Significance
The immune system defends against pathogens through a diverse population of T cells that display different antigen recognition surface receptor proteins. Receptor diversity is produced by an initial random gene recombination process followed by selection for a desirable range of peptide binding. Although recombination is well-understood, selection has not been quantitatively characterized. By combining high-throughput sequencing data with modeling, we quantify the selection pressure that shapes functional repertoires. Selection is found to vary little between individuals or between naive and memory repertoires. It reinforces the biases of the recombination process, meaning that sequences more likely to be produced are also more likely to pass selection. The model accounts for public sequences shared between individuals as resulting from pure chance.
Keywords: thymic selection, statistical inference, public repertoire, T cell
Abstract
The efficient recognition of pathogens by the adaptive immune system relies on the diversity of receptors displayed at the surface of immune cells. T-cell receptor diversity results from an initial random DNA editing process, called VDJ recombination, followed by functional selection of cells according to the interaction of their surface receptors with self and foreign antigenic peptides. Using high-throughput sequence data from the β-chain of human T-cell receptors, we infer factors that quantify the overall effect of selection on the elements of receptor sequence composition: the V and J gene choice and the length and amino acid composition of the variable region. We find a significant correlation between biases induced by VDJ recombination and our inferred selection factors together with a reduction of diversity during selection. Both effects suggest that natural selection acting on the recombination process has anticipated the selection pressures experienced during somatic evolution. The inferred selection factors differ little between donors or between naive and memory repertoires. The number of sequences shared between donors is well-predicted by our model, indicating a stochastic origin of such public sequences. Our approach is based on a probabilistic maximum likelihood method, which is necessary to disentangle the effects of selection from biases inherent in the recombination process.
The T-cell response of the adaptive immune system begins when receptor proteins on the surface of these cells recognize a pathogen peptide displayed by an antigen-presenting cell. The immune cell repertoire of a given individual is comprised of many clones, each with a distinct surface receptor. This diversity, which is central to the ability of the immune system to defeat pathogens, is initially created by a stochastic process of germline DNA editing (called VDJ recombination) that gives each new immune cell a unique surface receptor gene. This initial repertoire is subsequently modified by selective forces, including nonpathogen-related thymic selection against excessive (or insufficient) recognition of self proteins, which are also stochastic in nature. Because of this stochasticity and the large T-cell diversity, these repertoires are best described by probability distributions. In this paper, we apply a probabilistic approach to sequence data to obtain quantitative measures of the overall (not necessarily pathogenic) selection pressures that shape T-cell receptor repertoires.
New receptor genes are formed by randomly choosing alleles from a set of genomic templates for the subregions (V, D, and J) of the complete gene. Insertion and deletion of nucleotides in the junctional regions between the V and D and D and J genes greatly enhance diversity beyond pure VDJ combinatorics (1). The most variable region of the gene is between the last amino acids of the V segment and the beginning of the J segment; it codes for the Complementarity Determining Region 3 (CDR3) loop of the receptor protein, a region known to be functionally important in recognition (2). Previous studies have shown that immune cell receptors are not uniform in terms of VDJ gene segment use (3–6) or probability of generation (1) and that certain receptors are more likely than others to be shared by different individuals (4, 7). The statistical properties of the immune repertoire are, thus, rather complex, and their accurate determination requires sophisticated methods.
Recent advances in sequencing technology have made it possible to sample the T-cell receptor diversity of individual subjects in great depth (8). The availability of such data has, in turn, led to the development of sequence statistics-based approaches to the study of immune cell diversity (9, 10). In particular, we recently quantitatively characterized the preselection diversity of the human T-cell repertoire by learning the probabilistic rules of VDJ recombination from out-of-frame DNA sequences that cannot be subject to functional selection and whose statistics therefore reflect only the recombination process (1). After generation, T cells undergo a somatic selection process in the thymus (11) and later in the periphery (12). Cells that pass thymic selection enter the peripheral repertoire as naive T cells, and the subset of naive cells that eventually engage in an immune response will survive as a long-lived memory pool. Although we now understand the statistical properties of the initial repertoire of immune receptors (1) and despite some theoretical studies of thymic selection at the molecular level (13, 14), a quantitative understanding of how selection modifies those statistics to produce the naive and memory repertoires is lacking.
In this paper, we build on our understanding of the preselection distribution of T-cell receptors to derive a statistical method for identifying and quantifying selection pressures in the adaptive immune system. We apply this method to naive and memory DNA sequences of human T-cell β-chains obtained from peripheral blood samples of nine healthy individuals. Our goal is to characterize the likelihood that any given sequence, after it is generated, will survive selection for the ensemble of properties needed to pass into the peripheral repertoire(s). Our analysis reveals strong and reproducible signatures of selection on specific amino acids in the CDR3 sequence and on the usage of V and J genes. Most strikingly, we find significant correlation between the generation probability of a sequence and the probability that it will pass selection. This correlation suggests that natural selection, which acts on very long timescales to shape the generation mechanism itself, may have tuned it to anticipate somatic selection, which acts on single cells throughout the lifetime of an individual. The quantitative features of selection inferred from our model vary very little between donors, indicating that these features are universal. In addition, our measures of selection pressure on the memory and naive repertoires are statistically indistinguishable, consistent with the hypothesis that the memory pool is a random subsample of the naive pool.
