Abstract
Ursine bears are a mammalian subfamily that comprises six morphologically and ecologically distinct extant species. Previous phylogenetic analyses of concatenated nuclear genes could not resolve all relationships among bears, and appeared to conflict with the mitochondrial phylogeny. Evolutionary processes such as incomplete lineage sorting and introgression can cause gene tree discordance and complicate phylogenetic inferences, but are not accounted for in phylogenetic analyses of concatenated data. We generated a high-resolution data set of autosomal introns from several individuals per species and of Y-chromosomal markers. Incorporating intraspecific variability in coalescence-based phylogenetic and gene flow estimation approaches, we traced the genealogical history of individual alleles. Considerable heterogeneity among nuclear loci and discordance between nuclear and mitochondrial phylogenies were found. A species tree with divergence time estimates indicated that ursine bears diversified within less than 2 My. Consistent with a complex branching order within a clade of Asian bear species, we identified unidirectional gene flow from Asian black into sloth bears. Moreover, gene flow detected from brown into American black bears can explain the conflicting placement of the American black bear in mitochondrial and nuclear phylogenies. These results highlight that both incomplete lineage sorting and introgression are prominent evolutionary forces even on time scales up to several million years. Complex evolutionary patterns are not adequately captured by strictly bifurcating models, and can only be fully understood when analyzing multiple independently inherited loci in a coalescence framework. Phylogenetic incongruence among gene trees hence needs to be recognized as a biologically meaningful signal.
Keywords: species tree, introgressive hybridization, Ursidae, phylogenetic network, coalescence, multi-locus analyses
Introduction
Our understanding of evolutionary processes relies on a backbone of phylogenetic inferences from molecular data, but recombination imposes limits on the resolution that can be obtained from a single autosomal locus. High-resolution phylogenies can be obtained in multilocus analyses. In traditional phylogenetic analyses, several loci are concatenated and analyzed as one “superlocus.” However, incomplete lineage sorting (ILS), a process by which ancestral polymorphisms can persist through species divergences up to several million years, and gene flow across species boundaries caused by introgressive hybridization generate gene tree discordance, hampering species tree estimation (Tajima 1983; Pamilo and Nei 1988; Leaché et al. 2014). These evolutionary processes are not considered in phylogenetic analyses of concatenated data and can result in inconsistent phylogenetic estimates and high statistical support for an incorrect species tree topology (Kubatko and Degnan 2007).
Bears (Ursidae) are emerging as a prominent example of a mammalian family with a complex speciation history, showing discrepancies among mitochondrial and nuclear phylogenies (Yu et al. 2007; Krause et al. 2008; Nakagome et al. 2008; Pagès et al. 2008; Hailer et al. 2012, 2013; Miller et al. 2012; Cahill et al. 2013). Within bears, the ursine subfamily comprises the American and Asian black bear (Ursus americanus, U. thibetanus), sun bear (Helarctos malayanus), sloth bear (Melursus ursinus), brown bear (U. arctos), polar bear (U. maritimus), plus numerous extinct taxa. In addition, bears also include the giant panda (Ailuropoda melanoleuca) and spectacled bear (Tremarctos ornatus). In phylogenetic analyses of genes from the nuclear genome, the placement of the sun bear, sloth bear, and Asian black bear remained unclear (Yu et al. 2004; Nakagome et al. 2008; Pagès et al. 2008). These analyses were performed using a combination of intron and exon sequences, rendering it difficult to interpret whether nodes with low statistical support resulted from insufficient resolution or from actual conflict in evolutionary signals among loci. Moreover, in these studies only one (consensus) sequence per species was analyzed and data from several markers were concatenated, precluding the identification of paraphyletic relationships among species.
Recently, coalescence-based multilocus species tree approaches have been developed (e.g., Heled and Drummond 2010). These analytical advances make it possible to specifically model the complexity of lineage sorting and to incorporate intraspecific variation and heterozygosity within individuals. Accuracy of such multilocus species trees can be additionally improved by sampling several individuals per species, especially at shallow phylogenetic depths at which lineages are not completely sorted (Maddison and Knowles 2006). This is especially relevant in ursine bears, because the fossil record and dated phylogenies of mitochondrial genome sequences suggested a rapid radiation (Wayne et al. 1991; Yu et al. 2007; Krause et al. 2008), including time frames in which ILS is expected (Nichols 2001).
Another cause of gene tree discordance can be introgressive hybridization, resulting in gene flow across species boundaries, which can only be estimated when intraspecific variation is considered. Although ILS can be modeled in currently available species tree approaches, they cannot account for gene flow. A recent simulation study showed that gene flow can affect species tree inferences by decreasing posterior clade probabilities, underestimating divergence time estimates, and, in cases of high levels of gene flow, by altering the species tree topology (Leaché et al. 2014). Discordance among loci that differ in ploidy and inheritance mode can be explained by contrasting patterns of female and male gene flow (Chan and Levin 2005). In brown and polar bears, discordance between the mitochondrial gene tree and the nuclear species tree has been found (Hailer et al. 2012, 2013; Miller et al. 2012; Cronin et al. 2013), and explained with introgressive hybridization. Previous studies have also indicated phylogenetic discrepancies between mitochondrial and nuclear genes in American and Asian black bears (Yu et al. 2004; Nakagome et al. 2008; Pagès et al. 2008), suggesting that similar processes may have affected their evolution. To examine whether incongruences among nuclear loci and/or discordance between nuclear and mitochondrial phylogenies can be explained by introgression, coalescence-based multilocus gene flow analyses (e.g., Nielsen and Wakeley 2001; Hey 2010; Yu et al. 2012, 2013) can be used to complement species tree inferences. Thus, to more fully understand the evolutionary history of bears, it is crucial to analyze multiple independently inherited markers with a high resolution in several individuals per species. Such data sets need to be analyzed using coalescence models, tracing the evolutionary histories of individual alleles back in time, from extant individuals to their ancestral populations.
We here study the evolutionary history of bears, using a combination of coalescence-based species tree approaches and gene flow analyses. For this purpose, we generated sequence data of 14 independently inherited autosomal introns in 30 individuals and of 5.9 kb from the Y chromosome in 11 males from all eight extant bear species. We combine this with previous data into data sets comprising 29 kb of nuclear sequence and 10.8 kb of mitochondrial sequence to analyze the complexity of phylogenetic signals in bears through multilocus species tree and network analyses, and in statistical model comparisons. Further, we use coalescent-based gene flow analyses to specifically investigate whether remaining conflicts in phylogenetic signals in bears can be explained by introgressive hybridization.
Results
Basic Variability Statistics and Allele Sharing among Ursinae
We sequenced 14 autosomal introns from two to seven individuals per species yielding 7,991 bp, and nine markers from the Y chromosome yielding 5,907 bp in 11 male individuals, representing all extant bear species (supplementary table S1, Supplementary Material online). For giant panda, spectacled bear, sloth bear, sun bear, and Asian black bear, Y-chromosomal data were obtained from all available male individuals. Because of low intraspecific variability of Y chromosomes in brown, polar, and American black bears (Bidon et al. 2014), we included Y-chromosomal data from only one individual of each of these species.
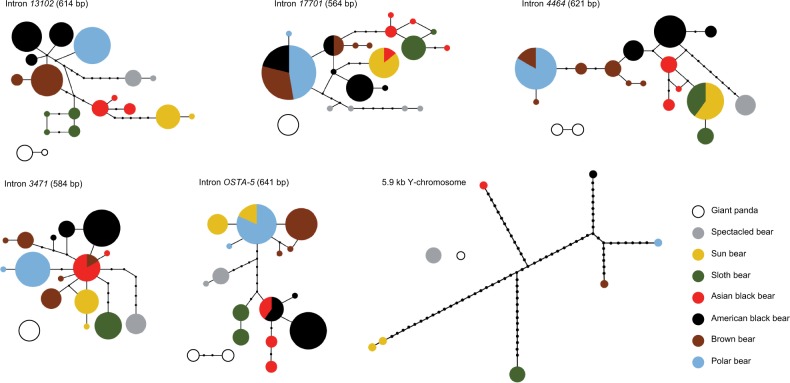
The number of variable sites was 515 across the 14 sequenced autosomal introns and 325 at Y-chromosomal sequence. The total sequence data generated in this study thus comprised 840 variable sites. In contrast, upon concatenation of the autosomal intron data, collapsing all variation within and among individuals into a 50% majority-rule consensus sequence per species, only 396 variable sites remained. Thus, intraspecific and intraindividual polymorphism contributed more than 30% to the phylogenetic signal in our autosomal data. Accordingly, interspecific p-distances of our autosomal introns including all phased individuals were on average 115% of the p-distances of the same 14 concatenated autosomal introns, and on average 178% of the p-distances of previously published autosomal sequences that did not consider intraspecific variability and that included both exon and intron sequences (Pagès et al. 2008; supplementary table S2, Supplementary Material online). High levels of shared polymorphisms were found between brown and Asian black bears, between American black and Asian black bears, and between brown and American black bears (supplementary table S3, Supplementary Material online). All ursine species pairs had similar mean genetic distances. Haplotype networks revealed various combinations of interspecific haplotype sharing for 12 of 14 autosomal introns (fig. 1, supplementary fig. S1 and table S4, Supplementary Material online). At eight introns, haplotypes were shared between closely related species, and at four introns, haplotypes were shared between more distantly related species. Across pairwise comparisons among species, the ratio of polymorphic sites to fixed differences increased toward shallower divergences (supplementary table S3, Supplementary Material online).
