Summary
ATP-dependent chromatin remodeling is involved in all DNA transactions and linked to numerous human diseases. We explored functions of chromatin remodelers during cellular aging. Deletion of ISW2, or mutations inactivating the Isw2 enzyme complex, extends yeast replicative lifespan. This extension by ISW2 deletion is epistatic to the longevity effect of calorie restriction (CR) and this mechanism is distinct from suppression of TOR signaling by CR. Transcriptome analysis indicates that isw2Δ partially mimics an up-regulated stress response in CR cells. In particular, isw2Δ cells show an increased response to genotoxic stresses, and the DNA repair enzyme Rad51 is important for isw2Δ-mediated longevity. We show that lifespan is also extended in C. elegans by reducing levels of athp-2, a putative ortholog of Itc1/ACF1, a critical subunit of the enzyme complex. Our findings demonstrate that the ISWI class of ATP-dependent chromatin remodeling complexes play a conserved role during aging and in calorie restriction.
Keywords: Chromatin remodeling, aging, calorie restriction, stress response, Isw2, yeast replicative aging, cellular senescence
Introduction
The eukaryotic genome is packaged into a highly organized and largely repressive structure of chromatin, hence DNA-based processes require remodeling of chromatin to expose or occlude DNA elements. ATP-dependent chromatin remodeling enzymes use the energy of ATP hydrolysis to alter chromatin states in chromatin assembly, DNA replication, recombination and repair, gene transcription and silencing, chromatin domain insulation, chromosomal dosage compensation, and chromosome segregation (Clapier and Cairns, 2009). Chromatin remodelers play critical roles in development, cell differentiation, and stem cell maintenance. Mutations in these enzymes contribute to cancer and congenital syndromes (Wu, 2012).
The four sub-families of the chromatin remodeling ATPases (SWI/SNF, ISWI, CHD/NURD, and INO80) have distinct domain organization and catalytic mechanisms (Clapier and Cairns, 2009; Hota and Bartholomew, 2011). The ISWI subfamily alters nucleosome positioning, catalyzes chromatin assembly, and leads to chromosome condensation (Corona and Tamkun, 2004). These enzymes regulate DNA replication, transcription regulation, and cell fate determination (Yadon and Tsukiyama, 2011). The ISWI subfamily has simple enzyme complex composition compared to other remodelers, but, nonetheless, form distinct complexes with different subunits for varied functions. Notably, chromatin remodeling ATPases are highly conserved and orthologs exist from yeast to humans (Clapier and Cairns, 2009). Despite considerable understanding, a role of ATPases in aging and lifespan regulation remains underexplored.
A hallmark of aging is the accumulation of deleterious cellular damage, including oxidized, misfolded and/or aggregated proteins, dysfunctional organelles, and damaged DNA and chromatin structures (Feser and Tyler, 2011; Guarente, 2008; Kourtis and Tavernarakis, 2011; Sahin and Depinho, 2010). Various mechanisms reduce insults and remove damaged components in normal young cells, including enzymes to remove reactive oxygen species (Landis and Tower, 2005), heat shock proteins to remove misfolded proteins (Koga et al., 2011; Kourtis and Tavernarakis, 2011), recycling of damaged organelles (Green et al., 2011; Koga et al., 2011), and DNA repair and check point systems to fix DNA damage prior to replication (Langerak and Russell, 2011). These mechanisms comprise the cellular stress response system, and genetic and environmental interventions often extend lifespan via enhanced stress responses (Kourtis and Tavernarakis, 2011). There are age-dependent changes in these stress response pathways (Gorbunova et al., 2007; Kourtis and Tavernarakis, 2011). However, the underlying mechanisms leading to altered stress responses during aging remain elusive.
Calorie restriction (CR), or more generally dietary restriction (DR), is the most robust and conserved intervention to extend lifespan (Mair and Dillin, 2008). Studies in model organisms indicate multiple pathways in mediating longevity and health benefits from DR, including reduced insulin-like growth factor (IGF) signaling (Mair and Dillin, 2008), down-regulated TOR signaling and ribosome abundance (Johnson et al., 2013), elevated sirtuin activity and reduced oxidative stress (Guarente, 2008), as well as improved DNA damage repair (Martins et al., 2011). For replicative aging of the budding yeast Saccharomyces cerevisiae, various CR conditions are proposed to mediate lifespan extension: enhancing Sir2 function through increased NAD/NADH ratio (Longo and Kennedy, 2006), repressing ribosome biogenesis through down-regulation of TOR pathway kinases Tor1 and Sch9 (Johnson et al., 2013), and enhancing mitochondrial function and oxidative stress response (Molin et al., 2011; Ristow and Schmeisser, 2011).
Recently, chromatin remodeler SWI/SNF was linked to DAF-16 mediated longevity in C. elegans (Riedel et al., 2013). Here we investigate whether other remodelers impact aging in S. cerevisiae. We discovered that the Isw2 enzyme complex regulates lifespan through stress response pathways. Further, ISW2 deletion results in a transcriptome and chromatin state that in part mimics CR conditions. Our findings reveal a novel CR pathway and suggest a conserved mechanism for regulation of longevity.
Results
Chromatin remodeling complex ISW2 is a novel aging regulator and effector of calorie restriction
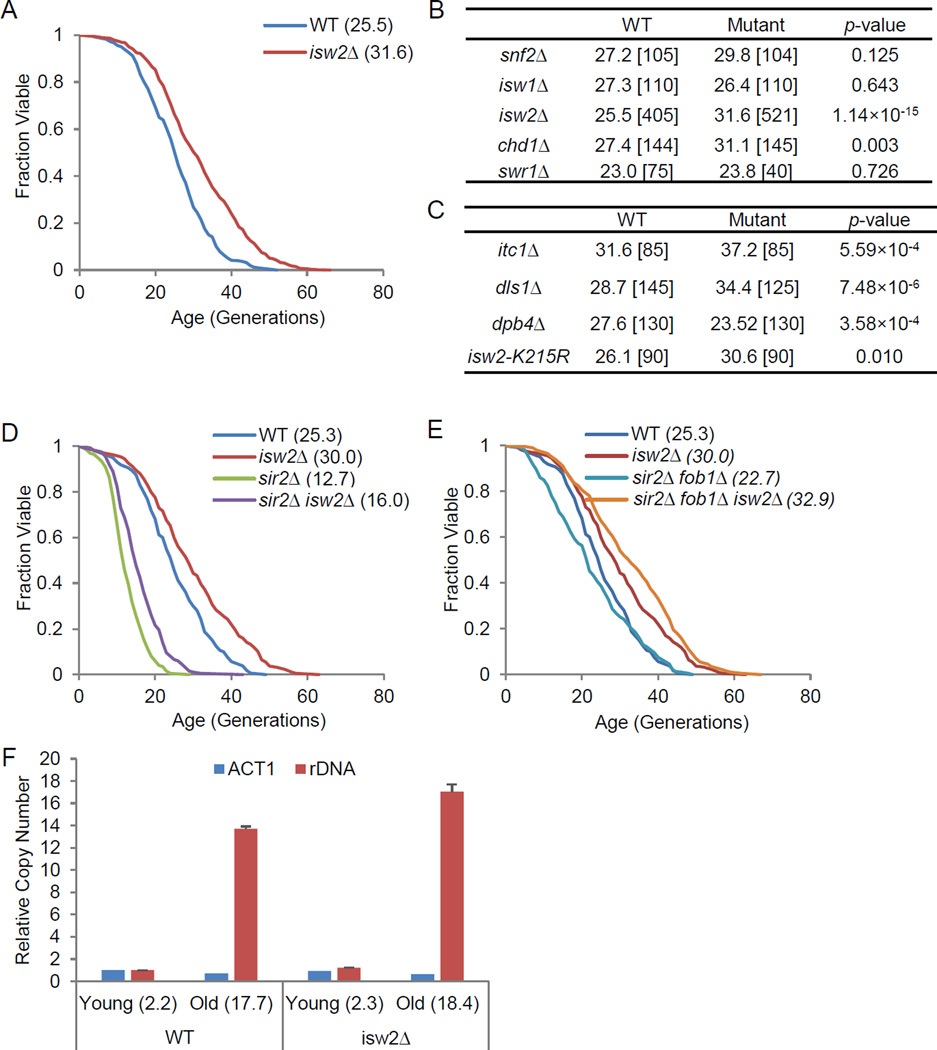
We screened chromatin regulators for lifespan alterations in the context of a large project quantifying replicative lifespan for single gene deletion strains from the yeast ORF deletion library (Kaeberlein et al., 2005a). Among nucleosome remodeling enzymes, only isw2Δ and chd1Δ were able to significantly extend lifespan, with isw2Δ robustly extending lifespan by 24% (Fig. 1A and B). Deletion of ISW2 extended both median and maximum lifespan (Fig. 1 A).
Figure 1. Disruption of ATP-dependent chromatin remodeling complex ISW2 extends yeast replicative lifespan.
(A) Replicative lifespan for wild-type (WT) and isw2Δ. Values in parenthesis (here and hereafter), are mean lifespan. (B) Mean replicative lifespan for strains deleted for chromatin remodeling ATPases. Values in brackets,are the number of cells tested. (C) Mean replicative lifespan for strains deleted for ISW2 complex subunits or via a catalytical mutant allele of ISW2. (D–E) Replicative lifespan for isw2Δ in combination with either sir2Δ or sir2Δ fob1Δ. (F) Quantitative real-time PCR analysis of rDNA copy number for young and old WT and isw2Δ cells. Values in parenthesis,are mean age. See also Figure S1 and Table S1.
