SUMMARY
MEDEA (MEA) is an Arabidopsis Polycomb group gene that is imprinted in the endosperm. The maternal allele is expressed and the paternal allele is silent. MEA is controlled by DEMETER (DME), a DNA glycosylase required to activate MEA expression, and METHYLTRANSFERASE I (MET1), which maintains CG methylation at the MEA locus. Here we show that DME is responsible for endosperm maternal-allele-specific hypomethylation at the MEA gene. DME can excise 5-methylcytosine in vitro and when expressed in E. coli. Abasic sites opposite 5-methylcytosine inhibit DME activity and might prevent DME from generating double-stranded DNA breaks. Unexpectedly, paternal-allele silencing is not controlled by DNA methylation. Rather, Polycomb group proteins that are expressed from the maternal genome, including MEA, control paternal MEA silencing. Thus, DME establishes MEA imprinting by removing 5-methylcytosine to activate the maternal allele. MEA imprinting is subsequently maintained in the endosperm by maternal MEA silencing the paternal allele.
INTRODUCTION
Alleles of imprinted genes are expressed differently depending on whether they are inherited from the male or female parent. Imprinting regulates a number of genes essential for normal development in mammals and angiosperms. In mammals, imprinted genes contribute to the control of fetal growth and placental development (Constancia et al., 2004). Human diseases are linked to mutations in imprinted genes or aberrant regulation of their expression (Constancia et al., 2004). Mechanisms of distinguishing maternal and paternal alleles have been extensively characterized in mammals. Imprinted genes reside in chromosomal clusters and are regulated by differentially methylated imprinting control regions (ICRs) (Reik and Walter, 2001). Differential DNA methylation is established during oogenesis or spermatogenesis by de novo methyltransferases and maintained somatically by the CG maintenance methyltransferase Dnmt1 (Li, 2002). ICRs are subject to differential histone modifications and in some instances can act as chromatin boundaries (Delaval and Feil, 2004). Other mechanisms to regulate allele-specific gene expression involve noncoding RNAs, including anti-sense transcripts and microRNAs (O’Neill, 2005). Polycomb group (PcG) proteins, which function in large complexes to methylate histones and modify chromatin (Cao and Zhang, 2004), maintain allele-specific silencing of some imprinted genes (Delaval and Feil, 2004).
The endosperm, one of the products of angiospermdouble fertilization, is an important site of imprinting in plants (Gehring et al., 2004) and has functions analogous to the placenta. In flowering plants, meiosis followed by mitosis produces the female and male gametophytes. Two cells of the female gametophyte, the haploid egg and the diploid central cell, are fertilized by two haploid sperm from the male gametophyte to form the diploid embryo and triploid endosperm, respectively. The endospermprovides nutrients to the embryo during seed development and, in Arabidopsis, is almost entirely consumed by the time embryo maturation is completed.
Molecular events that take place in the female gametophyte before fertilization have an essential role in endosperm gene imprinting. The imprinting of two genes, MEA and FWA, is regulated by DME, a helix-hairpin-helix DNA glycosylase (Choi et al., 2002; Kinoshita et al., 2004). DNA glycosylases function in the base-excision repair pathway by removing damaged or mismatched bases from DNA (Scharer and Jiricny, 2001). Bifunctional helix-hairpin-helix DNA glycosylases have both DNA glycosylase and apurinic/apyrimidinic (AP) lyase activities. The DNA glycosylase activity removes the damaged or mispaired base by cleaving the N-glycosylic bond, creating an abasic site, whereas the lyase activity nicks the DNA. An AP endonuclease generates a 3′-hydroxyl used by a DNA repair polymerase that inserts the proper nucleotide. A DNA ligase seals the nick to complete the repair process. DNA glycosylase/lyases have not been implicated in mammalian imprinting mechanisms.
Both MEA and FWA are expressed in the central cell before fertilization and in the endosperm, from the maternal allele, after fertilization (Kinoshita et al., 1999, 2004; Vielle-Calzada et al., 1999). In contrast, DME is expressed in the central cell of the female gametophyte but not in the endosperm (Choi et al., 2002). Expression of MEA and FWA in the central cell and early endosperm is dependent on DME (Choi et al., 2002; Kinoshita et al., 2004).
Though maternal expression of MEA and FWA is controlled by DME, there are important distinctions regarding the regulation of expression of these genes. FWA is silent in all vegetative and reproductive tissues except for expression of the maternal allele in the female gametophyte and endosperm (Kinoshita et al., 2004; Soppe et al., 2000). MEA is imprinted in the endosperm but is biallelically expressed in the embryo and in other sporophytic tissues (Kinoshita et al., 2004). Expression of MEA in the embryo is likely not under DME control as DME expression is not detected in the egg cell or embryo (Choi et al., 2002). Expression of FWA in the endosperm and elsewhere in the plant is associated with hypomethylation of repeats in the 5′ region of the gene (Kinoshita et al., 2004; Soppe et al., 2000). Paternal inheritance of met1 releases FWA paternal-allele silencing in the endosperm and embryo (Kinoshita et al., 2004). MET1 is the homolog of Dnmt1 (Bender, 2004).
DME, MEA, and MET1 genetically interact in the female gametophyte. MEA is an E(Z) homolog that functions in a PcG complex along with FIE (Kohler et al., 2003a), a homolog of Eed, to repress endosperm growth. Inheritance of mutant maternal dme or mea alleles causes endosperm overproliferation, embryo arrest, and seed abortion (Choi et al., 2002; Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999). Seed abortion caused by dme is suppressed by maternally inherited met1 if a wild-type maternal MEA allele is present (Xiao et al., 2003). Moreover, met1 can restore MEA expression in dme mutants (Xiao et al., 2003). The mechanism by which DME activates MEA is uncertain, although it is known that the glycosylase activity of DME is necessary for seed viability and activation of MEA transcription (Choi et al., 2004). DME could antagonize MET1 by specifically removing 5-methylcytosine from MEA in the central cell, allowing the maternal MEA allele to be expressed there before fertilization and in the endosperm after fertilization.
Here we report that, in wild-type seeds, the maternal endosperm allele was hypomethylated compared to the paternal endosperm allele. Hypomethylation was not observed in dme mutant endosperm, suggesting that, in the central cell, DME is responsible for the removal of MEA DNA methylation. Consistent with this hypothesis, DME with an active DNA glycosylase domain can excise 5-methylcytosine in vitro and when expressed in E. coli. We also found that paternal MEA allele expression is not subject to the same controls as the maternal MEA allele. Rather, Polycomb group proteins that are expressed from the maternal genome, including MEA, silence the paternal MEA allele, a novel example of self-imprinting.