Fig. 1.
Statistical parsimony networks for five autosomal intron markers and 5.9 kb of Y-chromosomal sequence in bears. Circle areas are proportional to haplotype frequencies and inferred intermediate states are shown as black dots. For some loci, spectacled bear and giant panda haplotypes were too divergent to be connected at the 95% credibility limit. Likewise, in the Y-chromosomal data set, sun bear haplotypes were connected at the 94% credibility limit. Haplotype networks for nine additional autosomal intron markers are shown in supplementary figure S1, Supplementary Material online.
Haplotype networks showed Y chromosomes from different species as clearly distinct from each other (fig. 1). In contrast to autosomal markers, no haplotype sharing was found. At marker 579.3C, a large insertion in sloth and sun bears (222 and 221 bp, respectively) was 93% identical to a transposable element from the giant panda (SINEC1_Ame). Mean pairwise distances between species were similar for the Y-chromosomal and autosomal data sets, when at least one of the compared species was giant panda or spectacled bear (supplementary table S3, Supplementary Material online). Within Ursinae, however, relatively fewer Y-chromosomal than autosomal substitutions were observed, a pattern also reported by Nakagome et al. (2008). We found a total of three pseudoheterozygous sites on the Y chromosome, all located within 119 bp of marker 403. The respective columns were removed from the alignment prior to any analysis. Pseudoheterozygous sites on the generally haploid Y chromosome can occur due to segmental duplications (Sachidanandam et al. 2001; Hallast et al. 2013).
Multilocus Species Tree Analyses
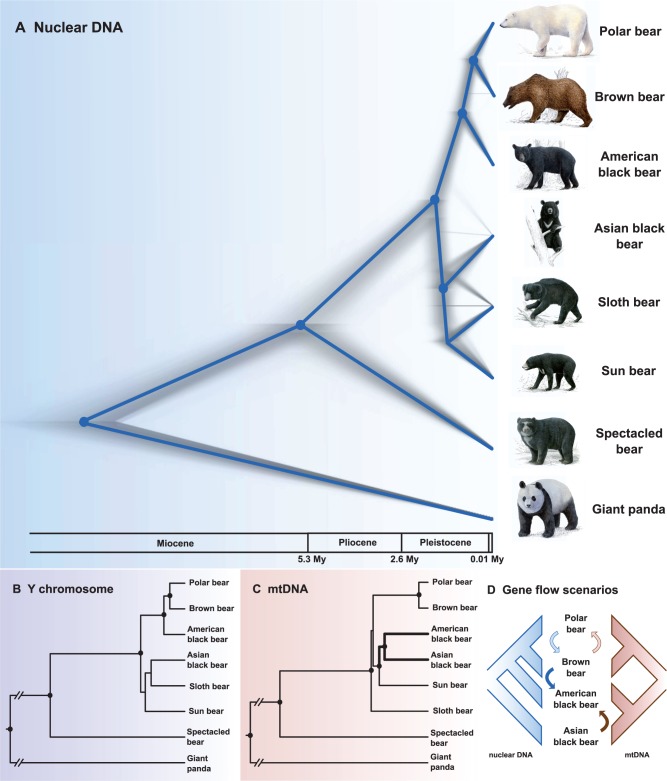
*BEAST, a multilocus coalescence approach, jointly estimates gene trees from independently inherited loci, as well as the species tree in which the gene trees are embedded. By including two phased haplotypes per individual and autosomal locus, and data from several individuals per species, variation among and within individuals could be explicitly considered. A multilocus analysis of all nuclear markers from this study yielded a topology placing the American black bear as sister-taxon to a brown/polar bear clade, which was supported by high posterior probability (fig. 2A). A clade consisting of Asian black, sun, and sloth bears was recovered with high statistical support. Topological uncertainty within this clade was represented in a cloudogram of species trees sampled from the posterior distribution (Bouckaert 2010) by lines (topologies) connecting the sloth bear with the Asian black bear, and a horizontal line indicating a placement of the sloth bear as sister-taxon to sun and Asian black bear (fig. 2A). A topology placing the Asian black bear as sister-taxon to the American black bear, brown bear, and polar bear was represented by faint lines in the cloudogram. Conflicting signals in our nuclear data were further illustrated in a consensus network of the 14 autosomal gene trees from all phased individuals (fig.3). Although there was a clear separation between an American black, brown, polar bear clade on the one side and an Asian black, sun, sloth bear clade on the other side, the topology deviated from a bifurcating tree. In particular, conflict among Asian black, sun, and sloth bears was depicted by a cuboid, and brown and American black bears were grouped closely together. Using a minimum estimate of 11.6 Ma for the divergence time of the giant panda from the other bear species resulted in a divergence time estimate of the ursine bears from the spectacled bear around the transition from the Miocene to the Pliocene (median: 5.88 Ma; fig. 2A, table 1). The divergence between the Asian black, sun, sloth bear clade and the American black, brown, polar bear clade was placed to the early Pleistocene (median: 1.78 Ma). Subsequent divergences within Ursinae occurred during the Pleistocene, within about 1.8 My. The average median posterior estimate of the substitution rate across loci obtained from our calibrated *BEAST analysis was 0.95 × 10−8 substitutions per site per generation, assuming an average generation time for bears of 7.2 years.
Fig. 2.
(A) Cloudogram of species trees from *BEAST analysis, based on 14 autosomal introns and 5.9 kb of Y-chromosomal sequence (90,000 species trees). The consensus tree of the most frequently occurring topology in the posterior distribution is superimposed onto the cloudogram in blue. Blue dots at nodes indicate posterior support >0.96 in the maximum-clade-credibility tree. Frequency of different topologies occurring in the posterior distribution is illustrated by width and intensity of grey branches. Variation in density along the x axis portrays variation in time estimates of divergences. (B) Gene tree of 5.9-kb Y-chromosomal sequence from BEAST. Note that in a *BEAST analysis of the 14 autosomal introns alone, the same topology was obtained, with low statistical support (P < 0.95) for a clade of Asian black bears, sun bears, and sloth bears (data not shown). (C) Gene tree of mitochondrial genome data (protein-coding regions, excluding ND6) from BEAST. Black dots at nodes indicate posterior support >0.95. (D) Schematic scenarios for interspecific gene flow that could explain discordance between mitochondrial and nuclear phylogenies. Blue arrows: Nuclear gene flow, brown arrows: Introgression of mtDNA. Light blue and light brown arrows indicate gene flow identified in previous studies (Hailer et al. 2012, 2013; Miller et al. 2012; Cahill et al. 2013; Liu et al. 2014). Note that IMa2 identified additional introgression signals from Asian black into sloth bears (supplementary fig. S3B, Supplementary Material online).
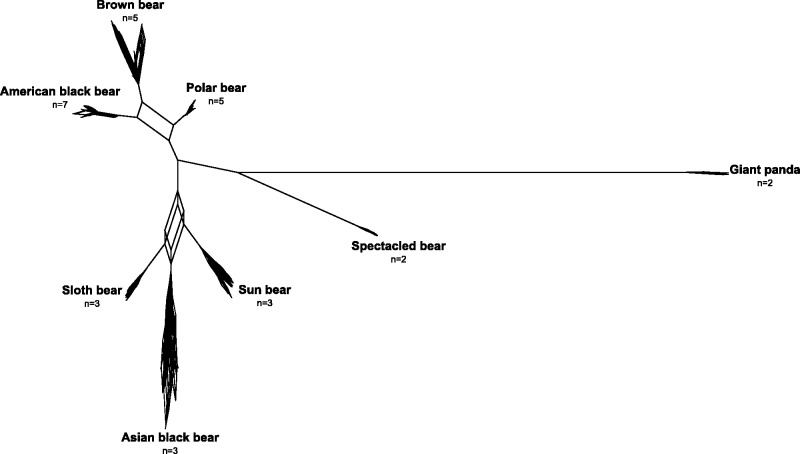
Fig. 3.
Consensus network of 14 autosomal gene trees obtained from a *BEAST analysis of 14 nuclear introns. All splits found in at least two gene trees (2/14, threshold = 0.14) are shown. n, number of individuals analyzed per species.
Table 1.
