Abstract
A genetic screen for mutants that alter circadian rhythms in Drosophila identified the first clock gene - the period (per) gene. The per gene is a central player within a transcriptional feedback loop that represents the core mechanism for keeping circadian time in Drosophila and other animals. The per feedback loop, or core loop, is interlocked with the Clock (Clk) feedback loop, but whether the Clk feedback loop contributes to circadian timekeeping is not known. A series of distinct molecular events are thought to control transcriptional feedback in the core loop. The time it takes to complete these events should take much less than 24h, thus delays must be imposed at different steps within the core loop. As new clock genes are identified, the molecular mechanisms responsible for these delays have been revealed in ever-increasing detail, and provide an in depth accounting of how transcriptional feedback loops keep circadian time. The phase of these feedback loops shift to maintain synchrony with environmental cycles, the most reliable of which is light. Although a great deal is known about cell-autonomous mechanisms of light-induced phase shifting by CRYPTOCHROME (CRY), much less is known about non-cell autonomous mechanisms. CRY mediates phase shifts through an uncharacterized mechanism in certain brain oscillator neurons, and carries out a dual role as a photoreceptor and transcription factor in other tissues. Here I will review how transcriptional feedback loops function to keep time in Drosophila, how they impose delays to maintain a 24h cycle, and how they maintain synchrony with environmental light:dark cycles. The transcriptional feedback loops that keep time in Drosophila are well conserved in other animals, thus what we learn about these loops in Drosophila should continue to provide insight into the operation of analogous transcriptional feedback loops in other animals.
I. Introduction
Research on the fruit fly Drosophila melanogaster has had a profound impact on our understanding of the circadian timekeeping mechanism. Groundbreaking studies by Ron Konopka and Seymour Benzer identified the first “clock gene”, period (per), in a screen for mutants with altered free-running (in constant darkness or DD) circadian periods in the rhythm of adult emergence (Konopka and Benzer, 1971). Since per mutants alter circadian period, per was thought to be integrally involved in keeping circadian time, but determining how per contributed to circadian timekeeping didn’t take off until the per gene was isolated at Brandeis University in the labs of Michael Rosbash and Jeff Hall and at Rockefeller University in Michael Young’s lab (Bargiello et al., 1984; Bargiello and Young, 1984; Reddy et al., 1984; Zehring et al., 1984). The PER protein sequence didn’t offer many clues about its role in the clock because its only distinguishing features were a stretch of Threonine-Glycine repeats, later shown to be involved in adapting to different thermal environments (Sawyer et al., 1997), and a region similar to a portion of the Drosophila SINGLE-MINDED (SIM) and mammalian aryl hydrocarbon receptor nuclear translocator (Arnt) proteins, termed the Per-Arnt-Sim (PAS) domain (Nambu et al., 1991), that mediates protein-protein interactions (Huang et al., 1993). However, the discovery that per mRNA and protein cycle in a circadian manner (Hardin et al., 1990; Siwicki et al., 1988), and that PER protein is required for cycling of per mRNA (Hardin et al., 1990), suggested that per contributes to circadian timekeeping via a feedback loop in which PER protein controls rhythms in per mRNA expression (Hardin et al., 1990). Studies demonstrating transcriptional control of per mRNA cycling and PER-dependent inhibition of per mRNA expression further refined the role of PER in this feedback loop as a transcriptional repressor (Hardin et al., 1992; Zeng et al., 1994). Subsequent studies not only support the view that this transcriptional feedback loop keeps circadian time in Drosophila, but show that similar feedback loops keep circadian time in diverse eukaryotic species including plants, fungi, and animals, the latter of which even shares critical feedback loop components such as per (for reviews see (Bell-Pedersen et al., 2005; Dunlap, 1999; Young and Kay, 2001).
Although per is an essential feedback loop component, many other genes are required to sustain the per feedback loop. In the first section of this review I will explain the roles other genes play within the per feedback loop, and describe how the per feedback loop relates to other interlocked feedback loops. Since the per feedback loop has many components and is inextricably linked to other feedbacks loops, I will refer to the per feedback loop as the ‘core loop’ and to the combined core and interlocked feedback loops as ‘circadian feedback loops’. The different steps required to construct a transcriptional feedback loop (such as the core loop) can be completed in far less than ~24h, indicating that potent mechanisms have evolved to impart delays in feedback regulation. The second section will focus on how post-transcriptional regulation of key feedback loop components produces delays in transcriptional feedback that set the pace of the circadian oscillator. Entrainment of circadian feedback loops to environmental light-dark cycles is critical for driving physiological, metabolic and behavioral rhythms at the appropriate time of day. Unlike the case in mammals, light can directly entrain circadian feedback loops in peripheral tissues from Drosophila, thus there is no intermediary “master” pacemaker in the Drosophila brain that relays light information to the periphery. However, light can entrain the network of Drosophila brain pacemaker neurons via multiple mechanisms depending on which cells detect the light. Section three will discuss how light entrains circadian feedback loops in different cells and tissues.
In the last section of this review I will summarize the main conclusions from each section, point out where there are gaps in our understanding, discuss how filling these gaps may explain how these circadian feedback loops account for basic features of circadian clock such as the 24h period and entrainment to environmental cycles. The general consensus is that circadian feedback loops sit at the heart of the circadian timekeeping mechanism in eukaryotes. However, experiments demonstrating that circadian rhythms in the phosphorylation/dephosphorylation of KaiC protein in cyanobacteria can be reconstituted in a test-tube (Nakajima et al., 2005), and rhythms in the oxidation of peroxiredoxins in a primitive eukaryotic alga Ostrecoccus tauri and red blood cells occurs in the absence of transcription (O’Neill and Reddy, 2011; O’Neill et al., 2011), demonstrate that other circadian timekeeping mechanisms exist in eukaryotes that don’t require transcriptional feedback. I will discuss the role of circadian feedback loops in circadian timekeeping in light of these new results.
II. The circadian feedback loops of Drosophila
The per feedback loop suggested a mechanism for keeping circadian time: Once per transcription is initiated during mid day the levels of per mRNA rise until early evening, when accumulating levels of PER protein repress per transcription, thereby reducing the levels of per mRNA until the early day, when PER protein is eliminated and the next round of per transcription begins. Although this feedback loop provided a framework for how per contributes to circadian timekeeping, it raised many questions. For instance, PER feeds back to repress transcription of its own gene, but does PER achieve this by binding DNA at specific sites to repress transcription, competing with per activators for DNA binding sites, or binding to per activators to inhibit their DNA binding? Does PER feed back to repress transcription alone, or are other factors involved in insuring that feedback occurs at the correct time of day? What genes are responsible for activating per transcription? A combination of approaches including genetic screens, molecular interaction assays, per promoter analysis, and molecular searches for clock gene orthologs were used to answer these questions.
Genetic screens uncovered many additional clock genes including timeless (tim) (Sehgal et al., 1994), Clock (Clk) (Allada et al., 1998), cycle (cyc) (Rutila et al., 1998), doubletime (dbt) (Kloss et al., 1998; Price et al., 1998), shaggy (sgg) (Martinek et al., 2001), casein kinase 2 (CK2) subunits (Akten et al., 2003; Lin et al., 2002a) and cryptochrome (cry) (Stanewsky et al., 1998). A screen for proteins that interact with PER also identified tim (Gekakis et al., 1995), which binds to the PAS domain of PER. Dissecting the per promoter for transcriptional regulatory elements identified a canonical E-box element (in this case 5’ CACGTG 3’) that is required to activate per transcription (Hao et al., 1997), and was later found to be a conserved ‘circadian’ regulatory element for many animal genes (reviewed in (Bell-Pedersen et al., 2005; Hardin, 2004; Young and Kay, 2001)). The first genetic screen for mouse clock genes identified Clock (King et al., 1997a; King et al., 1997b; Vitaterna et al., 1994), which is a member of the basic-helix-loop-helix-PAS (bHLH-PAS) transcription factor family that typically binds E-box elements to activate transcription. Protein coding sequences from mouse Clock were used to recover the Drosophila Clock ortholog from a cDNA library screen (Darlington et al., 1998), thereby complementing the identification of Clk via genetic screening (Allada et al., 1998). The discovery of additional clock genes and regulatory sequences not only support the per feedback loop model, but add to its mechanistic detail. I will briefly summarize clock gene function within the core feedback loop, and refer you to a number of reviews for more detailed descriptions (Allada and Chung, 2010; Hardin, 2005; Zheng and Sehgal, 2008).
