoenzymes may be more catalytically efficient than their Cys counterparts. Increased enzymatic efficiencies of the Sec variant range from four-fold to perhaps several hundred-fold (Stadtman, 1996, Hazebrouck et al., 2000, Kim et al., 2006). Second, the Sec variants may be significantly more resistant to oxidative inactivation (Hondal and Ruggles, 2011, Nauser et al., 2012).
Selenoproteins are especially prominent in methanogens. For example, Methanococcus maripaludis encodes ten Sec proteins and eight of these are involved in methanogenesis. These include subunits of both the F420-dependent and F420-independent hydrogenases, W-containing formyl-methanofuran dehydrogenase, Mo-containing formylmethanofuran dehydrogenase, heterodisulfide reductase and two formate dehydrogenases (Hohn et al., 2011). Despite this abundance of Sec-containing proteins, M. maripaludis is able to grow in the absence of Sec by the expression of alternate enzymes containing Cys in place of Sec. The only exception is the formate dehydrogenase, which requires Se for both isozymes. As a result, inactivation of Sec incorporation prevented growth on formate, but not growth by methanogenesis (Rother et al., 2003).
The substitute Cys-containing methanogenesis proteins are transcriptionally regulated by a Se-sensing regulatory protein, HrsM (Sun and Klein, 2004). This LysR family regulator represses expression in the presence of Se. Since growth of M. maripaludis in the absence of Sec synthesis requires deletion of either the selD gene (encoding selenophosphate synthase) or hrsM (encoding the repressor), it is hypothesized that selenophosphate may be the co-repressor for HrsM-mediated transcriptional repression (Hohn et al., 2011). The use of selenophosphate as corepressor is expedient, since synthesis of this intermediate is catalyzed by SelD, which is, remarkably, itself a selenoprotein (Stock et al., 2010). Thus, selenophosphate synthesis requires both Se and a functional Sec synthesis and incorporation pathway.
It is interesting that many, but not all, SelD homologs are Sec enzymes, which has previously led to the suggestion that this might serve a positive feedback mechanism (Guimaraes et al., 1996). We suggest instead that the Sec in SelD may be present not for its catalytic advantages, but as an indicator of the cell's ability to synthesize selenoproteins. Although the mechanisms are not yet defined, it has also been proposed that M. maripaludis may have a hierarchy for Sec-insertion into proteins with formate hydrogenase (which lacks a Cys-containing alternate) given priority under conditions of limited Sec synthetic capacity (Stock et al., 2011).
IV. Major Themes - Microbial Adaptations to Elemental Limitation
We noted at the outset the explosion of knowledge in the molecular life sciences occasioned by the convergence of biology and chemistry. The periodic table is, arguably, the greatest organizing theme of chemistry, while the theory of evolution similarly pervades biological thought (Figure 1). We have here considered several different microbial systems and the remarkable ways in which they have adapted to elemental limitations. Conceptually similar evolutionary responses have emerged in systems as distinct as: methanogens facing a Ni famine at the end of the Archean era; phytoplankton in the contemporary ocean's surface waters facing chronic limitations for P, Fe or Zn; and mammalian pathogens facing metal ion deprivation imposed by the innate immune system. Our scope has been very broad, and many of these same topics are reviewed (in cited references) in greater detail but with an emphasis on a single element, a single organism, or both. As is often the case in biology, comparisons between systems help reveal common themes, which we hope we have accomplished in this work.
Elemental limitations may be chronic in the environment, imposed by a proliferation of neighboring cells (of the same or a different species), or imposed by a host organism. The corresponding adaptations and acclimations include alterations of the elemental composition of macromolecules to reduce or eliminate the elemental requirement. Elimination of a requirement has occurred repeatedly in evolution: cells have learned to thrive in the absence of elements like Ni, Co, and even Fe (in rare examples). More commonly, cells develop strategies for reducing demand, particularly under times of duress, by altering the composition of abundant constituents such as proteins or membrane lipids. Often, eliminating or reducing the demand for one element increases demand for another. The process of substitution, in which one form of an enzyme or other macromolecule is replaced by an alternate form, is a widespread feature of elemental economy. In many cells, these changes are now fixed and the corresponding evolutionary pressures can only be inferred.
Perhaps the most widespread adaptation to elemental restriction is the development of acclimation mechanisms. Acclimation typically involves several complementary strategies that include: (i) high affinity acquisition mechanisms and pathways to access recalcitrant resources that may not be available to all organisms, (ii) mobilization of elements stored during times of relative abundance, (iii) elemental sparing responses, and (iv) elemental recycling. Elemental sparing often involves the synthesis of substitute enzymes that rely on different elements, but these are often catalytically inferior or less robust than the preferred isozymes. Collectively, these responses help to redirect elemental resources to the highest priority targets.
These same strategies, implemented daily by microbes, have numerous parallels throughout society. When a yeast cell is challenged by Cd, S is preferentially directed towards synthesis of the defensive molecule glutathione, while a widespread S-sparing response is implemented to suppress now lower priority uses. Analogously, when the United States entered World War II, the massive shift to synthesis of armaments required the introduction of widespread recycling programs (scrap drives) to recover valuable metals, while the "lower priority" needs of the civilian population were accommodated by substitution (a shift from metal-based consumer goods to those based on wood and other renewables). We are still in the early stages of dealing with our dependency on non-renewable resources including, in many cases, elemental resources. For example, much of modern computer-based technology relies on a family of rare earth metals with ~97% of today’s world supply controlled by a single country (Service, 2010) and our agricultural supply of phosphate is expected to run out by the end of this century (Gilbert, 2009). Ultimately, our dependence on elemental resources will require an ever increasing reliance on recycling rather than new acquisitions.
While work in microbial systems highlights these mechanisms of elemental economy, there is still much to be learned. The mechanisms that regulate the processes of sparing, recycling, and substitution are often poorly understood. There are numerous hints that organisms prioritize targets for synthesis as elemental availability declines, but the mechanisms are still obscure. A major challenge for the future is to define the complete elemental requirements for cell growth and how this varies with the environment and form of metabolism. This is presently a focus of numerous metallomics studies which seek to define the elemental quotas for various model organisms. It is remarkable that we, in most cases, are still unable to provide a satisfying answer to the seeming simple questions: which elements, and in which amounts, are required for life, and perhaps more important, why are they needed?
Acknowledgements
Work related to elemental economy in the authors' laboratories is supported by grants from the National Institutes of Health (GM059323 to JDH and GM042143 to SM) and the US Department of Energy (DE-FD02-04ER15529 to SM). We thank our colleagues for their insights and comments on this work.
LITERATURE CITED
- Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, Harrison PM, Guest JR, Andrews SC. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol. 1999;181:1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MM, Gomez-Garcia MR, Grossman AR, Bhaya D. Phosphorus deprivation responses and phosphonate utilization in a thermophilic Synechococcus sp. from microbial mats. J Bacteriol. 2008;190:8171–8184. doi: 10.1128/JB.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Africa LA, Murphy ER, Egan NR, Wigley AF, Wing HJ. The iron-responsive Fur/RyhB regulatory cascade modulates the Shigella outer membrane protease IcsP. Infect Immun. 2011;79:4543–4549. doi: 10.1128/IAI.05340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afting C, Kremmer E, Brucker C, Hochheimer A, Thauer RK. Regulation of the synthesis of H2-forming methylenetetrahydromethanopterin dehydrogenase (Hmd) and of HmdII and HmdIII in Methanothermobacter marburgensis. Arch Microbiol. 2000;174:225–232. doi: 10.1007/s002030000197. [DOI] [PubMed] [Google Scholar]
- Aguirre JD, Culotta VC. Battles with iron: manganese in oxidative stress protection. J Biol Chem. 2012 doi: 10.1074/jbc.R111.312181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn BE, Cha J, Lee EJ, Han AR, Thompson CJ, Roe JH. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol Microbiol. 2006;59:1848–1858. doi: 10.1111/j.1365-2958.2006.05065.x. [DOI] [PubMed] [Google Scholar]
- Akanuma G, Nanamiya H, Natori Y, Nomura N, Kawamura F. Liberation of zinc-containing L31 (RpmE) from ribosomes by its paralogous gene product, YtiA, in Bacillus subtilis. J Bacteriol. 2006;188:2715–2720. doi: 10.1128/JB.188.7.2715-2720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci U S A. 2002;99:3695–3700. doi: 10.1073/pnas.062526999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, Del Campo JA, Kropat J, Merchant SS. FEA-1, FEA-2, and FRE-1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX-1 and FTR-1 in iron-deficient Chlamydomonas reinhardtii. Eukaryot Cell. 2007a;6:1841–1852. doi: 10.1128/EC.00205-07. Epub 2007 Jul 27 PMC2043389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, Kropat J, Tottey S, Del Campo JA, Merchant SS. Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 2007b;143:263–277. doi: 10.1104/pp.106.088609. Epub 2006 Nov 3 PMC1761973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves R, Savageau MA. Evidence of selection for low cognate amino acid bias in amino acid biosynthetic enzymes. Mol Microbiol. 2005;56:1017–1034. doi: 10.1111/j.1365-2958.2005.04566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir A, Meshner S, Beatus T, Stavans J. Damped oscillations in the adaptive response of the iron homeostasis network of E. coli. Mol Microbiol. 2010;76:428–436. doi: 10.1111/j.1365-2958.2010.07111.x. [DOI] [PubMed] [Google Scholar]
- Andreini C, Bertini I, Rosato A. Metalloproteomes: a bioinformatic approach. Acc Chem Res. 2009;42:1471–1479. doi: 10.1021/ar900015x. [DOI] [PubMed] [Google Scholar]
- Andrews SC. Making DNA without iron - induction of a manganese-dependent ribonucleotide reductase in response to iron starvation. Mol Microbiol. 2011;80:286–289. doi: 10.1111/j.1365-2958.2011.07594.x. [DOI] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Anjem A, Varghese S, Imlay JA. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol. 2009;72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV. The life of diatoms in the world's oceans. Nature. 2009;459:185–192. doi: 10.1038/nature08057. [DOI] [PubMed] [Google Scholar]
- Arudchandran A, Bullerjahn GS. Expression of the petE gene encoding plastocyanin in the photosynthetic prokaryote, Prochlorothrix hollandica. Biochem. Biophys. Res. Commun. 1996;226:626–630. doi: 10.1006/bbrc.1996.1406. [DOI] [PubMed] [Google Scholar]
- Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K, Kanematsu S, Uchida K. Superoxide dismutases in photosynthetic organisms: absence of the cuprozinc enzyme in eukaryotic algae. Arch Biochem Biophys. 1977;179:243–256. doi: 10.1016/0003-9861(77)90109-6. [DOI] [PubMed] [Google Scholar]
- Askwith C, Kaplan J. An oxidase-permease-based iron transport system in Schizosaccharomyces pombe and its expression in Saccharomyces cerevisiae. J Biol Chem. 1997;272:401–405. doi: 10.1074/jbc.272.1.401. [DOI] [PubMed] [Google Scholar]
- Assuncao AG, Schat H, Aarts MG. Regulation of the adaptation to zinc deficiency in plants. Plant Signal Behav. 2010;5:1553–1555. doi: 10.4161/psb.5.12.13469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- Baek JH, Lee SY. Transcriptome analysis of phosphate starvation response in Escherichia coli. J Microbiol Biotechnol. 2007;17:244–252. [PubMed] [Google Scholar]
- Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol. 2002;45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Kenney GE, Rosenzweig AC. Dual pathways for copper uptake by methanotrophic bacteria. J Biol Chem. 2011;286:37313–37319. doi: 10.1074/jbc.M111.284984. Epub 2011 Sep 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basturea GN, Zundel MA, Deutscher MP. Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA. 2011;17:338–345. doi: 10.1261/rna.2448911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudouin-Cornu P, Surdin-Kerjan Y, Marliere P, Thomas D. Molecular evolution of protein atomic composition. Science. 2001;293:297–300. doi: 10.1126/science.1061052. [DOI] [PubMed] [Google Scholar]
- Behrenfeld MJ, Westberry TK, Boss ES, O'malley RT, Siegel DA, Wiggert JD, Franz BA, Mcclain CR, Feldman GC, Doney SC, Moore JK, Dall'olmo G, Milligan AJ, Lima I, Mahawold N. Satellite-detected fluorescence reveals global physiology of ocean phytoplankton. Biogeosciences. 2009;6:779–794. [Google Scholar]
- Bellenger JP, Wichard T, Xu Y, Kraepiel AM. Essential metals for nitrogen fixation in a free-living N-fixing bacterium: chelation, homeostasis and high use efficiency. Environ Microbiol. 2011;13:1395–1411. doi: 10.1111/j.1462-2920.2011.02440.x. [DOI] [PubMed] [Google Scholar]
- Benning C, Huang ZH, Gage DA. Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch Biochem Biophys. 1995;317:103–111. doi: 10.1006/abbi.1995.1141. [DOI] [PubMed] [Google Scholar]
