Significance
Vibrio cholerae classical (CL) biotype was isolated, along with biotype El Tor (ET) and altered ET carrying the cholera toxin (CTX) gene of CL biotype, during the 1991 cholera epidemic in Mexico, subsequently becoming endemic until 1997. Microbiological, molecular, and phylogenetic analyses of V. cholerae isolated from both clinical and environmental samples during 1998–2008 confirm important genetic events, namely predominance of ET over CL and altered ET in Mexico. Although altered ET is predominantly associated with cholera globally, progression of CTX+ V. cholerae ET with truncated CTX prophage to the predominant pathogen causing endemic cholera in Mexico may prove to be yet another key historical point in the global epidemiology of cholera.
Abstract
The seventh cholera pandemic caused by Vibrio cholerae O1 El Tor (ET) has been superseded in Asia and Africa by altered ET possessing the cholera toxin (CTX) gene of classical (CL) biotype. The CL biotype of V. cholerae was isolated, along with prototypic and altered ET, during the 1991 cholera epidemic in Mexico and subsequently remained endemic until 1997. Microbiological, molecular, and phylogenetic analyses of clinical and environmental V. cholerae isolated in Mexico between 1998 and 2008 revealed important genetic events favoring predominance of ET over CL and altered ET. V. cholerae altered ET was predominant after 1991 but not after 2000. V. cholerae strains isolated between 2001 and 2003 and a majority isolated in 2004 lacked CTX prophage (Φ) genes encoding CTX subunits A and B and repeat sequence transcriptional regulators of ET and CL biotypes: i.e., CTXΦ−. Most CTXΦ− V. cholerae isolated in Mexico between 2001 and 2003 also lacked toxin coregulated pili tcpA whereas some carried either tcpAET or a variant tcpA with noticeable sequence dissimilarity from tcpACL. The tcpA variants were not detected in 2005 after CTXΦ+ ET became dominant. All clinical and environmental V. cholerae O1 strains isolated during 2005–2008 in Mexico were CTXΦ+ ET, carrying an additional truncated CTXΦ instead of RS1 satellite phage. Despite V. cholerae CTXΦ− ET exhibiting heterogeneity in pulsed-field gel electrophoresis patterns, CTXΦ+ ET isolated during 2004–2008 displayed homogeneity and clonal relationship with V. cholerae ET N16961 and V. cholerae ET isolated in Peru.
The causative agent of cholera, Vibrio cholerae, is a genetically versatile bacterial species for which more than 200 serogroups have been identified and for which significant lateral transfer of genes has been demonstrated. Pandemic cholera is generally caused by toxigenic strains of V. cholerae serogroups O1 and O139. V. cholerae O1 has been divided into two biotypes, classical (CL) and El Tor (ET), differing primarily in phenotypic traits and distinct signature genome sequences (1). Of seven cholera pandemics recorded since 1817, the sixth and presumably the earlier pandemics have been caused by CL biotype (1). V. cholerae ET biotype was first recognized in 1905, but not until 1961 was it considered the causative agent of the seventh cholera pandemic, during which the CL biotype was no longer isolated from cholera cases in Asia.
Although cholera has been endemic in Asia for centuries and sporadic cases have been recorded in the Americas, the presence of V. cholerae CL biotype as the causative agent of cholera in the Americas has not been clarified. A massive epidemic of cholera occurred in South America during 1991–1992, first reported in Peru in January 1991, after which cholera appeared in other countries of Latin America, notably Mexico, by June 1991. Although the characteristic features of the Latin American strains of V. cholerae O1 biotype ET distinguished them from seventh pandemic V. cholerae ET isolated in Asia (2), clonal relatedness led some investigators to conclude that the 1991 Latin American cholera epidemic was simply an extension of the seventh pandemic from the Western hemisphere (3).
A significant recent development in cholera epidemiology has been the emergence of toxigenic variant strains of V. cholerae ET carrying traits of the CL biotype isolated in Asia and Africa (4, 5). Genetic changes in Latin American strains of V. cholerae have been described and the Peruvian V. cholerae O1 isolated between 1991 and 2003 has been shown to be similar to the ET of the seventh pandemic prototype, carrying a distinct signature in the VSP-II region that distinguished it from the Asian ET prototype (6). A recent study of V. cholerae isolated between 1991 and 1997 from diarrhea patients and surface water sources in Mexico showed both CL and ET biotype strains were present, along with the altered ET involved in epidemic cholera globally (7). In this study, V. cholerae isolated between 1998 and 2008 from diarrheal patients and the aquatic environment in Mexico was characterized, using microbiological, molecular, and phylogenetic techniques to elucidate events leading to the outbreaks of cholera in Mexico.
Results
Source and Distribution of Cholera Cases and V. cholerae in Mexico.
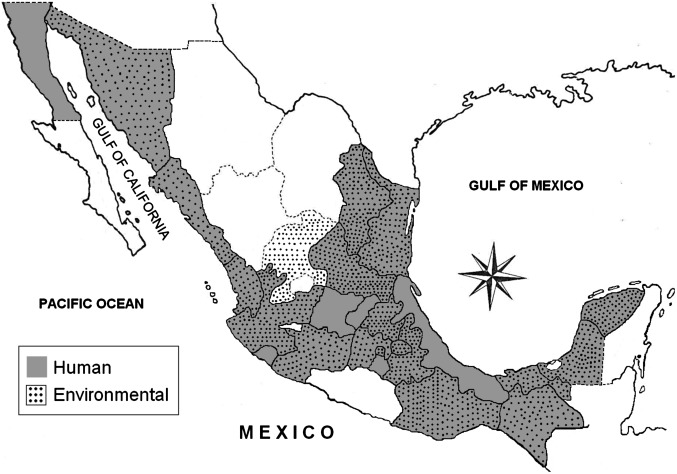
V. cholerae was isolated from hospitalized diarrheal patients and from surface water sources (Table 1) collected from nearly every province of Mexico, including those bordering the United States in the north and Guatemala in the south (Fig. 1). Of 91 V. cholerae isolates included in this study, all produced characteristic colonies on taurocholate tellurite gelatin agar, reacted to polyvalent O1 antisera, were agglutinated by either monovalent Inaba or Ogawa antisera, and amplified primers for the O-biosynthetic gene wbe (O1), confirming all to be serogroup O1. Among these 91 V. cholerae O1, 48 were Inaba and 43 Ogawa (Table 1). The ctxA gene was present in 58 strains, confirming CTX+ V. cholerae O1 (Table 1) and 33 were confirmed CTX−.
