Abstract
The controversy around sirtuins and their functions in aging has drawn in the past few years as much attention, if not more, from the scientific community and the public as they did when first proposed as the key conserved aging regulators in eukaryotes. With some of the basic observations on sirtuin longevity promoting functions being questioned in popular model systems, researchers are wondering if this family of conserved enzymes still holds strong potential as therapeutic targets. This review examines the several controversial issues around sirtuins and their functions in aging, calorie restriction, as well as age-related diseases in light of recent studies in mammalian systems and discusses whether modulators of sirtuins still hold the secret of life.
Introduction
The word “sirtuin” was coined from its founding member Sir2 in budding yeast Saccharomyces cerevisiae 1. SIR stands for Silent Information Regulator. Four SIR genes were identified through a genetic screen for mutations that suppressed transcription silencing 1. Among them, SIR2 is essential for silencing at all heterochromatin-like regions, including the ribosomal gene cluster (rDNA), telomeres, and the hidden mating type loci HML/HMR. The gene product Sir2 protein is an enzyme called histone deacetylase (HDAC, class III) that removes the acetyl group from acetylated lysines in histones. Very soon, many homologs of Sir2 were found from bacteria to humans, establishing a highly conserved class of enzymes 2.
Sirtuins are protein deacetylases and ADP-ribosyltransferases
Distinct from previously identified deacetylases (class I and II), sirtuins couple the deacetylation of lysine to the hydrolysis of NAD+ by transferring the acetyl group to the ADP-ribose moiety to form O-acetyl-ADP-ribose, releasing free nicotinamide (Fig. 1A). Because NAD+ is required for this reaction and the NAD+/NADH is determined by the nutritional state of the cell, sirtuins directly link cellular metabolic signaling to the state of protein post-translational modifications. In addition to the NAD+/NADH influence, this reaction is also regulated by one of its products, nicotinamide, which is a non-competitive inhibitor of sirtuins. The other reaction product, O-acetyl-ADP-ribose, has been suggested to facilitate heterochromatin formation and silencing, regulate ion channels, and modulate cellular redox state 3.
Figure 1.
Typical chemical reactions catalyzed by sirtuin enzymes. (A) Protein lysine deacetylation by sirtuins requires NAD+ as a cofactor, releasing deaceylated protein, nicotinamide, and 2’-O-acetyl-ADP-ribose. (B) Certain sirtuins are ADP-ribosyltransferases, attaching the ADP-ribose moiety to the ε-amine of lysine, releasing nicotinamide.
Some sirtuins are found to carry out reactions to remove larger PTM groups, such as malonyl and succinyl from their target proteins in a way very similar to deacetylation. Yet, other sirtuins do not exhibit the deacetylation activity, but rather act as ADP-ribosyltransferases, enzymes that add an ADP-ribosyl group to lysines 4 (Fig. 1B).
Sirtuins in simple model organisms
In budding yeast, Sir2 is recruited to silencing loci by other DNA binding factors such as Rap1 and removes acetyl group from histone H4 lysine 16 (H4K16), histone H3K56, and H3K45–7. It forms a heterotrimeric complex with Sir3 and Sir4 at HML/HMR and telomeres, and exists in a homotrimeric form with Net1 and Cdc14 at rDNA 8. Besides Sir2, there are four other sirtuins, Hst1, Hst2, Hst3, and Hst4, all of which are involved in transcription silencing 2, even though Hst2 predominantly localizes to the cytoplasm 9. The involvement of sirtuins in aging was discovered through a genetic screen that identified a long-lived mutant featuring a C-terminal truncation to Sir4 10. It turns out that this mutation causes the Sir2-Sir3-Sir4 complex to relocalize from telomeres to the rDNA cluster in the nucleolus and to suppress the rDNA recombination and the formation of extrachromosomal rDNA circles (ERCs), an aging factor unique to yeast 11. Deletion or inhibition of Sir2 shortens lifespan, whereas overexpression with a second integrated copy of the SIR2 gene extends the replicative lifespan, suggesting that Sir2 promotes longevity and is a limiting factor for lifespan 12. More recently, Sir2's function at telomeres is also shown to be important for longevity and that the Sir2 protein becomes much less abundant in old cells, providing more insights to the cause of yeast replicative aging 13.
In the nematode Caenorhabditis elegans, four sirtuins are identified with SIR-2.1 showing the highest similarity to yeast Sir2. Worm SIR-2.1 deacetylates H3K9 and is involved in transcription silencing and heterochromatin formation 14. It is shown to interact with non-histone targets, 14-3-3 and DAF-16, to activate DAF-16 target genes, which is proposed to be a longevity promoting mechanism under stress 15. Deletion of this gene moderately shortens lifespan 16. However, the effect of sir-2.1 overexpression has been recently disputed. The initially observed lifespan extension by either chromosomal duplication or transgene has been attributed to secondary mutations unrelated to sir-2.1 17,18.
In Drosophila melanogaster, five sirtuins have been found, with dSir2 being the closest homolog to yeast Sir2. dSir2 shows deacetylase activity in vitro and is required for heterochromatin silencing in vivo 19. Knocking out dSir2 shortens lifespan 20. Lifespan extension upon dSir2 overexpression has been found in a number of different configurations, including several different transgenes, different drivers, tissue-specific and adult-only inductions 21. However, several of these have also been challenged recently 18. These controversies will be discussed in detail later.