- Benoist BD, Mclean E, Egli I, Cogswell M. Worldwide prevalence of anaemia 1993–2005: WHO global database on anaemia. 20 Avenue Appia 1211 Geneva 27, Switzerland: WHO Press, World Health Organization; 2008. [Google Scholar]
- Bernal M, Casero D, Singh V, Wilson GT, Grande A, Yang H, Dodani SC, Pellegrini M, Huijser P, Connolly EL, Merchant SS, Krämer U. Transcriptome sequencing identifies SPL7-regulated Cu acquisition genes FRO4/FRO5 and the Cu dependence of Fe homeostasis in Arabidopsis. Plant Cell. 2012 doi: 10.1105/tpc.111.090431. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertilsson S, Berglund O, Karl DM, Chisholm SW. Elemental composition of marine Prochlorococcus and Synechococcus: Implications for the ecological stoichiometry of the sea. Limnol Oceanogr. 2003;48:1721–1731. [Google Scholar]
- Bertrand EM, Saito MA, Jeon YJ, Neilan BA. Vitamin B biosynthesis gene diversity in the Ross Sea: the identification of a new group of putative polar B biosynthesizers. Environ Microbiol. 2011a;13:1285–1298. doi: 10.1111/j.1462-2920.2011.02428.x. [DOI] [PubMed] [Google Scholar]
- Bertrand EM, Saito MA, Lee PA, Dunbar RB, Sedwick PN, Ditullio GR. Iron limitation of a springtime bacterial and phytoplankton community in the ross sea: implications for vitamin b(12) nutrition. Front Microbiol. 2011b;2:160. doi: 10.3389/fmicb.2011.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevers LE, Theil EC. Maxi- and mini-ferritins: minerals and protein nanocages. Prog Mol Subcell Biol. 2011;52:29–47. doi: 10.1007/978-3-642-21230-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar AP, Brown ED. Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol Microbiol. 2006;60:1077–1090. doi: 10.1111/j.1365-2958.2006.05169.x. [DOI] [PubMed] [Google Scholar]
- Bieleski RL. Phosphate pools, phosphate transport, and phosphate availability. Annu Rev Plant Physiol. 1973;24:225–252. [Google Scholar]
- Bini E. Archaeal transformation of metals in the environment. FEMS Microbiol Ecol. 2010;73:1–16. doi: 10.1111/j.1574-6941.2010.00876.x. [DOI] [PubMed] [Google Scholar]
- Bird AJ. Metallosensors, the ups and downs of gene regulation. Adv Microb Physiol. 2008;53:231–267. doi: 10.1016/S0065-2911(07)53004-3. [DOI] [PubMed] [Google Scholar]
- Bird AJ, Gordon M, Eide DJ, Winge DR. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 2006;25:5726–5734. doi: 10.1038/sj.emboj.7601453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AJ, Zhao H, Luo H, Jensen LT, Srinivasan C, Evans-Galea M, Winge DR, Eide DJ. A dual role for zinc fingers in both DNA binding and zinc sensing by the Zap1 transcriptional activator. EMBO J. 2000;19:3704–3713. doi: 10.1093/emboj/19.14.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop PE, Jarlenski DM, Hetherington DR. Evidence for an alternative nitrogen fixation system in Azotobacter vinelandii. Proc Natl Acad Sci U S A. 1980;77:7342–7346. doi: 10.1073/pnas.77.12.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, De Crecy-Lagard V. Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol Microbiol. 2011;79:700–715. doi: 10.1111/j.1365-2958.2010.07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby-Haas CE, Merchant SS. The ins and outs of algal metal transporters. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.04.010. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black R. Micronutrient deficiency — an underlying cause of morbidity and mortality. Bulletin of the World Health Organization. 2003;81:79. [PMC free article] [PubMed] [Google Scholar]
- Blanco NE, Ceccoli RD, Segretin ME, Poli HO, Voss I, Melzer M, Bravo-Almonacid FF, Scheibe R, Hajirezaei MR, Carrillo N. Cyanobacterial flavodoxin complements ferredoxin deficiency in knocked-down transgenic tobacco plants. Plant J. 2011;65:922–935. doi: 10.1111/j.1365-313X.2010.04479.x. [DOI] [PubMed] [Google Scholar]
- Bleackley MR, Macgillivray RT. Transition metal homeostasis: from yeast to human disease. Biometals. 2011;24:785–809. doi: 10.1007/s10534-011-9451-4. [DOI] [PubMed] [Google Scholar]
- Boal AK, Cotruvo JA, Jr, Stubbe J, Rosenzweig AC. Structural basis for activation of class Ib ribonucleotide reductase. Science. 2010;329:1526–1530. doi: 10.1126/science.1190187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boal AK, Rosenzweig AC. Structural biology of copper trafficking. Chem Rev. 2009;109:4760–4779. doi: 10.1021/cr900104z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohner H, Böhme H, Böger P. Reciprocal formation of plastocyanin and cytochrome c-553 and the influence of cupric ions on photosynthetic electron transport. Biochim Biophys Acta. 1980a;592:103–112. doi: 10.1016/0005-2728(80)90117-6. [DOI] [PubMed] [Google Scholar]
- Bohner H, Merkle H, Kroneck P, Böger P. High variability of the electron carrier plastocyanin in microalgae. Eur J Biochem. 1980b;105:603–609. doi: 10.1111/j.1432-1033.1980.tb04538.x. [DOI] [PubMed] [Google Scholar]
- Borghouts C, Osiewacz HD. GRISEA, a copper-modulated transcription factor from Podospora anserina involved in senescence and morphogenesis, is an ortholog of MAC1 in Saccharomyces cerevisiae. Mol Gen Genet. 1998;260:492–502. doi: 10.1007/s004380050922. [DOI] [PubMed] [Google Scholar]
- Borghouts C, Werner A, Elthon T, Osiewacz HD. Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina. Mol Cell Biol. 2001;21:390–399. doi: 10.1128/MCB.21.2.390-399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovok I, Gorovitz B, Schreiber R, Aharonowitz Y, Cohen G. Coenzyme B12 controls transcription of the Streptomyces class Ia ribonucleotide reductase nrdABS operon via a riboswitch mechanism. J Bacteriol. 2006;188:2512–2520. doi: 10.1128/JB.188.7.2512-2520.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella E, Hubner S, Hokamp K, Hansen A, Bisicchia P, Noone D, Powell L, Salzberg LI, Devine KM. Cell envelope gene expression in phosphate-limited Bacillus subtilis cells. Microbiology. 2011;157:2470–2484. doi: 10.1099/mic.0.049205-0. [DOI] [PubMed] [Google Scholar]
- Bottin H, Lagoutte B. Ferredoxin and flavodoxin from the cyanobacterium Synechocystis sp PCC-6803. Biochim Biophys Acta. 1992;1101:48–56. doi: 10.1016/0167-4838(92)90465-p. [DOI] [PubMed] [Google Scholar]
- Boulter D, Haslett BG, Peacock D, Ramshaw JAM, Scawen MD. Chemistry, function and evolution of plastocyanin. In: Northcote DH, editor. Int Rev Biochem. Baltimore: University Park Press; 1977. [Google Scholar]
- Bovy A, De Kruif J, De Vrieze G, Borrias M, Weisbeek P. Iron-dependent protection of the Synechococcus ferredoxin I transcript against nucleolytic degradation requires cis-regulatory sequences in the 5' part of the messenger RNA. Plant Mol Biol. 1993;22:1047–1065. doi: 10.1007/BF00028977. [DOI] [PubMed] [Google Scholar]
- Bovy A, De Vrieze G, Borrias M, Weisbeek P. Transcriptional regulation of the plastocyanin and cytochrome c553 genes from the cyanobacterium Anabaena species PCC 7937. Mol Microbiol. 1992;6:1507–1513. doi: 10.1111/j.1365-2958.1992.tb00871.x. [DOI] [PubMed] [Google Scholar]
- Bowler C, Vardi A, Allen AE. Oceanographic and biogeochemical insights from diatom genomes. Ann Rev Mar Sci. 2010;2:333–365. doi: 10.1146/annurev-marine-120308-081051. [DOI] [PubMed] [Google Scholar]
- Bragg JG, Thomas D, Baudouin-Cornu P. Variation among species in proteomic sulphur content is related to environmental conditions. Proc Biol Sci. 2006;273:1293–1300. doi: 10.1098/rspb.2005.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg JG, Wagner A. Protein material costs: single atoms can make an evolutionary difference. Trends Genet. 2009;25:5–8. doi: 10.1016/j.tig.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Briggs LM, Pecoraro VL, Mcintosh L. Copper-induced expression, cloning, and regulatory studies of the plastocyanin gene from the cyanobacterium Synechocystis sp. PCC-6803. Plant Mol Biol. 1990;15:633–642. doi: 10.1007/BF00017837. [DOI] [PubMed] [Google Scholar]
- Brouwer M, Hoexum Brouwer T, Grater W, Brown-Peterson N. Replacement of a cytosolic copper/zinc superoxide dismutase by a novel cytosolic manganese superoxide dismutase in crustaceans that use copper (haemocyanin) for oxygen transport. Biochem J. 2003;374:219–228. doi: 10.1042/BJ20030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulté L, Bennoun P. Translational accuracy and sexual differentiation in Chlamydomonas reinhardtii. Curr Genet. 1990;18:155–160. doi: 10.1007/BF00312603. [DOI] [PubMed] [Google Scholar]
- Busch A, Rimbauld B, Naumann B, Rensch S, Hippler M. Ferritin is required for rapid remodeling of the photosynthetic apparatus and minimizes photo-oxidative stress in response to iron availability in Chlamydomonas reinhardtii. Plant J. 2008;55:201–211. doi: 10.1111/j.1365-313X.2008.03490.x. Epub 2008 Mar 19. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Ball DA, Guerquin-Kern JL, Rouiller I, Wu TD, Downing KH, Vali H, Komeili A. Desulfovibrio magneticus RS-1 contains an iron- and phosphorus-rich organelle distinct from its bullet-shaped magnetosomes. Proc Natl Acad Sci U S A. 2010;107:12263–12268. doi: 10.1073/pnas.1001290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SL, Foito A, Hedley PE, Morris JA, Stewart D, Barth S. Early response mechanisms of perennial ryegrass (Lolium perenne) to phosphorus deficiency. Ann Bot. 2011;107:243–254. doi: 10.1093/aob/mcq234. Epub 2010 Dec 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon GC, Heinhorst S, Kerfeld CA. Carboxysomal carbonic anhydrases: Structure and role in microbial CO2 fixation. Biochim Biophys Acta. 2010;1804:382–392. doi: 10.1016/j.bbapap.2009.09.026. Epub 2009 Oct 8. [DOI] [PubMed] [Google Scholar]
- Carr HS, Winge DR. Assembly of cytochrome c oxidase within the mitochondrion. Acc Chem Res. 2003;36:309–316. doi: 10.1021/ar0200807. [DOI] [PubMed] [Google Scholar]
- Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Interplay of metal ions and urease. Metallomics. 2009;1:207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter EL, Tronrud DE, Taber SR, Karplus PA, Hausinger RP. Iron-containing urease in a pathogenic bacterium. Proc Natl Acad Sci U S A. 2011;108:13095–13099. doi: 10.1073/pnas.1106915108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castruita M, Casero D, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, Yan W, Cokus S, Loo JA, Benning C, Pellegrini M, Merchant SS. Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell. 2011;23:1273–1292. doi: 10.1105/tpc.111.084400. Epub 2011 Apr 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini G. Function and structure of complex II of the respiratory chain. Annu Rev Biochem. 2003;72:77–109. doi: 10.1146/annurev.biochem.72.121801.161700. [DOI] [PubMed] [Google Scholar]
- Cellier MF, Courville P, Campion C. Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect. 2007;9:1662–1670. doi: 10.1016/j.micinf.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Chai SC, Wang WL, Ye QZ. Fe(II) is the native cofactor for Escherichia coli methionine aminopeptidase. J Biol Chem. 2008;283:26879–26885. doi: 10.1074/jbc.M804345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazarreta-Cifre L, Martiarena L, De Mendoza D, Altabe SG. Role of ferredoxin and flavodoxins in Bacillus subtilis fatty acid desaturation. J Bacteriol. 2011;193:4043–4048. doi: 10.1128/JB.05103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Hsieh SI, Kropat J, Merchant SS. A ferroxidase encoded by FOX1 contributes to iron assimilation under conditions of poor iron nutrition in Chlamydomonas. Eukaryot Cell. 2008;7:541–545. doi: 10.1128/EC.00463-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Roberts E, Luthey-Schulten Z. Horizontal gene transfer of zinc and non-zinc forms of bacterial ribosomal protein S4. BMC Evol Biol. 2009;9:179. doi: 10.1186/1471-2148-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillappagari S, Seubert A, Trip H, Kuipers OP, Marahiel MA, Miethke M. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J Bacteriol. 2010;192:2512–2524. doi: 10.1128/JB.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Davidson VL. Cupredoxins--a study of how proteins may evolve to use metals for bioenergetic processes. Metallomics. 2011;3:140–151. doi: 10.1039/c0mt00061b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AK, Campbell D. Inactivation of the petE gene for plastocyanin lowers photosynthetic capacity and exacerbates chilling-induced photoinhibition in the cyanobacterium Synechococcus. Plant Physiol. 1996;112:1551–1561. doi: 10.1104/pp.112.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys JP, Havarstein LS. Cannibalism and fratricide: mechanisms and raisons d'etre. Nat Rev Microbiol. 2007;5:219–229. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- Collier JL, Grossman AR. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J Bacteriol. 1992;174:4718–4726. doi: 10.1128/jb.174.14.4718-4726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin RA, Holmes WR, Fontaine CP, Maret W. Cytosolic zinc buffering and muffling: their role in intracellular zinc homeostasis. Metallomics. 2010;2:306–317. doi: 10.1039/b926662c. [DOI] [PubMed] [Google Scholar]
- Cook JD, Penner-Hahn JE, Stemmler TL. Structure and dynamics of metalloproteins in live cells. Methods Cell Biol. 2008;90:199–216. doi: 10.1016/S0091-679X(08)00810-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- Cotruvo JA, Jr, Stubbe J. NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Proc Natl Acad Sci U S A. 2008;105:14383–14388. doi: 10.1073/pnas.0807348105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotruvo JA, Stubbe J. Escherichia coli class Ib ribonucleotide reductase contains a dimanganese(III)-tyrosyl radical cofactor in vivo. Biochemistry. 2011;50:1672–1681. doi: 10.1021/bi101881d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CD. Deferration of laboratory media and assays for ferric and ferrous ions. Methods Enzymol. 1994;215:315–329. doi: 10.1016/0076-6879(94)35150-3. [DOI] [PubMed] [Google Scholar]
- Crichton RR, Pierre JL. Old iron, young copper: from Mars to Venus. Biometals. 2001;14:99–112. doi: 10.1023/a:1016710810701. [DOI] [PubMed] [Google Scholar]
- Csáki R, Bodrossy L, Klem J, Murrell JC, Kovács KL. Genes involved in the copper-dependent regulation of soluble methane monooxygenase of Methylococcus capsulatus (Bath): cloning, sequencing and mutational analysis. Microbiology. 2003;149:1785–1795. doi: 10.1099/mic.0.26061-0. [DOI] [PubMed] [Google Scholar]