Since isw2Δ had a more significant effect, we chose to further investigate how it regulated lifespan, even though Chd1, a regulator of transcription elongation and chromatin assembly (Sims and Wade, 2011), might also be an interesting subject. Isw2 functions in a complex associated with three other subunits, Itc1, Dls1, and Dpb4, and we found that itc1Δ and dls1Δ also extended lifespan, by 18% and 20%, respectively (Fig. 1C and S1A–B). Deletion of DPB4 did not extend lifespan (Fig. 1C and S1C), likely because Dpb4 is also a subunit of DNA polymerase ε, and a defect in this enzyme may counterbalance any benefits from reduced Isw2 function (Iida and Araki, 2004). Isw2 shares a conserved ATPase domain with other members of the remodeler family; we found that a mutation in the conserved catalytic domain, K215R, also significantly extended lifespan by 15% (Fig. 1C and Fig. S1D).
Because Isw2 regulates chromatin accessibility, we tested whether lifespan extension by isw2Δ required the histone deacetylase Sir2. An important cause of aging in yeast is the accumulation of extra-chromosomal circles (ERCs) in old cells, which is normally suppressed by Sir2, and requires Fob1, a replication fork protein (Longo and Kennedy, 2006). Homologs of Sir2 are found in all eukaryotes, collectively called sirtuins. Previous studies with fob1Δ mutants have revealed ERC-independent mechanisms that involve Sir2 in promoting longevity, such as telomeric heterochromatic silencing (Dang et al., 2009; Kaeberlein et al., 1999). We found that isw2Δ significantly extended lifespan in both sir2Δ and sir2Δ fob1Δ backgrounds (Fig. 1D–E). The ability of isw2Δ to extend the short lifespan of sir2Δ cells is noteworthy, as it was recently reported that, of 33 gene deletions that extend lifespan, only fob1Δ had this property (Delaney et al., 2011a). This might suggest that effects of isw2Δ on lifespan are mediated by reduction of rDNA recombination. Hence, we further tested whether isw2Δ could reduce the accumulation of ERCs in old cells. Using quantitative real-time PCR targeting rDNA sequences, we observed increased rDNA copy number in old cells compared to young cells due to accumulation of ERCs. Interestingly, isw2Δ did not reduce the levels of ERCs in old cells compared to wild-type (WT) (Fig. 1F), suggesting that isw2Δ did not extend lifespan by suppressing the formation and accumulation of ERCs. This is consistent with the results that isw2Δ extended lifespan in sir2Δ fob1Δ cells. Hence, we conclude that Isw2 regulates aging through a Sir2-independent and ERC-independent pathway.
Isw2 functions in a distinct calorie restriction pathway
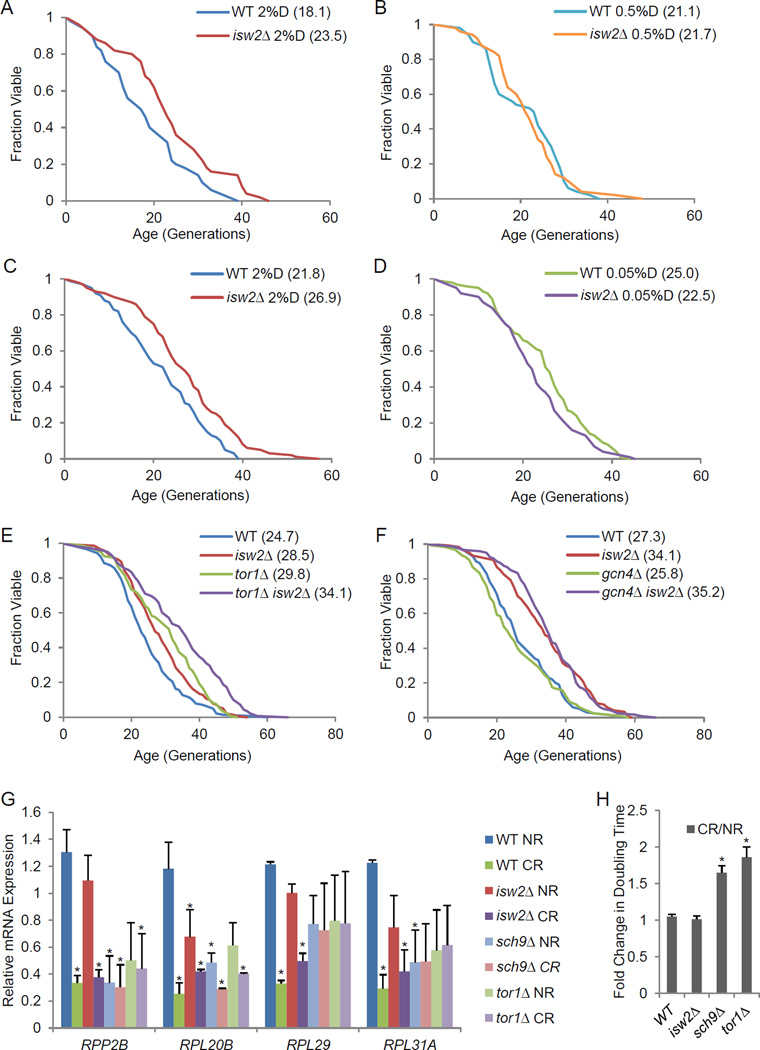
Since Isw2 requires ATP hydrolysis for its activity, we tested whether the altered cellular energy state in CR might be related to isw2Δ-mediated lifespan extension. As expected, lifespan was extended by limiting glucose concentrations to either 0.5% or 0.05%; however, these CR conditions were not able to extend lifespan when ISW2 was deleted (compare Fig. 2A to B, and 2C to D). Similarly, isw2Δ was unable to extend lifespan further under these CR conditions (Fig. 2B/D). This is similar to findings that tor1Δ and sch9Δ are epistatic to CR (Kaeberlein et al., 2005b). Our observation suggests that isw2Δ and CR may share a common pathway to confer lifespan extension.
Figure 2. Deletion of ISW2 extends lifespan through a novel mechanism exploited by calorie restriction.
(A–B) Replicative lifespan for isw2Δ under the moderate CR condition, YPD with 0.5% D-glucose (0.5%D), compared to the non-restricted (NR) condition, YPD with 2% D-glucose (2%D). (C–D) Replicative lifespan for isw2Δ under the severe CR condition, YPD with 0.05% D-glucose (0.05%D), compared to NR. (E–F) Replicative lifespan for isw2Δ in the tor1Δ (E) or gcn4Δ (F) background. (G) Gene expression analysis by RT-qPCR for selected 60S ribosome subunit genes. NR, SC with 2% glucose. CR, SC with 0.05% glucose. (H) Fold change in doubling time during exponential growth in SC media. * p values < 0.05 compared to WT NR. See also Table S1
Therefore, we next tested how isw2Δ is related to known effectors of CR. Tor1 and Sch9 are kinases in the TOR (target of rapamycin) signaling pathway that respond to nutrient availability and become inactivated in CR (Kaeberlein, 2010). A mechanism proposed for this pathway is that inactivation of TOR inhibits ribosome biogenesis and protein translation, which in turn induces the expression of Gcn4, a transcription factor activated in CR (Steffen et al., 2008). In order to determine if lifespan extension by isw2Δ is mediated by the same mechanism, we tested the lifespan of the isw2Δ tor1Δ double mutant. Interestingly, isw 2Δ tor1Δ had a lifespan significantly longer than either of the single mutants (Fig. 2E), suggesting that Isw2 and Tor1 regulate lifespan through distinct and parallel pathways. To further verify that the lifespan extension by isw2Δ is independent of the TOR pathway and Gcn4, we tested lifespan epistasis between isw2Δ and gcn4Δ. As predicted, gcn4Δ did not influence the longevity effect of isw2Δ (Fig. 2F), further supporting the idea that Isw2 functions in a pathway different from TOR.
To further confirm this finding, we compared effects of these mutants directly on ribosomal gene expression, as well as their growth phenotypes. Ribosome gene expression is strictly controlled by the cellular energy state (Shore et al., 2009). Nutrient deprivation can reduce ribosome gene expression and ribosome assembly. Likewise, treatment with the mTOR inhibitor rapamycin, a CR mimetic, has a similar effect on ribosome gene expression as CR (Jorgensen et al., 2004). As expected, sch9Δ and tor1Δ reduced gene expression of several ribosome large subunit proteins, however, isw2Δ did not show such an effect on ribosome gene expression (Fig. 2G), suggesting that isw2Δ did not reduce global mRNA translation and that isw2Δ extended lifespan through a mechanism distinct from that of ribosome regulation.
We then examined their growth effects. Deletion of SCH9 extends lifespan by reducing the expression of ribosomal proteins, thus reducing the rate of protein synthesis, resulting in significant growth defects (Delaney et al., 2011b). We found that both tor1Δ and sch9Δ showed stronger growth defects under CR than normal growth conditions (Fig. 2H); these mutants, which lack nutrient sensing capabilities, were apparently unable to cope with the nutrient deprivation stress. In contrast, both WT and isw2Δ showed no growth phenotype in CR compared to NR (Fig. 2H). These distinct responses to CR between isw2Δ and tor1Δ/sch9Δ provided further evidence that Isw2 and Tor1/Sch9 function in distinct pathways.
Isw2 regulates a cohort of stress response genes
Yeast Isw2 plays numerous important cellular roles, such as in retrotransposition, transcription, and DNA replication (Yadon and Tsukiyama, 2011). We next investigated functions of Isw2 relevant to lifespan extension under CR conditions.
Isw2 facilitates integration of retrotransposon Ty1 near tRNA genes (Gelbart et al., 2005), which could restrict lifespan by reducing tRNA levels. We tested Isw2 effects on Ty1, but did not uncover evidence that either isw2Δ or CR altered Ty1 genome copy number, Ty1 integration patterns, or adjacent tRNA expression (Fig. S2A–C). In addition, Bdp1 recruits Isw2 to loci of Ty1 integration (Bachman et al., 2005), however, disruption of Isw2 recruitment by Bdp1 mutants failed to extend lifespan (Fig. S2D), ruling out the possibility that lifespan extension by isw2Δ was mediated by Ty1 retrotransposon.
