Abstract
Non-homologous end joining (NHEJ) is the major pathway for repair of DNA double-strand breaks (DSBs) in human cells. NHEJ is also needed for V(D)J recombination and the development of T and B cells in vertebrate immune systems, and acts in both the generation and prevention of non-homologous chromosomal translocations, a hallmark of genomic instability and many human cancers. X-ray crystal structures, cryo-electron microscopy envelopes, and small angle X-ray scattering (SAXS) solution conformations and assemblies are defining most of the core protein components for NHEJ: Ku70/Ku80 heterodimer; the DNA dependent protein kinase catalytic subunit (DNA-PKcs); the structure-specific endonuclease Artemis along with polynucleotide kinase/phosphatase (PNKP), aprataxin and PNKP related protein (APLF); the scaffolding proteins XRCC4 and XLF (XRCC4-like factor); DNA polymerases, and DNA ligase IV (Lig IV). The dynamic assembly of multi-protein NHEJ complexes at DSBs is regulated in part by protein phosphorylation. The basic steps of NHEJ have been biochemically defined to require: 1) DSB detection by the Ku heterodimer with subsequent DNA-PKcs tethering to form the DNA-PKcs-Ku-DNA complex (termed DNA-PK), 2) lesion processing, and 3) DNA end ligation by Lig IV, which functions in complex with XRCC4 and XLF. The current integration of structures by combined methods is resolving puzzles regarding the mechanisms, coordination and regulation of these three basic steps. Overall, structural results suggest the NHEJ system forms a flexing scaffold with the DNA-PKcs HEAT repeats acting as compressible macromolecular springs suitable to store and release conformational energy to apply forces to regulate NHEJ complexes and the DNA substrate for DNA end protection, processing, and ligation.
1. Introduction
Non-homologous end joining (NHEJ, sometimes referred to as classical, or C- NHEJ) is the major pathway for repair of ionizing radiation (IR)-induced double-stranded DNA breaks (DSBs) in human cells [1, 2]. NHEJ is required for antigen receptor gene rearrangements via V(D)J recombination and the development of T and B cells in the vertebrate immune system [3], and it is implicated in both the generation and prevention of non-homologous chromosomal translocations, a major hallmark of genomic instability and many human cancers [4]. The main proteins required for NHEJ are the Ku70/Ku80 heterodimer, the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs), the endo/structure specific nuclease Artemis, the scaffolding protein XRCC4, DNA ligase IV (Lig IV) and XRCC4-like factor, XLF. NHEJ can be thought of as occurring in three distinct stages: (1) detection of the DSB by the Ku heterodimer, with subsequent tethering by DNA-PKcs to form the DNA-PKcs-Ku-DNA complex (termed DNA-PK), (2) processing of IR-induced lesions and (3) ligation of the DNA ends by Lig IV, that functions in complex with XRCC4 and XLF. DNA end processing is required to remove damaged DNA and non-ligatable end groups at the termini of the DSB to facilitate ligation. End processing in response to IR-induced damage may include a variety of enzymes including polynucleotide kinase/phosphatase (PNKP), aprataxin and PNKP related protein (APLF), DNA polymerases and Artemis [1, 5, 6] (Fig. 1).
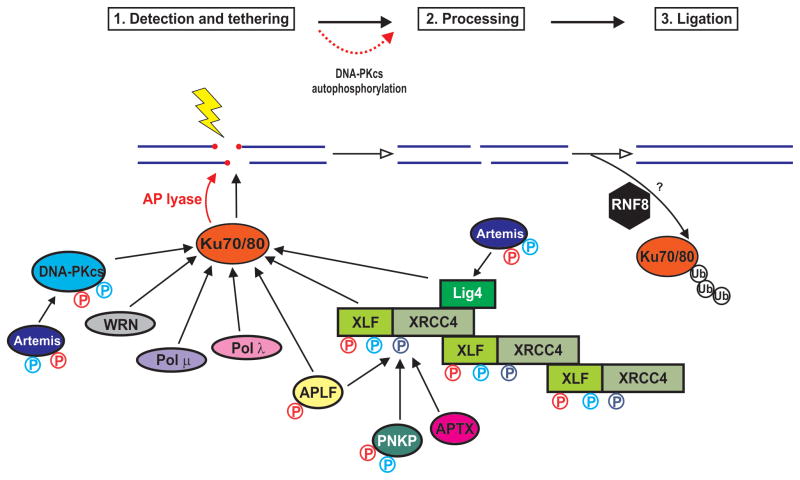
Figure 1. A model for non-homologous end joining.
NHEJ is proposed to occur in three stages, detection, and tethering, processing and, finally ligation. Central to NHE is recognition of the DSB by the Ku70/80 heterodimer. Ku interacts with multiple NHEJ proteins including DNA-PKcs, the Werner Syndrome helicase (WRN), DNA polymerases mu and lambda, APLF and XLF, contributing to their recruitment to DSBs. The catalytic activity of Ku is shown by the red arrows. XLF, LigIV, APLF, PNKP and APTX interact with XRCC4. Artemis interacts with DNA-PKcs and with LigIV. Together, these protein-protein interactions stabilize the dynamic NHEJ complex at DSBs. DNA-PK and ATM-dependent phosphorylation sites are indicated in blue and red, respectively. DNA-PKcs autophosphorylation regulates its association with the Ku-DNA complex, end processing and DSB pathway choice (dashed red line). See text for details. Figure adapted from [6].
Over the past few years it has become apparent that Ku plays multiple roles in NHEJ; not only does it recognize DSB ends, it has catalytic activity [7, 8] and is required for recruitment of multiple NHEJ proteins to the DSB (reviewed in [5, 6]). A picture is thus emerging of the dynamic assembly of a multi-protein NHEJ complex at DSBs, the function of which is regulated, at least in part, by protein phosphorylation [9]. Unraveling the multiple protein-protein and protein-DNA interactions within such dynamic protein-DNA assemblies presents formidable challenges for traditional approaches such as X-ray crystallography; however, small angle X-ray scattering (SAXS) has emerged as a powerful tool for elucidating overall shapes and conformations of protein-protein and protein-DNA complexes [10, 11].
In addition, we are learning more about the pathway and cross-pathway interactions between NHEJ and homologous recombination (HR) and its initiating complex MRE11-RAD50-NBS1 (MRN) [12]. During S and G2 phases of the cell cycle when NHEJ and HR are both active, the choice between which of the two pathways is used for repair is controlled in part by resection of DSBs, as the 3′ single-strand DNA (ssDNA) generated by extensive resection inhibits NHEJ but is required for Rad51 filament formation and strand invasion during HR [13]. We can now start to consider the structural implications of the complexes acting in the two DSB repair pathways for mechanisms of pathway choice. Here, we will review how structural biology on individual NHEJ components and, in some cases NHEJ complexes, is providing important insights into the mechanism of NHEJ, how MRN activities interface with NHEJ, and how this combined information may lead to identification of critical interfaces that may be targets for therapeutic intervention.
2. The Ku70/80 heterodimer
The Ku70/80 heterodimer was initially described as an autoantigen in the blood of patients with autoimmune disease [14]. Ku binds with high affinity to the ends of DSBs [15], targets DNA-PKcs to DSB ends [16, 17] and plays critical roles in NHEJ (reviewed in [5, 6]). The central DNA binding region of the heterodimer exists as a pre-formed ring that encircles dsDNA [18], allowing Ku to not only bind double stranded (ds) DNA in a largely sequence independent manner, but also to translocate away from the DSB end, allowing recruitment of DNA-PKcs, which interacts directly with the DNA termini [19, 20]. The Ku heterodimer is orientated such that Ku70 faces the DSB, whereas Ku80 is distal to the DSB [18, 20].
In addition to this central DNA binding core, Ku70 contains an acidic N-terminal region and a C-terminal SAP (SAF-A/B, Acinus and PIAS) domain [21, 22]. The extreme N-terminal region of Ku70 (the first 30 residues) contains twelve acidic amino acids (aspartic and glutamic acids) and Ku70 is phosphorylated on serine 6 in vitro by DNA-PKcs [23] and in vivo [24], providing additional negatively charged groups. The function of this region is unknown but its acidic nature suggests possible interaction with basic histones, or possibly DNA mimicry as proposed to involve such negative regions mimicking specific DNA structures rather than standard dsDNA [25]. At the C-terminus of Ku70, the SAP domain (residues 561–609) forms a well-defined helical structure that is linked to the DNA binding core by a highly flexible linker (residues 536–560). Both the linker and the SAP domain are reported to bind DNA in vitro [26, 27] suggesting that the SAP domain may assist in tethering the Ku heterodimer to chromatin.
