Abstract
The discovery that many intron-containing genes can be cotranscriptionally spliced has led to an increased understanding of how splicing and transcription are intricately intertwined. Cotranscriptional splicing has been demonstrated in a number of different organisms and has been shown to play roles in coordinating both constitutive and alternative splicing. The nature of cotranscriptional splicing suggests that changes in transcription can dramatically affect splicing, and new evidence suggests that splicing can, in turn, influence transcription. In this chapter, we discuss the mechanisms and consequences of cotranscriptional splicing and introduce some of the tools used to measure this process.
Keywords: Splicing, Cotranscriptional, RNA, Spliceosome, Transcription, Intron, RNA polymerase II
1 Early Indications of Cotranscriptional Splicing
The last decade has seen a rapid evolution of our understanding of the process of pre-messenger RNA splicing. While elegant genetics and biochemistry have provided a “parts list” of the components of the core splicing machinery and important insights into the well-conserved functions of these proteins and RNAs, it has also become clear that in vivo assembly of the spliceosome and at least some splicing catalysis occur cotranscriptionally, while the elongating RNA polymerase is still actively engaged with a chromatin template (Fig. 1). Furthermore, the cotranscriptional nature of splicing has important functional and regulatory implications.
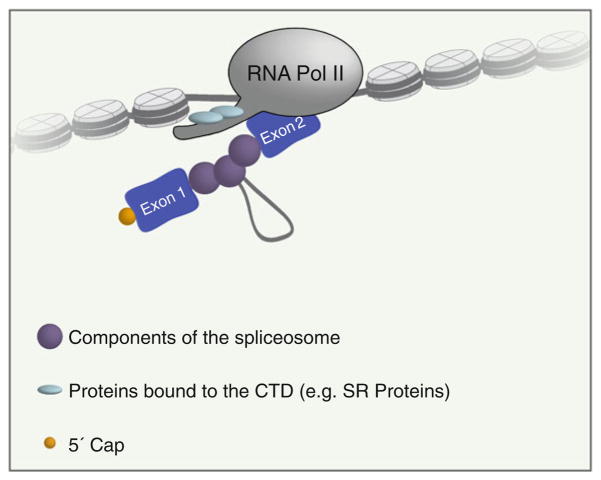
Fig. 1.
Coupling between pre-mRNA splicing and transcription. Components of the splicing machinery localize to the nascent RNA while transcription is occurring. SR proteins (blue ovals) facilitate cross talk between the CTD tail of RNA polymerase II and the splicing machinery. Cross talk also occurs between the splicing machinery and modified histones. Other RNA processing events, such as 5′ capping, also occur cotranscriptionally
Remarkably, within a decade of the discovery of split genes, elegant studies by Ann Beyer and Yvonne Osheim using electron microscopy to examine the highly transcribed Drosophila chorion genes showed spliceosomes associated at splice junctions of nascent transcripts [1]. These EM results nicely complemented Beyer’s earlier studies of nascent transcripts in dipteran embryos, which revealed formation of looped structures (lariats) on the nascent transcripts prior to their release [2], and similar phenomena were later reported on amphibian oocyte lampbrush chromosomes [3]. While these studies showed evidence of cotranscriptional splicing, it is important to point out that they did not address whether the transcription and splicing machineries are functionally coupled. Nonetheless, the ground was set early for studies to explore both the extent of cotranscriptional splicing and the mechanism by which it occurs.
2 Evidence of Widespread Cotranscriptional Splicing
These early studies suggest that both spliceosome assembly and catalysis of splicing can occur in a cotranscriptional manner. Assembly of the spliceosome has been shown to occur in a highly ordered and stepwise fashion in vitro (Chapter 1), and the same is true of spliceosome assembly that occurs on nascent transcripts [4]. In fact, chromatin immunoprecipitation (ChIP) experiments of experimentally tractable genes were the first to demonstrate that the stepwise assembly of the spliceosome in cotranscriptional splicing is akin to how the spliceosome is understood to assemble in in vitro experiments in yeast [5, 6] and in metazoans [7]. While ChIP experiments detect interactions between proteins and DNA, since nascent RNPs lie adjacent to the DNA axis [8], protein—nucleic acid interactions can illustrate cotranscriptional spliceosome assembly, and this method has become a useful proxy to study protein-RNA interactions in cotranscriptional splicing [9]. These analyses have begun to address the specific requirements for proper cotranscriptional spliceosome assembly. For example, studies utilizing ChIP to analyze spliceosome assembly on cotranscriptionally spliced genes have revealed that a histone acetyltransferase regulates the association of components of the U2 snRNP to nascent RNAs in S. cerevisiae [10].
