Abstract
It can be convenient to think of the genome as simply a string of nucleotides, the linear order of which encodes an organism’s genetic blueprint. However, the genome does not exist as a linear entity within cells where this blueprint is actually utilized. Inside the nucleus, the genome is organized in three-dimensional (3D) space, and lineage-specific transcriptional programs that direct stem cell fate are implemented in this native 3D context. Here, we review principles of 3D genome organization in mammalian cells. We focus on the emerging relationship between genome organization and lineage-specific transcriptional regulation, which we argue are inextricably linked.
Introduction
One of the most fundamental questions in human biology is how one genome sequence can give rise to so many different cell types. The answer to this question lies, at least in part, in the ability of distinct cell types to express genes at different levels and in different combinations. Much of the cell-type-specific (or “lineage-specific”) regulation of gene expression occurs at the level of transcription. Such lineage-specific transcriptional regulation is not simply a product of genome sequence because all cells in an individual have essentially the same genetic content. Thus, features of the genome beyond its primary nucleotide sequence must contribute to the lineage-specific gene regulation that underlies cellular identity.
Tremendous effort has been dedicated to the study of genomic features other than primary nucleotide sequence. To this end, biochemical assays and computational tools have been employed to map sites of active transcription, chromatin accessibility, transcription factor (TF) binding, and chemical modification to histones and to the DNA itself; culminating in the discovery of tens of thousands of transcription units and millions of potential cis-regulatory elements in the human genome (Consortium et al., 2012, Bernstein et al., 2010). These data provide myriad layers of information about the genome’s lineage-specific biochemical activity that can be superimposed on its primary nucleotide sequence, and these additional annotations have proven to be a valuable resource for biomedical researchers (Maurano et al., 2012, Hnisz et al., 2013, Weedon et al., 2014, Praetorius et al., 2013).
However, no linear representation of the human genome – no matter how well annotated with functional elements – can fully capture the molecular mechanisms responsible for lineage-specific transcriptional regulation. The process of transcriptional regulation is not carried out on a linear string of nucleotides. In vivo this string of nucleotides is wrapped around histones, divided into chromosomes, highly compacted, and enclosed within the crowded and non-uniform environment of the interphase nucleus. Transcriptional regulation depends on physical interactions between regulatory elements like enhancers and promoters that are often not adjacent in a linear sense. The role of non-linear interactions in transcriptional regulation is exemplified by two fundamental properties of metazoan enhancer function: 1) enhancers can direct the expression of target genes located far away in linear distance (i.e. number of intervening base pairs), and 2) the gene most heavily influenced by an enhancer is not always the gene that is closest by linear distance (for illustrative examples see Lettice et al., 2003, Sagai et al., 2005, Montavon et al., 2011, Benko et al., 2009). Mounting evidence suggests that this ostensibly “long-range” regulation is possible because enhancers are in close physical proximity to the promoters of their target genes in vivo, despite long stretches of intervening nucleotides (de Laat and Duboule, 2013). This physical proximity allows protein complexes bound at enhancers to interact with those bound at promoters, thereby influencing transcription of target genes.
For much of its history, the study of genome organization has relied on microscopy-based techniques, which lack the resolution necessary to observe individual physical interactions like those between an enhancer and promoter. However, researchers have overcome this limitation in recent years with a series of molecular techniques based on the concept of Chromatin Conformation Capture (3C) (de Laat and Dekker, 2012, de Wit and de Laat, 2012, Dekker et al., 2002, Dekker et al., 2013). Briefly, these 3C-derived technologies (collectively referred to here as “C-technologies”) have a common methodological underpinning in which chemical crosslinking is used to secure 3D contacts between genomic loci occurring in live cells. This cross-linked chromatin is then isolated, digested with a restriction enzyme, and re-ligated in extremely dilute solution so that only loci that were contacting each other in vivo (and thus fixed together by crosslinking) will be ligated together. Therefore, in theory each ligation product contains a pair of loci that were in contact in vivo at the time of crosslinking. These ligation products can then be assayed to determine the frequency of contacts between specific loci, albeit with varying scope and throughput. Collectively, data from C-technologies (which we refer to below as “C-data”) has allowed researchers to answer questions about genome organization that were previously beyond reach.
Here, we discuss recent findings related to 3D genome organization in mammalian cells, with a particular focus on how different levels of organization contribute to lineage-specific transcriptional regulation. As we are primarily focused on global principles, we rely heavily on evidence from genome-wide studies, although key findings at specific gene loci are also discussed where applicable. We begin our discussion with higher-order organizational features that are observed at the level of the whole genome or whole chromosome, and work progressively downward in scale to the level of interactions between individual genomic loci. Throughout the review, we highlight changes in genome organization that occur during the course of differentiation, and we conclude with a discussion of genome organization in pluripotent cells. In sum, we believe that recent developments firmly support the notion that genome organization plays an essential role in orchestrating the lineage-specific gene expression programs that underlie cellular identity.
Higher-order genome organization influences but does not determine transcriptional output
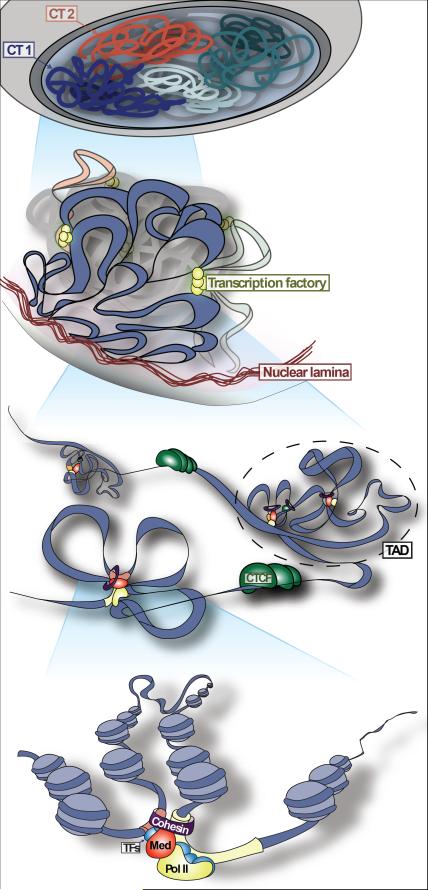
The genome is organized at many levels ranging from higher-order structures that are visible under the microscope down to smaller-scale structures that are detectable only by molecular techniques (Figure 1) (Gibcus and Dekker, 2013, Bickmore, 2013). Perhaps the most fundamental unit of higher-order genome organization is the chromosome. Each chromosome occupies its own sub-volume of the interphase nucleus, known as a Chromosome Territory (CT) (Cremer and Cremer, 2010). CTs can be visualized by Fluorescent in Situ Hybridization (FISH) using probes sets designed to paint entire chromosomes (Bolzer et al., 2005), and are also evident in C-data which demonstrate a consistent preference for intra-chromosomal over inter-chromosomal interactions (Lieberman-Aiden et al., 2009). Although CTs are spatially distinct, there is considerable intermingling between different chromosomes near the border of CTs (Branco and Pombo, 2006). The position of specific regions within their resident CT is non-random, and is correlated (albeit loosely) with transcriptional activity. Gene-rich regions tend to localize to the periphery of CTs (Boyle et al., 2011), which likely facilitates access to transcriptional machinery as well as sharing of this machinery between active genes on different chromosomes (Schoenfelder et al., 2010, Osborne et al., 2004). It has also been observed that specific regions can shift position from the CT interior to the CT periphery as genes in those regions become active during development (Morey et al., 2007, Chambeyron and Bickmore, 2004). While CT positioning correlates with transcriptional activity, the details of this relationship remain unclear. Notably, a shift in CT position is not always accompanied by a change in transcriptional activity, and active transcription is not limited to a specific zone of the CT (Morey et al., 2007, Zink et al., 2004). In this way, CT positioning exemplifies a common theme in higher-order genome organization: organization and transcriptional activity influence each other, but one does not strictly determine the other (Misteli, 2009, Cavalli and Misteli, 2013).
Figure 1. Different levels of genome organization.