The C-terminal region (CTR) of Ku80 (residues 592–709) contains a globular domain connected to the DNA binding core by a disordered, flexible linker [28, 29]. The CTR forms a flexible arm that extents from the DNA binding core, suitable for interaction with other proteins [30]. The extreme C-terminus of Ku80 contains twelve residues (EGGDVDDLLDMI) shown to interact directly with DNA-PKcs [31, 32]. This region is conserved in Ku80 from vertebrates but is not present in Arabidopsis thaliana, Caenorhabditis elegans or Saccharomyces cerevisiae, consistent with the absence of DNA-PKcs in these organisms [31, 33]. However, although cells lacking Ku80CTR are radiation sensitive and have low DNA-PK activity [34], Ku80CTR was not absolutely required for recruitment and activation of DNA-PKcs at DSBs [35]. Since DNA-PKcs only interacts with full-length Ku heterodimer in the presence of DNA [36], it seems likely that the interaction of DNA-PKcs and Ku on DNA is more complex, and involves more, than the extreme C terminal 14 amino acids of Ku80. The core, DNA-binding region of the Ku heterodimer is required for its interaction with XLF [37, 38] and residues L68, Y74 and I112 in the vonWillebrand (vWA) domain of Ku80 are required for interaction with residues 182–184 and 189 of APLF [39, 40]. Ku70 also interacts with XRCC4 but the region of interaction is unknown [41].
3. DNA-PKcs
DNA-PKcs is a member of the phosphatidyl inositol 3-kinase (PIKK) family of serine/threonine protein kinases [42]. Like the related protein kinases ataxia telangiectasia mutated (ATM) and ATM, Rad3-related (ATR), it is composed of a large N-terminal α-helical domain composed of multiple Huntington, Elongation factor 2, PP2A-A subunit, mTOR (HEAT) repeats separated by a flexible linker to the C-terminal FRAP, ATM and TRRAP (FAT) [43], kinase and FAT-C domains (Fig. 3). At over 4000 amino acids in length, elucidation of the structure of DNA-PKcs represents a considerable challenge for structural biology. Low resolution structures obtained from SAXS and cryo-electron microscopy (EM) revealed the overall shape and dimensions of DNA-PKcs, including head and palm regions separated by a large, central cavity with dimensions suitable for binding dsDNA [30, 44]. The presence of a smaller channel, possibly capable of binding single-stranded (ss) DNA, was proposed to occur in the head region of DNA-PKcs [44], consistent with the ability of DNA-PKcs to open or fray dsDNA ends [45]. The X-ray structure of DNA-PKcs (in complex with the Ku80CTR) at 6.6 Å confirmed the overall shape and dimensions of DNA-PKcs and revealed that the FAT, kinase and FATC domains are located in the head/crown of the molecule while the N-terminal HEAT repeat region forms the sides or arms of the molecule, which surround the central cavity [46]. Of note, a gap was detected at the base of the arms and the apex of the arms was predicted to be flexible, suggesting that DNA-PKcs may function like pincers, opening and closing around the central channel [9, 46] (Fig. 3).
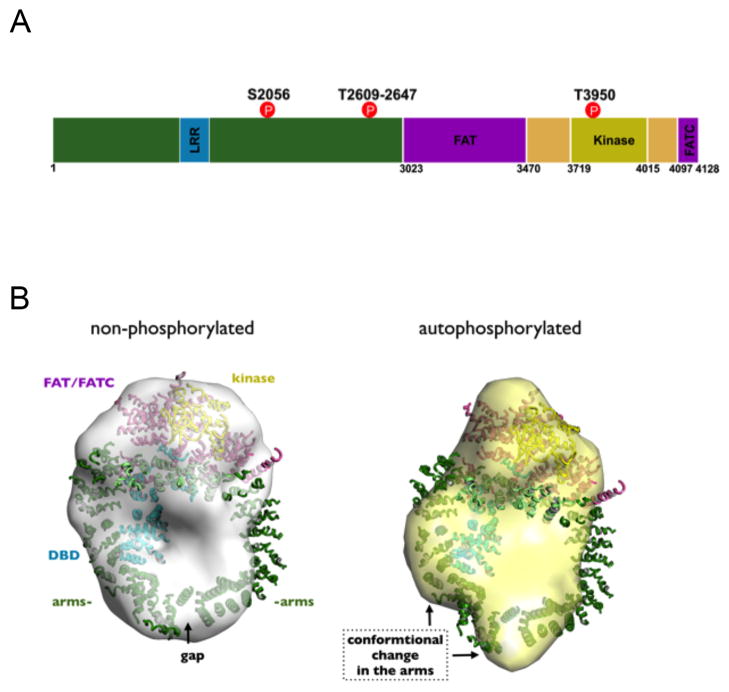
Figure 3. DNA-PKcs.
A. Schematic of DNA-PKcs showing putative domains and in vivo phosphorylation sites The N-terminal region of DNA-PKcs, extending from residues 1–3022 contains multiple HEAT repeats and other α-helical regions. A leucine rich region (LRR) spans residues 1503–1602 and has been proposed to be involved in DNA binding. The FAT domain encompasses residues 3023–3470 and is followed by PI3K-like kinase catalytic region (residues 3719–4015). The FATC domain spans residues 4097–4128. A PIKK regulatory domain (PRD) has been proposed between the kinase and FATC domains [57]. Well-characterized in vivo phosphorylation sites that function in NHEJ (S2056, T2609 and T3950) are indicated in red. See text for details.
B. Structure of non-phosphorylated and autophosphorylated DNAPKcs highlighting the autophosphorylation-induced conformational switch. Left: X-ray crystal structure of DNA-PKcs in complex with the Ku80CTR at 6.6 Å from (PDB ID 3KGV) showing the N-terminal HEAT/α-helical domains or arms (green), the FAT and FATC domains (magenta), and the kinase domain (yellow). A putative DNA binding domain (DBD) within the central cavity is shown in blue and the forehead in light green. The regions where the HEAT repeat/α-helical (green) arms abut the FAT/kinase/FATC domains (magenta) are predicted to exhibit conformational flexibility [46]. Phosphorylation within or close to this flexible region may induce conformational changes that regulate the interaction of DNA-PKcs with DNA (see [9] and text for details); Overlay of the DNA-PKcs crystal structure on the average SAXS envelope revealed hollow regions between the arms and bulky kinase region [30]. Right: Average SAXS envelopes for autophosphorylated DNA-PKcs overlaid with the X-ray crystal structure indicate the main conformational changes in the arm regions that open up the gap at the base [30].
DNA-PKcs is predicted to contain multiple intrinsically disordered regions, suggesting it has a high degree of flexibility [47]. Indeed, this conformational flexibility may account for subtle differences between the cryo-EM and low resolution X-ray structures, particularly at the base of DNA-PKcs. Interestingly, a recent study using cryo-EM suggests that dsDNA binds to the flexible region at the base of DNA-PKcs rather than directly in the large central cavity as suggested earlier [47]. Thus, further work is required to precisely localize the DNA binding regions within DNA-PKcs. Conformational flexibility is a theme conserved among other DNA damage response proteins, as RAD50 opens and closes in an ATP-dependent manner that may be somewhat analogous to DNA-PKcs [48].
The concave surface of the HEAT repeats of DNA-PKcs revealed in the recent X-ray structure [46] is reminiscent of the structure of PR65, the A subunit of protein phosphatase 2A (PP2A), which is composed entirely of HEAT repeats [49] that act like a molecular spring to transmit forces between the catalytic and regulatory subunits of PP2A [50]. It is therefore interesting to speculate that the HEAT repeats in DNA-PKcs and other members of the PIKK family may function in a similar manner.