Analysis
We analyzed human CD4+ T-cell β-chain DNA sequence reads (60- or 101-nucleotide long) centered around the CDR3 region. T cells were obtained from nine individuals and sorted into naive (CD45RO−) and memory (CD45RO+) subsets, yielding datasets of ∼200,000 unique naive and ∼120,000 unique memory sequences per individual on average. The datasets are the same as those used in ref. 1 and were obtained by previously described methods (15, 16).
In ref. 1, we used the out-of-frame sequences to characterize the receptor generation process. That analysis yielded an accurate model for the probability that a VDJ recombination event will produce a β-chain gene consistent with the sequence read (for any ). In this study, we focus instead on the in-frame sequences free of stop codons, with the goal of quantifying how their probability of occurrence, , differs from the preselection distribution . (We only consider the presence or absence of a sequence and not the size of its clone.) Here, we distinguish between the read and the entire β-chain sequence, which is characterized uniquely by the V and J gene choices (denoted by V and J) as well as the CDR3 region ; the latter is defined to run from a conserved Cys near the end of the V segment to the last amino acid of the read (we note that the last amino acid in the read is separated from a conserved Phe in the J gene by two variable amino acids). The CDR3 sequence can be uniquely read off from each sequence read; by contrast, the V and J may not be uniquely identifiable (because of the relatively short read length). Because V and J may play a role in selection outside the read , we must consider selection in terms of the full β-chain rather than the incomplete .
For each β-chain sequence , we define a selection factor Q = Ppost/Ppre that quantifies whether selection (thymic selection or subsequent selection in the periphery) has enriched or impoverished the frequency of that sequence compared with the preselection ensemble. Because Ppre varies over many orders of magnitude, such a relative enhancement factor is the only way to define selection strength. Our goal is to find a model for Q, such that the distribution gives a good account of an observed set of selected sequences. We cannot directly estimate from the data, but as we outline in Fig. 1A, we can use a reduced complexity model for Q to infer it (and therefore Ppost) from the data. Specifically, we will show that the following factorized model for Q captures the main features of selection:
| [1] |
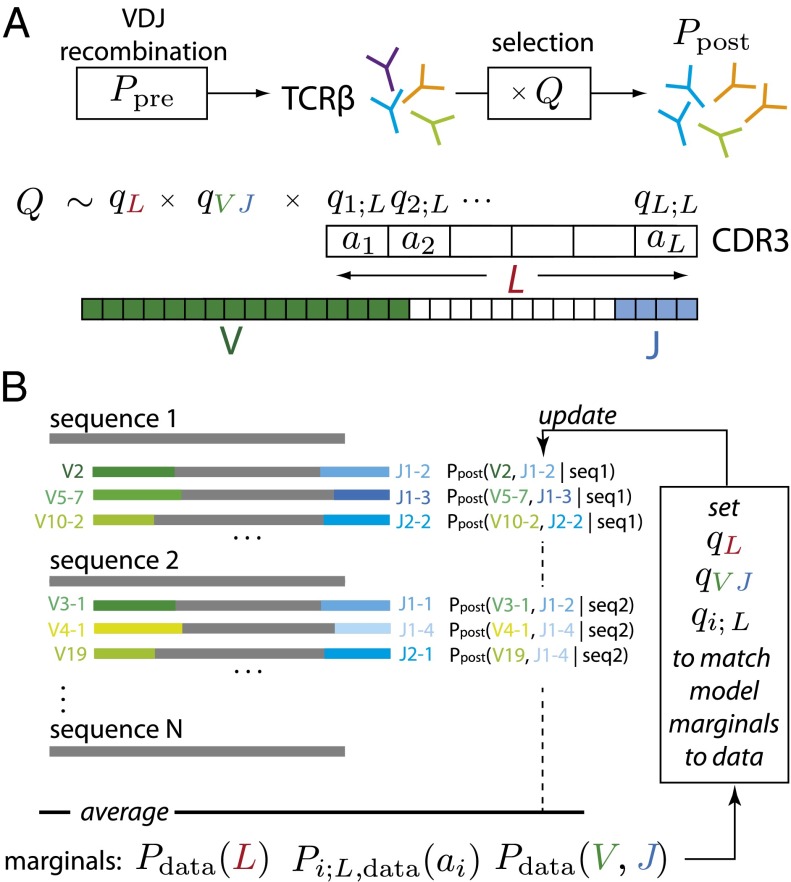
Fig. 1.
Graphical representation of our method. (A) T-cell receptor β-chain sequences are formed during VDJ recombination. Sequences from this probability distribution, described by Ppre, are then selected with a factor Q defined for each sequence, resulting in the observed Ppost distribution of receptor sequences. Selection is assumed to act independently on the V and J genes, the length of the CDR3 region, and each of the amino acids, ai, therein. (B) A schematic of the fitting procedure: the parameters are set so that Ppost fits the marginal frequencies of amino acids at each position, the distribution of CDR3 lengths, and VJ gene choices. Because the latter is not known unambiguously from the observed sequences, it is estimated probabilistically using the model itself in an iterative procedure.
where (a1, … , aL) is the amino acid sequence of the CDR3 (i.e., the translation of ), and L is its length. The factors qL, qi;L(a), and qVJ denote selective pressures on the CDR3 length, its composition, and the associated VJ identities, respectively. Note that the D segment is entirely included in this junctional region, and therefore, selection acting on it is encoded in the qi;L factors. Z enforces the model normalization condition .
