Significance
Defense mechanisms against predators, parasites, and pathogens are a hallmark of all multicellular life forms. A conserved defense mechanism is the production of toxic proteins. Because of the limited number of innate defense effectors in a specific host organism, the target epitopes of such toxins are usually highly conserved or occur in different molecular contexts to cover a large spectrum of antagonists. Because glycan epitopes are part of different surface-displayed glycoconjugates in different organisms, carbohydrate-binding proteins (lectins) are the prevailing type of protein toxins in many multicellular organisms. Here we provide evidence that defense lectins can be specific for secondary glycan modifications, such as O-methylation, thereby broadening the range of target organisms.
Abstract
Effector proteins of innate immune systems recognize specific non-self epitopes. Tectonins are a family of β-propeller lectins conserved from bacteria to mammals that have been shown to bind bacterial lipopolysaccharide (LPS). We present experimental evidence that two Tectonins of fungal and animal origin have a specificity for O-methylated glycans. We show that Tectonin 2 of the mushroom Laccaria bicolor (Lb-Tec2) agglutinates Gram-negative bacteria and exerts toxicity toward the model nematode Caenorhabditis elegans, suggesting a role in fungal defense against bacteria and nematodes. Biochemical and genetic analysis of these interactions revealed that both bacterial agglutination and nematotoxicity of Lb-Tec2 depend on the recognition of methylated glycans, namely O-methylated mannose and fucose residues, as part of bacterial LPS and nematode cell-surface glycans. In addition, a C. elegans gene, termed samt-1, coding for a candidate membrane transport protein for the presumptive donor substrate of glycan methylation, S-adenosyl-methionine, from the cytoplasm to the Golgi was identified. Intriguingly, limulus lectin L6, a structurally related antibacterial protein of the Japanese horseshoe crab Tachypleus tridentatus, showed properties identical to the mushroom lectin. These results suggest that O-methylated glycans constitute a conserved target of the fungal and animal innate immune system. The broad phylogenetic distribution of O-methylated glycans increases the spectrum of potential antagonists recognized by Tectonins, rendering this conserved protein family a universal defense armor.
Organisms relying solely on innate immunity face the problem that a limited set of effector proteins has to be effective against a large number of different antagonists. This problem is alleviated by targeting glycan rather than protein epitopes, as similar glycan epitopes can occur in various glycoconjugates and/or various organisms. In addition, glycans show much less adaptation to selective pressure than proteins, due to the fact that they are not primary but secondary gene products. Accordingly, glycan-binding proteins, commonly referred to as lectins, play a crucial role in innate (and adaptive) immunity. Binding of potential pathogens by lectins leads to phagocytosis, complement activation, and antigen processing but also regulation of adaptive immune functions (1). In addition to their role in pathogen recognition, some lectins act as direct defense effectors by intoxicating the antagonist upon binding. Examples are lectins from plants and fungi directed against herbivores and fungivores, as well as human galectins targeting bacterial pathogens (2–4).
One class of lectins that has been associated with innate immunity is the superfamily of Tectonin domain-containing proteins, commonly referred to as Tectonins. The Tectonin domain comprises a 33–37 amino acid consensus sequence that was first reported in Tectonin I and II proteins from the slime mold Physarum polycephalum (5). To date, Tectonins from various organisms including sponge, horseshoe crab, fish, and human have been characterized (6–12). Some of these proteins consist exclusively of Tectonin domain tandem repeats, whereas others comprise additional modules (Fig. 1B). Tandem repeats of Tectonin domains are predicted to form β-propeller structures, with each domain representing a blade formed by a four-stranded antiparallel β-sheet (5, 10). Several members of the Tectonin superfamily have been described as defense molecules or recognition factors in innate immunity based on antibacterial activity or bacteria-induced expression. Because most of them bind to bacterial lipopolysaccharide (LPS), these proteins were proposed to be lectins. Examples are the P. polycephalum Tectonins suggested to play a role in phagocytosis of bacteria (5), limulus lectin L6 (Tachylectin 1) from the Japanese horseshoe crab Tachypleus tridentatus, a Tectonin from the sponge Suberites domuncula shown to exhibit antibacterial activity (7, 9), and galactose-binding protein from the mangrove horseshoe crab Carcinoscorpius rotundicauda (Cr-GBP) and human Tectonin, suggested to mediate antimicrobial defense by interaction with host defense proteins as well as pathogenic bacteria (8, 10). Although binding to bacterial LPS has been demonstrated for all these proteins, their exact carbohydrate-binding specificity remained unclear. In contrast to these representatives of the Tectonin superfamily, Tachylectin-P (TL-P), a Tectonin isolated from perivitelline fluid of T. tridentatus, which is highly homologous to L6, did not bind to bacteria and was proposed to play a role in the completion of embryonic development of this organism by interacting with N-acetylhexosamines of endogenous glycoproteins (13).
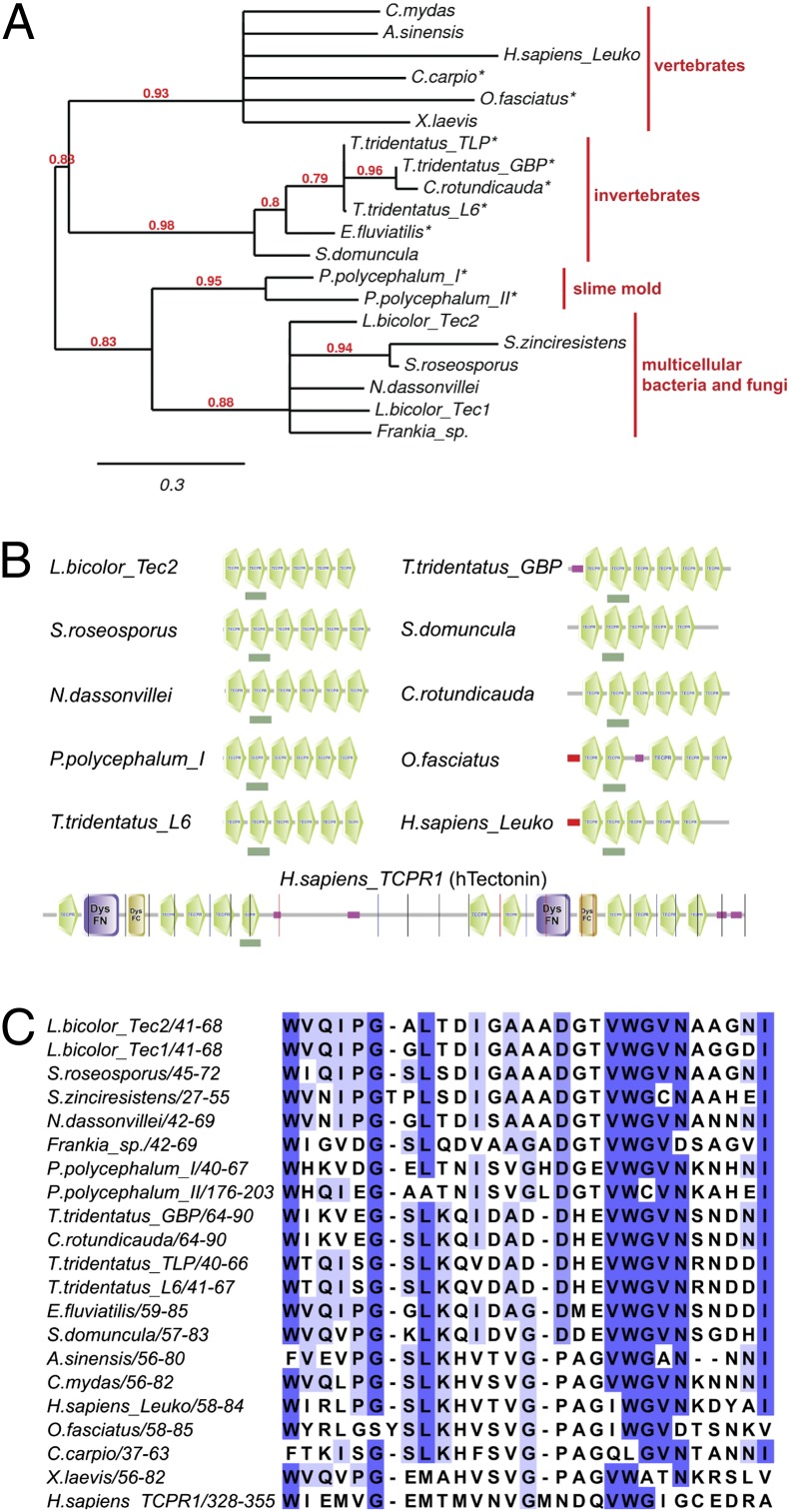
Fig. 1.
Phylogenetic analysis of Tectonin proteins. (A) Phylogenetic tree of previously characterized Tectonins (marked with an asterisk) and selected animal, fungal, and bacterial homologs. GenBank identifiers of the analyzed protein sequences are listed in SI Appendix, Table S1. Numbers at the nodes indicate the level of confidence for the branches as determined by bootstrap analysis. Branches with confidence lower than 0.6 were collapsed. (B) Domain architecture of selected Tectonins predicted by SMART. Tectonin domains are depicted in green. Dark green boxes below the schemes indicate the domains used for the alignment in C. DysFC, Dysferlin domain, C-terminal region; DysFN, Dysferlin domain, N-terminal region. Pink box, low-complexity region; red box, signal sequence for the classical secretory pathway. (C) Multiple sequence alignment of a selected Tectonin domain of the proteins included in the phylogenetic tree in A. The location of the aligned domain within the protein is indicated by the positions of the flanking amino acids. The darkness of the color of specific amino acid residues indicates the degree of conservation.
