Summary
Receptor down-regulation in the MVB pathway is mediated by the ESCRT complexes. ESCRT-III is composed of four protein subunits that are monomeric in the cytosol and oligomerize into a protein lattice only upon membrane binding. Recent studies have shown that the ESCRT-III protein Snf7 can form a filament by undergoing homo-oligomerization. To examine the role of membrane binding and of interactions with other ESCRT components in initiating Snf7 oligomerization, we used fluorescence spectroscopy to directly detect and characterize the assembly of the Snf7 oligomer on liposomes using purified ESCRT components. The observed fluorescence changes reveal an obligatory sequence of membrane-protein and protein-protein interactions that generate the active conformation of Snf7. Also, we demonstrate that ESCRT-III assembly drives membrane deformation. Furthermore, using an in vitro disassembly assay, we directly demonstrate that Vps24 and Vps2 function as adaptors in the ATP-dependent membrane disassembly of the ESCRT-III complex by recruiting the AAA ATPase Vps4.
Introduction
A number of seemingly unrelated biological processes such as Multivesicular body (MVB) biogenesis, cytokinesis and retroviral budding require the ESCRT (Endosomal Sorting Complex Required for Transport) machinery. In contrast to the formation of secretory and endocytic vesicles where membrane budding occurs into the cytosol, MVB formation requires membrane budding away from the cytosol. The cellular machinery responsible for MVB formation under normal and pathological conditions is comprised of a subset of vacuolar protein sorting (Vps) gene products that were first implicated in receptor downregulation via the MVB pathway (Hurley, 2008; Saksena et al., 2007; Williams & Urbe, 2007). The majority of these Vps proteins are constituents of five separate heteromeric protein complexes called ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III, and the Vps4 AAA-ATPase complex (Hurley, 2008; Williams & Urbe, 2007). These complexes are transiently recruited from the cytoplasm to the endosomal membrane where they function sequentially in sorting ubiquitinated transmembrane proteins into the MVB pathway.
During the process of MVB sorting, ESCRT-0, ESCRT-I and ESCRT-II are recruited to the endosomal membrane as stable hetero-oligomeric complexes from the cytosol. In contrast, the ESCRT-III proteins (Vps20, Snf7, Vps24 and Vps2) remain monomeric in the cytosol, and only upon membrane binding oligomerize into an ESCRT-III lattice of indeterminate stoichiometry. The fact that the ESCRT-III complex is comprised of four protein subunits that undergo a membrane-dependent monomer to hetero-oligomer transition raises a number of mechanistic questions, including (i) what is the order of membrane recruitment and assembly for each of the ESCRT-III proteins? and (ii) what are the molecular signals that prevent premature assembly of the ESCRT-III complex in the cytosol and direct assembly on the membrane?
Genetic and biochemical studies in yeast have provided some clues regarding the order of membrane recruitment of the individual ESCRT-III proteins. Vps20 is the first ESCRT-III protein to associate with ESCRT-II on the endosome. Endosomal recruitment of the Vps24-Vps2 subcomplex appears to be dependent on the Vps20-Snf7 subcomplex, thereby indicating that the two subcomplexes are recruited sequentially (Teis et al., 2008). Recycling of membrane-bound ESCRT complexes into the cytosol has been shown to involve interactions between Vps2 and the AAA+ATPase Vps4 (Obita et al., 2007; Stuchell-Brereton et al., 2007).
While it is challenging to determine the precise stoichiometry and molecular architecture of a membrane-bound protein lattice, studies on hSnf7-1 (CHMP4A) have shown that overexpressed hSnf7-1/CHMP4A spontaneously forms membrane-attached filaments 5 nm in width that curve and self associate to create circular arrays that can promote or stabilize negative membrane curvature and outward budding of plasma membrane tubules (Hanson et al., 2008). These data indicate that a single ESCRT-III protein can form an ESCRT-III lattice, presumably by Snf7 binding to itself. This possibility is further supported by observations that Snf7 is the most abundant ESCRT-III subunit in yeast (Teis et al., 2008), and that most Snf7 isoforms self-associate in yeast two hybrid analyses (reviewed in Saksena et al., 2008).
ESCRT-III assembly is spatially restricted to the endosomal membrane by the upstream ESCRT complexes (ESCRT-0, -I, and –II), which selectively bind ubiquitin-modified transmembrane cargo and the endosomally enriched lipid phosphatidylinositol 3-phosphate (PI3P). There is growing evidence that activation of ESCRT-III subunits for assembly into the membrane-bound ESCRT-III complex requires an activating conformational switch. All of the ESCRT-III subunits are similar in size (221–241 residues) and domain organization, with N-terminal basic and C-terminal acidic regions (Saksena et al., 2007). Hence, the crystal structure of hVps24/CHMP3 (residues 9–183) reveals structural elements that are probably common to all ESCRT-III subunits (Muziol et al., 2006). Five alpha helices (α1– α5) span most of the N-terminal two thirds of the protein and are connected by a relatively long linker to a sixth short predicted helix (α6) near the C-terminus (α6 and the loop linking α5 to α6 were not seen in the crystal structure indicating conformational flexibility). The most prominent structural element observed in the crystal structure is the N-terminal helical hairpin that is 70 Å long and composed of helices α1 and α2, which are critical for membrane binding and homo- or hetero-dimerizati on events (Muziol et al., 2006). The C-terminal region that includes helix α5, the microtubule interacting and transport (MIT)-interacting region (MIR), and α6 constitutes a putative autoinhibitory region that is thought to interact with helix α2 of the core and thereby block homo-or hetero-dimerization of ESCRT-III components (Shim et al., 2007). Such intramolecular interactions may therefore prevent ESCRT-III-ESCRT-III intermolecular association and thereby maintain the ESCRT-III proteins as metastable monomers in the cytosol in a ‘closed’ state. ESCRT-III protein assembly may then occur only upon recruitment to endosomes where the autoinhibitory intramolecular interaction is lost during a putative ‘closed’ to ‘open’ state transition that now permits ESCRT-III proteins to associate. The molecular trigger for the proposed release of this inhibition and ESCRT-III assembly on endosomal membranes remains unclear.
Here we have employed primarily fluorescence spectroscopy to directly examine the molecular requirements for the assembly and disassembly of a Snf7 (ESCRT-III) polymer in vitro using purified components. Probes at several different positions were monitored spectroscopically under near-native aqueous conditions to detect changes in probe environment and also their sensitivity to different ESCRT components and membranes. The observed spectral changes indicate that upon membrane binding: (i) the conformations of both Vps20 and Snf7 change; (ii) these conformations are further altered upon association with, respectively, Vps25 and Vps20; (iii) most of the surface of the Snf7 molecule is in contact with the membrane surface; (iv) the C-terminal end of Snf7 moves adjacent to the bilayer except in mutants with restricted conformational flexibility; (v) Snf7 molecules oligomerize only in the presence of both membranes and Vps20; and (vi) this oligomerization event is critical for membrane deformation. The combined data strongly suggest that these ESCRT-III interactions dictate an obligatory sequence of assembly reactions during formation of the ESCRT-III complex. Furthermore, we demonstrate that disassembly of the membrane-bound Snf7 oligomer requires Vps24 and Vps2, which function as adaptors for ESCRT-III disassembly by capping the Snf7 oligomer and providing an entry point for the AAA-ATPase Vps4.
Results
Experimental Strategy
To monitor the conformation, environment, and interactions of a single protein during the assembly and disassembly of the multi-component, membrane-bound ESCRT-III complex, the Cys-free Vps20 and Snf7 proteins were each modified by the replacement of a single amino acid with Cys. Cys substitutions were made at sites that lie within domains predicted to have functional importance (autoinhibitory loop, membrane binding etc.), and are solvent exposed (based on the hVps24 crystal structure) to enhance the efficiency of covalent modification. Importantly, the introduction of a single Cys at different locations in monocysteine mutants had no effect on the folding and function of Vps20 or Snf7 since cells expressing the different cysteine mutants were able to sort MVB cargo (GFP-CPS) to the vacuolar lumen (Fig. S1). A fluorescent 7-nitrobenz-2-oxa-1,3-diazole (NBD) dye was then covalently attached to the Cys in each derivative. NBD is small, uncharged, and its O and N atoms have sufficient polar character for the dye to be soluble and exist stably in both aqueous and non-aqueous environments. Moreover, NBD is environmentally sensitive and its spectral properties are dramatically different in aqueous and non-aqueous environments (Johnson, 2005; Shepard et al., 1998). When NBD moves from an aqueous milieu (e.g., the surface of a soluble protein) to a hydrophobic environment (e.g., the nonpolar interior of a membrane or protein), its emission intensity and fluorescence lifetime (τ) increase while its wavelength of maximum emission (λem max) shifts to the blue (but not always, see Supplement). To distinguish the nonpolar membrane interior from a nonpolar environment inside a folded protein or at the nonpolar interface of associated proteins or domains, one can diagnostically identify the former by collisions between NBD probes and nitroxide collisional quenching moieties that are restricted to locations within the nonpolar core of the bilayer. Thus, by continuously monitoring in real time the spectral properties of an NBD-labeled protein (emission intensity, fluorescence lifetime, accessibility to quenchers, anisotropy, FRET) before, during, and after it associates with other proteins or membranes, both kinetic and thermodynamic parameters of these interactions can be quantified directly. In addition, since each protein contains NBD at only a single location, this approach provides an opportunity to characterize the structural changes that occur at specific sites in Vps20 and Snf7. The different NBD-labeled and Rhodamine (Rh)-labeled mutants of Vps20 and Snf7 are denoted as Vps20/Snf7n-NBD/Rh, where n denotes the location of the cysteine residue modified with NBD or Rh.
