Abstract
Recent studies have identified rice (Oryza sativa) as a major dietary source of inorganic arsenic (As) and poses a significant human health risk. The predominant model for plant detoxification of heavy metals is complexation of heavy metals with phytochelatins (PCs), synthesized non-translationally by PC synthase (PCS) and compartmentalized in vacuoles. In this study, in order to restrict As in the rice roots as a detoxification mechanism, a transgenic approach has been followed through expression of phytochelatin synthase, CdPCS1, from Ceratophyllum demersum, an aquatic As-accumulator plant. CdPCS1 expressing rice transgenic lines showed marked increase in PCS activity and enhanced synthesis of PCs in comparison to non-transgenic plant. Transgenic lines showed enhanced accumulation of As in root and shoot. This enhanced metal accumulation potential of transgenic lines was positively correlated to the content of PCs, which also increased several-fold higher in transgenic lines. However, all the transgenic lines accumulated significantly lower As in grain and husk in comparison to non-transgenic plant. The higher level of PCs in transgenic plants relative to non-transgenic presumably allowed sequestering and detoxification of higher amounts of As in roots and shoots, thereby restricting its accumulation in grain.
Arsenic (As) is well known for its carcinogenic effect on humans1 and has been recognized as a group I human carcinogen by the International Agency for Research of Cancer2. Various studies have identified rice (Oryza sativa) as a major dietary source of As which poses a significant health risk3,4. High accumulation of As in rice has been correlated with the anaerobic conditions prevailing in paddy soil leading to As mobilization5,6. The role of silicic acid and phosphate uptake pathways has been suggested for the inadvertent uptake of As in rice7,8. It has been reported that rice accumulate up to 2 mg As kg−1 grains in areas where severe As contamination occurs10.
The inorganic forms of the arsenic, [arsenate (AsV) and arsenite (AsIII)], are more prominent in soil in comparison to the organic As species [Monomethyl arsenic acid (MMA), Dimethylarsonic acid (DMA) ]9. AsIII as arsenous acid resembles structurally with silicic acid and thus transported as neutral molecule through Lsi1 in rice root7. AsV being non-functional phosphate analog enters the root cells via plant phosphate transporters10,11. Apart from these transporters for the uptake of As from soil, role of Natural Resistance-Associated Macrophage Protein (NRAMP) has been suggested in xylem loading and root to shoot mobilization8. AsV interferes with phosphate metabolism (such as phosphorylation and ATP synthesis) while AsIII has high affinity towards sulphydryl groups and binds to proteins affecting their structures and catalytic functions9,12. Methylated species such as DMA and MMA are also taken up by rice plants but at a much slower rate than inorganic As13. In the rice root, the reduction of AsV leads to the formation of AsIII which is more toxic as compared to AsV6,13. AsIII enters the xylem via a silicic acid/AsIII effluxer7,13. Detoxification of AsIII takes place through complexation with thiol-rich peptides including phytochelatins (PCs) and glutathione (GSH) followed by sequestration into vacuoles13,14
PCs, non-translationally synthesized small polypeptides, have a general structure (γ-Glu-Cys)n-Gly where n = 2–11. PCs are synthesized by the transpeptidation of γ-glutamylcysteinyl dipeptides from GSH with the help of Phytochelatin Synthase (PCS)15,16. PCS is a constitutively expressed enzyme and is known for the post-translational activation in the presence of heavy metals17,18. It has been suggested that PCS and PCs play very important role in heavy metal detoxification19,20,21. In the last decade, genes encoding PCS have been cloned from Arabidopsis (AtPCS1), wheat (TaPCS1), Schizosaccharomyces pombe (SpPCS), Caenorhabditis elegans (CePCS1), and from other species22,23,24,25,26. Different groups overexpressed PCS to enhance plant heavy metal tolerance and accumulation but observed varying results11,27,28,29,30,31,32,33,34,35,36. It was presumed that these disparities in metal response in transgenic plants may be due to the functional differences prevailing among the PCS genes from various sources. In our previous study, we showed that regulation of PCS at transcriptional level does not play any significant role in As detoxification as no significant change in expression of PCS was observed in rice seedlings in the presence of As37. Therefore, it was hypothesized that use of PCS gene from a potential accumulator plant might help in enhancing metal accumulation in transgenic plants.
The aquatic plant, Ceratophyllum demersum is known for its ability to accumulate toxic metals from water. It is a potential accumulator of heavy metals38,39,40,41,42. The capacity of metal accumulation in C. demersum has been correlated to the coordination between the induction of both biosynthesis and consumption pathways of thiols as well as induction of PCs41,42. In our recent studies, we have shown that expression of CdPCS1 in tobacco and Arabidopsis enhances Cd and As accumulation in roots as well as there was coordinated increase in levels of PCs and other precursor non-protein thiols14,21.
The overall goal of this study was to develop a strategy to generate transgenic rice with low As accumulation in grain. Recently, it has been reported that PCs are involved in trapping inorganic As in roots which subsequently reduces movement of As in grain43. Therefore, it is speculated that expression of an efficient PCS in rice might accumulate As in root and restrict its movement to the aerial parts. In the present study, CdPCS1 has been expressed in rice under control of constitutive promoter CaMV35S. Developed transgenic lines showed marked increase in PCS activity in comparison to non-transgenic plant. Further, analysis suggested that CdPCS1 expression in rice enhanced accumulation of As in roots leading to significantly low accumulation in aerial parts including rice grain.
Results
Selection of CdPCS1 expressing rice transgenic lines
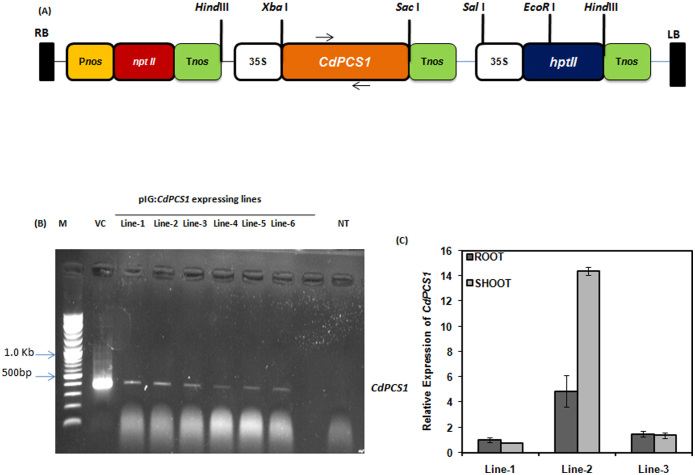
Transgenic lines (T0 generation) were confirmed for the presence of CdPCS1 by genomic DNA PCR using primers CdPCS1 RTF and CdPCS1 RTR. Genomic DNA amplification revealed that all the six transgenic lines selected on plates containing antibiotic harbour CdPCS1(Fig. 1A and B). Three independent transgenic lines of T4 generation were further selected for expression analysis in root as well as in shoot of CdPCS1 by real time PCR (Fig. 1C). Analysis revealed significant variability between individual transformants for CdPCS1 expression. The level of accumulation of a foreign gene in mRNA/protein level in a transgenic plant/tissue is dependent on many different factors. These include transgene integration at deferent chromosomal locations, the rate of transcription of the introduced gene and the stability of the resultant mRNA/protein44.