Here we demonstrate that Tectonin 2 from the ectomycorrhizal mushroom Laccaria bicolor (Lb-Tec2) agglutinates Gram-negative bacteria and is toxic for the model nematode Caenorhabditis elegans. Genetic analysis revealed that bacterial agglutination and nematotoxicity of the protein depend on its binding to bacterial LPS and C. elegans N-glycans, respectively. We provide biochemical evidence that Lb-Tec2 is specific for O-methylated mannose (and fucose) residues present on C. elegans N-glycans and some bacterial LPS. Intriguingly, the lectin L6 from the Japanese horseshoe crab T. tridentatus has the same specificity. Thus, we suggest that Tectonins constitute a previously unrecognized class of lectins specific for O-methylated glycans. Because glycans carrying this modification are present in bacteria, worms, and mollusks, this epitope represents a hitherto unknown and conserved target of fungal and animal defense strategies that allows the coverage of a wide range of antagonists using a single specificity. Finally, we used C. elegans genetics as a tool to study the largely unknown biosynthesis of methylated glycans in nematodes. Mutation of a C. elegans gene coding for a member of the major facilitator superfamily conferred resistance to Lb-Tec2 and resulted in the lack of methylated N- and O-glycans. We hypothesize that the encoded protein, termed SAMT-1, is required for the transport of the donor substrate for glycan O-methylation, S-adenosyl-methionine (SAM), from the cytoplasm to the Golgi lumen.
Results
Tectonins from L. bicolor Are Related to Animal Proteins Involved in Innate Immunity Against Bacteria.
The genome of the ectomycorrhizal mushroom L. bicolor encodes several predicted Tectonins (GenBank accession nos. XP_001877906.1, XP_001876432.1, XP_001876091.1, and XP_001875654.1). Of these, Tectonin 1 (Lb-Tec1) (XP_001877906.1) and Tectonin 2 (Lb-Tec2) (XP_001876432.1) each consists of six tandemly arranged Tectonin domains (Fig. 1B). Both proteins lack a signal sequence and are therefore predicted to be cytoplasmic. Transcription of the gene coding for Lb-Tec2 is 18-fold up-regulated in fruiting bodies compared with vegetative mycelium, whereas the expression of Lb-Tec1 is only 3-fold induced (14). In addition, transcription of Lb-Tec2 is up-regulated in L. bicolor vegetative mycelium upon challenge with several rhizobacteria (15). Similar expression patterns have been observed for lectins involved in fungal defense against predators and parasites (3).
Tectonins have been identified and characterized in slime molds as well as in animals ranging from invertebrates to humans (10), and genes coding for Tectonins have been discovered in the genomes of various, mostly multicellular (filamentous), bacteria. The L. bicolor Tectonins show highest homology to the predicted Tectonins of filamentous actinobacteria and to Tectonin I and II of the slime mold P. polycephalum (Fig. 1A). More distantly related homologs of fungal Tectonins are found in marine invertebrates (arthropods and sponges) as well as vertebrates (fishes, frogs, reptiles, and mammals). Among fungi, Tectonin-encoding genes occur mainly in the genomes of agaricomycetes, including the ectomycorrhizal species L. bicolor, Laccaria amethystina, Paxillus rubicundulus, Hebeloma cylindrosporum, Cortinarius glaucopus, and the saprobic species Galerina marginata, but are also found in the genome of the glomeromycete Rhizophagus irregularis (Glomus intraradices), an arbuscular mycorrhizal species. No homologous genes have been identified in ascomycetes, zygomycetes, or chytrids to date. Sequencing of further fungal genomes will clarify the distribution of Tectonins within the fungal kingdom. Like the characterized Tectonins from slime mold, sponge, and horseshoe crab (5, 7–9, 13, 16) and the predicted Tectonins from filamentous bacteria, fungal Tectonins consist of multiple Tectonin domains and lack a signal sequence for secretion (Fig. 1B).
A multiple sequence alignment of an individual Tectonin domain (underlined by a green box in Fig. 1B) of each protein included in our phylogenetic analysis is shown in Fig. 1C. The conserved amino acid residues are in accordance with previous reports (5, 10). In contrast to their animal homologs, L. bicolor and most bacterial Tectonins do not contain any cysteine residues. The repetitive sequences of Tectonins are most closely related to WD proteins, suggesting that they form β-propeller structures (17). Accordingly, Lb-Tec2 is predicted by the structure prediction program Phyre2 (18) to adopt a six-bladed β-propeller. However, neither is a crystal structure of a Tectonin available nor has lectin function of such a protein unequivocally been demonstrated to date.
Lb-Tec2 Is Toxic Toward C. elegans, and This Toxicity Depends on Binding to N-Glycans.
Lb-Tec2 shares several properties with fungal defense lectins, such as small size, predicted cytoplasmic localization, presumptive binding to carbohydrates, and induction in the fruiting body or upon bacterial challenge. We therefore assessed a potential function of Lb-Tec2 in fungal defense against insects and nematodes. For this purpose the corresponding cDNA was generated and cloned in an Escherichia coli expression vector, and soluble recombinant Lb-Tec2 was obtained upon expression in E. coli BL21 at 23 °C for 16 h. Attempts to purify His-tagged Lb-Tec2 via nickel-Sepharose revealed binding of the protein to the Sepharose CL-6B matrix, which is a cross-linked algal polysaccharide consisting of d-galactose β1,4-linked to 3,6-anhydro-l-galactose. Thus, Sepharose binding was a first indication of a carbohydrate-binding function of Lb-Tec2. Interestingly, binding to Sepharose CL-6B has also been reported for L6 and GBPs (7, 8, 16).
We next tested recombinant Lb-Tec2 for toxicity against Drosophila melanogaster, Aedes aegypti, and C. elegans using previously described assays (19–21). Recombinant Lb-Tec2 was not toxic for the tested insects (SI Appendix, Fig. S1) but displayed significant toxicity toward C. elegans N2, impairing the development of 100% of the larvae (Fig. 2A). In this assay, C. elegans L1 larvae (L) failed to develop to the L4 stage when fed E. coli BL21 expressing Lb-Tec2. To investigate whether the observed nematotoxicity was glycan-dependent, we studied the effect of Lb-Tec2 on established C. elegans N-glycosylation mutants (Fig. 2A). C. elegans fut-8(ok2558) as well as the double mutants fut-6(ok475)fut-1(ok892) and fut-6(ok475);fut-8(ok2558), lacking fucose residues linked to the GlcNAc units of the N-glycan core (22–24), were equally susceptible toward Lb-Tec2 as the wild-type strain. We therefore concluded that α1,3- or α1,6-linked core fucose was not required for Lb-Tec2–induced nematotoxicity. Similarly, C. elegans bre-3(ye26), lacking extended glycosphingolipids (25), was fully susceptible toward intoxication by Lb-Tec2, indicating that glycolipids are not targets of fungal Tectonin. In contrast, the C. elegans triple mutant gly-14(id48);gly-12(id47)gly-13(ok712) and the single mutant aman-2(tm1078) were still susceptible to Lb-Tec2 but showed a considerably higher fraction of L4 staged larvae. In accordance with these results, the hex-3(tm2725);hex-2(tm2530) double mutant did not show any apparent susceptibility to Lb-Tec2 intoxication. Reduced toxicity toward these C. elegans mutants displaying altered structures of N-glycan antennae was evidence for a carbohydrate-dependent function of Lb-Tec2. To further establish this observation, we tested additional glycosylation mutants of C. elegans with pmk-1(km25) background. Similar to the N2 wild-type strain, C. elegans pmk-1(km25), which was previously shown to be hypersensitive to different kinds of stress, including exposure to fungal lectins, due to a defective MAPK pathway (26), did not develop to the L4 stage when feeding on Lb-Tec2–expressing bacteria (Fig. 2B). As observed for the N2 background, Lb-Tec2 caused toxicity toward C. elegans fut-6(ok475)fut-1(ok892);pmk-1(km25). In agreement with these results, a pmk-1(km25);galt-1(op497) mutant lacking the Galβ1,4Fuc epitope on the innermost GlcNAc of the N-glycan core (27) was also susceptible to the toxin. Finally, we tested C. elegans pmk-1(km25)bre-1(op509) and ger-1(op499);pmk-1(km25), two strains defective in the fucose biosynthesis pathway (28, 29). Both strains exhibited significant susceptibility toward Lb-Tec2, but this susceptibility was reduced compared with the pmk-1(km25) strain, suggesting a role of fucose in nematotoxicity. As changes in fucosylation of the N-glycan core did not affect toxicity whereas the complete absence of fucose did reduce toxicity, we concluded that fucosylation of the antennae is at least partially required for susceptibility to Lb-Tec2. The enzyme transferring fucose to the antennae of N-glycans in C. elegans, however, is not known. Fucosyltransferases encoded by fut-2, fut-3, and fut-4 seem not to be involved in this process, as C. elegans single mutants in these genes were susceptible to Lb-Tec2 (SI Appendix, Fig. S2). However, redundancy of the enzymes cannot be excluded.
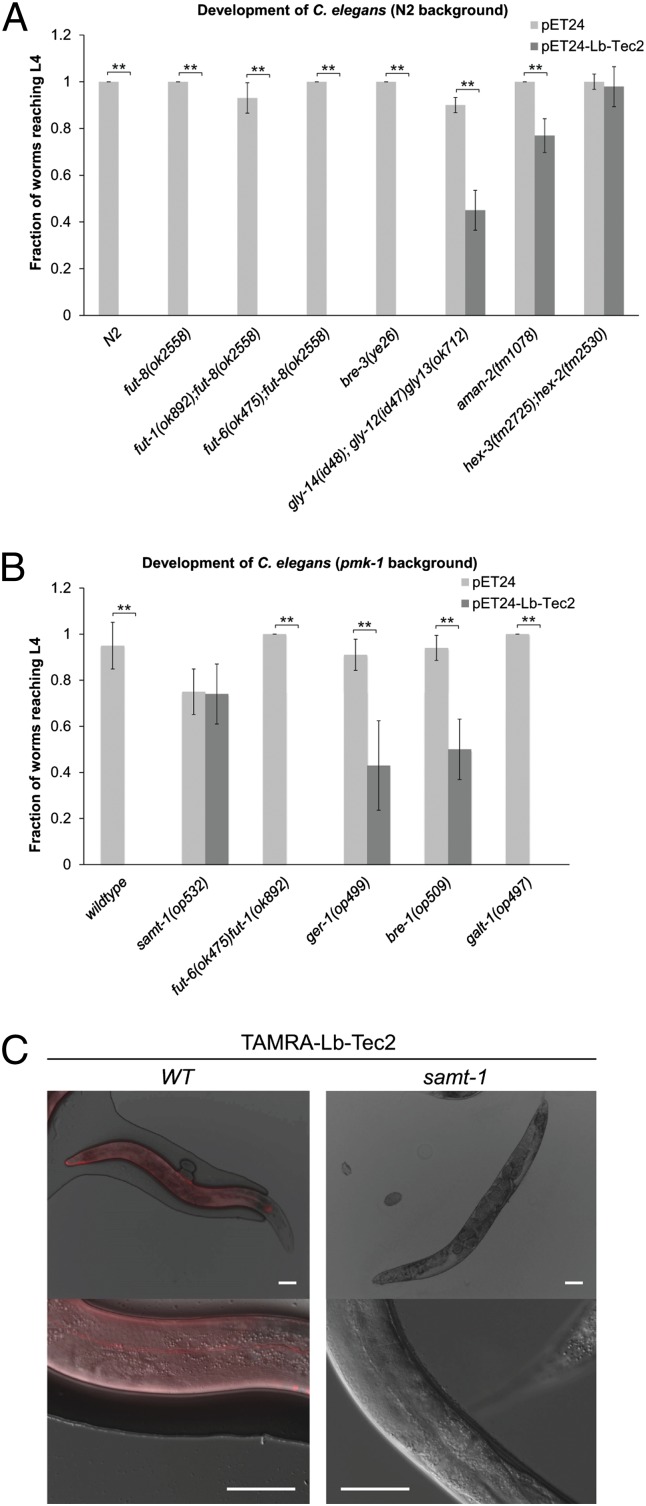
Fig. 2.