Vps20 Binding to Membranes
ESCRT-III assembly is initiated by membrane binding of Vps20 (Teis et al., 2008). To detect Vps20 binding to a membrane surface, NBD probes were positioned at four different sites in mono-cysteine mutants of Vps20: residue 7 in the unstructured N-terminal loop, 61 in the α2 helix, and residues 170 and 190 on either side of the α5 helix (Fig. 1F). The fluorescence emission of each of these NBD-labeled Vps20 derivatives was then characterized before and after the addition of liposomes comprised of phosphatidylcholine, phosphatidylserine, and phosphatidylinositol at molar ratios of 80:15:5 (hereafter termed PC/PS/PI). No change in emission was observed for probes located at position 7 upon exposure to PC/PS/PI (Fig. 1A). However, a ~6-fold increase in intensity and a 13-nm blue shift in the λem max was observed for the NBD probe at residue 61 following liposome addition (Fig. 1B). Such substantial changes show that the NBD dye at residue 61 moves from an aqueous to non-aqueous milieu upon addition of membranes, a conclusion verified by a large increase in its fluorescence lifetime (data not shown).
Fig. 1. Vps20 binding to membranes.
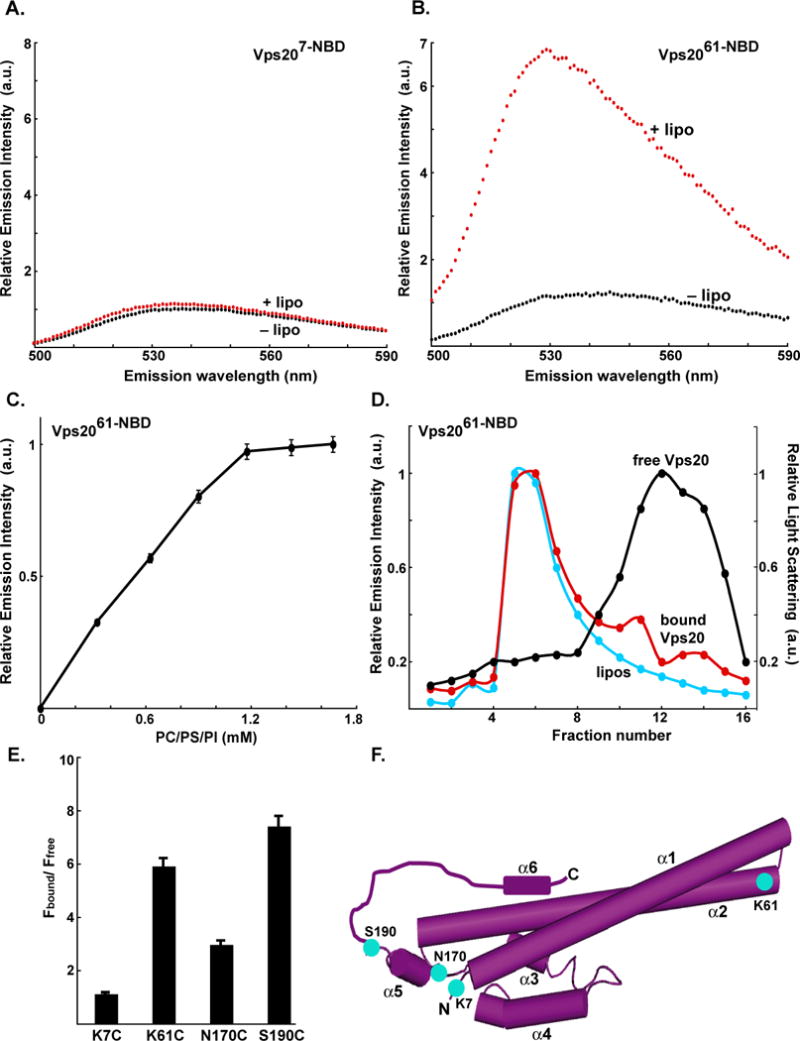
Fluorescence emission spectra of 750 nM Vps207-NBD (A) and 1 μM Vps2061-NBD (B) before (black) and after (red) addition of excess (1.6 mM; see Fig. 1C) PC/PS/PI. (C) Titration of 1 μM Vps2061-NBD with liposomes reveals that fluorescence-detected (λem = 530 nm) binding is complete after addition of 1.2 mM PC/PS/PI. (D) Sepharose CL-2B chromatography of 800 nM Vps2061-NBD that had been preincubated with either 1.5 mM PC/PS/PI (red) or no liposomes (black). Protein was detected by NBD emission intensity and liposomes by light scattering (cyan). The elution profile of liposomes that were not exposed to protein was identical to the profile shown in this and later figures (data not shown). (E) The ratios of NBD emission intensities at 530 nm are shown for each derivative with (Fbound) and without (Ffree) membranes. (F) Cartoon representation of the water-soluble hVps24 monomer. Based on the sequence similarities of the ESCRT-III proteins, Vps20, Snf7 and Vps2 are believed to share structural features with the solved crystal structure of hVps24. Helices α1–5 and the N- and C-termini of the protein are labeled. Residues that were mutated to cysteine for labeling with NBD are depicted in cyan and labeled accordingly. Helix α6 as well as the α5–α6 loop have been manually sketched.
When Vps2061-NBD was titrated with PC/PS/PI, the emission intensity increased and reached a plateau (Fig. 1C), thereby suggesting that all Vps20 had bound to the membrane. To test this conclusion by an independent technique, a sample of Vps2061-NBD was incubated with liposomes and then purified by gel filtration chromatography. Since essentially all of the NBD fluorescence was found to co-elute in the void volume with the liposomes (detected by light scattering) and very little eluted with free Vps20 assayed in the absence of liposomes (Fig. 1D), more than 95% of the Vps2061-NBD was bound to liposomes after elution. Thus, the membrane-dependent spectral changes do indeed result from Vps20 binding to a membrane surface.
Large increases in intensity were also observed when excess liposomes were added to Vps20 with an NBD probe at either residue 170 or 190 (Fig. 1E). When each of these derivatives was titrated with PC/PS/PI, the binding curves were similar to that of Fig. 1C (Fig. S2; data not shown). Thus, the similar PC/PS/PI-dependent spectral changes of probes at residues 61, 170, and 190 suggest that the probes near α2 and α5 are exposed to the nonpolar bilayer. This conclusion was confirmed directly when the intensity of each probe was reduced by collisions with quenchers restricted to the nonpolar membrane core (Fig. S3B–S3F) (Johnson, 2005). Interestingly, although the spectral changes for the probe at residue 7 in the unstructured Vps20 N-terminal domain are slight upon PC/PS/PI addition, the fluorescence lifetime of this probe is somewhat high (<τ> = ~4 ns; aqueous τ = 1–2 ns and nonpolar τ = 7–9 ns for NBD-labeled protein) and its λem max is somewhat low (538 nm; typical range = 520–546 nm for nonpolar to aqueous) even in the absence of membranes, which indicates that this probe is in a relatively nonpolar environment when Vps20 is free in solution.
Vps25 Binds to Membrane-bound Vps20
When either Vps2061-NBD or Vps20170-NBD was titrated with Vps25, a protein in the ESCRT-II complex that directly interacts with Vps20, little change in NBD emission intensity was observed (Fig. 2A–C). These small spectral changes reveal that the probes located at positions 61 and 170 in Vps20 do not experience significant changes in environment upon exposure to free Vps25 in solution and/or Vps20 and Vps25 associate only weakly in solution. Probes at residues 7 and 190 were also insensitive to Vps20 exposure to Vps25 (Fig. 2F, S4B). Similarly, when Vps20170-NBD was first bound to liposomes and then titrated with Vps25, the NBD fluorescence signal was unchanged by Vps25 (Fig. S4A;2E,F). However, when Vps25 was added to membrane-bound Vps2061-NBD, a large additional increase of NBD emission intensity was observed that was Vps25-dependent (Fig. 2D–F). These data strongly suggest that Vps25 and Vps20 associate on a membrane, but not in solution. Furthermore, the spectroscopic insensitivity of both free and membrane-bound Vps20170-NBD to the presence of Vps25 suggests that a significant conformational change alters the environment of a probe at residue 61, but not at 170, upon association with Vps25 on the membrane.
Fig. 2. Vps20 interactions with the Vps25 component of ESCRT-II.