Table 1.
Phenotypic and genotypic characteristics of V. cholerae O1 strains (n = 91) isolated from clinical and environmental samples collected in Mexico (1998–2008)
| Year of isolation | No. of isolates | Serotype | Source | Phenotypic properties | Genetic screening by PCR | Deduced biotype | |||||||||||
| Chick cell aggl | Poly B (50U) | Phage IV CL | Phage V ET | ompW | wbeO1 | ctxA | CtxB type* | tcpA | rtxC | hlyA | rstC | rstR type | |||||
| 1998 | 3 | Ogawa | Hum. | + | R | R | S | + | + | + | C | E | E | E | + | E,C | Alt-ET |
| 1999 | 4 | Ogawa | Hum. | + | R | R | S | + | + | + | C | E | E | E | + | E,C | Alt-ET |
| 5 | Ogawa | Hum. | + | R | R | S | + | + | − | − | E | E | E | − | − | ET | |
| 1 | Ogawa | Hum. | + | R | R | R | + | + | − | − | E | E | E | − | − | ET | |
| 1 | Inaba | Hum. | + | R | R | S | + | + | − | − | E | E | E | − | − | ET | |
| 2000 | 2 | Ogawa | Hum. | + | R | R | S | + | + | + | E | E | E | E | + | E | ET |
| 8 | Ogawa | Hum. | + | R | R | S | + | + | + | C | E | E | E | + | E,C | Alt-ET | |
| 1 | Ogawa | Hum. | + | R | R | R | + | + | + | C | E | E | E | + | E,C | Alt-ET | |
| 1 | Ogawa | Hum. | + | R | R | S | + | + | − | − | E | E | E | − | − | ET | |
| 4 | Inaba | Env. | + | R | R | S | + | + | − | − | − | E | E | − | − | ET | |
| 2001 | 1 | Ogawa | Hum. | + | R | R | S | + | + | − | − | E | E | E | − | − | ET |
| 2 | Ogawa | Env. | + | R | R | S | + | + | − | − | E | E | E | − | − | ET | |
| 2 | Ogawa | Env. | + | R | R | S | + | + | − | − | C | E | E | − | − | TCP-Var | |
| 3 | Ogawa | Env. | + | R | R | R | + | + | − | − | − | E | E | − | − | ET | |
| 1 | Inaba | Env. | + | R | R | R | + | + | − | − | − | E | E | − | − | ET | |
| 2002 | 1 | Ogawa | Hum. | + | R | R | S | + | + | − | − | − | E | E | − | − | ET |
| 1 | Ogawa | Hum. | + | R | R | S | + | + | − | − | E | E | E | − | − | ET | |
| 1 | Inaba | Env. | + | R | R | S | + | + | − | − | E | E | E | − | − | ET | |
| 2003 | 1 | Ogawa | Hum. | + | R | R | S | + | + | − | − | − | E | E | − | − | ET |
| 1 | Ogawa | Env. | + | R | R | S | + | + | − | − | − | E | E | − | − | ET | |
| 1 | Inaba | Env. | + | R | R | S | + | + | − | − | − | E | E | − | − | ET | |
| 1 | Ogawa | Env. | + | R | R | S | + | + | − | − | C | E | E | − | − | TCP-Var | |
| 2004 | 1 | Ogawa | Hum. | + | R | R | S | + | + | − | − | C | E | E | − | − | TCP-Var |
| 1 | Ogawa | Hum. | + | R | R | R | + | + | − | − | − | E | E | − | − | ET | |
| 3 | Ogawa | Env. | + | R | R | S | + | + | − | − | E | E | E | − | − | ET | |
| 4 | Inaba | Env. | + | R | R | S | + | + | + | E | E | E | E | − | E | ET | |
| 2005 | 10 | Inaba | Env. | + | R | R | S | + | + | + | E | E | E | E | − | E | ET |
| 2006 | 11 | Inaba | Env. | + | R | R | S | + | + | + | E | E | E | E | − | E | ET |
| 2007 | 1 | Inaba | Hum. | + | R | R | S | + | + | + | E | E | E | E | − | E | ET |
| 3 | Inaba | Hum. | + | R | R | R | + | + | + | E | E | E | E | − | E | ET | |
| 2 | Inaba | Env. | + | R | R | S | + | + | + | E | E | E | E | − | E | ET | |
| 2008 | 8 | Inaba | Env. | + | R | R | S | + | + | + | E | E | E | E | − | E | ET |
| 1 | Inaba | Env. | + | R | R | R | + | + | + | E | E | E | E | − | E | ET | |
| N16961 | ET | Inaba | Hum. | + | R | R | S | + | + | + | E | E | E | E | + | E | El Tor |
| O395 | CL | Ogawa | Hum. | − | S | S | R | + | + | + | C | C | − | C | − | C | Classical |
Alt, altered; C, classical; Chick cell aggl, chicken cell agglutination; CL, classical biotype; E, El Tor; Env., environment; ET, El Tor biotype; Hum., human; Poly B, polymixin B; R, resistant; S, sensitive; TCP-Var, TCP-variant; +, positive; −, negative.
Determined by mismatch amplification mutation assay (MAMA) PCR (38).
Fig. 1.
Geographic map of Mexico showing provinces and locations near the Gulf of Mexico where samples from human and environmental sources were collected and tested for V. cholerae O1 as part of a national surveillance between 1991 and 2008.
Phenotypic and Related Genotypic Characteristics.
