Abstract
Background
In Escherichia coli the genes involved in the acquisition of tetracycline resistance are mainly tet(A) and tet(B). In addition, tet(M) is the most common tetracycline resistance determinant in enterococci and it is associated with conjugative transposons and plasmids. Although tet(M) has been identified in E. coli, to our knowledge, there are no previous reports studying the linkage of the tet(M) gene in E. coli to different mobile genetic elements. The aim of this study was to determine the occurrence of tet(A), tet(B), and tet(M) genes in doxycycline-resistant E. coli isolates from pigs, as well as the detection of mobile genetic elements linked to tet(M) in E. coli and its possible transfer from enterococci.
Results
tet(A) was the most frequently detected gene (87.9%) in doxycycline-resistant isolates. tet(M) was found in 13.1% E. coli isolates. The tet(M) gene was detected in relation with conjugative transposons in 10 out of 36 enterococci isolates analyzed but not in any of E. coli isolates positive for tet(M). Southern blot showed that in E. coli and in most of the enterococci isolates the tet(M) gene was carried on a plasmid. According to the phylogenetic analysis, E. coli contained a new tet(M) allele grouping separately. Mating experiments revealed that tet(M) was carried on a mobile element successfully transferred between enterococci and between enterococci and E. coli.
Conclusions
The detection of tet(M) in E. coli isolates from pigs was higher than expected. In our study, tet(M) detected in E. coli seems not to have been transferred from enterococci, although it can not be ruled out that the horizontal transfer of this gene occurred from other intestinal tract bacteria.
Keywords: E. coli, Pigs, tet(M), Horizontal gene transfer
Background
Tetracyclines, including doxycycline, are a family of antimicrobial agents that are frequently used in veterinary medicine because of their broad-spectrum of activity and their relatively low cost [1]. Besides the therapeutic use of tetracyclines, they have also been administered as growth promoters in many countries [1]. The extensive use of tetracyclines have resulted in an emergence of resistant bacteria [1]. Thus, commensal and pathogenic Escherichia coli isolated from pigs are often resistant to tetracycline [2-4].
Tetracycline resistance usually results from the acquisition of genes that are involved mainly in three processes: antibiotic efflux through energy-dependent membrane-associated proteins, ribosomal protection, and enzymatic inactivation of tetracycline [1,5]. More than 40 different classes of tetracycline resistance genes have been identified [5-7]. In commensal and pathogenic E. coli, the genes involved mainly in the acquisition of tetracycline resistance are genes encoding efflux proteins, being tet(A) and tet(B) most frequently detected [2,4,8-10]. The ribosomal protection gene tet(M) was first reported in E. coli in 2004, when Bryan et al. detected a tet(M) gene in strains from chicken and pigs that shared a 98% identity over 386 bp to a tet(M) gene found in Enterococcus faecalis[8]. Since then, this gene has also been identified in an E. coli from a river basin [11] and in a small number of avian, porcine, and human E. coli isolates [12-15].
tet(M) has been identified in more than 40 genera of bacteria and it has become the widest host range of any tetracycline resistance gene [5]. This may be due, at least partially, to its association with conjugative transposons [5]. In enterococci, tet(M) is the most common tetracycline resistance determinant and it is mainly associated with the conjugative transposon Tn916[16-19], although it has also been found in another conjugative transposons (Tn5397 and Tn5801) and on plasmids [20-22]. To our knowledge, in E. coli, there are no previous reports studying the linkage of the tet(M) gene to different mobile genetic elements.
Enterococci and E. coli are natural inhabitants of the gastrointestinal tract of humans and animals. In previous studies, the in vivo transfer of resistance genes among intestinal tract bacteria has been showed. Thus, the transfer of resistance genes from E. faecalis to E. coli and between E. coli isolates in the gut has been demonstrated [23,24]. Therefore, it is possible that the finding of the tet(M) gene in E. coli strains it is due to a horizontal transfer of this gene from enterococci.
The aim of this study was to determine the occurrence of tet(A), tet(B), and tet(M) genes in doxycycline-resistant E. coli isolates from pigs, as well as the detection of mobile genetic elements linked to tet(M) in E. coli and its possible transfer from enterococci.