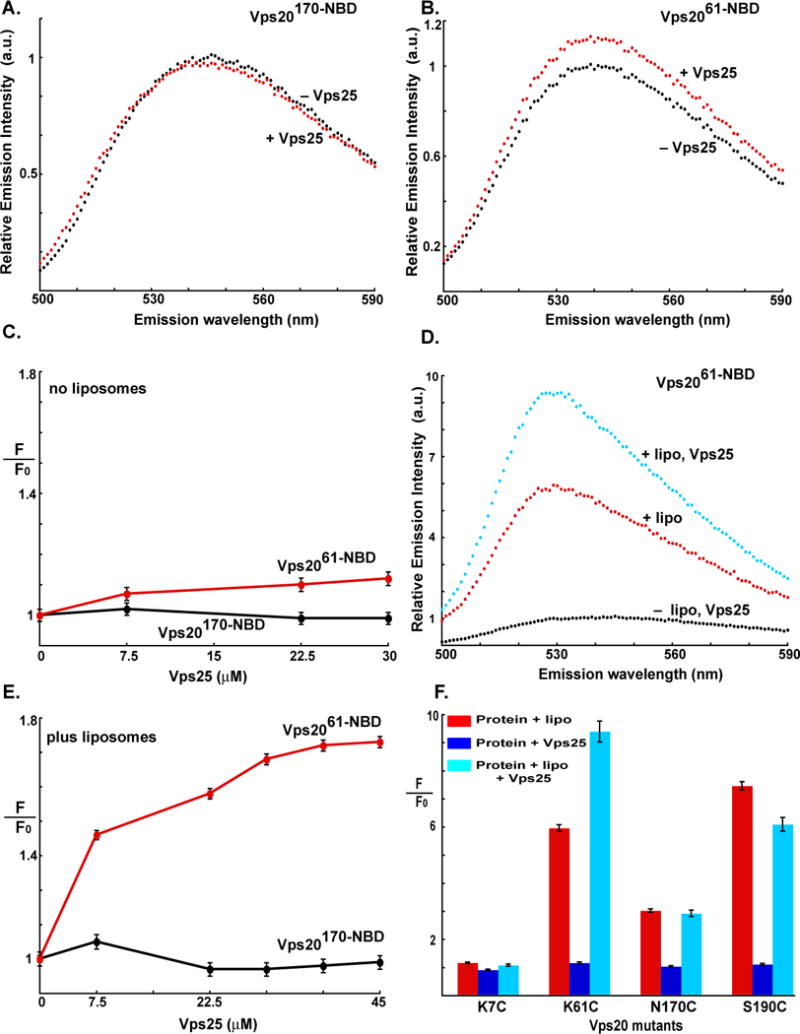
Fluorescence emission scans of 1 μM Vps20170-NBD D (A) and Vps2061-NBD (B) are shown in buffer (black) and after addition of 30 μM Vps25 (red). (C) The dependence of the emission intensities (F; λex = 468, λem = 530 nm) of 1 μM Vps20170-NBD (black) and Vps2061-NBD (red) on Vps25 addition are shown; F0 is the intensity in the absence of Vps25. (D) Emission scans of 1 μM Vps2061-NBD are shown in buffer (black), after addition of excess 1.5 mM liposomes (red), and after the addition of 30 μM Vps25 to Vps2061-NBD preincubated with 1.5 mM liposomes. (E) The dependence of the emission intensities (F; λex = 468, λem = 530 nm) of 1 μM Vps20170-NBD (black) and Vps2061-NBD (red) on Vps25 concentration is shown after the Vps20 derivatives were incubated with 1.5 mM liposomes for 30 min at 22 °C before Vps25 addition; F0 is the intensity in the absence of Vps25. (F) Summary of changes in the emission intensities of different NBD-labeled Vps20 mutants upon incubation with liposomes (red bar); Vps25 alone (dark blue bar); or liposomes and Vps25 (cyan). In each case, F0 represents emission intensity of the protein in buffer; and F represents emission intensity of the protein following incubation either with liposomes, Vps25 or liposomes and Vps25.
When excess Vps25 was added to membrane-bound Vps20190-NBD, a small emission intensity decrease was observed (Fig. S4C), in contrast to the large increase seen with Vps2061-NBD (Fig. 2D). In addition, little change was detected with the Vps207-NBD derivative (Fig. 2F). Thus, the use of multiple fluorescent probes reveals that the environments of Vps20 residues 61, 170, and 190 are altered upon membrane binding, and that the environments of the probes at both residues 61 and 190 are changed when Vps25 binds to the membrane-bound Vps20 (Fig. 2F). It must be emphasized that the experiments described above were carried out using concentrations of PC/PS/PI at which all of the NBD-labeled Vps20 is membrane-bound (Fig. 1C). Hence, the changes in NBD intensity upon Vps25 addition reflect changes in the local environment of NBD as a result of Vps25-Vps20 interactions rather than changes in the amount of membrane-bound Vps20. It must also be noted that the size of a spectral change does not necessarily correlate with the magnitude of the conformational change that altered dye environment. Thus, one cannot conclude that the membrane- and Vps25-induced structural changes in Vps20 were smaller near residue 170 than residue 61. However, the most important observation is that the binding of Vps25 to Vps20 changed its conformation at two sites that are well separated in the predicted Vps20 structure (based on the hVps24 crystal structure) (Fig. 1F). Furthermore, the simultaneous binding of both Vps25 and a membrane surface elicited a unique Vps20 conformation.
Snf7 Binding to Membranes
To characterize Snf7 binding to liposomes, nine mono-cysteine Snf7 mutants were prepared and examined as above with Vps20. With the exception of Snf714-NBD, addition of PC/PS/PI to a Snf7 derivative elicited a blue shift in its λem max and an increase in its τ (data not shown). Furthermore, collisional quenching experiments (Johnson, 2005) demonstrated that these spectral changes resulted from the exposure of the NBD probes (except at residue 14) to the nonpolar core of the bilayer (Fig. S5). However, as is evident by comparing emission scans of Snf781-NBD (Fig. 3A) and Snf7200-NBD (Fig. 3B) in the presence and absence of PC/PS/PI, the magnitudes of the membrane-dependent intensity increases varied considerably (Fig. 3C). Hence, the environments of probes at different locations, even those that are as spatially close as K14, K21 and K35 in the α1 helix, P191 and P200 in the α5– α6 loop and E81 and S93 in the α2 helix, differ for free and/or membrane-bound Snf7 (Fig. 3D). To ensure that the binding of protein to liposomes was complete in each of these samples, PC/PS/PI was titrated into the Snf7 proteins and intensities were measured only at liposome concentrations that gave maximal intensities (Fig. 3E, S6; data not shown). Thus, the different Fbound/Ffree ratios suggest that the membrane-dependent structural changes vary for different Snf7 domains and even for sites within the same domain. When examined using gel filtration, Snf7 binding to liposomes was found to differ from that of Vps20. Whereas more than 95% of the Vps20 remained bound to the PC/PS/PI liposomes after the chromatography (Fig. 1D), less than 50% of Snf781-NBD (Fig. 3F), and the other eight Snf7 derivatives (data not shown) remained bound to liposomes. This result was not due to partial inactivation of the Snf781-NBD proteins because the same proteins were completely bound to the same liposomes under different conditions (see below). Instead, the reduced Snf7 binding to liposomes following gel filtration indicates that the membrane association and dissociation rates are faster for Snf7 than for Vps20, even though maximal fluorescence-detected binding was obtained at similar PC/PS/PI concentrations for equivalent concentrations of Snf7 and Vps20.
Fig. 3. Snf7 binding to membranes.
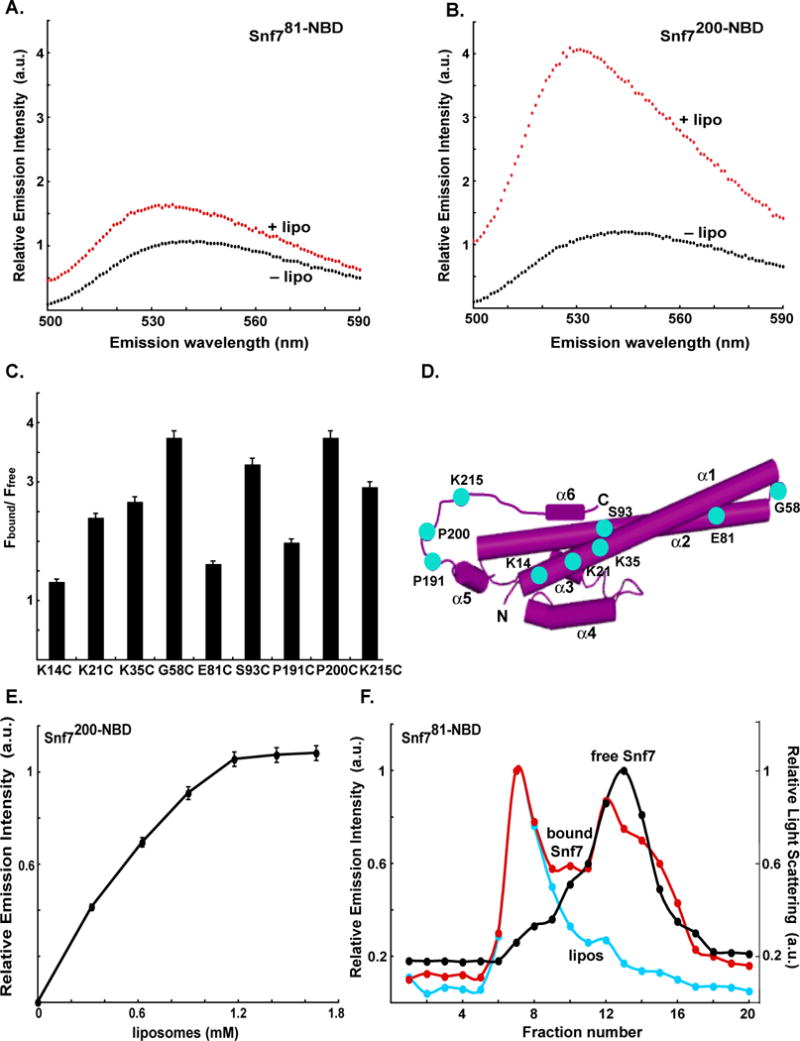
Fluorescence emission spectra of 540 nM Snf781-NBD (A) and 300 nM Snf7200-NBD (B) before (black) and after (red) addition of an excess (1.5 mM; see Fig. 1C) of PC/PS/PI. (C) The ratios of NBD emission intensities at 530 nm are shown for each derivative with (Fbound) and without (Ffree) membranes. (D) Cartoon representation of the water-soluble hVps24 monomer as in Fig. 1F, but here showing the locations of the probes used in the Snf7 experiments (cyan). (E) Titration of 300 nM Snf7200-NBD with liposomes reveals that fluorescence-detected (λem = 530 nm) binding is complete after addition of 1.2 mM PC/PS/PI. (F) Sepharose CL-2B chromatography of 540 nM Snf781-NBD that had been preincubated with either 1.5 mM PC/PS/PI (red) or no liposomes (black). Protein was detected by NBD emission intensity, and liposomes by light scattering (cyan).