Figure 1. Plasmid map of pIG121-CdPCS1 and molecular analysis of CdPCS1 expressing rice transgenic lines.
(A) Plasmid map of pIG121-CdPCS1. Horizontal arrows above the CdPCS1 gene in the plasmid represents the region selected for PCR amplification. (B) Confirmation of the presence of CdPCS1 in transgenic lines through genomic DNA PCR. Lane-M represent O' Range RulerTM 100 bp + 500 bp DNA ladder, VC represent vector control and NT represents non transgenic control. PCR amplification was carried out using gene-specific CdPCS1-RTF and CdPCS1-RTR primers. Full-length gel is presented in Figure S3. (C) Relative expression analysis of CdPCS1 in root and shoot of CdPCS1 transgenic lines through qRT-PCR analysis by normalizing to the actin level as a control. Data are shown as mean ± SD of 3 independent experiments.
Non-protein thiol (NPT), PC and PC synthase activity in transgenic lines
To further investigate the effect of expression of CdPCS1 on the pool of non-protein thiols and PCs, the NT and transgenic lines were grown in Hewitt media for 10 days followed by growth in Hewitt media supplemented with 100 μM of AsV for next 10 days. Accumulation of Cys was higher in shoots in comparison to roots in NT and transgenic lines. Accumulation of Cys was significantly increased in shoots of transgenic lines in comparison to NT (p < 0.05; Fig. 2A). Accumulation of GSH was found to be higher in roots in comparison to shoots in NT plant. While in transgenic lines, the accumulation of GSH was higher in shoots. Transgenic plants accumulated significantly lesser amount of GSH in roots in comparison to NT (p < 0.05; Fig. 2B, Fig. S1). Accumulation of GSH in shoots showed the same pattern of Cys. There was significant increased accumulation of GSH in shoots of transgenic lines in comparison to NT (p < 0.05; Fig. 2B, Fig. S2). Line-2 and 3 accumulated higher amount of GSH in shoot as compared to NT (p < 0.05; Fig. 2B, Fig. S1 and S2).
Figure 2. Comparison of Cys and GSH levels in roots and shoots of NT and CdPCS1 expressing rice seedlings.
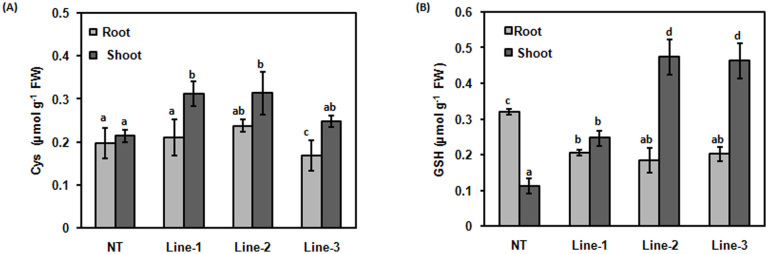
NT and transgenic lines of rice (Line-1, Line-2 and Line-3) were grown for 10 days on Hewitt media and then transferred in fresh Hewitt media containing 100 μM Na2AsO4. Levels of Cys (A) and GSH (B) were measured in root and shoot extracts (after 10 days of treatment) after derivatization with mBBr, separation using HPLC and fluorometric detection. Data are expressed as means ± SD of at least 3 independent experiments. Values marked with similar letters are not significantly (Duncan's test: p < 0.05) different.
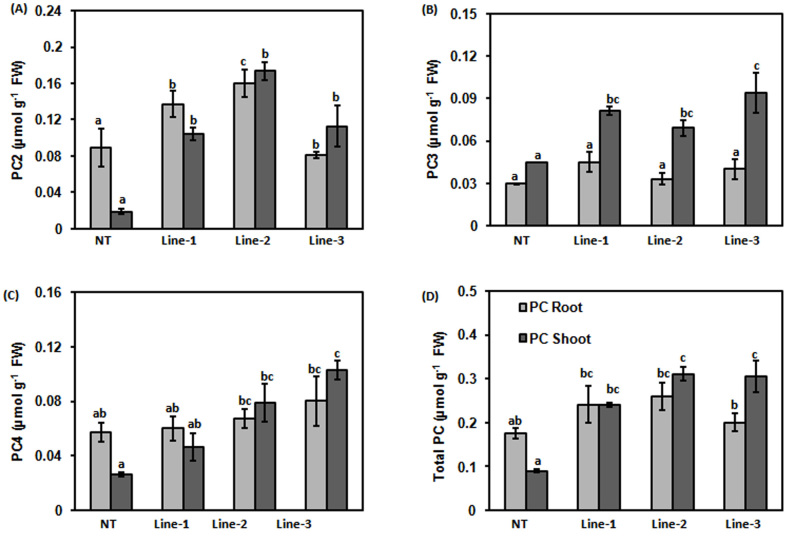
Transgenic lines (Line-1 and 2) accumulated significantly higher amount of PC2 in roots and shoots in comparison to NT. Line-3 accumulated significantly higher amount of PC2 in shoots in comparison to shoots of NT. In Line-2, 1.79 -fold and 9.21 -fold higher PC2 accumulation were observed in roots and shoots, respectively in comparison to NT. There was significantly higher accumulation of PC2 in shoots of all transgenic lines (Line-1, 2 and 3) in comparison to NT (p < 0.05; Fig. 3A, Fig. S1 and S2).
Figure 3. Comparison of PC levels in roots and shoots of NT and CdPCS1expressing rice seedlings.
NT and transgenic lines of rice (Line-1, Line-2 and Line-3) were grown for 10 days on Hewitt media and then transferred in fresh Hewitt media containing either 100 μM Na2AsO4. After 10 days of treatment the extract was used for accumulation of PCs. Levels of PC2 (A), PC3 (B), PC4 (C) and total PC (D) were measured in root and shoot extracts after derivatization with mBBr, separation in HPLC and fluorometric detection. Data are expressed as means ± SD of at least 3 independent experiments. Values marked with similar letters are not significantly (Duncan's test:p < 0.05) different.
Accumulation of PC3 was found to be higher in shoots of all transgenic lines in comparison to NT (Fig. 3B, Fig. S2). Line-3 had highest accumulation of PC3 in shoots in comparison to all transgenic lines as well as NT (p < 0.05; Fig. 3B, Fig. S2). However, there was no statistically significant difference was observed in roots of transgenic lines and NT (p < 0.05; Fig. 3B, Fig. S1). Roots and shoots of transgenic lines showed increased accumulation of PC4 when compared to NT. Line-2 and 3 accumulated significantly higher amount of PC4 in shoots in comparison to roots. On the contrary, there was no significant changes in accumulation of PC4 in roots of Line-1 as compared to NT (p < 0.05; Fig. 3C, Fig. S1).
In terms of total PCs, the transgenic plants accumulated more PCs in shoots as compared to shoots of NT. Roots of transgenic lines also showed higher accumulation of total PCs but the increase was more in shoots of Line-2 and Line-3 when compared with root (p < 0.05; Fig. 3D, Fig. S1 and S2).
