Abstract
Medicine is expected to benefit from combining usual cellular and molecular studies with high-throughput methods (genomics, transcriptomics, proteomics and metabolomics). These methods, collectively known as omics, permit the determination of thousands of molecules (variations within genes, RNAs, proteins, metabolites) within a tissue, cell or biological fluid. The utilization of these methods is very demanding in terms of the design of the study, acquisition, storage, analysis and interpretation of the data. When carried out properly, these studies can reveal new etiological pathways, help to identify patients at risk for disease, and predict the response to specific treatments. Here we review these omics methods and mention several applications in hepatology research.
Keywords: genomics, proteomics, metabolomics, lipidomics, microbiome
Introduction
There is a nautical chart attributed to Christopher Columbus, obviously drawn before he set sail on the voyage that would lead to the discovery of America, that stretches from the south of Scandinavia to the mouth of the river Congo showing all the Mediterranean ports of Europe and Africa in detail. The enormous space that Columbus dedicated to the Atlantic Ocean is conspicuously lacking in detail. In all probability, this huge blank space served not only to mark the frontier of the known world and therefore the potential expansion of world knowledge. It also opened up a route for the imagination and the adventure of sailing through it, a route travelled by numerous sixteenth and seventeenth-century explorers who, although in most cases were destined to remain anonymous, changed the world forever.
In the same way, sequencing the human genome opened up a new era in biomedical sciences that is being explored by a legion of scientists. Biomedical research has evolved from the analysis of the effects of individual genes to a more integrated view that examines whole ensembles of genes as they interact during a biological process. This has changed the way we look at human disease and understand better why specific therapies work or do not work. An example in hepatology is the use of a genetic variation near the IL28B gene that predicts the response to hepatitis C therapy (1). This way of thinking has given excessive value, however, to a way of carrying out research in biosciences that consists in measuring everything (genes, proteins, metabolites) in a biological system in the hope that upon analyzing this huge amount of information, new properties of the system will emerge that will allow an integral understanding. This holistic approach often forgets that in biology the interactions between molecules (DNA, RNA, proteins and metabolites) are characterized not by exclusivity, but by the multiplicity of possible interactions between some molecules and others. The problem is that it is not possible to discover how an organism works based on a model that incorporates hundreds of thousands of measurements of its internal components simply because there is no single solution, no predefined design, not an unique three-dimensional structure. From this perspective, health or disease cannot be viewed as the result of the fulfillment of a linear program, but the result of an open process in which a specific biological state springs from certain genetic information interacting with other information existing at that moment.
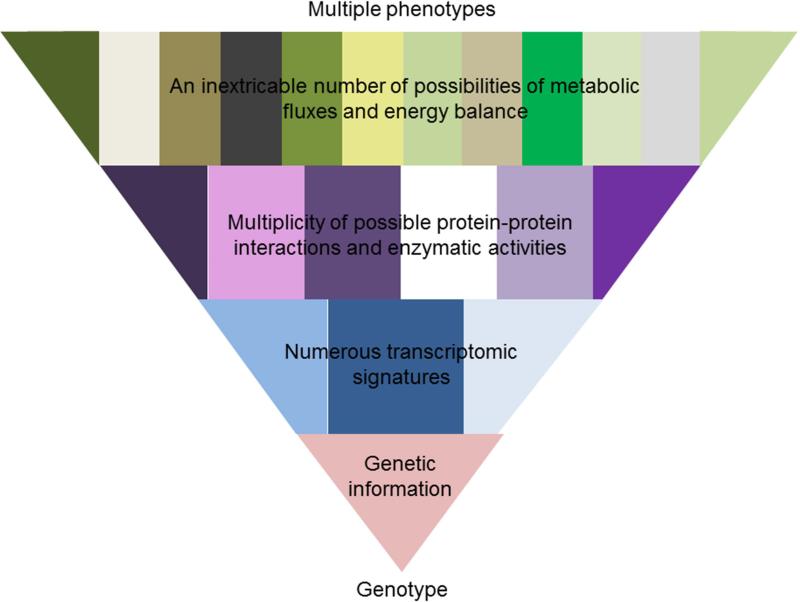
This vision helps to understand how multiple phenotypes can be formed from a single genome, and how environment and chance select, at each moment, one from among all possible phenotypes (Figure 1). Even in those diseases caused by mutations in a single gene, as in the case of phenylketonuria, although the genotype predicts well the biochemical phenotype (the concentration of phenylalanine in the blood), it does not predict the clinical phenotype (the appearance of intellectual disability) (2). It is important to emphasize, that clinical reasoning is basically Bayesian. In other words, the predictive value associated with a diagnostic test varies when it is applied to populations with indices of prevalence very different to those of the study condition. For example, in a person diagnosed of iron overload the presence of a mutation in the HFE gene is a highly reliable predictor of hereditary hemochromatosis. However, in a population that has not been preselected for iron overload, the presence of the same mutation confers only a slight risk of developing clinical symptoms (3). These results speak by themselves of the importance of interpreting the results of studies of genetic variations within an adequate medical context.