Results
Detection of tetracycline resistance genes
All of the analyzed E. coli isolates contained at least one of the three tetracycline resistance genes studied. The most frequently detected gene, tet(A), was found alone or combined with other genes in 87 of the 99 (87.9%) tetracycline-resistant isolates. tet(B) and tet(M) were detected in 42 (42.4%) and 13 (13.1%) of the E. coli isolates, respectively (Table 1).
Table 1.
Number (percentage) of tetracycline resistance genes in doxycycline-resistant E. coli isolates from pigs
| Resistance genes | E. coli isolates |
|---|---|
|
tet(A) |
46 (46.5) |
|
tet(B) |
12 (12.1) |
|
tet(A) + tet(B) |
28 (28.3) |
|
tet(A) + tet(M) |
11 (11.1) |
| tet(A) + tet(B) + tet(M) | 2 (2) |
Detection of Tn916-, Tn5397-, and Tn5801-like conjugative transposons
None of the 13 tet(M)-positive E. coli isolates carried the xis-Tn gene from Tn916, the tndX gene from Tn5397, or the int gene from Tn5801. Of the 36 tet(M)-positive enterococci isolates (28 E. faecalis, four Enterococcus faecium, and four Enterococcus hirae) selected from the pigs from which the tet(M)-positive E. coli were isolated, seven (five E. faecalis and two E. faecium) contained the xis-Tn gene, two (one E. faecium and one E. hirae) carried the tndX gene, and one (E. faecium) carried both the tndX and int genes. Twenty-six isolates (23 E. faecalis and three E. hirae) were negative for the xis-Tn, tndX, and int genes.
Southern blot
Hybridization to the tet(M) probe was obtained in the plasmid DNA from the four E. coli isolates tested (CICYT-268, CICYT-320, CICYT-332, and CICYT-348) and from three (CICYT-381, CICYT-436, and CICYT-453) of the four enterococci isolates analyzed. The approximate size of the plasmids from the E. coli and enterococci isolates is around 36 Kb.
Sequencing of the tet(M) gene and phylogenetic analysis
The upstream part of the tet(M) gene was amplified in all the E. coli and enterococci isolates analyzed in the study. However, the downstream part of the gene was only amplified in 12 out of the 36 enterococci isolates and it was not amplified in any E. coli isolate.
Comparison of the 11 tet(M) sequences selected from this study [1802 bp of the total tet(M) of 1920 bp] revealed five different sequence types and the pylogenetic analysis divided these into four phylogenetic groups (Figure 1). The phylogenetic tree (Figure 1) showed the plasmid-borne tet(M) (group 1) of the four E. coli isolates (CICYT-268, CICYT-320, CICYT-332, and CICYT-348) represent a new tet(M) allele. Four enterococci isolates negative for the xis-Tn, tndX, and int genes (CICYT-67, CICYT-381, CICYT-436, and CICYT-453) and one isolate positive for tndX from Tn5397 (CICYT-452) contained identical tet(M) genes (group 2) (Figure 1). The tet(M) gene from enterococci isolates containing xis-Tn from Tn916 or int from Tn5801 (CICYT-383 and CICYT-205, respectively) grouped with the tet(M) genes of the respective transposons (Figure 1, groups 3 and 4).
Figure 1.
