SUMMARY
Virus infection is sensed in the cytoplasm by retinoic acid-inducible gene I (RIG-I, also known as DDX58), which requires RNA and polyubiquitin binding to induce type I interferon (IFN), and activate cellular innate immunity. We show that the human IFN-inducible oligoadenylate synthetases-like (OASL) protein had antiviral activity and mediated RIG-I activation by mimicking polyubiquitin. Loss of OASL expression reduced RIG-I signaling and enhanced virus replication in human cells. Conversely, OASL expression suppressed replication of a number of viruses in a RIG-I-dependent manner and enhanced RIG-I-mediated IFN induction. OASL interacted and colocalized with RIG-I, and through its C-terminal ubiquitin-like domain specifically enhanced RIG-I signaling. Bone marrow derived macrophages from mice deficient for Oasl2 showed that among the two mouse orthologs of human OASL; Oasl2 is functionally similar to human OASL. Our findings show a mechanism by which human OASL contributes to host antiviral responses by enhancing RIG-I activation.
INTRODUCTION
Initiation of cellular innate immunity to virus infection is reliant upon sensing viral nucleic acid in the cytoplasm by the retinoic acid-inducible gene I (RIG-I)-like receptors (RLR) (Yoneyama et al., 2004). Upon binding to viral RNA, RIG-I or melanoma differentiation-associated gene 5 (MDA5) engage with their common adaptor mitochondrial antiviral-signaling protein (MAVS) (Loo and Gale, 2011)) on the mitochondrial surface, and initiate a signal transduction to transcriptionally induce a series of antiviral genes including type I interferons (IFN) (Loo and Gale, 2011; Yan and Chen, 2012). RLR signaling is tightly regulated by several mechanisms, such as protein expression and post-translational modifications, to prevent aberrant activation and IFN production that can be toxic to the cell. Ubiquitination or deubiquitination of critical RLR pathway components appears to be one such mode of regulation (Oshiumi et al., 2012). Although a number of cellular proteins associated with mitochondrial function have been found to be important for the successful signal transduction through this pathway (Moore and Ting, 2008), host cell factors that function as positive regulators of RIG-I signaling have been less characterized.
Number of structural studies (Civril et al., 2011; Jiang et al., 2011; Kowalinski et al., 2011; Luo et al., 2011) have indicated a two-step mechanism for RIG-I activation, involving sequential binding of 5’-triphosphate containing RNA and K63-linked polyubiquitin (pUb) (Zeng et al., 2010). The ubiquitin ligase TRIM25 has been described to be primarily responsible for the second step of RIG-I activation by ubiquitination or synthesis of short K63-linked pUb chains (Gack et al., 2007; Zeng et al., 2010). Another ubiquitin ligase RNF135 (also known as Riplet) is similarly involved in the positive regulation of RIG-I signaling, while RNF125 and the deubiquitinase CYLD (Oshiumi et al., 2012) are known to negatively regulate RIG-I signaling. In contrast, activation of MDA5 involves the formation of ATP sensitive MDA5 filaments along dsRNAs. As the CARDs (Caspase activation and recruitment domains) of MDA5 are accessible and aligned in these filaments, it is believed to lead to MAVS aggregation along these filaments and TRIM25 independent activation of MAVS (Wu et al., 2013)
Type I IFNs exert their pleiotropic effects through the induction of a variety of IFN stimulated genes (ISGs) (Sadler and Williams, 2008; Sarkar and Sen, 2004; Yan and Chen, 2012). Although the antiviral activities for a comprehensive list of ISGs have been recently described (Schoggins et al., 2011), the mechanism of action for the majority of these proteins remains largely unknown. Oligoadenylate synthetases (OAS) are a family of ISGs characterized by their ability to synthesize 2’-5’ oligoadenylates, which induce RNA degradation by activating RNaseL (Kristiansen et al., 2011). However, the recent identification of the cytoplasmic DNA sensor cyclic GMP-AMP synthase (cGAS), which is another member of OAS family, shows potential diverse functions of these proteins (Sun et al., 2013). Human OASL is related to the OAS family by its N-terminal OAS-like domain, but lacks 2’-5’ oligoadenylate synthetase activity because of characteristic changes in the active site. Furthermore, OASL contains two tandem ubiquitin-like domains (UBL) in the C-terminus, which are absent in other OAS proteins (Hartmann et al., 1998; Rebouillat et al., 1998). OASL is rapidly induced by virus infection via interferon regulatory factor 3 (IRF3) as well as by IFN signaling (Melchjorsen et al., 2009; Sarkar and Sen, 2004) and has been shown to have antiviral activities, which requires the UBL domain (Marques et al., 2008; Schoggins et al., 2011). However, in the absence of the catalytic activity, the mechanism of human OASL antiviral activity has remained elusive.
Here we describe a role for human OASL in enhancing RIG-I-mediated signaling through its UBL, and define a mechanism by which OASL induces viral resistance. Genetic targeting of OASL in human cells, as well as in mouse macrophages derived from Oasl2−/− mice reduced RIG-I activity, resulting in enhanced virus replication. We propose a model whereby OASL, induced after virus infection directly binds to RIG-I and mimic pUb in order to sensitize its activation by viral RNA, thus enhancing antiviral signaling.
RESULTS
Unlike mouse Oasl1, human OASL does not bind IRF7 mRNA and instead exhibits antiviral activity
The human OASL was discovered as a member of the OAS family genes from the EST database (Hartmann et al., 1998; Rebouillat et al., 1998). Subsequently, two orthologs were identified in mouse chromosome 5 and termed as Oasl1 and Oasl2 with respectively 70 and 48% amino acid sequence identity with human OASL (Eskildsen et al., 2003). A pseudogene, named ψhOASL2 has been found adjacent to human OASL with partial homology to the mouse Oasl2 gene, but contains only the first two exons and lacks protein-coding ability (Suppl. Fig. S1A)(Eskildsen et al., 2003).
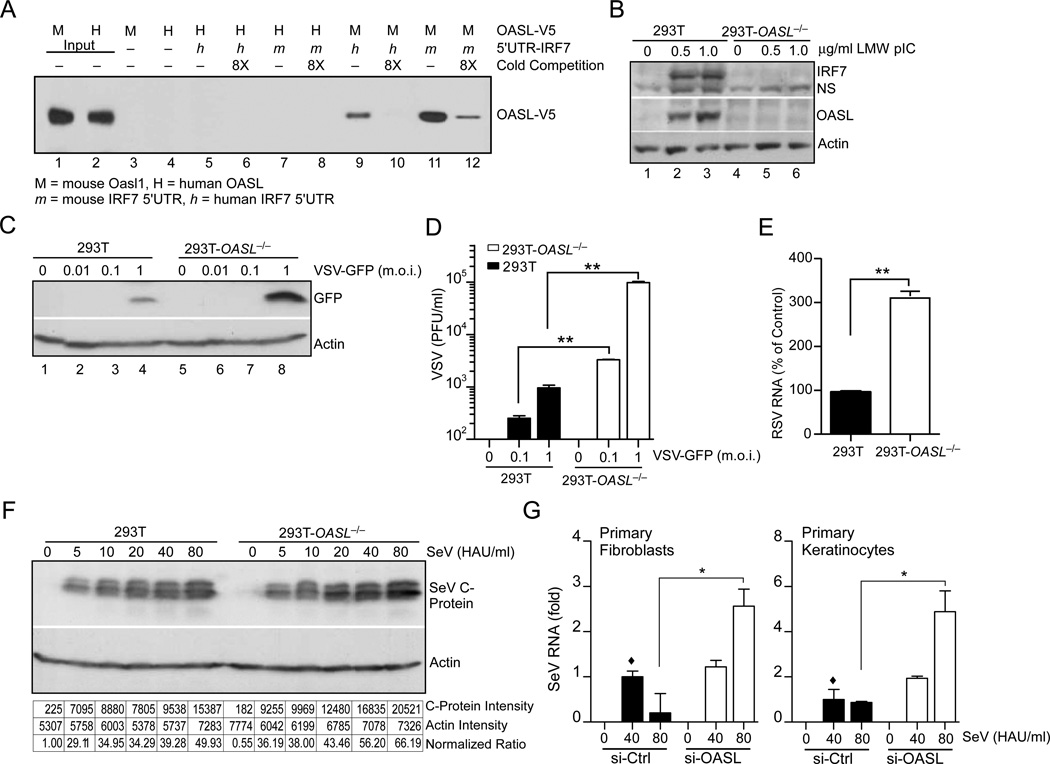
Human OASL protein has been reported to provide antiviral activity against RNA viruses (Marques et al., 2008; Schoggins et al., 2011). However, the mouse ortholog of human OASL, Oasl1, has been recently shown to inhibit IFN induction by binding to the 5’ UTR of IRF7 and inhibiting its translation. Consequently, targeted deletion of Oasl1 led to enhanced IFN induction and diminished viral replication in vivo (Lee et al., 2013). To determine whether human OASL is capable of binding to the IRF7 5’ UTR, we used a RNA pull-down assay (Fig. 1A). 3'-biotin labeled RNA corresponding to the 5'-UTR of human or mouse IRF7 was incubated with purified V5-tagged human or mouse OASL proteins in various combinations, the complexes were precipitated with Streptavidin-agarose beads, and analyzed by immunoblotting with V5 antibody. As observed before, mouse Oasl1 strongly bound to the 5’UTR of mouse IRF7 (Fig. 1A, Lane 11), but less strongly to the 5’UTR of human IRF7 (Fig. 1A, Lane 9). However, we did not detect appreciable binding of human OASL to either human or mouse IRF7 5’UTR (Fig. 1A, Lane 5–8). The binding specificity of each protein was demonstrated by competition with eight fold molar excess of corresponding unlabelled RNA. These results indicate that unlike mouse, human OASL does not bind to the IRF7 5’UTR to inhibit its translation and may exert unique biological activities.