Because V and J cannot always be inferred deterministically from the read , the V and J assignments of any given read will have to be treated as probabilistically defined hidden variables. In addition, because of correlations in Ppre, the q factors cannot be identified with marginal enrichment factors [therefore, for example, Pi;L,data(ai)/Pi;L,pre(ai) cannot be set equal to qi;L(ai)]. For these reasons, we must use a maximum likelihood procedure to learn the qL, qi;L, and qVJ factors of Eq. 1. We use an expectation maximization algorithm that iteratively modifies the q values until the observed marginal frequencies—CDR3 length distribution, amino acid usage as a function of CDR3 position, and VJ usage—in the data match those implied by the model distribution in Eq. 1, with the preselection distribution Ppre being taken as a fixed, known input. The procedure is schematically depicted in Fig. 1B (full details are in SI Appendix).
Our model for the selection factor Q assumes factorization on a small set of sequence features with no interactions between these features. This choice is in the same spirit as the classic position weight matrix method for identifying transcription factor binding sites (17). We have verified that such interactions are, in fact, not necessary to describe the data: Fig. 2B plots the covariances of amino acid pairs as predicted by Ppost vs. the observed values in the data, and SI Appendix, Fig. S10 displays a similar comparison of the covariances of (V, J) with L on the one hand and the (V, J) identity with amino acid choice on the other hand. All of the pairwise correlations in the data are well-predicted by the model, although Q does not model them directly. Nonzero pairwise correlations are, in fact, inherited from the preselection distribution, which has correlations of its own (shown by the green points in Fig. 2B).
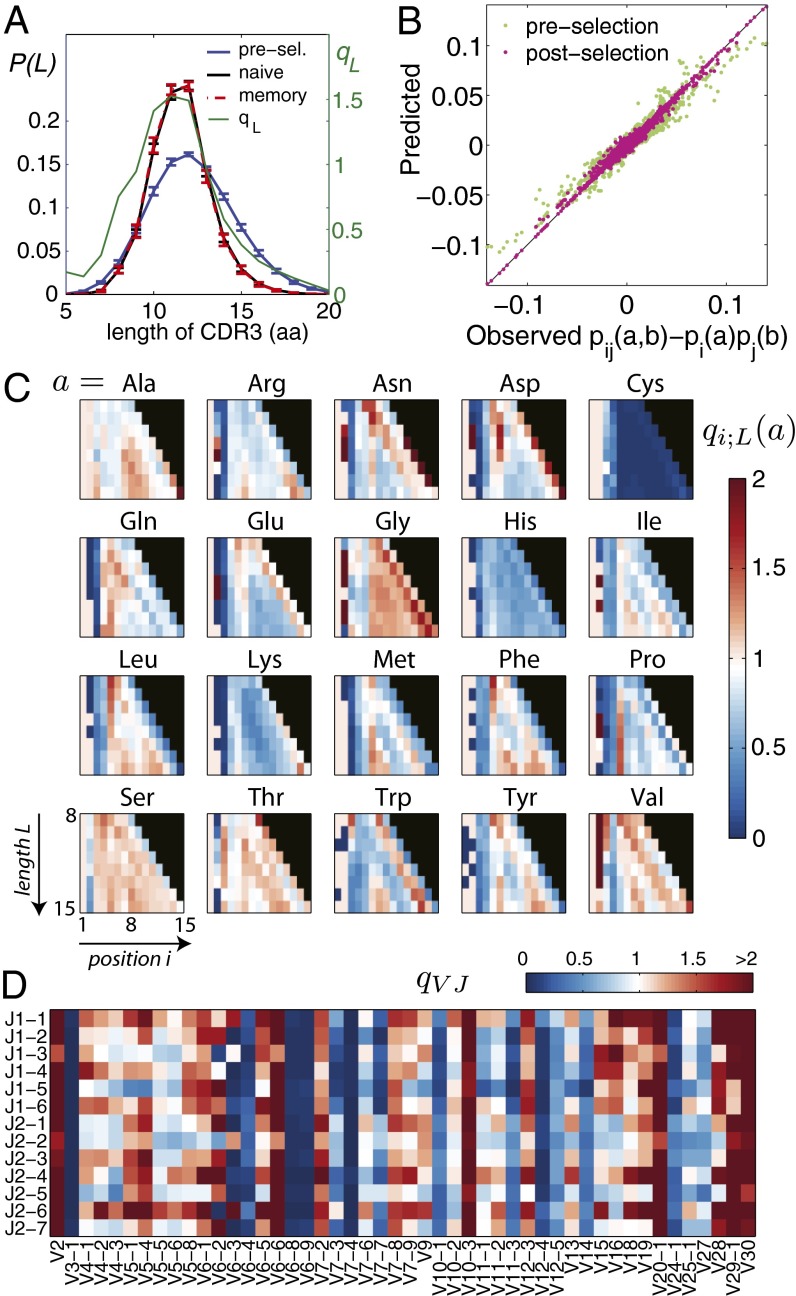
Fig. 2.
Characteristics of selection. (A) CDR3 length distributions pre- and postselection and the length selection factor qL (green). Selection makes the length distribution of CDR3 regions in the preselection repertoire more peaked for the naive and memory repertoires (overlapping). Error bars show standard deviation over nine individuals. (B) Comparison between data and the model of the connected pairwise correlation functions, which were not fitted by our model. The excellent agreement validates the inference procedure. As a control, the prediction from the preselection model (green) does not agree with the data as well. (C) Values of the inferred amino acid selection factors for each amino acid, ordered by length of the CDR3 region (ordinate) and position in the region (abscissa). (D) Values of the VJ gene selection factors.
