Significance
Ribosomes exist in every cell and are responsible for translation from mRNA to protein. The structure of the ribosomal common core is highly conserved in all living species, while the outer regions of the ribosome are variable. Ribosomal RNA of eukaryotes contains expansion segments accreted onto the surface of the core, which is nearly identical in structure to that in prokaryotic ribosomes. Comparing eukaryotic and prokaryotic ribosomes allows us to identify 3D insertion fingerprints of the expansion segments. Similar fingerprints allow us to analyze the common core and detect ancestral expansion segments within it. We construct a molecular model of ribosomal evolution starting from primordial biological systems near the dawn of life, culminating with relatively recent changes specific to metazoans.
Keywords: RNA evolution, C value, origin of life, translation, phylogeny
Abstract
The origins and evolution of the ribosome, 3–4 billion years ago, remain imprinted in the biochemistry of extant life and in the structure of the ribosome. Processes of ribosomal RNA (rRNA) expansion can be “observed” by comparing 3D rRNA structures of bacteria (small), yeast (medium), and metazoans (large). rRNA size correlates well with species complexity. Differences in ribosomes across species reveal that rRNA expansion segments have been added to rRNAs without perturbing the preexisting core. Here we show that rRNA growth occurs by a limited number of processes that include inserting a branch helix onto a preexisting trunk helix and elongation of a helix. rRNA expansions can leave distinctive atomic resolution fingerprints, which we call “insertion fingerprints.” Observation of insertion fingerprints in the ribosomal common core allows identification of probable ancestral expansion segments. Conceptually reversing these expansions allows extrapolation backward in time to generate models of primordial ribosomes. The approach presented here provides insight to the structure of pre-last universal common ancestor rRNAs and the subsequent expansions that shaped the peptidyl transferase center and the conserved core. We infer distinct phases of ribosomal evolution through which ribosomal particles evolve, acquiring coding and translocation, and extending and elaborating the exit tunnel.
The translation system, one of life’s universal processes, synthesizes all coded protein in living systems. Our understanding of translation has advanced over the last decade and a half with the explosion in sequencing data and by the determination of 3D structures (1–4). X-ray crystallography and cryoelectron microscopy (cryo-EM) have provided atomic resolution structures of ribosomes from all three domains of life. Eukaryotic ribosomal structures are now available from protists (5), fungi (6), plants (7), insects, and humans (8). Here we describe an atomic level model of the evolution of ribosomal RNA (rRNA) from the large ribosomal subunit (LSU). Our evolutionary model is grounded in patterns of rRNA growth in relatively recent ribosomal expansions, for which there is an extensive, atomic-resolution record.
The common core LSU rRNA (9, 10), which is approximated here by the rRNA of Escherichia coli, is conserved over the entire phylogenetic tree, in sequence, and especially in secondary structure (11) and 3D structure (12). By contrast, the surface regions and the sizes of ribosomes are variable (13, 14). Most of the size variability is found in eukaryotic LSUs (Fig. 1). The integrated rRNA size in the LSU follows the trend Bacteria ≤ Archaea < Eukarya. The added rRNA in eukaryotes interacts with eukaryotic-specific proteins (5, 8, 9) (SI Appendix, Fig. S1 and Dataset S1).
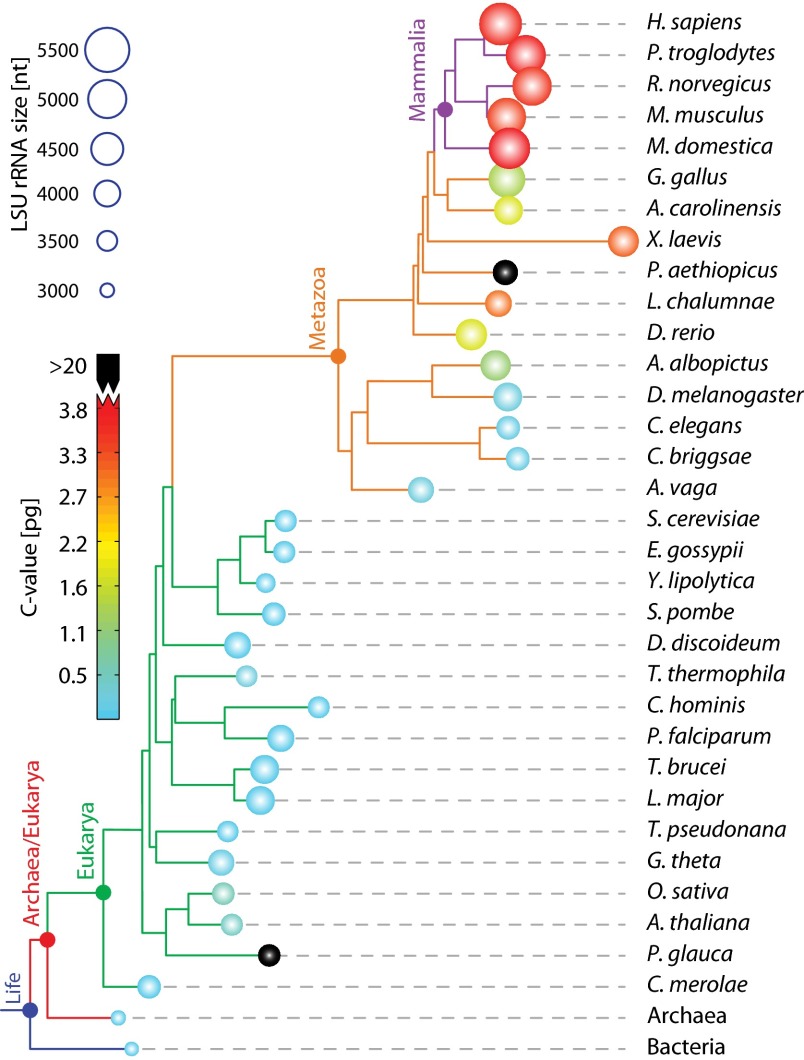
Fig. 1.