Mammalian Sirtuins
In mammals, there are seven sirtuins, named SIRT1 to SIRT7. They are involved in a much broader range of cellular processes and pathways with distinct cellular localization and molecular targets (Table 1).
Table 1.
Summary of sirtuin functions
| Localization | Activity | Target | Molecular and cellular function | ||
|---|---|---|---|---|---|
| Yeast | |||||
| Sir2 | Nucleus, nucleolus | Deacetylation | H4K16, H3K56, H3K4 | Gene silencing, heterochromatin | |
| Hst1 | Nucleus | Deacetylation | H3K4 | Transcription repression | |
| Hst2 | Cytoplasm | Deacetylation | H4K16 | Nucleolar and telomere silencing | |
| Hst3 | Nucleus | Deacetylation | H3K56 | DNA replication and repair | |
| Hst4 | Nucleus | Deacetylation | H3K56 | DNA replication and repair | |
| Worm | |||||
| sir-2.1 | Nucleus | Deacetylation | H3K9 | Transcription silencing, heterochomatin | |
| Fly | |||||
| dSir2 | Nucleus, cytoplasm | Deacetylation | Histone, Dmp53 | Transcription silencing, heterochomatin | |
| Mammal | |||||
| SIRT1 | Nucleus | Deacetylation | H1K26, H4K16, p53, PGClα, NF-κB, FOXO1, FOXO3, FOXO4, Notch, HIFlα, 14-3-3, PI3K, DNMT1, TORC1, HSF1, Ku70 | Transcription silencing, mitochondria regulation, insulin signaling, tumorigenesis, apoptosis, cell proliferation and survival, tissue regeneration, differentiation, stress response | |
| SIRT2 | Cytoplasm | Deacetylation | H4K16, Tubulin, PAR-3, FOXO1, FOXO3, CDH1, CDC20, PGClα | Mitosis, nerve myelination and regeneration, brain aging, adipocyte differentiation, genome integrity, oxidative catabolism | |
| SIRT3 | Mitochondria | Deacetylation | LCAD, IDH2, GDH, ACS2, SOD2 | Fatty acid oxidation, TCA cycle, oxidative phosphorylation, oxidative stress | |
| SIRT4 | Mitochondria | ADP-ribosylation | GDH | TCA cycle, fatty acid oxidation | |
| SIRT5 | Mitochondria | Deacetylation, demalonylation, desuccinylation | CPS1 | Urea cycle | |
| SIRT6 | Nucleus | Deacetylation ADP-ribosylation | H3K9, H3K56, PARP1 | Genome stability, telomere silencing | |
| SIRT7 | Nucleolus | Unknown | Unknown | rDNA transcription | |
SIRT1 is the closest mammalian homolog of the yeast Sir2 protein in sequence and is the most studied mammalian sirtuin. SIRT1 predominantly localizes to the nucleus and acts as a deacetylase for histones H1K26 and H4K16 22, as well as many non-histone targets (Table 1). Deacetylation of p53 by SIRT1 upon DNA damage and oxidative stress results in reduced apoptosis 23,24. SIRT1 activates PGC1α through deacetylation and regulates mitochondria biogenesis and activities 25. Acetylation state of FOXO transcription factors, regulated by SIRT1, is thought to selectively direct them to certain targets, representing another level of regulation of metabolism and stress response 4. Upon inhibition of insulin signaling, SIRT1 is actively shuttled out of the nucleus into the cytoplasm. Recently, more evidence suggested SIRT1 is very profoundly involved in cancers and apoptosis, which is reviewed in greater detail below. Other newly identified novel functions of SIRT1 include neuroprotection against various neurodegenerative diseases 26,27, promoting liver function and regeneration 28, stem cell differentiation and cell fate determination 29–31, and delaying replicative senescence in primary fibroblasts 32.
SIRT2 exists in the cytoplasm, deacetylates tubulin, and regulates skeletal muscle differentiation 4. Recently, SIRT2 has been found to accumulate in neurons in the central nervous system in aging brains and its microtubule deacetylase activity is linked to the pathology of brain aging and neurodegenerative diseases 33. In the peripheral nervous system, however, SIRT2 appears important for nerve myelination and regeneration by deacetylating Par-3, a critical regulator of cell polarity and myelin assembly in Schwann cells 34. In adipocytes, SIRT2 regulates metabolism through deacetylating FOXO1 and PCG1α 4,35. When FOXO3 is deacetylated by SIRT1 or SIRT2, it is targeted for polyubiquitination and degradation 36. Tissues from human breast cancers and hepatocellular carcinoma exhibit reduced levels of SIRT2, which regulates the anaphase-promoting complex (APC) activity in these tissues by deacetylating CDH1 and CDC20 to preserve genome integrity and to antagonize tumorigenesis 37.
