Abstract
Alkyl isocyanides (CNRs) identify pathways for diatomic ligand movement into and out of Mb, with their side chains acting as transition state analogs. The bound alkyl groups point either into the back of the distal pocket (in conformation, νCN ≈ 2070–2090 cm−1), which allows hydrogen bond donation from His64(E7) to the isocyano group, or toward solvent through an open His(E7) channel (out conformation, νCN ≈ 2110–2130 cm−1), which prevents polar interactions with the isocyano atoms. Fractions of the in conformer (Fin) were measured by FTIR spectroscopy for methyl through n-pentyl isocyanide bound to a series of 20 different distal pocket mutants of sperm whale myoglobin and found to be governed by the ease of rotation of the His(E7) side chain, distal pocket volume and steric interactions, and, for the longer isocyanides, the unfavorable hydrophobic effect of placing their terminal carbon atoms into the solvent phase in the out conformation. There are strong correlations between the fraction of in conformer, Fin, for long chain MbCNR complexes measured by FTIR spectroscopy, the fraction of geminate recombination of photodissociated O2, and the bimolecular rates of O2 entry into the distal pocket. These correlations indicate that alkyl isocyanides serve as transition state analogs for the movement of O2 into and out of the binding pocket of Mb.
Keywords: myoglobin, globin, alkyl isocyanide, isonitrile, transition state, FTIR, infrared, vibrational spectroscopy, geminate recombination, iron coordination
In the preceding paper, we used crystal structures of alkyl isocyanides bound to Mb (MbCNRs) to visualize the entry and exit channels involved in diatomic ligand binding (1). Isocyanides coordinate to the Mb heme iron, and the alkyl side chain (R-group) is relatively free to rotate and occupy the lowest free energy positions adjacent to the active site, which are presumably the spaces and channels most accessible to the movement of the physiologically relevant ligands, O2, CO, and NO. Two major isocyanide conformations are observed in the crystal structures of native and wild-type (wt) MbCNR complexes that correlate with the two major isocyanide stretching frequency bands observed in solution FTIR spectra (1, 2). The low frequency (~2075 cm−1) νCN band represents the in conformer, in which the bound alkyl chain is pointing toward the protein interior. In this orientation, the distal histidine gate is closed and forms a hydrogen bond to the bound isocyano group and decreases its bond order. The high frequency (~2125 cm−1) νCN band represents the out conformer, in which the ligand side chain is pointing toward solvent through an open E7 channel created by outward movement of His64 side chain. In this orientation, the bound isocyano group is in an apolar environment, which favors a larger bond order (2).
In the case of bound n-butyl isocyanide (CNC4), the fractions of in and out conformations are roughly equal in solution, indicating that the unfavorable free energy for packing the four butyl carbon atoms in the interior of the distal pocket is roughly equal to that for opening the distal histidine gate and filling the E7 channel (Fig. 1). In the previous paper we suggested that the inward- and outward-pointing positions of the butyl side chain represent, respectively, the initial and final pathways for the movement of photo- and thermally dissociated diatomic ligands.
Figure 1. Stereo view of amino acids selected for mutation near bound CNRs.
The binding pockets are shown for native (pH 7) and wt (pH 9) MbCNC4 (104m and 111m, respectively, (1)) and are globally aligned by their Cα atoms. Mutation of the residues shown in dark blue are expected to affect the free energy of CNC4 in the in conformation (slate blue) by increased or decreased steric hindrance, whereas those in brown should selectively affect the out conformer of CNC4 (orange). The heme groups are in white, and van der Waals spheres for CNC4 are shown to provide an indication of residue/ligand packing. The alkyl groups of bound CNRs may act as transitions state analogs for CO in the photodissociated B state or during entry into the binding pocket through the His64 “gate” (marked as CO not observable). CO in the dissociated B state has been observed directly in low temperature and time-resolved crystallography experiments, whereas, to date, ligand movement through the E7 gate has not been observed directly.
Olson, Gibson, Phillips, and coworkers (3, 4) have proposed that diatomic ligands enter mammalian Mbs and hemoglobins through the distal His(E7) gate and are then captured in the interior portion of the distal pocket and adjacent Xe binding cavities before binding to the iron atom. They suggested the metaphor of catching of a ball in a baseball glove. Opening of the glove by movement of the thumb is analogous to the outward movement of the His(E7) side chain, and catching in the web of the glove is analogous to ligand capture in the interior cavities of the globin.
Ligand dissociation represents a reversal of this process and can be examined directly by laser photolysis of Mb-ligand complexes. Gibson, Scott, and Olson (3, 4) described the internal ligand movements and geminate recombination reaction in terms of a side path scheme. Photolysis breaks the Fe-ligand bond causing the dissociated ligand to move into the initial B state, which is located directly above the interior pyrrole rings of the heme group. From there, the ligand can either move into the Xe4 and Xe1 sites (states C and D), rebind, or escape through the His(E7) gate (Fig. 1). Movement into the C and D sites represents a side path from which the ligand rarely leaves the protein. Instead, it returns to the distal pocket to either rebind to the iron (geminate recombination) or escape from the protein.
We hypothesize that the CNR alkyl C atoms in the in conformation simulate the initial trajectory taken by photodissociated diatomic ligands into the B-state site, and that the alkyl carbon atoms in the out conformation mimic O2, CO, and NO at points along their escape path through the E7 channel (Fig. 1; (4, 5)). To examine the correspondence between the side path scheme and the isocyanide conformation, we systematically measured the effects of key distal pocket mutations at positions 29, 45, 46, 68, and 107 (Fig. 1) on the fraction of in versus out CNR conformers as measured by FTIR spectroscopy. The results were then compared to previously measured rates of O2 entry and fractions of geminate rebinding for the same library of Mb variants (4). The resultant correlations between these independently measured parameters are striking and support strongly the idea that the positions of bound isocyanide side chains do identify the pathways for diatomic ligand movement in myoglobin.
MATERIALS AND METHODS
Detailed methods can be found in the first paper of this series (2). Recombinant Mbs were expressed in E. coli and purified as described by Springer and Sligar (6) and modified by Carver et al. (7). The CNRs were synthesized by our group or by Mark Hargrove’s group at Iowa State University using the methods of Casanova et al. (8–10).
Geminate recombination measurements were carried out for MbCNRs in N2-equilibrated, pH 7.0 buffer containing 100 mM potassium phosphate, 1 mM EDTA, 50–100 μM Mb and 100–1000 μM CNR. Rate constants for CNR binding were measured as described in (11). We chose not to determine all the rate parameters for the isocyanides because our goal was to correlate the position of the ligand side chain with the rate constants for diatomic ligand binding and not to study isocyanide binding per se as has been done previously for various naturally occurring globins (10, 12) and His64 mutants (11).
FTIR measurements were conducted by loading into a 40 μm CaF2 cell a ~20 μl sample of 2–5 mM Mb and 1X and 5X molar equivalents of CNR and sodium dithionite, respectively. All solutions contained N2-equilbrated, 100 mM potassium phosphate, 1 mM EDTA, pH 7.0 buffer. The FTIR spectra were collected on a Nicolet Nexus 470 FTIR spectrometer and have a 1 or 2 cm−1 resolution,
RESULTS
Effects of Mutations that Alter the Flexibility of His64(E7) Side Chain
Substitutions of His64 clearly alter ligand entry and exit rates. Scott et al. (4) observed a roughly linear dependence of the O2 entry rate on the size of the side chain at position E7 for the series of relatively apolar amino acids, Gly, Ala, Val, Leu, Phe, and Trp. They also observed that mutations at locations spatially near the distal histidine, Arg45(CD3) and Phe46(CD4), could cause significant increases in the rates of ligand entry and exit by enhancing the mobility of His64 side chain.
The Arg45(CD3) guanidinium group is part of a hydrogen bonding network that includes the Nδ of His64, the heme propionates, and water molecules when His64 is in the closed conformation (13, 14). Disruption of this hydrogen bonding network moderately enhances the rates of ligand entry and exit by destabilizing the closed conformation of the histidine gate. The Phe46(CD4) benzyl group sterically hinders outward rotation of the His64 side chain. In the crystal structure of F46V MbCO, His64 rotates upward to fill the space left by removal of the phenyl group and, in doing so, opens a pore between the binding pocket and the solvent phase near the heme propionates (15). Smith et al. (1) observed an identical opening of the His64 side chain in the F46V MbCNC4 crystal structure. In the latter case, the pore is filled with the butyl group of the bound isocyanide in the out conformation (Fig. 2C).