Another assumption of our model is that selection acts at the level of the amino acid sequence, regardless of the underlying codons. To test this, we learned more general models, where a represented one of 61 possible codons instead of one of 20 aa. We found that codons coding for the same residue had similar selection factors (SI Appendix, Fig. S2), except near the edges of the CDR3, where amino acids may actually come from genomic V and J segments and reflect their codon biases.
To compare the different donors, we learned a distinct model for each donor and cell type (memory or naive) as well as a universal model for all sequences of a given type from all donors taken together (details are in SI Appendix). We also learned models from random subsets of the sequence dataset to assess the effects of low-number statistical noise.
Results
Characteristics of Selection and Repertoire Diversity.
The length, single-residue, and VJ selection factors, learned from the naive datasets of all donors taken together, are presented in Fig. 2 A, C, and D. The qL factor (Fig. 2A) simply reflects the substantial reduction in variance in CDR3 lengths between the preselection ensemble and the observed sequence datasets. The qVJ factor shows that the different V and J genes are subject to a wide range of selection factors (note that these factors act in addition to the quite varied gene segment use probabilities in Ppre). The position-dependent amino acid selection factors qi;L(a) are also quite variable but have striking systematic features, such as uniform suppression (or enhancement) away from the CDR3 region boundaries. We looked for correlations between the qi;L(a) factors and a variety of amino acid biochemical properties (18): hydrophobicity, charge, pH, polarity, volume, and propensities to be found in α- or β-structures in turns at the surface of a binding interface, on the rim, or in the core (19) (details in SI Appendix). We found no significant correlations, except for a negative correlation with amino acid volume and α-helix association as well as a positive correlation with the propensities to be in turns or the core of an interacting complex (SI Appendix, Fig. S7).
To estimate differences between datasets, we calculated the correlation coefficients between the logs of the qVJ and qi;L(a) selection factors (SI Appendix, Fig. S4). Comparing naive vs. naive, memory vs. memory, or naive vs. memory between donors (Fig. 3 A–C shows an example for qi;L, and SI Appendix, Fig. S3 shows an example for qVJ) gave correlation coefficients of ∼0.9 in log qi;L, whereas the naive vs. memory repertoires of the same donor gave 0.95. To get a lower bound on small-number statistical noise, we also compared the factors inferred from artificial datasets obtained by randomly shuffling sequences between donors (SI Appendix), yielding an average correlation coefficient of 0.98. Repeating the analysis for log qVJ, we found correlation coefficients of ∼0.8 between datasets of different donors and 0.84 for the naive and memory dataset of the same donor, all of which must be compared with 0.94, which was obtained between shuffled datasets. We also calculated Jensen–Shannon divergences (SI Appendix) between the Ppost distributions of all donors and found them to be small—0.07 bits on average. Thus, the observed differences between donors of qi;L and qVJ are small and consistent with their expected statistical variability.
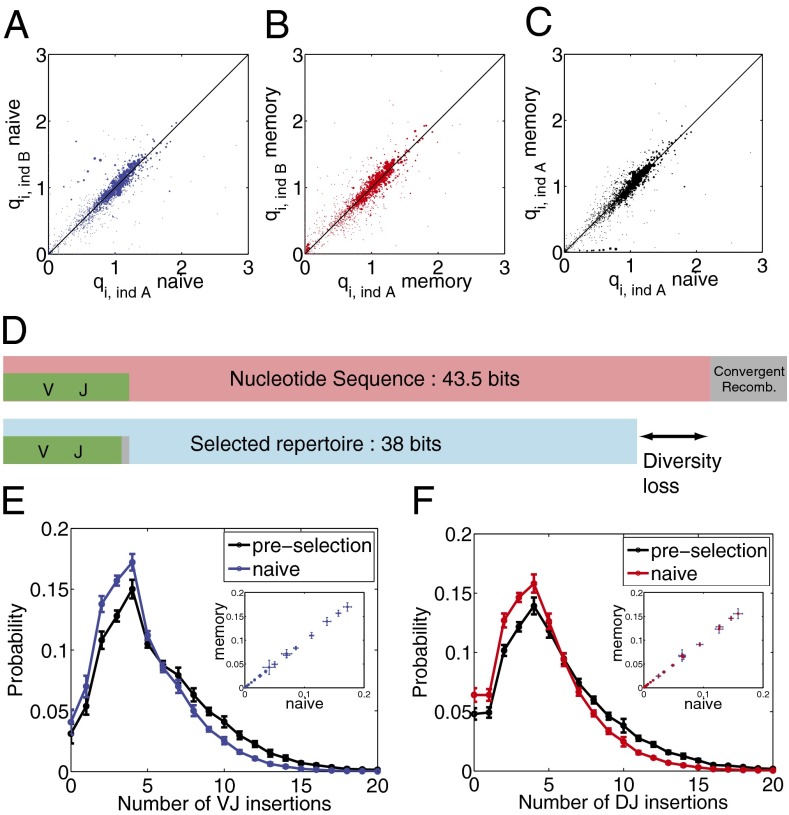
Fig. 3.
Repertoire diversity. (A–C) Variability between repertoires. The scatter between qi;L selection factors between two sample individuals A and B for (A) naive and (B) memory repertoires compared with that of (C) memory and naive repertoires for the same individual shows great similarity between them (SI Appendix, Fig. S4). (D) The entropy of the preselection repertoire (Upper) is reduced in the postselection repertoire (Lower). (E and F) Distribution of (E) VJ and (F) DJ insertions in the preselection and naive repertoires shows elimination of long insertions. Error bars show standard deviations over nine donors. The insertion distributions for the memory repertoire are the same as for the naive repertoire (scatter plots in Insets).