A. The Core Feedback Loop
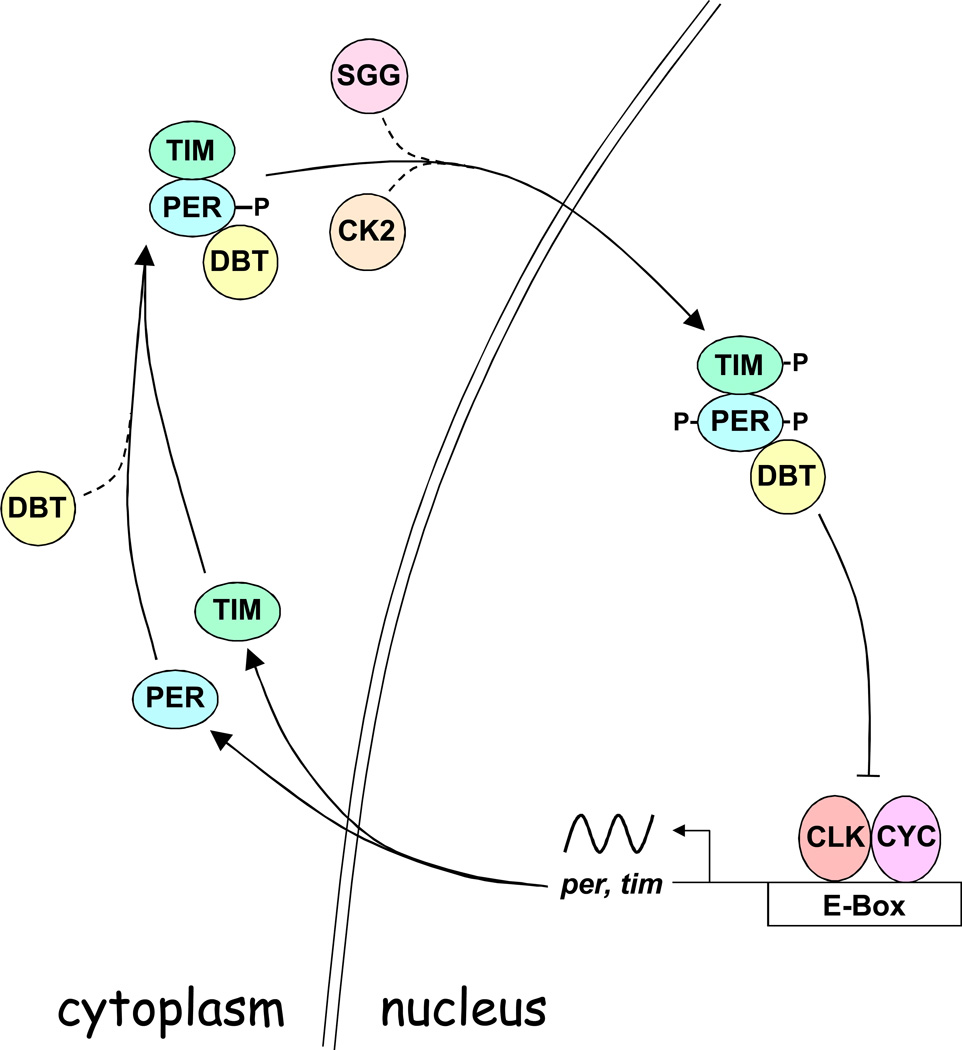
In the core feedback loop (Fig. 1), per and tim transcription are activated from ~ZT4 to ~ZT18 (Zeitgeber Time, or ZT, refers to time in hours during a light-dark cycle, where ZT0 is lights on and ZT12 is lights off), when CLK and its heterodimeric bHLH-PAS partner CYC bind E-boxes in the per and tim promoters (Allada et al., 1998; Darlington et al., 1998; Hao et al., 1997; Rutila et al., 1998). PER and TIM proteins start accumulating in the cytoplasm at ~ZT12, about 6–8hr after their respective mRNAs. This lag in PER and TIM protein accumulation is thought to be caused by the combined effects of phosphorylation-dependent degradation of PER by DBT, a homolog of mammalian casein kinase 1ɛ (Kloss et al., 1998; Kloss et al., 2001; Price et al., 1998), and stabilization of PER-DBT complexes by TIM (Price et al., 1995), which enables cytoplasmic accumulation of DBT-PER-TIM complexes (Curtin et al., 1995; Gekakis et al., 1995; Zeng et al., 1996). However, recent data (described below) suggest that translational regulation may mediate the delay in PER cytoplasmic accumulation rather than DBT-dependent destabilization of PER (Lim et al., 2011; Chiu et al., 2008). Phosphorylation of PER by CK2 and TIM by SGG, a homolog of mammalian Glycogen Synthase Kinase 3, promotes nuclear localization of PER-DBT and TIM (Akten et al., 2003; Lin et al., 2002a; Martinek et al., 2001). PER and TIM phosphorylation are counterbalanced by protein phosphatase 2a (PP2a) and protein phosphatase 1 (PP1) mediated dephosphorylation, respectively, which stabilize PER and TIM and alter their nuclear localization (Fang et al., 2007; Sathyanarayanan et al., 2004). Once in the nucleus, PER-DBT complexes (and/or PER-TIM-DBT complexes) then bind CLK, promote CLK phosphorylation, and release CLK-CYC from E-boxes to inhibit transcription from ~ZT18 to ~ZT4 (Bae et al., 2000; Lee et al., 1998, 1999; Menet et al., 2010; Yu et al., 2006). At ZT0 lights turn on and induce TIM degradation (Hunter-Ensor et al., 1996; Lee et al., 1996; Myers et al., 1996; Zeng et al., 1996), thus ‘deprotecting’ PER. Progressive phosphorylation of PER by DBT in the nucleus ultimately triggers binding of the E3 ubiquitin ligase SLIMB, which targets PER for degradation in the proteasome by ~ZT4 (Grima et al., 2002; Kloss et al., 2001; Ko et al., 2002). Once PER is degraded, hypophosphorylated CLK accumulates and CLK-CYC binds E-boxes to initiate another cycle of per and tim transcription.
Figure 1.
The core feedback loop. All gene, regulatory element, and protein names are as defined in the text. Double line, nuclear envelope; sinusoidal line, mRNA rhythm; solid arrows, steps in the pathway; blocked line, repression; dashed line, step employing protein activity; P, phosphorylation. See text for detailed description.
