SUMMARY
The role of lipids in the assembly, structure and function of hetero-oligomeric membrane protein complexes is poorly understood. The photosynthetic cytochrome b6f complex, a 16-mer of eight distinct subunits and 26 trans-membrane helices, catalyzes trans-membrane proton-coupled electron transfer for energy storage. Using a 2.5 Å crystal structure of the dimeric complex, 23 distinct lipid binding sites per monomer have been identified in the present study. Annular lipids are proposed to provide a connection for (i) super-complex formation with the photosystem-I reaction center, and (ii) the LHCII kinase enzyme for trans-membrane signaling. Internal lipids mediate (iii) cross-linking to stabilize the domain-swapped iron-sulfur protein subunit, (iv) dielectric heterogeneity within inter-monomer and intra-monomer electron transfer pathways, and (v) dimer stabilization through lipid-mediated inter-monomer interactions. This study provides the most complete structure-analysis thus far obtained of lipid-mediated functions in a multi-subunit membrane protein complex, and reveals lipid-sites at positions essential for assembly and function.
INTRODUCTION
Membrane proteins can assemble into large complexes and super-complexes (Iwai et al., 2010; Schagger and Pfeiffer, 2000), whose stability is dependent on association with lipids (Pfeiffer et al., 2003; Wenz et al., 2009). Specific lipid-protein interactions have been inferred from crystal structures of a number of membrane proteins (Galdiero and Gouaux, 2004; Hanson et al., 2008; Hite et al., 2010; Lee et al., 2005), including hetero-oligomeric complexes such as cytochrome c oxidase (Qin et al., 2006; Qin et al., 2008; Shinzawa-Itoh et al., 2007; Tiefenbrunn et al., 2011), cytochrome bc1 complex (Hunte and Richers, 2008; Lange et al., 2001; Palsdottir and Hunte, 2004), and the photosystem-II reaction center (Guskov et al., 2009). Despite the increase in resolved membrane protein structures (~450, http://blanco.biomol.uci.edu/membrane_proteins_xtal.html), hetero-oligomeric complexes constitute only a small fraction, <75 structures, of the depositions in the PDB, thereby limiting information concerning general principles that determine integral lipid functions.
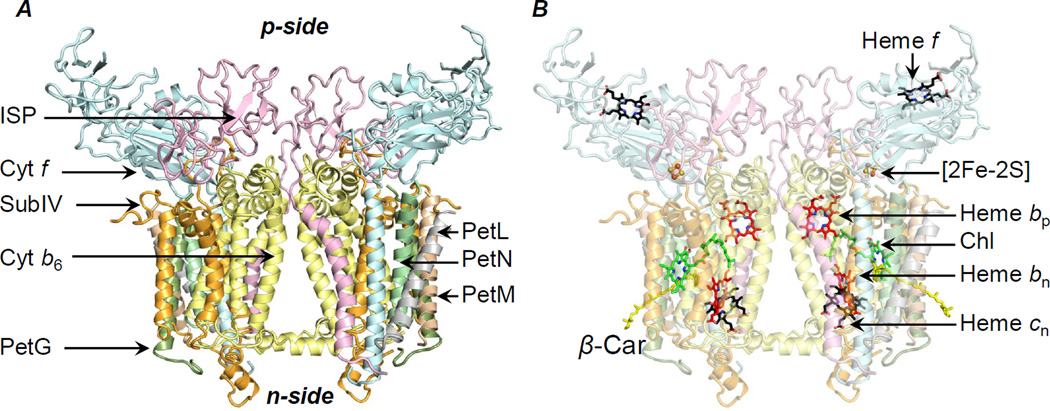
The hetero-oligomeric cytochrome bc complexes, b6f and bc1, catalyze proton-coupled quinone-redox reactions to generate a trans-membrane proton electrochemical gradient for energy storage, as summarized (Berry et al., 2000; Cramer et al., 2006). The b6f complex embedded in the thylakoid membrane consists of two monomers related by a 2-fold symmetry axis normal to the membrane (Figures 1A, B). Each monomer has at least eight distinct trans-membrane subunits (Figure 1A)- cytochrome b6 (cytochrome b) and subunit IV, respectively, with 4 (“A–D”) and 3 (“E–G”) trans-membrane helices, and the single helix subunits cytochrome f, iron-sulfur protein (ISP), and PetG, PetL, PetM and PetN, the latter four forming a “picket-fence” around the periphery of the complex (Kurisu et al., 2003; Stroebel et al., 2003). The cytochrome b subunit is linked non-covalently to the trans-membrane hemes, bp and bn respectively on the electrochemically positive (p) and negative (n) sides, and the covalently to heme cn (Figure 1B). The p-side extrinsic domains of ISP and cytochrome f respectively bind a [2Fe-2S] cluster and a c-type heme (heme f) (Figure 1B). Structural stability of the b6f complex dimer (Breyton et al., 1997) as well as formation of well-diffracting crystals (Zhang et al., 2003), depends on the presence of lipids. In the related bc1 complex, structure-function dependence on lipids has been demonstrated (Wenz et al., 2009), and functions suggested for specific lipid sites (Lange et al., 2001; Palsdottir and Hunte, 2004). Conservation of lipid-binding sites has been noted between the b6f and bc1 complexes (Hasan et al., 2011).
Figure 1.
Dimeric cytochrome b6f complex from Nostoc PCC 7120. (A) The monomer consists of eight polypeptides (ribbons)- cytochrome b6 (yellow, 4 helices), subunit IV (orange, 3 helices), cytochrome f (cyan, 1 helix), ISP (pink, 1 helix), PetG (dark green, 1 helix), PetL (gray, 1 helix), PetM (light brown, 1 helix), PetN (light green, 1 helix). (B) Prosthetic groups associated with the b6f complex- two non-covalently linked hemes, bp and bn, a covalently linked heme cn, a chlorophyll-a/β-carotene pair, a covalently linked heme (heme f) and a [2Fe-2S] cluster. n, p, electrochemically negative (stroma) and positive (lumen) sides.
