Abstract
SUMOylation is the covalent conjugation of SUMO polypeptides to cellular target proteins. Psmd1 is a subunit of the proteasomal 19S regulatory particle that acts as a docking site for Adrm1, another proteasome subunit that recruits ubiquitinated substrates for proteolysis. Here, we show that the SUMO deconjugating enzyme xSENP1 specifically interacts with Psmd1, and that disruption of xSENP1 targeting delays mitotic exit. Psmd1 becomes SUMOylated through the action of the SUMO E3 enzyme PIASy. We mapped SUMOylation sites within Psmd1, and find that SUMOylation of a critical lysine immediately adjacent to the Adrm1 binding domain regulates Adrm1 association with Psmd1. Together, our findings suggest that the interaction of Psmd1 with Adrm1 is controlled by SUMOylation in a manner that may alter proteasome composition and function. These findings demonstrate a new mechanism for regulation of ubiquitin-mediated protein degradation by ubiquitin-like proteins of the SUMO family.
Introduction
SUMOylation is the covalent conjugation of SUMO proteins (Small ubiquitin-related modifiers) to target proteins through the sequential action of E1 (Uba2/Aos1) and E2 (Ubc9) enzymes (Gareau and Lima, 2010). Most targets also require a SUMO ligase or E3 enzyme to facilitate their SUMOylation. SUMOylation is reversed by SUMO-specific deconjugating enzymes called Ulp/SENPs (Mukhopadhyay and Dasso, 2007). Yeast has two Ulp/SENPs, Ulp1p and Ulp2p. ULP1 is essential, and ulp1Δ strains arrest in mitosis (Li and Hochstrasser, 1999). There are four Ulp1p-like Ulp/SENPs in mammals: SENP1, SENP2, SENP3 and SENP5 (Mukhopadhyay and Dasso, 2007). SENP1 and SENP2 are most similar to each other; like Ulp1p, the vertebrate SENP1/SENP2 subfamily is important for mitosis (Cubenas-Potts et al., 2013; Era et al., 2012; Zhang et al., 2008).
Proteasomes are multi-subunit proteases that mediate the degradation of proteins that have been targeted for destruction by ubiquitination (Tomko Jr and Hochstrasser, 2013). Ubiquitinated degradation substrates are fed into the proteasome's catalytic 20S core particle (20S-CP) through the 19S regulatory particle (19S-RP). Psmd1 (Rpn2 in yeast) is the largest subunit of 19S-RP (Tomko Jr and Hochstrasser, 2013). Psmd1 plays a key structural role in the 19S-RP and acts as a docking site for other proteasome subunits, including Adrm1 (Rpn13 in yeast), a subunit that recruits ubiquitinated substrates to the 19S-RP. Adrm1 also recruits and activates UCH37, a deubiqitinating enzyme (Lee et al., 2011). Proteasomal subunits have been found in proteomic screens for SUMOylation substrates (Becker et al., 2013; Golebiowski et al., 2009), but no role of their modifications has been reported.
Taking advantage of the fact that the frog X. laevis has only one member of the SENP1/SENP2 subfamily, xSENP1 (Wang et al., 2009), we have investigated the mitotic function of SENP1/SENP2 proteases through manipulation of xSENP1 in Xenopus egg extracts (XEEs) (Maresca and Heald, 2006). We found that disruption of xSENP1 targeting caused defects in mitotic exit, and that xSENP1 associated strongly with Psmd1. We mapped SUMOylation sites within Psmd1, and found that modification of a critical lysine adjacent to the Adrm1 binding domain regulates Adrm1 association with Psmd1. Our findings suggest Psmd1 SUMOylation controls proteasome composition and function, providing a new mechanism for regulation of ubiquitin-mediated protein degradation through the SUMO pathway.
Results and Discussion
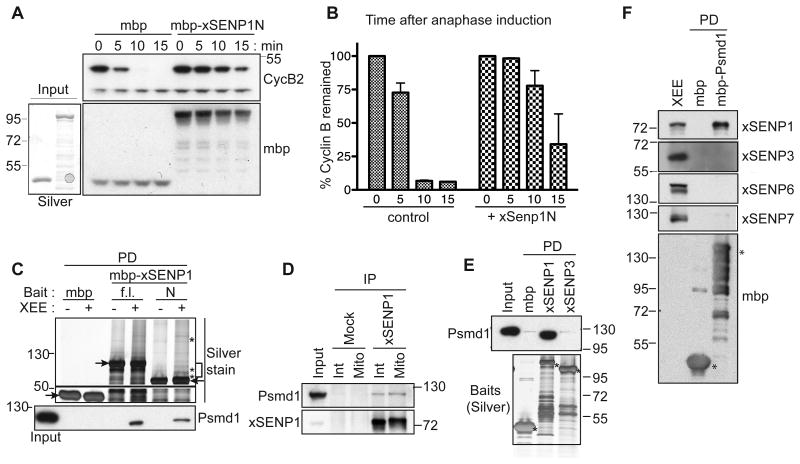
The N-terminal domains of SENPs determine their localization and contribute to their substrate specificity (Mukhopadhyay and Dasso, 2007). We reasoned that addition of a recombinant N-terminal xSENP1 fragment (xSENP1N) might act in a dominant negative manner by displacing endogenous xSENP1. We added MBP-fused xSENP1N to M-phase arrested XEEs (CSF-XEEs), followed by induction of anaphase (Figure 1A, 1B). As shown by the rate of Cyclin B protein destruction, the addition of xSENP1N delayed anaphase progression in comparison to control XEEs to which MBP was added, suggesting that xSENP1 function is important in some way for mitotic exit.