The core feedback loop nicely explains the regulation of cycling mRNAs that peak in abundance during the early evening. However, not all cycling gene expression peaks in the early evening. The first example of this in flies was Clk mRNA, which peaks during the early morning (Bae et al., 1998; Darlington et al., 1998). Rhythms in Clk mRNA levels are also dependent on the core feedback loop as these rhythms were abolished in per01 and tim01 null mutants (Bae et al., 1998). How these rhythms were abolished was intriguing; whereas per and tim mRNAs remain at relatively high levels in per01 and tim01 mutants (Hardin et al., 1990; Sehgal et al., 1995), Clk mRNA falls to constant low levels (Bae et al., 1998). Although PER and TIM function to repress per and tim transcription (Sehgal et al., 1994; Zeng et al., 1994), the low levels of Clk mRNA in per01 and tim01 flies suggested that PER and TIM somehow activate Clk transcription (Bae et al., 1998). Since per and tim expression is abolished in severe loss of function ClkJrk and cyc01 mutants (Allada et al., 1998; Rutila et al., 1998), Clk mRNA levels were expected to also be low in ClkJrk and cyc01 flies due to the loss of PER and TIM. However, Clk mRNA was expressed at peak levels in ClkJrk and cyc01 mutants, which suggested that CLK-CYC somehow represses Clk expression, in contrast to its traditional role as a transcription activator (Glossop et al., 1999). A model was developed to explain these puzzling results that invoked a second ‘Clk feedback loop’ that interlocked with the core feedback loop (Glossop et al., 1999). The subsequent identification of the PAR transcription factors vrille (vri) and PAR domain protein 1 ε and δ (Pdp1ε/δ) as components of the Clk feedback loop strongly support the interlocked feedback loop model (Cyran et al., 2003; Glossop et al., 2003; Zheng et al., 2009). I will briefly outline the important features of the Clk feedback loop, and refer you to other reviews for a more detailed description (Allada and Chung, 2010; Hardin, 2005; Zheng and Sehgal, 2008).
B. Interlocked Feedback Loops
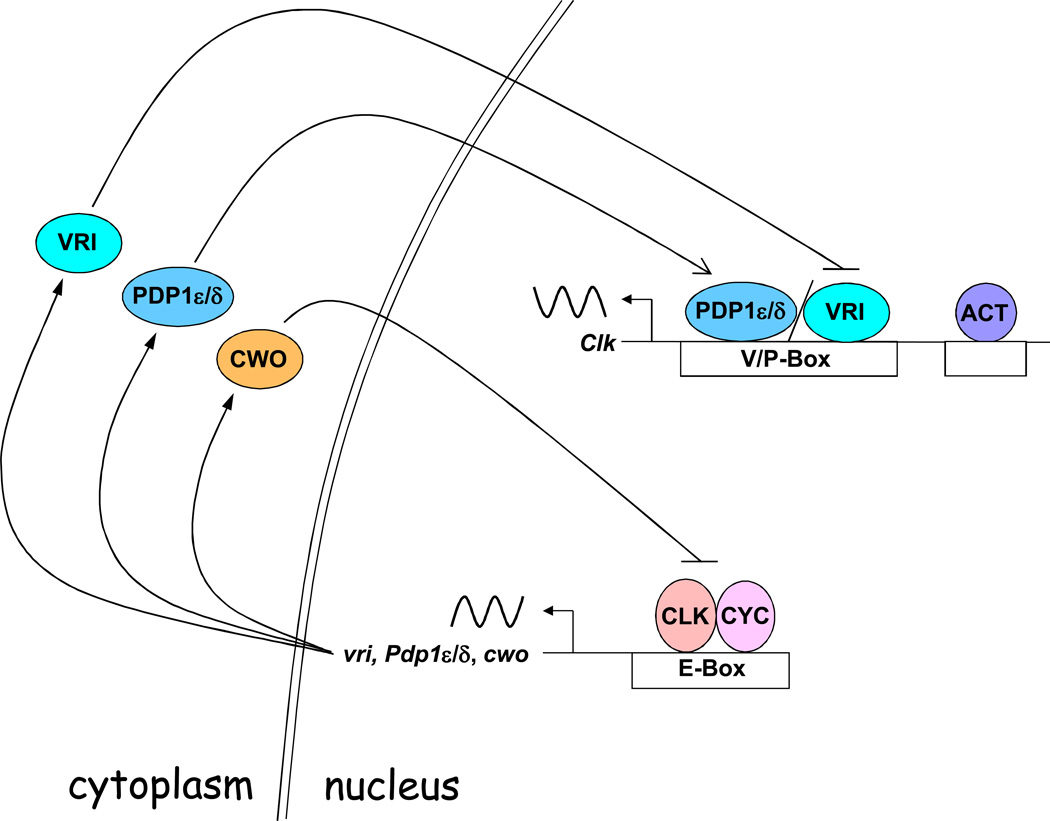
In the Clk loop (Fig. 2), CLK-CYC binds E-boxes to activate vri transcription between ~ZT4 and ZT16 (Blau and Young, 1999; Cyran et al., 2003). VRI protein accumulates in phase with vri mRNA, ultimately peaking in abundance at ~ZT14. As VRI levels rise, VRI binds VRI/PDP1-boxes (V/P-boxes) in the Clk promoter, thereby repressing Clk transcription (Cyran et al., 2003; Glossop et al., 2003). As PER-DBT and PER-TIM-DBT complexes feed back to inhibit CLK-CYC dependent transcription from ~ZT16 to ZT4, vri mRNA and protein decline to low levels, thus permitting activation of Clk transcription. Mutants that disrupt CLK-CYC transcriptional activity (e.g. ClkJrk, cyc01) exhibit constant high levels of Clk mRNA (Glossop et al., 1999), indicating that Clk is constitutively activated independent of circadian oscillator function. However, another PAR transcription factor, PDP1ε/δ, also plays a role in Clk activation (Benito et al., 2007; Cyran et al., 2003; Zheng et al., 2009). CLK-CYC binds E-boxes to activate Pdp1ε/δ between ZT4 and ZT16 (Cyran et al., 2003). PDP1ε/δ rises to peak levels ~ZT18, several hours after VRI levels peak, and binds V/P-boxes to activate Clk transcription (Cyran et al., 2003). The extent to which Pdp1ε/δ activates Clk transcription is debatable, as altering Pdp1ε/δ levels via RNA interference and overexpression has mild effects on Clk mRNA levels compared to mutants that eliminate Pdp1ε/δ specifically or all Pdp1 isoforms (Benito et al., 2007; Cyran et al., 2003; Zheng et al., 2009). Perhaps vri and Pdp1ε/δ function to enhance Clk mRNA amplitude once developmental activators establish the Clk expression pattern (Houl et al., 2008). Once PER complexes are degraded during mid-day, the next cycle of vri transcription and VRI-dependent repression is initiated.
Figure 2.
The Clk feedback loop. All gene, regulatory element, and protein names are as defined in the text. All symbols are as defined in Figure 1. ACT, Clk activator; open arrow, transcription activation; antiphase sinusoidal line, antiphase mRNA rhythm; backslash, binding by one or the other protein. See text for detailed description.
In addition to the per and Clk feedback loops, another feedback loop that is controlled by the bHLH-orange transcription inhibitor CLOCKWORK ORANGE (CWO) has been proposed (Fig. 2). In this feedback loop, CLK-CYC binds E-boxes to activate cwo transcription, and CWO then feeds back to inhibit CLK-CYC transcription (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007). Inhibition of CLK-CYC occurs through competition for binding E-boxes (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007), and thus independently reinforces inhibition by PER complexes. This model for CWO function is primarily based on in vitro and Drosophila Schneider 2 (S2) cell culture data, which contrasts with cwo mutant data showing period lengthening and lower levels of per, tim, vri and Pdp1ε/δ levels (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007; Richier et al., 2008). This in vivo data implies that cwo is necessary for high-level transcription of CLK-CYC activated genes. Additional studies that document the phase of CWO protein cycling and DNA binding will help to resolve the role of CWO in the circadian oscillator.