To understand the role of lipids in the biogenesis of hetero-oligomeric membrane protein complexes, it is essential to obtain high resolution crystallographic information that elucidates lipid-binding sites for targeted analysis by mutagenesis (de Vitry et al., 2004; Lange et al., 2001; Wenz et al., 2009). In the present study, 23 lipid-binding sites per monomer were identified in a 2.5 Å crystal structure of the dimeric b6f complex isolated from Nostoc PCC 7120. Specific functions are proposed for each lipid site: (i) formation of an interface around the β-carotene to facilitate super-complex formation; (ii) stabilization of the chlorophyll binding site inferred to have a role in trans-membrane signaling; (iii) a determinant of position of the ISP helix; and (iv) a determinant of quinone traffic in the inter-monomer cavity that modulates the intra-membrane electron-proton transfer. The study provides the most comprehensive description thus far available of lipid functions in a hetero-oligomeric membrane protein complex, partly as a consequence of the multiple pathways of electron-proton transfer in the cytochrome complex.
RESULTS
Lipid and Detergent Sites in the Cytochrome b6f Complex
Crystallographically ordered lipids and detergents indicate physiologically relevant lipid-binding sites in membrane proteins (Qin et al., 2007), with an average lipid density of approximately 10 lipid sites per 100 kDa (SI, Table T1). The 2.5 Å cytochrome b6f complex structure described in the present study (Table 1) has revealed the presence of 12 lipids, 6 partially ordered alkyl chains, and 5 detergents per monomer (SI, Tables T2, T3, Figures S1, and S2). Head-group atoms could be assigned to three n-side lipids- a sulfolipid and monogalactosyldiacylglycerol (MGDG), which are unique to the membranes of oxygenic photosynthesis, and the synthetic dioleoylphosphatidylcholine (DOPC). Other lipids were modeled as diacylglycerols (DAG). It is significant to note that thylakoid membranes, unlike other cellular membranes, consist of primarily neutral galactolipids (SI, Tables T4, T5). The 6 partially ordered alkyl chains represent loosely bound lipids.
Table 1.
Summary of crystallographic data. Values in parentheses correspond to the outer shell.
| Crystal | Cyt b6f (PDB ID 4OGQ) |
|---|---|
| Data Collection | |
| Space Group | P6122 |
| Cell Dimensions | |
| a, b, c (Å) | 159.3, 159.3, 365.9 |
| α, β, γ (Å) | 90, 90, 120 |
| Number of Molecules (A. U.) | 1 |
| Resolution (Ǻ) | 50.00-2.50 (2.54-2.50) |
| Rpim (%) | 4.8 (90.6) |
| CC* | 0.68 |
| <I>/<σI> | 16.7 (1.0) |
| Measured Reflections | 498,334 |
| Unique Reflections | 93,444 |
| Completeness (%) | 98.2 (89.9) |
| Redundancy | 5.3 (5.2) |
| Refinement | |
| Resolution (Ǻ) | 39.57-2.50 |
| Rwork/Rfree (%) | 20.0 (33.8)/23.2 (36.3) |
| Coordinate Error (M.L., Å) | 0.33 |
| Ramachandran statistics* | |
| Favored | 93.11 |
| Allowed | 6.14 |
| Unfavored | 0.75 |
| Rotamer Outliers (%) | 0.90 |
| Amino Acids (A.U.) | 949 |
| No. of atoms | |
| Protein | 7296 |
| Ligand/Ion | 1007 |
| Water | 93 |
| B-factors (Ǻ2,Average) | |
| Protein | 94.40 |
| Ligand/Ion | 108.60 |
| Water | 82.90 |
| RMS Deviation | |
| Bond lengths (Ǻ) | 0.003 |
| Bond angles (°) | 0.766 |
Data merged from two crystals; 5% of the reflections used for Rfree calculation; A.U., asymmetric unit; CC*, correlation coefficient; M.L., maximum-likelihood;
Molprobity statistics.
Boundary Lipids Adjacent to the β-Carotene
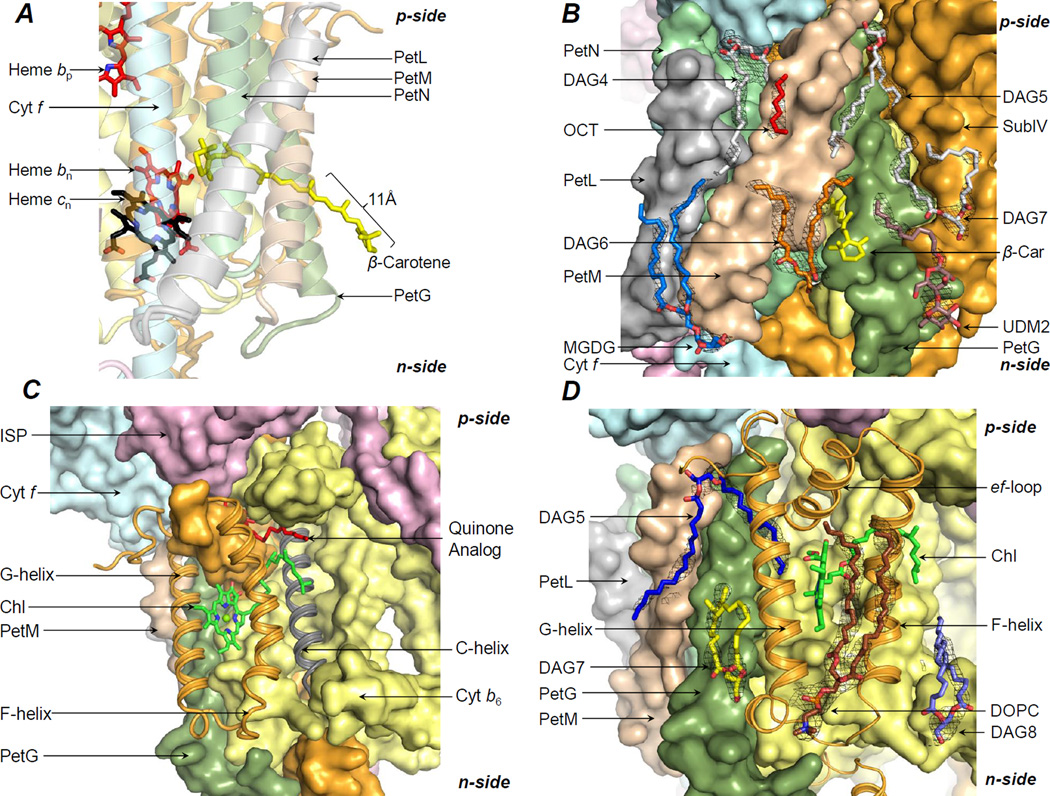
An integral β-carotene-chlorophyll pigment pair, separated by ~14 Å, is associated with the cytochrome b6f complex (Kurisu et al., 2003; Stroebel et al., 2003) (Figure 2A). The 11 Å exposed conjugated chain of β-carotene has been proposed to mediate super-complex formation with the photosystem-I complex (Hasan and Cramer, 2012). However, whether the β-carotene performs this “latch” function alone, or in combination with surface-bound lipids, remains unknown.