Divergence Time Estimates Obtained from *BEAST Based on 15 Nuclear Markers (14 autosomal introns and Y-chromosomal sequence).
| Prior | Estimated Divergence Time, Ma (95% HPD interval) |
||||||
|---|---|---|---|---|---|---|---|
| Giant Panda/Spect. Bear + Ursinae | Spect. Bear/Ursinae | Polar + Brown + Am. Black Bear/Asian Black + Sun + Sloth Bear | Asian Black Bear/Sun + Sloth Bear | Sun/Sloth Bear | Am. Black Bear/Polar + Brown Bear | Polar/ Brown Bear | |
| Root height min. 11.6 Ma | 12.46 | 5.88 | 1.78 | 1.56 | 1.42 | 0.94 | 0.62 |
| (Abella et al. 2012) | (11.6–14.48) | (4.67–7.18) | (1.42–2.2) | (1.2–1.96) | (1.04–1.81) | (0.67–1.25) | (0.38–0.89) |
In a *BEAST analysis of the 14 autosomal introns alone (data not shown), and in a BEAST analysis of the Y-chromosomal sequences alone (fig. 2B), the same topology was obtained as in the combined species tree analysis (fig. 2A), but with lower statistical support for an Asian black, sun, sloth bear clade. Phylogenetic analyses of concatenated nuclear data were conducted for comparison and are described in the supplementary material, Supplementary Material online. A *BEAST analysis of a combined data set including our data and previously published sequences (29 kb from 30 nuclear markers; supplementary table S5, Supplementary Material online, lists all analyzed data sets) did not converge within 2 × 109 generations, likely due to incongruent signals among loci. In a cloudogram of this analysis (results not shown), the three most frequent topologies were the same as obtained from our 15 loci data set (fig. 2A), but the third most common topology, which was identical to those previously published by Nakagome et al. (2008) and Pagès et al. (2008), was represented by thick lines placing the Asian black bear as sister-taxon to the American black, brown, polar bear clade. Thus, this third topology occurred more often in the data from previous studies than in our own intron and Y chromosome data, illustrating the heterogeneity of phylogenetic signals in bears.
Contrasting Signals from Nuclear and Mitochondrial DNA
When reanalyzing mitochondrial genomes from all eight extant bear species in BEAST, we obtained a topology with the sloth bear as sister-taxon to all other ursines with limited support and the sun bear as sister-taxon to an American and Asian black bear clade (fig. 2C; Yu et al. 2007; Krause et al. 2008). This topology differed from nuclear phylogenies (fig. 2A and B).
We evaluated the phylogenetic signal from the mitochondrial data set and two Y-chromosomal data sets (supplementary tables S5 and S6, Supplementary Material online) for their fit on 105 different tree topologies that can be built for five operational taxonomical units. In these topologies, polar and brown bears were constrained to be sister taxa, the spectacled bear as sister-taxon to all ursines, and the giant panda as outgroup.
In approximately unbiased (AU) tests of mitochondrial data, all three possible positions of the sloth bear in this phylogeny obtained high probability (P) values, with low differences in the log-likelihood values (ΔlogL) relative to the best tree (supplementary table S6A, Supplementary Material online). All topologies obtained from analyses of nuclear DNA in this and in previous studies (Nakagome et al. 2008; Pagès et al. 2008) were incompatible with the mitochondrial data set (P < 0.01; supplementary table S6A, Supplementary Material online). Conversely, all three mitochondrial topologies were incompatible with the Y-chromosomal data, regardless whether our Y-chromosomal sequences were analyzed alone, or when combining Y-chromosomal sequences from this study with Y-linked markers from Nakagome et al. (2008) and Pagès et al. (2008) (supplementary table S6B and C, Supplementary Material online). For both these Y-chromosomal data sets, the highest P value was observed for the topology that was also reconstructed in BEAST using our own Y-chromosomal data set (fig. 2B) Additional topologies could not be rejected (P ≥ 0.05), including the species tree topology (fig. 2A). Topologies from previous publications were characterized by large ΔlogL values, and some were incompatible (P < 0.05).
To perform statistical comparisons of the mitochondrial and the nuclear species tree topologies, we conducted analyses of our nuclear data in *BEAST, in which we constrained the species tree topology to either the mitochondrial topology (fig. 2C) or the species tree topology (fig. 2A), respectively. The latter analysis was carried out to ensure that constraining per se did not affect the analysis. To test the two hypotheses, posterior probabilities were compared using Bayes factors (BF) (Kass and Raftery 1995; Suchard et al. 2005), the Bayesian analog of likelihood ratio (LLR) tests. Considering a log10(BF) >2 (or BF >100) as “decisive” (Kass and Raftery 1995), the nuclear species tree topology was favored over the mitochondrial gene tree topology with high statistical support (log10[BF] = 4.2, or BF = 15,811).
Gene Flow and Demographic Analyses
Multilocus coalescence approaches such as *BEAST can efficiently accommodate ILS, but they do not model gene flow, although the latter can significantly impact phylogenetic inferences (Leaché et al. 2014). We therefore used IMa2, which is based on an isolation-with-migration model and jointly estimates six demographic parameters, including population migration rates between populations since their divergence from a common ancestral population. We analyzed species pairs where conflict between mitochondrial and species tree topologies was found (brown bear–American black bear, American black bear–Asian black bear), or based on shared haplotypes between distantly related species (polar bear–sun bear). Pairs of Asian bear species (Asian black bear–sun bear, Asian black bear–sloth bear, sloth bear–sun bear) were selected to investigate whether past introgression may explain the uncertain branching order among these species (P = 0.67; fig. 2A).
IMa2 analyses indicated significant unidirectional gene flow from the brown bear into the American black bear lineage (table 2 and supplementary fig. S3A, Supplementary Material online), irrespective of the upper prior boundaries chosen. This was also evident from haplotype sharing between brown and American black bears, which shared four haplotypes at three introns (fig. 1, supplementary fig. S1 and table S4, Supplementary Material online). Between American and Asian black bears, two haplotypes were shared at two introns, but multilocus analyses in IMa2 revealed no significant gene flow between these two species. The posterior distribution for gene flow from American into Asian black bears showed a peak at 0.03 migrants per generation, but the 95% highest posterior density (HPD) interval included zero (table 2 and supplementary fig. S3A, Supplementary Material online). The same applied to sun and polar bears, which also shared two haplotypes at two introns. Although the 95% HPD interval for gene flow from sun into polar bears also included zero, the posterior distribution had a clear peak at 0.01 migrants per generation.
Table 2.
Demographic Parameters (modal values; 95% HPD interval in parentheses) from Analyses of Bear Species Pairs in IMa2, Based on 14 Autosomal Introns.
| Species 1 | Species 2 | Ne1 | Ne2 | 2N1M1 | 2N2M2 |
|---|---|---|---|---|---|
| American black bear | Asian black bear | 21,432 (8,664–44,233) | 44,233 (18,696–94,394) | 0 (0–0.16) | 0.03 (0–0.38) |
| American black bear | Brown bear | 20,178 (8,550–37,963) | 43,435 (24,282–76,267) | 0.08a (0.01–0.24) | 0 (0–0.12) |
| Polar bear | Sun bear | 3,967 (1,231–11,355) | 16,279 (6,703–33,517) | 0.01 (0–0.06) | 0 (0–0.09) |
| Asian black bear | Sun bear | 46,969 (21,432–89,834) | 19,608 (7,752–44,233) | 0.03 (0–0.23) | 0 (0–0.12) |
| Asian black bear | Sloth bear | 46,969 (22,344–88,922) | 4,104 (1,368–16,872) | 0 (0–0.18) | 0.03a (0–0.1) |
| Sloth bear | Sun bear | 1,368 (0–10,488) | 4,104 (1,368–16,872) | 0.01 (0–0.07) | 0.04 (0–0.16) |
Ne1 and Ne2, effective population sizes for species 1 and 2, respectively; 2N1M1, population migration rate into species 1 from species 2 per generation; 2N2M2, population migration rate into species 2 from species 1 per generation. Posterior probability distributions for parameters are shown in supplementary figure S3, Supplementary Material online.
aMigration rates that are significantly different from zero at the P < 0.05 level in LLR tests (Nielsen and Wakeley 2001; Hey 2010).
In IMa2 analyses of Asian bear species pairs, significant unidirectional gene flow was detected from the Asian black bear lineage into the sloth bear lineage at a rate of 0.03 migrants per generation (table 2 and supplementary fig. S3B, Supplementary Material online), consistent with shared variation between the two species (supplementary table S3, Supplementary Material online). Neither between Asian black and sun bears, nor between sloth and sun bears, significant signals of gene flow were detected (table 2), although in both cases, two haplotypes were shared at two introns (fig. 1, supplementary fig. S1 and table S4, Supplementary Material online). The posterior distributions for gene flow from sun into Asian black bears, from sloth into sun bears, and from sun into sloth bears showed clear peaks at 0.01–0.04 migrants per generation, but the 95% HPD intervals included zero (table 2 and supplementary fig. S3B, Supplementary Material online).