Figure 1. Psmd1 binds xSENP1 specifically in XEE.
(A) 5 μM MBP-tagged N-terminal xSENP1 fragment (mbp-xSENP1N; amino acids 1-420) or MBP were added to CSF-XEE in the presence of sperm chromatin. Anaphase was induced with 0.6 mM Ca2+ (time = 0 min), and samples were taken periodically for analysis by Western blotting with anti-Cyclin B and anti-MBP. Lower left panel, shows silver stain of input proteins (mbp and mbp-xSENP1N).
(B) Mean Cyclin B levels from three independent experiments performed as in (A), quantified using ImageJ. Error bars = standard deviation.
(C) Pull-down (PD) samples from XEE using MBP, MBP-tagged full length xSENP1 (f.l.) or MBP-xSENP1N (N), subjected to SDS-PAGE and silver staining. Arrowheads and asterisks indicate bait and binding proteins, respectively. Proteins within the bracket were analyzed by mass spectrometry. Psmd1 association to xSENP1 was confirmed by Western blotting (lower panel). Input: 2.5%
(D) immunoprecipitates (IP) from interphase (Int) or mitotic (Mito) XEEs using either IgG (Mock) or anti-xSENP1 antibodies were analyzed by Western blotting with the indicated antibodies. Input: 1%
(E) Pull-down samples from XEE using MBP, MBP-xSENP1 or MBP-xSENP3 were analyzed by Western blotting with anti-Psmd1 (upper panel) or silver staining (lower panel). Asterisks indicate bait. Input: 2.5%
(F) Reciprocal Pull-down assays samples from XEE using MBP or MBP-Psmd1 were analyzed by Western blotting for the indicated proteins. Asterisks indicate bait. Input: 5%
To understand xSENP1's function, we performed pull-down assays from XEE (Figure 1C), and observed several proteins on silver stained gels that bound xSENP1 and xSENP1N but not MBP. These proteins were excised from a Coomassie blue stained gel (bracket) and analyzed by mass spectrometry. Psmd1 was among the most prominent proteins identified, and Western blotting confirmed its association to both full-length xSENP1 and xSENP1N (Figure 1C, bottom panel). Psmd1 was present in anti-xSENP1 immunoprecipitates from interphase and mitotic XEEs (Figure 1D), indicating that this association occured throughout the cell cycle.
We examined Psmd1 binding to other SENPs in two ways: First, we performed pull-down experiments comparing MBP-xSENP1 to MBP-xSENP3, the other Ulp1p-like SENP present in XEEs (Wang et al., 2009) (Figure 1E). While Psmd1 bound strongly to MBP-xSENP1, its binding to MBP-xSENP3 was negligible. Second, we performed reciprocal pull-down experiments using MBP-Psmd1, which showed strong interaction with xSENP1 but not xSENP3, xSENP6 or xSENP7 (Figure 1F). Additionally, we observed co-precipitation of bacterially expressed Psmd1 with purified xSENP1, indicating that they associate in the absence of any other XEE components (Figure S1). Together, our data suggest that Psmd1 binds xSENP1 in a direct, specific fashion.