Figure 2. Effect of mutating residues Arg45 and Phe46.
A. The νCN peaks in the FTIR spectra of methyl through pentyl isocyanides (CNC1-CNC5) bound to wt, R45K, R45E, and F46V Mbs. B. The fraction of in conformers (Fin) for each mutant in panel A as a function of CNR size. These values indicate that the distal histidine is pushed outward more easily in the order wt < R45K < R45E ≈ F46V by the alkyl tail of each ligand. C. Structures of native (orange) and F46V (slate blue) MbCNC4 (104M and 101M, respectively; (1)), globally aligned by their Cα atoms. The smaller Val46 side chain allows the His64 imidazole to rotate away from the terminal carbon of the ligand and relieve steric hindrance of the CNR in the out conformation.
The FTIR measurements shown in Fig. 2 support the idea that the R45K, R45E, and F46V mutations allow a greater ease of opening the E7 gate. Although, the conservative R45K substitution retains a positively charged amino acid at position 45(CD3), it causes small increases in the amplitudes of the νCN out peaks in all of the R45K MbCNR spectra (Fig. 2A). Loss of the three N atoms of the arginine guanidinium partially disrupts the hydrogen bonding network that stabilizes His64 in the closed conformation and allows more of the bound ligands to point outward. The larger increase in the amount of out conformer for the R45E mutant is due both to disruption of the hydrogen bond network and to introduction of a negative charge near His(E7). The net change in charge of −2 due to the R45E mutation increases the pKa of the His64 side chain, its extent of protonation at pH 7, and the amount of outward rotation of the resulting imidazolium cation. Carver et al. (13) studied a similar mutant, K45E pig Mb, and found that CNC3 and CNC4 had 10-fold increases in affinity over the wt protein (Table 1, last column). This effect is most likely caused by selective relief of steric hindrance on the bound CNR in the out conformation.
Table 1.
FTIR spectroscopy and ligand binding kinetics parameters measured for MbCNR complexes with R45 mutations E, and K, and F46V.a
| FTIR spectra data | CNR binding data | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| MbCNR complex | in νCN cm−1 | out νCN cm−1 | Fin | Fgem | k′ μM−1s−1 | k s−1 | Ka μM−1 |
| wt | |||||||
| CNC1 | 2083 | 2139 | 0.60 | 0.80 | 0.12b | 4.3b | 0.028b |
| CNC2 | 2065 | 2106 | 0.82 | 0.95 | 0.074b | 0.27b | 0.27b |
| CNC3 | 2077 | 2112 | 0.69 | 0.79 | 0.043b | 0.33b | 0.13b |
| CNC4 | 2083 | 2113 | 0.47 | 0.63 | 0.029b | 0.60b | 0.048b |
| CNC5 | 2081 | 2131 | 0.68 | 0.89 | 0.030c | 0.44c | 0.069c |
| R45E | |||||||
| CNC1 | 2083 | 2139 | 0.16 | 0.59d | 0.23d | 4.7d | 0.049d |
| CNC2 | 2077 | 2112 | 0.61 | 0.29d | 1.0d | 0.28d | |
| CNC3 | 2077 | 2118 | 0.29 | 0.23d | 0.46d | 0.51d | |
| CNC4 | 2077 | 2118 | 0.20 | 0.18d | 0.36d | 0.51d | |
| CNC5 | 2081 | 2121 | 0.29 | ||||
| R45K | |||||||
| CNC1 | 2082 | 2139 | 0.45 | 0.78d | 0.11d | 4.4d | 0.025d |
| CNC2 | 2071 | 2107 | 0.76 | 0.098d | 0.63d | 0.15d | |
| CNC3 | 2082 | 2116 | 0.55 | 0.063d | 0.70d | 0.090d | |
| CNC4 | 2078 | 2116 | 0.36 | 0.040d | 0.50d | 0.081d | |
| CNC5 | 2085 | 2122 | 0.57 | ||||
| F46V | |||||||
| CNC1 | 2083 | 2131 | 0.07 | 0.47 | |||
| CNC2 | 2079 | 2110 | 0.52 | 0.54 | |||
| CNC3 | 2075 | 2110 | 0.37 | 0.42 | |||
| CNC4 | 2074 | 2112 | 0.26 | 0.30 | 0.22e | 0.63e | 0.36e |
| CNC5 | 2074 | 2118 | 0.16 | ||||
The infrared spectra for methyl through pentyl isocyanide bound to F46V Mb are also shown in Fig. 2A. The fractions of in conformers for F46V Mb are roughly 2 to 10-fold lower than their corresponding wt Mb values (Fin, Fig. 2B). Clearly, the histidine gate in this mutant is easily opened (Fig. 2C). Although the ligand binding data is limited to CNC4, the out conformer in the F46V mutant is less hindered than either conformer in wt Mb, and as a result, the affinity of F46V Mb for CNC4 is roughly 10-fold higher than that for wt Mb and similar to that for K45E pig Mb (Table 1).
For wt Mb and the R45K and R45E mutants, a plot of Fin versus CNR size has an undulating up-down-up shape with CNC2 and CNC4 at the vertices (Fig 2B, tops of the bars for each variant). Blouin and Olson (2) postulated that the increase in Fin for CNC2 compared to CNC1 is due to the inductive effect of the ethyl group, which favors more unpaired electron density on the N atom for interaction with His64. The space occupied by the ligand Cβ is unhindered and is in the same location as the initial position of photodissociated ligands (Fig. 1). The decrease in Fin for CNC2 through CNC4 is due to steric constraints on the Cγ and Cδ atoms of the alkyl side chain, which begin to push the n-propyl and n-butyl groups out of the pocket and into the E7 channel. CNC5 is long enough to access solvent directly outside the binding pocket in the out conformer, where it disrupts the water hydrogen-bonding lattice. Thus, for the longest ligands, the hydrophobic effect becomes dominate and the fraction in becomes higher again for both CNC5 (Fig. 2) and CNC6 (see Fig. 6C).
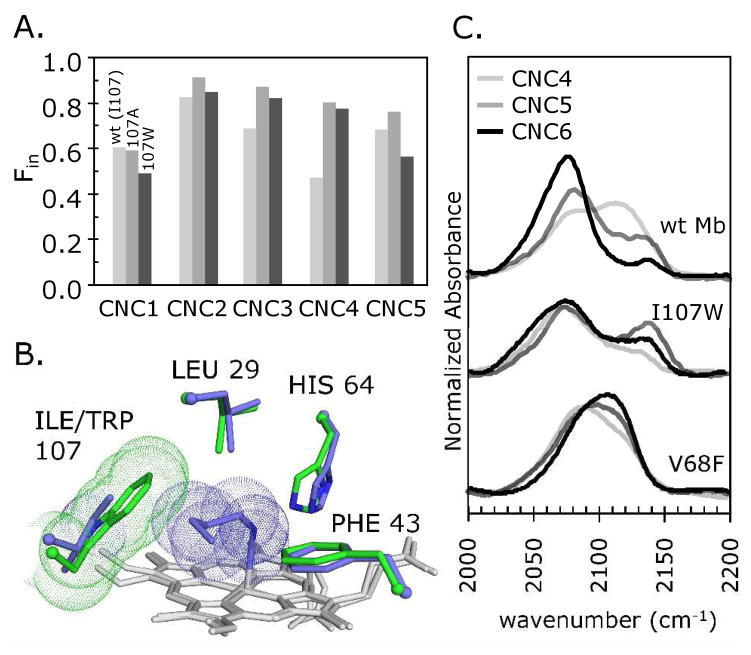
Figure 6. Effect of side chain size at residue 107 and blocking the Xe4 pocket.