As discussed elsewhere, DNA-PKcs function is highly regulated by phosphorylation [1, 9, 51], and over 40 phosphorylation sites have been reported in vitro and/or in vivo [9]. The solution SAXS structure of autophosphorylated DNA-PKcs revealed extensive conformational changes and a substantial increase in the maximum dimension (Dmax) of DNA-PKcs [30]. We have interpreted the overall SAXS results as indicating that DNA-PKcs autophosphorylation, possibly at the flexible regions that we expect contains the cluster of autophosphorylation sites around threonine 2609 (also called the ABCDE cluster), induces opening of the arms, thus facilitating dissociation of DNA-PKcs from DNA and Ku [9] (Fig. 3). However, a more detailed understanding of the regulation of DNA-PKcs function by phosphorylation awaits a higher resolution structure.
The recent 3.2 Å crystal structure of the FAT, kinase and FAT-C domains of the related PIKK, mTOR, provides the first glimpse of the catalytic mechanism of the PIKK family of protein kinases [52]. Like the N-terminal domain of DNA-PKcs, the FAT domain of mTOR is composed of multiple α-helices that form a C-shaped solenoid. Importantly, both the FAT domain and the FAT-C domain of mTOR are integrally linked to the catalytic domain, suggesting that they play critical roles in regulating catalytic activity. The kinase domain of mTOR adopts the two-lobed structure typical of the eukaryotic protein kinase family, as exemplified by the cAMP-dependent protein kinases and phosphatidyl inositol 3 kinase p110 catalytic subunit itself [53, 54], suggesting that the PIKKs share a similar catalytic mechanism to other serine/threonine and lipid kinases. Intriguingly, DNA-PKcs was shown to contain a putative FKBP12-rapamycin-binding (FRB) domain [52] raising the possibility that DNA-PKcs could be regulated by rapamycin and/or mTOR inhibitors, many of which are in clinical trials [55].
The region of DNA-PKcs that interacts with Ku was originally mapped to the C-terminal ~1000 amino acids [56] and residues 4043–4090 have been suggested to act as a PIKK regulatory domain (PRD) similar to the ATR region required for interaction with ATRIP [57], however, the precise functional interface between DNA-PKcs and Ku within the DNA-PK complex remains under investigation.
4. Artemis
Artemis is an endonuclease that plays an essential role in processing DNA hairpins formed during V(D)J recombination, and acts in the repair of a subset of DNA DSBs by NHEJ that have “complex” ends and require further end-processing [58]. Artemis has an N-terminal nuclease domain that belongs to the metallo-β-lactamase and β-CASP families [59] and a C-terminal regulatory domain that is highly phosphorylated in vivo and predicted to be unstructured [60–62] (Fig. 4). Artemis interacts with DNA-PKcs [63] and LigIV [64] through motifs contained in the C-terminal region. Residues 399–404 of Artemis, which are adjacent to the nuclease domain, interact with DNA-PKcs [65] although the corresponding interaction region in DNA-PKcs is not known. Recently, Artemis residues 485–495 were shown to interact with LigIV, with this interaction being essential for V(D)J recombination [64, 66]. Two groups independently solved crystal structures of this motif in complex with an open conformation of LigIV [67], or with the LigIV-DNA binding domain (DBD) only [66]. These structures show that the Artemis motif forms a three-helix bundle with the first two helices of the LigIV DBD (Fig. 4B). Artemis Trp489 and Phe493 residues bind to a hydrophobic pocket in LigIV to anchor the interaction (Fig. 4C).
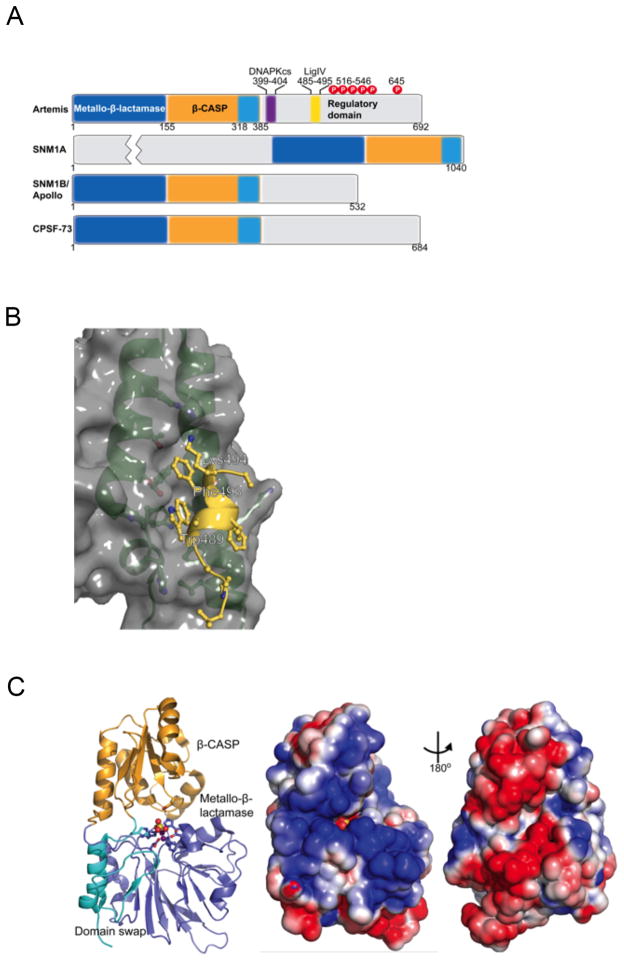
Figure 4. Structural insights of Artemis.
A. Schematic of the domain structure of Artemis showing DNA-PKcs and LigIV interacting regions, C-terminal phosphorylation sites, and domain similarities to SNM1A, SNM1B/Apollo and CPSF-73.
B. Structure of the Artemis-LigIV interaction interface (PDB 3W1G). Artemis residues 485–495 (yellow) complete a 3-helix bundle with the first two helices of LigIV (green). The DNA LigIV surface reveals a hydrophobic pocket into which Artemis residues Trp489 and Phe493 insert.
C. The SNM1B/Apollo (PDB 3ZDK) structure as a model for Artemis. (Left) The structure of SNM1B/Apollo reveals the overall fold of the nuclease domain, which is conserved with Artemis. The β-CASP (orange) and metallo-β-lactamase (blue) subdomains form two distinct lobes, with a domain swap (cyan) from the C-terminal region of the nuclease domain capping the metallo-β-lactamase subdomain. Zn and sulfate ions bound in the active site are shown as spheres, with neighboring residues within 4 Å shown as sticks. (Middle and right) Electrostatic surface of the two faces of SNM1B/Apollo reveal an electropositive surface (middle) that is likely to bind DNA.
The structure of the Artemis nuclease domain has remained elusive. Fortunately, crystal structures of related family members have recently been solved, and these provide interesting insights relevant to the structure and function of Artemis. Structures of the inter-strand crosslink repair nuclease SNM1A and Apollo (SNM1B), which share sequence identity of ~30% to each other and 26% to Artemis, are deposited in the protein databank (PDB IDs 4B87 and 3ZDK respectively), although details of these structures are unpublished. The SNM1A and Apollo nucleases superimpose well with each other (root mean square deviation [RMSD] = 2.2 Å over Cα residues), and the pre-mRNA-3′-end-processing endonuclease CPSF-73 (RMSD ~3 Å) [68] which shares <20% sequence identity with SNM1A/Apollo/Artemis.
These structures reveal that the nuclease separates into metallo-β-lactamase and β-CASP subdomains, with a surprising domain swap where the C-terminal region of the nuclease, which is annotated as being part of β-CASP subdomain based on sequence, folds with the metallo-β-lactamase subdomain, capping a beta sheet formed in this region (Fig. 4B). Interestingly, this domain swap positions a conserved histidine (H319 in Artemis), which has been proposed to be catalytically important for CSPF-73 [68] into the active site. A histidine in the Mre11 nuclease acts in opening the dsDNA for access to the active site that is capped and restricted in access analogously to the Artemis family [69]. One intriguing possibility for how Artemis activity is regulated is that when DNA-PKcs binds to and/or phosphorylates the Artemis C-terminal domain, this may be transmitted to the domain swap region, which is only 15 residues away from the DNA-PKcs interaction motif, to alter the conformation of key catalytic residues in the active site. For the Mre11-Rad50 end-processing complex, ATP binding and hydrolysis by Rad50 drives the conformational change [12].