Fig. 1. Unlike mouse Oasl1, human OASL does not bind IRF7 mRNA, and its loss enhances replication of a number of RNA viruses.
(A) Comparison between human and mouse IRF7 5’-UTR binding to human OASL or mouse Oasl1 proteins. Biotin labeled IRF7 5’-UTR RNA were incubated with the partially purified V5 tagged OASL or Oasl proteins and pulled down by streptavidin beads. Following extensive wash, bead-bound proteins were analyzed by immunoblotting (IB) using V5 antibody. (B) Reduced IRF7 induction in 293T-OASL−/− cells. 293T-OASL−/− and control 293T cells were transfected with LMW for 24 h, and analyzed by IB. NS, Non-specific. (C–D) VSV replication in 293T-OASL−/− cells detected by IB using anti-GFP antibody (C) or by plaque assay (D) 24 h post infection. (E) RSV replication in 293T-OASL−/− cells. Cells were infected with RSV at 3 m.o.i. for 18 h followed by total RNA extraction and detection by qRT-PCR. (F) SeV replication in 293T-OASL−/− cells detected by IB using SeV C protein antibody 24 h post infection. Densitometric analysis of the immunoblot is shown below. (G) OASL silencing results in higher SeV replication in primary human cells. Primary human foreskin fibroblasts or keratinocytes were transfected using lipfectamine RNAi MAX with either 50 nM OASL siRNA or control siRNA (ON-TARGETplus, SMARTpool, Thermo Scientific) for 48 h. The cells were then infected with SeV at indicated doses for another 24 h followed by detection of SeV specific RNA by qRT-PCR.
To further examine the antiviral function of human OASL, we generated OASL-deficient 293T cells (293T-OASL−/−) through TALEN-mediated genome editing (Schmid-Burgk et al., 2013). In contrast to mouse Oasl1−/− cells, which exhibited enhanced IRF7 induction, human OASL−/− cells showed a severe reduction in IRF7 induction upon activation of RLR signaling by low molecular weight p(I):p(C) (LMW) transfection (Fig. 1B). Further, human 293T-OASL−/− cells showed enhanced viral replication when infected with various RNA viruses including vesicular stomatitis virus (VSV), respiratory syncytial virus (RSV), and Sendai virus (SeV) (Fig. 1C–F). We validated the antiviral role of OASL by silencing OASL expression with siRNA in primary human fibroblasts and keratinocytes by siRNA and observed enhanced SeV replication (Fig. 1G). Taken together, these results indicated unique antiviral functions for human OASL compared to the mouse Oasl1, which was found to enhance viral replication.
OASL expression leads to resistance to infection by a number of DNA and RNA viruses
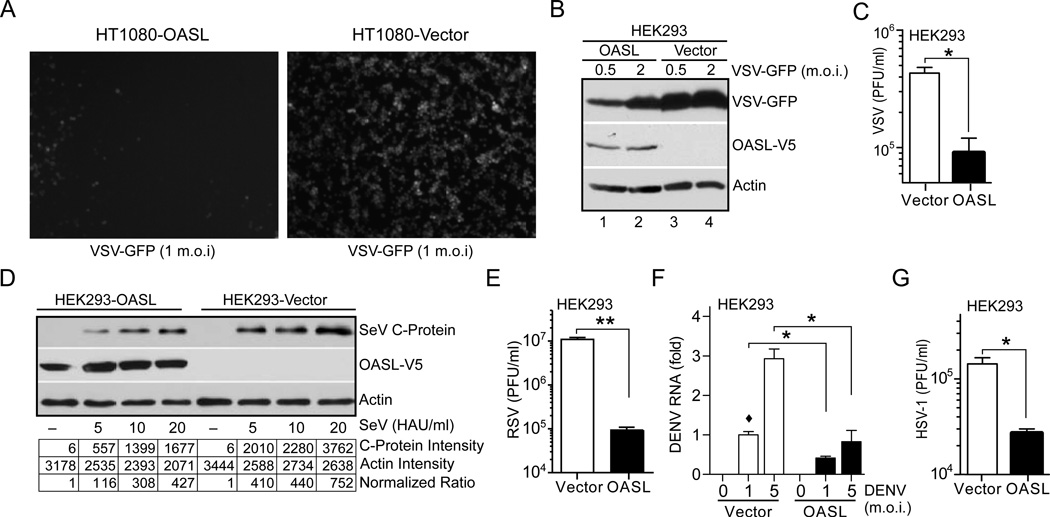
As a typical ISG, OASL protein is not expressed in naïve cells. To further clarify these apparent contradictory functions of human OASL and mouse Oasl1 genes, we used stable expression of V5-tagged human OASL in various cell lines and examined their antiviral activity against a broad range of viruses. The physiologically relevant amount of ectopic OASL expression was confirmed by comparing the amounts of exogenous OASL expression with that of endogenous OASL induction after IFN-treatment (Suppl. Fig. S1B). OASL-expressing HT1080 cells infected with VSV exhibited lower levels of viral replication (as measured by GFP fluorescence) compared to HT1080-Vector cells (Fig. 2A). Similarly, HEK293-OASL cells showed reduced VSV replication by immunoblotting (Fig. 2B) and plaque assay (Fig. 2C). SeV infection of these cells showed reduced SeV RNA (Suppl. Fig. S1C), and C protein (Fig. 2D). Markedly reduced virus replication in HEK293-OASL cells was observed with RSV (Fig. 2E), and dengue virus type 2 (DENV) (Fig. 2F) indicating antiviral activity of human OASL. We examined the antiviral activity of OASL against the herpes simplex virus-1 (HSV-1), and observed reduced virus production in HEK293-OASL cells (Fig. 2G). However, OASL did not provide protection against encephalomyocarditis virus (EMCV)(Suppl. Fig. S1D). Furthermore, siRNA-mediated silencing of OASL did not enhance EMCV replication in primary human keratinocytes (Suppl. Fig. S1E) or fibroblasts (Suppl. Fig. S1F). These results show that unlike mouse Oasl1, human OASL provides antiviral activity against specific RNA and DNA viruses.
Fig. 2. OASL expression provides cellular antiviral activity.
(A) OASL expression inhibits VSV infection. Cells were infected with VSV-GFP at 1 m.o.i. GFP florescence was observed under florescence microscope 8 h post-infection. Representative micrograph from at least three separate experiments is shown. (B–C) Expression of OASL reduces VSV replication. Cells were infected with VSV-GFP at the indicated m.o.i for 8 h followed by IB using GFP antibody (B). Supernatants from similarly infected cells (5 m.o.i) were used for plaque assay on BHK21 cells (C). (D) OASL expression inhibits SeV infection. SeV infected (24 h) cells were analyzed by IB using antibody against SeV C protein. Densitometric analysis of the protein bands are shown below. (E–G) OASL expression inhibits RSV, DENV and HSV-1 replication. Cells were infected with RSV (3 m.o.i) for 48 h, followed by plaque assay on Hep2 cells (E). Cells were infected with type 2 dengue virus (DENV) at indicated doses for 48 h followed by qRT-PCR for DENV specific RNA (F). Cells were infected with HSV-1 at 5 m.o.i. for 24 h followed by plaque assay on Vero cells (G).