A. Fin plotted against ligand size for methyl through pentyl isocyanides (CNC1-CNC5) bound to wt, I107A and I107W Mbs. B. Native MbCNC4 (slate blue; pH 9 structure; PDB ID 105m; (1)) and I107W Mb (green; ferric form; PDB ID 2ohb; (5)) globally aligned by their Cα atoms, with van der Waals spheres shown for CNC4 and the Trp107 side chain. The Fin value for the actual I107W MbCNC4 complex is unexpectedly high (panel A). C. FTIR spectra of CNC4-CNC6 bound to wt, I107W and V68F Mbs. Mutations that block access to the Xe4 site inhibit inward movement of longer CNRs, as shown by increases in the high-frequency out conformation band at ~2125 cm−1.
However, for F46V Mb, Fin continues to decrease for CNC5 indicating that despite the unfavorable hydrophobic effect, the open conformation is more favorable than packing the five alkyl carbon atoms in the interior of the distal pocket. Superimposition of the native MbCNC4 (P21, pH 7) and F46V MbCNC4 (P6, pH 9) crystal structures (1) shows that, although the distal histidine in both structures is rotated into an open conformation, the space left by the removal of the Phe46 phenyl group allows the His64 imidazole to rotate further away from the ligand alkyl atoms in the out conformation. As a result, there is a clearance of 3.8 Å between the closest atom of the His64 side chain (Cδ) and the terminal carbon of the CNC4 alkyl group in F46V Mb. In contrast, this distance is 3.3 Å in the native Mb structure (Fig. 2C).
Correlations between Fin and Rates of O2 and NO Entry for Mutations at the E7 Gate
The values of Fin for the R45E, R45K, and F46V MbCNC5 complexes show the greatest variation (Fig. 2B, last set of bars) and were compared to previously determined rate constants for O2 entry, k′entry,O2, and fraction of O2 geminate recombination, Fgem,O2, for the same set of Mb variants (Fig. 3). As shown in Fig. 3, there is a strong linear correlation (R2 values ≥ 0.9) between Fin for CNC5 measured by FTIR and Fgem,O2 (4). The positive slopes confirm that larger Fin values, which indicate a higher free energy barrier to E7 gate opening, are associated with larger Fgem values for O2, which indicate greater trapping of diatomic ligands in the distal pocket after photolysis.
Figure 3. The CNR out conformation acts as a transition state analog for diatomic ligand entry.
A. Correlations between the fraction of geminate recombination of photolyzed O2, Fgem,O2, and the fraction of bound CNC5 that adopts the in conformation, Fin,CNC5, for Mb mutants with substitutions near the His(E7) gate, including R45K, R45E and F46V. Fgem,O2 values were taken from (4). B. Correlations for wt, R45K, R45E, and F46V Mbs between the logs of k′entry,O2 or k′NO and Fin,CNC5.
The conclusion that His(E7) in Mb controls ligand access to the Mb distal pocket is supported by the inverse correlation of the fraction of in CNC5 conformer with both the computed rates of ligand entry for O2 binding, k′entry, and the directly measured association rate constants for NO binding, k′NO, for the position 45 and 46 mutants (Fig. 3B). For NO, the fraction of internal binding versus escape is always ≥ 0.99 due to the high reactivity of the NO radical, and thus the rate limiting step for NO binding is entry into the distal pocket (4). This inverse correlation demonstrates that a lower barrier to opening the histidine gate, indicated by a smaller Fin,CNC5, predicts significantly higher bimolecular rates of diatomic ligand entry.
Similar, though less strong, correlations of Fin for the CNC1 through CNC4 complexes of these mutants with Fgem,O2, k′entry,O2, and k′NO are observed (data not shown). The close proximity of His64 to the Mb heme iron allows any bound CNR, regardless of its size, to act as a probe of the stability of its closed versus open conformations. Longer CNRs are required to study the effects of mutations at positions deeper in the binding pocket and to indicate the importance of capture volume in the reactions of diatomic ligands with Mb.
Effects of Val68(E11) Mutations: Decreasing and Enhancing Direct Steric Hindrance and Blocking Access to the Xe4 Pocket
The distal valine(E11) has multiple effects on ligand binding to Mb including: (a) sterically hindering access to the heme iron (17–19), (b) affecting the position of the distal histidine in its closed conformation, (c) lining a portion of both the E7 channel, and (d) being part of the short path between the initial geminate or B state and the more interior Xe4 cavity (Fig. 1). Thus, mutations at the E11 position have complex, sometimes competing effects on bimolecular and geminate rebinding and on the orientation of bound isocyanides.
The V68A mutation in Mb results in a large decrease in steric pressure on all bound CNRs, as judged by the 10 to 100-fold increases in overall affinity (Ka values in Table 2). However, the effects of this release of hindrance are different for the short versus the long straight-chained ligands (see the spectra in Fig. 4A, and the first two bars of each set in Fig. 4B). The Fin values for CNC1, CNC2 and CNC3 bound to the V68A mutant are all smaller than those for wt Mb, indicating that opening up of the E7 channel due to loss of the adjacent valine γ2 methyl group is the dominant effect. In contrast, the Fin values for CNC4 and CNC5 bound to V68A Mb are larger than the wt values (Fig. 4B; Table 2), indicating that loss of the γ1 methyl group creates more space in the back of the distal pocket for a direct connection to the Xe4 cavity and accommodation of the terminal carbon atoms of the n-butyl and n-pentyl side chains (Fig. 1). This increase in space allows the hydrophobic effect to push the side chains of the longer ligands back into the protein interior (Fig. 4B; Table 2). These differences roughly correlate with the differences in the fractions of geminate recombination observed for the rebinding of these ligands to wt and V68A Mb after laser photolysis. The short chain isocyanides show smaller while the longer ligands show larger Fgem and Fin values (Fig. 4B; Table 2).
Table 2.
FTIR spectroscopy and ligand binding kinetics parameters measured for MbCNR complexes with V68 mutations A, I, F, L and T.a
| FTIR spectra data | CNR binding data | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| MbCNR complex | in νCN cm−1 | out νCN cm−1 | Fin | Fgem | k′ μM−1s−1 | k s−1 | Ka μM−1 |
| wt | |||||||
| CNC1 | 2083 | 2139 | 0.60 | 0.80 | 0.12b | 4.3b | 0.028b |
| CNC2 | 2065 | 2106 | 0.82 | 0.95 | 0.074b | 0.27b | 0.27b |
| CNC3 | 2077 | 2112 | 0.69 | 0.79 | 0.043b | 0.33b | 0.13b |
| CNC4 | 2083 | 2113 | 0.47 | 0.63 | 0.029b | 0.60b | 0.048b |
| CNC5 | 2081 | 2131 | 0.68 | 0.89 | 0.030c | 0.44c | 0.069c |
| CNC6 | 2078 | 2137 | 0.90 | 0.97 | 0.037c | 0.15c | 0.25c |
| V68A | |||||||
| CNC1 | 2100 | 2137 | 0.38 | 0.66 | 0.38d | 0.76d | 0.50d |
| CNC2 | 2064 | 2106 | 0.69 | 0.83 | 0.18d | 0.07d | 2.6d |
| CNC3 | 2089 | 2121 | 0.61 | 0.97 | 0.11d | 0.022d | 5.0d |
| CNC4 | 2079 | 2121 | 0.60 | 0.99 | 0.14d | 0.011d | 13d |
| CNC5 | 2065 | 2121 | 0.75 | ||||
| V68I | |||||||
| CNC1 | 2091 | 2138 | 0.21 | 0.57e | 0.05d | 21d | 0.0024d |
| CNC2 | 2065 | 2095 | 0.53 | 0.63e | 0.047d | 3.4d | 0.014d |
| CNC3 | 2073 | 2116 | 0.37 | 0.016d | 1.5d | 0.011d | |
| CNC4 | 2092 | 2115 | 0.39 | 0.018d | 1.5d | 0.012d | |
| CNC5 | ND | 2118 | 0.00 | ||||
| V68F | |||||||
| CNC1 | 2098 | 2139 | 0.59 | 0.99 | 0.013d | 0.03d | 0.43d |
| CNC2 | 2064 | 2100 | 0.86 | 0.99 | 0.0061d | 0.0035d | 1.7d |
| CNC3 | 2079 | 2108 | 0.71 | 0.96 | 0.004d | 0.0019d | 2.1d |
| CNC4 | 2079 | 2108 | 0.53 | 0.95 | 0.0058d | 0.0077d | 0.75d |
| CNC5 | 2079 | 2107 | 0.46 | 0.44 | |||
| CNC6 | 2079 | 2107 | 0.40 | 0.72 | |||
| V68L | |||||||
| CNC1 | 2140 | ~0.0 | 0.36 | 1.7 | 0.21 | ||
| CNC2 | 2065 | 2100 | ~0.8 | 0.19 | 0.55 | 0.35 | |
| CNC3 | 2090 | 0.26 | 0.34 | 0.76 | |||
| CNC4 | 2090 | 0.22 | 0.13 | 1.7 | |||
| CNC5 | 2100 | ||||||
| V68T | |||||||
| CNC1 | 2118 | 2139 | 0.64 | 0.63f | 0.039f | 1.2f | 0.033f |
| CNC2 | 2085 | 2109 | 0.65 | 0.67f | 0.018f | 0.092f | 0.20f |
| CNC3 | 2095 | 2121 | 0.64 | 0.0095f | 0.082f | 0.12f | |
| CNC4 | 2080 | 2127 | 0.67 | 0.0088f | 0.16f | 0.056f | |
| CNC5 | 2074 | 2142 | 0.86 | ||||
Figure 4. Effect of mutating residue V68.