While assembly of the spliceosome during transcription is a key aspect of cotranscriptional splicing, a key question is whether splicing catalysis occurs cotranscriptionally, before the termination of transcription [11]. The results showing that spliceosome assembly, from the early steps involving the U1 and U2 snRNPs to the later steps involving the U4/U6-U5 tri-snRNP, occurs on nascent RNA support the notion of cotranscriptional splicing catalysis. However, there is still some question about the proportion of introns that are removed cotranscriptionally. Early studies utilizing chromatin immunoprecipitation methods posited that although spliceosome assembly could begin cotranscriptionally, most genes in Saccharomyces cerevisiae are spliced posttranscriptionally [12]. The rationale was that yeast genes are too short for cotranscriptional catalysis, since the polymerase would be expected to terminate transcription before exon ligation could occur. However, more recent studies applying global analysis of nascent RNA support the argument that most intron-containing genes in S. cerevisiae are indeed spliced cotranscriptionally. Although terminal exons are short, these data show evidence that the polymerase pauses at the terminal exon, effectively allowing time to splice (this phenomenon is explained in greater detail below) [13, 14] (Fig. 2a). Our current understanding of the breadth of cotranscriptional splicing in other organisms continues to evolve, though it has been reported that the majority of intron-containing genes are at least partially spliced cotranscriptionally in Drosophila [15], as well as in human tissues and cell lines [16 – 20], and these transcripts remain associated with chromatin until fully spliced [21]. Nonetheless, it will be important to understand the potential biological significance of posttranscriptional splicing when it occurs.
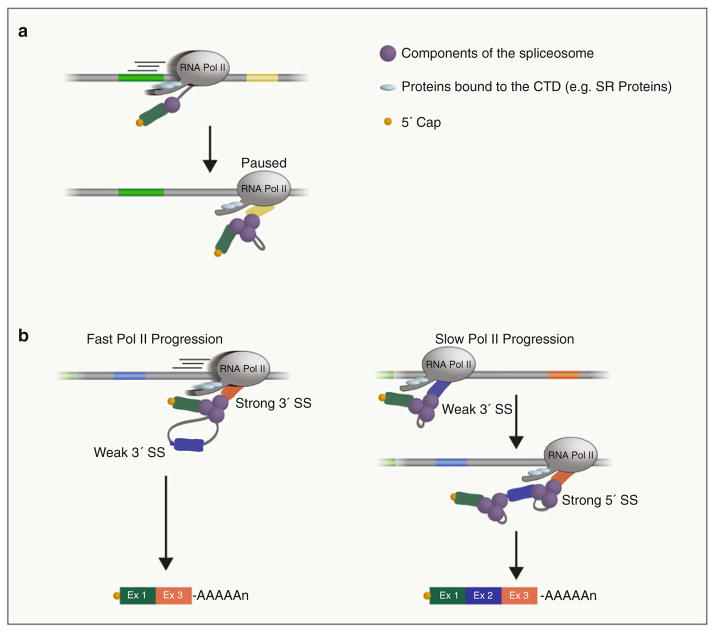
Fig. 2.
Kinetic Model of Cotranscriptional Splicing. (a) In Saccharomyces cerevisiae, RNA Pol II pauses at the terminal exon and/or 3′ SS to facilitate cotranscriptional splicing. (b) RNA Pol II rate of elongation modulates cotranscriptional alternative splicing. Fast elongation rate of transcription (left) favors skipping of exons with “weak” upstream 3′ splice sites (blue exon). Slower Pol II elongation rates (right) favor inclusion of exons with weak 3′ SS sites. Constitutive exons (containing strong 3′ SS) are included independently of RNA Pol II elongation rate (not shown)
3 Constitutive and Alternative Cotranscriptional Splicing and the Transcriptional Machinery
It has become clear that cotranscriptional splicing is spatially and temporally linked to transcription, and a key player in coordinating transcription with splicing is the RNA polymerase itself. RNA polymerase II, the polymerase responsible for transcribing intron-containing pre-mRNAs, is distinguished from the other eukaryotic RNA polymerases by the presence of a C-terminal “tail” made up of numerous heptad repeats (YSPTSPS), the number of which roughly correlates with organism complexity. For example, the CTD consists of 26 repeats in the yeast Saccharomyces cerevisiae and 52 repeats in humans. Posttranslational modifications on the CTD tail play a key role in the regulation of Pol II activity, and modifications of the CTD, particularly phosphorylation, help couple transcription and numerous RNA processing events (reviewed in refs. 22 – 24). Serines 2 and 5 of the CTD have been identified as major phosphorylated residues [25, 26]. Serine 5 is phosphorylated by the basal transcription factor TFIIH at the initiation of transcription [27]. Subsequent to initiation, promoter clearance and transcriptional elongation occur, during which serine 2 of the CTD is phosphorylated by the transcriptional elongation factor P-TEFb. This shift from serine 5 to serine 2 phosphorylation of the CTD during transcriptional elongation may play an important role in the regulation of cotranscriptional splicing [28, 29].