Antiviral activity of OASL is reduced in the absence of RIG-I
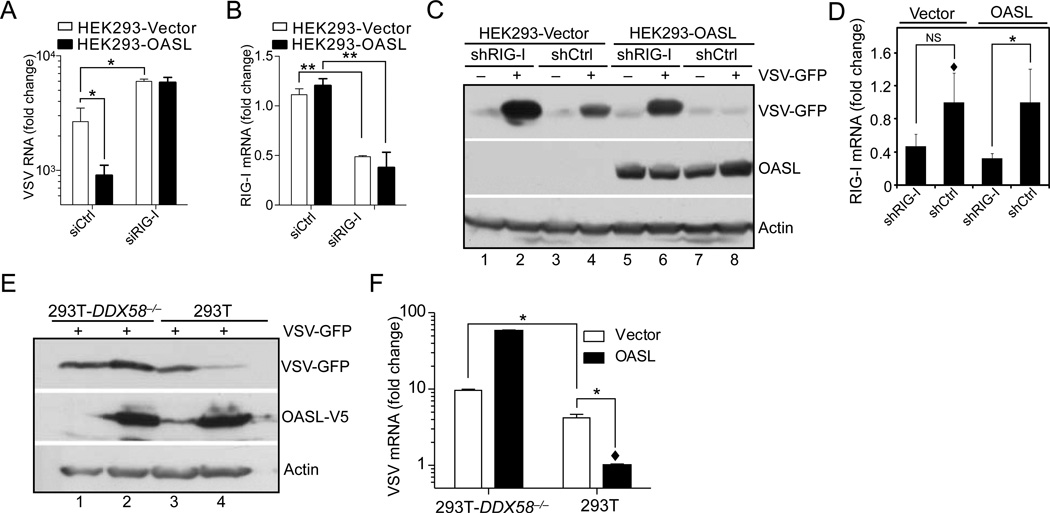
RIG-I is the primary sensor of viral RNA for VSV, SeV and RSV infection (Loo and Gale, 2011). In conjunction with RNA polymerase III, it also participates in sensing HSV-1 infection in HEK293 cells (Ablasser et al., 2009; Chiu et al., 2009). In contrast, MDA5 has been implicated in the sensing of picornaviruses such as EMCV (Kato et al., 2006; Loo and Gale, 2011). Since, OASL showed antiviral activity against VSV, SeV, RSV and HSV-1, but not against EMCV, we examined the dependence of this antiviral activity on RIG-I by three approaches: 1) transient silencing of RIG-I in HEK293 vector or OASL stable cells by siRNA; 2) establishment of an OASL expressing HEK293 stable cell line where RIG-I has been partially silenced by shRNA expression; and 3) expression of OASL in RIG-I-deficient 293T cells (293T-DDX58−/−). As expected, targeting of RIG-I increased VSV replication (assessed by quantitation of VSV RNA by qRT-PCR (Fig. 3A), immunoblotting with anti-GFP (Fig. 3C, lane 2 and 4)), while OASL expression reduced VSV replication (Fig. 3A, and 3C, lane 4 and 8). However, in the absence of RIG-I, there was no antiviral activity of OASL (Fig. 3A and 3C, lane 2 and 6). The steady state levels of RIG-I mRNA are shown in Fig. 3B and 3D, demonstrating more than 50% reduction of RIG-I mRNA with siRNA or shRNA targeted to RIG-I. In parallel, we expressed OASL in 293T-DDX58−/− and 293T cells. OASL expressing 293T cells inhibited VSV replication (Fig. 3E, lane 3 and 4), while OASL expressing 293T-DDX58−/− cells did not (Fig. 2E, lane 1 and 2, Fig. 3F). Since, RIG-I is the primary sensor for VSV, IFN and ISG induction by VSV infection is mostly absent in the RIG-I null cells. However, to exclude the possible role of any residual ISG induced in 293T-DDX58−/− cells via IFN receptor (IFNAR) signaling, we silenced IFNAR1 expression in these cells. As shown in the Suppl. Fig. S1G, despite functional reduction of IFN signaling indicated by the reduced ISG60 induction (Suppl. Fig S1G, lane 5, 6, 7 and 8), VSV replication in 293T-DDX58−/− cells remained unaffected by OASL either in the presence or absence of IFNAR signaling (Suppl. Fig S1G, lane 1, 2, 3 and 4). Results from the above experiments indicate that OASL-mediated antiviral activity against VSV is dependent on RIG-I.
Fig. 3. OASL antiviral activity is dependent on RIG-I.
(A–B) Loss of RIG-I expression reduces OASL antiviral activity against VSV. Cells were transfected with 50 nM RIG-I or control siRNA (Dharmacon) for 48 h, followed by VSV infection (1 m.o.i., 24 h). Total RNA from the infected cells were extracted and analyzed for VSV replication (A) and RIG-I mRNA (B) by qRT-PCR. (C) Indicated cells were transfected with psiRNA-Ctrl or psiRNA-hRIG-I plasmids (Invivogene) followed by Zeocin selection. VSV infected (1 m.o.i) cells were immunobloted with indicated antibodies. (D) Levels of RIG-I silencing were estimated by qRT-PCR. (E–F) Expression of OASL in RIG-I null cells does not protect them from VSV. OASL expressing 293T-DDX58−/− and control cells were infected with 1 m.o.i VSV for 24 h followed by IB with indicated antibodies (E), and qRT-PCR to detect VSV replication (F).
Expression of OASL enhances sensitivity of RIG-I signaling
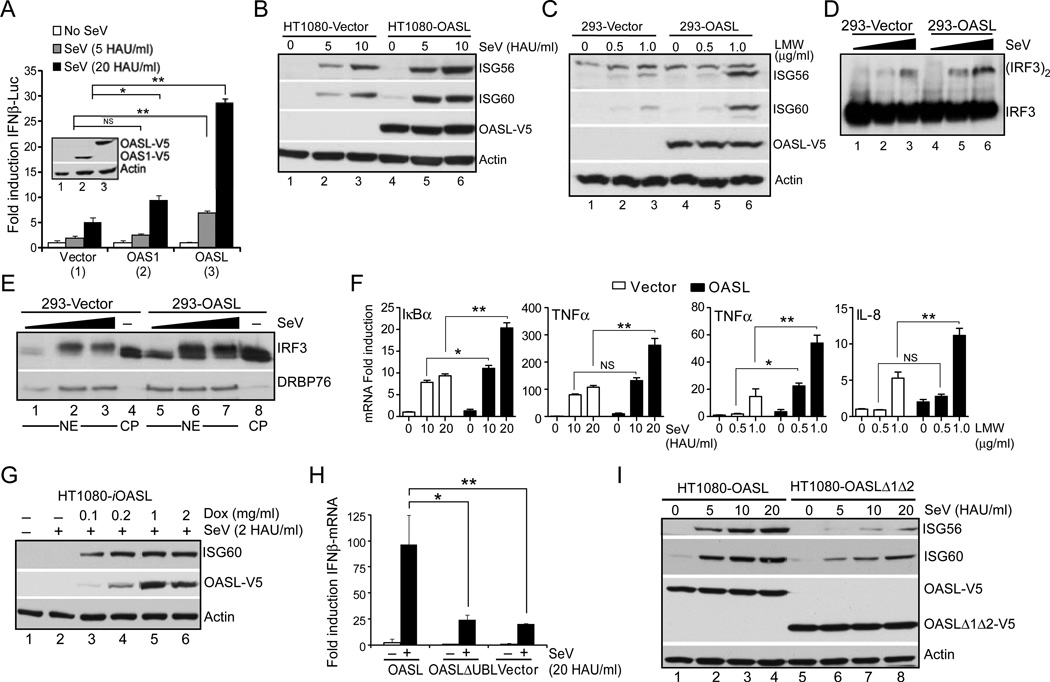
RIG-I initiates downstream signaling through its adaptor MAVS, and induce transcription of type I IFNs and ISGs. Since, OASL antiviral activity was dependent on RIG-I, in the next series of experiments we tested whether OASL modulated gene induction via the RIG-I pathway. OASL led to a substantial enhancement of IFNβ and ISG56 promoter activity at various doses of SeV, whereas another OAS family member, OAS1, did not (Fig. 4A and Suppl. Fig. S2A). Similarly, OASL-expressing HEK293 or HT1080 cells demonstrated higher levels of endogenous ISG56, ISG60 and IFN induction following SeV infection, or RIG-I specific LMW transfection, indicating enhanced RIG-I pathway activation in the presence of OASL (Fig. 4B, 4C Suppl. Fig. S2B, S2C). Early events of RIG-I signaling, such as IRF3 dimerization (Fig. 4D) and nuclear translocation (Fig. 4E) were also enhanced in presence of OASL. Induction of RIG-I-mediated NF-κB-regulated genes was also enhanced in OASL expressing cells (Fig. 4F). Presence of OASL did not alter the kinetics of RIG-I-mediated ISG induction, but the magnitude was substantially enhanced (Suppl. Fig. S2D). To further understand the nature of this enhancement in RIG-I signaling by OASL, we created HT1080 cell line with doxycycline inducible OASL expression (HT1080-iOASL) and infected them with sub-threshold doses of SeV (2 HAU/ml, 8 h). As shown in Fig. 4G, there was no detectable ISG60 induction in response to this sub-threshold SeV infection in cells (Fig. 4G, lane 2). However, upon doxycycline treatment, ISG60 was upregulated in an OASL expression-dependent manner. Taken together, these results indicate that OASL expression enhances viral RNA-induced RIG-I signaling and that the increases in IFN and ISG induction in presence of OASL are due to enhanced sensitivity of the RIG-I pathway, not due to changes in the kinetics of induction.
Fig. 4. OASL enhances RIG-I signaling.