Phylogram indicating the sizes of LSU rRNAs and the sizes of genomes. Circle radii are proportional to total length of LSU rRNAs. Circles are colored by C value, which is genome size measured in picograms. Two species here have anomalously high C values and are colored in black (Protopterus aethiopicus: C-value 133 pg, and Picea glauca: C-value 24 pg). The sizes of archaeal and bacterial LSU rRNAs are highly restrained, so they are represented by just one species each. The phylogram was computed using sTOL (37) and visualized with ITOL (38). Three species (P. aethiopicus, Adineta vaga, P. glauca) were manually added to the phylogram, because the genomes are not sufficiently annotated for sTOL analysis.
Bacterial and archaeal LSU rRNAs are composed entirely of the common core, with only subtle deviations from it. By contrast, eukaryotic LSU rRNAs are expanded beyond the common core. Sacccharomyces cerevisiae LSU rRNAs are around 650 nucleotides larger than the common core rRNA. Drosophila melanogaster LSU rRNAs are larger than those of S. cerevisiae by 524 nucleotides. Homo sapiens LSU rRNAs are larger than those of D. melanogaster by 1,149 nucleotides. The correlation of general level of biological complexity with LSU rRNA size (Fig. 1) could have profound implications for the nature and definition of complexity in biological systems. The C value, a measure of the genome size, does not correlate well with complexity (15). LSU size reaches a maximum in modern metazoans, with immense rRNA polymers of tremendous complexity, many proteins (8, 9), and a total atomic mass of well over 4 MDa. The differences in the small ribosomal subunit (SSU) components are more modest, with 69 additional nucleotides in the H. sapiens SSU rRNA over S. cerevisiae and 258 additional nucleotides in S. cerevisiae over E. coli (SI Appendix, Table S1).
Variation in rRNA structure across species provides information on ribosomal evolution. Mutation frequencies are greater in helices than in loops (16, 17). While examining archaeal 5S rRNAs, Luehrsen et al. made the first observation of an rRNA insertion (18). Comparisons of rRNA secondary structures between bacteria and eukaryotes led to the discovery of expansion segments in eukaryotic rRNAs (13, 14, 19, 20). As confirmed by recent structural studies, expansion segments are constrained to the periphery of the LSU, far from the peptidyl transferase center (PTC). The locations of the sites of expansion of S. cerevisiae and H. sapiens are indicated by arrows on the secondary structures in Fig. 2. In general, rRNA expansion does not perturb the common core or other ancestral rRNA: essentially all secondary structural helices of the E. coli rRNA are intact within the (larger) S. cerevisiae rRNA. Likewise, nearly all secondary structural helices of the S. cerevisiae rRNA are intact within the (larger) H. sapiens rRNA.
Fig. 2.
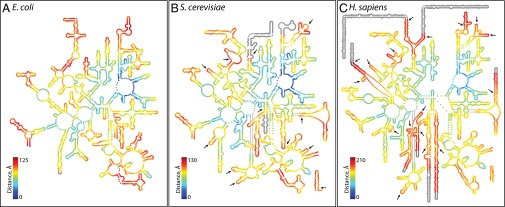
LSU rRNA secondary structures. (A) E. coli, (B) S. cerevisiae, and (C) H. sapiens. The color indicates the proximity in three dimensions to the site of peptidyl transfer. Blue is close to the site of peptidyl transfer and red is remote. In the secondary structures, the sites of expansion from E. coli to S. cerevisiae and from S. cerevisiae to H. sapiens are marked by arrows. Nucleotides that were not experimentally resolved in three dimensions are gray on the secondary structures.
Moving from secondary structures to 3D structures reveals that sites of expansion are associated with distinctive rRNA structures that we call “insertion fingerprints.” Comparison of pre- and postexpanded rRNAs in three-dimensions clearly reveals these insertion fingerprints, composed of ancestral helical trunks joined to more recent helical branches. The junctions are highly localized and do not perturb surrounding rRNA. New rRNA is added to old rRNA with preservation of the local conformation and of structural integrity of the old rRNA. Conformation and base pairing of ancestral helices are preserved upon expansion. Here, taking the conservative assumption that expansions within the common core followed the same rules as recent rRNA expansions, we use insertion fingerprints to infer a stepwise building up of the common core.
Results and Discussion
Steps in rRNA expansion can be “observed” by comparing 3D structures of serially increasing size (Fig. 3). This approach incorporates an assumption that the common ancestor of a pair of ribosomes is best approximated by the subset of rRNA that is present in both species. This subset of rRNA is, typically, most similar to the smaller rRNA. The general pattern is that as eukaryotic organisms increase in overall complexity, the rRNA becomes longer. However, during evolution of individual species, some expansion segments decrease in size. The reduction of rRNA also occurs during the evolution of some archaeal or bacterial species. The distribution of rRNA size mapped onto the phylogenetic tree is shown in Fig. 1.
Fig. 3.
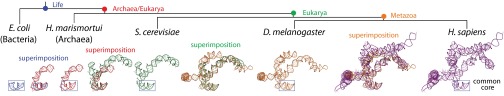
The evolution of helix 25/ES 7 shows serial accretion of rRNA onto a frozen core. This image illustrates at the atomic level how helix 25 of the LSU rRNA grew from a small stem loop in the common core into a large rRNA domain in metazoans. Each accretion step adds to the previous rRNA core but leaves the core unaltered. Common ancestors, as defined in Fig. 1, are indicated. Pairs of structures are superimposed to illustrate the differences and to demonstrate how new rRNA accretes with preservation of the ancestral core rRNA. Each structure is experimentally determined by X-ray diffraction or Cryo-EM.
A “movie” of rRNA growth is exemplified by the lineage of expansion segment 7 (ES 7), as shown in Fig. 3. A stem loop of rRNA (helix 25) and its progeny rRNAs present a multistep model of evolution of an rRNA domain (ES 7), at high resolution in three dimensions. ES 7 originates with a short 22-nucleotide stem loop in the last universal common ancestor, which is approximated here by E. coli. This stem loop grows to an 80-nucleotide bent helix in the common ancestor of Archaea and Eukarya. The common ancestor of Archaea and Eukarya is approximated by the archaeon Haloarcula marismortui. In the next step, ES 7 grows to a branched 210-nucleotide structure in the common ancestor of eukaryotes, which is approximated by S. cerevisiae. In the next step, ES 7 grows to a 342-nucleotide structure in the common ancestor of metazoans (approximated by the arthropod D. melanogaster). Mammalian rRNA grows further, exemplified by the 876 nucleotide ES 7 domain in H. sapiens.
In this series, one can observe accretion at the atomic level. The foundational helix 25 is fully intact in all larger rRNAs (Fig. 3) and was structurally conserved during a long evolutionary process. In general, each expansion step builds on preexisting rRNA, without substantially perturbing its 3D structure. This process has consistently been ongoing as the rRNA nearly doubled in size over 3.5 billion years of evolution, using the prokaryotic LSU as a foundation for the massive metazoan LSU.
Insertion Fingerprints.
The available structures allow us to make direct comparisons of pre- and postexpanded rRNA, and to observe rRNA conformation at sites where expansion elements join common core rRNA. We call the patterns observed at these sites insertion fingerprints.
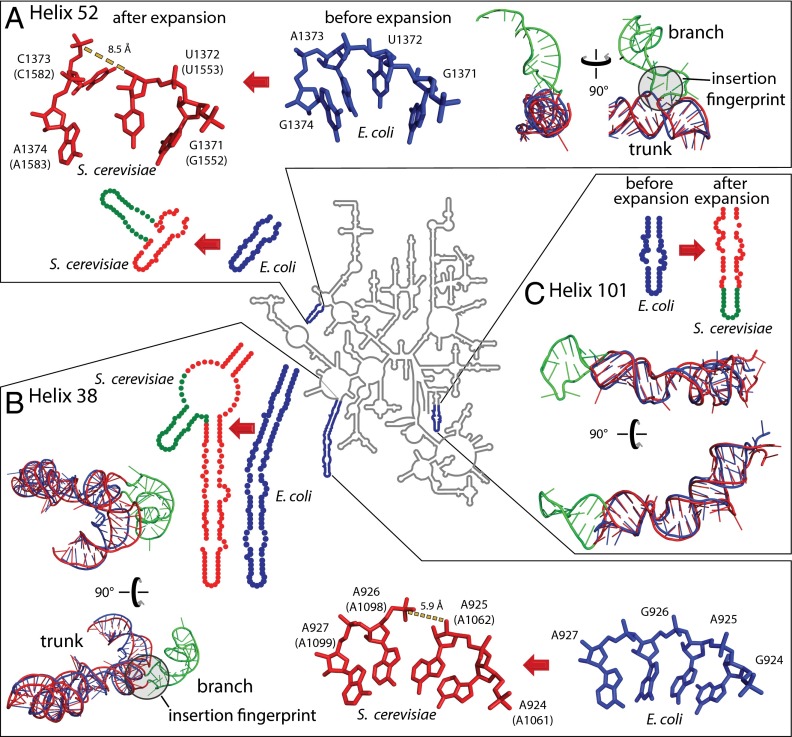
The predominant insertion fingerprint is a helical trunk joined to a secondary branching helix at a highly localized three- or four-way junction (21) that minimally perturbs the trunk helix. At most, a few base pairs of the trunk rRNA are disrupted or unstacked at the site of insertion. These atomic-level fingerprints are seen by comparing many pre- and postinserted expansion sites. For example, helix 52 (Fig. 4A and SI Appendix, Fig. S2) and helix 38 (Fig. 4B and SI Appendix, Fig. S3) are common core trunks in E. coli that have grown branches in the rRNA of S. cerevisiae. The E. coli rRNA shows trunk helices 38 and 52 before insertion of the branching helices, whereas the S. cerevisiae rRNA shows trunk helices sporting branch helices after insertion. A second type of expansion is elongation of a previous helix. Helix 101 of E. coli is elongated in S. cerevisiae (Fig. 4C and SI Appendix, Fig. S4) to form a continuous stack within the previous helical element. Helix elongations do not leave distinctive structural fingerprints. Comparisons of pre- and postexpanded rRNAs reveal that helix insertions or elongations occurred within the common core in helices 25, 30, 38, 52, 54, 63, 79, 98, and 101 of the LSU rRNA. Each of the expansion sites of the LSU, obtained by comparing pre- and postexpanded rRNA crystal structures of E. coli and S. cerevisiae, are shown in three dimensions and annotated in SI Appendix, Table S2.
Fig. 4.
rRNA expansion elements in two and three dimensions. (A) Helix 52 is expanded by insertion. (B) Helix 38 is expanded by insertion. (C) Helix 101 is expanded by elongation. The secondary structure of the LSU common core rRNA, represented by that of E. coli (34), is a gray line at the center of the figure. Selected regions where the E. coli rRNA has been expanded to give the S. cerevisiae rRNA are enlarged. In the enlargements, the rRNA is blue for E. coli and red for S. cerevisiae, except that expansion elements of S. cerevisiae rRNA are green. These observed expansion processes, from blue rRNA to red/green rRNA, are symbolized by red arrows. Superimposed pre- and postexpanded rRNAs indicate trunk (old) and branch (new) elements. Insertion fingerprints, where trunk meets branch, are highlighted by gray circles. E. coli nucleotide numbers are provided, with S. cerevisiae numbering in parentheses.
The patterns of conformation at sites of rRNA expansion suggests the reverse process, which is excision of inserted helices followed by religation to generate the ancestral RNA (SI Appendix, Fig. S5). The expansion is predicted to be conformationally facile and readily reversible in silico. We have tested this prediction. In general, a branching helix at an insertion fingerprint can be computationally excised, and the trunk rRNA can be religated by subtle shifts in the positions of a few nucleotides or even a single phosphate group. In all cases examined here, the religation can be achieved by gentle energy minimization with a shift of local atomic positions by a few ångströms and minimal perturbation of the trunk rRNA. Our modeling demonstrates how rRNA can be expanded (and contracted) with preservation of the ancestral core. Highly localized insertions have been reported in mRNAs (22).
Ancestral Insertion Fingerprints.
Here, we step back in time and reconstruct the growth of the common core rRNA, assuming that the common core developed in accordance with the patterns of expansion observed in eukaryotic rRNAs. We recapitulate the building up of the common core by stepwise additions of ancestral expansion segments (AESs) to a growing rRNA core, at sites marked by insertion fingerprints. We observe insertion fingerprints deeply buried within the common core of the LSU. These ancestral insertion fingerprints appear identical in form to modern insertion fingerprints of eukaryotic expansions. The observation of ancestral insertion fingerprints suggests that addition of eukaryotic expansion segments followed patterns established in biological antiquity. The ancestral insertion fingerprints within the common core point to some of the oldest imaginable evolutionary events and imply a method to work backward in time, to identify pathways of expansion during formation of the common core.
Building Up the Peptidyl Tranferase Center.
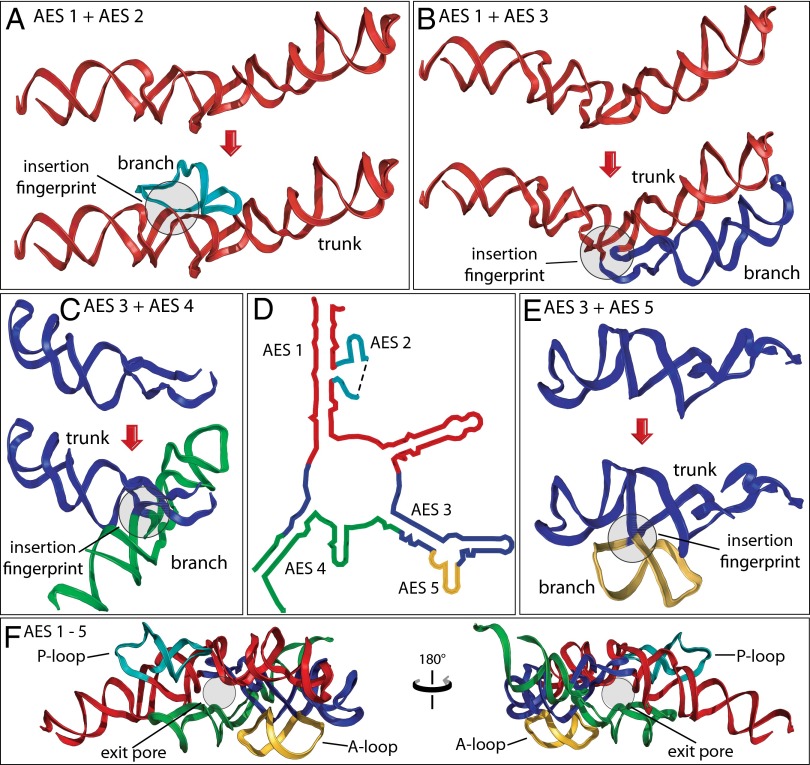
The PTC is an essential component of the ribosome, responsible for peptide bond formation. The PTC is thought to predate coded protein (23, 24) and is believed to be among the oldest polymeric elements of biological systems. The rRNA that forms the PTC (Fig. 5D) contains four insertion fingerprints (Table 1). A single continuous trunk helix (red) with a defect at the base of the P region appears to be the ultimate ancestor of the PTC. This rRNA fragment, denoted as ancestral expansion segment 1 (AES 1), is joined by AES 2 (the P loop) at an insertion fingerprint (Fig. 5A and SI Appendix, Fig. S6). AES 1 and AES 2 together comprise the P region. AES 1 is also joined by AES 3 at a second insertion fingerprint (Fig. 5B and SI Appendix, Fig. S7). The temporal ordering of the additions of AES 2 and AES 3 to AES 1 is undetermined.
Fig. 5.
Origins and evolution of the PTC. Trunk rRNA is shown before and after insertion of branch helix. (A) AES 1 (red) is expanded by insertion of AES 2 (teal). (B) AES 1 is expanded by insertion of AES 3 (blue). (C) AES 3 is expanded by insertion of AES 4 (green). (D) The secondary structure of AESs 1–5, which form the PTC and the exit pore (helices 74, 80, 89, 90, and 91–93). The ends of AES 2 are located in direct proximity to each other in three dimensions, indicated by a dashed line in the secondary structure. (E) AES 3 is expanded by insertion of AES 5 (gold). (F) The 3D structure of AESs 1–5, colored as in A–E. In each case, the before state was computationally modeled by removing the branch helix and sealing the trunk using energy minimization protocols. Positions of the P loop, the A loop, and the exit pore are marked. Enlarged and more detailed representations of the structures of AESs 1–5 are available in SI Appendix, Figs. S6–S9.
Table 1.
AESs within the PTC
| Expansion segments | Helices | Nucleotide numbers (E. coli) |
| AES 1 | H74, H75, H89 | 2061–2092; 2226–2245; 2435–2501 |
| AES 2 | H80 | 2246–2258; 2427–2434 |
| AES 3 | H90, H91 | 2053–2060; 2502–2546; 2567–2576 |
| AES 4 | H73, H93 | 2043–2052; 2577–2629 |
| AES 5 | H93 | 2547–2566 |
AES 3 appears to be expanded in turn by the addition of AES 4 (Fig. 5C and SI Appendix, Fig. S8) at one insertion fingerprint and the addition of AES 5 (the A loop, Fig. 5E, and SI Appendix, Fig. S9) at a second insertion fingerprint. AESs 3–5 form the A region of the PTC and the exit pore, which is the entrance to the exit tunnel. By the method of Steinberg (25), AESs 4 and 5 appear to be added after AESs 2 and 3. In our model, AES 1 and four expansion segments (AES 2–AES 5) together form not only the A and P regions but also a pore that, with later expansions, develops into the exit tunnel (Fig. 5F). In sum, we have a well-grounded model for evolution of some of the oldest polymeric elements in all of biology.
Building Up the Common Core and Eukaryotic LSUs.
The approach described here is readily extended, leading to a stepwise model of evolution of the common core and beyond (Fig. 6A and SI Appendix, Fig. S10). We propose that functional elements of the LSU emerge in a specific progressive ordering, in a series of distinct phases (Fig. 6B and SI Appendix, Fig. S11).
Phase 1: Folding, rudimentary binding, and catalysis. AESs 1–2 is a branched duplex with a defect that forms the P loop. This defect may confer catalytic activity (26) and/or ability to bind specifically to small molecules.
Phase 2: Maturation of the PTC and formation of an exit pore. Inclusion of AESs 3–5 adds the A region to the P region, in concert with formation of an exit pore (27).
Phase 3: Early tunnel extension. Inclusion of AESs 6–10 extends the exit pore, creating a short tunnel. The stability and rigidity of the tunnel are increased by buttressing.
Phase 4: Acquisition of the SSU interface. AESs 11–28 are included. AESs 11, 12, and 15 form the LSU interface for association with the SSU. The other segments enhance the stability and efficiency of the LSU by embracing the PTC and further extending the exit tunnel.
Phase 5: Acquisition of translocation function. Inclusion of AESs 29–39 adds essential components of the modern energy-driven translational machinery: the L7/L12 stalk and central protuberance (28, 29), and binding site (sarcin–ricin loop) for elongation factors G and Tu (28, 29). The tunnel is further extended.
Phase 6: Late tunnel extension. Further expansion of the LSU by inclusion of AESs 40–59 results in the maturation of common core of the LSU. In the final phase of prokaryotic ribosomal evolution (Fig. 2A and SI Appendix, Fig. S12A), the exit tunnel is extended. A majority of elements added here are located at the ribosomal surface and interact with ribosomal proteins.
Phase 7: Encasing the common core (simple eukaryotes). Eukaryotic expansion segments are acquired and previous AESs are elongated. This eukaryotic-specific rRNA combines with eukaryotic-specific proteins (9) (Fig. 2B and SI Appendix, Fig. S12B) to form a shell around the common core.
Phase 8: Surface elaboration (complex eukaryotes). Metazoan ribosomes are decorated with “tentacle-like” rRNA elements that extend well beyond the subunit surfaces (8). These tentacles (Fig. 2C and SI Appendix, Fig. S12C), are fundamentally different in structure and function than common core rRNA. Metazoan expansions appear to enable elaborate control, delivery, and complexity, and are thought, for example, to enable communication between the mRNA exit in the SSU and the exit tunnel terminus in the LSU, and to facilitate interactions with eukaryotic-specific factors involved in membrane localization.
Fig. 6.
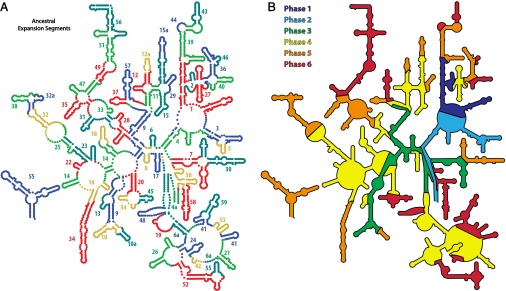
rRNA evolution mapped onto the LSU rRNA secondary structure. The common core is built up in six phases, by stepwise addition of ancestral expansion segments at sites marked by insertion fingerprints. (A) Each AES is individually colored and labeled by temporal number. AES colors are arbitrary, chosen to distinguish the expansions, such that no AES is the color of its neighbor. (B) Accretion of ancestral and eukaryotic expansion segments is distributed into eight phases, associated with ribosomal functions. Phase 1, rudimentary binding and catalysis (dark blue); phase 2, maturation of the PTC and exit pore (light blue); phase 3, early tunnel extension (green); phase 4, acquisition of the SSU interface (yellow); phase 5, acquisition of translocation function (orange); phase 6, late tunnel extension (red). Some AESs appear to be discontinuous on the secondary structure and so are labeled twice. A description of each AES and their partitioning into phases is given in SI Appendix, Table S3. The 3D structure of each phase is shown in SI Appendix, Fig. S11.