SIRT3, SIRT4, and SIRT5 localize primarily to mitochondria. SIRT3 is the major mitochondrial deacetylase, targeting numerous enzymes playing critical roles in maintaining metabolic homeostasis 38. In the fatty acid oxidation pathway, SIRT3 deacetylates the long-chain acyl CoA dehydrogenase (LCAD). Among enzymes involved in the TCA cycle, both isocitrate dehydrogenase (IDH2) and glutamate dehydrogenase (GDH) are deacetylation targets of SIRT3. Furthermore, SIRT3 deacetylates components in each step of the electron transport chain in oxidative phosphorylation 39. The important mitochondrial enzyme required for generating acetyl-coA, AceCS2 is also a deacetylation target of SIRT3 38. In addition to these metabolic enzymes, SIRT3 provides protection against oxidative stress by deacetylation and activation of SOD2, an important mitochondrial antioxidant enzyme 38. SIRT4 is primarily an ADP-ribosylase targeting GDH 4. More recently, SIRT4 has been shown to negatively regulate fatty acid oxidation in liver and muscle cells, an apparent opposing role to SIRT3 4. SIRT5 targets carbamoyl phosphate synthetase (CPS1) to remove malonyl or succinyl groups in a fashion very similar to deacetylation 4.
SIRT6 localizes in the nucleus and deacetylates histone H3K9 and H3K56 to maintain genome stability and telomere function 40. More recently, SIRT6 is also found to be an ADP-ribosylase for PARP1 under oxidative stress and stimulates PARP1 function to support DNA repair 41.
SIRT7 exists in nucleolus and activates RNA polymerase I transcription 4. Although several of its interaction partners have been identified, including RNA polI, rDNA transcription factor UBF, and chromatin remodeling complex WICH (consisting of WSTF and SNF2h) 42, SIRT7's catalytic activity and targets remain elusive.
Controversy 1: Sirtuins mediates effects of calorie restriction
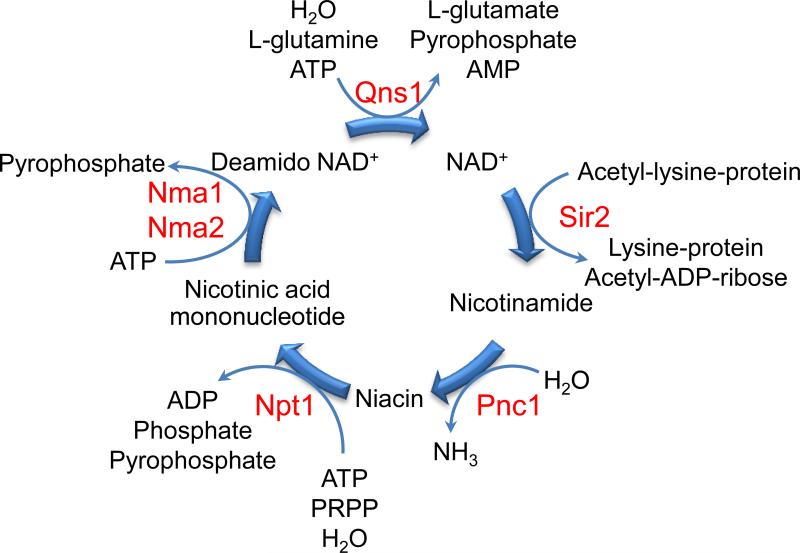
Calorie restriction (CR) is the most robust way to extend lifespan in most model organisms studied so far, from yeast to primates. Yet, its molecular mechanism remains elusive 43. After the discovery of sirtuins as a class of conserved aging regulators, genetic studies in simple organisms, such as yeast and fruit fly, suggested sirtuins mediates the effects of CR 44. Specifically, in yeast, CR increases NAD+/NADH ratio through elevated level of respiration. Since the Sir2 deacetylation reaction consumes NAD+, an increased NAD+/NADH ratio would favor the reaction. Meanwhile, CR enhances the salvage pathway of NAD+ synthesis by up-regulating nicotinamidase Pnc1, an enzyme that converts nicotinamide to niacin (Fig. 2). Since nicotinamide is a product of the NAD+-dependent deacetylation reaction and an inhibitor of sirtuins, activation of the salvage pathway under CR is thought to be another piece of important evidence supporting the activation of sirtuin under CR 44. Most interesting evidence came from genetic experiments, where deletion of SIR2 gene in yeast was reported to block the lifespan extension by CR. Similar results were also found in worms 45 and fruit flies 46. In mammals, however, direct evidence for sirtuin's function in CR in other model systems remains to be evaluated. Nevertheless, in several cases, CR associated with increased sirtuin levels or activities in various mouse tissues, suggesting that sirtuins are involved in CR 47,48.
Figure 2.
NAD+ salvage pathway recycles nicotinamide from the sirtuin catalyzed deacetylation reaction and regenerates NAD+. Activation of this pathway may play an important role in modulating sirtuin activities by removing the sirtuin inhibitor nicotinamide and generating the cofactor NAD+. Key yeast enzymes and metabolites in this pathway are shown.
