There is a known hierarchy of selenoprotein mRNA expression when selenium is absent. This article shows that this hierarchy is determined by the susceptibility of different selenoprotein mRNAs to degradation by the NMD pathway.
Keywords: selenium, nonsense-mediated decay, UPF1, SMG1
Abstract
Selenoproteins contain the unique amino acid selenocysteine (Sec), which is encoded by the triplet UGA. Since UGA also serves as a stop codon, it has been postulated that selenoprotein mRNAs are targeted for degradation by the nonsense-mediated mRNA decay pathway (NMD). Several reports have observed a hierarchy of selenoprotein mRNA expression when selenium (Se) is limiting, whereby the abundance of certain transcripts decline while others do not. We sought to investigate the role of NMD in this hierarchical response that selenoprotein mRNAs exhibit to environmental Se status. Selenoprotein mRNAs were categorized as being predicted sensitive or resistant to NMD based on the requirements held by the current model. About half of the selenoprotein transcriptome was predicted to be sensitive to NMD and showed significant changes in mRNA abundance in response to cellular Se status. The other half that was predicted to be resistant to NMD did not respond to Se status. RNA immunoprecipitation with essential NMD factor UPF1 revealed that the mRNAs that were the most sensitive to Se status were also the most enriched on UPF1 during Se deficiency. Furthermore, depletion of SMG1, the kinase responsible for UPF1 phosphorylation and NMD activation, abrogated the decline in transcript abundance of Se-responsive transcripts. Lastly, mRNA decay rates of Se-responsive transcripts were altered upon the addition of Se to resemble the slower decay rates of nonresponsive transcripts. Taken together, these results present novel evidence in support of a crucial role for the NMD pathway in regulating selenoprotein mRNA levels when Se is limiting.
INTRODUCTION
Selenium (Se) is an essential trace element that exerts its function predominantly through selenoproteins, which are characterized by the presence of the unusual amino acid selenocysteine (Sec). The evolutionary preservation of selenoproteins as well as their conservation across phyla is just one indication of their importance to life. Maintenance of Se homeostasis in the body promotes optimal function of key antioxidant selenoenzymes that are essential to preserving the integrity of the organism, and Se deficiency has been linked to several conditions including male infertility, cancer, improper thyroid function, and improper immune function (Foresta et al. 2002; Tinggi 2008). If environmental constraints limit the availability of Se, living systems would likely adapt in a way to preserve those selenoproteins that are more vital to maintaining homeostasis. Several studies have reported a hierarchy of selenoprotein mRNA expression when Se is limiting, whereby the levels of certain selenoprotein transcripts are decreased while others are maintained. However none of these studies have elucidated an underlying mechanism (Bermano et al. 1996; Sun et al. 2001; Sunde et al. 2009).
The stop codon UGA serves redundantly as the triplet for Sec, and incorporation of Sec into protein depends on the coordinated action of unique cis and trans acting factors. These include, but are not limited to, a Sec-specific elongation factor (EFsec), a secondary structure present in the 3′ UTR of all selenoprotein mRNAs termed a Sec insertion sequence (SECIS) element, and a SECIS-binding protein (SBP2). These factors are thought to facilitate the placement of a Sec-specific tRNA into the A site of the ribosome during translation (Hubert et al. 1996; Fagegaltier et al. 2000; Tujebajeva et al. 2000; Small-Howard et al. 2006). The nonsense-mediated mRNA decay (NMD) pathway is generally assumed to play an influential role in selenoprotein expression because it targets aberrant transcripts with premature termination codons (PTCs) for degradation; and since UGA encodes Sec, this would include selenoprotein mRNAs. The NMD pathway is well studied, and much has been uncovered regarding elements that would subject a transcript for decay. The current model for mammalian NMD holds that if a PTC is situated at least 50–55 nucleotides upstream of an exon junction, it will render that particular transcript vulnerable to NMD (Nagy and Maquat 1998; for review, see Maquat 2004; Chang et al. 2007). The reason for this is due to the deposition of a multimeric complex of proteins just upstream of exon–exon junctions during splicing (Le Hir et al. 2000). These exon junction complexes (EJCs) are displaced by the first translocating ribosome during the first round of translation (Ishigaki et al. 2001; Gehring et al. 2009); and in the case of a PTC, the ribosome will stall leaving one or more EJCs on the transcript. Remaining EJCs have the capacity to initiate decay based on a spatially dependent ability to associate with a PTC-stalled ribosome. NMD initiation has been proposed to follow a release factor binding the PTC-stalled ribosome and subsequent bridging of the ribosome-EJC gap by the RNA helicase UPF1. Phosphorylation of UPF1 by its kinase, SMG1, then signals mRNP remodeling and initiates RNA decay (Yamashita et al. 2001; Lejeune et al. 2003; Kashima et al. 2006; Isken et al. 2008; for review, see Schweingruber et al. 2013). Despite the widespread presumption that NMD plays a significant role in the regulation of selenoproteins at the RNA level, there has been little experimental evidence to support this.
In order to better understand the relationship between NMD and selenoprotein mRNAs, we first characterized the selenoprotein transcriptome in terms of its predicted susceptibility to the NMD pathway and assessed changes in selenoprotein mRNA levels in response to Se deficiency. We then performed RNA immunoprecipitation (RIP) with UPF1 to assess selenoprotein mRNA enrichment under the same conditions of limiting Se. We also combined Se deficiency with knockdown of SMG1. Lastly, mRNA decay rates were measured under conditions of varying Se. We show that selenoprotein mRNAs predicted to be sensitive to the current model of NMD show significant changes in response to Se deficiency, whereas those predicted to be resistant do not. Further, the mRNA abundance profile observed in our assessment of NMD susceptibility is almost identical to the abundance profile observed after immunoprecipitation with UPF1. We also show that the decline in transcript abundance of the RNAs in response to Se deficiency is abrogated with knockdown of SMG1. Finally, mRNA decay rate profiles show that the addition of Se altered the decay rates of selenoprotein mRNAs in conjunction with predictions of NMD susceptibility. Taken together, these results strongly support the influence of the NMD pathway on selenoprotein mRNAs when Se is limiting. This study is the first to directly evaluate a long-assumed relationship between the two and provides novel insight into a mechanism of selenoprotein regulation at the molecular level.