IMa2 showed small effective population sizes (Ne) for polar bears and sloth bears, and much larger values for brown and Asian black bears (table 2 and supplementary fig. S3C, Supplementary Material online), consistent with current nucleotide diversity levels (supplementary table S7, Supplementary Material online). In all IMa2 runs, the posterior distributions of ancestral population size had a clear peak, but for some species pairs, the upper tails did not approach zero, even in runs based on much wider priors (supplementary fig. S3D, Supplementary Material online). The right tails of the posterior distributions of the time since population splitting also did not converge on zero, so this parameter could not be estimated with certainty for any species pair (supplementary fig. S3E, Supplementary Material online). However, when restricting the prior for the splitting time to the minimum age of the youngest Ursavus fossil (ca. 7.1 My; Fortelius 2003), the genus that is believed to have given rise to the Ursus lineage (Kurtén 1968), the highest peaks of the posterior distributions coincided with the geological ages of time estimates inferred in *BEAST (table 1 and supplementary fig. S3E, Supplementary Material online). In summary, our gene flow analyses thus indicated that besides ILS, introgression also played a role during the evolutionary history of bears.
Discussion
Introgression and ILS both lead to variation in the phylogenetic signal among loci and individuals from the same species, causing gene tree discordance. Especially in rapidly diverged species such as ursine bears, disentangling the effects of ILS and introgression remains challenging. Because concatenation approaches cannot model or portray either of these processes, we instead used coalescent-based multilocus methods to analyze multiple independently inherited loci sequenced in several individuals from each extant bear species.
We first reconstructed phylogenetic trees based on nuclear data. Next, we specifically investigated whether gene flow could explain observed incongruences among nuclear loci, and the conflict between the nuclear species tree and the mitochondrial phylogeny. This approach provided a more comprehensive understanding of the evolutionary process than by simply aiming at a fully resolved bifurcating tree. By explicitly considering intraspecific and intraindividual variation, we demonstrate that both ILS and introgression have shaped the evolutionary history of ursine bears.
Species Tree Inferences in the Presence of ILS and Introgression
The multilocus species tree of autosomal introns and Y-chromosomal sequence from this study (fig. 2A) is similar, but not identical, to phylogenetic trees reconstructed in previous studies based on concatenated nuclear data. In contrast to the concatenation approach, however, ILS is specifically considered and modeled in our species tree estimation. We obtained high posterior support for a placement of the giant panda and the spectacled bear outside the variation of all Ursinae, and for a brown, polar, and American black bear clade. A previous study placed the Asian black bear as sister-taxon to the brown, polar, and American black bear with high statistical support (Pagès et al. 2008). In our species tree, however, sun, sloth, and Asian black bear, the three species whose current distributions are limited to Asia, form a highly supported clade. The sun, sloth, and Asian black bear clade is distinct from the brown, polar, and American black bear clade also in our consensus network of autosomal gene trees (fig. 3). Because the sun and sloth bear are currently not included in the Ursus genus, our findings render Ursus, as it is currently defined, paraphyletic.
The exact branching order within the clade of Asian bear species is complex, however, as illustrated by a cuboid connecting Asian black bears, sun bears, and sloth bears in the consensus network. Some support for a sister relationship between the sun bear and the sloth bear comes from a sequence insertion in sun and sloth bears in the Y chromosome, which is 93% identical to a transposable element from the giant panda (SINEC1_Ame). We note, however, that more insertions are required to obtain statistical significance (Waddell et al. 2001). Low statistical support for a sister relationship of sun and sloth bears in the species tree (fig. 2A) can result from introgression, as *BEAST does not model gene flow. A recent simulation study showed that even low levels of gene flow between nonsister species reduce statistical support for the true sister species clade in species tree inferences using *BEAST (Leaché et al. 2014). Indeed, we detect weak, but significant unidirectional gene flow from the Asian black bear lineage into the sloth bear lineage (table 2 and supplementary fig. S3B, Supplementary Material online). This is consistent with low statistical support for a sun and sloth bear clade, and with alternative topologies in the cloudogram of species trees showing Asian black and sloth bears as sister species. Thus, a combination of phylogenetic and gene flow estimation approaches suggests that sun and sloth bears may be sister species that have been impacted by introgression from a bear lineage related to extant Asian black bears.
Due to their haploid nature and uniparental inheritance, mitochondrial and Y-chromosomal loci are expected to sort more rapidly than biparentally inherited autosomal loci. In contrast to mtDNA, intraspecific variation on the Y chromosome is low in many mammals (Hellborg and Ellegren 2004), but differences are predicted to accumulate quickly among lineages (Petit et al. 2002). Furthermore, the Y chromosome lacks recombination over most of its length. Therefore, it constitutes a high-resolution record of evolutionary history. Accordingly, the Y chromosome shows haplotypes from different species as clearly distinct (fig. 1). Despite differences in the pattern of haplotype sharing and in the mean distances between pairs of ursine species, the Y-chromosomal gene tree and the autosomal species tree show congruent phylogenetic signals, and both marker systems contrast with the phylogenetic signal of mtDNA with high statistical confidence (fig. 2 and supplementary table S6, Supplementary Material online).
Rapid Speciation and ILS in Ursine Bears
Several lines of evidence suggest extensive ILS for autosomal loci in ursine bears. A large number of polymorphic sites within species compared with the number of fixed differences between ursine species pairs confirm that intraspecific polymorphism makes a major contribution to the overall phylogenetic signal on autosomal loci—a signal that needs to be considered. However, this is not possible in noncoalescence-based phylogenetic analyses of concatenated data. We show that haplotype sharing in bears occurs most frequently between closely related species. Neither haplotypes nor polymorphic sites are shared between giant pandas, spectacled bears, and ursine bears. Our divergence time estimates indicate that speciation events in ursine bears occurred within only about 1.8 My. Assuming an average Ne of 28,000 individuals for brown and polar bears (Miller et al. 2012; Hailer et al. 2013; Nakagome et al. 2013) and a generation time of 10 years (Tallmon et al. 2004; Cronin et al. 2009), lineage sorting for most autosomal loci in bears requires 1.1–2.0 My, based on coalescence theory (corresponding to 4–7 Ne generations; Nichols 2001). Considering the rapid radiation of ursine bears, ILS is thus expected to be common in the autosomal part of their genome.
Ursine bears descended directly from U. minimus (Kurtén 1968), a species known from the fossil record. Thus, modern ursine bears most likely radiated after the last occurrence of this species in the fossil record. Indeed, our time estimate for the onset of the ursine radiation is younger than the youngest U. minimus fossil, which was dated to 2.6–3.4 Ma (Fortelius 2003). Our estimation places the onset of the radiation of Ursinae to the early Pleistocene, and the most recent speciation event, the polar/brown bear divergence, to the mid Pleistocene. In contrast to divergence time estimates based on mitochondrial genomes (Yu et al. 2007; Krause et al. 2008), our estimated time frame excludes the Miocene. Our polar/brown bear divergence time estimate is similar to other recent estimates from nuclear data (Edwards et al. 2011; Hailer et al. 2012; Cahill et al. 2013; Liu et al. 2014), but younger than the 4–5 Ma proposed by Miller et al. (2012). We note that our estimates may underestimate the actual divergence times, and that the incorporation of sequence data from ancient bear specimens as fossil tip calibration points will likely allow for more refined divergence time estimates. The average substitution rate across all loci obtained from our calibrated *BEAST analyses of 0.95 × 10−8 substitutions per site per generation is lower than a rate estimated for primates (2.5 × 10−8 substitutions per site per generation; Nachman and Crowell 2000). Applying the faster rate from primates would lead to even younger divergence time estimates for bears. Regardless of the exact timing, the Plio-/Pleistocene epoch was characterized by climatic fluctuations, dramatic changes in habitat characteristics and habitat fragmentation, promoting population differentiation and speciation but also allowing for secondary contact.
Our study shows that the rapid radiation of bears did not allow for complete lineage sorting on their autosomes. This is reflected in the high degree of shared polymorphic sites and haplotypes between ursine species, in our network analyses, and in the short internal branches found in the present and in previous phylogenetic analyses of ursines (Yu et al. 2007; Krause et al. 2008; Nakagome et al. 2008; Pagès et al. 2008). These findings highlight that the extent of ILS on the autosomes of species with similar population sizes and speed of speciation as ursine bears is not to be underestimated.
Accounting for ILS was only possible because we consider intraspecific variability within a coalescence framework. In contrast, previous phylogenetic studies of the bear family analyzed concatenated sequences of only one (consensus) individual per species, without being able to specifically model the genealogical history of intraspecific variation, which was made possible by recent methodological developments. A recent simulation study demonstrated that sampling effort in terms of number of individuals and markers had a large effect on species tree accuracy, especially when lineage sorting was incomplete (Lanier and Knowles 2012). In that study, accurate species tree estimates were obtained by sampling three individuals per species and nine independent loci, suggesting that our sampling scheme should yield reliable results. Thus, by extending the available data on bears with sequences of high resolution from several individuals per species, and by using an advanced coalescence multilocus approach that specifically models ILS, complemented by multilocus gene flow analyses, our data set allows for the estimation of a statistically robust species tree of bears, including divergence time estimates.
Haplotype networks of autosomal introns further illustrate the effect of sampling several individuals per species. For example, depending on which Asian black bear individual is chosen for phylogenetic analysis, the signal would be altered, as each Asian black bear individual shares different haplotypes with different other bear species. Moreover, data sets analyzed in previous studies contained less than half of the number of variable sites of our data set, highlighting that a considerable amount of genealogical information resides within species, including the variation found among individuals, as well as intraindividual variability (heterozygous sites).