We use Shannon entropy, , to quantify the diversity of the naive and memory distributions. Entropy is a diversity measure that accounts for nonuniformity of the distribution, and it is additive in independent components. Because when there are Ω equally likely outcomes, the diversity index 2S can be viewed as an effective number of states. The entropy of the naive repertoire according to the model is 38 bits (corresponding to a diversity of ∼2.7⋅1011), which is down from 43.5 bits in the preselection repertoire (Fig. 3D). The majority of this 5.5-bits (or 50-fold) reduction in diversity comes from insertions and deletions, which accounted for most of the diversity in the preselection repertoire. The entropies of the memory and naive repertoires are the same, indicating that selection in the periphery does not further reduce diversity.
Knowing the postselection distribution of sequences, we can ask how different features of the recombination scenario fare in the face of selection. We do not mean to imply that somatic selection acts on the scenarios themselves—it acts on the final product—but it is an a posteriori assessment of the fitness of particular rearrangements. For example, the distributions of insertions at VD and DJ junctions in the postselection ensemble have shorter tails (Fig. 3 E and F), whereas the distribution of deletions at the junctions seems little affected by selection (SI Appendix, Fig. S5), although large numbers of deletions are selected against.
Selection Factor Q as a Measure of Fitness.
The selection factor Q is a proxy for the probability of an in-frame sequence after it is generated by recombination to survive the different forms of selection to which it is subjected: the proper folding of the T-cell receptor (TCR) protein, appropriate binding to self peptides, etc. One can think of Q as an intrinsic physical property of the β-chain, and it is instructive to compare Q distributions of the various sequence repertoires of interest: preselection model Ppre, postselection model Ppost, and postselection observed sequences; for each of these repertoires, we assign a Q value to each sequence using the inferred model and create Q-value histograms, denoted by Ppre(Q), Ppost(Q), and Pdata(Q), respectively (SI Appendix, Eqs. S34–S37 shows details of the calculations).
We observe that the data sequences are enriched in large Q values compared with preselection sequences (Fig. 4 A, Inset and B, Inset), consistent with the interpretation of Q as a selection factor. Furthermore, because the definition of Ppost implies that Ppost(Q) = QPpre(Q), we expect Pdata(Q)/Ppre(Q) = Q if the selection model accurately describes the data. This ratio is plotted in Fig. 4, and we see that, for Q ≤ 5 (accounting for more than 91% of the data sequences), this ratio is, indeed, equal to Q, whereas for Q > Qmax ∼ 7 (accounting for less than 3% of the data sequences), the ratio plateaus. Thus, only the small population of high-Q (fittest) data sequences fails to satisfy this stringent model prediction.
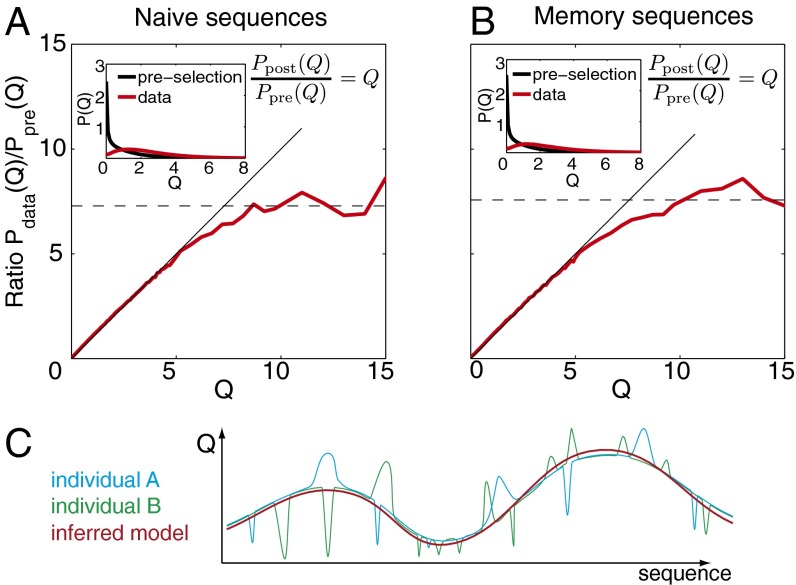
Fig. 4.
Probability of passing selection. (A and B) Ratio of the distributions of sequence-wide selection factors Q between the observed sequences and the preselection ensemble (red line), plotted as a function of Q for (A) naive and (B) memory repertoires. The model prediction Ppost(Q)/Ppre(Q) = Q is shown in black, and the preselection and observed distributions of Q are shown in Insets. The selection ratio saturates around approximately seven, which may be interpreted as the maximum probability of being selected. Naive and memory repertoires show similar behaviors. (C) A cartoon of the effective selection landscape captured by our model (red line). Our method does not capture localized selection pressures (such as avoiding self) specific to each individual but captures general global properties.
The approach of projecting genotypes onto a single phenotypic variable and using the distribution of that variable to identify selection effects has previously been used to characterize the fitness landscape of transcription factor binding sites (20, 21). Although in that problem, the phenotypic variable, equivalent to our log Q, is simply the binding affinity of the sequence to the transcription factor, we have (so far) not been able to identify a simple physical quantity linked to Q.