In the active sites, one zinc ion is seen for SNM1A, with two zinc ions and one sulfate ion seen for Apollo and CSPF-73. The sulfate has been postulated to mimic the scissile phosphate, and a mechanism proposed for how CSPF-73 cleaves nucleic acid [68]. The metal ions are found in a protein cleft that is accessible from a very electropositive face of the enzyme (Fig. 4B/C). The reverse side of the protein is electronegative, thus it is likely that DNA substrates bind across the electropositive face of these proteins with the scissile phosphate entering the active site through the cleft. Interestingly, these enzymes all show distinct substrate preferences. While CSPF-73 binds to and endonucleolytically cleaves 3′-ends of mRNA [68]. SNM1A, Apollo and Artemis all show specificity for DNA. SNM1A and Apollo both have exonuclease activity and can act on ss and dsDNA, with SNM1A showing a preference for ssDNA and processivity on long DNA substrates not seen for Apollo [70]. In contrast, Artemis is an endonuclease that cuts in a structure-specific manner with a preference for hairpins [63, 71] which resembles Mre11 specificity [72]. It will be interesting to understand how these enzymes bind their nucleic acid substrates, and what structural features define substrate preferences; however this knowledge awaits determination of co-complex structures and further biochemical analyses. Currently, comparative analyses of the DNA-PK and MRN systems may provide insights for experimental design and interpretation.
5. The XLF-XRCC4-DNA Ligase IV complex
XRCC4 is a critical NHEJ scaffolding protein. It can interact with itself to form multimeric complexes, as well as with LigIV, XLF, PNKP, APTX, APLF, Ku and DNA (see [1, 6, 73]). Examination of full-length XRCC4 and its complexes with LigIV tandem BRCT domains and XLF in solution by SAXS revealed XRCC4 forms tetramers via a head-to-head interface supported by the XRCC4 C-terminal region which folds back on itself [74]. Combined crystallography and SAXS shows that the head domain of XRCC4 forms a hydrophobic pocket for specific interaction with the head domain of XLF via L115 [75]. The resulting dimers of XRCC4 and XLF interact to form long, super-helical filaments suitable to bridge DSB ends for ligation (Figure 6A) [73, 75]. These structures suggest how human XLF and XRCC4 interact to bind DNA and help explain the mechanistic consequences of patient mutations [75].
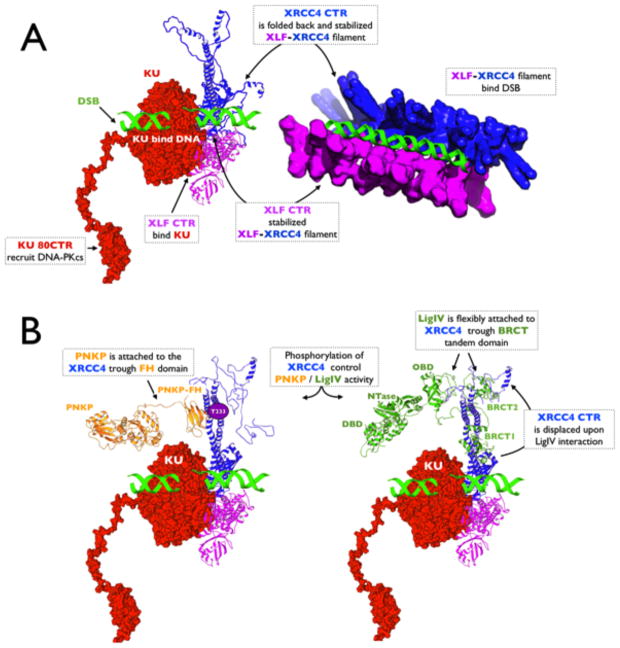
Figure 6. Towards a structure of NHEJ multi-protein-DNA complexes.
A. Left: Atomistic model of full length Ku (red surface) bound to DNA (green) indicating the extended and flexible Ku80-CTR domain [30] reported to recruit DNA-PKcs to the DSB [31]. Superimposition of the crystal structure of XLF1–224 -XRCC41–140 [75] and solution atomistic model of full length XLF (magenta) / full length XRCC4 (blue) [74, 75] was used to build a hypothetical assembly of Ku-XLF-XRCC4. The XLF CTR region is required for recruitment of XLF to Ku at the DSB, stabilizing the interaction with XRCC4 [38]. Right: The initial Ku-XLF-XRCC4 complex may initiate XLF-XRCC4 filament formation [30, 75, 135, 136] and create an extended grooved scaffold to stabilize DSBs [75].
B. Left: Phosphorylation of XRCC4 promotes a high affinity interaction with the FHA domain of PNKP (orange). The atomistic model of flexibly tethered PNKP was built based on the PNKP crystal structure [104], SAXS models of PNKP [105] and full length XRCC4 (blue) [74]. The model shows a far-reaching enzyme in the process of interacting with a DSB.
Right: An atomistic model of XRCC4-LigIV was built based on the known crystal structures of XRCC4 in complex with the LigIV tandem BRCT domain [77, 78], a SAXS model of full length XRCC4 [74], the crystal structure of DNA Ligase III [82] and the assumption that LigIV is flexibly attached to XRCC4 [86]. The model shows the far-reaching LigIV catalytic core, composed of DBD, NTase and OBD, in the process of DSB ligation.
LigIV is composed of N-terminal DNA binding and catalytic domains connected to a C-terminal tandem BRCT domain separated by a short linker [76] that interacts directly with the coiled-coil stalk domain of XRCC4 [77, 78]. The BRCT domains of many DNA damage response proteins bind phospho-proteins [79], however, no phosphoprotein binding partners have been identified for the tandem BRCT domains of LigIV. Recently, the LigIV BRCT domains were shown to interact directly with poly ADP ribose polymerase (PAR), the product of DNA damage activated poly ADP ribose polymerase (PARP) [80]. Moreover, PAR was responsible for the rapid recruitment of LigIV to sites of DNA damage in vivo [80], providing a novel mechanism for the recruitment of the XRCC4-LigIV complex to DSBs. Other PAR binding motifs include the FHA domains of PNKP, and Aprataxin (APTX) and the FHA-BRCT domain of NBS1, indicating that this mode of recruitment is common to other DNA damage response proteins.
The recently solved structure of LigIV (residues 1–609) [67] revealed three catalytic domains (the DNA-binding domain [DBD]; the nucleotidyltransferase domain [Ntase or NTD], and an OB-fold domain [OBD]) with similarities to those of human DNA ligases I [81] and III [82]. The LigIV structure is in a relatively open conformation, and we speculate that the DBD-Ntase-OBD domains are flexible relative to each other, as shown in the solution X-ray scattering studies of LigI [83] and LigIII [82] in the absence of DNA. In LigI and III, DNA binding leads to encircling of DNA by the DBD-Ntase-OBD domains, and a similar arrangement is expected for LigIV.
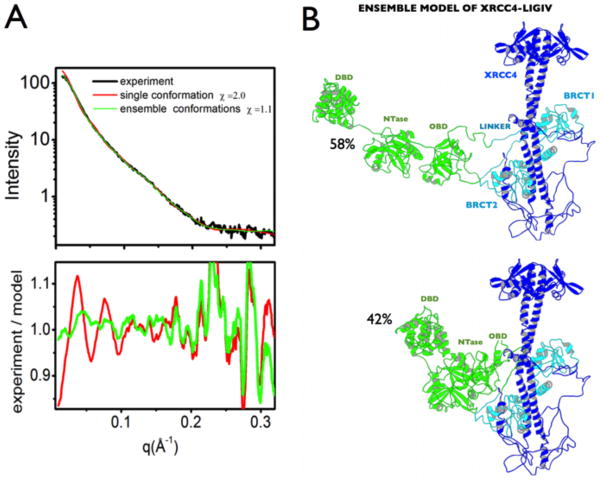
The flexible linker region between the catalytic core of LigIV and the tandem BRCT domain is predicted to provide LigIV with considerable range of motion [84]. Although the cryo-EM structure of XRCC4-LigIV positions the catalytic domain of LigIV near the XRCC4 head domain [85], our solution X-ray scattering studies of the XRCC4-LigIV complex suggest flexibility of the catalytic region such that the catalytic domains are in proximity to the BRCT domain (Figure 5). Together with advanced ensemble modeling, solution scattering also suggests internal conformational disorder between the individual DBD-Ntase-OBD domains (Figure 5). Similar conclusions have been postulated from the solution studies of LigIV in complex with truncated and modified XRCC4ΔCTD;CtoA, where the XRCC4 C-terminal domain (residues 214–334) is omitted and all cysteines are mutated to alanines [86]. Together, these studies suggest that the LigIV catalytic domain adopts dynamic conformations capable of reaching towards DNA substrates (Figure 5B).