(A) OASL expression enhances IFNβ-reporter activity. HEK293 cells were co-transfected either with control pcDNA3 (1), pcDNA3-OAS1 (2) or pcDNA3-OASL (3) in combination with IFNβ-luciferase reporter and β-actin Renilla luciferase reporter for 24 h. One part of the transfected cells was used for IB to detect protein expression (inset) and the other part was stimulated with SeV for 16 h and luciferase activities were measured. (B–C) OASL expression enhances RIG-I signaling. Cells were infected with SeV for 16 h followed by IB using ISG56, ISG60, V5 (OASL) and actin antibodies (B). Cells were transfected with LMW for 24 h followed by IB using indicated antibodies (C). (D–E) IRF3 dimerization and nuclear localization is enhanced in OASL expressing cells. Indicated cells were stimulated with SeV for 12h. Cell lysates were analyzed in native-PAGE followed by IB with IRF3 antibody (D). Nuclear extracts (NE) from similarly stimulated cells were analyzed along with control uninfected cytoplasm (CP) by IB for IRF3 and nuclear marker DRBP76 respectively (E). (F) NF-κB-dependent gene inductions are also enhanced in OASL expressing cells. Indicated cells were either transfected with LMW or infected with SeV for 24 h followed by qRT-PCR analysis using indicated primers to detect NF-κB-controlled gene induction. (G) Dose-dependent enhancement of RIG-I-signaling sensitivity by OASL. HT1080-iOASL cells were stimulated with Doxycycline at indicated concentrations for 48h followed by infection with SeV (2 HAU/ml) for 8 h. Cell lysates were analyzed by IB as indicated. (H) UBL deleted OASL mutant does not enhance SeV mediated IFNβ induction. Cells were either mock infected or infected with 20 HAU/ml SeV for 16 h. Total RNA from each well were extracted followed by qRT-PCR using IFNβ specific primers. (I) OASL mediated enhancement of ISG induction by SeV is dependent on its UBL. Various OASL expressing HT1080 cells were infected with SeV as indicated for 16 h followed by IB with indicated antibodies as before.
The UBL domain of OASL has been implicated in its antiviral activity (Marques et al., 2008). We confirmed the contribution of the UBL in enhancing RIG-I signaling in cell lines expressing UBL deleted OASL (OASLΔUBL). Cells expressing OASLΔ1Δ2 were not protected from VSV infection (Suppl. Fig. S2E), and did not show enhanced induction of IFNβ, ISG56 and ISG60 mRNA (Fig. 4H; Suppl. Fig. S2F and S2G) and proteins (Fig. 4I) with SeV. However, expression of the UBL of OASL alone did not cause enhancement of RIG-I signaling (Suppl. Fig. 2H). These results indicate that the OASL UBL is necessary, but not sufficient, to enhance RIG-I signaling.
Loss of OASL expression reduces ISG induction and enhances virus replication
Next, we investigated the involvement of OASL in RIG-I signaling by ablating OASL expression in different systems. We tested various cell lines and found that the colon cancer cell line HCT-116 showed detectable, albeit low, basal OASL expression, that was strongly induced by SeV infection or IFNα treatment (Suppl. Fig. S3A and S3B). Multiple shRNAs targeting OASL were used to establish stable cell lines (HCT-116 and HEK293) in which basal and SeV or dsRNA-inducible endogenous OASL expression was partially ablated (Fig. 5A, panel 3, and Suppl. Fig. S3C and S3F). Upon SeV infection or LMW transfection of these cells ISG60, ISG56 and IFN induction was substantially reduced compared to control (Fig. 5A, panel 1 and 2; Fig. 5B; Fig. 5C and Suppl. Fig. S3D, S3E, and S3F). Consequently, loss of OASL in HCT-116-shOASL cells substantially increased VSV and SeV replication (Suppl. Fig. S3G–H). For further validation homozygous short deletions in the genomic OASL locus were introduced using TALENs. The resulting HCT-116-OASL−/− and 293T-OASL−/− cells showed reduced RIG-I signaling as measured by ISG induction (Fig. 5D, 5E and 5F, lane 4–6). As expected, the control 293T-DDX58−/− cells also showed complete loss of ISG induction (Fig. 5F, lane 7–9). In addition, confirming the role of OASL in RIG-I-mediated NF-κB activation, 293T-OASL−/− cells also showed diminished NF-κB-regulated gene induction (Fig. 5G). Similar results were obtained in primary human keratinocytes upon OASL silencing by siRNA (Fig. 5H and 5I). Taken together, these results confirm the positive role of human OASL in RIG-I signaling and antiviral responses.
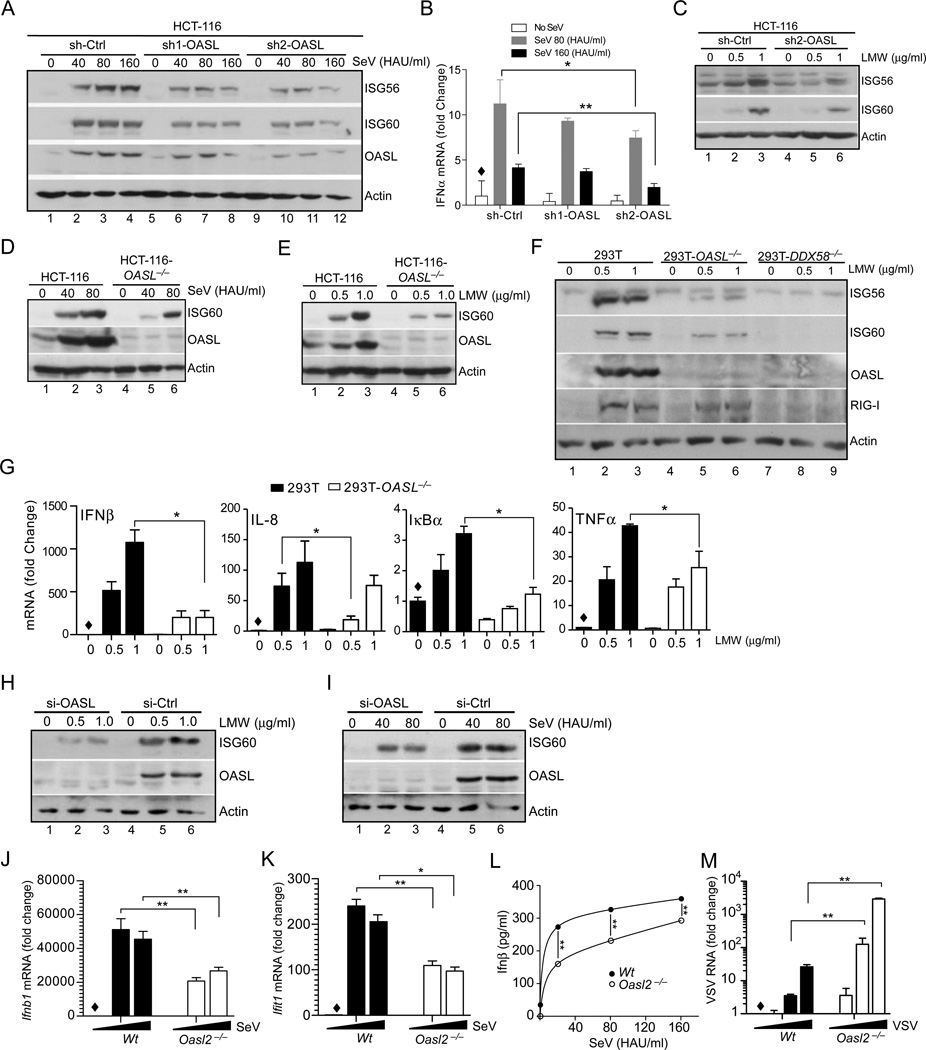
Fig. 5. Loss of human OASL and mouse Oasl2 reduces RIG-I signaling and enhances virus replication.
(A–C) Inhibition of RIG-I signaling in HCT-116 cells following OASL silencing. HCT-116 cells stably expressing OASL shRNA (two different shRNA, sh1-OASL and sh2-OASL targeting two different regions of OASL mRNA) or control vector stimulated with SeV as indicated for 24h followed by IB with indicated antibodies (A). Indicated cells were stimulated with SeV as in (A), and the cellular RNA were analyzed for IFNα mRNA by qRT-PCR (B). Same HCT-116 OASL shRNA and control cells were transfected with LMW for 24 h followed by IB with the indicated antibodies (C). (D–E) Reduction of RIG-I-mediated ISG induction in HCT-116-OASL−/− cells. OASL−/− HCT-116 and wild type HCT-116 cells were either infected with SeV (D) or transfected with LMW (E) as indicated, followed by IB for ISG60, OASL and actin antibodies respectively. (F) ISG induction in OASL and RIG-I targeted 293T cells. Wild type 293T, 293T-OASL−/− and 293T-DDX58−/− cells were transfected with LMW for 24 h, followed by IB for the indicated proteins. (G) Effect of OASL loss on RIG-I-mediated induction of NF-κB target genes. Cells were treated similarly as in (F) followed by qRT-PCR analysis of indicated NF-κB-regulated genes. (H–I) OASL silencing reduces ISG60 induction by RIG-I signaling in primary human keratinocytes. Primary keratinocytes were transfected either with OASL or control siRNA for 48 h, followed by either transfection with LMW or infection with SeV at the indicated doses for another 24 h and analysis by IB. (J–K) Downregulation of RIG-I-mediated Ifnb1 (IFNβ) and Ifit1 (ISG56) mRNA induction in Oasl2−/− macrophages. BMDM from wild type (Wt) or Oasl2−/− mice were infected with SeV for 8 h followed by detection of Ifnb1 (J) and Ifit1 (K) mRNA by qRT-PCR. (L) Secreted IFNβ was measured in the supernatants from SeV infected (8h) Wt (●) and Oasl2−/− (○) BMDM using the mouse IFNβ ELISA kit. (M) Increased VSV replication in BMDM from Oasl2−/− mice. BMDM from Wt or Oasl2−/− mice were infected with VSV for 24 h followed by quantitation of VSV RNA by qRT-PCR.