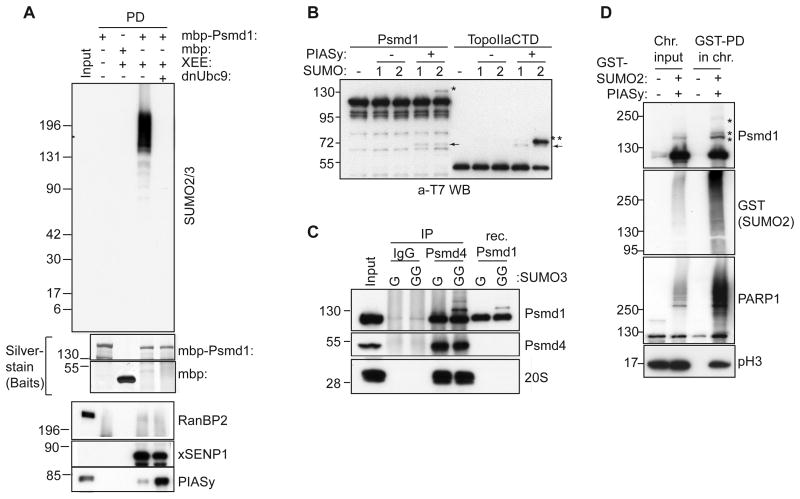
Western blotting of isolated mitotic spindles formed in CSF-XEEs indicated that both xSENP1 and Psmd1 are concentrated on spindles (Figure S2A). A smaller amount of these proteins associated with chromosomes purified from nocodazole-treated CSF-XEEs. We determined the distribution of Psmd1 and xSENP1 on mitotic chromosomes by immunofluorescent staining. Psmd1 and xSENP1 concentrated at centromeres (Figure S2B). SUMOylated species are abundantly concentrated on mitotic centromeres in XEEs (Ryu and Azuma, 2010), and we speculated that Psmd1 might be a SUMOylation target. To test this idea, MBP-Psmd1 was incubated in XEE, re-isolated and analyzed by Western blotting. Antibodies against MBP and SUMO2 both detected a smear migrating more slowly than MBP-Psmd1 that was abolished in reactions containing a dominant negative form of the SUMO E2 enzyme (dnUbc9) (Figure S2C and Figure 2A). No corresponding smear was observed when we blotted the same samples with anti-SUMO1 antibodies. These data suggest that Psmd1 is a paralog-specific target for conjugation to SUMO2/3 in mitotic XEE.
Figure 2. Psmd1 is modified by SUMO2/3 in XEE and in vitro.
(A) MBP or MBP-Psmd1 were incubated under the indicated conditions, and pulled down using Amylose resin. Where indicated, dominant negative E2 (dnUbc9) was included to inhibit SUMOylation. The samples were analyzed by Western blotting with antibodies against the indicated proteins. Silver stain shows bait proteins. Input: 5%
(B) T7-tagged Psmd1 or T7-tagged TopoIIαCTD were subjected to in vitro SUMOylation with or without PIASy, and analyzed by Western blotting with anti-T7 antibodies. 1 and 2 indicate SUMO1 and SUMO2, respectively. Asterisks and arrows indicate SUMO-conjugated species and PIASy, respectively.
(C) Immunoprecipitations (IP) from XEE using IgG and anti-Psmd4 antibodies were subjected to in vitro reactions as in (B), except that SUMO3 replaced the other paralogs. GG indicates that the mature form of SUMO3 was used, while G indicates use of a truncated, non-conjugatable form. Recombinant (Rec.) Psmd1 was concurrently subjected to in vitro SUMOylation. The samples were analyzed by Western blotting, as indicated. Antibodies against subunit C2 were used to detect 20S proteasome. Input: 5%
(D) Sperm chromatin was incubated for 60 min in XEE in the absence or presence of GST-SUMO2 and PIASy. The isolated chromosome fractions (Chr.) were processed and pulled down for GST-SUMO2 (see experimental procedures), followed by Western blotting with the indicated antibodies. Asterisks indicate SUMO-modified forms of Psmd1. pH3 indicates phosphohistone H3. Input: 6%
PIASy, a major mitotic SUMO E3 ligase in XEE (Azuma et al., 2005), was among the proteins that were pulled down from XEE with Psmd1 (Figure 2A). However, we did not detect RanBP2, another SUMO ligase reported to associate with Psmd1 (Yi et al., 2007). PIASy binding to Psmd1 was enhanced upon dnUbc9 addition. This phenomenon may be analogous to ‘substrate trapping’, wherein dominant negative mutant enzymes form stabilized complexes with their substrates (Flint et al., 1997). We tested whether PIASy catalyzed Psmd1 SUMOylation within in vitro assays that also contained E1 and E2 at concentrations similar to those in XEE. We observed PIASy-dependent Psmd1 SUMOylation (Figure 2B), which occurred specifically with SUMO2, as we had observed in XEE. Moreover, PIASy SUMOylated Psmd1 in the context of the intact 19S-RP: we immunoprecipitated proteasomes from XEE using anti-Psmd4 antibodies, and subjected them to in vitro SUMOylation as in Figure 2B. The efficiency of Psmd1 SUMOylation within the 19S-RP was comparable to that observed for recombinant Psmd1 (Figure 2C).
To determine whether chromosome-associated Psmd1 becomes SUMOylated, we isolated chromosomes formed in CSF-XEE containing sperm chromatin in the presence or absence of exogenous PIASy and GST-SUMO2. GST-SUMO2-conjugated proteins were isolated from the chromosomal fractions by affinity chromatography, and we found that a portion of Psmd1 was SUMOylated in the mitotic chromosomal fraction (Figure 2D). Together, our data indicated that Psmd1 is a substrate for PIASy-dependent conjugation to SUMO2/3, and that it can become SUMOylated in the context of intact proteasomes and on mitotic chromosomes.
