Summary
The existence of sub-populations of cells in cancers with increased tumor initiating capacities and self-renewal potential, often termed ‘cancer stem cells’, is a much discussed and key area of cancer biology. Such cellular heterogeneity is very important due to its impact on therapy and especially states of treatment resistance. A major question is whether there is plasticity for evolution of these cell states during tumorigenesis which can involve movement between cell populations in a reversible fashion. In this review, we discuss the possible role of epigenetic abnormalities, as well as genetic alterations in such dynamics, and in the creation of cellular heterogeneity in cancers of all types.
Introduction
Cellular heterogeneity is a well-recognized attribute of both normal and neoplastic tissues. The difference is that in the former there is an ordered developmental program underlying the heterogeneity. This order dictates that from a single genome, or the “hard drive” of DNA, without base sequence changes, multiple cell types can be generated through the “software packages” of proper co-ordination of dynamic signal transduction, and subsequently, long term maintenance of gene expression patterns through epigenetic mechanisms (Allis et al., 2008). These control processes ensure proper balance between cells capable of continued self-renewal, or being maintained in stem cell-like states, and their generation of progeny cells committed to tissue lineages and differentiation. By contrast, disorder characterizes cancer cell populations. One driving factor for this is obviously genetic instability through which mutations alter gene function such that cells either do not exit self-renewal states and/or commit properly to tissue lineage and differentiation (Stratton et al., 2009; Vogelstein et al., 2013). In this review, we visit the possibility that aberrations of epigenetic control may also significantly contribute to the disorder of cancer. If so, the consequences are profound since reversal of abnormalities for therapy strategies is difficult in terms of correcting mutations but much more promising in terms of reversing epigenetic abnormalities. Also, and related to therapy strategies, the points we will make are key because the dynamic variability, or heterogeneity, of cell populations provides the driving force for tumors to utilize selection pressures to evolve. While progressive mutations certainly do play a role in such evolution, we will emphasize that epigenetic changes are also key factors and may be especially important to the emergence of, and plasticity for, formation of the most tumor initiating cell sub-populations in cancer. Such cells may also be key for treatment resistance; indeed, they may be the major factor in therapy failures that plague the management of the most common cancers and those with the highest mortality statistics.
Inherent to the above concept of cancer cell heterogeneity as it contributes to tumor initiation and progression, is the “cancer stem cell” hypothesis. However one frames this concept, most cancer biologists accept that, at any given time in a cancer, there are populations of cells with cancer cell renewal and tumor initiating properties (Beck and Blanpain, 2013; Nguyen et al., 2012; Wang and Dick, 2008). Their frequency may differ from tumor to tumor, ranging from virtually all the cells to small populations. Also, arguments continue as to whether there is a hierarchical arrangement for such populations versus their less tumorigenic counterparts, or whether there is plasticity in which such stem-cell like populations can always be generated, especially under stress situations, from other cells in the population (Meacham and Morrison, 2013). Whatever the exact situation, in addressing the biology of the heterogeneity and evolution of cancer stem cell sub-populations, both genetic and epigenetic dynamics must be considered. In this review, we discuss the possibility that during cancer evolution, and during tumor initiation from cancer risk states, such as inflammation, that predispose cells to undergo transformation a cellular plasticity may exist allowing dynamic shifts of more and less virulent cells differing in their tumor initiation and therapeutic resistance capabilities. We will specifically address the potential importance of epigenetic abnormalities, which may underlie such plasticity in cell phenotypes and their link to processes by which, from cancer risk states through tumor progression, cells survive stress to create cancer cell populations. We will also consider how the epigenetic molecular profiles of cancers may reflect the cell sub-compartments in normal cell renewal systems from which cancers arise – and, in turn, how these issues frame the molecular and cell phenotype subpopulations of self-renewing cells in tumors.
Clonal evolution of tumors and evolution of cell heterogeneity
From a histologic and cell population standpoint phenotypic heterogeneity in tumor tissues has long been observed (Beck and Blanpain, 2013; Nguyen et al., 2012; Wang and Dick, 2008). Whether this heterogeneity contributes to functional sub-populations, such as those with metastatic capabilities or stem cell-like properties as discussed above, has been the subject of intense study. Despite the heterogeneity, most likely the diverse cancer cell subpopulations arise within the clone originally giving rise to the cancer. However, it is increasingly being recognized at a molecular level that clonal subpopulations evolve in tumors along courses which may vary considerably by tumor type (Garraway and Lander, 2013; Vogelstein et al., 2013; Yachida et al., 2010). Here we describe the concepts of tumor cell heterogeneity in the context of genetic alterations and aim to distinguish these from those due to epigenetic alterations.
Contribution to tumor heterogeneity from mutations
Recent deep sequencing exercises are providing invaluable insights into the evolutionary dynamics of cancer cell populations and contributions to drug resistance and tumor relapse. Importantly, these studies have reinforced some early hypotheses about the stem-cell origins in the natural history of cancers (Cairns, 1975) and the clonal evolution of cancers through Darwinian selection (Nowell, 1976). Thus, analysis of genome copy number at the level of single cell and genetic mutations from different sites in a tumor and metastasis from the same patient have revealed genetic heterogeneity within the same tumor tissue (Anderson et al., 2011) (Navin et al., 2011) (Gerlinger et al., 2012). Most recently this phenomenon has been observed in subpopulation differences between the primary tumor and distant metastases (Gerlinger et al., 2012). These studies have revealed that there may be transitions, arising either from sub-clonal evolution of heterogeneous cell populations present at the earliest points in tumor evolution or through the new appearance of such cells, which manifest as a linear evolutionary process with branching points (Gerlinger et al., 2012) (Fig 1). In this concept, independent sub-clones arise in bursts of sub-clonal expansion (Navin et al., 2011) which may well arise in response to pressures from Darwinian selection – including those imposed on cancers from therapeutic intervention (Mullighan et al., 2008). A prime example from deep sequencing studies is renal cancer in which the key early mutations in the VHL gene are constant from early time points on, while other mutations arise along a branching pattern consistent with emerging subclones (Gerlinger et al., 2012). From a therapy point of view, such spatial genetic heterogeneity within the same tumor and metastases translates into variegated diagnostic signatures that will have important implications for targeted therapeutic approaches (Anderson et al., 2011; Yang et al., 2012). However, to date, most therapeutically targeted mutations are those that are dominant in the tumor, most likely to have occurred in the founder clone. Certainly, new mutations in the targeted gene do evolve with therapy resistance (Sierra et al., 2010), and recent studies indicate that these may have been present in tumor subclones when therapy was initiated (Turke et al., 2010).