RESULTS
Selenoprotein mRNAs predicted to be sensitive to NMD exhibit significant changes in response to Se status, whereas those predicted to be resistant remain stable
Selenoprotein mRNAs were categorized as being predicted sensitive or resistant to NMD based on the location of the Sec UGA relative to exon–exon junctions. In accordance with the current model for mammalian NMD, Sec codons in the last exon are immune to NMD due to the displacement of all upstream EJCs by the first translocating ribosome. Interestingly, about half of selenoprotein mRNAs have their Sec codon in the last exon, whereas the other half are in either the second or third exon. Selenoprotein K (SelK) and Thioredoxin reductase 2 (Txnrd2) are exceptions, as they have their Sec codons in the penultimate exon. However, they are both categorized as being predicted resistant to NMD due to the proximity of the Sec codon to the last exon junction. Cellular Se status directly influences Sec-tRNA biosynthesis and thus its availability (Hatfield et al. 1991; Chittum et al. 1997; Moustafa et al. 2001; Jameson and Diamond 2004; for review, see Hatfield et al. 2006; Carlson et al. 2009). We hypothesized that conditions of limiting Se would alter the availability of the Sec-tRNA and shift any existing equilibrium between termination and Sec insertion toward the former. Since NMD initiates with the binding of a release factor to a stop codon, it would presumably ensue if a release factor were to bind a Sec UGA codon.
To test this hypothesis, we modeled conditions of low Se and assessed mRNA levels based on our categorization of NMD susceptibility. HEK293T cells were cultured under standard conditions until confluent and then switched to Se-deficient (3 nM Se) or Se-supplemented (60 nM Se) media. After 48 h, cells were collected, and qRT-PCR was performed on 18 selenoprotein mRNAs that are detectable in this cell line (Supplemental Fig. S1). Of the transcripts predicted to be sensitive to NMD, selenoprotein W (SelW) was the most responsive to Se status, showing a fourfold reduction in 3 nM versus 60 nM Se. Selenoprotein P (SelP), glutathione peroxidase 1 (GPx1), glutathione peroxidase 3 (GPx3), selenoprotein M (SelM), and selenoprotein H (SelH) also showed a statistically significant response to low Se albeit to a lesser degree than SelW. Glutathione peroxidase 4 (GPx4) and selenoprotein N (SelN) did not show a statistically significant response to Se status. The observation that GPx4 remains stable under conditions of varying Se has previously been reported (Bermano et al. 1996; Sun et al. 2001; Sunde et al. 2009) and will be subsequently discussed further. Of all the transcripts predicted resistant to NMD, none were decreased in 3 nM versus 60 nM Se (Fig. 1A). Only SPS2 mRNA abundance changed, and this change was inversely related to Se concentration.
FIGURE 1.
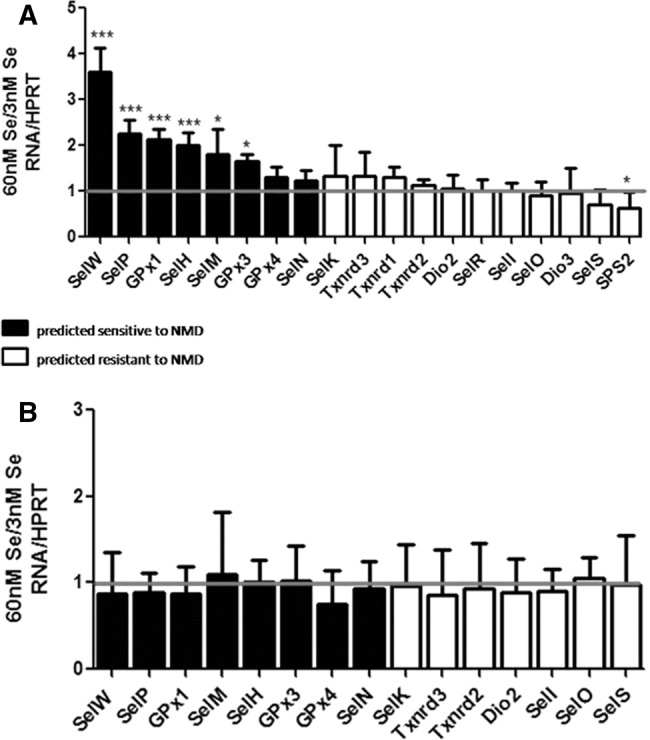
Fold change of a subset of selenoprotein mRNAs and pre-mRNAs in high versus low Se. (A) Levels of selenoprotein mRNAs in 60 nM Se versus 3 nM Se in HEK293T cells. (B) Same as A for pre-mRNAs. RNA levels are normalized to HPRT. Error bars represent standard deviation of the mean; n = 6; (*) P < 0.05; (***) P < 0.001.
Selenoprotein pre-mRNAs are not affected by Se status, indicating changes are post-transcriptional
To help clarify the nature of the observed changes in mRNA levels, we sought to address the role transcription could be playing within the context of our model. It has previously been shown that changes in GPx1 mRNA levels in response to Se are not due to transcription, but similar studies have not been reported for other selenoprotein mRNAs (Christensen and Burgener 1992; Moriarty et al. 1998). To examine this, an intron-based method for transcription analysis was utilized (Ponzio et al. 2007). Primers were designed to introns of a representative sample of transcripts from each category, and qRT-PCR was performed. Regardless of their predicted sensitivity to NMD, none of the pre-mRNA transcripts responded to changes in Se (Fig. 1B), suggesting the observed effects in Figure 1A to be post-transcriptional.