Early studies investigating the role of the CTD in pre-mRNA splicing proposed that the CTD interacts directly with RNA splicing proteins to recruit them to the nascent transcript [30], as truncation or mutation of the CTD of RNAP II leads to changes in splicing in vitro and in vivo [31, 32]. For example, the splicing protein U2AF65 interacts directly with the phosphorylated CTD [33] to promote its association with the pre-mRNA [33, 34]. There is also evidence of interactions between the CTD and serine-arginine (SR) proteins required for constitutive and alternative splicing in metazoans [35] (Fig. 1), although the precise consequence3s of these interactions are still poorly understood. Interestingly, the SR protein SRSF3 negatively regulates the inclusion of the EDI exon of the fibronectin gene in a manner that is dependent on the presence of the CTD of RNA Pol II [36]. A number of other members of the SR family of proteins have been shown to functionally interact with the RNA Pol II CTD to affect pre-mRNA splicing [37].
As further evidence of an important role for the CTD in cotranscriptional splicing, in vitro transcription/splicing systems show that the presence of the CTD enhances the rate of splicing, as in vitro T7 RNA polymerase-transcribed RNAs are spliced less efficiently than those transcribed with the CTD-containing RNAP II [38, 39], and this requires CTD phosphorylation [32]. Posttranslational modifications of the CTD may mediate physical interactions between the elongating RNAP II and the splicing machinery by creating a binding platform for splicing factors (bound directly to the CTD or indirectly with other CTD associated proteins), that can then be transferred to the nascent RNA [40].
These interactions between the CTD and splicing proteins represent examples of a “recruitment model” of cotranscriptional splicing, which posits that there are physical contacts between the transcriptional and splicing machineries, and perturbing these alters cotranscriptional splicing. In addition to interactions between the CTD and splicing proteins, interactions between spliceosomal snRNP complexes and transcription elongation factors are likely to be very important for the coupling of these two processes [41, 42], as are interactions between chromatin marks (or proteins associated with chromatin marks) and the spliceosome, which will be explored further in a subsequent chapter.
Posttranslational modifications on the CTD, as well as other factors that affect the rate of elongation of transcription, influence splice site recognition, spliceosome assembly, and splicing patterns [43 – 45] through kinetic coupling of these two processes. This model has been termed the “kinetic model” of cotranscriptional splicing. For example, as described above, changes in Pol II elongation, specifically Pol II pausing, couple splicing with transcriptional elongation in Saccharomyces cerevisiae [13, 14] (Fig. 2a). Thus, genes that are predicted to be spliced posttranscriptionally are in fact spliced cotranscriptionally. While it is unclear what the precise mechanism of pausing is or to what extent this phenomenon occurs in other species, this terminal exon pausing represents a functional coupling between transcription and splicing.
Some of the most compelling evidence of the importance of the rate of Pol II elongation on splicing outcomes comes from analyses of splicing of RNAs containing alternative 3′ splice sites [46], as changes in the kinetics of RNA Pol II elongation can markedly affect splice site selection in alternatively spliced genes (Fig. 2b). Studies in both yeast and mammalian cells expressing constructs in which an intron-containing gene contains a strong 3′ SS downstream of a weak 3′ SS support this model [46]. A decrease in the elongation rate of the polymerase or pausing by Pol II favors the inclusion of exons possessing the weak 3′ splice site, whereas Pol II with a normal elongation rate, or without pausing during elongation, favors the exclusion of these exons [43, 47, 48]. Furthermore, an exon containing a suboptimal 3′ SS that is normally not utilized in a reporter minigene is included in the transcribed mRNA in Drosophila cells expressing a mutant form of Pol II which transcribes at a slower elongation rate [49]. One particularly intriguing possible mechanism is that nucleosomes, which can form a natural barrier to the transcribing polymerase [50, 51], may alter polymerase elongation rates to facilitate inclusion of weak splice sites. Consistent with this, exons flanked by weak splice sites are more enriched with nucleosomes compared with those containing strong splice sites, and exon inclusion levels correlate with nucleosome occupancy [52, 53]. It is important to note that the elongation rate may also influence the ability of splicing regulators (both positive and negative) to bind to sequences in the nascent RNA, which could also affect exon inclusion and skipping.