Although the per and Clk feedback loops control mRNA cycling in opposite phases of the circadian cycle, they are not equally important for circadian oscillator function (Hardin, 2006). The per loop is required for Clk loop function since CLK-CYC dependent activation and PER-TIM-DBT dependent repression control rhythms in vri transcription. However, the Clk loop is not necessary for per loop function since reversing Clk mRNA cycling has little affect on molecular or behavioral rhythms (Kim et al., 2002), and rhythms in Clk mRNA levels don’t drive rhythms in CLK levels or transcriptional activity (Yu et al., 2006). If Clk mRNA rhythms are not important for core feedback loop function, then what is the importance of the Clk feedback loop? Microarray studies show that ~10% of Drosophila head mRNAs are rhythmically expressed (Ceriani et al., 2002; Claridge-Chang et al., 2001; Keegan et al., 2007; Lin et al., 2002b; McDonald and Rosbash, 2001; Ueda et al., 2002; Wijnen et al., 2006), and a large fraction of those are expressed with Clk-like peak times near dawn. Reducing, eliminating or increasing Pdp1ε/δ expression abolishes behavioral rhythms (Benito et al., 2007; Zheng et al., 2009), which suggests that Pdp1ε/δ controls output gene expression. Indeed, Pdp1ε/δ controls rhythmic expression of takeout (to), which regulates courtship behavior (Dauwalder et al., 2002), but such regulation appears to be indirect as no canonical V/P-boxes are present near the to promoter (Benito et al., 2010). Although vri overexpression abolishes oscillator function due to repression of Clk transcription (Cyran et al., 2003; Glossop et al., 2003), it has not been possible to test whether vri is required for oscillator function because vri null mutants are lethal (Cyran et al., 2003; George and Terracol, 1997). Perhaps vri, like Pdp1ε/δ, also is required to control rhythmic transcription of output genes that peak near dawn. Given the importance of the core loop, it is imperative that the mechanisms governing rhythmic transcription within this loop are defined.
III. Post-Transriptional Regulation of Rhythmic Transcription
To keep circadian time, the various molecular events within the core feedback loop must be completed in ~24h. However, the transcriptional activation and elongation, transcript processing and transport to the cytosol, protein synthesis and accumulation, nuclear localization, transcriptional repression, and repressor degradation that mediate feedback loop function collectively take much less than 24h to complete. Consequently, delays must be imposed at one or more steps in the core loop to achieve a 24h cycle. As described in the last section, there is good evidence that PER stability, nuclear localization, and transcriptional repression is controlled at the post-transcriptional level by TIM binding and kinases and phosphatases that control the phosphorylation state of PER and TIM. In this section I will review recent studies that begin to reveal the molecular consequences of PER-TIM interactions and site specific PER phosphorylation by different kinases. In addition, I will discuss the regulation and function of rhythms in CLK phosphorylation and other forms of post-transcriptional regulation that govern timekeeping by the core loop.
A. PER Phosphorylation and Translational Control
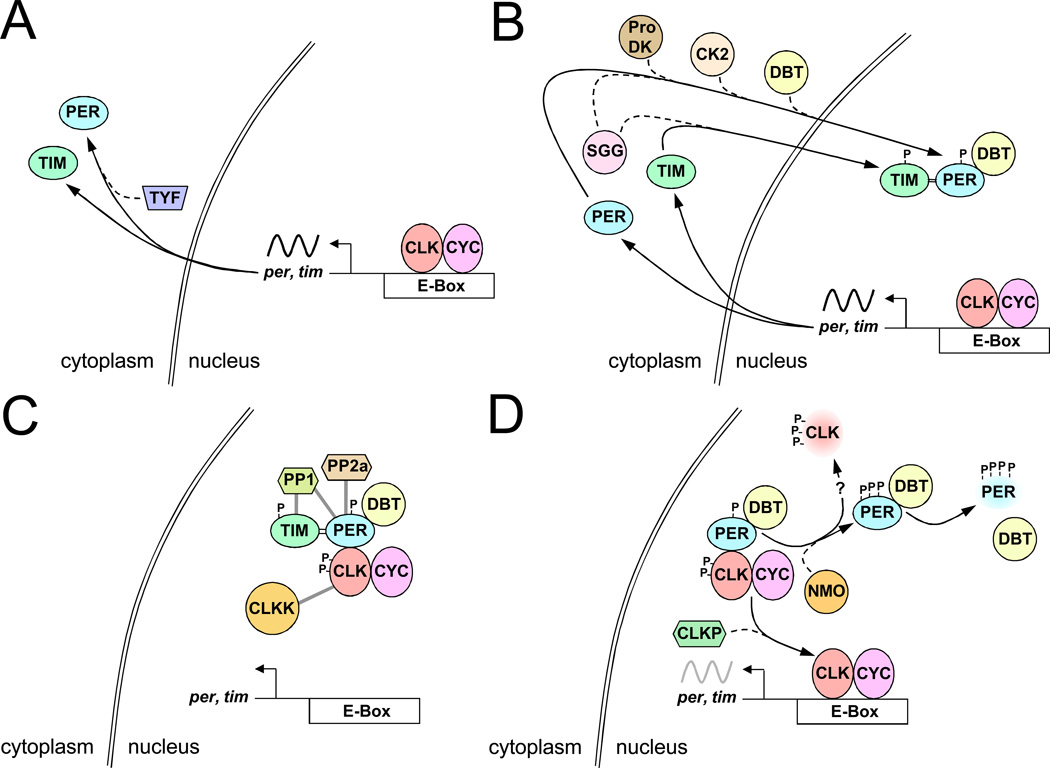
PER phosphorylation increases as PER accumulates during the night, and peaks as PER is degraded in the proteasome a few hours after dawn (Edery et al., 1994; Naidoo et al., 1999). DBT binds to PER and promotes PER degradation (Kim et al., 2007; Kloss et al., 1998; Kloss et al., 2001; Price et al., 1998), whereas TIM binds to PER and prevents PER degradation (Kloss et al., 1998; Kloss et al., 2001; Ko et al., 2002; Price et al., 1998). Much progress has been made in understanding the regulation of PER degradation after light-induced removal of TIM (Fig. 3c). At this time of the circadian cycle, PER is complexed with DBT and CLK-CYC to repress transcription (Kloss et al., 2001; Lee et al., 1999; Yu and Hardin, 2006). PER is progressively phosphorylated during this timeframe, but its SLIMB F-box protein (homolog of mammalian B-TrCP) induced degradation is delayed for several hours even in the absence of TIM (Grima et al., 2002; Ko et al., 2002). The reason for this delay is that DBT phosphoryation at PER serine 47 (S47) is the final step in a series of DBT phosphorylation events that produce an atypical SLIMB binding site (Chiu et al., 2008). Once SLIMB binds, PER is ubiquitinated and rapidly degraded (Chiu et al., 2008), thereby releasing transcriptional repression.
Figure 3.
Post-transcriptional regulatory steps within the core feedback loop. All gene, regulatory element, and protein names are as defined in the text. All symbols are as defined in Figure 1. A. Delay in PER synthesis from ~ZT6-ZT12. B. Movement of PER and TIM into the nucleus from ~ZT16-20. C. Stabilization of nuclear PER as PER complexes repress CLK-CYC transcription from ~ZT16-ZT0. D. Release of PER repression and reactivation of CLK-CYC transcription from ~ZT3-ZT6. Double bar, stabilizing activity; CLKK, CLK kinase; ProDK, proline-directed kinase; gray bars, kinase and phosphatase targets; faded protein symbols, protein degradation; ?, putative direct effect; gray sinusoidal line, initiation of transcription. See text for detailed description.