Selenoprotein mRNA abundance on UPF1 correlates with mRNA responses to Se status
Although the data from Figure 1 suggests a relationship between NMD and selenoprotein mRNAs, they lack a mechanistic link. In order to investigate a potential mechanism, we directed our focus to key NMD mediator protein, UPF1. UPF1 belongs to superfamily I of RNA helicases, and studies have identified it as the central regulator of NMD (Hwang et al. 2010; Chakrabarti et al. 2011). We reasoned that if the transcripts that decreased in low Se were undergoing NMD, then they should similarly be enriched on UPF1. The same experiment modeling conditions of low Se was repeated, and RNA immunoprecipitation was performed with UPF1. The cytoplasmic nucleoprotein complexes were coimmunoprecipitated with an anti-UPF1 antibody, followed by quantification of coimmunoprecipitated RNAs by qRT-PCR. A representative set of RNAs from each category of NMD susceptibility was analyzed, including those most responsive to Se status (SelW, SelP, GPx1), as well as GPx4, which has been consistently unresponsive to Se status in past research. Interestingly, all of the selenoprotein mRNAs analyzed coimmunoprecipitated with UPF1 to some degree in agreement with UPF1's general role in translation (Ghosh et al. 2010). A strikingly similar pattern to the profile in Figure 1A was observed in response to Se status. In this case, the RNAs predicted resistant to NMD showed no difference in their abundance on UPF1 with respect to Se. The RNAs that were the most responsive to Se status however, were also the most abundantly bound to UPF1 under conditions of low Se (Fig. 2A). Importantly, UPF1 did not change with Se, ruling out the possibility that changes in immunoprecipitated RNAs could have been due to changes in total UPF1 protein (Fig. 2B,C). Overall, the degree of mRNA responsiveness to Se status directly correlated with the degree of mRNA abundance on UPF1, suggesting the involvement of NMD in the regulation of selenoprotein mRNAs during Se deficiency.
FIGURE 2.
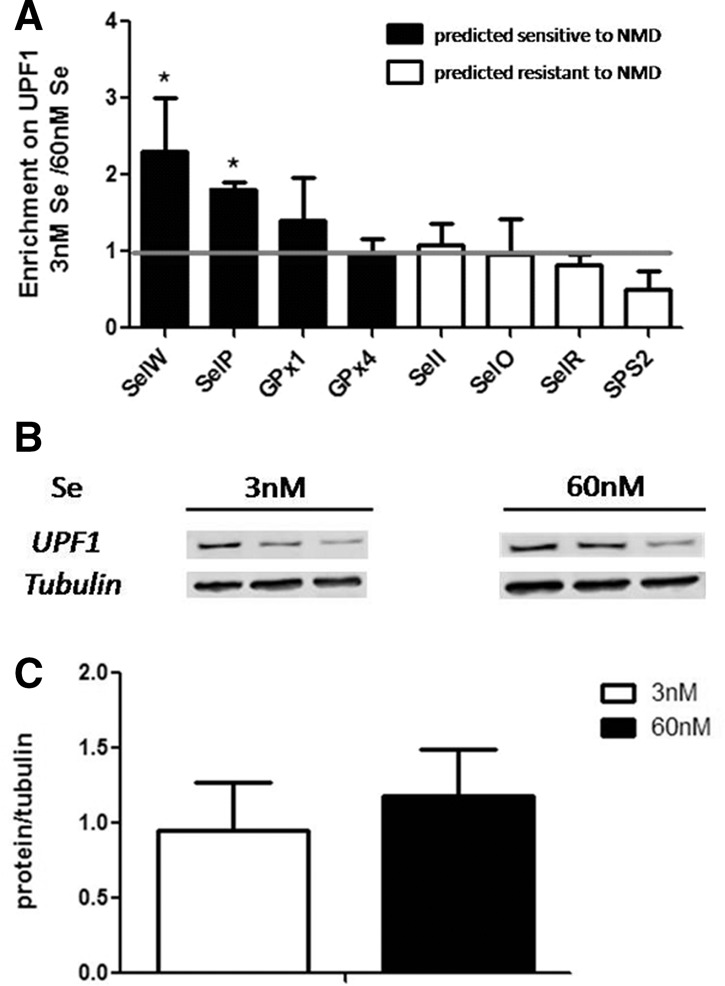
Fold enrichment of selenoprotein mRNAs on UPF1 in low versus high Se. (A) Levels of selenoprotein mRNAs that coprecipitated with UPF1, following culturing HEK293T cells in 3 nM Se or 60 nM Se. Transcript levels are normalized to a synthetic in vitro transcribed RNA and then to the value of the corresponding sample from the total RNA extraction (see Methods; Supplemental Fig. S3). (B) UPF1 protein levels in 3 nM Se or 60 nM Se. (C) Quantitation of B. Error bars represent standard deviation of the mean; n = 4 for A, and n = 6 for B and C; (*) P < 0.05.