Vps20 Appears to Bind to One End of Snf7
When NBD-labeled Snf7 was titrated with Vps20 in solution, probes positioned at opposite ends of Snf7 (Fig. 3D) responded very differently. The emission intensity of an NBD at residue 200 was completely unaffected by the addition of Vps20 (Fig. S7A). In contrast, the intensity of the probe at residue 58 increased substantially (Fig. 4A), either by Vps20 allosterically altering the local Snf7 conformation near residue 58 and/or by Vps20 binding and covering the surface of Snf7 near residue 58. Thus, while one probe is spectroscopically insensitive to Vps20 binding to Snf7, the dependence of the Snf758-NBD spectral change on Vps20 concentration shows that Vps20 and Snf7 associate in solution (Fig. 4B). Such an association is not thought to occur between Vps20 and Snf7 in solution, but the spectral data show that such binding does occur at the non-physiological protein concentrations used in our experiments.
Fig. 4. Vps20 binding to membrane-bound Snf7.
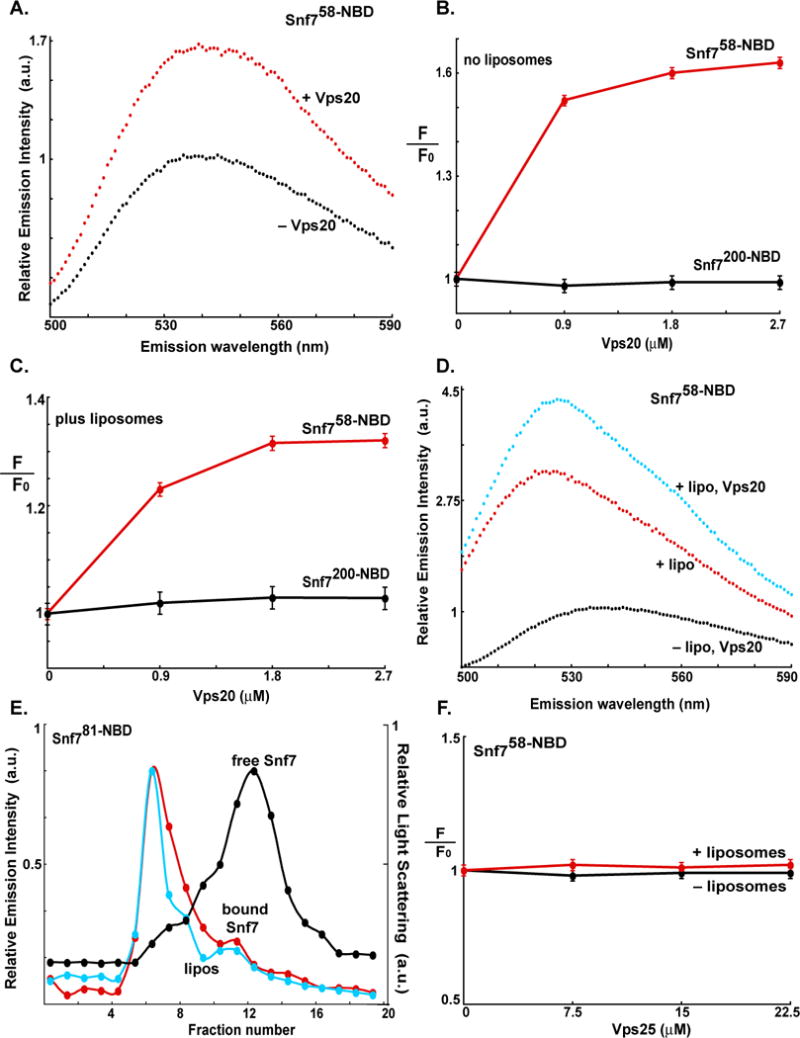
Fluorescence emission spectra of 300 nM Snf7200-NBD (A) and 2.2 μM Snf758-NBD (B) before (black) and after (red) addition of Vps20. Titration (λem = 530 nm) of 300 nM Snf7200-NBD (black) and 2.2 μM Snf758-NBD (red) with Vps20 either before (C) or after incubation with 1.5 mM PC/PS/PI (D). (E) Emission spectra of 2.2 μM Snf758-NBD bound to 1.5 mM PC/PS/PI before (red) and after (cyan) addition of 2.7 μM Vps20. For comparison, the spectrum of 2.2 μM Snf758-NBD in buffer is shown (black). (F) Sepharose CL-2B chromatography of samples containing 540 nM Snf781-NBD and 2.7 μM Vps20 that had been preincubated with either 1.5 mM PC/PS/PI (red) or no liposomes (black). Protein was detected by NBD emission intensity, and liposomes by light scattering (cyan).
Interestingly, the spectral data indicate that Vps20 binds similarly to both free and membrane-bound Snf7. Vps20 addition did not significantly alter emission of both free (Fig. 4B) and membrane-bound (Fig. 4C) Snf7200-NBD, even though the probe at residue 200 was very sensitive to Snf7 binding to the membrane (Fig. 3B, C). The probe at residue 58 was also sensitive to Snf7 binding to the membrane (Fig. 3C), but its environment was further altered by the binding of Vps20 to membrane-bound Snf7 (Fig. 4C, D). Given the similarities in the magnitudes of the Snf758-NBD intensity changes and their dependence on Vps20 concentration (Fig. 4B, C), it appears that the extent of Vps20 binding to free and membrane-bound Snf7 is similar. Since Vps20 binding to Snf7 appears to be unaffected by whether or not Snf7 is bound to a liposome, it suggests that Snf7 interactions with Vps20 and membranes are uncoupled and non-cooperative. However, a Vps20 effect on Snf7 binding to liposomes was revealed by gel filtration. When Snf781-NBD was incubated with both liposomes and Vps20 and then chromatographed, essentially all (>95%) of the Snf7 was found bound to the liposomes (Fig. 4E). Thus, Vps20 markedly stabilizes Snf7 binding to membranes, as is clearly evident by comparing the elution profiles of Snf7 in liposomal samples that either contain (Fig. 4E) or lack (Fig. 3F) Vps20. These results demonstrate that Vps20 functions to stabilize Snf7 binding to the membrane, presumably by changing the Snf7 conformation and reducing its rate of dissociation from the membrane.
As was true with Snf7200-NBD (Fig. 4B), titration of Snf735-NBD with Vps20 had little effect on NBD emission intensity both in the presence and absence of PC/PS/PI (Fig. S7C,D). Hence, the probe at residue 35 was not especially sensitive to Vps20 binding to Snf7. However, Vps20-dependent emission intensity increases were detected for both free and membrane-bound Snf781-NBD, though the spectral changes were small especially for membrane-bound Snf781-NBD (Fig. S7B,C,D).
The probes between α5 and α6 (residue 200) and near the middle of the α1 helix (residues 35 and 21; data not shown) detected Snf7 binding to membranes, but not to Vps20. In contrast, probes at residue 58, at the other end of the protein (according to the hVps24 crystal structure), and at residue 81 in α2 were sensitive to Snf7 binding to both membranes and Vps20. These data strongly suggest that the short Snf7 α1–α2 loop and the α2 helix are sensitive to Vps20 binding both in solution and on the membrane surface, while Vps20 binding has no detectable effect on the conformation or environment of the α5–α6 loop or of α1 for either free or membrane-bound Snf7.
Vps25 Does Not Detectably Bind to Snf7
Since Vps25 binding to Vps20 on the membrane alters Vps20 conformation (Fig. 2D–F), we also determined whether Vps25 binding to Snf7 could be detected spectroscopically. No Vps25-dependent emission intensity changes were detected for both free and membrane-bound Snf758-NBD (Fig 4F) Similarly no change in emission was detected when the NBD probe was placed at any of the other eight probe locations in Snf7 (data not shown). Since Snf7 labeled at multiple sites did not detectably interact with Vps25, it therefore appears that, as suggested by genetic studies, the protein-protein interactions that lead to the formation of the ESCRT assemblies occur in an obligatory sequence: Vps25 in ESCRT-II interacts with Vps20 on the membrane, and then Vps20 in turn promotes Snf7 binding to the membrane.