RESULTS
The Maternal MEA Allele Is Hypomethylated in Wild-Type Endosperm
Four regions around the MEA locus were previously shown to be methylated: a helitron DNA transposon element (Kapitonov and Jurka, 2001), AtREP2, about 4 kb 5′ of the start site (Xiao et al., 2003); CG sites 3 kb and 500 bp upstream (Xiao et al., 2003); and seven ~182 bp direct repeats 3′ of the gene, termed MEA-ISR (Cao and Jacobsen, 2002). Here we show that bisulfite sequencing covering 91% of the CG sites in the MEA coding region did not reveal any additional methylated cytosines (Figure 1A). To see whether DME antagonizes MET1 by removing MEA DNA methylation in the central cell, we compared the methylation of maternal and paternal alleles in the embryo and endosperm of seeds dissected between 7 and 8 days after pollination (DAP). Allele-specific methylation was determined in reciprocal crosses between the accessions Columbia with the glabrous mutation (Col-gl) and RLD and between Landsberg with the erecta mutation (Ler) and RLD. This allowed us to discount any methylation effects due to natural variation or the direction of the cross. Maternal and paternal alleles could be distinguished after sequencing because of polymorphisms between RLD and Col-gl/Ler near the regions of methylation. The three accessions used in this study have similar levels of MEA methylation in leaves with one exception: Ler consistently has much lower levels of CG methylation in the −500 bp region (Table S1).
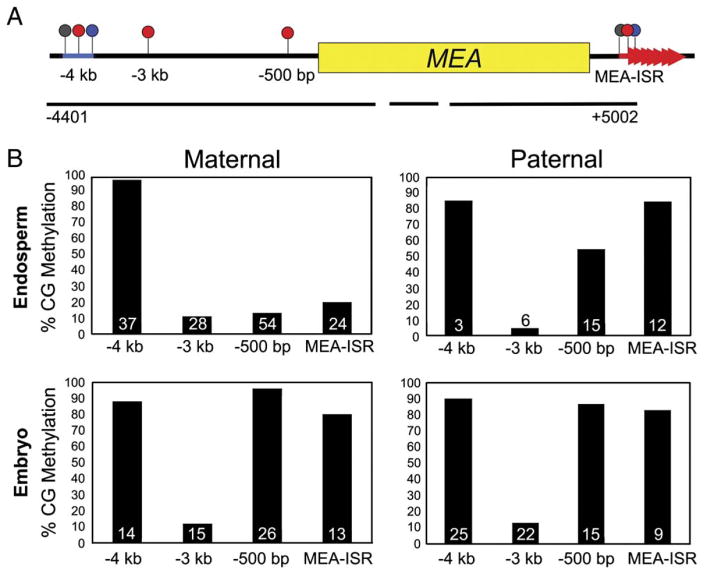
Figure 1. MEA Methylation in Dissected Seeds.
(A) MEA is methylated in four regions. Numbers are relative to the translation start site.
(B) CG methylation of maternal and paternal embryo and endosperm alleles from a Col-gl female crossed to a RLD male. The number of clones sequenced is given at the base of each column. Black lines, sequences assayed by bisulfite sequencing; blue bar, helitron transposon element; red arrowheads, 182 bp direct repeats; lollipops, sites of DNA methylation (red, CG; blue, CNG; gray, CNN).
In a cross between a Col-gl female and a RLD male, the −4 kb transposon element was highly methylated on both maternal and paternal embryo and endosperm alleles (Figure 1B). The −3 kb region exhibited low levels of methylation on all alleles (Figure 1B). However, the maternal endosperm allele was hypomethylated at the −500 bp (13% CG) region compared to the paternal endosperm allele (54%) and the maternal (96%) and paternal (87%) embryo alleles (Figure 1B). The same relationship was observed at the MEA-ISR. The maternal endosperm allele had 20% CG methylation compared to the paternal endosperm allele, which had 83%, and maternal and paternal embryo alleles, with 80% and 85% CG methylation, respectively (Figure 1B). The −500 bp region and MEA-ISR were also maternally hypomethylated in the endosperm of the reciprocal cross with RLD as the female and Col-gl as the male (Table S2).
Maternal endosperm alleles also had low levels of methylation at the −500 bp region and MEA-ISR in reciprocal crosses between Ler and RLD (Table S2). However, consistent with the data from rosette leaves (Table S1), the −500 bp region had low levels of methylation on any allele inherited from the Ler background, whether from the male or female parent, embryo, or endosperm (Table S2). Thus, though there are differences in MEA DNA methylation attributable to the accession (Tables S1 and S2), each accession examined had at least one region of allele-specific DNA hypomethylation in the endosperm (Figure 1B and Table S2).
Maternal MEA Is Not Hypomethylated in dme Endosperm
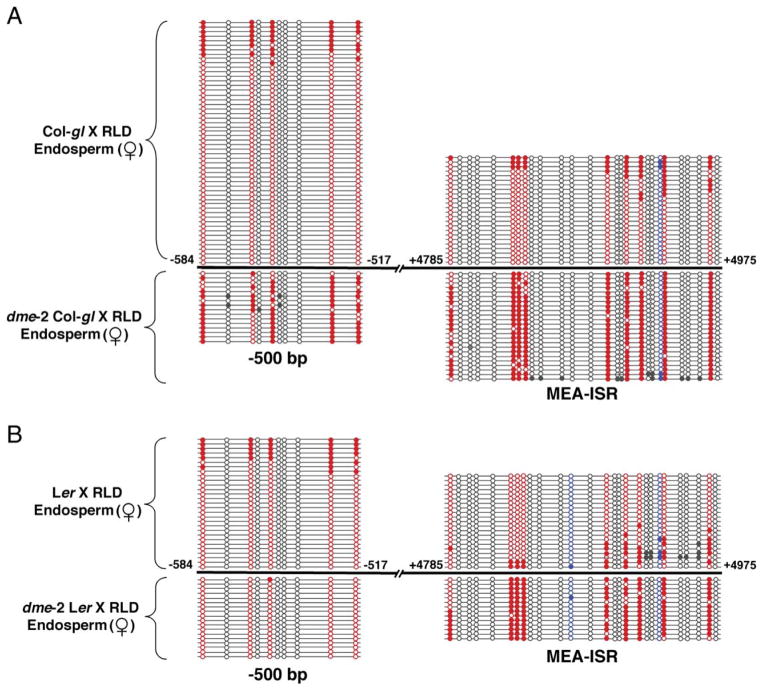
If DME is responsible for hypomethylation of MEA in the female gametophyte, then dme mutant endosperm should, in comparison, inherit hypermethylated maternal MEA alleles from dme central cells. We crossed dme-2 heterozygous mutant females in both the Col-gl and Ler backgrounds to wild-type RLD males and analyzed methylation of maternal and paternal alleles from dme mutant endosperm 9 or 10 DAP. Compared to maternal-allele methylation in wild-type endosperm, we found a substantial increase in maternalallele CG methylation in both the −500 bp (76% versus 13% for wild-type) and MEA-ISR (89% versus 20% for wild-type) regions in crosses with dme in a Col-gl background (Figure 2A). In crosses with dme in a Ler background, methylation on the maternal allele increased at the MEA-ISR (84% versus 18% for wild-type) but not in the −500 bp region (1% versus 22% for wild-type) (Figure 2B). We expected no change for the −500 bp region in the dme Ler mutant because there is very little methylation there for DME to act on in wild-type (Tables S1 and S2). We conclude that, in wild-type, DME DNA glycosylase is responsible for hypomethylation of the maternal endosperm allele observed at the MEA-ISR in the Col-gl, Ler, and RLD backgrounds and for hypomethylation of the −500 bp region in Col-gl and RLD.