Figure 2.
Lipidic-interface for super-complex formation. (A) The β-carotene molecule (yellow sticks) has been proposed to serve as a “latch” for interactions with the PSI complex. (B) On the n-side, the lipids DAG6 (orange/red sticks), DAG7 (white/red sticks), MGDG (blue/red sticks) and the partially ordered detergent UDM2 (brown/red sticks), and on the p-side, the lipids DAG4 and DAG5 (white/red sticks), and an 8-carbon OCT (red) form the interface. (C) A chlorophyll (green/red/blue sticks) is inserted between the F and G-helices of subunit IV (orange ribbons). C-helix shown in gray. A quinone analog (red sticks, from PDB ID 2E76) is shown for reference. (D) A DOPC lipid (brown/blue/red/yellow sticks) is associated with the F and G-helices. Further stabilization is provided by the lipids DAG7 (yellow/red sticks) and DAG8 (light blue/red sticks) that interact, respectively, with the G and F-helices of subunit IV. On the p-side, an acyl tail of lipid DAG5 (blue/red sticks) further stabilizes the chlorin-ring. Black mesh, 2Fo-Fc map at 1.1 σ.
Seven lipidic sites occupied by lipids, detergents, and an 8-carbon chain were assigned around the b6f β-carotene pigment (Figure 2B). (i) The acyl tails of the lipid DAG6 (Figure 2B; SI, Figure S1A) interact with the subunits PetG and PetM (SI, Tables T2, T3). (ii) The detergent UDM2 (Figure 2B; SI, Figure S1B) is observed within interaction distance of the PetG subunit, and the n-side subunit IV fg-loop (SI, Tables T2, T3). (iii) The lipid DAG7 interacts with the subunit IV G-helix and PetG (Figure 2B; SI, Table T2, T3, Figure S1C). (iv) The lipid MGDG (Figure 2B; SI, Figure S1D) is located on the n-side, proximal to the subunits PetL and PetM (SI, Tables T2, T3). (v) The lipid DAG4 (Figure 2B; SI, Figure S1E) is associated with the core helices A, B and E (SI, Table T2). One acyl tail of DAG4 is exposed on the b6f periphery, proximal to PetL, PetM, PetG and PetN (Figure 2B; SI, Table T2, T3). (vi) The lipid DAG5 (Figure 2B; SI, Figure S1F) interacts with the core helices B and E (SI, Table T2, T3). The surface-proximal DAG5 acyl chain is associated with the protein periphery, in contact with the subunits PetG and PetM (Figure 2B). (vii) An 8-carbon chain (OCT, Figure 2B; SI, Figure S1G) was modeled on the p-side, between the peripheral acyl chains of the lipids DAG4 and DAG5.
Stabilization of the Chlorophyll-Binding Niche
A chlorophyll molecule is associated with the p-side quinone-binding (Qp) site of the b6f complex (Figure 2C), and has been proposed to be involved in mediating photosynthetic state-transitions to maintain redox balance (de Lacroix de Lavalette et al., 2008; Hasan et al., 2013b; Pierre et al., 1997). The chlorophyll phytyl-tail, which is inserted into the Qp-portal between the F and C-helices, may function as a “sensor” for quinone/quinol traffic (Hasan et al., 2013b). The chlorophyll chlorin-ring is inserted between the F and G-helices, and is exposed to the lipid bilayer on the edge (Baniulis et al., 2009; Kurisu et al., 2003; Stroebel et al., 2003) (Figure 2C). In the cyanobacterial b6f complex structures, the exposed chlorin-ring edge interacts with a DOPC lipid molecule (Figure 2D; SI, Figure S1H).
The chlorophyll binding site is stabilized by lipid. (i) The n-side lipid DAG8 interacts with the subunit IV F-helix (Figure 2D; SI, Tables T2, T3, Figure S1I). (ii) The n-side lipid DAG7 (Figure 2C; SI, Figure S1C) is associated with the subunit IV G-helix and PetG (SI, Tables T2, T3). (iii) The p-side lipid DAG5 (Figure 2D; SI, S1F) interacts with the subunit IV ef-loop and G-helix (SI, Tables T2, T3), thus forming an extensive association with the chlorophyll niche.