Phenotypic and related genetic characteristics of V. cholerae O1 strains are presented in Table 1. All V. cholerae O1 strains were resistant to CL biotype-specific phage IV, all but 11 were responsive to El Tor (ET)-specific phage V, and all showed ET-specific phenotypic traits, including the ability to agglutinate chicken blood cells (CCA) and resistance to polymyxin B (poly-B). The V. cholerae strains amplified primers for repeat in toxin rtxC, an ET-specific marker gene, and ET-specific hemolysin hlyAET (Table 1). All but 13 amplified primers for tcpA, 74 amplified primers for tcpAET, and 4 for tcpACL (Table 1). Of 58 CTX+ strains, 16 amplified primers for tcpAET, ctxBCL, rstRET, and rstRCL, indicating V. cholerae altered (variant) ET, as reported previously (7). All altered ET and two ET (prototype) strains amplified primers specific for phage encoded antirepressor rstC of the RS1 satellite phage flanking ET CTXΦ. Forty-two CTX+ strains amplified primers for rstRET and ctxBET, confirming V. cholerae ET of seventh pandemic prototype, of which 40 failed to amplify rstC of the RS1 element. Thirty-three V. cholerae ET strains lacked ctxA, ctxB, and rstR, suggesting major genetic truncation, of which 13 lacked tcpA, whereas the remaining strains carried either ET-specific tcpA (tcpAET) or CL-specific tcpA (tcpACL) in their ET biotype background.
Year-wise analysis showed that most of the clinical V. cholerae strains isolated between 1998 and 2004 were Ogawa (Table 1), with Inaba dominant among the environmental isolates. Inaba was the only serotype isolated in Mexico between 2005 and 2008. The three V. cholerae strains isolated in 1998 and four in 1999 were CTXΦ+ altered ET whereas the remaining seven isolated in 1999 were CTXΦ− ET. V. cholerae CTXΦ+ altered ET was isolated in 2000 in relatively larger proportions, together with V. cholerae ET strains with and without the CTXΦ, but, thereafter, CTXΦ+ V. cholerae was not consistently isolated from clinical cases (Table 1). V. cholerae O1 biotype ET isolated between 2001 and 2003 lacked CTXΦ. However, most remarkable was the isolation in 2004 of CTXΦ+ V. cholerae ET, and, thereafter, between 2005 and 2008, all Mexican isolates were the CTXΦ+ V. cholerae ET seventh pandemic prototype (Table 1).
Chronologically, the majority of CTXΦ− V. cholerae ET isolated in 1999 and 2000 possessed ET biotype tcpA (Table 1), and all ET strains isolated during 2001 and thereafter were CTXΦ−. The majority of CTXΦ− V. cholerae isolated in Mexico between 2001 and 2003 lacked tcpA whereas some CTXΦ− V. cholerae carried the tcpA allele of either ET or CL type in an ET host; the latter were designated TCP variant. Only V. cholerae CTXΦ− ET was isolated from environmental sources between 2001 and 2003. V. cholerae CTXΦ− ET was no longer isolated in Mexico after 2004, indicating clonal switching. During 2005–2008, CTXΦ+ V. cholerae O1 ET Inaba was the dominant serotype (Table 1). The Mexican CTXΦ+ V. cholerae O1 ET isolated during 2005–2008 exhibited phenotypic and molecular traits of the seventh pandemic prototype.
Nucleotide Sequencing of Targeted Genes.
Data on the ctxB (460 bp) sequence of CTXΦ+ strains from each year class (1998–2008) showed that altered ET strains isolated before 2004 presented amino acid sequences identical to CL biotype CT (ctxB genotype 1), with histidine and threonine at positions 39 and 68, respectively. However, ctxB sequences of ET strains isolated in 2004 and thereafter matched ET biotype CT (ctxB genotype 3): i.e., the cholera toxin type of seventh cholera pandemic strains.
Genome Organization of CTXΦ and Flanking Region.
The Mexican CTXΦ+ ET strains isolated between 2004 and 2008 failed to amplify rstC of the RS1 element. To follow up on this finding, representative ET biotype-specific ctxBET and rstRET were tested for genetic arrangements in CTXΦ and its flanking region by primer walking. Results showed that, unlike ET reference strain N16961, none of the rstC-negative strains amplified the ∼1.6 kb and 2.1 kb targeted RS1 sequences primed by rstA-F/rstC-R and rstRET-F/rstC-R, respectively (Table S1), confirming the absence of RS1 in the genome of the Mexican V. cholerae ET strains.
Thus, the Mexican V. cholerae ET lacked RS1, a characteristic of the CL biotype observed in ET variant strains from Bangladesh and Mozambique (8). A PCR assay was performed using primers CIIF and CIIR targeting dif2 (9) to determine the presence of CTXΦ in the small chromosome (Chr II). As shown in Table S1, all strains possessed an ∼747-bp amplicon, suggesting the absence of CTXΦ in Chr II. Because the ET strains were CTXΦ+, primer sets ctxAF/RTX5R and TLC3F/rstAR were used to locate the exact position of CTXΦ. As shown in Table S1, amplification of ∼1.4-kb and 1.8-kb DNA fragments, respectively, confirmed CTXΦ to be present in the large chromosome (Chr I). The presence or absence of CTXΦ tandem repeat was determined using the primer pair ctxBF/cepR. This amplicon was not found using template DNA prepared from the Mexican strains (Table S1), indicating that a tandem repeat of CTXΦ was not present in either chromosome.
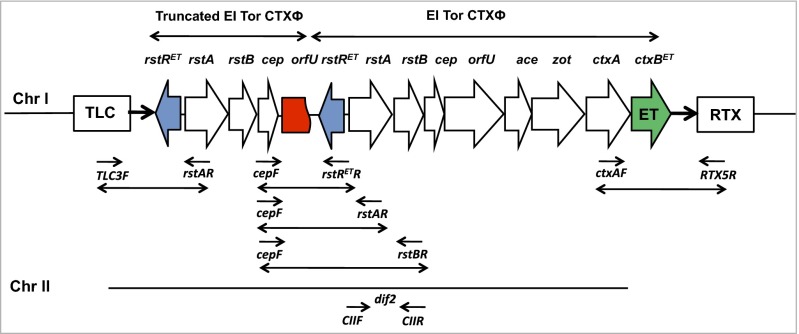
Primer sets cepF/rstRETR, cepF/rstAR, and cepF/rstBR were used to detect truncated CTXΦ at the upstream region of CTXΦ for strains lacking RS1. All yielded amplicons, ∼1.2 kb, 2.2 kb, and 2.7 kb for cepF/rstRETR, cepF/rstAR, and cepF/rstBR primers, respectively, as did CL reference strain O395, confirming the presence of a truncated CTXΦ at the upstream region of CTX. DNA sequence analysis of the ∼1.2-kb amplicon, using cepF/rstRETR primers, confirmed the presence of a truncated orfU subunit (453 bp) within the truncated CTXΦ. Thus, the results showed that V. cholerae O1 ET strains isolated in Mexico between 2004 and 2008 had a truncated CTXΦ, in addition to CTX prophage, in the large chromosome, with neither an RS1 element nor a tandem array of CTX prophage. A schematic genetic map displaying the deduced chromosomal localization of CTXΦ and its flanking genetic region is provided in Fig. 2.