Figure 5. Flexibility of XRCC4-LigIV in solution.
A. Comparison of experimental data and calculated scattering profiles for human XRCC4-LigIV. Experimental data are represented by the black line. Red and green lines represent the theoretical scattering curves of the single best-fit conformation (red), and the ensemble model determined by BILBOMD – MES [134]. Residuals calculated as I(q)experimental/I(q)model are shown below the scattering curves. The single best model (not shown) fits the data with χ=2.0; a MES model with both open (52%) and closed (48%) conformations significantly improved the fit (χ=1.1) and indicated flexibly tethered LigIV.
B. The Ensemble model of XRCC4-LigIV is shown. The sample had been prepared by size exclusion chromatography immediately prior to SAXS data collection with a final protein concentration of approximately 1.0 mg/mL. SAXS data were collected and processed at the Advanced Light Source Beamline 12.3.1 SIBYLS, Lawrence Berkeley National Laboratory as described [89].
6. Towards a structural and mechanistic understanding of NHEJ multi-protein-DNA complexes
As discussed above, considerable progress has been made towards determining the structures of individual NHEJ components and their subdomains. The next challenge is to elucidate structures of multi protein complexes assembled together on their target, dsDNA ends, to link complexes and conformations to biology. To deal with the size and conformational plasticity of these multi-component complexes, approaches such as small angle x-ray scattering (SAXS) that can characterize the assembly, morphology and the conformational states [87–89] are likely to be particularly informative. With new methods, SAXS can comprehensively define protein and DNA conformational states in solution by objective quantitative measurements, as seen for the mismatch repair ATPase complexes with DNA [90, 91]. Such comprehensive measurements may be needed to fully understand NHEJ pathway interactions.
What should we look for to gain a mechanistic and predictive knowledge of NHEJ biology at the molecular level? Discoveries made from the structural biochemistry of other DNA repair pathways, such as DNA base excision repair, suggest that protein complexes and DNA distortions can create platforms for pathway progression and coordination [92–94]. Structures of DNA repair complexes with substrate and product DNA also reveal principles of negative design that suggest how cells avoid reactions that would cause a loss of genetic integrity, as seen for the structure-specific nuclease FEN1 [95]. Consideration of the key importance of coordination of DSB repair steps while maintaining protection of DNA ends suggests that NHEJ will employ both protein and DNA conformational controls by analogy with other repair machinery. As discussed in Section 5, recent results have revealed that NHEJ components form remarkable scaffolding architectures at DSBs, with important roles for XRCC4 and XLF in this process. The implications of these scaffolds in promoting NHEJ are vast, thus an important unanswered question in the field is whether such scaffolds do indeed form in vivo (discussed in [73]).
Given that human cells contain more XRCC4 than LigIV [96], it seems unlikely that each subunit of XRCC4 in the filament contains a bound LigIV molecule. XRCC4 filaments may represent a transient storage form that dissociates into dimers upon interaction with the LigIV-BRCT domain. Indeed, the binding constant for XRCC4 tetramerization is weaker (KD 22 μM) than that of XRCC4 binding to the LigIV·BRCT domain (estimated KD ~1 nM) [97] and is reversible. Thus, the interaction of XRCC4 with LigIV will be dominant over XRCC4 oligomerization and shift the formation of filaments towards the dimeric form [97]. The BRCT domain probably promotes dissociation of XRCC4 filaments by the displacement of the XRCC4 C-terminus from the head region, which destabilizes the head-to-head interface [74]. We propose that LigIV may only be present at the ends of the filaments, where it can directly carry out ligation (Figure 6B). The LigIV tandem BRCT domain disrupted the large protein-DNA networks observed for XLF-XRCC4-DNA and disrupted DNA end-bridging by XLF-XRCC4 [98, 99] or XRCC4 [74] in vitro. Thus, we hypothesize that XRCC4 homodimers directly involved in end joining might be complexed to LigIV (Figure 6B), while XRCC4 molecules involved in alignment of DNA ends by XLF-XRCC4 filaments may be free of LigIV (Figure 6A). This would provide a mechanism for forming the XRCC4-LigIV complex only at the ends of filaments in the proximity of Ku, and support a model in which LigIV is not present within the filaments (Figure 6B).
The flexible XRCC4 scaffolding also interacts with PNKP [100] and the related proteins APLF [101] and APTX [102] providing possible mechanisms to recruit proteins with DNA-end processing activities to DSBs. PNKP is a 3′-DNA phosphatase, 5′-DNA kinase that directly removes non-ligatable groups from DNA termini and replaces them with ligatable 5′-phosphates and 3′-hydroxyl groups [103]. The X-ray structure of PNKP reveals an N-terminal fork-head associated (FHA) domain attached by a flexible linker to the catalytic phosphatase-kinase domains [104, 105]. The flexible PNKP linker is phosphorylated by ATM after DNA damage, regulating PNKP function [106, 107]. The PNKP-FHA domain interacts with CK2-phosphorylated XRCC4 (Figure 6B), through phosphorylated T233 [100], a site in the CTR that is constitutively phosphorylated in vivo [96]. Thus, it has long been thought that PNKP is recruited to DSBs through its interaction with XRCC4. However, the FHA domain of PNKP also interacts with PAR and this interaction facilitates the rapid recruitment of PNKP to sites of DNA damage in vivo [80]. Mechanisms that regulate binding of phospho-XRCC4 and PAR to the PNKP FHA domain remain to be determined.
Like PNKP, APLF also contains an N-terminal FHA domain that interacts directly with T233-phosphorylated XRCC4 [108, 109]. Indeed, APLF has emerged as an important scaffolding protein in NHEJ, interacting with Ku80 as well as XRCC4 and stimulating NHEJ by enhancing ligation [39, 40, 110]. APLF has also been reported to have exonuclease activity [101, 111], thus may contribute to processing of DNA termini prior to ligation. APLF also interacts with PAR, but via its C-terminal PBZ domains [112, 113] rather than its FHA domain [80], providing a mechanism for its recruitment to chromatin in vivo. APLF also interacts with histones, and can act as a histone chaperone [114], revealing additional roles in the DNA damage response. Aprataxin (APTX), which is mutated in ataxia with oculomotor apraxia 1 (AOA1), removes AMP, formed from abortive ligation reactions, from DNA termini [102, 115, 116]. Like APLF and PNKP, APTX also interacts with T233-phosphorylated XRCC4 suggesting roles in DSB repair [102], however, to date, no defects in DSB repair have been reported in APTX defective cells (discussed in [117]).
The effects of CK2 mediated phosphorylation of XRCC4 on filament structure and how the interaction of phosphorylated XRCC4 with these various FHA domain-containing proteins affects filament assembly and dynamics are important questions that need to be addressed. However, the flexible attachment of the FHA domain to the PNKP catalytic segment, elucidated by SAXS [105], suggests that PNKP is capable of reaching distal DNA substrates that are bound by Ku, as proposed for LigIV (Figure 6B). Like LigIV, PNKP, APLF and APTX carry out their enzymatic functions at DNA ends, therefore, as discussed for LigIV and PNKP, the flexible scaffold-like arrangement of BRCT and FHA domains may provide mechanisms to control the access of these enzymes to DSB ends rather than place them throughout the filaments. The ability of the FHA and BRCT domains of these enzymes to interact with PAR may also contribute to their localization to DSB ends [80, 112].
The Ku heterodimer plays a critical role in recruiting multiple NHEJ factors to DSBs [118]. Both XRCC4 and XLF interact with Ku [37, 41, 119, 120] (Figure 6A), and Ku is required to recruit XRCC4 and XLF to sites of laser-induced DNA damage in vivo [37]. Similarly, Ku enhances recruitment of XRCC4-LigIV and XLF to DNA in vitro [37, 120]. Surprisingly, XRCC4 was not required to recruit XLF to sites of damage, although it did help to stabilize/retain XLF at DSBs [37]. Understanding how filaments coordinate with Ku will therefore be important to understanding their broader functions. As a first step towards this goal, we determined the overall arrangement of the Ku-DNA-XLF-XRCC4 assembly in solution using SAXS [75]. Our results suggest that the Ku-DNA complex is positioned on one side of the XLF-XRCC4 assembly (Figure 6A), providing a possible mechanistic basis for the Ku-DNA dependent recruitment of the XLF-XRCC4 complex to DSBs in vivo [37]. However, whether Ku binds to DNA ends then translocates inwards allowing the filaments to form on the free end of the DNA (i.e. proximal to the break), or remains bound to the DNA termini, allowing filaments to form internal to DNA-bound Ku (i.e. distal to the break) (Figure 6A), remains to be determined.