A. FTIR spectra of methyl through pentyl isocyanides (CNC1-CNC5) bound to wt and V68A, V68F, and V68I Mbs. B. Fin for each mutant-isocyanide pair as a function of ligand size. C. Native (orange) and wt (slate blue) MbCNC4 and V68I MbCO (green) globally aligned by their Cα atoms (PDB IDs 104m, 111m (1) and 1mlm (18), respectively). The V68I MbCO structure has two Ile68 χ2 rotomers (A and B). Hemes are in white. D. The same structural overlay as in panel C, except V68I MbCO is replaced by V68F MbCNC4 (PDB ID 107m; (1)).
In contrast to the V68A mutation, which reduces steric hindrance of the bound ligands, the V68I mutation increases direct hindrance significantly and decreases the affinities for O2, CO, and the alkyl isocyanides CNC1 through CNC4 by 4-fold to 20-fold (Table 2; (17)). The Cδ methyl group of isoleucine is in van der Waals contact with bound CO in the X-ray structure of V68I MbCO and is found in two rotomers indicating significant hindrance and disorder in the liganded complex (Fig. 4C, green sticks; (18)).
The IR spectra of V68I MbCNRs are complex with two to three broad peaks, reflecting the conformational heterogeneity observed in the V68I MbCO crystal structure. The three νCN bands are assignable based on their alignments with the wt MbCNR peaks. One low frequency peak probably represents an in conformation, and the two high frequency peaks probably represent out conformations. The splitting of the out conformer peak may be due to the presence of both Ile68 rotomers shown in Fig. 4C. Despite this complexity, it is clear from the spectra in Fig. 4A that the dominant effect of the V68I mutation is to push the alkyl isocyanides into an out conformation, which opens the His(E7) gate. This effect is most dramatic for the longer isocyanides where ≥60% and 100% of CNC4 and CNC5 conformers, respectively, are pointing outward.
The V68F mutation offers a test of the ability of longer alkyl isocyanides to access the distal Xe4 cavity. In the crystal structures of V68F Mb, the benzene ring of Phe68 partially fill and block access to the Xe4 cavity (Fig. 4D, green sticks). The lack of an effect of the Phe68 mutation on Fin for CNC1, CNC2 and CNC3 demonstrates that, as in the crystal structure, the 68(E11) phenyl group does not significantly intrude into the distal pocket and hinder bound ligands (Fig. 4B; Table 2). Fin for CNC4 is also unchanged by this mutation, although in the crystal structure, the Phe68 side chain sterically hinders the terminal carbon of the CNC4 butyl group (Fig. 4D). However, the V68F mutation does significantly decrease Fin for CNC5 from 0.68 in wt Mb to 0.46 in the mutant (Fig. 4B; Table 2). The increase in steric hindrance on CNC5 in the in conformation due to blockage of the Xe4 cavity by Phe68 partially overcomes the unfavorable hydrophobic forces on the ligand in the out conformation (2). The effect of decreasing Fin by blocking the Xe4 cavity is even larger for the longer CNC6 ligand, as shown in Fig. 6.
The IR spectra of the V68L MbCNR complexes are difficult to interpret because, except for CNC1, a single broad band is observed with a peak maximum midway between that for the normal in and out peaks of the wt complexes (Table 2; spectra not shown). Consequently Fin values could not be determined or used in the more global analyses described in DISCUSSION (Table 2). In the crystal structure of V68L metMb, deoxyMb, and MbCO, the isobutyl side chain occupies multiple conformations, which alternatively fill the back of the distal pocket or the space directly above the iron atom, displacing distal pocket water from the unliganded protein (18). As a result, an ensemble of isocyanide conformers is most likely present that cannot be described in terms of only one in and one out orientation.
The effect of increasing the distal pocket polarity was also examined by replacing Val68 with Thr. Following the work of Smerdon et al. (21), Li et al. found the V68T mutation causes a 17 cm−1 blue shift of the major νCO band of pig and sw MbCO and attributed this effect to the proximity of the negative dipole of the 68T hydroxyl group to the bound ligand atoms (22). There are similar large blue-shifts for the low frequency in peaks of CNC1, CNC2, and CNC3 bound to V68T Mb, but not for CNC4 and CNC5 (Table 2; spectra not shown).
Effects of Leu29(B10) Mutations
In globins, the B-helix forms part of the roof of the distal binding pocket above the heme, and the B10 side chain extends down toward the bound ligand (Figs. 1 and 5C). In mammalian Mbs and Hbs, the native Leu(B10) directs the movement of dissociated ligands either into the back of the distal pocket or out through the E7 gate and, if increased in size to Phe or Trp, can directly interact with bound diatomic ligands (7, 23–25). Substitution of Leu(B10) with the smaller Ala residue significantly increases the binding pocket volume available to bound isocyanides, causing the marked increases in ligand affinities, particularly for the larger isocyanides (Table 3). In contrast, the Phe(B10) replacement causes decreases in isocyanide affinity. However, remarkably, the Fin values for L29A and L29F MbCNR complexes determined by νCN measurements are similar to each other and to those for the wt MbCNRs.
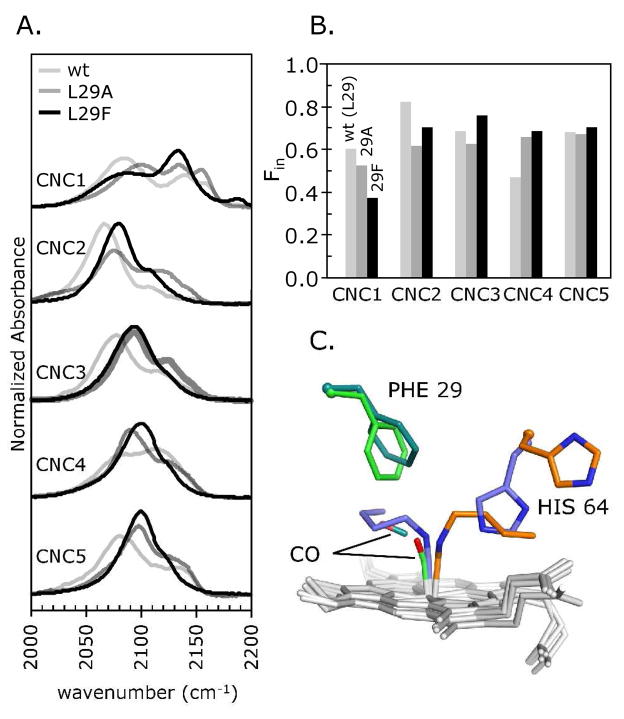
Figure 5. Effect of mutating residue Leu29.
A. FTIR spectra of methyl through pentyl isocyanides (CNC1-CNC5) bound to wt, L29A, and L29F Mbs. B. Fin for each mutant-isocyanide pair as a function of ligand size. C. Structures of native (orange) and wt (slate blue) MbCNC4, and L29F MbCO (light green) and L29F Mb:CO photoproduct (dark green), globally aligned by their Cα atoms (PDB IDs 104m, 111m (1), 2g0r and 2g0v (23), respectively; hemes in light gray). The benzyl side chain at residue 29 rotates to accommodate photodissociated CO in the B-state. A similar rotation may explain how the large CNR alkyl groups are accommodated within the L29F Mb binding pocket.