When DBT is co-expressed with PER in S2 cells, DBT phosphorylates PER at many sites before the ultimate phosphorylation event on S47 (Chiu et al., 2008; Kivimae et al., 2008). However, PER is phosphorylated by other kinases in the presence or absence of DBT in S2 cells (Chiu et al., 2008; Kivimae et al., 2008). For instance, PER is phosphorylated by CK2 at S149, S151 and S153 to promote nuclear localization of PER complexes (Chiu et al., 2008; Lin et al., 2005) (Fig. 3b). PER is also phosphorylated at multiple consensus proline-directed kinase target sites (Chiu et al., 2008), in which a proline follows the phosphorylated serine or threonine. One such proline-directed site at S661 is phosphorylated by an as yet unidentified kinase, but rather than promoting PER degradation, S661 phosphorylation primes phosphorylation of S657 by SGG to promote PER nuclear localization (Ko et al., 2010). SGG was thought to promote nuclear localization of PER complexes by phosphorylating TIM (Martinek et al., 2001), but SGG directly interacts with and stabilizes CRY, which in turn stabilizes TIM (Stoleru et al., 2007), and consequently PER, thus providing increased CK2 and SGG substrate to effect PER nuclear localization. Moreover, CK2 and SGG are found predominantly in the cytoplasm (Lin et al., 2002a; Yuan et al., 2005), consistent with their role in promoting PER nuclear localization. SGG phosphorylation promotes PER nuclear localization but not PER degradation (Ko et al., 2010), which implies that these nuclear localization and degradation are programmed by independent phosphorylation cascades. The (or at least a) lynchpin for PER nuclear localization is the proline-directed kinase that acts in the cytoplasm to prime SGG phosphorylation by phosphorylating S661. Identifying this proline-directed kinase and determining the relationship between SGG and CK2 dependent phosphorylation will provide a clearer picture of how PER nuclear localization is regulated.
PER is phosphorylated at S661 and several additional consensus proline-directed kinase sites (Chiu et al., 2008; Kivimae et al., 2008). One such site at S596 is situated within the “short period domain” of PER, a region spanning amino acids S585 to Y601 that contains mutants that shorten circadian period including perS and perT (Baylies et al., 1987; Baylies et al., 1992; Konopka et al., 1994; Rutila et al., 1992; Yu et al., 1987). Disrupting the only proline-directed phosphorylation site in this region by mutating Y597 results in short period rhythms (Baylies et al., 1992), suggesting that phosphorylation at this site normally acts to lengthen (or delay) the circadian cycle. Indeed, recent work demonstrates that NEMO (NMO) kinase phosphorylates S596 (Chiu et al., 2011), and nmo mutant and nmo RNAi knockdown flies show short period rhythms (Chiu et al., 2011; Yu et al., 2011) (Fig. 3d). Mutating S595 to A also shortens circadian period, and gates the phosphorylation of nearby residues S589 (the original perS mutant site), S595 and S593 by DBT (Chiu et al., 2011). Phosphorylation of PER by NMO at S596 delays the phosphorylation of S47 by DBT (Chiu et al., 2011), and thus PER degradation, consistent with a delay in PER degradation in the morning when NMO is overexpressed (Yu et al., 2011). A model proposed by Edery and colleagues postulates that phosphorylation of PER by NMO and DBT within the “short period domain” alters the conformation of PER, thereby inhibiting the phosphorylation of sites required for SLIMB binding and delaying PER degradation (Chiu et al., 2011).
In addition to the delay in PER degradation, PER protein accumulation lags 6–8h behind per mRNA accumulation. This lag was thought to be produced by DBT induced PER degradation and protection by TIM. However, PER is not phosphorylated at the key S47 site until after dawn (Chiu et al., 2008), thus arguing against the same SLIMB-dependent mechanism functioning in the cytoplasm. The delayed accumulation of PER in the cytoplasm could be achieved by accelerating PER degradation or impeding PER synthesis (Fig. 3a). DBT associates with PER as PER accumulates in the cytoplasm, thus it is possible that cytoplasmically localized F-box proteins could bind phosphorylated PER and promote degradation. Alternatively, there may be a delay in the translation of per mRNA. Recent work on the twenty-four (tyf) gene suggests that PER accumulation is translationally controlled (Lim et al., 2011). TYF associates with the 5’ cap binding complex, polyA binding protein and per mRNA, and loss of tyf reduces PER accumulation in ventralateral neurons (LNvs) in the fly brain, which are necessary and sufficient for locomotor activity rhythms (Frisch et al., 1994; Grima et al., 2004; Renn et al., 1999; Stoleru et al., 2004), suggesting that TYF promotes PER translation in LNvs (Lim et al., 2011). This loss of tyf function result implies that per mRNA is poorly translated and/or under active translational repression, and that TYF enhances translation efficiency or removes the translation block. The extent to which regulation of per mRNA translation contributes to the delay in PER cytoplasmic accumulation is not known, but such regulation could play a major role in determining the pace of the circadian oscillator. TYF appears to function only in LNvs (Lim et al., 2011), thus translational regulation by different factors or other post-transcriptional regulatory processes act to delay the cytoplasmic accumulation of PER in other oscillator cells. Translational regulation of other oscillator components is mediated by microRNAs. The microRNA bantam targets sequences in the 3’UTR of Clk mRNA, and mutating these bantam target sites greatly decreases rescue of behavioral rhythms by Clk transgenes (Kadener et al., 2009). Translation of vri and cwo mRNAs are also thought to be regulated by microRNAs (Kadener et al., 2009), and a number of microRNAs are rhythmically expressed (Yang et al., 2008).
As PER accumulates it is phosphorylated and enters the nucleus, where TIM plays an important role in stabilizing phosphorylated PER (Fig. 3b). TIM likely stabilizes PER by inhibiting phosphorylation at sites that promote SLIMB binding and degradation. How TIM inhibits PER phosphorylation at these sites is not well characterized, but recent analysis suggests that TIM may inhibit PER phosphorylation by delivering protein phosphatase 1 (PP1) to PER (Fang et al., 2007). PER is also dephosphorylated by protein phosphatase 2a (PP2a) (Sathyanarayanan et al., 2004), but dephosphorylation of PER by PP2a is not mediated by TIM (Fang et al., 2007). Nevertheless, PP2a dependent dephosphorylation of PER would further delay PER phosphorylation and degradation (Sathyanarayanan et al., 2004). Since phosphorylation enhances PER’s ability to repress transcription (Nawathean and Rosbash, 2004), TIM-dependent stabilization of phosphorylated PER likely plays a key role in effectively repressing CLK-CYC transcription. It is not clear how PER phosphorylation enhances transcriptional repression, but it doesn’t appear to act by enabling PER-CLK binding (Yu et al., 2009). Thus, delays in the synthesis, nuclear localization and degradation of PER contribute to the determination of circadian period.
B. CLK Phosphorylation
CLK is phosphorylated coincident with the entry of PER repression complexes into the nucleus (Yu et al., 2006). CLK phosphorylation coincides with transcriptional repression (Kim and Edery, 2006; Menet et al., 2010; Yu et al., 2006; Yu et al., 2009), but whether CLK phosphorylation is required for repression is not known (Fig. 3c). PER carries DBT into the nucleus (Kloss et al., 2001), and DBT is required for CLK phosphorylation (Kim and Edery, 2006; Yu et al., 2006; Yu et al., 2009). However, DBT doesn’t phosphorylate CLK directly, though DBT (whether catalytically active or inactive) must be present in the PER repression complex to mediate CLK phosphorylation by one or more other kinases (Yu et al., 2009). The kinase(s) responsible for most CLK phosphorylation has not been identified, but one kinase implicated in CLK phosphorylation is NMO (Fig. 3d). Loss of nmo function increases CLK levels and shortens circadian period, whereas increasing nmo function decreases CLK levels and lengthens circadian period (Yu et al., 2011), consistent with period changes associated with increased or decreased CLK copy number (Kadener et al., 2008). These results suggest that NMO promotes CLK degradation to slow the pace of the circadian cycle (Yu et al., 2011), but whether NMO lowers CLK levels directly by phosphorylating CLK or indirectly by phosphorylating PER is not known.