Snf7 Oligomerization Requires Both Membranes and Vps20
The association of Snf7 into polymeric assemblies was detected directly by using a variation of the fluorescence resonance energy transfer (FRET) technique involving two samples of Snf7 E81C, one labeled with an NBD donor dye and the other with a rhodamine (Rh) acceptor dye. When Snf781-NBD and Snf781-Rh were mixed in solution at final concentrations of 320 nM and 4.2 μM, respectively, no FRET was detected (Table S1). Similarly, no FRET was observed with equimolar mixtures of Snf721-NBD and Snf721-Rh or of Snf721-NBD and Snf781-Rh (Table S1). Thus, no dimers or higher oligomers of Snf7 were detected in solution at this concentration.
However, when liposomes were added to the sample, some FRET was observed between each of these pairs of labeled proteins (Table S1). Thus, upon binding to the liposomal surface, some donor-labeled Snf7 are positioned sufficiently close to acceptor-labeled Snf7 for FRET to be detected. But when Vps20 was added to these samples, a large increase in FRET was observed in each case (Table S1). The maximum FRET efficiencies (E) differed for Snf7 pairs labeled at different sites (Table S1), as would be expected. These differences were not due to incomplete binding of Snf7 to the membrane because the Vps20, Snf7, and liposome concentrations used in the FRET experiments were in excess of that required to maximize Snf7 binding (Fig. 4D,3E). It is therefore clear that Vps20 strongly promotes the oligomerization of Snf7 on the membrane surface, and that both membranes and Vps20 are required for maximal association of Snf7 molecules. This conclusion is dramatically supported by the absence of FRET when the Vps20PW mutant, a variant of Vps20 that is conformationally restricted at the C-terminus (see Snf7PW results below), is added to the sample instead of wild-type Vps20 (Table S1).
Proper determination of E in a membrane-containing sample always requires at least four samples. Emission spectra for donor-containing (D), donor- and acceptor-containing (DA), acceptor-containing (A), and blank (B) samples containing Vps20, liposomes, and Snf781-C labeled with NBD, Rh, or nothing are shown in Fig. 5A. E was quantified by comparing the donor emission intensities at 530 nm in the normalized net D and DA spectra (Fig. 5B). In the absence of Vps20, the donor emission intensity was nearly the same in the normalized net D and DA spectra (Fig. S8). The much higher FRET efficiency observed in the presence of Vps20 (Table S1) is probably due to Vps20-dependent conformational changes in Snf7 that stabilize its binding to the membrane surface (cf. Fig. 4E) and/or stabilize its binding to itself to form multimeric assemblies.
Fig. 5. Oligomerization of Snf7.
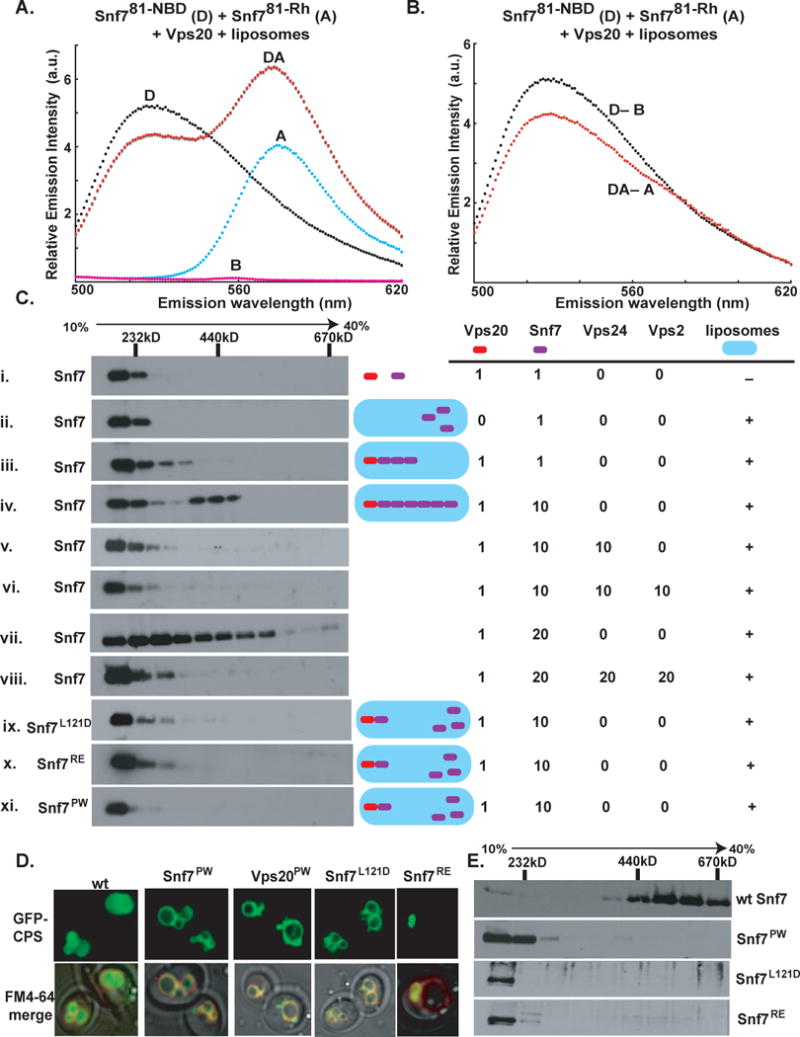
(A) Emission spectrum (λex = 478 nm) of the DA sample (red) containing 320 nM Snf781-NBD, 4.2 μM of Snf781-Rh, 0.5 μM Vps20, and 1.5 mM PC/PS/PI. Unlabeled Snf781-C was substituted for one or both of the fluorescent derivatives to form the D (black), A (cyan), and B (magenta) samples. (B) The normalized net D (black) and net DA (red) emission spectra obtained from the samples in A. (C) Purified ESCRT-III proteins were mixed in different combinations and using different molar ratios (as indicated) in the presence or absence of PC/PS/PI. The liposomal fraction was isolated by centrifugation, detergent-solubilized, and analyzed by velocity sedimentation. (D) In wild-type (wt) cells, GFP-tagged carboxypeptidase-S (GFP-CPS) accumulates in the vacuolar lumen, while the FM4-64 dye stains the limiting membrane of the vacuole. In snf7Δ cells expressing the different mutant forms of Snf7 or in vps20Δ cells expressing the Vps20PW mutant, GFP-CPS co-localizes either with the limiting membrane of the vacuole or the class E compartment. (E) Endosome (P13) fractions prepared from vps4Δsnf7Δ cells expressing either wt Snf7 (top panel) or the different Snf7 mutants were detergent solubilized, and analyzed using glycerol velocity gradients.
The requirements of Vps20 and membranes for inducing Snf7 oligomerization were also observed in a non-spectroscopic assay that detected Snf7 oligomerization in vitro using velocity sedimentation centrifugation on 10%–40% glycerol gradients. The majority of Snf7 sediments in a low molecular weight fraction (Mr<156kD) upon incubation with soluble Vps20 or liposomes alone (Fig. 5Ci,ii). Consistent with the FRET data, we found that addition of Vps20 to a mixture of Snf7 and liposomes triggered Snf7 oligomerization that was exquisitely sensitive to the molar ratio of Vps20 to Snf7. When equimolar Vps20 and Snf7 were mixed in the presence of liposomes, a small amount of Snf7 sedimented in the high molecular weight fractions (Fig. 5Ciii). Since the majority of Snf7 sedimented in the low molecular weight fraction under these conditions, equimolar Vps20 was inefficient in nucleating Snf7 oligomerization, presumably because each Snf7 monomer was bound to Vps20, and no Snf7 monomers are available for oligomerization.
The Vps20:Snf7 molar ratio in yeast cells is ~1:10 (Teis et al., 2008). When Vps20 and Snf7 were mixed at the physiological ratio in the presence of liposomes and sedimented, Snf7 was found in low molecular weight fractions containing monomeric Snf7 and/or Vps20-bound Snf7, and also in high molecular weight fractions (~440kD) that contain Snf7 polymers (Fig. 5Civ). Since a 1:10 molar ratio drove Snf7 oligomerization, a 1:20 Vps20:Snf7 molar ratio was tested. Under these conditions, instead of two distinct species of membrane-bound Snf7 (monomer & oligomer), Snf7 was smeared over the gradient in what appears to be a heterogeneous mixture of different sized Snf7 polymers (Fig. 5Cvii). Thus, Vps20 nucleates Snf7 oligomerization on membranes, but the size of the oligomers is dictated by the relative concentrations of Vps20 and Snf7.
The α5–α6 Loop and Key Residues In and Around α4 are Involved in Snf7 Oligomerization and Function
To test if displacement of the autoinhibitory loop (α5–α6 loop) upon membrane binding (indicated by collisional quenching data; Fig. S5) is a prerequisite for Snf7 oligomerization; we generated the Snf7PW mutant (details in Supplement S.iv). We predicted that bulky tryptophans introduced in the α5–α6 loop would restrict conformational rearrangements of the loop and the movement of α6, thus making the Snf7PW mutant defective in oligomerization. As expected, the Snf7PW mutant did not oligomerize in vitro (Fig. 5Cxi, Table S1). Furthermore, an in vivo experiment performed using vps4Δsnf7Δ cells expressing the Snf7PW mutant revealed that the mutant form of Snf7 does not oligomerize on endosomes (Fig. 5E); whereas wild type Snf7 expressed in the same cells oligomerizes into a 440–600 kDa complex (Fig. 5E). Also, snf7Δ cells expressing the Snf7PW mutant were defective in the vacuolar sorting of GFP-CPS (Fig. 5D). The combined results reveal that the α5–α6 loop is required for Snf7 oligomerization, and this oligomerization is essential for the sorting function of Snf7.