[From top to bottom] Level 1: Chromosomes occupy distinct sub-regions of the nucleus known as chromosome territories (CTs). Individual chromosomes are indicated by different colors. Level 2: Transcriptionally inactive regions are enriched at the nuclear periphery where they contact the nuclear lamina (red). Actively transcribed genes often co-localize at RNA polymerase II transcription factories (yellow). These and other instances of co-localization between regions with similar transcriptional activity may provide the physical basis for the observations of A and B compartments in C-data. Level 3: Topological domains, or Topologically-Associating Domains (TADs) are regions of frequent local interactions separated by boundaries across which interactions are less frequent. CTCF binding sites and other sequence features (TSS, SINEs; not depicted here) are enriched at TAD boundaries. Note that CTCF also binds within TADs. Cohesin is often present at TAD boundaries, although it is not shown here. Level 4: Transcriptional regulation depends on long-range Interactions between cis-regulatory elements such as enhancers (light red) and promoters (light yellow). These cis-regulatory interactions are facilitated by proteins including Transcription Factors (“TFs”; blue), co-factors such as Mediator (“Med”; red) and Cohesin (purple ring), and RNA Polymerase II (“Pol II”; yellow).
The position of a given CT within the nucleus is highly stable through interphase, but a reshuffling of chromatin occurs during mitosis such that neighboring CTs can vary between mother and daughter cells, and thus also between cells within an ostensibly homogenous population (Thomson et al., 2004, Walter et al., 2003, Nagano et al., 2013, Parada et al., 2003). Despite this cell-to-cell variation, there are several features of genome organization above the level of the CT that are consistent across a population of cells. One such feature is that genomic regions tend to contact other regions with similar transcriptional activity. High-throughput C-technologies have demonstrated that regions showing characteristics of transcriptional activity (including accessible chromatin, activating histone modifications, high gene density, and high expression levels) most frequently interact in space with other active loci (Lieberman-Aiden et al., 2009, Simonis et al., 2006). Similarly, regions that lack characteristics of transcriptional activity tend to interact with other inactive regions. This tendency of regions with similar transcriptional activity to contact each other extends beyond a single chromosome, as the same trend is readily apparent in C-data even when only trans contacts are considered. These distinct active and inactive networks of co-interaction are referred to in the literature as the A and B compartments, respectively. It is not yet clear how these compartments (active compartment A and inactive B compartments) are established, but they presumably reflect a global tendency of euchromatin and heterochromatin to segregate in space. The concept that regions with similar transcriptional activity can co-localize in nuclear space is well established. For example, rRNA gene clusters from different chromosomes co-localize at the nucleolus where they undergo transcription by RNA polymerase I. Genes transcribed by RNA polymerase II also co-localize at foci of transcriptional activity known as transcription factories (Figure 1), although there can be hundreds or thousands of such factories in a single nucleus (Papantonis and Cook, 2013, Edelman and Fraser, 2012). In addition, transcriptionally inactive regions are enriched at the nuclear periphery in most cell types, leading to the classic appearance of a dense ring of heterochromatin just under the inner nuclear membrane in electron micrographs (Padeken and Heun, 2014).
Genomic regions at the nuclear periphery have been studied in further detail using the DamID method (van Steensel and Henikoff, 2000). DamID can identify regions that come into contact with proteins of the nuclear lamina, a filamentous network of proteins abutting the inner nuclear membrane (Pickersgill et al., 2006, Guelen et al., 2008). Genomic regions that contact the nuclear lamina, which are known as Lamin Associated Domains (LADs), are characterized by low levels of transcriptional activity, low gene density, and repressive histone modifications including H3K27me3 and H3K9me2 (Guelen et al., 2008, Kind et al., 2013, Akhtar et al., 2013). These observations suggest a link between transcriptional silencing and the nuclear lamina. Consistent with this theory, the association of specific genes with the nuclear lamina often coincides with their transcriptional silencing during differentiation (Peric-Hupkes et al., 2010). Examples include the key pluripotency genes Oct4, Nanog, and Klf4. Conversely, loss of association with the lamina and re-positioning away from the nuclear periphery often coincides with transcriptional activation (Kosak et al., 2002, Williams et al., 2006, Peric-Hupkes et al., 2010). Forcing the localization of specific genomic regions to the nuclear periphery via tethering to the lamina or other proteins of the inner nuclear membrane leads to a loss of transcriptional activity (Finlan et al., 2008, Reddy et al., 2008). It is important to note, however, that not all genes are equally affected by such forced relocation, and localization to the nuclear periphery is not incompatible with active transcription. Moreover, a given region is not found exclusively at either the nuclear periphery or the interior in a population of cells. The specific regions associated with the lamina can differ considerably within a population of cells, and even between mother and daughter cells (Kind et al., 2013). Thus, nuclear localization can influence transcriptional activity, but it does not determine the transcription level of any given gene. We suspect that this pattern – of widespread but non-definitive influence on transcriptional activity – is likely to be characteristic of many features of higher-order genome organization.
Topological domains coordinate regulatory influences
At increasing resolution, below the scale of an individual CT, another major feature of genome organization is observed: chromosomes are comprised of structural units called Topological Domains, also known as Topologically-Associating Domains (TADs) (Nora et al., 2012, Dixon et al., 2012). TADs are regions of high local contact frequency, which are separated by sharp boundaries across which contacts are relatively infrequent (Figures 1, 2). Mammalian genomes contain roughly 2000 TADs covering more than 90% of the mapable genome, and varying in size from a few hundred kilobases (kb) to several megabases with an average size of approximately 1 Mb. TADs are too small to study comprehensively with current microscopy-based methods, but visual evidence obtained by FISH is generally consistent with C-data (Dixon et al., 2012, Nora et al., 2012, Sofueva et al., 2013). It has also been suggested that TADs correspond to chromatin structures of roughly the same size that were previously observed in micrographs (Gibcus and Dekker, 2013).
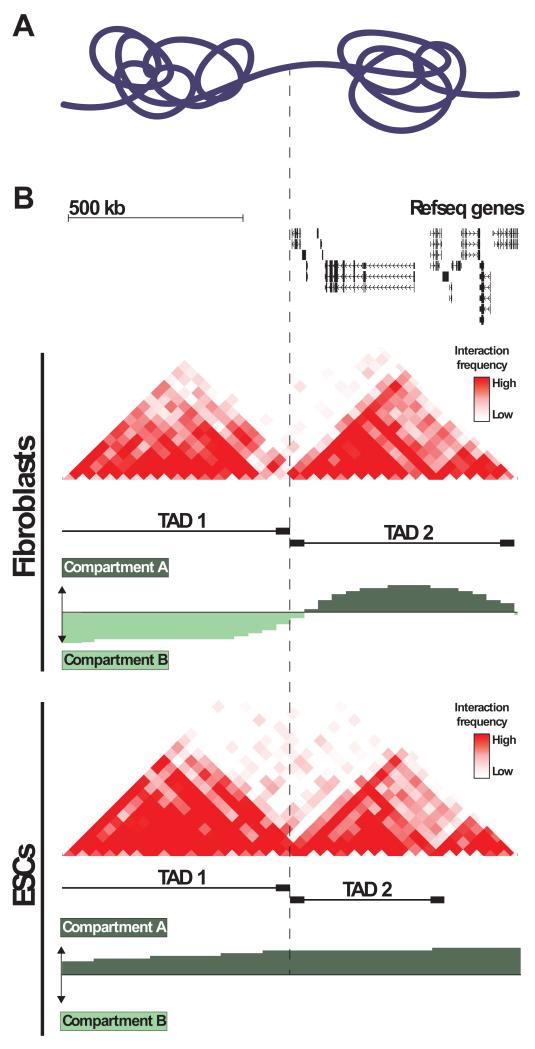
Figure 2. TADs and A/B compartments.