Figure 1. Metabolic fluxes are the best representation of the phenotype of an organism.
Health or disease cannot be viewed as the result of the fulfillment of a linear program, but as the result of an open process in which a specific biological state or phenotype springs from certain genetic information interacting with other information existing at that moment. This vision helps to understand how multiple phenotypes can be formed from a single genome, and how environment and chance select, at each moment, one from among all possible phenotypes.
Genomics
When carried out within the adequate medical context, genetic screens are powerful tools for identifying new genes and variations within genes that are involved in specific physiopathological processes. For example, one single nucleotide polymorphism (SNP) that is consistently associated with nonalcoholic fatty liver disease (NAFLD) is a nonsynonymous substitution (a mutation in which a single nucleotide change results in a codon that codes for a different amino acid) in the PNPLA3 gene (4). To identify this gene variant, a genome-wide association study (GWAS) of 9,299 nonsynonymous sequence variations was carried out in a population of 2,111 individuals from three different ethnic groups, in which hepatic fat content was measured by proton magnetic resonance spectroscopy (5). The substitution of an isoleucine by a methionine at position 148 (I148M) of PNPLA3 was found to associate strongly with the accumulation of fat across the three ethnic groups studied with an overall P value of 5.9E-10. Recently, this variant was associated with NAFLD progression to nonalcoholic steatohepatitis (NASH), alcoholic fatty liver disease, and hepatocellular carcinoma (HCC) (6-8). The experience of identifying PNPLA3 teaches us that this hypothesis-free approach to the identification of new genes and variations within genes involved in a pathological process needs to be statistically sound and requires a large sample size of clinically well-characterized patients.
Through December 2013, 2,034 GWAS have been published that have led to the identification of several hundreds of disease associated gene variants (9). GWAS are approaches that aim to identify potential associated genes, at whole genome level, based on the statistical significance of the differential occurrence of common SNPs when comparing populations with distinct traits such as disease and health or drug responders and non-responders (10). Interestingly the majority of these SNPs are not in gene coding regions, which suggests that these variants affect regulatory elements of the genome that have key functions in the development of complex diseases, such as those of the liver. This agrees with the results of the Encyclopedia of DNA Elements (ENCODE) consortium, an ambitious project that aims to identify and characterize all functional elements in the human genome (11), whose principal conclusion is that the majority of the genome, although not coding for proteins, is active and plays important regulatory functions (12).
Transcriptomics and proteomics
Genomic technologies have made feasible to investigate the expression of thousands of genes at a time using large sets of samples. This technology has often been used with the aim to develop tests that more reliably diagnose diseases and predict the response to specific treatments. However, the clinical application of these diagnostic and prognostic gene expression signatures has been delayed due to three main reasons. First, complex diseases, like NASH, cirrhosis or HCC, likely involve a large number of different genes and biological pathways and are very heterogeneous in terms of clinical manifestations, genomic alterations, and gene expression patterns (13-15). Therefore, large cohorts of well-characterized patients are necessary to obtain genomic signatures of clinical relevance. Second, diagnostic and prognostic gene signatures contain a large number of genes and the prediction algorithms are complex and not easy to use routinely in clinics (16). Third, the development of complex molecular tests based on DNA, RNA, proteins or metabolite profiles carries a series of problems inherent to all high-throughput techniques where large datasets are analyzed (17). Selecting the statistically significant results from a large dataset containing also nonsignificant data is challenging, because when multiple significance tests are calculated the probability that at least one reaches by chance significance increases with the number of tests performed (18). It is therefore critical to control this multiplicity problem as well as to use one or more model validation technique for assessing how the results of a statistical analysis will generalize to an independent dataset (18). However, in the race to apply genomics technology, genomics works are too frequently published in which massive quantities of data containing avoidable errors are handled (17). Yet, when used correctly microarray technologies may be translated into score systems that can reproducibly predict clinical outcomes. An example in hepatology is the development of a simple risk score classifier based on the expression of a small number of genes that can predict in a reproducible manner overall survival of patients after surgical resection for HCC (19). In a recent report, the Institute of Medicine identified best practices for future research and development of omics-based tests (20). These practices include the use of rigorous statistical methods, bioinformatics and data management, and open access to the datasets and algorithms used to develop the test. The application of these best practices should be reinforced by all research organizations (21).