A Lipidic-Cage around the ISP-Cytochrome f Trans-Membrane Helices
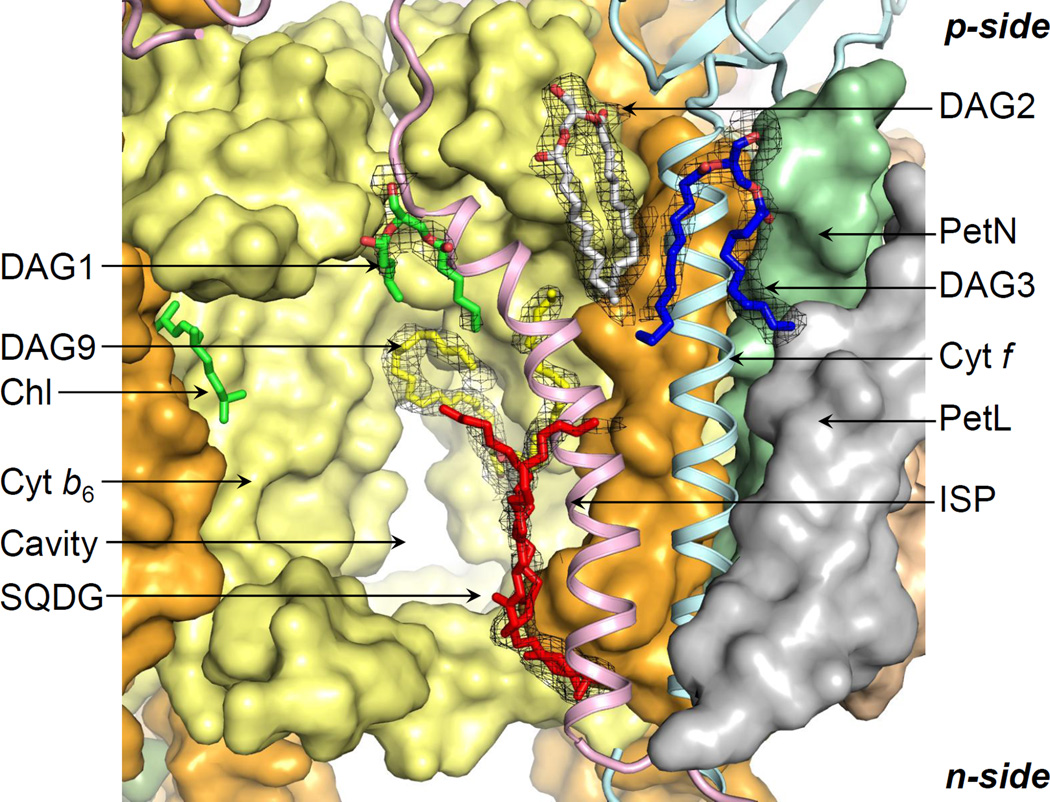
The ISP p-side extrinsic domain undergoes motion from the p-side quinone-binding site to the cytochrome c1 (bc1) or cytochrome f (b6f) extrinsic domain for electron transfer (Yamashita et al., 2007; Zhang et al., 1998). Previously, a UDM molecule was modeled at the p-side interface between the cytochrome f and ISP helices in the cyanobacterial b6f structures, and a 20-carbon chain in the algal b6f structure. However, in the 2.7 Å b6f structure from Nostoc PCC 7120 (PDB ID 4H44), the 2.8 Å structure from M. laminosus (PDB ID 4I7Z), and the 2.5 Å structure described in the present study (PDB ID 4OGQ), a lipid DAG2 (Figure 3; SI, Figure S1J) was modeled in the electron density. The lipid position is conserved in the related cytochrome bc1 complex (Hasan et al., 2011). The DAG2 acyl tails interact with the cytochrome f and ISP helices, and with the E-helix of subunit IV (SI, Tables T2, T3). Additional interaction of the ISP helix is observed with the lipid DAG1 (Figure 3; SI, Figure S1K, Tables T2, T3). On the n-side, it has previously been shown that the sulfolipid mediates interactions between the ISP and cytochrome f helices (de Vitry et al., 2004) (Figure 3; SI Figure S1M). In the present study, an additional n-side lipid DAG9 interacts with the ISP helix through one of its acyl tails to enhance contacts of the ISP subunit with the b6f core (Figure 3; SI, Tables T2, T3, Figure S1N).
Figure 3.
p-Side lipid cage around ISP-cytochrome f. Lipid DAG2 (white/red sticks) enhances the stabilization of the ISP helix (pink ribbon) by cross-linking with the cytochrome f helix (cyan ribbon). The lipid DAG1 (green/red) connects the ISP helix to the cytochrome b polypeptide (yellow surface). The cytochrome f helix (cyan ribbon) is stabilized by the lipid DAG3 (blue/red sticks). On the n-side, interactions of the ISP helix with the b6f core are mediated by the sulfolipid (red sticks, SQDG) and lipid DAG9 (yellow/red sticks). Black mesh, 2Fo-Fc map at 1.1 σ.
On the p-side, the cytochrome f helix is stabilized by interactions with the lipids DAG2 and DAG3 (Figure 3; SI, Tables T2, T3, Figure S1L).
Nature of the Inter-Monomer Cavity
The b6f complex dimer encloses a large 30Å × 15Å × 25Å cavity, which functions in quinone/quinol exchange with the bulk membrane (Cramer et al., 2006). The contents of the cavity have not been resolved in the previously published lower resolution b6f structures. In the present study, a total of 6 lipidic sites were assigned in the cavity per monomer (SI, Table T2). (i) A lipid DAG9 was modeled proximal to heme cn along the cavity walls defined by the helices A, D and E (Figure 4A; SI, Tables T2, T3 and Figure S1N). (ii) A partially ordered UDM5 detergent was modeled close to the axial position of heme cn (Figure 4A; SI, Tables T2, T3 and Figure S1P). However, the 11-carbon UDM tail does not sufficiently occupy the long tubular density, which extends further toward the p-side. (iii) Hence, an 18-carbon chain (8K6-4) was also modeled in the electron density (Figure 4A; SI, Tables T2, T3, and Figure S1P). (iv, v) Two n-side 18-carbon chains (8K6-2, and 8K6-3) were modeled at the boundary/cavity interface of the b6f dimer, proximal to the DAG9 lipid (Figure 4B; SI, Tables T2, T3 and Figure S1Q). (vi) A lipid, DAG8 (SI, Figure S1I) was found to be located proximal to 8K6-2 and 8K6-3 (Figure 4B). The DAG8 glycerol moiety interacts with the N-terminal surface helix of the cytochrome b subunit in one monomer, while the DAG8 acyl tails contact the cytochrome b C-helix, and the subunit IV F-helix in the other monomer (SI, Tables T2, T3).
Figure 4.