The high-Q plateau suggests that sequences with Q > Qmax all have the same selective advantage within the resolution of the model. We can use this line of reasoning to put bounds on the probability for rearranged TCR sequences to pass selection. If we assume that Q is proportional to the probability for sequence to be selected, then . Because Psel cannot exceed unity, Q cannot exceed α−1 or . The mean probability that a sequence produced by VDJ rearrangement will pass selection is (as follows from the normalization condition on Ppost). Thus, an upper limit on the average fraction of rearranged TCRs to pass selection is . This limit is consistent with existing estimates (2) for passing positive and negative thymic selection: 10–30% for positive selection only and ∼5% for both together. Our analysis only includes the β-chain, and including the α-chain could further reduce our estimate.
The saturation phenomenon indicates that our model is too coarse-grained to describe the very fit (high-Q) sequences. Because of its factorized structure, our model can only account for the coarse features of selection and may not capture very individual-specific traits, such as avoidance of self (corresponding to Q ≪ 1 in localized regions of the sequence space) or response to pathogens (Q ≫ 1 for particular sequences). This individual-dependent ruggedness of the fitness landscape Q, schematized in Fig. 4C, is probably ignored by our description and may be hard to model in general. To check that the saturation does not affect our inference procedure, we relearned our model parameters from simulated data, where sequences were generated from Ppre and then selected with probability min(Q/Qmax, 1) (details in SI Appendix). We found that essentially the same model was recovered (SI Appendix, Fig. S6).
Natural Selection Anticipates Somatic Selection.
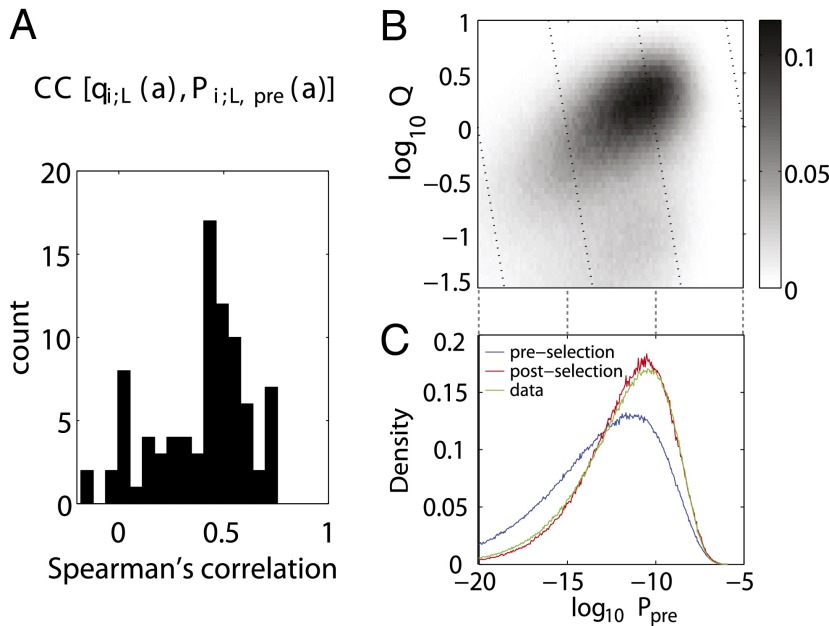
Comparing the pre- and postselection length distributions in Fig. 2A shows that the CDR3 lengths that were the most probable to be produced by recombination are also more likely to be selected. Formally, the Spearman rank correlation coefficient between Ppre(L) and qL is 0.76, showing good correlation between the probability of a CDR3 length and the corresponding selection factor. We asked whether this correlation was also present in the other sequence features. The histogram of Spearman correlations between the selection factors qi;L(a) and the preselection amino acid use Pi;L,pre(a) for different lengths and positions (i, L) (Fig. 5A) shows a clear majority of positive correlations. Likewise, the selection factors qVJ are positively correlated with the preselection VJ use PVJ,pre (Spearman rank correlation = 0.3, P < 2⋅10−20).
Fig. 5.
Correlations between the pre- and postselection repertoires. (A) A histogram of Spearman correlation coefficient (CC) values between the qi;L(a) selection factors in the CDR3 region and their generation probabilities Pi:L,pre(a) for all i, L shows an abundance of positive correlations. (B) Heat map of the joint distribution of the preselection probability distribution Ppre and selection factors Q for each sequence shows that the two quantities are correlated. (C) Sequences in the observed selected repertoire (green line) had a higher probability to have been generated by recombination than unselected sequences (blue line). Agreement between the postselection model (red line) and data distribution (green line) is a validation of the model.
The correlations observed for each particular feature of the sequence (CDR3 length, amino acid composition, and VJ use) combine to create a global correlation between the probability that a sequence was generated by recombination and its propensity to be selected (Spearman rank correlation = 0.4, P = 0) (Fig. 5B). Consistent with this observation, the postselection repertoire is enriched in sequences that have a high probability to be produced by recombination (Fig. 5C). This enrichment is well-predicted by the model, providing another validation of its predictions at the sequence-wide level.
Taken together, these results suggest that the mechanism of VDJ recombination has evolved to preferentially produce sequences that are more likely to be selected by thymic or peripheral selection.
Shared Sequences Between Individuals.
The observation of unique sequences that are shared between different donors has suggested that these sequences make up a public repertoire common to many individuals that is formed through convergent evolution or a common source. However, it is also possible that these common sequences are just statistically more frequent (6) and likely to be randomly recombined in two individuals independently, as discussed by Venturi et al. (7, 22). In other words, public sequences could just be chance events. Here, we revisit this question by asking whether the number of observed shared sequences between individuals is consistent with random choice from our inferred sequence distribution Ppost.