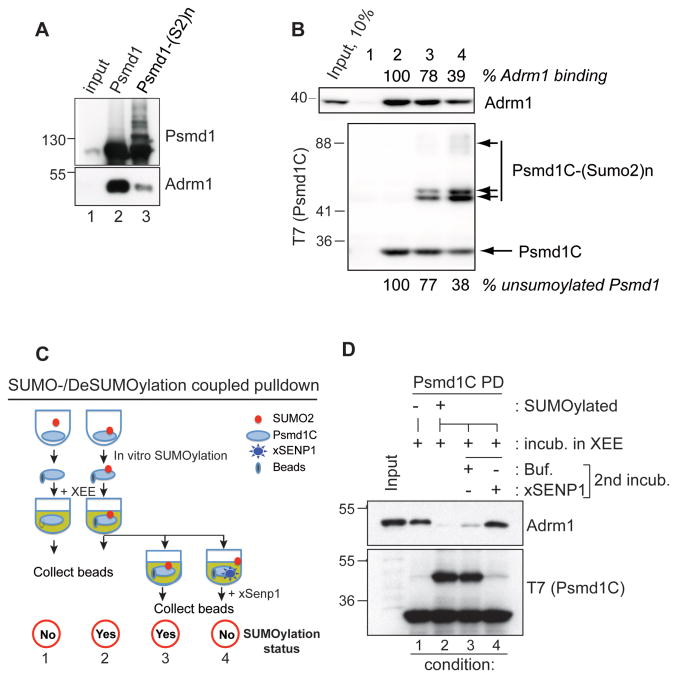
We wished to determine the sites of Psmd1 SUMOylation and the consequences of this modification. A SUMOylation site prediction program, SUMOsp 2.0, indicated that potential SUMO acceptor lysines lie mainly within the Psmd1 C-terminal domain. Human Adrm1 associates to the proteasome through the C-terminus of Psmd1 (He et al., 2012); Xenopus Adrm1 likewise bound the C-terminus of Psmd1 (Figure S3), while xSENP1 bound to the middle domain of Psmd1 (PC domain, a.a. 348-782). We hypothesized that Psmd1 SUMOylation might alter Adrm1 binding. To test this idea, we SUMOylated Psmd1 in vitro using elevated concentrations of enzymes to enhance its modification. Psmd1 was isolated on beads, which were introduced to XEE to allow Adrm1 binding. After re-isolation and washing, SUMOylated Psmd1 beads showed correspondingly less co-precipitating Adrm1 than those from a mock reaction lacking ATP (Figure 3A), indicating that SUMOylation compromises Psmd1 binding to Adrm1.
Figure 3. SUMOylation of the Psmd1 C-terminus negatively regulates its interaction with Adrm1.
(A) Full length MBP-Psmd1 was either mock-treated (lane 2) or subjected to in vitro SUMOylation (lane 3), followed by incubation with XEE. The samples were subjected to affinity chromatography, followed by Western blotting of the bound fractions with anti-Psmd1 (upper panel; bracket indicates SUMOylated Psmd1) or anti-Adrm1 (lower panel). Input: 5% of mock-treated input reaction.
(B) A T7-tagged C-terminal fragment of Psmd1 (Psmd1C) was subjected to in vitro SUMOylation for 0 (lane 2), 20 (lane 3) and 60 min (lane 4). The beads were incubated in CSF-XEE, re-isolated and washed. Bound proteins were analyzed by Western blotting with anti-Adrm1 (upper panel) or anti-T7 (lower panel). Lane 1 shows a control sample with empty beads. The amounts unSUMOylated T7-Psmd1C (below lower panel) and of Adrm1 bound to the beads (above upper panel) were quantitated for reactions containing T7-Psmd1C, and normalized relative to levels in lane 2. Input: 10%
(C) Schematic of experiment Figure 3D. In vitro SUMOylation reactions of T7-Psmd1C containing one volume (1x) or three volumes (3x) were incubated without or with ATP, respectively, followed by proportional addition of CSF-XEE and further incubation for 30 min at 23°C. T7-Psmd1C-bound proteins were isolated from the first reaction (-ATP) on beads. The latter (3x) was split into three equal portions; T7-Psmd1C-bound proteins were isolated on beads from the first portion without further manipulation. The second and third portions were supplemented with buffer or xSENP1 catalytic domain, respectively, and incubated for 30 min at 23°C, followed by capture of T7-Psmd1C-bound proteins on beads. All samples were eluted with 1x sample buffer.