Currently we know Ku recruits DNA-PKcs to DNA ends [16, 19], so the location of Ku relative to the filaments is critical for understanding how DNA-PKcs accesses DNA ends and functions at DSBs. Upon recruitment, DNA-PKcs undergoes autophosphorylation-dependent conformational changes (Figure 3B) that regulate its interaction with DNA ends [9, 30, 121]. The timing of DNA-PKcs recruitment and release, how it is coordinated with filament formation, and how it might regulate the function of the filaments are all unknown and will be exciting to decipher. Interestingly, both DNA-PKcs and XLF appear to protect DNA ends from resection [51, 122–125]. Thus, for NHEJ it is possible that the XRCC4-XLF filaments do not form until DNA-PKcs has been released and, at this point, the filaments function to restrict DNA end resection. This would be consistent with the observation that XLF promotes retention of 3′ overhangs in vitro [126] and that end resection during V(D)J recombination seems to be restricted by XLF [122, 124, 127, 128]. For MRE11-RAD50, the complex evidently moves away from the DNA ends to make a nick internal to the DSB that allows resection toward and away from the break [129]. It will be interesting to see how Ku and XRCC4-XLF protects ends but also allow access for repair while somehow competing with and regulating Mre11-Rad50 initiation of excision for homologous recombination. An intriguing possibility that merits more attention is that direct interactions may provide functional regulation of pathway choice for DSB repair, such as the interaction between Ku70 and Mre11 proposed from two-hybrid and other data [130]. However we have not seen this interaction in pull-down experiments. Another area of interest with be the licensing of NHEJ versus HR pathways, which appears to be controlled in part by the MRE11 nuclease, as well are regulation of MRE11 nuclease end processing and end protection at stalled forks [131, 132]. One key lesson so far that is underscored by both the MRN complex [12, 133] and the complexes of Ku-DNA-XLF-XRCC4 is that both major DSB repair machines employ flexing scaffolds to allow functional activities, regulation and protection of DSB ends. In this context, the DNA-PKcs and ATM kinases, composed of multiple HEAT repeats, appear suitable to act as springs that can store conformational energy, be regulated by post-translational modifications and apply forces to regulate the flexible scaffold complexes. As we approach answers to questions about the mechanisms regulating DSB repair, the structures of components and complexes are providing useful information that will be complemented by a growing understanding of dynamic complexes including their overall super assembly architecture and conformational transitions.
Figure 2. The Ku70/80 heterodimer.
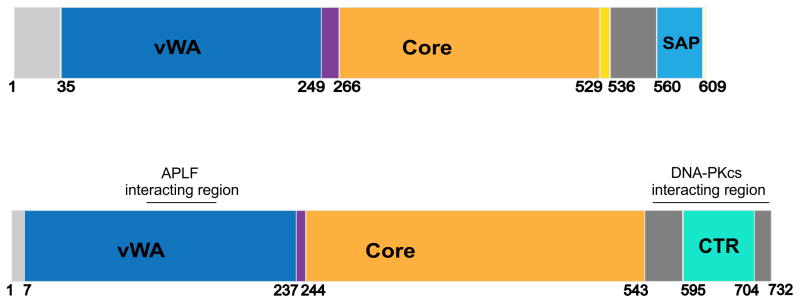
Schematics of Ku70 (upper) and Ku80 (lower) showing regions of interaction with XLF, APLF and DNA-PKcs. The N-terminal region of Ku70 (amino acids 1–33) is disordered and contains 12 highly acidic amino acids. Ku70 contains N-terminal von Willebrand domain (vWA) (amino acids 35–249), a core domain (amino acids 266–529) and a C-terminal SAP domain (amino acids 561–609), which is linked to the core domain via a disordered linker region (amino acids 536–560). The SAP domain has been proposed to function in DNA binding and chromatin organization. Ku80, like Ku70, shows similar domain organization with a vWA domain (amino acids 7–237), a core domain (amino acids 244–543) and a unique C-terminal region (amino acids 544–732). Amino acids 544–594 are predicted to be disordered and are followed by a globular region (amino acids 595–704) followed by another disordered region (amino acids 705–732). The Ku80CTR region has been proposed to be required for the recruitment of DNA-PKcs. See text for details.
Acknowledgments
Work in the authors’ laboratories is funded by NCI-NIH P01 grant CA92584 to JAT and SPLM and CA84442 to DAR.
Footnotes
Conflict of interest
The authors declare there are no conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helmink BA, Sleckman BP. The response to and repair of RAG-mediated DNA double- strand breaks. Annu Rev Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer. 2013;13:443–454. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Lees-Miller SP. Detection and Repair of Ionizing Radiation-Induced DNA Double Strand Breaks: New Developments in Nonhomologous End Joining. Int J Radiat Oncol Biol Phys. 2013;86:440–449. doi: 10.1016/j.ijrobp.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radhakrishnan SK, Jette N, Lees-Miller SP. Nonhomologous end joining: new and emerging themes. DNA Repair (Amst) doi: 10.1016/j.dnarep.2014.01.009. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strande N, Roberts SA, Oh S, Hendrickson EA, Ramsden DA. Specificity of the dRP/AP lyase of Ku promotes nonhomologous end joining (NHEJ) fidelity at damaged ends. J Biol Chem. 2012;287:13686–13693. doi: 10.1074/jbc.M111.329730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA- PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 11.Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr Opin Struct Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 2010;9:1299–1306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 14.Mimori T, Akizuki M, Yamagata H, Inada S, Yoshida S, Homma M. Characterization of a high molecular weight acidic nuclear protein recognized by autoantibodies in sera from patients with polymyositis-scleroderma overlap. J Clin Invest. 1981;68:611–620. doi: 10.1172/JCI110295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mimori T, Hardin JA. Mechanism of interaction between Ku protein and DNA. J Biol Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 16.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 17.Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci U S A. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 19.Yoo S, Dynan WS. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999;27:4679–4686. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo S, Kimzey A, Dynan WS. Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J Biol Chem. 1999;274:20034–20039. doi: 10.1074/jbc.274.28.20034. [DOI] [PubMed] [Google Scholar]
- 21.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 22.Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan DW, Ye R, Veillette CJ, Lees-Miller SP. DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry. 1999;38:1819–1828. doi: 10.1021/bi982584b. [DOI] [PubMed] [Google Scholar]
- 24.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 25.Putnam CD, Tainer JA. Protein mimicry of DNA and pathway regulation. DNA Repair (Amst) 2005;4:1410–1420. doi: 10.1016/j.dnarep.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Zhu L, Lin D, Chen F, Chen DJ, Chen Y. The three-dimensional structure of the C-terminal DNA-binding domain of human Ku70. J Biol Chem. 2001;276:38231–38236. doi: 10.1074/jbc.M105238200. [DOI] [PubMed] [Google Scholar]
- 27.Hu S, Pluth JM, Cucinotta FA. Putative binding modes of Ku70-SAP domain with double strand DNA: a molecular modeling study. J Mol Model. 2012;18:2163–2174. doi: 10.1007/s00894-011-1234-x. [DOI] [PubMed] [Google Scholar]
- 28.Harris R, Esposito D, Sankar A, Maman JD, Hinks JA, Pearl LH, Driscoll PC. The 3D solution structure of the C-terminal region of Ku86 (Ku86CTR) J Mol Biol. 2004;335:573–582. doi: 10.1016/j.jmb.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Hu W, Cano L, Lee TD, Chen DJ, Chen Y. Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions. Structure (Camb) 2004;12:495–502. doi: 10.1016/j.str.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, Abolfath RM, Chen DJ, Lees-Miller SP, Tainer JA. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gell D, Jackson SP. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 1999;27:3494–3502. doi: 10.1093/nar/27.17.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 33.Tamura K, Adachi Y, Chiba K, Oguchi K, Takahashi H. Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA double-strand breaks. Plant J. 2002;29:771–781. doi: 10.1046/j.1365-313x.2002.01258.x. [DOI] [PubMed] [Google Scholar]