- Cvetkovic A, Menon AL, Thorgersen MP, Scott JW, Poole FL, 2nd, Jenney FE, Jr, Lancaster WA, Praissman JL, Shanmukh S, Vaccaro BJ, Trauger SA, Kalisiak E, Apon JV, Siuzdak G, Yannone SM, Tainer JA, Adams MW. Microbial metalloproteomes are largely uncharacterized. Nature. 2010;466:779–782. doi: 10.1038/nature09265. [DOI] [PubMed] [Google Scholar]
- Daly MJ. A new perspective on radiation resistance based on Deinococcus radiodurans. Nat Rev Microbiol. 2009;7:237–245. doi: 10.1038/nrmicro2073. [DOI] [PubMed] [Google Scholar]
- Danchin A. Science and Arsenic Fool's Gold: A Toxic Broth. Journal of Cosmology. 2010;13:3617–3620. [Google Scholar]
- Danielli A, Roncarati D, Delany I, Chiarini V, Rappuoli R, Scarlato V. In vivo dissection of the Helicobacter pylori Fur regulatory circuit by genome-wide location analysis. J Bacteriol. 2006;188:4654–4662. doi: 10.1128/JB.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PC, Benner SA, Cleland CE, Lineweaver CH, Mckay CP, Wolfe-Simon F. Signatures of a shadow biosphere. Astrobiology. 2009;9:241–249. doi: 10.1089/ast.2008.0251. [DOI] [PubMed] [Google Scholar]
- Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J Bacteriol. 2005;187:4005–4014. doi: 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bivort BL, Perlstein EO, Kunes S, Schreiber SL. Amino acid metabolic origin as an evolutionary influence on protein sequence in yeast. J Mol Evol. 2009;68:490–497. doi: 10.1007/s00239-009-9218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Domenico I, Ward DM, Kaplan J. Specific iron chelators determine the route of ferritin degradation. Blood. 2009;114:4546–4551. doi: 10.1182/blood-2009-05-224188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nicola R, Hazelwood LA, De Hulster EA, Walsh MC, Knijnenburg TA, Reinders MJ, Walker GM, Pronk JT, Daran JM, Daran-Lapujade P. Physiological and transcriptional responses of Saccharomyces cerevisiae to zinc limitation in chemostat cultures. Appl Environ Microbiol. 2007;73:7680–7692. doi: 10.1128/AEM.01445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany I, Rappuoli R, Scarlato V. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol Microbiol. 2004;52:1081–1090. doi: 10.1111/j.1365-2958.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- Desnoyers G, Morissette A, Prevost K, Masse E. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J. 2009;28:1551–1561. doi: 10.1038/emboj.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci. 2009;85:369–391. doi: 10.1016/S0079-6603(08)00809-X. [DOI] [PubMed] [Google Scholar]
- Dingler C, Kuhla J, Wassink H, Oelze J. Levels and activities of nitrogenase proteins in Azotobacter vinelandii grown at different dissolved oxygen concentrations. J Bacteriol. 1988;170:2148–2152. doi: 10.1128/jb.170.5.2148-2152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2004;2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- Dobbs BJT. The Foundations of Newton's Alchemy. Cambridge University Press; 1983. [Google Scholar]
- Domaille DW, Que EL, Chang CJ. Synthetic fluorescent sensors for studying the cell biology of metals. Nat Chem Biol. 2008;4:168–175. doi: 10.1038/nchembio.69. [DOI] [PubMed] [Google Scholar]
- Dos Santos PC, Dean DR. Co-ordination and fine-tuning of nitrogen fixation in Azotobacter vinelandii. Mol Microbiol. 2011;79:1132–1135. doi: 10.1111/j.1365-2958.2011.07541.x. [DOI] [PubMed] [Google Scholar]
- Downie JA, Garland PB. An antimycin A- and cyanide-resistant variant of Candida utilis arising during copper-limited growth. Biochem J. 1973;134:1051–1061. doi: 10.1042/bj1341051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozd M, Gangaiah D, Liu Z, Rajashekara G. Contribution of TAT system translocated PhoX to Campylobacter jejuni phosphate metabolism and resilience to environmental stresses. PLoS One. 2011;6:e26336. doi: 10.1371/journal.pone.0026336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond JT, Matthews RG. Cobalamin-dependent and cobalamin-independent methionine synthases in Escherichia coli: two solutions to the same chemical problem. Adv Exp Med Biol. 1993;338:687–692. doi: 10.1007/978-1-4615-2960-6_142. [DOI] [PubMed] [Google Scholar]
- Dubrac S, Touati D. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J Bacteriol. 2000;182:3802–3808. doi: 10.1128/jb.182.13.3802-3808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrac S, Touati D. Fur-mediated transcriptional and post-transcriptional regulation of FeSOD expression in Escherichia coli. Microbiology. 2002;148:147–156. doi: 10.1099/00221287-148-1-147. [DOI] [PubMed] [Google Scholar]
- Dühring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci U S A. 2006;103:7054–7058. doi: 10.1073/pnas.0600927103. Epub 2006 Apr 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont CL, Barbeau K, Palenik B. Ni uptake and limitation in marine Synechococcus strains. Appl Environ Microbiol. 2008a;74:23–31. doi: 10.1128/AEM.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont CL, Butcher A, Valas RE, Bourne PE, Caetano-Anolles G. History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc Natl Acad Sci U S A. 2010;107:10567–10572. doi: 10.1073/pnas.0912491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont CL, Neupane K, Shearer J, Palenik B. Diversity, function and evolution of genes coding for putative Ni-containing superoxide dismutases. Environ Microbiol. 2008b;10:1831–1843. doi: 10.1111/j.1462-2920.2008.01604.x. [DOI] [PubMed] [Google Scholar]
- Duran RV, Hervas M, De La Rosa MA, Navarro JA. The efficient functioning of photosynthesis and respiration in Synechocystis sp. PCC 6803 strictly requires the presence of either cytochrome c6 or plastocyanin. J Biol Chem. 2004;279:7229–7233. doi: 10.1074/jbc.M311565200. [DOI] [PubMed] [Google Scholar]
- Eide DJ. Homeostatic and adaptive responses to zinc deficiency in S. accharomyces cerevisiae. J Biol Chem. 2009;284:18565–18569. doi: 10.1074/jbc.R900014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Bonnett S, Anderson JN, Swairjo MA, Iwata-Reuyl D, De Crecy-Lagard V. Discovery of a new prokaryotic type I GTP cyclohydrolase family. J Biol Chem. 2006;281:37586–37593. doi: 10.1074/jbc.M607114200. [DOI] [PubMed] [Google Scholar]
- Elango N, Radhakrishnan R, Froland WA, Wallar BJ, Earhart CA, Lipscomb JD, Ohlendorf DH. Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci. 1997;6:556–568. doi: 10.1002/pro.5560060305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elser JJ, Acquisti C, Kumar S. Stoichiogenomics: the evolutionary ecology of macromolecular elemental composition. Trends Ecol Evol. 2011;26:38–44. doi: 10.1016/j.tree.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Moseley JL, Tottey S, Del Campo JA, Quinn JM, Kim Y, Merchant S. Genetic dissection of nutritional copper signaling in Chlamydomonas distinguishes regulatory and target genes. Genetics. 2004;168:795–807. doi: 10.1534/genetics.104.030460. PMC1448816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann B, Guler S, Narang RA, Linke D, Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnelid H, Andersson AF, Bertilsson S, Al-Soud WA, Hansen LH, Sorensen S, Steward GF, Hagstrom A, Riemann L. Nitrogenase gene amplicons from global marine surface waters are dominated by genes of non-cyanobacteria. PLoS One. 2011;6:e19223. doi: 10.1371/journal.pone.0019223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchon M, Lagniel G, Aude JC, Lombardia L, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol Cell. 2002;9:713–723. doi: 10.1016/s1097-2765(02)00500-2. [DOI] [PubMed] [Google Scholar]
- Faulkner MJ, Ma Z, Fuangthong M, Helmann JD. Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J Bacteriol. 2011 doi: 10.1128/JB.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fee JA. Regulation of sod genes in Escherichia coli: relevance to superoxide dismutase function. Mol Microbiol. 1991;5:2599–2610. doi: 10.1111/j.1365-2958.1991.tb01968.x. [DOI] [PubMed] [Google Scholar]
- Feingersch R, Philosof A, Mejuch T, Glaser F, Alalouf O, Shoham Y, Beja O. Potential for phosphite and phosphonate utilization by Prochlorococcus. ISME J. 2011 doi: 10.1038/ismej.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M, Golyshina OV, Beloqui A, Golyshin PN, Timmis KN. The cellular machinery of Ferroplasma acidiphilum is iron-protein-dominated. Nature. 2007;445:91–94. doi: 10.1038/nature05362. [DOI] [PubMed] [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- Finney LA, O'Halloran TV. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol. 2006;2:132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- Fischer E, Sauer U. Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat Genet. 2005;37:636–640. doi: 10.1038/ng1555. [DOI] [PubMed] [Google Scholar]
- Frangipani E, Haas D. Copper acquisition by the SenC protein regulates aerobic respiration in Pseudomonas aeruginosa PAO1. FEMS Microbiol Lett. 2009;298:234–240. doi: 10.1111/j.1574-6968.2009.01726.x. [DOI] [PubMed] [Google Scholar]
- Frangipani E, Slaveykova VI, Reimmann C, Haas D. Adaptation of aerobically growing P seudomonas aeruginosa to copper starvation. J Bacteriol. 2008;190:6706–6717. doi: 10.1128/JB.00450-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frausto Da Silva J, Williams R. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. Oxford, UK: Oxford University Press; 2001. [Google Scholar]
- Frere F, Reents H, Schubert WD, Heinz DW, Jahn D. Tracking the evolution of porphobilinogen synthase metal dependence in vitro. J Mol Biol. 2005;345:1059–1070. doi: 10.1016/j.jmb.2004.10.053. [DOI] [PubMed] [Google Scholar]
- Frey AG, Bird AJ, Evans-Galea MV, Blankman E, Winge DR, Eide DJ. Zinc-regulated DNA binding of the yeast Zap1 zinc-responsive activator. PLoS One. 2011;6:e22535. doi: 10.1371/journal.pone.0022535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden E. New perspectives on the essential trace elements. J Chem Ed. 1985;62:917–923. [Google Scholar]
- Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, Helmann JD. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci U S A. 2008;105:11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Wang T, Ye RW, Helmann JD. Functional analysis of the Bacillus subtilis Zur regulon. J Bacteriol. 2002;184:6508–6514. doi: 10.1128/JB.184.23.6508-6514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel C, Bittinger MA, Maier RJ. Cytochrome aa3 gene regulation in members of the family Rhizobiaceae: comparison of copper and oxygen effects in Bradyrhizobium japonicum and Rhizobium tropici. Appl Environ Microbiol. 1994;60:141–148. doi: 10.1128/aem.60.1.141-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SE, Helmann JD. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J Bacteriol. 2009;191:6116–6122. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SE, Miyagi F, Gaballa A, Helmann JD. Regulation of the Bacillus subtilis yciC gene and insights into the DNA-binding specificity of the zinc-sensing metalloregulator Zur. J Bacteriol. 2008;190:3482–3488. doi: 10.1128/JB.01978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaby JC, Buckley DH. A global census of nitrogenase diversity. Environ Microbiol. 2011;13:1790–1799. doi: 10.1111/j.1462-2920.2011.02488.x. [DOI] [PubMed] [Google Scholar]
- Gangeswaran R, Eady RR. Flavodoxin 1 of Azotobacter vinelandii: characterization and role in electron donation to purified assimilatory nitrate reductase. Biochem J. 1996;317:103–108. doi: 10.1042/bj3170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T. Iron in innate immunity: starve the invaders. Curr Opin Immunol. 2009;21:63–67. doi: 10.1016/j.coi.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattis SG, Hernick M, Fierke CA. Active site metal ion in UDP-3-O-((R)-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase (LpxC) switches between Fe(II) and Zn(II) depending on cellular conditions. J Biol Chem. 2010;285:33788–33796. doi: 10.1074/jbc.M110.147173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger O, Rohrs V, Weissenmayer B, Finan TM, Thomas-Oates JE. The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti. Mol Microbiol. 1999;32:63–73. doi: 10.1046/j.1365-2958.1999.01325.x. [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Wong B, Ferreira F, Markley JL, Straus NA. Cloning, sequencing and transcriptional studies of the genes for cytochrome c-553 and plastocyanin from Anabaena sp. PCC 7120. Microbiology. 1994;140:1151–1159. doi: 10.1099/13500872-140-5-1151. [DOI] [PubMed] [Google Scholar]
- Gilbert N. Environment: The disappearing nutrient. Nature. 2009;461:716–718. doi: 10.1038/461716a. [DOI] [PubMed] [Google Scholar]
- Giordano M, Beardall J, Raven JA. CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol. 2005;56:99–131. doi: 10.1146/annurev.arplant.56.032604.144052. [DOI] [PubMed] [Google Scholar]
- Giorgio AJ, Cartwright GE, Wintrobe MM. Preparation of a Copper-Deficient Medium for Yeast Growth. J Bacteriol. 1963;86:1037–1040. doi: 10.1128/jb.86.5.1037-1040.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goñi G, Serrano A, Frago S, Hervás M, Peregrina JR, De La Rosa MA, Gómez-Moreno C, Navarro JA, Medina M. Flavodoxin-mediated electron transfer from photosystem I to ferredoxin-NADP+ reductase in Anabaena: role of flavodoxin hydrophobic residues in protein-protein interactions. Biochemistry. 2008;47:1207–1217. doi: 10.1021/bi7017392. Epub 2008 Jan 5. [DOI] [PubMed] [Google Scholar]
- González-Ballester D, Casero D, Cokus S, Pellegrini M, Merchant SS, Grossman AR. RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell. 2010;22:2058–2084. doi: 10.1105/tpc.109.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE. Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbiol Rev. 2011;35:415–424. doi: 10.1111/j.1574-6976.2010.00253.x. [DOI] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci U S A. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AI, Hunt S, Stokes SL, Bramall N, Bunch J, Cox AG, Mcleod CW, Poole RK. Severe zinc depletion of Escherichia coli: roles for high affinity zinc binding by ZinT, zinc transport and zinc-independent proteins. J Biol Chem. 2009;284:18377–18389. doi: 10.1074/jbc.M109.001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DW, Chaudhary JA, Hanson RS, Arnold RG. Factors affecting competition between Type I and Type II methanotrophs in two-organism, continuous-flow reactors. Microb Ecol. 1993;25:1–17. doi: 10.1007/BF00182126. [DOI] [PubMed] [Google Scholar]