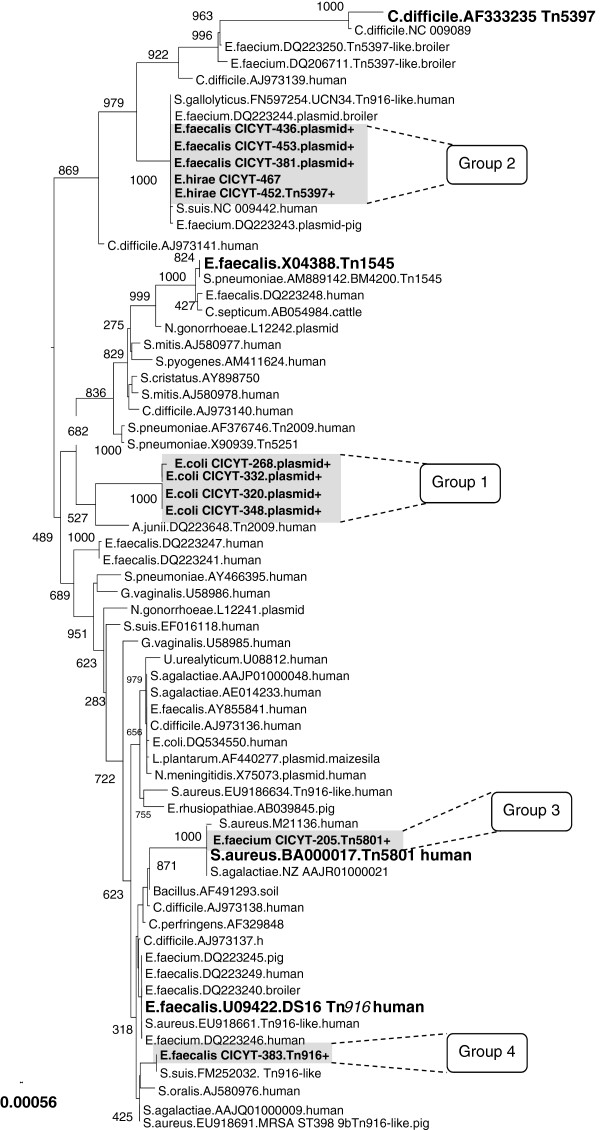
Phylogenetic gene tree of tet(M). Bootstrap values are indicated at branch points (out of 1000 generated NJ trees).
Conjugative transfer of tet(M) in filter mating experiments
Conjugal transfer of tet(M) gene between donor (three E. faecalis and one E. hirae) and recipient (E. faecium BM4105 and E. faecalis JH2-2) enterococci was observed, except from E. hirae to E. faecium BM4105, and from E. faecalis CICYT-381 to E. faecalis JH2-2 (Table 2). tet(M) was successfully transferred from all the four donor enterococci strains to the recipient E. coli CICYT70-Ri. No transfer of tet(M) gene was detected from E. coli to E. faecium or E. faecalis.
Table 2.
Results of the mating experiments
| Donor strains* | Species | Transfer frequency to E. faecalis JH2-2 (tc/dn) | Transfer frequency to E. faecium BM4105 (tc/dn) | Transfer frequency to E. coli CYCIT-70-Ri (tc/dn) |
|---|---|---|---|---|
| CICYT-381 |
E. faecalis |
ND (<1 × 10−9)** |
1.1 × 10−9 |
3.1 × 10−8 |
| CICYT-436 |
E. faecalis |
4.5 × 10−9 |
7 × 10−9 |
3.1 × 10−8 |
| CICYT-453 |
E. faecalis |
2.8 × 10−8 |
1.3 × 10−8 |
3.8 × 10−8 |
| CICYT-467 |
E. hirae |
3.6 × 10−6 |
ND (<0.7 × 10−9) |
2.2 × 10−8 |
| CICYT-268 |
E. coli |
ND (<1.1 × 10−10) |
ND (<1.7 × 10−10) |
|
| CICYT-320 |
E. coli |
ND (<1.5 × 10−10) |
ND (<1.4 × 10−10) |
|
| CICYT-332 |
E. coli |
ND (<1.3 × 10−10) |
ND (<1.5 × 10−10) |
|
| CICYT-348 | E. coli | ND (<2.3 × 10−10) | ND (<1.8 × 10−10) |
tc/dn transconjugans/donor, ND no transconjugants detected. *All the strains were positive for the tet(M) gene but negative for the Tn916, Tn5397, and Tn5801 transposons. **(): detection limit.
Discussion
In the present study, tet(A) was the tetracycline resistance gene detected most frequently, which is in agreement with a previous study carried out on E. coli isolated from healthy pigs [10]. On the contrary, in other studies tet(B) was detected more frequently than tet(A) in E. coli isolated from healthy pigs [9,25]. A negative association between the presence of tet(A) and tet(B) in E. coli has been described in previous studies [10,25]. It has been suggested that this negative association is probably caused by plasmid incompatibilities [26]. However, in the present study, 28 of the 99 (28.3%) E. coli isolates tested carried both tet(A) and tet(B) (Table 1).