CdPCS1 expression leads to enhanced PCS activity in transgenic lines
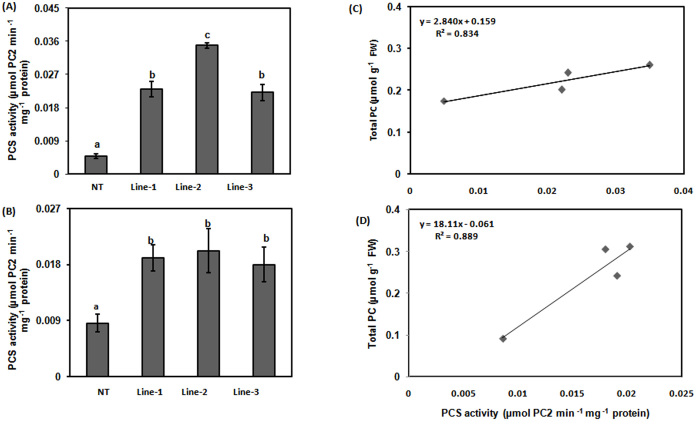
To find the reason of increased accumulation of PCs, activity of PCS enzyme in roots and shoots of both NT as well as transgenic lines were analyzed. The roots and shoots of transgenic lines showed increased enzyme activity in comparison to roots and shoots of NT (p < 0.05; Fig. 4A, B). Roots of Line-2 showed around 8-fold increase in enzyme activity in comparison to roots of NT (p < 0.05; Fig. 4A). Shoots of all transgenic lines showed around 2-fold increased enzyme activity in comparison to NT (p < 0.05; Fig. 4B). Increased PCS enzyme activity and accumulation of total PCs in transgenic lines suggested that there could be a positive correlation between increased enzyme activity and production of PCs. To check this hypothesis, correlation between enzyme and total accumulation of PCs was studied. These were positively correlated with coefficient of determination R2 = 0.839 in root (Fig. 4C) while in shoots at R2 = 0.834 (Fig. 4D). However, no correlation was observed in PCS activity and mRNA level in transgenic lines in shoots as well as roots. It seems that due to expression through 35S promoter, expression was enhanced several fold whereas PCS activity was enhanced at a certain level depending upon enzyme characteristics. Activation of enzyme and expression are two different phenomenon and one may not get a direct correlation all the time.
Figure 4. Phytochelatin synthase activity in roots and shoots in NT and CdPCS1expressing rice seedlings.
NT and transgenic lines of rice (Line-1, Line-2 and Line-3) were grown for 10 days on Hewitt media and then transferred in fresh Hewitt media containing 100 μM Na2AsO4. After 10 days of treatment, PCS activity in roots (A) and shoots (B) was measured as the production of PC2 min−1 mg−1 protein−1. Levels of PC2 were measured in root and shoot extracts after derivatization with mBBr, separation in HPLC and fluorometric detection. Correlation between PCS activity and accumulation of PCs in root (C) and shoot (D) of NT and CdPCS1expressing rice seedlings. Data are expressed as means ± SD of at least 3 independent experiments. Values marked with similar letters are not significantly (Duncan's test: p < 0.05) different.
Arsenic sensitivity and enhanced accumulation in transgenic lines under hydroponic condition
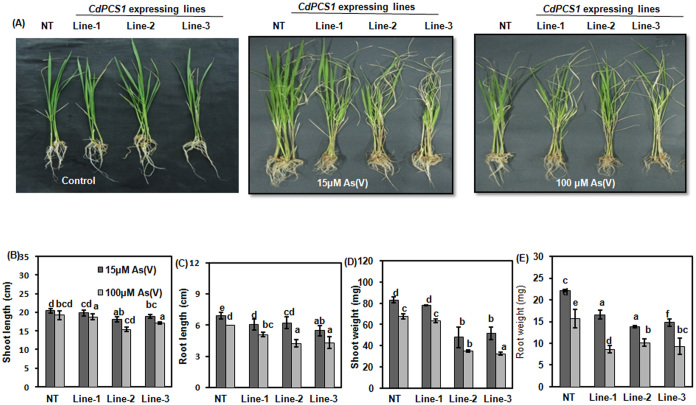
To check response of the transgenic lines to As stress, seeds of NT and transgenic lines (Line-1, 2 and 3) were grown in Hewitt media for 10 days and then transferred to Hewitt media supplemented with 15 and 100 μM AsV for next 10 days. In control, AsV was not supplemented; however, other growth conditions were maintained similar for all the seedlings. After 10 days of the growth, various growth parameters were recorded. Analysis suggests that transgenic lines were sensitive to As stress. Phenotypically, leaves of transgenic plants turned yellow after As treatment at higher concentration and roots also turned brown as the sign of heavy metal toxicity (Fig. 5A). Though these effects were also observed in NT plants however, transgenic lines were more sensitive to As stress.
Figure 5. Effect of As on growth of CdPCS1 expressing lines and NT in hydroponic condition.
(A) NT and transgenic lines of rice (Line-1, Line-2and Line-3, were grown for 10 days on Hewitt media and then transferred in fresh Hewitt media containing 15 μM and 100 μM Na2AsO4. (B) shoot length, (C) root length, (D) shoot weight and (E) root weight were recorded after 10 days of treatment. Data are expressed as means ± SD of at least 3 independent experiments. Values marked with similar letters are not significantly (Duncan's test: p < 0.05) different.
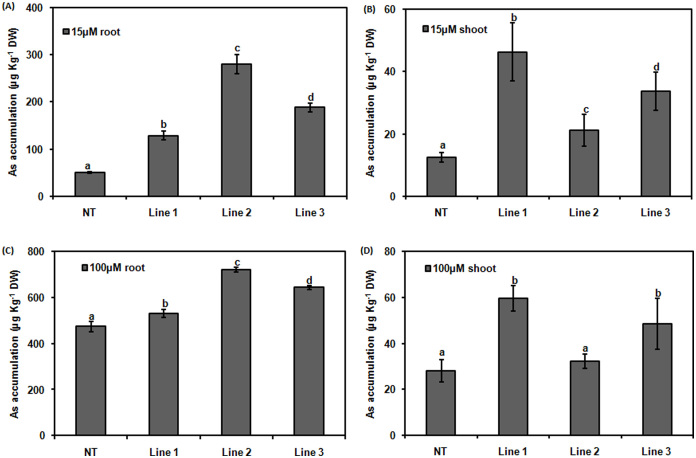
The shoot growth and root growth in terms of length and weight were affected more in transgenic lines when exposed to As. The effect was more prominent in root. There was no significant difference in the shoot length of transgenic lines and NT at 15 μM of AsV while at 100 μM of AsV, there was 25% reduction in shoot length of Line-2 in comparison to NT (p < 0.05; Fig. 5B). Root was affected more as these are the primary sight of defense. Even at 15 μM of AsV, the decrease in root length was significant in transgenic lines but this decrease was more at 100 μM of AsV. The maximum decrease in root length was observed in Line-2 under 100 μM of AsV which was 19.81% of NT (p < 0.05; Fig. 5C). There was 38% decrease in shoot weight of Line-2 and Line-3 at 15 μM of AsV in comparison to NT. At higher concentration, 47.74 and 51.78% reduction in the shoot weight of Line-2 and Line-3, respectively were observed (p < 0.05; Fig. 5D). Root weight of all the transgenic line decreased significantly. Line-1 showed 31% of reduction while 45% of reduction was observed in Line-2 and Line-3 at 15 μM of AsV in comparison to NT. At 100 μM of AsV, there was 44.87, 35.25 and 40.38% of reduction in root weight of Line-1, 2 and 3 in comparison to NT, respectively (p < 0.05; Fig. 5E).To compare As accumulation in transgenic lines and NT, As content was measured in NT and transgenic lines grown under AsV exposure of 15 and 100 μM for 10 days. All the transgenic lines accumulated significantly higher amount of As. Transgenic lines accumulated more As in roots as well as shoots in comparison to NT (Fig. 6A, B, C and D). Line-2 accumulated more As in roots as compared to all other transgenic lines (Fig. 6A).