(D) Proteins prepared as in (C) were analyzed by Western blotting with anti-Adrm1 and anti-T7. Buf. = buffer. Input: 5%
We prepared a Psmd1 C-terminal fragment (Psmd1C; a.a. 783-951) that harbors most of the predicted SUMO acceptor lysines and the Adrm1 binding motif. Psmd1C was incubated with SUMO2, E1, E2 and PIASy in the presence or absence of ATP (Figure S4). We incubated SUMOylated or mock-treated Psmd1C with XEE, followed by isolation and detection of Adrm1 by Western blotting. Adrm1 binding was lost in close correlation with increasing levels of Psmd1C SUMOylation (Figure 3B). We predicted that if SUMOylation occludes Adrm1 binding to the C-terminus of Psmd1, its deSUMOylation should restore binding. To test this idea, a deSUMOylation step was included in the SUMOylation-coupled pull-down (Figure 3C). As before, Psmd1C SUMOylation decreased Adrm1 binding (Figure 3D, conditions 1 and 2), but the deconjugation of SUMOylated Psmd1C by an exogenous xSENP1 catalytic fragment restored Adrm1 interaction (conditions 2, 3, and 4), indicating that SUMOylation indeed blocks Adrm1 association to the C-terminal domain of Psmd1.
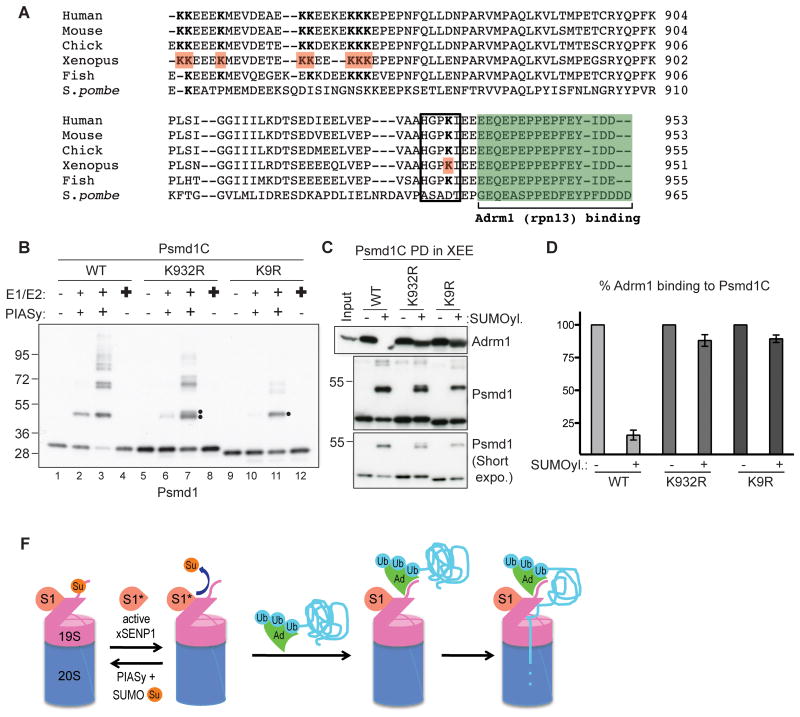
We used LC/MS/MS to map acceptor lysines of in vitro SUMOylated full length Psmd1 and Psmd1C, in combination with a candidate approach. We identified nine lysines near the Adrm1 binding motif as bona fide SUMO acceptors (a.a. 848, 849, 853, 861, 862, 865, 866, 867, and 932 of Xenopus Psmd1; Figure 4A, B). A Psmd1C mutant in which these lysines were substituted with arginine, Psmd1C-K9R, showed dramatically reduced SUMOylation (compare lane 2 and 10). Psmd1C-K9R showed modest SUMOylation at elevated SUMO enzyme concentrations, although it remained below the level of SUMOylation observed for wild type Psmd1C (lane 3 and 11). One predicted SUMOylation site, K932, lies immediately adjacent to the Adrm1 binding site and has an excellent “ΨKXE/D” SUMOylation consensus motif (Yang et al., 2006) (Figure 4A). Mutation of K932 caused a substantial reduction of SUMOylation (Figure 4B, lanes 2 and 6), although other lysines were still modified at high enzyme concentrations (lane 3 and 7). Notably, PIASy was essential for efficient SUMOylation of Psmd1C (Figure 4B, lanes 4, 8, 12).
Figure 4. SUMOylation on Lys 932 of Psmd1 is critical to inhibit Adrm1 binding.
(A) C-terminal sequences of human, mouse, chicken, frog, fish and yeast Psmd1 protein, aligned using ClustalW2 program. The green box indicates Adrm1-binding motif. Lysines identified as SUMO acceptors in X. laevis are in red and bold. The black box shows a SUMOylation consensus motif. Note that SUMO acceptor lysines are conserved among higher eukaryotes.
(B) Psmd1C wild type (WT), K932R, and K9R were subjected to in vitro reactions that contain various concentrations of SUMO E1, E2, PIASy. Small “+”reactions contain enzyme concentrations similar to XEE endogenous levels: 15 nM E1, 30 nM E2 and 10 nM PIASy. Medium “+” reactions contain double the level of SUMO enzymes. Bold “+” reactions contain 150 nM E1 and 300 nM E2, but no PIASy. Note that PIASy is essential for Psmd1C SUMOylation. The double dot indicates alternatively SUMOylated forms. The single dot indicates residually SUMOylated forms of Psmd1C observed after nine lysines were mutated to arginine.