Figure 2. Hypermethylation of Maternal MEA in dme Mutant Endosperm.
Maternal-allele methylation in the −500 bp and MEA-ISR regions in endosperm from crosses between dme-2 heterozygous females and RLD males compared to maternal endosperm allele methylation from crosses between wild-type females and RLD males.
(A) dme-2 heterozygous Col-gl crossed to RLD.
(B) dme-2 heterozygous Ler crossed to RLD. Mutant endosperm was collected at 9 DAP from seeds with the dme endosperm overproliferation phenotype. Numbers are from the translation start site. To determine the pattern of DNA methylation, DNA was treated with bisulfite, PCR amplified, cloned, and sequenced. Circles connected by lines represent the results from determining the DNA sequence of one clone. Filled circle, methylated cytosine; open circle, unmethylated cytosine; red circle, CG site; blue circle, CNG site; gray circle, CNN site.
DME with a Wild-Type DNA Glycosylase/Lyase Domain Excises 5-Methylcytosine In Vitro
DME is related to DNA glycosylases (Choi et al., 2002) that catalyze the first steps in the base-excision DNA repair pathway (Scharer and Jiricny, 2001). The reaction mechanism of bifunctional DNA glycosylases is well known (Scharer and Jiricny, 2001). A conserved aspartic acid acquires a proton from a conserved lysine residue that attacks the C1′ carbon of the deoxyribose ring, creating a covalent DNA-enzyme intermediate (Figure 3A). β or δ elimination reactions release the enzyme from the DNA and cleave one of the phosphodiester bonds (Figure 3A). Cleavage 5′ to the abasic site of the β or δ elimination produced by an AP endonuclease generates a 3′-hydroxyl used by a DNA repair polymerase that inserts the proper nucleotide, and a DNA ligase seals the nick.
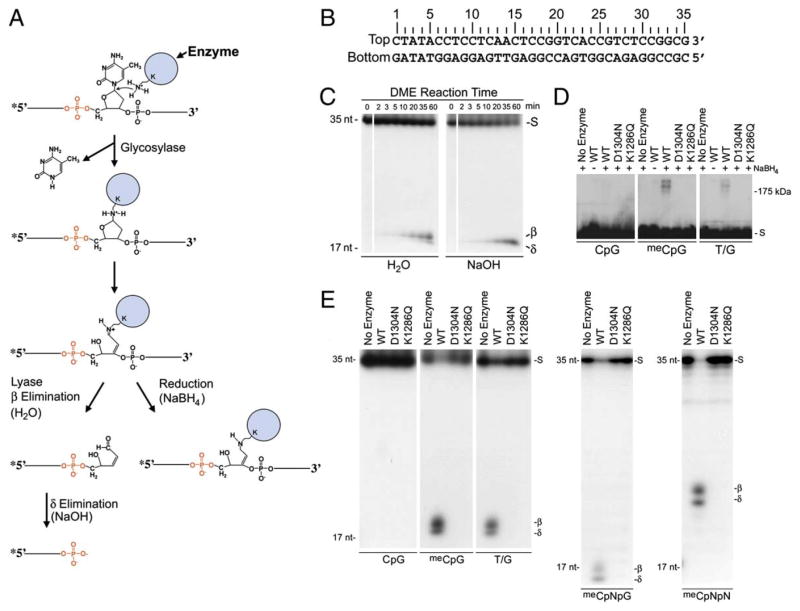
Figure 3. DME In Vitro Activity.
(A) Schematic mechanism of bifunctional DNA glycosylases.
(B) DNA substrate sequence. Base pair positions relative to the 5′ end of the top DNA strand are shown. Double-stranded DNA oligonucleotide substrates in (C)–(E) were labeled at the 5′ end of the top strand. DNAs in (C) had 5-methylcytosine at position 18 in the top strand. The top strand for (D) and (E) has: CpG, C at position 18; meCpG, 5-methylcytosine at position 18; T/G, T at position 18; meCpNpG, 5-methylcytosine at position 17; meCpNpN, 5-methylcytosine at position 15. All reactions were for 1 hr.
(C) Reaction products of DME. Products were treated with either water or NaOH as indicated, denatured, and analyzed on 15% polyacrylamide gels with 7.5 M urea.
(D) Covalent crosslinking of DME to DNA. Reaction products were treated with NaBH4, denatured, and analyzed on a 10% SDS-polyacrylamide gel.
(E) Substrate specificity of DME. Reaction products were denatured and analyzed on 15% polyacrylamide gels with 7.5 M urea. Both β and δ elimination products are observed because reactions were not treated with NaOH before gel electrophoresis. S, uncleaved substrate; β, predicted β elimination product; δ, predicted δ elimination product; 35 nt, 35 nucleotide size marker; 17 nt, 17 nucleotide size marker.
We expressed in E. coli an 1192 amino acid portion of DME that lacks 537 amino-terminal amino acids (Δ537DME) but includes the predicted DNA glycosylase domain. Δ537DME was fused to the maltose binding protein (MBP). MBP-Δ537DME was purified over an amylose column (Figure S1) and is referred to as wild-type DME. For control experiments, we expressed and purified mutant forms of DME where the invariant aspartic acid at position 1304 was converted to asparagine (D1304N) or the lysine at position 1286 was converted to glutamine (K1286Q). Both mutations reduce DNA glycosylase activity while preserving enzyme structure and stability (Fromme et al., 2004; Norman et al., 2003).
We incubated DME with various double-strand oligonucleotides (Figure 3B) to understand its biochemical mechanism. DME breaks the phosphodiester linkage on the 3′ side of a 5-methylcytosine residue (hemimethylated substrate) and generates end-labeled DNAs that migrate on denaturing polyacrylamide gels at the predicted position for β elimination products (Figure 3C). The subsequent cleavage of the phosphodiester linkage on the 5′ side yields δ elimination products through the same mechanism found in related DNA glycosylases (Bhagwat and Gerlt, 1996). Treatment of products with strong base (NaOH) prior to gel electrophoresis confirmed the δ elimination process at the predicted position (Figure 3C). Consistent with the reaction mechanism for a bifunctional DNA glycosylase/lyase (Figure 3A), products treated with a reducing agent (NaBH4) migrated in the predicted region for trapped enzyme-DNA complexes (~200 kDa), suggesting that the Schiff base intermediate between DME and a ring-opened sugar is covalently reduced (Figure 3D). No lyase activity (Figure 3E) or covalent trapping (Figure 3D) was detected when DME was incubated with nonmethylated oligonucleotides or when hemimethylated substrate was incubated with no enzyme or mutant enzymes (D1304N or K1286Q). Plants have 5-methylcytosine in the three sequence contexts: CpG, CpNpG, and CpNpN (Bender, 2004). DME has activity on 5-methylcytosine in each of these sequence contexts (Figure 3E). We detected no DME activity when single-stranded oligonucleotides with 5-methylcytosine were used in the reaction (data not shown). These results show that DME is a bifunctional DNA glycosylase/lyase with activity on 5-methylcytosine substrates.