In addition to transcription elongation, cotranscriptional alternative splicing can be influenced by promoters and transcriptional activators or repressors. Promoter-swapping experiments indicate that changes in the structure of these sequences result in a change in alternative splice site selection [54]. There is evidence that this promoter-driven effect on splice site selection may occur through interactions between the transcription and splicing machineries modulated by transcriptional activators such as PGC-1 [55]. Recently, the mediator complex has also been implicated in cross talk between transcription and alternative splicing through its ability to link transcriptional activators or repressors that interact with splicing silencers or enhancers (such as hnRNPs and SR proteins) with transcription factors associated with core promoters [56]. These data show that while elongation influences splicing outcomes, early transcriptional events can also affect splicing.
While many of the studies mentioned above appear to support either the “recruitment” or the “kinetic” model of cotranscriptional splicing, these mechanisms are by no means mutually exclusive. It is becoming increasingly clear that both the recruitment of splicing and splicing-associated factors by the transcriptional machinery, as well as the kinetics of the transcription machinery, play critical roles in regulating pre-mRNA splicing.
4 Cotranscriptional Splicing and Its Effects on Transcription
An obvious implication of the close spatial and temporal proximity of the splicing and transcription machineries is that the relationship between transcription and splicing could work both ways, namely, that splicing and splicing factors could also influence transcription. Indeed there is a growing body of evidence indicating that this is the case.
Some of the earliest indications of this came from work as far back as the late 1980s and early 1990s in which it was shown that the presence of an intron increases expression of mouse transgenes [57 – 59]. In the subsequent years there have been a number of important discoveries that have shed light on the mechanisms by which introns can exert a positive effect on transcription. One of the first was the striking observation that interactions between U snRNPs and the transcription elongation factor TAT-SF1 stimulated RNA polymerase II elongation. More specifically, the authors proposed that stimulation of Pol II elongation was the result of TAT-SF1 interaction with the positive transcription elongation factor b (P-TEFb), which phosphorylates the CTD of RNA polymerase II [42]. This study hinted at a central role for P-TEFb in mediating communication between components of the splicing machinery and the RNA polymerase—a role supported by subsequent studies.
As previously described, SR proteins associate cotranscriptionally with the RNA polymerase during active transcription. In vivo depletion of either of the SR proteins SRSF1 or SRSF2 decreases nascent RNA production, with dramatic effects on transcription elongation seen upon SRSF2 depletion [29]. SRSF2 co-IPs with both P-TEFb and TAT-SF1, and its depletion correlates with defective P-TEFb recruitment. Moreover, in these cells, Pol II accumulates in the body of genes and Ser-2 CTD phosphorylation is abrogated, indicative of defective transcription elongation. One intriguing model is that SR proteins such as SRSF2 dynamically associate with Pol II and enhance elongation by stimulating P-TEFb, and at emerging splice sites, the SR proteins disembark and bind to the appropriate RNA signals. Interestingly, SR proteins have also been shown to bind directly to histones [60], raising the possibility that SR protein binding to histones may affect the state of the chromatin and, in turn, affect transcription. Moreover, SR proteins’ interactions with chromatin may facilitate their roles in splicing; since it has been shown that nucleosomes are enriched in exons (discussed elsewhere in this issue), the ability of SR proteins to bind to histones may facilitate their association in exonic RNA sequences. It should be noted that SR proteins are also found associated with intronless genes [61, 62], so the presence of an intron may not be a prerequisite for SR protein effects on transcription. Nonetheless, SR proteins appear to play a central role in mediating the bidirectional relationship between transcription and splicing.
In addition to SR proteins, other proteins involved in RNA processing in general and RNA splicing in particular may affect CTD phosphorylation. In fact, the cap-binding complex, which binds to the 5′ cap structure of pre-mRNAs and has long been known to interact with the core splicing machinery to affect spliceosome assembly, also interacts with P-TEFb. Moreover, the CBC is required for P-TEFb-dependent alternative splicing [63]. This example of the CBC again illustrates the strong bidirectional relationship between splicing and transcription: RNA processing factors can affect transcription, which in turn affects RNA processing. The effect of the CBC on RNA Pol II CTD phosphorylation appears to be conserved, as the yeast cap-binding complex interacts with the yeast ortholog of P-TEFb and stimulates transcription elongation [64].