Discordance between Mitochondrial and Nuclear Phylogenies of Bears
We find evidence for ILS among ursine bear species and gene flow from Asian black bears into sloth bears, causing incongruences among genealogical histories of nuclear loci. Similarly, discordances between mitochondrial and nuclear phylogenies in bears have been reported previously, but without explicitly testing alternative hypotheses considering ILS or introgression. We show that the nuclear species tree of ursine bears conflicts with the mitochondrial gene tree topology using statistical model comparisons in a coalescence framework, and that the Y-chromosomal and the mitochondrial gene tree are mutually exclusive using likelihood-based statistical tests, both with high statistical significance. Such discordance can be explained by differences in ploidy and inheritance mode of the maternally inherited mtDNA, the paternally inherited Y chromosome, and the biparentally inherited autosomal loci, which capture different aspects of evolutionary history. Therefore, comparing differentially inherited loci allows for the identification of possibly contrasting patterns of female and male gene flow, and of introgression events.
Discordance between the mitochondrial gene tree on the one side and the autosomal species tree and the Y-chromosomal gene tree on the other side has already been documented for brown bears and polar bears (Hailer et al. 2012, 2013; Miller et al. 2012; Cahill et al. 2013; Bidon et al. 2014). This pattern was explained with introgressive hybridization between the two species and the replacement of the polar bear mitochondrial genome (mitochondrial capture; fig. 2D). Hybridization between different bear species has been observed in zoos and in the wild (Gray 1972; Kelly et al. 2010). The discordant placement of the American black bear in the nuclear species tree and in the mitochondrial gene tree (fig. 2A–D), and the detection of unidirectional gene flow from the brown bear into the American black bear lineage suggest a similar process for American black, Asian black, and brown bears.
Two hybridization scenarios could explain the incongruent placement of the American black bear in the nuclear species compared with the mitochondrial gene tree (fig. 2D): A) The replacement of the original American black bear mtDNA by an Asian black bear-like lineage through introgressive hybridization (mitochondrial capture), leading to a matrilineal sister-relationship of the two species. Alternatively B), nuclear swamping of the American black bear genome by genetic material from the brown bear through male-mediated introgressive hybridization, causing the placement of the American black bear with the brown/polar bear clade in the nuclear species tree (see Leaché et al. 2014).
Mitochondrial capture (scenario A) would require hybridization between Asian and American black bears (fig. 2D). The current distribution of Asian and American black bears is allopatric. However, the Bering land bridge connected eastern Asia and North America several times for long time periods during the Pleistocene (Hoffecker and Elias 2007). Today, populations from both species occur proximal to this region: Asian black bears in eastern Russia, the Korean Peninsula and Japan, and American black bears in Alaska and Yukon, Canada (Servheen et al. 1990). The Bering land bridge may thus have provided opportunity for sympatry of American and Asian black bears in former times. Asian and American black bears share two haplotypes at two intron loci, and are polymorphic for the same variants at four sites (fig. 1, supplementary fig. S1 and tables S3 and S4, Supplementary Material online), but we find no significant multilocus signal of gene flow between the two species under the isolation-with-migration model. mtDNA was shown to introgress more easily than paternally or biparentally inherited genetic material (Chan and Levin 2005). Numerous cases of mitochondrial introgression across species boundaries have been documented, often with lower levels or without introgression of nuclear DNA, for example in polar and brown bears (Hailer et al. 2012), elephants (Roca et al. 2005), chipmunks (Good et al. 2008), colobine monkeys (Roos et al. 2011), hares (Melo-Ferreira et al. 2012), and in black rats (Pagès et al. 2013). Thus, mitochondrial capture can explain our observations.
Several other observations argue for nuclear swamping (scenario B). Such a forceful process could result from male-biased gene flow from brown into American black bears, with physically larger male brown bears mating with female black bears, without mtDNA passing the species boundary. Such gene flow must have stopped at some time in the past to explain the level of differentiation observed between brown bear and American black bear Y chromosomes. Indeed, we find significant, but weak signals of gene flow from the brown bear lineage into the American black bear lineage (table 2 and supplementary fig. S3A, Supplementary Material online), consistent with three haplotypes and three polymorphic sites shared between brown and American black bears (fig. 1, supplementary fig. S1 and tables S3 and S4, Supplementary Material online). Similarly, Miller et al. (2012) observed gene flow between brown and American black bears since their speciation, lasting until the late Pleistocene. Scenario B postulates that the mitochondrial gene tree reflects the speciation history of American and Asian black bears. Indeed, there is paleontological evidence for a sister-species relationship between American and Asian black bears (Kurtén and Anderson 1980). Remains of the ancestral nuclear genome, from times prior to introgression of brown bear genes into the American black bear lineage should still be detectable in American black bears. These ancestral remains may be represented by two haplotypes and four polymorphisms shared between American and Asian black bears. There is evidence for nuclear swamping affecting the genomes of brown and polar bears (fig. 2D): At the mitochondrial genome, polar bears were found to be closely related to brown bears from the Alaskan ABC (Admiralty, Baranof, and Chichagof) islands and from Ireland (now extinct) (Cronin et al. 1991; Edwards et al. 2011). At the nuclear genome, unidirectional gene flow has been detected from polar bears into North American brown bears, including ABC island brown bears (Cahill et al. 2013; Liu et al. 2014). Based on these findings, ABC island brown bears have been suggested to carry a mitochondrial haplotype that derives from an initial polar bear ancestry, whereas extensive male-biased gene flow from mainland brown bears has replaced much of the original polar bear-like genome with genetic material from immigrant brown bears (Cahill et al. 2013; Bidon et al. 2014). Considering these observations from different bear species, nuclear swamping is a reasonable explanation for the different placement of the American black bear lineage in nuclear and mitochondrial phylogenies.
Both hypotheses regarding American black bears appear rather drastic. Another source of conflict between nuclear and mitochondrial phylogenies can be the faster lineage sorting of the mitochondrial genome compared with autosomal DNA, due to the smaller effective population size of mtDNA (Funk and Omland 2003; McKay and Zink 2010). However, ILS was accounted for in our statistical comparisons of mitochondrial and nuclear topologies in a coalescence framework, rendering differences in lineage sorting an unlikely cause for the observed discrepancies between mitochondrial and nuclear phylogenies. Nonetheless, a scenario including several hybridization events during the evolutionary history of ursine bears is conceivable, involving ancient hybridization of American and Asian black bears, gene flow from Asian black bears into sloth bears, and/or male-biased gene flow from brown bears into American black bears. Extended population-level and/or genome-wide studies and analytical approaches that incorporate both ILS and introgression into species tree estimation will be required to fully understand the evolutionary processes leading to the observed discrepancies between nuclear and mitochondrial phylogenies in these species.
Capturing the Complexity of Evolutionary Processes
Charles Darwin pointed out that many closely related species are not completely reproductively isolated (Darwin 1859), and in recent decades, molecular studies have identified introgressive hybridization as a pervasive evolutionary process (Schwenk et al. 2008). At least 10% of animal species hybridize with closely related species in well-studied taxa (Gray 1972; Mallet 2005). In addition, based on predictions from coalescence theory, lineage sorting of autosomal genes should be completed within about four to seven Ne generations (Nichols 2001). Thus, ILS spans time scales of up to several million years, often covering longer time frames than required for speciation in mammals. ILS has been shown to affect a large proportion of the genomes of humans and their closest relatives (Hobolth et al. 2011; Prüfer et al. 2012; Scally et al. 2012), but only few studies have specifically examined both ILS and gene flow in vertebrates that diverged several million years ago. Notably, many species have a larger population size than bears and great apes, so their genomes will be even more affected by ILS.
Initially, when technological advances made it feasible to sequence multiple loci, phylogenetic methods developed for single loci were used to analyze a concatenated superlocus. This approach ignored the heterogeneity of the phylogenetic signal among loci, and disregarded the vast amount of phylogenetic information that resides within individuals and species by including only one individual per species. Indeed, simulation studies have shown that the concatenation procedure can provide high statistical support for an incorrect species tree, because lineage sorting processes are not modeled (Kubatko and Degnan 2007). Finally, branch length estimates are affected when heterozygous sites are excluded from phylogenetic analyses (Lischer et al. 2014), which was common practice in phylogenetic analyses of concatenated autosomal data. Conceptual advances and recently developed coalescence-based multilocus species tree approaches now provide a means to infer overall phylogenetic relationships (species trees), against which individual gene trees can be contrasted to identify the underlying evolutionary processes. Although species tree approaches such as *BEAST (Heled and Drummond 2010) do not model gene flow, coalescence-based gene flow analyses can be used to complement phylogenetic inferences of evolutionary history: For example, in orioles (Jacobsen and Omland 2012), hares (Melo-Ferreira et al. 2012), and gibbons (Chan et al. 2013). By comparing marker systems with different inheritance modes and ploidy, sex-biased mechanisms and introgression events can be identified. To depict the complexity of evolutionary processes, networks of individual loci and multilocus networks (Holland et al. 2004; Bapteste et al. 2013) are better suited than bifurcating trees, because the latter may obscure evolutionary signals (Morrison 2005; Hallström and Janke 2010; Bapteste et al. 2013). In summary, advanced phylogenetic studies that aim to capture the full complexity of the evolutionary process need to consider “phylogenetic incongruence [as] a signal, rather than a problem” (Nakhleh 2013).