We then considered potential effects of Isw2 on anti-sense transcription (Whitehouse et al., 2007). We found that levels of anti-sense transcription were not globally altered in old cells compared to young (Fig. S2E). In addition, we note that a global increase in anti-sense transcription via reduced Isw2 is not a plausible pathway to provide longevity benefits to cells.
We then considered effects on Isw2’s function to promote DNA replication fork progression in parallel with the INO80 complex (Vincent et al., 2008). Given our recent findings of a correlation between short lifespan and S phase arrest in yeast terminal state (Delaney et al., 2013), it seems highly unlikely that a defect in DNA replication via reduced Isw2 would promote longevity.
Having ruled out these previously identified Isw2 functions as plausible explanations for the longevity effect of isw2Δ, we turned our attention to the characterized role of Isw2 in creating regular nucleosome spacing, leading to transcriptionally repressive chromatin (Fazzio et al., 2001). However, previous microarray analysis showed that only 35 genes were derepressed more than two-fold by ISW2 deletion (Fazzio et al., 2001). Hence, we reexamined this dataset with a 1.5-fold cutoff, which was less stringent but still statistically relevant. Among the 281 genes derepressed more than 1.5-fold in isw2Δ, a majority of them, significantly greater than by chance, were also bound by Isw2 (Whitehouse et al., 2007) (Fig. S2F). Using gene ontology (GO) analysis for these Isw2-regulated genes, we found a significant enrichment for stress response pathways (Fig. S2G and Table S2). Examination of the published ChIP-chip dataset (Whitehouse et al., 2007) confirmed that Isw2 localizes to these genes and promoters; response to abiotic stimulus/stress was again the second most significant GO cluster among Isw2-bound genes (Fig. S2H). In summary, stress response genes are the most significant group both bound by Isw2 and derepressed in isw2Δ.
Activation of homologous recombination-based DNA damage repair promotes longevity
We next examined which Isw2-regulated stress response pathway might be crucial for aging. RAD51 was among the most derepressed genes in isw2Δ (Fazzio et al., 2001); genome-wide ChIP-chip showed a specific localization of Isw2 to the RAD51 promoter (Whitehouse et al., 2007). We were able to reproduce these results for RAD51 by RT-qPCR and ChIP-qPCR (Fig. S3A–B).
We then turned to the specific function of Rad51, to unravel the role of Isw2. Rad51 is required for homologous recombination (HR) (Symington, 2002). Although both HR and nonhomologous end joining (NHEJ) are major DNA double-strand break repair mechanisms (Polo and Jackson, 2011), HR, but not NHEJ, has been implicated in replicative lifespan in yeast (Kaeberlein et al., 1999; Park et al., 1999). Hence, we investigated whether the HR-based DNA repair pathway was responsible for the effects of isw2Δ.
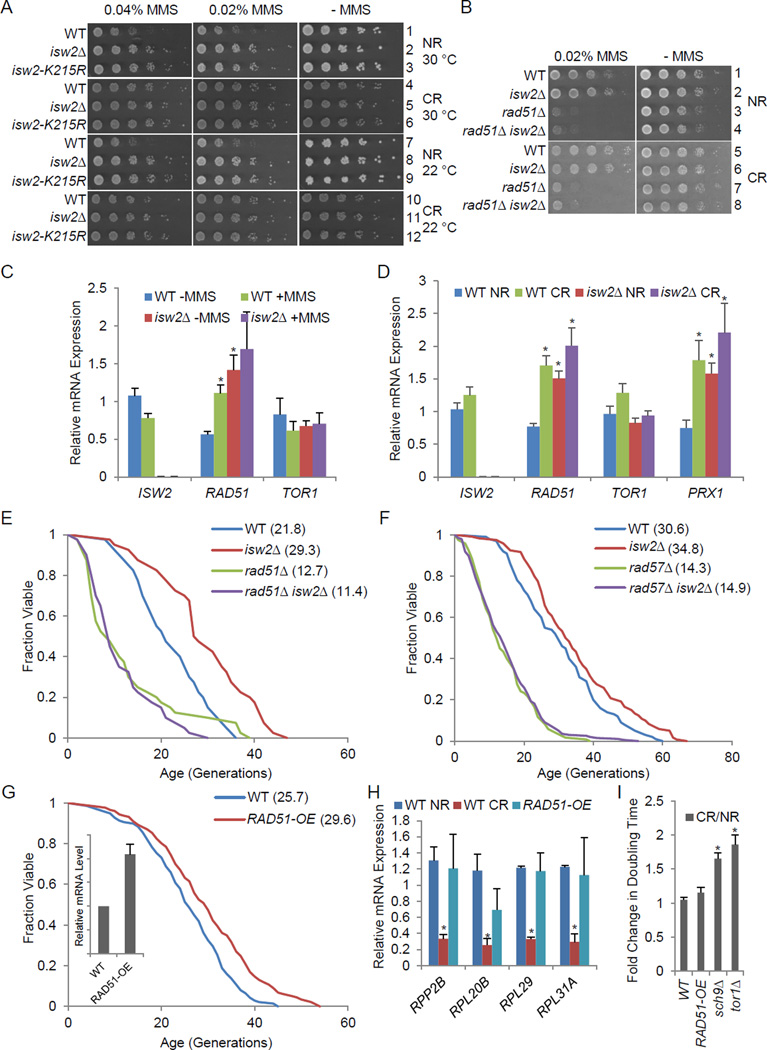
We found that inactivation of ISW2 improved the cellular response to genotoxic stress induced by DNA damaging agents. Specifically, either isw2Δ or a catalytic mutation improved resistance to methyl methanesulfonate (MMS) and camptothecin (CPT) (Fig. 3A rows 1–3 and S3C–D). Interestingly, CR also improved resistance to such stresses, and isw2Δ did not further increase the resistance, again suggesting epistasis between isw2Δ and CR (Fig. 3 A rows 4–6, compare to rows 1–3). Notably, resistance to MMS under CR was not due simply to slow growth since isw2Δ still improved MMS growth when assayed at a lower temperature (Fig. 3 A rows 7–12). Deletion of RAD51 caused hypersensitivity to MMS and CPT; however, additional deletion of ISW2 did not restore resistance to these agents (Fig. 3B and S3D), indicating that Rad51 was required for elevated resistance to genotoxic stress in ISW2 mutants.
Figure 3. Isw2 mediates effects of calorie restriction through homologous recombination (HR)-based DNA repair.
(A–B) Designated strains were 5-fold serial diluted and spotted on SC with 2% glucose (NR) and SC with 0.05% glucose (CR). (C–D) Gene expression by qPCR for WT and isw2Δ cells with or without MMS (C) and in NR and CR conditions (D). * p<0.05 compared to WT NR. (E– F) Replicative lifespan for isw2Δ in the rad51Δ (E) or rad57Δ (F) backgrounds. (G) Replicative lifespan for RAD51 overexpression strain (RAD51-OE). Inset, relative RAD51 expression level. (H) Gene expression analysis by RT-qPCR for selected 60S ribosome subunit genes. NR, SC with 2% glucose; CR, SC with 0.05% glucose. (I) Fold change in doubling time during exponential growth in SC. * p <0.05 compared to WT NR. See also Figures S2–S3 and Tables S1–S2.
To better understand the molecular mechanism underlying the improved genotoxic response, we further investigated RAD51 expression under these conditions. In cells either treated with MMS or deleted for ISW2, RAD51 expression levels were significantly increased (Fig. 3C); little further induction was seen when isw2Δ cells were treated with MMS, suggesting that isw2Δ alone was sufficient to mimic the elevated RAD51 expression induced by genotoxic stress. In contrast, no significant change in TOR1 expression was observed under these conditions (Fig. 3C), further distinguishing these pathways. Since isw2Δ showed epistasis to CR, we then compared the effect of RAD51 induction between isw2Δ cells and cells grown under CR conditions. Again, either CR or isw2Δ significantly induced RAD51 expression, while there was insignificant further induction when growing isw2Δ cells in CR conditions, and, again, there was no significant change of TOR1 expression (Fig. 3D). These observations are consistent with the epistasis between isw2Δ and CR. Since Isw2 regulates a cohort of stress response genes, we examined another gene showing the most upregulation in isw2Δ: the PRX1 gene encodes the mitochondria peroxiredoxin, whose expression is activated by oxidative stress and is derepressed in isw2 by nearly two folds (Fazzio et al., 2001). Similar to RAD51, PRX1 expression was also significantly elevated under CR, to an extent similar to isw2Δ cells (Fig. 3D).
We next examined whether epistasis between isw2Δ and rad51Δ also occurs in the context of aging. As shown previously, lifespan was shortened by deletion of RAD51 (Fig. 3E), and we found that isw2Δ was unable to extend the rad51Δ short lifespan (Fig. 3E), indicating that Rad51 was required for longevity regulated by Isw2. Rad57 is a critical facilitator protein for HR that forms a heterodimer with Rad55 and promotes the assembly of Rad51 at sites of DNA double strand breaks (Symington, 2002). To further verify that it was the HR-based DNA damage repair that was required for the longevity extension by isw2Δ, we tested epistasis between isw2Δ and rad57Δ. Consistent with the rad51Δ results, isw2Δ was not able to extend lifespan in the rad57Δ (Fig. 3F), confirming that isw2Δ required a functional HR pathway to extend lifespan.
Since RAD51 expression was elevated in isw2Δ cells, we next tested the longevity effect of a strain carrying an extra copy of the RAD51 gene integrated in its genome. Overexpression of RAD51 is known to suppress certain mutations in the DNA double strand break repair pathways and does not show deleterious effects in WT cells (Klein, 2008). Strikingly, lifespan was extended 24% by overexpressing RAD51 (Fig. 3G). These data showed that lifespan extension by isw2Δ required RAD51, and that up-regulation of RAD51 alone could promote longevity.