One of the most important objectives of genomic research is also to associate transcriptomic data with the molecular pathways that underlie disease. However, gene expression changes in complex diseases, such as those of the liver, often reflect processes that are secondary to the pathological process. To overcome this problem, transcriptional networks have been developed based on the assumption that gene products that are causative of a disease process and whose expression is altered in a pathological condition have similar expression patterns (co-expressed genes) (22). An impressive example of this approach is the use of an integrative genomic method, based on the analysis of transcriptional networks in human brain, to identify a new molecular pathway linking late-onset Alzheimer's disease, aging, and APOE4 (23). The same principle applies to proteomic research where the concentration of hundreds to thousands of proteins is determined simultaneously in a cell type or tissue. An application of this method in hepatology is the demonstration that knocking out the liver-specific prohibitin-1 gene (Phb1) in mice results in the spontaneous development of severe liver disease and HCC (24) after identification, using proteomics, that liver PHB1 content was decreased in an experimental model of NASH (25).
One of the best ways to learn about the function of a gene is to generate a mouse with a deletion in that specific gene. Hence, the function of around 7,300 mouse genes has been described utilizing this approach (26). In general, these knockout mice have been generated towards genes previously studied and where a phenotype was anticipated, as in the case of Phb1 mentioned above. Interestingly, in many occasions no obvious phenotype is observed, probably because only the expected traits are investigated. As a result of this, the full biological function of many genes for which knockout mice are available is not known. Genome-wide mouse programs aiming to generate knockout mice with mutations in all protein coding genes are underway (26,27). Over 900 knockout mice, many with phenotypic data, have been made openly available for further analysis (27). It is important to note that even when the deletion of a known gene is associated to a phenotype, elucidating the molecular mechanism by which mutation of that gene leads to a particular phenotype is not obvious at first glance and requires extensive experimental work to elucidate it.
The coordinated changes in gene expression patterns associated with the molecular pathways that underlie disease are controlled at multiple levels. Examples include, nucleosome remodeling, non-coding RNAs, histone variants and modifications (e.g. by acetylation or methylation) (28). MicroRNAs and RNA-binding proteins play a critical role in the post-transcriptional regulation of global changes in gene expression (11,12). DNA methylation is another key epigenetic modulator of gene expression that is generally associated with transcriptional repression (11,12). Large-scale DNA methylation mapping studies have provided important insights in the gene regulation and the development of various diseases, particularly in tumorigenesis (29). One of the most important objectives of DNA-methylation mapping research is to link DNA methylation changes with the expression of genes pathways that underlie disease. Here, similarly to genome research, a variety of software have been released to facilitate the identification of differentially methylated regions (DMRs), classification of DMRs into enriched genomic regions, and comparison of DNA methylation, and gene expression changes. Recently, this technique has elegantly been applied to identify differences in DNA methylation that could distinguish patients with advanced versus mild NAFLD (30), and that led to the identification of MAT1A (methionine adenosyltransferase 1A) as one of the principal down-regulated genes in NASH (31). These findings agree with earlier work demonstrating that MAT1A expression and MAT activity are markedly reduced in human liver cirrhosis (32,33), and that MAT1A expression is silenced in human HCC (34). Furthermore, deletion of Mat1a in mice causes NASH and HCC (35,36), which support the concept that from NASH to HCC MAT1A may be a therapeutic target (37).
Metabolomics
Metabolomics, the high-throughput identification and quantification of small sized (<1,500Da) molecules, is the last branch of omics-based technology incorporated into biomedical research. While in other omics fields thousands of targets are routinely analyzed at a time, till recently few studies had identified and quantified more than 100 metabolites at a time in a large set of samples. Two factors have made it feasible to determine the concentration of hundreds of metabolites at a time using large sets of samples. First, the release of electronic database equivalent to GenBank or UniProt, like the Human Metabolome Database; and second, the development of modern high-resolution NMR spectroscopy and of mass spectrometry (MS) technologies, such as ultraperformance liquid chromatography-MS (UPLC-MS) and gas chromatography-MS, for the identification and quantification of thousands of metabolites at a time in as little as a few minutes per sample.