- Gralla EB, Kosman DJ. Molecular genetics of superoxide dismutases in yeasts and related fungi. Adv Genet. 1992;30:251–319. doi: 10.1016/s0065-2660(08)60322-3. [DOI] [PubMed] [Google Scholar]
- Gray HB, Malmström BG, Williams RJ. Copper coordination in blue proteins. J Biol Inorg Chem. 2000;5:551–559. doi: 10.1007/s007750000146. [DOI] [PubMed] [Google Scholar]
- Grossman AR, Schaefer MR, Chiang GG, Collier JL. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol Rev. 1993;57:725–749. doi: 10.1128/mr.57.3.725-749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes MJ, Peterson D, Vicari A, Cocks BG, Copeland NG, Gilbert DJ, Jenkins NA, Ferrick DA, Kastelein RA, Bazan JF, Zlotnik A. Identification of a novel selD homolog from eukaryotes, bacteria, and archaea: is there an autoregulatory mechanism in selenocysteine metabolism? Proc Natl Acad Sci U S A. 1996;93:15086–15091. doi: 10.1073/pnas.93.26.15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güler S, Seeliger A, Härtel H, Renger G, Benning C. A null mutant of Synechococcus sp. PCC7942 deficient in the sulfolipid sulfoquinovosyl diacylglycerol. J Biol Chem. 1996;271:7501–7507. doi: 10.1074/jbc.271.13.7501. [DOI] [PubMed] [Google Scholar]
- Guss JM, Freeman HC. Structure of oxidized poplar plastocyanin at 1.6 Ǻ resolution. J Mol Biol. 1983;169:521–563. doi: 10.1016/s0022-2836(83)80064-3. [DOI] [PubMed] [Google Scholar]
- Gutu A, Alvey RM, Bashour S, Zingg D, Kehoe DM. Sulfate-driven elemental sparing is regulated at the transcriptional and posttranscriptional levels in a filamentous cyanobacterium. J Bacteriol. 2011;193:1449–1460. doi: 10.1128/JB.00885-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas CE, Rodionov DA, Kropat J, Malasarn D, Merchant SS, De Crecy-Lagard V. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics. 2009;10:470. doi: 10.1186/1471-2164-10-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakemian AS, Rosenzweig AC. The biochemistry of methane oxidation. Annu Rev Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- Haley KP, Skaar EP. A battle for iron: host sequestration and Staphylococcus aureus acquisition. Microbes Infect. 2011 doi: 10.1016/j.micinf.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer ND, Skaar EP. The impact of metal sequestration on Staphylococcus aureus metabolism. Curr Opin Microbiol. 2011 doi: 10.1016/j.mib.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 2003;132:578–596. doi: 10.1104/pp.103.020941. Epub 2003 May 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanikenne M, Merchant SS, Hamel PP. Transition metal nutrition: a balance between deficiency and toxicity. In: Stern D, editor. The Chlamydomonas sourcebook. 2nd ed. San Diego: Academic Press; 2009. http://www.chem.ucla.edu/dept/Faculty/merchant/pdf/hanikenne.pdf. [Google Scholar]
- Hanke GT, Kimata-Ariga Y, Taniguchi I, Hase T. A post genomic characterization of Arabidopsis ferredoxins. Plant Physiol. 2004;134:255–264. doi: 10.1104/pp.103.032755. Epub 2003 Dec 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol Gen Genet. 1987;210:135–139. doi: 10.1007/BF00337769. [DOI] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr Opin Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Hao Z, Reiske HR, Wilson DB. Characterization of cadmium uptake in Lactobacillus plantarum and isolation of cadmium and manganese uptake mutants. Appl Environ Microbiol. 1999;65:4741–4745. doi: 10.1128/aem.65.11.4741-4745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happe T, Mosler B, Naber JD. Induction, localization and metal content of hydrogenase in the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 1994;222:769–774. doi: 10.1111/j.1432-1033.1994.tb18923.x. [DOI] [PubMed] [Google Scholar]
- Havaux M, Guedeney G, Hagemann M, Yeremenko N, Matthijs HC, Jeanjean R. The chlorophyll-binding protein IsiA is inducible by high light and protects the cyanobacterium Synechocystis PCC6803 from photooxidative stress. FEBS Lett. 2005;579:2289–2293. doi: 10.1016/j.febslet.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Hazebrouck S, Camoin L, Faltin Z, Strosberg AD, Eshdat Y. Substituting selenocysteine for catalytic cysteine 41 enhances enzymatic activity of plant phospholipid hydroperoxide glutathione peroxidase expressed in Escherichia coli. J Biol Chem. 2000;275:28715–28721. doi: 10.1074/jbc.M004985200. [DOI] [PubMed] [Google Scholar]
- He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, Korsmeyer S, Packer M, May HI, Hill JA, Virgin HW, Gilpin C, Xiao G, Bassel-Duby R, Scherer PE, Levine B. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012 doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol. 2011;28:2921–2933. doi: 10.1093/molbev/msr124. [DOI] [PubMed] [Google Scholar]
- Hensley MP, Gunasekera TS, Easton JA, Sigdel TK, Sugarbaker SA, Klingbeil L, Breece RM, Tierney DL, Crowder MW. Characterization of Zn(II)-responsive ribosomal proteins YkgM and L31 in E. coli. J Inorg Biochem. 2011 doi: 10.1016/j.jinorgbio.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert SK, Samson G, Fork DC, Laudenbach DE. Characterization of damage to photosystems I and II in a cyanobacterium lacking detectable iron superoxide dismutase activity. Proc Natl Acad Sci U S A. 1992;89:8716–8720. doi: 10.1073/pnas.89.18.8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JA, George SJ, Rubio LM. Molybdenum trafficking for nitrogen fixation. Biochemistry. 2009;48:9711–9721. doi: 10.1021/bi901217p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás M, Navarro JA, Molina-Heredia FP, De La Rosa MA. Ther reaction mechanism of photosystem I reduction by plastocyanin and cytochrome c6 follows two different kinetic models in the cyanobacterium Pseudoanabaena sp. PCC6903. Photosyn Res. 1998;57:93–100. [Google Scholar]
- Hewson I, Poretsky RS, Beinart RA, White AE, Shi T, Bench SR, Moisander PH, Paerl RW, Tripp HJ, Montoya JP, Moran MA, Zehr JP. In situ transcriptomic analysis of the globally important keystone N2-fixing taxon Crocosphaera watsonii. ISME J. 2009;3:618–631. doi: 10.1038/ismej.2009.8. [DOI] [PubMed] [Google Scholar]
- Hill KL, Merchant S. In vivo competition between plastocyanin and a copper-dependent regulator of the Chlamydomonas reinhardtii cytochrome c6 gene. Plant Physiol. 1992;100:319–326. doi: 10.1104/pp.100.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KL, Merchant S. Coordinate expression of coproporphyrinogen oxidase and cytochrome c6 in the green alga Chlamydomonas reinhardtii in response to changes in copper availability. EMBO J. 1995;14:857–865. doi: 10.1002/j.1460-2075.1995.tb07067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille R. Molybdenum and tungsten in biology. Trends Biochem Sci. 2002;27:360–367. doi: 10.1016/s0968-0004(02)02107-2. [DOI] [PubMed] [Google Scholar]
- Ho KK, Krogmann DW. Electron-Donors to P700 in Cyanobacteria and Algae - an Instance of Unusual Genetic-Variability. Biochim, Biophys. Acta. 1984;766:310–316. [Google Scholar]
- Hohn MJ, Palioura S, Su D, Yuan J, Soll D. Genetic analysis of selenocysteine biosynthesis in the archaeon Methanococcus maripaludis. Mol Microbiol. 2011;81:249–258. doi: 10.1111/j.1365-2958.2011.07690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondal RJ, Ruggles EL. Differing views of the role of selenium in thioredoxin reductase. Amino Acids. 2011;41:73–89. doi: 10.1007/s00726-010-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondorp ER, Matthews RG. Oxidation of cysteine 645 of cobalamin-independent methionine synthase causes a methionine limitation in Escherichia coli. J Bacteriol. 2009;191:3407–3410. doi: 10.1128/JB.01722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard JA, Hughes MN, Poole RK. Effects of copper concentration in continuous culture on the aa3-type cytochrome oxidase and respiratory chains of Paracoccus denitrificans. Arch Microbiol. 1989;151:300–306. [Google Scholar]
- Hulett FM. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol Microbiol. 1996;19:933–939. doi: 10.1046/j.1365-2958.1996.421953.x. [DOI] [PubMed] [Google Scholar]
- Hutber GN, Hutson KG, Rogers LJ. Effect of iron deficiency on levels of two ferredoxins and flavodoxin in a cyanobacterium. FEMS Microbiol Lett. 1977;1:193–196. [Google Scholar]
- Ihrig J, Hausmann A, Hain A, Richter N, Hamza I, Lill R, Muhlenhoff U. Iron regulation through the back door: iron-dependent metabolite levels contribute to transcriptional adaptation to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2010;9:460–471. doi: 10.1128/EC.00213-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- Irihimovitch V, Yehudai-resheff S. Phosphate and sulfur limitation responses in the chloroplast of Chlamydomonas reinhardtii. FEMS Microbiol Lett. 2008;283:1–8. doi: 10.1111/j.1574-6968.2008.01154.x. [DOI] [PubMed] [Google Scholar]
- Ivanov AG, Park Y-I, Miskiewicz E, Raven JA, Huner NP, Öquist G. Iron stress restricts photosynthetic intersystem electron transport in Synechococcus sp. PCC 7942. FEBS Lett. 2000;485:173–177. doi: 10.1016/s0014-5793(00)02211-0. [DOI] [PubMed] [Google Scholar]
- Jacques JF, Jang S, Prevost K, Desnoyers G, Desmarais M, Imlay J, Masse E. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol Microbiol. 2006;62:1181–1190. doi: 10.1111/j.1365-2958.2006.05439.x. [DOI] [PubMed] [Google Scholar]
- Jaffe EK. An unusual phylogenetic variation in the metal ion binding sites of porphobilinogen synthase. Chem Biol. 2003;10:25–34. doi: 10.1016/s1074-5521(02)00296-x. [DOI] [PubMed] [Google Scholar]
- Jain R, Hao B, Liu RP, Chan MK. Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal. J Am Chem Soc. 2005;127:4558–4559. doi: 10.1021/ja0503074. [DOI] [PubMed] [Google Scholar]
- Janagama HK, Kumar S, Bannantine JP, Kugadas A, Jagtap P, Higgins L, Witthuhn B, Sreevatsan S. Iron-sparing Response of Mycobacterium avium subsp. paratuberculosis is strain dependent. BMC Microbiol. 2010;10:268. doi: 10.1186/1471-2180-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyasingh PD, Weider LJ. Fundamental links between genes and elements: evolutionary implications of ecological stoichiometry. Mol Ecol. 2007;16:4649–4661. doi: 10.1111/j.1365-294X.2007.03558.x. [DOI] [PubMed] [Google Scholar]
- Jiang W, Metcalf WW, Lee KS, Wanner BL. Molecular cloning, mapping, and regulation of Pho regulon genes for phosphonate breakdown by the phosphonatase pathway of Salmonella typhimurium LT2. J Bacteriol. 1995;177:6411–6421. doi: 10.1128/jb.177.22.6411-6421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger RD, Bishop PE. Bacterial alternative nitrogen fixation systems. Crit Rev Microbiol. 1988;16:1–14. doi: 10.3109/10408418809104465. [DOI] [PubMed] [Google Scholar]
- Jung YS, Kwon YM. Small RNA ArrF regulates the expression of sodB and feSII genes in Azotobacter vinelandii. Curr Microbiol. 2008;57:593–597. doi: 10.1007/s00284-008-9248-z. [DOI] [PubMed] [Google Scholar]
- Kaluarachchi H, Chan chung KC, Zamble DB. Microbial nickel proteins. Nat Prod Rep. 2010;27:681–694. doi: 10.1039/b906688h. [DOI] [PubMed] [Google Scholar]
- Kaplan J, Mcvey ward D, Crisp RJ, Philpott CC. Iron-dependent metabolic remodeling in S. cerevisiae. Biochim Biophys Acta. 2006;1763:646–651. doi: 10.1016/j.bbamcr.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Kaplan R, Apirion D. The fate of ribosomes in Escherichia coli cells starved for a carbon source. J Biol Chem. 1975;250:1854–1863. [PubMed] [Google Scholar]
- Kathuria S, Martiny AC. Prevalence of a calcium-based alkaline phosphatase associated with the marine cyanobacterium Prochlorococcus and other ocean bacteria. Environ Microbiol. 2011;13:74–83. doi: 10.1111/j.1462-2920.2010.02310.x. [DOI] [PubMed] [Google Scholar]
- Katoh S, Takamiya A. A new leaf copper protein 'plastocyanin', a natural Hill oxidant. Nature. 1961;189:665–666. doi: 10.1038/189665a0. [DOI] [PubMed] [Google Scholar]
- Kehl-fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl-fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Kidd MJ, Penner-hahn JE, Pakrasi HB. A light-dependent mechanism for massive accumulation of manganese in the photosynthetic bacterium Synechocystis sp. PCC 6803. Biochemistry. 2002;41:15085–15092. doi: 10.1021/bi026892s. [DOI] [PubMed] [Google Scholar]
- Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci U S A. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hirai MY, Hayashi H, Chino M, Naito S, Fujiwara T. Role of O-acetyl-l-serine in the coordinated regulation of the expression of a soybean seed storage-protein gene by sulfur and nitrogen nutrition. Planta. 1999;209:282–289. doi: 10.1007/s004250050634. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Graham DW, Dispirito AA, Alterman MA, Galeva N, Larive CK, Asunskis D, Sherwood PM. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science. 2004;305:1612–1615. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- Kim HY, Fomenko DE, Yoon YE, Gladyshev VN. Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry. 2006;45:13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Kang SO, Lee JK. The protein complex composed of nickel-binding SrnQ and DNA binding motif-bearing SrnR of Streptomyces griseus represses sodF transcription in the presence of nickel. J Biol Chem. 2003;278:18455–18463. doi: 10.1074/jbc.M211740200. [DOI] [PubMed] [Google Scholar]
- King AL, Sanudo-wilhelmy SA, Leblanc K, Hutchins DA, Fu F. CO2 and vitamin B12 interactions determine bioactive trace metal requirements of a subarctic Pacific diatom. ISME J. 2011;5:1388–1396. doi: 10.1038/ismej.2010.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC. Zinc: an essential but elusive nutrient. Am J Clin Nutr. 2011;94:679S–684S. doi: 10.3945/ajcn.110.005744. Epub 2011 Jun 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp CW, Fowle DA, Kulczycki E, Roberts JA, Graham DW. Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proc Natl Acad Sci U S A. 2007;104:12040–12045. doi: 10.1073/pnas.0702879104. Epub 2007 Jul 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight E, JR, D'eustachio AJ, Hardy RW. Flavodoxin: a flavoprotein with ferredoxin activity from Clostrium pasteurianum. Biochim Biophys Acta. 1966;113:626–628. doi: 10.1016/s0926-6593(66)80025-5. [DOI] [PubMed] [Google Scholar]