Fig. 2.
Genetic mapping of CTXΦ lacking RS1 in V. cholerae O1 biotype El Tor strains isolated in Mexico during 2004–2008. The deduced genetic organization shows integration of an additional truncated CTXΦ in the upstream region of the CTXΦ in the large chromosome (Chr I) of V. cholerae whereas the dif2 site in the small chromosome did not harbor CTXΦ. Arrows indicate transcription directions of each of the genes in the CTXΦ and flanking regions. The map is without a scale bar. rstRET (blue arrow), El Tor type rtsR; ctxBET (green arrow), El Tor type ctxB; orfU (red box), truncated orfU subunit; TLC, toxin-linked cryptic; RTX, repeat in toxin. Locations of primers are also shown.
Sequencing and Phylogenetic Analysis of tcpA.
The results of DNA sequencing of the tcpAET allele of the Mexican ET strains possessing only ET type tcpA showed that the sequences were identical to reference ET N16961(accession no. AF536868.1). The nucleotide sequences of tcpACL of four V. cholerae ET strains (TCP variant ET, Mex-2174, Mex-3358, Mex-3065, and Mex-2058) were identical, with 81% sequence homology with reference CL and 74% with ET biotype tcpA alleles. The nucleotide sequence of the tcpACL allele of the Mexican V. cholerae ET strains matched exactly the tcpA sequence in GenBank for nontoxigenic V. cholerae O1 ZJ59 (accession no. EU622531) and LN93094 (accession no. AF512422) reported from China. The deduced amino acid sequences revealed substitutions at positions 74 [valine to isoleucine (V→I)], 99 [proline to serine (P→S)], 145 [alanine to glycine (A→G)], and 189 [threonine to alanine (T→A)], suggesting genetic differences from CL, ET, and other types of tcpA alleles described previously (10).
A phylogenetic tree was constructed using tcpA sequences of the two Mexican strains (Mex-2058 and Mex-3065) together with tcpA gene sequences from GenBank (Fig. S1). The Mexican and Chinese strains with analogous tcpA sequences formed a separate cluster to the 10 clusters for tcpA alleles reported to date (11). Overall, results of the analysis clearly indicate that tcpA of Mex-2058 and Mex-3065 have a lineage distinct from the seventh pandemic ET.
PFGE and Cluster Analysis.
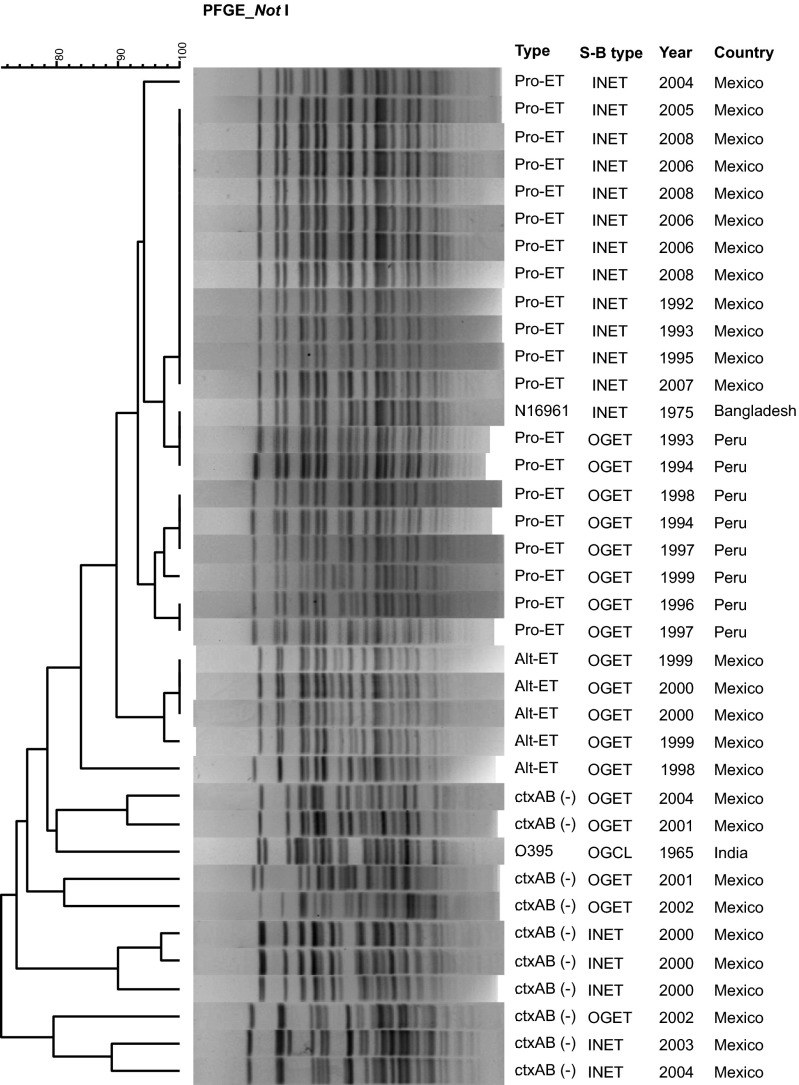
The results of genomic fingerprinting determined by pulsed-field gel electrophoresis (PFGE) showed Mexican V. cholerae ET strains carrying tcpAET, and ET biotype-specific CTX-prophage marker genes yielded an overall banding pattern characteristic of seventh pandemic ET N16961 (Fig. 3). V. cholerae CTXΦ− ET and TCP variants differed significantly from the signature PFGE banding patterns of the CTXΦ+ ET isolated in Mexico between 1998 and 2008.
Fig. 3.