However, sirtuin's critical role in CR in yeast, one of the best defined genetic systems, has been challenged. These results suggested that yeast Sir2 is not required for CR to function in yeast as deletion of SIR2 does not block the lifespan extension effect of CR when generation of extrachromosomal RNA circles (ERCs) was prevented through the FOB1 deletion 49. Moreover, CR still extended lifespan when SIR2 and two of its paralogs HST1 and HST2 had all been deleted under such a condition 50. Meanwhile, the TOR nutrient sensing kinase signaling pathway emerged as a conserved mechanism behind CR since disruption of TOR1 mimics and is epistatic to CR 51. Consistently, Sir2 is also not required for the lifespan extension by TOR1 deletion, a proposed CR mimetic. Others have also found that the effect Sir2 activation, monitored by gene silencing, upon CR is moderate or undetectable, much less than previous suggested 52. In worms, it is also controversial: one study found sir-2.1 required for CR mediated longevity 45; while others showed sir-2.1 to be dispensable for CR effects 53,54. These studies have casted serious doubt on the fundamentals of sirtuin's role in CR.
Many questions remain unanswered amongst the debates around this controversy. In yeast, it seems clear that Sir2 is not required for CR when ERCs are blocked. However, Sir2 activity is activated by the metabolic (NAD+/NADH ratio) changes associated with CR conditions. Since Sir2 promotes longevity as overexpression of SIR2 gene reproducibly extends lifespan in yeast, does activated Sir2 contribute to the longevity effect of CR? Interestingly, although sirtuin's role in CR has been challenged in yeast and worms, the evidence for Sir2 being critical for CR in flies seem plentiful 21. In mammals, although direct genetic tests remain difficult, accumulating evidence suggest that sirtuins are regulated in various tissues at both expression levels and metabolic states. The results from changes in sirtuin abundance and activity can be at multitudes of levels, given the broad substrates of SIRT1 and mitochondria-specific sirtuins 47. With the intimate regulatory relationship between sirtuins and various metabolic pathways regulated by AMPK, FOXO, and AceCS2, etc, it is an intriguing hypothesis that sirtuins mediate at least some of the effects of CR in mammals. However, the exact contributions of sirtuins to animal health, fitness, and potentially longevity under CR conditions remain to be explored.
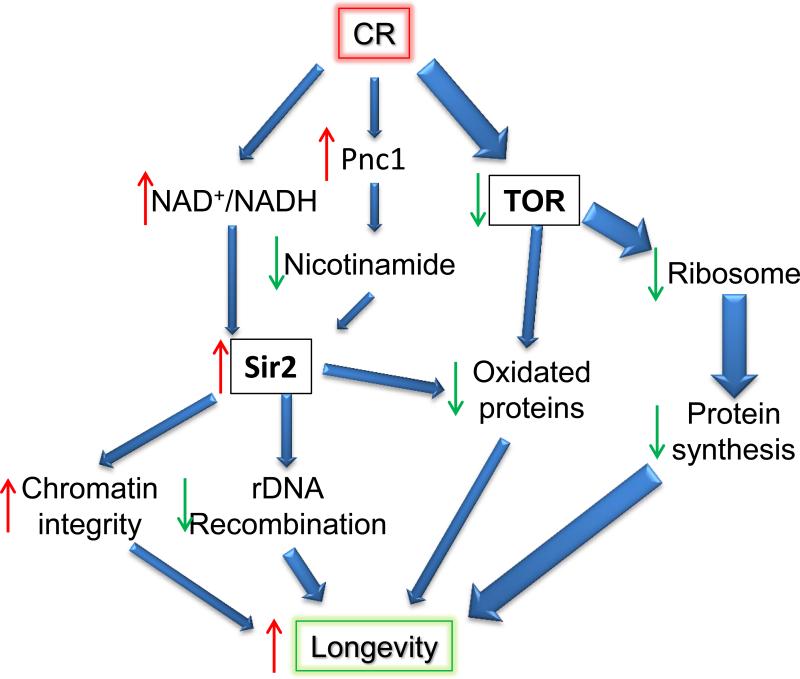
A model could at least partially reconcile the controversy about sirtuins’ role in CR. That is the activation of sirtuins, either through increase NAD+/NADH ration or increased expression level, is one of the many anti-aging mechanisms exploited by CR (Fig. 3). Since it is a pathway further downstream of nutrient sensing and is not a common pathway node shared by other CR mechanisms, disruption of it does not block the majority effect of CR. Although sirtuins are not the central controllers of the longevity effects of CR, accumulating evidence does suggest their contributions to the full benefits of CR 4,55.
Figure 3.
A proposed mechanism of both sirtuins and TOR regulated pathways are modulated by the availability of nutrition states. The red “up” and green “down” arrows indicate increased and decreased activity or abundance, respectively. The thickness of the blue arrows indicates the relative impact to the downstream targets.
Controversy 2: Resveratrol is a sirtuin activator
Soon after sirtuins emerged as a class of conserved aging regulators, efforts were made to identify chemical inhibitors and potential activators of these enzymes. It was especially intriguing to search for activators since overexpression of sirtuins was shown to have longevity benefits. A screen using an in vitro deacetylation assay featuring a fluorophore labeled acetylated p53 peptide identified a number of inhibitors and activator for mammalian SIRT1. Among them, a compound found most enriched in red wines, resveratrol, activated the reaction as much as 13 fold by increasing the substrate binding affinity 56, and was proposed to mimic the calorie restriction in vivo 57. Resveratrol extends lifespan for several model organisms, including yeast, worms, flies, and fish 58. In mice, although the lifespan of mice fed on regular diet was not affected by resveratrol, it showed significant longevity and overall health benefits for mice fed on a high fat diet 59.