- Konhauser KO, Pecoits E, Lalonde SV, Papineau D, Nisbet EG, Barley ME, Arndt NT, Zahnle K, Kamber BS. Oceanic nickel depletion and a methanogen famine before the Great Oxidation Event. Nature. 2009;458:750–753. doi: 10.1038/nature07858. [DOI] [PubMed] [Google Scholar]
- Kosman DJ. Molecular mechanisms of iron uptake in fungi. Mol Microbiol. 2003;47:1185–1197. doi: 10.1046/j.1365-2958.2003.03368.x. [DOI] [PubMed] [Google Scholar]
- Kraepiel AM, Bellenger JP, Wichard T, Morel FM. Multiple roles of siderophores in free-living nitrogen-fixing bacteria. Biometals. 2009;22:573–581. doi: 10.1007/s10534-009-9222-7. [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Kropat J, Hong-hermesdorf A, Casero D, Ent P, Castruita M, Pellegrini M, Merchant SS, Malasarn D. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 2011;66:770–780. doi: 10.1111/j.1365-313X.2011.04537.x. Epub 2011 Mar 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J, Tottey S, Birkenbihl RP, Depège N, Huijser P, Merchant S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci U S A. 2005;102:18730–18735. doi: 10.1073/pnas.0507693102. Epub 2005 Dec 13 PMC1311908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Gladyshev VN. The prokaryotic selenoproteome. EMBO Rep. 2004;5:538–543. doi: 10.1038/sj.embor.7400126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunert K-J, Böger P. Absence of plastocyanin in the alga Bumilleriopsis and its replacement by cytochrome 553. Z. Naturforsch. 1975;30c:190–200. [Google Scholar]
- La fontaine S, Quinn JM, Nakamoto SS, Page MD, Gohre V, Moseley JL, Kropat J, Merchant S. Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot Cell. 2002;1:736–757. doi: 10.1128/EC.1.5.736-757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La roche J, Boyd PW, Mckay RML, Geider RJ. Flavodoxin as an in situ marker for iron stress in phytoplankton. Nature. 1996;382:802–804. [Google Scholar]
- Labbé S, Peña MM, Fernandes AR, Thiele DJ. A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. J Biol Chem. 1999;274:36252–36260. doi: 10.1074/jbc.274.51.36252. [DOI] [PubMed] [Google Scholar]
- Lafontaine DL. A 'garbage can' for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem Sci. 2010;35:267–277. doi: 10.1016/j.tibs.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Lahooti M, Harwood CR. Transcriptional analysis of the Bacillus subtilis teichuronic acid operon. Microbiology. 1999;145(Pt 12):3409–3417. doi: 10.1099/00221287-145-12-3409. [DOI] [PubMed] [Google Scholar]
- Lane TW, Morel FM. A biological function for cadmium in marine diatoms. Proc Natl Acad Sci U S A. 2000a;97:4627–4631. doi: 10.1073/pnas.090091397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane TW, Morel FM. Regulation of carbonic anhydrase expression by zinc, cobalt, and carbon dioxide in the marine diatom Thalassiosira weissflogii. Plant Physiol. 2000b;123:345–352. doi: 10.1104/pp.123.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane TW, Morel FM. Regulation of carbonic anhydrase expression by zinc, cobalt, and carbon dioxide in the marine diatom Thalassiosira weissflogii. Plant Physiol. 2000c;123:345–352. doi: 10.1104/pp.123.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane TW, Saito MA, George GN, Pickering IJ, Prince RC, Morel FM. Biochemistry: a cadmium enzyme from a marine diatom. Nature. 2005;435:442. doi: 10.1038/435042a. [DOI] [PubMed] [Google Scholar]
- Lang WK, Glassey K, Archibald AR. Influence of phosphate supply on teichoic acid and teichuronic acid content of Bacillus subtilis cell walls. J Bacteriol. 1982;151:367–375. doi: 10.1128/jb.151.1.367-375.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudenbach DE, Reith ME, Straus NA. Isolation, sequence analysis, and transcriptional studies of the flavodoxin gene from Anacystis nidulans R2. J Bacteriol. 1988;170:258–265. doi: 10.1128/jb.170.1.258-265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson RJ, Von wachenfeldt C, Haq I, Perkins J, Munro AW. Expression and characterization of the two flavodoxin proteins of Bacillus subtilis YkuN and YkuP: biophysical properties and interactions with cytochrome P450 BioI. Biochemistry. 2004;43:12390–12409. doi: 10.1021/bi049131t. [DOI] [PubMed] [Google Scholar]
- Lee BC, Lobanov AV, Marino SM, Kaya A, Seravalli J, Hatfield DL, Gladyshev VN. A 4-selenocysteine, 2-selenocysteine insertion sequence (SECIS) element methionine sulfoxide reductase from Metridium senile reveals a non-catalytic function of selenocysteines. J Biol Chem. 2011;286:18747–18755. doi: 10.1074/jbc.M111.229807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- Leigh JA, Albers SV, Atomi H, Allers T. Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol Rev. 2011;35:577–608. doi: 10.1111/j.1574-6976.2011.00265.x. [DOI] [PubMed] [Google Scholar]
- Leonhardt K, Straus NA. An iron stress operon involved in photosynthetic electron transport in the marine cyanobacterium Synechococcus sp. PCC 7002. J Gen Microbiol. 1992;138:1613–1621. doi: 10.1099/00221287-138-8-1613. [DOI] [PubMed] [Google Scholar]
- Li HH, Merchant S. Two metal-dependent steps in the biosynthesis of Scenedesmus obliquus plastocyanin. Differential mRNA accumulation and holoprotein formation. J. Biol. Chem. 1992;267:9368–9375. [PubMed] [Google Scholar]
- Li HH, Merchant S. Degradation of plastocyanin in copper-deficient Chlamydomonas reinhardtii. J Biol Chem. 1995;270:23504–23510. doi: 10.1074/jbc.270.40.23504. [DOI] [PubMed] [Google Scholar]
- Li HH, Quinn J, Culler D, Girard-bascou J, Merchant S. Molecular genetic analysis of plastocyanin biosynthesis in Chlamydomonas reinhardtii. J Biol Chem. 1996;271:31283–31289. doi: 10.1074/jbc.271.49.31283. [DOI] [PubMed] [Google Scholar]
- Li L, Chen OS, Mcvey ward D, Kaplan J. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem. 2001;276:29515–29519. doi: 10.1074/jbc.M103944200. [DOI] [PubMed] [Google Scholar]
- Li X, Yakunin AF, Mckay RML. Fe-responsive accumulation of redox proteins ferredoxin and flavodoxin in a marine cryptomonad. Eur J Phycol. 2004;39:73–82. [Google Scholar]
- Li Y, Zamble DB. Nickel homeostasis and nickel regulation: an overview. Chem Rev. 2009;109:4617–4643. doi: 10.1021/cr900010n. [DOI] [PubMed] [Google Scholar]
- Liang W, Malhotra A, Deutscher MP. Acetylation regulates the stability of a bacterial protein: growth stage-dependent modification of RNase R. Mol Cell. 2011;44:160–166. doi: 10.1016/j.molcel.2011.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman RL, Rosenzweig AC. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005;434:177–182. doi: 10.1038/nature03311. Epub 2005 Jan 26. [DOI] [PubMed] [Google Scholar]
- Lindahl PA, Holmes-hampton GP. Biophysical probes of iron metabolism in cells and organelles. Curr Opin Chem Biol. 2011;15:342–346. doi: 10.1016/j.cbpa.2011.01.007. Epub 2011 Feb 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jin W, Theil EC. Opening protein pores with chaotropes enhances Fe reduction and chelation of Fe from the ferritin biomineral. Proc Natl Acad Sci U S A. 2003;100:3653–3658. doi: 10.1073/pnas.0636928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov AV, Hatfield DL, Gladyshev VN. Eukaryotic selenoproteins and selenoproteomes. Biochim Biophys Acta. 2009;1790:1424–1428. doi: 10.1016/j.bbagen.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell. 2010;22:2219–2236. doi: 10.1105/tpc.110.074096. Epub 2010 Jul 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-lara IM, Gao JL, Soto MJ, Solares-pérez A, Weissenmayer B, Sohlenkamp C, Verroios GP, Thomas-oates J, Geiger O. Phosphorus-free membrane lipids of Sinorhizobium meliloti are not required for the symbiosis with alfalfa but contribute to increased cell yields under phosphorus-limiting conditions of growth. Mol Plant Microbe Interact. 2005;18:973–982. doi: 10.1094/MPMI-18-0973. [DOI] [PubMed] [Google Scholar]
- Luo H, Benner R, Long RA, Hu J. Subcellular localization of marine bacterial alkaline phosphatases. Proc Natl Acad Sci U S A. 2009;106:21219–21223. doi: 10.1073/pnas.0907586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Gabriel SE, Helmann JD. Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res. 2011;39:9130–9138. doi: 10.1093/nar/gkr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, Smith I, Palu G, Riccardi G, Manganelli R. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J Bacteriol. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Ponomarev VA, Koonin EV. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-research0033. RESEARCH 0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmström BG, Leckner J. The chemical biology of copper. Curr Opin Chem Biol. 1998;2:286–292. doi: 10.1016/s1367-5931(98)80071-9. [DOI] [PubMed] [Google Scholar]
- Martin JE, Imlay JA. The alternative aerobic ribonucleotide reductase of Escherichia coli NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol. 2011;80:319–334. doi: 10.1111/j.1365-2958.2011.07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NC, Chiang KS, Goodenough UW. Turnover of chloroplast and cytoplasmic ribosomes during gametogenesis in Chlamydomonas reinhardi. Dev Biol. 1976;51:190–201. doi: 10.1016/0012-1606(76)90137-8. [DOI] [PubMed] [Google Scholar]
- Martin P, Van mooy BA, Heithoff A, Dyhrman ST. Phosphorus supply drives rapid turnover of membrane phospholipids in the diatom Thalassiosira pseudonana. ISME J. 2011;5:1057–1060. doi: 10.1038/ismej.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Tyson GW, Delong EF. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol. 2010;12:222–238. doi: 10.1111/j.1462-2920.2009.02062.x. [DOI] [PubMed] [Google Scholar]
- Masepohl B, Drepper T, Paschen A, Gross S, Pawlowski A, Raabe K, Riedel KU, Klipp W. Regulation of nitrogen fixation in the phototrophic purple bacterium Rhodobacter capsulatus. J Mol Microbiol Biotechnol. 2002;4:243–248. [PubMed] [Google Scholar]
- Masepohl B, Hallenbeck PC. Nitrogen and molybdenum control of nitrogen fixation in the phototrophic bacterium Rhodobacter capsulatus. Adv Exp Med Biol. 2010;675:49–70. doi: 10.1007/978-1-4419-1528-3_4. [DOI] [PubMed] [Google Scholar]
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara T, Frunzke K, Zumft WG. Modulation by copper of the products of nitrite respiration in Pseudomonas perfectomarinus. J Bacteriol. 1982;149:816–823. doi: 10.1128/jb.149.3.816-823.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D, Marlière P. Adaptive eradication of methionine and cysteine from cyanobacterial light-harvesting proteins. Nature. 1989;341:245–248. doi: 10.1038/341245a0. [DOI] [PubMed] [Google Scholar]
- Mckay RM, Geider RJ, Laroche J. Physiological and Biochemical Response of the Photosynthetic Apparatus of Two Marine Diatoms to Fe Stress. Plant Physiol. 1997;114:615–622. doi: 10.1104/pp.114.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcrae R, Bagchi P, Sumalekshmy S, Fahrni CJ. In situ imaging of metals in cells and tissues. Chem Rev. 2009;109:4780–4827. doi: 10.1021/cr900223a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier B, Sehn AP, Schininà ME, Barra D. In vivo incorporation of copper into the iron-exchangeable and manganese-exchangeable superoxide dismutase from Propionibacterium shermanii Amino acid sequence and identity of the protein moieties. Eur J Biochem. 1994;219:463–468. doi: 10.1111/j.1432-1033.1994.tb19960.x. [DOI] [PubMed] [Google Scholar]
- Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA. A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J Bacteriol. 2007;189:3686–3694. doi: 10.1128/JB.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S. Synthesis of metalloproteins involved in photosynthesis: plastocyanin and cytochromes. In: Rochaix J-D, Goldschmidt-clermont M, Merchant S, editors. The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Kluwer Academic Publishers; 1998. [Google Scholar]
- Merchant S, Bogorad L. Rapid degradation of apoplastocyanin in Cu(II)-deficient cells of Chlamydomonas reinhardtii. J Biol Chem. 1986a;261:15850–15853. [PubMed] [Google Scholar]
- Merchant S, Bogorad L. Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardtii. Mol Cell Biol. 1986b;6:462–469. doi: 10.1128/mcb.6.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S, Bogorad L. Metal ion regulated gene expression: use of a plastocyanin-less mutant of Chlamydomonas reinhardtii to study the Cu(II)-dependent expression of cytochrome c-552. EMBO J. 1987a;6:2531–2535. doi: 10.1002/j.1460-2075.1987.tb02540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S, Bogorad L. The Cu(II)-repressible plastidic cytochrome c Cloning and sequence of a complementary DNA for the pre-apoprotein. J Biol Chem. 1987b;262:9062–9067. [PubMed] [Google Scholar]
- Merchant S, Hill K, Howe G. Dynamic interplay between two copper-titrating components in the transcriptional regulation of cyt c6. EMBO J. 1991;10:1383–1389. doi: 10.1002/j.1460-2075.1991.tb07658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Müller J, Lippard SJ. Dioxygen Activation and Methane Hydroxylation by Soluble Methane Monooxygenase: A Tale of Two Irons and Three Proteins. Angew Chem Int Ed Engl. 2001;40:2782–2807. doi: 10.1002/1521-3773(20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Messerschmidt A, Huber R. The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships. Eur J Biochem. 1990;187:341–352. doi: 10.1111/j.1432-1033.1990.tb15311.x. [DOI] [PubMed] [Google Scholar]
- Mey AR, Craig SA, Payne SM. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect Immun. 2005;73:5706–5719. doi: 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin DE, Abdolrahimzadeh H, Baddiley J. Replacement of acidic phosphates by acidic glycolipids in Pseudomonas diminuta. Nature. 1974;249:268–269. doi: 10.1038/249268a0. [DOI] [PubMed] [Google Scholar]
- Miramar MD, Inda LA, Saraiva LM, Peleato ML. Plastocyanin/cytochrome c6 interchange in Scenedesmus vacuolatus. J Plant Physiol. 2003;160:1483–1486. doi: 10.1078/0176-1617-01009. [DOI] [PubMed] [Google Scholar]
- Moisander PH, Beinart RA, Hewson I, White AE, Johnson KS, Carlson CA, Montoya JP, Zehr JP. Unicellular cyanobacterial distributions broaden the oceanic N2 fixation domain. Science. 2010;327:1512–1514. doi: 10.1126/science.1185468. [DOI] [PubMed] [Google Scholar]