The human serum metabolome is composed, with today's technology, of around 4,200 metabolites half of which are phospholipids and over a thousand glycerolipids (triglycerides (TG), diglycerides, and monoacylglycerols) (38). In other words, around three quarters of the known human serum metabolome are lipids. Amino acids, peptides, carbohydrates, amines, and carboxylic acids complete the list of the serum metabolome. Thousands of different lipids seem much more than the 4 bases utilized by DNA to encode the genetic information of an organism, much more than the 23 amino acids that are the building blocks of proteins, much more than the hundreds of carbohydrates and carboxylic acids that form the central carbon metabolism. But ultimately this many thousands of lipids make sense, if we think for instance, that an average car has over 10,000 moving parts. From the storage of energy and the establishment of the permeability barrier for cells and cell organelles, to the regulation of membrane-associated processes –such as oxidative phosphorylation, intracellular trafficking, cell growth, apoptosis- and the facilitation of membrane protein folding in a manner similar to protein molecular chaperones, lipids play an essential biological function.
Metabolic dysfunction has been implicated in a wide variety of human diseases, such as obesity, NAFLD, diabetes, inborn errors of metabolism and cancer, just to mention a few (39). Results are consistent with an important contribution of metabolic disbalance, that is the rerouting of the metabolic fate of lipids, carbohydrates and amino acids through the intermediary metabolism, to the initiation and/or progression of these and other diseases. Consequently, there is an increased interest in understanding what are the metabolic differences between normal and diseased tissues that can lead to development of more selective and effective treatments. Cellular metabolism consists of a multitude of enzymatic reactions, inextricably interconnected, that are involved in two functions: one, the conversion of thousands of molecules into building blocks for macromolecular biosynthesis; and two, the reactions that ensure the constant supply of energy via ATP and redox equivalents (NADPH). The concentrations of the metabolites in a cell are the result of the fluxes in the metabolic reactions, which ultimately depend on the conditions of the moment such as the available nutrients, hormonal and neural factors, the properties of the enzymes involved and the levels of the metabolites themselves, as they exert important feed-back and feed-forward regulation on the system (40). Notoriously, the liver parenchyma shows a zonal distribution of key metabolic enzymes and metabolism, which indicates that there are different types of hepatocytes in the liver. For instance, oxidative phosphorylation, glucose output, urea synthesis, and bile acid synthesis is higher in the periportal area, whereas glucose uptake, glutamine formation, and xenobiotic metabolism are greater in the perivenous area (41). Metabolic zonation is altered in liver steatosis (42), but whether this reflects processes that are secondary to the pathological process is an open question. From this perspective, it is clear that the metabolic fluxes represent the final outcome of cellular regulation at many different levels, and hence they are the best representation of the phenotype of an organism (Figure 1).
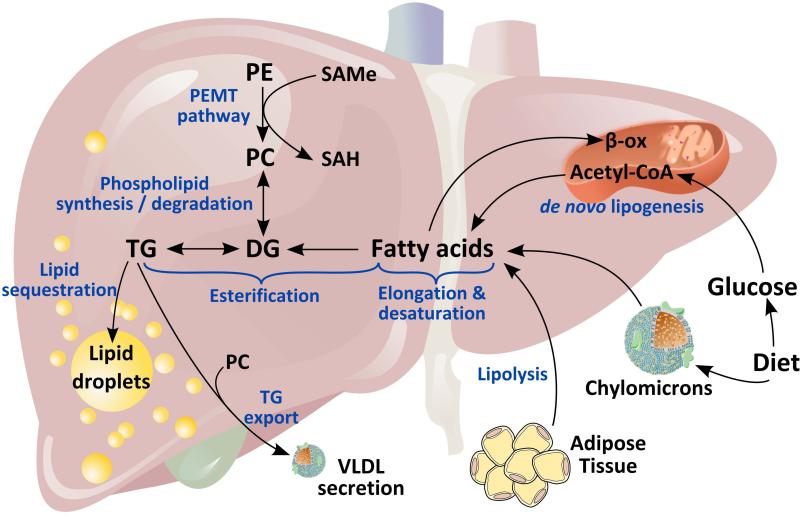
A consequence of this convoluted network of enzymatic reactions that integrate cellular metabolism is that it is not possible to conclude that a certain metabolic pathway is altered in a cell or tissue simply by measuring the steady state concentration of its metabolites at one single time point. For example, the three major sources of fatty acids (FA) used by the liver to synthesize TG are the diet, de novo lipogenesis, and the adipose tissue; and the four major fates of hepatic FA are mitochondrial β-oxidation, biosynthesis of other lipid classes, esterification and storage as TG into lipid droplets, and assembly as TG into very low-density lipoproteins and export into blood (Figure 2). Processes that lead to an imbalance between the intake and biosynthesis of TG and the export and catabolism of TG cause NAFLD. Elucidating which of all these potential mechanisms are responsible of hepatic TG accumulation under a specific condition requires careful measurements of metabolic fluxes, using labeled tracers, as well as the determination of the content of dozens to hundred of metabolites and activities of key enzymes. Unfortunately, studies are too frequently published in which a pathological process