Figure 6. As accumulation in roots (A) and shoots (B) of NT and CdPCS1 expressing transgenic lines.
The NT and transgenic lines of rice (Line-1, Line-2 and Line-3) were grown for 10 days on Hewitt media and then transferred in fresh Hewitt media containing 15 μM and 100 μM Na2AsO4. After 10 day treatment roots and shoots of NT and transgenic lines were washed, dried and accumulation of As was measured from 100 mg dried tissue. Data are expressed as means ± SD of at least 3 independent experiments. Values marked with similar letters are not significantly (Duncan's test: p < 0.05) different.
Arsenic uptake and accumulation in transgenic rice lines under simulated pot condition
In hydroponic experiment, transgenic plants were sensitive to As which might be due to enhanced accumulation of metal. To study response of transgenic lines in the field, pot experiment was designed under simulated condition. Different growth parameters such as plant height, tiller number, panicle length, grain number/panicle were observed before harvesting. All transgenic lines showed increase in tiller number, and panicle length in comparison to NT (Table 1).
Table 1. Relative growth parameters of transgenic lines as compared to NT plant in simulated condition.
| Line-1 | Line-2 | Line-3 | ||||
|---|---|---|---|---|---|---|
| Trait | -As(V) | +As(V) | -As(V) | +As(V) | -As(V) | +As(V) |
| Plant Height | 101.35 ± 0.521 | 106.31 ± 0.2022 | 98.32 ± 0.694 | 110.34±3.96 | 100.13 ± 0.99 | 108.70 ± 0.232 |
| Tiller number | 96.24 ± 2.61 | 122.50 ± 1.36 | 100.15 ± 2.37 | 177.46 ± 13.72 | 102.47 ± 3.13 | 169.99 ± 0.562 |
| Panicle length | 96.39 ± 3.61 | 117.28 ± 8.85 | 99.29 ± 3.08 | 124.22 ± 3.03 | 96.72 ± 2.77 | 103.51 ± 4.26 |
| Grain number/panicle | 97.67 ± 2.58 | 97.61 ± 2.27 | 101.10 ± 3.51 | 104.76 ± 2.61 | 99.94 ± 2.437 | 103.96 ± 3.14 |
| Germination | 100 | 100 | 100 | 100 | 100 | 100 |
| Grain yield (g/panicle) | 95.26 ± 4.03 | 166.81 ± 47.71 | 103.13 ± 8.829 | 136.64 ± 59.97 | 100.92 ± 2.71 | 134.03 ± 50.122 |
| 1000 Grain weight | 101.34 ± 6.24 | 93.86 ± 0.998 | 99.80 ± 4.066 | 114.81 ± 0.678 | 96.81 ± 3.41 | 110.01 ± 2.26 |
Relative values have been calculated as change in different parameters in transgenic lines in comparison to NT. NT has been considered as 100 for all the traits.
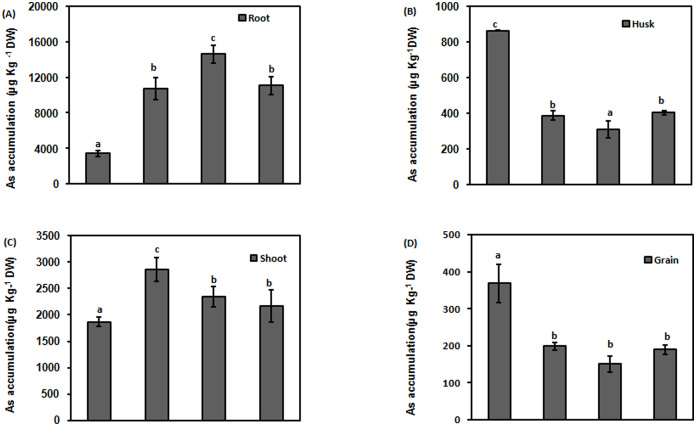
To observe the partitioning of As in different parts of rice plants, As accumulation was measured in roots, shoots, husk and grain of transgenic lines and NT plants. Transgenic lines accumulated significantly decreased level of As in husk in comparison to NT plants (p < 0.05; Fig. 7B). All the transgenic lines showed more than 50% reduction in the accumulation of As in husk in comparison to husk of NT (Supplementary Table 1). Grain of transgenic lines also accumulated significantly lesser amount of As in comparison to NT (p < 0.05; Fig. 7D). Transgenic lines accumulated 50% reduced level of As in grain in comparison to NT (Supplementary Table 1). On the contrary, transgenic lines accumulated significantly increased accumulation of As in shoot (p < 0.05; Fig. 7C; Supplementary Table-1). Transgenic lines also showed significantly enhanced accumulation of As in root (p < 0.05; Fig. 7A). The accumulation was at least 200% more in all the transgenic lines (Line 2 with 327%) in comparison to NT (Supplementary Table-1).
Figure 7. Effect of As in simulated condition on growth and As accumulation in different parts of NT and CdPCS1 expressing transgenic lines.
Grains were surface sterilized with 0.1% HgCl2 for 1 min, allowed to germinate in moist conditions. AsV was supplied as a solution of Na2HAsO4·7H2O in distilled water, in concentrations (15 ppm) until the rice grain was ripe. Applications of As ceased 10 days before harvest. As accumulation in root (A), husk (B), shoot (C) and root (D) of NT and CdPCS1 expressing transgenic lines. Data are expressed as means ± SD of at least 3 independent experiments. Values marked with similar letters are not significantly (Duncan's test: p < 0.05) different.
Discussion
PCs are Cys rich small peptides having general structure as (γ-glu-cys)ngly and are known for their protective role during different heavy metal stresses45. These molecules form complexes with heavy metals and sequester inside the vacuole. Synthesis of PCs takes place non-translationally by the transpeptidation of γ-glutamyl cysteine moiety of GSH with the help of PCS enzyme46. PCS is a constitutively expressed enzyme but its activity is regulated at post-translation level through binding to heavy metals27. It was hypothesized that by expressing the PCS in homologous or heterologous systems, the production of PCs can be manipulated which can lead to heavy metal tolerance and enhanced accumulation of heavy metals in plants27,32,47. But this hypothesis did not provide expected results. Overexpression of AtPCS1, TaPCS1 and SpPCS1 have been achieved in different plants that resulted in enhanced production of PC peptides in transgenic lines. However the results from these studies were contradictory in terms of metal accumulation and plant tolerance towards metal tolerance. Expression of AtPCS1 in Escherichia coli45 and Saccharomyces cerevisiae25 led to enhanced Cd tolerance and accumulation, however, overexpression in Arabidopsis resulted in Cd-hypersensitivity despite the enhanced PC production11,47. At the same time, Nicotiana tabacum expressing AtPCS1 displayed enhanced cadmium tolerance and accumulation32. However, expression of the same gene (AtPCS1) in another plant Brassica juncea led to higher Cd and Zn tolerance, but significantly lower accumulation of these elements in both root and shoot tissues27. Transgenic tobacco (Nicotiana glauca) expressing TaPCS1 (Wheat PCS) had shown increased Cd and Pb tolerance and accumulation28. These disparities in transgenic plants expressing PCS might have arisen due to differential PCS activity in source genes and nature of plant species selected for transformation. To understand response of PCS genes isolated from different organisms in one model organism, Wojas et al. (2008) expressed AtPCS1 and CePCS1 in tobacco concluded that not all PCS genes would be suitable for the transformation of all plant species for the phytoremediation purposes33. Unfortunately, none of the plants from which PCS genes were used to raise transgenic plants were potential hyperaccumulator of the heavy metals. Therefore, we thought that use of PCS gene from a potential accumulator plant might help in enhancing metal accumulation in transgenic plants. In this study, we investigated the effect of expression of CdPCS1 in rice for As response and accumulation. We hypothesized that CdPCS1 expression may increase production of PCs which will form complex with As for its detoxification. Enhanced PC synthesis in root might lead to increased accumulation of As in vacuoles. As a consequence of enhanced accumulation in root tissue, As movement to the above ground tissues including grains might decrease leading to development of low As accumulating rice varieties.