(C) Psmd1C WT and mutants treated as in Panel B, under conditions without (lanes 1, 5, 9) or with SUMOylation (lanes 3, 7, 11), were used for pull down assays in XEE. Bound proteins were analyzed by Western blotting with anti-Adrm1 and anti-Psmd1. Input: 5%
(D) Two independent experiments performed as in (C) were quantified using ImageJ. The graph shows Adrm1 levels bound to SUMOylated Psmd1C WT or mutants normalized to Adrm1 bound to the same forms of Psmd1 without prior SUMOylation. Error bar shows standard deviation.
(F) Model: PIASy conjugates SUMO2/3 (Su) to the C-terminus of Psmd1 (extension from 19S-RP), occluding the Adrm1 (Ad) docking site. Active xSENP1 (S1*) antagonizes this modification, allowing Adrm1 recruitment. The balance of conjugation and deconjugation might be regulated, perhaps through conversion of xSENP1 between inactive (S1) and active forms, with deconjugation favoring proteasome activity. Ub: ubiquitinated targets of Adrm1.
We subjected Psmd1C WT, K932R, and K9R to in vitro SUMOylation or mock treatment, followed by their introduction to XEE to analyze Adrm1 binding as in Figure 3A. The capacity of WT Psmd1C to bind Adrm1 decreased by 85% after SUMOylation (Figure 4C and 4D). However, SUMOylation of either mutant protein caused less than a 10% decrease in Adrm1 binding in comparison to the mock-treated control samples. Taken together, our results suggest that K932 is a major SUMO acceptor whose conjugation regulates Adrm1 binding, while SUMOylation of nearby lysines may help to modulate Adrm1 recruitment.
Collectively, our data suggest a model in which Psmd1 becomes SUMO2/3-modified by PIASy, preventing Adrm1 docking (Figure 4E). We propose that xSENP1 removes SUMOylation from Psmd1, allowing Adrm1 loading and the degradation of key proteasomal targets. Under circumstances when xSENP1 is inhibited, this pathway would be disrupted, causing an inability to degrade Adrm1-dependent substrates. While our data indicate that PIASy and xSENP1 mediate Psmd1 SUMOylation and deSUMOylation, respectively, precisely how and when their activities are regulated remains to be elucidated. We do not know the identity of the protein(s) whose degradation might be controlled in this manner, although clearly this will be another important point for future investigation. Notably, Psmd1 associates with xSENP1 throughout the cell cycle (Figure 1D), so this mechanism could operate in other contexts.
Disruption of xSENP1 targeting in XEEs delays mitotic exit (Figure 1A, B). SENP1 depletion from cultured mammalian cells likewise delays sister chromatid segregation and anaphase onset (Cubenas-Potts et al., 2013), suggesting that SENP1 function is conserved among vertebrates. However, the bulk of Psmd1 remains un-SUMOylated in XEE (Figure 2). Thus, only a small fraction of proteasomes should be inhibited through Psmd1 SUMOylation, making it difficult to rationalize how such a marginal loss of proteasomal activity could slow mitotic progression. These issues might be reconciled in two ways: First, deSUMOylation of a protein other than Psmd1 could be necessary, and the delay caused by xSENP1N might reflect failure to deSUMOylate this substrate. Alternatively, there might be a SUMO-regulated proteasome sub-population that is essential for the proteolysis of key proteins. For example, if ubiquitination of mitotic targets were both spatially regulated and closely coupled to degradation, local regulation of proteasomes could also modulate their destruction. This is an attractive idea, particularly because activation of the anaphase promoting complex, a major mitotic ubiquitin ligase, is coupled to chromosome localization (Sivakumar et al., 2014), where we likewise observe SUMOylated Psmd1 (Figure S2). Further work will clearly be needed to test these possibilities.
There are a number of ways in which Psmd1 SUMOylation could impact the degradation of proteasomal targets: Changes in Psmd1-Adrm1 interactions could modulate the recruitment of ubiquitinated proteins to the 19S-RP. Proteasomes bind ubiquitinated substrates through Adrm1 and Rpn10 (Tomko Jr and Hochstrasser, 2013), which show distinct substrate recognition profiles. Genetic analysis shows that these two recognition pathways are not functionally redundant (Elangovan et al., 2010; Fatimababy et al., 2010), and some substrates are particularly dependent upon Adrm1 for their degradation, including Cyclin B (Chen et al., 2010). Additionally, Adrm1 mediates the recruitment and activation of UCH37, an enzyme that antagonizes the degradation of some ubiquitinated species (Lee et al., 2011). Changes in Adrm1 binding are thus strongly predicted to modulate the stability of these proteins. Finally, association to the 19S-RP places xSENP1 in an ideal location to cleave SUMO chains from proteins that are targeted for proteasomal degradation by SUMO-targeted ubiquitin ligases (STUbLs) (Geoffroy and Hay, 2009), and thus to modulate the destruction of STUbL substrates.