- 34.Singleton BK, Torres-Arzayus MI, Rottinghaus ST, Taccioli GE, Jeggo PA. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol Cell Biol. 1999;19:3267–3277. doi: 10.1128/mcb.19.5.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weterings E, Verkaik NS, Keijzers G, Florea BI, Wang SY, Ortega LG, Uematsu N, Chen DJ, van Gent DC. The Ku80 carboxy terminus stimulates joining and artemis-mediated processing of DNA ends. Mol Cell Biol. 2009;29:1134–1142. doi: 10.1128/MCB.00971-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suwa A, Hirakata M, Takeda Y, Jesch SA, Mimori T, Hardin JA. DNA-dependent protein kinase (Ku protein-p350 complex) assembles on double-stranded DNA. Proc Natl Acad Sci U S A. 1994;91:6904–6908. doi: 10.1073/pnas.91.15.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yano KI, Morotomi-Yano K, Lee KJ, Chen DJ. Functional significance of the interaction with Ku in DNA double-strand break recognition of XLF. FEBS Lett. 2011;585:841–846. doi: 10.1016/j.febslet.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grundy GJ, Rulten SL, Zeng Z, Arribas-Bosacoma R, Iles N, Manley K, Oliver A, Caldecott KW. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013;32:112–125. doi: 10.1038/emboj.2012.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirodkar P, Fenton AL, Meng L, Koch CA. Identification and functional characterization of a Ku-binding motif in aprataxin polynucleotide kinase/phosphatase-like factor (APLF) J Biol Chem. 2013;288:19604–19613. doi: 10.1074/jbc.M112.440388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, Houtsmuller AB, van Gent DC. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, Admon A, Lees-Miller SP, Anderson CW, Jackson SP. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 43.Bosotti R, Isacchi A, Sonnhammer EL. FAT: a novel domain in PIK-related kinases. Trends Biochem Sci. 2000;25:225–227. doi: 10.1016/s0968-0004(00)01563-2. [DOI] [PubMed] [Google Scholar]
- 44.Williams DR, Lee KJ, Shi J, Chen DJ, Stewart PL. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals alpha helices and insight into DNA binding. Structure. 2008;16:468–477. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. Embo J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villarreal SA, Stewart PL. CryoEM and image sorting for flexible protein/DNA complexes. J Struct Biol. 2013 doi: 10.1016/j.jsb.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 48.Williams RS, Tainer JA. Learning our ABCs: Rad50 directs MRN repair functions via adenylate kinase activity from the conserved ATP binding cassette. Mol Cell. 2007;25:789–791. doi: 10.1016/j.molcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 49.Groves MR, Hanlon N, Turowski P, Hemmings BA, Barford D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell. 1999;96:99–110. doi: 10.1016/s0092-8674(00)80963-0. [DOI] [PubMed] [Google Scholar]
- 50.Grinthal A, Adamovic I, Weiner B, Karplus M, Kleckner N. PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis. Proc Natl Acad Sci U S A. 2010;107:2467–2472. doi: 10.1073/pnas.0914073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meek K, Dang V, Lees-Miller SP. DNA-PK: the means to justify the ends? Adv Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 52.Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature. 2013;497:217–223. doi: 10.1038/nature12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walker EH, Perisic O, Ried C, Stephens L, Williams RL. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature. 1999;402:313–320. doi: 10.1038/46319. [DOI] [PubMed] [Google Scholar]
- 54.Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 55.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 56.Jin S, Kharbanda S, Mayer B, Kufe D, Weaver DT. Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J Biol Chem. 1997;272:24763–24766. doi: 10.1074/jbc.272.40.24763. [DOI] [PubMed] [Google Scholar]
- 57.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 59.Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L, Morio T, Minegishi Y, Nakada S, Nagasawa M, Komatsu K, Chessa L, Villa A, Lecis D, Delia D, Mizutani S. Ataxia-telangiectasia-mutated dependent phosphorylation of Artemis in response to DNA damage. Cancer Sci. 2005;96:134–141. doi: 10.1111/j.1349-7006.2005.00019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poinsignon C, de Chasseval R, Soubeyrand S, Moshous D, Fischer A, Hache RJ, de Villartay JP. Phosphorylation of Artemis following irradiation-induced DNA damage. Eur J Immunol. 2004;34:3146–3155. doi: 10.1002/eji.200425455. [DOI] [PubMed] [Google Scholar]
- 62.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. Embo J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 64.Malu S, De Ioannes P, Kozlov M, Greene M, Francis D, Hanna M, Pena J, Escalante CR, Kurosawa A, Erdjument-Bromage H, Tempst P, Adachi N, Vezzoni P, Villa A, Aggarwal AK, Cortes P. Artemis C-terminal region facilitates V(D)J recombination through its interactions with DNA Ligase IV and DNA-PKcs. J Exp Med. 2012;209:955–963. doi: 10.1084/jem.20111437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soubeyrand S, Pope L, De Chasseval R, Gosselin D, Dong F, de Villartay JP, Hache RJ. Artemis phosphorylated by DNA-dependent protein kinase associates preferentially with discrete regions of chromatin. J Mol Biol. 2006;358:1200–1211. doi: 10.1016/j.jmb.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 66.De Ioannes P, Malu S, Cortes P, Aggarwal AK. Structural basis of DNA ligase IV-Artemis interaction in nonhomologous end-joining. Cell Rep. 2012;2:1505–1512. doi: 10.1016/j.celrep.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ochi T, Gu X, Blundell TL. Structure of the catalytic region of DNA ligase IV in complex with an Artemis fragment sheds light on double-strand break repair. Structure. 2013;21:672–679. doi: 10.1016/j.str.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, Tainer JA. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sengerova B, Allerston CK, Abu M, Lee SY, Hartley J, Kiakos K, Schofield CJ, Hartley JA, Gileadi O, McHugh PJ. Characterization of the human SNM1A and SNM1B/Apollo DNA repair exonucleases. J Biol Chem. 2012;287:26254–26267. doi: 10.1074/jbc.M112.367243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 72.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem Cell Biol. 2013;91:31–41. doi: 10.1139/bcb-2012-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hammel M, Yu Y, Fang S, Lees-Miller SP, Tainer JA. XLF Regulates Filament Architecture of the XRCC4.Ligase IV Complex. Structure. 2010;18:1431–1442. doi: 10.1016/j.str.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney B, Weinfeld M, Schriemer DC, Lees-Miller SP, Tainer JA. XRCC4 interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double-strand break repair. J Biol Chem. 2011;286:32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol. 2001;8:1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- 78.Wu PY, Frit P, Meesala S, Dauvillier S, Modesti M, Andres SN, Huang Y, Sekiguchi J, Calsou P, Salles B, Junop MS. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol Cell Biol. 2009;29:3163–3172. doi: 10.1128/MCB.01895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reinhardt HC, Yaffe MB. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat Rev Mol Cell Biol. 2013;14:563–580. doi: 10.1038/nrm3640. [DOI] [PubMed] [Google Scholar]
- 80.Li M, Lu LY, Yang CY, Wang S, Yu X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013;27:1752–1768. doi: 10.1101/gad.226357.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 82.Cotner-Gohara E, Kim IK, Hammel M, Tainer JA, Tomkinson AE, Ellenberger T. Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry. 2010;49:6165–6176. doi: 10.1021/bi100503w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, Tomkinson AE, Tainer JA, Ellenberger T. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol Cell. 2006;24:279–291. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 84.Perry JJ, Cotner-Gohara E, Ellenberger T, Tainer JA. Structural dynamics in DNA damage signaling and repair. Curr Opin Struct Biol. 2010 doi: 10.1016/j.sbi.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Recuero-Checa MA, Dore AS, Arias-Palomo E, Rivera-Calzada A, Scheres SH, Maman JD, Pearl LH, Llorca O. Electron microscopy of Xrcc4 and the DNA ligase IV-Xrcc4 DNA repair complex. DNA Repair (Amst) 2009;8:1380–1389. doi: 10.1016/j.dnarep.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 86.Ochi T, Wu Q, Chirgadze DY, Grossmann JG, Bolanos-Garcia VM, Blundell TL. Structural insights into the role of domain flexibility in human DNA ligase IV. Structure. 2012;20:1212–1222. doi: 10.1016/j.str.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rambo RP, Tainer JA. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annual Review of Biophysics. 2013;42:415–441. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- 89.