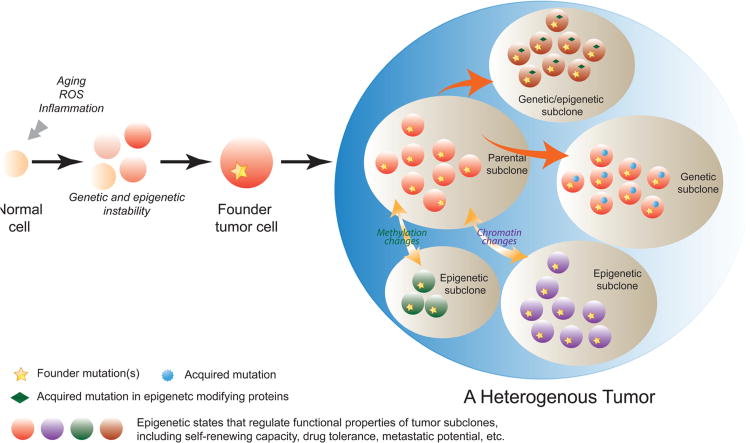
Figure 1. Genetic and epigenetic contributions to tumor heterogeneity.
During oncogenesis, environmental stress such as chronic inflammation, accumulating reactive oxygen species (ROS) or aging, may promote clonal expansion of cells with genetic or epigenetic abnormalities. These cells then acquire further mutations or epigenetic alterations and become founder tumor cells that initiate pre-cancer or cancer. The evolution of tumor clones continues as tumors develop. In an established tumor, the parental subclone may acquire new driver or passenger mutations (genetic subclone), or undergo epigenetic alterations on the levels of chromatin or DNA methylation, or both (epigenetic subclones). Some subclones may acquire mutations in epigenetic modifying proteins resulting in emergence of epigenetic changes (genetic/epigenetic subclones). Either genetic or epigenetic subclones can exhibit different functional attributes from the parental clone in terms of self-renewing capacity, drug tolerance or metastatic potential. Thus, both genetic and epigenetic mechanisms contribute to the intra-tumoral heterogeneity.
In the context of tumor “stem cell” populations, a widely held model for the evolution of cancer is that initial oncogenic mutations result in transformation of a tissue stem or progenitor cell (Greaves and Maley, 2012). This cell then acquires the aberrant property of unrestricted growth giving rise to patches of genetically altered cells in the normal tissue/organ that becomes the substrate for tumor progression via selection due to genetic variation (Braakhuis et al., 2003). Certainly, the driver mutations such as those for K-RAS in multiple tumor types, APC in colorectal cancer and VHL in renal cancers likely are central forces that induce appearance of cells with tumor initiating and self-renewal properties. We will discuss later the concepts of shifts in self-renewing cell sub-populations in cancers that may appear with time, and in response to various survival pressures, and the plasticity of the cell populations involved.
Contribution to tumor heterogeneity from epigenetic alterations
Brief overview of the concept of epigenetics
Before considering evidence for the role of epigenetics, it is important to strictly define this term versus other non-genetic processes, such as signal transduction, which serve to change and maintain cell phenotypes (Ptashne, 2007). The original definition of the term, coined by Waddington, referred to stable changes in cell phenotype based on other than genetic changes (Waddington, 2012). Today's definition encompasses the concept that epigenetic control means a stable, or heritable change in gene expression, without any changes in DNA sequence (Bird, 2007). Certainly the consequences of the gene expression changes are what contribute to the phenotype. Strictly speaking, an epigenetic event implies that the constituent molecular determinants should be heritable through every round of cell replication, thereby, acting to maintain a stable pattern of gene expression subserving a given cell state. This pattern can be preserved even if the original transcription stimuli for the initiation of involved gene states are no longer present. The most agreed upon parameter which strictly fits this definition is DNA methylation, which is the covalent modification of DNA by a methyl group, occurring in humans at predominantly cytosines in a CpG context (Bestor, 1990). During cell division, DNA methylation is maintained on the parent DNA strand and copied on the daughter strand. The patterns of DNA methylation at any point in time can help maintain transcriptional activities of the genome, both for canonical genes and non-coding RNA's, and potentially in enhancer regions, generally serving as a component of transcription repression (Baylin and Jones, 2011; Encode Project Consortium et al., 2012). However, when present in the body of genes, the relationship can be the inverse wherein gene expression may be enhanced in the presence of DNA methylation which may facilitate transcriptional elongation (Shenker and Flanagan, 2012) or alternate promoter usage (Maunakea et al., 2010). DNA methylation cannot, alone, modulate transcription but must work in association with repressive chromatin modifications (Allis et al., 2008; Baylin and Jones, 2011). It is less well proven whether these histone marks remain associated with DNA during replication. However, in a holistic sense if they are re-established heritably after DNA replication, they serve to maintain the stable patterns of transcription for maintenance of cell phenotypes, and thus are essential to epigenetic control of the genome. Thus, histone modifications, in addition to DNA methylation are vital to control of normal cellular states, and vis a vis the focus of this review, cancer cell populations.
Key to each of the above processes is their interaction with how nucleosomes are positioned linearly, and three dimensionally, in the context of DNA, thus providing the “packaging” component of epigenetics (Korber and Becker, 2010). The critical end result of this positioning, in association with DNA methylation and chromatin modifications, is the establishment in normal cells with proper boundaries, which separate the tightly packaged and repressive nucleosome domains from the more spaced and loosely configured nucleosome arrangements. The former facilitate active or permissive transcriptional states (Fan et al., 2005; Thurman et al., 2007). Early in tumor progression, there is increasing evidence that these boundaries break down (Baylin and Jones, 2011; Hansen et al., 2011) (Berman et al., 2011). The result is alteration of structural control of DNA replication and a cancer “epigenome” in which hundreds of coding and non-coding regions have altered transcription states. For the most studied abnormalities, those of DNA methylation, both widespread losses and more focal gains often occur simultaneously within defined megabase regions (Berman et al., 2011) (Hon et al., 2012). The gains are often cancer-specific and involve normally non-DNA methylated CpG islands located in proximal promoter regions of hundreds of genes (Baylin and Jones, 2011) (Berman et al., 2011). This change can be associated with loss of gene expression, which can serve as an alternative to mutations for abolition of tumor suppressor gene function through associated gene silencing. In addition to DNA methylation abnormalities, chromatin changes, such as those driven by over-activity of the long term gene silencing protein complex, polycomb group proteins (PcG), are frequently observed in multiple tumor types (You and Jones, 2012).