To investigate whether the longevity benefit of RAD51 overexpression was mediated by Tor1/Sch9, we again examined both ribosome gene expression and growth phenotype. We found RAD51 overexpression did not cause a significant decrease in ribosome protein expression, as opposed to the case for CR (Fig. 3H) and for tor1Δ and sch9Δ (Fig. 2G). In addition, RAD51 overexpression did not lead to a slow growth phenotype under CR, again differing from tor1Δ and sch9Δ (Fig. 3I). These observations provided further evidence that the extended lifespan by RAD51-OE was not the result of an altered Tor1/Sch9 signaling pathway.
ISW2 deletion partially mimics CR effects and potentiates stress response in old cells
For several phenotypes described above, isw2Δ showed genetic epistasis to CR, and stress response pathways appeared to be similarly up-regulated by isw2Δ and CR treatment. To obtain a broader view, we compared transcriptome changes between isw2Δ and CR cells. Two biological replicates of RNA-seq experiments were carried out for WT cells grown in synthetic complete (SC) media containing 2% glucose (non-restricted, NR) or 0.05% glucose (calorie-restricted, CR). There was strong agreement between the two replicates, with Pearson correlation coefficients of 0.97 and 0.91 for NR and CR samples, respectively. For all 6437 annotated protein-coding genes with RNA-seq data for all samples, we plotted the log2(CR/NR) data in a histogram, which followed a normal distribution (Fig. S4). Genes that had a change in log2(CR/NR) greater than 1 SD (standard deviation) from the mean were selected for GO analysis, which included 775 up-regulated and 853 down-regulated genes. This 1-SD cutoff represented over 2 fold change in either direction (Table S3).
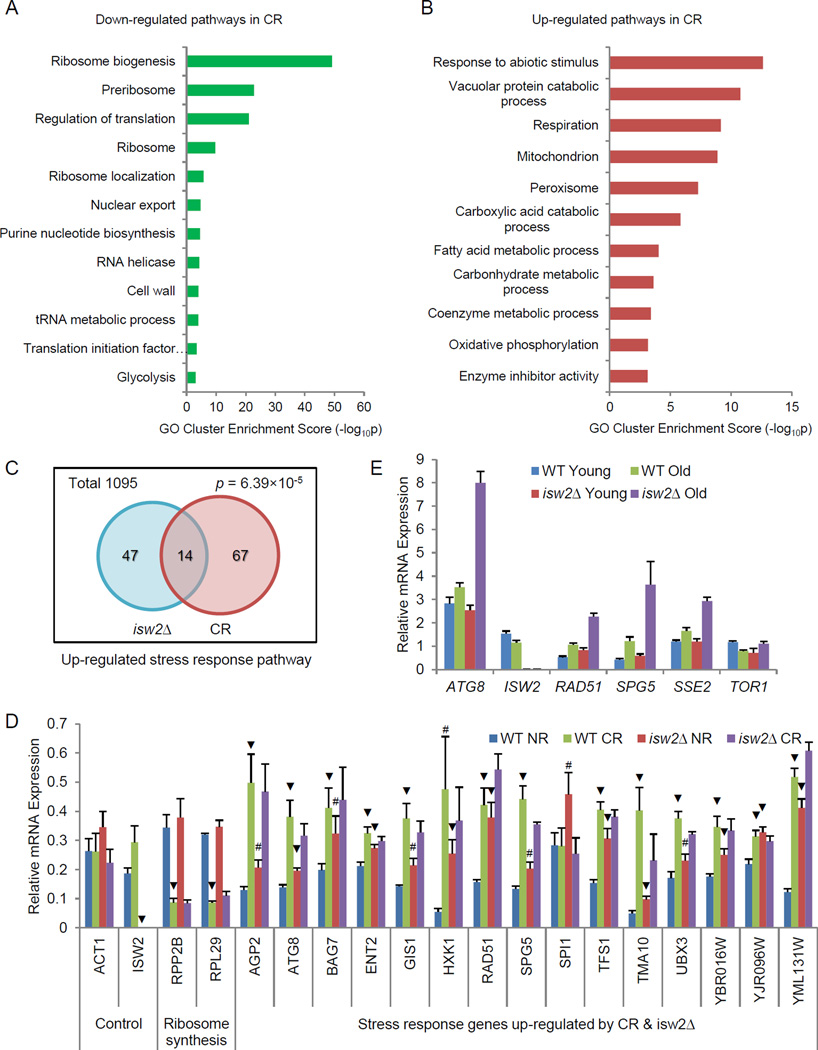
GO clustering analysis for these genes showed characteristic changes expected for calorie restricted cells (Lee and Lee, 2008; Sharma et al., 2011; Steffen et al., 2008; Wang et al., 2010). These changes included down-regulated ribosome biogenesis and protein translation, purine nucleotide biosynthesis, and glycolysis, as well as up-regulated respiration, mitochondria function, and carbohydrate, fatty acid and protein metabolic processes (Fig. 4A – B). Notably, response to abiotic stimulus, which largely overlaps with response to stress, was one of the most significantly enriched functional clusters among up-regulated genes (Fig. 4B).
Figure 4. ISW2 deletion partially mimics calorie restriction and potentiates stress response in old cells.
(A–B) GO clustering analysis for down-regulated (A) and up-regulated (B) genes in CR. GO categories with p <0.001 were included in enrichment score. (C) Venn diagram showing statistically significant overlap between up-regulated stress/abiotic stimulus response pathways in isw2Δ cells (61 genes) and in cells grown in CR conditions (81 genes). (D) Validation of gene expression by qPCR for overlapped genes. # p<0.05, ▼ p <0.01, compared to WT NR. (E) Gene expression for young and old, WT and isw2Δ cells. See also Figure S4 and Tables S3–S4.
Since the response to abiotic stimulus/stress cluster was up-regulated in both isw2Δ cells and CR-treated cells, we compared the lists of genes present in this functional cluster from the two datasets. Among all genes in the stress response cluster, there was a statistically significant overlap (14 genes) between those up-regulated in CR cells (81 genes) and in isw2Δ cells (61 genes) (Fig. 4C and Table S4). These data suggested that isw2Δ cells partially resembled CR-treated cells at the transcriptome level. By RT-qPCR, we validated the transcriptome analysis for a majority of 14 genes elevated in both isw2Δ and CR treated cells (Fig. 4D). In addition, we found that isw2Δ did not alter the expression of ribosome genes, as was the case for CR and tor1Δ (Fig. 4D and Fig. 2G). Taken together, we conclude that isw2Δ and CR similarly increase expression of a cohort of stress response genes, and this pathway is distinct from the Tor1 pathway that regulates protein synthesis.
To further evaluate the contribution of isw2Δ to the stress response specifically associated with aged cells, we compared expression changes by RT-qPCR for selected stress response genes in WT and isw2Δ cells during aging. We found that a number of stress response genes were moderately up-regulated in aged WT cells, compared to young cells (Fig. 4E), consistent with elevated stress levels in old cells. Interestingly, the induction of stress response genes in the aged population was much greater in isw2Δ cells (Fig. 4E). Hence, it is likely that derepression of stress response pathways in young isw2Δ cells establishes a responsive state poised to antagonize age-associated elevation of cellular stress. Hormesis of this nature has been previously proposed with respect to aging (Martins et al., 2011).