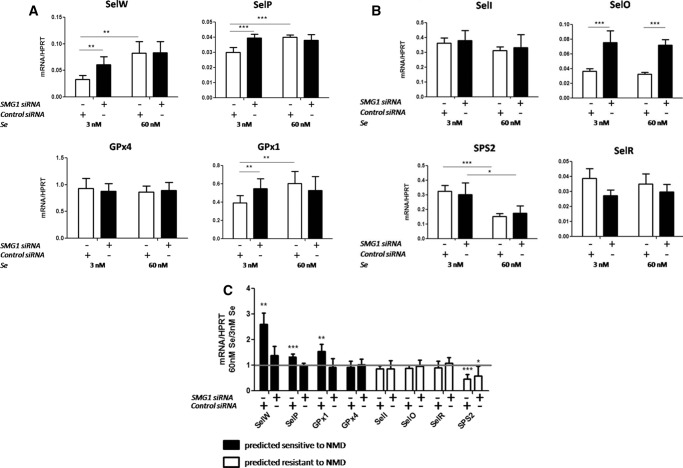
SMG1 knockdown abrogates Se responsiveness of mRNAs predicted sensitive to NMD
To further investigate the mechanistic link between NMD and selenoprotein mRNAs, expression of SMG1 was knocked down under the same Se conditions. SMG1 is a phosphatidylinositol-kinase-related kinase that phosphorylates UPF1 to initiate the decay cascade and has been shown to be necessary for NMD (Yamashita et al. 2001; Usuki et al. 2013). If NMD is involved in the regulation of Se-responsive transcripts, such as SelW and SelP, then knocking down SMG1 should alter their response. HEK293T cells were again cultured under standard conditions to ∼40% confluence and transfected with SMG1 siRNA using a two-hit protocol (see Materials and Methods). Two days after the second transfection, the media was changed to Se-deficient or Se-supplemented as in the previous two experiments. qRT-PCR was performed on the same representative transcripts from each category of predicted NMD susceptibility. SMG1 did not change with Se status, and protein quantification after the final 48 h of incubation showed almost 90% knockdown regardless of Se conditions (Fig. 3). Of those transcripts that were predicted sensitive to NMD and were responsive to Se, SMG1 knockdown restored SelW, SelP, and GPx1 mRNAs to levels that were no longer statistically different from Se-supplemented levels. GPx1 and SelP mRNAs in 3 nM Se were increased with SMG1 knockdown to levels nearly identical to the levels in 60 nM Se. SelW mRNA was also increased with SMG1 knockdown in 3 nM Se albeit not to levels equivalent to that of 60 nM Se. The levels of these NMD-sensitive mRNAs in 60 nM Se were unchanged with SMG1 knockdown, suggesting that under conditions of Se-supplementation, they are not undergoing NMD (Fig. 4A). SMG1 knockdown did not have an effect on the Se response of the transcripts predicted to be NMD resistant (Fig. 4B). Surprisingly, SMG1 knockdown significantly up-regulated SelO mRNA independent of Se concentration. This result is unexpected and will require further research to elucidate the mechanism underlying this effect.
FIGURE 3.
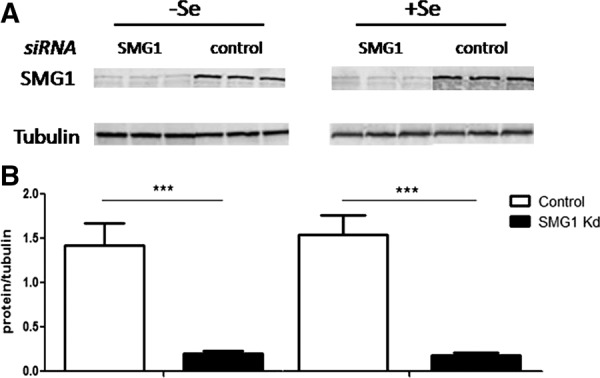
SMG1 knockdown in HEK293T cells. (A) Western blot of SMG1 protein after knockdown with SMG1 siRNA or nonspecific control siRNA. α-Tubulin was used as a loading control. (B) Quantitation of A. Error bars represent standard deviation of the mean; n = 5; (***) P < 0.001.
FIGURE 4.
Effect of SMG1 knockdown on selenoprotein mRNAs in high versus low Se. (A) mRNAs predicted sensitive to NMD. (B) mRNAs predicted resistant to NMD. (C) Fold change representation of data from A and B. Transcript levels are normalized to HPRT. Error bars represent a combined standard deviation of the compared means; n = 5; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
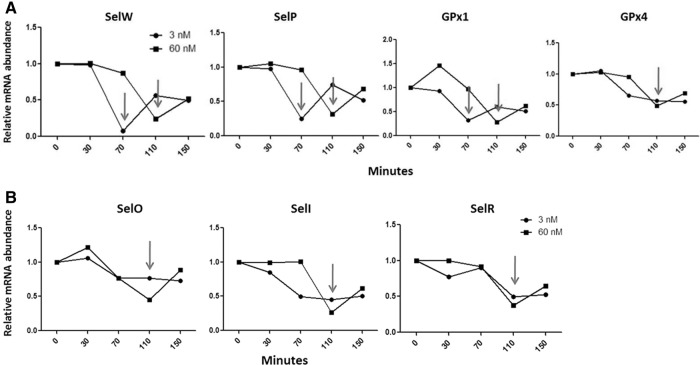
Se alters mRNA decay rates in conjunction with predictions of NMD susceptibility
To further examine the effect of Se on selenoprotein mRNAs predicted sensitive and resistant to NMD, mRNA decay rates were assessed using actinomycin D and qRT-PCR (Scherrer et al. 1963; Sobell 1985; Leclerc et al. 2002). Actinomycin D was used to block transcription, and the decay rates of selenoprotein mRNAs were measured using the same model of Se-deficiency previously described. In agreement with predictions of NMD susceptibility, the decay rates of the predicted-NMD-sensitive and Se-responsive mRNAs (SelW, SelP, and GPx1) were altered with the addition of Se (Fig. 5A). Under conditions of limiting Se, the abundance of these transcripts reaches a minimum at 70 min while the actinomcyin treatment runs its course. When Se is sufficient, however, the reduction in transcript abundance is delayed, reaching a minimum at 110 min. In addition, the predicted-NMD-resistant transcripts that did not respond to Se or SMG1 knockdown also showed no difference in their decay rates with respect to Se. The abundance of these transcripts declined less rapidly and resembled the Se-responsive transcripts under high Se, reaching a minimum at 110 min (Fig. 5B). Consistent with previous results, the decay rate of GPx4 mRNA was also not influenced by Se. In summary, this data suggests that when Se is limiting, Se-responsive selenoprotein mRNAs that were predicted to be sensitive to NMD decay more rapidly than the transcripts that did not respond to Se and were predicted resistant to NMD. Furthermore, the addition of Se altered the mRNA decay rates of the Se-responsive mRNAs to resemble those of the nonresponsive transcripts. This change in decay rate due to Se provides further support contributing to the involvement of NMD in the regulation of selenoprotein mRNAs during Se deficiency.