Epigenetic alterations in tumor progression
Primal to understanding the contribution of epigenetic changes in tumor progression is the key issue of how do epigenetic changes track with the different progression stages of cancers. Although such studies are far fewer than those for mutations, it is very clear that abnormalities of DNA methylation occur quite early during tumorigenesis frequently manifesting in pre-malignant cells (Suzuki et al., 2004) (Esteller et al., 2000; Greenspan et al., 2006; Licchesi et al., 2008), and in the context of field cancerization (Shen et al., 2005) (Mehrotra et al., 2008; Nonn et al., 2009) (Baylin and Jones, 2011; Ushijima, 2007). (Fig 1) These alterations include the above discussed widespread losses and promoter-focal gains of DNA methylation. These focal gains can involve well identified tumor suppressor genes like CDKN2A, which displays increasing frequency of methylation during progression from lung airway basal cell hyperplasia (17%) to squamous metaplasia (24%) to carcinoma in situ (50%) (Belinsky et al., 1998). Anti-Wnt activity genes become silenced and hypermethylated in early colon neoplasia and evidence indicates they may fundamentally complement key WNT pathway mutations, such as for the APC gene, for colon tumor initiation and/or early progression (Suzuki et al., 2004). With respect to these above early changes, it is important to note that many of the same parameters linked to occurrence of genetic changes are now being linked to causation of epigenetic aberrancies. Thus, aging (Ahuja et al., 1998; Maegawa et al., 2014), chronic inflammation (Niwa and Ushijima, 2010), and environmental exposures (Liu et al., 2010) are now well juxtaposed factors which alter the epigenome. Examples include a rat model of lung carcinogenesis in which a tobacco- specific carcinogen (4-methylnitrosamino-1-(3-pyridyl)-1-butanone) causes frequent hypermethylation of CDKN2A promoter in the early lesions (hyperplastic lesions and adenomas), and this change is then observed at a very high frequency (>90%) in the adenocarcinomas that later emerge (Belinsky et al., 1998). Further, CDKN2A promoter methylation is observed in rat lung tumors induced by exposure to various mutagenic agents (X-rays, plutonium-239 oxide, beryllium metal, cigarette smoke) indicating that the methylation induction can occur in response to different types of environmental insults. Epigenetic changes have been observed in various models of inflammation in mice (Ushijima, 2007) (Hahn et al., 2008), cigarette smoke exposure (Liu et al., 2010) and other environmental carcinogens (like cobalt) (Li et al., 2009). Aging is a leading scenario linked to increasing frequency of promoter, CpG island DNA hypermethylation of many genes, best outlined to date for normal colon over a time span that well tracks with risk for colon carcinoma (Ahuja et al., 1998; Toyota et al., 1999). For chronic inflammatory changes, not only the occurrences of DNA methylation abnormalities have now been chronicled, but the molecular parameters linked to boundary shifts which could trigger simultaneous losses and gains of DNA methylation are beginning to be deciphered, as will be re-visited below (O'Hagan et al., 2011). As for genetic changes, epigenetic alterations are now being mechanistically associated with various types of DNA damage ongoing in chronic inflammatory environments (Tili et al., 2011).
One of the most exciting developments in recent years has been the tying of epigenetic changes to genetic alterations in virtually all tumor types. Thus, very frequent mutations in the genes that encode for chromatin, nucleosome remodeling, and DNA methylation modifying proteins have been observed (Shen and Laird, 2013; You and Jones, 2012). As yet, the precise ramifications of most of these mutations are yet to be delineated; we will outline below their potential implications for understanding initiation and maintenance of stem cell-like subpopulations in specific cancer types.
Epigenetic regulation of cancer stem-like cell subpopulations
Once past the fact that epigenetic abnormalities are prevalent in early tumorigenesis, much less is known than for genetic changes as to how the cancer epigenome evolves with, and contributes functionally to, heterogeneity of tumor cell populations within the original tumor clone. As mentioned earlier in the case of renal cancer, genetic changes can be mapped along a branching evolutionary tree during tumor progression (Gerlinger et al., 2012). Such patterns are not well delineated for epigenetic alterations. A concept now emerging, however, is that from very early tumorigenesis steps on, at least as monitored by DNA methylation, the epigenome is in a state of chaotic shifting (Hansen et al., 2011). In this scenario, the widespread losses seem quite random, and non-clonal while the focal gains in promoter regions are far more clonal (Aryee et al., 2013). This is consistent with earlier studies of individual genes in tumors wherein the focal promoter gains were constant between more putative stem cell populations and their progeny (Yi et al., 2008). This conservation of epigenetic changes indicates that the aberrant hypermethylation that occurs early on is maintained, and that at least some of the changes potentially arise as a result of selection during subsequent steps of evolution. Defining which genes function as driver versus passenger events for such selection is currently a major question in cancer epigenetics research. In this regard, a recent study indicated that in a colon cancer cell line with engineered deletion of two major DNMT's, and 95% or more loss of DNA methylation, a group of genes still retain promoter hypermethylation - and that their loss of function may be essential for survival of the cells (De Carvalho et al., 2012).
How, in the above dynamics, do epigenetic abnormalities contribute to key cell sub-populations in cancers and particularly those with stem-like function? A key to their potential importance comes from the nature of genes with focal gains in promoter CpG islands and associated transcriptional silencing. Several groups, including ours have documented that a large proportion of genes with this cancer specific change are those with a history of a specific pattern of chromatin regulation in embryonic stem cells (Schlesinger et al., 2007) (Widschwendter et al., 2007) (Ohm et al., 2007) and which we have also traced back to adult stem cells (Easwaran et al., 2012). This chromatin, termed “bivalent chromatin” (Azuara et al., 2006) (Bernstein et al., 2006), is defined by simultaneous presence of the repressive histone marks, H3K27me3, and active, H3K4me3, and normally involves genes with non-DNA methylated, promoter CpG islands in embryonic and adult stem cells. These genes, when in a bivalent state, often have a low, poised level of transcription and generally switch to either an active state with predominantly promoter H3K4me3, or a repressed state, with predominantly H3K27me3, during differentiation (Bernstein et al., 2006). The majority of promoter DNA hypermethylated genes in cancer come from this pool and many of these control the balance between maintaining self renewal and commitment to differentiation in embryogenesis (Easwaran et al., 2012).
Although the existence of the bivalent state is now well established in terms of a promoter zone in which there is existence of both the H3K27me3 and H3K4me3 marks surrounding transcription start, some changes concerning the concept are emerging. First, the term bivalency refers to a zonal distribution of the marks and only recently is it established that there are individual nucleosomes that can carry both marks (Voigt et al., 2012). Second, there are suggestions that bivalency emerges to follow states of transcription rather than setting them. In this view, the degree of the H3K27me3 occupancy builds by default when transcription is low (Di Croce and Helin, 2013). Thus the initially proposed role for bivalent chromatin in maintaining developmental genes in a poised state for later activation or suppression might be an oversimplified. However, there is still no doubt that many genes marked by bivalent promoters do reside in relatively low expression states, and that during differentiation undergo activation or silencing in association with conversion to H3K4me3 or H3K27me3 marks. It then follows that the transcriptionally “off” state mediated by H3K27me3 can be modified by signal transduction for a range of generally low level transcription which is reflected by varying, simultaneous levels of the active H3K4me3 mark (De Gobbi et al., 2011). Such a model is consistent with studies showing that bivalent promoters mostly have paused RNA Pol II (Brookes et al., 2012; Min et al., 2011). In such a state, these genes may be primed to be activated or silenced in response to the right environmental cue and presence of transcription factors. Importantly, when DNA hypermethylated in cancer, these genes with potential roles in differentiation are less responsive to such cues and are fully suppressed due to a lack of, or markedly reduced, H3K4me3 and H3K27me3 (Easwaran et al., 2012) (Fig 2).