DME Excises Thymine from a T/G Mismatch
5-methylcytosine is mutagenic because it spontaneously deaminates to form thymine, generating a T/G mismatch. Deamination can also occur enzymatically by cytosine deaminase, a process that may play a role in mammalian epigenetic reprogramming and cell plasticity (Morgan et al., 2004). Specific DNA glycosylases initiate DNA repair by excising T from T/G mispairs (Scharer and Jiricny, 2001). We found that DME also is a thymine DNA glycosylase. DME activity on T/G mispairs is somewhat less than its activity on meC/G base pairs (Figure 3E and data not shown). DME also forms a trapped enzyme-DNA complex with DNA containing a T/G base pair (Figure 3D).
DME could cause hypomethylation of the maternal MEA alleles in the endosperm using two different mechanisms. DME might excise 5-methylcytosine, leading to its replacement with unmethylated cytosine, or DME might excise thymine from a T/G mismatch formed from deamination of 5-methylcytosine. To distinguish between these two mechanisms, we sequenced DNA from dme mutant endosperm. If DME excised thymine instead of 5-methylcytosine, we expected to find numerous C→T transitions at CG sites in the −500 bp region and MEA-ISR, which are hypomethylated in wild-type endosperm (Figure 1 and Figure 2). However, no C→T transitions were found (Figure S2). Thus, the thymine DNA glycosylase activity of DME is likely not responsible for maternal MEA allele hypomethylation.
DME Is Toxic in E. coli with 5-Methylcytosines
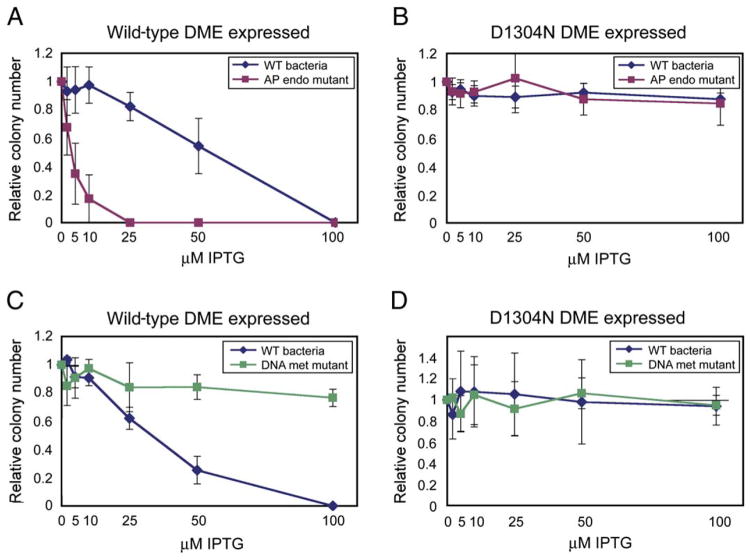
When expressing DME from an IPTG-inducible promoter, we found that DME was toxic to E. coli K-12 strains in an IPTG-concentration-dependent manner (Figures 4A and 4C). The toxicity of DME expression was significantly increased in a strain bearing mutations in two AP endonuclease genes (xth and nfo) (Cunningham et al., 1986), which remove abasic sites and trim the 3′ structure of nicks. This result suggests that DME DNA glycosylase and/or lyase activity is toxic, perhaps due to the formation of mutagenic abasic sites and/or nicks in the E. coli genome. Indeed, expression of inactive DME(D1304N) was nontoxic in xth nfo mutants or the isogenic wild-type background (Figure 4B).
Figure 4. DME Functions as a 5-methylcytosine DNA Glycosylase in E. coli.
Relative colony number; number of colonies on plate divided by the number of colonies obtained when plate has no IPTG inducer. The relative colony number at each concentration of IPTG was determined three times. Error bars represent the standard deviation from the mean.
(A and B) wt bacteria, AB1157; AP endo mutant RPC501 (Cunningham et al., 1986) isogenic to AB1157, with mutations in two AP endonuclease genes (xth, nfo).
(C and D) wt bacteria, GM30; DNA met mutant GM31 (Palmer and Marinus, 1994) isogenic to GM30, with a mutation in the dcm DNA methyltransferase.
DME has in vitro 5-methylcytosine activity (Figures 3C–3E), and E. coli K-12 strains have 5-methylcytosine in their genomes. Perhaps DME produces deleterious abasic sites in E. coli by excising 5-methylcytosine at a genome-wide level. We tested this hypothesis by expressing DME in a dcm mutant strain (Palmer and Marinus, 1994), which has no 5-methylcytosine in its genome. DME expression was not toxic to dcm bacteria compared to expression in the isogenic wild-type strain (Figure 4C). Expression of inactive DME(D1304N) had no effect on either strain (Figure 4D). This suggests that 5-methylcytosine is a substrate for DME in E. coli K-12 bacteria.
Base Excision Inhibits Further Excision by DME on the Opposite DNA Strand
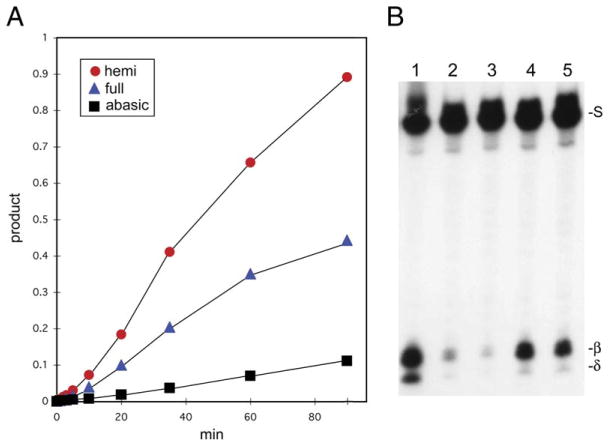
Excision of 5-methylcytosine from fully methylated meCpG/GpmeC sequences by DME would generate nicks 1 nucleotide apart on opposing DNA strands, which could lead to deleterious double-stranded breaks in the DNA (Hanai et al., 1998). A similar problem occurs when DNA glycosylases encounter clustered lesions on opposing DNA strands, where it has been shown that abasic sites and/or nicks on one DNA strand inhibit glycosylase-mediated excision of nearby lesions on the opposing strand (David-Cordonnier et al., 2001; Weinfeld et al., 2001). Consistent with this mechanism, we found that DME is more active on a specific 5-methylcytosine when it is in the hemimethylated state compared to the fully methylated state (Figure 5A). Moreover, an abasic site on the opposite strand (~pG/GpmeC, where ~ represents the abasic site) reduced the reaction rate approximately 10-fold compared to DME activity on hemimethylated DNA (Figures 5A and 5B, lane 2). A similar inhibitory effect was observed when an abasic site was in a hemimethylated CpNpG context(~pNpG/GpNpmeC) (Figure 5B, lane 3). By contrast, there is significantly less inhibition of DME activity when the abasic site is shifted 4 (Figure 5B, lane 4) or 7 nucleotides (Figure 5B, lane 5) away from the 5-methylcytosine. These results indicate that the abasic site created by excision of 5-methylcytosine from fully methylated CpG or CpNpG DNA specifically inhibits subsequent excision of 5-methylcytosine on the opposite strand. This would allow AP endonuclease, DNA polymerase, and ligase to complete the base-excision DNA repair pathway on one DNA strand before excising 5-methylcytosine on the opposite strand, thereby avoiding a double-strand break.