Expression of CdPCS1 in rice enhanced the synthesis of PCs (Fig. 4) as well as accumulation of As in roots and shoots of transgenic line as compared to NT (Fig. 7). However, growth was compromised as compare to NT (Fig. 5). While in simulated pot condition, the growth of transgenic lines was better in comparison to NT (Table 1). The possible explanation for this contrasting behaviour in the same lines could be that in hydroponic experiment, 10d old seedlings were continuously grown in liquid media supplemented with 100 μM AsV (phosphate analog) rendering the plants to easily uptake the As from the media. While in pot experiment, they are first grown in soil and then exposed to AsV in periodic interval. In soil-based studies, redox conditions and pH significantly affected the availability and consequent phytotoxicity of As as these parameters of the soil has a major influence on As speciation (inorganic and organic) and solubility. It is therefore not surprising that soil parameters influence the toxicity of As species due to altered availability (solubility or mobility). Apart from redox condition and pH, soil microorganism also plays an important role for its bioavailability following the reduction of AsV to AsIII and may potentially be further metabolized to methylated species. The presence of microorganisms and formation of iron plaques in the rhizosphere also affects bioavailability of As to the plants from the soil48. There are many reports about effects of mycorrhizal fungi as well as bacteria on As acquisition by plant. On the other hand, in hydroponic system As was supplied as AsV which acts as a phosphate analogue and is transported across the plasma membrane via phosphate cotransport systems. Therefore bioavailability of As is too high in this system. Phosphate is competing with AsV during its uptake by the roots of 10 d old rice seedlings. Once it enters inside the root cells, AsV is converted into AsIII, immediately chelate with PCs and sequestere inside the vacuole by the transgenic lines expressing CdPCS1. So a large amount of AsV enters through phosphate transporters and thereby reducing the phosphate uptake and a reduction of growth was observed in transgenic lines. In NT plants, the chelation is slow as production of PCs is low as compared to the transgenic lines. Therefore the uptake of phosphate is not hampered in NT plants as compared to transgenic lines. These results are in the agreement of the previous work in which PCSs from different plant sources were used to develop transgenic plants with higher metal accumulation and tolerance14,21,28.
Our analysis suggest that the increased accumulation of As was positively correlated with increased accumulation of PCs. In transgenic lines expressing CdPCS1, there was enhanced production of PCs and this increased level of PCs was responsible for enhanced sequestration of As9,15,21.
In the present study, modulated accumulation of GSH and Cys was also observed in the shoots of transgenic lines (Fig. 2). It could be explained on the basis of increased demand of these precursors for the synthesis of PCs. There was more accumulation of Cys, GSH and PCs in shoot tissue of transgenic lines in comparison to NT plants. This result can be explained on the basis of As accumulation data. Root accumulated more As in comparison to shoot (Fig. 6). For accumulation of As, enhanced synthesis of the PCs is required. This enhanced synthesis requires higher substrate flux as GSH and Cys. As the roots accumulated more As, the PCs which formed in the roots were utilised for more sequestration of As. Therefore in roots, there was enhanced accumulation of PCs but the GSH accumulation was decreased as GSH was utilised for PCs biosynthesis. As most of the As was accumulated in roots, there was lesser amount of As remained free for the transportation in shoots. As CdPCS1 was expressed under the CaMV35S promoter, there was increased PCS activity in roots as well as shoots. At the same time, roots of transgenic lines had higher PCS activity in comparison to shoots (Fig. 4). There was more demand of GSH and Cys in roots of transgenic lines in comparison to shoots and thus GSH and Cys content was enhanced in shoots while decrease amount of these compound were observed in roots of transgenic lines. Our results are in the agreement of the previous studies where increased amount of PCs was related with decrease amount in GSH as well as Cys11,21,47.
To study the accumulation of As in different parts of transgenic lines, simulated pot experiment was performed. Enhanced accumulation of As was observed in roots (327%) and shoots (53%) of transgenic lines while husk and grain accumulated significantly lower As in comparison to NT plants. As most of the As which entered the plant was sequestered inside the root, there was lesser As for transportation in shoots. Even then, the transgenic lines showed increased accumulation in shoots but this increase was only 29% of NT shoot tissue. Therefore, the accumulation of As was least in grains of transgenic lines.
In conclusion, among the transgenic lines analyzed for low grain As, Line-2 was the most effective to restrict the As level in roots. It showed the lowest As content in grains under stress conditions compared to the NT plants and other transgenic lines. It provides a good approach to generate transgenic rice with low grain As.
Methods
Plant materials and growth conditions
Mature, dehusked seeds of the rice variety IR-64, (Oryza sativa L. ssp. Indica) were used for callus induction. Callus were induced on MS basal medium49, supplemented with 2 mg l−1 2,4-dichlorophenoxyacetic acid (2,4-D), 30 g l−1 maltose, 0.3 g l−1 casein hydrolysate, 0.5 g l−1 proline and 4 g l−1 phytagel, and incubated at 26°C in the dark50. After 3–4 weeks, the proliferating calli were subcultured onto the same medium and cultured for another 3–4 weeks. White friable embryogenic calli dedveloped were subcultured onto the same medium 10 days before infection with Agrobacterium. Seeds of transgenic lines and non-transgenic line (NT) were used to study the morphological variability in response to As stress. Seeds were disinfected in 0.1% HgCl2 solution for 30 s followed by thorough washing with deionized water and soaking in milli-Q water for 24 h. These seeds were transferred to Petri dishes and kept in a culture room for 3–4 days at 26°C in the dark for proper germination. The plants were grown in hydroponic medium48 for 10 days before the treatment with different concentrations of AsV. The hydroponic culture and all experiments were conducted inside a controlled environment growth chamber under the following conditions: 16-h light period with a light intensity of 350 μmol m−2 s−1; 25/20°C day/night temperatures; and 60% relative humidity.