Carbohydrate binding-dependent toxicity of Lb-Tec2 toward C. elegans. (A) Development of C. elegans wild type (N2) and various glycosylation mutants feeding on Lb-Tec2–expressing E. coli (dark gray) or empty pET24 vector containing E. coli (light gray) (n = 5). Error bars indicate SD. Comparisons between Lb-Tec2 and vector control for each C. elegans strain were performed using the Mann–Whitney U test (**P < 0.01). (B) Development of C. elegans pmk-1(km25) strain (wild type) and various glycosylation mutants with pmk-1(km25) background feeding on Lb-Tec2–expressing E. coli (dark gray) or empty pET24 vector containing E. coli (light gray) (n = 5). Characteristics of the studied mutants are summarized in SI Appendix, Table S3. Error bars indicate SD. Comparisons between Lb-Tec2 and vector control for each C. elegans strain were performed using the Mann–Whitney U test (**P < 0.01). (C) Binding of TAMRA–Lb-Tec2 to the cuticle of C. elegans pmk-1(km25) (WT) and samt-1(op532)pmk-1(km25) (samt-1) at lower (Upper) and higher magnification (Lower). (Scale bars, 60 μm.)
In addition to C. elegans toxicity assays with Lb-Tec2–expressing bacteria, we performed feeding experiments with purified, fluorescently labeled Lb-Tec2. 5(6)-Carboxytetramethylrhodamine (TAMRA)–Lb-Tec2 bound to the intestine of C. elegans N2 (SI Appendix, Fig. S3), displaying a staining pattern that was previously observed with several nematotoxic fungal lectins (20, 26). Unlike any other of these lectins, TAMRA–Lb-Tec2 also bound to the surface of adult C. elegans, lighting up specific structures, such as annuli and alae, of the nematode (Fig. 2C). Interestingly, surface staining correlated with impaired movement of the nematodes that was previously described as skiddy phenotype (30) (Movies S1 and S2).
Deficiency in a Transporter of the Major Facilitator Superfamily Leads to Lb-Tec2 Resistance of C. elegans.
To identify additional genes involved in the biosynthesis of the Lb-Tec2 target epitope in C. elegans, we performed a forward genetic screen for Lb-Tec2–resistant mutants. For this purpose, a Mos1-mediated transposon mutagenesis was carried out on a population of C. elegans pmk-1(km25) and mutant nematodes were screened for absence of Lb-Tec2–induced skiddy movement. Indeed, one C. elegans mutant, carrying a Mos1 insertion (op532) in gene Y54G2A.4 (samt-1) encoding a protein of the major facilitator superfamily (MFS1), was unaffected in its movement by exposure to Lb-Tec2. In addition, this mutant was completely resistant for Lb-Tec2–induced toxicity (Fig. 2B) and was not stained with TAMRA-labeled Lb-Tec2 (Fig. 2C). We concluded that the samt-1 gene product was necessary for the biosynthesis of the Lb-Tec2 target epitope on C. elegans N-glycan antennae. However, as the function of the identified MFS1 transporter was not known, the target epitope of Lb-Tec2 remained unclear.
Lb-Tec2 Binds to N-Glycans of C. elegans Proteins In Vitro.
To confirm N-glycans as the possible target of Lb-Tec2 by other means, we took a biochemical approach. For this purpose, recombinant Lb-Tec2 was purified by affinity chromatography using Sepharose CL-6B as a matrix and 0.4 M GlcNAc as an eluent according to procedures that had previously been described for L6 and GBPs (7, 8, 16).
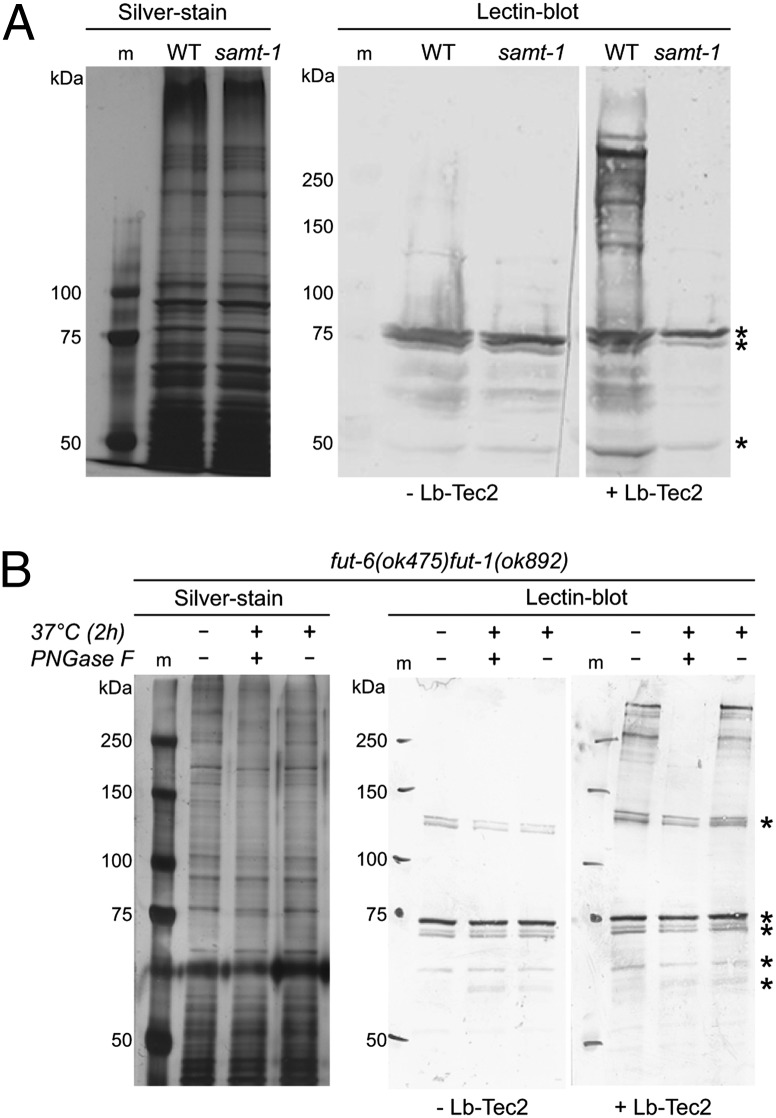
To assay binding of Lb-Tec2 to nematode N-glycoproteins, protein extracts from C. elegans pmk-1(km25) and samt-1(op532)pmk-1(km25) were probed with biotinylated Lb-Tec2 in a lectin blot (Fig. 3A). In this assay, Lb-Tec2 bound to several proteins from C. elegans pmk-1(km25) migrating at around 250 kDa by SDS/PAGE. In contrast, no binding was observed in the case of proteins extracted from the resistant samt-1(op532)pmk-1(km25) strain. A prominent band at around 73 kDa was also present in the negative control not incubated with the biotinylated lectin and therefore did not arise from Lb-Tec2 binding. Based on these results, we concluded that Lb-Tec2 recognizes a samt-1–dependent modification that is present on many C. elegans proteins. To confirm the genetic data suggesting N-glycans as targets of Lb-Tec2, we assessed binding to C. elegans proteins that had been deprived of N-glycans enzymatically. As peptidyl-N-glycosidase (PNGase) F is inhibited by α1,3-fucosylation of the N-glycan core, a modification highly abundant in nematodes, we took advantage of the C. elegans double mutant fut-6(ok475)fut-1(ok892) lacking α1,3-core fucosylation (20). In agreement with the genetic data, in vitro binding of Lb-Tec2 to C. elegans proteins was not affected by this alteration in N-glycan structure (Fig. 3B). However, de–N-glycosylation of fut-6(ok475)fut-1(ok892) proteins by PNGase F resulted in the loss of Lb-Tec2 binding. The absence of binding was not due to protein degradation, as a mock treatment of the protein extract without PNGase F did not affect binding of Lb-Tec2.
Fig. 3.
Binding of Lb-Tec2 to C. elegans N-glycans. (A) Silver-stained SDS/PAGE gel and lectin blot of protein extracts from C. elegans pmk-1(km25) (wild type) and samt-1(op532)pmk-1(km25) (samt-1). (B) Silver-stained SDS/PAGE gel and lectin blot of protein extracts from C. elegans fut-6(ok475)fut-1(ok892) after PNGase F digest. Dashes indicate negative control not incubated with the lectin. Asterisks indicate unspecific bands. m, marker.
Despite the partial resistance of C. elegans N-glycosylation mutants to Lb-Tec2 (Fig. 2A), glycan array analysis of purified Lb-Tec2 did not reveal binding to any structure present on the mammalian glycan array of the Consortium for Functional Glycomics (Dataset S1). We concluded that Lb-Tec2 targeted a modification of N-glycans on C. elegans proteins that was not present on mammalian N-glycans and that the samt-1 gene product was required for the biosynthesis of this modification.