FIGURE 5.
mRNA decay profiles during high and low Se. (A) mRNAs predicted sensitive to NMD. (B) mRNAs predicted resistant to NMD. Transcript levels are normalized to HPRT and then fold-change ratios were calculated by comparing levels of RNA in cells treated with actinomcycin D to those not treated.
DISCUSSION
Se-dependent regulation of selenoprotein mRNAs has long been observed, and NMD is generally regarded as a factor contributing to this (Sun et al. 2001; Sun and Maquat 2002; Sunde et al. 2009; Banerjee et al. 2012; Howard et al. 2013). Since all selenoprotein mRNAs essentially have at least one PTC, it is logical to assume that NMD would target these transcripts for decay. In accordance with the mammalian model of NMD, our categorization of selenoprotein mRNAs as being predicted sensitive or resistant depends on the location of the Sec codon relative to exon junctions. Since almost half of selenoprotein mRNAs have their Sec codon in the last exon, they are automatically considered immune to being degraded by NMD. This distribution of NMD susceptibility may be an evolutionary development that could represent the relative importance of each selenoprotein when environmental constraints limit the availability of Se. When we model conditions of low Se, we see that those transcripts predicted resistant to NMD remain stable, whereas those predicted sensitive respond to varying degrees (Fig. 1). SelW responds to the greatest extent, followed by SelP, GPx1, and SelH. SelW has similarly been reported by others to be the most sensitive to changes in Se status, and the biological significance of this extreme sensitivity is worth investigation (Sunde et al. 2009; Howard et al. 2013). Conversely, GPx4 is predicted to be sensitive to NMD, but it remains steady with changes in Se status. Of the selenoprotein mRNAs predicted to be resistant to NMD, none responded to changes in Se except SPS2. SPS2 mRNA is the only one that increases under conditions of low Se, which suggests autoregulation, possibly linked to the fact that it is an essential factor in Sec-tRNA biosynthesis (Ehrenreich et al. 1992; Veres et al. 1994; Itoh et al. 2009). These data are very similar to those published by Sunde et al. (2009), which show selenoprotein mRNA responses to Se in vivo. The agreement between these data and those of the in vivo model provides support for the validity of this in vitro model.
When selenoprotein mRNAs were immunoprecipitated with NMD factor UPF1, all transcripts analyzed were observed to coprecipitate to some degree. UPF1 is a member of the DEAD-box family of RNA helicases; and in addition to its role in NMD, it has also been implicated in several other cellular functions, such as translation stabilization, E3 ubiquitin ligation, cell cycle progression, HIV RNA metabolism, Staufen-mediated decay, telomere stability, and splicing (Kim et al. 2005; Azzalin and Lingner 2006; Ajamian et al. 2008; Takahashi et al. 2008; Ghosh et al. 2010; Chawla et al. 2011; De Turris et al. 2011; for review, see Imamachi et al. 2012). UPF1 has been shown to associate with unspliced pre-mRNAs in the nucleus in addition to mature mRNAs in the cytoplasm (De Turris et al. 2011), and it likely binds a large variety of cellular mRNAs. When conditions of low Se were modeled, the selenoprotein mRNAs that responded the most to Se status were also the ones that were the most abundantly bound to UPF1 (Fig. 2A). Although the magnitude of response to Se status in Figure 1 is not identical to the degree of abundance on UPF1 in low Se, the overall profile of the two results taken together provides a strong correlation between NMD and selenoprotein mRNA levels when Se is limiting.
Knocking down SMG1 under conditions of low Se restored the mRNAs of the Se-responsive transcripts to levels comparable to those at 60 nM Se (Fig. 4C), strongly suggesting a role for NMD in regulating specific transcript levels during Se deficiency. The knockdown had no effect on the mRNAs when Se was supplemented, suggesting that when Se is readily available, the mRNAs bypass NMD. This is further corroborated by the observation that the mRNA decay rates of the Se-responsive transcripts were altered by the addition of Se to resemble the decay rates of the nonresponsive transcripts (Fig. 5A,B). The availability of Se retards the decrease in transcript abundance only for the selenoprotein mRNAs that are predicted NMD targets and Se-responsive. These results strongly suggest that the stability of the mRNA is largely influenced by the location of the Sec UGA codon, which, depending on the availability of Se, predisposes a selenoprotein mRNA to NMD. Se-responsive selenoprotein mRNAs that are predicted NMD targets are therefore inherently less stable than selenoprotein mRNAs predicted resistant to NMD. This agrees with the current understanding of the NMD pathway taken together with selenoprotein biosynthesis. These two are constantly in competition with each other because release factors are proposed to initiate NMD, and the Sec-tRNA theoretically competes with release factors for binding at Sec UGA codons. Since Se status directly influences the abundance of the Sec-tRNA, when Se is sufficient, Se-responsive transcripts are more stable because the Sec-tRNA is readily available to bind Sec UGA codons, thereby preventing a release factor from binding and initiating NMD (Fig. 6). Furthermore, these transcripts are sensitive to conditions of limiting Se, because in addition to the various levels of control imposed on most mRNAs, the unique architecture of these selenoprotein mRNAs predisposes them to further control by NMD.
FIGURE 6.