Dendrogram of PFGE patterns of V. cholerae O1 prototype ET and transitional CTX− ET possessing TCP of ET or CL biotype. S-B type, sero-biotype. The tight clustering of Mexican V. cholerae O1 prototype ET strains indicates clonality whereas CTX− ET and transitional TCP variants of ET demonstrate heterogeneous patterns, indicating divergence. Peruvian prototype ET strains clustered differently from Mexican prototype V. cholerae ET (not identical because differentiating regional signatures were observed in the patterns). Alt-ET, altered ET; INET, Inaba ET; OGET, Ogawa ET; Pro-ET, prototype ET.
Cluster analysis, performed using PFGE patterns of NotI-digested genomic DNA, separated genetically heterogeneous CTXΦ− ET and TCP variants from pre- and post-2004 (1991 and 2008) V. cholerae ET, the latter comprising a cluster with Peruvian V. cholerae ET and V. cholerae N16961. The Mexican CTXΦ+ V. cholerae ET strains (1991–2008) formed a tight cluster including ET N16961 and closely linked to V. cholerae ET strains from Peru.
Discussion
Reports of the emergence and pandemic spread of an altered V. cholerae El Tor (ET) suggest that the pattern of global cholera is changing, a phenomenon that appears to be occurring in Mexico, where the V. cholerae CL biotype of pre-1991 cholera has been isolated, as well as altered V. cholerae ET, the latter dominant among V. cholerae CL and ET biotype progenitors associated with cholera outbreaks between 1991 and 1997 (7). To date, the CL biotype may no longer be associated with human disease in Mexico; and altered ET, previously considered the major cause of endemic cholera (7), was no longer isolated after the emergence in 2000 and predominance of the CTXΦ− population, including a unique TCP variant of V. cholerae ET. Characterization of V. cholerae population in subsequent years (2004–2008) suggests another major clonal shift, with the emergence and predominance of a CTX+ V. cholerae O1 ET lacking the RS1 element, with a truncated CTXΦ in the upstream region of the CTXΦ in the large chromosome.
Although the V. cholerae ET strains varied in PCR primer amplification for ctxB, tcpA, and rstR biotype-specific alleles (12, 13), overall serobiotyping results confirmed that the strains were a V. cholerae O1 ET seventh pandemic prototype. The genetic transitions in Asia from V. cholerae biotypes CL to ET (14) and ET prototype to altered ET are considered chronological events occurring in the 1980s and in 2001, respectively (4, 15, 16). In Mexico, both V. cholerae CL and ET biotype progenitors were isolated during cholera outbreaks, together with altered ET, dominant between 1991 and 1997 (7). The consistent association of V. cholerae CL, ET, and altered ET with local cholera outbreaks in the Americas between 1991 and 1997 and their occurrence in the natural aquatic environment, together with non-O1 phenotypically and genetically O1 CL, suggest that the emergence of altered V. cholerae ET has occurred locally in Mexico (7, 17) and is not clonal expansion from endemic regions of Asia (14), as has been proposed for Africa (5). According to our data, V. cholerae altered ET was not isolated in Mexico after 2000 nor was the CL biotype reported in Mexico until 1997 (7). However, V. cholerae altered ET continues to be routinely isolated from clinical cholera cases in Asia and Africa (4, 5, 15, 18), and both are reported to be capable of causing a more severe disease (19) and of spreading globally (20). The clonal shift observed in this study may well have implications for global cholera, considering that altered ET strains, such as those isolated from cholera cases in Asia and Africa, are dominant pandemic pathogens (5, 18, 20, 21).
The toxin coregulated pili (TCP), a receptor for lysogenic CTXΦ (22), is encoded by a 40-kb gene cluster, Vibrio pathogenicity island (VPI) (23). Like CTXΦ, TCP is biotype-specific, with two distinct alleles of tcpA encoding the major protein “pilin” (10, 11). In Mexico, the TCP variant of V. cholerae O1 strains carries a tcpA sequence that is different from tcpA types reported to date, including CL and ET (10, 11), although an analog of the Mexican type tcpA deposited in GenBank is from a nontoxigenic O1, reportedly isolated in 2006 from China. V. cholerae O1 strains from Mexico carrying the variant tcpA are nontoxigenic. Thus, nontoxigenic O1 strains with this tcpA can occur in geographically different ecosystems, namely Mexico and China.
As in the case of lysogenic CTXΦ that propagate by infecting susceptible nontoxigenic strains of both V. cholerae and Vibrio mimicus (22, 23), converting the host to CTXΦ+, excision can also occur, transforming the host to CTXΦ− (24). In the present study, V. cholerae O1 strains isolated in Mexico during 2000–2004 were predominantly CTXΦ−, TCP−. Although VPI is known to be horizontally transferred in V. cholerae (25), it is not generally considered a virion. In any case, CTXΦ−, TCP− V. cholerae in Mexico can be considered to have originated from a toxigenic progenitor via excision of both CTXΦ and VPI.
Bacterial clonal switching in V. cholerae can have a profound epidemiological influence (14, 19). Factors playing a role in clonal selection can be biotic, abiotic, or both, in natural aquatic ecosystems where V. cholerae is an autochthonous presence (24, 26, 27). In the study reported here, a unique clonal shift was observed whereby toxigenic V. cholerae O1 in Mexico was replaced, starting in 2001, with CTXΦ−, TCP− V. cholerae and superseded by toxigenic V. cholerae in 2005. It can be hypothesized that nontoxigenic V. cholerae in the aquatic environment will outnumber pathogenic strains when fecal–oral transmission is brought under control by application of stringent public health measures, as was done following the 1991–1997 epidemics in Mexico (7, 17, 28). Also important to note is that many bacterial pathogens including V. cholerae become nonculturable after release from human host into the aquatic environment, becoming noncompetitive with environmental V. cholerae, a phenomenon that may have occurred in Mexico between 2001 and 2004 (26, 27). An alternative hypothesis is that the predominance of a toxigenic or nontoxigenic clone may occur if phages provide selective advantage to a V. cholerae subpopulation (27, 29).