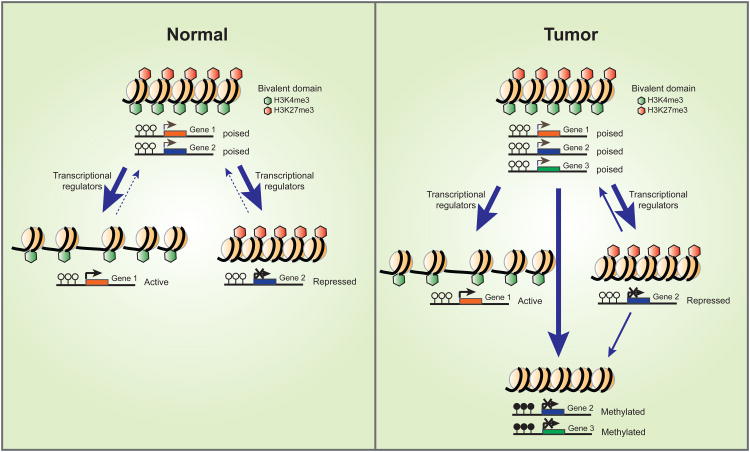
Figure 2. Epigenetic plasticity in normal and cancer cells.
In normal embryonic and adult stem cell states, many developmental genes maintain both active (H3K4me3) and repressed(H3K27me3) marks, or bivalent chromatin, at their promoter regions. This state helps maintain genes gene in poised states for transcription. During normal differentiation, bivalent domains will resolve into either active marks (H3K4me3) attendant to active transcription of certain differentiation-related genes, or repressive marks (H3K27me3) marks which accompany silencing of stem cell-related genes. Under certain circumstances, the resolution of bivalent domains can be reversed and cells may undergo dedifferentiation. Also, bivalency of genes may arise in cell populations distal to stem cell states. In a cancer cell, these chromatin shifts may be vital to cellular plasticity. Some of the bivalent genes may assume cancer-specific promoter DNA hypermethylation and, thus, be locked in a more permanent silenced state.
We suggest that in cancer, for gene promoters where DNA methylation replaces zones of bivalent marking, abnormal silencing for individual, or groups, of such genes, may help select events in initiation and/or progression of the cancer. We have hypothesized that their silencing, perhaps for different groups in different tumor types, can contribute mechanisms for how cancer cell subpopulations are held in an abnormal state of self-renewal potential at the expense of proper capacity to respond to lineage commitment and differentiation cues (Easwaran et al., 2012). This state is a fundamental defect of cancer cells and particularly those termed cancer “stem cells”. Recent studies illustrate how epigenetic abnormalities may drive these cell properties and serves as another illustration of how they can collaborate with genetic changes. Mutations in the isocitric dehydrogenase genes (IDH) have been linked to the genes that are bivalent in embryonic stem cells and DNA hypermethylated in subsets of glioblastomas (Noushmehr et al., 2010; Parsons et al., 2008), chondrosarcomas (Amary et al., 2011), and acute myelogenous leukemias (AML) (Figueroa et al., 2010). The molecular underpinning appears to be a combination of abnormal depletion of the Krebs cycle constituent α-keto-glutarate at the expense of marked increases in D-2-hydroxyglutarate (D-2-HG) which has been termed an “oncometabolite” (Dang et al., 2009). This oncometabolite inhibits various Fe(II)/2-oxoglutarate-dependent dioxygenases (Xu et al., 2011), including various histone demethylases that protect against DNA methylation by diminishing chromatin marks that attract DNA methylation, and the TET family of enzymes that catalyze DNA demethylation by converting 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC). Mice with genetic knock-in of IDH mutations have abnormally increased numbers of early haematopoietic progenitors and blockage of lineage commitment (Cairns and Mak, 2013; Sasaki et al., 2012). A similar phenotype has been seen for engineered mouse cells (Lu et al., 2013) and for patient-derived glioma xenografts with an IDH1 mutation (Borodovsky et al., 2013; Turcan et al., 2013). Critically, the build up of abnormal DNA methylation in the genes under discussion has been seen in each of the above scenarios, and treatment of cells with DNA demethylating agents can restore cell ability for induction of differentiation (Borodovsky et al., 2013) (Turcan et al., 2013). A challenge in all of this work is to directly identify whether, and which, genes that are bivalently marked in embryonic and adult stem cells may be responsible for abnormal retention of the cancer stem cell-like properties.
There is also growing evidence that cancer stem cells can harbor key epigenetic states defined by not just DNA methylation alterations but also solely by the histone modifications discussed above. For example, in AML, the AML-CSCs and non-CSCs differ in their histone modification patterns (H3K4me3 and H3K27me3) but not DNA methylation patterns (Yamazaki et al., 2013). In glioblastoma (GBM), the GBM-CSCs have aberrant activation of multiple transcription factors due to loss of the polycomb mark H3K27me3 from their promoters (Rheinbay et al., 2013). Aberrant expression of one of these transcription factors, ASCL1, causes Wnt activation, which is required for maintenance of the CSC state and its tumorigenecity. A dramatic instance of such loss of H3K27me3, and again an example of interactions between genetics and epigenetics, occurs in a type of pediatric brain tumor. Although these tumors harbor a mutation in the K27 residue of histone H3 in only one of multiple alleles encoding this histone (Shen and Laird, 2013), this change appears to act as a dominant negative effect inhibiting all EZH2 activity resulting in loss of H3K27me3 (Lewis et al., 2013). This in turn probably results in abnormal target gene activation, which might initiate and drive these tumors.
Epigenetic regulation of cancer cell plasticity
Our understanding of the clonal evolution of epigenetic alterations regulating gene expression is very limited. The median frequency of methylated gene promoters per tumor is in the range of 250 to 800 depending on the tumor type, which is far higher compared to the frequency of non-synonymous gene mutations per tumor, which maximally is in the range of 150-170 (Vogelstein et al., 2013). Further, compared to the spontaneous mutation rate in normal and cancer cells, which is about 10−10 mutations/nucleotide base pair/division (Jones et al., 2008), the error rate for gaining or losing methylation is far higher, estimated at 2 × 10−5 per CpG site per division (Yatabe et al., 2001). Therefore, despite the fact that focal promoter DNA hypermethylation is consistent between primary tumors and their metastatic counterparts (Aryee et al., 2013), the high error rate for replicating DNA methylation may indicate far more epigenetic than genetic variability occurs as tumors evolve. The precise ramifications of this epigenetic variability remain to be fully resolved. However, epigenetic changes, and especially DNA methylation, are heritable and can directly influence gene function when strategically located. They may, then contribute to clonal selection and thus be a factor in creating tumor cell heterogeneity.