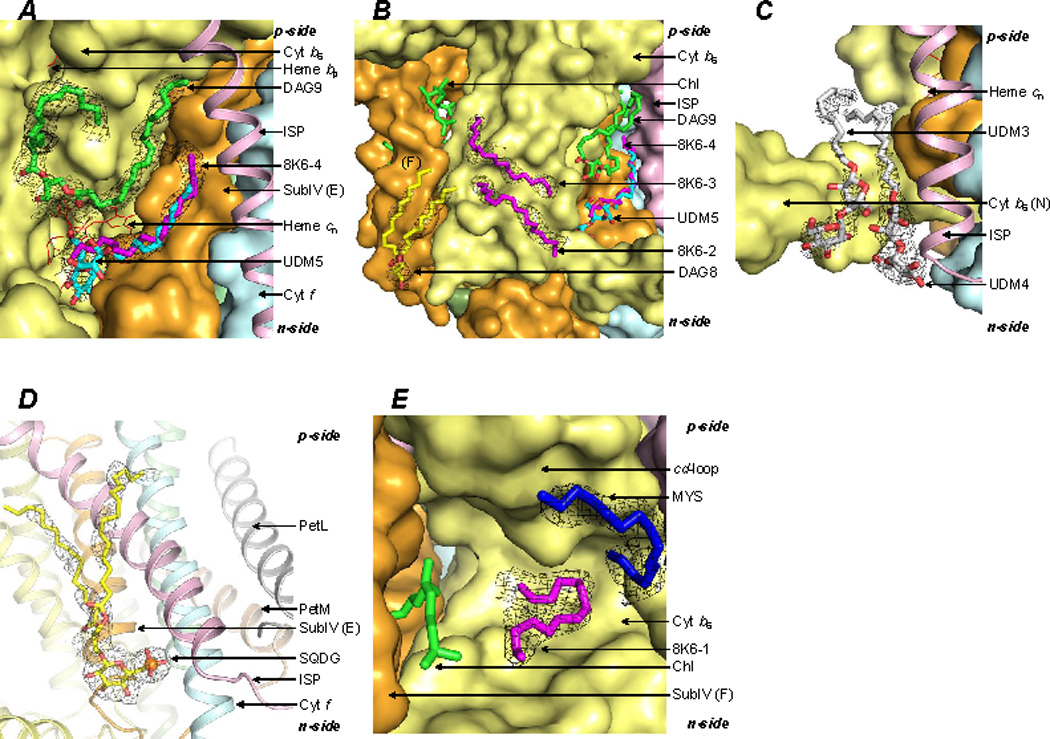
Lipidic sites in the inter-monomer cavity of the b6f complex. (A) Lipids located proximal to heme cn. The lipid DAG9 (green/red) interacts with the A, D and E-helices. The ligand bound to heme cn (red sticks) is modeled as a partially ordered detergent UDM5 (cyan/red). An alternate ligand is modeled as the 18-carbon 8K6-4 (magenta). (B) Two 18-carbon chains, 8K6-2 and 8K6-3 (magenta sticks), are modeled proximal to the C and D-helices. A lipid, DAG8 (yellow/red sticks), provides a connection between the inter-monomer cavity and the lipid bilayer. (C) The detergents, UDM3 and UDM4 (white/red sticks) are observed at the n-side lipid-water interface. ISP helix shown as a pink ribbon. (D) The sulfolipid (SQDG, yellow/red sticks) provides an n-side connection between the ISP (pink ribbon) and cytochrome f (cyan ribbon) helices. (E) An 18-carbon 8K6-1 (magenta sticks) and 15-carbon MYS (blue) chains were modeled in proximity to the Qp-site, marked by the chlorophyll phytyl-tail. Black mesh, 2Fo-Fc map at 1.1 σ.
The periphery of the cytochrome b6f complex inter-monomer cavity was observed to contain 5 lipidic sites. (i, ii) On the n-side, two partially ordered UDM molecules (UDM3 and UDM4) have been previously reported to be bound to the cytochrome b N-terminal surface helix (Figure 4C; SI, Tables T2, T3, Figure S1R). (iii) The sulfolipid acyl tails, located proximal to the cavity, were previously reported to be disordered (Baniulis et al., 2009; Stroebel et al., 2003; Yamashita et al., 2007). In the present study, electron density was assigned to portions of the acyl tails, one of which is wrapped around the ISP helix (Figure 4D). (iv, v) On the p-side of the b6f complex, an 18-carbon 8K6-1 and a 15-carbon MYS chain (Figure 4E; SI, Figure S1S, S1T) were assigned within the cavity (SI, Table T2, T3).
It is noted that the lipid binding sites described in the present study are abundant in aromatic residues (SI, Table T3). The relatively more polar tyrosine and tryptophan residues are located mainly near the polar lipid head groups, while the phenylalanine residues are found closer to the lipid acyl tails (SI, Figures S2A–S2T).
DISCUSSION
Specific interactions with lipids have been implicated in regulation of topology, assembly and oligomerization of membrane protein assemblies (de Vitry et al., 2004; Dowhan and Bogdanov, 2009; Hasan et al., 2011; Hasan et al., 2013b; Remy et al., 1982). Models have been proposed to describe the insertion and assembly of simple membrane proteins in lipid bilayers (Egea and Stroud, 2010; Rapoport et al., 2004; White and von Heijne, 2004). However, the sequence of assembly-related events relevant to multi-subunit membrane proteins, especially the hetero-oligomeric complexes remains unknown. It is inferred that insertion of lipids must occur during assembly of membrane protein complexes. The 23 lipid-binding sites identified in the present study provide clues about additional necessary steps in the assembly of hetero-oligomeric complexes and super-complexes. The association of the cytochrome b6f complex with lipids occurs mainly through the lipid acyl chains as the head-group electron density is poorly defined for nine of the 12 lipids reported to be bound to the b6f complex.
Lipids in the Vicinity of the Unique β-Carotene
In the present study, 7 lipid sites were identified on the b6f complex surface, close to the β-carotene (Figure 2B). Due to their surface location, lipids may provide cross-linking interactions that stabilize a b6f super-complex formed with the PSI reaction center complex. Hence, in contrast to information derived from lower resolution b6f structures (Hasan and Cramer, 2012; Hasan et al., 2011; Stroebel et al., 2003), the present study shows that stabilization of a b6f-PSI super-complex may be mediated by the β-carotene and supplemented through lipid contacts (Figure 2B), thereby providing a lipidbased mechanism of super-complex formation that is conserved between the respiratory and photosynthetic electron transfer chains.