Conclusions
Here, we analyze changes in ribosomal size, structure, and complexity over the course of ribosomal evolution. We observe distinct patterns in conformation and interactions of rRNA where expansion elements of S. cerevisiae join the common core. We tabulate the expansions and analyze the rRNA structure of each site. The analysis reveals patterns of rRNA conformation that we call insertion fingerprints. We then extrapolate backward, by identifying insertion fingerprints within the common core. Identification of insertion fingerprints within the common core allows us to construct a stepwise model of the evolution of the common core. Ultimately, this approach allows us to infer some of the earliest evolutionary steps in the formation of the peptidyl transferase center, at the very dawn of ribosomal evolution.
In our model, the LSU has evolved in distinct phases. This process started with the formation of the P site, possibly in an RNA world, and continues today in eukaryotes. A unifying theme of LSU evolution is the continuous extension, stabilization, and elaboration of exit tunnel structure and function. The exit tunnel is formed, extended, stabilized, and elaborated continuously in nearly all phases of ribosomal evolution.
The model of LSU origins and evolution described here is more fine grained than previous models but is in essential agreement with them, despite different assumptions and types of input data. Harvey and coworkers compared secondary structures and sequences across multiple species, identifying the RNA components of the “minimal ribosome” (11). Fox analyzed density of molecular interactions and interconnectivities (24). Bokov and Steinberg developed a powerful model by analyzing A-minor interactions (25). Williams and coworkers treated the LSU as a growing onion (12). Where they overlap, our stepwise model here corresponds well with each of these previous models, although it provides a more rigorous definition of the ancestral expansion segments and addresses the origin of the PTC. The cumulative effect of the first four initial expansions (Fig. 5) gives a structure that is strikingly similar to an ancestral PTC proposed independently by Yonath and coworkers (30, 31). Those investigators suggested rRNA components of the PTC as an ancient catalytic heart of the common core. Some of the AESs proposed here correspond to rRNA “elements” that were used to construct the ribosome in the Bokov–Steinberg model (25).
In our model, rRNA has evolved by analogous processes throughout its history, from the origin of the PTC, through the common core, to highly expanded rRNAs in complex metazoans. We also show that the size of the LSU rRNA correlates better with biological complexity than does genome size (C value), however complexity is defined. We suggest that the size of the LSU rRNA might be a universal proxy of biological complexity.
Materials and Methods
Alignments and Phylogenetic Trees.
We aligned complete LSU rRNA sequences from 135 organisms intended to represent the broadest sparse sampling of the phylogenetic tree of life, including all three domains of life. The alignment is provided in FASTA format (SI Appendix, Dataset S2). The phylogenetic tree was generated from sTOL.
Secondary Structures.
Secondary structures of LSU and SSU rRNAs are taken from our public gallery (http://apollo.chemistry.gatech.edu/RibosomeGallery/) and data are mapped by RiboVision (32–34).
Three-Dimensional Structures.
Three-dimensional structures of ribosomal particles were obtained from the Protein Data Bank (PDB) database [PDB IDs 1JJ2 (2), 3R8S, 4GD1 (35), 3U5B, 3U5C, 3U5D, 3U5E (25), 3J38, 3J3C, 3J39, 3J3E (8), 3J3A, 3J3B, 3J3D, and 3J3F]. Local and global superimpositions were performed using the built-in cealign functionality of PyMOL (36). Details are available in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We would like to acknowledge Dr. Eric Gaucher for helpful discussions. This work was supported by the National Aeronautics and Space Administration Astrobiology Institute (NNA09DA78A).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407205111/-/DCSupplemental.