Once PER is degraded, the levels of hypophosphorylated CLK increase along with CLK-CYC transcription. Since overall CLK levels are essentially constant (Yu et al., 2006), the increase in hypophosphorylated CLK levels could be due to the replacement of degraded hyperphosphoylated CLK with newly synthesized CLK, the dephosphorylation of hyperphosphorylated CLK, or both. Although Clk mRNA levels peak around dawn (Bae et al., 1998), Clk mRNA levels only vary ~3-fold over the circadian cycle, thus increased synthesis is less likely to account for the almost complete conversion of hyperphosphorylated to hypophosphorylated CLK over 3–6h. Phosphatases that would dephosphorylate CLK have not been identified, though PP1 and PP2a function within the core oscillator and PP2a regulatory subunit mRNAs cycle in abundance (Fang et al., 2007; Sathyanarayanan et al., 2004). Dephosphorylation of CLK after PER degradation may impose another delay within the feedback loop that is necessary to convert CLK-CYC to a transcriptionally active form. This is likely the case for CLOCK in mammals since CLK is phosphorylated upon interaction with PER-CRY repressor complexes and returned to a dephosphorylated form when CLOCK-BMAL1 activates transcription (Lee et al., 2001; Spengler et al., 2009; Yoshitane et al., 2009).
IV. Light entrainment of the Drosophila circadian feedback loops
In animals, environmental cycles of light, temperature and/or social cues set the phase of (i.e. entrain) circadian oscillators so that overt rhythms in physiology, metabolism and behavior occur at the appropriate time of day. The most potent and reliable environmental cue is light, which mediates entrainment via different mechanisms depending on tissue type and species. In mammals, light is detected by melanopsin in retinal ganglion cells in the eye, which transmit signals to the “master clock” in the suprachiasmatic nucleus (SCN) through the retinohypothalamic tract (reviewed in (Golombek and Rosenstein, 2010)). These signals entrain circadian oscillators in the SCN, which relay information about their new phase via humoral signals to entrain oscillators in peripheral tissues (reviewed in (Dibner et al., 2010)). This two-stage entrainment process (one stage for the SCN and a second for peripheral oscillators) differs from that in Drosophila because almost all Drosophila oscillator cells either detect light directly or receive light information from photoreceptor cells. The cell-autonomous nature of circadian light entrainment is best illustrated in flies that use a per-promoter driven luciferase reporter gene (per-luc) to drive rhythms in bioluminescence in isolated wings, and antennae and probosci that can be entrained by light (Plautz et al., 1997). Thus, there is no “master clock” in Drosophila that entrains peripheral oscillators (and no closed circulatory system to efficiently send entrainment signals through even if there were a master clock).
Nevertheless, like the SCN clock, pacemaker cells in the Drosophila brain control locomotor activity rhythms, which can be monitored with great precision to detect light-induced shifts in oscillator phase. Light alters the phase of activity differently depending on when it is detected during the circadian cycle (Pittendrigh and Minis, 1964). A light pulse applied during the early night delays the phase of activity by up to 4h, a light pulse applied during the day (or subjective day if the flies are in kept in constant darkness) has little effect on activity phase, and a light pulse applied during the late night advances the phase of activity up to 3h (Myers et al., 1996; Saunders et al., 1994). Light induces phase shifts by delaying or advancing the phase of the core feedback loop. This section will focus primarily on the molecular mechanism that governs light-induced phase shifts, but will also discuss how light integrates with other environmental cues to shift circadian phase.
A. TIM Degradation, CRY and Other Circadian Photoreceptors
In 1996 three groups discovered that light induces the degradation of TIM in fly heads (Hunter-Ensor et al., 1996; Myers et al., 1996; Zeng et al., 1996). Light drastically reduces TIM levels within 30 minutes, thereby destabilizing PER and shifting the phase of the core feedback loop. Although light’s effect on TIM levels is unidirectional, how reduced levels of TIM are interpreted by the clock depends on when the light pulse is applied. Light-induced degradation of TIM during the early evening produces a phase delay because tim mRNA levels are high, and new synthesis can replenish TIM levels within a few hours. During late night, light-induced degradation of TIM produces a phase advance because tim mRNA levels are low, thus premature loss of TIM resets the core loop to its normal state near dawn. Little or no phase shifts are seen during the actual or subjective day because TIM levels are normally very low and can’t be further reduced. Thus, light-dependent degradation of TIM initiates a phase shift, and the levels of tim mRNA and protein determine the direction of the phase shift through their effect on the core loop. Light induces TIM degradation in the proteasome (Naidoo et al., 1999). Upon light exposure, TIM is phosphorylated by a tyrosine kinase based on pharmacological experiments (Naidoo et al., 1999), but this kinase has not been identified.
Though TIM is rapidly degraded after light exposure, TIM is not itself a photoreceptor. Loss of external eyes and ocelli or eliminating opsin-based photoreception via vitamin A depletion or phototransduction mutants reduces light sensitivity of the clock but doesn’t abolish entrainment of locomotor activity rhythms by light (Blaschke et al., 1996; Hu et al., 1978; Ohata et al., 1998; Pearn et al., 1996), which suggests that other photoreceptors function to entrain circadian oscillators in brain pacemaker cells. A screen for mutants that disrupted rhythms in bioluminescence produced by per-luc identified CRYPTOCHROME (CRY) (Emery et al., 1998; Stanewsky et al., 1998), which is an ortholog of blue light photoreceptor cryptochromes in plants (Ahmad and Cashmore, 1993; Lin et al., 1998). The severely hypomorphic cryb mutant abolishes bioluminescence rhythms in whole flies during and after entrainment in 12h light:12h dark (LD) cycles, but not after entrainment to temperature cycles, suggesting a defect in the light entrainment pathway (Stanewsky et al., 1998). Despite the loss of bioluminescence rhythms, cryb flies are behaviorally rhythmic during and after LD entrainment, indicating that pacemaker cells continue to receive light information. However, cryb flies can’t be phase shifted by brief (e.g. 10 minute) pulses of light (Stanewsky et al., 1998), and remain rhythmic in constant light after entrainment in LD cycles (Emery et al., 2000a), in contrast to wild type flies that become arrhythmic in constant light (Konopka et al., 1989). Defects in behavioral rhythms due to cryb can be rescued by expressing CRY in brain pacemaker neurons, demonstrating that CRY functions cell-autonomously (Emery et al., 2000b). Taken as a whole, these studies demonstrate that CRY is a cell-autonomous photoreceptor that resets the phase of behavioral rhythms in Drosophila to short pulses of light, but is not required for light entrainment to LD cycles.
Loss of phototransduction in external eyes (i.e. compound eyes and ocelli) by removing the NORP-A phosholipase C together with CRY further weakens, but doesn’t abolish, light entrainment of behavioral rhythms (Stanewsky et al., 1998), indicating that other photoreceptors and/or phototransduction cascades function to entrain locomotor activity rhythms to light. The remaining photoreceptors capable of mediating light entrainment of locomotor activity are a pair of extraretinal eyes called the Hofbauer-Buchner (H-B) eyelet; eliminating all external photoreceptors, the H-B eyelet and CRY renders flies unentrainable to light (Helfrich-Forster et al., 2001). Eliminating phototransduction in external eyes along with CRY abolishes core feedback loop function in all brain clock neurons except the LNvs and dorsal neuron 1s (DN1s), which implies that the H-B eyelet relays light information to this subset of clock neurons to mediate behavioral rhythms (Helfrich-Forster et al., 2001). Thus, behavioral rhythms can be entrained by independent light input pathways in external eyes, the H-B eyelet, and CRY, but light resetting to short pulses is mediated by CRY.