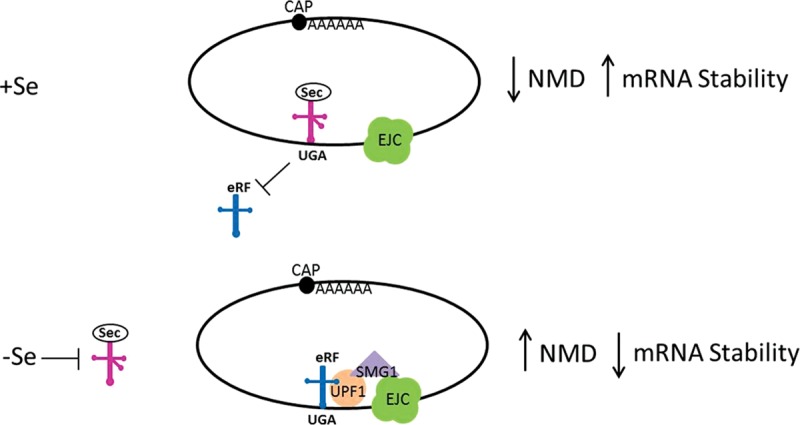
Se-responsive mRNAs are targeted by NMD when Se is limiting. When Se is sufficient, SectRNA will bind at Sec UGA codons, preventing potential release factor initiation of NMD. When Se is limiting, the abundance of SectRNA is decreased, in turn increasing the likelihood of a release factor binding to Sec UGA codons and up-regulating NMD of vulnerable transcripts.
GPx1 mRNA has been shown to be subject to NMD in other reports (Moriarty et al. 1998; Sun et al. 2000; Usuki et al. 2010), and it would not be surprising that other selenoprotein mRNAs would similarly be targeted. What remains puzzling is how GPx4, whose transcript is a predicted NMD target, remains stable regardless of Se status or SMG1 knockdown (Figs. 1A, 2A, 4A). One possible explanation would be that it is under the influence of some other mode of regulation that takes precedence over NMD. A recent study posits that under Se deficiency, eIF4A3, a core component of the EJC, is up-regulated and binds the SECIS element of GPx1 but not that of GPx4, competing for binding of the Sec-incorporation factor SBP2 and ultimately inhibiting Sec incorporation (Budiman et al. 2009). Although this is a plausible theory, it conflicts with a previous study (Weiss and Sunde 1998), showing that the GPx4 3′ UTR is not able to stabilize the Se responsiveness of GPx1 mRNA. Another explanation could relate to the relative abundance of each of two isoforms of the Sec-tRNA. The Sec-tRNA exists as two isoforms that differ in the methylation state of the wobble base, U34. The abundance of each isoform has been shown to correlate with Se status, and this could be an intermediate factor in susceptibility to NMD. The methylated isoform has been shown to be abundant when Se is sufficient and diminished when Se is limiting (Jameson and Diamond 2004; Carlson et al. 2009; Howard et al. 2013). Further, the synthesis of stress-related selenoproteins, such as SelW and GPx1, requires the methylated isoform, whereas the synthesis of housekeeping selenoproteins, such as GPx4 and Txnrd3, does not (Carlson et al. 2009). SelW and GPx1 are predicted to be sensitive to NMD, whereas Txnrd3 is predicted to be resistant. Degree of sensitivity to Se status is thus potentially a result of both mRNA architecture in predisposing transcripts to NMD as well as tRNA isoform specificity. This means that when Se is limiting, the abundance of the methylated isoform is decreased, and those mRNAs that require this isoform and are also sensitive to NMD will be rapidly degraded (e.g., SelW, GPx1). In the case of GPx4, although it is predicted susceptible to NMD, it does not require the methylated tRNA for its synthesis. Since the nonmethylated tRNA is abundant under conditions of low Se, it could potentially facilitate GPx4 mRNA translation and prevent degradation of the mRNA via NMD. This combination of tRNA isoform abundance and NMD susceptibility likely contributes significantly to the observed hierarchy of selenoprotein mRNA abundance when Se is limiting. In summary, this study is the first to present evidence supporting the involvement of the NMD pathway in the Se-dependent regulation of selenoprotein mRNAs.
MATERIALS AND METHODS
Selenium treatment, RNA extraction, cDNA synthesis, and qRT-PCR
HEK293T cells were plated in flat-bottom 35-mm six-well tissue culture plates (Corning, Inc.) and were grown to confluence in Dulbecco's modified eagle medium (DMEM) (Invitrogen) with 10% fetal bovine serum (FBS). FBS lots are routinely measured for Se content, and at 10%, the final Se concentration due to FBS is ∼30 nM. Media was then switched to 1% FBS with or without the addition of sodium selenite (final concentration of 3 nM Se or 60 nM Se) for 48 h. Cells were washed with phosphate buffered saline (PBS), and RNA was extracted with Trizol (Invitrogen), according to the manufacturer's instructions. RNA quantity and quality were measured using a ND1000 spectrophotometer (NanoDrop Technologies). For cDNA synthesis, 1 µg RNA was used in a total reaction volume of 20 µL using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). cDNA was diluted 5×, and for qRT-PCR, a volume of 0.5 µL was used per final 5 µL reaction volume. One µM specific primer pair and PerfeCTa SYBR Green FastMix were used (Quanta Biosciences) according to the manufacturer's instructions on a Light Cycler 480 II thermal Cycler (Roche). The sequences of all primers used in this study are shown in Supplemental Table S1.
RNA immunoprecipitation (RIP)
UPF1 antibody (anti-RENT1, A301-902A, Bethyl Laboratories, Inc.) was conjugated to magnetic Dynabeads (Invitrogen) using 5 mM BS3 (Proteochem), the reaction was quenched with Tris-HCl pH 7.5, washed twice with cold PBS, and resuspended in cytosolic lysis buffer (50 mM HEPES, 5 M NaCl, 75 mM NaF, 10 mM iodoacetamide, 0.05% Triton X-100, 1× Protease Inhibitor Cocktail set II [EMD Biosciences], 240 Units RNAse Inhibitor per sample [Applied Biosystems]).