The tet(M) gene is one of the most frequently detected tetracycline resistance determinant in enterococcal strains [16-19]. However, tet(M) is uncommon in Gram-negative coliforms such as E. coli[8,11-15]. In the present study, tet(M) was detected in 13 of the 99 (13.1%) tetracycline-resistant E. coli isolates tested. This may indicate a possible transfer of this gene from other intestinal tract bacteria, most likely from enterococci, to E. coli.
In enterococci tet(M) is often associated with conjugative transposons Tn916, Tn5397, and Tn5801[20,22]. Therefore, the presence of these transposons was determined in the 13 tet(M)-positive E. coli isolates and in 36 enterococci isolates selected from the pigs from which the tet(M)-positive E. coli were isolated. None of the E. coli isolates and only 10 of the enterococci carried some of these transposons. In contrast, Agersø et al.[20] detected Tn916-like in a high percentage (85%) of E. faecium isolated from pigs, although this percentage of detection was lower (53%) in E. faecalis strains from the same source. However, these authors [20] did not detect Tn5397-like among enterococci isolated from pigs, while in this work it was detected in 3 of the 36 (8.3%) isolates tested.
The absence of transposons in E. coli and in most of the enterococci isolates in the present study suggests that the tet(M) gene of these isolates is carried on a plasmid. Southern blot was performed in order to show the possible plasmid location of tet(M) and a positive hybridization with a tet(M) probe was obtained in the plasmid DNA from all the E. coli isolates and three of the four enterococci isolates tested.
The phylogenetic analysis shown in Figure 1 revealed a new tet(M) allele presents in the E. coli isolates which grouped separately and were only distantly related to the enterococcal tet(M) sequences detected in this study. Thus the origin of the plasmid-born tet(M) from the E. coli isolates is unknown, though is probably transferred from other bacteria in the intestinal tract. The tet(M) gene carried on a plasmid in E. faecalis isolates of this study was identical to tet(M) plasmid-borne from E. faecium (DQ223243) and E. faecium (DQ223244) isolated from pigs and broilers, respectively. In E. faecium CICYT-205 a Tn5801-like tet(M) gene identical to the sequence described in Tn5801 from Staphylococcus aureus of human origin (BA000017) was identified. To our knowledge, this is the first report of Tn5801-like tet(M) detection in E. faecium and this suggests the horizontal transfer of Tn5801 between different Gram-positive bacteria.
The mobility of tet(M) was investigated in filter mating experiments. The results confirmed that tet(M) in our enterococci isolates was linked to a mobile genetic element that could be transferred in vitro between enterococci, from enterococci to E. coli, but not from E. coli to enterococci. Thus, tetracycline-resistant transconjugants were obtained in all the mating experiments using E. faecalis as a donor and E. faecium BM4105 as a recipient. When E. faecalis JH2-2 was used as a recipient, the transfer of tet(M) was detected from only three of the four donor strains. Despite this result, the transfer rates for tet(M) between E. faecalis obtained in the present study were higher than those reported previously [18,19]. tet(M) was also transferred from E. hirae to the recipient strain E. faecalis JH2-2, but not to E. faecium BM4501. To the best of our knowledge, horizontal transfer of tet(M) from E. hirae to E. faecalis has not been reported previously.
Conclusions
In conclusion, the detection of tet(M) in E. coli isolates from healthy pigs was higher than expected. Our results suggest that the presence of tet(M) in the E. coli isolates may be the result of the transfer of this tetracycline resistance gene from another bacteria in the intestinal tract. However, in the present study, tet(M) detected in E. coli isolates was shown to be a new allele type carried on a plasmid of unknown origin. Nevertheless, it can not be ruled out that this plasmid was transferred from other bacteria in the intestinal tract, since it is known that a gene flow between bacteria belonging to different genera occurs.
Methods
Doxycycline-resistant E. coli isolates
The doxycycline-resistant E. coli isolates were obtained in an ongoing research project carried out in Spain designed to evaluate the effect of the oral administration of different doses of colistin on the frequency of resistance to different antimicrobials among E. coli and enterococci isolates from healthy pigs. In this project, 12 healthy weaned piglets, which were obtained from the same farm and without previous exposure to antimicrobials, were examined. Animals were randomly distributed into three groups of four pigs. Groups received different doses of colistin in drinking water for 5 days. Samples of ileal content were collected at three different times. From each sample, 10 E. coli isolates were chosen randomly. A total of 300 E. coli isolates were obtained, 204 of which were doxycycline-resistant. Because of the high number of doxycycline-resistant E. coli isolates, a sample of 99 was randomly selected for this study.