- Moore GR, Pettigrew GW. Cytochromes c: Evolutionary, structural and physicochemical aspects. New York: Springer-Verlag; 1990. [Google Scholar]
- Morel FM. The co-evolution of phytoplankton and trace element cycles in the oceans. Geobiology. 2008;6:318–324. doi: 10.1111/j.1472-4669.2008.00144.x. [DOI] [PubMed] [Google Scholar]
- Morel FM, Price NM. The biogeochemical cycles of trace metals in the oceans. Science. 2003;300:944–947. doi: 10.1126/science.1083545. [DOI] [PubMed] [Google Scholar]
- Moroney JV, Ynalvez RA. Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot Cell. 2007;6:1251–1259. doi: 10.1128/EC.00064-07. Epub 2007 Jun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J, Grossman AR. Phosphate metabolism and responses to phosphorus deficiency. In: Stern DB, editor. The Chlamydomonas Sourcebook. San Diego, CA: Academic Press; 2009. [Google Scholar]
- Moseley J, Quinn J, Eriksson M, Merchant S. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. Embo J. 2000;19:2139–2151. doi: 10.1093/emboj/19.10.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JL, Allinger T, Herzog S, Hoerth P, Wehinger E, Merchant S, Hippler M. Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 2002;21:6709–6720. doi: 10.1093/emboj/cdf666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota CS, Valette O, Gonzalez PJ, Brondino CD, Moura JJ, Moura I, Dolla A, Rivas MG. Effects of molybdate and tungstate on expression levels and biochemical characteristics of formate dehydrogenases produced by Desulfovibrio alaskensis NCIMB 13491. J Bacteriol. 2011;193:2917–2923. doi: 10.1128/JB.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller SI, Valdebenito M, Hantke K. Salmochelin, the long-overlooked catecholate siderophore of Salmonella. Biometals. 2009;22:691–695. doi: 10.1007/s10534-009-9217-4. [DOI] [PubMed] [Google Scholar]
- Murrell JC, Gilbert B, Mcdonald IR. Molecular biology and regulation of methane monooxygenase. Arch Microbiol. 2000a;173:325–332. doi: 10.1007/s002030000158. [DOI] [PubMed] [Google Scholar]
- Murrell JC, Mcdonald IR, Gilbert B. Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 2000b;8:221–225. doi: 10.1016/s0966-842x(00)01739-x. [DOI] [PubMed] [Google Scholar]
- Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Yamagishi M, Yoshizaki F, Sugimura Y. The syntheses of plastocyanin and cytochrome c-553 are regulated by copper at the pre-translational level in a green alga, Pediastrum boryanum. J Biochem. 1992;111:219–224. doi: 10.1093/oxfordjournals.jbchem.a123740. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Yoshizaki F, Sugimura Y. Accumulation of plastocyanin mRNA lacking 5' region in the green alga Pediastrum boryanum grown under copper-deficient conditions. Plant Cell Physiol. 2000;41:33–41. doi: 10.1093/pcp/41.1.33. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Hoaki T, Hanada S, Maruyama A, Kamagata Y, Fuse H. Soluble and particulate methane monooxygenase gene clusters in the marine methanotroph Methylomicrobium sp. strain NI. FEMS Microbiol Lett. 2007;277:157–164. doi: 10.1111/j.1574-6968.2007.00953.x. [DOI] [PubMed] [Google Scholar]
- Nanamiya H, Akanuma G, Natori Y, Murayama R, Kosono S, Kudo T, Kobayashi K, Ogasawara N, Park SM, Ochi K, Kawamura F. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol Microbiol. 2004;52:273–283. doi: 10.1111/j.1365-2958.2003.03972.x. [DOI] [PubMed] [Google Scholar]
- Nanamiya H, Kawamura F. Towards an elucidation of the roles of the ribosome during different growth phases in Bacillus subtilis. Biosci Biotechnol Biochem. 2010;74:451–461. doi: 10.1271/bbb.90859. [DOI] [PubMed] [Google Scholar]
- Nandal A, Huggins CC, Woodhall MR, Mchugh J, Rodriguez-Quinones F, Quail MA, Guest JR, Andrews SC. Induction of the ferritin gene (ftnA) of Escherichia coli by Fe(2+)-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol Microbiol. 2010;75:637–657. doi: 10.1111/j.1365-2958.2009.06977.x. [DOI] [PubMed] [Google Scholar]
- Nath K, Koch AL. Protein degradation in Escherichia coli. II. Strain differences in the degradation of protein and nucleic acid resulting from starvation. J Biol Chem. 1971;246:6956–6967. [PubMed] [Google Scholar]
- Natori Y, Nanamiya H, Akanuma G, Kosono S, Kudo T, Ochi K, Kawamura F. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol Microbiol. 2007;63:294–307. doi: 10.1111/j.1365-2958.2006.05513.x. [DOI] [PubMed] [Google Scholar]
- Naumann B, Busch A, Allmer J, Ostendorf E, Zeller M, Kirchhoff H, Hippler M. Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics. 2007;7:3964–3979. doi: 10.1002/pmic.200700407. [DOI] [PubMed] [Google Scholar]
- Nauser T, Steinmann D, Koppenol WH. Why do proteins use selenocysteine instead of cysteine? Amino Acids. 2012;42:39–44. doi: 10.1007/s00726-010-0602-7. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Ingraham JL, Schaechter M. Physiology of the Bacterial Cell: A Molecular Approach. Sunderland, MA: Sinauer Associates; 1990. [Google Scholar]
- Nguyen AV, Thomas-Hall SR, Malnoë A, Timmins M, Mussgnug JH, Rupprecht J, Kruse O, Hankamer B, Schenk PM. Transcriptome for photobiological hydrogen production induced by sulfur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryot Cell. 2008;7:1965–1979. doi: 10.1128/EC.00418-07. Epub 2008 Aug 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HH, Elliott SJ, Yip JH, Chan SI. The particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a novel copper-containing three-subunit enzyme. Isolation and characterization. J Biol Chem. 1998;273:7957–7966. doi: 10.1074/jbc.273.14.7957. [DOI] [PubMed] [Google Scholar]
- Nguyen KT, Wu JC, Boylan JA, Gherardini FC, Pei D. Zinc is the metal cofactor of Borrelia burgdorferi peptide deformylase. Arch Biochem Biophys. 2007;468:217–225. doi: 10.1016/j.abb.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhoffer EC, Naranjo CM, Bradley KL, Fee JA. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J Bacteriol. 1990;172:1930–1938. doi: 10.1128/jb.172.4.1930-1938.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AK, Gerdes K, Murrell JC. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol Microbiol. 1997;25:399–409. doi: 10.1046/j.1365-2958.1997.4801846.x. [DOI] [PubMed] [Google Scholar]
- Nyhus K, Jacobson ES. Oxy2 as a transcriptional activator gene for copper uptake in Cryptococcus neoformans. Med Mycol. 2004;42:325–331. doi: 10.1080/13693780310001658757. [DOI] [PubMed] [Google Scholar]
- O'halloran TV, Culotta VC. Metallochaperones, an intracellular shuttle service for metal ions. J Biol Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- Oglesby-Sherrouse AG, Vasil ML. Characterization of a heme-regulated non-coding RNA encoded by the prrF locus of Pseudomonas aeruginosa. PLoS One. 2010;5:e9930. doi: 10.1371/journal.pone.0009930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol. 2006;188:3664–3673. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphan VJ, House CH. Geobiological investigations using secondary ion mass spectrometry: microanalysis of extant and paleo-microbial processes. Geobiology. 2009;7:360–372. doi: 10.1111/j.1472-4669.2009.00201.x. [DOI] [PubMed] [Google Scholar]
- Osiewacz HD, Nuber U. GRISEA, a putative copper-activated transcription factor from Podospora anserina involved in differentiation and senescence. Mol Gen Genet. 1996;252:115–124. doi: 10.1007/BF02173211. [DOI] [PubMed] [Google Scholar]
- Outten CE, O'halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- Owen GA, Pascoe B, Kallifidas D, Paget MS. Zinc-responsive regulation of alternative ribosomal protein genes in Streptomyces coelicolor involves zur and sigmaR. J Bacteriol. 2007;189:4078–4086. doi: 10.1128/JB.01901-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MD, Allen MD, Urzica EI, Kropat J, Karpowicz S, Merchant SS. Two mechanisms for maintenance of superoxide dismutase capacity in iron-deficient Chlamydomonas. 2012 [Google Scholar]
- Palenik B, Morel FM. Amine oxidases of marine phytoplankton. Appl Environ Microbiol. 1991;57:2440–2443. doi: 10.1128/aem.57.8.2440-2443.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AE, Qin Y, Park JG, Mccombs JE. Design and application of genetically encoded biosensors. Trends Biotechnol. 2011;29:144–152. doi: 10.1016/j.tibtech.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panina EM, Mironov AA, Gelfand MS. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc Natl Acad Sci U S A. 2003;100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee AB. Purification and properties of a sulfate-binding protein from Salmonella typhimurium. J Biol Chem. 1966;241:5886–5892. [PubMed] [Google Scholar]
- Pardo MB, Gómez-Moreno C, Pelesto MI. Effect of iron-deficiency on ferredoxin levels in Anabaena variabilis PCC 6309. Arch Microbiol. 1990;153:528–530. [Google Scholar]
- Park H, Song B, Morel FM. Diversity of the cadmium-containing carbonic anhydrase in marine diatoms and natural waters. Environ Microbiol. 2007;9:403–413. doi: 10.1111/j.1462-2920.2006.01151.x. [DOI] [PubMed] [Google Scholar]
- Park YI, Sandström S, Gustafsson P, Öquist G. Expression of the isiA gene is essential for the survival of the cyanobacterium Synechococcus sp. PCC 7942 by protecting photosystem II from excess light under iron limitation. Mol Microbiol. 1999;32:123–129. doi: 10.1046/j.1365-2958.1999.01332.x. [DOI] [PubMed] [Google Scholar]
- Partensky F, Garczarek L. Prochlorococcus: Advantages and limits of minimalism. Annu Rev Marine Sci. 2010;2:303–331. doi: 10.1146/annurev-marine-120308-081034. [DOI] [PubMed] [Google Scholar]
- Pedro-Segura E, Vergara SV, Rodriguez-Navarro S, Parker R, Thiele DJ, Puig S. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J Biol Chem. 2008;283:28527–28535. doi: 10.1074/jbc.M804910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G, Price NM. A role for manganese in superoxide dismutases and growth of iron-deficient diatoms. Limnol. Oceanogr. 2004;49:1774–1783. [Google Scholar]
- Peers G, Price NM. Copper-containing plastocyanin used for electron transport by an oceanic diatom. Nature. 2006;441:341–344. doi: 10.1038/nature04630. [DOI] [PubMed] [Google Scholar]
- Pettigrew GW, Moore GR. Cytochromes c. Biological aspects. Berlin: Springer-Verlag; 1987. [Google Scholar]
- Petrucco S, Bolchi A, Foroni C, Percudani R, Rossi GL, Ottonello S. A maize gene encoding an NADPH binding enzyme highly homologous to isoflavone reductases is activated in response to sulfur starvation. Plant Cell. 1996;8:69–80. doi: 10.1105/tpc.8.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott CC, Protchenko O. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:20–27. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piir K, Paier A, Liiv A, Tenson T, Maivali U. Ribosome degradation in growing bacteria. EMBO Rep. 2011;12:458–462. doi: 10.1038/embor.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Cohu CM, Ravet K, Abdel-Ghany SE, Gaymard F. Essential transition metal homeostasis in plants. Curr Opin Plant Biol. 2009;12:347–357. doi: 10.1016/j.pbi.2009.04.011. Epub 2009 May 27. [DOI] [PubMed] [Google Scholar]
- Pomowski A, Zumft WG, Kroneck PM, Einsle O. N2O binding at a [4Cu:2S] copper-sulphur cluster in nitrous oxide reductase. Nature. 2011;477:234–237. doi: 10.1038/nature10332. [DOI] [PubMed] [Google Scholar]
- Posey JE, Gherardini FC. Lack of a role for iron in the Lyme disease pathogen. Science. 2000;288:1651–1653. doi: 10.1126/science.288.5471.1651. [DOI] [PubMed] [Google Scholar]
- Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Masse E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- Privalle CT, Fridovich I. Iron specificity of the Fur-dependent regulation of the biosynthesis of the manganese-containing superoxide dismutase in Escherichia coli. J Biol Chem. 1993;268:5178–5181. [PubMed] [Google Scholar]
- Priya B, Premanandh J, Dhanalakshmi RT, Seethalakshmi T, Uma L, Prabaharan D, Subramanian G. Comparative analysis of cyanobacterial superoxide dismutases to discriminate canonical forms. BMC Genomics. 2007;8:435. doi: 10.1186/1471-2164-8-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Puig S, Vergara SV, Thiele DJ. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 2008;7:555–564. doi: 10.1016/j.cmet.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Hulett FM. Role of Pho-P in transcriptional regulation of genes involved in cell wall anionic polymer biosynthesis in Bacillus subtilis. J Bacteriol. 1998;180:4007–4010. doi: 10.1128/jb.180.15.4007-4010.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Q, Helmann JD. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol. 2000;35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- Quigg A, Finkel ZV, Irwin AJ, Rosenthal Y, Ho TY, Reinfelder JR, Schofield O, Morel FM, Falkowski PG. The evolutionary inheritance of elemental stoichiometry in marine phytoplankton. Nature. 2003;425:291–294. doi: 10.1038/nature01953. [DOI] [PubMed] [Google Scholar]
- Quigg A, Irwin AJ, Finkel ZV. Evolutionary inheritance of elemental stoichiometry in phytoplankton. Proc Biol Sci. 2011;278:526–534. doi: 10.1098/rspb.2010.1356. Epub 2010 Sep 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JM, Barraco P, Eriksson M, Merchant S. Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J Biol Chem. 2000;275:6080–6089. doi: 10.1074/jbc.275.9.6080. [DOI] [PubMed] [Google Scholar]
- Quinn JM, Eriksson M, Moseley J, Merchant S. Oxygen Deficiency-Responsive Gene Expression in Chlamydomonas reinhardtii Through a Copper-Sensing Signal Transduction Pathway. Plant Physiol. 2002;128:463–471. doi: 10.1104/pp.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JM, Merchant S. Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell. 1995;7:623–638. doi: 10.1105/tpc.7.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JM, Merchant S. Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 1998;297:263–279. doi: 10.1016/s0076-6879(98)97020-3. [DOI] [PubMed] [Google Scholar]
- Ragsdale SW. Nickel-based Enzyme Systems. J Biol Chem. 2009;284:18571–18575. doi: 10.1074/jbc.R900020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale SW, Ljungdahl LG. Characterization of ferredoxin, flavodoxin, and rubredoxin from Clostridium formicoaceticum grown in media with high and low iron contents. J Bacteriol. 1984;157:1–6. doi: 10.1128/jb.157.1.1-6.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA, Evans MCW, Korb RE. The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosyn. Research. 1999;60:111–150. [Google Scholar]