Nontoxigenic V. cholerae O1 is associated with a mild but rather broad spectrum of human disease. A clinical link for nontoxigenic V. cholerae O1 ET was first reported by the US Centers for Disease Control and Prevention (CDC) between 1977 and 1991 (30). During the past three decades, there have been reports of isolation of nontoxigenic V. cholerae O1 from clinical, sewage, oyster, and surface-water samples collected in several countries, including Bangladesh, Guam, Brazil, Peru, Japan, England, the United States, and Mexico (7, 30). CT− V. cholerae O1 strains occur sporadically as the causative agent of cholera outbreaks, but not at as high a frequency as in Mexico between 2000 and 2004. It is important to note that nontoxigenic V. cholerae O1 was associated with a large cluster of cases of cholera in India (31). In addition to V. cholerae O1 CT− strains, currently available data suggest that a significant proportion of non-O1 strains possessing CT or non–O1-specific heat-stable enterotoxin (NAG-ST) are associated with cases of diarrhea (32).
In toxigenic V. cholerae O1 ET and O139 Bengal strains, the CTXΦ genome often is flanked by satellite phage RS1 carrying genes rstA, rstB, rstC, and rstR that determine integration of plasmids transporting portions of CTXΦ into the bacterial genome (23). The RS2 genetic element also is a satellite phage but differs from RS1 in lacking rstC (22). V. cholerae O1 CL biotype CTX prophages lack RS elements and exist either as a solitary prophage or as arrays of two truncated, fused prophages (8). V. cholerae O1 ET and O139 Bengal strains carrying RS1 generally yield infectious CTXΦ, but CL biotype strains do not. V. cholerae altered ET carrying the ctxB allele of the CL biotype recently was reported to have been isolated in Bangladesh (Matlab variant) and Mozambique (Mozambique variant), lacking RS1 (8, 18). Pathogenic V. cholerae ET strains isolated in Mexico between 2004 and 2008 lack RS1, but, unlike the Matlab and Mozambique variants (8, 18), the ctxB allele of these strains is an ET biotype. In addition, ET biotype strains isolated in Mexico during 2004–2008 exhibited a novel genetic array, with a truncated CTX prophage instead of RS1 element in the upstream region of the ET-specific CTXΦ located in the large chromosome (Chr I) of V. cholerae. It has been proposed that gene capture via plasmids or phages contributes to rapid adaptation and evolution in V. cholerae (33), a probable situation in Mexico, with clonal CTX+ ET displacing all other preexisting subtypes (7). Although the epidemiological significance of the newly emerged V. cholerae CTX+ ET in Mexico is yet to be understood, the truncated CTXΦ upstream could surrogate RS elements in facilitating replication of the lysogenic CTXΦ genome, producing infectious phage particles.
An aquatic reservoir of V. cholerae in the Americas has previously been documented, including association of V. cholerae with plankton in coastal waters of Peru and Mexico (26, 28, 34). Although the V. cholerae ET isolates causing epidemic cholera in Latin America in 1991 were initially considered to be homogeneous (3), divergence was demonstrated soon thereafter (2, 35, 36). Divergence was clearly evident among V. cholerae ET in Peru (6) and Mexico, and V. cholerae associated with the 1991 epidemic and subsequent endemic cholera in Mexico comprises a diverse population of serogroup O1 strains, including biotypes CL and ET, together with altered variant ET (7). V. cholerae O1 endemic cholera along the Gulf of Mexico coast after 1997 recently has been confirmed, with changes in ribosomal patterns separating strains into two distinct chronological groups, those isolated before and after 1997. Results presented here are in agreement with those of Lizarraga-Partida et al. (28), who described a new CTX prophage− V. cholerae lineage emerging shortly after 1997 in Mexico and continuing to displace preexisting V. cholerae O1 CL and ET strains, including altered ET strains (7). The CTX+ V. cholerae ET with a truncated CTX prophage isolated during endemic cholera in Mexico may prove to be a historical point in the global epidemiology of cholera.
Materials and Methods
Bacterial Strains.
Vibrio cholerae O1 strains included in the present study (n = 91) together with source, location, and year of isolation (Table 1), were provided by the Department of Public Health, Faculty of Medicine, National Autonomous University of Mexico (UNAM) and Centro de Investigación Científica y de Educación Superior de Ensenada. The strains were isolated from cholera patients (n = 36) and from surface-water samples (n = 55) as part of a nationwide cholera surveillance program conducted between 1998 and 2008. These isolates were identified as V. cholerae using a combination of biochemical and molecular procedures, as described previously (24).
Serogroup Analysis.
V. cholerae strains were screened serologically by slide agglutination, using polyvalent antisera specific for V. cholerae O1 or O139, followed by monoclonal antibody specific for each serogroup (1). Serogroups were reconfirmed using polyvalent O1 and monovalent Inaba and Ogawa antisera (7).
Biotype Analysis.
V. cholerae O1 strains were screened for chicken erythrocyte agglutination, sensitivity to polymyxin B, and Mukerjee classical (CL)-specific phage IV, and Mukerjee El Tor (ET)-specific phage V (1).
Storage.
All V. cholerae strains were subcultured on gelatin agar (GA), and a single representative colony from each GA plate was inoculated into T1N1 broth (1% Trypticase and 1% NaCl), incubated at 37 °C for 4–6 h, and stored at −80 °C with 15% (vol/vol) glycerol for future use.
Complementation of Serogrouping and Biotyping Results by PCR Assays.
Genomic DNA was extracted from each of the V. cholerae strains following previously described methods (7). V. cholerae strains identified primarily by phenotypic characteristics were reconfirmed by PCR for species-specific ompW (7). Serogroup-specific O biosynthetic wbeO1, wbfO139, and cholera toxin subunit A encoding ctxA were also examined by multiplex PCR (37). Furthermore, PCR targeting biotype-specific genetic markers, including tcpA (CL or ET), hlyA (CL or ET), and rstR (CL or ET) (13), were performed to complement phenotypic test results. Mismatch amplification mutation assay (MAMA)-PCR was performed to determine biotype-specific ctxB, as described previously (38).
DNA Sequencing of ctxB and tcpA.
Nucleotide sequencing of ctxB and tcpA of randomly selected representative strains was carried out using an ABI PRISM BigDye Terminator Cycle Sequencing Reaction kit (Perkin-Elmer Applied Biosystems) on an ABI PRISM 310 automated sequencer, as described previously (4). Nucleotide sequences were compared with the corresponding sequences of V. cholerae ET N16961 (GenBank accession no. NC_002505.1), and V. cholerae CL O395 (GenBank accession no. CP001235.1), retrieved from GenBank by Basic Local Alignment Search Tool (BLAST).