Figure 5. Inhibition of DME Activity by Abasic Sites.
(A) Rate of DME activity. Labeled (5′ end of the bottom strand) double-stranded oligonucleotides (Figure 3B) were used with the following sequences: hemi, 5-methylcytosine at position 19 (bottom strand); full, 5-methylcytosine at positions 19 (bottom strand) and 18 (top strand); abasic, 5-methylcytosine at position 19 (bottom strand) and an abasic site at 18 (top strand). Reactions were performed, terminated by addition of NaOH, boiled, and subjected to electrophoresis. Gels were exposed to a phosphorimager screen to determine the amount of product.
(B) Effect of abasic-site position on DME activity. Double-stranded oligonucleotides (Figure 3B) were labeled at the 5′ end of the bottom strand and had 5-methylcytosine at position 19 of the bottom strand (lane 1). In addition, abasic sites were in the top strand at position 18 (lane 2), position 17 (lane 3), position 15 (lane 4), and position 12 (lane 5).
A Hypomethylated Paternal Genome Does Not Release Paternal MEA Silencing
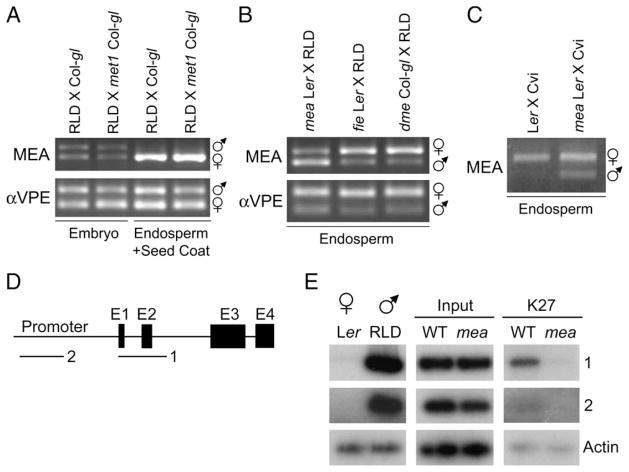
The silent paternal endosperm allele is hypermethylated compared to the expressed maternal allele (Figure 1B). Would inheritance of a hypomethylated paternal genome release silencing of the paternal allele in the endosperm? We crossed a wild-type female to a met1-6 homozygous mutant male and analyzed allele-specific expression in embryo and endosperm plus seed-coat fractions by RT-PCR. Expression was indistinguishable from wild-type crosses, indicating no change in MEA paternal-allele silencing (Figure 6A). We tested a variety of other mutations (Bender, 2004) that affect DNA methylation in various sequence contexts for their ability to alter imprinting in the endosperm. Paternal inheritance of ddm1-2, drm1 drm2 cmt3-7, ago4-1, rdr2-1, or dcl3-1 did not result in paternal-allele expression in the endosperm (data not shown).
Figure 6. Regulation of MEA Paternal-Allele Silencing.
(A) Paternal MEA silencing is not affected by a hypomethylated paternal genome. Expression of MEA in the embryo and endosperm/seed coat of crosses between a RLD female and Col-gl male and a RLD female and a met1-6 homozygous Col-gl male. Seeds were dissected 7 DAP.
(B) MEA expression in mutant endosperm of crosses between mea-3 homozygous Ler, fie-1 heterozygous Ler, and dme-2 heterozygous Col-gl females and RLD males, dissected 9 DAP.
(C) MEA expression in endosperm of crosses between Ler and mea-3 homozygous Ler females and Cvi males, dissected 7 and 8 DAP, respectively, at the torpedo stage of embryogenesis. VPE is a control for biallelic expression.
(D) Genomic structure of ArabidopsisMEAandregions examinedbyChIP.E1 throughE4;exons 1 through4.Regions amplified are shown bybars labeled 1 and 2.
(E) ChIP with anti-dimethylH3K27 comparing amplification ofMEA in wt Ler X RLD andmutant Lermea X RLD siliques 7 DAP. LNA primers were used to amplify regions 1 and 2 and not the actin control DNA.
Polycomb Group Proteins Maintain Paternal-Allele Silencing
What, then, is the mechanism for maintaining silencing of the paternal allele in the endosperm? In insects, mammals, and plants, PcG proteins maintain repressed states of gene transcription. PcG proteins are involved in a variety of epigenetic processes, including maintenance of X inactivation and of allele-specific silencing of a subset of imprinted genes in mammals (Cao and Zhang, 2004). We tested whether PcG genes are involved in MEA imprinting and found that endosperm paternal-allele silencing is lost when mutations in Polycomb group genes are inherited maternally.
In a cross between Ler mea-3 (Kiyosue et al., 1999) homozygous mutant females and wild-type RLD males, almost all seeds undergo endosperm overproliferation, embryo arrest, and seed abortion. We collected the mutant endosperm before seed abortion and analyzed allele-specific expression. Expression from both maternally and paternally inherited alleles was detected, indicating a loss of imprinting (Figure 6B).
Paternal-allele expression was also observed in endosperm from seeds that lack maternal MEA but do not abort. When Ler mea/mea plants are pollinated by the Cvi accession, the seed-abortion phenotype is suppressed and 95% viable seeds are produced (M.G., T. Kinoshita, and R.L.F, unpublished data). Endosperm allele-specific gene expression in seeds dissected at the torpedo stage of embryogenesis was compared in crosses between Ler and Cvi and Ler mea/mea and Cvi. In the wild-type cross, only maternal-allele expression was detected in the endosperm. When Ler mea/mea was the female in the cross, expression from both maternal and paternal alleles was observed (Figure 6C). Thus, MEA paternal-allele silencing is lost in both viable (Figure 6C) and aborting (Figure 6B) seeds when maternal MEA is not made.
FIE is a PcG gene homolog of Drosophila Esc and mammalian Eed, and fie mutants have a seed-abortion phenotype like mea (Ohad et al., 1999). FIE and MEA interact in a PcG complex (Kohler et al., 2003a). Loss of imprinting was also observed when fie-1 heterozygous females were crossed to wild-type males (Figure 6B). These results suggest that silencing of the paternal allele in the endosperm is maintained by maternally expressed Polycomb group proteins that likely act at the paternal MEA locus.
Paternal MEA Is Enriched in H3K27 Methylation
Polycomb group complexes modify histones. In Drosophila and mammals, ESC-E(Z) and EED-EZH2 PcG complexes methylate histone H3 at K27 (Czermin et al., 2002; Muller et al., 2002). H3K27 methylation is also a likely Polycomb mark in Arabidopsis. Expression of the FLC gene is regulated by vernalization (exposure to cold), which causes an increase in H3K27 dimethylation at the locus (Bastow et al., 2004; Sung and Amasino, 2004). This change is dependent on VRN2, a Polycomb group gene that maintains vernalization-induced downregulation of FLC expression (Bastow et al., 2004; Sung and Amasino, 2004).