A) Diagrammatic representation of two neighboring TADs. B) UCSC genome browser view of the region chr11:115,470,000-116,770,000 which contains two adjacent TADs. [Top] Scale bar and refseq genes. [Middle] C-data from IMR90 fibroblasts. Tracks show pairwise interaction frequencies (red; 40 kb bins), TADs (black bars), and compartments A and B (green). Dashed line marks the boundary between TAD 1 and TAD 2. Note that TAD 2 contains far more genes than TAD 1, and is in the inactive compartment A in IMR90, while TAD 1 is in the inactive compartment A. [Bottom] C-data from human ESCs. Tracks are arranged as above for IMR90. Note that overall TAD structure and location of TAD boundaries do not differ significantly between IMR90 and ESCs. However, TAD1 is in the active compartment A in ESCs. Association of this gene-poor TAD with compartment A in ESCs may be related to the global pervasiveness of open chromatin in pluripotent cells (see section “Genome organization and pluripotency” for further discussion). All C-data taken from Dixon et al. (2012).
A growing body of evidence suggests that TADs are a fundamental unit of genome organization. TADs have now been described in every mouse and human cell type in which they have been scrutinized (Dixon et al., 2012, Zuin et al., 2014, Jin et al., 2013, Naumova et al., 2013, Nora et al., 2012, Sofueva et al., 2013), as well as in Drosophila (TAD size is considerably smaller in Drosophila at ~100 kb on average) (Sexton et al., 2012). The boundaries between TADs are strikingly consistent across cell types. Roughly 50-90% of TAD boundaries overlap in pairwise comparisons between cell types (Dixon et al., 2012). In cases where boundaries do not overlap, it is often due to the lack of a precise definition of what constitutes a TAD boundary rather than gross changes in patterns of local interactions (although in isolated cases such gross changes are observed). The locations of TAD boundaries are also highly conserved between mouse and human, indicating that both the existence and location of TADs have functional significance that is under selective pressure. In addition, TADs are not detectable during mitosis (Naumova et al., 2013), suggesting that their function is specific to interphase when transcription is most active.
The transcriptional regulation of genes within the same TAD appears to be coordinated in a number of ways. TADs frequently overlap with regions demarcated by other functional annotations related to transcriptional activity including histone modifications (e.g. H3K9me2, H3K27me3), replication timing, and association with the nuclear lamina (Dixon et al., 2012, Nora et al., 2013, Nora et al., 2012). Transitions between compartment A and compartment B also frequently occur at TAD boundaries (Dixon et al., 2012). That is, a given TAD tends to be all in the active compartment A, or all in the inactive compartment B. Notably, TADs in the active compartment A tend to contain a higher density of internal interactions (Sofueva et al., 2013), as might be expected given the role of interactions between cis-regulatory elements in transcriptional activity. The same TAD can be found in different compartments (i.e. A or B) in different cell types (Figure 2). Such a shift between compartments is often accompanied by a respective gain or loss of internal interactions. TADs can also gain or lose association with the nuclear lamina during differentiation (Nora et al., 2012). Taken together, these data suggest that TADs represent structural units on which broad (but non-definitive) regulatory influences can be applied domain-wide.
Particular attention has been paid to the boundaries between adjacent TADs. By definition, cis interactions across TAD boundaries are infrequent, suggesting that these boundaries may limit the potential target genes of a given enhancer, or vice versa limit the potential enhancers of a given target gene. Indeed, promoters and enhancers within the same TAD often show coordinated activity (Shen et al., 2012). Moreover, the insertion of a reporter construct designed to act as a regulatory sensor into different locations within the same TAD yields highly similar patterns of reporter gene expression in transgenic mouse embryos (Symmons et al., 2014), further supporting a role for TAD boundaries in demarcating zones of enhancer influence. It has also been noted that well-described cases of long-range regulation involve a promoter and distal enhancer that lie within the same TAD (Smallwood and Ren, 2013). While TAD boundaries seem to play a general role in constraining interactions, they are likely to influence transcriptional regulation in a variety ways. In one example, the HOXD gene cluster straddles the border between two TADs, and is influenced by distal regulatory elements from those different TADs at different stages in development (Andrey et al., 2013).
Evidence suggests that specific sequence features at TAD boundaries contribute to their formation. Several sequence features, including binding sites for the protein CTCF, are highly enriched at TAD boundaries (Dixon et al., 2012). The function of CTCF is multifaceted and will be discussed in further detail below, but for the sake of this discussion it is important to note that CTCF can function as a transcriptional insulator in certain contexts by blocking enhancer-promoter interactions and/or preventing the spread of epigenetic marks (Bell et al., 1999, Cuddapah et al., 2009). In one study, deletion of a specific TAD boundary containing CTCF binding sites led to an increase in interactions between adjacent TADs (Nora et al., 2012). However, the deleted region encompasses more than 50 kb, making it hard to attribute boundary activity at this locus to any specific sequence element. From a global perspective, knockdown of CTCF leads to an increase in interactions between adjacent domains (so-called “inter-domain interactions”), though not complete abrogation of TAD boundaries (Zuin et al., 2014). Loss of Cohesin (recruited by CTCF and present at many TAD boundaries) also leads to an increase in inter-domain interactions (Sofueva et al., 2013). However, Cohesin loss appears to have a lesser impact on inter-domain interactions than does loss of CTCF (Zuin et al., 2014). Although CTCF clearly plays some role in maintaining TAD boundaries, the relationship between CTCF and TAD boundaries is not clear-cut. The majority of CTCF binding sites in the genome occur within TADs rather than at their boundaries, and nearly a quarter of TAD boundaries show no evidence of CTCF binding at all (Dixon et al., 2012). TAD boundaries are also enriched for SINE elements and Transcriptional Start Sites (TSSs, particularly those of so-called “housekeeping” genes), but the requirement of these elements for boundary activity has not been explored in as much detail.
While TAD boundaries appear sharp when viewed at Mb scale, at higher resolution it is not exactly clear where one TAD ends and another begins. TAD boundaries range in size from tens of kb to more than 100 kb. The lack of precise boundary locations may be due in part to limited resolution of the C-technologies used to identify TAD boundaries (currently between ~10-40 kb), but it is almost certainly also due to the physical nature and scale of the TAD boundaries themselves. We suspect that in most cases the formation of a TAD boundary requires more than one sequence element – for example, the combination of several CTCF binding sites, and perhaps housekeeping TSSs and SINEs, spread over several kb. Interestingly, a recent study has identified sub-TADs, which are similar to TADs but are roughly one tenth of the size on average (Phillips-Cremins et al., 2013). We theorize that while it may take multiple CTCF binding sites and other sequence elements to create a full TAD boundary, one or two such elements may be sufficient to create a sub-TAD boundary. Additional experiments, particularly those involving further genetic manipulation of TAD boundaries, are likely to reveal much more about the mechanisms involved in the formation of TADs and their boundaries.
Interactions between cis-regulatory elements direct lineage-specific transcription
To this point we have mainly discussed regulatory influences imposed by higher-order genome organization, which can be applied at a domain-wide level through TADs, but these influences account for only a small portion of transcriptional regulation from the perspective of a single gene. Much of the additional influence on a gene’s transcription comes from cis-regulatory elements such as enhancers and promoters, and it is well established that 3D interactions between these elements are integral to their function (Visel et al., 2009, de Laat and Duboule, 2013). Specific interactions usually cannot be observed under the microscope, particularly when they occur in cis (as most do). However, a rich landscape of interactions between specific genomic loci is readily detectable using C-technologies. The probability of contact between two loci is governed to some extent by random collision, which is influenced heavily by the linear distance between those loci (Lieberman-Aiden et al., 2009, Bornfleth et al., 1999, Marshall et al., 1997). However, certain contacts occur far more often than expected by chance based on the linear distance between the loci involved. Hereafter, we use the term “interaction” to describe the relationship between loci that are in contact more frequently than would be expected based on linear distance. The term “looping” is sometimes used to describe such interactions, but we avoid using that term here because we feel that “looping” more accurately describes the sequence between interacting loci, which may or may not adopt the shape of a loop.