Thus, epigenetic alterations arising stochastically, or as part of an aberrant transcriptional program, can potentially impart selective advantage just like genetic aberrations, such as inducing silencing of key tumor suppressor genes or inducing dysfunction in DNA repair genes. Such epigenetic variation might be one of the changes contributing to a cellular plasticity for formation of tumor initiating sub-populations during tumor evolution. This would be a factor in countering the strictly hierarchical theory for existence of cancer stem cells as a stable population with unrestricted tumor initiating potential that always give rise to progeny cells lacking, or reduced for, the tumor initiating property (Greaves and Maley, 2012; Gupta et al., 2011). By either theory or combinations thereof, tumors will be inherently heterogeneous in their cell populations based on their epigenetic makeup.
As mentioned, the question regarding a strict hierarchical model of tumors versus the plasticity model is still debated. However, evidence for tumor cell populations that can reversibly shift between functional states, including between stem-like and more committed cells, and a role in epigenetic shifts, is emerging. A key example is the concept that epithelial to mesenchymal transition (EMT) in cancer occurs along a spectrum found for normal cell transitions that are active from model organisms to man and which allow the proper control of organogenesis (Tam and Weinberg, 2013; Thiery et al., 2009). In the concept of EMT, the mesenchymal cells are more stem-like and have properties allowing for continued self-renewal which preserves stemlike populations in renewing systems including for migratory capabilities. These activities are fundamental for tissue formation during development and also in adult cell renewal. Inherent to this process, both in normal settings and cancer, is that with various cues cells can bi-directionally slide along this spectrum going from mesenchymal to epithelial characteristics and reverse this course as situations warrant (Fig 3). This has been experimentally seen for both normal and cancer-related mammary epithelial renewal. Although transitions occur even in normal cells, such switching occurs more efficiently in transformed cells (Chaffer et al., 2011). Thus, for breast cancer, the data indicate that more stem-like or mesenchymal phenotype cells can appear, and shift, at any time during tumor evolution leading to mixed cell populations in tumors with respect to more epithelial cell properties. (Chaffer et al., 2011; Marjanovic et al., 2013)
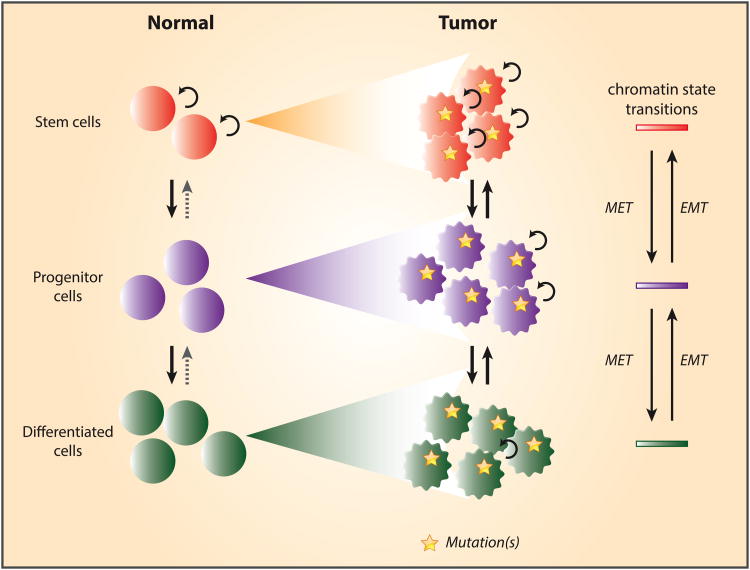
Figure 3. Cancer may derive from different compartments during normal cell differentiation and have plasticity for movement of cell subtypes.
During normal cell differentiation, self-renewing tissue stem cells undergo a series of chromatin state transitions and give rise to progenitor and differentiated cells. Cellular transformation may take place at different stages during normal differentiation, and give rise to malignant cells that carry similar, but abnormal chromatin states relative to their normal counterparts. Notably, tumor cells may exhibit a cellular plasticity through which they can switch between more stem cell-like states and more differentiated cell states through mesenchymal-epithelial transition (MET) or epithelial-mesenchymal transition (EMT).
How do epigenetic properties specifically enter the picture for controlling EMT plasticity? It is postulated that for EMT states at any given point in time, chromatin regulation of key transcription factors and their downstream targets is critical (Chaffer et al., 2011; Marjanovic et al., 2013). In this regulation, WNT activation, TGF-beta activation, and regulation of E-cadherin are all key pathway controls. For the chromatin regulation, gene silencing may be particularly important as controlled by the PcG proteins and the attendant H3K27me3 mark, and particularly in the setting of promoter bivalency(Chaffer et al., 2011; Marjanovic et al., 2013). Thus, especially for the mesenchymal or stem-like cells, such bivalency may hold genes like E-cadherin in a poised state that would otherwise allow epithelial characteristics to accrue if activated and suppress factors that allow WNT or TGF-beta activation. In contrast, and key for the concept of plasticity, some genes may similarly be held in bivalent states in more epithelial cells, which if more active might otherwise trigger EMT.
Overall, we hypothesize that a molecular progression along the types of chromatin regulation discussed above, and inherent to the earlier outlined relationships between bivalent chromatin and abnormal promoter DNA methylation, is key for cellular plasticity existing in cancers (Figs. 2 and 3). Further, our own studies indicate that cellular stresses affecting early events in tumor progression, probably during pre-malignant stages, are key to initiating progressive chromatin alterations in cancer. These epigenetic changes may allow survival of cell clones associated with increased cancer risk states, such as those induced by chronic inflammation (O'Hagan et al., 2011). In these dynamics, protein complexes with interaction between PcG proteins and DNMT's can rapidly move into promoter, CpG islands and associate with transcriptional silencing of the involved genes (O'Hagan et al., 2011). These shifts normally are probably transient events which serve to protect these promoter regions from errors of DNA damage repair that might accrue if transcription were not halted. However, during continued environmental insults, such as in chronic inflammation, low gene transcription associated with chromatin states such as bivalency, and/or typical of PcG marked genes, might render associated promoters vulnerable to retention of the above protein complexes and induce onset of time dependent accrual of abnormal DNA methylation (O'Hagan et al., 2011). Indeed, initiation of such methylation in a time frame as short as 30 minutes can be observed after exposure of cells to H2O2 exposure (O'Hagan et al., 2011). The deeper silencing of stem cell related genes with this abnormal DNA methylation and/or transcriptionally repressive chromatin, could reduce the plasticity of cell population changes and help lock in cell clones with abnormal retention of self-renewal capacity at the expense of capacity for differentiation (Easwaran et al., 2012). These types of chromatin alterations could also contribute to emergence of sub-clones in tumors for phenotypic heterogeneity with respect to plasticity for evolution and/or reversibility of tumor initiating and self-renewal properties (Fig. 2). Subclones with the least plasticity for reversion of stem-like properties could come to dominate the cell populations of a cancer at any given point in time of tumor progression.