C. elegans pmk-1(km25);samt-1(op532) Mutant Lacks O-Methylation of Glycans.
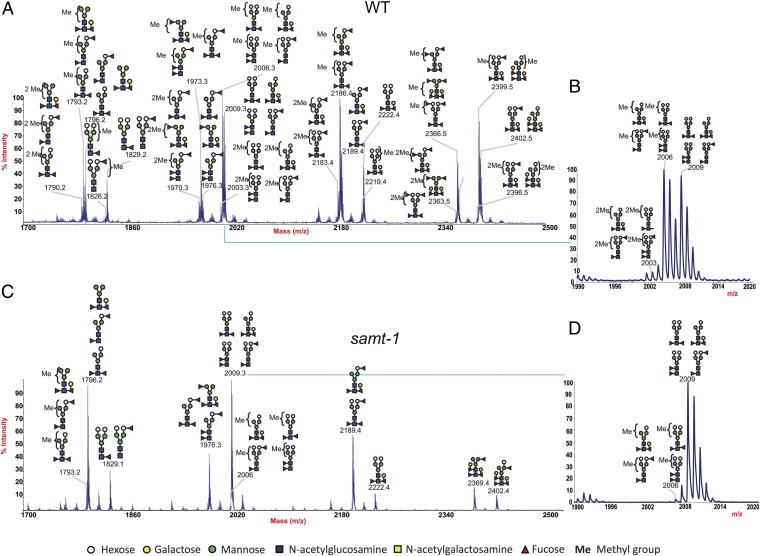
To identify this modification, we made use of the identified Lb-Tec2–resistant C. elegans mutant. First, we performed a structural analysis of N- and O-glycans of C. elegans pmk-1(km25) and samt-1(op532)pmk-1(km25) strains by MALDI-TOF-MS and TOF-TOF-MS/MS. To be able to detect glycan modifications, including the previously described O-methylation of C. elegans N-glycans (31), we perdeuteromethylated the glycans before analysis. Intriguingly, we found that PNGase A-released N-glycans from C. elegans pmk-1(km25) were highly modified with methyl groups (Fig. 4). In contrast, a much lower level of methylation was observed in N-glycans from samt-1(op532)pmk-1(km25). Detailed MS/MS analysis revealed the presence of up to two methylated residues on the antennae of C. elegans pmk-1(km25) N-glycans. However, it was not possible to deduce the exact position of the methyl group on the sugar residues and the position of the methylated sugar within the antennae. Our data yet provided evidence that core fucose residues were not modified with methyl groups. The N-glycan fraction released by PNGase F before PNGase A of both analyzed strains contained fewer glycan structures due to the inability of PNGase F to release α1,3-core–fucosylated N-glycans (SI Appendix, Fig. S5). Moreover, methylation of N-glycans occurred mainly on the antennae of α1,3-core–fucosylated glycans, as methyl groups were more abundant in PNGase A-released N-glycans of pmk-1(km25) compared with the PNGase F-released fraction. Similar to N-glycans, high amounts of methylated O-glycans were present in C. elegans pmk-1(km25), whereas very low levels of methylated O-glycan structures were detected in samt-1(op532)pmk-1(km25) (SI Appendix, Fig. S6). These results suggested that the samt-1–encoded MFS1 transporter was required for methylation of N- and O-glycans in C. elegans.
Fig. 4.
MALDI-TOF spectra of deuteromethylated wild-type and mutant C. elegans N-glycans released by PNGase A digestion. N-glycans were released from C. elegans tryptic glycopeptides by digestion with PNGase A after an initial PNGase F digest (see also SI Appendix, Fig. S5). Released glycans were deuteromethylated before analysis; all molecular ions are [M+Na]+. Structural assignments are based on monosaccharide composition, MS/MS fragmentation analyses, and knowledge of the glycan biosynthetic pathways. (A) N-glycans released from C. elegans pmk-1(km25) (WT), m/z 1,700–2,500. (B) N-glycans released from C. elegans pmk-1(km25), m/z 1,990–2,020. (C) N-glycans released from C. elegans samt-1(op532)pmk-1(km25) (samt-1), m/z 1,700–2,500. (D) N-glycans released from C. elegans samt-1(op532)pmk-1(km25), m/z 1,990–2,020.
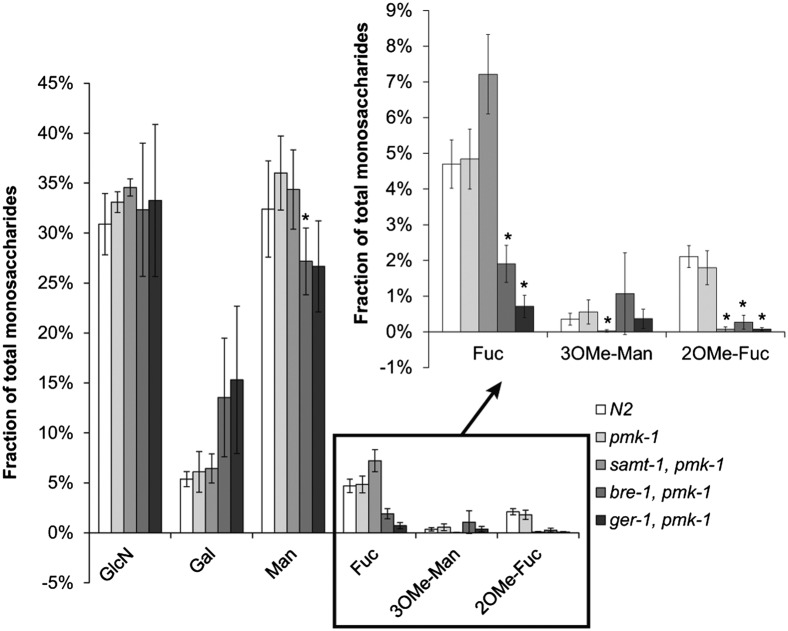
To confirm the results, we performed a monosaccharide analysis of C. elegans N-glycans. N-glycans were released from trypsinized protein extracts using PNGase A, and monosaccharide composition was determined by HPLC analysis in comparison with standard monosaccharides (Fig. 5 and SI Appendix, Fig. S4). We observed minimal amounts of 3-O-methyl-mannose in C. elegans N2 and pmk-1(km25) N-glycans. In addition, we detected small amounts of 2-O-methyl-fucose, as had previously been reported (31). Consistent with the results of the structural analysis, Lb-Tec2–resistant samt-1(op532)pmk-1(km25) did not contain any 3-O-methyl-mannose or 2-O-methyl-fucose. The increase of fucose in samt-1(op532)pmk-1(km25) compared with the wild-type strain was in accordance with the absence of the methylated monosaccharide, indicating that fucose biosynthesis itself was not affected by the mutation. Residual amounts of fucose were present in pmk-1(km25)bre-1(op509) and ger-1(op499);pmk-1(km25), possibly due to residual gene function or utilization of fucose of bacterial origin in these mutants.
Fig. 5.
Monosaccharide composition of N-glycans from Lb-Tec2–sensitive and –resistant C. elegans strains. N-glycans from C. elegans N2, pmk-1(km25)(pmk-1), samt-1(op532)pmk-1(km25) (samt-1, pmk-1), pmk-1(km25)bre-1(op509) (bre-1, pmk-1), and ger-1(op499);pmk-1(km25) (pmk-1, ger-1) were isolated and analyzed for their monosaccharide composition by HPLC (n = 4). Error bars indicate SD. Comparisons between C. elegans pmk-1(km25) and each mutant strain for each monosaccharide were performed using the nonparametric Mann–Whitney U test (*P < 0.05). GlcN, glucosamine (GlcNAc is deacetylated during analysis).
Taken together, these results strongly suggested that the nematotoxicity of Lb-Tec2 depended on the binding of the lectin to O-methylated mannose or fucose residues on the antennae of C. elegans N-glycans.
Lb-Tec2 Binds to 3-O-Methylated Mannose and 2-O-Methylated Fucose.
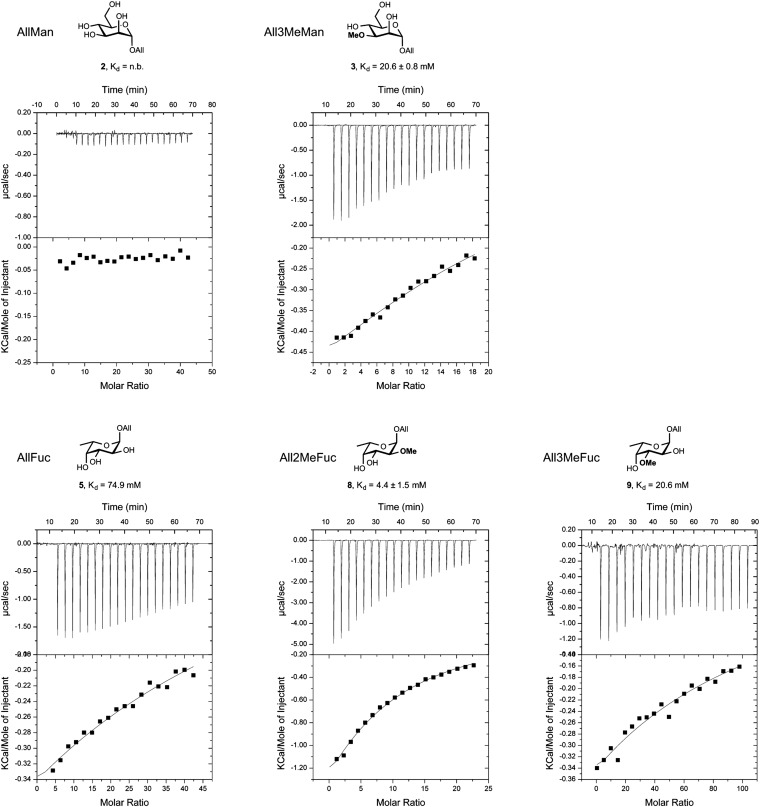
To obtain direct evidence for Lb-Tec2 binding to methylated glycans, microcalorimetry with methylated allyl-monosaccharides in comparison with nonmethylated monosaccharides was performed. Due to the low solubility (<350 μM) and low affinity of Lb-Tec2, titrations with low c values were performed. These titrations required a fixed N value and yielded reliable Kd values, whereas enthalpy and entropy values were of reduced reliability (32). Based on the six-bladed propeller structure predicted by Phyre2 and the fact that β-propeller lectins usually possess one binding site per blade, six binding sites were assumed in the fit of a one-binding site model. Lb-Tec2 bound to allyl 3-O-methyl-mannoside (3) and allyl 2-O-methyl-fucoside (8) with an average Kd value of 21 and 4 mM, respectively (Fig. 6). No binding was observed to allyl mannoside (2), whereas low binding (Kd value of 75 mM) was detected to allyl fucoside (5). A fivefold higher Kd value was observed for allyl 3-O-methyl-fucoside (9) compared with allyl 2-O-methyl-fucoside (8), demonstrating a certain regiospecificity of Lb-Tec2. The obtained microcalorimetry titration data are summarized in SI Appendix, Table S2.