Calorie restriction produces a chromatin conformation similar to isw2Δ cells
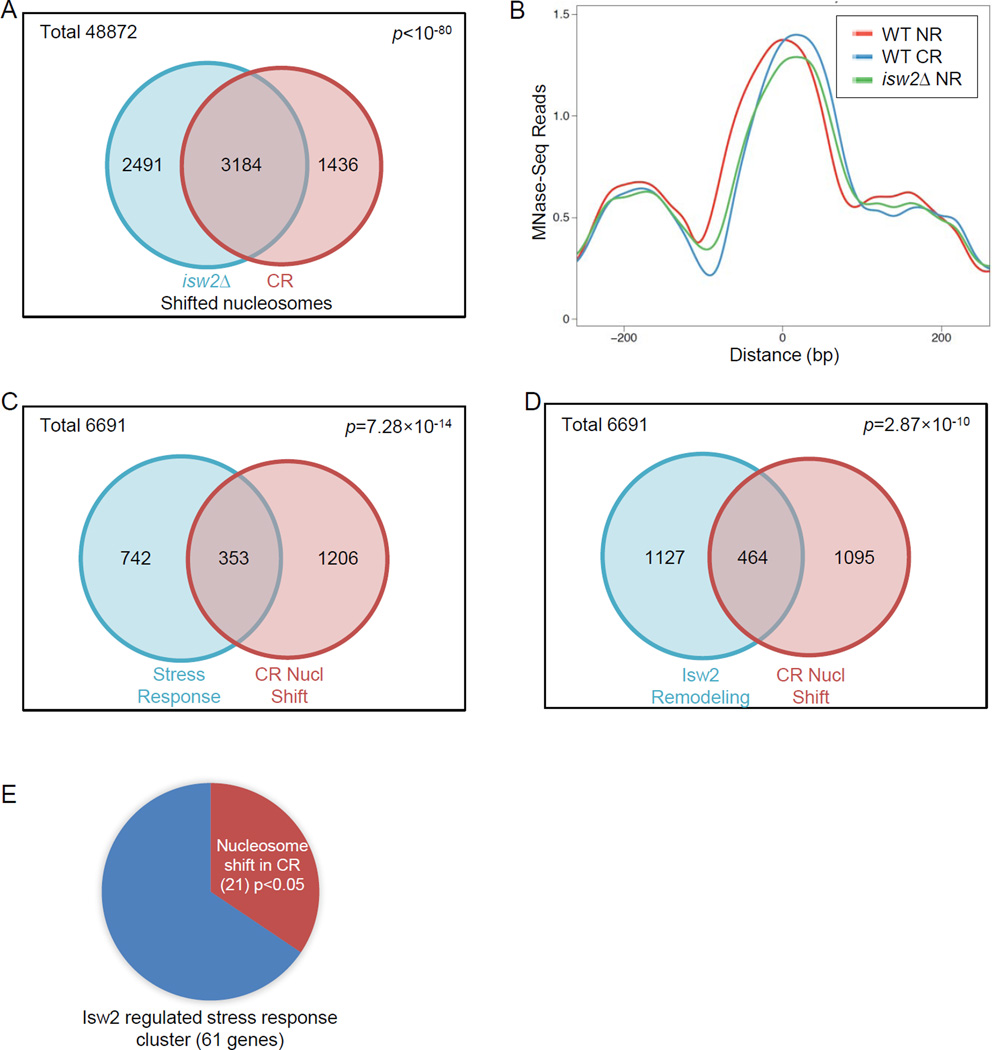
Isw2 alters nucleosome positioning in vivo (Whitehouse and Tsukiyama, 2006). Published datasets showed that isw2Δ caused nucleosome positioning changes at 1187 gene loci across the genome, more than half of which were specifically bound by Isw2 (Whitehouse et al., 2007). Since we found that CR led to transcriptional induction of stress response genes similar to those induced in isw2Δ cells, we then tested whether comparable chromatin conformation changes occur in CR cells and isw2Δ cells. Using a similar method of MNase/ExoIII digestion of chromatin (Whitehouse et al., 2007) followed by next generation sequencing, we found nucleosome positioning shifts (20–100 bp) at 1291 genes in cells grown under CR conditions, compared to NR. Similar shifts were also seen in isw2Δ cells grown under NR conditions. Specifically, of 4,620 nucleosomes showing changed positioning at promoters and ORFs in these 1291 genes in WT CR cells, a significant portion of them, 3,814 nucleosomes also shifted in isw2Δ cells under NR conditions (Fig. 5A). On average, these nucleosomes altered their positions by 40.3 bp (Fig. 5B). GO analysis showed a strong enrichment for stress response genes (Fig. 5C). We then compared genes showing nucleosome shifts in CR with genes containing the previously defined Isw2 remodeling regions (Whitehouse et al 2007), and found a significant overlap (Fig. 5D). To further focus on the 61 stress response genes up-regulated in isw2Δ (Fig. 4C), we found that a significant portion (21 genes, 34%) also showed nucleosome positioning shifts under CR conditions (Fig. 5E). Examples of similarly altered nucleosomes in CR as in isw2Δ NR are shown from (1) stress response genes up-regulated in both CR and isw2Δ cells identified in our study (Fig. S5A), (2) Isw2-regulated genes (Fazzio et al., 2001) (Fig. S5B), and (3) genes showing Isw2-dependent chromatin remodeling (Whitehouse et al., 2007) (Fig. S5C). These observations suggest that CR generated a chromatin conformation partially resembling the alterations caused by isw2Δ, supporting the model that Isw2 is one of the downstream effectors of calorie restriction.
Figure 5. Genome-wide nucleosome mapping for calorie restricted cells shows nucleosome positioning shifts also found in isw2Δ cells.
(A) Venn diagram showing statistically significant shifted promoter and ORF nucleosomes in both isw2Δ NR (SC with 2% glucose) cells and WT CR (SC with 0.05% glucose) cells. (B) Profiles of 3,184 nucleosomes at promoters and ORFs shifted in both WT CR cells and isw2Δ NR cells. (C) Statistically significant enrichment for response to stress/abiotic stimulus GO cluster (left circle; 1095 genes) in genes showing nucleosome positioning shifts in CR (right circle; 1559 genes). (D) Statistically significant overlap between genes showing nucleosome positioning shifts in CR (left circle; 1591 genes) and genes with Isw2 remodeling activities (right circle; 1559 genes). (E) Pie chart showing the fraction of Isw2-regulated stress response genes also bearing shifted nucleosomes in CR. See also Figure S5.
Implications of a conserved longevity mechanism in complex eukaryotes
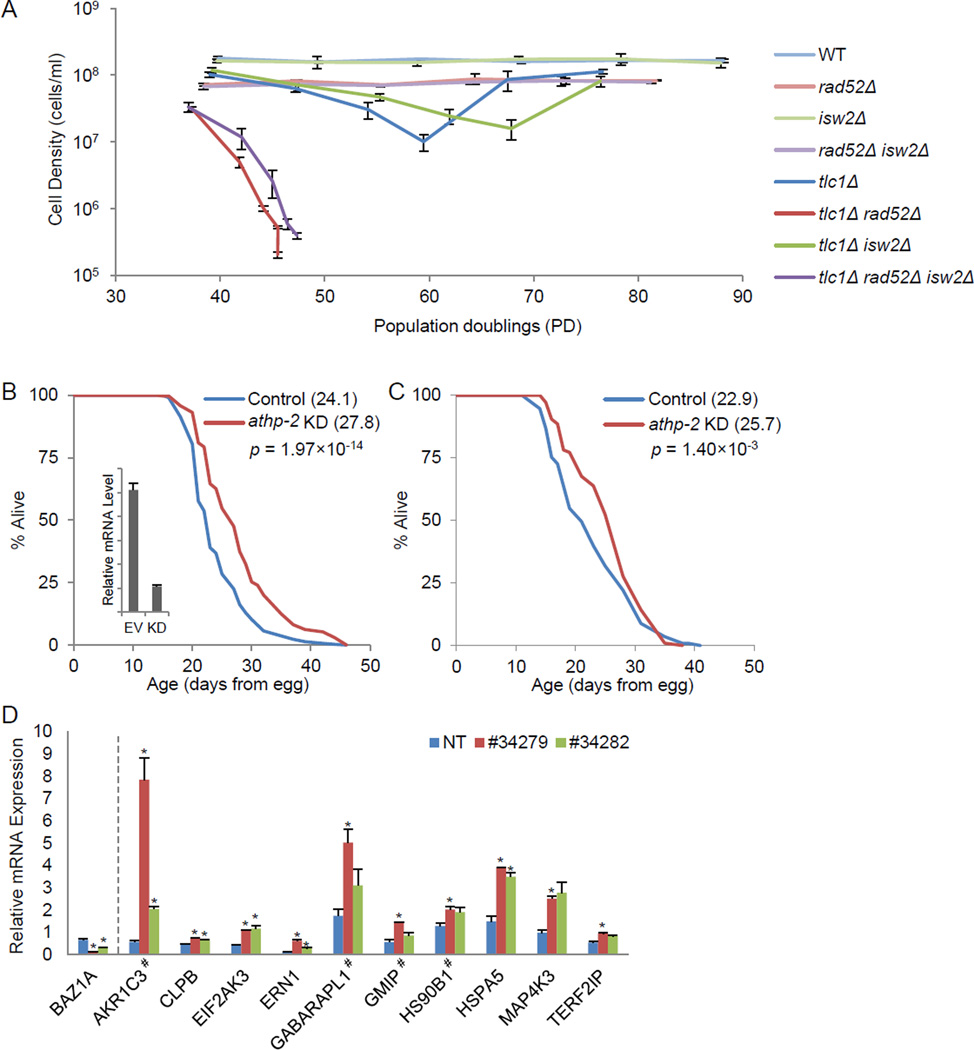
We next investigated the effect of Isw2 on aging and stress response pathways in other eukaryotes. A dramatic difference between yeast replicative aging and human aging is that yeast telomeres do not shorten with replicative age (Laun et al., 2007). However, yeast telomerase mutants (e.g. tlc 1Δ and est2Δ) senesce in response to progressive telomere shortening, providing a model for human cellular senescence (Teixeira, 2013). Hence, we tested whether isw2Δ could delay replicative senescence in tlc1Δ cells. Telomere shortening in tlc 1Δ mutants leads to a crisis involving the death of most cells, followed by emergence of rare survivors of the crisis, which use telomere recombination to re-lengthen chromosome ends (Fig. 6A, dark blue). Intriguingly, isw2Δ delayed replicative senescence in tlc1Δ cells, extending the crisis from 60 PD to 70 PD (Fig. 6A, dark green, also S6A–B). Deletion of RAD52, a gene facilitating Rad51 and required for HR-based DNA repair (Symington, 2002), led to much faster crisis, at 45 PD, as reported previously (Fig. 6A, dark red) (Le et al., 1999) and the more severe crisis in rad52Δ was not significantly extended by isw2Δ (Fig. 6A, dark purple), similar to the findings in the replicative aging model (Fig. 3E). These results indicate that the mechanism of lifespan extension conferred by isw2Δ also has a role in a genetic model where telomere length limits lifespan, and thus suggests possible relevance in mammalian cellular senescence.
Figure 6. Functional conservation in other eukaryotic model systems.
(A) Yeast replicative senescence assay. Cells of the indicated genotypes were sporulated from a TLC1/tlc1Δ ISW2/isw2Δ RAD52/rad52Δ diploid. The mean values for the indicated number of spore products are shown: n=2 for wild-type (WT), rad52Δ, isw2Δ, and isw2Δ rad52Δ; n=6 for tlc 1Δ, tlc 1Δ isw2Δ, and tlc1Δ rad52Δ; n=9 for tlc 1Δ isw2Δ rad52Δ. Significance test: tlc 1Δ isw2Δ vs. tlc1Δ, p = 0.04; rad52Δ tlc 1Δ vs. rad52Δ tlc1Δ isw2Δ, p = 0.37. (B–C) Mean lifespan (in parenthesis) for C. elegans treated with RNAi targeting athp-2 (KD) or empty vector control (EV). Experiments were performed with (B) or without (C) FUdR. Inset in (B) shows knockdown efficiency tested by RT-qPCR. (D) Relative mRNA expression for selected stress response genes in human fibroblast IMR90 (harvested at estimated population doublings of 38–40) treated with lentiviruses carrying either non-targeting control (NT), or shRNA (#34279 and #34282) targeting BAZ1A. * p<0.05, # genes homologous to yeast stress response genes up-regulated in both isw2Δ and CR. See also Figure S6 and Tables S4–S6.