While there has been a great deal of focus on the effect of splicing factors on transcription elongation, it has also been established that introns can affect early steps of transcription. A functional 5′ SS enhances pre-initiation complex (PIC) formation and stimulates recruitment of general transcription initiation factors [65]. The precise mechanism by which the 5′ SS influences early transcription complex formation is not yet clear; nonetheless, it is likely to involve protein and/or RNA interactions at the 5′ SS. Intriguingly, the U1 snRNA has been shown to associate with TFIIH and regulate transcriptional initiation in a reconstituted transcription system, and promoter proximal 5′ SS recognition by U1 snRNA stimulates TFIIH dependent reinitiation of transcription [66]. Consistent with this, removal of promoter proximal splice signals from a mammalian gene leads to a significant reduction in nascent transcription [67].
While the topic is discussed in more detail elsewhere, it is clear that splicing can influence transcription through its effects on chromatin. The Hu proteins are a family of mammalian RNA binding proteins that act as splicing regulators. Hu proteins are recruited to their RNA-binding sites and interact with histone deacetylase 2 (HDAC2) to inhibit its activity, alter histone acetylation, and, as a consequence, alter RNA polymerase elongation [68]. Several recent studies in mammalian cells demonstrate that histone H3K36me3, a mark of active transcription, is directly influenced by splicing [69, 70]. Moreover, histone H3K4me3 and H3K9ac, both marks of active transcription, are enriched at the first 5′ SS. Removal of endogenous introns or inhibition of splicing using the splicing inhibitor spliceostatin leads to a reduction in the overall H3K4me3 signal [71]. The next exciting challenge will be to determine whether specific components of the splicing machinery interact with the histone-modifying machinery to direct effects on chromatin and, if so, to identify these factors and their modes of action.
Finally, as described above and shown in Fig. 2, elegant yeast studies demonstrate polymerase pausing around the 3′ SS and/or in the 3′ exon, suggesting a model in which the splicing-induced polymerase pausing provides a checkpoint to allow time for splice site recognition and splicing catalysis. It is possible that components of the spliceosome involved in splicing events near the 3′ splice site feedback on the polymerase to induce pausing—either through changes in the chromatin, changes to the RNA polymerase itself (e.g., through CTD phosphorylation), or interactions with components of the transcription elongation machinery. Ongoing studies are aimed at understanding how splicing and/or specific splicing factors provide this feedback to the polymerase to affect pausing.
5 Cotranscriptional Splicing in Disease and Development
Given the requirement of precise expression of genes for normal cellular processes, it is not surprising that dysregulation of gene expression due to defects in pre-mRNA splicing can result in disease. In fact, it is likely that at least 30 % of mutations that cause disease do so by disrupting splicing, through cis-acting or trans - acting mechanisms [72 – 74]. Since a significant amount, if not most, of pre-mRNA splicing in humans occurs cotranscriptionally, it would be expected that defects in cotranscriptional splicing would lead to disease as well as defects in development. Consistent with this, genes that are highly cotranscriptionally alternatively spliced in the fetal brain have also been implicated in critical neurodevelopmental processes, suggesting that dysregulation of cotranscriptionally alternatively spliced genes may impair neural development [16]. Mutations in the autoimmune regulator (AIRE) transcription factor lead to autoimmune-polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED), likely due to a decrease in cotranscriptional splicing of the AIRE target genes [75]. Some of the genes that undergo dysregulated cotranscriptional alternative splicing are currently being investigated as targets for antisense oligonucleotide (AON) therapy. For example, antisense oligonucleotides, which induce exon skipping in the DMD gene, have shown promise as therapeutic intervention in Duchenne muscular dystrophy [76]. Further analysis to better understand which genes are cotranscriptionally spliced and the mechanisms of cotranscriptional splicing will likely uncover many genes whose dysregulation leads to disease or defects in development.
6 Tools of the Trade: Studying Cotranscriptional Splicing
Understanding cotranscriptional spliceosome assembly and splicing catalysis has evolved as more varied tools have been applied to study these processes. As described above, early EM studies provided direct visual evidence of cotranscriptional splicing, and more recent studies provide insights into the ordered nature of cotranscriptional spliceosome assembly, the extent of cotranscriptional splicing, and the roles played by specific transcription proteins as well as chromatin in directing cotranscriptional splicing.
Our grasp of spliceosome assembly has been largely informed by in vitro studies of the formation of splicing complexes using non-denaturing gel systems. These studies portrayed a picture of a spliceosome that assembled in an ordered, stepwise manner onto the pre-messenger RNA. It was an exciting surprise when it was shown using chromatin immunoprecipitation studies in yeast that, in vivo, the spliceosome followed a similar, stepwise pattern of assembly [5, 6, 9]. This approach allows inference of the kinetics of spliceosome assembly, based on the assumption that distance travelled by RNA Pol II is equivalent to time, as specific snRNPs localize to specific regions of the transcribed gene (e.g., U1 snRNP localizes to the 5′ SS as measured by ChIP) [77]. This is still an indirect measure of cotranscriptional splicing, and therefore, multiple caveats, such as accessibility of epitopes, should be considered when ChIP is used as a tool to measure cotranscriptional splicing [78]. Nonetheless, this approach has proven to be a powerful tool for measuring chromatin-associated RNA-binding proteins such as snRNPs [10, 78].
Advances in live-cell imaging have allowed for in vivo investigation of cotranscriptional splicing that was previously not feasible. For example, photobleaching experiments measuring mobility and distribution of spliceosomal proteins, as well as direct, real-time imaging of fluorescently tagged snRNP components, have provided significantly more insight into the kinetics of cotranscriptional spliceosome assembly in human cells [79 – 81].
The development of in vitro transcription-splicing coupled systems to study cotranscriptional splicing has also led to a further increase in our understanding of the interactions between the transcription machinery and the nascent transcript, as well as the effect that cotranscriptional splicing has on pre-mRNA stability and splicing efficiency [38, 82, 83]. However, a major drawback of these in vitro systems has been their inability to recapitulate the chromatin setting of cotranscriptionally spliced genes. However, as the technological challenges of in vitro splicing from in vitro assembled chromatin templates are addressed, this assay will certainly yield important mechanistic insights.
As next-generation deep sequencing of genomes and transcriptomes has become less costly, the role of these technologies in examining cotranscriptional splicing has also increased. The sequencing of nascent RNAs using high-density tiling arrays has previously shown that splicing catalysis occurs cotranscriptionally in S. cerevisiae [13]. More recently, studies utilizing RNA-Seq of nascent and chromatin-associated RNAs have revealed widespread cotranscriptional splicing in Drosophila and human cells [15 – 17]. The majority of these studies support widespread cotranscriptional splicing across species; however, there are reports suggesting otherwise [20]. The disparity between these observations may be due to cell-type differences and the conditions to which the cells are exposed. Furthermore, different methods used to calculate the frequency of cotranscriptional splicing may also result in a disparity between studies, particularly when assessing cotranscriptional splicing on an intron-to-intron versus entire gene basis (see also ref. 84). Therefore, it is necessary to use a technique such as RT-qPCR to validate high-throughput cotranscriptional splicing results. Even newer methods of deep sequencing and the availability of high-quality databases of transcriptomes will likely provide even further insight into the extent of cotranscriptional splicing across species.
Though newer high-throughput technologies such as RNA-seq have increased our knowledge of the breadth of cotranscriptional splicing, traditional methods such as classical yeast genetics still play a critical role in determining the underlying mechanisms of cotranscriptional splicing, particularly in S. cerevisiae. For example, genetic analyses have been instrumental in showing interactions between splicing factors and other cellular machineries, such as histone-modifying machinery and mRNA export factors [78, 85, 86]. The combined use of these tools will lead to a heightened understanding of the mechanisms and spectrum of cotranscriptional splicing.
Acknowledgments
We would like to thank members of the Johnson Lab for critical reading of the manuscript and apologize to any colleagues whose work is not referenced due to unintentional oversight or space constraints. Funding was provided by the National Institutes of General Medical Sciences (GM085474), the National Science Foundation (MCB-1051921), and an IRACDA fellowship to E.C.M. (K12 GM068524).