Fig. 6.
In vitro binding of Lb-Tec2 to O-methylated mannose and fucose. Indicated nonmethylated and O-methylated (Me) allyl(All)-mannoside (Man) and -fucoside (Fuc) were tested for binding to purified Lb-Tec2 by isothermal titration calorimetry. n.b., no binding observed. Errors indicate deviation of three independent titrations.
Lb-Tec2 Agglutinates E. coli O8 Cells Comprising LPS with Terminal 3-O-Methylated Mannose.
To further confirm the proposed carbohydrate specificity, we tested the lectin for its capacity to agglutinate a set of bacterial strains exhibiting defined LPS structures with regard to methylation (SI Appendix, Fig. S7). In many bacteria, the core oligosaccharide extending from lipid A is capped with a repeating unit glycan polymer known as the O-polysaccharide, or O-antigen. O-antigen structures within a given species can vary significantly with regard to composition of the repeating unit and the glycosidic linkages. The polymannan O-polysaccharide of E. coli O8 has a nonreducing terminal 3-O-methyl group (33); E. coli O9a comprises a methyl-phosphate group on the 3 position of the nonreducing terminal mannose of the O-antigen repeating unit (34). Intriguingly, Lb-Tec2 agglutinated E. coli O8:K− cells, whereas no agglutination was observed in the case of E. coli O8−:K− lacking the O-antigen. Agglutination was also absent for E. coli O9a:K− carrying LPS with terminal 3-O-methyl-phosphate-mannose. Therefore, we concluded that Lb-Tec2 specifically bound to 3-O-methylated mannose.
T. tridentatus L6 Has the Same Specificity as Lb-Tec2.
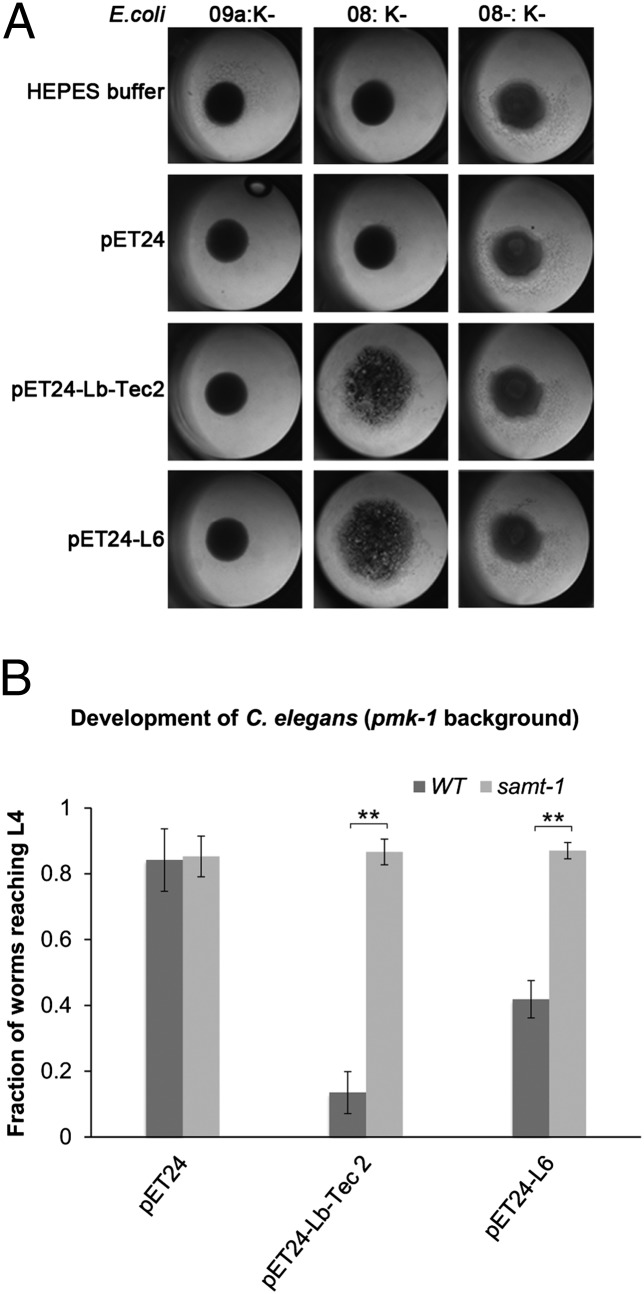
Finally, we set out to test whether the distantly related Tectonin L6 from the Japanese horseshoe crab T. tridentatus has the same specificity as Lb-Tec2. For this purpose, we synthesized the L6-coding region and expressed the protein in the E. coli cytoplasm analogous to Lb-Tec2. Due to solubility issues, we used the E. coli strain ArcticExpress and low temperature for expression. As a positive control for the E. coli agglutination tests and the C. elegans toxicity assays, we expressed Lb-Tec2 under the same conditions. The E. coli agglutination assays were performed with soluble extracts instead of purified proteins because L6 could not easily be purified from the chaperones of the ArcticExpress strain. Under the conditions used, both L6- and Lb-Tec2–containing extracts specifically agglutinated O8:K− cells, whereas no agglutination was observed with extracts of empty vector-containing bacteria or any of the other E. coli strains (Fig. 7A). Accordingly, L6- and Lb-Tec2–expressing ArcticExpress cells showed significant toxicity toward C. elegans pmk-1(km25) but not to samt-1(op532)pmk-1(km25) mutant worms (Fig. 7B). These results implied that the animal Tectonin L6 had a similar activity and specificity as the fungal Tectonin Lb-Tec2.
Fig. 7.
Conservation of O-methylation–dependent bacterial agglutination and nematotoxicity between Lb-Tec2 and limulus lectin L6. (A) Soluble extract of E. coli ArcticExpress expressing either Lb-Tec2 (as positive control) or L6 was added to E. coli O9a:K−, 08:K−, or O8−:K− in a 96-well plate. The soluble extract of E. coli ArcticExpress harboring the empty vector plasmid and Hepes buffer was used as negative controls. (B) Larval development of C. elegans pmk-1(km25) (wild type) and samt-1(op532)pmk-1(km25) (samt-1) was assayed as previously described but using E. coli ArcticExpress containing the empty vector (pET24a) or expressing Lb-Tec2 (pET24-Lb-Tec2) or limulus lectin L6 (pET24-L6) as food (n = 5). Error bars indicate SD. Comparisons between the two C. elegans strains for each plasmid-containing bacterium were performed using the Mann–Whitney U test (**P < 0.01).
Discussion
The domain architecture of Tectonins is well-conserved throughout the different phyla (Fig. 1B), and Tectonins from both invertebrates and vertebrates have been associated with innate immunity. Lectin functions have been ascribed to this protein family, but this functionality was primarily based on Tectonin binding to complex glycans including Sepharose and LPS. A clear carbohydrate-binding specificity has been missing to date.
In this study, we demonstrate that two rather distantly related members of this protein family, Tectonin 2 of the mushroom L. bicolor and L6 of the horseshoe crab T. tridentatus, both bind specifically to O-methylated glycans. These results confirm the lectin function of this protein family and suggest that not only the domain architecture but also the carbohydrate-binding specificity is conserved among the different family members. This conservation can be explained by a strong selection pressure or a rather recent horizontal transfer of the corresponding genes from animals to fungi (and bacteria). In agreement with the latter hypothesis, only a few fungal (and bacterial) genomes appear to contain Tectonin-encoding genes.
The relatively low affinity of Lb-Tec2 to allyl-3-O-methyl-mannose and allyl-2-O-methyl-fucose in vitro indicates that these monosaccharides are not sufficient for efficient binding of the lectin. The latter might be achieved by additional contacts of the carbohydrate-binding pocket of the lectin to subterminal glycan residues or by the presence of multiple methylated carbohydrate residues on the bound glycan. The importance of subterminal glycan residues has been demonstrated for many lectins, such as the fungal defense lectin CCL2, which was recently shown to bind with high affinity to the trisaccharide GlcNAc-β1,4-(Fuc-α1,3-)GlcNAc but had no detectable affinity to the monosaccharide fucose in isothermal titration calorimetry (20). Multivalency of the ligand, on the other hand, is highly relevant for the affinity of Tectonins to glycans, as these β-propeller lectins are expected to have multiple carbohydrate-binding sites. Accordingly, the affinity of another horseshoe crab β-propeller lectin, Tachylectin 2, which does not belong to the Tectonin superfamily, for BSA carrying multiple immobilized GlcNAc residues exceeded the affinity for free GlcNAc by three orders of magnitude (35, 36).
The lack of appropriate subterminal glycan residues might also explain the lack of Lb-Tec2 binding to PNGase F-treated C. elegans protein extracts (Fig. 3B) despite the presence of methylated O-glycans on these proteins (SI Appendix, Fig. S6). Alternatively, O-methylation of C. elegans O-glycans may occur on monosaccharides with a different configuration of the hydroxyl groups around the O-methylated position. In fact, 3-O-methyl-mannose and 2-O-methyl-fucose are very similar in this regard, which may explain why this lectin is able to bind to two different O-methylated monosaccharides. Testing of additional methylated monosaccharides, such as 4-O-methyl-mannose, for binding may reveal the pattern of hydroxyl groups recognized by Lb-Tec2. The molecular basis of the observed specificity should be resolved by the structural analysis of Lb-Tec2–ligand complexes. It will be interesting to see, for example, whether the coordination of this epitope occurs analogous to Anguilla anguilla agglutinin, where the methyl groups of either fucose or 3-O-methyl-galactose are bound via a hydrophobic pocket (37).