In summary, SUMO conjugation and deconjugation of Psmd1 by PIASy and xSENP1 provides a new mechanism to regulate proteasomal composition, as well as a novel and important point of cross-talk between ubiquitin-like modifier pathways.
Experimental Procedures
XEE preparation
Metaphase XEE (CSF-XEE) and sperm chromatin were prepared as described (Azuma, 2009). Interphase XEE was prepared by the addition of 0.6 mM CaCl2 to CSF-XEE and incubation for 60 min at 23°C. Unless otherwise specified, sperm chromatin was added at a concentration of 1000 nuclei per μl of final reaction.
Pull-down assays and immunoprecipitation
For pull-down assays, bacterially expressed His6-tagged proteins were bound to Talon affinity resin (Clontech), and MBP-tagged proteins were bound to Amylose resin (Biolabs) overnight at 4°C. The saturated resins we re blocked with 5% gelantin before mixing with 1:10 diluted XEE. The resins were incubated between 30 min and 2 hour at 23°C, washed three times with 1x PBS-T, and eluted in 1x SDS sample buffer. For immunoprecipitations (Figure 1D), anti-xSENP1 antibodies were bound to protein A-Dyna beads (Invitrogen) overnight at 4 °C and cross- linked using DMP (Thermo Scientific). The antibody-linked beads were incubated in 1:10 diluted XEE, washed with 1X PBS-T, and eluted in 1X SDS sample buffer. In Figure 2D, chromosomes were formed in CSF-XEE plus 8000 sperm nuclei per μl, with or without PIASy (50 nM) and GST-SUMO2 (5 μM). The isolated chromosome pellet was sonicated on ice and incubated for 30 min with DNaseI (Sigma) at 4°C. Th e samples were centrifuged at 10000g for 10 min, and the supernatants were subjected to GST-SUMO2 affinity chromatography over Glutathione-Sepharose. After elution with 1x SDS sample buffer, the samples were resolved on 4∼12% or 4∼20% tris-glycine gradient SDS-PAGE gels (NOVEX). Unless otherwise indicated, the input lane of each pull-down experiment using XEEs was loaded with a volume of XEE equivalent to the indicated percentage of the total reaction volume.
In vitro SUMOylation assays
Unless otherwise specified, in vitro SUMOylation assays were performed in the presence of 15 nM of E1, 30 nM of E2, 10 nM of PIASy, 5 μM SUMO paralogs, 0.5 μM substrates and 2.5 mM ATP. The reaction buffer contained 5 mM MgCl2, 100∼120 mM NaCl, 20 mM HEPES pH 7.8, 5% Glycerol, and 0.05% Tween20. Reactions were incubated in 27°C for one hour and stopped with 1x SDS sample buffer. For in vitro SUMOylation coupled pull-down assays (Figure 3 and 4, exclusive of Figure 3B), 10 μg of MBP-tagged Psmd1C was incubated with 150 nM E1, 300 nM E2, 100 nM PIASy, and 10 μM SUMO2GG at 27°C for 2 hours, with or without 5 mM ATP. The reactions were diluted 10-fold and incubated with Amylose resin for 90 min. The beads were added to CSF-XEE that had been diluted 10-fold with CSF-XB buffer, and incubated for 30 min at 23°C to allow Adrm1 binding. Finally, the beads were collected, washed three times with 1x PBS-T, and eluted in 1x SDS sample buffer. Where indicated, xSENP1 catalytic domain (amino acid 300-618) was added at a final concentration of 50∼100 nM.
For the in vitro SUMOylation coupled pull-down assay (Figure 3B), S-tagged Psmd1C was expressed in E. coli (BL21DE3Star) cultures containing 5% glycerol and 3% ethanol at 16°C for 40 hours, and purified using Ni-NTA beads followed by Superdex 200HR and Mono Q columns. 2 μg of S-tagged Psmd1C was incubated at 27°C for 0, 10, 20 or 60 min in a reaction buffer B (40 mM Tris, 100mM NaCl, 0.05% Tween-20, 5% glycerol, 2 mM DTT, 4 mM MgCl2, 2 mM ATP) that contained 150 nM E1, 200 nM E2, 50 nM PIASy, and 10 μM SUMO2GG. The reaction was diluted 20-fold with buffer B containing 10 mM EDTA and incubated with S-protein resin (EMD Millipore) for 90 min at 4°C. The beads were then mixed with CSF-XEE that ha d been diluted 10-fold in CSF-XB buffer, containing 10 ng/ml SUMO2-vinyl sulfone (Mukhopadhyay et al., 2006) to block SUMO isopeptidases and incubated for 90 min at 4°C. The beads were retrieved by centrifugation at 400 g for 10 s, washed 3 times in CSF-XB buffer, containing 0.05% Tween-20 and eluted in 1x SDS sample buffer.