Genetic depletion of mouse Oasl2 reduces RIG-I signaling and enhances virus replication
Since mouse Oasl1 did not share the antiviral properties of human OASL, we examined the properties of mouse Oasl2, and tested their comparative binding abilities to the 5’UTR regions of IRF7 mRNA. In a similar experiment to Fig. 1A, human OASL and mouse Oasl2 did not bind to human or mouse IRF7 5’ UTR (Suppl. Fig. S3I, lane 15–18). Next, we compared their effects on RIG-I signaling using isoform specific shRNA in mouse embryonic fibroblasts (MEFs). Specific silencing was validated by measuring the loss of SeV induced induction of either Oasl1 or Oasl2 mRNA (Suppl. Fig. S3J, bottom panels). In the same experiment, silencing of Oasl2 partially reduced SeV-mediated IFNβ (Ifnb1) and Isg56 (Ifit1) mRNA induction indicating that mouse Oasl2 might be functionally equivalent to human OASL. To confirm these results and provide evidence for the antiviral activity of Oasl2 in vivo, we used Oasl2-deficient mice. These mice were generated by inserting a gene trapping cassette in the Oasl2 locus, where the third exon was spliced into a LacZ cassette, thus abrogating the expression of the endogenous locus (Suppl. Fig. S3K–L), and the same resource had been successfully used to define cGAS-deficient mice phenotype (Li et al., 2013; Schoggins et al., 2013). Bone marrow derived macrophages (BMDM) from Oasl2−/− mice showed expected loss of Oasl2 expression compared to wild type upon SeV infection (Suppl. Fig. S3M). Subsequent analysis of Ifnb1 and Ifit1 mRNA induction by SeV also showed substantial reduction in Oasl2−/− BMDM (Fig. 5J and 5K), which were further corroborated by analyzing mouse IFNβ protein by ELISA (Fig. 5L). Finally, the Oasl2−/− macrophages demonstrated significant upregulation in VSV replication compared to wild type (Wt) establishing the antiviral role mouse Oasl2, which was similar to human OASL (Fig. 5M). These results suggest that mouse Oasl2 is functionally similar to human OASL and exerts a positive role in antiviral signaling.
OASL specifically activates RIG-I signaling upstream of MAVS
To understand the molecular mechanism and the possible stage at which OASL regulated RIG-I pathway, we overexpressed components of RLR signaling and assessed the effects of OASL expression. 293T-DDX58−/− cells were used to avoid any feed-forward effect from RIG-I on resulting ISG induction. As expected, transient overexpression of MAVS and constitutively active IRF3-5D caused ISG56 induction in 293T-DDX58−/− cells indicating the activation of the RLR signaling downstream of RIG-I (Fig. 6A, lanes 1–3, 6B, lanes 1–3). However, the presence of OASL did not affect ISG56 induction in these conditions (Fig. 6A, lanes 4–6, 6B, lanes 4–6), indicating that OASL affected the RLR signaling upstream of MAVS. To further define the RIG-I-specific activity of OASL, we tested the effect of OASL on MDA5 signaling using both OASL expressing and OASL−/− cells. Induction of ISG60 by full-length MDA5 overexpression was not affected by the presence of OASL at two different doses (Fig. 6C). Similarly, induction of either ISG60 or IRF7 by constitutively active MDA5 CARD (MDA5(N)) were not affected in 293T-OASL−/− cells compared to 293T cells (Fig. 6D). OASL silencing had no effect on EMCV-mediated induction of IFNβ in human primary keratinocytes (Fig. 6E). It has been demonstrated that transfection of replicating viral RNA into U2OS cells prepared from EMCV infected 293T cells induces IFNβ in a MDA5 dependent manner, while that from SeV or VSV infected cells activates RIG-I signaling (Jiang et al., 2012). We verified the ISG induction by replicating EMCV viral RNA in U2OS cells (Suppl. Fig. S4A) and tested the effect of OASL silencing on ISG induction. As shown in Fig. 6F, ISG60 and IRF7 induction by RNA prepared from SeV-infected cells was reduced by OASL silencing (lane 2, and lane 5) while that using RNA from EMCV-infected cells was not affected (lane 3, lane 6). Taken together, these results indicated OASL specifically enhanced RIG-I signaling upstream of MAVS.
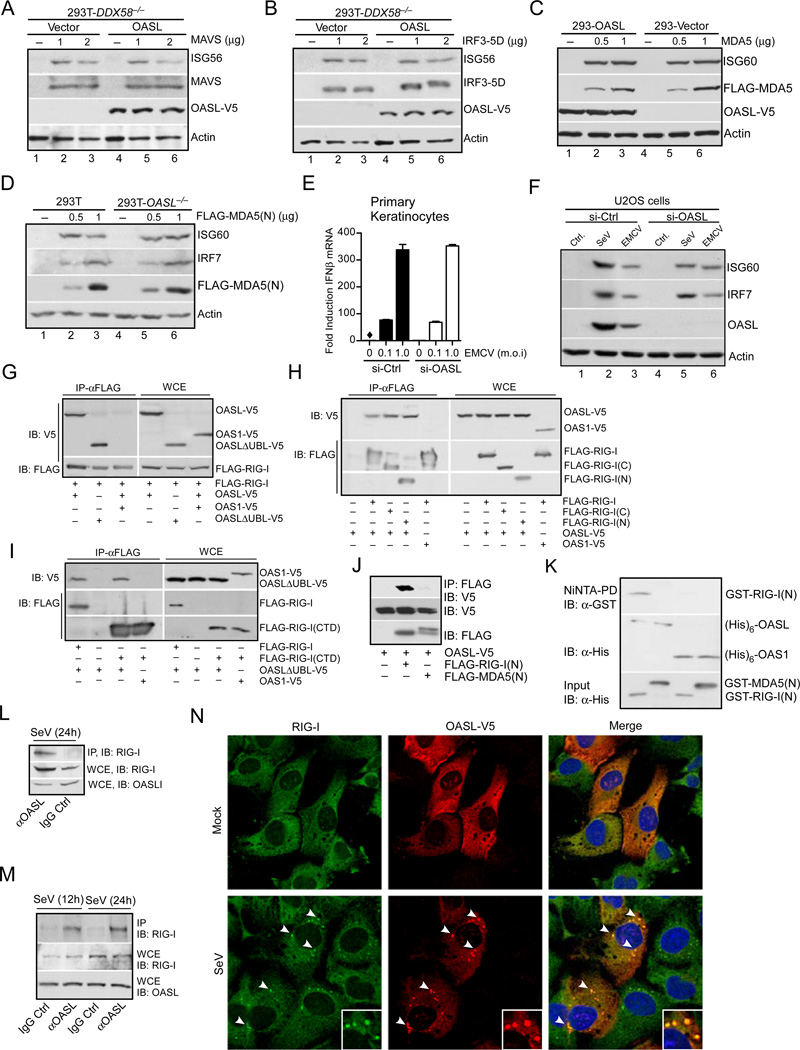
Fig. 6. OASL binds to and specifically enhances RIG-I signaling.