The predicted cytoplasmic localization of Lb-Tec2 is in accordance with its role in fungal defense, as fungivorous nematodes feed on the cytoplasm of fungal cells using a syringe-like stylet (38). Once ingested by the nematode, the lectin interacts with glycoproteins in the digestive tract of the fungivorous nematode. The relevance of the target glycan on the surface of the nematode is not clear, because the protein would have to be released from the fungal cytoplasm to bind to this target. The same issue applies to a possible role of Lb-Tec2 in fungal defense against bacteria. Thus far, this assumption is based on induction of Lb-Tec2 in vegetative mycelium of L. bicolor upon confrontation with an antagonistic soil bacterium and a mycorrhiza helper bacterium (15), agglutination of the E. coli O8:K− strain by Lb-Tec2 (Fig. 7A), and bacterial agglutination and growth inhibition reported for animal Tectonins including limulus lectin L6 (7). Lb-Tec2 would certainly be in place for the interaction with bacteria invading the fungal cytoplasm. To agglutinate noninvasive bacteria, however, the protein would have to be released, for example upon lysis of the fungal cell by bacterial enzymes. In contrast, limulus lectin L6 is located in large granules of hemocytes in the horseshoe crab despite the lack of classical secretory signal in its protein sequence (7). Similar to the induction of Lb-Tec2 by bacteria, a fish Tectonin has been shown to be induced upon challenge with pathogenic bacteria (11).
The dual phenotype of C. elegans upon Lb-Tec2 treatment, skiddy movement and inhibition of larval development, is intriguing. The underlying mechanisms are unclear at the moment and were not addressed in this study. Based on preliminary results, we hypothesize that the skiddy phenotype is caused by cross-linking of the outer surface of the worm cuticle with the agar surface by the lectin. Movement of the worm then leads to stripping of the outermost layer of the cuticle, namely the surface coat, and concomitant loss of traction of the worm on the agar surface. Inhibition of larval development, on the other hand, is likely to be caused by cross-linking of glycoproteins on the luminal side of the intestinal epithelium, which leads to destruction of its microvillar structure and, as a consequence, loss of its function in nutrient uptake.
Our study sheds light not only on the function of Tectonins but also on the structure and biosynthesis of O-methylated glycans in nematodes. In C. elegans, O-methylated carbohydrate residues were shown to be present on N-glycans as 3- or 4-O-methyl-mannose and 2-O-methyl-fucose (31). We demonstrate that O-methylated carbohydrate residues are also present on O-glycans, although these residues were not bound by Lb-Tec2 (Fig. 3B and SI Appendix, Fig. S6). The physiological role of glycan methylation in nematodes is not known. The C. elegans samt-1(op532)pmk-1(km25) mutant lacking methylated N- and O-glycans did not display any phenotype under laboratory conditions. Preliminary results indicated a possible regulation of surface but not intestinal Lb-Tec2 target epitope expression upon developmental and environmental signals. It is therefore possible that the target epitopes on the cuticle and the intestine are different. Interestingly, methylated glycans of Toxocara canis have been associated with antigenicity of this parasitic nematode, as this epitope is absent from mammals. The excretory–secretory antigens of this parasitic nematode contain 2-O-methyl-fucose and 4-O-methyl-galactose (39).
Biosynthesis of methylated glycans is proposed to take place in the Golgi, where a methyl group is transferred from the substrate SAM to the mature glycan. However, detailed studies of this process are missing. The products of this reaction are the O-methylated glycan and S-adenosyl-homocysteine. SAM, the donor substrate for various methylation reactions within the cell, is synthesized in the cytosol. Thus, availability of SAM in the lumen of the Golgi would require a specific transporter, as is the case for some nucleotide-activated monosaccharides. However, neither a Golgi SAM transporter nor a methyltransferase acting on glycans has been identified to date. A C. elegans forward genetic screen based on Lb-Tec2–induced skiddy movement and toxicity allowed for the identification of an MFS1 transporter required for methylation of glycans. The MFS family 1 is referred to as the sugar porter family, as substrates of many family members are mono- and disaccharides. However, also transporters of nonsugar molecules such as quinate, organocations, and inositols belong to this protein family (40). We propose that the identified transporter in C. elegans represents a Golgi SAM transporter. The respective gene was therefore named samt-1. Esterification of glucuronic acid residues with methyl groups in plant pectins is another SAM-dependent process taking place in the Golgi (41). Indeed, homologs of SAMT-1 are found encoded in plant genomes and may represent essential players for this common modification. SAM-dependent methyltransferases acting on animal glycans are still to be identified, as to date only a few bacterial and plant examples are known. The toxicity of methylation-specific Lb-Tec2 toward C. elegans may provide a tool for the identification of these enzymes in invertebrates.
O-methylated glycans have been reported in different phyla including bacteria (mainly as part of their LPS), fungi, amoebae, algae, plants, nematodes, and snails (42). Interestingly, methylated glycans have so far not been reported for insects or vertebrates. Methylation of glycans may, however, be more widespread than currently anticipated, as this modification cannot be detected when applying permethylation protocols during glycan analysis. Binding of Lb-Tec2 and its homologs to Sepharose is in accordance with reports of partially methylated galactose and galactans (2-O-, 4-O-, and 6-O-methyl-galactose residues as well as 3,6-anhydro-2-O-methyl-l-galactose) in this polysaccharide isolated from red seaweed (43).
So far, only a few proteins recognizing a specific modification of a glycan have been described, all of which have endogenous roles: P-type lectin recognizes terminal Man-6-P and has an important role in lysosomal targeting of glycoproteins (44), whereas Langerin binds to Gal-6-SO4 in sulfated proteoglycans and is involved in the modulation of innate immunity (45). In contrast, fungal Lb-Tec2 is a defense effector protein targeting a specific modification of exogenous glycans. The broad distribution of O-methylated glycans makes this epitope a perfect target of an innate immune system, as it allows a single type of effector to cope with a diversity of antagonists.
Materials and Methods
Strains and Cultivation Conditions.
E. coli strain DH5α was used for cloning and plasmid amplification. For protein purification and biotoxicity assays, proteins were expressed in E. coli strain BL21(DE3) and E. coli ArcticExpress (DE3). E. coli strains O8:K−, O8−:K−, O9a:K−, and O9a−:K− were kindly provided by Chris Whitfield (University of Guelph, Guelph, Canada) (33, 34). E. coli was cultivated on LB medium as described (46). C. elegans strains were grown on nematode growth medium (NGM) plates or in liquid medium as described previously (47). Nematodes were harvested by sedimentation, washed with S-basal, and stored at −80 °C. The Bristol isolate N2 was used as the wild-type strain. Strains with genotypes pmk-1(km25), bre-3(ye26), aman-2(tm1078), fut-1(ok892), fut-2(gk360), fut-2(ok509), fut-3(gk103), fut-4(gk111), fut-6(ok475), and fut-8(ok2558) were obtained from the Caenorhabditis Genetics Center at the University of Minnesota. The double mutants fut-6(ok475)fut-1(ok892), fut-6(ok475);fut-8(ok2558), fut-1(ok892);fut-8(ok2558), and hex-3(tm2725)hex-2(tm2530) were kindly provided by Iain Wilson [University of Natural Resources and Life Sciences (BOKU), Vienna]. The triple mutant gly-14(id48);gly-12(id47)gly-13(ok712) was kindly provided by Harry Schachter (University of Toronto, Toronto). The triple mutant fut-6(ok475)fut-1(ok892);pmk-1(km25) was generated by standard genetic crossing. Strains with genotype pmk-1(km25)bre-1(op509), ger-1(op499);pmk-1(km25), and pmk-1(km25);galt-1(op497) were generated by Mos1 mutagenesis of pmk-1(km25) worms as previously described (26). The samt-1(op532)pmk-1(km25) mutant was generated in this study by Mos1 mutagenesis as previously described (26).
Cloning and Expression.
The ORF encoding for Lb-Tec2 was amplified from genomic DNA of L. bicolor strain S238N using the primers TectNdeIfwd 5′-GGGGGGCATATGCCTTGGAAAGGAATCTCTGG-3′ and TectSalIrev 5′-GGGGGGGTCGACCTAATCACGAATCCAACGGTAAATACC-3′. The resulting fragment was ligated into the pET24a vector (Invitrogen) using the introduced restriction sites. For bacterial expression, E. coli BL21(DE3) transformed with the pET24-Lb-Tec2 construct was grown in LB medium supplemented with 50 µg/mL kanamycin. Expression was induced with 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) at an OD600 of 1.0, and the culture was incubated at 23 °C for 16 h.
cDNA encoding for limulus lectin L6 (7) was ordered from GenScript with two introduced restriction sites, an NdeI site at the 5′ end and a NotI site at the 3′ end. Using these restriction sites, the cDNA was cloned into the pET24a expression vector (Invitrogen). For expression of proteins in E. coli ArcticExpress (DE3) (Invitrogen), cells were transformed with either pET24a-L6 or pET24a-Lb-Tec2 and grown on LB medium according to the manufacturer’s protocol. Expression was induced with 0.5 mM IPTG at an OD600 of 0.6 for 24 h at 10 °C. Expression and solubility of recombinant proteins were checked as previously described (19).
Purification of Recombinant Lb-Tec2.
For purification of recombinant Lb-Tec2, induced cells were collected by centrifugation (15 min, 16,000 × g, 4 °C), washed with water, and shock-frozen in liquid nitrogen. The bacteria were resuspended in 0.1 M Hepes buffer (pH 7.5) containing 1 mM phenylmethylsulfonyl fluoride and lysed in a French press (SLM Aminco; SLM Instruments). The lysate was cleared by centrifugation (30 min, 38,000 × g, 4 °C) and the supernatant was applied to a Sepharose CL-6B (GE Healthcare) column equilibrated with 0.1 M Hepes (pH 7.5). After binding for 1 h at 4 °C, the flow-through was collected and the column was washed with 10 column volumes of 0.1 M Hepes (pH 7.5). Bound Lb-Tec2 was eluted with 0.4 M GlcNAc (Alfa Aesar) in Hepes buffer and desalted on a PD-10 column (Amersham Biosciences) or by dialysis. Purity of the protein was verified by SDS/PAGE, and the protein concentration was determined by measuring the absorbance at 280 nm, taking into account the extinction coefficient of Lb-Tec2. The purified protein was stable in Hepes (pH 7.5) and Tris (pH 8).