In order to gain more insights into the role of the ISWI class of enzymes in aging of metazoans, we determined how the lifespan of C. elegans was affected by RNAi-mediated knockdown of a key subunit of the putative complex homologous to the yeast ISW2 complex (Fig. 6B, insert). The worm ISWI class gene, isw-1, has been characterized genetically, together with nurf-1, an ortholog of NURF301 in the fruit fly D. melanogaster (Andersen et al., 2006). However, knocking down isw-1 did not affect worm lifespan (Curran et al., 2009). In the yeast ISW2 complex, the auxiliary subunit Itc1 is required for its in vivo function (Gelbart et al., 2001) and is homologous to ACF1 in the Drosophila CHRAC/ACF complex. A search for worm homologs of yeast Itc1 and fruit fly ACF1 led us to the worm gene athp-2 (H20J04.2), which shares a maximum of 42% identity (62% similarity) to ACF1 and 28% identity (47% similarity) to Itc1 in amino acid sequence, with the best E-values of 3× 10−34 and 9× 10−6, respectively. Multiple alignment of worm ATHP-2 with yeast Itc1, fly ACF1, and mammalian BAZ1A confirms their homology (Fig. S6C). We hypothesized that, similar to fruit fly, worm ISW-1 also forms multiple complexes, including an orthologous NURF complex with NURF-1 and an ACF/CHRAC complex with ATHP-2. We therefore focused on the specific orthologous complex, and knocked down athp-2 with the RNAi clone H20J04.f (Fig. 6B, insert shows mRNA levels were reduced compared to control). We found that athp-2 knockdown significantly extended worm lifespan by 15% (Fig. 6B), when the DNA synthesis inhibitor FUdR (5-fluorodeoxyuridine) was used in the worm lifespan assay to prevent the growth of progeny (Gandhi et al., 1980). Since FUdR has documented effects on lifespan for certain mutants (Aitlhadj and Stürzenbaum, 2010), we performed worm lifespan assays without FUdR, and observed consistent lifespan extension (Fig. 6C). These results suggested that the function of ISW2/ACF/CHRAC in aging regulation might be evolutionarily conserved.
To investigate possible functional conservation of the ISW2/ACF/CHRAC orthologous complex in a mammalian system in repressing stress response genes, we knocked down the human Itc1/ACF1 ortholog BAZ1A in IMR90 cells (primary fetal lung fibroblasts) by lentivirus based shRNA (Fig. 6D, left side). Using two validated shRNAs, #34279 and #34282, compared to a non-targeting control (NT), we tested expression changes for a list of 23 stress response genes. These genes included 10 human homologs of yeast stress response genes up-regulated in both isw2Δ and CR (Table S4), as well as 13 stress response genes whose promoters are bound by the catalytic subunit SMARCA5 (hSNF2H). These 13 genes were selected from a significantly enriched stress response GO cluster consisting of 54 genes from a GO analysis for all gene promoters bound by SMARCA5 (Table S6). We found that 4 out of 10 human homologs of yeast stress response genes (Fig. 6C marked with #, S6D and Table S4), as well as 6 out of the 13 SMARCA5-bound stress response genes, showed significantly elevated expression in the BAZ1A knock-down cells compared to the control, as measured by RT-qPCR (Fig. 6C, S6E, and Table S6). Overall, among the 23 tested genes, 10 were significantly activated upon BAZ1A knockdown, corresponding to a false discovery rate (FDR) of 0.115. This observation provides further evidence for a possible conserved role of the Isw2 orthologous complex in repressing stress response pathways.
Discussion
Using the yeast replicative aging model, we demonstrate a novel longevity regulation mechanism mediated by the ATP-dependent chromatin remodeling enzyme Isw2. Deletion or enzymatic inactivation of Isw2 extends lifespan. This longevity effect is the result of derepressing a cohort of stress response genes, in particular, RAD51 in the HR-mediated DNA damage repair pathway. Further, we find that these changes in transcriptome partially mimic the stress response state of calorie restricted cells.
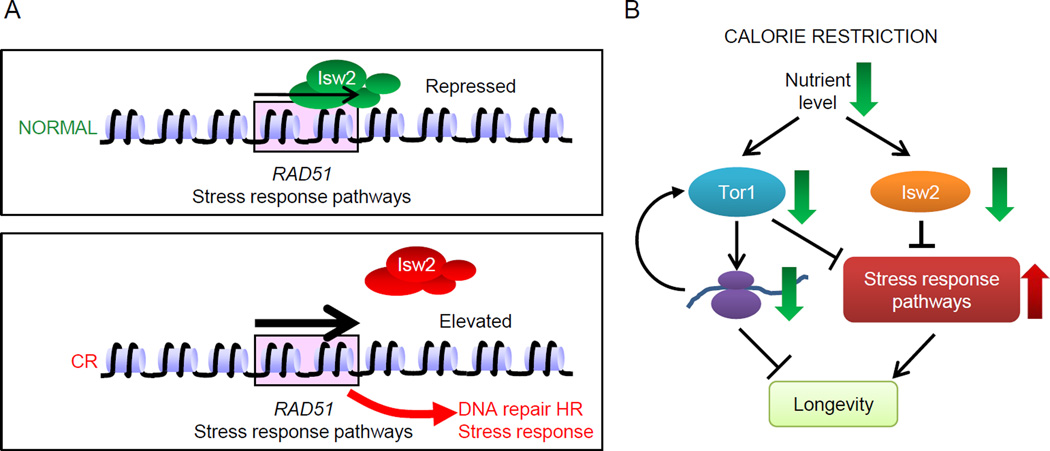
Our findings provide one of the first lines of evidence that ATP-dependent chromatin remodeling complexes play direct roles in aging regulation. Indeed, as this manuscript was in preparation, Riedel et al. showed that the SWI-SNF complex acts as a transcription cofactor for DAF-16/FOXO in C. elegans and is essential for DAF-16 mediated stress response and longevity (Riedel et al., 2013). Here, we demonstrate that the distinct ISWI subfamily of the ATP-dependent remodeling enzymes negatively regulates longevity through stress response pathways and contributes to the longevity effect of calorie restriction (Fig. 7A–B). In addition, we also provide evidence that this aging modulation mechanism might be evolutionarily conserved among eukaryotes. Since deletion of ISW2, or inhibition of enzymatic activity, provided longevity benefits, Isw2 could potentially be further explored as a pharmaceutical target for clinical applications.
Figure 7. Models of the Isw2 regulated pathway during calorie restriction.
(A) Effect of ISW2 complex on its stress response gene targets in non-restricted (upper) and in calorie-restricted cells (lower). We found that Isw2 represses a cohort of stress response genes, including RAD51, which are induced either by isw2Δ or CR. (B) During CR, reduced nutrient levels are signaled to TOR-signaling pathway and the Isw2 regulated pathway, resulting in down regulation of both pathways. Decreased TOR-signaling results in reduced ribosome biosysthesis and protein translation. Down-regulated Isw2 causes derepression of stress response genes, including RAD51, as shown in 7A. Both reduced protein synthesis and elevated stress response promote longevity.
Inactivation of Isw2 potentiates stress response during aging
After ruling out a number of possible Isw2 functions that may extend lifespan when removed, we found that Isw2 moderately represses a cohort of stress response genes (Fig. S2F–G). Thus, upon inactivation of Isw2, these genes become activated, rendering cells in a constitutive moderately stress-responsive state (Fig. 7A, lower panel). Pre-exposing cells to mild stress states has been shown to be beneficial for surviving future stronger stresses, such as heat shock, osmotic, oxidative, and genotoxic stresses, which is a phenomenon known as hormesis (Rattan, 2008). Our findings are consistent with the idea that reduction of Isw2 activity induces an elevated stress response state to mimic a hormetic effect, providing increased resistance to more severe genotoxic stresses during aging (Fig. 4E).
We show in this study that isw2Δ increases resistance to genotoxic stresses by activation of the HR-based DNA repair mechanism (Fig. 3 and S3), which in turn promotes longevity (Fig. 7B). In addition, Isw2 represses a number of other stress response pathways, including oxidative stress, heat shock, osmotic stress, autophagy, etc (Table S2). However, no phenotype was obvious for these stress conditions (data not shown); thus, either the appropriate test condition was not found, or the elevated response for other stress pathways by isw2Δ was subtle. Indeed, the increased resistance to MMS and CPT by isw2Δ was modest (Fig. 3A – B and S3C–D), providing a potential explanation as to why the phenotype was not observed in previous tests (Vincent et al., 2008). Hence, it appears that loss of Isw2 results in slight constitutive elevation of numerous stress pathways to extend lifespan.
Robust cellular stress response capability has been associated with longevity. An excellent example is the naked mole rat, which lives for nearly 30 years in captivity and is the longest-lived rodent. Fibroblasts obtained from these animals show much stronger resistance to various stressors compared to mouse (Lewis et al., 2012). In support of this paradigm, our results show that isw2Δ extends yeast replicative lifespan by activating cellular stress response pathways, especially HR-based DNA damage repair. Furthermore, old isw2Δ cells exhibit much higher expression of stress response genes when compared to old WT cells (Fig. 4E). This suggests that their derepression resulting from inactivation of Isw2 can lead to a more potent response to stress when needed during aging, hence conferring longevity benefits.
Isw2 regulates stress response pathways exploited by CR
Down-regulation of the TOR signaling pathway by CR through Tor1 and Sch9, and the downstream ribosome biogenesis has been extensively studied (Fig. 7B) (Steffen et al., 2008). In fact, yeast has evolved a sophisticated ribosome biogenesis regulation system called ribi regulon that precisely adjusts ribosome abundance to availability of nutrients (Shore et al., 2009). Thus, TOR signaling plays a critical role in mediating the effects of CR. One important experiment highlighting this mechanistic pathway is the epistasis of tor1Δ to CR, that is, tor1Δ cells under CR conditions show the same lengthened lifespan as either tor1Δ or CR alone (Kaeberlein, 2010). Our study shows that isw2Δ displays a similar relationship to CR (Fig. 2A–D), suggesting that Isw2 regulates an aging pathway downstream of CR.