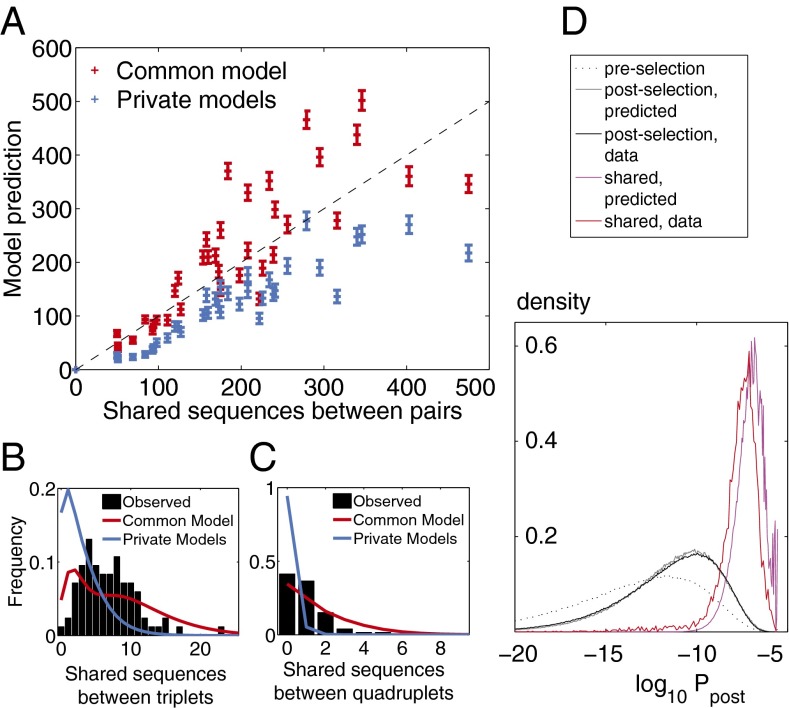
We estimated the expected number of shared sequences between groups of donors in two ways: (i) by assuming that each donor had its own private model learned from his own sequences or (ii) by assuming that sequences are drawn from a universal model learned from all sequences together (details on how these estimates are obtained from the models are in SI Appendix). Although the latter ignores small but perhaps, significant differences between the donors, the former may exaggerate them where statistics are poor. In Fig. 6A, we plot, for each pair of donors, the expected number of shared nucleotide sequences in their naive repertoires under assumptions i and ii vs. the observed number. The number is well-predicted under both assumptions: the universal model assumption gives a slight overestimate, and the private model gives a slight underestimate. We repeat the analysis for sequences that are observed to be common to at least three or four donors (Fig. 6 B and C). The universal model predicts their number better than the private models, although it still slightly overestimates it.
Fig. 6.
Shared sequences between individuals. (A) The mean number of shared sequences between any pair of individuals compared with the number expected by chance (model prediction) for one common model for all individuals (red crosses) and private models learned independently for each individual (blue crosses). Error bars are standard deviations from distributions over pairs. The distribution of shared sequences between (B) triplets and (C) quadruplets of individuals for the data (black histogram) from common (red line) and private (blue line) models. (D) The shared sequences are most likely to be generated and selected: comparison of the Ppost postselection distribution for sequences from the preselection (dotted line) and postselection repertoires (according to the model in gray and the data in black) as well as the sequences shared by at least two donors (model prediction in magenta and data in red).
These results suggest that shared sequences are, indeed, the result of pure chance. If that is so, shared sequences should have a higher occurrence probability than average; specifically, the model predicts that the sequences that are shared between at least two donors are distributed according to (SI Appendix). We test this prediction by plotting the distribution of Ppost for regular sequences as well as pairwise-shared sequences according to the model and in the naive datasets (Fig. 6D), and we find excellent agreement. In general, sequences that are shared between at least n individuals by chance should be distributed according to . For triplets and quadruplets, this model prediction is not as well-verified (SI Appendix, Fig. S8). This discrepancy may be explained by the fact that such sequences are outliers with very high occurrence probabilities and may not be well-captured by the model, which was learned on typical sequences.
We repeated these analyses for sequences shared between the memory repertoires of different individuals with very similar conclusions, except for donors 2 and 3 and donors 2 and 7, who shared many more sequences than expected by chance (SI Appendix, Fig. S9). We conclude that the vast majority of shared sequences occurs by chance and is well-predicted by our model of random recombination and selection.
Discussion
We have introduced and calculated a selection factor that serves as a measure of selection acting on a given receptor sequence in the somatic evolution of the immune repertoire. Using this measure, we show that the observed repertoires have undergone significant selection starting from the initial repertoire produced by VDJ recombination.
We find little difference between the naive and memory repertoires, which is in agreement with recent findings showing no correlation between TCR sequence and T-cell fate (23). We also find little difference between the repertoires of different donors, which is perhaps surprising, because the donors have distinct HLA types and could, therefore, experience markedly different selective pressures. Also, memory sequences have undergone additional selection compared with the naive ones—pathogen recognition—and could show different signatures of selection. A possible interpretation of both findings is that our model only captures coarse and universal features of selection related to the general fitness of receptors and not fine-grained, individual-specific selective pressures, such as avoidance of self, or recognition of particular pathogen epitopes, as illustrated schematically in Fig. 4C. A strategy for incorporating these highly specific effects in our analysis has yet to be defined. In other words, our selection factors may smooth out the complex landscapes of specific repertoires and fail to capture individual-specific tall peaks or deep valleys in the landscape of selection factors. To really probe these fine-grained individual-specific details, we need to develop methods based on accurate sequence counts. Another interesting future direction would be to see whether, at this global level, the signatures of selection are similar between (relatively) isolated populations. Lastly, comparing data from different species (mice and fish), particularly where inbred individuals with the same HLA type can be compared, would be an interesting avenue for addressing these issues.