HEK293T cells were plated in 10-cm tissue culture plates, cultured, and treated with sodium selenite as described above. After the 48 h incubation with 1% FBS in the presence or absence of Se (60 nM Se or 3 nM Se), cells were washed twice with cold PBS. Cold cytosolic lysis buffer was then added directly to each dish. The dishes containing lysis buffer were gently rocked for 2.5 min, and then incubated for another 2.5 min. The supernatant was aspirated, and cellular debris was removed by snap centrifugation on a tabletop microcentrifuge. The remaining lysate was collected and split in half. Trizol was added directly to one half for total RNA analysis (Supplemental Fig. S2), and the other half was used for the RIP. Protein was quantified using the ND1000 spectrophotometer. Lysate was added to the antibody–magnetic bead conjugate described above, followed by incubation on ice with rotation for 1 h. Samples were then split in half into individual tubes and placed on the DynaMag-2 magnet (Invitrogen) for collection. RNA/bead conjugates were washed twice with cold PBS, and one half was used for SDS-PAGE and Western blot (Supplemental Fig. S3). Trizol was added directly to the other half for RNA extraction. A random mouse DNA plasmid was used for in vitro RNA synthesis, and this RNA was added to the Trizol to serve as an internal control for the RNA extraction (Labeled mG1 in Supplemental Table S1). RNA extraction was then carried out according to the manufacturer's instruction, and cDNA was synthesized followed by qRT-PCR as previously described.
SMG1 knockdown
HEK293T cells were cultured in DMEM and 10% FBS until ∼40% confluence and then transfected with SMG1 siRNA (Qiagen) at a final concentration of 20 nM using RNAimax reagent (Invitrogen) according to the manufacturer's instructions for a forward transfection. After 24 h, cells were washed with PBS and transfected with SMG1 siRNA again under the same conditions as the previous day. Another 24 h later, the media was changed to fresh DMEM with 10% FBS. Twenty-four hours later, media was changed to DMEM plus 1% FBS with or without the addition of sodium selenite, and cells were cultured for 48 h. Cells were then washed with PBS and harvested for RNA and protein analysis. Protein was extracted using Cell Lytic MT Cell Lysis Reagent (Sigma-Aldrich) according to the manufacturer's instructions, and RNA was extracted with Trizol according to the manufacturer's instructions. RNA quantity and quality were measured using a ND1000 spectrophotometer. cDNA synthesis and qRT-PCR were carried out as previously described.
SDS-PAGE and Western Blot
Protein extracted using the preceding method was added to reduced Laemmli buffer (Bio-Rad), boiled for 5 min, and loaded into 4%–20% gradient polyacrylamide gels (Bio-Rad). Following electrophoresis, gel contents were transferred to a PVDF membrane (Millipore) and blocked with Odyssey blocking buffer (Li-Cor Biosciences) for 30 min. Membranes were then probed for proteins with the following primary antibodies: SMG1 (Dilution: 1:500; A300-394A, Bethyl Laboratories, Inc.) UPF1 (dilution: 1:1000; anti-RENT1, A301-902A, Bethyl Laboratories, Inc.), Grb2 (dilution: 1:1000; Upstate Cell Signaling), α-Tubulin (1:10,000; Novus Biologicals); and with the following secondary antibodies: goat anti-rabbit IRDye 680 (dilution: 1:10,000, Li-Cor Biosciences), goat anti-mouse IRDye 680 (dilution: 1:10,000, Li-Cor Biosciences), goat anti-mouse IRDye 800 (dilution: 1:10,000, Li-Cor Biosciences). All protein quantification was carried out using Odyssey's Image Studio version 3.0 (Li-Cor Biosciences).
mRNA decay rates in response to varying Se concentrations
HEK293T cells were plated in 12-well tissue culture plates and grown to confluence in DMEM and 10% FBS. Once confluent, media was changed to DMEM and 1% FBS with and without the addition of sodium selenite as previously described and left for 48 h. To stay consistent with our model, the 48-h mark was considered time point zero. At this point, the first set of samples was collected, and media was changed again. Actinomycin D (or water as vehicle control) was added to the media (DMEM and 1% FBS, with and without sodium selenite) to a final concentration of 5 µM, and samples were sequentially collected at the indicated time points. RNA was harvested, cDNA was synthesized, and qRT-PCR was performed as previously described.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Christy Gilman and Lucia Seale for their contributions to experimental design. We thank Arjun Raman for insightful discussion and Jeffrey Squires for valuable methodological suggestions. This research was supported by the National Institutes of Health grants R01-DK47320 and G12-MD007601.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.043463.113.
REFERENCES
- Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, Gehring NH, Mouland AJ 2008. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 14: 914–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J 2006. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol 16: 433–439 [DOI] [PubMed] [Google Scholar]
- Banerjee S, Yang S, Foster CB 2012. A luciferase reporter assay to investigate the differential selenium-dependent stability of selenoprotein mRNAs. J Nutr Biochem 23: 1294–1301 [DOI] [PubMed] [Google Scholar]
- Bermano G, Arthur JR, Hesketh JE 1996. Selective control of cytosolic glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase mRNA stability by selenium supply. FEBS Lett 387: 157–160 [DOI] [PubMed] [Google Scholar]
- Budiman ME, Bubenik JL, Miniard AC, Middleton LM, Gerber CA, Cash A, Driscoll DM 2009. Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol Cell 35: 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BA, Yoo MH, Tsuji PA, Gladyshev VN, Hatfield DL 2009. Mouse models targeting selenocysteine tRNA expression for elucidating the role of selenoproteins in health and development. Molecules 14: 3509–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E 2011. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 41: 693–703 [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF 2007. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Chawla R, Redon S, Raftopoulou C, Wischnewski H, Gagos S, Azzalin CM 2011. Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication. EMBO J 30: 4047–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittum HS, Baek HJ, Diamond AM, Fernandez-Salguero P, Gonzalez F, Ohama T, Hatfield DL, Kuehn M, Lee BJ 1997. Selenocysteine tRNA[Ser]Sec levels and selenium-dependent glutathione peroxidase activity in mouse embryonic stem cells heterozygous for a targeted mutation in the tRNA[Ser]Sec gene. Biochemistry 36: 8634–8639 [DOI] [PubMed] [Google Scholar]
- Christensen MJ, Burgener KW 1992. Dietary selenium stabilizes glutathione peroxidase mRNA in rat liver. J Nutr 122: 1620–1626 [DOI] [PubMed] [Google Scholar]