The first demonstration that 3D interactions between cis-regulatory elements contributes to transcriptional regulation came from studies of the β-globin gene cluster in mammals (Noordermeer and de Laat, 2008). Using C-technologies and other molecular techniques it was revealed that the promoters of active β-globin genes interact with an upstream regulatory sequence known as the Locus Control Region (LCR), despite more than 40 kb of intervening sequence (Carter et al., 2002, Tolhuis et al., 2002). These interactions were not observed in cell types where β-globin genes are silent, suggesting a role for the interactions in the lineage-specific regulation of β-globin genes. Similar cis-regulatory interactions (i.e. interactions between cis-regulatory elements such as promoters and enhancers) have now been described at many other genes (Montavon et al., 2011, Smemo et al., 2014, Phillips-Cremins et al., 2013, Visser et al., 2012). Researchers have also employed higher-throughput C-technologies to study interactions in parallel across many loci or even genome-wide (Dostie et al., 2006, Phillips-Cremins et al., 2013, Sanyal et al., 2012, Jin et al., 2013, Kieffer-Kwon et al., 2013, Li et al., 2012, Fullwood et al., 2009, Handoko et al., 2011, Zhang et al., 2013b, de Wit et al., 2013, Denholtz et al., 2013, Hughes et al., 2014). These studies consistently demonstrate that reproducible interactions are common in mammalian genomes, and that interacting loci are highly enriched for characteristics of cis-regulatory elements. One recent C-study detected more than a million interactions genome-wide between loci that are on average separated by roughly 100 kb, including approximately 30,000 interactions between active promoters and putative enhancers (as identified by epigenetic signature) (Jin et al., 2013). Notably, the vast majority of these interactions did not cross a TAD boundary, consistent with the role of TAD boundaries in constraining 3D interactions.
Several additional characteristics of cis-regulatory interactions are now apparent. First, interacting partners are not readily predicted by linear distance. Strikingly, within the ENCODE pilot regions (covering roughly 1% of the human genome) fewer than 10% of all interactions between TSSs and distal regions involved the closest TSS by linear distance (Sanyal et al., 2012). Second, enhancers and promoters do not interact in a simple 1:1 relationship. Studies have uncovered complex webs of interactions in which one promoter often interacts with multiple enhancers, one enhancer often interacts with multiple promoters, promoters often interact with other promoters, and enhancers often interact with other enhancers (Jin et al., 2013, Sanyal et al., 2012, Zhang et al., 2013b, Li et al., 2012, Kieffer-Kwon et al., 2013, Clowney et al., 2012). As further illustration of this regulatory complexity, the deletion of either of two long-range enhancers of the Aicda gene leads to decreased expression of both Aicda and Apobec1 despite more than 50 kb separating their TSSs. These enhancer deletions also cause a reduction in RNA polymerase II binding at numerous regulatory elements throughout a region of nearly 100 kb containing the two genes (Kieffer-Kwon et al., 2013). A third general principle of cis-regulatory interactions is that they often vary between cell types, which is particularly true for interactions between promoters and putative enhancers. For example, most interactions involving a TSS in the ENCODE pilot regions were found to be specific to one of three cell types examined (GM12878, K562 and HeLa-S3) (Sanyal et al., 2012). Fourth, the presence of putative enhancer-promoter interactions is highly correlated with a gene’s transcriptional activity (Sanyal et al., 2012, Jin et al., 2013, Kieffer-Kwon et al., 2013). One interesting exception to this trend is housekeeping genes, which tend to be highly expressed but not involved in interactions with putative enhancers (Jin et al., 2013, Li et al., 2012). These observations support the notion that lineage-specific genes are particularly dependent on long-range regulatory interactions. It has been observed, however, that some broadly-expressed genes (e.g. Myc) interact with distinct sets of enhancers in different cell types (Kieffer-Kwon et al., 2013).
Although there is a strong connection between transcriptional activity and enhancer-promoter interactions, it can be difficult to determine whether these interactions are a cause or a consequence of transcriptional activity. Here again, studies of the β-globin locus have been groundbreaking. Interactions between the LCR and β-globin genes are not simply a consequence of transcription, because inhibition of transcription by treatment with RNA polymerase II inhibitors does not disrupt these interactions, despite a drastic reduction in β-globin transcription (Palstra et al., 2008). Another line of evidence supporting a causal relationship between LCR-promoter interactions and transcriptional output is that forced ectopic interactions between the LCR and β-globin promoter (i.e. the creation of LCR-promoter interactions in cells where such an interaction is not naturally present) stimulates β-globin transcription. In a key study, Deng and colleagues (2012) created an ectopic interaction between the β-globin promoter and LCR in the pro-erythroblast cell line GE1, which does not normally express β-globin nor display an interaction between promoter and LCR. Creation of this ectopic interaction caused a dramatic increase in β-globin expression, albeit still below levels that would be considered full β-globin transcriptional activity (Deng et al., 2012).
Genome-wide evidence is consistent with the above findings, and demonstrates that enhancer-promoter interactions often exist prior to the onset of transcription. In one recent study, Jin and colleagues (2013) treated IMR90 fibroblasts with TNFα and then used high throughput C-technology to study changes in interactions that occur after treatment. As might be expected, TNFα treatment led to the induction of hundreds of genes, and these genes interacted with putative enhancers that also became active upon treatment with TNFα. More surprisingly, the TNFα-responsive enhancers were already involved in physical interactions with their target promoters prior to treatment and subsequent upregulation. Similar pre-induction interactions have been observed in the context of other cell types and other stimuli (Eijkelenboom et al., 2013, Hakim et al., 2011). Large-scale changes in interactions also precede transcriptional changes during somatic cell reprogramming to the induced pluripotent state (Apostolou et al., 2013). These data support a model in which cells are primed to respond to a specific set of developmental or environmental stimuli through pre-existing 3D interactions, which are likely anchored by pioneer transcription factors (Jin et al., 2011). We speculate that as differentiation proceeds, cells gain priming interactions for stimuli that are important at later stages of differentiation, while losing priming interactions required at earlier stages.
cis-regulatory interactions are secured by TFs and architectural proteins
Central to any discussion of cis-regulatory interactions is a consideration of how, at the molecular level, these interactions are established and maintained. At the sequence level both promoters and enhancers are composed of binding sites for TFs. We use the term TF here to refer to sequence-specific DNA binding factors, differentiating them from cofactors that are recruited to cis-regulatory elements but do not bind DNA in a sequence-specific manner. A classic model posits that promoters bind a core set of General Transcription Factors (GTFs), and these GTFs in turn recruit RNA polymerase II and additional cofactors (in the case of most protein-coding genes) (Fuda et al., 2009). The repertoire of TFs that bind at enhancers is more contingent on the cell type in question, but a common feature is that these enhancer-bound TFs recruit widely-expressed cofactors, including p300/CBP, Mediator, and Cohesin (Visel et al., 2009, Carlsten et al., 2013). In this classic model, enhancer-promoter communication involves a physical interaction between the factors bound at the enhancer and those bound at the promoter (Carlsten et al., 2013, Visel et al., 2009, Fuda et al., 2009). Recent studies have further illuminated the roles of many of these factors in establishing and maintaining the 3D interactions between regulatory elements.
The involvement of TFs in cis-regulatory interactions is not surprising given that TFs are essential to the function of regulatory elements, which in turn is facilitated by interactions. There is a clear correspondence between putative sites of TF binding and sites involved in 3D interactions (Jin et al., 2013, Fullwood et al., 2009, Li et al., 2012). In ESCs, binding sites for the pluripotency factors Oct4, Sox2, and Nanog are highly enriched at interacting loci (de Wit et al., 2013, Apostolou et al., 2013). Loss-of-function experiments further support the necessity of TFs in cis-regulatory interactions. Knockdown of either of the key pluripotency TFs Oct4 or Nanog in mouse ESCs results in specific loss of interactions anchored by these TFs (de Wit et al., 2013, Levasseur et al., 2008). Likewise, TFs bound at the β-globin promoter and LCR are required for LCR-to-promoter interactions in erythroid progenitors (Vakoc et al., 2005). Naturally occurring sequence variation in TF binding sites can also disrupt cis-regulatory interactions. The SNP rs12913832 is located in an enhancer that directs the expression of OCA2, a gene required for synthesis of the pigment melanin. The minor allele at rs12913832 interferes with TF binding to this enhancer, resulting in decreased enhancer-promoter interaction, and reduced OCA2 expression (Visser et al., 2012). Notably, this SNP has been associated by GWAS with a number of human pigment phenotypes, demonstrating that sequence variation that disrupts cis-regulatory interactions can have phenotypic consequences at the level of the whole organism.