We hypothesized that the maternal MEA-FIE complex methylates H3K27 at the paternal MEA allele in the endosperm. By a chromatin immunoprecipitation (ChIP) assay, we compared paternal-allele H3K27 dimethylation patterns in siliques from crosses between Ler females and RLD males and between Ler mea/mea females and RLD males. We took advantage of MEA sequence polymorphisms between Ler and RLD to specifically amplify paternal DNA by using PCR primers containing high-affinity DNA analogs known as locked nucleic acids (LNA) (Koshkin et al., 1998). The last base of each primer contains a LNA base analog that will pair with the RLD base at a much higher affinity than the Ler base. Primer sets for the MEA promoter and coding region (Figure 6D) amplified RLD (male parent) genomic DNA well but Ler (female parent) very poorly (Figure 6E).
The vast majority of silique DNA is of maternal origin, from the maternal silique and seed-coat tissue and the contributions of the maternal genome to the embryo and endosperm. The only paternal DNA in siliques is from the embryo and endosperm. Since paternal DNA is a small fraction of the total DNA, radioactive nucleotides were used to increase the sensitivity of the assay. As shown in Figure 6E, we found that, after ChIP with antibodies specific to H3 dimethyl K27, paternal MEA DNA was enriched in wild-type siliques compared to maternal mea siliques for the coding region from −5 to +440 (region 1). By contrast, little if any paternal MEA DNA was detected in MEA 5′ sequences from −947 to −547 (region 2). We cloned the −5 to +440 wild-type and mea PCR products, sequenced across an internal Ler/RLD polymorphism, and verified that almost all of the clones were from paternal RLD DNA (21 of 22 wild-type clones and 22 of 22 mea clones). Although paternal embryo and endosperm alleles cannot be distinguished, these results indicate that wild-type maternal MEA is required for paternal MEA H3 K27 dimethylation.
Paternal Silencing Is Lost in dme Mutants
Because dme mutants lack MEA expression in the female gametophyte (Choi et al., 2002), we looked at the effect of dme on paternal MEA expression in the endosperm. Paternal-allele expression was detected when dme-2 heterozygous plants were crossed as females to wild-type males (Figure 6B). This is consistent with our finding that maternal MEA expression in the female gametophyte, activated by DME, is required for paternal-allele silencing. The expressed paternal allele in dme endosperm is as highly methylated as the silent paternal allele from wild-type endosperm (Figure 1B and Table S2). In a cross between dme-2 Col-gl females and RLD males, expressed paternal endosperm alleles had 100% and 94% CG methylation in the −500 bp region and MEA-ISR, respectively (3 and 11 clones sequenced). In a cross between dme-2 Ler females and RLD males, expressed paternal endosperm alleles had 54% and 93% CG methylation in the −500 bp region and MEA-ISR (7 and 5 clones sequenced). This suggests, in agreement with results presented in Figure 6A, that the presence or absence of DNA methylation is not relevant to MEA paternal-allele silencing in the endosperm.
We also detected expression of the highly methylated maternal MEA allele (Figure 2A) in dme endosperm (Figure 6B). Previously, we showed that DME is required for MEA expression before fertilization (Choi et al., 2002). These results suggest that, although hypomethylation via DME is required for MEA expression in the central cell before fertilization and possibly during early endosperm development (Choi et al., 2002), it is not required for maternal MEA expression in the endosperm by 9 DAP.
DISCUSSION
Activation of Maternal MEA Allele Expression by DME
We have found that the expressed maternal endosperm allele of the imprinted MEA gene is hypomethylated in specific 5′ and 3′ regions (Figure 1 and Table S2). DME is required for MEA expression in the central cell (Choi et al., 2002) and for hypomethylation of the maternal MEA allele inherited from the central cell (Figures 1 and 2). Thus, expression of the maternal MEA allele is associated with removal of DNA methylation by a DNA glycosylase. This in vivo data suggests that one DNA repair function of DME is to excise 5-methylcytosine from CG contexts, leading to its replacement with cytosine. This is supported by DME excision of 5-methylcytosine in vitro (Figure 3) as well as DME activity on 5-methylcytosine in the base-excision repair pathway in E. coli (Figure 4). Another DME family member, ROS1, also has activity on 5-methylcytosine in vitro (Gong et al., 2002).
Excision of symmetric 5-methylcytosine is predicted to cause deleterious double-strand DNA breaks. However, this might be mitigated by the inhibition of DME activity by abasic sites (Figure 5). The mechanism for the inhibition is not known. DME has little lyase activity on abasic sites (data not shown), so it is likely to be the abasic site, not a nick in the DNA, that inhibits DME. One possibility is that DME binds to the abasic site and physically hinders other DME molecules from excising 5-methylcytosine on the opposite strand. Alternatively, an abasic site near the active site of a DME enzyme may inhibit an essential step of the base-excision reaction mechanism for the 5-methylcytosine on the opposing strand.
Several aspects of the activation of MEA by DME remain unclear. Do reduced levels of DNA methylation directly lead to expression of MEA in the central cell? Or does an accompanied change induced by the act of DNA repair render the locus transcriptionally competent? Unlike the maternal MEA allele in the central cell, paternal-allele expression in the endosperm is not affected by changes in DNA methylation (Figure 6A). Instead, paternal silencing is lost when the function of maternal MEA-FIE PcG complexes is perturbed (Figure 6B). This is associated with decreased H3K27 methylation on the paternal allele (Figure 6E).
Central-Cell-Specific Interpretation of MEA DNA Methylation
Our data show that removal of CG methylation is required for MEA expression in the central cell but not in the embryo or during later stages of endosperm development. A hypomethylated paternal genome does not affect MEA imprinting (Figure 6A). Furthermore, in dme endosperm, the expressed maternal and paternal alleles are highly methylated in the −500 bp region and MEA-ISR. Hypomethylation of MEA is only required for expression in the central cell and perhaps during early endosperm development at a stage prior to dme seed dissection. This conclusion is supported by embryo methylation data from wild-type crosses (Figure 1 and Table S2). MEA is expressed biallelically in the embryo (Figure 3; Kinoshita et al., 1999). Yet we found that the expressed embryo alleles are as highly methylated as the silent paternal endosperm allele and are hypermethylated compared to the expressed maternal endosperm allele (Figure 1 and Table S2). Differences in methylation between the maternal embryo and maternal endosperm alleles hearken back to the distinct origins of these alleles in the female gametophyte, which arise from the egg and central cell, respectively. Due to the exclusive expression of DME in the central cell, only the maternal endosperm allele, and not the maternal embryo allele, has been exposed to DME. Removal of DNA methylation at the maternal MEA allele in the central cell represents the first case in angiosperms in which changing the methylation status of a gene is an integral part of an essential developmental program, the formation of viable seeds.
The limited regulation of MEA expression by the removal of DNA methylation is in contrast to the imprinted gene FWA, where there is a strong correlation between DNA methylation and gene expression not only in the endosperm but also in the embryo and throughout the entire plant (Kinoshita et al., 2004; Soppe et al., 2000). FWA is not expressed vegetatively and is highly methylated on promoter repeats. These repeats are hypomethylated in mutants that ectopically express the gene (Soppe et al., 2000). Additionally, endosperm imprinting is lost when FWA is inherited from a met1 pollen parent (Kinoshita et al., 2004). Our results (Figure 1, Figure 6A, and Table S2) suggest that for MEA there is a high degree of specificity in the interpretation of DNA methylation. Methylation status is only relevant in the central cell. Thus, while maternal expression of both MEA and FWA is regulated by DNA methylation and DME in the central cell, additional distinct mechanisms, discussed below, control silencing of the paternal MEA allele.