Lipidic Environment of the Chlorophyll-Binding Site
The b6f complex chlorophyll (Figure 2C) can perform a quinone/quinol sensing function in state transitions in which the p-side quinol-binding site acts as a redox sensor (Vener et al., 1997; Zito et al., 1999). The interaction of the bound quinone with the chlorophyll phytyl-tail may generate a signal that activates the enzyme LHCII kinase for state transitions (Hasan et al., 2013b; Lemeille and Rochaix, 2010). Insertion of an α-helix proximal to the chlorophyll-binding site inhibits state transitions indicating a specific role of chlorophyll-binding niche in trans-membrane signaling (Zito et al., 2002). In the b6f structure described in the present study, a total of 4 lipids have been identified in the vicinity of the chlorophyll-binding niche (Figure 2D). The synthetic lipid DOPC has previously been reported to occupy an n-side site between the F and G-helices of subunit IV (Baniulis et al., 2009; Yamashita et al., 2007). The DOPC position is occupied by the cytochrome b terminal H-helix in the related bc1 complex (Hasan et al., 2011). Additional stabilization is provided by the lipids DAG5, DAG8, and DAG7 (Figure 2D). It is significant that the lipidic sites are located on the surface, proximal to the chlorophyll, thereby providing an adaptable surface for interaction with the LHCII kinase through lipid-mediated contacts.
Lipid-Mediated Stabilization of the Iron-Sulfur Protein
Enhanced flexibility of the b6f ISP subunit, which would require additional stabilization, is inferred from a comparative analysis of the biochemical and structural properties of the ISP subunit in the b6f and bc1 complexes (Hasan et al., 2013a) including, (i) a disordered flexible poly-Gly hinge in the b6f ISP subunit in crystal structures (Kurisu et al., 2003; Stroebel et al., 2003), and, (ii) resistance of b6f ISP poly-Gly hinge mutagenesis to loss of function (de Vitry et al., 2004; Yan and Cramer, 2003). Here, it is significant that b6f delipidation is correlated with ISP loss (Breyton et al., 1997; Zhang and Cramer, 2005). In the present study, multiple lipid sites were observed proximal to the ISP helix. On the p-side, the lipids DAG1 and DAG2 act as cross-linkers to enhance the stability of the ISP subunit (Figure 3). On the n-side, the sulfolipid headgroup augments interactions of the ISP with cytochrome f (de Vitry et al., 2004) (Figures 3, 4D). One of the sulfolipid acyl tails is inferred to interact with the ISP helix (Figure 4D). Hence, an extensive lipid-based network is involved in ISP stabilization in the b6f complex.
Role of Lipids in the Inter-Monomer Cavity
In the present study, five lipidic sites (SI, Figure S3A, B) have been identified that connect the Qp-portal to the inter-monomer cavity. The acyl tails of the lipid DAG8 (Figure 4B) point toward the Qp-portal. Additional sites along the Qp-portal are occupied by the 15-carbon chain MYS (Figure 4E), and the 18-carbon chains 8K6-1 (Figure 4E), 8K6-2 (Figure 4B), and 8K6-3 (Figure 4B). The location of the lipidic sites may be significant for quinone/quinol traffic. The quinone molecule has a long 45-carbon hydrophobic tail, which can potentially interact via Van der Waals’ interactions with sites other than the quinone binding site. The lipidic sites enhance tight packing of the b6f complex hydrophobic core, and occupy potentially superfluous quinone binding sites. It is expected that the efficiency of quinone/quinol traffic is enhanced in the presence of lipids (SI, Figures S3A, B), which reduce the number of quinone binding sites that may result in unproductive quinone-protein interactions.
It is relevant to note that delipidation has been implicated in monomerization and inactivation of the b6f complex (Breyton et al., 1997). In the present study, 11 distinct lipid sites (Figures 4A–E) per b6f monomer associated with the cavity have been identified. Recent studies on the cytochrome bc1 complex have defined inter-monomer electron transfer (Lanciano et al., 2011; Swierczek et al., 2010). The presence of lipids in the cavity may influence inter-monomer interactions relevant to electron transfer, by altering the dielectric constant between the inter-monomer heme pairs. In contrast, the intra-monomer heme pair is embedded in a protein environment. Therefore, lipids within the cavity may modulate the function of cytochrome bc complexes.
In summary, the present study shows that intrinsic lipids perform crucial functions as bridging molecules that mediate contacts between subunits of polytopic, multi-subunit membrane protein complexes, and thus enhance subunit-subunit interactions while providing an interface for super-complex formation. This study poses questions about the presently unknown mechanism by which lipids are inserted into critical internal sites within hetero-oligomeric complexes and super-complexes at the inter-subunit interface during translocon-dependent assembly.
EXPERIMENTAL PROCEDURES
Isolation and Crystallization of the Cyanobacterial Cytochrome b6f Complex
Purification and crystallization of the dimeric cytochrome b6f complex from Nostoc PCC 7120 have been described (Baniulis et al., 2009; Baniulis et al., 2011). The isolated b6f dimer complex was concentrated to 135 µM (monomer concentration) in the buffer Tris-HCl (30 mM, pH 7.5, at room temperature), NaCl (50 mM), sucrose (10%), analytical grade UDM (3 mM, Anatrace, Maumee OH) and the lipid DOPC (1 mM, Avanti Polar Lipids Inc., Alabaster AL), and crystallized as described (Baniulis et al., 2011). The concentration of the lipid DOPC is close to saturation in the protein buffer, which is essential for stabilization of the isolated dimeric b6f complex. Following cryo-protection, the cytochrome b6f complex crystals were frozen by plunging in liquid nitrogen.