Supplementary Material
Highlights.
The 19S regulatory particle (19S-RP) subunit Psmd1 is a SUMO2/3 conjugation target.
PIASy and xSENP1 mediate Psmd1 SUMOylation and deSUMOylation, respectively.
Psmd1 SUMOylation controls its binding to Adrm1, a ubiquitin-receptor of the 19S-RP.
Acknowledgments
We thank Kara Lukasiewicz for critical reading of this manuscript, and thank Woong Kim for the mass spectrometry analysis of Psmd1 SUMOylation sites. HR, AA and MD were supported by NICHD project # HD001902. YA was supported by NIH/NIGMS RO1 # GM80278.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azuma Y. Analysis of SUMOylation of topoisomerase IIalpha with Xenopus egg extracts. Methods Mol Biol. 2009;582:221–231. doi: 10.1007/978-1-60761-340-4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. The EMBO journal. 2005;24:2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Barysch SV, Karaca S, Dittner C, Hsiao HH, Berriel Diaz M, Herzig S, Urlaub H, Melchior F. Detecting endogenous SUMO targets in mammalian cells and tissues. Nature structural & molecular biology. 2013;20:525–531. doi: 10.1038/nsmb.2526. [DOI] [PubMed] [Google Scholar]
- Chen X, Lee BH, Finley D, Walters KJ. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Molecular cell. 2010;38:404–415. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubenas-Potts C, Goeres JD, Matunis MJ. SENP1 and SENP2 affect spatial and temporal control of sumoylation in mitosis. Molecular biology of the cell. 2013;24:3483–3495. doi: 10.1091/mbc.E13-05-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elangovan M, Oh C, Sukumaran L, Wojcik C, Yoo YJ. Functional differences between two major ubiquitin receptors in the proteasome; S5a and hRpn13. Biochemical and biophysical research communications. 2010;396:425–428. doi: 10.1016/j.bbrc.2010.04.108. [DOI] [PubMed] [Google Scholar]
- Era S, Abe T, Arakawa H, Kobayashi S, Szakal B, Yoshikawa Y, Motegi A, Takeda S, Branzei D. The SUMO protease SENP1 is required for cohesion maintenance and mitotic arrest following spindle poison treatment. Biochemical and biophysical research communications. 2012;426:310–316. doi: 10.1016/j.bbrc.2012.08.066. [DOI] [PubMed] [Google Scholar]
- Fatimababy AS, Lin YL, Usharani R, Radjacommare R, Wang HT, Tsai HL, Lee Y, Fu H. Cross-species divergence of the major recognition pathways of ubiquitylated substrates for ubiquitin/26S proteasome-mediated proteolysis. The FEBS journal. 2010;277:796–816. doi: 10.1111/j.1742-4658.2009.07531.x. [DOI] [PubMed] [Google Scholar]
- Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substratetrapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nature reviews. Molecular cell biology. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nature reviews. Molecular cell biology. 2009;10:564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Science signaling. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- He J, Kulkarni K, da Fonseca PC, Krutauz D, Glickman MH, Barford D, Morris EP. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric alpha-helical rings. Structure. 2012;20:513–521. doi: 10.1016/j.str.2011.12.015. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Molecular & cellular proteomics : MCP. 2011;10:R110 003871. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Heald R. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol Biol. 2006;322:459–474. doi: 10.1007/978-1-59745-000-3_33. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, Azuma Y, Wilkinson KD, Dasso M. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. The Journal of cell biology. 2006;174:939–949. doi: 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends in biochemical sciences. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Ryu H, Azuma Y. Rod/Zw10 complex is required for PIASy-dependent centromeric SUMOylation. The Journal of biological chemistry. 2010;285:32576–32585. doi: 10.1074/jbc.M110.153817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Daum JR, Tipton AR, Rankin S, Gorbsky GJ. The spindle and kinetochore-associated (Ska) complex enhances binding of the anaphase-promoting complex/cyclosome (APC/C) to chromosomes and promotes mitotic exit. Molecular biology of the cell. 2014;25:594–605. doi: 10.1091/mbc.E13-07-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ, Jr, Hochstrasser M. Molecular Architecture and Assembly of the Eukaryotic Proteasome. Annual review of biochemistry. 2013 doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mukhopadhyay D, Mathew S, Hasebe T, Heimeier RA, Azuma Y, Kolli N, Shi YB, Wilkinson KD, Dasso M. Identification and developmental expression of Xenopus laevis SUMO proteases. PloS one. 2009;4:e8462. doi: 10.1371/journal.pone.0008462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. The EMBO journal. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Friedman JL, Ferreira PA. The cyclophilin-like domain of Ranbinding protein-2 modulates selectively the activity of the ubiquitin-proteasome system and protein biogenesis. J Biol Chem. 2007;282:34770–34778. doi: 10.1074/jbc.M706903200. [DOI] [PubMed] [Google Scholar]
- Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Molecular cell. 2008;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.