The sequence-specific DNA binding factor CTCF stands apart from other TFs with respect to genome organization. A comprehensive discussion of the myriad functions of CTCF is beyond the scope of review, and this topic has been reviewed thoroughly elsewhere (Ong and Corces, 2014). However, one common theme in CTCF function appears to be that regions bound by CTCF are frequently engaged in physical interactions with themselves as well as with other regions (Handoko et al., 2011, Jin et al., 2013, Phillips-Cremins et al., 2013). Such observations have led to the description of CTCF as a “master weaver of genome” (Phillips and Corces, 2009), and more recently, as an “architectural protein” (Ong and Corces, 2014). CTCF is ubiquitously expressed, and binds to tens of thousands of sites throughout the genome (Kim et al., 2007, Barski et al., 2007, Chen et al., 2008). Many CTCF binding sites are consistent between cell types, and do not overlap classically defined enhancers or promoters. This suggests that at least part of CTCF’s function is to establish a structural framework that is similar between cell types (as seems true for the role of CTCF in establishing TAD boundaries). Within this framework, other factors may contribute more directly to lineage-specific transcriptional regulation (Shen et al., 2012, Neph et al., 2012). On the other hand, many other CTCF binding sites are not consistent between cell types, and do overlap enhancers and promoters. Lineage-specific binding of CTCF could be partly due to CpG methylation, which blocks CTCF binding when present in its recognition motif (Chen et al., 2012, Bell and Felsenfeld, 2000, Hark et al., 2000). In these instances, CTCF may be more directly involved in lineage-specific regulation. In different contexts, CTCF can act as a transcriptional activator, repressor, or insulator (Klenova et al., 1993, Bell et al., 1999, Cuddapah et al., 2009, Merkenschlager and Odom, 2013, Handoko et al., 2011). Thus, while it seems that the involvement of CTCF in 3D interactions is integral to its function, the impact of CTCF binding on transcription depends on the locus and cell type in question.
Despite apparent differences between CTCF and other TFs, they share the ability to recruit cofactors that are also involved in the formation of cis-regulatory interactions. One such cofactor is the Cohesin complex. Cohesin is well known for its role in holding sister chromatids together until anaphase when they are separated and migrate to opposite spindle poles. However, it has become increasingly clear that Cohesin is also a major player in transcriptional regulation during interphase, and that this regulatory function is largely independent of its role in sister chromatid cohesion (Merkenschlager and Odom, 2013, Remeseiro et al., 2013). Cohesin is commonly found at enhancers, where it acts together with the Mediator complex to maintain physical interaction between promoters and enhancers (Kagey et al., 2010). Mediator can directly interface with factors bound at enhancers and those bound at promoters, facilitating communication between them (Carlsten et al., 2013, Ebmeier and Taatjes, 2010, Lariviere et al., 2012). Cohesin is also present at CTCF binding sites, many of which are outside of traditional enhancers and lack Mediator binding (Wendt et al., 2008, Parelho et al., 2008, Rubio et al., 2008). In fact, it seems that in a given cell type (including ESCs) the majority of Cohesin binding falls into one of two categories: 1) sites that are co-occupied by Mediator and multiple TFs, or 2) sites that co-occupied by CTCF (Kagey et al., 2010, Yan et al., 2013, Faure et al., 2012, Hnisz et al., 2013).
The extensive role of Cohesin in 3D interactions is exemplified by a recent study that employed ChIA-PET (a C-technology that uses an immunoprecipitation step to specifically identify interactions that involve a particular protein of interest) to identify Cohesin-mediated interactions in developing mouse limbs (DeMare et al., 2013). This method identified more than 2,000 such interactions, including over 1000 interactions involving sites bound by CTCF, and 680 interactions between promoters and putative enhancers (although this study did not directly examine Mediator occupancy). Another recent study provides additional insight into the roles of Cohesin, Mediator, and CTCF in cis-regulatory interactions. Phillips-Cremins et al. (2103) used C-technology to study interactions within six ~1-2 Mb regions in murine ESCs and Neuronal Precursor Cells (NPCs), identifying roughly 500 such interactions. Strikingly, they found that more than 80% of these interactions involved loci bound by some combination of Cohesin, Mediator, and/or CTCF, leading the authors to label these factors as “architectural proteins.” More specifically, properties of these interactions varied depending on which architectural proteins were involved. Interactions between loci bound by Cohesin-Mediator or Mediator alone were often specific to either ESCs or NPCs, consistent with the view that these interactions involve lineage-specific regulatory elements. In contrast, interactions between sites bound by Cohesin-CTCF or CTCF alone were often constant between these cell types. Cohesin-mediator interactions also occurred over shorter distances (mean <100 kb) than did Cohesin-CTCF interactions (mean >1 Mb).
The observations that Cohesin is widely involved in 3D interactions, and is a common thread between interactions with different properties, suggest that Cohesin may function as a general stabilizer of these interactions (Kagey et al., 2010). In further support of this theory, studies examining the effects of Cohesin loss have consistently demonstrated a requirement for Cohesin in maintaining interactions. Cohesin depletion leads to the loss of interactions at several individual gene loci including INFG, β-globin, Olig1, Nanog, OCT4, and Tcra (Chien et al., 2011, Phillips-Cremins et al., 2013, Apostolou et al., 2013, Seitan et al., 2011, Zhang et al., 2013a, Hadjur et al., 2009). The global effects of Cohesin depletion have also been investigated. Deletion of the Cohesin subunit Rad21 in non-cycling thymocytes leads to global misregulation of gene expression, and a loss of contacts between Cohesin-bound sites throughout the genome (Seitan et al., 2013). Notably, the set of misregulated genes is enriched for GO terms related to lineage-specific processes such as “hematopoiesis” and “lymphocyte activation.” In HEK293 cells, Zuin et al. (2014) observed a global loss of intra-TAD interactions (particularly those between Cohesin binding sites) upon depletion of Cohesin using an alternative approach in which a recognition site for the Human rhinovirus 3C (HRV) protease is introduced into RAD21, allowing for its rapid cleavage after HRV expression. A third study also found widespread perturbation of interactions and gene expression upon deletion of Rad21 in post-mitotic astrocytes (Sofueva et al., 2013). This study reported a loss of both intra-TAD and inter-TAD interactions after Cohesin loss. Together, these data support a model in which Cohesin provides structural stability to a range of interactions with a variety of potential regulatory consequences.
The above findings paint a complex picture in which a number of trans factors including lineage-specific TFs, CTCF, Mediator, and Cohesin are involved in anchoring different types of cis-regulatory interactions (including, but not limited to, interactions between promoters and enhancers). We have focused here on several of the factors involved in cis-regulatory interactions, but it is also important to note that many additional factors contribute to genome organization and transcriptional regulation in important ways that were not elaborated on here (Jones et al., 2000, Cai et al., 2006, Soler et al., 2010). Despite the complexity, common themes have emerged that may characterize the molecular machinery involved in nearly all cis-regulatory interactions (Figure 3). First, interactions are anchored by factors that recognize DNA in a sequence-specific manner, thereby determining which specific loci are most likely to participate in stable interactions. Examples of such factors mentioned above include TFs, GTFs, and CTCF. Second, these DNA binding factors in turn recruit cofactors such as Cohesin and Mediator, which further promote and stabilize the interactions. One interesting twist on these themes is a newly-described class of non-coding RNA (ncRNA-a), which can direct the transcriptional upregulation of other genes in cis, thus functioning analogously to classically-defined enhancer elements (Lai et al., 2013, Orom et al., 2010, Orom and Shiekhattar, 2013). As ncRNA-a are transcribed, they engage in physical interactions with their target promoters, and these interactions are dependent on the recruitment of Mediator by the nascent ncRNA-a. This fascinating discovery indicates that, like TFs, ncRNA-a can anchor cis-regulatory interactions and recruit cofactors to further stabilize these interactions.