We recently showed that soluble Vps24 forms filament-like structures in vitro, and identified key residues in α4 and the α3–α4 loop that mediate monomer-monomer interactions (Ghazi-Tabatabai et al., 2008). Snf7 contains identical or similar residues at these sites (Fig. S9). To examine the role of these key residues in Snf7 oligomerization, two point mutants of Snf7 (Snf7L121D and Snf7RE; details in Supplement S.iv) were generated. As with the Snf7PW mutant, the two point mutants were impaired in oligomerization both in vitro (Fig. 5Cix,x) and in vivo (Fig. 5E), and consequently, in sorting GFP-CPS to the vacuolar lumen (Fig. 5D).
Vps24 limits Snf7 Oligomerization
We recently showed that deletion of Vps24 results in unrestricted growth of the Snf7 polymer on endosomes in yeast cells (Teis et al., 2008). To test directly if Vps24 is sufficient to limit the growth of Snf7 polymers assembled in vitro, Snf7 was mixed with liposomes, Vps20, Vps24, and sized using glycerol gradients. To ensure that Vps24 was not limiting, Snf7 and Vps24 were mixed at a non-physiological equimolar ratio in the sample. Strikingly, the addition of Vps24 either by itself (Fig. 5Cv) or in equimolar combination with Vps2 (Snf7:Vps24:Vps2 molar ratio of 1:1:1) (Fig. 5Cvi) resulted in almost complete disappearance of the ~440kD Snf7 oligomer. Vps24 therefore effectively blocks Snf7 oligomerization. Vps24 also blocked the formation of the heterogeneous mixture of Snf7 polymers assembled using a Vps20:Snf7 molar ratio of 1:20 (Fig. 5Cviii). While the exact mechanism of Vps24 prevention of Snf7 oligomerization is not yet clear, it seems likely that Vps24 ‘caps’ or prevents polymerization by binding to Snf7. The prevention of Snf7 oligomerization appears to be solely a function of Vps24 because in the absence of Vps24, equimolar (to Snf7) Vps2 did not block Snf7 oligomerization (data not shown).
Vps24, Vps2 and Vps4 Mediate Disassembly of Membrane-bound Snf7 Oligomers
To monitor disassembly of the membrane-bound Snf7 oligomer spectroscopically, six parallel reactions (A–F) containing Snf781-NBD were mixed with Vps20 and NBD emission was monitored. After 600 sec, PC/PS/PI was added to each sample to initiate Vps20-induced Snf7 oligomerization. The resulting intensity changes were equivalent for samples A–F and are shown by a single black trace in Fig. 6A. At 1200 sec, the samples received different combinations of purified proteins. After a 37°C, 45 min incubation, little to no change in NBD intensity was observed in samples lacking Vps24 (A), Vps2 (B), or a functional Vps4 (C). In contrast, the NBD intensity increases were nearly reversed in a sample containing Vps24, Vps2, Vps4, and ATP (E), while a much smaller decrease in NBD emission was observed when ADP replaced ATP (D). Similar results were obtained when samples were prepared with Snf721-NBD, Snf735-NBD, or Snf7215-NBD instead of Snf781-NBD (data not shown).
Fig. 6. ESCRT-III assembly, disassembly and liposome deformation.
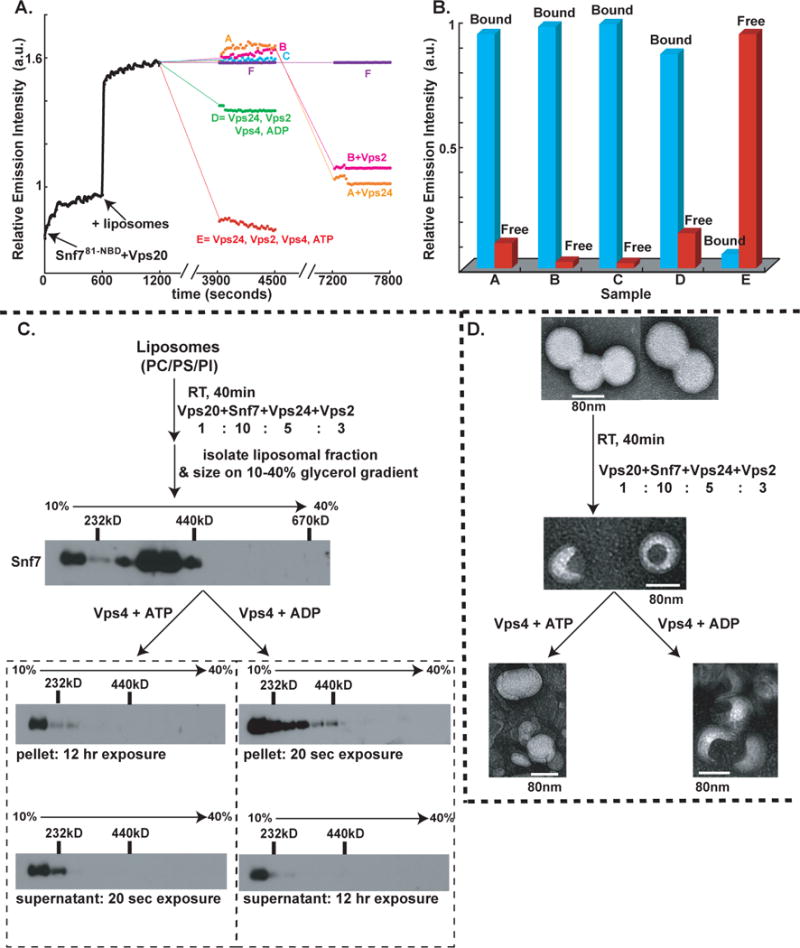
(A) Time- and component-dependent emission intensity profiles. Each reaction sample (A–F) contained 540 nM Snf781-NBD mixed with Vps20 at time 0. At 600 sec, 1.5mM PC/PS/PI was added to each sample. At 1200 sec, additions were made as follows: sample A (orange trace) to 180 nM Vps2, 1 μM Vps4, 1mM ATP; B (magenta) to 270 nM Vps24, 1 μM Vps4, 1mM ATP; C (cyan) to 270 nM Vps24, 180 nM Vps2, 1 μM Vps4 (E233Q), 1mM ATP; D (green) to 270 nM Vps24, 180 nM Vps2, 1 μM Vps4, 1mM ADP; E (red) to 270 nM Vps24, 180 nM Vps2, 1 μM Vps4, 1mM ATP; and F (purple) received only buffer. After incubation (37°C, 45 min), emission was re-measured from 3900–4500 sec at 22°C. Samples A, B, and F then received 270 nM Vps24, 180 nM Vps2, or buffer, respectively, were incubated (37°C, 45 min), and emission was re-measured from 7200–7800 sec. (B) In some experiments, samples were analyzed by Sepharose CL-2B gel filtration chromatography after completing the first 45 min incubation. Protein was detected by NBD emission intensity, and liposomes by light scattering (cyan). The red and blue bars indicate the relative amounts of free Snf781-NBD and liposome-bound Snf781-NBD respectively. (C) Outline of the in vitro ESCRT-III disassembly assay. ESCRT-III complex was assembled on liposomes using purified ESCRT-III proteins and centrifuged to separate the liposome-bound complex The liposome-bound ESCRT-III complex was treated with either (i) Vps4 and ATP or (ii) Vps4 and ADP for 45 min at 37°C. Following incubation, the samples were centrifuged to separate the liposome-bound proteins (“pellet”) from free ESCRT-III proteins (“supernatant”), and then further analyzed by velocity sedimentation. (D) Negative stain EM analyses of PC/PS/PI liposomes, ESCRT-III bound liposomes, ESCRT-III-bound liposomes treated with 1μM Vps4 + 1mM ATP, and ESCRT-III-bound liposomes treated with 1 μM Vps4 + 1mM ADP.
To determine whether the large drop in NBD emission intensity reflects Vps4-mediated release of NBD-labeled Snf7 from the membrane into the aqueous milieu, samples were analyzed by gel filtration after the 37°C incubation. Nearly all of the NBD emission in the complete sample (E) co-eluted with liposome-free Snf7 (red bar in Fig. 6B), and very little co-eluted in the void volume with the liposomes (detected by light scattering; blue bar in Fig. 6B), thereby showing efficient Snf7 release from the liposomes. In contrast, when the other samples (A–D) were analyzed by gel filtration, nearly all of the NBD emission co-eluted with liposomes in the void volume (Fig. 6B; data not shown).
Since membrane-bound Snf7 was not released into the aqueous milieu in samples lacking only Vps24 or Vps2, the missing proteins were then added to A and B, respectively. After 45 min at 37°C, the lowered NBD intensity shows that Vps4-mediated Snf7 disassembly now occurred in each sample (Fig. 6A; the reduced intensity decrease compared to sample E presumably results from ATP hydrolysis during the second 37°C incubation). Thus, efficient release/disassembly of membrane-bound Snf7 requires Vps24, Vps2, the ATPase activity of Vps4, and ATP.