Agrobacterium-mediated transformation
Transformation was carried out using Agrobacterium tumefaciens strain EHA101 containing engineered binary vector in pIG121Hm51. Engineered plasmid contains CdPCS1 gene (NCBI Accession No. HM855235) in place of uidA and hygromycin resistance (hptII) gene as selectable marker. The calli were immersed in bacterial suspension for 15–20 min, and the excess bacterial suspension was removed by blotting on sterile tissue paper. Infected calli were transferred onto an MS co-cultivation medium51. After co-cultivation, infected calli were washed 2 times with sterile distilled water followed by aqueous solution containing cefotaxime (250 mg l−1) and carbenicillin (250 mg l−1), blotted on sterile tissue paper, and transferred to MS selection medium (MS callus induction medium containing 40 mg l−1 hygromycin).
After 5 rounds of selection, actively growing pieces of calli were transferred to MS regeneration medium [MS basal medium supplemented with 1 mg l−1 1-naphthaleneacetic acid (NAA), 3 mg l−1 6-benzyl adenine (6-BA), 1 mg l−1 thidiazuron (TDZ), 0.3 g l−1 casein hydrolysate, 30 g l−1 maltose and 40 mg l−1 hygromycin] and kept under a 16-h-light (110–130 μmol m−2 s−1) and 8-h-dark photoperiod51. Shoot regeneration was observed after 4 weeks. The regenerated shoots were transferred to the rooting medium (half-strength MS medium without growth regulator containing 40 mg l−1 hygromycin). After 4 weeks, rooted plants were kept in Soilrite for 4 weeks for hardening. The hardened plants were transferred to plastic pots in the greenhouse till flowering and seed setting.
Analysis of transgenic lines
Three rice homozygous transgenic lines (Lines-1, 2 and 3) expressing CdPCS1 of T4 generation were used for further analysis. In this study, only AsV was used to study response of transgenic lines as this species is predominant in the aerobic soil. Studies using soil and pure Fe hydroxides generally agree that AsV solubility increases upon pH increase within pH-ranges commonly found in soil (pH 3–8), whereas AsIII tends to follow the opposite pattern51,52. NT and transgenic rice lines were grown in hydroponic medium for 10 days followed by in hydroponic media supplemented with AsV (15 and 100 μM) for 10 days under standard physiological conditions of 16 h light (115 μmol m−2 s−1) and 8 h dark photoperiod at 25 ± 2°C53,54,55,56. Na2HAsO4 (Merck, India) was used to prepare AsV stock solution. The root length, shoot length, root weight as well as shoot weight were measured after 10 days of treatment. All the samples were ground in liquid N2 and stored at −80°C.
Molecular analysis of transgenic lines
PCR analysis was used to confirm presence of the CdPCS1 transgene in T4 generation. Primers CdPCS-RTF and CdPCS-RTR were designed to confirm the presence of CdPCS1 in transgenic lines.
Approximately, 5 μg RNase free DNase treated total RNA isolated from root and shoot of rice was reverse-transcribed using SuperScriptII (Fermentas, USA), following the manufacturer's recommendation. Real Time PCR was performed in 25 μl reaction volume using CdPCS1 specific primers (CdPCS1RTF,TGCTCGATTCAAGTATCCTCCACA; CdPCS1RTR, CTTGCCGTTCTCAGTACATCTTC) using Power SYBR Green PCR Master Mix (ABI, USA) and Fast Real Time PCR System (Model 7500; ABI, USA). Oligonucleotide primers for rice actin gene (F, GAGTATGATGAGTCGGGTCCAG; R-ACACCAACACCAACAATCCCAAACAGAG) were used to calculate the relative expression of the gene in independent lines. The reactions were performed using the following cycle conditions, an initial 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and the final 5 min extension at 72°C. After obtaining ct value for each reaction, the relative expression was calculated by using Delta-Delta ct method57.
HPLC analysis of non-protein thiols
For the analysis of non-protein thiols (PCs, GSH and Cys), seeds of NT and transgenic lines were grown in Hewitt-media hydroponicaly for 10 days followed by growth in same media supplemented with 100 μM of AsV for next 10 days. Sample were prepared by crushing 100 mg of rice roots in liquid nitrogen with the help of extraction buffer which contain 6.3 mM diethylenetriamine-penta-acetic acid (DTPA) and 0.1% TFA (Trifluoroacetic acid). Samples were kept in −20°C until further processing. Extraction and derivetization of peptides was done with the help of monobromobimane (mBBr)21. Fluorescence HPLC detection method was utilized for analysis of peptides as described21 The analytical data were integrated and quantified using Empower Software (Waters, USA) using standard curves made by running mBBr-labeled NPT standards. The NPT (Cys, GSH, PC2, PC3 and PC4) standards are commercially available from Sigma-Aldrich (St. Louis, MO, U.S.A. and Link-biotech, India).
Phytochelatin synthase activity
For analysis of phytochelatin synthase (PCS; EC 2.5.2.15) activity, extraction was performed following Dave et al. (2013)16. Ten gram of root and shoots of transgenic and NT plant was frozen and ground in liquid nitrogen. The powdered material was homogenized in 20 ml of chilled Tris buffer (50 mM; pH 8.0) containing 10 mM β-mercaptoethanol (BME), 1 mM phenylmethyl sulphonyl fluoride (PMSF) and 14% (v/v) glycerol at 40°C. The homogenate was centrifuged at 10,000 × g for 10 min and the supernatant obtained was used for PCS assay. Briefly, the reaction mixture contained 100 mM Tris-HCl (pH 8.0), 1 mM BME, 100 μM AsV 5 mM GSH, and 200 μL enzyme extract, in a final volume of 1 ml. The reaction was terminated after 30 min by addition of 150 μL of 5% TFA. The reaction mixture was cleared by centrifugation and filtered, and 50 μL was injected on the HPLC for analyzing the PCs following pre-column derivatization with mBBr as mentioned above. Only the synthesis of PC2 could be detected whose amount was calculated in terms of GSH equivalents. Enzyme activity is expressed as μmoles PC2 min−1 mg−1 protein.
Simulated pot experiment
The simulated pot experiment was conducted in transgenic glass house. Grains were surface sterilized with 0.1% HgCl2 for 1 min and allowed to germinate in moist conditions. The seedlings were transplanted to one liter plastic pots (having no hole) filled with 1 kg of soil [alluvial soil and compost (in ratio 3:1)] (pH: 8.4, As content: 0.013 mg kg−1). The pots were placed in a transgenic greenhouse under natural sun light and 90% humidity with overall temperature fluctuation between 20°C and 30°C. After transplantation, the plants were grown under flooded conditions, saturation to permanent immersion of the soil under up to 3–4 cm of solution. For soil fertility, nitrogen was supplied as urea (76 mg kg−1) in four equal splits, phosphorous as Triple supper phosphate (TSP) (14.3 mg kg−1) and potassium as Murate of potash (MOP) (28.6 mg kg−1) were applied once before transplantation of the rice seedlings.
AsV was supplied as a solution of Na2HAsO4·7H2O in distilled water, in concentrations (15 ppm) until the rice grains were ripe (110 + 15 days after germination). The amount of water used for irrigation was between 600 and 800 ml pot−1 week−1 to maintain flooded condition. Applications of AsV was ceased 10 days before harvest. After harvesting the rice plants, the soil pH (7.6) and arsenic content (10.43 mg/kg) were measured. After measuring plant height (panicle top to level of soil in the pot) and tiller numbers, rice plants were harvested by cutting at 4 cm above the soil. Plant height, tiller number and panicle length were recorded at the time of harvesting. Grain number per panicle was calculated by counting the number of filled spikelet per panicle manually. Thousand-grain weight (the mass of 1000 unhusked rice grains) was measured for each transgenic line and NT. Rice grains were separated from husks using a pestle and mortar. Root, shoot, husk and grain were collected and stored for the estimation of AsV accumulation. Root and shoot were dried in oven before storage.