Our results suggest that natural selection has refined the VDJ recombination process over evolutionary timescales to produce a preselection repertoire that anticipates the downstream actions of somatic selection: sequences that are likely to fail selection are not very likely to be produced in the first place. Because of this rich become richer effect, selection reduces the diversity of the repertoire by a factor of 50 in terms of diversity index. This reduction in diversity does not mean that only 2% of the sequences pass selection: our results are consistent with an acceptance ratio as large as 15%. This paradoxical result is possible because selection, by preferentially keeping clones that were more likely to be generated, gets rid of the many rare clones that are responsible for the large initial sequence diversity. We do not have a mechanistic understanding of how the VDJ recombination process has evolved to achieve this result. Exploration of this question would require an analysis of data on multiple species in different environments.
To summarize, our work has provided the first, to our knowledge, quantitative statistical description of the way that thymic selection and later, peripheral selection modify the TCR sequence repertoire that emerges from VDJ recombination. These results provide a detailed characterization of the background against which one would have to work to detect sequence signatures of more subtle selection effects, such as those associated with autoimmunity and pathogen response.
Supplementary Material
Acknowledgments
The work of Y.E., T.M., and A.M.W. was supported, in part, by European Research Council Starting Grant 306312. The work of C.G.C. was supported, in part, by National Science Foundation Grants PHY-0957573 and PHY-1305525 and the W. M. Keck Foundation Award (dated December 15, 2009).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409572111/-/DCSupplemental.
References
- 1.Murugan A, Mora T, Walczak AM, Callan CG., Jr Statistical inference of the generation probability of T-cell receptors from sequence repertoires. Proc Natl Acad Sci USA. 2012;109(40):16161–16166. doi: 10.1073/pnas.1212755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janeway C. Immunobiology, the Immune System in Health and Disease. New York: Garland; 2005. [Google Scholar]
- 3.Weinstein JA, Jiang N, White RA, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324(5928):807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ndifon W, et al. Chromatin conformation governs T-cell receptor Jβ gene segment usage. Proc Natl Acad Sci USA. 2012;109(39):15865–15870. doi: 10.1073/pnas.1203916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mora T, Walczak AM, Bialek W, Callan CG., Jr Maximum entropy models for antibody diversity. Proc Natl Acad Sci USA. 2010;107(12):5405–5410. doi: 10.1073/pnas.1001705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quigley MF, et al. Convergent recombination shapes the clonotypic landscape of the naive T-cell repertoire. Proc Natl Acad Sci USA. 2010;107(45):19414–19419. doi: 10.1073/pnas.1010586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol. 2008;8(3):231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- 8.Baum PD, Venturi V, Price DA. Wrestling with the repertoire: The promise and perils of next generation sequencing for antigen receptors. Eur J Immunol. 2012;42(11):2834–2839. doi: 10.1002/eji.201242999. [DOI] [PubMed] [Google Scholar]
- 9.Six A, et al. The past, present, and future of immune repertoire biology – the rise of next-generation repertoire analysis. Front Immunol. 2013;4:1–16. doi: 10.3389/fimmu.2013.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robins H. Immunosequencing: Applications of immune repertoire deep sequencing. Curr Opin Immunol. 2013;25(5):646–652. doi: 10.1016/j.coi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Yates AJ. Theories and quantification of thymic selection. Front Immunol. 2014;5:13. doi: 10.3389/fimmu.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jameson SC. Maintaining the norm: T-cell homeostasis. Nat Rev Immunol. 2002;2(8):547–556. doi: 10.1038/nri853. [DOI] [PubMed] [Google Scholar]
- 13.Detours V, Mehr R, Perelson AS. A quantitative theory of affinity-driven T cell repertoire selection. J Theor Biol. 1999;200(4):389–403. doi: 10.1006/jtbi.1999.1003. [DOI] [PubMed] [Google Scholar]
- 14.Kosmrlj A, Jha AK, Huseby ES, Kardar M, Chakraborty AK. How the thymus designs antigen-specific and self-tolerant T cell receptor sequences. Proc Natl Acad Sci USA. 2008;105(43):16671–16676. doi: 10.1073/pnas.0808081105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robins H, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2(47):47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg OG, von Hippel PH. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J Mol Biol. 1987;193(4):723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- 18.Stryer L, Berg JM, Tymoczko JL. Biochemistry. 5th Ed. New York: Freeman; 2002. [Google Scholar]
- 19.Martin J, Lavery R. Arbitrary protein-protein docking targets biologically relevant interfaces. BMC Biophysics. 2012;1(5):7. doi: 10.1186/2046-1682-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mustonen V, Lässig M. Evolutionary population genetics of promoters: Predicting binding sites and functional phylogenies. Proc Natl Acad Sci USA. 2005;102(44):15936–15941. doi: 10.1073/pnas.0505537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mustonen V, Kinney J, Callan CG, Jr, Lässig M. Energy-dependent fitness: A quantitative model for the evolution of yeast transcription factor binding sites. Proc Natl Acad Sci USA. 2008;105(34):12376–12381. doi: 10.1073/pnas.0805909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venturi V, et al. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proc Natl Acad Sci USA. 2006;103(49):18691–18696. doi: 10.1073/pnas.0608907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci USA. 2010;107(4):1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.