Maternally and Paternally Silent Alleles of Imprinted Genes Are Maintained by Polycombs
The mouse Polycomb group protein EED, a homolog of FIE, is required to maintain silencing of some imprinted autosomal genes (Delaval and Feil, 2004; Lewis et al., 2004; Umlauf et al., 2004). Certain paternally silent alleles in the placenta are associated with repressive histone H3K27 methylation regulated by the Polycomb complex EED-EZH2 (Lewis et al., 2004; Umlauf et al., 2004). Some of these genes are also imprinted in the embryo. However, unlike in the embryo, placental repression takes place in the absence of the promoter DNA methylation (Lewis et al., 2004; Umlauf et al., 2004). Köhler et al. (2003b, 2005) showed that maternal MEA PcG complexes repress maternal expression of the MADS-box gene PHERES1 (PHE1). PHE1 is an example of a gene imprinted oppositely to MEA and FWA, such that the maternal allele is largely silent and the paternal allele is expressed in the endosperm (Kohler et al., 2005). MEA PcG complexes likely assemble at the maternal PHE1allele in the central cell before fertilization (Kohler et al., 2005). We found that maternal MEA PcG complexes maintain silencing of the paternal MEA allele (Figures 6B, 6C, and 6E). The paternal MEA allele is enriched in H3K27 dimethylation when the maternal MEA allele is wild-type compared to when the maternal mea allele is mutant (Figure 6E). This suggests that maternal MEA Polycomb group complexes play a direct role in regulating the chromatin structure at the paternal MEA allele. Paternal-allele silencing is maintained even if the paternal genome is hypomethylated (Figure 6A). Thus, maternal MEA functions in maintaining both maternally (e.g., PHE1) and paternally (e.g., MEA) silenced alleles of imprinted genes. It remains unknown how PcG complexes are directed to the paternal MEA locus or how the silent state is initially established. Our data indicate that the PcG complex is one means by which the maternal genome modifies the activity of the paternal genome. This emphasizes the prominent role the maternal genome has in controlling endosperm imprinting and development.
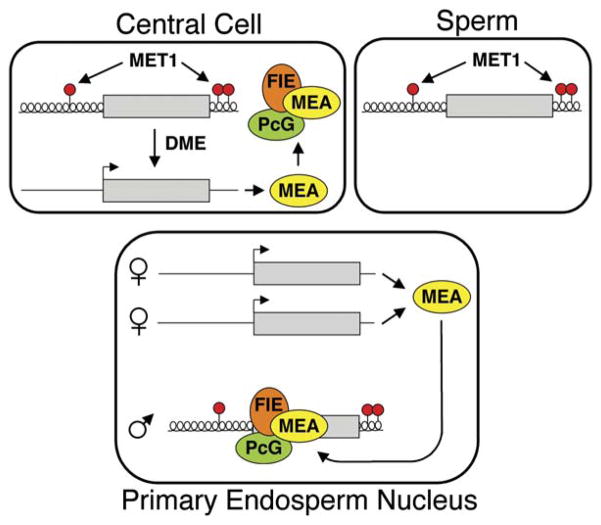
Model for the Regulation of MEA Imprinting
We propose the following model for MEA imprinting (Figure 7). DME is expressed in the central cell of the female gametophyte and removes MEA DNA methylation by excising 5-methylcytosine. The hypomethylated maternal MEA allele is expressed, producing MEA protein. Shortly after fertilization, FIE-MEA PcG complexes assemble at the paternal MEA allele, maintaining its previously established silent state. Thus, DME-mediated methylation changes that take place in the central cell before fertilization control both aspects of MEA imprinting—maternal-allele expression and subsequent paternal-allele silencing. Imprinting is lost in maternal mea and dme mutant endosperm because maternal MEA protein is not present at the time of fertilization. Methylation does not inhibit maternal MEA expression in dme endosperm during later stages of endosperm development (by 9 DAP), but, by this time, the paternal MEA allele has already lost its silent state. Maintenance of MEA silencing by MEA represents a unique instance of a Polycomb group gene regulating its own imprinting.
Figure 7. Model for Regulation of MEA Imprinting.
MEA methylation is maintained by MET1. In the central cell, DME removes methylation at the −500 bp region and MEA-ISR. MEA protein is produced and forms PcG complexes. After fertilization, MEA-FIE PcG complexes target the paternal allele to maintain its silent state. Maternal MEA continues to be expressed in the endosperm. Gray box, MEA gene; red circles, DNA methylation; helical line, nontranscribed compacted chromatin; straight line, transcribed open chromatin.
EXPERIMENTAL PROCEDURES
Plant Material
Seeds were plated on 0.5× Murashige and Skoog salts (Caisson Laboratories, Inc.), 1× Gamborg’s Vitamins (Sigma), and 2% sucrose; stratified at 4ºC for 2 days; grown in continuous light in a growth chamber for 10 days; and then transplanted to soil and grown in greenhouse conditions (16 hr light). For crosses, flowers were emasculated 2 days before pollination. met1-6 homozygous plants were obtained from a self-pollinated met1-6 heterozygote that had never been homozygous. Ler mea/mea plants were the F3 generation.
Bisulfite DNA Sequencing
Seeds at the mid- to late torpedo stage of embryogenesis (7 to 8 DAP) were dissected into embryo, endosperm, and seed-coat fractions in 0.3 M sorbitol, 5 mM MES (pH 5.7) on a slide under a dissecting microscope. Endosperm tissue was ground in CTAB to isolate DNA. Embryos were washed to remove contaminating endosperm. Bisulfite treatment and sequencing were performed as described (Xiao et al., 2003). Primer sequences for PCR amplification are in the Supplemental Experimental Procedures.
DME Activity
5′-labeled oligonucleotide substrates (13.3 nM) were incubated with DME protein (250 nM) in a 15 μl reaction with 40mMHEPES-KOH (pH 8.0), 0.1 MKCl, 0.1mMEDTA, 0.5mMdithiothreitol, and 200 μg/ml BSA at 37º for 1 hr. The reaction was terminated with 15 μl of 95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF and boiled for 5 min. To induce δ elimination, NaOH was added at a final concentration of 0.1 M and the reaction was boiled for 7 min. Products were fractionated on a 15% polyacrylamide gel containing 7.5 M urea and 1× TBE. Electrophoresis was done at 1000V for 4 hr with a Hoefer SQ3 gel apparatus. The gel was exposed to Kodak BioMax MR film at −80ºC. Methods for purification of recombinant DME, oligonucleotide substrates, NaBH4 trapping, and toxicity in E. coli are in the Supplemental Experimental Procedures.