Arsenic estimation
Shoots and roots from hydroponic media grown rice seedlings as well as roots, shoots, husk and grain from simulated pot experiment were dried in hot air oven at 40°C to reach constant weight. Dried plant tissues (100 mg) were digested in HNO3 and H2O2, 3:1, through microwave digestion, in CEM-MDS 2000 Microwave digester58. This digested solution was diluted from which 10 μl aliquot was quantitatively analyzed for As through atomic absorption spectrometry (Perkin–Elmer; AAnalyst 600) fitted with graphite furnace. Reference standard for calibration was made using 1000 mg/ml (AA03N-5) standard supplied by Accustandard, USA.
Stastiscal analysis
Each experiment was carried out under a completely randomized design with three independent experiments with atleast three replications. The data were analyzed by two way analysis of variance to confirm the variability and validity of results, and Duncan's multiple range test (DMRT) was performed to determine significant difference between treatments.
Author Contributions
M.S., R.D., D.S., R.K. and S.D. performed experiments. M.S., D.C., R.D., R.D.T. and P.K.T. discussed the results. M.S., P.K.T. and D.C. wrote the manuscript.
Supplementary Material
Supplementary info
Acknowledgments
Authors acknowledge Director, CSIR-National Botanical Research Institute for providing facilities and support during the study. The authors acknowledge the financial assistance from CSIR-Network project (BSC-107). MS acknowledge CSIR for providing SRF. This work is a part of AcSIR Ph.D program. Authors have no conflict of interest.
References
- Tsuji J. S., Yost L. J., Barraj L. M., Scrafford C. G. & Mink P. J. Use of background inorganic arsenic exposures to provide perspective on risk assessment results. Regul. Toxicol. Pharmacol. 48, 59–68 (2007). [DOI] [PubMed] [Google Scholar]
- IARC. Some drinking-water disinfectants and contaminants, including arsenic. IARC Monogr. Eval. Carcinog. Risks Hum. 84, 1–477 (2004). [PMC free article] [PubMed] [Google Scholar]
- Tuli R., Chakrabarty D., Trivedi P. K. & Tripathi R. D. Recent advances in arsenic accumulation and metabolism in rice. Mol. Breed. 26, 307–323 (2010). [Google Scholar]
- Banerjee M. et al. High arsenic in rice is associated with elevated genotoxic effects in humans. Sci. Rep. 3, 1–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y. et al. Arsenic behavior in paddy fields during the cycle of flooded and non-flooded periods. Environ. Sci. Technol. 38, 1038–1044 (2004). [DOI] [PubMed] [Google Scholar]
- P N. et al. Greatly enhanced arsenic shoot assimilation in rice leads to elevated grain levels compared to wheat and barley. Environ. Sci. Technol. 41, 6854–6859 (2007). [DOI] [PubMed] [Google Scholar]
- Ma J. F. et al. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Nat. Acad. Sci. U.S.A. 105, 9931–9935(2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari M. et al. Expression in Arabidopsis and cellular localization reveal involvement of rice NRAMP, OsNRAMP1, in arsenic transport and tolerance. Plant Cell Environ. 37, 140–152 (2014). [DOI] [PubMed] [Google Scholar]
- Tripathi R. D. et al. Arsenic hazards: strategies for tolerance and remediation by plants. Trends Biotechnol. 25, 158–165 (2007). [DOI] [PubMed] [Google Scholar]
- Clark G. T., James D. & Phung H. T. Phosphate absorption by Arabidopsis thaliana: interactions between phosphorus status and inhibition by arsenate. Aus. J. Plant Physiol. 27, 959–965 (2000). [Google Scholar]
- Lee S. et al. Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiol. 131, 656–663 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F. J. et al. The role of the rice aquaporin Lsi1 in arsenite efflux from roots. New Phytol. 186, 392–399 (2010). [DOI] [PubMed] [Google Scholar]
- Abedin M. J., Feldmann J. & Meharg A. A. Uptake kinetics of arsenic species in rice plants. Plant Physiol. 128, 1120–1128 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla D., Tiwari M., Tripathi R. D., Nath P. & Trivedi P. K. Synthetic phytochelatins complement a phytochelatin-deficient Arabidopsis mutant and enhance the accumulation of heavy metal(loid)s. Biochem. Biophys. Res. Commu. 434, 664–669 (2013). [DOI] [PubMed] [Google Scholar]
- Grill E., Loffler S., Winnacker E. L. & Zenk M. H. Phytochelatins, the heavy metal-binding peptides of plants are synthesized from glutathione by a specific γ-glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase). Proc. Natl. Acad. Sci. U.S.A. 86, 6838–6842 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave R. et al. Arsenite Tolerance is Related to Proportional Thiolic Metabolite Synthesis in Rice (Oryza sativa L.). Arch. Environ. Contam. Toxicol 64, 235–242 (2013). [DOI] [PubMed] [Google Scholar]
- Vatamaniuk O. K. et al. Phytochelatin synthase, a dipeptidyltransferase that undergoes multisite acylation with γ-glutamylcysteine during catalysis. J. Biol. Chem 279, 22449–22460 (2004). [DOI] [PubMed] [Google Scholar]
- Rea P. A. Phytochelatin synthase: of a protease a peptide polymerase made. Physiol. Plant. 145, 154–164 (2012). [DOI] [PubMed] [Google Scholar]
- Glaeser H. et al. Glutathione metabolism and heavy metal detoxification in Schizosaccharomyces pombe. Curr. Genet. 19, 207–213 (1991). [Google Scholar]
- Mishra S., Srivastava S., Tripathi R. D. & Trivedi P. K. Thiol metabolism and antioxidant systems complement each other during arsenate detoxification in Ceratophyllum demersum L. Aquat. Toxicol. 31, 205–215 (2008). [DOI] [PubMed] [Google Scholar]
- Shukla D. et al. Expression of phytochelatin synthase from aquatic macrophyte Ceratophyllum demersum L. enhances cadmium and arsenic accumulation in tobacco. Plant Cell Rep. 31, 1687–1699 (2012). [DOI] [PubMed] [Google Scholar]
- Clemens S., Kim E. J., Neumann D. & Schroeder J. L. Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 18, 3325–3333 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S., Schroeder J. I. & Degenkolb T. Caenorhabditis elegans expresses a functional phytochelatin synthase. Eur. J Biochem. 268, 3640–3643 (2001). [DOI] [PubMed] [Google Scholar]
- Ha S. B. et al. Phytochelatin synthase genes from Arabidopsis and the yeast, Schizosaccharomyces pombe. Plant Cell. 11, 1153–1164 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk O. K., Mari S., Lu Y. P. & Rea P. A. AtPCS1, a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proc. Natl. Acad. Sci. U.S.A. 96, 7110–7115 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk O. K., Bucher E., Ward J. T. & Rea P. A. A new pathway for heavy metal detoxification in animals. J. Biol Chem. 276, 20817–20820 (2001). [DOI] [PubMed] [Google Scholar]