References
- 1.Osheim YN, Miller OL, Jr, et al. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 1985;43(1):143–151. doi: 10.1016/0092-8674(85)90019-4. [DOI] [PubMed] [Google Scholar]
- 2.Beyer AL, Bouton AH, Miller OL., Jr Correlation of hnRNP structure and nascent transcript cleavage. Cell. 1981;26(2 Pt 2):155–165. doi: 10.1016/0092-8674(81)90299-3. [DOI] [PubMed] [Google Scholar]
- 3.Wu ZA, Murphy C, Callan HG, et al. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres, and snurposomes. J Cell Biol. 1991;113(3):465–483. doi: 10.1083/jcb.113.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36(2):178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gornemann J, Kotovic KM, Hujer K, et al. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19(1):53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ ss base pairing in yeast. Mol Cell. 2005;19(1):65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13(9):815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 8.Wetterberg I, Zhao J, Masich S, et al. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 2001;20(10):2564–2574. doi: 10.1093/emboj/20.10.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotovic KM, Lockshon D, Boric L, et al. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol. 2003;23(16):5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5(10):e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrillo Oesterreich F, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21(6):328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast premRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24(6):917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40(4):571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Alexander RD, Innocente SA, Barrass JD, et al. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40(4):582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khodor YL, Rodriguez J, Abruzzi KC, et al. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25(23):2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ameur A, Zaghlool A, Halvardson J, et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18(12):1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 17.Tilgner H, Knowles DG, Johnson R, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22(9):1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girard C, Will CL, Peng J, et al. Posttranscriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat Commun. 2012;3:994. doi: 10.1038/ncomms1998. [DOI] [PubMed] [Google Scholar]
- 19.Windhager L, Bonfert T, Burger K, et al. Ultrashort and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res. 2012;22(10):2031–2042. doi: 10.1101/gr.131847.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatt DM, Pandya-Jones A, Tong AJ, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150(2):279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandya-Jones A, Bhatt DM, Lin CH, et al. Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression. RNA. 2013;19(6):811–827. doi: 10.1261/rna.039081.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26(19):2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Almeida SF, Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS lett. 2008;582(14):1971–1976. doi: 10.1016/j.febslet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 24.Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20(3):260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15(10):383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 26.West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140(4):1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36(4):541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barboric M, Lenasi T, Chen H, et al. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci USA. 2009;106(19):7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin S, Coutinho-Mansfield G, Wang D, et al. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15(8):819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mortillaro MJ, Blencowe BJ, Wei X, et al. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93(16):8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCracken S, Fong N, Yankulov K, et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385(6614):357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 32.Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13(10):1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.David CJ, Boyne AR, Millhouse SR, et al. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011;25(9):972–983. doi: 10.1101/gad.2038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu B, Eick D, Bensaude O. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res. 2012;41(3):1591–1603. doi: 10.1093/nar/gks1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuryev A, Patturajan M, Litingtung Y, et al. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93(14):6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13(11):973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 37.Das R, Yu J, Zhang Z, et al. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26(6):867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh S, Garcia-Blanco MA. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA. 2000;6(9):1325–1334. doi: 10.1017/s1355838200992537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das R, Dufu K, Romney B, et al. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev. 2006;20(9):1100–1109. doi: 10.1101/gad.1397406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23(13):2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115 (Pt 20):3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 42.Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414(6866):929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 43.de la Mata M, Alonso CR, Kadener S, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12(2):525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Munoz MJ, Perez Santangelo MS, Paronetto MP, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137(4):708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Ip JY, Schmidt D, Pan Q, et al. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011;21(3):390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dujardin G, Lafaille C, Petrillo E, et al. Transcriptional elongation and alternative splicing. Biochimica et Biophysica Acta. 2012;1829(1):134–140. doi: 10.1016/j.bbagrm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 47.Kornblihtt AR, de la Mata M, Fededa JP, et al. Multiple links between transcription and splicing. RNA. 2004;10(10):1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howe KJ, Kane CM, Ares M., Jr Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9(8):993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Chafin D, Price DH, et al. Drosophila RNA polymerase II mutants that affect transcription elongation. J Biol Chem. 1996;271(11):5993–5999. [PubMed] [Google Scholar]