Protein Gel Analysis
Protein purity was determined by staining gels with Code Blue reagent (Pierce). Gels were blotted on nitrocellulose membranes (Bio-Rad) and reacted with anti-MBP monoclonal antibody (New England Biolabs) as described by the manufacturer. Goat anti-mouse IgG-AP-conjugated antibody (Bio-Rad) and the AP Conjugate Substrate Kit (Bio-Rad) were used for colorimetric detection. Goat anti-mouse IgG-HRP-conjugated antibody (Bio-Rad) and SuperSignal Substrate (Pierce) were used for chemiluminescent detection. Reacted membranes were exposed to Kodak BioMax MS film for 5 to 10 min.
Expression Analysis
RNA was isolated using an RNAqueous Kit with Plant RNA Isolation Aid (Ambion, Inc.), and treated with DNase I (Invitrogen) before reverse transcription. For Figures 6A and 6B, the 72ºC amplification step for PCR was 10 s. For Figure 6C, 533 bp of MEA RNA from exons 3 to 6 was amplified with primers SR12 (5′-CAGAGGATGATAATGGAGGAGA-3′) and UCB3SR8 (5′-GCTTGAGTTCATTGTATCTTTCC-3′) for 40 cycles with a 40 s amplification step. An XbaI site is present in exon 3 in Cvi and not in Ler. After XbaI digestion, Cvi is cut into 395 and 138 bp pieces. For αVPE, primers for first amplification were VPE2912 (5′-ACAA CTTTCCCACTTCCTCCT-3′) and VPEdSal (5′-TCGCCGGATCCAGCG GATACTGGAATTGTCG-3′). Primers for a second amplification were VPE2679 (5′-GATTCTCCTCGTTCTCCGCA-3′) and VPEdSal. Digestion of VPE with SalI restriction endonuclease cut the RLD allele.
ChIP Assay
Siliques were collected 7–8 DAP, slit, and fixed in 1% formaldehyde. Tissue (0.4 g) was used for ChIP with anti-dimethyl histone H3 (Lys27) (Upstate Biotechnology). After immunoprecipitation, protein A bound immunocomplexes were washed as described (Johnson et al., 2002). ChIP PCR reactions (25 μl) were performed with 35 or 45 amplification cycles for Actin and MEA, respectively. The amount of immunoprecipitate was quantified so that equal amounts of ACTIN were amplified from wt and mea. The annealing temperature was 61ºC for Actin, 58ºC for MEA region 1, and 60ºC for MEA region 2. LNA primer sequences are in the Supplementary Experimental Procedures.
Supplementary Material
Acknowledgments
We thank S. Mitra, T. Hazra, and S. Linn for helpful discussions. We thank E. Richards (ddm1-2), S. Jacobsen (drm1 drm2 cmt3-7), and J. Carrington (dcl3-1, rdr2-1) for Arabidopsis mutants. We thank R.P. Cunningham (AB1157, RPC501) and M.G. Marinus (GM30, GM31) for E. coli strains. We thank R. Uzawa for technical assistance. R.L.F. received grants from the NIH (GM069415), USDA (2005-02355), BARD (IS-3604-04CR), and Ceres, Inc. (B97062). M.G. received an NSF Graduate Research Fellowship, and J.H.H. received a fellowship from the National Institute for International Education Development, South Korea.
Footnotes
Supplemental Data include Supplemental Experimental Procedures, Supplemental References, two tables, and two figures and can be found with this article online at http://www.cell.com/cgi/content/full/124/3/495/DC1/.
References
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- Bender J. DNA methylation and epigenetics. Annu Rev Plant Biol. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- Bhagwat M, Gerlt JA. 3′- and 5′-strand cleavage reactions catalyzed by the Fpg protein from Escherichia coli occur via successive β- and δ-elimination mechanisms, respectively. Biochemistry. 1996;35:659–665. doi: 10.1021/bi9522662. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA. 2002;99:16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- Choi Y, Harada JJ, Goldberg RB, Fischer RL. An invariant aspartic acid in the DNA glycosylase domain of DEMETER is necessary for transcriptional activation of the imprinted MEDEA gene. Proc Natl Acad Sci USA. 2004;101:7481–7486. doi: 10.1073/pnas.0402328101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constancia M, Kelsey G, Reik W. Resourceful imprinting. Nature. 2004;432:53–57. doi: 10.1038/432053a. [DOI] [PubMed] [Google Scholar]
- Cunningham RP, Saporito SM, Spitzer SG, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- David-Cordonnier MH, Boiteux S, O’Neill P. Efficiency of excision of 8-oxo-guanine within DNA clustered damage by XRS5 nuclear extracts and purified human OGG1 protein. Biochemistry. 2001;40:11811–11818. doi: 10.1021/bi0112356. [DOI] [PubMed] [Google Scholar]
- Delaval K, Feil R. Epigenetic regulation ofmammalian genomic imprinting. Curr Opin Genet Dev. 2004;14:188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- Gehring M, Choi Y, Fischer RL. Imprinting and seed development. Plant Cell. 2004;16:S203–S213. doi: 10.1105/tpc.017988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JJ. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a Polycomb-group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Hanai R, Yazu M, Hieda K. On the experimental distinction between ssbs and dsbs in circular DNA. Int J Radiat Biol. 1998;73:475–479. doi: 10.1080/095530098142013. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Cao X, Jacobsen SE. Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci USA. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Yadegari R, Harada JJ, Goldberg RB, Fischer RL. Imprinting of the MEDEA Polycomb gene in the Arabidopsis endosperm. Plant Cell. 1999;11:1945–1952. doi: 10.1105/tpc.11.10.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. Published online November 20, 2003. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada J, Goldberg RB, Fischer RL. Control of fertilization-independent endosperm development by the MEDEA Polycomb gene in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 2003a;22:4804–4814. doi: 10.1093/emboj/cdg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 2003b;17:1540–1553. doi: 10.1101/gad.257403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Page DR, Gagliardini V, Grossniklaus U. The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet. 2005;37:28–30. doi: 10.1038/ng1495. [DOI] [PubMed] [Google Scholar]
- Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Norman DP, Chung SJ, Verdine GL. Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase. Biochemistry. 2003;42:1564–1572. doi: 10.1021/bi026823d. [DOI] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL. Mutations in FIE, a WD Polycomb group gene, allow endosperm development without fertilization. Plant Cell. 1999;11:407–415. doi: 10.1105/tpc.11.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill MJ. The influence of non-coding RNA on allele-specific gene expression in mammals. Hum Mol Genet. 2005;14:R113–R120. doi: 10.1093/hmg/ddi108. [DOI] [PubMed] [Google Scholar]
- Palmer BR, Marinus MG. The dam and dcm strains of Escherichia coli - a review. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Scharer OD, Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Soppe WJJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJM. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. Published online October 31, 2004. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Thomas J, Spillane C, Coluccio A, Hoeppner MA, Grossniklaus U. Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 1999;13:2971–2982. doi: 10.1101/gad.13.22.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinfeld M, Rasouli-Nia A, Chaudhry A, Britten RA. Response of base excision repair enzymes to complex DNA lesions. Radiat Res. 2001;156:584–589. doi: 10.1667/0033-7587(2001)156[0584:robere]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL. Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell. 2003;5:891–901. doi: 10.1016/s1534-5807(03)00361-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.