Genetic Analysis of CTXΦ and Flanking Region.
Mexican V. cholerae ET strains possessing ctxBET and rstRET and unable to amplify rstC of the RS1 element were further analyzed using primers rstAF/rstCR targeting the RS1 element (39). V. cholerae N16961, possessing RS1 downstream of CTXΦ, and V. cholerae O395, lacking RS1, were used as controls, considering that N16961 has CTXΦ only in the large chromosome, whereas O395 possesses CTXΦ in both the large and small chromosomes. The genetic structure of CTXΦ and its flanking region in representative El Tor strains harboring ctxBET and rstRET was analyzed using similar primers and primer walking and sequencing as described previously (39, 40). Primers cepF/rstRETR, cepF/rstAR, and cepF/rstB were used to detect truncated CTXΦ in strains lacking RS1 element and tandem repeat of CTX prophage. The ∼1,200-bp amplicon for the cepF/rstRETR primers was sequenced for confirmation of truncated CTXΦ and subsequently was deposited in GenBank under accession no. KC952008.
Phylogenetic Analysis of tcpA.
To determine the genetic relatedness of tcpA in the Mexican V. cholerae O1 strains, tcpA of representative ET, CL, and TCP variants was sequenced, using primers and PCR conditions as described previously (11). Multiple sequence alignment of trimmed tcpA (600 bp, encoding mature protein) and phylogenetic analysis of tcpA alleles were done using the ClustalW multiple alignment program and the Molecular Evolutionary Genetic Analysis program, version 5.05 (MEGA5), respectively. TcpA sequences of two Mexican strains (Mex-2058 and Mex-3065) were used for construction of a phylogenetic tree with 33 tcpA sequences, of which 31 were of serogroup O1 and non-O1/non-O139 from a previous study (11), and two of nontoxigenic O1 strains from China (ZJ59 and LN93097, accession nos. EU622531 and AF512425, respectively, in GenBank). A neighbor-joining tree with 1,000 bootstrap was generated to determine the phylogenetic relationships on the basis of tcpA sequences.
Pulsed-Field Gel Electrophoresis.
Agarose-embedded genomic DNA was prepared from each of the V. cholerae strains. Genomic DNA was digested by NotI restriction enzyme (GIBCO-BRL). Salmonella braenderup DNA digested by XbaI served as molecular size markers. PFGE was carried out using a contour-clamped homogeneous electrical field (CHEF-DRII) apparatus (Bio-Rad), following procedures described elsewhere (6, 41). Gel fingerprint patterns were analyzed using the Bionumeric Software Package (Applied Maths). After background subtraction and gel normalization, fingerprint patterns were subjected to typing based on banding similarity and dissimilarity, using Dice similarity coefficient and Unweighted Pair Group Method with Arithmetic Mean (UPGMA) clustering, as recommended by the manufacturer. Results are graphically represented as a dendrogram.
Supplementary Material
Acknowledgments
This research was supported in part by National Institutes of Health Grant 1R01A13912901, under collaborative agreements between the Johns Hopkins Bloomberg School of Public Health, the University of Maryland, College Park, and the International Center for Diarrheal Disease Research, Bangladesh (icddr,b); by the National Institute of Infectious Diseases (NIID), Tokyo; and by National Oceanic and Atmospheric Administration (NOAA) Grant SO660009. The icddr,b acknowledges the following donors who provide unrestricted support to the center’s research efforts: the Australian Agency for International Development (AusAID), the government of Bangladesh, the Canadian International Development Agency (CIDA), the Swedish International Development Cooperation Agency (SIDA), and the United Kingdom Department for International Development (DFID).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper of the ∼1,200-bp amplicon for the cepF/rstRETR primers has been deposited in the GenBank database (accession no. KC952008).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323408111/-/DCSupplemental.
References
- 1.Kaper JB, Morris JG, Jr, Levine MM. Cholera. Clin Microbiol Rev. 1995;8(1):48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evins GM, et al. The emerging diversity of the electrophoretic types of Vibrio cholerae in the Western Hemisphere. J Infect Dis. 1995;172(1):173–179. doi: 10.1093/infdis/172.1.173. [DOI] [PubMed] [Google Scholar]
- 3.Wachsmuth IK, et al. The molecular epidemiology of cholera in Latin America. J Infect Dis. 1993;167(3):621–626. doi: 10.1093/infdis/167.3.621. [DOI] [PubMed] [Google Scholar]
- 4.Nair GB, et al. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol. 2006;44(11):4211–4213. doi: 10.1128/JCM.01304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Safa A, Sultana J, Dac Cam P, Mwansa JC, Kong RY. Vibrio cholerae O1 hybrid El Tor strains, Asia and Africa. Emerg Infect Dis. 2008;14(6):987–988. doi: 10.3201/eid1406.080129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nusrin S, et al. Peruvian Vibrio cholerae O1 El Tor strains possess a distinct region in the Vibrio seventh pandemic island-II that differentiates them from the prototype seventh pandemic El Tor strains. J Med Microbiol. 2009;58(Pt 3):342–354. doi: 10.1099/jmm.0.005397-0. [DOI] [PubMed] [Google Scholar]
- 7.Alam M, et al. Cholera between 1991 and 1997 in Mexico was associated with infection by classical, El Tor, and El Tor variants of Vibrio cholerae. J Clin Microbiol. 2010;48(10):3666–3674. doi: 10.1128/JCM.00866-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun J, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA. 2009;106(36):15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd EF. Efficiency and specificity of CTXphi chromosomal integration: Dif makes all the difference. Proc Natl Acad Sci USA. 2010;107(9):3951–3952. doi: 10.1073/pnas.1000310107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukhopadhyay AK, Chakraborty S, Takeda Y, Nair GB, Berg DE. Characterization of VPI pathogenicity island and CTXphi prophage in environmental strains of Vibrio cholerae. J Bacteriol. 2001;183(16):4737–4746. doi: 10.1128/JB.183.16.4737-4746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P, Thulaseedharan A, Chowdhury G, Ramamurthy T, Thomas S. Characterization of novel alleles of toxin co-regulated pilus A gene (tcpA) from environmental isolates of Vibrio cholerae. Curr Microbiol. 2011;62(3):758–763. doi: 10.1007/s00284-010-9774-3. [DOI] [PubMed] [Google Scholar]