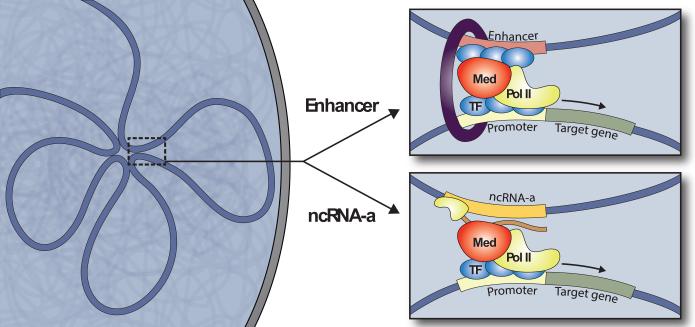
Figure 3. Molecular machinery of cis-regulatory interactions.
[Top] Enhancer-promoter interactions. TFs (blue) bind to enhancers and promoters. RNA polymerase II (yellow) is recruited to the promoter, and Mediator (red) and Cohesin (purple ring) are recruited to the enhancer. Cohesin stabilizes the interaction, perhaps by forming a ring around interacting loci (although there is little experimental evidence to support this at present). [Bottom] ncRNA-mediated interactions. ncRNA-a (orange) have enhancer-like function: they upregulate genes in cis, dependent on Mediator and coincident with 3D interactions between the ncRNA locus and target gene promoter. Involvement of Cohesin in these interactions has not been demonstrated to date.
Genome organization and pluripotency
Pluripotent cells have been integral to the study of genome organization. Indeed, many of the studies cited above examine ESCs and their derivatives. ESCs have many of the same features of genome organization as differentiated cells, including A/B compartments, LADs, TADs, and cis-regulatory interactions. However, there are other organizational features that appear to be somewhat unique to pluripotent cells (Denholtz and Plath, 2012, Apostolou and Hochedlinger, 2013). One such feature is that chromatin is generally less condensed and more loosely organized in pluripotent cells than in lineage committed cells (Melcer and Meshorer, 2010, Meshorer et al., 2006, Gaspar-Maia et al., 2011). Correspondingly, histone modifications that mark heterochromatin expand during lineage commitment to cover a substantially larger portion of the genome in differentiated cells than in ESCs (Hawkins et al., 2010, Zhu et al., 2013). More recently, analysis of C-data revealed that transcriptionally inactive regions tend to participate in fewer specific long-range interactions in ESCs than in non-ESCs (de Wit et al., 2013). These results are all consistent with a chromatin conformation that is particularly malleable in pluripotent cells, and which may function to maintain a state of permissiveness for the different transcriptional programs required for lineage specification. Although condensed heterochromatin is less prevalent in ESCs than in other cell types, transcriptional repression is still important to the pluripotent state. The repression of many genes associated with lineage commitment requires Polycomb group (PcG) proteins. Genomic regions enriched for PcG binding and/or its associated repressive histone modification H3K27me3 contact each other at high frequency in C-data generated from ESCs, and this contact is dependent on the Eed subunit of the Polycomb PRC2 complex (Denholtz et al., 2013). These data are consistent with the observation that PcG proteins form visible foci in the nuclei of ESCs (Eskeland et al., 2010, Isono et al., 2013), and with a large body of work linking Polycomb-mediated silencing to genome organization in Drosophila (reviewed in Cheutin and Cavalli, 2014). However, PcG foci and PcG-mediated chromatin interactions have also been observed in non-pluripotent cells, suggesting that while PcG-mediated silencing is critical to pluripotency it is probably also important in differentiated cells (Isono et al., 2013, Tiwari et al., 2008).
Another unique feature of higher-order genome organization in pluripotent cells is that regions with a high density of binding sites for the key pluripotency TFs Oct4, Sox2, and Nanog (together abbreviated as OSN) tend to co-localize in nuclear space. In ESCs, OSN binding is the strongest determinant of genome-wide contact frequency other than transcriptional activity (i.e. A/B compartments) (Denholtz et al., 2013). Furthermore, an examination of long-range contacts in ESCs (in this case between loci separated by more than 5 Mb) revealed that clustered binding of OSN is highly enriched at regions involved in such contacts (de Wit et al., 2013). These observations suggest that OSN are directly involved in higher-order genome organization in ESCs, which is further supported by the demonstration that loss of either Oct4 or Nanog diminishes long-range contacts between OSN-bound regions. Moreover, binding of a Nanog-LacR fusion protein to a LacO array inserted into the genome was sufficient to create long-range contacts between this ectopic array and endogenous regions bound by OSN. Surprisingly, binding of CTCF and Cohesin is not enriched at long-range contact sites in ESCs, suggesting that the role of OSN in shaping higher-order structure of the pluripotent genome is independent of these so-called architectural proteins. Although OSN seem to facilitate long-range contacts independently of Cohesin, OSN also anchor short-range cis-regulatory interactions that do require Cohesin as discussed in the previous section (Kagey et al., 2010, Phillips-Cremins et al., 2013, Nitzsche et al., 2011).
The OSN proteins are clearly involved in organizing the pluripotent genome, but another important aspect of the relationship between OSN and genome organization is that the OSN genes are highly regulated and are thus influenced by genome organization themselves in many of the ways discussed throughout this review. For example, OSN and other key pluripotency genes are in contact with the silencing environment of the nuclear lamina less frequently in ESCs than in differentiated derivates (Peric-Hupkes et al., 2010). In addition, interactions between the promoters of different pluripotency genes can be detected In ESCs both in cis and in trans (de Wit et al., 2013, Kieffer-Kwon et al., 2013, Apostolou et al., 2013), indicating that they colocalize in the pluripotent nucleus, perhaps at shared RNA Polymerase II transcription factories. Like many lineage-specific genes, the transcription of pluripotency genes is heavily influenced by interactions with distal regulatory elements. One particularly well-characterized example is Oct4, the expression of which is dependent on interactions between the Oct4 promoter and an upstream enhancer bound by multiple TFs (Oct4, Sox2, and Klf4), Mediator, and Cohesin (Wei et al., 2013, Zhang et al., 2013a, Huang et al., 2013). Knockdown of either Klf4 or of Cohesin abrogates the enhancer-promoter interaction, leading to a reduction in Oct4 transcription and subsequent disruption of the pluripotent state. The importance of cis-regulatory interactions to pluripotency is further underscored by the results of a shRNA-based screen of thousands of genes in ESCs to identify regulators of the pluripotent state (Kagey et al., 2010). As one might expect, this screen identified well-known pluripotency factors such as OSN. However, this screen also revealed that architectural proteins – including all four Cohesin subunits and more than 10 Mediator subunits – are critical regulators of the pluripotent state. Knockdown of either Mediator or Cohesin in ESCs causes reduced expression of pluripotency genes, and increased expression of genes associated with differentiation (Nitzsche et al., 2011, Tutter et al., 2009). Knockdown of Mediator or Cohesin also reduces the efficiency of somatic cell reprograming to induced pluripotent stem cells (iPSCs) (Apostolou et al., 2013, Zhang et al., 2013a). Thus, the impact of genome organization on pluripotency manifests at many levels.
Conclusions and future perspectives
It has been difficult to move beyond a linear model of the genome to a more comprehensive view of the genome as a 3D entity. The linear model has proven sufficient for a long time in part because many of the functional modules in the genome are arranged in linear fashion. For example, exons are always transcribed in linear order, and promoters are always located immediately upstream of the transcription unit. The linear arrangement of these modules is reflective of the transcriptional machinery on which their function depends. This machinery is processive – that is, it moves along a stretch of DNA in a line – and thus the functional modules on which the machinery acts are arranged in linear fashion in the genome. While a linear conception of the genome may be sufficient for predicting the direction of transcription, it is becoming increasingly inadequate for describing transcriptional regulation. Unlike the exons of a gene, the enhancers that regulate a gene’s transcription are often not arranged in a linear fashion with respect to the gene in question. Rather, enhancers can be found upstream or downstream of the genes they regulate, can act over large linear distances, and can skip over intervening genes. Just as the linear arrangement of exons reflects the processive nature of the transcriptional machinery, the non-linear arrangement of cis-regulatory elements reflects the nonprocessive nature of the cis-regulatory machinery (e.g. many of TFs and cofactors discussed above). We suggest that the machinery of transcriptional regulation is structural – that is, it relies on 3D interactions between modules that may be separated by considerable linear distance. In other words, just as the processive transcriptional machinery must traverse a DNA molecule to carry out its function, the structural regulatory machinery must establish and maintain 3D interactions to carry out its function.