In the above hypotheses, altered chromatin patterns during cancer development would be a factor for evolution of cellular heterogeneity by helping to lock in tumor phenotypes, which may reflect the cell compartments from which the tumors arise (Fig. 3). The type of chromatin abnormalities would, in turn, also reflect such timing and be dependent upon the degree of molecular progression from the more plastic expression states of bivalency or PcG control to the tightest form of abnormal gene silencing associated with promoter CpG island DNA hypermethylation. Perhaps these proposed dynamicsmay have roots in relationships between given cancers and the normal stem cell compartments in which they arise. Such relationships could be determinants of which genes, relative to their initial chromatin patterns during cell stresses in cancer risk states, undergo epigenetic abnormalities such as abnormal promoter DNA methylation (Easwaran et al., 2012). Breast cancers may be a key example of these possibilities. Multiple groups (Fang et al., 2011), including ours (Easwaran et al., 2012), have reported that the more differentiated luminal phenotype breast cancers with epithelial features, have far more genes with promoter CpG island DNA hypermethylation than do the basal or triple negative tumors, which are more stem-like with mesenchymal phenotype. These latter, proposed to derive from a more primitive, and/or less committed normal breast epithelial compartment (Lo and Sukumar, 2008), might arise faster with cell phenotypes more dependent on retention of PcG driven chromatin control than its conversion to DNA methylation. In contrast, development of luminal phenotype tumors may evolve over a longer time course with more chromatin evolution from bivalency/PcG control to promoter DNA methylation. The latter change may reflect, and/or help lock in, subpopulations of self-renewing cells reflective of the origin of luminal tumors in more committed stem/progenitor cell compartments in normal breast epithelium. Plasticity of the resultant populations for self renewing properties may still be present, but the EMT phenotypes involved might be of the more mixed EMT-epithelial types proposed recently in the range of breast cancer cell phenotypes (Tam and Weinberg, 2013).
The high frequency of DNA methylation patterns in luminal breast cancer, termed as a CpG island hypermethylation phenotype or CIMP, and differences from basal and triple negative tumors, has great relevance for other cancer types as well. Classic CIMP has been defined in colorectal cancer, and again is a pattern separating one group of tumors arising in the right side of the colon from the more common ones arising on the left side (Toyota et al., 1999) (Weisenberger et al., 2006). We have mentioned earlier that tumors with IDH mutations, first recognized in brain tumors, are accompanied by a CIMP pattern again involving genes with a history of bivalency (Turcan et al., 2012) and having a phenotype of being locked in a self-renewal state resistant to differentiation induction (Lu et al., 2013; Lu et al., 2012). The cells of origin in this scenario may be particularly relevant. These mutations are confined almost totally to a form of low-grade gliomas occurring in a far younger group of patients than the more typical gliomas (Parsons et al., 2008; Yan et al., 2009). Also, they arise in a proneural, or more committed neural stem cell than the glial cell origins of typical gliomas and are much slower growing with a far better prognosis (Noushmehr et al., 2010; Parsons et al., 2008).
The implications of epigenetic abnormalities for cancer treatment and a role in treatment resistance
In addition to their importance for understanding basic facets of tumor biology, epigenetic alterations are increasingly being recognized as being integral for new approaches to cancer therapies - and for their possible role in treatment resistance, the major barrier to successful therapies for advanced malignancies (Fig. 4). Potential for preventing, delaying, or reversing such resistance with epigenetic therapies, such as use of small molecules that can reverse DNA methylation and histone deacetylation, have been the subjects of recent reviews as have implications for targeting other chromatin regulatory proteins (Azad et al., 2013) (Baylin and Jones, 2011). A background context for this is the excitement surrounding recent data indicating a role for chromatin changes in linking simultaneous emergence of stem cell populations and their role in the evolution of treatment resistance to multiple therapy types (Sharma et al., 2010). Certainly, and especially for targeted therapies, such resistance can be due to new mutations arising directly in the targeted molecule, or in genes in the direct downstream pathways, or complementing pathways (Redmond et al., 2008)(Fig. 4). However, it may be that epigenetic alterations could play just as important a role in such resistance. As we have pointed out, every cancer has multiple genes with epigenetic alterations in every key pathway fundamental to tumor development. In fact, experimentally, a reversible type of resistance has been induced in cancer cells (Fig. 4). This resistance, reversible with drug withdrawal and with treatment by low doses of histone deacetylase inhibitors, correlates with selection and/or induction of stem-like cancer cells from heterogeneous tumor cell populations. In turn, a key association in these cell subpopulations is very high levels in these cells of the H3K4me3 demethylase, JARID1A and decreased overall H3K4me3 (Roesch et al., 2010; Sharma et al., 2010). A close relative of this protein, JARID1B, has been associated with maintenance of tumor initiating cells (Roesch et al., 2010). All of these data, again, juxtapose cancer cell subpopulation plasticity with respect to stem-like phenotypes, and chromatin alterations. Notably, the reversibility of epigenetic abnormalities in cancer subpopulations with inherent drug tolerance also provides a potentially exciting therapeutic strategy. Epigenetic therapy could be utilized as a priming therapy, which may sensitize cancer cells, which are otherwise drug-resistant, to conventional/targeted chemotherapy (Fig. 4).
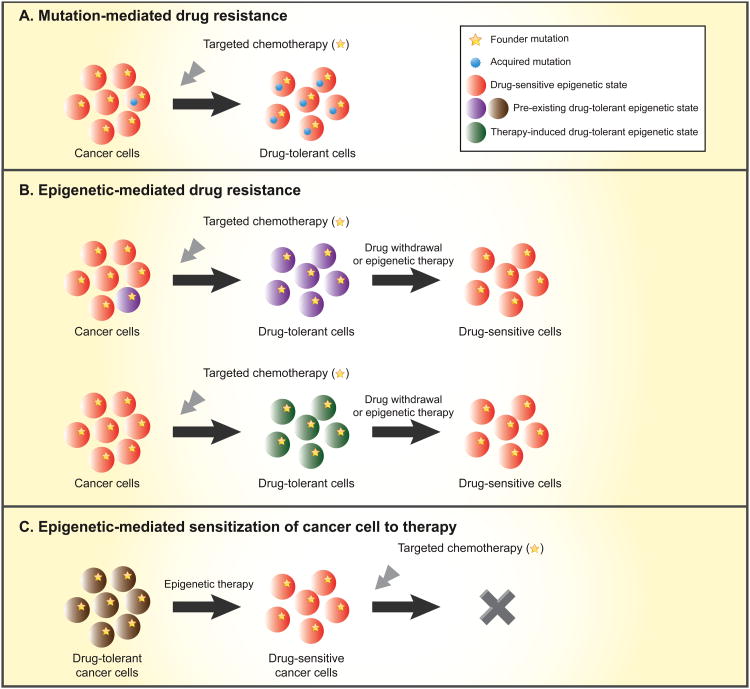
Figure 4. Genetic and epigenetic mechanisms for drug resistance and implications of epigenetic therapy.