- 12.Keasler SP, Hall RH. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341(8861):1661 (lett). doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 13.Kimsey HH, Nair GB, Ghosh A, Waldor MK. Diverse CTXphis and evolution of new pathogenic Vibrio cholerae. Lancet. 1998;352(9126):457–458. doi: 10.1016/S0140-6736(05)79193-5. [DOI] [PubMed] [Google Scholar]
- 14.Siddique AK, et al. Survival of classic cholera in Bangladesh. Lancet. 1991;337(8750):1125–1127. doi: 10.1016/0140-6736(91)92789-5. [DOI] [PubMed] [Google Scholar]
- 15.Nair GB, et al. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol. 2002;40(9):3296–3299. doi: 10.1128/JCM.40.9.3296-3299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raychoudhuri A, et al. Classical ctxB in Vibrio cholerae O1, Kolkata, India. Emerg Infect Dis. 2009;15(1):131–132. doi: 10.3201/eid1501.080543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alam M, et al. Vibrio cholerae classical biotype strains reveal distinct signatures in Mexico. J Clin Microbiol. 2012;50(7):2212–2216. doi: 10.1128/JCM.00189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ansaruzzaman M, et al. Mozambique Cholera vaccine Demonstration Project Coordination Group Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis. 2004;10(11):2057–2059. doi: 10.3201/eid1011.040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siddique AK, et al. El Tor cholera with severe disease: A new threat to Asia and beyond. Epidemiol Infect. 2010;138(3):347–352. doi: 10.1017/S0950268809990550. [DOI] [PubMed] [Google Scholar]
- 20.Chin CS, et al. The origin of the Haitian cholera outbreak strain. N Engl J Med. 2011;364(1):33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Islam MS, et al. Phenotypic, genotypic, and antibiotic sensitivity patterns of strains isolated from the cholera epidemic in Zimbabwe. J Clin Microbiol. 2011;49(6):2325–2327. doi: 10.1128/JCM.00432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272(5270):1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 23.Faruque SM, Rahman MM, Asadulghani, Nasirul Islam KM, Mekalanos JJ. Lysogenic conversion of environmental Vibrio mimicus strains by CTXPhi. Infect Immun. 1999;67(11):5723–5729. doi: 10.1128/iai.67.11.5723-5729.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alam M, et al. Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc Natl Acad Sci USA. 2007;104(45):17801–17806. doi: 10.1073/pnas.0705599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karaolis DKR, et al. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95(6):3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colwell R, Spira W. In: The ecology of Vibrio cholerae. Cholera. Barua D, Greenough WI, editors. New York: Plenum; 1992. pp. 107–127. [Google Scholar]
- 27.Nelson EJ, Harris JB, Morris JG, Jr, Calderwood SB, Camilli A. Cholera transmission: The host, pathogen and bacteriophage dynamic. Nat Rev Microbiol. 2009;7(10):693–702. doi: 10.1038/nrmicro2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lizárraga-Partida ML, et al. Association of Vibrio cholerae with plankton in coastal areas of Mexico. Environ Microbiol. 2009;11(1):201–208. doi: 10.1111/j.1462-2920.2008.01753.x. [DOI] [PubMed] [Google Scholar]
- 29.Nelson EJ, et al. Transmission of Vibrio cholerae is antagonized by lytic phage and entry into the aquatic environment. PLoS Pathog. 2008;4(10):e1000187. doi: 10.1371/journal.ppat.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodrigue DC, Popovic T, Wachsmuth IK (1994) Vibrio Cholerae and Cholera: Molecular to Global Perspectives, eds Wachsmuth IK, Blake PA, Olsvik Ø (ASM Press, Washington, DC), pp 69–76.
- 31.Saha PK, et al. Nontoxigenic Vibrio cholerae 01 serotype Inaba biotype El Tor associated with a cluster of cases of cholera in southern India. J Clin Microbiol. 1996;34(5):1114–1117. doi: 10.1128/jcm.34.5.1114-1117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagchi K, et al. Epidemic of diarrhea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp in Thailand. J Clin Microbiol. 1993;31(5):1315–1317. doi: 10.1128/jcm.31.5.1315-1317.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manning PA, Clark CA, Focareta T. Gene capture in Vibrio cholerae. Trends Microbiol. 1999;7(3):93–95. doi: 10.1016/s0966-842x(99)01464-x. [DOI] [PubMed] [Google Scholar]
- 34.Gil AI, et al. Occurrence and distribution of Vibrio cholerae in the coastal environment of Peru. Environ Microbiol. 2004;6(7):699–706. doi: 10.1111/j.1462-2920.2004.00601.x. [DOI] [PubMed] [Google Scholar]
- 35.Dalsgaard A, et al. Molecular evolution of Vibrio cholerae O1 strains isolated in Lima, Peru, from 1991 to 1995. J Clin Microbiol. 1997;35(5):1151–1156. doi: 10.1128/jcm.35.5.1151-1156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beltrán P, et al. Genetic diversity and population structure of Vibrio cholerae. J Clin Microbiol. 1999;37(3):581–590. doi: 10.1128/jcm.37.3.581-590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoshino K, et al. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol. 1998;20(3):201–207. doi: 10.1111/j.1574-695X.1998.tb01128.x. [DOI] [PubMed] [Google Scholar]
- 38.Morita M, et al. Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol Immunol. 2008;52(6):314–317. doi: 10.1111/j.1348-0421.2008.00041.x. [DOI] [PubMed] [Google Scholar]
- 39.Mohapatra SS, Mantri CK, Turabe Fazil MH, Singh DV. Vibrio cholerae O1 biotype El Tor strains isolated in 1992 from Varanasi, India harboured El Tor CTXΦ and classical ctxB on the chromosome-I and classical CTXΦ and classical ctxB on the chromosome-II. Environ Microbiol Rep. 2011;3(6):783–790. doi: 10.1111/j.1758-2229.2011.00287.x. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen BM, et al. Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J Clin Microbiol. 2009;47(5):1568–1571. doi: 10.1128/JCM.02040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cameron DN, Khambaty FM, Wachsmuth IK, Tauxe RV, Barrett TJ. Molecular characterization of Vibrio cholerae O1 strains by pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32(7):1685–1690. doi: 10.1128/jcm.32.7.1685-1690.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.