X-ray Data Collection, Data Reduction, and Electron Density Map Calculation
Diffraction data sets were previously collected at 100 K for the cytochrome b6f complex crystals (PDB ID 2ZT9), at beamline 19-ID-B of the Advanced Photon Source, Argonne IL. Based on the correlation-coefficient method of diffraction data merging (Karplus and Diederichs, 2012), the X-ray diffraction data (PDB ID 2ZT9) (Baniulis et al., 2009) were re-processed in HKL2000 (Otwinowski and Minor, 1997) to a resolution of 2.5 Å (Table 1). Rigid-body refinement of the b6f complex atomic model (PDB ID 4H44) was performed in Phenix (Adams et al., 2010) against the 2.5 Å diffraction data-set, using a single rigid-body. Model building was performed in COOT (Emsley et al., 2010). Simulated annealing of torsion angles and Cartesian coordinates was performed, along with Translation-Libration-Screw refinement. The fitting of coordinates of the Rieske iron-sulfur protein extrinsic domain (chain D, Thr54-Ser179) into the electron density map was improved by manually modeling the coordinates on the cyanobacterial iron-sulfur protein extrinsic domain crystal structure (PDB ID 3AZC) (Veit et al., 2012), followed by restrained refinement. Lipid and detergent sites were located using the Fo-Fc and 2Fo-Fc maps, respectively contoured at 3.0–3.5 σ and 1.0–1.5 σ. The ligands were built manually into the electron density in COOT, followed by restrained refinement.
Analysis of Lipid-Protein Contacts
A model of the dimeric b6f complex was analyzed in PyMol (www.pymol.org). A cut-off distance of 4.0 Å was used to identify the amino acids which interact with each lipid. The residues are listed in SI Table T3.
Supplementary Material
Highlights.
Presently, understanding of lipid-based biogenesis of membrane proteins is limited
23 distinct lipid sites revealed per monomer in 2.5Å structure of b6f complex dimer
Lipids located at inter-subunit interface, mediate complex & super-complex formation
ACKNOWLEDGEMENTS
The authors thank D. Baniulis for b6f purification and crystallization, E. Yamashita for X-ray diffraction data collection, staff at beam line APS-19-ID-B for technical assistance, J. P. Whitelegge, M. G. Rossmann and V. M. Prasad for discussions, and members of the Phenix team and CCP4 workshop-2010 for advice on structure solution. The studies were supported by NIH-GM-038323 and the Henry Koffler Distinguished Professorship (WAC).
ABBREVIATIONS
- 8K6
18-carbon chain
- Car
β-carotene
- Chl
chlorophyll
- Cyt
cytochrome
- DAG
diacylglycerol lipid
- DOPC
dioleoylphosphatidylcholine lipid
- Heme cn
covalently linked heme on n-side of cytochrome b polypeptide, serves as quinone binding site
- ISP
iron-sulfur protein, bound to high-potential [2Fe-2S] cluster
- MGDG
monogalactosyldiacylglycerol lipid
- MYS
15-carbon chain
- n, p-sides
electrochemically negative and positive side of membrane and cytochrome bc complexes
- OCT
8-carbon chain
- PDB
Protein Data Bank
- PSI
photosystem-I reaction centre complex
- Qn, Qp-sites
quinone reduction-protonation (n-side) and quinol deprotonation-oxidation sites (p-side)
- SQDG
sulfoquinovosyldiacylglycerol lipid
- SubIV
subunit IV of b6f complex
- TMH
trans-membrane helix
- UDM
n-undecyl-β-D-maltopyranoside
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
SSH performed crystallographic data reduction, model building and refinement, analyzed results, wrote manuscript; WAC analyzed results, wrote manuscript.
ACCESSION NUMBER
The Protein Data Bank accession number for the structure reported in this paper is 4OGQ.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniulis D, Yamashita E, Whitelegge JP, Zatsman AI, Hendrich MP, Hasan SS, Ryan CM, Cramer WA. Structure-function, stability, and chemical modification of the cyanobacterial cytochrome b6f complex from Nostoc sp. PCC 7120. J. Biol. Chem. 2009;284:9861–9869. doi: 10.1074/jbc.M809196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniulis D, Zhang H, Yamashita E, Zakharova T, Hasan SS, Cramer WA. Purification and crystallization of the cyanobacterial cytochrome b6f complex. In: Carpentier R, editor. Methods Mol. Biol. (Photosyn. Res. Protoc.) Totowa, NJ: Humana Press Inc; 2011. pp. 65–77. [DOI] [PubMed] [Google Scholar]
- Berry EA, Guergova-Kuras M, Huang L-S, Crofts AR. Structure and function of cytochrome bc complexes. Ann. Rev. Biochem. 2000;69:1005–1075. doi: 10.1146/annurev.biochem.69.1.1005. [DOI] [PubMed] [Google Scholar]
- Breyton C, Tribet C, Olive J, Dubacq JP, Popot JL. Dimer to monomer conversion of the cytochrome b6f complex. Causes and consequences. J. Biol. Chem. 1997;272:21892–21900. doi: 10.1074/jbc.272.35.21892. [DOI] [PubMed] [Google Scholar]
- Cramer WA, Zhang H, Yan J, Kurisu G, Smith JL. Transmembrane traffic in the cytochrome b6f complex. Ann. Rev. Biochem. 2006;75:769–790. doi: 10.1146/annurev.biochem.75.103004.142756. [DOI] [PubMed] [Google Scholar]
- de Lacroix de Lavalette A, Finazzi G, Zito F. b6f-Associated chlorophyll: structural and dynamic contribution to the different cytochrome functions. Biochemistry. 2008;47:5259–5265. doi: 10.1021/bi800179b. [DOI] [PubMed] [Google Scholar]
- de Vitry C, Ouyang Y, Finazzi G, Wollman FA, Kallas T. The chloroplast Rieske iron-sulfur protein. At the crossroad of electron transport and signal transduction. J. Biol. Chem. 2004;279:44621–44627. doi: 10.1074/jbc.M406955200. [DOI] [PubMed] [Google Scholar]
- Dowhan W, Bogdanov M. Lipid-Dependent Membrane Protein Topogenesis. Ann. Rev. Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea PF, Stroud RM. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl. Acad. Sci. USA. 2010;107:17182–17187. doi: 10.1073/pnas.1012556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero S, Gouaux E. High resolution crystallographic studies of α-hemolysin-phospholipid complexes define heptamer-lipid head group interactions: implication for understanding protein-lipid interactions. Protein Sci. 2004;13:1503–1511. doi: 10.1110/ps.03561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Cramer WA. Lipid functions in cytochrome bc complexes: An odd evolutionary transition in a membrane protein structure. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:3406–3411. doi: 10.1098/rstb.2012.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Stofleth JT, Yamashita E, Cramer WA. Lipid-induced conformational changes within the cytochrome b6f complex of oxygenic photosynthesis. Biochemistry. 