(A–B) OASL does not affect RLR signaling downstream of MAVS. 293T-DDX58−/− cells stably expressing OASL or vector control were transfected with indicated plasmids to express either MAVS (A) or constitutively active IRF3-5D (B) for 24 h followed by immunoblot analysis. (C) OASL does not enhance MDA5 signaling. Cells were transfected either with vector or two different concentrations of FLAG-MDA5 plasmid for 24 h followed by IB. (D) Loss of OASL expression does not affect MDA5 signaling. Cells were transfected for 24 h with indicated amounts of MDA5(N) plasmid and analyzed by IB. (E) OASL silencing does not affect IFNβ induction stimulated by EMCV infection in keratinocytes. Primary human keratinocytes were transfected with 50 nM OASL siRNA and control siRNA for 48 h, followed by EMCV infection for another 24 h. The OASL silencing (Suppl. Fig. S1E), IFNβ production (E) and EMCV replication (Suppl. Fig. S1E) were analyzed by specific qRT-PCR. (F) OASL does not affect the gene ISG60 and IRF7 induction triggered by a physiological MDA5 agonist. 293T cells were infected either with EMCV, VSV and SeV or mock infected (Ctrl.) and extracted for total RNA as described before (Jiang et al., 2012). U2OS cells were first transfected with control or OASL siRNA as described in Fig. 1 for 48 h followed by transfection with 1mg of total RNA prepared form infected cells as indicated. Cell lysates were analyzed by IB. (G) RIG-I interacts with OASL. 293T cells were co-transfected for 24 h with FLAG-RIG-I and OASL-V5, OASL-ΔUBL-V5 or OAS1-V5 plasmids, immunoprecipitated (IP) with FLAG antibody, followed by IB with V5 antibody. Whole cell extracts (WCE) from transfected cells were similarly analyzed for expression levels. (H) Both N-terminal CARD and C-terminal Helicase-CTD domains of RIG-I interact with OASL. 293T cells were either individually transfected with OASL-V5 or co-transfected as before with indicated plasmids followed by IP analysis. (I) OAS domain of OASL interacts with RIG-I CTD domain. 293T cells were transfected as indicated, followed by similar IP analysis. (J) RIG-I CARD domain (RIG-I(N)) specifically interacts with OASL, while MDA5 CARD domain (MDA5(N)) does not. 293T cells were transfected as indicated followed by IP analysis. (K) Specific in vitro interaction of RIG-I(N) with OASL. GST-RIG-I(N) and GST-MDA5(N) proteins purified from bacteria were incubated on ice as indicated with (His)6-OASL or with (His)6-OAS1 purified from insect cells followed by Ni-NTA pull down and IB. (L) Interaction of endogenous OASL and RIG-I. HEK293 cells were infected with SeV (100 HAU/ml) for 24 h. Cell lysates were immunoprecipitated either with OASL antibody or control IgG followed by IB with RIG-I antibody. Control WCE was analyzed to show expression. (M) Interaction of endogenous RIG-I and OASL in primary human fibroblsts. Human primary foreskin fibroblasts (HFF) were stimulated with SeV (160 HAU/ml) for 12 and 24 h respectively, followed by IP analysis as indicated. (N) OASL co-localizes with RIG-I in virus infected cells. HT1080-OASL cells were either mock infected or infected with 100 HAU/ml SeV for 8 h immuno-stained with RIG-I and V5 antibodies followed by confocal microscopy. The arrows denote the puntae formation, and insets show magnified puntaes.
OASL specifically binds and colocalizes with RIG-I
To understand the biochemical mechanism of RIG-I signaling enhancement by OASL, we investigated their interaction. V5-tagged OASL was co-expressed with FLAG-tagged RIG-I in HEK293 cells (schematic, Suppl. Fig. S4B). Imunoprecipitation with FLAG antibody showed OASL co-precipitated with RIG-I, while another OAS family member, OAS1, did not (Fig. 6G). Additionally, UBL-deleted OASL (OASLΔUBL) also interacted with RIG-I, indicating that the OAS domain of OASL is sufficient to mediate RIG-I interaction (Fig. 6G). To map the interaction domain in RIG-I, we used various RIG-I mutants with deleted domains (schematic, Suppl. Fig. S4B). Surprisingly, both N-terminal CARD deleted (RIG-I(C)) and helicase plus C-terminal domain (CTD) deleted RIG-I (RIG-I(N)) interacted with OASL (Fig. 6H), indicating that both domains of RIG-I interact with full-length OASL. However, when the C-terminal domain of RIG-I (RIG-I(CTD)) was co-expressed with either OASLΔUBL or OAS1, only the OAS-like domain of OASL (OASLΔUBL) specifically interacted with RIG-I(CTD) (Fig. 6I). This showed that OAS-like domain of OASL specifically interacted with the CTD of RIG-I, allowing further interaction of RIG-I CARDS with OASL, presumably with the UBL (see below). To understand the biochemical basis of OASL to specifically enhance RIG-I and not MDA5 signaling, we compared interaction of OASL with RIG-I and MDA5 CARDs. As shown in Fig. 6J, RIG-I(N), not MDA5(N), specifically co-immunoprecipitated with OASL. Furthermore, in an in vitro binding assay using purified RIG-I(N) or MDA5(N) and OASL or OAS1, we found that it is only RIG-I(N), which specifically bound to OASL (Fig. 6K). Endogenous OASL and RIG-I showed interaction in co-immunoprecipitation studies in HEK293 cells (Fig. 6L) and primary human fibroblasts (Fig. 6M). Due to the undetectable levels of OASL in unstimulated cells, we infected cells with SeV (100 HAU/ml) to induce OASL expression. As shown in Fig. 6L and 6M, endogenous RIG-I co-precipitated with endogenous OASL. The specificity of this interaction was demonstrated by testing OASL-MAVS co-precipitation, which was negative irrespective of the presence of RIG-I (Suppl. Fig. S4C). To examine the ligand or virus infection dependence of RIG-I and OASL interaction, we infected HEK293 cells co-expressing RIG-I and OASL with different amounts of SeV and immunoprecipitated RIG-I with anti-FLAG. As shown in Suppl. Fig. S4D, RIG-I-OASL interaction was detected even in the absence of SeV infection, indicating that this interaction was not dependent on the presence of viral RNA.
Finally, to establish the physiological relevance of the RIG-I-OASL interaction, and to examine if these proteins exist within the same cellular compartments we performed immunofluorescence microscopy for endogenous RIG-I and ectopically-expressed OASL in uninfected and SeV-infected HT1080 cells. In uninfected cells, both RIG-I and OASL exhibited diffuse staining throughout the cytoplasm (Fig 6N, top). However, upon infection with SeV, RIG-I and OASL translocated to discrete punctae localized throughout the cytoplasm (Fig 6N, bottom). RNA virus infection has been shown to translocate RIG-I to stress-granules appearing as discrete punctae (Onomoto et al., 2012). We found that OASL was relocalized to stress granules in response to SeV infection, and not in mitochondria, as confirmed by its colocalization with a GFP-tagged marker protein for stress granules, G3BP (Suppl. Fig. S4E and S4F). Taken together, our results show that OASL interacts with RIG-I to positively modulate its function and relocalizes to RIG-I-containing stress granules in response to specific virus infection.
OASL specifically enhances RIG-I signaling by mimicking polyubiquitin
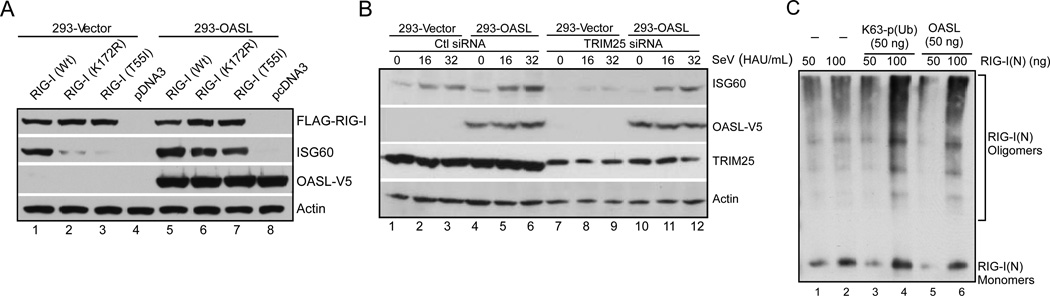
RIG-I is K63-ubiquitinated in its CARDs by the ubiquitin ligases TRIM25 and Riplet, which is essential for its activation (Gack et al., 2007; Oshiumi et al., 2012). However, K63-linked pUb chains synthesized by TRIM25 are sufficient for the complete activation of RIG-I signaling upon ligand binding (Zeng et al., 2010). Two mutations in the RIG-I CARD, T55I and K172R, abolish this ubiquitination and inhibit pUb binding to RIG-I, thereby rendering it inactive. Due to the presence of UBL, and its specific interaction with RIG-I, we hypothesized that the UBL of OASL might interact with RIG-I and mimic the K63-linked pUb to activate RIG-I. To test this, we created full-length and RIG-I(N) carrying T55I and K172R mutations, and measured their OASL binding, and signaling abilities with or without OASL. In co-immunoprecipitation assay all the mutants and wild type RIG-I bound to OASL with equal efficiencies (Suppl. Fig. S5A). Expression of wild-type RIG-I in HEK293 cells caused ISG60 induction and IFNβ reporter activation that was enhanced by OASL expression (Fig. 7A, lane 1 and lane 5; Suppl. Fig. 5B). As expected, both the T55I and K172R mutants of RIG-I were inactive when expressed in HEK293-Vector cells (Fig. 7A, lane 2 and lane 3; Suppl. Fig. S5B). However, when the same mutants were expressed in HEK293 cells expressing OASL, they were functional and induced near-wild-type amounts of ISG60 and IFNβ reporter activity (Fig. 7A, lane 6 and lane 7; Suppl. Fig. S5B). Similar results were obtained for the K172R mutant in the RIG-I(N) context (Suppl. Fig. S5C). Thus, OASL expression rescued RIG-I mutants that failed to interact with K63-linked pUb and are thus defective in signaling. To provide additional evidence, we silenced TRIM25 expression by siRNA. Reduction of TRIM25 expression caused inhibition of SeV-mediated ISG60 induction in HEK293-Vector cells (Fig. 7B, lane 1–3, lane 7–9). In contrast, silencing of TRIM25 had no effect on ISG60 induction in OASL expressing cells (Fig. 7B, lane 4–6, lane 10–12). Recently, it has been shown that following long (≤112 bp) RNA binding, full length RIG-I can be activated in vitro without pUb by ATP dependent oligomerization (Patel et al., 2013; Peisley et al., 2011). However, pUb binding is necessary for RNA-independent oligomerization of RIG-I(N); and with full length RIG-I and short dsRNA (<60 bp) indicating synergistic parallel mechanisms (Peisley et al., 2011). To investigate how OASL affect RIG-I oligomerization, we used purified RIG-I(N) at two different protein concentrations and compared its oligomerization in presence or absence of pUb and OASL using native gel electrophoresis (Patel et al., 2013). As shown in Fig. 7C, both 50 and 100 ng RIG-I(N) showed monomeric and oligomeric complexes in the absence of RNA or ATP (lane 1, lane 2). However, with 100 ng RIG-I, the oligomeric fraction was increased by pUb as well as by OASL (lane 4, lane 6). In a similar experiment oligomeric fraction for MDA5(N) was neither affected by pUb nor by OASL (Suppl. Fig. S5D). This suggested that OASL binding to RIG-I may help RIG-I-CARD oligomerization, possibly through its C-terminal UBL, which is required for MAVS binding and signal propagation.
