Abstract
Genome-scale models are used for an ever-widening range of applications. Although there has been much focus on specifying the stoichiometric matrix, the predictive power of genome-scale models equally depends on reaction directions. Two-thirds of reactions in the two eukaryotic reconstructions Homo sapiens Recon 1 and Yeast 5 are specified as irreversible. However, these specifications are mainly based on biochemical textbooks or on their similarity to other organisms and are rarely underpinned by detailed thermodynamic analysis. In this study, a to our knowledge new workflow combining network-embedded thermodynamic and flux variability analysis was used to evaluate existing irreversibility constraints in Recon 1 and Yeast 5 and to identify new ones. A total of 27 and 16 new irreversible reactions were identified in Recon 1 and Yeast 5, respectively, whereas only four reactions were found with directions incorrectly specified against thermodynamics (three in Yeast 5 and one in Recon 1). The workflow further identified for both models several isolated internal loops that require further curation. The framework also highlighted the need for substrate channeling (in human) and ATP hydrolysis (in yeast) for the essential reaction catalyzed by phosphoribosylaminoimidazole carboxylase in purine metabolism. Finally, the framework highlighted differences in proline metabolism between yeast (cytosolic anabolism and mitochondrial catabolism) and humans (exclusively mitochondrial metabolism). We conclude that network-embedded thermodynamics facilitates the specification and validation of irreversibility constraints in compartmentalized metabolic models, at the same time providing further insight into network properties.
Introduction
Genome-scale metabolic models (GeMs) have been reconstructed for many model organisms, including Escherichia coli, Saccharomyces cerevisiae, Mus musculus, Homo sapiens, and Arabidopsis thaliana (1–5). These models enable a systematic approach to investigating genotype-phenotype relationships at the metabolic level (6), and they are applied in a variety of research fields, such as metabolic engineering (7).
Each reconstruction in principle captures the complete metabolic capabilities of a given organism. These metabolic capabilities are explored by flux balance analysis (FBA) using the topological constraints (e.g., reaction connectivity and reversibility) contained within the reconstruction (8). The flux balances greatly reduce the dimension of the solution space, but GeMs still have hundreds of degrees of freedom, and directional constraints are critical to the predictive powers of the models. The quality of predictions improves as more constraints are specified (9). Conversely, erroneous irreversibility constraints can result in incorrect flux predictions (10).
Reaction directionality can be specified from the Gibbs energy of reaction (ΔrG) using the second law of thermodynamics. The range of ΔrG can be calculated from estimates of standard Gibbs energy and physiological ranges of metabolite concentrations. The range of ΔrG has been used to assign directionality to individual reactions in E. coli iJR904 (11), E. coli iAF1260 (12), and H. sapiens Recon 1 (13).
Through the network topology, individual irreversible reactions may dictate the direction of otherwise reversible reactions and the range of feasible metabolite concentrations. Accordingly, thermodynamic constraints can propagate widely in the network. Network thermodynamics was first exploited in the 1990s to analyze glycolysis (14). More recently, network-embedded thermodynamic analysis (NET analysis) (15) has enabled analysis of large-scale networks using nonlinear optimization.
NET analysis was developed to assess whether experimentally derived metabolite concentrations are feasible with respect to the reaction directionalities. In this study, we use NET analysis in reverse to assign reaction directionalities based on physiological ranges of metabolite concentrations and thermodynamic constraints in two eukaryotic GeMs, namely, H. sapiens Recon 1 (3) and Yeast 5 (16). Erroneous irreversibility constraints were found in both models, whereas several new constraints revealed unconnected loops in the models. The analysis also enabled resolution of the different compartmentalization of L-proline metabolism in humans and yeast. A new open-source software, NExT, was developed to implement correct estimations of Gibbs energy for transport reactions.
Materials and Methods
Calculation of Gibbs energy (ΔG)
The estimation of Gibbs energy of formation (ΔfG) was adjusted for physiological conditions of pH and ionic strength. For reactions coupled to transport (e.g., ATP synthase), the differences in pH and redox potential were taken into account (17). The full description of the ΔG calculations can be found in the Supporting Material.
NET analysis
For biological systems where constant temperature and pressure applies, the second law of thermodynamics states that spontaneity is defined by negative Gibbs free energy (ΔG < 0). Therefore, for biochemical reactions, the reaction occurs only in the direction where ΔrG is negative. ΔrG depends on metabolite concentrations and the standard Gibbs energy of reaction (ΔrG0), the latter of which is determined from the standard Gibbs energy of formation of the components (ΔfG0) and the stoichiometric coefficients.
NET analysis (15) applies thermodynamic principles to a complete metabolic network simultaneously to check the thermodynamic feasibility of a metabolite data set, estimate feasible ranges for unmeasured metabolites, and assign reaction directions. The algorithm integrates the second law of thermodynamics and ΔfG′0 with a stoichiometric model (defined by a stoichiometric matrix S and reaction directionalities):
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
By minimizing and maximizing ΔrG' of each reaction k (Eq. 1), the range of ΔrG'k is estimated for all reactions. Equations 2 and 3 impose thermodynamic constraints for irreversible reactions, and Eqs. 4 and 5 describe the calculation of ΔrG'. The measured metabolite concentrations and the physiological range of concentrations are expressed in Eq. 6; the range of cofactor ratios can also be incorporated as constraints. In an analogous process, the feasible range of metabolite concentrations can be calculated (15).
If a reaction specified to be reversible in the model has its maximum ΔrG calculated to be negative, the reaction operates strictly in the forward direction. Conversely, if the minimum ΔrG is positive, the reaction operates strictly in the reverse direction. No direction can be inferred when the ΔrG minimum is negative and the maximum is positive. Our Matlab implementation of NET analysis, NExT, is available from http://web.aibn.uq.edu.au/cssb/Resources.html.
Workflow to thermodynamically curate a metabolic model
A novel workflow for the thermodynamic curation of GeMs was developed (Fig. 1). The following information and data are required as inputs:
-
•
a metabolic model (which represents the metabolic reactions with their directions and the specific compartment where each reaction takes place);
-
•
biophysical properties of cellular compartments (pH, ionic strength, etc.);
-
•
thermodynamic properties (standard free energy of formation, charge, and number of hydrogen atoms of the reactants); and
-
•
physiological metabolite concentrations.
Figure 1.