We also show that isw2Δ and tor1Δ have a combinatorial effect on lifespan extension (Fig. 2E), suggesting parallel pathways. This would seem to be conflicting with the epistasis of either tor1Δ or isw2Δ with CR. To explain this, we hypothesize that tor1Δ and isw2Δ phenocopy two separate events, each promoted by CR, and that CR may exert an effect from each pathway to a lesser extent compared to the gene deletion. Thus both pathways act downstream of CR to mediate lifespan extension in yeast. In addition, although tor1Δ and isw2Δ each mimic CR, the presence of both TOR1 and ISW2 genes may be important to realize the benefits of CR. Hence, deletion of either one blocks the CR longevity effect.
It has been well established that CR provides cellular protection and stress resistance (Lee and Longo, 2011). Since isw2Δ showed a partial overlap with CR-regulated stress response, we compared the correlation at the chromatin level and found similar nucleosome positioning shifts between isw2Δ and CR for a gene set that is also enriched for stress response (Fig. 5A–C), a significant fraction of which appear to be attributable to changes in Isw2 activity under CR conditions (Fig. 5D). However, not all Isw2-regulated genes showed similar nucleosome positioning shifts under CR (Fig. S5), which is likely due to additional transcription regulation for Isw2 target genes. Indeed, isw2Δ alone has a very modest effect on gene transcription in general and other transcription repressors, such as Rpd3 and Fkh2, are known to work in parallel with Isw2 (Fazzio et al., 2001; Sherriff et al., 2007) and to regulate stress responses (Postnikoff et al., 2012; Ruiz-Roig et al., 2010). However, neither rpd3Δ nor fkh2Δ extended replicative lifespan (data not shown), suggesting that Isw2 is important in conferring the longevity effect. Overall, our evidence supports the model that regulation of stress response by Isw2 is one of the downstream effects of CR that contributes to longevity.
Implications to aging in higher eukaryotes
Yeast Isw2 and the ISWI class of chromatin remodeling ATPases are evolutionarily conserved. Enzyme complexes homologous to ISW2 have been characterized in plants, worms, flies, mice, and humans. However, the degree of diversity and complexity associated with this class of enzymes also increases significantly, hence, genetic and longevity investigations in these more complex organisms are difficult. We sought to use several eukaryotic model systems to test functional conservation of Isw2 orthologous enzyme complexes.
The nematode C. elegans is a popular animal model for aging reseerch due to its short lifespan, easy lifespan determination, and the availability of RNAi libraries. Studies in worm aging have led to discoveries of many aging regulation and CR pathways, such as insulin/IGF signaling (IIS), autophagy, mitochondria, and histone methylation (Tissenbaum, 2012). Thus, we tested the worm aging phenotype. The observed lifespan extension by knocking down the Itc1/ACF1 ortholog athp-2 suggests that the ISW2/ACF/CHRAC complex may function in a conserved mechanism to regulate aging in eukaryotes (Fig. 6B).
Although mammalian cellular senescence has long been thought to be an cancer suppression mechanism, recent evidence also links senescence to aging (Campisi, 2013). Therefore, it is intriguing to test whether elevated stress responses in isw2Δ cells can mitigate the stress associated with cellular senescence. The yeast tlc1Δ telomerase mutant is valuable as a cellular senescence model to initially test this idea. Our data shows that isw2Δ delays replicative senescence, and is also dependent on HR (Fig. 6A). This result suggests that the elevated stress response in isw2Δ antagonizes senescence caused by telomere shortening, leading to the hypothesis that a similar pathway might function in mammalian senescence and aging.
Homozygous knockout of mammalian SMARCA5 (SNF2H), the closest homolog to yeast Isw2, is embryonic lethal (Skoultchi, PNAS, 2003). Adding to this complexity, SMARCA5 is a component of, at minimum, five distinct complexes. Hence, we instead focused on the BAZ1A gene, which encodes a subunit orthologous to yeast Itc1 and fruit fly ACF1, and knocked down expression in human primary lung fibroblasts IMR90 in their early passages. The moderate increase in several stress response genes upon BAZ1A knockdown (Fig. 6D) supports the hypothesis of a conserved mechanism of stress response regulation by ISW2/ACF/CHRAC chromatin remodeling enzyme complexes. Future studies will establish whether this pathway may be one of the anti-aging mechanisms in mammals that can be exploited by dietary restriction and, potentially, clinical intervention in age-associated human diseases.
Experimental Procedures
Yeast strains and media
All yeast mutant strains were derived from BY4741/2, except as noted. Strains YWD781/2 were made by integrating plasmid pWD210, containing the RAD51 gene and flanking sequences. See Table S7 for strain details. Standard Synthetic Complete (SC) solid or liquid media were used in yeast experiments except as noted.
Yeast replicative and senescent lifespan
Assays were performed as described (Kaeberlein et al., 1999; Kozak et al., 2010).
Yeast phenotype assay
Equal amounts of cells were serially-diluted, spotted on SC agar media with 2% (NR) or 0.05% (CR) D-glucose and indicated compounds, and incubated at 30°C or 22°C for 2–5 days.
Isolation of young and old cells
Young and old cells were purified as described (Dang et al., 2009) with the following changes: SC media with 2% (NR) or 0.05% (CR) D-glucose were used; cell density was maintained within logarithmic growth phase (OD600 <2.0 for NR and <0.2 for CR); bud scars were stained with WGA Alexa Fluor 488 (Life Technologies).
Worm lifespan assay with athp-2 RNAi knockdown
RNAi feeding bacteria were obtained from the Ahringer RNAi library and verified by sequencing. C. elegans strain N2 was used in lifespan experiments conducted at 20°C as described (Sutphin, G. L., Kaeberlein, 2009). Worms prepared in parallel were harvested for RNA analysis. In experiments without FUdR, adult animals were transferred to fresh RNAi plates at each scoring when eggs or larvae were present.
IMR90 culturing and lentivirus knockdown
IMR90 cells were cultured in standard conditions with 3% O2. Control (non-mammalian target) and knockdown (TRC #34279 and #34282 targeting human BAZ1A) shRNA lentiviruses were produced by standard methods. Infected cells were selected with puromycin and harvested after two passages.
RNA extraction, RT-PCR and quantitative real-time PCR analysis
Yeast cells were lysed in QIAzol (QIAGEN) by beadbeating with 0.5 mm dia. zirconia/silica beads (BioSpec) four cycles (1-min beating, 2-min pause on ice). Total RNA was purified with miRNeasy Mini Kit (QIAGEN). Worm RNA was prepared similarly, except using 1.0 mm dia. beads. IMR90 cell pellets were lysed with QIAshredder (QIAGEN) and purified with RNeasy Mini Kit (QIAGEN). TaqMan Reverse Transcription Reagents (Life Technologies) were used to synthesize cDNA from 1 µg purified RNA with random hexamers. Relative changes in mRNA levels were determined by quantitative real-time PCR. Yeast gene expression data were normalized to ACT1; worm data were normalized to the ribosomal RNA rrn-1.1; human data were normalized to the average of β-actin and GAPDH. See real-time PCR primers in Table S8.
Ribosomal DNA copy number
Yeast genomic DNA was extracted by standard methods and diluted 10 fold before realtime PCR. Relative copy number was estimated by normalizing to an intergenic region in chromosome V.
Preparation of RNA-seq and nucleosome-seq libraries and next-gen sequencing
Poly-adenylated RNA was enriched from 10 µg purified total RNA with Poly(A)Purist Kit (Life Technologies) and treated with Fragmentation Reagents (Life Technologies). Strand-specific RNA-seq libraries were prepared with Small RNA Sample Preparation Kit (Illumina). Sequencing was done in 50-bp single-end format on Illumina HiSeq 2000.
For nucleosome-seq, chromatin was digested with MNase and ExoIII as described (Whitehouse et al., 2007) and mononucleosome-sized DNA was purified by agarose gel electrophoresis. Sequencing libraries were prepared with NEBNext DNA Library Prep Master Mix Set for Illumina (NEB) and Multiplexing Sample Preparation Oligonucleotide Kit (Illumina). Sequencing was done in 100-bp paired-end format on Illumina HiSeq 2000.
Bioinformatics and Data Analysis
Sequencing read mapping and quantification of gene expression in FPKM by RNA-seq was performed with Bowtie and Cufflink, respectively, with default settings. Subsequent RNA-seq analysis was based on the geometric mean of FPKM from two biological replicates, and included genes that scored in both replicates.
An upper and lower 1-SD cutoff was used to generate gene lists for GO analysis by DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/). The Functional Annotation Clustering option was used to group similar categories. An enrichment score for each cluster was given as a mean of -log10p of member GO categories with p < 0.01 or 0.001.Nucleosome positions were called from the nucleosome-seq data using NucPosSimulator (Schöpflin et al., 2013). Nucleosomes were called that significantly shifted positions in WT_CR from WT_NR (20 to 100 bps). The average shift of nucleosomes was calculated over the promoter and gene body regions. Metanucleosome analysis included only shifts found in both WT_CR and isw2Δ.
Fisher’s exact test was used to assess the null hypothesis, i.e. that the proportion of genes in common for two independent observations is less than or equal to random chance. Yeast replicative lifespan and worm lifespan p-values were determined by the Wilcoxon rank-sum test. Statistical significance p-values for non-high throughput data and non-lifespan data were calculated by unpaired two-tailed student’s t-test. All error bars are standard error of the mean.
RNA-seq and nucleosome-Seq data in this study can be access at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53721.