Materials and Methods
Samples and DNA Extraction
Samples were obtained from one giant panda, two spectacled bears, three sloth bears, three sun bears, three Asian black bears, one American black bear, two brown bears, and three polar bears (supplementary table S1, Supplementary Material online). All samples originated from zoo individuals or from animals legally hunted for purposes other than this study. Total DNA was extracted from muscle, skin, and blood samples using a standard salt extraction protocol (Crouse and Amorese 1987), or a standard phenol–chloroform extraction protocol (Sambrook and Russell 2000).
Amplification and Sequencing
We used primer pairs for 14 independently inherited autosomal markers (Hailer et al. 2012) to amplify intron sequences with flanking exon sequences in 15 individuals. We amplified nine Y-chromosomal markers in 11 male individuals (supplementary table S8, Supplementary Material online), using primers that were either described in Bidon et al. (2014), or newly designed (322, 389, 403) based on the polar bear genome (Liu et al. 2014), or based on male giant panda reads (Zhao et al. 2013) mapped against the polar bear genome. Polymerase chain reactions (PCRs) were performed using 5–15 ng of genomic DNA, and each PCR setup contained no-template controls. For amplification of Y-chromosomal markers, female DNA controls were included to ensure male-specificity throughout all experiments. PCR conditions and primers are listed in supplementary table S8, Supplementary Material online. PCR products were detected using standard agarose gel electrophoresis, and cycle sequenced with BigDye 3.1 chemistry (Applied Biosystems, Foster City, CA) in both directions according to the manufacturer’s recommendation, and detected on an ABI 3100 instrument (Applied Biosystems). Electropherograms were checked manually. For autosomal introns, sequence data were included from Hailer et al. (2012) and from the giant panda genome assembly (Li et al. 2010), the final data set comprised 30 individuals. The Y-chromosomal data set included sequence data from Bidon et al. (2014). Therefore, American black bear and polar bear individuals differed between this and the autosomal intron data set. Accession numbers are listed in supplementary table S1, Supplementary Material online. Sequences were aligned using ClustalW implemented in Geneious 5.6.6 and 6.1.6 (Biomatters, Auckland, New Zealand; Drummond et al. 2012). We compared Y-chromosomal sequences from our single male giant panda individual with the mapped panda reads. Although this genome’s Y-chromosomal sequence could not be included in our analyses because of some missing data, we found that all panda-specific divergent sites that were covered by both individuals were identical.
Data Analyses
We resolved heterozygous indels at autosomal markers using Champuru (Flot 2007) and Indelligent (Dmitriev and Rakitov 2008). Haplotypes were deduced using PHASE implemented in the software DnaSP v5.0 (Librado and Rozas 2009), based on alignments containing all available unphased sequences from the present and from a previous study (Hailer et al. 2012), allowing for recombination within haplotypes and using a cutoff value of 0.6 (Harrigan et al. 2008; Garrick et al. 2010). Twelve heterozygous sites could not be resolved and respective alignment columns were discarded from analyses. Sites containing floating indels, gaps, or missing data (N) were deleted from the alignments. In the Y-chromosomal alignment, three pseudoheterozygous sites were removed. Sequence diversity and differentiation statistics were calculated in Arlequin 3.5 (Excoffier and Lischer 2010), MEGA 5.2.2 (Tamura et al. 2011), and DnaSP v5.0 (Librado and Rozas 2009). To investigate the heterogeneity among different loci, statistical parsimony networks were reconstructed using TCS 1.21 (Clement et al. 2000). For this analysis, indels were treated as single mutational events, and gaps as a fifth character state. Longer gaps were treated as single mutational changes. The connection probability limit was set to 0.95 (autosomal loci) or 0.94 (Y-chromosomal sequence).
We reconstructed multilocus species trees from different data sets (supplementary table S5, Supplementary Material online), using *BEAST 1.7.5 (Drummond et al. 2012). Recombination is not modeled in *BEAST, but sampling effort (number of loci, number of individuals) has a much larger effect on species tree accuracy than the error introduced by recombination (Lanier and Knowles 2012). Hence, by reducing an alignment to its largest nonrecombining section, abundant phylogenetic information is discarded. We therefore used the total sequence length of the 14 autosomal introns (8 kb) in all *BEAST analyses. *BEAST was run applying a Yule prior on the species tree and a normal prior of 0.001 ± 0.001 (mean ± SD) on the substitution rates. We used a strict clock, because a relaxed, uncorrelated lognormal clock approach (Drummond et al. 2006) showed no significant departure from the strict clock model for our data. Models of sequence evolution were used as indicated by jModeltest (Posada 2008) and *BEAST was run for 2 × 109 generations, sampling every 10,000th iteration. Convergence was checked in Tracer with effective sampling sizes (ESS) >200. Two runs with identical settings were combined in LogCombiner v1.7.5 using a burnin of 10%, and a maximum clade credibility tree was constructed using TreeAnnotator.
For divergence time estimates, we assumed a minimum age of 11.6 My for the divergence of the giant panda from other bears, based on the oldest described fossil from the subfamily Ailuropodinae (Abella et al. 2012). Generation time for American black bears has been estimated at 6.27 years (Onorato et al. 2004) and 10 years for brown and polar bears (Tallmon et al. 2004; Cronin et al. 2009). For spectacled, sloth, sun, Asian black bears, and giant pandas, no adequate data were available, but as generation time is correlated with body size in mammals (Bonner 1965), we used the estimate of 6.27 years for American black bears also for these species. Based on the arithmetic mean of these generation time estimates, we assumed an overall generation time of 7.2 years to transform per-year estimates of ursid mutation rates from *BEAST into per-generation values. For statistical comparisons of the mitochondrial and the species tree topologies, we performed *BEAST analyses of autosomal introns and Y-chromosomal data combined. The species tree topology was either constrained to the mitochondrial topology (monophyly of American black bear and Asian black bear, and monophyly of American black bear, Asian black bear, and sun bear), or to the species tree topology (monophyly of polar bear, brown bear, and American black bear). BF were estimated in Tracer based on likelihood traces of the two constrained analyses (Suchard et al. 2005), using 1,000 bootstrap replicates.
To illustrate the extent of phylogenetic conflict in the nuclear signal, DensiTree (Bouckaert 2010) was used to generate a cloudogram of the posterior distribution of species trees from *BEAST, and a consensus network (Holland et al. 2004) was generated using SplitsTree4 (Huson and Bryant 2006). For the latter, *BEAST maximum clade credibility gene trees from the 14 autosomal introns were used as input gene trees, displaying splits that occurred in at least 2 of the 14 gene trees (edge threshold: 0.14).
For phylogenetic analyses of concatenated mitochondrial and Y-chromosomal data, we reconstructed different data sets (supplementary table S5, Supplementary Material online) from sequence data generated in the present and in previous studies (Jameson et al. 2003; Nakagome et al. 2008; Pagès et al. 2008). Pagès et al. (2008) published a consensus sequence of several individuals per species, with intraspecific polymorphisms coded by ambiguity codes. Alignment columns with these sites were disregarded in all analyses. Protein-coding regions from the mitochondrial genomes of all eight bear species (excluding ND6) were obtained from OGRe (Jameson et al. 2003) (for accession numbers, see supplementary table S1, Supplementary Material online), and aligned and concatenated in Geneious 5.6.6. For each data set, the optimal model of sequence evolution was determined using jModeltest (Posada 2008). Concatenated Y-chromosomal data (present study) and mitochondrial sequences were analyzed in BEAST 1.7.5 (Drummond et al. 2012) using a Yule prior on the species tree and a normal prior of 0.001 ± 0.001 on the substitution rates. BEAST was run for 1 × 109 generations, sampling every 10,000th iteration. Convergence was checked in Tracer (ESS > 200) and maximum-clade credibility trees were reconstructed in TreeAnnotator using a burnin of 10%. The AU test (Shimodaira 2002) was performed in Treefinder (Jobb et al. 2004) with 50,000 bootstrap replicates each, using mitochondrial and two Y-chromosomal data sets. Likelihoods and tree statistics were calculated in Treefinder in an exhaustive search among all 105 topologies that are possible for five operational taxonomical units. The giant panda served as outgroup, with the spectacled bear as sister-taxon to all ursines, and the polar and brown bear were constricted to be sister lineages.