However, activation of sirtuins by resveratrol and other similar activators was quickly disputed because the covalently attached fluorophore was required for the enhanced substrate binding and stimulated deacetylation by SIRT1 60–62 and the activation of sirtuins cannot be observed in vivo 62. The lifespan extension effect of resveratrol in worms and flies also seems dependent on strain backgrounds 63.
As the controversy unfolds, the interests in resveratrol did not subside, because many in vivo experiments continue to show its CR mimetic effects 64, as well as age related health benefits. These benefits include protection against cancer, cardiovascular diseases, insulin resistance, and cognitive problems 65. Meanwhile, many reports showed that resveratrol mimics the effects of SIRT1 overexpression in various mammalian cell cultures 64; many metabolic and signaling pathways activated by resveratrol also seemed to be dependent on the presence of sirtuins in various cell lines, tissues and mouse models 58. Most interestingly, several clinic trials have shown encouraging results for resveratrol: reduced oxidative stress and inflammation; improved cardiovascular function; enhanced cognitive function; inhibition of cancerous growth; decreased insulin resistance in diabetic patients; and improved metabolic profiles for obese and aged individuals 66.
Are sirtuins the critical targets of resveratrol for all these health benefits? The exact molecular mechanism remains to be explored. In some cases, the answer might be “yes”; in others, the effect is likely mediated by other pathways. After all, resveratrol can activate AMPK through an upstream kinase LKB1 in the absence of SIRT1 58. Direct inhibition of p300 activity can be another mechanism independent of sirtuins 67. For the situations where sirtuins do seem to mediate the health benefits, they may not be the direct targets of resveratrol, but rather being in the downstream pathways. To reconcile some apparent sirtuin-dependent resveratrol effects and the requirement of the artificial fluorophore for resveratrol stimulated sirtuin activation in biochemical assays, it is hypothesized that fluorophore labeled peptides are a closer mimic of the in vivo substrates of sirtuins because of their hydrophobicity 58. However, biochemical assays with the full length SIRT1 substrate AceCS1 didn't support this hypothesis 61. Obviously, the debate will continue until a satisfying molecular mechanism is identified.
Controversy 3: Overexpression of sirtuins extends lifespan
Extension of lifespan by overexpression of sirtuins was first reported for yeast Sir2 12. This overexpression was achieved by inserting another copy of the SIR2 gene in the genome. This SIR2 double copy strain showed a lifespan 30% greater than the wild-type control. Soon, similar effects were also shown in worms and flies.
In worms, the results came from a screen for long-lived mutants in strains containing partial genome duplications 68. The strain with a genome duplication carrying sir-2.1 extended lifespan by 10%; while two other strains with a duplication of the nearby regions excluding sir-2.1 shortened lifespan. Three independent sir-2.1 extrachromosomal transgenic lines showed lifespan extension by 20-50%. Lastly, three sir-2.1 integrated strains also showed similar extensions. It is further suggested that SIR-2.1 functions in the same DAF-2 and DAF-16 regulated insulin signaling pathway. Later, an integrated low-copy sir-2.1 transgenic strain also showed a lifespan extension of 26% 69.
In flies, three independent strains were constructed to express a GAL4 transcription activator driver and to carry the native dSir2 gene with a GAL4 binding site. They had an over 4 fold increase in dSir2 expression and showed a lifespan extension of up to 57% 46. This lifespan extension appeared to be dose-dependent, since a 10% dSir2 increase did not extend lifespan. Increase in lifespan was also observed when dSir2 was overexpressed in neurons, the type of cells normally having the highest dSir2 expression in adult flies.
Interestingly, the results for both worms and flies have been disputed by a study conducted among several labs in Europe and in the US 18. First, they examined one of the three worm strains carrying an integrated sir-2.1 transgene. Although it was long-lived compared to the wild-type, outcrossing 5 times to the wild-type strains abrogated the longevity effect. It was then found that a secondary mutation was responsible for the increase lifespan in the original sir-2.1 transgenic strain. Then, a similar experiment was repeated for the integrated low-copy strain. After outcrossing 6 times, the mutant strain again failed to show lifespan extension. Lastly, upon sir-2.1 RNAi knockdown, the original long-lived genome duplication strain still lived long, suggesting the longevity was not due to the increased sir-2.1 expression. In flies, outcross of the long-lived dSir2 overexpression strain was still long-lived compared to the wild-type control, but the GAL4 driver only strain (no dSir2 overexpression) also had a similar long lifespan. Therefore, the longevity effect of the strain did not appear to be conferred by dSir2 overexpression, but rather the GAL4 driver itself. An independently created strain with a higher dSir2 overexpression level did not extend lifespan.