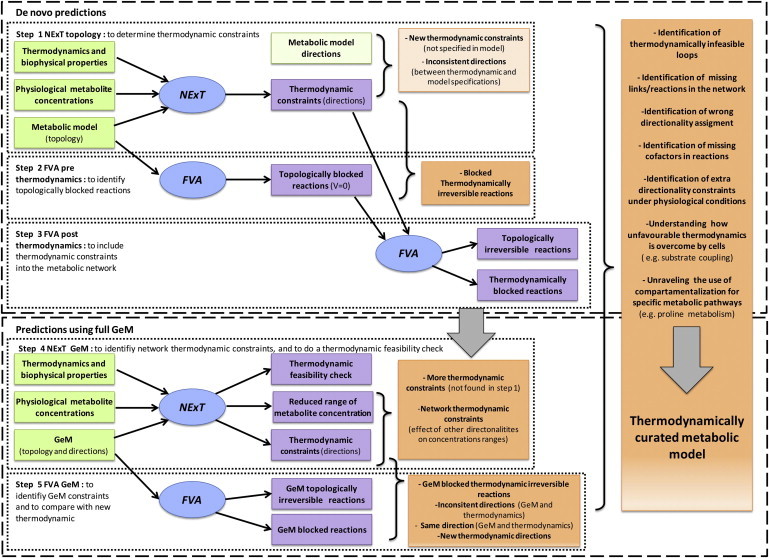
Methodology workflow. A five-steps process is used to generate a thermodynamically curated metabolic model. The first step (NExT topology) identifies thermodynamic constraints, which are compared with the model specifications. Step 2 (FVA prethermodynamics) identifies blocked reactions caused solely by network topology; together with the results from step 1, blocked thermodynamically irreversible reactions are thus identified. Step 3 (FVA post thermodynamics) identifies the reduction of the solution space due to thermodynamic constraints. Step 4 (NExT GeM) identifies new thermodynamic constraints that are a consequence of network-specified directionality constraints; the metabolite concentration ranges are also thermodynamically constrained. Step 5 (FVA GeM) identifies the solution space of the model using network-specified directionality constraints only, which are compared with results of step 4 to identify the new thermodynamic constraints that will reduce the flux solution space. Inputs to the process are in green boxes, outputs in purple boxes, and information inferred in orange boxes; simulations are in blue ovals.
The workflow is divided into two stages: de novo predictions without using directional specifications from the GeM and predictions using the full GeM. In the first stage, NExT identifies purely thermodynamic constraints (Fig. 1, Step 1). These constraints are compared to the directionality constraints specified in the GeM to identify new constraints and correct any inconsistent constraints. In the second step, flux variability analysis (FVA) is used to identify topologically blocked reactions (Fig. 1, Step 2). Finally, a second FVA, including the thermodynamic constraints from Step 1, is used to identify additional irreversible reactions, which we call topologically irreversible reactions (Fig. 1, Step 3). For example, reactions belonging to a linear pathway become topologically irreversible when one reaction of the pathway is thermodynamically irreversible.
The new, reconciled thermodynamic constraints are carried forward to the second stage, where the directionality constraints from the GeM are integrated in NExT (Fig. 1, Step 4). Using the full set of constraints, metabolite concentration ranges are narrowed, and consequently further thermodynamic constraints are identified. A final FVA using the GeM directionality constraints is used to identify reactions that are either blocked or irreversible due to the GeM topology (Fig. 1, Step 5). A final comparison between thermodynamic constraints and FVA predictions is used to identify needs for further curation.
Metabolic models and assumptions
The human GeM H. sapiens Recon 1 (3) and Yeast 5 GeM (16) were modified as follows to make them suitable for thermodynamic calculations. 1), Reactions that are catalyzed by different enzymes in either direction were lumped together, otherwise a thermodynamic equilibrium state with a zero Gibbs energy of reaction would be considered for both of them. 2), The species HCO3−, CO32−, CO2, and H2CO3 were aggregated as the reactant CO2tot, since carbon dioxide is distributed among these species in aqueous phase (H2O was added to the other side of the reaction to balance oxygen atoms) (18). 3), The oxidized and reduced FAD of mitochondria were replaced by the oxidized and reduced FADenz to represent the enzyme-bound FAD cofactor (18).
Biophysical compartment properties
Literature values for compartment-specific biophysical properties (pH, redox potential, ionic strength, and size) were used in the calculations of ΔrG to represent real physiological conditions (Table 1 and Table S1 in the Supporting Material). Mitochondrial and cytosolic accurate biophysical properties were found in the literature (15,19–23); and for the remaining compartments, the same properties were assumed as for the cytosol (with the exception of size), keeping the model stoichiometrically intact.
Table 1.
Biophysical properties and concentration ranges
| Human | Yeast | |
|---|---|---|
| pH, cytosol | 7.4a | 7b |
| Redox potential, cytosol | −315 mVc | −286 mVd |
| pH, mitochondria | 7.98a | 7.4b |
| Redox potential, mitochondria | −360 mVe | −296 mVd |
| pH, IMS | 6.7f | 6.3f |
| Redox potential, IMS | −242g | −213d |
| Ionic strength | 0.15 Mb | |
| Oxygen concentration (mM) | [10−4;0.1] | |
| [CO2], [Pi], [Ppi] (mM) | [1;100]b | |
| Other reactants (mM) | [10−4,10]b,h | |
| Adenylate energy charge (AEC) | [10−4,10]b | |
| NADH/NAD ratio | [10−4,10] | |
| NADPH/NADP ratio | [10−4,10] | |
Values from Llopis et al. (21).
Values from Kummel et al. (15).
Values from Björnberg et al. (19).
Values from Hu et al. (20).
Value from Hanson et al. (23).
Values from Porcelli et al. (22), considering 0.7 units lower than cytosolic pH.
The variation between redox potential in cytosol and IMS of yeast was used as a reference.
Value from Haraldsdóttir et al. (13).
Physiological range of metabolite concentrations
The concentrations for all metabolites were assumed to be between 0.0001 mM and 10 mM to represent the range of observed physiological concentrations (13,15). Exceptions were made for oxygen, carbon dioxide, and inorganic phosphate (Table 1). In addition, an arbitrarily wide range of concentration ratios were specified for the adenylate energy charge (AEC) and the redox cofactors (i.e., NAD+/NADH and NADP+/NADPH) (Table 1).
Physical and chemical metabolite properties
To calculate the Gibbs energy of reactions, the following information was needed for all reactants in the network: standard Gibbs energy of formation (ΔfG0), number of hydrogen atoms, and charge. The thermodynamic properties for 186 metabolites were obtained from published data (24). For metabolites for which we found no published ΔfG0 values, a separate algorithm based on the group contribution method was used to estimate ΔfG0 (25,26). In addition, the number of hydrogen atoms and the charge of metabolites at physiological conditions were collected from the literature (15,26–29). The final database contains information for 860 metabolites (Table S2).
Results and Discussion
Software implementation of NET analysis
NET analysis has previously been implemented in the software anNET (30). A new software, NExT, was developed to implement corrections regarding how transport reactions are handled. First, when calculating the ΔrG of reactions coupled to transport (e.g., ATP synthase), anNET only considers the transport part (e.g., proton transport) and not the reaction part (e.g., ATP synthesis). H. sapiens Recon 1 has almost 50 transport coupled reactions, so it was important to correct the erroneous calculations. Second, we made the necessary corrections for the transport of weak acids and bases, i.e., reactants composed of multiple proton-bound species in rapid equilibrium (e.g., lactate and lactic acid), allowing for the different equilibria in compartments that differ in pH (17). For a given external concentration, the corrections significantly affect the feasible internal concentrations of many transported weak acids and bases.
ATP synthase was affected by both modifications as an intercompartment enzyme that produces ATP using the energy generated from proton motive force. Although protons are exchanged between the mitochondrial matrix and the mitochondrial intermembrane space (IMS), this exchange is commonly treated as protons being transported to and from the cytoplasm. Given the moderate differences in pH and redox potential between the cytoplasm and mitochondria observed for both yeast and humans (Table 1), the reaction ΔrG'0 becomes positive (10.57 and 25.15 KJ/mol for the human and yeast models, respectively (Table S3)). This value translates into a nonphysiological, low maximum mitochondrial ATP concentration (0.014 and 0.00004 mM for human and yeast cells, respectively). This was not apparent in anNET, since it ignores ATP production when estimating the ΔrG'0 of ATP synthase (see Eq. S11) (30), which is the thermodynamically unfavorable part of the reaction (ΔrG'0compartment values for producing ATP are 41.20 and 38.14 KJ/mol for the human and yeast models, respectively (Table S3)).