Detection of tetracycline resistance genes
The presence of the tetracycline resistance genes tet(A), tet(B), and tet(M) was determined in doxycycline-resistant E. coli isolates by PCR using the primers described in Table 3. The following strains were used as positive controls: E. coli Co228 [tet(A)], E. coli Co71 [tet(B)], and E. faecalis CG110 [tet(M)].
Table 3.
Primers used in this study
| Primer use and primer | Sequence (5′-3′) | Reference |
|---|---|---|
|
Detection
tet
(A) | ||
| Tet(A)-F |
GCTACATCCTGCTTGCCTTC |
[27] |
| Tet(A)-R |
CATAGATCGCCGTGAAGAGG |
[27] |
|
Detection
tet
(B) | ||
| Tet(B)-F |
TTGGTTAGGGGCAAGTTTTG |
[27] |
| Tet(B)-R |
GTAATGGGCCAATAACACCG |
[27] |
|
Detection
tet
(M) | ||
| Tet(M)-1 (266) |
GTTAAATAGTGTTCTTGGAG |
[16] |
| Tet(M)-2 (267) |
CTAAGATATGGCTCTAACAA |
[16] |
|
Detection Tn
916
-like (
xis
-
Tn
) | ||
| Tn916-1 (327) |
GCCATGACCTATCTTATA |
[20] |
| Tn916-2 (328) |
CTAGATTGCGTCCAA |
[20] |
|
Detection Tn
5397
-like (
tndX
) | ||
| Tn5397-tndX-1 (864) |
ATGATGGGTTGGACAAAGA |
[20] |
| Tn5397-tndX-2 (865) |
CTTTGCTCGATAGGCTCTA |
[20] |
|
Detection Tn
5801
-like (
int
) | ||
| intcw459-1 (1811) |
CCGATATTGAGCCTATTGATGTG |
[22] |
| intcw459-2 (1812) |
GTCCATACGTTCCTAAAGTCGTC |
[22] |
|
Amplification and sequencing
tet
(M) | ||
| TetM-upstream (526) |
TTGAATGGAGGAAAATCAC |
[20] |
| TetM-up (323) |
CTGGCAAACAGGTTC |
[20] |
| TetM sequence-1 (525) |
TACTTTCCCTAAGAAAGAAAGT |
[20] |
| TetM sequence-3 (540) |
GCAGAAATCAGTAGAATTGC |
[20] |
| TetM sequence-6 (709) |
TCGAGGTCCGTCTGAAC |
[22] |
| Reverse TetM-2 (307) |
TTGTTAGAGCCATATCTTAG |
[20] |
| TetM sequence-9 (1756) |
AACAGTAAAATGTATAGAGGTG |
[22] |
| F2R (1837) |
GTGTCTTATACCATGGAAGGA |
[22] |
| TetM-down (324) |
TAGCTCATGTTGATGC |
[22] |
| Tet(M)-1 (266) | GTTAAATAGTGTTCTTGGAG | [16] |
Detection of Tn916-, Tn5397-, and Tn5801-like conjugative transposons
The presence of Tn916-, Tn5397-, and Tn5801-like transposons was first analyzed by PCR in the E. coli isolates that carried the tet(M) gene. Later, the occurrence of these transposons was also determined in 36 tet(M)-positive enterococci isolates selected from the four pigs belonging to the three groups studied from which tet(M)-positive E. coli were isolated (nine isolates from each animal).
Tn916-, Tn5397-, and Tn5801-like were detected by amplifying the xis-Tn, tndX, and int genes, respectively, using the primers shown in Table 3. PCR reactions were performed as described previously [20] using E. faecalis CG110 (Tn916), Bacillus subtilis CU2189 (Tn5397), and S. aureus Mu50 (Tn5801) as positive controls.