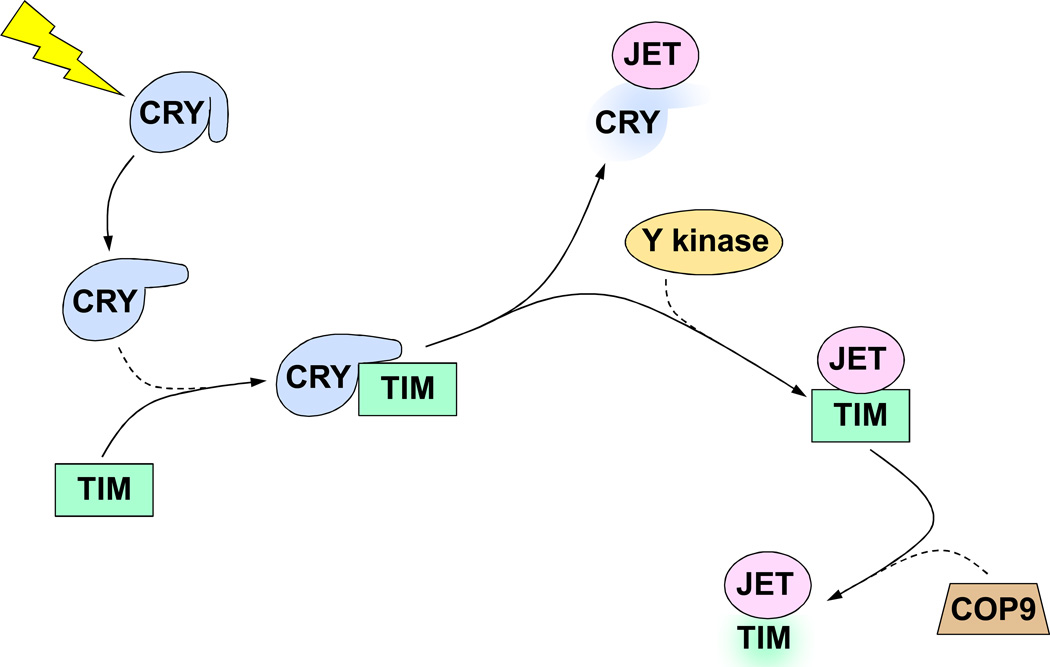
Since TIM is degraded in response to short pulses of light, photodetection by CRY must initiate a cell-autonomous pathway leading to TIM degradation (Fig. 4). Indeed, CRY binds directly to TIM in a light-dependent manner, which irreversibly commits TIM to degradation in the proteasome (Busza et al., 2004; Ceriani et al., 1999; Dissel et al., 2004; Naidoo et al., 1999). CRY is also degraded in the proteasome upon activation by light, but CRY degradation occurs more slowly than TIM degradation (Busza et al., 2004; Dissel et al., 2004; Lin et al., 2001). Although light-induced TIM and CRY degradation were thought to be controlled independently (Busza et al., 2004; Sathyanarayanan et al., 2008), more recent evidence suggests that they are linked (Peschel et al., 2009). Light-dependent degradation of both TIM and CRY is mediated by the F-box protein JETLAG (JET) (Koh et al., 2006; Peschel et al., 2009; Peschel et al., 2006). However, TIM and CRY are degraded sequentially due to a higher affinity of JET for TIM than CRY (Peschel et al., 2009). Thus, when CRY is activated by light it becomes a substrate for JET binding, and binding of activated CRY to TIM makes TIM an even higher affinity substrate for JET than CRY. Light-induced degradation of TIM requires the COP9 signalosome (Knowles et al., 2009), which apparently acts downstream of JET to promote TIM (and maybe CRY) degradation in the proteasome. Mutants in cry, jet and the COP9 signalosome are all rhythmic in constant light (Emery et al., 2000a; Knowles et al., 2009; Koh et al., 2006; Peschel et al., 2006), which suggests that these genes function in the same pathway to mediate light-dependent resetting of circadian oscillators in pacemaker cells that control rhythmic activity.
Figure 4.
Light-induced phase resetting mechanism. All gene, regulatory element, and protein names are as defined in the text. All symbols are as defined in Figures 1 and 3. Y kinase, tyrosine kinase. See text for detailed description.
In contrast to CRY mediated phase resetting to light, the molecular and cellular mechanisms by which the H-B eyelet and external photoreceptors in the compound eye and ocelli have only just begun to be characterized. The H-B eyelet projects to the large subset of ventrolateral neurons (lLNvs), which are necessary for light-induced phase resetting at dawn and increase their firing rate when stimulated by light (Shang et al., 2008; Sheeba et al., 2008). The lLNvs project throughout the optic lobe, including the vicinity of the small subset of ventrolateral neurons (sLNvs) (Helfrich-Forster et al., 2007), suggesting that light information from the H-B eyelet may be sent to lLNvs, and then on to the sLNvs to mediate phase resetting near dawn (Shang et al., 2008; Sheeba et al., 2010). How compound eyes and ocelli contribute to phase resetting is not known.
B. Resetting Behavioral Rhythms – Cellular and Sensory Integration
Since TIM is rapidly degraded in fly heads after light exposure, it has been assumed that light leads to TIM degradation uniformly in all oscillator cells including the LNv pacemaker neurons. Although TIM is degraded in all brain oscillator neurons including the LNvs when phase advancing light pulses are applied at ZT21, TIM is not degraded in LNvs (but is degraded in dorsal brain oscillator neurons) when phase delaying light pulses are applied at ZT15 (Tang et al., 2010). Overexpressing JET in the LNvs of a jet null mutant enables light-induced TIM degradation at ZT15, but doesn’t produce a phase delay in behavior (Tang et al., 2010), thus TIM degradation in LNvs is neither necessary nor sufficient for phase delays. If CRY expression is knocked down in only LNvs, light-induced phase delays and advances are both blocked, indicating that LNvs are necessary for behavioral phase shifts even though TIM is not degraded by a light pulse at ZT15 in these cells (Tang et al., 2010).
These results suggest that light-sensitive dorsal brain oscillator neurons signal through LNvs to promote light-induced phase delays. A good candidate for dorsal neurons that initiate light-induced phase delays are the DN1s because a subset of these neurons are CRY positive (Benito et al., 2008; Yoshii et al., 2008), light pulses at ZT15 consistently induce TIM degradation in these neurons (Tang et al., 2010), and LNvs signal to DN1s via PDF while DN1s may signal LNvs via projections to LNv cell bodies (Helfrich-Forster et al., 2007; Kaneko and Hall, 2000; Zhang et al., 2010a). Whether DN1s or other dorsal oscillator neurons mediate light-dependent phase delays via LNvs, it is clear that the standard cell-autonomous CRY-dependent degradation of TIM paradigm for light-induced phase resetting doesn’t explain how light induces phase delays in locomotor activity.
During LD conditions Drosophila display morning and evening peaks in activity that anticipate the lights-on and lights-off transitions, respectively. These activity peaks are controlled by separate sets of brain oscillator neurons, where morning activity is controlled by LNvs that express the PDF neuropeptide and evening activity is controlled by LNds and the PDF negative 5th sLNv (Grima et al., 2004; Stoleru et al., 2004). The phase of these morning and evening activity peaks adjusts to seasonal differences in photoperiod and temperature. When photoperiod is long and temperature is high in the summer, flies are more active in before dawn and after dusk, but when photoperiod is short and temperatures are low, flies are more active after dawn and before dusk (Majercak et al., 1999). This behavioral plasticity is influenced by molecular mechanisms that act to adjust oscillator phase including temperature sensitive splicing of per intron 8 (Collins et al., 2004; Low et al., 2008; Majercak et al., 2004; Majercak et al., 1999), temperature sensitive splicing of the last tim intron (Boothroyd et al., 2007), and light-induced transcription of tim only at low temperatures (Chen et al., 2006). Communication between morning cells that serve as dominant clocks in the dark and evening cells that serve as dominants clocks in light (along with a subset of DN1s) is also important for controlling seasonal differences in activity (Cusumano et al., 2009; Murad et al., 2007; Stoleru et al., 2007; Zhang et al., 2009). Integration of light and temperature information was recently shown to occur in the DN1s, which alter the phase and amplitude of morning and evening activity peaks depending on temperature and light intensity (Zhang et al., 2010a; Zhang et al., 2010b). How temperature and light information is integrated in DN1s to modulate morning and evening activity will no doubt be the subject of future studies.