- Gasic K. & Korban S. S. Expression of Arabidopsis phytochelatin synthase in Indian mustard (Brassica juncea) plants enhances tolerance for Cd and Zn. Planta 225, 1277–1285 (2007). [DOI] [PubMed] [Google Scholar]
- Gisbert C. et al. Plant genetically modified that accumulates Pb is especially promising for phytoremediation. Biochem. Biophys. Res. Commun. 303, 440–445 (2003). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Overexpression of phytochelatin synthase in Arabidopsis leads to enhanced arsenic tolerance and cadmium hypersensitivity. Plant Cell Physiol. 45, 1787–1797 (2004). [DOI] [PubMed] [Google Scholar]
- Liu G. Y., Zhang Y. X. & Chai T. Y. Phytochelatin synthase of Thlaspi caerulescens enhanced tolerance and accumulation of heavy metals when expressed in yeast and tobacco. Plant Cell Rep. 30, 1067–1076 (2011). [DOI] [PubMed] [Google Scholar]
- Martinez M. et al. An engineered plant that accumulates higher levels of heavy metals than Thlaspi caerulescens, with yields of 100 times more biomass in mine soils. Chemosphere 64, 478–485 (2006). [DOI] [PubMed] [Google Scholar]
- Pomponi M. et al. Overexpression of Arabidopsis phytochelatin synthase in tobacco plants enhances Cd(+2) tolerance and accumulation but not translocation to the shoot. Planta. 223, 180–189 (2006). [DOI] [PubMed] [Google Scholar]
- Wojas S. et al. Overexpression of phytochelatin synthase in tobacco: distinctive effects of AtPCS1 and CePCS genes on plant response to cadmium. J. Exp. Bot. 92, 1–15 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojas S., Clemens S., Sklodowska A. & Maria Antosiewicz D. Arsenic response of AtPCS1- and CePCS-expressing plants— effects of external As(V) concentration on As-accumulation pattern and NPT metabolism. J Plant Physiol 167, 169–175 (2010a). [DOI] [PubMed] [Google Scholar]
- Wojas S., Ruszczynska A., Bulska E., Clemens S. & Antosiewicz D. M. The role of subcellular distribution of cadmium and phytochelatins in the generation of distinct phenotypes of AtPCS1- and CePCS3-expressing tobacco. J. Plant Physiol. 167, 981–988 (2010b). [DOI] [PubMed] [Google Scholar]
- Brunetti P. et al. Cadmium tolerance and phytochelatin content of Arabidopsis seedlings over-expressing the phytochelatin synthase gene AtPCS1. J Exp Bot 62, 5509–5519 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty D. et al. Comparative transcriptome analysis of arsenate and arsenite stresses in rice seedlings. Chemosphere 74, 688–702 (2009). [DOI] [PubMed] [Google Scholar]
- Bunluesin S. et al. Plant screening and comparison of Ceratophyllum demersum and Hydrilla verticillata for cadmium accumulation. Bull. Environ. Contam. Toxicol. 73, 591–598 (2004). [DOI] [PubMed] [Google Scholar]
- Keskinkan O., Goksu M. Z., Basibuyuk M. & Forster C. F. Heavy metal adsorption properties of a submerged aquatic plant (Ceratophyllum demersum). Bioresour. Technol. 92, 197–200 (2004). [DOI] [PubMed] [Google Scholar]
- Robinson B. et al. Arsenic hyperaccumulation by aquatic macrophyte in the Taupo volcanic zone NZ. Environ. Exp. Bot. 58, 206–215 (2006). [Google Scholar]
- Mishra S. et al. Thiol metabolism play significant role during cadmium detoxification by Ceratophyllum demersum L. Bioresour. Technol. 100, 2155–2161 (2009). [DOI] [PubMed] [Google Scholar]
- Sun Q., Liu W. B. & Wang C. Different response of phytochelatins in two aquatic macrophytes exposed to cadmium at environmentally relevant concentrations. Afr. J. Biotechnol. 10, 6292–6299 (2011). [Google Scholar]
- Batista B. L. et al. Identification and quantification of phytochelatins in roots of rice to long-term exposure: evidence of individual role on arsenic accumulation and translocation. J. Exp. Bot. 65, 1467–1479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M., Mol J. N. M. & Kooter J. M. The Silence of Genes in Transgenic Plants. Ann. Bot. 79, 3–12 (1997). [Google Scholar]
- Hall J. L. Cellular mechanism for heavy metal detoxification and tolerance. J. Exp. Bot. 53, 1–11 (2001). [PubMed] [Google Scholar]
- Sauge-Merle S. et al. Enhanced toxic metal accumulation in engineered bacterial cells expressing Arabidopsis thaliana phytochelatin synthase. Appl Environ. Microbiol. 69, 490–494 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Overexpression of phytochelatin synthase in Arabidopsis leads to enhanced arsenic tolerance and cadmium sensitivity. Plant Cell Physiol. 45, 1787–1797 (2004). [DOI] [PubMed] [Google Scholar]
- Liu W. J., Zhu Y. G., Smith F. A. & Smith S. E. Do phosphorus nutrition and iron plaque alter arsenate (As) uptake by rice seedlings in hydroponic culture? New Phytol. 162, 481–488 (2004). [Google Scholar]
- Murash I. T. & Skoog F. A revised mediurn for rapid growth and bio-assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497 (1962). [Google Scholar]
- Chakrabarty D. et al. Differential transcriptional expression following thidiazuron-induced callus differentiation developmental shifts in rice. Plant Biol. 12, 46–59 (2010). [DOI] [PubMed] [Google Scholar]
- Jain A. & Loeppert R. H. Effect of competing anions on the adsorption of arsenate and arsenite by ferrihydrite. J. Environ. Qual. 29, 1422–1430 (2000). [Google Scholar]
- Walter J. F. & Walter W. W. Arsenic transformations in the soil–rhizosphere–plant system: fundamentals and potential application to phytoremediation. J. Biotechnol. 99, 259–278 (2002). [DOI] [PubMed] [Google Scholar]
- Dubey S. et al. Transcriptomic and metabolomic shifts in rice roots in response to Cr(VI) stress. BMC Genomics. 11, 648 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Asif M. H., Chakrabarty D., Tripathi R. D. & Trivedi P. K. Differential expression and alternative splicing of rice sulphate transporter family members regulate sulphur status during plant growth, development and stress conditions. Funtc. Integr. Genomics 11, 259–273 (2011). [DOI] [PubMed] [Google Scholar]
- Kumar S. et al. Differential expression of rice Lambda class GST gene family members during plant growth, development, and in response to stress conditions. Plant Mol. Biol. Rep. 31, 569–580 (2013). [Google Scholar]
- Kumar S. et al. Expression of a rice Lambda class of Glutathione S-transferase, OsGSTL2, in Arabidopsis provides tolerance to heavy metal and other abiotic stresses. J. Hazard. Mater. 249, 228–237 (2013). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. Enhanced transgene expression in rice following selection controlled by weak promoters. BMC Biotechnol. 13, 29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury T., Uchino T., Toluanga H. &. Ando M. Survey of arsenic in food composites from arsenic affected area of West Bengal, India. Food Chem. Toxicol. 40, 1611–1621 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary info