Abstract
Purpose.
Myasthenia gravis demonstrates a distinct predilection for involvement of the extraocular muscles (EOM), and we have hypothesized that this may be due to a unique immunological environment. To assess this hypothesis, we took an unbiased approach to analyze RNA expression profiles in EOM, diaphragm, and extensor digitorum longus (EDL) in rats with experimentally acquired myasthenia gravis (EAMG).
Methods.
Experimentally acquired myasthenia gravis was induced in rats by intraperitoneal injection of antibody directed against the acetylcholine receptor (AChR), whereas control rats received antibody known to bind the AChR but not induce disease. After 48 hours, animals were killed and muscles analyzed by RNA expression profiling. Profiling results were validated using qPCR and immunohistochemical analysis.
Results.
Sixty-two genes common among all muscle groups were increased in expression. These fell into four major categories: 12.8% stress response, 10.5% immune response, 10.5% metabolism, and 9.0% transcription factors. EOM expressed 212 genes at higher levels, not shared by the other two muscles, and a preponderance of EOM gene changes fell into the immune response category. EOM had the most uniquely reduced genes (126) compared with diaphragm (26) and EDL (50). Only 18 downregulated genes were shared by the three muscles. Histological evaluation and disease load index (sum of fold changes for all genes) demonstrated that EOM had the greatest degree of pathology.
Conclusions.
Our studies demonstrated that consistent with human myasthenia gravis, EOM demonstrates a distinct RNA expression signature from EDL and diaphragm, which is based on differences in the degree of muscle injury and inflammatory response.
Keywords: myasthenia gravis, acetylcholine receptor, autoimmunity, complement, skeletal muscle
The preferential involvement of extraocular muscle by myasthenia gravis is clinically well known. Here we provide evidence that the muscle is a unique immunological environment that contributes to its targeting by myasthenia gravis.
Introduction
Myasthenia gravis (MG) is an autoimmune neuromuscular transmission disorder that shows a distinct propensity to compromise the extraocular muscles (EOMs). Several hypotheses have been considered.1,2 The least esoteric is that patients come to clinical attention with even slight visual disturbance produced by EOM weakness, whereas minor reductions of limb or bulbar muscle power are not as immediately appreciated. Other explanations can be divided into two categories: one suggests anatomic-physiological properties make the ocular motor system more susceptible to the neuromuscular transmission defect2 and the other is that the EOM possesses a unique immunological environment,1 which places the tissue at risk for antibody-mediated attack. The two hypotheses are not mutually exclusive; however, distinguishing between these considerations has therapeutic implications. Verification that immune properties target the EOM for attack suggests that immune-based treatments could be devised to reverse ocular involvement of MG.
The basis for the differential involvement of muscle groups by neuromuscular disease, in general, is an open question for muscle biology. Despite advances in understanding the primary pathophysiology of many nerve and muscle disorders, little is known as to why individual diseases target or spare specific muscle groups. For example, Duchenne muscular dystrophy, caused by a mutation in dystrophin, which is expressed in all skeletal muscles, produces weakness of quadriceps before deltoid, and the tibialis anterior is compromised more severely than the gastrocnemius.3 Dystrophies related to mutations in dysferlin, which is involved in muscle membrane repair, produce dramatically different clinical presentations among patients,4 and dysferlin-deficient mice have relative sparing of proximal muscles.5 Extraocular muscle, even late in the disease course, is spared in most human muscular dystrophies and animal models.6–8 Inflammatory and endocrine muscle diseases tend to involve proximal muscles and spare EOM and other cranial nerve–innervated muscles, and for these disorders no studies of differential pathology have been performed. In contrast,9 Graves' ophthalmopathy and orbital myositis can be isolated to orbital tissue supporting a unique immunological environment for the EOM, and more generally the supporting orbital tissue.10,11
Myasthenia gravis is caused primarily by antibodies directed against the skeletal muscle acetylcholine receptor (AChR).12,13 The muscle-specific kinase and lipoprotein receptor protein-4 has been identified, as other autoantigens and patients with antibodies to these proteins may have ocular manifestations of MG. Experimentally acquired MG (EAMG) may be induced in animals by administration of antibodies directed against the AChR, which mimics the final effector mechanism of disease pathology observed in humans.14 The passive transfer model of EAMG offers the advantage of a single mechanism of injury to the neuromuscular junction, which is applied in a uniform fashion among animals at a single time point. We administered a monoclonal antibody, which has been used by us and others15–20 to evaluate differential RNA expression profiles of EOM, diaphragm (DIA), and extensor digitorum longus (EDL) to assess the potential differential response of these muscles to injury and susceptibility to injury.
Methods
Animals
Eight- to 10-week-old female Lewis rats weighing 125 to 150 g (Harlan, Indianapolis, IN, USA) were used for the study. Animals were maintained in the Case Western Reserve University animal facility. Animals were housed in isolator cages in a pathogen-free environment, and rodent chow and water were provided ad libitum. A veterinarian was on staff and observed the health of the animals throughout the study. All experiments were conducted in accordance with the principles and procedures established by the National Institutes of Health and the Association for Assessment of Laboratory Animal Care and in the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Experimentally Acquired MG Induction
Eight- to 10-week-old female Lewis rats weighing 125 to 150 g (Harlan) received 1.56 μg per gram of body weight intraperitoneal injections of rat anti-mouse muscle AChR monoclonal IgG2b isotype antibody McAb-3, which binds the skeletal muscle AChR's extracellular region. McAb-1 was used as an isotype control that binds the AChR but does not induce EAMG (both gifts of Vanda Lennon, Mayo Clinic, Rochester, MN, USA).15 The dose of McAb-3 consistently produces mild weakness (Class 1) in 24 hours and moderate weakness (Class 2) in 48 hours by a generally accepted grading scale.21 Rats were killed 48 hours after antibody administration. To qualify for analysis, rats receiving McAb-3 must have developed Class 2 weakness, but not greater. McAb-1–treated rats never demonstrated evidence of weakness.
Tissue Preparation
Extraocular rectus muscles, and DIA and EDL muscles were dissected from rats 48 hours after antibody administration. Tissues were snap frozen in liquid nitrogen and stored at −80°C until use. Muscles were pooled from three rats for each of three independent replicate groups. This procedure served to limit interanimal and interexperiment variability.
DNA Microarray
Isolation of RNA and preparation of labeled cRNA followed methods described previously.22,23 Labeled cRNA was hybridized to Affymetrix GeneChip rat genome 430 v2.0 (RAE230 2.0) microarrays (Santa Clara, CA, USA), which evaluates 31,000 probe sets representing 30,000 unique transcripts and variants from more than 28,000 substantiated rat genes. The manufacturer's standard posthybridization wash, double-stain, and scanning protocols used an Affymetrix GeneChip Fluidics Station 400 and GeneChip Scanner 3000.
Microarray Data Analysis
Affymetrix Microarray Suite (MAS) software version 5.0 was used for initial data processing and fold ratio analyses. The MAS evaluates sets of perfect match and mismatch probe sequences to obtain both hybridization signal values and present/absent calls for each transcript. The MAS filter was used to exclude transcripts that were absent from all samples for further analysis. Present calls were as indicated for EOM (57.4% ± 2.6%), DIA (53.4% ± 4.7%), and EDL (52.9% ± 2.2%). Any transcripts with expression intensity below 400 (five times of background level) across all the samples were also excluded because distortion of fold difference values results when expression levels are low and may be within the level of background noise. Pairwise comparisons were used. For McAb-1 and McAb-3, transcripts defined as differentially regulated met the criteria of the following: nine of nine increase/decrease call each McAb-3 versus McAb-1 in three replicates with nine comparisons, and absolute value of the mean fold difference value greater than or equal to 2.0. Hierarchical clustering was performed by using the clustering function in GeneSpring software (Agilent Technologies, Santa Clara, CA, USA). All differentially regulated genes as a gene list and their expression levels were organized by both individual samples as well as groups.
Affymetrix transcript annotations were replaced with official gene nomenclature and functions were assigned using information in National Center for Biotechnology Information Entrez Gene, UniGene, PubMed, and Affymetrix NetAffx and Weizmann Institute of Science GeneCards databases. To track disease-dependent changes in EAMG mice, we used an aggregate measure designated as the disease load index.24 The disease load index is a unitless measure, representing the sum of the fold change absolute values of all differentially regulated transcripts of a given muscle.
Quantitative Real-Time PCR (qPCR)
Select transcripts were reanalyzed by qPCR, using the same samples as in the microarray studies. The qPCR used SYBR green PCR core reagent with an Applied Biosystems PRISM 7000 Sequence Detection System (Foster City, CA, USA), as described previously.22,23 Rat glyceraldehyde 3-phosphate dehydrogenase was used as an internal positive loading control. For primer sequences, see Supplementary Table S1. Fold change values represent averages from triplicate measurements, by using the 2−ΔΔCT method.25
Histological Assessment
As an additional validation of genomic profiling observations, we performed standard histological examination. Muscle tissues were mounted on cork with 8% tragacanth (Sigma, St. Louis, MO, USA) and immediately frozen in liquid nitrogen-cooled 2-methylbutane, sectioned at 8 μm, stained with hematoxylin and eosin, and viewed under a bright field microscope (Diaphot Nikon Instruments, Inc., Melville, NY, USA).
Immunohistochemistry
The EOMs were dissected and frozen in liquid N2-cooled 2-methybutane and stored at −80°C until use. Ten-micrometer cryosections were fixed for 5 minutes with ice cold acetone and rinsed with PBS (pH 7.4) before they were blocked with 3% normal goat serum for at least 1 hour. The sections were then incubated in diluted primary antibody ED1 (mouse anti-rat CD68; Serotec, Raleigh, NC, USA) or ED2 (mouse anti-rat CD163; Serotec) or at a dilution of 1:200 or for 1 hour at room temperature and washed with PBS before application of secondary antibody Alexa 488 goat anti-mouse (1:500 dilution; Invitrogen, Grand Island, NY, USA). Images were captured with a digital camera (Spot; Diagnostic Instruments, Sterling Heights, MI, USA) and software (Spot Advanced; Diagnostic Instruments) before processing with image-management software (Adobe Photoshop; Adobe Systems, Inc., San Jose, CA, USA).
Results
Expression Profile Analysis
To identify global patterns of gene expression in rat muscles induced by EAMG, triplicate RNA samples from EOM, DIA, and EDL were prepared and further processed for microarray hybridization from McAb-3 antibody–induced EAMG rats and McAb-1–injected control rats. The expression profiling showed 331, 276, and 164 increased and 164,103, and 67 decreased (Supplementary Table S2) transcripts in EOM, EDL, and DIA, respectively, of EAMG-induced rats compared with those injected with control antibody with a total of 787 differentially regulated genes among the three muscle groups. Hierarchical cluster analysis of these 787 transcripts (Fig. 1) showed a distinct expression pattern of the genes found to be differentially influenced by induction of EAMG in rats. Approximately two-thirds of the differentially regulated genes were increased (528 of 787), whereas one-third of the differentially regulated genes were reduced (259 of 787). Comparison of the differentially regulated transcripts across all three muscle groups (Fig. 2) identified 62 upregulated transcripts (Table 1) and 18 downregulated transcripts (Table 2) common to all three muscle groups. All transcripts that showed differential expression were classified into basic categories of function: response to stress/stimulus, immune response, transcription factor, metabolism, cell adhesion/migration, cell-cell signaling/signal transduction, cell growth/maintenance, regeneration related, muscle related, cell death/apoptosis, transcription factor related, protein synthesis/modification, receptor/ion channel plus some unclassified molecules and EST. The percentage of transcripts by category among EOM, DIA, and EDL are listed in Table 3. The functional classification demonstrated that highly expressed transcripts among all muscles were predominantly in the areas of immune response, response to stress, and metabolism.
Figure 1.

Hierarchical clustering of the 787 gene probes identified as differentially expressed in EAMG rats compared to control rats among EOM, DIA, and EDL muscle groups. Transcripts identified are those at the intersection of data obtained with the Affymetrix MAS and RMA algorithms. The three independent replicates of each group are represented. The scale at the top right denotes normalized expression levels (red, high expression; blue, low expression).
Figure 2.

Venn diagram showing the numbers of differentially expressed transcripts in EAMG muscles compared with control rats shared by or unique to EOM, DIA, and EDL.
Table 1.
Common Gene Transcript Increases Common to All Muscles
|
Accession No. |
Gene |
Mean Fold Difference |
||
|
EOM |
Diaphragm |
EDL |
||
| Immune response | ||||
| AB001382 | Osteopontin (Spp1) | 33.0 | 11.2 | 30.2 |
| NM_133298 | Glycoprotein (transmembrane) nmb (Gpnmb) | 22.0 | 6.3 | 12.4 |
| AA945643 | Chitinase 3-like 1 (RGD:620874) | 14.0 | 5.2 | 17.5 |
| BI284255 | FK506 binding protein 5 (predicted) (RGD:1309155) | 9.0 | 13.3 | 9.7 |
| AW534837 | FK506 binding protein 5 (predicted) (RGD:1309155) | 7.6 | 11.3 | 8.4 |
| BF555488 | CD163 antigen (predicted) (RGD:1310382) | 4.6 | 2.90 | 3.3 |
| NM_031832 | Lectin, galactose binding, soluble 3 (Lgals3) | 4.2 | 2.0 | 2.9 |
| NM_012580 | Heme oxygenase (decycling) 1 (Hmox1) | 3.7 | 4.5 | 3.8 |
| BF419200 | CCAAT/enhancer binding protein (C/EBP), delta (Cebpd) | 2.9 | 6.3 | 6.1 |
| NM_017020 | Interleukin 6 receptor (Il6r) | 2.8 | 3.1 | 5.2 |
| BI276554 | Transcribed locus | 2.3 | 3.0 | 3.4 |
| BI284739 | LPS-induced TNF-alpha factor (RGD:69294) | 2.3 | 2.0 | 2.1 |
| NM_080903 | Tripartite motif-containing 63 (Trim63) | 2.2 | 5.3 | 4.5 |
| NM_013154 | CCAAT/enhancer binding protein (C/EBP), delta (Cebpd) | 2.1 | 5.7 | 6.5 |
| AB025017 | Zinc finger protein 36 (Zfp36) | 2.0 | 3.2 | 4.4 |
| Metabolism | ||||
| NM_053551 | Pyruvate dehydrogenase kinase, isoenzyme 4 (Pdk4) | 4.8 | 5.3 | 9.0 |
| M29853 | Cytochrome P450, family 4, subfamily b, polypeptide 1 (Cyp4b1) | 4.0 | 2.5 | 2.1 |
| U92069 | Uncoupling protein 3 (Ucp3) | 3.6 | 3.8 | 3.2 |
| BI274605 | Uncoupling protein 3 (Ucp3) | 3.5 | 4.1 | 3.5 |
| NM_031544 | Adenosine monophosphate deaminase 3 (Ampd3) | 3.1 | 3.9 | 15.6 |
| NM_053433 | Flavin containing monooxygenase 3 (RGD:619761) | 3.0 | 4.0 | 3.7 |
| BI279069 | Diacylglycerol O-acyltransferase homolog 2 (mouse) (Dgat2) | 2.4 | 3.1 | 2.5 |
| NM_012792 | Flavin containing monooxygenase 1 (Fmo1) | 2.3 | 2.1 | 2.4 |
| NM_017154 | Xanthine dehydrogenase (Xdh) | 2.1 | 2.1 | 2.5 |
| AF394783 | Sulfotransferase family 1A, phenol-preferring, member 1 (Sult1a1) | 2.0 | 2.4 | 2.3 |
| NM_133521 | F-box only protein 32 (Fbxo32) | 2.0 | 3.6 | 4.8 |
| Response to stress/stimulus | ||||
| AB001382 | Osteopontin (Spp1) | 33.0 | 11.2 | 30.2 |
| AF411318 | Metallothionein (Mt1a) | 26.2 | 32.1 | 64.7 |
| AA849895 | Ankyrin repeat domain 2 (stretch responsive muscle) (predicted) (RGD:1305104) | 8.1 | 5.4 | 5.2 |
| NM_024127 | Growth arrest and DNA-damage-inducible 45 alpha (Gadd45a) | 4.9 | 4.2 | 8.1 |
| NM_080906 | DNA-damage-inducible transcript 4 (Ddit4) | 4.3 | 12.1 | 10.6 |
| NM_012580 | Heme oxygenase (decycling) 1 (Hmox1) | 3.7 | 4.5 | 3.8 |
| U92069 | Uncoupling protein 3 (Ucp3) | 3.6 | 3.8 | 3.2 |
| U24174 | Cyclin-dependent kinase inhibitor 1A (Cdkn1a) | 3.4 | 4.1 | 15.8 |
| AI406908 | Similar to arsenite inducible RNA associated protein (predicted) (RGD1310991_predicted) | 3.0 | 4.7 | 7.0 |
| AI169756 | Similar to Mitogen-inducible gene 6 protein homolog (Mig-6) (Gene 33 polypeptide) | 2.7 | 5.0 | 8.4 |
| AB025017 | Zinc finger protein 36 (Zfp36) | 2.0 | 3.2 | 4.4 |
| Transcription factor | ||||
| NM_013220 | Ankyrin repeat domain 1 (cardiac muscle) (Ankrd1) | 65.5 | 31.3 | 20.7 |
| NM_017325 | Runt related transcription factor 1 (Runx1) | 8.0 | 4.1 | 6.8 |
| BM392224 | TG interacting factor (predicted) (RGD:1310517) | 3.4 | 5.2 | 9.7 |
| BF419200 | CCAAT/enhancer binding protein (C/EBP), delta (Cebpd) | 2.9 | 6.3 | 6.1 |
| AI411375 | V-ets erythroblastosis virus E26 oncogene homolog 2 (avian) (Ets2) | 2.5 | 2.4 | 2.8 |
| NM_080903 | Tripartite motif-containing 63 (Trim63) | 2.2 | 5.3 | 4.5 |
| NM_013154 | CCAAT/enhancer binding protein (C/EBP), delta (Cebpd) | 2.1 | 5.7 | 6.5 |
| AB025017 | Zinc finger protein 36 (Zfp36) | 2.0 | 3.2 | 4.4 |
| Cell adhesion/migration | ||||
| AB001382 | Osteopontin (Spp1) | 33.0 | 11.0 | 30.0 |
| AA818262 | Angiopoietin-like protein 4 (Angptl4) | 12.5 | 13 | 17.3 |
| AI548856 | Poliovirus receptor (PVR) | 8.5 | 5.9 | 11.2 |
| NM_031832 | Lectin, galactose binding, soluble 3 (Lgals3) | 4.2 | 2.0 | 2.9 |
| Muscle | ||||
| NM_013220 | Ankyrin repeat domain 1 (cardiac muscle) (Ankrd1) | 65.5 | 31.3 | 20.7 |
| NM_017325 | Runt related transcription factor 1 (Runx1) | 8.0 | 4.1 | 6.8 |
| AI411375 | V-ets erythroblastosis virus E26 oncogene homolog 2 (avian) (Ets2) | 2.5 | 2.4 | 2.8 |
| NM_133521 | F-box only protein 32 (Fbxo32) | 2.0 | 3.6 | 4.8 |
| Regeneration | ||||
| NM_013220 | Ankyrin repeat domain 1 (cardiac muscle) (Ankrd1) | 65.5 | 31.3 | 20.7 |
| AB001382 | Osteopontin (Spp1) | 33.0 | 11.2 | 30.2 |
| NM_012792 | Flavin containing monooxygenase 1 (Fmo1) | 2.3 | 2.1 | 2.4 |
| Cell growth/maintenance | ||||
| BI288701 | B-cell translocation gene 2, anti-proliferative (Btg2) | 3.5 | 2.1 | 3.6 |
| U24174 | Cyclin-dependent kinase inhibitor 1A (Cdkn1a) | 3.4 | 4.1 | 15.8 |
| AI179988 | Ectodermal-neural cortex 1 (RGD:1303152) | 2.1 | 4.1 | 5.3 |
| Cell-cell signaling/signal transduction | ||||
| NM_133298 | Glycoprotein (transmembrane) nmb (Gpnmb) | 22.0 | 6.3 | 12.4 |
| NM_053551 | Pyruvate dehydrogenase kinase, isoenzyme 4 (Pdk4) | 4.8 | 5.3 | 9.0 |
| NM_053338 | Ras-related associated with diabetes (Rrad) | 2.1 | 3.6 | 28.6 |
| Cell death/apoptosis | ||||
| NM_024127 | Growth arrest and DNA-damage-inducible 45 alpha (Gadd45a) | 4.9 | 4.2 | 8.1 |
| NM_012580 | Heme oxygenase (decycling) 1 (Hmox1) | 3.7 | 4.5 | 3.8 |
| Protein synthesis/modification | ||||
| None | ||||
| Receptor/ion channel | ||||
| None | ||||
| Other | ||||
| AI010427 | Ultraviolet B radiation-activated UV96 mRNA, partial sequence | 2.3 | 3.4 | 10.8 |
| NM_053819 | Tissue inhibitor of metalloproteinase 1 (Timp1) | 23.2 | 6.0 | 8.5 |
| Unclassified | ||||
| None | ||||
| EST | ||||
| BM383531 | Transcribed locus | 25.2 | 30.8 | 31.4 |
| BI274903 | Similar to RIKEN cDNA 2310057H16 (predicted) (RGD1305887_predicted) | 6.7 | 2.9 | 9.8 |
| BI294706 | Similar to MS4A6B protein (MGC94557) | 5.3 | 2.0 | 2.3 |
| AA859079 | Transcribed locus | 4.6 | 5.4 | 4.8 |
| BM385061 | Transcribed locus | 3.9 | 2.4 | 3.7 |
| AA956555 | Transcribed locus, strongly similar to XP_232342.2 similar to macrophage hemoglobin scavenger receptor CD163 precursor (Rattus norvegicus) | 3.6 | 2.8 | 3.0 |
| BG371620 | Transcribed locus | 3.1 | 2.4 | 2.5 |
| AA850715 | Transcribed locus | 2.9 | 2.4 | 3.0 |
| AI170665 | Transcribed locus | 2.9 | 4.3 | 2.2 |
| BI278547 | Hypothetical LOC300027 (LOC300027) | 2.9 | 2.1 | 2.1 |
| AI172302 | Transcribed locus | 2.9 | 2.6 | 6.9 |
| BF565188 | Transcribed locus | 2.7 | 2.2 | 2.4 |
| BF396512 | Transcribed locus | 2.7 | 3.6 | 3.1 |
| AA925542 | Transcribed locus | 2.5 | 2.7 | 3.7 |
| BF565188 | Transcribed locus | 2.2 | 2.2 | 2.4 |
| BE115141 | Transcribed locus | 2.1 | 2.2 | 2.3 |
| BI282296 | Transcribed locus | 2.1 | 2.2 | 2.0 |
| AI045724 | Transcribed locus | 2.1 | 3.5 | 4.1 |
| AI412174 | Similar to hypothetical protein MGC34760 (MGC94799) | 2.1 | 3.0 | 3.4 |
Table 2.
Common Gene Transcript Reductions Across All Muscle Groups
|
Accession No. |
Gene |
Mean Fold Difference |
||
|
EOM |
Diaphragm |
EDL |
||
| Immune response | ||||
| AI406939 | Putative lymphocyte G0/G1 switch gene | −3.1 | −2.1 | −3.9 |
| BG381127 | Ly6/neurotoxin 1 (predicted) (RGD:1312017) | −2.0 | −2.5 | −2.0 |
| Metabolism | ||||
| AA945624 | NAD(P)H dehydrogenase, quinone 2 (RGD:1303320) | −2.2 | −2.3 | −2.3 |
| Response to stress/stimulus | ||||
| NM_031750 | Heat shock 27kD protein family, member 3 (Hspb3) | −2.7 | −2.9 | −2.3 |
| Transcription factor | ||||
| BE111791 | CDNA clone MGC:95090 IMAGE:7126503, complete cds | −2.4 | −2.3 | −2.0 |
| Cell adhesion/migration | ||||
| AA799521 | Similar to RIKEN cDNA 9830160G03 (predicted) (RGD1305167_predicted) | −4.2 | −3.5 | −4.3 |
| Muscle | ||||
| NM_017130 | Neuraminidase 2 (Neu2) | −4.6 | −5.3 | −5.2 |
| AA799521 | Similar to RIKEN cDNA 9830160G03 (predicted) (RGD1305167_predicted) | −4.2 | −3.5 | −4.3 |
| NM_031750 | Heat shock 27kD protein family, member 3 (Hspb3) | −2.7 | −2.9 | −2.3 |
| U22830 | Purinergic receptor P2Y, G-protein coupled 1 (P2ry1) | −2.3 | −2.7 | −3.2 |
| AA945624 | NAD(P)H dehydrogenase, quinone 2 (RGD:1303320) | −2.2 | −2.3 | −2.3 |
| Regeneration | ||||
| NM_017130 | Neuraminidase 2 (Neu2) | −4.6 | −5.3 | −5.2 |
| BE107450 | Transcribed locus, strongly similar to NP_835197.1 neuronal regeneration related protein (Rattus norvegicus) | −3.3 | −3.2 | −2.6 |
| U22830 | Purinergic receptor P2Y, G-protein coupled 1 (P2ry1) | −2.3 | −2.7 | −3.2 |
| Cell growth/maintenance | ||||
| AI406939 | Putative lymphocyte G0/G1 switch gene | −3.1 | −2.1 | −3.9 |
| Cell-cell signaling/signal transduction | ||||
| NM_053703 | Mitogen-activated protein kinase kinase 6 (RGD:620666) | −2.9 | −3.2 | −2.8 |
| NM_022618 | A kinase (PRKA) anchor protein 6 (Akap6) | −2.4 | −2.1 | −2.1 |
| U22830 | Purinergic receptor P2Y, G-protein coupled 1 (P2ry1) | −2.3 | −2.7 | −3.2 |
| Cell death/apoptosis | ||||
| BM389225 | Angiopoietin-like 2 (Angptl2) | −2.1 | −3.2 | −2.9 |
| Protein synthesis/modification | ||||
| None | ||||
| Receptor/ion channel | ||||
| None | ||||
| Other | ||||
| None | ||||
| Unclassified | ||||
| BI289329 | Similar to hypothetical protein DKFZp434H2010 | −3.1 | −2.4 | −2.8 |
| BI289459 | Hypothetical LOC300061 (LOC300061) | −2.6 | −2.2 | −2.8 |
| AI013568 | Similar to 1300013J15Rik protein | −2.5 | −2.9 | −3.0 |
| EST | ||||
| BE111310 | Transcribed locus | −6.1 | −10.1 | −7.1 |
| BF283398 | Transcribed locus | −3.2 | −2.4 | −2.6 |
| AI070365 | Transcribed locus | −2.2 | −2.5 | −3.6 |
Table 3.
Transcript Change in Percent by Category
|
Shared |
EOM |
DIA |
EDL |
|
| Response to stress/stimulus | 12.8 | 3.8 | 6.4 | 5.4 |
| Immune response | 10.5 | 15.7 | 7.2 | 7.8 |
| Metabolism | 10.5 | 9.8 | 12.1 | 9.3 |
| Cell adhesion/migration | 3.8 | 2.5 | 2.7 | 2.2 |
| Cell-cell signaling/signal transduction | 4.5 | 5.2 | 5.2 | 5.2 |
| Cell growth/maintenance | 3.0 | 2.7 | 3.3 | 3.9 |
| Regeneration related | 5.3 | 1.5 | 3.9 | 2.6 |
| Muscle related | 6.8 | 4.3 | 5.8 | 4.1 |
| Cell death/apoptosis | 2.3 | 2.0 | 3.3 | 2.2 |
| Transcription factor related | 9.0 | 5.5 | 7.6 | 8.5 |
| Protein synthesis/modification | 1.5 | 2.5 | 2.7 | 3 .2 |
| Receptor/ion channel related | 1.5 | 1.8 | 2.7 | 2.6 |
| Other | 1.5 | 3.5 | 3.6 | 2.4 |
| Unclassified function | 4.5 | 7.5 | 3.9 | 4.8 |
| ESTs | 22.6 | 31.5 | 30 | 35.6 |
The percentage of transcripts in each category was calculated among EOM, DIA, and EDL separately and calculated in the commonly shared transcript list.
A disease load index was calculated in a similar fashion to a study of muscular dystrophy24 (Fig. 3). Both increased and decreased transcripts contribute to the RNA expression assessment of the disease impact and summing their absolute fold change values provides a single transcriptional index of EAMG pathology. The higher number of induced/repressed transcripts in the EOM versus EDL and DIA is consistent with greater disease pathology affecting the EOM. The disease load index based on the increased gene expression levels for EOM was substantially higher, as was the disease load index of reduced gene levels, to a lesser degree, in comparison with the other two muscles.
Figure 3.
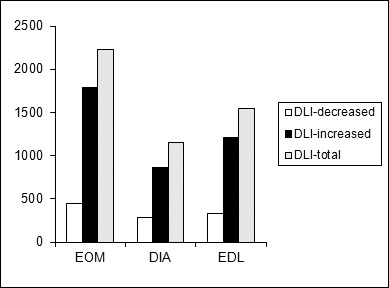
Aggregate of disease load index (DLI) plots for EOM, DIA, and EDL in EAMG rats, illustrating disease severity in different muscles by summing the absolute values of fold changes of differentially regulated in each muscle.
Validation of RNA Profile
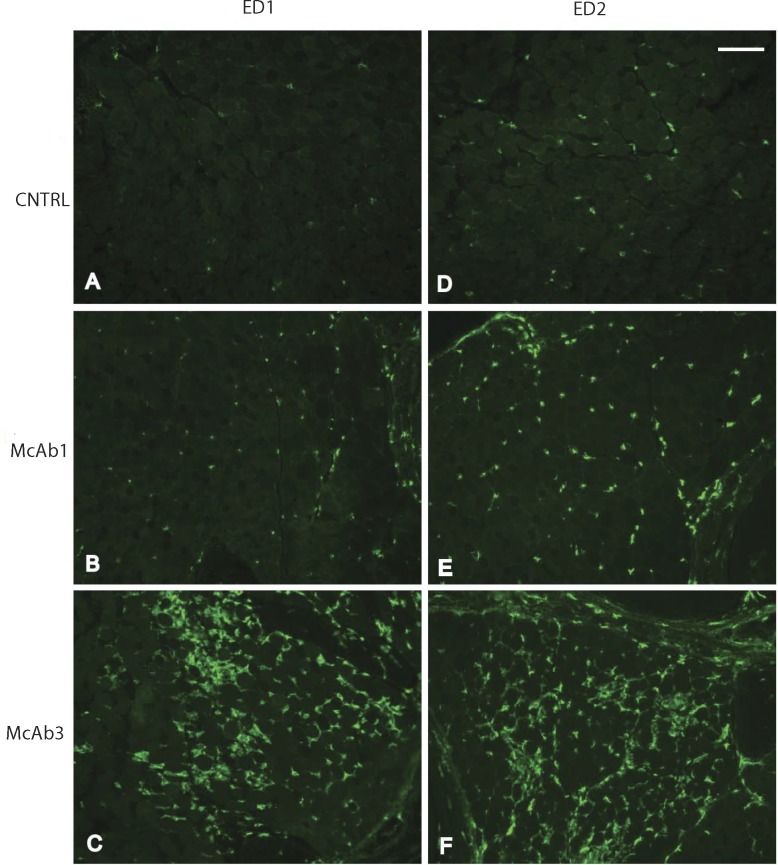
The qPCR and histological assessment validated results of RNA profiling. The expression levels of eight upregulated genes (Timp1, Spp1, Runx1, Myog1, Mt1a, Gpnmb, Gadd45a, Cebpd) and four downregulated genes (P2ry1, Neu2, fibronectin, Ly6) assessed by qPCR correlated well with the profiling analysis (Table 4). These specific genes were selected for validation because of their specific involvement in muscle, immune system, or cellular repair function. In keeping with a large percentage of immune response gene alterations, histological assessment demonstrated cellular infiltration of muscle, consistent with inflammatory infiltrates, which was more prominent in EOM (Fig. 4). Among the immune response genes identified, CD68 and CD163 are expressed in the infiltrating and residential macrophages. Figure 5 shows that in the untreated normal rat EOM, there are rare positive cells for ED1 (which recognizes CD68 antigen, Fig. 5A) and for CD163 (which recognizes CD163 antigen, Fig. 5B). The number of positive cells increased only slightly in McAb-1–injected control rat EOM (Figs. 5C, 5D); however, EAMG produced by administration of McAb-3 induced an obvious increase of CD68- and CD163-positive cells (Figs. 5E, 5F).
Table 4.
Fold Change Comparison Between Real-Time PCR and Microarray
|
Gene Symbol |
Gene Title |
EDL |
EOM |
DIA |
|||
|
qPCR |
Array |
qPCR |
Array |
qPCR |
Array |
||
| Timp1 | Tissue inhibitor of metalloproteinase 1 | 8.9 ± 2.9 | 8.5 | 20 ± 3.0 | 23 | 9.0 ± 2.3 | 6.0 |
| Spp1 | Secreted phosphoprotein 1 | 30 ± 1.7 | 30 | 82 ± 0.3 | 33 | 22 ± 1.8 | 11 |
| Runx1 | Runt-related transcription factor 1 | 18 ± 3.5 | 6.8 | 12 ± 2.0 | 7.9 | 1.4 ± 0.3 | 4.1 |
| Myog | Myogenin | 15 ± 1.6 | 6.4 | 22 ± 2.6 | 9.4 | 3.0 ± 1.2 | 2.3 |
| Mt1a | Metallothionein | 262 ± 3.2 | 65 | 54 ± 0.8 | 26 | 50 ± 1.7 | 32 |
| Gpnmb | Glycoprotein (transmembrane) nmb | 20 ± 4.0 | 12 | 31 ± 3.0 | 22 | 2.0 ± 1.2 | 6.2 |
| Gadd45a | Growth arrest and DNA-damage–inducible 45 alpha | 30 ± 7.2 | 8.1 | 41 ± 10 | 4.9 | 18 ± 4.5 | 4.2 |
| Cebpd | CCAAT/enhancer binding protein (C/EBP), delta | 22 ± 1.5 | 6.5 | 5.5 ± 0.1 | 2.9 | 6.2 ± 0.6 | 6.3 |
| P2ry1 | Purinergic receptor P2Y, G-protein coupled 1 | −3.5 ± 0.1 | −3.2 | −1.6 ± 0.1 | −2.3 | −2.1 ± 0.1 | −2.7 |
| Neu2 | Neuraminidase 2 | −8.7 ± 1.2 | −5.2 | −2.9 ± 0.4 | −4.6 | −3.6 ± 1.4 | −5.3 |
| 218860 | Fibronectin, type III /// SPla/RYanodine receptor SPRY | −4.3 ± 1.2 | −4.3 | −11 ± 3.6 | −4.2 | −4.3 ± 2.2 | −3.5 |
| 216965 | Ly6/neurotoxin 1 (predicted)/CD59 antigen | 1.6 ± 0.1 | −2.0 | −1.8 ± 0.1 | −2.0 | −2.3 ± 0.3 | −2.5 |
Figure 4.
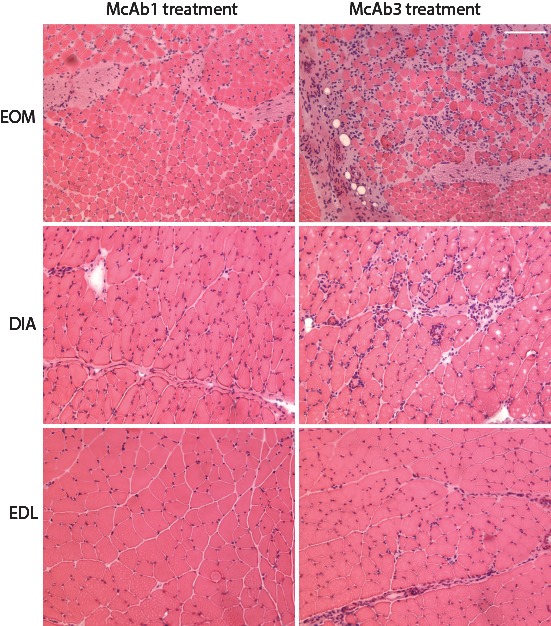
Histological sections stained with hematoxylin and eosin and viewed at low magnification to provide a representative view from EOM, EDL, and DIA from control and McAb-3 injected mice. Note more extensive inflammatory inflammation in EOM compared with other muscles.
Figure 5.
Validation by immunohistochemistry of increased expression of CD163 and CD68 in EAMG EOM compared with control EOM. Ten-micron EOM cross sections from McAb-3–injected EAMG rats, McAb-1–injected control rats, and PBS-injected control rats were immunostained with mouse anti-rat CD68 ED1 (A, B, C) and mouse anti-rat with CD163 ED2 (D, E, F) muscle. Scale bar: 100 μm.
Discussion
The RNA expression profiling indicates that EOM has a differential response gene expression compared with diaphragm and a leg muscle to EAMG produced by administration of antibody directed toward the AChR. We calculated a disease load index to assess the degrees to which the gene expression of each muscle diverged from the nondiseased state, which offers a means to compare the muscles' global response to injury. The EOM had the greatest disease load index, indicating that EAMG had provoked the greatest alteration of RNA expression in EOM, suggesting a greater degree of injury. The greater extent of inflammatory infiltration of EOM also indicates greater involvement of EOM by EAMG. Therefore, as in humans with MG, the EOMs appear to be preferentially involved by antibody-mediated attack of the neuromuscular junction. Finding that a large proportion of gene transcript alterations were related to immune function supports our hypothesis that the EOM, and more broadly the supportive tissue, are a unique immunological environment compared with other skeletal muscles.11,23,26 We suspect that intrinsic properties make the EOM particularly susceptible to passively induced EAMG,11,23 as appears to be the case for the preferential sparing of EOM by muscular dystrophies.6,10,27
Our conclusions are subject to some caveats. The EOMs are highly vascular,28 and therefore delivery of pathogenic antibody could be greater than other muscle. This could contribute to their greater damage, which would not be related directly to the intrinsic immune environment of the EOMs. In addition, the innervational ratio of EOM is higher than other muscles29 and, therefore, could be subject to greater antibody-induced injury from an AChR-specific antibody per area of muscle. This explanation is unlikely, as the amount of antibody infused is extremely high and likely to bind a large proportion on AChR at all neuromuscular junctions. The greater disease load index of EOM could represent a greater adaptive response to injury than other muscle, but this would not change our conclusion of EOM having a divergent response to EAMG compared with the other muscles.
Expression Profiling Similarities Across Muscles
The three muscles shared an increase in gene expression involved in regenerative processes, and several of these genes are known to be expressed in both muscle and cells of the immune system. Osteopontin gene transcripts were dramatically increased more than 30-fold in EOM and EDL and 10 times in DIA. Osteopontin, which is secreted from myoblasts and various immune cells, is involved in recruitment of inflammatory cells and muscle regeneration.30,31 Osteoactivin (GPNMB) is a type I transmembrane glycoprotein that is expressed in various cell types, including skeletal muscle and macrophages. Osteoactivin has immunosuppressive effects32 and promotes maintenance of innervation33; each of these properties would be particularly beneficial for recovery from injury induced by antibody attack of the neuromuscular junction. The FK506 binding protein 5 is a member of the immunophilin family, which is involved in calcineurin inhibition, protein folding and trafficking, and immune regulation. In common with osteopontin and osteoactivin, the gene is expressed in normal skeletal muscle and several component lines of the immune system. The MG sera and passive EAMG compromises muscle contractility beyond that expected by the neuromuscular transmission defect.34,35 The increase of FK506-binding protein, which moderates calcium activation of the ryanodine receptor, could explain this phenomenon.
Assessment by immunohistochemistry identified both CD68- and CD163-positive cells within EOM. The CD68 marker is generally accepted to identify M1 macrophages with a proinflammatory phenotype, whereas the CD163 marker identifies M2 macrophages, which are primarily involved in tissue repair.36 The CD163 RNA expression was markedly elevated in the RNA profile. It should be appreciated that neither marker definitively differentiates pro- and anti-inflammatory macrophages.36 Taken together, the data indicate at this early stage of antibody-induced injury both populations of macrophages are present in EOM, indicating that proinflammatory and tissue repair is occurring simultaneously, which is consistent with the entire RNA profile.
The genes Runx1 and Ankrd1, which were most likely exclusively muscle derived, were increased in each muscle group; Runx1 has been found to increase in response to denervation and prevent atrophy of skeletal muscle.37,38 The genes Spp1 and Rrad are influenced by Runx1 expression, and Ankrd1 is a member of a conserved gene family, referred to as muscle ankyrin repeat proteins (MARPs). The expression of MARPs is induced on injury and hypertrophy, stretch or denervation, and during recovery following starvation, suggesting that they are involved in muscle stress response pathways.39 Increase in Runx1 and Ankrd1 is also observed with axotomy and mouse models of motor neuron disease, which has been found to have early neuromuscular junction degeneration. The Lgals3 gene (galectin), which we also found increased, is elevated in expression also in these other disease models.40,41 The CCAAT/enhancer binding protein (C/EBP), delta (Cebpd) is a transcription factor found in immune cells and several other tissues,42 which is increased with motor neuron degeneration and food deprivation, in which it enhances myostatin expression.43 Deranged neuromuscular transmission or the reduction of animal weight, presumably caused by reduced food intake, could have led to Cebpd increase.
Chitinase 3–like 1, a member of a family of genes that are induced at sites of infection, was elevated across all three muscles. These genes are part of innate antipathogen responses and appear to minimize oxidative damage and thereby reduce tissue injury. They increase the adaptive immune response to cancer and infection to eradicate pathogen invasion.44 Chitinase proteins have been shown to enhance tissue healing and fibrosis.
The Ly6/neurotoxin 1 was the only one of two immune-related genes found to be reduced in all three muscles; the other was a gene whose function is as yet to be characterized (see Table 2). In addition to being a cell surface marker of lymphocytes, Ly6 activates nicotinic acetylcholine receptors in various tissues, which may lead to cell injury.45,46 Its downregulation may serve a protective effect from potential synapse injury from depolarization and subsequent calcium influx, as observed in slow channel syndrome.47 There was a general lack of alterations in interleukins and other cytokines observed to be elevated in circulation or circulating cells in MG, although there was a small increase in IL-6 receptor gene expression.
In addition to osteopontin, three cell adhesion–related genes were found to be increased across each of the three muscles. Angiopoietin-like protein 4 promotes angiogenesis in the context of wound healing and is activated during induction of systemic inflammation.48,49 The poliovirus receptor gene is a transmembrane glycoprotein and is part of the immunoglobulin superfamily with functional domains that mediate cell attachment to vitronectin and an intracellular domain that binds dynein. Its increased expression is most likely related to immune cell infiltration, in which it is known to function.50 Lgals3 is a member of the lectin family and is involved in many processes, including inflammation and tissue repair.51,52
Myogenin, the myogenic regulatory factor, was increased among all muscles, which would also support repair of injured muscle and activation of satellite cells. In contrast, Myf6 was increased only in EDL, and DIA and MyoD transcripts were only elevated EOM. These observations point to fundamental differences in response to injury among the muscles. These observations support the postulate that EOM is unique in its repair mechanism. McLoon and colleagues53,54 proposed that EOM is under a constant active state of remodeling, which explains its sparing by most muscular dystrophies.
Altered Metabolism
Nearly a quarter of the differentially expressed genes common to all muscles were related to cellular metabolism and an upregulation of transcripts occurred rather than a suppression. This observation is surprising, as the EOM, EDL, and DIA differ markedly in constitutive metabolic properties. Extraocular muscle does not have glycogen stores, has extremely high mitochondrial content, and relies on glucose and lactate as metabolic substrates.55 In contrast, EDL is relatively poor in mitochondrial content and is primarily glycolytic, whereas DIA has a mixture of fiber types, which rely on oxidative and glycolytic metabolism. The gene responsible for synthesis of the key metabolic enzyme, pyruvate dehydrogenase kinase (Pdk4), was increased, which would be expected to lead to conservation of glucose and reduce free radical production through the electron transport chain. The gene Dgat2 catalyzes the conversion from diacylglycerol to triglyceride and its elevated expression would be expected to enhance lipid synthesis.56 The Cyp4b1 gene promotes omega-hydroxylation of medium-chain fatty acids and is generated with acute inflammation.57–59 An increase in uncoupling protein 3 (Ucp3) occurs when fatty acid delivery to mitochondria exceeds capacity for oxidation, which appears to be occurring based on the described gene changes and its activity leads to reduced reactive oxygen species formation.60 Taken together, these relatively focused gene transcript alterations would favor oxidative metabolism and reduce free radical generation, perhaps to limit further tissue injury. Consistent with this conclusion, hemoxygenase-1 was elevated, which would be expected to reduce tissue injury with a potent anti-inflammatory influence as well as the promotion of wound healing.61
EOM-Specific RNA Expression Alterations
In addition to the overall disease load index being highest for EOM, immune-related gene expression changes were most prominent for the EOM compared with the other muscles. Part of this can be explained by the greater infiltration of immune cells, as genes specific for these cells (Chemokine receptor 2, Leukocyte peptidase receptor 2, Sectm1, CD18, Igsf6, major histocompatibility related genes, CD68, Ccr1, CD44) or chemotactic for lymphocytes (Ccl20 and other chemokine-ligands) are predominant in the EOM genomic profile. Three complement pathway genes (C5r1, C2, C1qb) were elevated only in EOM; C5r1 is likely acting to resolve acute inflammation as has been observed in other conditions,62 and C2 and C1qb are known to be expressed by myoblasts63,64 and cardiomyocytes,65 which may lead to enhanced inflammation or clearance of cellular debris. Consistent with our previous investigations, transcript levels of the intrinsic complement inhibitor, Daf, were reduced in response to EAMG in EOM, but not the other muscles.20
There is considerable evidence for skeletal muscle group heterogeneity from DNA microarray and proteomic studies. These all support the existence of broad-based, inter- and intra-allotype diversity.26,66–71 Genomic profiles support the notion that despite similarities in morphology (i.e., myotubes formed by a syncitium of myoblasts and striated appearance), EOM and skeletal muscles have significant differences in transcriptomes,68–70 which warrant EOM classification as a distinct allotype, as cardiac and smooth muscle. Comparing results of the present RNA profile with those previously done of the wild-type state,68–70 indicates that transcripts that are differentially expressed in EOM, for example, several myosin heavy chain isoforms, are not identified in the present profile. This observation indicates that induction of EAMG alters constitutive transcription.
In keeping with the baseline differences in the normal state, our results demonstrate that the inherent targeting of EOM by antibody attack is greater in EOM and the muscle responds in a fundamentally different way. This is consistent with observations of other muscle disorders, such as muscular dystrophies and motor neuron disease.6,10,27
Significance for Human Disease
The administration of antibody against the AChR mimics the final effector mechanism of human MG, and the antibody used in this study activates complement, which is a primary mechanism of disease induction in humans.13 However, it must be appreciated that the human disorder does not demonstrate inflammation,72 as is observed in the passive transfer model of MG used here, and therefore, the present results may be most relevant to inflammatory disorders that affect EOM. We demonstrated that EOMs have inherent properties that make them more susceptible to injury, as demonstrated by the greater disease load index. Essentially all patients with MG have ocular muscle involvement and a large minority has disease isolated to the ocular muscles. Therefore, the underlying autoimmune pathology appears not to be the determining factor, but rather inherent properties of muscle. The results demonstrate that a relatively limited alteration of global RNA expression of muscle occurs in response to EAMG induced by antibody transfer. Muscles activate genes that are regenerative. Gene profiles indicate that metabolism is altered to limit generation of reactive oxygen species, which may produce further injury. EOM has a much greater increase in gene expression, which suggests that either there is a greater degree of injury or a particularly robust response to a similar level of injury to all three muscles evaluated. It is impossible to absolutely distinguish these possibilities; however, the greater infiltration of inflammatory cells does suggest a greater degree of injury to us. Continued work defining the immune environment of EOM may lead to targeted immunotherapy for orbital diseases, such as ocular myasthenia, orbital myositis, and Graves' ophthalmopathy.6,10,27
Acknowledgments
Supported by National Institutes of Health Grant R24EY014837 (HJK, LK).