is associated to changes in a certain metabolite or group of metabolites based simply in their steady-state concentration, or quantification of mRNA and/or protein of specific enzymes. Moreover, it is important to remember that changes in the concentration of metabolites often reflect processes that are secondary to the pathological process. However, when used correctly metabolic studies may lead to the identification of the rate-limiting step responsible of a pathological process. An example in hepatology is the identification that an excess of hepatic S-adenosylmethionine (SAMe), which occurs in the setting of impaired glycine N-methyltransferase-mediated catabolism, reroutes phosphatidylethanolamine (PE) metabolism towards the biosynthesis of phosphatidylcholine (PC), via activation of the enzyme PE methyltransferase (Figure 2). The excess PC thus generated is used by the hepatocyte to synthesize TG that accumulate into lipid droplets causing NAFLD (43).
Figure 2. Fatty acid metabolism in liver.
The three major sources of fatty acids (FA) used by the liver to synthesize triglyceride (TG) are the diet, de novo lipogenesis, and the adipose tissue; and the four major fates of hepatic FA are mitochondrial β-oxidation, biosynthesis of other lipid classes (such as phospholipids, cholesterol esters and sphingolipids), esterification and storage as TG into lipid droplets (LD), and assembly as TG into very low-density lipoproteins (VLDL) and export into blood. Hepatic TG can be synthesized by desaturation, elongation and esterification of FA, or by the phosphatidylethanolamine N-methyltransferase (PEMT) pathway that converts phosphatidylethanolamine (PE) to phosphatidylcholine (PC). TG export via VLDL requires incorporation of PC synthesized by the PEMT pathway.
The human metabolome is an ocean full of biomarkers. Accordingly, a central objective in metabolomics research is the discovery of specific metabolic profiles (in serum, urine, feces, sweet, tears, saliva, tissues) that associate with disease or the response to specific treatments. The development of metabolomic-based diagnostic and prognostic tests has the same problems inherent to all high-throughput techniques, namely the detection of statistically significant relationships between a group of metabolites and disease while minimizing the risk of false positive associations (18,44). An additional complication in metabolomics, as compared to other omics-based methods, is the preparation and storage of the samples due to large differences in solubility and stability among metabolites. When used correctly metabolomics is a powerful novel approach for biomarker identification. For example, a serum lipidomic signature associated with NAFLD progression has been identified (45). To obtain this signature, 540 serum metabolites were determined by UPLC-MS in a population of 467 biopsied individuals with different body mass index (45,46).
Microbiome
In addition to the 22,000 or so protein-coding genes of the human genome, the collective genome of the human gut flora is guessed to contain 100 to 200-times that number (47). This collective genome, the microbiome, provides us with an additional and extraordinary metabolic capacity that modulates host energy and lipid metabolism (48), whose importance in health and disease we are beginning to understand. Thus, gut microbiota alterations have been associated to the susceptibility of developing certain diseases such as obesity, diabetes, celiac disease, cardiovascular disease and NASH (48). An example of this complex relationship between the gut microbes and the host metabolism is the discovery of a new pathway for gut flora mediated generation of the pro-atherosclerotic metabolite trimethylamine N-oxide (TMAO) from dietary PC (49). An example that illustrates the complex relationship between gut microbiota and liver disease is the demonstration that bile acid metabolism by intestinal bacteria has a key role in obesity-associated HCC development (50). These authors analyzed the serum metabolites of high fat-diet and normal-diet fed mice by UPLC/MS and observed an increase in the levels of deoxycholic acid (DCA), a secondary bile acid solely produced by hydroxylation of primary bile acids by gut bacteria. DCA is known to cause DNA damage and enhance liver cancer. Interestingly, lowering DCA levels in obese mice treated with the carcinogen dimethylbenz(a)anthracene decreased HCC development (50). These results speak for themselves of the complex relationship between the gut flora and the host metabolism and the importance to assess medical risks, monitor, diagnose and treat patients according to their specific metabolic phenotype.