We have limited our focus here to the connection between genome organization and lineage-specific transcriptional regulation, but genome organization plays a role in myriad other processes including DNA repair, DNA replication, and X chromosome inactivation (Engreitz et al., 2013, Xu et al., 2007, Zhang et al., 2012, Roukos et al., 2013, Cavalli and Misteli, 2013). The study of genome organization also informs our understanding of human disease in several ways. First, mutations in genes that encode genome and nuclear architectural components (including subunits of the Cohesin complex, Mediator complex, and Nuclear Lamins) can result in severe developmental phenotypes (Misteli, 2010, Carlsten et al., 2013). Second, genes encoding Mediator subunits, Cohesin subunits, and CTCF are also mutated at significant frequency in cancer, raising questions about the potential contribution of defects in genome organization to malignancy (Lawrence et al., 2014). Third, SNPs that are linked to human disease by Genome-Wide Association Study (GWAS) are commonly found within enhancers, suggesting that perturbation of long-range regulation is the mechanism behind a sizable portion of pathogenic sequence variation (Maurano et al., 2012, Hindorff et al., 2009). In order to establish the target gene of a given enhancer it is often necessary to demonstrate a 3D interaction between the enhancer and the target gene’s promoter (Smemo et al., 2014).
Our understanding of 3D genome organization is evolving rapidly. Despite tremendous progress, some of which has been covered here, many questions remain unanswered. These include important questions about the mechanisms that establish genome organization, and how at the molecular level each layer of genome organization influences transcriptional regulation. Molecular techniques are often performed on populations of cells, which can make it difficult to extrapolate about mechanisms that operate within an individual cell. If a single promoter is found to interact with multiple enhancers in population-derived data, do all of these interactions occur simultaneously within an individual cell, or do these multiple interactions only become apparent when data is merged from multiple cells? Likewise, how can we reconcile the cell-to-cell variability in higher-order genome organization with the reproducible correlations between gene expression and higher-order organization at the population level? Recent breakthroughs allowing for the application of C-technologies and DamID to single cells have started to close these gaps, and promise to yield additional insight in the future (Kind et al., 2013, Nagano et al., 2013). Advances in microscopy-based techniques and in computational modeling have also been trailblazing in this regard (Boyle et al., 2011, Song et al., 2014, Muller et al., 2012). In particular, one recent study used population-based C-data to train a biophysical model of TAD structure that could account for variation between cells in a population. This model predicted substantial variation in TAD structure between cells, and also identified key DNA sequences responsible for establishing TAD structures, both of which were validated by super-resolution DNA FISH (Giorgetti et al., 2014). Progress has also been made in distinguishing between homologous chromosomes in C-data (Selvaraj et al., 2013), currently a limitation of many C-studies. Perhaps the most pressing challenge is to continue to lower the cost and increase the resolution of these technologies so that they can be applied to additional cell types and experimental systems. As the complexities of lineage-specific gene expression continue to be unraveled, and as functional elements in expansive mammalian genomes continue to be delineated, we anticipate that the importance of studying the genome from a 3D perspective will only continue to increase.
Acknowledgements
We thank members of the Ren lab for helpful discussion and feedback – particularly Tinting Du, Anthony Schmitt, Andrea Local, Chloe Rivera, Fulai JIn, and Jesse Dixon. We also thank our reviewers for their valuable input. We apologize to the many authors whose work we were unable to highlight in this manuscript. This work was supported by funds from the Ludwig Institute for Cancer Research and NIH (grants U01ES017166, U01HL107442, P50GM085764 and U54 HG006997) to B.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhtar W, De Jong J, Pindyurin AV, Pagie L, Meuleman W, De Ridder J, Berns A, Wessels LF, Van Lohuizen M, Van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–27. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, Kharchenko PV, Park PJ, Hochedlinger K. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–71. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–5. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–96. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Benko S, Fantes JA, Amiel J, Kleinjan DJ, Thomas S, Ramsay J, Jamshidi N, Essafi A, Heaney S, Gordon CT, Mcbride D, Golzio C, Fisher M, Perry P, Abadie V, Ayuso C, Holder-Espinasse M, Kilpatrick N, Lees MM, Picard A, Temple IK, Thomas P, Vazquez MP, Vekemans M, Roest Crollius H, Hastie ND, Munnich A, Etchevers HC, Pelet A, Farlie PG, Fitzpatrick DR, Lyonnet S. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41:359–64. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA. The spatial organization of the human genome. Annu Rev Genomics Hum Genet. 2013;14:67–84. doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornfleth H, Edelmann P, Zink D, Cremer T, Cremer C. Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy. Biophys J. 1999;77:2871–86. doi: 10.1016/S0006-3495(99)77119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Rodesch MJ, Halvensleben HA, Jeddeloh JA, Bickmore WA. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. 2011;19:901–9. doi: 10.1007/s10577-011-9245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–88. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Carlsten JO, Zhu X, Gustafsson CM. The multitalented Mediator complex. Trends Biochem Sci. 2013;38:531–7. doi: 10.1016/j.tibs.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–6. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–9. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–30. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Tian Y, Shu W, Bo X, Wang S. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One. 2012;7:e41374. doi: 10.1371/journal.pone.0041374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cheutin T, Cavalli G. Polycomb silencing: from linear chromatin domains to 3D chromosome folding. Curr Opin Genet Dev. 2014;25C:30–37. doi: 10.1016/j.gde.2013.11.016. [DOI] [PubMed] [Google Scholar]
- Chien R, Zeng W, Kawauchi S, Bender MA, Santos R, Gregson HC, Schmiesing JA, Newkirk DA, Kong X, Ball AR, Jr., Calof AL, Lander AD, Groudine MT, Yokomori K. Cohesin mediates chromatin interactions that regulate mammalian beta-globin expression. J Biol Chem. 2011;286:17870–8. doi: 10.1074/jbc.M110.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, Legros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–37. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laat W, Dekker J. 3C-based technologies to study the shape of the genome. Methods. 2012;58:189–91. doi: 10.1016/j.ymeth.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- De Wit E, Bouwman BA, Zhu Y, Klous P, Splinter E, Verstegen MJ, Krijger PH, Festuccia N, Nora EP, Welling M, Heard E, Geijsen N, Poot RA, Chambers I, De Laat W. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–31. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- De Wit E, De Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Demare LE, Leng J, Cotney J, Reilly SK, Yin J, Sarro R, Noonan JP. The genomic landscape of cohesin-associated chromatin interactions. Genome Res. 2013;23:1224–34. doi: 10.1101/gr.156570.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–44. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M, Bonora G, Chronis C, Splinter E, De Laat W, Ernst J, Pellegrini M, Plath K. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13:602–16. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M, Plath K. Pluripotency in 3D: genome organization in pluripotent cells. Curr Opin Cell Biol. 2012;24:793–801. doi: 10.1016/j.ceb.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeier CC, Taatjes DJ. Activator-Mediator binding regulates Mediator-cofactor interactions. Proc Natl Acad Sci U S A. 2010;107:11283–8. doi: 10.1073/pnas.0914215107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman LB, Fraser P. Transcription factories: genetic programming in three dimensions. Curr Opin Genet Dev. 2012;22:110–4. doi: 10.1016/j.gde.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Eijkelenboom A, Mokry M, De Wit E, Smits LM, Polderman PE, Van Triest MH, Van Boxtel R, Schulze A, De Laat W, Cuppen E, Burgering BM. Genome-wide analysis of FOXO3 mediated transcription regulation through RNA polymerase II profiling. Mol Syst Biol. 2013;9:638. doi: 10.1038/msb.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, Mcdonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–64. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, Ramsay RG, Odom DT, Flicek P. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res. 2012;22:2163–75. doi: 10.1101/gr.136507.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–92. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–82. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, Tiana G, Heard E. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–63. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, De Klein A, Wessels L, De Laat W, Van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–3. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Sung MH, Voss TC, Splinter E, John S, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, De Laat W, Hager GL. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 2011;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan Y, Wei CL. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–8. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–9. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–91. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Wei Z, Lu W. Genome organization by Klf4 regulates transcription in pluripotent stem cells. Cell Cycle. 2013;12:3351–2. doi: 10.4161/cc.26577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Roberts N, Mcgowan S, Hay D, Giannoulatou E, Lynch M, De Gobbi M, Taylor S, Gibbons R, Higgs DR. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet. 2014;46:205–12. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- Isono K, Endo TA, Ku M, Yamada D, Suzuki R, Sharif J, Ishikura T, Toyoda T, Bernstein BE, Koseki H. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell. 2013;26:565–77. doi: 10.1016/j.devcel.2013.08.016. [DOI] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–4. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Ren B, Natarajan R. PU.1 and C/EBP(alpha) synergistically program distinct response to NF-kappaB activation through establishing monocyte specific enhancers. Proc Natl Acad Sci U S A. 2011;108:5290–5. doi: 10.1073/pnas.1017214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DO, Cowell IG, Singh PB. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays. 2000;22:124–37. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, Van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, Vian L, Nelson S, Zare H, Hakim O, Reyon D, Yamane A, Nakahashi H, Kovalchuk AL, Zou J, Joung JK, Sartorelli V, Wei CL, Ruan X, Hager GL, Ruan Y, Casellas R. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–20. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, De Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, Van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–92. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, Neiman PE, Lobanenkov VV. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–24. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–62. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere L, Seizl M, Cramer P. A structural perspective on Mediator function. Curr Opin Cell Biol. 2012;24:305–13. doi: 10.1016/j.ceb.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, De Beer P, Oostra BA, Goode D, Elgar G, Hill RE, De Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–35. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Levasseur DN, Wang J, Dorschner MO, Stamatoyannopoulos JA, Orkin SH. Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes Dev. 2008;22:575–80. doi: 10.1101/gad.1606308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G, Wei CL, Ge W, Wang H, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan Y. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7:930–9. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcer S, Meshorer E. Chromatin plasticity in pluripotent cells. Essays Biochem. 2010;48:245–62. doi: 10.1042/bse0480245. [DOI] [PubMed] [Google Scholar]
- Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–97. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–16. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Self-organization in the genome. Proc Natl Acad Sci U S A. 2009;106:6885–6. doi: 10.1073/pnas.0902010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, De Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–45. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–19. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- Muller WG, Heymann JB, Nagashima K, Guttmann P, Werner S, Rehbein S, Schneider G, Mcnally JG. Towards an atlas of mammalian cell ultrastructure by cryo soft X-ray tomography. J Struct Biol. 2012;177:179–92. doi: 10.1016/j.jsb.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–53. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, Maurano MT, Humbert R, Rynes E, Wang H, Vong S, Lee K, Bates D, Diegel M, Roach V, Dunn D, Neri J, Schafer A, Hansen RS, Kutyavin T, Giste E, Weaver M, Canfield T, Sabo P, Zhang M, Balasundaram G, Byron R, Maccoss MJ, Akey JM, Bender MA, Groudine M, Kaul R, Stamatoyannopoulos JA. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, De Vries I, Ding L, Huebner N, Mann M, Stunnenberg HG, Buchholz F. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS One. 2011;6:e19470. doi: 10.1371/journal.pone.0019470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, De Laat W. Joining the loops: beta-globin gene regulation. IUBMB Life. 2008;60:824–33. doi: 10.1002/iub.129. [DOI] [PubMed] [Google Scholar]
- Nora EP, Dekker J, Heard E. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays. 2013;35:818–28. doi: 10.1002/bies.201300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, Van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Bluthgen N, Dekker J, Heard E. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–46. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154:1190–3. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–71. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Padeken J, Heun P. Nucleolus and nuclear periphery: Velcro for heterochromatin. Curr Opin Cell Biol. 2014;28C:54–60. doi: 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, De Laat W. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One. 2008;3:e1661. doi: 10.1371/journal.pone.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantonis A, Cook PR. Transcription factories: genome organization and gene regulation. Chem Rev. 2013;113:8683–705. doi: 10.1021/cr300513p. [DOI] [PubMed] [Google Scholar]
- Parada LA, Roix JJ, Misteli T. An uncertainty principle in chromosome positioning. Trends Cell Biol. 2003;13:393–6. doi: 10.1016/s0962-8924(03)00149-1. [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–33. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, Van Lohuizen M, Reinders M, Wessels L, Van Steensel B. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–13. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, Bland MJ, Wagstaff W, Dalton S, Mcdevitt TC, Sen R, Dekker J, Taylor J, Corces VG. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–95. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, De Wit E, Talhout W, Fornerod M, Van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–14. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Praetorius C, Grill C, Stacey SN, Metcalf AM, Gorkin DU, Robinson KC, Van Otterloo E, Kim RS, Bergsteinsdottir K, Ogmundsdottir MH, Magnusdottir E, Mishra PJ, Davis SR, Guo T, Zaidi MR, Helgason AS, Sigurdsson MI, Meltzer PS, Merlino G, Petit V, Larue L, Loftus SK, Adams DR, Sobhiafshar U, Emre NC, Pavan WJ, Cornell R, Smith AG, Mccallion AS, Fisher DE, Stefansson K, Sturm RA, Steingrimsson E. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell. 2013;155:1022–33. doi: 10.1016/j.cell.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–7. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Losada A. Cohesin in development and disease. Development. 2013;140:3715–8. doi: 10.1242/dev.090605. [DOI] [PubMed] [Google Scholar]
- Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341:660–4. doi: 10.1126/science.1237150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–14. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Faure AJ, Zhan Y, Mccord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, Flicek P, Dekker J, Merkenschlager M. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–77. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–71. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj S, J RD, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol. 2013;31:1111–8. doi: 10.1038/nbt.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, Mccleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–20. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, De Wit E, Van Steensel B, De Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr Opin Cell Biol. 2013;25:387–94. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung HK, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui CC, Gomez-Skarmeta JL, Nobrega MA. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–5. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–29. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, De Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, Van Ijcken W, Hou J, Steinhoff C, Rijkers E, Lenhard B, Grosveld F. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010;24:277–89. doi: 10.1101/gad.551810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu RM, Zhu P, Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–80. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson I, Gilchrist S, Bickmore WA, Chubb JR. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol. 2004;14:166–72. doi: 10.1016/j.cub.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Tiwari VK, Cope L, Mcgarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18:1171–9. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, De Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Tutter AV, Kowalski MP, Baltus GA, Iourgenko V, Labow M, Li E, Kadam S. Role for Med12 in regulation of Nanog and Nanog target genes. J Biol Chem. 2009;284:3709–18. doi: 10.1074/jbc.M805677200. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–62. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–8. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser M, Kayser M, Palstra RJ. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22:446–55. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol. 2003;160:685–97. doi: 10.1083/jcb.200211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Cebola I, Patch AM, Flanagan SE, De Franco E, Caswell R, Rodriguez-Segui SA, Shaw-Smith C, Cho CH, Lango Allen H, Houghton JA, Roth CL, Chen R, Hussain K, Marsh P, Vallier L, Murray A, International Pancreatic Agenesis, C. Ellard S, Ferrer J, Hattersley AT. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat Genet. 2014;46:61–4. doi: 10.1038/ng.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, Mcqueen P, Misteli T, Merkenschlager M, Fisher AG. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–40. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–6. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, Taipale J. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801–13. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- Zhang H, Jiao W, Sun L, Fan J, Chen M, Wang H, Xu X, Shen A, Li T, Niu B, Ge S, Li W, Cui J, Wang G, Sun J, Fan X, Hu X, Mrsny RJ, Hoffman AR, Hu JF. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell. 2013a;13:30–5. doi: 10.1016/j.stem.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mccord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–21. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wong CH, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, Mulawadi FH, Sung WK, Nicolis S, Ahituv N, Ruan Y, Wei CL. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013b;504:306–10. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, Bennett DA, Houmard JA, Muoio DM, Onder TT, Camahort R, Cowan CA, Meissner A, Epstein CB, Shoresh N, Bernstein BE. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–54. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, Rosenecker J, Schindelhauer D. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–25. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, Van Der Reijden MI, Ye Z, Kolovos P, Brouwer RW, Van De Corput MP, Van De Werken HJ, Knoch TA, Van IWF, Grosveld FG, Ren B, Wendt KS. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]