- Reddi AR, Jensen LT, Culotta VC. Manganese homeostasis in Saccharomyces cerevisiae. Chem Rev. 2009;109:4722–4732. doi: 10.1021/cr900031u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redinbo MR, Yeates TO, Merchant S. Plastocyanin: Structural and functional analysis. J. Bioenerg. Biomembr. 1994;26:49–66. doi: 10.1007/BF00763219. [DOI] [PubMed] [Google Scholar]
- Regulski EE, Moy RH, Weinberg Z, Barrick JE, Yao Z, Ruzzo WL, Breaker RR. A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol Microbiol. 2008;68:918–932. doi: 10.1111/j.1365-2958.2008.06208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle C, Coosemans N, Jans F, Hanikenne M, Motte P, Cardol P. Knock-down of the COX3 and COX17 gene expression of cytochrome c oxidase in the unicellular green alga Chlamydomonas reinhardtii. Plant Mol Biol. 2010;74:223–233. doi: 10.1007/s11103-010-9668-6. Epub 2010 Aug 11. [DOI] [PubMed] [Google Scholar]
- Riccardi G, Milano A, Pasca MR, Nies DH. Genomic analysis of zinc homeostasis in Mycobacterium tuberculosis. FEMS Microbiol Lett. 2008;287:1–7. doi: 10.1111/j.1574-6968.2008.01320.x. [DOI] [PubMed] [Google Scholar]
- Ridge PG, Zhang Y, Gladyshev VN. Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS One. 2008;3:e1378. doi: 10.1371/journal.pone.0001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof WR, Ruckle ME, Lydic TA, Sears BB, Benning C. The sulfolipids 2'-O-acyl-sulfoquinovosyldiacylglycerol and sulfoquinovosyldiacylglycerol are absent from a Chlamydomonas reinhardtii mutant deleted in SQD1. Plant Physiol. 2003;133:864–874. doi: 10.1104/pp.103.029249. Epub 2003 Sep 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts K, Granum E, Leegood RC, Raven JA. Carbon acquisition by diatoms. Photosynth Res. 2007;93:79–88. doi: 10.1007/s11120-007-9172-2. Epub 2007 May 12. [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. Epub 2003 Jul 17. [DOI] [PubMed] [Google Scholar]
- Rohlin L, Gunsalus RP. Carbon-dependent control of electron transfer and central carbon pathway genes for methane biosynthesis in the Archaean, Methanosarcina acetivorans strain C2A. BMC Microbiol. 2010;10:62. doi: 10.1186/1471-2180-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen BP, Ajees AA, Mcdermott TR. Life and death with arsenic. Arsenic life: an analysis of the recent report "A bacterium that can grow by using arsenic instead of phosphorus". Bioessays. 2011;33:350–357. doi: 10.1002/bies.201100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother M, Mathes I, Lottspeich F, Bock A. Inactivation of the selB gene in Methanococcus maripaludis: effect on synthesis of selenoproteins and their sulfur-containing homologs. J Bacteriol. 2003;185:107–114. doi: 10.1128/JB.185.1.107-114.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc Natl Acad Sci U S A. 2010;107:16184–16189. doi: 10.1073/pnas.1009513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito MA. Biogeochemistry: Less nickel for more oxygen. Nature. 2009;458:714–715. doi: 10.1038/458714a. [DOI] [PubMed] [Google Scholar]
- Saito MA, Bertrand EM, Dutkiewicz S, Bulygin VV, Moran DM, Monteiro FM, Follows MJ, Valois FW, Waterbury JB. Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc Natl Acad Sci U S A. 2011;108:2184–2189. doi: 10.1073/pnas.1006943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon E, Keren N. Manganese limitation induces changes in the activity and in the organization of photosynthetic complexes in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 2011;155:571–579. doi: 10.1104/pp.110.164269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvail H, Lanthier-Bourbonnais P, Sobota JM, Caza M, Benjamin JA, Mendieta ME, Lepine F, Dozois CM, Imlay J, Masse E. A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proc Natl Acad Sci U S A. 2010;107:15223–15228. doi: 10.1073/pnas.1007805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvail H, Masse E. Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip Rev RNA. 2012;3:26–36. doi: 10.1002/wrna.102. [DOI] [PubMed] [Google Scholar]
- Sandmann G, Böger P. Copper-induced exchange of plastocyanin and cytochrome c-533 in cultures of Anabaena variabilis and Plectonema boryanum. Plant Sci Lett. 1980;17:417–424. [Google Scholar]
- Sandmann G, Malkin R. Iron-Sulfur Centers and Activities of the Photosynthetic Electron Transport Chain in Iron-Deficient Cultures of the Blue-Green Alga Aphanocapsa. Plant Physiol. 1983;73:724–728. doi: 10.1104/pp.73.3.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann G, Malkin R. Light inhibition of respiration is due to a dual function of the cytochrome b6-f complex and the plastocyanin/cytochrome c-553 pool in Aphanocapsa. Arch Biochem Biophys. 1984;234:105–111. doi: 10.1016/0003-9861(84)90329-1. [DOI] [PubMed] [Google Scholar]
- Sandmann G, Peleato ML, Fillat MF, Lázaro MC, Gómez-Moreno C. Consequences of the iron-dependent formation of ferredoxin and flavodoxin on photosynthesis and nitrogen fixation on Anabaena strains. Photosyn Res. 1990;26:119–125. doi: 10.1007/BF00047083. [DOI] [PubMed] [Google Scholar]
- Sandmann G, Reck H, Kessler E, Boger P. Distribution of plastocyanin and soluble plastidic cytochrome c in various classes of algae. Arch Microbiol. 1983;134:23–27. [Google Scholar]
- Sandy M, Butler A. Microbial iron acquisition: marine and terrestrial siderophores. Chem Rev. 2009;109:4580–4595. doi: 10.1021/cr9002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran B, Bonnett SA, Shah K, Gabriel S, Reddy R, Schimmel P, Rodionov DA, De Crecy-Lagard V, Helmann JD, Iwata-Reuyl D, Swairjo MA. Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB. J Bacteriol. 2009;191:6936–6949. doi: 10.1128/JB.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanvisens N, Bano MC, Huang M, Puig S. Regulation of ribonucleotide reductase in response to iron deficiency. Mol Cell. 2011;44:759–769. doi: 10.1016/j.molcel.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan J, Dumont MG, Murrell JC. Involvement of MmoR and MmoG in the transcriptional activation of soluble methane monooxygenase genes in Methylosinus trichosporium OB3b. FEMS Microbiol Lett. 2009;301:181–187. doi: 10.1111/j.1574-6968.2009.01816.x. Epub 2009 Oct 10. [DOI] [PubMed] [Google Scholar]
- Scheckhuber CQ, Grief J, Boilan E, Luce K, Debacq-Chainiaux F, Rittmeyer C, Gredilla R, Kolbesen BO, Toussaint O, Osiewacz HD. Age-related cellular copper dynamics in the fungal ageing model Podospora anserina and in ageing human fibroblasts. PLoS One. 2009;4:e4919. doi: 10.1371/journal.pone.0004919. Epub 2009 Mar 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid AK, Pan M, Sharma K, Baliga NS. Two transcription factors are necessary for iron homeostasis in a salt-dwelling archaeon. Nucleic Acids Res. 2011;39:2519–2533. doi: 10.1093/nar/gkq1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneegurt MA, Sherman DM, Nayar S, Sherman LA. Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp. strain ATCC 51142. J Bacteriol. 1994;176:1586–1597. doi: 10.1128/jb.176.6.1586-1597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönheit P, Brandis A, Thauer RK. Ferredoxin degradation in growing Clostridium pasteurianum during periods of iron deprivation. Arch Microbiol. 1979;120:73–76. doi: 10.1007/BF00413277. [DOI] [PubMed] [Google Scholar]
- Schwarz G, Mendel RR, Ribbe MW. Molybdenum cofactors, enzymes and pathways. Nature. 2009;460:839–847. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- Sebastian M, Ammerman JW. The alkaline phosphatase PhoX is more widely distributed in marine bacteria than the classical PhoA. ISME J. 2009;3:563–572. doi: 10.1038/ismej.2009.10. [DOI] [PubMed] [Google Scholar]
- Seguin A, Ward DM, Kaplan J. Regulation of Ribonucleotide Reductase during Iron Limitation. Mol Cell. 2011;44:683–684. doi: 10.1016/j.molcel.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seravalli J, Ragsdale SW. Expanding the biological periodic table. Chem Biol. 2010;17:793–794. doi: 10.1016/j.chembiol.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service RF. Nations Move to Head Off Shortages of Rare Earths. Science. 2010;327:1596–1597. doi: 10.1126/science.327.5973.1596. [DOI] [PubMed] [Google Scholar]
- Shatzman AR, Kosman DJ. The utilization of copper and its role in the biosynthesis of copper-containing proteins in the fungus, Dactylium dendroides. Biochim Biophys Acta. 1978;544:163–179. doi: 10.1016/0304-4165(78)90220-9. [DOI] [PubMed] [Google Scholar]
- Shatzman AR, Kosman DJ. Biosynthesis and cellular distribution of the two superoxide dismutases of Dactylium dendroides. J Bacteriol. 1979;137:313–320. doi: 10.1128/jb.137.1.313-320.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiller AM, Boyle E. Dissolved Zinc in Rivers. Nature. 1985;317:49–52. [Google Scholar]
- Shin JH, Oh SY, Kim SJ, Roe JH. The zinc-responsive regulator Zur controls a zinc uptake system and some ribosomal proteins in Streptomyces coelicolor A3(2) J Bacteriol. 2007;189:4070–4077. doi: 10.1128/JB.01851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S, Phung LET. Novel expansion of living chemistry or just a serious mistake? FEMS Microbiol Lett. 2011;315:79–80. doi: 10.1111/j.1574-6968.2010.02202.x. [DOI] [PubMed] [Google Scholar]
- Simm C, Lahner B, Salt D, Lefurgey A, Ingram P, Yandell B, Eide DJ. Saccharomyces cerevisiae vacuole in zinc storage and intracellular zinc distribution. Eukaryot Cell. 2007;6:1166–1177. doi: 10.1128/EC.00077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Kaur N, Kosman DJ. The metalloreductase Fre6p in Fe-efflux from the yeast vacuole. J Biol Chem. 2007;282:28619–28626. doi: 10.1074/jbc.M703398200. [DOI] [PubMed] [Google Scholar]
- Singh BK, Bardgett RD, Smith P, Reay DS. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol. 2010;8:779–790. doi: 10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- Smaldone G, Antelmann H, Gaballa A, Helmann JD. The FsrA sRNA and FbpB protein mediate the iron-dependent induction of the Bacillus subtilis LutABC iron-sulfur containing oxidases. J Bacteriol. 2012 doi: 10.1128/JB.05567-11. (in revision). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL. The physiological role of ferritin-like compounds in bacteria. Crit Rev Microbiol. 2004;30:173–185. doi: 10.1080/10408410490435151. [DOI] [PubMed] [Google Scholar]
- Sohm JA, Webb EA, Capone DG. Emerging patterns of marine nitrogen fixation. Nat Rev Microbiol. 2011;9:499–508. doi: 10.1038/nrmicro2594. [DOI] [PubMed] [Google Scholar]
- Sohrin Y, Bruland KW. Global status of trace elements in the ocean. Trends in Analytical Chemistry. 2011;30:1291–1307. [Google Scholar]
- Sommer F, Kropat J, Malasarn D, Grossoehme NE, Chen X, Giedroc DP, Merchant SS. The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell. 2010;22:4098–4113. doi: 10.1105/tpc.110.080069. Epub 2010 Dec 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo L, Toro I, D'orazio M, O'neill P, Pedersen JZ, Carugo O, Rotilio G, Battistoni A, Djinovic-Carugo K. Unique features of the sodC-encoded superoxide dismutase from Mycobacterium tuberculosis a fully functional copper-containing enzyme lacking zinc in the active site. J Biol Chem. 2004;279:33447–33455. doi: 10.1074/jbc.M404699200. Epub 2004 May 23. [DOI] [PubMed] [Google Scholar]
- Sprott GD, Bakouche L, Rajagopal K. Identification of sulfoquinovosyl diacylglycerol as a major polar lipid in Marinococcus halophilus and Salinicoccus hispanicus and substitution with phosphatidylglycerol. Can J Microbiol. 2006;52:209–219. doi: 10.1139/w05-112. [DOI] [PubMed] [Google Scholar]
- Stadtman TC. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- Steunou AS, Jensen SI, Brecht E, Becraft ED, Bateson MM, Kilian O, Bhaya D, Ward DM, Peters JW, Grossman AR, Kühl M. Regulation of nif gene expression and the energetics of N2 fixation over the diel cycle in a hot spring microbial mat. Isme J. 2008;2:364–378. doi: 10.1038/ismej.2007.117. Epub 2008 Mar 6. [DOI] [PubMed] [Google Scholar]
- Stock T, Selzer M, Connery S, Seyhan D, Resch A, Rother M. Disruption and complementation of the selenocysteine biosynthesis pathway reveals a hierarchy of selenoprotein gene expression in the archaeon Methanococcus maripaludis. Mol Microbiol. 2011;82:734–747. doi: 10.1111/j.1365-2958.2011.07850.x. [DOI] [PubMed] [Google Scholar]
- Stock T, Selzer M, Rother M. In vivo requirement of selenophosphate for selenoprotein synthesis in archaea. Mol Microbiol. 2010;75:149–160. doi: 10.1111/j.1365-2958.2009.06970.x. [DOI] [PubMed] [Google Scholar]
- Stoof J, Breijer S, Pot RG, Van Der Neut D, Kuipers EJ, Kusters JG, Van Vliet AH. Inverse nickel-responsive regulation of two urease enzymes in the gastric pathogen Helicobacter mustelae. Environ Microbiol. 2008;10:2586–2597. doi: 10.1111/j.1462-2920.2008.01681.x. [DOI] [PubMed] [Google Scholar]
- Strzepek RF, Harrison PJ. Photosynthetic architecture differs in coastal and oceanic diatoms. Nature. 2004;431:689–692. doi: 10.1038/nature02954. [DOI] [PubMed] [Google Scholar]
- Subramanian P, Rodrigues AV, Ghimire-Rijal S, Stemmler TL. Iron chaperones for mitochondrial Fe-S cluster biosynthesis and ferritin iron storage. Curr Opin Chem Biol. 2011;15:312–318. doi: 10.1016/j.cbpa.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Sato N, Tsuzuki M. Utilization of a chloroplast membrane sulfolipid as a major internal sulfur source for protein synthesis in the early phase of sulfur starvation in Chlamydomonas reinhardtii. FEBS Lett. 2007;581:4519–4522. doi: 10.1016/j.febslet.2007.08.035. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Tsuzuki M, Sato N. Regulation of synthesis and degradation of a sulfolipid under sulfur-starved conditions and its physiological significance in Chlamydomonas reinhardtii. New Phytol. 2010;185:676–686. doi: 10.1111/j.1469-8137.2009.03115.x. [DOI] [PubMed] [Google Scholar]
- Sukdeo N, Honek JF. Pseudomonas aeruginosa contains multiple glyoxalase I-encoding genes from both metal activation classes. Biochim Biophys Acta. 2007;1774:756–763. doi: 10.1016/j.bbapap.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Sun J, Klein A. A lysR-type regulator is involved in the negative regulation of genes encoding selenium-free hydrogenases in the archaeon Methanococcus voltae. Mol Microbiol. 2004;52:563–571. doi: 10.1111/j.1365-2958.2004.03998.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Braby CE, Grossman AR. Sulfur economy and cell wall biosynthesis during sulfur limitation of Chlamydomonas reinhardtii. Plant Physiol. 2001;127:665–673. [PMC free article] [PubMed] [Google Scholar]