Conclusions
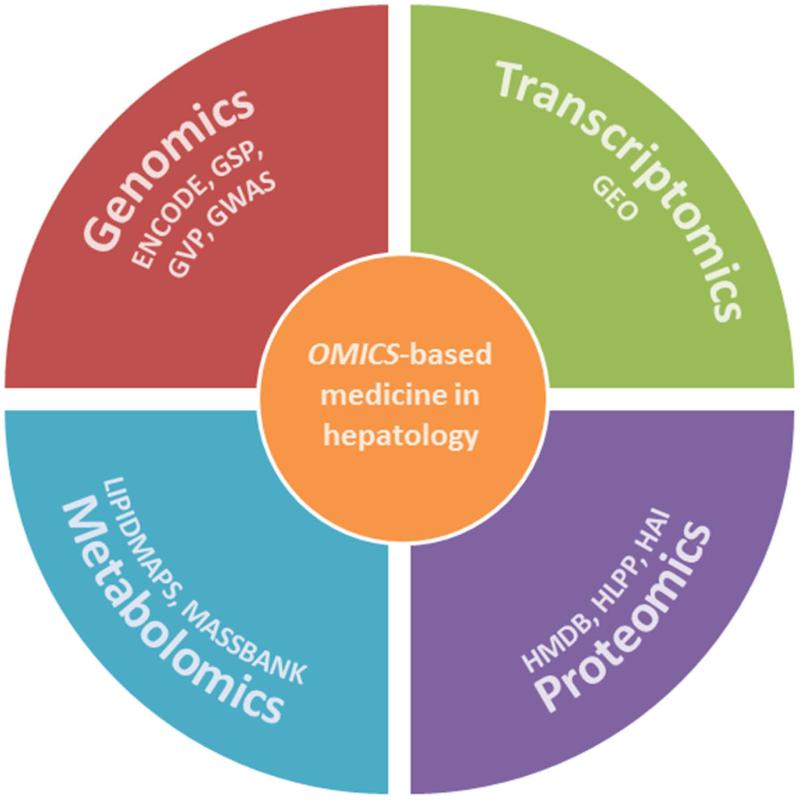
The ultimate aim of omics-based research in hepatology is to translate this knowledge into useful results that improve our understanding of complex biological processes, make reliable predictions in silico of human liver drug toxicity, and provide clinically relevant tests (Figure 3). However, several problems need to be overcome to ensure the successful translation of these technologies. One is adopting protocols that yield consistent results in different laboratories so that data can be built into a single repository. Another problem is the integration of all the data generated by omics-based screens (such as RNAs, proteins, metabolites, protein-protein interactions, protein-lipid interactions, protein-nucleic acid interactions, and so on). Once these two problems are solved, the translation of omics-based results into clinically useful products will be within reach.
Figure 3. Omics-based medicine.
The ultimate aim of omics-based medicine is to translate human genomics, transcriptomics, proteomics and metabolomics results into clinically useful products. To help researchers achieve this goal several freely accessible initiatives have been established, such as the Genome Sequencing Program (GSP), the Encyclopedia of DNA Elements (ENCODE), the Genetic Variation Program (GVP), or the Genome-Wide Associations Studies (GWAS) of the National Human Genome Research Institute (http://www.genome.gov/). In transcriptomics, the Gene Expression Omnibus (GEO) provides a public repository that archives and freely distributes (http://www.ncbi.nlm.nih.gov/geo/info/overview.html) microarrays and other functional genomics data. In proteomics, the Human Proteome Organization (HUPO, http://www.hupo.org/initiatives/) sponsors several initiatives such as the Human Liver Protein Project (HLPP) or the Human Antibody Initiative (HAI); and in metabolomics the Human Metabolome Database (HMDB, http://www.hmdb.ca/) and other related resources such as KEGG, LipidMaps, and MassBank, contain freely available information about metabolites found in the human body,
Acknowledgments
Grant support: NIH RO1AT1576 (M.L.M-C., S.C.L., J.M.M.), RO1DK051719 (S.C.L., J.M.M.), Spanish Plan Nacional I+D SAF 2011-29851 (J.M.M.), ETORTEK-2010 Gobierno Vasco (M.L.M.-C, J.M.M.), PI11/01588, Sanidad Gobierno Vasco 2008, Educación Gobierno Vasco 2011 (M.L.M.-C), 2012 (J.M.M.). Ciberehd is funded by ISCiii.