References
- 1.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285(5436):2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 2.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 3.Harms J, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107(5):679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 4.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 5.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334(6058):941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 6.Jenner L, et al. Crystal structure of the 80S yeast ribosome. Curr Opin Struct Biol. 2012;22(6):759–767. doi: 10.1016/j.sbi.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Armache JP, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. Proc Natl Acad Sci USA. 2010;107(46):19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anger AM, et al. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497(7447):80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 9.Melnikov S, et al. One core, two shells: Bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19(6):560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 10.Michot B, Qu LH, Bachellerie JP. Evolution of large-subunit rRNA structure. The diversification of divergent D3 domain among major phylogenetic groups. Eur J Biochem. 1990;188(2):219–229. doi: 10.1111/j.1432-1033.1990.tb15393.x. [DOI] [PubMed] [Google Scholar]
- 11.Mears JA, et al. Modeling a minimal ribosome based on comparative sequence analysis. J Mol Biol. 2002;321(2):215–234. doi: 10.1016/s0022-2836(02)00568-5. [DOI] [PubMed] [Google Scholar]
- 12.Hsiao C, Mohan S, Kalahar BK, Williams LD. Peeling the onion: Ribosomes are ancient molecular fossils. Mol Biol Evol. 2009;26(11):2415–2425. doi: 10.1093/molbev/msp163. [DOI] [PubMed] [Google Scholar]
- 13.Gerbi SA. Expansion segments: Regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In: Zimmermann RA, Dahlberg AE, editors. Ribosomal RNA—Structure, Evolution, Processing, and Function in Protein Synthesis. Boca Raton, FL: CRC; 1996. pp. 71–87. [Google Scholar]
- 14.Hassouna N, Michot B, Bachellerie JP. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984;12(8):3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doolittle WF. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci USA. 2013;110(14):5294–5300. doi: 10.1073/pnas.1221376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van de Peer Y, Neefs JM, De Rijk P, De Wachter R. Reconstructing evolution from eukaryotic small-ribosomal-subunit RNA sequences: Calibration of the molecular clock. J Mol Evol. 1993;37(2):221–232. doi: 10.1007/BF02407359. [DOI] [PubMed] [Google Scholar]
- 17.Gillespie JJ. Characterizing regions of ambiguous alignment caused by the expansion and contraction of hairpin-stem loops in ribosomal RNA molecules. Mol Phylogenet Evol. 2004;33(3):936–943. doi: 10.1016/j.ympev.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Luehrsen KR, Nicholson DE, Eubanks DC, Fox GE. An archaebacterial 5S rRNA contains a long insertion sequence. Nature. 1981;293(5835):755–756. doi: 10.1038/293755a0. [DOI] [PubMed] [Google Scholar]
- 19.Michot B, Bachellerie JP. Comparisons of large subunit rRNAs reveal some eukaryote-specific elements of secondary structure. Biochimie. 1987;69(1):11–23. doi: 10.1016/0300-9084(87)90267-7. [DOI] [PubMed] [Google Scholar]
- 20.Bachellerie JP, Michot B. Evolution of large subunit rRNA structure. The 3′ terminal domain contains elements of secondary structure specific to major phylogenetic groups. Biochimie. 1989;71(6):701–709. doi: 10.1016/0300-9084(89)90086-2. [DOI] [PubMed] [Google Scholar]
- 21.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12(1):83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19(10):1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woese CR. On the evolution of cells. Proc Natl Acad Sci USA. 2002;99(13):8742–8747. doi: 10.1073/pnas.132266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fox GE. Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol. 2010;2(9):a003483. doi: 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bokov K, Steinberg SV. A hierarchical model for evolution of 23S ribosomal RNA. Nature. 2009;457(7232):977–980. doi: 10.1038/nature07749. [DOI] [PubMed] [Google Scholar]
- 26.Wolf YI, Koonin EV. On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol Direct. 2007;2:14. doi: 10.1186/1745-6150-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox GE, Tran Q, Yonath A. An exit cavity was crucial to the polymerase activity of the early ribosome. Astrobiology. 2012;12(1):57–60. doi: 10.1089/ast.2011.0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114(1):123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 29.Lancaster L, Lambert NJ, Maklan EJ, Horan LH, Noller HF. The sarcin-ricin loop of 23S rRNA is essential for assembly of the functional core of the 50S ribosomal subunit. RNA. 2008;14(10):1999–2012. doi: 10.1261/rna.1202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krupkin M, et al. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos Trans R Soc Lond B Biol Sci. 2011;366(1580):2972–2978. doi: 10.1098/rstb.2011.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belousoff MJ, et al. Ancient machinery embedded in the contemporary ribosome. Biochem Soc Trans. 2010;38(2):422–427. doi: 10.1042/BST0380422. [DOI] [PubMed] [Google Scholar]
- 32.Petrov AS, et al. Secondary structures of rRNAs from all three domains of life. PLoS ONE. 2014;9(2):e88222. doi: 10.1371/journal.pone.0088222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernier C, et al. FD169: RiboVision: Visualization and analysis of ribosomes. Faraday Discussions. 2014 doi: 10.1039/c3fd00126a. 10.1039/C3FD00126A. [DOI] [PubMed] [Google Scholar]
- 34.Petrov AS, et al. Secondary structure and domain architecture of the 23S and 5S rRNAs. Nucleic Acids Res. 2013;41(15):7522–7535. doi: 10.1093/nar/gkt513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrödinger LLC. 2014. The PyMOL Molecular Graphics System, version 1.5.0.4.
- 37.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313(4):903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 38.Letunic I, Bork P. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39(Web Server issue):W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.