To allow the ATP synthase reaction to function as expected, it must be written with the consideration of proton flux from the IMS rather than the cytoplasm. The IMS itself has a much lower pH and a more oxidizing environment (Table 1) compared to the cytoplasm (20,22). With this modification, the concentration of mitochondrial ATP is not constrained by ATP synthase, thus rendering the model thermodynamically feasible (see Table S3 for the new ΔrG'0 when protons are transported from the IMS). The change from compartmentalization of protons in the cytoplasm to compartmentalization in the IMS was performed on all reactions involved in oxidative phosphorylation to maintain consistency of the model. Transporters involved in solute transport between the cytoplasm and mitochondria (e.g., mitochondrial pyruvate transporter) are written as such without an intermediate IMS stage.
The NExT software is provided in an open-source format to enable facile modification and integration with other workflows by the community. The software and corrected GeMs are available from http://web.aibn.uq.edu.au/cssb/Resources.html.
Prediction of irreversible reactions in H. sapiens Recon 1
This work uses the H. sapiens Recon 1 GeM (3) rather than the recently published Recon 2 (31), since initial releases of the latter contained a number of errors. All of the observations made below, however, have been cross-checked against the current database, Recon X (http://humanmetabolism.org/), and, unless otherwise stated, remain true as of September 1, 2013. The IMS compartment was introduced in Recon 1 for oxidative phosphorylation. Additional minor modifications to equations were made to enable accurate calculations (see Materials and Methods). Excluding artificial boundary exchange reactions, the modified model contains 1160 reversible reactions and 2149 reactions defined as irreversible. Biophysical properties were specified based on literature values, whereas broad concentration ranges and ratio ranges for adenylate energy charge and redox factors were used (Table 1).
De novo predictions
The novel framework used for curation of GeMs, introduced in Materials and Methods, is summarized in Fig. 1. We first used NExT to identify irreversible reactions without using the 2149 irreversibilities already specified in Recon 1 (Fig. 1, Step 1). Using broad concentration ranges (Table 1), a total of 319 reactions were found to be irreversible under physiological conditions. Of these irreversible reactions, 306 were among the 2149 specified in Recon 1, whereas the remaining 13 are new irreversibility constraints not previously specified in the GeM (Table 2).
Table 2.
Reactions identified as irreversible by thermodynamic constraints in Recon1 that were not specified as such in the model
| ID | Reaction | ΔrG min (KJ/mol) | ΔrG max (KJ/mol) | Direction | Pathway |
|---|---|---|---|---|---|
| P450SCC1ma | [m]chsterol + h + nadph + o2 = 20ahchsterol + h2o+nadp | −427.91 | −302.34 | Forward | Cholesterol metabolism |
| AIRCrb | [c]air + co2tot = 5aizc + h + h2o | 3.17 | 71.67 | Reverse | IMP biosynthesis |
| G5SADrm | [m]glu5sa = 1pyr5c + h + h2o | −94.29 | −37.21 | Forward | Arginine and proline metabolism |
| PHCHGSm | [m]1p3h5c + h + h2o = 4hglusa | 44.75 | 101.83 | Reverse | Arginine and proline metabolism |
| GTHP | [c] (2)gthrd + h2o2 = gthox + (2)h2o | −316.29 | −202.13 | Forward | Glutathione metabolism |
| GTHPe | [e] (2)gthrd + h2o2 = gthox + (2)h2o | −316.29 | −202.13 | Forward | Glutathione metabolism |
| GTHPm | [m] (2)gthrd + h2o2 = gthox + (2)h2o | −316.29 | −202.13 | Forward | Glutathione metabolism |
| DPGM | [c]13dpg = 23dpg + h | −70.55 | −13.47 | Forward | Glycolysis /gluconeogenesis |
| DCMPDA | [c]dcmp + h + h2o = dump + nh4 | −87.76 | −2.14 | Forward | Pyrimidine catabolism |
| DCT | [c]L_dpchrm = 56dihindlcrbxlt | −119.48 | −62.40 | Forward | Tyrosine metabolism |
| DOPAQNISO1 | [c]dopaqn = 2c23dh56dhoxin + h | −86.28 | −29.20 | Forward | Tyrosine metabolism |
| PDX5PO | [c]o2 + pdx5p = h2o2 + pydx5p | −162.89 | −65.86 | Forward | Vitamin B6 metabolism |
| PYAM5POr | [c]h2o + o2 + pyam5p = h2o2 + nh4 + pydx5p | −193.98 | −68.41 | Forward | Vitamin B6 metabolism |
See Supporting Material for analysis of errors in estimated values.
Linked to singleton metabolite and cannot carry flux.
AIRCr is treated in a separate section.
Even in the absence of irreversibility constraints, 525 of 3711 reactions in the human model are linked to singleton (dead-end) metabolites, i.e., metabolites unable to carry any flux (Fig. 1, Step 2). Of the 319 irreversible reactions predicted, 77, including one of the 13 new irreversible reactions (P450SCC1m), cannot carry flux. Thermodynamically constraining these 77 reactions will not further reduce the solution space.
Introducing the remaining 242 irreversibility constraints caused an additional 27 reactions to lose the ability to carry flux, whereof 17 were also unable to carry flux in the constrained Recon 1 (see Table S4) (Fig. 1, Step 3). The additional 10 reactions unable to carry flux belonged to the purine metabolism pathway producing inosine monophosphate (IMP) from 5-phosphoribosyl-1-pyrophosphate (PRPP) and glutamine. In the model, flux through this essential pathway is prevented by the reaction catalyzed by phosphoribosylaminoimidazole carboxylase (AIRCr) being thermodynamically constrained to operate in the reverse direction, which contradicts both the essential requirement for flux through the pathway and the thermodynamic direction of the reaction glutamine phosphoribosyldiphosphate amidotransferase. AIRCr will be discussed in a separate section below, where it will become clear that it is not thermodynamically constrained. Hence, the prediction will be excluded from further analysis.
Apart from preventing flux through 27 reactions, the introduction of 242 thermodynamic constraints within the network caused an additional 307 reactions to be topologically irreversible (Fig. 1, Step 3). These reactions include two that are a direct consequence of the new thermodynamic constraints added into the model; the directions of mitochondrial L-1-pyrroline-3-hydroxy-5-carboxylate dehydrogenase (PHCDm) and mitochondrial L-4-hydroxyglutamate semialdehyde dehydrogenase (4HGLSDm) were fixed by the direction of mitochondrial L-1-pyrroline-3-hydroxy-5-carboxylate (PHCHGSm), since the three reactions form an unbranched pathway.
Predictions using the full GeM
Using NET analysis without including any prior irreversibility constraints is equivalent to testing each reaction in the network independently using the metabolite ranges specified in Table 1. NET analysis uses separately specified irreversibility constraints implicitly to further constrain the metabolite range, thereby potentially identifying additional irreversible reactions. Such an approach must be used with care, since errors in ΔfG estimates or irreversibility assumptions can propagate widely. Moreover, the algorithm uses constraints from all reactions, whether or not they are active under specific growth conditions.