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hura GL, Tsai CL, Claridge SA, Mendillo ML, Smith JM, Williams GJ, Mastroianni AJ, Alivisatos AP, Putnam CD, Kolodner RD, Tainer JA. DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals. Proc Natl Acad Sci U S A. 2013;110:17308–17313. doi: 10.1073/pnas.1308595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hura GL, Budworth H, Dyer KN, Rambo RP, Hammel M, McMurray CT, Tainer JA. Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nat Methods. 2013;10:453–454. doi: 10.1038/nmeth.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 93.Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, McGown G, Thorncroft M, Santibanez-Koref MF, Millington C, Arvai AS, Kroeger MD, Peterson LA, Williams DM, Fried MG, Margison GP, Pegg AE, Tainer JA. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 95.Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, Cooper PK, Grasby JA, Tainer JA. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mani RS, Yu Y, Fang S, Lu M, Fanta M, Zolner AE, Tahbaz N, Ramsden DA, Litchfield DW, Lees-Miller SP, Weinfeld M. Dual modes of interaction between XRCC4 and polynucleotide kinase/phosphatase: implications for nonhomologous end joining. J Biol Chem. 2010;285:37619–37629. doi: 10.1074/jbc.M109.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Modesti M, Junop MS, Ghirlando R, van de Rakt M, Gellert M, Yang W, Kanaar R. Tetramerization and DNA ligase IV interaction of the DNA double-strand break repair protein XRCC4 are mutually exclusive. J Mol Biol. 2003;334:215–228. doi: 10.1016/j.jmb.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 98.Andres SN, Junop MS. Crystallization and preliminary X-ray diffraction analysis of the human XRCC4-XLF complex. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:1399–1402. doi: 10.1107/S1744309111033549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Andres SN, Vergnes A, Ristic D, Wyman C, Modesti M, Junop M. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 2012;40:1868–1878. doi: 10.1093/nar/gks022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. Embo J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kanno S, Kuzuoka H, Sasao S, Hong Z, Lan L, Nakajima S, Yasui A. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. Embo J. 2007;26:2094–2103. doi: 10.1038/sj.emboj.7601663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, Brenner C, Moreira MC, Taylor AM, Caldecott KW. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 2004;3:1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 103.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci. 2011;36:262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galicia S, Koch CA, Cass CE, Durocher D, Weinfeld M, Glover JN. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 105.Bernstein NK, Hammel M, Mani RS, Weinfeld M, Pelikan M, Tainer JA, Glover JN. Mechanism of DNA substrate recognition by the mammalian DNA repair enzyme, Polynucleotide Kinase. Nucleic Acids Res. 2009;37:6161–6173. doi: 10.1093/nar/gkp597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zolner AE, Abdou I, Ye R, Mani RS, Fanta M, Yu Y, Douglas P, Tahbaz N, Fang S, Dobbs T, Wang C, Morrice N, Hendzel MJ, Weinfeld M, Lees-Miller SP. Phosphorylation of polynucleotide kinase/ phosphatase by DNA-dependent protein kinase and ataxia-telangiectasia mutated regulates its association with sites of DNA damage. Nucleic Acids Res. 2011;39:9224–9237. doi: 10.1093/nar/gkr647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Segal-Raz H, Mass G, Baranes-Bachar K, Lerenthal Y, Wang SY, Chung YM, Ziv-Lehrman S, Strom CE, Helleday T, Hu MC, Chen DJ, Shiloh Y. ATM-mediated phosphorylation of polynucleotide kinase/phosphatase is required for effective DNA double-strand break repair. EMBO Rep. 2011;12:713–719. doi: 10.1038/embor.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Iles N, Rulten S, El-Khamisy SF, Caldecott KW. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol Cell Biol. 2007;27:3793–3803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Macrae CJ, McCulloch RD, Ylanko J, Durocher D, Koch CA. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair (Amst) 2008;7:292–302. doi: 10.1016/j.dnarep.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 110.Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 111.Li S, Kanno S, Watanabe R, Ogiwara H, Kohno T, Watanabe G, Yasui A, Lieber MR. Polynucleotide kinase and aprataxin-like forkhead-associated protein (PALF) acts as both a single-stranded DNA endonuclease and a single-stranded DNA 3′ exonuclease and can participate in DNA end joining in a biochemical system. J Biol Chem. 2011;286:36368–36377. doi: 10.1074/jbc.M111.287797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 113.Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, Ahel I, Neuhaus D. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nat Struct Mol Biol. 2010;17:241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mehrotra PV, Ahel D, Ryan DP, Weston R, Wiechens N, Kraehenbuehl R, Owen-Hughes T, Ahel I. DNA repair factor APLF is a histone chaperone. Mol Cell. 2011;41:46–55. doi: 10.1016/j.molcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rass U, Ahel I, West SC. Molecular mechanism of DNA deadenylation by the neurological disease protein aprataxin. J Biol Chem. 2008;283:33994–34001. doi: 10.1074/jbc.M807124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reynolds JJ, El-Khamisy SF, Katyal S, Clements P, McKinnon PJ, Caldecott KW. Defective DNA ligation during short-patch single-strand break repair in ataxia oculomotor apraxia 1. Mol Cell Biol. 2009;29:1354–1362. doi: 10.1128/MCB.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rulten SL, Caldecott KW. DNA strand break repair and neurodegeneration. DNA Repair (Amst) 2013;12:558–567. doi: 10.1016/j.dnarep.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 118.Gu J, Lieber MR. Mechanistic flexibility as a conserved theme across 3 billion years of nonhomologous DNA end-joining. Genes Dev. 2008;22:411–415. doi: 10.1101/gad.1646608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Costantini S, Woodbine L, Andreoli L, Jeggo PA, Vindigni A. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair (Amst) 2007;6:712–722. doi: 10.1016/j.dnarep.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 120.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Neal JA, Meek K. Choosing the right path: does DNA-PK help make the decision? Mutat Res. 2011;711:73–86. doi: 10.1016/j.mrfmmm.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dai Y, Kysela B, Hanakahi LA, Manolis K, Riballo E, Stumm M, Harville TO, West SC, Oettinger MA, Jeggo PA. Nonhomologous end joining and V(D)J recombination require an additional factor. Proc Natl Acad Sci U S A. 2003;100:2462–2467. doi: 10.1073/pnas.0437964100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu X, Jiang W, Dubois RL, Yamamoto K, Wolner Z, Zha S. Overlapping functions between XLF repair protein and 53BP1 DNA damage response factor in end joining and lymphocyte development. Proc Natl Acad Sci U S A. 2012;109:3903–3908. doi: 10.1073/pnas.1120160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oksenych V, Alt FW, Kumar V, Schwer B, Wesemann DR, Hansen E, Patel H, Su A, Guo C. Functional redundancy between repair factor XLF and damage response mediator 53BP1 in V(D)J recombination and DNA repair. Proc Natl Acad Sci U S A. 2012;109:2455–2460. doi: 10.1073/pnas.1121458109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, Dubois RL, Alt FW. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2011;469:250–254. doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci U S A. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 128.Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol Cell. 2008;31:631–640. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, Ismail A, Ismalaj E, Petricci E, Neale MJ, Bristow RG, Masson JY, Wyman C, Jeggo PA, Tainer JA. DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities. Mol Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Goedecke W, Eijpe M, Offenberg HH, van Aalderen M, Heyting C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet. 1999;23:194–198. doi: 10.1038/13821. [DOI] [PubMed] [Google Scholar]
- 131.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 134.Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal Structure of Human XLF: A Twist in Nonhomologous DNA End-Joining. Mol Cell. 2007;28:1093–1101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 136.Ropars V, Drevet P, Legrand P, Baconnais S, Amram J, Faure G, Marquez JA, Pietrement O, Guerois R, Callebaut I, Le Cam E, Revy P, de Villartay JP, Charbonnier JB. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc Natl Acad Sci U S A. 2011;108:12663–12668. doi: 10.1073/pnas.1100758108. [DOI] [PMC free article] [PubMed] [Google Scholar]