The debate quickly heated up. The original authors of the worm work responded by admitting the existence of the secondary mutation in one of the original long-lived sir-2.1 transgenic strains, but maintained that their own outcrossing (6-8 times) resulted in strains still 10-15% longer-lived than the control strains 17. In addition, another research group also confirmed that the lifespan extension of the integrated low-copy sir-2.1 transgenic strain can be abrogated by RNAi knockdown of sir-2.1 expression 70.
Debates on the longevity effect of sirtuin overexpression in these two important model systems are likely to continue until all the discrepancies are resolved. Better communication and prompt exchange of strains and protocols may be helpful in eliminating some technical disparities that might have contributed to the apparent discrepancy in results. The overexpression dosage of sirtuins may be an important factor to determine lifespan; hence it is prudent to compare a series of strains with different expression levels of sirtuins in lifespan experiments. The neuronal-specific dSir2 overexpression strain should also be outcrossed and evaluated the same way.
While the sirtuin's lifespan extension effect is disputed, works in mice have shown promising progress. Overexpression of the SIRT6 transgene extended lifespan for male mice, but not female mice 71. The longevity effect appeared to be mediated by the insulin-like growth factor 1 (IGF1) signaling pathway, a mechanism consistent with findings in the previous worm studies. Yet, much more work remains to address whether activation of sirtuins can directly provide longevity benefits. Many new results in mice suggest a still promising future.
Controversy 4: Sirtuins are oncogenes or tumor suppressors
Not surprisingly, in mammals, many sirtuins targets are involved in cancer. SIRT1's pro-survival effects seem to promote oncogenesis and malignancy. It inactivates the p53 tumor suppressor through deacetylation and inhibits p53-dependent apoptosis upon DNA damage and oxidative stress 23,24. SIRT1 interacts with E2F1, a cell cycle regulator, and interferes with its apoptotic function during DNA damage response 72. DBC1 (Deleted in Breast Cancer 1), a tumor suppressor, binds to SIRT1 and inhibits its activity 73. In many types of cancers, SIRT1 is found to be overexpressed 74; knocking down SIRT1 in these cancer cells restores radiation induced apoptosis 75. SIRT2 is up-regulated in acute myeloid leukemia (AML), deacetylating and activating Akt 76. These observations and many others seem to support SIRT1 being an oncogene.
However, SIRT1 also clearly shows functions of a tumor suppressor in many other situations. Overexpression of SIRT1 in intestines reduces the incidence of colon cancer by deacetylation of oncogene β-catenin, preventing its localization to the nucleus 73. SIRT1 also functions in DNA damage repair as it relocates to sites of DNA damage and promotes genome integrity 77. Reduced SIRT1 expression levels associate with increase sarcoma and lymphoma in p53 heterozygous mice 78. SIRT2 targets the APC to regulate cell cycle. Mice with disrupted SIRT2 develop cancers 37. Interestingly, SIRT1 and SIRT2 levels are reduced in some types of cancers 73. SIRT6 maintains telomere and genome stability 79 and its overexpression induces apoptosis in cancer cells 80.
How can sirtuins function as both oncogenes and tumor suppressors? At first, the situation seems extremely confusing, and sirtuin's functions in cancer appear dependent on the type of tissue or cancer. However, it does seem that their tumor suppressing roles are consistent with their anti-aging functions in animals as well as various tissues. It is likely that certain types of cancers have evolved a mechanism to usurp the general pro-survival properties of sirtuins in order to circumvent apoptosis. It remains to be investigated how such a mechanism works and if the tumor suppressor functions can be restored in such a case.
Despite controversies, can sirtuins be therapeutic targets in future medicine?
The answer is most likely affirmative for most researchers studying sirtuins and their functions. This is because, despite so many controversies, mammalian sirtuins have been clearly shown as a class of critical factors regulating many cellular processes, playing important functions in diverse tissues and systems. Sirtuin functions have been described in the central/peripheral nerve system, cardiovascular system, immune system, liver, bone, skeletal muscles, stem cells, and tissue regeneration. They have also been associated with most major diseases, such as cardiovascular diseases, cancer, metabolic disorders, neurodegenerative diseases, arthritis, and osteoporosis, all of which are age-related. For instance, in several types of cancers, knocking down SIRT1 sensitizes cancer cells to radiation and chemotherapies 73,81. However, the complexity of activities of sirtuins, and their widespread roles and activities, increases the difficulties in determining how best to modulate them therapeutically.
Sirtuins are a class of epigenetic regulators that modulate the activity of their targets by removing covalently attached acetyl groups. Small molecule regulators targeting sirtuins would provide a robust, rapid, and yet reversible cellular response. Indeed, two HDAC inhibitor drugs have already been approved to treat a certain type of lymphoma, and many more are under clinical trials for several types of cancers.
It should be noted that current HDAC inhibitor drugs all target the class I and II HDACs, not sirtuins, the class III HDAC. Nevertheless, thanks to the interest in screening for small molecular modulators for sirtuins, many sirtuin-specific inhibitors have been discovered and characterized, several of which have been tested to treat cancer in mouse models 81. Although resveratrol's function as a sirtuin activator is under debate, the compound rising with the fame of sirtuins did show many health benefits in clinical trials 66.
Current controversies around sirtuins will eventually lead us to a much better understanding for this class of enzymes, which should provide a clearer perspective for their use as targets in future medicine.