- De Turris V, Nicholson P, Orozco RZ, Singer RH, Mühlemann O 2011. Cotranscriptional effect of a premature termination codon revealed by live-cell imaging. RNA 17: 2094–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich A, Forchhammer K, Tormay P, Veprek B, Böck A 1992. Selenoprotein synthesis in E. coli. Purification and characterisation of the enzyme catalysing selenium activation. Eur J Biochem 206: 767–773 [DOI] [PubMed] [Google Scholar]
- Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A 2000. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J 19: 4796–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresta C, Flohé L, Garolla A, Roveri A, Ursini F, Maiorina M 2002. Male fertility is linked to the selenoprotein phospholipid hydroperoxide glutathione peroxidase. Biol Reprod 67: 967–971 [DOI] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW 2009. Disassembly of exon junction complexes by PYM. Cell 137: 536–548 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Ganesan R, Amrani N, Jacobson A 2010. Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA 16: 1832–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D, Lee BJ, Hampton L, Diamond AM 1991. Selenium induces changes in the selenocysteine tRNA[Ser]Sec population in mammalian cells. Nucleic Acids Res 19: 939–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN 2006. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog Nucleic Acid Res Mol Biol 81: 97–142 [DOI] [PubMed] [Google Scholar]
- Howard MT, Carlson BA, Anderson CB, Hatfield DL 2013. Translational redefinition of UGA codons is regulated by selenium availability. J Biol Chem 288: 19401–19413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert N, Walczak R, Carbon P, Krol A 1996. A protein binds the selenocysteine insertion element in the 3′-UTR of mammalian selenoprotein mRNAs. Nucleic Acids Res 24: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE 2010. UPF1 Association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell 39: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamachi N, Tani H, Akimitsu N 2012. Up-frameshift protein 1 (UPF1): multitalented entertainer in RNA decay. Drug Discov Ther 6: 55–61 [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617 [DOI] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE 2008. UPF1 phosphorylation triggers translational repression and nonsense-mediated mRNA decay. Cell 133: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Sekine S, Matsumoto E, Akasaka R, Takemoto C, Shirouzu M, Yokoyama S 2009. Structure of selenophosphate synthetase essential for selenium incorporation into proteins and RNAs. J Mol Biol 385: 1456–1469 [DOI] [PubMed] [Google Scholar]
- Jameson RR, Diamond A 2004. A regulatory role for Sec tRNA[Ser]Sec in selenoprotein synthesis. RNA 10: 1142–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S 2006. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Furic L, DesGroseillers L, Maquat LE 2005. Mammalian staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195–208 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc GJ, Leclerc GM, Barredo JC 2002. Real-time PCR analysis of mRNA decay: half-life of β-actin mRNA in human leukemia CCRF-CEM and Nalm-6 cell lines. Cancer Cell Int 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12: 675–687 [DOI] [PubMed] [Google Scholar]
- Maquat LE 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Moriarty PM, Reddy CC, Maquat LE 1998. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol 18: 2932–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa ME, Carlson BA, El-Saadani MA, Kryukov GV, Sun QA, Harney JW, Hill KE, Combs GF, Feigenbaum L, Mansur DB, et al. 2001. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol Cell Biol 21: 3840–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE 1998. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 23: 198–199 [DOI] [PubMed] [Google Scholar]
- Ponzio TA, Chunmei Y, Gainer H 2007. An intron-based real-time PCR method for measuring vasopressin gene transcription. J Neurosci Methods 164: 149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer K, Latham H, Darnell JE 1963. Demonstration of an unstable RNA and of a precursor to ribosomal RNA in HeLa cells. Proc Natl Acad Sci 49: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O 2013. Nonsense-mediated mRNA decay—mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta 1829: 612–623 [DOI] [PubMed] [Google Scholar]
- Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, Berry MJ 2006. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol 26: 2337–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell HM 1985. Actinomycin and DNA transcription. Proc Natl Acad Sci 83: 5328–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Maquat LE 2002. Nonsense-mediated decay: assaying for effects on selenoprotein mRNAs. Methods Enzymol 347: 49–57 [DOI] [PubMed] [Google Scholar]
- Sun X, Moriarty PM, Maquat LE 2000. Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position. EMBO J 19: 4734–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Li X, Moriarty PM, Henics T, LaDuca JP, Maquat LE 2001. Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide glutathione peroxidase is detectable in cultured cells but masked or inhibited in rat tissues. Mol Biol Cell 12: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde RA, Raines AM, Barnes KM, Evenson JK 2009. Selenium status highly regulates selenoprotein mRNA levels for only a subset of the selenoproteins in the selenoproteome. Biosci Rep 29: 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Araki Y, Ohya Y, Sakuno T, Hoshino S, Kontani K, Nishina H, Katada T 2008. Upf1 potentially serves as a RING-related E3 ubiquitin ligase via its association with Upf3 in yeast. RNA 14: 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinggi U 2008. Selenium: its role as antioxidant in human health. Environ Health Prev Med 13: 102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, Berry MJ 2000. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep 1: 158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuki F, Yamashita A, Fujimura M 2010. Post-transcriptional defects of antioxidant selenoenzymes cause oxidative stress under methylmercury exposure. J Biol Chem 286: 6641–6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuki F, Yamashita A, Shiraishi T, Shiga A, Onodera O, Higuchi I, Ohno S 2013. Inhibition of SMG-8, a subunit of SMG-1 kinase, ameliorates nonsense-mediated mRNA decay-exacerbated mutant phenotypes without cytotoxicity. Proc Natl Acad Sci 110: 15037–15042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres Z, Kim IY, Scholz TD, Stadtman TC 1994. Selenophosphate synthetase. Enzyme properties and catalytic reaction. J Biol Chem 269: 10597–10603 [PubMed] [Google Scholar]
- Weiss SL, Sunde RA 1998. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA 4: 816–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev 15: 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]