Table 3.
FTIR spectroscopy and ligand binding kinetics parameters measured for MbCNR complexes with L29 mutations A, and F.a
| FTIR spectra data | CNR binding data | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| MbCNR complex | in νCN cm−1 | out νCN cm−1 | Fin | Fgem | k′ μM−1s−1 | k s−1 | Ka μM−1 |
| wt | |||||||
| CNC1 | 2083 | 2139 | 0.60 | 0.80 | 0.12b | 4.3b | 0.028b |
| CNC2 | 2065 | 2106 | 0.82 | 0.95 | 0.074b | 0.27b | 0.27b |
| CNC3 | 2077 | 2112 | 0.69 | 0.79 | 0.043b | 0.33b | 0.13b |
| CNC4 | 2083 | 2113 | 0.47 | 0.63 | 0.029b | 0.60b | 0.048b |
| CNC5 | 2081 | 2131 | 0.68 | 0.89 | 0.030c | 0.44c | 0.069c |
| L29A | |||||||
| CNC1 | 2098 | 2154 | 0.53 | 0.62 | 0.58d | 7.7d | 0.075d |
| CNC2 | 2075 | 2119 | 0.62 | 0.97 | 0.70d | 0.63d | 1.1d |
| CNC3 | 2093 | 2123 | 0.63 | 0.97 | 0.45 | 0.023 | 20 |
| CNC4 | 2089 | 2126 | 0.66 | 0.98 | 2.0 | 0.020 | 100 |
| CNC5 | 2098 | 2134 | 0.68 | ||||
| L29F | |||||||
| CNC1 | 2084 | 2133 | 0.38 | 0.29 | 0.19d | 57d | 0.0033d |
| CNC2 | 2079 | 2108 | 0.71 | 0.44 | 0.090d | 4.2d | 0.021d |
| CNC3 | 2093 | 2127 | 0.76 | 0.42 | |||
| CNC4 | 2099 | 2123 | 0.69 | 0.28 | |||
| CNC5 | 2098 | 2124 | 0.71 | ||||
The FTIR results in Table 3 and Fig. 5 show that an increase or a decrease in the size of the B10 amino acid causes only small changes in Fin, even though ligand affinities change dramatically (Figs. 5B; Table 3). The in νCN peak is shifted to higher frequencies for both the L29A and L29F mutants suggesting a more upright conformation in both cases, and the amount of in conformer for the larger CNC4 ligands is significantly greater than for wt Mb. The latter result is easy to understand for the L29A mutant in which the internal distal cavity is much larger than that in wt Mb; however, the increase in Fin for Phe(B10) Mb is more difficult to rationalize. Aranda et al. (23) have shown that Phe(B10) can move away from photodissociated CO in time resolved X-ray crystallographic studies of L29F MbCO and does not appear to significantly obstruct the B-state site, which is readily occupied by photodissociated CO (Fig. 5C). The FTIR data also indicate that the long alkyl side chains of bound isocyanides are accommodated in the interior distal pocket of L29F MbCNR.
Effects of Ile107(G8) Mutations and Blocking Access to the Xe4 Cavity for Bound CNC5 and CNC6
The Ile107(G8) side chain is located at the back of the distal pocket and serves as a partial barrier to ligand movement into the Xe4 pocket (4). Substitution of alanine for isoleucine at G8 enhances the stability of ethyl through pentyl isocyanides in the in conformation, with CNC4 giving the greatest Fin increase from 0.47 in wt Mb to 0.80 in I107A Mb (Fig. 6A; Table 4; spectra not shown). Inspection of the wt MbCNC4 crystal structure shows that the Ile107 Cδ atom contacts the C3 and C4 atoms of the ligand butyl group along its C1-C4 path from the B state near the heme iron to the Xe4 pocket (Fig. 1). The structural and FTIR data indicate that crowding by the Ile107 side chain significantly contributes to the large free energy penalty of adding a fourth carbon to the CNR n-alkyl group in the in conformation, and that the further addition of a fifth carbon is energetically neutral because it is located within the Xe4 cavity.
Table 4.
FTIR spectroscopy and ligand binding kinetics parameters measured for MbCNR complexes with I107 mutations A, and W.a
| FTIR spectra data | CNR binding data | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| MbCNR complex | in νCN cm−1 | out νCN cm−1 | Fin | Fgem | k′ μM−1s−1 | k s−1 | Ka μM−1 |
| wt | |||||||
| CNC1 | 2083 | 2139 | 0.60 | 0.80 | 0.12b | 4.3b | 0.028b |
| CNC2 | 2065 | 2106 | 0.82 | 0.95 | 0.074b | 0.27b | 0.27b |
| CNC3 | 2077 | 2112 | 0.69 | 0.79 | 0.043b | 0.33b | 0.13b |
| CNC4 | 2083 | 2113 | 0.47 | 0.63 | 0.029b | 0.60b | 0.048b |
| CNC5 | 2081 | 2131 | 0.68 | 0.89 | 0.030c | 0.44c | 0.069c |
| CNC6 | 2078 | 2137 | 0.90 | 0.97 | 0.037c | 0.15c | 0.25c |
| I107A | |||||||
| CNC1 | 2089 | 2141 | 0.59 | 0.73 | 0.091 | 2.3 | 0.040 |
| CNC2 | 2067 | 2147 | 0.91 | 0.92 | 0.12 | 0.15 | 0.80 |
| CNC3 | 2083 | 2141 | 0.87 | 0.99 | 0.13 | 0.025 | 5.2 |
| CNC4 | 2081 | 2143 | 0.80 | 0.99 | 0.19 | 0.0088 | 22 |
| CNC5 | 2079 | 2144 | 0.76 | 0.99 | |||
| I107W | |||||||
| CNC1 | 2096 | 2143 | 0.49 | 0.87 | 0.18 | 0.21 | 0.86 |
| CNC2 | 2066 | 2147 | 0.85 | 0.94 | 0.16 | 0.025 | 6.4 |
| CNC3 | 2077 | 2139 | 0.82 | 0.96 | 0.017 | 0.029 | 0.59 |
| CNC4 | 2077 | 2141 | 0.78 | 0.87 | 0.062 | 0.044 | 1.4 |
| CNC5 | 2073 | 2137 | 0.57 | 0.91 | |||
| CNC6 | 2074 | 2136 | 0.68 | 0.97 | |||
Surprisingly, the I107W mutation appears to expand the distal pocket. The Trp107 mutant more readily accommodates moderately sized isocyanides, CNC2 to CNC4, as judged by significantly larger Fin values compared to those for wt Mb (Fig. 6A; Table 4). This result is consistent with the 30-fold greater Ka and the larger fraction of geminate recombination for CNC4 binding to I107W Mb than to wt Mb. The structure of I107W metMb shows that the Trp107 indole is sterically restricted by the edge of the heme group from rotating into the B-state associated with the initial CO photoproduct (Fig. 6B). However, this mutation does significantly lower the Fin values for bound CNC5 and CNC6 compared to their much higher wild-type values (Fig. 6A; Table 4). As is the case for V68F Mb (Figs. 4B and 4D), the indole ring of Trp107 appears to block access to the Xe4 cavity, and sterically hinders the ends of the longer CNRs as they wrap around the back of the distal pocket.
Complexes of n-hexyl isocyanide, CNC6, bound to wt Mb, V68F Mb and I107W Mb serve as tests of the ease of movement into the Xe4 cavity, which is described as state C in most photodissociation schemes. Figure 6C shows comparisons of the νCN spectra for these complexes and those for CNC4 and CNC5 bound to the same proteins. As described in the first paper of this series (2), there is a progressive increase in the inward-pointing population for CNC4, CNC5, and CNC6 bound to wt Mb, with Fin values of 0.47, 0.68 and 0.90, respectively (Fig. 6C, top set of spectra; Table 4). The size-dependent increase of the in conformation for these three CNRs is due to the terminal alkyl carbon atoms being driven out of the solvent phase and into the protein interior by a hydrophobic effect. There appear to be only small increases in steric hindrance for the C5 and C6 carbon atoms in the in conformation because they can be accommodated within the empty Xe4 cavity (2). This interpretation is supported by the effects of placing a tryptophan at 107(G8) or phenylalanine at 68(E11), which block access to the Xe4 cavity and partially reverse the trend to higher Fin values for the CNC4 to CNC6 series (Fig. 6C; Table 4). Thus, when access to the protein interior is sterically obstructed, there is a roughly 40% decrease in the inward-pointing population of the long chain isocyanides. Under these conditions, the hydrophobic effect driving inward movement roughly equals the steric hindrance caused by forcing the terminal carbon atoms into the protein interior.
Correlations between Fin and O2 binding parameters as function of distal pocket size
Scott et al. (4) analyzed the reactions of O2 with 90 mutants of Mb and found that the bimolecular entry rate for O2, defined as k′entry=k′overall/Fgem, was dependent on the volume available for ligand capture within the distal pocket and in the Xe4 cavity. To examine this idea further, we compared the measured values of Fin for MbCNR mutants containing small (Ala) and large (Trp or Phe) amino acids at the Leu29(B10), Val68(E11), and Ile107(G8) positions with the fractions of geminate O2 recombination (Fgem,O2), the microscopic geminate rate constants k′entry, kbond, and kescape for O2 binding, and the bimolecular association rate constants for NO binding, k′NO, taken from Scott et al. (4). The oxygen binding parameters only show significant correlations with the Fin values for the larger isocyanides, particularly CNC5, presumably because the bound smaller isocyanides cannot sense volume changes near the back of the distal pocket (see Figs. 1, 4D and 6B).
Remarkably, there is a significant inverse correlation between Fgem,O2 and Fin,CNC5 (R2 = 0.62; Fig. 7A). The fraction of geminate O2 recombination reflects the competition between internal ligand rebinding and escape to solvent on nanosecond timescales. The internal distal pocket mutations have little effect on O2 escape (Fig. 7D) but do markedly affect the speed of internal bond formation (Fig 7C). Reducing the volume sequesters the O2 molecule close to the iron atom increasing kbond markedly, whereas increasing the volume available to dissociated diatomic ligands decreases kbond due an increased entropic barrier to return to the heme center (4). Thus, the extent of geminate O2 recombination in I107W and V68F MbO2 is large, whereas the Fgem,O2 values for L29A and I107A MbO2 is small. The situations for L29F and V68A are more complex because of increased and decreased, respectively, direct hindrance of bound ligands.
Figure 7. Correlations between the Fin values for MbCNC5 complexes and parameters describing the binding kinetics of O2 for internal distal pocket mutants of Mb.
Fgem and the logarithms of k′entry and kbond, but not kescape, measured for the reaction of O2 with Leu29, Val68, and Ile107 Mb mutants show moderate correlations with the Fin values for bound CNC5. k′NO (gray circles, upper right panel) serves as an independent measure of the entry rate of diatomic gases. The O2 and NO kinetics data were taken from Scott et al. (4).
There is a strong linear correlation between Fin,CNC5 and log(k′entry) for diatomic ligands, as measured by k′NO or calculated from k′O2/Fgem,O2 (R2 = 0.81, Fig. 7B). The increase in k′entry with increasing accessible distal pocket volume measured by Fin for MbCNC5 complexes indicates that the net rate of entry is governed by the capture volume available for the incoming ligands as well as the ease of opening of the His(E7) gate. A larger volume increases the residence time of the ligand in the pocket, allowing the His(E7) gate to close before the ligand can return to solvent, increasing the net rate of ligand capture. In contrast, there is no correlation between Fin,CNC5 and kescape (Fig. 7, lower right), which is governed primarily by the extent and frequency of opening of the His(E7) gate and not the volume of the distal pocket.
DISCUSSION
The results presented here, in Blouin et al. (2), and in Smith et al. (1) demonstrate that bound straight-chain alkyl isocyanides act as probes of the spaces and pathways used by ligands to bind to myoglobin. The fraction of in conformer obtained from FTIR measurements with wt MbCNR complexes correlates strongly and directly with the fraction of geminate recombination of bound methyl through n-hexyl isocyanide (CNC1-6) after nanosecond laser photolysis (2). Photodissociated alkyl isocyanides originally oriented in the in conformation are trapped and efficiently rebind, but those oriented in the out conformation with the His(E7) gate open are much more likely to escape completely to solvent. The original evidence in support of His64(E7) regulating CNR entry and escape from Mb is shown in Fig. 8. The logarithms of the measured overall bimolecular rate constants for O2, methyl, ethyl, n-propyl, and n-butyl isocyanide binding are plotted versus the size of the amino acid at the 64(E7) position. Most of the data were taken from Rohlfs et al. (11) and Scott et al. (4), and new parameters for CNR binding to H64W Mb were measured for this work. There is an ~10-fold decrease in k′O2 when the size of the E7 amino acid is increased from Gly to Trp, but the increase in the size of the E7 amino acid has a much more dramatic effect on k′CNR, which decreases from ~100 μM-1s-1 to ≤ 0.1 μM−1s−1 for the same set of mutations (Fig. 8).
Figure 8. The bimolecular association rate (k′) for O2 and CNC1-4 binding to Mb mutants with varied residue sizes at the E7 gate.

There is only a small dependence of k′O2 on the size of the residue at 64(E7). The association rate constants for the larger CNRs, however, are highly dependent on the amino acid size in the channel normally occupied by the E7 gate. The k′ values are from this work (W64), Rohlfs et al. (11) and Olson et al. (26).
The complex correlations between the O2 binding parameters, k′entry,O2, and Fgem,O2 and the fraction of in MbCNR conformer shown in Figs. 3 and 7 support our idea that the alkyl side chains of isocyanide ligands act as transition state analogues for both O2 entry and O2 capture in the back of the distal pocket. The crystal structures of MbCNC4 taken from the second paper of this series are shown in Fig. 9 and can be used to visualize diatomic ligand pathways. Two key caveats to using the isocyanides as probes of ligand channels are: (1) the isocyanide alkyl chain is less mobile than a freely diffusing diatomic gas because it is tethered to the iron atom, and (2) the connectivity of the alkyl side chain may require displacement of several amino acids simultaneously rather than just one or two side chains. However, the flexibility of the straight-chained CNRs allows more accurate sensing of steric constraints and the relative free energies required for opening potential pathways than the more rigid aromatic rings of bound imidazole and phenyl ligands.
Figure 9.

Left panel – structure of the native out MbCNC4 conformer (PDB ID 104m; (1)) with CO atoms placed at the terminal carbon atoms of the alkyl side chain. Right panel – structure of wt in MbCNC4 conformer (PDB ID 111m; (1)) with CO placed in the position found in the structure of the room temperature B state photoproduct of wt MbCO (PDB ID 1abs; (27)). All structures were globally aligned by Cα atoms and rendered in the same orientation in Pymol. The right panel represents ligand entry through the E7 channel and the left panel, represents ligand capture in the side path model of ligand binding first described in detail by (4).
In the E7 gate mechanism, ligands enter Mb when the imidazole side chain of His64 has rotated outward opening a direct channel to the distal pocket. This conformation is seen for the out conformer of native MbCNC4 P21 crystals with the n-butyl chain occupying an open E7 channel with enough space for a diatomic gas molecule to enter the protein (Fig. 9, left panel). This interpretation is strongly supported by both the inverse and direct correlations, respectively, of log(k′entry,O2) and Fgem,O2 with the fraction of CNC5 in conformers, Fin,CNC5, measured by FTIR spectroscopy for a series of mutations at the 45 (CD3) and 46 (CD4) positions that alter the flexibility of the His64(E7) side chain (Fig. 3). In both cases, smaller values of Fin for n-pentyl isocyanide indicate a lower barrier to opening of the E7 channel, which increases the rate of bimolecular entry and at the same time decrease geminate recombination after laser photolysis by facilitating escape.
The side path or baseball glove mechanism for O2 binding also predicts that the rate of ligand entry depends on the size of the distal pocket. The larger the volume available to incoming O2, the greater will be the chance of ligand capture, just as a large pocket in a baseball glove makes it easier to catch a ball. This interpretation is supported by the direct correlation between Fin for bound CNC5 and log(k′entry,O2) for mutations in which the volume of the interior of the distal pocket is increased or decreased by Ala or Phe/Trp mutations, respectively. An increase in the fraction of in CNC5 conformer indicates more free space in the interior of the distal pocket and this increase in volume correlates directly with greater rates of O2 entry (Fig. 7B). This interpretation is shown in the right panel of Fig. 9, where the structure of the in conformer of wt MbCNC4 is compared to the structure of the initial B-state structure of photodissociated CO. The terminal atom of the n-butyl side chain is pointing toward the Xe4 pocket which may also be part of the capture volume and is the location of state C for dissociated CO (4, 23, 24, 28–31).
There is debate over the functional significance of the interior Xe cavities that were characterized at high resolution in Mb by Tilton et al. (32). Their original observations were followed by molecular dynamics simulations that suggested a “multiple pathways” model in which ligands can freely migrate from one Xe site to another in the interior of Mb and then exit from these positions through several transient openings to solvent, with the distal histidine gate being only a very minor pathway (33). Even the most recent molecular dynamics simulations (34–36) and energy mapping computations (37) still consistently find multiple apolar pathways for ligand entry and exit that lead from the distal pocket through the protein interior to solvent.
In 1994, Huang and Boxer (38) carried out a random mutagenesis study of point mutations of recombinant Mb. In their experiments, changes in O2 and CO rebinding time courses were measured after laser photolysis of bacterial cell lysates without purifying the Mb variants. Most of the mutations that markedly affected ligand binding rates were located at or near the His(E7) position but a number of replacements remote from the heme pocket also gave large effects. The latter observations led Huang and Boxer (38) to propose that their results supported the multiple pathway model.
Building on the work of Huang and Boxer (38), Scott et al. (3) used a rational mutagenesis strategy to systematically introduce small or large amino acids at positions along all the proposed ligand entry and exit pathways in sperm whale Mb. In their work, all the variants were expressed, purified to homogeneity, and both geminate and overall association and dissociation rate parameters were measured. They only found significant effects on rates of ligand entry and escape for substitutions near His64 and the E7 channel and suggested that ~80% of diatomic gases enter and exit through this pathway. Some of the remote mutations reported to have large effects by Huang and Boxer (38) were examined and found to be false positives (3). Many of the other remote mutations identified in the screen of bacterial lysates involved proline substitutions in the middle of helices or other replacements that are predicted to markedly decreased Mb stability. As a result, the lysates probably contained significant amounts of partially denatured protein and free heme, which would give large changes in the time courses for CO and O2 rebinding after laser photolysis.
Time resolved X-ray crystallography has also provided strong experimental evidence in favor of the side path/E7 gate model proposed by Scott et al. (3). In crystals of wt Mb, the electron density for photolyzed CO is found to persist in the Xe1 cavity on the proximal side of the heme group with a life time of ~10–20 μs and then disappears as the ligand moves out into solvent (30). This observation first led workers to propose that CO escapes directly to solvent from the Xe1 cavity, thus supporting the multiple paths model. However, blocking access to the distal pocket from these internal cavities by L29F and L29W mutations traps CO within the Xe1 cavity for even longer times, ~300 and ~1,500 μs, respectively (24). If ligands could escape directly from the Xe sites, inhibiting their return to the distal pocket by the large Phe(B10) and Trp(B10) side chains should not have affected the life times of ligand electron density in the Xe1 cavity. The simplest interpretation of these time resolved crystallographic data is that ligands have to return to the active site to escape through the E7 channel as predicted by the side pathway model. Thus, there is still a strong disconnect between theoretical simulations that consistently predict multiple pathways and the experimental effects of mutagenesis on ligand binding that strongly indentify the E7 gate and channel as the primary route for entry and exit.
In this work, we have taken another experimental approach, used CNRs to probe the spaces and paths near the Mb binding pocket, and found correlations that also indicate that ligands primarily enter and exit the protein through the His(E7) gate. An alternative explanation for the direct correlation between Fin,CNC5 and log(k′entry,O2) is that when the connection between the Xe4 cavity and the distal pocket is blocked by the V68F and I107W mutations, ligand entry and escape through the multiple interior pathways will be markedly inhibited. However this argument and the multiple interior pathway mechanism cannot explain the lack of correlation of Fin,CNC5 with kescape,O2 for the “pocket volume” mutants including V68F and I107W (Fig. 7D). In addition, the multiple pathway model cannot readily explain the decrease in k′ with increasing size of the E7 amino acid shown in Fig. 8. Thus, in our view, the E7gate/baseball glove model provides the best structural explanation of ligand binding to Mb, and bound alkyl isocyanides serve as excellent probes of the flexibility of the distal histidine gate and the capture volume in the interior of the active site, as shown in Figs. 1 and 9.
Abbreviations
- Mb
myoglobin
- wt
wild-type
- CNR
alkyl isocyanide, where R = C1, C2, C3, C4, C5 and C6 for methyl, ethyl, n-propyl, n-butyl, n-pentyl and n-hexyl groups
- FTIR
Fourier-transform infrared spectroscopy
- νCN
isocyano group stretching frequency
- Fin
fraction of CNRs that point into the Mb binding pocket as measured by FTIR
- k′
association rate
- k
dissociation rate
- Ka
equilibrium association constant calculated as k′/k
- Fgem
fraction of geminate recombination
Footnotes
Supported by U.S. Public Health Service Grants GM 35649 (J.S.O.) and HL 47020 (J.S.O.), and Grant C-612 (J.S.O.) from the Robert A. Welch Foundation. R.L.S. and G.C.B. were recipients of traineeships from The Houston Area Molecular Biophysics Predoctroral Training Grant GM08280.
References
- 1.Smith RD, Johnson KA, Blouin GC, Phillips GN, Jr, Olson JS. Straight-Chain Alkyl Isocyanides Open the Distal Histidine Gate in Crystal Structures of Myoglobin. Biochemistry. 2010 doi: 10.1021/bi1001739. to be co-published with this article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blouin GC, Olson JS. The Stretching Frequencies of Bound Alkyl Isocyanides Indicate Two Distinct Ligand Orientations within the Distal Pocket of Myoglobin. Biochemistry. 2010 doi: 10.1021/bi100172c. to be co-published with this article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott EE, Gibson QH. Ligand migration in sperm whale myoglobin. Biochemistry. 1997;36:11909–11917. doi: 10.1021/bi970719s. [DOI] [PubMed] [Google Scholar]
- 4.Scott EE, Gibson QH, Olson JS. Mapping the pathways for O2 entry into and exit from myoglobin. J Biol Chem. 2001;276:5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]
- 5.Olson JS, Soman J, Phillips GN., Jr Ligand pathways in myoglobin: a review of Trp cavity mutations. IUBMB Life. 2007;59:552–562. doi: 10.1080/15216540701230495. [DOI] [PubMed] [Google Scholar]
- 6.Springer BA, Sligar SG. High-level expression of sperm whale myoglobin in Escherichia coli. Proc Natl Acad Sci U S A. 1987;84:8961–8965. doi: 10.1073/pnas.84.24.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carver TE, Brantley RE, Jr, Singleton EW, Arduini RM, Quillin ML, Phillips GN, Jr, Olson JS. A novel site-directed mutant of myoglobin with an unusually high O2 affinity and low autooxidation rate. J Biol Chem. 1992;267:14443–14450. [PubMed] [Google Scholar]
- 8.Casanova J, Jr, Schuster RE, Werner ND. Synthesis of aliphatic isocyanides. Journal of the Chemical Society. 1963:4280–4281. [Google Scholar]
- 9.Reisberg PI, Olson JS. Equilibrium binding of alkyl isocyanides to human hemoglobin. J Biol Chem. 1980;255:4144–4130. [PubMed] [Google Scholar]
- 10.Mims MP, Porras AG, Olson JS, Noble RW, Peterson JA. Ligand binding to heme proteins. An evaluation of distal effects. J Biol Chem. 1983;258:14219–14232. [PubMed] [Google Scholar]
- 11.Rohlfs RJ, Mathews AJ, Carver TE, Olson JS, Springer BA, Egeberg KD, Sligar SG. The effects of amino acid substitution at position E7 (residue 64) on the kinetics of ligand binding to sperm whale myoglobin. J Biol Chem. 1990;265:3168–3176. [PubMed] [Google Scholar]
- 12.Stetzkowski F, Cassoly R, Banerjee R. Binding of alkylisocyanides with soybean leghemoglobin. Comparisons with sperm whale myoglobin. J Biol Chem. 1979;254:11351–11356. [PubMed] [Google Scholar]
- 13.Carver TE, Olson JS, Smerdon SJ, Krzywda S, Wilkinson AJ, Gibson QH, Blackmore RS, Ropp JD, Sligar SG. Contributions of residue 45(CD3) and heme-6-propionate to the biomolecular and geminate recombination reactions of myoglobin. Biochemistry. 1991;30:4697–4705. doi: 10.1021/bi00233a009. [DOI] [PubMed] [Google Scholar]
- 14.Lecomte JT, La Mar GN. 1H NMR study of labile proton exchange in the heme cavity as a probe for the potential ligand entry channel in myoglobin. Biochemistry. 1985;24:7388–7395. doi: 10.1021/bi00346a054. [DOI] [PubMed] [Google Scholar]
- 15.Lai HH, Li T, Lyons DS, Phillips GN, Jr, Olson JS, Gibson QH. Phe-46(CD4) orients the distal histidine for hydrogen bonding to bound ligands in sperm whale myoglobin. Proteins. 1995;22:322–339. doi: 10.1002/prot.340220404. [DOI] [PubMed] [Google Scholar]
- 16.Smith RD. Biochemistry & Cell Biology, Doctoral Thesis. Rice University; Houston, TX: 1999. p. 203. [Google Scholar]
- 17.Egeberg KD, Springer BA, Sligar SG, Carver TE, Rohlfs RJ, Olson JS. The role of Val68(E11) in ligand binding to sperm whale myoglobin. Site-directed mutagenesis of a synthetic gene. J Biol Chem. 1990;265:11788–11795. [PubMed] [Google Scholar]
- 18.Quillin ML, Li T, Olson JS, Phillips GN, Jr, Dou Y, Ikeda-Saito M, Regan R, Carlson M, Gibson QH, Li H, et al. Structural and functional effects of apolar mutations of the distal valine in myoglobin. J Mol Biol. 1995;245:416–436. doi: 10.1006/jmbi.1994.0034. [DOI] [PubMed] [Google Scholar]
- 19.De Angelis F, Jarzecki AA, Car R, Spiro TG. Quantum chemical evaluation of protein control over heme ligation: CO/O2 discrimination in myoglobin. J Phys Chem B. 2005;109:3065–3070. doi: 10.1021/jp0451851. [DOI] [PubMed] [Google Scholar]
- 20.Carver TE, Rohlfs RJ, Olson JS, Gibson QH, Blackmore RS, Springer BA, Sligar SG. Analysis of the kinetic barriers for ligand binding to sperm whale myoglobin using site-directed mutagenesis and laser photolysis techniques. J Biol Chem. 1990;265:20007–20020. [PubMed] [Google Scholar]
- 21.Smerdon SJ, Dodson GG, Wilkinson AJ, Gibson QH, Blackmore RS. Distal pocket polarity in ligand binding to myoglobin: structural and functional characterization of a threonine68(E11) mutant. Biochemistry. 1991;30:6252–6260. doi: 10.1021/bi00239a025. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Quillin ML, Phillips GN, Jr, Olson JS. Structural determinants of the stretching frequency of CO bound to myoglobin. Biochemistry. 1994;33:1433–1446. doi: 10.1021/bi00172a021. [DOI] [PubMed] [Google Scholar]
- 23.Aranda Rt, Levin EJ, Schotte F, Anfinrud PA, Phillips GN., Jr Time-dependent atomic coordinates for the dissociation of carbon monoxide from myoglobin. Acta Crystallogr D Biol Crystallogr. 2006;62:776–783. doi: 10.1107/S0907444906017318. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt M, Nienhaus K, Pahl R, Krasselt A, Anderson S, Parak F, Nienhaus GU, Srajer V. Ligand migration pathway and protein dynamics in myoglobin: a time-resolved crystallographic study on L29W MbCO. Proc Natl Acad Sci U S A. 2005;102:11704–11709. doi: 10.1073/pnas.0504932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirota S, Li T, Phillips GN, Jr, Olson JS, Mukai M, Kitagawa T. Perturbation of the Fe-O2 Bond by Nearby Residues in Heme Pocket: Observation of .nu.Fe-O2 Raman Bands for Oxymyoglobin Mutants. Journal of the American Chemical Society. 1996;118:7845–7846. [Google Scholar]
- 26.Olson JS, McKinnie RE, Mims MP, White DK. Mechanisms of ligand binding to pentacoordinate protoheme. Journal of the American Chemical Society. 1983;105:1522–1527. [Google Scholar]
- 27.Schlichting I, Berendzen J, Phillips GN, Jr, Sweet RM. Crystal structure of photolysed carbonmonoxy-myoglobin. Nature. 1994;371:808–812. doi: 10.1038/371808a0. [DOI] [PubMed] [Google Scholar]
- 28.Brunori M, Vallone B, Cutruzzola F, Travaglini-Allocatelli C, Berendzen J, Chu K, Sweet RM, Schlichting I. The role of cavities in protein dynamics: crystal structure of a photolytic intermediate of a mutant myoglobin. Proc Natl Acad Sci U S A. 2000;97:2058–2063. doi: 10.1073/pnas.040459697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostermann A, Waschipky R, Parak FG, Nienhaus GU. Ligand binding and conformational motions in myoglobin. Nature. 2000;404:205–208. doi: 10.1038/35004622. [DOI] [PubMed] [Google Scholar]
- 30.Srajer V, Ren Z, Teng TY, Schmidt M, Ursby T, Bourgeois D, Pradervand C, Schildkamp W, Wulff M, Moffat K. Protein conformational relaxation and ligand migration in myoglobin: a nanosecond to millisecond molecular movie from time-resolved Laue X-ray diffraction. Biochemistry. 2001;40:13802–13815. doi: 10.1021/bi010715u. [DOI] [PubMed] [Google Scholar]
- 31.Bourgeois D, Schotte F, Brunori M, Vallone B. Time-resolved methods in biophysics. 6. Time-resolved Laue crystallography as a tool to investigate photo-activated protein dynamics. Photochem Photobiol Sci. 2007;6:1047–1056. doi: 10.1039/b704249c. [DOI] [PubMed] [Google Scholar]
- 32.Tilton RF, Jr, Kuntz ID, Jr, Petsko GA. Cavities in proteins: Structure of a metmyoglobin-xenon complex solved to 1.9 Å. Biochemistry. 1984;23:2849–2857. doi: 10.1021/bi00308a002. [DOI] [PubMed] [Google Scholar]
- 33.Elber R, Karplus M. Enhanced sampling in molecular dynamics: use of the time dependent Hartree approximation for simulation of carbon monoxide diffusion through myoglobin. J Am Chem Soc. 1990;112:9161–9175. [Google Scholar]
- 34.Ruscio JZ, Kumar D, Shukla M, Prisant MG, Murali TM, Onufriev AV. Atomic level computational identification of ligand migration pathways between solvent and binding site in myoglobin. Proc Natl Acad Sci U S A. 2008;105:9204–9209. doi: 10.1073/pnas.0710825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golden SD, Olsen KW. Use of the conjugate peak refinement algorithm for identification of ligand-binding pathways in globins. Methods Enzymol. 2008;437:417–437. doi: 10.1016/S0076-6879(07)37021-3. [DOI] [PubMed] [Google Scholar]
- 36.Elber R, Gibson QH. Toward quantitative simulations of carbon monoxide escape pathways in myoglobin. J Phys Chem B. 2008;112:6147–6154. doi: 10.1021/jp0769779. [DOI] [PubMed] [Google Scholar]
- 37.Cohen J, Arkhipov A, Braun R, Schulten K. Imaging the migration pathways for O2, CO, NO, and Xe inside myoglobin. Biophys J. 2006;91:1844–1857. doi: 10.1529/biophysj.106.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X, Boxer SG. Discovery of new ligand binding pathways in myoglobin by random mutagenesis. Nat Struct Biol. 1994;1:226–229. doi: 10.1038/nsb0494-226. [DOI] [PubMed] [Google Scholar]