Disassembly of liposome-bound Snf7 oligomers was also examined by glycerol gradient sizing assays. ESCRT-III complexes were assembled on liposomes in vitro by combining Vps20, Snf7, Vps24 and Vps2 at their previously-determined physiological molar ratio of 1:10:5:3, and their ~400 kD mass [as judged by immunoblotting with Snf7 (Fig. 6C) or Vps24 (data not shown) antibodies] was very similar to the masses of ESCRT-III complexes that assemble on endosomes in vivo (Teis et al., 2008; no upstream ESCRT components were required for assembly presumably because of the high ESCRT-III protein concentrations used in the assay). The liposome fraction was isolated by centrifugation and split in two. One half was mixed with Vps4 and ATP, while the other half received equivalent amounts of Vps4 and ADP. After another 45 min at 37°C incubation, the liposomes were separated from soluble proteins by centrifugation, and both the pellet and supernatant fractions were further analyzed by glycerol gradient sizing. As shown in Fig. 6C, the liposome-bound ~400kD ESCRT-III complex was disassembled by exposure to Vps4 and ATP. Most Snf7 was detected in the supernatant-derived fractions by immunoblotting with Snf7 antibodies (20 sec exposure), while Snf7 could only be detected in the pellet-derived fractions after long exposure (12 hr). Thus Vps4- and ATP-mediated release of Snf7 from the liposomes into the supernatant was very efficient. Interestingly, Vps4 appears to disassemble the membrane-bound Snf7 oligomers into monomers because Snf7 present in the supernatant was found in the low molecular weight fractions of the glycerol gradient (Fig. 6C).
The ATP dependence of the disassembly reaction was shown by the inability of the ATPase inactivated Vps4 E233Q to disassemble the membrane-bound Snf7 oligomer even in the presence of ATP (data not shown). Also, Vps4-mediated disassembly of membrane-bound Snf7 oligomers was much less efficient with ADP than with ATP since the majority of Snf7 was found in the pellet-derived fractions (20 sec exposure); only a small amount of Snf7 was found in the supernatant after a 12-hr exposure (Fig. 6C). The size of the membrane-bound ESCRT-III complex was slightly smaller after than before treatment with Vps4 and ADP. We attribute this to one round of Vps4-mediated disassembly that utilizes the ATP that may remain bound to Vps4 during purification. Such an effect would also explain the small drop in NBD emission observed in sample D in our spectroscopic Snf7 disassembly assay (Fig. 6A). Consistent with the spectral data, no Vps4- and ATP-mediated disassembly of the membrane-bound Snf7 oligomer was observed in the absence of either Vps24 or Vps2 (data not shown).
ESCRT-III Assembly Induces Membrane Deformation
To determine if the assembly and disassembly of the Snf7 oligomer induces membrane deformation, we analyzed liposome morphology at different stages of the ESCRT-III disassembly assay using negative stain electron microscopy (EM). Untreated PC/PS/PI liposomes prepared by extrusion have a uniform, spherical appearance with a diameter of 80–100 nm (Fig. 6D). Upon exposure to the ESCRT-III complex (Vps20, Snf7, Vps24, Vps2), a distinct change in morphology was observed for ~90% of the liposomes. ESCRT-III-bound liposomes had an inward invaginated appearance, and the size of the invagination was ~40 nm in diameter. This deformation was observed only when liposomes were treated with all four ESCRT-III proteins: incubation with individual ESCRT-III proteins did not induce any detectable liposome deformation (data not shown). Also, no liposome morphology changes were observed when mutants of Snf7 that do not oligomerize (Snf7PW, Snf7L121D and Snf7RE) were mixed with Vps20, Vps24 and Vps2 (data not shown). These data reveal that Snf7 oligomerization is critical for membrane invagination.
Importantly, when ESCRT-III-bound liposomes were exposed to Vps4 and ATP, the near normal spherical morphology of the liposomes was restored (Fig. 6D). In contrast, ESCRT-III-bound liposomes retained their invaginated appearance when treated with Vps4 and ADP. The reversibility of ESCRT-III-induced liposome deformation upon treatment with Vps4 and ATP, coupled with the other data presented above, demonstrate directly that liposome morphology is dictated by the assembly and disassembly of the ESCRT-III complex.
Discussion
The ordered [ESCRT-II (Vps25)-Vps20-Snf7-Vps24-Vps2] assembly and disassembly of the ESCRT-III complex has been reconstituted in vitro using purified proteins, fluorescent-labeled derivatives of those proteins, liposomes comprised of PC/PS/PI, and multiple independent fluorescence and other techniques. Specifically, our studies reveal that: (i) Vps25 binds to Vps20, but not to Snf7; (ii) both Vps20 and Snf7 bind to a membrane surface; (iii) Vps25 binding to membrane-bound Vps20 alters its conformation; (iv) Vps20 binds the N-terminal end of membrane-bound Snf7, and this association both stabilizes Snf7 binding to the membrane surface and alters its conformation; (v) Vps20 nucleates Snf7 oligomerization on the membrane surface; (vi) Vps20 stabilizes and/or changes the conformation of the Snf7 oligomers; (vii) Vps24 caps membrane-bound Snf7 oligomers; (viii) Snf7 oligomerization is blocked by mutations just C-terminal of the α5 helix and by mutation of critical residues in and around helix α4; (ix) membrane-bound ESCRT-III complex is disassembled by Vps4 in a process that requires Vps24, Vps2 and ATP; (x) liposome-bound ESCRT-III complex induces membrane deformation, and Snf7 oligomerization is critical for this deformation. When considered in toto, these results suggest that the ESCRT-III functional cycle consists of an ordered multistep assembly involving three distinct stages (filament/lattice nucleation, polymerization, and capping), followed by an ATP-dependent disassembly reaction. These complex, coupled, and regulated structural changes ultimately mediate membrane deformation, cargo sorting, and MVB vesicle formation (Fig. 7).
Fig. 7. Speculative model for the ESCRT-III reaction cycle.
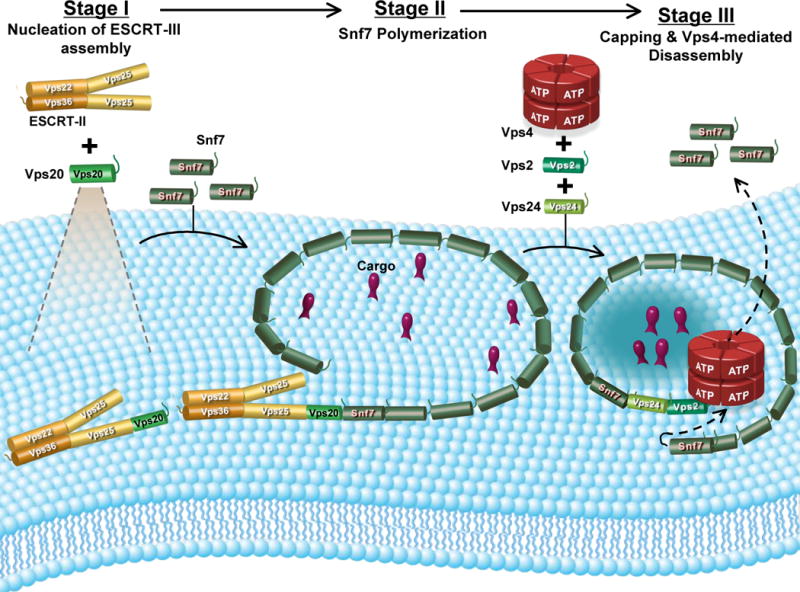
ESCRT-III assembly is initiated when Vps20 binds to Vps25 on the membrane surface. In the second stage, Snf7 binds to the membrane-bound Vps25-Vps20 complex, and this triggers the formation of a membrane-bound Snf7 oligomer. The Snf7 oligomer traps cargo into a localized sorting domain and induces membrane deformation. Vps24 caps the Snf7 oligomer, and interactions between Vps2 and Vps4 initiate membrane disassembly of the ESCRT-III complex (see Discussion).
ESCRT-III assembly involves an obligatory sequence of protein-protein interactions & conformational rearrangements
The fluorescence-detected conformational changes in the α5–α6 loop strongly suggest that they are required to nucleate the assembly of the ESCRT-III complex. Such a conformational change was proposed earlier to mediate the association of ESCRT-III subunits by an intramolecular autoinhibition of ESCRT-III polymer formation (Shim et al., 2007). The autoinhibition model proposes that ESCRT-III subunits exist as metastable ‘closed’ monomers in the cytosol because the C-terminal autoinhibitory region of the molecule (including α5 and α6) binds to α2 of the core and thus prevents hetero- and homo-dimerization. Consistent with this, our results indicate that Vps25 has no affinity for Vps20 in solution (Fig. 2A–C;S4B), there is no energy transfer between NBD- and Rh-labeled Snf7 molecules in solution (Table S1), and no oligomerization of Snf7 in solution (Fig. 5Ci). Implicit in the autoinhibition model is the idea that membrane binding and/or interactions with ESCRT-II generate the ‘open’ state of the ESCRT-III subunit that is active for assembly into the ESCRT-III lattice/filament. The spectral data presented here show that NBD probes incorporated within the α5–α6 loop of both Vps20 and Snf7 exhibit a dramatic increase in NBD emission intensity upon membrane binding (Fig. 1E,3B,3C), and – more importantly – that NBD emission from these sites can be efficiently quenched by membrane-restricted quenchers (Fig. S3C,S5B,S3F). These data therefore directly show that membrane binding of an ESCRT-III subunit is accompanied by the movement of its C-terminal α5–α6 loop adjacent to the membrane surface. Rearrangement of the C-terminal end of both Vps20 and Snf7 upon membrane binding may expose α2 in each protein for homo- and hetero-dimerization interactions, and thereby activate the ESCRT-III subunit (‘closed’ to ‘open’ state transition) for continued assembly into the ESCRT-III lattice. Consequently, mutations that block conformational flexibility around the α5–α6 loop impair Snf7 oligomerization and function (Fig. 5Cxi,5D,5E,Table S1).