Top panel - cancers may consist of different subclones that carry a founder mutation(s) alone, or additional acquired mutations, that confer drug resistant states. When treated with targeted therapies, a pre-existing drug tolerant clone may remain unaffected and through outgrowth can come to dominate the entire cancer population. Middle panel - cancer subclones with epigenetic-mediated drug tolerance may exist in the original caner population or develop as a result of targeted chemotherapy. The drug tolerance may be reversed and cells may return to the sensitive epigenetic state after a prolonged drug withdrawal or after the tumor is treated with epigenetic therapy, such as low dose histone deacetylase (HDAC) inhibitors and/or DNA demethylating agents. Lower panel - epigenetic therapy may convert cancer cells in a drug-resistance state to a drug-sensitive state, thereby sensitizing the cells to targeted chemotherapy that was not effective for the original cancer population.
Conclusion and future directions
We have reviewed data to suggest that, beginning in cancer risk states and moving forward through tumor progression, epigenetic abnormalities may play key roles in determining patterns of cellular heterogeneity in cancers. These may vary at any steady state point in time during tumor evolution. We have defined the concept of molecular progression for epigenetic changes occurring even before frank tumor appearance, which may be induced by agents predisposing to cancer. These events could modulate evolution of non-hierarchical sub-populations of cancer stem-like and tumor initiating cells. We have also raised the hypothesis that a key underlying mechanism for how such chromatin patterns arise may be inherent to responses of cells to chronic stress which may select for, or induce, stem-like cell populations which otherwise would not survive and have the capacity to maintain cell renewal. In turn, we suggest that the initial chromatin make up of such cells may set the stage for patterns of epigenetic abnormalities that might help maintain self renewal properties at the expense of normal responses to lineage commitment and differentiation cues. Thus, the cell compartments from which a tumor arises, and the duration of time to tumor appearance, may dictate the extent of molecular progression of epigenetic abnormalities and the degree of cell plasticity. Various technical challenges have to be overcome in studies aimed at further understanding the role of epigenetic alterations in tumor heterogeneity. Firstly, measuring the variations in epigenetic alterations within a tumor is very challenging. Although with the advent of deep sequencing this could be tackled in the case of DNA methylation, this remains a challenge for histone modifications by the sheer nature of these modifications. Secondly, important to this endeavor will be efforts to sort out the direct and indirect gene and molecular pathway targets of the genes subject to tumor-specific epigenetic alterations, and to define their driver versus passenger roles in tumorigenesis. Finally, we have stressed that any tumor cell constitution represents interplay between the above dynamics and the genetic abnormalities arising at each stage of tumor progression from initiation onwards. Particularly intriguing are the frequent mutations that are being increasingly recognized for genes encoding major determinants of epigenomes and how these dictate the phenotypic make up of cancers. For the future, investigation strategies will be key for not only determining the role of these mutations in determining patterns of epigenetic abnormalities in cancer, but for exploring facets of all the dynamics we have been discussing in this review.
In addition to advancing our understanding of cancer biology, the data reviewed and the concepts derived are extremely important for their translational potential. There is a rapidly evolving enterprise for targeting epigenetic abnormalities as a therapeutic strategy for cancer. Attempts are ongoing to use older drugs in newer more targeted ways, and efforts are building to develop new drugs that target a myriad of proteins which regulate the epigenome (Cole, 2008; Dawson and Kouzarides, 2012; Vedadi et al., 2011). All of these endeavors will benefit immensely from considering the implications of the cellular and molecular biology we have reviewed and some of the hypotheses put forth. Particularly important may be the patterns of epigenetic abnormalities we have discussed for different tumor types and for sub-types within tumor categories. Targeting of therapies to fit the precise epigenetic abnormalities contributing to the phenotype of a given cancer may, in the end, determine the success or failure of such strategies. Epigenetic therapies may eventually work best not only in combination with other treatment modalities, but especially may benefit from following biochemically driven hypotheses for combining drugs targeting different regulators of the cancer epigenome. Already in this regard, use of older drugs targeting the epigenetic machinery, like DNA demethylating agents and histone deactylase inhibitors, are showing very promising signs of efficacy by potentially sensitizing to other therapies (Juergens et al., 2011). The future of the epigenetic therapy lies in following up on the potential leads for using epigenetic drugs to reverse and/or delay resistance to current cancer therapies. As we learn more about relationships between epigenetics and their contribution to cellular composition of cancers, the future appears very bright for leveraging its translational potential.
Acknowledgments
Portions of the authors' writing is supported by National Cancer institute (NCI) CA043318, National Institutes of Environmental Health Sciences (NIEHS) ES011858, National Institutes of Health (NIH) HL099775 and the Hodson Endowment Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- 2.Allis CD, Jenuwein T, Reinberg D. Epigenetics. 1st. Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- 3.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O'Donnell P, Grigoriadis A, Diss T, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 4.Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 5.Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC, Esopi D, Irizarry RA, Getzenberg RH, Nelson WG, et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med. 2013;5:169ra110. doi: 10.1126/scitranslmed.3005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azad N, Zahnow CA, Rudin CM, Baylin SB. The future of epigenetic therapy in solid tumours--lessons from the past. Nat Rev Clin Oncol. 2013;10:256–266. doi: 10.1038/nrclinonc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 8.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13:727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- 10.Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2011;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 13.Bestor TH. DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos Trans R Soc Lond B Biol Sci. 1990;326:179–187. doi: 10.1098/rstb.1990.0002. [DOI] [PubMed] [Google Scholar]
- 14.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 15.Borodovsky A, Salmasi V, Turcan S, Fabius AW, Baia GS, Eberhart CG, Weingart JD, Gallia GL, Baylin SB, Chan TA, et al. 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget. 2013;4:1737–1747. doi: 10.18632/oncotarget.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 17.Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell. 2012;10:157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 19.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3:730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 20.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole PA. Chemical probes for histone-modifying enzymes. Nat Chem Biol. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 24.De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, Yang X, Liang G, Jones PA. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Gobbi M, Garrick D, Lynch M, Vernimmen D, Hughes JR, Goardon N, Luc S, Lower KM, Sloane-Stanley JA, Pina C, et al. Generation of bivalent chromatin domains during cell fate decisions. Epigenetics Chromatin. 2011;4:9. doi: 10.1186/1756-8935-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nature structural & molecular biology. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 27.Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, Aryee MJ, Joyce P, Ahuja N, Weisenberger D, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 2012;22:837–849. doi: 10.1101/gr.131169.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Encode Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Gonzalez S, Tarafa G, Sidransky D, Meltzer SJ, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. [PubMed] [Google Scholar]