2013a;52:2649–2654. doi: 10.1021/bi301638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Yamashita E, Cramer WA. Trans-membrane signaling and assembly of the cytochrome b6f-lipidic charge transfer complex. Biochim. Biophys. Acta. 2013b;1827:1295–1308. doi: 10.1016/j.bbabio.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Yamashita E, Ryan CM, Whitelegge JP, Cramer WA. Conservation of lipid functions in cytochrome bc complexes. J. Mol. Biol. 2011;414:145–162. doi: 10.1016/j.jmb.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite RK, Li Z, Walz T. Principles of membrane protein interactions with annular lipids deduced from aquaporin-0 2D crystals. EMBO J. 2010;29:1652–1658. doi: 10.1038/emboj.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C, Richers S. Lipids and membrane protein structures. Curr. Opin. Struct. Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Iwai M, Takizawa K, Tokutsu R, Okamuro A, Takahashi Y, Minagawa J. Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature. 2010;464:1210–1213. doi: 10.1038/nature08885. [DOI] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: Tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- Lanciano P, Lee DW, Yang H, Darrouzet E, Daldal F. Intermonomer electron transfer between the low-potential b hemes of cytochrome bc1. Biochemistry. 2011;50:1651–1663. doi: 10.1021/bi101736v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Nett JH, Trumpower BL, Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc. Natl. Acad. Sci. USA. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeille S, Rochaix JD. State transitions at the crossroad of thylakoid signalling pathways. Photosynth. Res. 2010;106:33–46. doi: 10.1007/s11120-010-9538-8. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. Macromol. Crystallogr. A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim. Biophys. Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- Pierre Y, Breyton C, Lemoine Y, Robert B, Vernotte C, Popot JL. On the presence and role of a molecule of chlorophyll-a in the cytochrome b6f complex. J. Biol. Chem. 1997;272:21901–21908. doi: 10.1074/jbc.272.35.21901. [DOI] [PubMed] [Google Scholar]
- Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Mills DA, Buhrow L, Hiser C, Ferguson-Miller S. A conserved steroid binding site in cytochrome c oxidase. Biochemistry. 2008;47:9931–9933. doi: 10.1021/bi8013483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Sharpe MA, Garavito RM, Ferguson-Miller S. Conserved lipid-binding sites in membrane proteins: A focus on cytochrome c oxidase. Curr. Opin. Struct. Biol. 2007;17:444–450. doi: 10.1016/j.sbi.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA, Goder V, Heinrich SU, Matlack KE. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 2004;14:568–575. doi: 10.1016/j.tcb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Remy R, Tremolieres A, Duval JC, Ambardbretteville F, Dubacq JP. Study of the Supramolecular Organization of Light-Harvesting Chlorophyll Protein (Lhcp) - Conversion of the Oligomeric Form into the Monomeric One by Phospholipase-A2 and Reconstitution with Liposomes. FEBS Lett. 1982;137:271–275. [Google Scholar]
- Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D, Choquet Y, Popot JL, Picot D. An atypical haem in the cytochrome b6f complex. Nature. 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- Swierczek M, Cieluch E, Sarewicz M, Borek A, Moser CC, Dutton PL, Osyczka A. An electronic bus bar lies in the core of cytochrome bc1. Science. 2010;329:451–454. doi: 10.1126/science.1190899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiefenbrunn T, Liu W, Chen Y, Katritch V, Stout CD, Fee JA, Cherezov V. High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment. PloS One. 2011;6:e22348. doi: 10.1371/journal.pone.0022348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit S, Takeda K, Tsunoyama Y, Rexroth D, Rogner M, Miki K. Structure of a thermophilic cyanobacterial b6f-type Rieske protein. Acta Crystallogr. D Biol. Crystallogr. 2012;68:1400–1408. doi: 10.1107/S0907444912034129. [DOI] [PubMed] [Google Scholar]
- Vener AV, van Kan PJ, Rich PR, Ohad I, Andersson B. Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: Thylakoid protein kinase deactivation by a single-turnover flash. Proc. Natl. Acad. Sci. USA. 1997;94:1585–1590. doi: 10.1073/pnas.94.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Hielscher R, Hellwig P, Schagger H, Richers S, Hunte C. Role of phospholipids in respiratory cytochrome bc1 complex catalysis and supercomplex formation. Biochim. Biophys. Acta. 2009;1787:609–616. doi: 10.1016/j.bbabio.2009.02.012. [DOI] [PubMed] [Google Scholar]
- White SH, von Heijne G. The machinery of membrane protein assembly. Curr. Opin. Struct. Biol. 2004;14:397–404. doi: 10.1016/j.sbi.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Yamashita E, Zhang H, Cramer WA. Structure of the cytochrome b6f complex: quinone analogue inhibitors as ligands of heme cn. J. Mol. Biol. 2007;370:39–52. doi: 10.1016/j.jmb.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Cramer WA. Functional insensitivity of the cytochrome b6f complex to structure changes in the hinge region of the Rieske iron-sulfur protein. J. Biol. Chem. 2003;278:20925–20933. doi: 10.1074/jbc.M212616200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cramer WA. Problems in obtaining diffraction-quality crystals of heterooligomeric integral membrane proteins. J. Struct. Funct. Genomics. 2005;6:219–223. doi: 10.1007/s10969-005-1912-y. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kurisu G, Smith JL, Cramer WA. A defined protein-detergent-lipid complex for crystallization of integral membrane proteins: The cytochrome b6f complex of oxygenic photosynthesis. Proc. Nat. Acad. Sci. USA. 2003;100:5160–5163. doi: 10.1073/pnas.0931431100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- Zito F, Finazzi G, Delosme R, Nitschke W, Picot D, Wollman FA. The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J. 1999;18:2961–2969. doi: 10.1093/emboj/18.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito F, Vinh J, Popot JL, Finazzi G. Chimeric fusions of subunit IV and PetL in the b6f complex of Chlamydomonas reinhardtii: Structural implications and consequences on state transitions. J. Biol. Chem. 2002;277:12446–12455. doi: 10.1074/jbc.M110914200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.