PCR amplification of full-length tet(M)
For all E. coli isolates and all enterococci isolates, but one, full-length tet(M) gene was amplified using the strategy suggested by Agersø et al. [20]. To amplify the upstream part of tet(M) by PCR, primers TetM-up (323), TetM sequence-1 (525), TetM-upstream (526), TetM sequence-3 (540), and TetM sequence-6 (709) were used (Table 3). The downstream part was amplified using the primers Reverse TetM-2 (307) and TetM sequence-9 (1756) (Table 3). One E. faecium isolate (CICYT-205) was suspected to contain two tet(M) genes. Therefore a long PCR product (4780 bp) containing the Tn5801-like tet(M) genes was amplified using primer pair TetM-upstream (526) and F2R (1837) with Phusion™ High-Fidelity DNA Polymerase (Finzymes). PCR conditions were 30 s at 98°C followed by 30 cycles of 10 s at 98°C, 30 s at 60°C and 145 s at 72°C, and a final extension for 10 min at 72°C. The sequencing primers TetM sequence-3 (540), TetM-upstream (526), TetM down (324), TetM sequence-1 (525), Tet(M)-1 (266), Reverse TetM-2 (307), and TetM sequence-9 (1756) were used (Table 3).
Phylogenetic analysis of tet(M)
GenBank was searched for full length tet(M) genes based on the definition that tet(M) genes share ≥80% similarity on the amino acid level [1] and 58 nucleotide sequences were selected for the phylogenetic analysis. Eleven unique gene sequences from this study were selected to represent E. coli and enterococci: four of E. coli and four of enterococci negative for transposons [one E. coli and one enterococcus from each of the four animals from which tet(M)-positive E. coli were isolated] and three from enterococci positive to each of the transposons studied. A neighbor-joining (NJ) tree based on a multiple alignment of the 11 tet(M) sequences obtained in this study and 58 sequences from GenBank [1802 bp of the total tet(M) gene of 1920 bp] was constructed in Clustal X [28] and visualized by MEGA 4.0 [29]. The tree was rooted with the tet(O) gene (GenBank/EMBL/DDBJ accession no. Y07780) as outgroup.
Filter mating experiments
Mating experiments were performed as described previously [30]. The conjugal transfer of tet(M) was analyzed in three different assays, using: enterococci as donor and recipient; E. coli as donor and enterococci as recipient; and enterococci as donor and E. coli as recipient. As donors, we selected four tet(M)-positive enterococci (three E. faecalis and one E. hirae) strains in which no transposons had been detected and four E. coli that carried the tet(M) gene. As recipients, E. faecium BM4105, E. faecalis JH2-2 (both resistant to rifampicin and fusidic acid), and E. coli CICYT70-Ri (rifampicin resistant) were used.
In the mating experiment between enterococci, transconjugants were selected on brain heart infusion (BHI) agar that contained tetracycline (8 μg/ml), rifampicin (12.5 μg/ml), and fusidic acid (12.5 μg/ml). When E. coli was used as a donor, transconjugants were selected in the same BHI agar, except that polymyxin B (32 μg/ml) was included instead of fusidic acid to avoid the growth of E. coli donors in the selection media. To select transconjugants in the mating experiment between enterococci and E. coli, BHI agar with rifampicin (50 μg/ml) and tetracycline (4 μg/ml) was used. Transconjugants were restricted on selective media that contained tetracycline and confirmed by the tet(M)-PCR screen (Table 3).
Southern blot
Total DNA and plasmid DNA from the four E. coli and four enterococci used as donors in the mating experiments were purified (QIAmp DNA mini Kit and QIAGEN Tip-100, Qiagen). Southern blot was performed using the total DNA and the plasmid DNA from E. coli and enterococci isolates after separation by electrophoresis in 0.8% agarose gel. A specific tet(M) probe made from the PCR product of the TetM sequence-1 (525) and Reverse TetM-2 (307) primers was used in Southern analysis.
Nucleotide sequence accession numbers
The sequences of the tet(M) gene from the E. coli isolates CICYT-332, CICYT-268, CICYT-320, and CICYT-348 have been deposited into GenBank under the accession numbers KJ55873-KJ55876, respectively.
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
SJR and YA participated in the experiments and helped to draft the manuscript. LEV participated in the experiments. RF and JARQS designed and coordinated the study and helped to draft and write the manuscript. JAO helped to draft and write the manuscript. All authors critically read and approved the final manuscript.