Supplementary Material
Highlights.
ISW2 deletion extends yeast replicative lifespan and is epistatic to CR.
Isw2 regulates lifespan through stress response pathways.
ISW2 deletion partially mimics CR effects on stress response pathways.
Isw2-mediated longevity is TOR-independent and likely conserved in eukaryotes.
Acknowledgements
We thank Dr. Matteo Cesaroni in the RS lab and Dr. Wei Liu in the WD lab for technical assistance. We also thank members of the BKK and MK labs who participated in yeast lifespan experiments. University of Pennsylvania PGFI provided Next-Gen Sequencing service. The Wistar Institute Molecular Screening Facility provided shRNA lentivirus. This work was supported by NIH grants (P01AG031862 to SLB, FBJ; K99AG037646 to WD; R01AG039390 to MK; R01AG043080 to BKK; and T32AG000057 for GLS), and an Ellison Medical Foundation grant (SLB).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aitlhadj L, Stürzenbaum SR. The use of FUdR can cause prolonged longevity in mutant nematodes. Mech. Ageing Dev. 2010;131:364–365. doi: 10.1016/j.mad.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Andersen EC, Lu X, Horvitz HR. C. elegans ISWI and NURF301 antagonize an Rb-like pathway in the determination of multiple cell fates. Development. 2006;133:2695–2704. doi: 10.1242/dev.02444. [DOI] [PubMed] [Google Scholar]
- Bachman N, Gelbart ME, Tsukiyama T, Boeke JD. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev. 2005;19:955–964. doi: 10.1101/gad.1299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Corona DFV, Tamkun JW. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim. Biophys. Acta. 2004;1677:113–119. doi: 10.1016/j.bbaexp.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature. 2009;459:1079–1084. doi: 10.1038/nature08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JR, Sutphin GL, Dulken B, Sim S, Kim JR, Robison B, Schleit J, Murakami CJ, Carr D, An EH, et al. Sir2 deletion prevents lifespan extension in 32 long-lived mutants. Aging Cell. 2011a;10:1089–1091. doi: 10.1111/j.1474-9726.2011.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JR, Murakami CJ, Olsen B, Kennedy BK, Kaeberlein M. Quantitative evidence for early life fitness defects from 32 longevity-associated alleles in yeast. Cell Cycle. 2011b;10:156–165. doi: 10.4161/cc.10.1.14457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JR, Chou A, Olsen B, Carr D, Murakami C, Ahmed U, Sim S, An EH, Castanza AS, Fletcher M, et al. End-of-life cell cycle arrest contributes to stochasticity of yeast replicative aging. 2013 doi: 10.1111/1567-1364.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 2001;21:6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Tyler J. Chromatin structure as a mediator of aging. FEBS Lett. 2011;585:2041–2048. doi: 10.1016/j.febslet.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S, Santelli J, Mitchell DH, Stiles JW, Sanadi DR. A simple method for maintaining large, aging populations of Caenorhabditis elegans. Mech. Ageing Dev. 1980;12:137–150. doi: 10.1016/0047-6374(80)90090-1. [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 2001;21:2098. doi: 10.1128/MCB.21.6.2098-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19:942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Mao Z, Hine C. Changes in DNA repair during aging. Nucleic Acids Res. 2007;35:7466–7474. doi: 10.1093/nar/gkm756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Mitochondria--a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota SK, Bartholomew B. Diversity of operation in ATP-dependent chromatin remodelers. Biochim. Biophys. Acta. 2011;1809:476–487. doi: 10.1016/j.bbagrm.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T, Araki H. Noncompetitive counteractions of DNA polymerase ε and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:217–227. doi: 10.1128/MCB.24.1.217-227.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. 2013 doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Rupeš I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech. Ageing Dev. 2005a;126:491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005b;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 2008;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res. Rev. 2011;10:205–215. doi: 10.1016/j.arr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N, Tavernarakis N. Cellular stress response pathways and ageing: intricate molecular relationships. EMBO J. 2011;30:2520–2531. doi: 10.1038/emboj.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak ML, Chavez A, Dang W, Berger SL, Ashok A, Guo X, Johnson FB. Inactivation of the Sas2 histone acetyltransferase delays senescence driven by telomere dysfunction. EMBO J. 2010;29:158–170. doi: 10.1038/emboj.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis GN, Tower J. Superoxide dismutase evolution and life span regulation. Mech. Ageing Dev. 2005;126:365–379. doi: 10.1016/j.mad.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Langerak P, Russell P. Regulatory networks integrating cell cycle control with DNA damage checkpoints and double-strand break repair. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2011;366:3562–3571. doi: 10.1098/rstb.2011.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laun P, Bruschi CV, Dickinson JR, Rinnerthaler M, Heeren G, Schwimbersky R, Rid R, Breitenbach M. Yeast mother cell-specific ageing, genetic (in)stability, and the somatic mutation theory of ageing. Nucleic Acids Res. 2007;35:7514–7526. doi: 10.1093/nar/gkm919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S, Moore JK, Haber JE, Greider CW. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Longo VD. Fasting vs dietary restriction in cellular protection and cancer treatment: from model organisms to patients. Oncogene. 2011;30:3305–3316. doi: 10.1038/onc.2011.91. [DOI] [PubMed] [Google Scholar]
- Lee Y, Lee C. Transcriptional response according to strength of calorie restriction in Saccharomyces cerevisiae. Mol. Cells. 2008;26:299–307. [PubMed] [Google Scholar]
- Lewis KN, Mele J, Hornsby PJ, Buffenstein R. Stress resistance in the naked mole-rat: the bare essentials - a mini-review. Gerontology. 2012;58:453–462. doi: 10.1159/000335966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- Martins I, Galluzzi L, Kroemer G. Hormesis, cell death and aging. Aging (Albany. NY) 2011;3:821–828. doi: 10.18632/aging.100380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin M, Yang J, Hanzén S, Toledano MB, Labarre J, Nyström T. Life span extension and H(2)O(2) resistance elicited by caloric restriction require the peroxiredoxin Tsa1 in Saccharomyces cerevisiae. Mol. Cell. 2011;43:823–833. doi: 10.1016/j.molcel.2011.07.027. [DOI] [PubMed] [Google Scholar]
- Park PU, Defossez PA, Guarente L. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:3848–3856. doi: 10.1128/mcb.19.5.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postnikoff SDL, Malo ME, Wong B, Harkness TAA. The yeast forkhead transcription factors fkh1 and fkh2 regulate lifespan and stress response together with the anaphase-promoting complex. PLoS Genet. 2012;8:e1002583. doi: 10.1371/journal.pgen.1002583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattan SIS. Hormesis in aging. Ageing Res. Rev. 2008;7:63–78. doi: 10.1016/j.arr.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Riedel CG, Dowen RH, Lourenco GF, Kirienko NV, Heimbucher T, West JA, Bowman SK, Kingston RE, Dillin A, Asara JM, et al. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 2013;15:491–501. doi: 10.1038/ncb2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Schmeisser S. Extending life span by increasing oxidative stress. Free Radic. Biol. Med. 2011;51:327–336. doi: 10.1016/j.freeradbiomed.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Ruiz-Roig C, Viéitez C, Posas F, de Nadal E. The Rpd3L HDAC complex is essential for the heat stress response in yeast. Mol. Microbiol. 2010;76:1049–1062. doi: 10.1111/j.1365-2958.2010.07167.x. [DOI] [PubMed] [Google Scholar]
- Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöpflin R, Teif VB, Müller O, Weinberg C, Rippe K, Wedemann G. Modeling nucleosome position distributions from experimental nucleosome positioning maps. Bioinformatics. 2013;29:2380–2386. doi: 10.1093/bioinformatics/btt404. [DOI] [PubMed] [Google Scholar]
- Sharma PK, Agrawal V, Roy N. Mitochondria-mediated hormetic response in life span extension of calorie-restricted Saccharomyces cerevisiae. Age (Dordr) 2011;33:143–154. doi: 10.1007/s11357-010-9169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherriff JA, Kent NA, Mellor J. The Isw2 chromatin-remodeling ATPase cooperates with the Fkh2 transcription factor to repress transcription of the B-type cyclin gene CLB2. Mol. Cell. Biol. 2007;27:2848–2860. doi: 10.1128/MCB.01798-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D, Lempia H, Lempiäinen H. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Sims JK, Wade PA. SnapShot: Chromatin remodeling: CHD. Cell. 2011;144:626.e1–626.e1. doi: 10.1016/j.cell.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutphin GL, Kaeberlein M. Measuring Caenorhabditis elegans Life Span on Solid Media. J. Vis. Exp. 2009;27:e1152. doi: 10.3791/1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 Epistasis Group Genes in Homologous Recombination and Double-Strand Break Repair Role of RAD52 Epistasis Group Genes in Homologous Recombination and Double-Strand Break Repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT. Saccharomyces cerevisiae as a Model to Study Replicative Senescence Triggered by Telomere Shortening. Front. Oncol. 2013;3:101. doi: 10.3389/fonc.2013.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA. Genetics, life span, health span, and the aging process in Caenorhabditis elegans. J. Gerontol. A. Biol. Sci. Med. Sci. 2012;67:503–510. doi: 10.1093/gerona/gls088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat. Struct. Mol. Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jiang JC, Jazwinski SM. Gene regulatory changes in yeast during life extension by nutrient limitation. Exp. Gerontol. 2010;45:621–631. doi: 10.1016/j.exger.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat. Struct. Mol. Biol. 2006;13:633 –640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Wu J. Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta Biochim. Biophys. Sin. (Shanghai) 2012;44:54–69. doi: 10.1093/abbs/gmr099. [DOI] [PubMed] [Google Scholar]
- Yadon AN, Tsukiyama T. SnapShot: Chromatin remodeling: ISWI. Cell. 2011;144:453.e1–453.e1. doi: 10.1016/j.cell.2011.01.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.