- Terauchi AM, Lu SF, Zaffagnini M, Tappa S, Hirasawa M, Tripathy JN, Knaff DB, Farmer PJ, Lemaire SD, Hase T, Merchant SS. Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J Biol Chem. 2009;284:25867–25878. doi: 10.1074/jbc.M109.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi AM, Peers G, Kobayashi MC, Niyogi KK, Merchant SS. Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth Res. 2010;105:39–49. doi: 10.1007/s11120-010-9562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Kaster AK, Goenrich M, Schick M, Hiromoto T, Shima S. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu Rev Biochem. 2010;79:507–536. doi: 10.1146/annurev.biochem.030508.152103. [DOI] [PubMed] [Google Scholar]
- Tortell PD, Price NM. Cadmium toxicity and zinc limitation in centric diatoms of the genus Thalassiosira. Mar Ecol Prog Ser. 1996;138:245–254. [Google Scholar]
- Tottey S, Harvie DR, Robinson NJ. Understanding how cells allocate metals using metal sensors and metallochaperones. Acc Chem Res. 2005;38:775–783. doi: 10.1021/ar0300118. [DOI] [PubMed] [Google Scholar]
- Tottey S, Waldron KJ, Firbank SJ, Reale B, Bessant C, Sato K, Cheek TR, Gray J, Banfield MJ, Dennison C, Robinson NJ. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature. 2008;455:1138–1142. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trefry JH, Metz S, Trocine RP, Nelsen TA. A decline in lead transport by the Mississippi river. Science. 1985;230:439–441. doi: 10.1126/science.230.4724.439. [DOI] [PubMed] [Google Scholar]
- Tripp BC, Bell CB, 3rd, Cruz F, Krebs C, Ferry JG. A role for iron in an ancient carbonic anhydrase. J Biol Chem. 2004;279:6683–6687. doi: 10.1074/jbc.M311648200. [DOI] [PubMed] [Google Scholar]
- Tripp HJ, Bench SR, Turk KA, Foster RA, Desany BA, Niazi F, Affourtit JP, Zehr JP. Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature. 2010;464:90–94. doi: 10.1038/nature08786. [DOI] [PubMed] [Google Scholar]
- Tu WY, Pohl S, Gray J, Robinson NJ, Harwood CR, Waldron KJ. Cellular Iron Distribution in Bacillus anthracis. J Bacteriol. 2011 doi: 10.1128/JB.06195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- Van Bakel H, Strengman E, Wijmenga C, Holstege FC. Gene expression profiling and phenotype analyses of S. cerevisiae in response to changing copper reveals six genes with new roles in copper and iron metabolism. Physiol Genomics. 2005;22:356–367. doi: 10.1152/physiolgenomics.00055.2005. Epub 2005 May 10. [DOI] [PubMed] [Google Scholar]
- Van Mooy BA, Fredricks HF, Pedler BE, Dyhrman ST, Karl DM, Koblízek M, Lomas MW, Mincer TJ, Moore LR, Moutin T, Rappé MS, Webb EA. Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature. 2009;458:69–72. doi: 10.1038/nature07659. [DOI] [PubMed] [Google Scholar]
- Van Mooy BA, Rocap G, Fredricks HF, Evans CT, Devol AH. Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc Natl Acad Sci U S A. 2006;103:8607–8612. doi: 10.1073/pnas.0600540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese S, Wu A, Park S, Imlay KR, Imlay JA. Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol Microbiol. 2007;64:822–830. doi: 10.1111/j.1365-2958.2007.05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara SV, Thiele DJ. Post-transcriptional regulation of gene expression in response to iron deficiency: co-ordinated metabolic reprogramming by yeast mRNA-binding proteins. Biochem Soc Trans. 2008;36:1088–1090. doi: 10.1042/BST0361088. [DOI] [PubMed] [Google Scholar]
- Vinkenborg JL, Koay MS, Merkx M. Fluorescent imaging of transition metal homeostasis using genetically encoded sensors. Curr Opin Chem Biol. 2010;14:231–237. doi: 10.1016/j.cbpa.2009.11.022. [DOI] [PubMed] [Google Scholar]
- Wakeman CA, Skaar EP. Metalloregulation of Gram-positive pathogen physiology. Curr Opin Microbiol. 2011 doi: 10.1016/j.mib.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- Wang D, Hurst TK, Thompson RB, Fierke CA. Genetically encoded ratiometric biosensors to measure intracellular exchangeable zinc in Escherichia coli. J Biomed Opt. 2011a;16:087011. doi: 10.1117/1.3613926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pantopoulos K. Regulation of cellular iron metabolism. Biochem J. 2011;434:365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Duanmu D, Spalding MH. Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: inorganic carbon transport and CO2 recapture. Photosynth Res. 2011b;109:115–122. doi: 10.1007/s11120-011-9643-3. [DOI] [PubMed] [Google Scholar]
- Wang Y, Secco D, Poirier Y. Characterization of the PHO1 gene family and the responses to phosphate deficiency of Physcomitrella patens. Plant Physiol. 2008;146:646–656. doi: 10.1104/pp.107.108548. Epub 2007 Nov 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZQ, Lawson RJ, Buddha MR, Wei CC, Crane BR, Munro AW, Stuehr DJ. Bacterial flavodoxins support nitric oxide production by Bacillus subtilis nitric-oxide synthase. J Biol Chem. 2007;282:2196–2202. doi: 10.1074/jbc.M608206200. [DOI] [PubMed] [Google Scholar]
- Warner DF, Savvi S, Mizrahi V, Dawes SS. A riboswitch regulates expression of the coenzyme B12-independent methionine synthase in Mycobacterium tuberculosis: implications for differential methionine synthase function in strains H37Rv and CDC1551. J Bacteriol. 2007;189:3655–3659. doi: 10.1128/JB.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg ED. The Lactobacillus anomaly: total iron abstinence. Perspect Biol Med. 1997;40:578–583. doi: 10.1353/pbm.1997.0072. [DOI] [PubMed] [Google Scholar]
- Wen J, Chen X, Bowie JU. Exploring the allowed sequence space of a membrane protein. Nat Struct Biol. 1996;3:141–148. doi: 10.1038/nsb0296-141. [DOI] [PubMed] [Google Scholar]
- Wennerhold J, Bott M. The DtxR regulon of Corynebacterium glutamicum. J Bacteriol. 2006;188:2907–2918. doi: 10.1128/JB.188.8.2907-2918.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerhold J, Krug A, Bott M. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J Biol Chem. 2005;280:40500–40508. doi: 10.1074/jbc.M508693200. [DOI] [PubMed] [Google Scholar]
- White AE. New insights into bacterial acquisition of phosphorus in the surface ocean. Proc Natl Acad Sci U S A. 2009;106:21013–21014. doi: 10.1073/pnas.0912475107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C, Lee J, Kambe T, Fritsche K, Petris MJ. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PJ, Sowa NA, Fitzgerald DJ, Fitzgerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci U S A. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildner GF, Hauska G. Localization of the reaction site of cytochrome 552 in chloroplasts from Euglena gracilis Cytochrome content and photooxidation in different chloroplast preparations. Arch Biochem Biophys. 1974;164:127–135. doi: 10.1016/0003-9861(74)90014-9. [DOI] [PubMed] [Google Scholar]
- Williams R. Chemical selection of elements by cells. Coordination Chemistry Reviews. 2001;216–217:583–595. [Google Scholar]
- Winge DR. Copper-regulatory domain involved in gene expression. Prog Nucleic Acid Res Mol Biol. 1998;58:165–195. doi: 10.1016/s0079-6603(08)60036-7. [DOI] [PubMed] [Google Scholar]
- Wintjens R, Noël C, May AC, Gerbod D, Dufernez F, Capron M, Viscogliosi E, Rooman M. Specificity and phenetic relationships of iron- and manganese-containing superoxide dismutases on the basis of structure and sequence comparisons. J Biol Chem. 2004;279:9248–9254. doi: 10.1074/jbc.M312329200. Epub 2003 Dec 12. [DOI] [PubMed] [Google Scholar]
- Wolfe-Simon F, Davies PCW, Anbar AD. Did nature also choose arsenic? International Journal of Astrobiology. 2009;8:69–74. [Google Scholar]
- Wolfe-Simon F, Starovoytov V, Reinfelder JR, Schofield O, Falkowski PG. Localization and role of manganese superoxide dismutase in a marine diatom. Plant Physiol. 2006;142:1701–1709. doi: 10.1104/pp.106.088963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe-Simon F, Switzer Blum J, Kulp TR, Gordon GW, Hoeft SE, Pett-Ridge J, Stolz JF, Webb SM, Weber PK, Davies PC, Anbar AD, Oremland RS. A bacterium that can grow by using arsenic instead of phosphorus. Science. 2011;332:1163–1166. doi: 10.1126/science.1197258. [DOI] [PubMed] [Google Scholar]
- Wood PM. Interchangeable copper and iron proteins in algal photosynthesis. Studies on plastocyanin and cytochrome c-552 in Chlamydomonas. Eur J Biochem. 1978;87:9–19. doi: 10.1111/j.1432-1033.1978.tb12346.x. [DOI] [PubMed] [Google Scholar]
- Wu CH, Jiang W, Krebs C, Stubbe J. YfaE, a ferredoxin involved in diferric-tyrosyl radical maintenance in Escherichia coli ribonucleotide reductase. Biochemistry. 2007;46:11577–11588. doi: 10.1021/bi7012454. [DOI] [PubMed] [Google Scholar]
- Wu WF, Urbanowski ML, Stauffer GV. Role of the MetR regulatory system in vitamin B12-mediated repression of the Salmonella typhimurium metE gene. J Bacteriol. 1992;174:4833–4837. doi: 10.1128/jb.174.14.4833-4837.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtmann EJ, Wolin SL. A role for a bacterial ortholog of the Ro autoantigen in starvation-induced rRNA degradation. Proc Natl Acad Sci U S A. 2010;107:4022–4027. doi: 10.1073/pnas.1000307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc Natl Acad Sci U S A. 1999;96:15336–15341. doi: 10.1073/pnas.96.26.15336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Degui T, Shaked Y, Morel FMM. Zinc, cadmium, and cobalt interreplacement and relative use efficiencies in the coccolithophore Emiliania huxleyi. Limnol Oceanogr. 2007;52:2294–2305. [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell. 2009;21:347–361. doi: 10.1105/tpc.108.060137. Epub 2009 Jan 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D, Morel FMM. In vivo substitution of zinc by cobalt in carbonic anhydrase of a marine diatom. Limnol Oceanogr. 1996;41:573–577. [Google Scholar]
- Yehudai-Resheff S, Zimmer SL, Komine Y, Stern DB. Integration of chloroplast nucleic acid metabolism into the phosphate deprivation response in Chlamydomonas reinhardtii. Plant Cell. 2007;19:1023–1038. doi: 10.1105/tpc.106.045427. Epub 2007 Mar 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 1996;10:835–844. doi: 10.1046/j.1365-313x.1996.10050835.x. [DOI] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci U S A. 2002;99:5732–5737. doi: 10.1073/pnas.082696499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaheer R, Morton R, Proudfoot M, Yakunin A, Finan TM. Genetic and biochemical properties of an alkaline phosphatase PhoX family protein found in many bacteria. Environ Microbiol. 2009;11:1572–1587. doi: 10.1111/j.1462-2920.2009.01885.x. [DOI] [PubMed] [Google Scholar]
- Zahn JA, Bergmann DJ, Boyd JM, Kunz RC, Dispirito AA. Membrane-associated quinoprotein formaldehyde dehydrogenase from Methylococcus capsulatus Bath. J Bacteriol. 2001;183:6832–6840. doi: 10.1128/JB.183.23.6832-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaleta-Pastor M, Sohlenkamp C, Gao JL, Guan Z, Zaheer R, Finan TM, Raetz CR, Lopez-Lara IM, Geiger O. Sinorhizobium meliloti phospholipase C required for lipid remodeling during phosphorus limitation. Proc Natl Acad Sci U S A. 2010;107:302–307. doi: 10.1073/pnas.0912930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr JP. Nitrogen fixation by marine cyanobacteria. Trends Microbiol. 2011;19:162–173. doi: 10.1016/j.tim.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Zehr JP, Bench SR, Mondragon EA, Mccarren J, Delong EF. Low genomic diversity in tropical oceanic N2-fixing cyanobacteria. Proc Natl Acad Sci U S A. 2007;104:17807–17812. doi: 10.1073/pnas.0701017104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr JP, Jenkins BD, Short SM, Steward GF. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol. 2003;5:539–554. doi: 10.1046/j.1462-2920.2003.00451.x. [DOI] [PubMed] [Google Scholar]
- Zehr JP, Kudela RM. Nitrogen cycle of the open ocean: from genes to ecosystems. Ann Rev Mar Sci. 2011;3:197–225. doi: 10.1146/annurev-marine-120709-142819. [DOI] [PubMed] [Google Scholar]
- Zhang L, Mcspadden B, Pakrasi HB, Whitmarsh J. Copper-mediated regulation of cytochrome c553 and plastocyanin in the cyanobacterium Synechocystis 6803. J Biol Chem. 1992;267:19054–19059. [PubMed] [Google Scholar]
- Zhang Y, Gladyshev VN. Molybdoproteomes and evolution of molybdenum utilization. J Mol Biol. 2008;379:881–899. doi: 10.1016/j.jmb.2008.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gladyshev VN. General trends in trace element utilization revealed by comparative genomic analyses of Co, Cu, Mo, Ni, and Se. J Biol Chem. 2010;285:3393–3405. doi: 10.1074/jbc.M109.071746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gladyshev VN. Comparative genomics of trace element dependence in biology. J Biol Chem. 2011;286:23623–23629. doi: 10.1074/jbc.R110.172833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rodionov DA, Gelfand MS, Gladyshev VN. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics. 2009;10:78. doi: 10.1186/1471-2164-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Stubbe J. Bacillus subtilis class Ib ribonucleotide reductase is a dimanganese(III)-tyrosyl radical enzyme. Biochemistry. 2011;50:5615–5623. doi: 10.1021/bi200348q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Gibbons J, Zhang S, Williamson PR. Copper-mediated reversal of defective laccase in a Δvph1 avirulent mutant of Cryptococcus neoformans. Mol Microbiol. 2003;47:1007–1014. doi: 10.1046/j.1365-2958.2003.03340.x. [DOI] [PubMed] [Google Scholar]
- Zumft WG, Spiller H. Characterization of a flavodoxin from the green alga Chlorella. Biochem Biophys Res Commun. 1971;45:112–118. doi: 10.1016/0006-291x(71)90057-x. [DOI] [PubMed] [Google Scholar]
- Zundel MA, Basturea GN, Deutscher MP. Initiation of ribosome degradation during starvation in Escherichia coli. RNA. 2009;15:977–983. doi: 10.1261/rna.1381309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurbriggen MD, Tognetti VB, Carrillo N. Stress-inducible flavodoxin from photosynthetic microorganisms. The mystery of flavodoxin loss from the plant genome. IUBMB Life. 2007;59:355–360. doi: 10.1080/15216540701258744. [DOI] [PubMed] [Google Scholar]