We used IMa2 (Hey 2010) on the 14 autosomal introns to assess the level of gene flow among species. This software is based on an isolation-with-migration model and estimates effective population sizes (present and ancestral), splitting times, and population migration rates using Markov chain Monte Carlo (MCMC) simulations. As the isolation-with-migration model assumes no recombination within and free recombination between markers (Hey and Nielsen 2004), the nonrecombining sections of the 14 autosomal introns (in total 5.1 kb) were used as reconstructed in IMgc (Woerner et al. 2007). Substitution rates per marker per year were estimated from the average divergence (DXY = 2Tµ) between the giant panda and polar bear, assuming a divergence time (T) of 12 Ma (Abella et al. 2012), and the Hasegawa–Kishino–Yano model of sequence evolution. We assumed a generation time of 8 years for the pairwise comparisons brown bear—American black bear and polar bear—sun bear, and a generation time of 6 years for the other pairwise comparisons. Generation times were based on estimates of 6.27 years for American black bears (Onorato et al. 2004) and of 10 years for brown and polar bears (Tallmon et al. 2004; Cronin et al. 2009). Preliminary runs were performed to evaluate various prior settings, heated chain conditions, and the necessary MCMC lengths. To set an upper bound for the splitting time, we assumed that time since divergence could not be older than the minimum age of the youngest Ursavus fossil (ca. 7.1 My; Fortelius 2003), the genus from which the Ursus lineage is thought to have descended (Kurtén 1968). For effective population sizes, we defined an upper bound for the prior by multiplying the arithmetic mean of θπ (Tajima 1983) of each species pair by approximately nine, allowing for larger population sizes in the past (Miller et al. 2012). Four independent runs, each with different starting seeds, were performed with optimized priors and heating schemes, using 40 Markov chains. After a burnin period with stationary already reached, 25,000 genealogies were saved. Convergence was assessed based on ESS >50, stable parameter trend plots, and similar parameter estimates from the first and the second half of the runs. Marginal posterior probability density estimates and LLR tests to assess whether migration rates were significantly different from zero were calculated in “L mode” of IMa2, using 100,000 sampled genealogies from each of the four independent runs.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by LOEWE Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz, the Arthur und Aenne Feindt-Stiftung, Hamburg, and the RISE Research Internships in Science and Engineering (RISE) program of the German Academic Exchange Service (DAAD). The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the US Fish and Wildlife Service. The authors thank U. Arnason, S.B. Hagen, H.-G. Eiken N. Lecomte, M. Onucsán, and B. Steck for providing samples, J.B. Hlíðberg (www.fauna.is) for the artwork, and the editor and reviewers for helpful comments on a previous version of the manuscript.
References
- Abella J, Alba DM, Robles JM, Valenciano A, Rotgers C, Carmona R, Montoya P, Morales J. Kretzoiarctos gen. nov., the oldest member of the giant panda clade. PLoS One. 2012;7:e48985. doi: 10.1371/journal.pone.0048985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste E, van Iersel L, Janke A, Kelchner S, Kelk S, McInerney JO, Morrison DA, Nakhleh L, Steel M, Stougie L, et al. Networks: expanding evolutionary thinking. Trends Genet. 2013;29:439–441. doi: 10.1016/j.tig.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Bidon T, Janke A, Fain SR, Eiken HG, Hagen SB, Saarma U, Hallström BM, Lecomte N, Hailer F. Brown and polar bear Y chromosomes reveal extensive male-biased gene flow within brother lineages. Mol Biol Evol. 2014;31:1353–1363. doi: 10.1093/molbev/msu109. [DOI] [PubMed] [Google Scholar]
- Bonner JT. Size and cycles: an essay on the structure of biology. Princeton (NY): Princeton University Press; 1965. [Google Scholar]
- Bouckaert RR. DensiTree: making sense of sets of phylogenetic trees. Bioinformatics. 2010;26:1372–1373. doi: 10.1093/bioinformatics/btq110. [DOI] [PubMed] [Google Scholar]
- Cahill JA, Green RE, Fulton TL, Stiller M, Jay F, Ovsyanikov N, Salamzade R, St. John J, Stirling I, Slatkin M, et al. Genomic evidence for island population conversion resolves conflicting theories of polar bear evolution. PLoS Genet. 2013;9:e1003345. doi: 10.1371/journal.pgen.1003345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KMA, Levin SA. Leaky prezygotic isolation and porous genomes: rapid introgression of maternally inherited DNA. Evolution. 2005;59:720–729. [PubMed] [Google Scholar]
- Chan Y-C, Roos C, Inoue-Murayama M, Inoue E, Shih C-C, Pei KJ-C, Vigilant L. Inferring the evolutionary histories of divergences in Hylobates and Nomascus gibbons through multilocus sequence data. BMC Evol Biol. 2013;13:82. doi: 10.1186/1471-2148-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Cronin MA, Amstrup SC, Garner GW, Vyse ER. Interspecific and intraspecific mitochondrial DNA variation in North American bears (Ursus) Can J Zool. 1991;69:2985–2992. [Google Scholar]
- Cronin MA, Amstrup SC, Talbot SL, Sage GK, Amstrup KS. Genetic variation, relatedness, and effective population size of polar bears (Ursus maritimus) in the southern Beaufort Sea, Alaska. J Hered. 2009;100:681–690. doi: 10.1093/jhered/esp061. [DOI] [PubMed] [Google Scholar]
- Cronin MA, McDonough MM, Huynh HM, Baker RJ. Genetic relationships of North American bears (Ursus) inferred from amplified fragment length polymorphisms and mitochondrial DNA sequences. Can J Zool. 2013;91:626–634. [Google Scholar]
- Crouse J, Amorese D. Ethanol precipitation: ammonium acetate as an alternative to sodium acetate. Focus. 1987;19:13–16. [Google Scholar]
- Darwin CR. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- Dmitriev DA, Rakitov RA. Decoding of superimposed traces produced by direct sequencing of heterozygous indels. PLoS Comput Biol. 2008;4:e1000113. doi: 10.1371/journal.pcbi.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M. Geneious v5.6. 2012 [Internet]. [2012 Mar]. Available from: http://www.geneious.com. [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CJ, Suchard MA, Lemey P, Welch JJ, Barnes I, Fulton TL, Barnett R, O’Connell TC, Coxon P, Monaghan N, et al. Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr Biol. 2011;21:1251–1258. doi: 10.1016/j.cub.2011.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Flot J-F. champuru 1.0: a computer software for unraveling mixtures of two DNA sequences of unequal lengths. Mol Ecol Notes. 2007;7:974–977. [Google Scholar]
- Fortelius M (coordinator). 2003. New and old worlds database of fossil mammals (NOW). University Helsinki. [cited 2014 Jan]. Available from: http://www.helsinki.fi/science/now.