Disclosure: Y. Zhou, None; H.J. Kaminski, None; B. Gong, None; G. Cheng, None; J.M. Feuerman, None; L. Kusner, None
References
- 1. Kaminski HJ, Li Z, Richmonds C, Ruff RL, Kusner L. Susceptibility of ocular tissues to autoimmune diseases. Ann N Y Acad Sci. 2003; 998: 362–374 [DOI] [PubMed] [Google Scholar]
- 2. Serra A, Ruff R, Kaminski H, Leigh RJ. Factors contributing to failure of neuromuscular transmission in myasthenia gravis and the special case of the extraocular muscles. Ann N Y Acad Sci. 2011; 1233: 26–33 [DOI] [PubMed] [Google Scholar]
- 3. Johnson M, Polgar J, Weightman D, Appleton D. Data on the distribution of fibre types in thirty-six human muscles: an autopsy study. J Neurol Sci. 1973; 18: 111–129 [DOI] [PubMed] [Google Scholar]
- 4. Bansal D, Campbell KP. Dysferlin and the plasma membrane repair in muscular dystrophy. Trends Cell Biol. 2004; 14: 206–213 [DOI] [PubMed] [Google Scholar]
- 5. von der Hagen M, Laval SH, Cree LM, et al. The differential gene expression profiles of proximal and distal muscle groups are altered in pre-pathological dysferlin-deficient mice. Neuromuscul Disord. 2005; 15: 863–877 [DOI] [PubMed] [Google Scholar]
- 6. Kallestad KM, Hebert SL, McDonald AA, Daniel ML, Cu SR, McLoon LK. Sparing of extraocular muscle in aging and muscular dystrophies: a myogenic precursor cell hypothesis. Exp Cell Res. 2011; 317: 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lewis C, Ohlendieck K. Proteomic profiling of naturally protected extraocular muscles from the dystrophin-deficient mdx mouse. Biochem Biophys Res Comm. 2010; 396: 1024–1029 [DOI] [PubMed] [Google Scholar]
- 8. Pertille A, de Carvalho CL, Matsumura CY, Neto HS, Marques MJ. Calcium-binding proteins in skeletal muscles of the mdx mice: potential role in the pathogenesis of Duchenne muscular dystrophy. Int J Exp Pathol. 2010; 91: 63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harrison AR, Lee MS, McLoon LK. Effects of elevated thyroid hormone on adult rabbit extraocular muscles. Invest Ophthalmol Vis Sci. 2010; 51: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pedrosa Domellof F. Extraocular muscles: response to neuromuscular diseases and specific pathologies. In: McLoon LK, Andrade F, eds Craniofacial Muscles: A New Framework for Understanding the Effector Side of Craniofacial Muscle Control. New York: Springer-Verlag; 2013: 75–90 [Google Scholar]
- 11. Soltys J, Gong B, Kaminski HJ, Zhou Y, Kusner LL. Extraocular muscle susceptibility to myasthenia gravis: unique immunological environment? Ann N Y Acad Sci. 2008; 1132: 220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vincent A, Leite MI. Neuromuscular junction autoimmune disease: muscle specific kinase antibodies and treatments for myasthenia gravis. Curr Opin Neurol. 2005; 18: 519–525 [DOI] [PubMed] [Google Scholar]
- 13. Conti-Fine BM, Milani M, Kaminski HJ. Myasthenia gravis: past, present, and future. J Clin Invest. 2006; 116: 2843–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christadoss P, Poussin M, Deng C. Animal models of myasthenia gravis. Clin Immunol. 2000; 94: 75–87 [DOI] [PubMed] [Google Scholar]
- 15. Lennon VA, Lambert EH. Monoclonal autoantibodies to acetylcholine receptors: evidence for a dominant idiotype and requirement of complement for pathogenicity. Ann N Y Acad Sci. 1981; 377: 77–96 [DOI] [PubMed] [Google Scholar]
- 16. Morgan BP, Chamberlain-Banoub J, Neal JW, Song W, Mizuno M, Harris CL. The membrane attack pathway of complement drives pathology in passively induced experimental autoimmune myasthenia gravis in mice. Clin Exp Immunol. 2006; 146: 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin F, Kaminski H, Conti-Fine B, Wang W, Richmonds C, Medof M. Enhanced susceptibility to experimental autoimmune myasthenia gravis in the absence of decay-accelerating factor protection. J Clin Invest. 2002; 110: 1269–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruff RL, Lennon V. End-plate voltage-gated sodium channels are lost in clinical and experimental myasthenia gravis. Ann Neurol. 1998; 43: 370–379 [DOI] [PubMed] [Google Scholar]
- 19. Kaminski HJ, Kusner LL, Richmonds C, Medof ME, Lin F. Deficiency of decay accelerating factor and CD59 leads to crisis in experimental myasthenia. Exp Neurol. 2006; 202: 287–293 [DOI] [PubMed] [Google Scholar]
- 20. Kaminski HJ, Li Z, Richmonds C, Lin F, Medof ME. Complement regulators in extraocular muscle and experimental autoimmune myasthenia gravis. Exp Neurol. 2004; 189: 333–342 [DOI] [PubMed] [Google Scholar]
- 21. Lennon VA, Lambert EH, Leiby KR, Okama TB, Talib S. Recombinant human acetylcholine receptor alpha-subunit induces chronic experimental autoimmune myasthenia gravis. J Immunol. 1991; 146: 2245–2248 [PubMed] [Google Scholar]
- 22. Zhou Y, Gong B, Kaminski HJ. Genomic profiling reveals pitx2 controls expression of mature extraocular muscle contraction-related genes. Invest Ophthalmol Vis Sci. 2012; 53: 1821–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kusner LL, Young A, Tjoe S, Leahy P, Kaminski HJ. Perimysial fibroblasts of extraocular muscle, as unique as the muscle fibers. Invest Ophthalmol Vis Sci. 2010; 51: 192–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Porter JD, Merriam AP, Leahy P, Gong B, Khanna S. Dissection of temporal gene expression signatures of affected and spared muscle groups in dystrophin-deficient (mdx) mice. Hum Mol Gen. 2003; 12: 1813–1821 [DOI] [PubMed] [Google Scholar]
- 25. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 26. Porter JD, Merriam AP, Leahy P, et al. Temporal gene expression profiling of dystrophin-deficient (mdx) mouse diaphragm identifies conserved and muscle group-specific mechanisms in the pathogenesis of muscular dystrophy. Hum Mol Gen. 2004; 13: 257–269 [DOI] [PubMed] [Google Scholar]
- 27. McDonald AA, Kunz MD, McLoon LK. Dystrophic changes in extraocular muscles after gamma irradiation in mdx:utrophin(+/–) mice. PLoS One. 2014; 9: e86424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oh SY, Poukens V, Cohen MS, Demer JL. Structure-function correlation of laminar vascularity in human rectus extraocular muscles. Invest Ophthalmol Vis Sci. 2001; 42: 17–22 [PubMed] [Google Scholar]
- 29. Porter JD, Baker R. Muscles of a different “color”: The unusual properties of the extraocular muscles may predispose or protect them in neurogenic and myogenic disease. Neurol. 1996; 46: 30–37 [DOI] [PubMed] [Google Scholar]
- 30. Uaesoontrachoon K. Wasgewatte Wijesinghe DK, Mackie EJ, Pagel CN. Osteopontin deficiency delays inflammatory infiltration and the onset of muscle regeneration in a mouse model of muscle injury. Dis Model Mech. 2013; 6: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vetrone SA, Montecino-Rodriguez E, Kudryashova E, et al. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-beta. J Clin Invest. 2009; 119: 1583–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwarzbich MA, Gutknecht M, Salih J, et al. The immune inhibitory receptor osteoactivin is upregulated in monocyte-derived dendritic cells by BCR-ABL tyrosine kinase inhibitors. Cancer Immunol Immunother. 2012; 61: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Furochi H, Tamura S, Takeshima K, et al. Overexpression of osteoactivin protects skeletal muscle from severe degeneration caused by long-term denervation in mice. J Med Invest. 2007; 54: 248–254 [DOI] [PubMed] [Google Scholar]
- 34. Imai T, Tsuda E, Hozuki T, et al. Anti-ryanodine receptor-positive antibody acetylcholine receptor negative myasthenia gravis: evidence of impaired excitation-contraction coupling. Muscle Nerve. 2011; 43: 294–295 [DOI] [PubMed] [Google Scholar]
- 35. van Lunteren E, Moyer M, Kaminski HJ. Adverse effects of myasthenia gravis on rat phrenic diaphragm contractile performance. J Appl Physiol. 2004; 97: 895–901 [DOI] [PubMed] [Google Scholar]