- 50.Hodges C, Bintu L, Lubkowska L, et al. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325(5940):626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469(7330):368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16(9):990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 53.Tilgner H, Nikolaou C, Althammer S, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16(9):996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 54.Cramer P, Pesce CG, Baralle FE, et al. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA. 1997;94(21):11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monsalve M, Wu Z, Adelmant G, et al. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell. 2000;6(2):307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 56.Huang Y, Li W, Yao X, et al. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell. 2012;45(4):459–469. doi: 10.1016/j.molcel.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brinster RL, Allen JM, Behringer RR, et al. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA. 1988;85(3):836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi T, Huang M, Gorman C, et al. A generic intron increases gene expression in transgenic mice. Mol Cell Biol. 1991;11(6):3070–3074. doi: 10.1128/mcb.11.6.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmiter RD, Sandgren EP, Avarbock MR, et al. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci USA. 1991;88(2):478–482. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loomis RJ, Naoe Y, Parker JB, et al. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33(4):450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7(4):899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 62.Pozzoli U, Riva L, Menozzi G, et al. Over-representation of exonic splicing enhancers in human intronless genes suggests multiple functions in mRNA processing. Biochem Biophys Res Commun. 2004;322(2):470–476. doi: 10.1016/j.bbrc.2004.07.144. [DOI] [PubMed] [Google Scholar]
- 63.Lenasi T, Peterlin BM, Barboric M. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb) J Biol Chem. 2011;286(26):22758–22768. doi: 10.1074/jbc.M111.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hossain MA, Chung C, Pradhan SK, et al. The yeast cap binding complex modulates transcription factor recruitment and establishes proper histone H3K36 trimethylation during active transcription. Mol Cell Biol. 2013;33(4):785–799. doi: 10.1128/MCB.00947-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Damgaard CK, Kahns S, Lykke-Andersen S, et al. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell. 2008;29(2):271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 66.Kwek KY, Murphy S, Furger A, et al. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol. 2002;9(11):800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 67.Furger A, O’Sullivan JM, Binnie A, et al. Promoter proximal splice sites enhance transcription. Genes Dev. 2002;16(21):2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou HL, Hinman MN, Barron VA, et al. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc Natl Acad Sci USA. 2011;108(36):E627–E635. doi: 10.1073/pnas.1103344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim S, Kim H, Fong N, et al. PremRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci USA. 2011;108(33):13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Almeida SF, Grosso AR, Koch F, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18(9):977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 71.Bieberstein NI, Carrillo Oesterreich F, Straube K, et al. First exon length controls active chromatin signatures and transcription. Cell Rep. 2012;2(1):62–68. doi: 10.1016/j.celrep.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 72.Lopez-Bigas N, Audit B, Ouzounis C, et al. Are splicing mutations the most frequent cause of hereditary disease? FEBS lett. 2005;579(9):1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 73.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136(4):777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17(4):419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 75.Zumer K, Plemenitas A, Saksela K, et al. Patient mutation in AIRE disrupts P-TEFb binding and target gene transcription. Nucleic Acids Res. 2011;39(18):7908–7919. doi: 10.1093/nar/gkr527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pramono ZA, Wee KB, Wang JL, et al. A prospective study in the rational design of efficient antisense oligonucleotides for exon skipping in the DMD gene. Hum Gene Ther. 2012;23(7):781–790. doi: 10.1089/hum.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nilsen TW. Spliceosome assembly in yeast: one ChIP at a time? Nat Struct Mol Biol. 2005;12(7):571–573. doi: 10.1038/nsmb0705-571. [DOI] [PubMed] [Google Scholar]
- 78.Gunderson FQ, Merkhofer EC, Johnson TL. Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc Natl Acad Sci USA. 2011;108(5):2004–2009. doi: 10.1073/pnas.1011982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmidt U, Basyuk E, Robert MC, et al. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. J Cell Biol. 2011;193(5):819–829. doi: 10.1083/jcb.201009012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huranova M, Ivani I, Benda A, et al. The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells. J Cell Biol. 2010;191(1):75–86. doi: 10.1083/jcb.201004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rino J, Carvalho T, Braga J, et al. A stochastic view of spliceosome assembly and recycling in the nucleus. PLoS Comput Biol. 2007;3(10):2019–2031. doi: 10.1371/journal.pcbi.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu Y, Das R, Folco EG, et al. A model in vitro system for co-transcriptional splicing. Nucleic Acids Res. 2010;38(21):7570–7578. doi: 10.1093/nar/gkq620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hicks MJ, Yang CR, Kotlajich MV, et al. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 2006;4(6):e147. doi: 10.1371/journal.pbio.0040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brugiolo M, Herzel L, Neugebauer KM. Counting on co-transcriptional splicing. F1000Prime Rep. 2013;5:9. doi: 10.12703/P5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilmes GM, Bergkessel M, Bandyopadhyay S, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32(5):735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moehle EA, Ryan CJ, Krogan NJ, et al. The yeast SR-like protein Npl3 links chromatin modification to mRNA processing. PLoS Genet. 2012;8(11):e1003101. doi: 10.1371/journal.pgen.1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]