Acknowledgements
I thank Jean Dorsey and Rocco Perry for proofreading the manuscript. The work is supported by NIH grant 5K99AG037646.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The author has no conflict of interest to declare.
References
- 1.Guarente L. Diverse and dynamic functions of the Sir silencing complex. Nature genetics. 1999;23:281–5. doi: 10.1038/15458. [DOI] [PubMed] [Google Scholar]
- 2.Brachmann CB, et al. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes & Development. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 3.Denu JM. The Sir 2 family of protein deacetylases. Current opinion in chemical biology. 2005;9:431–40. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nature Reviews Molecular Cell Biology. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 6.Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Molecular cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guillemette B, et al. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS genetics. 2011;7:e1001354. doi: 10.1371/journal.pgen.1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cubizolles F, Martino F, Perrod S, Gasser SM. A homotrimer-heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Molecular cell. 2006;21:825–36. doi: 10.1016/j.molcel.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Perrod S, et al. A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. The EMBO journal. 2001;20:197–209. doi: 10.1093/emboj/20.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy B, Jr NA, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- 11.Sinclair DA, Guarente L. Extrachromosomal rDNA circles-a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 12.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Development. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–7. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wirth M, et al. HIS-24 linker histone and SIR-2.1 deacetylase induce H3K27me3 in the Caenorhabditis elegans germ line. Molecular and cellular biology. 2009;29:3700–9. doi: 10.1128/MCB.00018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berdichevsky A, Viswanathan M, Horvitz HR, Guarente LC. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–77. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 16.Pan KZ, et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. doi:10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1–2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- 18.Burnett C, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annual review of biochemistry. 2004;73:417–35. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 20.Åström S, Cline T, Rine J. The Drosophila melanogaster sir2+ gene is nonessential and has only minor effects on position-effect variegation. Genetics. 2003;163:931. doi: 10.1093/genetics/163.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frankel S, Ziafazeli T, Rogina B. dSir2 and longevity in Drosophila. Experimental gerontology. 2011;46:391–6. doi: 10.1016/j.exger.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 23.Vaziri H, et al. hSIR2 SIRT1 functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 24.Luo J. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 25.Sepharose PG, et al. Nutrient control of glucose homeostasis through a complex of PGC-1 a and SIRT1. Nature. 2005;434:3–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava S, Haigis MC. Role of sirtuins and calorie restriction in neuroprotection: implications in Alzheimer's and Parkinson's diseases. Current pharmaceutical design. 2011;17:3418–33. doi: 10.2174/138161211798072526. [DOI] [PubMed] [Google Scholar]
- 27.Donmez G, et al. SIRT1 Protects against α-Synuclein Aggregation by Activating Molecular Chaperones. The Journal of Neuroscience. 2012;32:124–132. doi: 10.1523/JNEUROSCI.3442-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Jin J, Iakova P, Jiang Y, Medrano EE, Timchenko N. a The reduction of SIRT1 in livers of old mice leads to impaired body homeostasis and to inhibition of liver proliferation. Hepatology (Baltimore, Md.) 2011;54:989–98. doi: 10.1002/hep.24471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Wang J, Chen G, Fan D, Deng M. Inhibition of Sirt1 promotes neural progenitors toward motoneuron differentiation from human embryonic stem cells. Biochemical and biophysical research communications. 2011;404:610–4. doi: 10.1016/j.bbrc.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 30.Han M-K, et al. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell stem cell. 2008;2:241–51. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chae H-D, Broxmeyer HE. SIRT1 deficiency downregulates PTEN/JNK/FOXO1 pathway to block reactive oxygen species-induced apoptosis in mouse embryonic stem cells. Stem cells and development. 2011;20:1277–85. doi: 10.1089/scd.2010.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita S, et al. SIRT1 prevents replicative senescence of normal human umbilical cord fibroblast through potentiating the transcription of human telomerase reverse transcriptase gene. Biochemical and biophysical research communications. 2012;417:630–4. doi: 10.1016/j.bbrc.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 33.Maxwell MM, et al. The Sirtuin 2 microtubule deacetylase is an abundant neuronal protein that accumulates in the aging CNS. Human molecular genetics. 2011;20:3986–96. doi: 10.1093/hmg/ddr326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beirowski B, et al. Sir-two-homolog 2 (Sirt2) modulates peripheral myelination through polarity protein Par-3/atypical protein kinase C (aPKC) signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E952–61. doi: 10.1073/pnas.1104969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishnan J, et al. Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes & development. 2012;26:259–70. doi: 10.1101/gad.180406.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F, et al. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene. 2012;31:1546–57. doi: 10.1038/onc.2011.347. [DOI] [PubMed] [Google Scholar]