Contributor Information
Sonia Jurado-Rabadán, Email: sjurado@vet.ucm.es.
Ricardo de la Fuente, Email: rifuente@vet.ucm.es.
José A Ruiz-Santa-Quiteria, Email: ruizsanta@vet.ucm.es.
José A Orden, Email: jaorden@vet.ucm.es.
Lisbeth E de Vries, Email: ledv90@hotmail.com.
Yvonne Agersø, Email: yvoa@food.dtu.dk.
Acknowledgements
This study was funded by grants from the Spanish Ministry of Science and Innovation (AP2004-4242 and AGL2004-08139) and in part by a grant from the Danish Research Council for Technology and Production Sciences (274-05-0117).
The authors thank Dr. I. Badiola (CReSA, UAB-IRTA, Spain) for generously providing the enterococci isolates used in this study and Dr. C. Torres (Universidad de la Rioja, Spain) for the positive controls of tet(A) and tet(B) genes.
References
- Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz R, Kuhnert P, Boerlin P. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet Microbiol. 2003;91:73–84. doi: 10.1016/s0378-1135(02)00263-8. [DOI] [PubMed] [Google Scholar]
- Lim SK, Lee HS, Nam HM, Cho YS, Kim JM, Song SW, Park YH, Jung SC. Antimicrobial resistance observed in Escherichia coli strains isolated from fecal samples of cattle and pigs in Korea during 2003–2004. Int J Food Microbiol. 2007;116:283–286. doi: 10.1016/j.ijfoodmicro.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Maynard C, Fairbrother JM, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, Masson L, Lariviere S, Harel J. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob Agents Chemother. 2003;47:3214–3221. doi: 10.1128/AAC.47.10.3214-3221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Kazimierczak KA, Rincon MT, Patterson AJ, Martin JC, Young P, Flint HJ, Scott KP. A new tetracycline efflux gene, tet(40), is located in tandem with tet(O/32/O) in a human gut firmicute bacterium and in metagenomic library clones. Antimicrob Agents Chemother. 2008;52:4001–4009. doi: 10.1128/AAC.00308-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SA, Maani EV, Lindell AH, King CJ, McArthur JV. Novel tetracycline resistance determinant isolated from an environmental strain of Serratia marcescens. Appl Environ Microbiol. 2007;73:2199–2206. doi: 10.1128/AEM.02511-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan A, Shapir N, Sadowsky MJ. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl Environ Microbiol. 2004;70:2503–2507. doi: 10.1128/AEM.70.4.2503-2507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumai Y, Suzuki Y, Tanaka Y, Shima K, Bhadra RK, Yamasaki S, Kuroda K, Endo G. Characterization of multidrug-resistance phenotypes and genotypes of Escherichia coli strains isolated from swine from an abattoir in Osaka, Japan. Epidemiol Infect. 2005;133:59–70. doi: 10.1017/s0950268804003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengeløv G, Halling-Sørensen B, Aarestrup FM. Susceptibility of Escherichia coli and Enterococcus faecium isolated from pigs and broiler chickens to tetracycline degradation products and distribution of tetracycline resistance determinants in E. coli from food animals. Vet Microbiol. 2003;95:91–101. doi: 10.1016/s0378-1135(03)00123-8. [DOI] [PubMed] [Google Scholar]
- Hu J, Shi J, Chang H, Li D, Yang M, Kamagata Y. Phenotyping and genotyping of antibiotic-resistant Escherichia coli isolated from a natural river basin. Environ Sci Technol. 2008;42:3415–3420. doi: 10.1021/es7026746. [DOI] [PubMed] [Google Scholar]