Fig. 7. OASL specifically enhances RIG-I-signaling by mimicking pUb to activate RIG-I.
(A) Ubiuitination or p(Ub) binding mutants of RIG-I can be activated in presence of OASL. Cells were transfected with wild type (Wt) or mutant RIG-I constructs for 24 h and analyzed by IB with indicated antibodies. (B) OASL is capable of promoting RIG-I activation in the absence of TRIM25. Cells were transfected with 50 nM TRIM25 siRNA (Zeng et al., 2010) or control siRNA for 48 h followed by SeV infection for 16 h and IB analysis. (C) OASL enhances RIG-I(N) oligomerizaiton similar to pUb. 50 or 100 ng of purified RIG-I(N) (GST tagged) were incubated either with K63-linked polyubiquitin chain (mixture of (pUb)>3, Boston Biochem) or with purified OASL for 1h at 30°C in the binding buffer (20 mM HEPES-KOH pH 7.4; 5 mM MgCl2 and 1X proteinase inhibitor cocktails). The incubated proteins were analyzed by native PAGE followed by IB with anti-GST antibody.
DISCUSSION
We described a mechanism explaining the antiviral activity of human OASL whereby OASL bound to RIG-I, and specifically enhanced RIG-I-mediated antiviral signaling. We showed that similar to human OASL, mouse Oasl2 had antiviral activity and loss of these proteins resulted in reduced IFN signaling and enhanced RNA virus replication. Very recently, Lee et al. (Lee et al., 2013) has reported that mouse Oasl1 binds to the IRF7 5’ UTR to inhibit its translation, thereby downregulating IFN induction and enhancing viral replication. However, human OASL has been reported to have antiviral properties (Marques et al., 2008; Schoggins et al., 2011). Furthermore, SNP in human OASL has been linked to altered susceptibility to Hepatitis C and West Nile virus infection (Li et al., 2008; Yakub et al., 2005). Here using human cells either with targeted OASL deletion or ectopic OASL expression, and Oasl2−/− macrophages, we established the antiviral property of OASL and provide a mechanistic model consistent with previous observation. In contrast to mouse Oasl1, human OASL and mouse Oasl2 did not bind to human and mouse IRF7 5’UTR, neither did human OASL suppress IRF7 protein induction. As noted recently the innate immune responses can be quite unique between mice and humans (Seok et al., 2013). Specifically, the rodent OAS system is unique, with a total of 12 OAS genes and at least one previously described function specific to rodents. The mouse Oas1b is critical for resistance to Flavivirus in mice but a similar mechanism does not exist in humans (Perelygin et al., 2002). Therefore, this study establishes the unique antiviral activity of human OASL and provides a mechanistic basis of its function.
Type I IFNs induce hundreds of genes in different cells collectively known as ISGs. Although the antiviral activities of a large number of ISGs have been tested against a number of viruses (Liu et al., 2012; Schoggins et al., 2013; Schoggins et al., 2011), the biochemical and cellular basis of the antiviral property for only a handful of them has been delineated (Sadler and Williams, 2008; Sarkar and Sen, 2004). Our results therefore demonstrate a unique mechanism by which an ISG modulates antiviral signaling and provide important insights into the molecular events that mediate the antiviral effects of human OASL, a commonly expressed ISG.
From a number of biochemical and structural studies (Civril et al., 2011; Jiang et al., 2011; Kowalinski et al., 2011; Luo et al., 2011), a model for RIG-I activation has been proposed as follows: without RNA, full-length RIG-I adopts an autoinhibited conformation. In this state, intramolecular interactions between the CARDs and the helicase domain inhibit pUb binding to the CARDs, while the CTD remains relatively free to bind . Upon binding to viral RNA through the CTD, the helicase domain changes conformation, thereby enabling it to hydrolyze ATP and further interact with RNA, thus disrupting the CARD-helicase interaction (termed the ‘competent state’). The CARDs then bind to pUb, converting RIG-I to an active competent state, which is followed by CARD-mediated MAVS aggregation and signaling (Jiang and Chen, 2011; O'Neill and Bowie, 2011). Recent observations also suggest RNA dependent RIG-I aggregation in case of longer RNA to promote MAVS activation (Patel et al., 2013; Peisley et al., 2013). Although, for larger dsRNA the strict requirement of pUb for RIG-I activation has been a topic of debate, in most cases RIG-I activation is strongly regulated by a two-step mechanism requiring simultaneous binding of two ligands – RNA and pUb – likely to avoid aberrant activation of antiviral innate immunity and IFN induction. Based upon our studies, we propose an additional model of RIG-I activation (Suppl. Fig. S5E). In this model, OASL would be present at very low levels in naive cells. Upon virus infection, RIG-I would be activated by viral RNA and pUb, resulting in quick induction of IFN and ISGs like OASL through IRF3. Following the initial viral infection and OASL induction (which is equivalent to our OASL expressing cells) in the infected and the surrounding cells through IFN signaling, OASL will bind to RIG-I and mimic pUb. This will likely make RIG-I activation more sensitive requiring just one ligand, viral RNA, and lead to enhanced IFN induction. The OASL bound RIG-I may well represent a distinct intermediate state of RIG-I, with increased RNA sensitivity and isreadily activated by lower amount of viral RNA.
Our results also demonstrate a unique specificity of human OASL in enhancing the RIG-I pathway. We found that OASL did not enhance MDA5 activity, which is supported by our data showing the inability of OASL to protect from EMCV and Coxsackievirus B infection (data not shown), which are both detected by MDA5 (Loo and Gale, 2011). Although there has been no evidence that MDA5 requires ubiquitination for its activation, recent results suggest that MDA5 also binds pUb, which is important for its function (Jiang et al., 2012). However, when we tested the interaction of MDA5 CARDs (MDA5(N)) with OASL and compared it with that of RIG-I, it was only the RIG-I CARDs (RIG-I(N)) that interacted with OASL providing an explanation for the specific nature of OASL-mediated RIG-I activation. Additionally, OASL also enhanced RIG-I(N) oligomerization without affecting that for MDA5(N). This demonstrates a highly specific function of OASL as an alternative ligand and activator of the RIG-I pathway.
The expression of RIG-I and a number of RIG-I-associated pathway proteins is generally maintained at low levels and controlled through IRF3 and/or IFN signaling. Thus, a critical aspect of host cell defense is to induce potent and rapid antiviral signaling upon sensing of a virus infection. The rapid induction of OASL in uninfected cells via IFN signaling could thereby increase both the sensitivity of RIG-I signaling and induce a more potent antiviral response. Another possible advantage for OASL-mediated RIG-I activation is that the pUb chains can be labile in the presence of cellular or viral deubiquitinase activity, which may limit the quantities available for RIG-I activation during the course of a viral infection. Furthermore, this could serve as an additional mechanism utilized by the host cell to activate RIG-I in the absence of pUb as a means to escape viral antagonism. For example, several viruses directly target RIG-I and/or regulators of RIG-I signaling as a mechanism to attenuate host innate immune signaling. (Bowie and Unterholzner, 2008; Versteeg and Garcia-Sastre, 2010). Of particular interest, a number of viruses target ubiquitin ligases or encode for deubiquitinases (DUBs) and DUB-like molecules that function to alter the cellular ubiquitin machinery in order to facilitate their replication or to evade host innate immunity. For example, the influenza virus non-structural protein NS1 targets TRIM25 to inhibit RIG-I-mediated type I IFN induction (Gack et al., 2009). Thus, the activation of RIG-I by OASL in the absence of pUb may provide the host cell with a critical layer of defense that might serve to escape virus-mediated antagonism. In summary, we describe a mechanism by which human OASL, an evolutionary conserved ISG, and its functional mouse homolog Oasl2, confer an added layer of antiviral signal regulation to host cells in order to efficiently combat viral infections.