NExT was used to identify new irreversible reactions when including 2032 of the 2149 irreversibilities already specified in Recon 1 (Fig. 1, Step 4). To avoid unreal constraints, the 118 irreversible transport reactions between the compartments with assumed biophysical properties equal to those of cytoplasm were set to be reversible. First, the ΔrG range was determined for 926 of 2032 irreversible reactions for which sufficient information was available, i.e., where ΔfG was available for all reactants. All 926 specified reaction directions were thermodynamically feasible under the broad conditions specified in Table 1. Of 1277 reactions specified as reversible in Recon 1, 701 could be thermodynamically assessed. In addition to the 12 reactions (ignoring AIRCr, as discussed above) already found to be irreversible given broad concentration constraints (Table 2), an additional 18 reactions labeled as reversible were identified as thermodynamically irreversible when the Recon 1 constraints were included (Fig. 1, Step 4).
The new irreversibility constraints are caused by surrounding irreversibility constraints limiting substrate or product concentration ranges. The sources of these constraints were further analyzed to establish whether they came from a reliable source. The initial list of new irreversible reactions included four enzymes normally considered reversible (three nucleotide phosphatases (NPs) and a phosphopentomutase). All four were made irreversible through the model specification that NP1 (another NP activity) is irreversible, which dictates a minimal ribose-1-phosphate (R1P) concentration of 2 mM and a maximum inorganic phosphate concentration of 5 mM. The source for the irreversibility specification for NP1 is unclear; it is not found in any standard database, including Recon X. Nicotinate incorporation in the NAD salvage pathway is normally reported to use NAPRT1 (EC 2.4.2.11), which uses PRPP instead of R1P. Moreover, the exact same enzyme (EC 2.4.2.1) is known to work in the opposite direction for other substrates (purine nucleosides, nicotinate-D-riboside). Given the unlikely high predictions for the minimal R1P concentration and direction of other nucleotide phosphorylases, NExT was rerun without the NP1 specification, leading to the identification of 13 irreversible reactions (Table 3).
Table 3.
Reactions that were identified as irreversible by NExT considering directionality constraints in Recon1 that were not already specified in the model
| ID | Reaction | ΔrG min (KJ/mol) | ΔrG max (KJ/mol) | Direction | Pathway |
|---|---|---|---|---|---|
| 4HGLSDm | [m]e4hglu + (2)h + nadh = 4hglusa + h2o + nad | 44.75 | 100.95 | Reverse | Arginine and proline metabolism |
| L_LACtm | h[c] + lac_L[c] = h[m] + lac_L[m] | −31.85 | −7.66 | Forward | Transport, mitochondrial |
| GTHRDt | atp[c] + gthrd[c] + h2o[c] = adp[c] + gthrd[m] + h[c] + pi[c] | −108 | −8.72 | Forward | Glutathione metabolism |
| RADH | [c]h2o + nad + retinal = (2)h + nadh + retn | −109.87 | −29.39 | Forward | Vitamin A metabolism |
| RADH2 | [c]h2o + nadp + retinal = (2)h + nadph + retn | −86.86 | −15.97 | Forward | Vitamin A metabolism |
| RADH3 | [c]h2o + nad + retinal_cis_13 = 13_cis_retn + (2)h + nadh | −109.87 | −29.39 | Forward | Vitamin A metabolism |
| RADH4 | [c]h2o + nadp + retinal_cis_13 = 13_cis_retn + (2)h + nadph | −86.86 | −15.97 | Forward | Vitamin A metabolism |
| 3M4HDXPAC | [c]3mox4hpac + h2o + nad = (2)h + homoval + nadh | −91.83 | −26.06 | Forward | Tyrosine metabolism |
| 34DHPLACOX_NADP | [c]34dhpac + h2o + nadp = 34dhpha + (2)h + nadph | −83.54 | −12.65 | Forward | Tyrosine metabolism |
| 34DHXMANDACOX_NADP | [c]34dhmald + h2o + nadp = 34dhoxmand + (2)h + nadph | −83.54 | −12.65 | Forward | Tyrosine metabolism |
| 3MOX4HOXPGALDOX_NADP | [c]3m4hpga + h2o + nadp= 3mox4hoxm + (2)h + nadph | −83.54 | −12.65 | Forward | Tyrosine metabolism |
| 4HOXPACDOX_NADP | [c]4hoxpacd + h2o + nadp = 4hphac + (2)h + nadph | −83.54 | −12.65 | Forward | Tyrosine metabolism |
| FACOAL161 | [c]atp + coa + hdcea = amp + hdcoa + ppi | −119.89 | −0.65 | Forward | Fatty acid metabolism |
See Supporting Material for analysis of errors in estimated values.
The causes for the 10 new irreversible reactions were further analyzed. The 4HGLSDm reaction was predicted to be irreversible. This reaction was also identified by Haraldsdóttir et al. (13), and the same enzyme catalyzes other irreversible reactions, including those of 1-pyrroline-5-carboxylate dehydrogenase (P5CD) and PHCD. Indeed, NExT predicts irreversibility of 4HGLSDm on the basis of PHCDm constraining the mitochondrial NADH/NAD ratio.
Mitochondrial L-lactate transport through the proton symport (L-LACtm) is made irreversible by the inclusion of an irreversible facilitated transport reaction for lactate into mitochondria through a broad selectivity neutral solute channel (AQP9). Although the transport specification in Recon 1 is incorrect (AQP9 only transports the neutral acid form), the correct transporter would have the same effect if irreversibility is accepted. There is, however, limited basis for the latter considering that the existence of mitochondrial lactate oxidation is still subject to debate (32).
Glutathione transport into mitochondria (GTHRDt) becomes irreversible because the estimated minimum cytosolic ATP concentration is ∼0.5 mM. This estimate is well below the reported minimum value of 1.29 mM (13) and thus is a reasonable constraint. It should be noted that glutathione is only transported to mitochondria to establish the glutathione pool; redox cycling is performed internally.
Reaction fatty-acid-CoA ligase hexadecenoate (FACOAL161) becames irreversible due to a reduced range of AEC, caused by other irreversible reactions. Finally, a number of cytosolic aldehyde dehydrogenases used in vitamin A and tyrosine metabolism were found to be irreversible in the oxidation direction. The constraints are caused by a reduced range of cytosolic redox ratios due to other irreversible dehydrogenases. The irreversibility of retinal dehydrogenase was reported previously (13), using a narrower initial concentration range for the dinucleotides. The four NADP-linked aldehyde dehydrogenases are all catalyzed by ALDH3 (EC 1.2.1.5), and one of these (4HOXPACDOX_NADP) is topologically constrained to operate only in the forward direction; thus, it is reasonable to assume that all four reactions are thermodynamically constrained in this direction under normal conditions.