Abbreviations used in alphabetical order
- DCA
deoxycholic acid
- DMRs
differentially methylated regions
- ENCODE
Encyclopedia of DNA Elements
- FA
fatty acids
- GC
gas chromatography
- GWAS
genome-wide association study
- HCC
hepatocellular carcinoma
- MAT1A
methionine adenosyltransferase 1A
- MS
mass spectrometry
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PHB1
prohibtin-1
- SAMe
S-adenosylmethionine
- SNP
single nucleotide polymorphism
- TG
triglycerides
- TMAO
trimethylamine N-oxide
- UPLC
ultraperformance liquid chromatography
Footnotes
* None of the authors have conflict of interest to declare
REFERENCES
- 1.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 2.Okano Y, Eisensmith RC, Güttler F, Lichter-Konecki U, Konecki DS, Trefz FK, et al. Molecular basis of phenotypic heterogeneity in phenylketonuria. N Engl J Med. 1991;324:1232–1238. doi: 10.1056/NEJM199105023241802. [DOI] [PubMed] [Google Scholar]
- 3.Adams PC, Reboussin DM, Barton JC, McLaren CE, Eckfeldt JH, McLaren GD, et al. Hemochromatosis and Iron Overload Screening (HEIRS) Study Research Investigators. Hemochromatosis and iron-overload screening in a racially diverse population. N Engl J Med. 2005;352:1769–1778. doi: 10.1056/NEJMoa041534. [DOI] [PubMed] [Google Scholar]
- 4.Daly AK, Ballestri S, Carulli L, Loria P, Day CP. Genetic determinants of susceptibility and severity in nonalcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol. 2011;5:253–263. doi: 10.1586/egh.11.18. [DOI] [PubMed] [Google Scholar]
- 5.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 7.Stickel F, Buch S, Lau K, Meyer H, Berg T, et al. Variation in the PNPLA3 gene is associated with alcoholic liver injury in caucasians. Hepatology. 2011;53:86–95. doi: 10.1002/hep.24017. [DOI] [PubMed] [Google Scholar]
- 8.Guyot E, Sutton A, Rufat P, Laguillier C, Mansouri A, Moreau R, et al. PNPLA3 rs738409, hepatocellular carcinoma occurrence and risk model prediction in patients with cirrhosis. J Hepatol. 2013;58:312–318. doi: 10.1016/j.jhep.2012.09.036. [DOI] [PubMed] [Google Scholar]
- 9.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehta G, Jalan R, Mookerjeee RP. Cracking the ENCODE: from transcription to therapeutics. Hepatology. 2013;57:2532–2535. doi: 10.1002/hep.26449. [DOI] [PubMed] [Google Scholar]
- 13.Villanueva A, Minguez B, Forner A, Reig M, Llovet JM. Hepatocellular carcinoma: novel molecular approaches for diagnosis, prognosis, and therapy. Annu Rev Med. 2010;61:317–328. doi: 10.1146/annurev.med.080608.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo HG, Park ES, Thorgeirsson SS, Kim YJ. Exploring genomic profiles of hepatocellular carcinoma. Mol Carcinog. 2011;50:235–243. doi: 10.1002/mc.20691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loomba R, Abraham M, Unalp A, Wilson L, Lavine J, Doo E, et al. Nonalcoholic Steatohepatitis Clinical Research Network. Association between diabetes, family history of diabetes, and risk of nonalcoholic steatohepatitis and fibrosis. Hepatology. 2012;56:943–951. doi: 10.1002/hep.25772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 17.Reich ES. Cancer trial errors revealed. Nature. 2011;469:139–140. doi: 10.1038/469139a. [DOI] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 19.Nault JC, De Reyniès A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176–187. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 20.Micheel CM, Nass SJ, Omenn GS. Evolution of translational omics: lessons learned and the path to follow. http://www.iom.edu/Reports/2012/Evolution-of-Translational-Omics.aspx. [PubMed]
- 21.Wadman M. NIH mulls rules for validating key results. Nature. 2013;500:14–16. doi: 10.1038/500014a. [DOI] [PubMed] [Google Scholar]
- 22.Heyer LJ, Kruglyak, Yooseph S. Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhinn H, Fujita R, Qiang L, Cheng R, Lee JH, Abeliovich A. Integrative genomics identifies APOE ε4 effectors in Alzheimer's disease. Nature. 2013;500:45–50. doi: 10.1038/nature12415. [DOI] [PubMed] [Google Scholar]