- Funk DJ, Omland KE. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 2003;34:397–423. [Google Scholar]
- Garrick RC, Sunnucks P, Dyer RJ. Nuclear gene phylogeography using PHASE: dealing with unresolved genotypes, lost alleles, and systematic bias in parameter estimation. BMC Evol Biol. 2010;10:118. doi: 10.1186/1471-2148-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Hird S, Reid N, Demboski JR, Steppan SJ, Martin-Nims TR, Sullivan J. Ancient hybridization and mitochondrial capture between two species of chipmunks. Mol Ecol. 2008;17:1313–1327. doi: 10.1111/j.1365-294X.2007.03640.x. [DOI] [PubMed] [Google Scholar]
- Gray A. Mammalian hybrids. A check-list with bibliography. Slough (United Kingdom): Commonwealth Agricultural Bureaux; 1972. [Google Scholar]
- Hailer F, Kutschera VE, Hallström BM, Fain SR, Leonard JA, Arnason U, Janke A. Response to comment on “Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage”. Science. 2013;339:1522. doi: 10.1126/science.1228066. [DOI] [PubMed] [Google Scholar]
- Hailer F, Kutschera VE, Hallström BM, Klassert D, Fain SR, Leonard JA, Arnason U, Janke A. Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage. Science. 2012;336:344–347. doi: 10.1126/science.1216424. [DOI] [PubMed] [Google Scholar]
- Hallast P, Balaresque P, Bowden GR, Ballereau S, Jobling MA. Recombination dynamics of a human Y-chromosomal palindrome: rapid GC-biased gene conversion, multi-kilobase conversion tracts, and rare inversions. PLoS Genet. 2013;9:e1003666. doi: 10.1371/journal.pgen.1003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallström BM, Janke A. Mammalian evolution may not be strictly bifurcating. Mol Biol Evol. 2010;27:2804–2816. doi: 10.1093/molbev/msq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrigan RJ, Mazza ME, Sorenson MD. Computation vs. cloning: evaluation of two methods for haplotype determination. Mol Ecol Resour. 2008;8:1239–1248. doi: 10.1111/j.1755-0998.2008.02241.x. [DOI] [PubMed] [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellborg L, Ellegren H. Low levels of nucleotide diversity in mammalian Y chromosomes. Mol Biol Evol. 2004;21:158–163. doi: 10.1093/molbev/msh008. [DOI] [PubMed] [Google Scholar]
- Hey J. Isolation with migration models for more than two populations. Mol Biol Evol. 2010;27:905–920. doi: 10.1093/molbev/msp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobolth A, Dutheil JY, Hawks J, Schierup MH, Mailund T. Incomplete lineage sorting patterns among human, chimpanzee, and orangutan suggest recent orangutan speciation and widespread selection. Genome Res. 2011;21:349–356. doi: 10.1101/gr.114751.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffecker JF, Elias SA. The human ecology of Beringia. New York: Columbia University Press; 2007. [Google Scholar]
- Holland BR, Huber KT, Moulton V, Lockhart PJ. Using consensus networks to visualize contradictory evidence for species phylogeny. Mol Biol Evol. 2004;21:1459–1461. doi: 10.1093/molbev/msh145. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jacobsen F, Omland KE. Extensive introgressive hybridization within the northern oriole group (Genus Icterus) revealed by three-species isolation with migration analysis. Ecol Evol. 2012;2:2413–2429. doi: 10.1002/ece3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson D, Gibson AP, Hudelot C, Higgs PG. OGRe: a relational database for comparative analysis of mitochondrial genomes. Nucleic Acids Res. 2003;31:202–206. doi: 10.1093/nar/gkg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- Kelly BP, Whiteley A, Tallmon D. The Arctic melting pot. Nature. 2010;468:891. doi: 10.1038/468891a. [DOI] [PubMed] [Google Scholar]
- Krause J, Unger T, Nocon A, Malaspinas A-S, Kolokotronis S-O, Stiller M, Soibelzon L, Spriggs H, Dear PH, Briggs AW, et al. Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene-Pliocene boundary. BMC Evol Biol. 2008;8:220. doi: 10.1186/1471-2148-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubatko LS, Degnan JH. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst Biol. 2007;56:17–24. doi: 10.1080/10635150601146041. [DOI] [PubMed] [Google Scholar]
- Kurtén B. Pleistocene mammals of Europe. Chicago (IL): Aldine; 1968. [Google Scholar]
- Kurtén B, Anderson E. Pleistocene mammals of North America. New York: Columbia University Press; 1980. [Google Scholar]
- Lanier HC, Knowles LL. Is recombination a problem for species-tree analyses? Syst Biol. 2012;61:691–701. doi: 10.1093/sysbio/syr128. [DOI] [PubMed] [Google Scholar]
- Leaché AD, Harris RB, Rannala B, Yang Z. The influence of gene flow on species tree estimation: a simulation study. Syst Biol. 2014;63:17–30. doi: 10.1093/sysbio/syt049. [DOI] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lischer HEL, Excoffier L, Heckel G. Ignoring heterozygous sites biases phylogenomic estimates of divergence times: implications for the evolutionary history of Microtus voles. Mol Biol Evol. 2014;31:817–831. doi: 10.1093/molbev/mst271. [DOI] [PubMed] [Google Scholar]
- Liu S, Lorenzen ED, Fumagalli M, Li B, Harris K, Xiong Z, Zhou L, Korneliussen TS, Somel M, Babbitt C, et al. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell. 2014;157:785–794. doi: 10.1016/j.cell.2014.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Knowles LL. Inferring phylogeny despite incomplete lineage sorting. Syst Biol. 2006;55:21–30. doi: 10.1080/10635150500354928. [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- McKay BD, Zink RM. The causes of mitochondrial DNA gene tree paraphyly in birds. Mol Phylogenet Evol. 2010;54:647–650. doi: 10.1016/j.ympev.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Melo-Ferreira J, Boursot P, Carneiro M, Esteves PJ, Farelo L, Alves PC. Recurrent introgression of mitochondrial DNA among hares (Lepus spp.) revealed by species-tree inference and coalescent simulations. Syst Biol. 2012;61:367–381. doi: 10.1093/sysbio/syr114. [DOI] [PubMed] [Google Scholar]
- Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, Zhao F, Lim Kim H, Burhans RC, Drautz DI, Wittekindt NE, et al. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci U S A. 2012;109:E2382–E2390. doi: 10.1073/pnas.1210506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DA. Networks in phylogenetic analysis: new tools for population biology. Int J Parasitol. 2005;35:567–582. doi: 10.1016/j.ijpara.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagome S, Mano S, Hasegawa M. Comment on “Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage”. Science. 2013;339:1522. doi: 10.1126/science.1227339. [DOI] [PubMed] [Google Scholar]
- Nakagome S, Pecon-Slattery J, Masuda R. Unequal rates of Y chromosome gene divergence during speciation of the family Ursidae. Mol Biol Evol. 2008;25:1344–1356. doi: 10.1093/molbev/msn086. [DOI] [PubMed] [Google Scholar]
- Nakhleh L. Computational approaches to species phylogeny inference and gene tree reconciliation. Trends Ecol Evol. 2013;28:719–728. doi: 10.1016/j.tree.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols R. Gene trees and species trees are not the same. Trends Ecol Evol. 2001;16:358–364. doi: 10.1016/s0169-5347(01)02203-0. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorato DP, Hellgren EC, Bussche RA, van D, Doan-Crider DL. Phylogeographic patterns within a metapopulation of black bears (Ursus americanus) in the American southwest. J Mammal. 2004;85:140–147. [Google Scholar]
- Pagès M, Bazin E, Galan M, Chaval Y, Claude J, Herbreteau V, Michaux J, Piry S, Morand S, Cosson J-F. Cytonuclear discordance among Southeast Asian black rats (Rattus rattus complex) Mol Ecol. 2013;22:1019–1034. doi: 10.1111/mec.12149. [DOI] [PubMed] [Google Scholar]
- Pagès M, Calvignac S, Klein C, Paris M, Hughes S, Hänni C. Combined analysis of fourteen nuclear genes refines the Ursidae phylogeny. Mol Phylogenet Evol. 2008;47:73–83. doi: 10.1016/j.ympev.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Pamilo P, Nei M. Relationships between gene trees and species trees. Mol Biol Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- Petit E, Balloux F, Excoffier L. Mammalian population genetics: why not Y? Trends Ecol Evol. 2002;17:28–33. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Prüfer K, Munch K, Hellmann I, Akagi K, Miller JR, Walenz B, Koren S, Sutton G, Kodira C, Winer R, et al. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca AL, Georgiadis N, O’Brien SJ. Cytonuclear genomic dissociation in African elephant species. Nat Genet. 2005;37:96–100. doi: 10.1038/ng1485. [DOI] [PubMed] [Google Scholar]
- Roos C, Zinner D, Kubatko LS, Schwarz C, Yang M, Meyer D, Nash SD, Xing J, Batzer MA, Brameier M, et al. Nuclear versus mitochondrial DNA: evidence for hybridization in colobine monkeys. BMC Evol Biol. 2011;11:77. doi: 10.1186/1471-2148-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk K, Brede N, Streit B. Introduction. Extent, processes and evolutionary impact of interspecific hybridization in animals. Philos Trans R Soc B Biol Sci. 2008;363:2805–2811. doi: 10.1098/rstb.2008.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servheen C. The status and conservation of the bears of the world. Int Conf Bear Res and Manage Monogr Series. 1990;2:1–32. [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Suchard MA, Weiss RE, Sinsheimer JS. Models for estimating Bayes factors with applications to phylogeny and tests of monophyly. Biometrics. 2005;61:665–673. doi: 10.1111/j.1541-0420.2005.00352.x. [DOI] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of Dna sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallmon DA, Bellemain E, Swenson JE, Taberlet P. Genetic monitoring of Scandinavian brown bear effective population size and immigration. J Wildl Manag. 2004;68:960–965. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell PJ, Kishino H, Ota R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 2001;12:141–154. [PubMed] [Google Scholar]
- Wayne RK, Van Valkenburgh B, O’Brien SJ. Molecular distance and divergence time in carnivores and primates. Mol Biol Evol. 1991;8:297–319. doi: 10.1093/oxfordjournals.molbev.a040651. [DOI] [PubMed] [Google Scholar]
- Woerner AE, Cox MP, Hammer MF. Recombination-filtered genomic data sets by information maximization. Bioinformatics. 2007;23:1851–1853. doi: 10.1093/bioinformatics/btm253. [DOI] [PubMed] [Google Scholar]
- Yu L, Li Q-W, Ryder OA, Zhang Y-P. Phylogeny of the bears (Ursidae) based on nuclear and mitochondrial genes. Mol Phylogenet Evol. 2004;32:480–494. doi: 10.1016/j.ympev.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Yu L, Li Y-W, Ryder OA, Zhang Y-P. Analysis of complete mitochondrial genome sequences increases phylogenetic resolution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evol Biol. 2007;7:198. doi: 10.1186/1471-2148-7-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Barnett RM, Nakhleh L. Parsimonious inference of hybridization in the presence of incomplete lineage sorting. Syst Biol. 2013;62:738–751. doi: 10.1093/sysbio/syt037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Degnan JH, Nakhleh L. The probability of a gene tree topology within a phylogenetic network with applications to hybridization detection. PLoS Genet. 2012;8:e1002660. doi: 10.1371/journal.pgen.1002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Zheng P, Dong S, Zhan X, Wu Q, Guo X, Hu Y, He W, Zhang S, Fan W, et al. Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat Genet. 2013;45:67–71. doi: 10.1038/ng.2494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.