Reaction directionalities obtained from thermodynamics (Tables 2 and 3) were compared with the direction of fluxes generated by FVA (Fig. 1, Step 5). Six reactions (PHCHGSm, GTHPm, DPGM, DCT, 4HOXPACDOX_NADP, and DOPAQNISO1) were found to be GeM topologically irreversible in the same direction as the thermodynamic constraints. On the other hand, five reactions (AIRCr, 4HGLSDm, 34DHPLACOX_NADP, 34DHXMANDACOX_NADP, and 3MOX4HOXPGALDOX_NADP) showed inconsistency in terms of the directionality calculated by FVA and thermodynamics (see below). Another three reactions (P450SCC1m, GTHRDt, and 3M4HDXPAC) were linked to singleton (dead-end) metabolites and cannot carry any flux. For the remaining 12 reactions (GTHP, GTHPe, G5SADrm, DCMPDA, L_LACtm, PDX5PO, PYAM5POr, RADH, RADH2, RADH3, RADH4, and FACOAL161), however, the thermodynamic constraints represent novel information that can be used to constrain the feasible flux space.
The reactions showing inconsistency between FVA and thermodynamics highlight problems in the model. When 4HGLSDm is constrained, no flux is possible through either PHCHGSm or PHCDm; further analysis revealed that Recon 1 lacks mitochondrial import for 4-hydroxyproline (a degradation product of collagen and other extracellular matrix proteins) and that flux is restored when this link is introduced. The three NADP-linked ALDH3 reactions only function in isolated loops with their irreversible NAD-linked counterparts (Table S5). The thermodynamic constraints conflict with the GeM topological constraints (Fig. 1, Step 5) and no flux is possible through these loops. These isolated loops highlight that the conventional emphasis during curation on ensuring that reactions carry flux may be flawed, and a more accurate approach would be to incorporate thermodynamics to help curate reactions when biochemical evidence is lacking. AIRCr is caused by another problem and will be discussed in a separate section.
Reduced range of metabolite concentration
Initially, a broad range of 10−4 to 10 mM was allocated for the concentration of all metabolites in the model. If a metabolite is present in more than one compartment, then this concentration range is applied to the total metabolite concentration from all compartments, since metabolite concentrations are typically measured for the whole cell. Using thermodynamics together with the directions in Recon 1, NExT narrowed the compartmentalized concentration ranges of 44 metabolites (Table S6), including six from central carbon metabolism (3-phosphoglycerate, ATP, L-Lactate, NAD, NADH, and PPi).
Comparison of reactions found in both compartments
Of the total 110 reactions recorded in both the cytosol and mitochondria in Recon 1, 33 were found to operate differently between the cytoplasm and mitochondria (Table S7). Of these 33 reactions, 30 carry flux only in one cell compartment due to the network topology, including L-glutamate 5-semialdehyde dehydratase (G5SAD), a spontaneous irreversible reaction (Table 2), which can carry flux only in mitochondria. Thermodynamically reversible L-aminoadipate-semialdehyde dehydrogenase (AASAD3) is topologically constrained in the cytosol due to a link to the thermodynamically irreversible PPD2CSPp reaction, whereas the mitochondrial version is part of a different pathway and unconstrained. Reaction D-glucuronolactone NAD oxidoreductase was specified in the model as irreversible in both compartments; however, thermodynamic analysis only specifies the mitochondrial reaction as irreversible. Finally, thermodynamically reversible glutathione oxidoreductase is topologically free in the cytosol but topologically constrained in the direction of oxidation of NADPH in mitochondria. This direction is imposed by the thermodynamically irreversible mitochondrial glutathione peroxidase (GTHPm) reaction. The cytoplasm isoform of glutathione peroxidase is also thermodynamically irreversible, but in the cytoplasm there are other reactions that are able to produce oxidized glutathione, removing the substrate constraint to the glutathione oxidoreductase reaction.
Prediction of irreversible reactions in Yeast 5 model
In this study, the most recently published version of the yeast consensus GeM, Yeast 5 (16), was selected. Yeast 5 was modified as discussed for the human model, i.e., introduction of IMS, and some minor modifications of equations to enable accurate calculations. The modified model contains a total of 1891 reactions, whereof 630 are reversible. Biophysical properties were specified based on literature values, whereas broad concentration ranges and ratio ranges for adenylate energy charge and redox factors were used (Table 1).
De novo predictions
Using NExT without the irreversibility constraints specified in Yeast 5, 124 reactions were identified as irreversible under physiological conditions (Fig. 1, Step 1). Of these reactions, 109 were already specified among the 1261 irreversible reactions in Yeast 5. Another three reactions were specified as irreversible in Yeast 5, but in the opposite direction to that determined by NExT (Table 4, bold reactions). The remaining 12 reactions were not specified in Yeast 5 (Table 4). Eight of the fifteen reactions with new directions were also found to be thermodynamically irreversible in Recon 1 (Tables 2 and 3) linked to arginine and proline metabolism (r_1871 and r_1888), glutathione metabolism (r_0483 and r_0484), glycolysis (r_0356), pyrimidine catabolism (r_0326), vitamin B6 metabolism (r_0955), and IMP biosynthesis (r_0911). As for the human model, AIRCr (r_0911) was predicted to operate in the incorrect direction and will be discussed later. One of the new thermodynamically irreversible reactions, DPGM (r_0356), belongs to the 474 reactions in Yeast 5 that are linked to singleton metabolites and hence cannot carry flux (Fig. 1, Step 2).
Table 4.
Reactions that were identified as irreversible by thermodynamic constraints in Yeast 5 that were not previously specified, or were specified with the opposite direction in the model
| ID | Reaction | ΔrGmin (KJ/mol) | ΔrGmax (KJ/mol) | Direction | Pathway | ID hGeM |
|---|---|---|---|---|---|---|
| r_1871 | [m]1p3h5c + h + h2o = 4hglusa | 41.44 | 98.52 | Reverse | Arginine and proline metabolism | PHCHGSm |
| r_1887 | [c]glu5sa = 1pyr5c + h + h2o | −88.7 | −31.62 | Forward | Arginine and proline metabolism | G5SADs |
| r_1888 | [m]glu5sa = 1pyr5c + h + h2o | −90.98 | −33.9 | Forward | Arginine and proline metabolism | G5SADrm |
| r_0483 | [c] (2)gthrd + h2o2 = gthox + (2)h2o | −316.29 | −202.13 | Forward | Glutathione metabolism | GTHP |
| r_0484 | [m] (2)gthrd + h2o2 = gthox + (2)h2o | −316.29 | −202.13 | Forward | Glutathione metabolism | GTHPm |
| r_0356 | [c]13dpg = 23dpg + h | −71.96 | −14.88 | Forward | Glycolysis/gluconeogenesis | DPGM |
| r_0911 | [c]air + co2tot = 5aizc + h + h2o | 3.34 | 71.84 | Reverse | IMP biosynthesis | AIRCr |
| r_0073 | [c]4ppmip + h2o = (2)h + minohp + pi | −69.16 | −0.67 | Forward | Inositol phosphate metabolism | NA |
| r_0092 | [c]6ppmip + h2o = (2)h + minohp + pi | −69.16 | −0.67 | Forward | Inositol phosphate metabolism | NA |
| r_0767 | [n]h2o + NAD = ADP-rib + h + ncam | −89.44 | −3.82 | Forward | NAD metabolism | NADN |
| r_0204 | [c]ap4a + h2o = (2)adp + (2)h | −102.97 | −17.35 | Forward | Nucleotides metabolism | ∼AP4AH1 |
| r_0326 | [c]dcmp + h + h2o = dump + nh4 | −90.02 | −4.4 | Forward | Pyrimidine catabolism | DCMPDA |
| r_0110 | [c]ac + coa + h = accoa + h2o | 15.52 | 101.14 | Reverse | Pyruvate metabolism | ACOAHi |
| r_0953 | [c]nh4 + (0.5)o2 + pydx + (2)h2o = (2)h2o2 + pydam | 306.21 | 454.62 | Reverse | Vitamin B6 metabolism | NA |
| r_0955 | [c]o2 + pdx5p = h2o2 + pydx5p | −162.89 | −65.86 | Forward | Vitamin B6 metabolism | PDX5PO |
Reactions in bold are those specified with the opposite direction in the model. See the Supporting Material for analysis of errors in estimated values. NA, not applicable.