Spectroscopic characterization of Vps25 interactions with NBD-labeled Vps20 mutants revealed that the binding of Vps25 to Vps20 changed its conformation at both the N- and C-terminus. Thus, although the N-terminal domain of Vps20 undergoes a larger spectral change and perhaps a larger conformational change upon association with Vps25 (Fig. 2D–F;S4A,C), Vps25 binding clearly elicits a long-range conformational change in Vps20 that extends throughout its entire length (see Supplement). Similarly, spectroscopic examination of Snf7 mutants containing NBD probes at multiple locations and domains within the molecule allowed us to determine possible sites on Snf7 that bind to Vps20. Our data indicate that the short Snf7 α1–α2 loop and the α2 helix are sensitive to Vps20 binding both in solution and on the membrane (Fig. 4A–D;S7B–D). In contrast, Vps20 binding had no detectable effect on the probe environment and presumably conformation of the C-terminal α5–α6 loop or of α1 for either free or membrane-bound Snf7 (Fig. 4B,C;S7A,C,D). These results strongly indicate that the hetero-dimerization interactions during the assembly of the ESCRT-III lattice are mediated by interactions involving the α2 helix and the short α1–α2 loop.
As was true with Vps20, Snf7 exhibited low affinity for Vps25 in solution (Fig. 4F). However, in contrast to Vps20, we also did not detect binding of Vps25 to any of the membrane-bound Snf7 monocysteine mutants (Fig. 4F; data not shown). Specifically worth noting is the probe at residue 58 which is sensitive to Snf7 binding both to membranes (Fig. 3C) and to Vps20 (Fig. 4A–D) but remains insensitive to Vps25 both in solution and when liposome-bound (Fig. 4F). When combined, these data indicate that an obligatory sequence of protein-protein interactions mediates assembly of the ESCRT-III complex (Fig. 7).
How then does the system regulate interactions between the structurally similar ESCRT-III proteins that associate via a common surface? The most likely explanation is that individual interactions elicit unique conformational changes, and that the next component in the assembly pathway binds only to a specific ‘active/open’ conformation. In this way, one can ensure that ESCRT-III assembly occurs in the proper order because a component can only associate with the precursor complex that has the necessary conformation. The spectral changes detected along the entire length of the Snf7 molecule upon membrane binding and interactions with Vps20 correlate with alterations in protein conformation that shift the functional/structural state of the protein from one state to another (‘closed’ to ‘open’ state transition) and thereby activate it for assembly into the ESCRT-III lattice (Fig. 7).
Specific function for each protein in the ESCRT-III reaction cycle
The assembly of ESCRT-III on a membrane surface has striking mechanistic analogies to the assembly of filaments like actin or tubulin, where ESCRT-III appears to form a polarized filament that assembles in a sequential fashion with distinct nucleation and capping sites (Fig. 7). Our results indicate that Vps20 plays a pivotal role in the assembly of a membrane-bound Snf7 polymer by stabilizing the binding of Snf7 derivatives to the membrane surface (Fig. 4E) and by nucleating Snf7 oligomerization either by stabilizing the binding of Snf7 to itself to form multimeric assemblies and/or by altering the conformation of membrane-bound Snf7 to allow the assembly of an oligomer (Fig. 5A–E;S8;Table S1). Interestingly, the ability of Vps20 to nucleate Snf7 oligomerization in vitro (Fig. 5A–C) and in vivo (Fig. S10) appears to be independent of its N-terminal myristoylation.
Vps20-induced Snf7 oligomerization on the membrane appears to be critical for membrane deformation as judged by the fact that Snf7 mutants that do not oligomerize (in vitro and in vivo) are defective in inducing changes in liposome morphology in vitro and in MVB sorting in vivo (Fig. 5Cix–xi,5D,5E;data not shown). How ESCRT-III induced liposome deformation in vitro relates to MVB formation in vivo remains an open question. However, the strong dependence of the observed liposome deformation on Snf7 oligomerization (Fig. 5C,data not shown), and the Vps4-mediated reversibility of the deformation (Fig. 6D) suggest a direct link between Snf7 oligomerization and membrane deformation. In addition to inducing membrane deformation, Snf7 oligomers could form rings, similar to the ring-like spiral filaments generated at the plasma membrane in mammalian cells over-expressing hSnf7 (Hanson et al., 2008). A Snf7 membrane-associated ring/spiral could encircle cargo molecules and keep them committed to MVB sorting by confining the integral membrane cargo proteins to a sorting domain from which they cannot escape (Snf7 filaments tightly associate with the membrane surface) (Fig. 7). Recent work has shown that Snf7 oligomers (filaments) associated with the endosomal membrane may play a direct role in cargo sequestration possibly by restricting lateral movement of cargo molecules (Teis et al., 2008).
Our data suggest that Vps24 and Vps2 serve as adaptors for Vps4-mediated disassembly of the Snf7 oligomer (Fig. 6A). The data presented here (Fig. 5C,6) and results of in vivo studies (Teis et al., 2008; Obita et al., 2007; Stuchell-Brereton et al., 2007) are consistent with a model in which Vps24 terminates oligomerization, possibly by capping Snf7 oligomers and/or by recruiting Vps2 to the Snf7 oligomer in order to initiate Vps4-mediated disassembly (Fig. 7). The Vps24-Vps2 ‘capping’ complex may itself be in the form of a short mixed oligomer (Lata et al., 2008), thereby providing multiple binding sites (Vps2 MIM domain) for Vps4 recruitment via the Vps4 MIT domain (Obita et al., 2007). Although not addressed by the present study, we speculate that the Vps4 AAA ATPase may drive the sequential release/disassembly of Snf7 molecules from one end of the Snf7 filament ring/spiral. This would result in closure of the Snf7 ring/spiral and concentration of cargo encircled by the ring (Fig. 7). The Vps4 dodecamer could stabilize the ever-shrinking Snf7 ring via a combination of strong interactions with Vps2, and weak interactions with Snf7 mediated by the N-terminal MIT domain on each Vps4 subunit (Obita et al., 2007). The closure of the Snf7 ring/spiral may also drive invagination and fission of the sorting domain/vesicle thereby directly coupling ESCRT-III disassembly to cargo sorting and vesicle formation (purse string model). Clearly, this speculative hypothesis will require considerable new experimentation. However, it does offer a framework that could explain the mechanism by which ESCRT-III can sort cargo (even after the Ub sorting signal has been cleaved), deform the membrane, and ultimately release the invaginated MVB vesicle into the lumen of the endosome.
Experimental Procedures
Purification & labeling of ESCRT-III proteins
Recombinant Vps20 and Snf7 were expressed in C41(DE3) cells and purified using a combination of Ni2+-affinity and ion exchange chromatography. Purified proteins were labeled with NBD as described earlier (Shepard et al., 1998). Details of the protein purification and NBD-modification procedures are presented in the Supplement.
Liposome preparation
Liposomes containing 100 mol% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC; Avanti) or 80 mol% POPC, 15 mol% egg-phosphatidyl serine (PS), and 5 mol% egg-phosphatidyl inositol (PI) were prepared by extrusion through a Liposofast extruder using 100-nm pore polycarbonate membranes as before (Shepard et al., 1998).
Fluorescence spectroscopy
All intensity and anisotropy measurements were carried out in 50mM HEPES (pH 7.5), 100mM NaCl at 22°C using the same steady-state instrumentation and procedures described previously (Shepard et al., 1998). Emission spectra for NBD-labeled proteins were recorded in 4 mm × 4 mm quartz cuvettes coated with POPC to minimize adsorption. See Supplement for further details on fluorescence spectroscopy, fluorescence microscopy, and FRET experiments.
Glycerol velocity gradients
Endosome-enriched P13 fractions were prepared and solubilized with 0.5% Tween-20 and analyzed using a glycerol velocity gradient as before (Teis et al., 2008). Analysis details are given in the Supplement.
Supplementary Material
Acknowledgments
We are grateful to Y. Miao, Y. Shao, S. Weys for technical assistance; B. Judson for EM expertise; and E.V. MacGurn for assistance with graphics. S.S. and D.T. were supported by fellowships from American Heart Association (AHA 0826060D) and Human Frontier Science Program (LT00634/2006-L) respectively. This work was supported by NIH grant GM26494 and the Robert A. Welch Foundation (Chair Grant BE-0017) (A.E.J.) and funds from the Weill Institute for Cell and Molecular Biology (S.D.E.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, Emr SD, Egelman EH, Williams RL. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AE. Fluorescence approaches for determining protein conformations, interactions and mechanisms at membranes. Traffic. 2005;6:1078–1092. doi: 10.1111/j.1600-0854.2005.00340.x. [DOI] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzioł T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Göttlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Shepard LA, Heuck AP, Hamman BD, Rossjohn J, Parker MW, Ryan KR, Johnson AE, Tweten RK. Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: an alpha-helical to beta-sheet transition identified by fluorescence spectroscopy. Biochemistry. 1998;37:14563–14574. doi: 10.1021/bi981452f. [DOI] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