- 24.Ko KS, Tomasi ML, Iglesias-Ara A, French BA, French SW, Ramani K, et al. Liver-specific deletion of prohibitin 1 results in spontaneous liver injury, fibrosis, and hepatocellular carcinoma in mice. Hepatology. 2010;52:2096–2108. doi: 10.1002/hep.23919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santamaria E, Avila MA, Latasa MU, Rubio A, Martin-Duce A, Lu SC, et al. Functional proteomics of nonalcoholic steatohepatitis: mitochondrial proteins as targets of S-adenosylmethionine. Proc Natl Acad Sci U S A. 2003;100:3065–3070. doi: 10.1073/pnas.0536625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White JK, Gerdin A- K, Karp NA, Ryder E, Buljan M, Bussell JN, et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Lyer V, et al. A conditional knockout resource for genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker PB, Workman JL. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol. 2013 Sep 1;5(9) doi: 10.1101/cshperspect.a017905. doi:pii: a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, et al. Relationship between the methylome and transcriptome in patients with non-alcoholic fatty liver disease. Gastroenterology. 2013;145:1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moylan CA, Pang H, Dellinger A, Suzuki A, Garrett ME, Guy CD, et al. Hepatic gene expression profiles differentiate pre-symptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology. 2013 doi: 10.1002/hep.26661. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duce AM, Ortíz P, Cabrero C, Mato JM. S-adenosyl-L-methionine synthetase and phospholipid methyltransferase are inhibited in human cirrhosis. Hepatology. 1988;8:65–68. doi: 10.1002/hep.1840080113. [DOI] [PubMed] [Google Scholar]
- 33.Avila MA, Berasain C, Torres L, Martín-Duce A, Corrales FJ, Yang H, et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. 2000;33:907–914. doi: 10.1016/s0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 34.Cai J, Sun W, Hwang JJ, Stain S, Lu SC. Changes in S-adenosylmethionine synthetase in human liver cancer: molecular characterization and significance. Hepatology. 1996;24:1090–1097. doi: 10.1002/hep.510240519. [DOI] [PubMed] [Google Scholar]
- 35.Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci U S A. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martínez-Chantar ML, Corrales FJ, Martínez-Cruz LA, García-Trevijano ER, Huang ZZ, Chen L, et al. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 37.Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, et al. The human serum metabolome. PLoS One. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeBernardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–1144. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen J. It is all about metabolic fluxes. J Bacteriol. 2003;185:7031–7035. doi: 10.1128/JB.185.24.7031-7035.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jungermann K, Kietzmann T. Oxygen: modulator of metabolic zonation and disease of the liver. Hepatology. 2000;31:255–260. doi: 10.1002/hep.510310201. [DOI] [PubMed] [Google Scholar]
- 42.Debois D, Bralet MP, Le Naour F, Brunelle A, Laprévote O. In situ lipidomic analysis of nonalcoholic fatty liver by cluster TOF-SIMS imaging. Analytical Chemistry. 2009;81:2823–2831. doi: 10.1021/ac900045m. [DOI] [PubMed] [Google Scholar]
- 43.Martínez-Uña M, Varela-Rey M, Cano A, Fernández-Ares L, Beraza N, Aurrekoetxea I, et al. Excess S-adenosylmethionine reroutes phosphatidylethanolamine towards phosphatidylcholine and triglyceride synthesis. Hepatology. 2013;58:1296–1305. doi: 10.1002/hep.26399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chadeau-Hyam M, Ebbels TM, Brown IJ, Chan Q, Stamler J, Huang CC, et al. Metabolic profiling and the metabolome-wide association study: significance level for biomarker identification. J Proteome Res. 2010;9:4620–4627. doi: 10.1021/pr1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barr J, Caballería J, Martínez-Arranz I, Domínguez-Díez A, Alonso C, Muntané J, et al. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J Proteome Res. 2012;1:2521–2532. doi: 10.1021/pr201223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barr J, Vázquez-Chantada M, Alonso C, Pérez-Cormenzana M, Mayo R, Galán A, et al. Liquid chromatography-mass spectrometry-based parallel metabolic profiling of human and mouse model serum reveals putative biomarkers associated with the progression of nonalcoholic fatty liver disease. J Proteome Res. 2010;9:4501–4512. doi: 10.1021/pr1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 48.de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev. 2012;70(Suppl 1):S45–56. doi: 10.1111/j.1753-4887.2012.00505.x. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]