One of the three reactions (r_1871) for which the direction was specified differently between thermodynamics analysis and the model was found in the thermodynamically correct direction in the human model. If present, acetyl-CoA hydrolase (r_0110) must operate in the hydrolysis direction. Acetyl-CoA biosynthesis from CoA and acetate requires ATP and is covered by a separate reaction encoded by acetyl-CoA synthetase (r_0112). Cytosolic acetyl-CoA hydrolase was deleted in the most recent online version, Yeast 7 GeM, and the same was done in this study. The reaction pyridoxal oxidase (r_0953) has an extremely high ΔrG, and it is thus highly improbable that the model-specified direction is correct.
Incorporating the 112 thermodynamic constraints into the network, FVA identified 21 blocked reactions (Table S8), including four of the new thermodynamically constrained reactions (r_0484, r_0955, r_0767, and r_1888) and one of the reactions with contradicting directions between thermodynamics and model specifications (r_0953) (Fig. 1, Step 3). Mitochondrial oxidation of glutathione (r_0484) was blocked with another four mitochondrial reactions linked to hydrogen peroxide degradation. Evidently, Yeast 5 lacks hydrogen-peroxide-forming reactions in mitochondria. The main source of hydrogen peroxide in mitochondria is from disproportionation of superoxide (formed as a byproduct of electron transport chain reactions) by manganese-dependent superoxide dismutase (Mn-SOD) (33); neither the byproduct formation reaction nor Mn-SOD is included in Yeast 5. The constraints in reactions r_0953 and r_0955 together prevent flux through these reactions and another four reactions for vitamin B6 metabolism (r_0954, r_0956, r_2025, and r_2029). These six reactions (r_0953, r_0955, r_0954, r_0956, r_2025, and r_2029) are part of a closed loop in Yeast 5 (Table S8), which can be resolved by connecting pyridoxal-5-phosphate to the network. Cytosolic and nuclear NAD nucleases (r_1955 and r_0767) are thermodynamically irreversible in the same direction; these reactions and three more (r_1646, r_2097, and r_1966) belong to an intercompartment closed loop. Therefore, the six reactions became blocked. The consequences of the null flux of r_1888 in the proline metabolism will be discussed later.
Three new thermodynamically irreversible reactions were neither blocked nor found irreversible in the human model. The two depyrophosphorylation reactions (r_0073 and r_0092) are catalyzed by (EC 3.6.1.52). The same enzyme, diphosphoinositol-polyphosphate diphosphatase catalyzes reaction r_0082 (converting 5PP-IP5 to IP6), which is reported as irreversible in both Yeast 5 and Recon 1. The analogous human reaction to Ap4A hydrolase (r_0204) was specified in Recon 1 as irreversible, supporting the direction assigned based on thermodynamics.
Predictions using the full GeM
The original reversibility constraints of the model were checked using NExT. Analogous to what was done with the human model, 54 irreversible intercompartment reactions were assumed reversible. The ΔrG values for 493 of 1205 remaining irreversible reactions were estimated. Resolving the three directional conflicts already identified (Table 4) produced a thermodynamically feasible model. One new thermodynamically irreversible reaction was identified in the constrained model, namely fatty-acid-CoA ligase (hexadecenoate) (r_0403) with a maximum ΔrG of −0.66 KJ/mol (Fig. 1, Step 4). The new direction is a consequence of the reduction of the cytoplasmic AEC range and the coenzyme A concentration range by other reactions specified as irreversible in the network.
The 13 new thermodynamic constraints (r_0403 and those listed in Table 4) were compared to FVA using the Yeast 5 directionality constraints only (Fig. 1, Step 5). The analysis revealed that three reactions are blocked irrespective of thermodynamics (r_0356, r_0484, and r_0955), whereas six reactions have conflicting directions between thermodynamics and FVA (r_0073, r_0092, r_0326, r_0767, r_0911, and r_1888). The conflicts point to a need for further curation of these reactions. Thermodynamics and FVA agreed on the directions specified for three reactions (r_0204, r_0483, and r_1887), and only the new thermodynamic constraint on fatty-acid-CoA ligase (hexadecenoate) adds a new flux constraint to Yeast 5.
Reduced range of metabolite concentration
NExT was used to calculate thermodynamically feasible concentration ranges of metabolites. The concentration range of 40 metabolites was significantly narrower than the allocated physiological range (Table S9). Among these metabolites, nine also had significantly reduced ranges in the human model (Table S5), including the highly connected cofactors NADH[c], NAD[c], and ATP[c], which themselves can constrain many reaction directions.
Comparison of reactions in both compartments
There are 73 reactions in Yeast 5 that are present in both the cytoplasm and mitochondria. Using the network topology and/or thermodynamic constraints, 19 of these reactions were predicted to behave differently between the cytoplasm and mitochondria (Table S10). Network topology alone prevents flux in one cellular compartment in 13 of the 19 reactions.
Of the remaining six reactions, glutamate 5-semialdehyde dehydratase will be discussed under proline metabolism in another section, whereas four of the six reactions are linked to hydrogen peroxide metabolism. As mentioned above, mitochondrial glutathione peroxidase (r_0484) is blocked once thermodynamic irreversibility is considered, since no hydrogen-peroxide generating reactions have been included in mitochondria. As a result, several linked irreversible mitochondrial reactions are unable to carry flux, namely, hydrogen peroxide reductase (r_0551), thioredoxin reductase (NADPH) (r_1039), and glutathione oxidoreductase (r_0482). In the cytosol, other sources of hydrogen peroxide exist, and the corresponding reactions (r_0481, r_0483, r_0550, and r_1038) can all carry flux.
The final reaction is aldehyde dehydrogenase, which in Yeast 5 was listed as irreversible in both compartments. Although thermodynamics analysis indicates that only the mitochondrial aldehyde dehydrogenase (r_0175) is thermodynamically irreversible, the cytoplasmic enzyme is reversible due to the different compartmental conditions (pH and redox potential).
Phosphoribosylaminoimidazole carboxylase (AIRCr and ADE2)
Phosphoribosylaminoimidazole carboxylase (EC 4.1.1.21) is an essential enzyme in purine synthesis in both humans and yeast (Fig. 2). The estimated ΔrG, however, contradicts the expected flux direction (Tables 2 and 4). In Recon 1, the pathway to produce IMP from PRPP cannot carry flux as a result of the reverse direction, whereas in Yeast 5 the pathway can still carry flux but in the opposite direction.
Figure 2.