C. elegans Biotoxicity Assays.
Liquid toxicity assays with C. elegans feeding on E. coli BL21(DE3) or E. coli ArcticExpress (DE3) expressing Lb-Tec2 or limulus lectin L6 or containing the empty vector were performed as described (20). Fractions were calculated from five independent experiments.
TAMRA Labeling of Lb-Tec2 and Fluorescence Microscopy of C. elegans.
Purified Lb-Tec2 was labeled with TAMRA succinimidyl ester or Alexa Fluor 488 5-sulfodichlorophenol (SDP) ester (Molecular Probes) according to the manufacturer’s instructions and desalted on a PD-10 column (Amersham Biosciences). Nematodes were harvested from NGM plates, washed with S-basal (47), and incubated for 10 min in 50 µL S-basal containing 2 μg/mL TAMRA-labeled protein in the dark. Worms were washed three times with S-basal for 10 min each, anesthetized with levamisol (10 mM), and transferred to a glass slide coated with 1% agarose for microscopy. Phase-contrast and fluorescence images were acquired on a Zeiss Axiophot microscope using AxioVision 4.8 with a 10× or 20× objective.
Protein Extraction from C. elegans.
Three hundred milligrams of nematodes was resuspended in 300 µL of ice-cold extraction buffer (PBS containing 1% Nonidet P-40 and Roche proteinase inhibitor mixture) in a 2-mL screw-cap tube. Two hundred microliters of acid-washed 0.5-mm glass beads (Sigma-Aldrich) was added and the worms were disrupted in a FastPrep FP120 (Thermo Savant) tissue and cell homogenizer (three times for 45 s at level 6, cooled on ice in-between). Samples were incubated for 30 min at 4 °C on a rotating wheel before collecting the supernatant by centrifugation for 15 min at 12,000 × g at 4 °C.
Enzymatic Removal of N-Glycans from C. elegans Proteins.
C. elegans proteins were extracted as described above, denatured, and digested with PNGase F (New England BioLabs) according to the manufacturer’s instructions. Samples with and without PNGase F were incubated for 2 h at 37 °C while shaking.
Biotinylation of Lb-Tec2 and Lectin Blot.
Purified Lb-Tec2 was biotinylated with EZ-Link Sulfo-NHS-SS-Biotin (Pierce) according to the manufacturer’s instructions and desalted on a PD-10 column (Amersham Biosciences). For lectin blots, protein extracts were separated on a 7% (wt/vol) SDS/PAGE gel and transferred to a nitrocellulose membrane by semidry blotting. The membrane was blocked with 5% BSA in PBS containing 0.1% Tween-20 (PBS-T) for 16 h at 4 °C. After rinsing with PBS-T the membrane was incubated with biotinylated Lb-Tec2 in PBS-T at a concentration of 5 µg/mL for 1 h at room temperature. A negative control was incubated in PBS-T without lectin. The membrane was washed three times for 20 min with PBS-T before incubation with streptavidin-linked alkaline phosphatase. This solution was obtained by diluting reagents A and B of a Vectastain ABC-Alkaline Phosphatase Kit (AK-5000; Vector Laboratories) 150-fold in PBS-T. After a 1-h incubation at room temperature the membrane was washed three times for 5 min in PBS-T. Finally, the membrane was overlaid with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium substrate (B1911; Sigma-Aldrich) and developed to the desired intensity. The color reaction was stopped with two washes in water and the membrane was air-dried. SDS/PAGE gels were stained by Coomassie blue or EZBlue (Sigma-Aldrich). Silver staining was performed as described (48).
Bacterial Agglutination Assay.
For assays shown in SI Appendix, Fig. S7, E. coli strains O8:K−, O8−:K−, O9a:K−, and O9a−:K− were grown in LB medium at 37 °C to an OD600 of 0.7–1, washed with PBS, and resuspended to an OD600 of 2.5. Agglutination assays were performed in a U-shaped 96-well microtiter plate. Ten microliters of bacteria was mixed with 40 µL Lb-Tec2 at a final protein concentration of 15–500 µg/mL. BSA was used as a negative control. Images were taken after a 10–20 min incubation at room temperature using a Leica MZ125 stereomicroscope.
For assays shown in Fig. 7, 50-mL cultures of E. coli ArcticExpress (DE3) expressing Lb-Tec2 or limulus lectin L6 or containing an empty vector pET24a plasmid were washed in 0.1 M Hepes buffer (pH 7.5) supplemented with 0.15 M NaCl and resuspended in 1 mL to an OD600 of 20. Cells were lysed with 1 g of 0.5-mm glass beads in four consecutive homogenization steps with a FastPrep FP120 (Thermo Savant) for 45 s at level 6.5. The homogenate was centrifuged at 12,000 × g for 30 min, and 40 µL of the supernatant was mixed with 10 µL bacteria. ArcticExpress (DE3) harboring an empty vector pET24a plasmid and 0.1 M Hepes buffer (pH 7.5) served as negative controls. Images were taken after a 1-h incubation at room temperature using a Leica MZ125 stereomicroscope.
Structural Analysis of C. elegans N- and O-Glycans.
Details of the isolation and analysis of C. elegans N- and O-glycans by mass spectrometry are described in SI Appendix.
Monosaccharide Analysis of C. elegans N-Glycans.
Details of the monosaccharide analysis of C. elegans N-glycans by HPLC are described in the SI Appendix.
Microcalorimetry.
Isothermal titration calorimetry was performed on a MicroCal iTC200 (General Electric) and the data were analyzed using MicroCal Origin software. Lb-Tec2 (130–328 μM) in Hepes buffer (0.1 M Hepes, 150 mM NaCl, pH 7.5, 0.1% NaN3) was placed in the sample cell at 25 °C. Details of the individual titrations are described in SI Appendix.
Phylogenetic Analysis.
Amino acid sequences of L. bicolor Tectonin 2 homologs were retrieved from the National Center for Biotechnology Information. Sequences were aligned using MUSCLE (49), multiple sequence alignments were curated by applying Gblocks (50), a phylogenetic tree was constructed with ProtDist/FastDist + Neighbor (PHYLIP) (51) with 1,000 bootstrap replicates under the Jones–Taylor–Thornton matrix, and tree rendering was performed with TreeDyn (52). All software was used at the Phylogeny.fr server (53). Domain prediction was performed with SMART (54, 55). Tectonin domains were aligned by MUSCLE, and the multiple sequence alignment was edited using Jalview (56). Structure prediction was performed on the Phyre2 server (18). Distribution of Tectonin proteins among fungi was analyzed using BLAST on the Joint Genome Institute fungal genome portal (57).
Chemical Synthesis.
General and specific experimental details of the chemical synthesis of the various allyl-mannosides and -fucosides used in this study are described in SI Appendix. NMR spectra of synthetic glycosides are shown in Dataset S2.
Statistical Analysis.
The statistical significance of the toxicity assays and the monosaccharide analysis was evaluated by pairwise comparisons using the nonparametric Mann–Whitney U test.
Supplementary Material
Acknowledgments
We thank Chris Whitfield for providing E. coli strains O8:K−, O8−:K−, O9a:K−, and O9−:K− and Iain B.H. Wilson and Harry Schachter for providing the C. elegans strains. Simon Flückiger is acknowledged for cultivation of C. elegans, protein purification, and bacterial agglutination assays. Our thanks go to Silvia Bleuler-Martínez and Ramon Sieber for performing the insect biotoxicity assays. We are grateful to the Protein-Glycan Interaction Resource of the Consortium for Functional Glycomics for glycan array analysis. Valentin Wittmann is acknowledged for access to the microcalorimeter and Holger Bußkamp for acquisition of the high-resolution mass spectrometry spectra. This work was supported by the National Institute of General Medical Sciences (Grant R24 GM098791), the European Commission Marie Curie Program (EuroGlycoArrays Initial Training Network), the Swiss National Science Foundation (Grant 31003A-130671), ETH Zürich, the Zukunftskolleg and the Graduate School Chemical Biology at the University of Konstanz, and the Fonds der Chemischen Industrie. The mass spectrometry work was supported by the Biotechnology and Biological Sciences Research Council (BBF0083091).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 9669.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401176111/-/DCSupplemental.