EXPERIMENTAL PROCEDURES
Cells, reagents, viruses and mice
HT1080, HEK293 and 293T cells were cultured in DMEM with 10% FBS and Penicillin/Streptomycin; HCT-116 cells were in McCoy's 5a Medium (Life Technologies, Grand Island, NY) with 10% FBS and antibiotics. BMDM were obtained by differentiating bone marrow cells from 4 week old Oasl2−/− and control Wt mice for one week using M-CSF (30 ng/mL) containing medium as described before. Low molecular weight p(I):p(C), was from InvivoGen (San Diego, CA), and high molecular weight p(I):p(C) was from GE Life Science (Pittsburgh, PA). Fugene 6 and GFP antibody were purchased from Roche (Indianapolis, IN). V5 monoclonal, FITC conjugated anti-rabbit and Alexa Fluor 594 conjugated anti-mouse antibodies from Life Technologies. FLAG antibody and antibody conjugated beads, OASL shRNA pLKO.1 constructs were from Sigma-Aldrich (St Louis, MO). RIG-I antibody was from Cell Signaling Technology (Danvers, MA) and OASL antibody from Abgent (San Diego, CA). ISG56, ISG60, actin, and Sendai virus C protein antibodies have been described before (Zhu et al., 2011). Mouse IFNβ ELISA kit was from PBL interferon source (Piscataway, NJ). Sendai virus (SeV, Cantell strain) was purchased from Charles River Laboratories (Wilmington, MA). EMCV, (ATCC, Manassas, VA), EGFP-tagged VSV and GFP tagged HSV1 (KOS strain) have been described before (Zhu et al., 2011). Long strain A serotype RSV that has been described before (Goswami et al., 2013). DENV-2 (16681) was from BEI Resources and expanded as described (Vasilakis et al., 2008). Oasl2−/− mice (Strain: C57BL/6N) embryos were obtained from International Knockout Mouse Consortium and rederived at the Jackson Lab (Bar Harbor, Maine), while the control mice (Strain: C57BL/6J) were purchased from Jackson Lab. All the mice experiments were carried out according to University of Pittsburgh IACUC guidelines. For all the experiments representative results are shown from minimum three biological repeats. Wherever applicable, plots show mean with standard error bars, where ♦ set as 1 for comparison. Statistical analyses were carried out using GraphPad Prism. * P < 0.05, ** P < 0.01 and NS = not statistically significant by two-tailed Student’s t test analysis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Zijian J. Chen for sharing crucial reagents and expert advice. We thank Drs. Prashant Desai, Asit Pattnaik, Kate Fitzgerald and Toni Cathomen for reagents. This work was supported in part by AI082673 from NIAID/NIH (SNS), and University of Pittsburgh Cancer Institute Startup funds (SNS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and Supplemental Experimental Procedures and can be found at
AUTHOR CONTRIBUTIONS
JZ and YZ spearheaded the project, where YZ performed initial, and JZ carried out later experiments. AG was responsible for Oasl2−/− mouse experiments. RAC and AF respectively carried out immunofluorescence and virus titration experiments. JD along with SB performed RNA binding and RSV replication experiments. MSI and RH provided various OASL constructs and purified proteins. JLS, TS along with VH helped create all the genome edited human cells. TF provided with crucial RIG-I antibody. SNS with initial input from MKG conceived and directed the project. CBC provided crucial reagents and intellectual inputs. SNS with help from CBC, RH, SB and VH wrote the manuscript.
REFERENCES
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Bennett M, Moldt M, Deimling T, Witte G, Schiesser S, Carell T, Hopfner KP. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen S, Justesen J, Schierup MH, Hartmann R. Characterization of the 2'-5'-oligoadenylate synthetase ubiquitin-like family. Nucleic acids research. 2003;31:3166–3173. doi: 10.1093/nar/gkg427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Goswami R, Majumdar T, Dhar J, Chattopadhyay S, Bandyopadhyay SK, Verbovetskaya V, Sen GC, Barik S. An RNA viral degradasome hijacks mitochondria to suppress innate immunity. Cell Res. 2013;23:1025–1042. doi: 10.1038/cr.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R, Olsen HS, Widder S, Jorgensen R, Justesen J. p59OASL, a 2'-5' oligoadenylate synthetase like protein: a novel human gene related to the 2'-5' oligoadenylate synthetase family. Nucleic acids research. 1998;26:4121–4128. doi: 10.1093/nar/26.18.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang QX, Chen ZJ. Structural insights into the activation of RIG-I, a nanosensor for viral RNAs. EMBO reports. 2011;13:7–8. doi: 10.1038/embor.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. Ubiquitin-Induced Oligomerization of the RNA Sensors RIG-I and MDA5 Activates Antiviral Innate Immune Response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- Lee MS, Kim B, Oh GT, Kim YJ. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat Immunol. 2013;14:346–355. doi: 10.1038/ni.2535. [DOI] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wan D, Wei W, Su J, Cao J, Qiu X, Ou C, Ban K, Yang C, Yue H. Candidate genes responsible for human hepatocellular carcinoma identified from differentially expressed genes in hepatocarcinogenesis of the tree shrew (Tupaia belangeri chinesis) Hepatol Res. 2008;38:85–95. doi: 10.1111/j.1872-034X.2007.00207.x. [DOI] [PubMed] [Google Scholar]
- Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci U S A. 2012;109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques J, Anwar J, Eskildsen-Larsen S, Rebouillat D, Paludan SR, Sen G, Williams BR, Hartmann R. The p59 oligoadenylate synthetase-like protein possesses antiviral activity that requires the C-terminal ubiquitin-like domain. J Gen Virol. 2008;89:2767–2772. doi: 10.1099/vir.0.2008/003558-0. [DOI] [PubMed] [Google Scholar]
- Melchjorsen J, Kristiansen H, Christiansen R, Rintahaka J, Matikainen S, Paludan SR, Hartmann R. Differential regulation of the OASL and OAS1 genes in response to viral infections. J Interferon Cytokine Res. 2009;29:199–207. doi: 10.1089/jir.2008.0050. [DOI] [PubMed] [Google Scholar]
- Moore CB, Ting JP. Regulation of mitochondrial antiviral signaling pathways. Immunity. 2008;28:735–739. doi: 10.1016/j.immuni.2008.05.005. [DOI] [PubMed] [Google Scholar]
- O'Neill LA, Bowie AG. The powerstroke and camshaft of the RIG-I antiviral RNA detection machine. Cell. 2011;147:259–261. doi: 10.1016/j.cell.2011.09.027. [DOI] [PubMed] [Google Scholar]
- Onomoto K, Jogi M, Yoo J-S, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, Matsumiya T, et al. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One. 2012 doi: 10.1371/journal.pone.0043031. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H, Matsumoto M, Seya T. Ubiquitin-mediated modulation of the cytoplasmic viral RNA sensor RIG-I. J Biochem. 2012;151:5–11. doi: 10.1093/jb/mvr111. [DOI] [PubMed] [Google Scholar]
- Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO reports. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, Hur S. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci U S A. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Molecular cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Perelygin AA, Scherbik SV, Zhulin IB, Stockman BM, Li Y, Brinton MA. Positional cloning of the murine flavivirus resistance gene. Proc Natl Acad Sci U S A. 2002;99:9322–9327. doi: 10.1073/pnas.142287799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebouillat D, Marie I, Hovanessian AG. Molecular cloning and characterization of two related and interferon-induced 56-kDa and 30-kDa proteins highly similar to 2'-5' oligoadenylate synthetase. Eur J Biochem. 1998;257:319–330. doi: 10.1046/j.1432-1327.1998.2570319.x. [DOI] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar SN, Sen GC. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol Ther. 2004;103:245–259. doi: 10.1016/j.pharmthera.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol. 2013;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Macduff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2013 doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Fokam EB, Hanson CT, Weinberg E, Sall AA, Whitehead SS, Hanley KA, Weaver SC. Genetic and phenotypic characterization of sylvatic dengue virus type 2 strains. Virology. 2008;377:296–307. doi: 10.1016/j.virol.2008.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg GA, Garcia-Sastre A. Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol. 2010;13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- Yakub I, Lillibridge KM, Moran A, Gonzalez OY, Belmont J, Gibbs RA, Tweardy DJ. Single nucleotide polymorphisms in genes for 2'-5'-oligoadenylate synthetase and RNase L inpatients hospitalized with West Nile virus infection. J Infect Dis. 2005;192:1741–1748. doi: 10.1086/497340. [DOI] [PubMed] [Google Scholar]
- Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Coyne CB, Sarkar SN. PKC alpha regulates Sendai virus-mediated interferon induction through HDAC6 and beta-catenin. EMBO J. 2011;30:4838–4849. doi: 10.1038/emboj.2011.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.