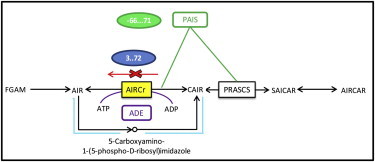
AIRCr. The human enzyme PAIS catalyzes both the AIRCr and PRASCS reactions (green). The yeast enzyme ADE2 catalyzes the AIRCr reaction coupled with ATP hydrolysis (purple). The parallel pathway utilized by bacteria is shown in light blue. These are the three different ways that different organisms overcome the energy demand to produce CAIR from AIR.
Prokaryotes (e.g., E. coli) use the reactions 5-(carboxyamino)imidazole ribonucleotide synthase (EC 6.3.4.18) and 5-(carboxyamino)imidazole ribonucleotide mutase (EC 5.4.99.18) to bypass phosphoribosylaminoimidazole carboxylase (Fig. 2). Although the bypass does not exist in eukaryotes, two other strategies are used to overcome the unfavorable thermodynamics. In humans, AIRCr is carried out by a dual-function enzyme (PAIS) that also catalyzes the thermodynamically favored reaction PRASCS, which has a ΔrG range of −97.42 to 27.68 KJ/mol under physiological conditions. This observation suggests that substrate channelling is being used to couple the unfavorable AIRCr reaction with the favorable PRASCS reaction and that the two reactions should be replaced with a single combined reaction (Fig. 2).
In yeast, the two enzymes are not coupled. Instead, yeast uses a variant of AIRCr (ADE2) that hydrolyzes ATP in stoichiometric proportion to AIR conversion (34). Since no new EC number has been created for the reaction, many metabolic models including KEGG and Yeast 5 erroneously describe the reaction without ATP, whereas the Biocyc database for yeast correctly describes it with ATP. This error has been corrected in the two latest online Yeast models, 6 and 7.
Proline synthesis and degradation pathways
Using NExT, it was possible to unravel a putative role of yeast compartmentalization for separation of proline synthesis (cytoplasm) and degradation (mitochondria), and the use of P5CD as a unique degradation pathway. The reaction G5SAD has an important role in proline metabolism (Fig. 3). This spontaneous reaction is thermodynamically irreversible under physiological conditions for both the human and yeast models. For yeast, the enzyme pyrroline-5-carboxylate reductase (P5CR) is only found in the cytoplasm (35); thus, proline synthesis occurs exclusively in cytoplasm. Initially, without considering the fixed direction for G5SAD, there are two possible degradation pathways for proline in the yeast GeM model: direct degradation of 1-pyrroline-5-carboxylate to glutamate, or through L-glutamate 5-semialdehyde. Due to the thermodynamically determined direction for G5SAD (for both compartments), yeast can only degrade proline using P5CD. Since this enzyme is strictly mitochondrial for yeast (35), the degradation of proline takes place in mitochondria in yeast.
Figure 3.
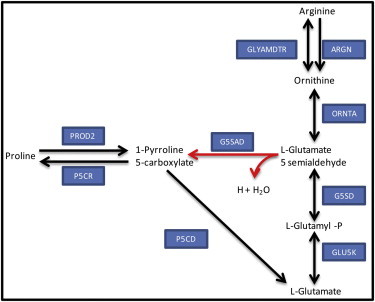
Proline synthesis and degradation pathways. The spontaneous reaction L-glutamate 5-semialdehyde dehydratase (red arrows) has a fixed direction due to thermodynamic constraints under physiological conditions for both of the eukaryotic models studied. ARGN, arginase; G5SAD, L-glutamate 5-semialdehyde dehydratase; G5SD, L-glutamate 5-semialdehyde dehydratase; GLU5K, glutamate 5-kinase; GLYAMDTR, glycine amidinotransferase; ORNTA, ornithine transaminase; P5CD, 1-pyrroline-5-carboxylate dehydrogenase; P5CR, pyrroline-5-carboxylate reductase; PROD2, proline dehydrogenase.
In contrast to yeast, human cells use only one compartment (mitochondria) for proline synthesis and degradation. In the human model, mitochondrial G5SAD (G5SADm) can be active due to the presence of ornithine transaminase (ORNTA) or L-glutamate 5-semialdehyde dehydratase (G5SD) in the mitochondrial compartment (36–38). Since ORNTA and G5SD are not present in the cytoplasm, the cytoplasmic G5SAD is unable to carry flux. In a similar way, the enzyme P5CD required for proline catabolism is only localized in mitochondria (36,37). Thus, for human cells, proline metabolism takes place exclusively in mitochondria.
Comparing with previous approaches
The range of estimated ΔrG values for physiological ranges of metabolite concentrations have been used to assign directions to individual reactions in GeMs (11–13). A recent implementation of this approach is the von Bertalanffy package (39,40). This corresponds to the first step in the workflow developed (Fig. 1). As seen in this study, only a small fraction of the directional constraints can be identified when using broad metabolite concentrations. Using NET analysis through NExT and FVA on the full model, it is possible to evaluate the wider impact of the directional constraints in the model.
NET analysis is well suited for open-ended network analysis, where the gross metabolic phenotype is not specified, i.e., when we are evaluating and refining a GeM in generic form. For a specific metabolic study where the direction of exchange reactions are specified, thermodynamics-based metabolic flux analysis (TMFA) is a superior algorithm that directly estimates thermodynamically feasible metabolic flux distributions from a set of directionality constraints and a reduced model (25). For eukaryotic models, TMFA is limited to the use of compartment-specific metabolite concentration ranges.
Conclusion
Thermodynamics analysis is a potent tool for validation of existing irreversibility constraints and identification of new ones in multicompartment metabolic networks.
We developed an open-source software package for NET analysis, NExT, which implements correct Gibbs energy estimations for transport reactions. NExT revealed that proton transport in the electron transport chain must occur in the IMS rather than the cytoplasm, and ATP synthase was modified to use IMS protons.
In two well-curated eukaryotic models, NExT revealed a few erroneous irreversibility constraints, as well as some thermodynamically infeasible internal loops. Despite the use of a broad range of metabolite concentrations, additional irreversibility constraints were identified, including some supported by the literature. Improved curation, together with the new thermodynamic constraints will provide more accurate flux estimations.
The analysis further highlighted the alternative approaches used by different organisms to overcome thermodynamic irreversibility of phosphoribosylaminoimidazole carboxylase using, for example, a parallel pathway (E. coli), ATP coupling (yeast), and substrate channeling (human). The analysis also highlighted differences in proline metabolism between yeast (cytosolic anabolism and mitochondrial catabolism) and humans (exclusively mitochondrial metabolism).