References
- 1.Vasta GR, Ahmed H, Tasumi S, Odom EW, Saito K. Biological roles of lectins in innate immunity: Molecular and structural basis for diversity in self/non-self recognition. Adv Exp Med Biol. 2007;598:389–406. doi: 10.1007/978-0-387-71767-8_27. [DOI] [PubMed] [Google Scholar]
- 2.Vandenborre G, Smagghe G, Van Damme EJ. Plant lectins as defense proteins against phytophagous insects. Phytochemistry. 2011;72(13):1538–1550. doi: 10.1016/j.phytochem.2011.02.024. [DOI] [PubMed] [Google Scholar]
- 3.Bleuler-Martínez S, et al. A lectin-mediated resistance of higher fungi against predators and parasites. Mol Ecol. 2011;20(14):3056–3070. doi: 10.1111/j.1365-294X.2011.05093.x. [DOI] [PubMed] [Google Scholar]
- 4.Stowell SR, et al. Innate immune lectins kill bacteria expressing blood group antigen. Nat Med. 2010;16(3):295–301. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huh CG, et al. Cloning and characterization of Physarum polycephalum tectonins. Homologues of limulus lectin L-6. J Biol Chem. 1998;273(11):6565–6574. doi: 10.1074/jbc.273.11.6565. [DOI] [PubMed] [Google Scholar]
- 6.Galliano M, et al. Structural and biochemical characterization of a new type of lectin isolated from carp eggs. Biochem J. 2003;376(Pt 2):433–440. doi: 10.1042/BJ20030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito T, Kawabata S, Hirata M, Iwanaga S. A novel type of limulus lectin-L6. Purification, primary structure, and antibacterial activity. J Biol Chem. 1995;270(24):14493–14499. doi: 10.1074/jbc.270.24.14493. [DOI] [PubMed] [Google Scholar]
- 8.Low DH, et al. Molecular interfaces of the galactose-binding protein Tectonin domains in host-pathogen interaction. J Biol Chem. 2010;285(13):9898–9907. doi: 10.1074/jbc.M109.059774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schröder HC, et al. Emergence and disappearance of an immune molecule, an antimicrobial lectin, in basal metazoa. A tachylectin-related protein in the sponge Suberites domuncula. J Biol Chem. 2003;278(35):32810–32817. doi: 10.1074/jbc.M304116200. [DOI] [PubMed] [Google Scholar]
- 10.Low DH, et al. A novel human tectonin protein with multivalent beta-propeller folds interacts with ficolin and binds bacterial LPS. PLoS ONE. 2009;4(7):e6260. doi: 10.1371/journal.pone.0006260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim BS, et al. Molecular characterisation and expression analysis of a fish-egg lectin in rock bream, and its response to bacterial or viral infection. Fish Shellfish Immunol. 2011;31(6):1201–1207. doi: 10.1016/j.fsi.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 12.Funayama N, et al. Isolation of Ef silicatein and Ef lectin as molecular markers for sclerocytes and cells involved in innate immunity in the freshwater sponge Ephydatia fluviatilis. Zoolog Sci. 2005;22(10):1113–1122. doi: 10.2108/zsj.22.1113. [DOI] [PubMed] [Google Scholar]
- 13.Nagai T, Kawabata S, Shishikura F, Sugita H. Purification, characterization, and amino acid sequence of an embryonic lectin in perivitelline fluid of the horseshoe crab. J Biol Chem. 1999;274(53):37673–37678. doi: 10.1074/jbc.274.53.37673. [DOI] [PubMed] [Google Scholar]
- 14.Martin F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452(7183):88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 15.Deveau A, et al. The mycorrhiza helper Pseudomonas fluorescens BBc6R8 has a specific priming effect on the growth, morphology and gene expression of the ectomycorrhizal fungus Laccaria bicolor S238N. New Phytol. 2007;175(4):743–755. doi: 10.1111/j.1469-8137.2007.02148.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen SC, Yen CH, Yeh MS, Huang CJ, Liu TY. Biochemical properties and cDNA cloning of two new lectins from the plasma of Tachypleus tridentatus: Tachypleus plasma lectin 1 and 2+ J Biol Chem. 2001;276(13):9631–9639. doi: 10.1074/jbc.M008414200. [DOI] [PubMed] [Google Scholar]
- 17.Chen CK, Chan NL, Wang AH. The many blades of the β-propeller proteins: Conserved but versatile. Trends Biochem Sci. 2011;36(10):553–561. doi: 10.1016/j.tibs.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 19.Künzler M, et al. Biotoxicity assays for fruiting body lectins and other cytoplasmic proteins. Methods Enzymol. 2010;480:141–150. doi: 10.1016/S0076-6879(10)80007-2. [DOI] [PubMed] [Google Scholar]
- 20.Schubert M, et al. Plasticity of the β-trefoil protein fold in the recognition and control of invertebrate predators and parasites by a fungal defence system. PLoS Pathog. 2012;8(5):e1002706. doi: 10.1371/journal.ppat.1002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabotič J, et al. Structural basis of trypsin inhibition and entomotoxicity of cospin, serine protease inhibitor involved in defense of Coprinopsis cinerea fruiting bodies. J Biol Chem. 2012;287(6):3898–3907. doi: 10.1074/jbc.M111.285304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paschinger K, Rendic D, Lochnit G, Jantsch V, Wilson IB. Molecular basis of anti-horseradish peroxidase staining in Caenorhabditis elegans. J Biol Chem. 2004;279(48):49588–49598. doi: 10.1074/jbc.M408978200. [DOI] [PubMed] [Google Scholar]
- 23.Paschinger K, Staudacher E, Stemmer U, Fabini G, Wilson IB. Fucosyltransferase substrate specificity and the order of fucosylation in invertebrates. Glycobiology. 2005;15(5):463–474. doi: 10.1093/glycob/cwi028. [DOI] [PubMed] [Google Scholar]
- 24.Yan S, Serna S, Reichardt NC, Paschinger K, Wilson IB. Array-assisted characterization of a fucosyltransferase required for the biosynthesis of complex core modifications of nematode N-glycans. J Biol Chem. 2013;288(29):21015–21028. doi: 10.1074/jbc.M113.479147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffitts JS, et al. Glycolipids as receptors for Bacillus thuringiensis crystal toxin. Science. 2005;307(5711):922–925. doi: 10.1126/science.1104444. [DOI] [PubMed] [Google Scholar]
- 26.Butschi A, et al. Caenorhabditis elegans N-glycan core beta-galactoside confers sensitivity towards nematotoxic fungal galectin CGL2. PLoS Pathog. 2010;6(1):e1000717. doi: 10.1371/journal.ppat.1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Titz A, et al. Molecular basis for galactosylation of core fucose residues in invertebrates: Identification of Caenorhabditis elegans N-glycan core alpha1,6-fucoside beta1,4-galactosyltransferase GALT-1 as a member of a novel glycosyltransferase family. J Biol Chem. 2009;284(52):36223–36233. doi: 10.1074/jbc.M109.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marroquin LD, Elyassnia D, Griffitts JS, Feitelson JS, Aroian RV. Bacillus thuringiensis (Bt) toxin susceptibility and isolation of resistance mutants in the nematode Caenorhabditis elegans. Genetics. 2000;155(4):1693–1699. doi: 10.1093/genetics/155.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhomberg S, et al. Reconstitution in vitro of the GDP-fucose biosynthetic pathways of Caenorhabditis elegans and Drosophila melanogaster. FEBS J. 2006;273(10):2244–2256. doi: 10.1111/j.1742-4658.2006.05239.x. [DOI] [PubMed] [Google Scholar]
- 30.Gravato-Nobre MJ, et al. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2005;171(3):1033–1045. doi: 10.1534/genetics.105.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altmann F, Fabini G, Ahorn H, Wilson IB. Genetic model organisms in the study of N-glycans. Biochimie. 2001;83(8):703–712. doi: 10.1016/s0300-9084(01)01297-4. [DOI] [PubMed] [Google Scholar]
- 32.Turnbull WB, Daranas AH. On the value of c: Can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc. 2003;125(48):14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 33.Clarke BR, Cuthbertson L, Whitfield C. Nonreducing terminal modifications determine the chain length of polymannose O antigens of Escherichia coli and couple chain termination to polymer export via an ATP-binding cassette transporter. J Biol Chem. 2004;279(34):35709–35718. doi: 10.1074/jbc.M404738200. [DOI] [PubMed] [Google Scholar]
- 34.Clarke BR, et al. In vitro reconstruction of the chain termination reaction in biosynthesis of the Escherichia coli O9a O-polysaccharide: The chain-length regulator, WbdD, catalyzes the addition of methyl phosphate to the non-reducing terminus of the growing glycan. J Biol Chem. 2011;286(48):41391–41401. doi: 10.1074/jbc.M111.295857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beisel HG, Kawabata S, Iwanaga S, Huber R, Bode W. Tachylectin-2: Crystal structure of a specific GlcNAc/GalNAc-binding lectin involved in the innate immunity host defense of the Japanese horseshoe crab Tachypleus tridentatus. EMBO J. 1999;18(9):2313–2322. doi: 10.1093/emboj/18.9.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawabata S, Tsuda R. Molecular basis of non-self recognition by the horseshoe crab tachylectins. Biochim Biophys Acta. 2002;1572(2-3):414–421. doi: 10.1016/s0304-4165(02)00322-7. [DOI] [PubMed] [Google Scholar]
- 37.Bianchet MA, Odom EW, Vasta GR, Amzel LM. A novel fucose recognition fold involved in innate immunity. Nat Struct Biol. 2002;9(8):628–634. doi: 10.1038/nsb817. [DOI] [PubMed] [Google Scholar]
- 38.Fisher JM, Evans AAF. Penetration and feeding by Aphelenchus avenae. Nematologica. 1967;13(3):425–428. [Google Scholar]
- 39.Khoo KH, et al. Characterization of nematode glycoproteins: The major O-glycans of Toxocara excretory-secretory antigens are O-methylated trisaccharides. Glycobiology. 1991;1(2):163–171. doi: 10.1093/glycob/1.2.163. [DOI] [PubMed] [Google Scholar]
- 40.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62(1):1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibar C, Orellana A. The import of S-adenosylmethionine into the Golgi apparatus is required for the methylation of homogalacturonan. Plant Physiol. 2007;145(2):504–512. doi: 10.1104/pp.107.104679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staudacher E. Methylation—An uncommon modification of glycans. Biol Chem. 2012;393(8):675–685. doi: 10.1515/hsz-2012-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zibetti RG, et al. Galactans from Cryptonemia species. Part II: Studies on the system of galactans of Cryptonemia seminervis (Halymeniales) and on the structure of major fractions. Carbohydr Res. 2009;344(17):2364–2374. doi: 10.1016/j.carres.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Song X, et al. Glycan microarray analysis of P-type lectins reveals distinct phosphomannose glycan recognition. J Biol Chem. 2009;284(50):35201–35214. doi: 10.1074/jbc.M109.056119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tateno H, et al. Dual specificity of Langerin to sulfated and mannosylated glycans via a single C-type carbohydrate recognition domain. J Biol Chem. 2010;285(9):6390–6400. doi: 10.1074/jbc.M109.041863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 47.Stiernagle T. 2006. Maintenance of C. elegans. WormBook, ed. The C. elegans Research Community, WormBook. 10.1895/wormbook.1.101.1.
- 48.Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8(2):93–99. [Google Scholar]
- 49.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 51.Retief JD. Phylogenetic analysis using PHYLIP. Methods Mol Biol. 2000;132:243–258. doi: 10.1385/1-59259-192-2:243. [DOI] [PubMed] [Google Scholar]
- 52.Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dereeper A, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci USA. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Letunic I, Doerks T, Bork P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40(Database issue):D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grigoriev IV, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40(Database issue):D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.