- 36. Barros MH, Hauck F, Dreyer JH, Kempkes B, Niedobitek G. Macrophage polarisation: an immunohistochemical approach for identifying M1 and M2 macrophages. PLoS One. 2013; 8: e80908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X, Blagden C, Fan J, et al. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 2005; 19: 1715–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu CL, Kandarian SC, Jackman RW. Identification of genes that elicit disuse muscle atrophy via the transcription factors p50 and Bcl-3. PLoS One. 2011; 6: e16171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller MK, Bang ML, Witt CC, et al. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J Mol Biol. 2003; 333: 951–964 [DOI] [PubMed] [Google Scholar]
- 40. Takaku S, Yanagisawa H, Watabe K, et al. GDNF promotes neurite outgrowth and upregulates galectin-1 through the RET/PI3K signaling in cultured adult rat dorsal root ganglion neurons. Neurochem Internat. 2013; 62: 330–339 [DOI] [PubMed] [Google Scholar]
- 41. Barrette B, Calvo E, Vallieres N, Lacroix S. Transcriptional profiling of the injured sciatic nerve of mice carrying the Wld(S) mutant gene: identification of genes involved in neuroprotection, neuroinflammation, and nerve regeneration. Brain Behav Immun. 2010; 24: 1254–1267 [DOI] [PubMed] [Google Scholar]
- 42. Tsukada J, Yoshida Y, Kominato Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011; 54: 6–19 [DOI] [PubMed] [Google Scholar]
- 43. Allen DL, Cleary AS, Hanson AM, Lindsay SF, Reed JM. CCAAT/enhancer binding protein-delta expression is increased in fast skeletal muscle by food deprivation and regulates myostatin transcription in vitro. Am J Physiol Regul Integr Comp Physiol. 2010; 299: R1592–R1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee CG, Da Silva CA, Dela Cruz CS, et al. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol. 2011; 73: 479–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miwa JM, Stevens TR, King SL, et al. The prototoxin lynx1 acts on nicotinic acetylcholine receptors to balance neuronal activity and survival in vivo. Neuron. 2006; 51: 587–600 [DOI] [PubMed] [Google Scholar]
- 46. Ibanez-Tallon I, Miwa JM, Wang HL, et al. Novel modulation of neuronal nicotinic acetylcholine receptors by association with the endogenous prototoxin lynx1. Neuron. 2002; 33: 893–903 [DOI] [PubMed] [Google Scholar]
- 47. Gomez CM, Maselli R, Gundeck JE, et al. Slow-channel transgenic mice: a model of postsynaptic organellar degeneration at the neuromuscular junction. J Neurosci. 1997; 17: 4170–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lu B, Moser A, Shigenaga JK, Grunfeld C, Feingold KR. The acute phase response stimulates the expression of angiopoietin like protein 4. Bioch Biophys Research Com. 2010; 391: 1737–1741 [DOI] [PubMed] [Google Scholar]
- 49. Raschke S, Eckel J. Adipo-myokines: two sides of the same coin—mediators of inflammation and mediators of exercise. Med Inflam. 2013; 2013: 320724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lozano E, Joller N, Cao Y, Kuchroo VK, Hafler DA. The CD226/CD155 interaction regulates the proinflammatory (Th1/Th17)/anti-inflammatory (Th2) balance in humans. J Immunol. 2013; 191: 3673–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Henderson NC, Sethi T. The regulation of inflammation by galectin-3. Immunol Rev. 2009; 230: 160–171 [DOI] [PubMed] [Google Scholar]
- 52. Liu FT, Hsu DK. The role of galectin-3 in promotion of the inflammatory response. Drug News Perspect. 2007; 20: 455–460 [DOI] [PubMed] [Google Scholar]
- 53. McLoon LK, Rowe J, Wirtschafter J, McCormick KM. Continuous myofiber remodeling in uninjured extraocular myofibers: myonuclear turnover and evidence for apoptosis. Muscle Nerve. 2004; 29: 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McLoon LK, Wirtschafter J. Activated satellite cells in extraocular muscles of normal adult monkeys and humans. Invest Ophthalmol Vis Sci. 2003; 44: 1927–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Spencer RF, Porter JD. Biological organization of the extraocular muscles. Prog Brain Res. 2005; 151: 43–80 [DOI] [PubMed] [Google Scholar]
- 56. Levin MC, Monetti M, Watt MJ, et al. Increased lipid accumulation and insulin resistance in transgenic mice expressing DGAT2 in glycolytic (type II) muscle. Am J Physiol Endocrinol Metab. 2007; 293: E1772–E1781 [DOI] [PubMed] [Google Scholar]
- 57. Mezentsev A, Mastyugin V, Seta F, et al. Transfection of cytochrome P4504B1 into the cornea increases angiogenic activity of the limbal vessels. J Pharmacol Exp Ther. 2005; 315: 42–50 [DOI] [PubMed] [Google Scholar]
- 58. Baer BR, Rettie AE. CYP4B1: an enigmatic P450 at the interface between xenobiotic and endobiotic metabolism. Drug Metab Rev. 2006; 38: 451–476 [DOI] [PubMed] [Google Scholar]
- 59. Stoilov I, Krueger W, Mankowski D, et al. The cytochromes P450 (CYP) response to allergic inflammation of the lung. Arch Biochem Biophys. 2006; 456: 30–38 [DOI] [PubMed] [Google Scholar]
- 60. Toime LJ, Brand MD. Uncoupling protein-3 lowers reactive oxygen species production in isolated mitochondria. Free Radic Biol Med. 2010; 49: 606–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Patil K, Bellner L, Cullaro G, Gotlinger KH, Dunn MW, Schwartzman ML. Heme oxygenase-1 induction attenuates corneal inflammation and accelerates wound healing after epithelial injury. Invest Ophthalmol Vis Sci. 2008; 49: 3379–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nishiura H. The alternative C5a receptor function. Adv Exp Med Biol. 2013; 735: 111–121 [DOI] [PubMed] [Google Scholar]
- 63. Gasque P, Morgan BP, Legoedec J, Chan P, Fontaine M. Human skeletal myoblasts spontaneously activate allogeneic complement but are resistant to killing. J Immunol. 1996; 156: 3402–3411 [PubMed] [Google Scholar]
- 64. Legoedec J, Gasque P, Jeanne JF, Scotte M, Fontaine M. Complement classical pathway expression by human skeletal myoblasts in vitro. Mol Immunol. 1997; 34: 735–741 [DOI] [PubMed] [Google Scholar]
- 65. Yasojima K, Schwab C, McGeer EG, McGeer PL. Human heart generates complement proteins that are upregulated and activated after myocardial infarction. Circ Res. 1998; 83: 860–869 [DOI] [PubMed] [Google Scholar]
- 66. Campbell WG, Gordon SE, Carlson CJ, Pattison JS, Hamilton MT, Booth FW. Differential global gene expression in red and white skeletal muscle. Am J Physiol Cell Physiol. 2001; 280: C763–C768 [DOI] [PubMed] [Google Scholar]
- 67. Cheng G, Porter JD. Transcriptional profile of rat extraocular muscle by serial analysis of gene expression. Invest Ophthalmol Vis Sci. 2002; 43: 1048–1058 [PubMed] [Google Scholar]
- 68. Fischer MD, Budak MT, Bakay M, et al. Definition of the unique human extraocular muscle allotype by expression profiling. Physiol Genomics. 2005; 22: 283–291 [DOI] [PubMed] [Google Scholar]
- 69. Fischer MD, Gorospe JR, Felder E, et al. Expression profiling reveals metabolic and structural components of extraocular muscles. Physiol Genomics. 2002; 9: 71–84 [DOI] [PubMed] [Google Scholar]
- 70. Porter JD, Khanna S, Kaminski HJ, et al. Extraocular muscle is defined by a fundamentally distinct gene expression profile. Proc Natl Acad Sci U S A. 2001; 98: 12062–12067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Okumura N, Hashida-Okumura A, Kita K, et al. Proteomic analysis of slow- and fast-twitch skeletal muscles. Proteomics. 2005; 5: 2896–2906 [DOI] [PubMed] [Google Scholar]
- 72. Nakano S, Engel AG. Myasthenia gravis: quantitative immunocytochemical analysis of inflammatory cells and detection of complement membrane attack complex at the end-plate in 30 patients. Neurol. 1993; 43: 1167–1172 [DOI] [PubMed] [Google Scholar]