- 30.Fan HY, Trotter KW, Archer TK, Kingston RE. Swapping function of two chromatin remodeling complexes. Molecular cell. 2005;17:805–815. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 31.Fang F, Turcan S, Rimner A, Kaufman A, Giri D, Morris LG, Shen R, Seshan V, Mo Q, Heguy A, et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med. 2011;3:75ra25. doi: 10.1126/scitranslmed.3001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenspan EJ, Jablonski MA, Rajan TV, Levine J, Belinsky GS, Rosenberg DW. Epigenetic alterations in RASSF1A in human aberrant crypt foci. Carcinogenesis. 2006;27:1316–1322. doi: 10.1093/carcin/bgi373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 38.Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–10289. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, Traulsen A, Nowak MA, Siegel C, Velculescu VE, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korber P, Becker PB. Nucleosome dynamics and epigenetic stability. Essays Biochem. 2010;48:63–74. doi: 10.1042/bse0480063. [DOI] [PubMed] [Google Scholar]
- 44.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q, Ke Q, Costa M. Alterations of histone modifications by cobalt compounds. Carcinogenesis. 2009;30:1243–1251. doi: 10.1093/carcin/bgp088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Licchesi JD, Westra WH, Hooker CM, Herman JG. Promoter hypermethylation of hallmark cancer genes in atypical adenomatous hyperplasia of the lung. Clin Cancer Res. 2008;14:2570–2578. doi: 10.1158/1078-0432.CCR-07-2033. [DOI] [PubMed] [Google Scholar]
- 47.Liu F, Killian JK, Yang M, Walker RL, Hong JA, Zhang M, Davis S, Zhang Y, Hussain M, Xi S, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene. 2010;29:3650–3664. doi: 10.1038/onc.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo PK, Sukumar S. Epigenomics and breast cancer. Pharmacogenomics. 2008;9:1879–1902. doi: 10.2217/14622416.9.12.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu C, Venneti S, Akalin A, Fang F, Ward PS, Dematteo RG, Intlekofer AM, Chen C, Ye J, Hameed M, et al. Induction of sarcomas by mutant IDH2. Genes Dev. 2013;27:1986–1998. doi: 10.1101/gad.226753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maegawa S, Gough SM, Watanabe-Okochi N, Lu Y, Zhang N, Castoro RJ, Estecio MR, Jelinek J, Liang S, Kitamura T, et al. Age-related epigenetic drift in the pathogenesis of MDS and AML. Genome Res. 2014 doi: 10.1101/gr.157529.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marjanovic ND, Weinberg RA, Chaffer CL. Cell plasticity and heterogeneity in cancer. Clin Chem. 2013;59:168–179. doi: 10.1373/clinchem.2012.184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehrotra J, Varde S, Wang H, Chiu H, Vargo J, Gray K, Nagle RB, Neri JR, Mazumder A. Quantitative, spatial resolution of the epigenetic field effect in prostate cancer. Prostate. 2008;68:152–160. doi: 10.1002/pros.20675. [DOI] [PubMed] [Google Scholar]
- 56.Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, Downing JR. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 60.Niwa T, Ushijima T. Induction of epigenetic alterations by chronic inflammation and its significance on carcinogenesis. Adv Genet. 2010;71:41–56. doi: 10.1016/B978-0-12-380864-6.00002-X. [DOI] [PubMed] [Google Scholar]
- 61.Nonn L, Ananthanarayanan V, Gann PH. Evidence for field cancerization of the prostate. Prostate. 2009;69:1470–1479. doi: 10.1002/pros.20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 64.O'Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 68.Redmond KM, Wilson TR, Johnston PG, Longley DB. Resistance mechanisms to cancer chemotherapy. Front Biosci. 2008;13:5138–5154. doi: 10.2741/3070. [DOI] [PubMed] [Google Scholar]
- 69.Rheinbay E, Suva ML, Gillespie SM, Wakimoto H, Patel AP, Shahid M, Oksuz O, Rabkin SD, Martuza RL, Rivera MN, et al. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep. 2013;3:1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 73.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan PS, Krouse RS, Prasad AR, Einspahr JG, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 76.Shenker N, Flanagan JM. Intragenic DNA methylation: implications of this epigenetic mechanism for cancer research. Br J Cancer. 2012;106:248–253. doi: 10.1038/bjc.2011.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sierra JR, Cepero V, Giordano S. Molecular mechanisms of acquired resistance to tyrosine kinase targeted therapy. Mol Cancer. 2010;9:75. doi: 10.1186/1476-4598-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 80.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 82.Thurman RE, Day N, Noble WS, Stamatoyannopoulos JA. Identification of higher-order functional domains in the human ENCODE regions. Genome Res. 2007;17:917–927. doi: 10.1101/gr.6081407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tili E, Michaille JJ, Wernicke D, Alder H, Costinean S, Volinia S, Croce CM. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci U S A. 2011;108:4908–4913. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Turcan S, Fabius AW, Borodovsky A, Pedraza A, Brennan C, Huse J, Viale A, Riggins GJ, Chan TA. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT Inhibitor Decitabine. Oncotarget. 2013;4:1729–1736. doi: 10.18632/oncotarget.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ushijima T. Epigenetic field for cancerization. J Biochem Mol Biol. 2007;40:142–150. doi: 10.5483/bmbrep.2007.40.2.142. [DOI] [PubMed] [Google Scholar]
- 89.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, Dimaggio PA, Wasney GA, Siarheyeva A, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Voigt P, LeRoy G, Drury WJ, 3rd, Zee BM, Son J, Beck DB, Young NL, Garcia BA, Reinberg D. Asymmetrically modified nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Waddington CH. The epigenotype. Int J Epidemiol. 2012;41:10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 93.Wang JCY, Dick JE. Cancer: Principles & Practice of Oncology. Cancer Stem Cells. (Eighth) 2008 [Google Scholar]
- 94.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 95.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 96.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamazaki J, Estecio MR, Lu Y, Long H, Malouf GG, Graber D, Huo Y, Ramagli L, Liang S, Kornblau SM, et al. The epigenome of AML stem and progenitor cells. Epigenetics. 2013;8:92–104. doi: 10.4161/epi.23243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang J, Luo H, Li Y, Li J, Cai Z, Su X, Dai D, Du W, Chen T, Chen M. Intratumoral heterogeneity determines discordant results of diagnostic tests for human epidermal growth factor receptor (HER) 2 in gastric cancer specimens. Cell Biochem Biophys. 2012;62:221–228. doi: 10.1007/s12013-011-9286-1. [DOI] [PubMed] [Google Scholar]
- 101.Yatabe Y, Tavare S, Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc Natl Acad Sci U S A. 2001;98:10839–10844. doi: 10.1073/pnas.191225998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yi JM, Tsai HC, Glockner SC, Lin S, Ohm JE, Easwaran H, James CD, Costello JF, Riggins G, Eberhart CG, et al. Abnormal DNA methylation of CD133 in colorectal and glioblastoma tumors. Cancer research. 2008;68:8094–8103. doi: 10.1158/0008-5472.CAN-07-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]