Supporting Material
References
- 1.Reed J.L., Vo T.D., Palsson B.O. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR) Genome Biol. 2003;4:R54. doi: 10.1186/gb-2003-4-9-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Oliveira Dal’Molin C.G., Quek L.E., Nielsen L.K. AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 2010;152:579–589. doi: 10.1104/pp.109.148817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duarte N.C., Becker S.A., Palsson B.O. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrgård M.J., Swainston N., Kell D.B. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat. Biotechnol. 2008;26:1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quek L.E., Nielsen L.K. On the reconstruction of the Mus musculus genome-scale metabolic network model. Genome Inform. 2008;21:89–100. [PubMed] [Google Scholar]
- 6.Feist A.M., Palsson B.O. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat. Biotechnol. 2008;26:659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curran K.A., Alper H.S. Expanding the chemical palate of cells by combining systems biology and metabolic engineering. Metab. Eng. 2012;14:289–297. doi: 10.1016/j.ymben.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Varma A., Palsson B.O. Metabolic flux balancing: basic concepts, scientific and practical use. Nat. Biotechnol. 1994;12:994–998. [Google Scholar]
- 9.Thiele I., Palsson B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faria J.P., Rocha M., Henry C.S. Analysis of the effect of reversibility constraints on the predictions of genome-scale metabolic models. Adv. Bioinform. 4th Int. Workshop Pract. Appl. Comput. Biol. Bioinform. 2010;74:209–215. [Google Scholar]
- 11.Henry C.S., Jankowski M.D., Hatzimanikatis V. Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophys. J. 2006;90:1453–1461. doi: 10.1529/biophysj.105.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming R.M., Thiele I., Nasheuer H.P. Quantitative assignment of reaction directionality in constraint-based models of metabolism: application to Escherichia coli. Biophys. Chem. 2009;145:47–56. doi: 10.1016/j.bpc.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haraldsdóttir H.S., Thiele I., Fleming R.M.T. Quantitative assignment of reaction directionality in a multicompartmental human metabolic reconstruction. Biophys. J. 2012;102:1703–1711. doi: 10.1016/j.bpj.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mavrovouniotis M.L. Identification of localized and distributed bottlenecks in metabolic pathways. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1993;1:275–283. [PubMed] [Google Scholar]
- 15.Kummel A., Panke S., Heinemann M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol. Syst. Biol. 2006;2:2006.0034. doi: 10.1038/msb4100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heavner B.D., Smallbone K., Walker L.P. Yeast 5—an expanded reconstruction of the Saccharomyces cerevisiae metabolic network. BMC Syst. Biol. 2012;6:55. doi: 10.1186/1752-0509-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jol S.J., Kümmel A., Heinemann M. Thermodynamic calculations for biochemical transport and reaction processes in metabolic networks. Biophys. J. 2010;99:3139–3144. doi: 10.1016/j.bpj.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alberty R.A. Wiley; Hoboken, NJ: 2003. Thermodynamics of Biochemical Reactions. [Google Scholar]
- 19.Björnberg O., Ostergaard H., Winther J.R. Measuring intracellular redox conditions using GFP-based sensors. Antioxid. Redox Signal. 2006;8:354–361. doi: 10.1089/ars.2006.8.354. [DOI] [PubMed] [Google Scholar]
- 20.Hu J., Dong L., Outten C.E. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J. Biol. Chem. 2008;283:29126–29134. doi: 10.1074/jbc.M803028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llopis J., McCaffery J.M., Tsien R.Y. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. USA. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porcelli A.M., Ghelli A., Rugolo M. pH difference across the outer mitochondrial membrane measured with a green fluorescent protein mutant. Biochem. Biophys. Res. Commun. 2005;326:799–804. doi: 10.1016/j.bbrc.2004.11.105. [DOI] [PubMed] [Google Scholar]
- 23.Hanson G.T., Aggeler R., Remington S.J. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 24.Alberty R.A. Wiley-Interscience; New York: 2006. Biochemical Thermodynamics: Applications of Mathematica. [PubMed] [Google Scholar]
- 25.Henry C.S., Broadbelt L.J., Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophys. J. 2007;92:1792–1805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jankowski M.D., Henry C.S., Hatzimanikatis V. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys. J. 2008;95:1487–1499. doi: 10.1529/biophysj.107.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wishart D.S., Jewison T., Scalbert A. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M., Goto S., Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hastings J., de Matos P., Steinbeck C. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res. 2013;41:D456–D463. doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zamboni N., Kümmel A., Heinemann M. anNET: a tool for network-embedded thermodynamic analysis of quantitative metabolome data. BMC Bioinformatics. 2008;9:199. doi: 10.1186/1471-2105-9-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiele I., Swainston N., Palsson B.O. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gladden L.B. Is there an intracellular lactate shuttle in skeletal muscle? J. Physiol. 2007;582:899. doi: 10.1113/jphysiol.2007.138487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luk E., Yang M., Culotta V.C. Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:22715–22720. doi: 10.1074/jbc.M504257200. [DOI] [PubMed] [Google Scholar]
- 34.Meyer E., Leonard N.J., Smith J.M. Purification and characterization of the purE, purK, and purC gene products: identification of a previously unrecognized energy requirement in the purine biosynthetic pathway. Biochemistry. 1992;31:5022–5032. doi: 10.1021/bi00136a016. [DOI] [PubMed] [Google Scholar]
- 35.Brandriss M.C., Magasanik B. Subcellular compartmentation in control of converging pathways for proline and arginine metabolism in Saccharomyces cerevisiae. J. Bacteriol. 1981;145:1359–1364. doi: 10.1128/jb.145.3.1359-1364.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muzio G., Maggiora M., Canuto R.A. Aldehyde dehydrogenases and cell proliferation. Free Radic. Biol. Med. 2012;52:735–746. doi: 10.1016/j.freeradbiomed.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 37.Marchitti S.A., Deitrich R.A., Vasiliou V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol. Rev. 2007;59:125–150. doi: 10.1124/pr.59.2.1. [DOI] [PubMed] [Google Scholar]
- 38.Wang G., Shang L., Wang X. Diazonamide toxins reveal an unexpected function for ornithine δ-amino transferase in mitotic cell division. Proc. Natl. Acad. Sci. USA. 2007;104:2068–2073. doi: 10.1073/pnas.0610832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleming R.M., Thiele I. von Bertalanffy 1.0: a COBRA toolbox extension to thermodynamically constrain metabolic models. Bioinformatics. 2011;27:142–143. doi: 10.1093/bioinformatics/btq607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noor E., Haraldsdóttir H.S., Fleming R.M.T. Consistent estimation of Gibbs energy using component contributions. PLOS Comput. Biol. 2013;9:e1003098. doi: 10.1371/journal.pcbi.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamilton J.J., Dwivedi V., Reed J.L. Quantitative assessment of thermodynamic constraints on the solution space of genome-scale metabolic models. Biophys. J. 2013;105:512–522. doi: 10.1016/j.bpj.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alberts B., Wilson J., Hunt T. Garland Science; New York: 2008. Molecular Biology of the Cell. [Google Scholar]
- 43.Dewey W.C., Fuhr M.A. Quantification of mitochondria during the cell cycle of Chinese hamster cells. Exp. Cell Res. 1976;99:23–30. doi: 10.1016/0014-4827(76)90675-3. [DOI] [PubMed] [Google Scholar]
- 44.Jorgensen P., Edgington N.P., Futcher B. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell. 2007;18:3523–3532. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maskow T., von Stockar U. How reliable are thermodynamic feasibility statements of biochemical pathways? Biotechnol. Bioeng. 2005;92:223–230. doi: 10.1002/bit.20572. [DOI] [PubMed] [Google Scholar]
- 46.Vojinović V., von Stockar U. Influence of uncertainties in pH, pMg, activity coefficients, metabolite concentrations, and other factors on the analysis of the thermodynamic feasibility of metabolic pathways. Biotechnol. Bioeng. 2009;103:780–795. doi: 10.1002/bit.22309. [DOI] [PubMed] [Google Scholar]
- 47.Walker G.M. Wiley; Chichester, United Kingdom: 1998. Yeast Physiology and Biotechnology. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