- Hölzel CS, Harms KS, Bauer J, Bauer-Unkauf I, Hörmansdorfer S, Kämpf P, Mölle G, Oehme C, Preikschat P, Schwaiger K. Diversity of antimicrobial resistance genes and class-1-integrons in phylogenetically related porcine and human Escherichia coli. Vet Microbiol. 2012;160:403–412. doi: 10.1016/j.vetmic.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Jones CH, Tuckman M, Murphy E, Bradford PA. Identification and sequence of a tet(M) tetracycline resistance determinant homologue in clinical isolates of Escherichia coli. J Bacteriol. 2006;188:7151–7164. doi: 10.1128/JB.00705-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger K, Hölzel C, Bauer J. Resistance gene patterns of tetracycline resistant Escherichia coli of human and porcine origin. Vet Microbiol. 2010;142:329–336. doi: 10.1016/j.vetmic.2009.09.066. [DOI] [PubMed] [Google Scholar]
- Zhang T, Wang CG, Lv JC, Wang RS, Zhong XH. Survey on tetracycline resistance and antibiotic-resistant genotype of avian Escherichia coli in North China. Poult Sci. 2012;91:2774–2777. doi: 10.3382/ps.2012-02453. [DOI] [PubMed] [Google Scholar]
- Aarestrup FM, Agersø Y, Gerner-Smidt P, Madsen M, Jensen LB. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn Microbiol Infect Dis. 2000;37:127–137. doi: 10.1016/s0732-8893(00)00130-9. [DOI] [PubMed] [Google Scholar]
- Cauwerts K, Decostere A, De Graef EM, Haesebrouck F, Pasmans F. High prevalence of tetracycline resistance in Enterococcus isolates from broilers carrying the erm(B) gene. Avian Pathol. 2007;36:395–399. doi: 10.1080/03079450701589167. [DOI] [PubMed] [Google Scholar]
- Hummel A, Holzapfel WH, Franz CM. Characterisation and transfer of antibiotic resistance genes from enterococci isolated from food. Syst Appl Microbiol. 2007;30:1–7. doi: 10.1016/j.syapm.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Huys G, D'Haene K, Collard JM, Swings J. Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl Environ Microbiol. 2004;70:1555–1562. doi: 10.1128/AEM.70.3.1555-1562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agersø Y, Pedersen AG, Aarestrup FM. Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J Antimicrob Chemother. 2006;57:832–839. doi: 10.1093/jac/dkl069. [DOI] [PubMed] [Google Scholar]
- Roberts AP, Johanesen PA, Lyras D, Mullany P, Rood JI. Comparison of Tn5397 from Clostridium difficile, Tn916 from Enterococcus faecalis and the CW459tet(M) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology. 2001;147:1243–1251. doi: 10.1099/00221287-147-5-1243. [DOI] [PubMed] [Google Scholar]
- de Vries LE, Christensen H, Skov RL, Aarestrup FM, Agersø Y. Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J Antimicrob Chemother. 2009;64:490–500. doi: 10.1093/jac/dkp214. [DOI] [PubMed] [Google Scholar]
- Hart WS, Heuzenroeder MW, Barton MD. A study of the transfer of tetracycline resistance genes between Escherichia coli in the intestinal tract of a mouse and a chicken model. J Vet Med B. 2006;53:333–340. doi: 10.1111/j.1439-0450.2006.00967.x. [DOI] [PubMed] [Google Scholar]
- Trobos M, Lester CH, Olsen JE, Frimodt-Moller N, Hammerum AM. Natural transfer of sulphonamide and ampicillin resistance between Escherichia coli residing in the human intestine. J Antimicrob Chemother. 2009;63:80–86. doi: 10.1093/jac/dkn437. [DOI] [PubMed] [Google Scholar]
- Boerlin P, Travis R, Gyles CL, Reid-Smith R, Janecko N, Lim H, Nicholson V, McEwen SC, Friendship R, Archambault M. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl Environ Microbiol. 2005;71:6753–6761. doi: 10.1128/AEM.71.11.6753-6761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CS, Osborne DJ, Stanley J. Enterobacterial tetracycline resistance in relation to plasmid incompatibility. Mol Cell Probes. 1992;6:313–317. doi: 10.1016/0890-8508(92)90007-k. [DOI] [PubMed] [Google Scholar]
- Ng LK, Martin I, Alfa M, Mulvey M. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes. 2001;15:209–215. doi: 10.1006/mcpr.2001.0363. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Hammerum AM, Jensen LB, Aarestrup FM. Detection of the satA gene and transferability of virginiamycin resistance in Enterococcus faecium from food-animals. FEMS Microbiol Lett. 1998;168:145–151. doi: 10.1111/j.1574-6968.1998.tb13267.x. [DOI] [PubMed] [Google Scholar]