- 37.Kim H-S, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer cell. 2011;20:487–99. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell EL, Guarente L. The SirT3 divining rod points to oxidative stress. Molecular cell. 2011;42:561–8. doi: 10.1016/j.molcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jing E, et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14608–13. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tennen RRI, Chua KF. Chromatin regulation and genome maintenance by mammalian SIRT6. Trends in biochemical sciences. 2011;36:39–46. doi: 10.1016/j.tibs.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao Z, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science (New York, N.Y.) 2011;332:1443–6. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai Y-C, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional Proteomics Establishes the Interaction of SIRT7 with Chromatin Remodeling Complexes and Expands Its Role in Regulation of RNA Polymerase I Transcription. Molecular & cellular proteomics: MCP. 2012;11:60–76. doi: 10.1074/mcp.A111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annual review of biochemistry. 2008;77:727–54. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 44.Guarente L, Picard F. Calorie restriction--the SIR2 connection. Cell. 2005;120:473–82. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, et al. C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mechanisms of ageing and development. 2006;127:741–7. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. PNAS. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu X, Brown KV, Moran Y, Chen D. Sirtuin regulation in calorie restriction. Biochimica et biophysica acta. 2010;1804:1576–83. doi: 10.1016/j.bbapap.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 48.Guarente L. Sirtuins, Aging, and Metabolism. Cold Spring Harbor symposia on quantitative biology. 2011 doi: 10.1101/sqb.2011.76.010629. doi:10.1101/sqb.2011.76.010629. [DOI] [PubMed] [Google Scholar]
- 49.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS biology. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaeberlein M, et al. Comment on “HST2 Mediates SIR2-Independent Life-Span Extension by Calorie Restriction. Science. 2006;312:5–6. doi: 10.1126/science.1124608. [DOI] [PubMed] [Google Scholar]
- 51.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science (New York, N.Y.) 2005;310:1193–6. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 52.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–9. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee GD, et al. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging cell. 2006;5:515–24. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hansen M, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 55.Guarente L. Sirtuins and calorie restriction. Nature reviews. Molecular cell biology. 2012;13:4288. doi: 10.1038/nrm3308. [DOI] [PubMed] [Google Scholar]
- 56.Howitz KKT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 57.Wood JG, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–9. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 58.Baur J. a Resveratrol, sirtuins, and the promise of a DR mimetic. Mechanisms of ageing and development. 2010;131:261–9. doi: 10.1016/j.mad.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baur J. a, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. The Journal of biological chemistry. 2005;280:17187–95. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 61.Pacholec M, et al. SRT1720 , SRT2183 , SRT1460 , and Resveratrol Are Not Direct Activators of SIRT1. Journal of Biological Chemistry. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaeberlein M, et al. Substrate-specific activation of sirtuins by resveratrol. The Journal of biological chemistry. 2005;280:17038–45. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 63.Bass TM, Weinkove D, Houthoofd K, Gems D, Partridge L. Effects of resveratrol on lifespan in Drosophila melanogaster and Caenorhabditis elegans. Mechanisms of ageing and development. 2007;128:546–52. doi: 10.1016/j.mad.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 64.da Cunha FM, Demasi M, Kowaltowski AJ. Aging and calorie restriction modulate yeast redox state, oxidized protein removal, and the ubiquitin-proteasome system. Free radical biology & medicine. 2011;51:664–70. doi: 10.1016/j.freeradbiomed.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 65.Baur J. a. Biochemical effects of SIRT1 activators. Biochimica et biophysica acta. 2010;1804:1626–34. doi: 10.1016/j.bbapap.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Timmer S, Auwerx J, Schrauwen P. The journey of resveratrol from yeast to human. Aging. 2012;4:146–58. doi: 10.18632/aging.100445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huber JL, McBurney MW, Distefano PS, McDonagh T. SIRT1-independent mechanisms of the putative sirtuin enzyme activators SRT1720 and SRT2183. Future medicinal chemistry. 2010;2:1751–9. doi: 10.4155/fmc.10.257. [DOI] [PubMed] [Google Scholar]
- 68.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 69.Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Developmental cell. 2005;9:605–15. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 70.Rizki G, et al. The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS genetics. 2011;7:e1002235. doi: 10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kanfi Y, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;05:3–6. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 72.Rhind N. DNA replication timing: random thoughts about origin firing. Nature cell biology. 2006;8:1313–1316. doi: 10.1038/ncb1206-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang BL. Sirt1's systemic protective roles and its promise as a target in antiaging medicine. Translational research: the journal of laboratory and clinical medicine. 2011;157:276–84. doi: 10.1016/j.trsl.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 74.Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer research. 2009;69:1702–5. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 75.Chang C-J, et al. Enhanced radiosensitivity and radiation-induced apoptosis in glioma CD133-positive cells by knockdown of SirT1 expression. Biochemical and biophysical research communications. 2009;380:236–42. doi: 10.1016/j.bbrc.2009.01.040. [DOI] [PubMed] [Google Scholar]
- 76.Dan L, et al. The role of sirtuin 2 activation by nicotinamide phosphoribosyltransferase in the aberrant proliferation and survival of myeloid leukemia cells. Haematologica. 2012;97:551–9. doi: 10.3324/haematol.2011.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–18. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang R-H, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer cell. 2008;14:312–23. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 80.Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell cycle (Georgetown, Tex.) 2011;10:3153–8. doi: 10.4161/cc.10.18.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carafa V, Nebbioso A, Altucci L. Sirtuins and disease: the road ahead. Frontiers in pharmacology. 2012;3:4. doi: 10.3389/fphar.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]