Social interactions among flies also alters locomotor activity rhythms. Flies entrained to a particular circadian phase influence the activity of flies entrained to a different phase (Levine et al., 2002b), and courtship interactions between males and females shift activity to the night and away from dusk (Fujii et al., 2007). In both of these cases olfactory cues are responsible for the activity changes (Fujii et al., 2007; Krupp et al., 2008; Levine et al., 2002b), which indicates that olfactory signaling, which is also under circadian control (Krishnan et al., 1999; Krishnan et al., 2008; Tanoue et al., 2008; Tanoue et al., 2004), is likely integrated with light and temperature to determine the timing of activity. Several excellent reviews have been published that provide a more in-depth description of the discovery, function and modulation of morning and evening oscillators (Allada and Chung, 2010; Choi and Nitabach, 2010; Dubruille and Emery, 2008), and insight into the mechanisms by which temperature entrains circadian oscillators to effect seasonal adaptation (Chen et al., 2007; Dubruille and Emery, 2008; Glaser and Stanewsky, 2007).
C. Other Functions of Drosophila CRY
Bioluminescence rhythms in cryb flies bearing per-luc can be entrained by temperature cycles, but not LD cycles (Stanewsky et al., 1998). Since the vast majority of per-luc expression is from peripheral oscillators, this result implies that CRY is also required for light entrainment of circadian oscillators in peripheral tissues. While this may be the case, CRY is also required for oscillator function per se in at least some peripheral tissues including the eye, the antenna, Malpighian tubules (the fly kidney equivalent), and forelegs (Collins et al., 2006; Ivanchenko et al., 2001; Krishnan et al., 2001; Levine et al., 2002a). The evidence that CRY is required for oscillator function differs depending on the peripheral tissue. In antenna, rhythmic output in the form of rhythmic electroantennagram (EAG) responses to odors is severely impaired or abolished in cryb mutants during and after entrainment to LD or temperature cycles (Krishnan et al., 2001). This loss of EAG rhythms in cryb flies is due to the disruption of rhythmic expression within the core feedback loop, as reported by per-luc and tim-luc in antennae cultured during and after LD entrainment and after temperature entrainment (Krishnan et al., 2001; Levine et al., 2002a). This result is reminiscent of the arrhythmicity seen in mice lacking mCRY1 and mCRY2, which repress CLOCK-BMAL1 mediated transcription (Griffin et al., 1999; Kume et al., 1999; Lee et al., 2001; Preitner et al., 2002).
These experiments show that CRY is required for oscillator function in antennae, but it is difficult to know whether CRY also serves to entrain antennae to light cycles since EAGs and gene expression in cryb flies were arrhythmic during LD. Loss of rhythmic per-luc and tim-luc expression in forelegs from cryb flies during and after LD entrainment implies that CRY also functions within the oscillator, in contrast to the situation in wing where per-luc and tim-luc expression is rhythmic in cryb flies during and after LD entrainment (Levine et al., 2002a). In Malpighian tubules from cryb flies, loss of TIM degradation to light pulses suggests that CRY is required for phase resetting, but cycling of PER and TIM levels during LD cycles suggests that entrainment occurs through some other mechanism (Ivanchenko et al., 2001). Rhythms in tim-luc and PER and TIM protein cycling were abolished in cryb mutants during DD, demonstrating that CRY is necessary for oscillator function (Ivanchenko et al., 2001). The role CRY plays within fly peripheral oscillators was investigated in fly heads, where >80% of clock gene expression is from peripheral oscillators in the eye (Glossop and Hardin, 2002). In cryb flies, expression of CLK-CYC activated genes was constantly near peak levels, suggesting that CRY represses CLK-CYC transcription (Collins et al., 2006). Consistent with this result, CRY collaborates with PER to inhibit CLK-CYC transcription in eyes and S2 cells, but not in sLNvs (Collins et al., 2006). If CRY is indeed required for rhythmic transcription within peripheral oscillators, it is difficult to understand how oscillator function persists in cryb fly heads after temperature entrainment (Stanewsky et al., 1998). Despite this inconsistency, CRY and PER dependent repression of CLK-CYC transcription is completely in line with mCRY1 and mCRY2 function in mammals, which repress CLOCK-BMAL1 transcription in collaboration with mPER2 (Griffin et al., 1999; Kume et al., 1999; Lee et al., 2001; Preitner et al., 2002). How Drosophila CRY carries out independent functions in brain pacemaker cells and peripheral oscillators is not known, though other insects (e.g. Monarch butterflies, mosquito, moth) carry out photoreceptor and transcriptional repressor function using two independent cry genes (Yuan et al., 2007; Zhu et al., 2008).
V. Summary and Conclusions
In this review I have covered several topics related to the interlocked feedback loops that keep circadian time in Drosophila. There are several take home points from this review. First, many feedback loop components have been identified in Drosophila that carry out specific roles in regulating time-dependent transcription including the CLK and CYC activators and PER and TIM repressors. Although the per and Clk feedback loops drive rhythmic transcription in opposite phases of the circadian cycle, the per loop controls the Clk loop and functions to keep circadian time. Second, time delays are built into multiple steps of the core feedback loop to achieve a period of ~24h. These delays are controlled post-transcriptionally via regulated synthesis, accumulation, nuclear localization, degradation, and activity of transcriptional activators and repressors. Third, light shifts the Drosophila circadian oscillator to a new circadian phase by promoting TIM degradation. In most oscillator cells CRY acts as a cell-autonomous photoreceptor that binds TIM to induce JET-dependent degradation of both TIM and CRY in the proteasome after light exposure, but also functions as a transcriptional repressor to support core feedback loop function in many peripheral oscillator cells. Retinal photoreceptors, extraretinal photoreceptors and CRY all contribute to the entrainment of locomotor activity pacemaker cells to light-dark cycles, but other environmental factors such as temperature and olfactory cues also integrate with light information to modulate fly activity according to the season and social context. Understanding how the circadian clock is entrained by environmental cues and maintains a ~24h period under constant conditions in a simple model organism such as Drosophila will no doubt continue to provide important insights into the basic mechanisms that control circadian clock function in mammals.
Despite the progress that has been made in identifying components of the circadian feedback loops, major gaps in our understanding of how these components contribute to circadian timekeeping remain. The Clk feedback loop controls rhythmic transcription that peaks near dawn, but is dependent on the core loop. Though rhythmic outputs including locomotor activity are dependent on Pdp1ε/δ within the Clk loop (Benito et al., 2008; Benito et al., 2010; Zheng et al., 2009), we don’t know if this feedback loop contributes to circadian timekeeping. The Neurospora interlocked feedback loop contributes to oscillator stability (Cheng et al., 2001), whereas the mammalian interlocked loop influences period length and phase shifting to light (Preitner et al., 2002). Manipulating components of the Clk loop such as vri and Clk activators will provide the tools to test whether the Clk loop contributes to timekeeping function. Identifying Clk activators would fill a gap in our understanding of the Clk loop since we would be able to determine whether these activators both determine clock cell identity and maintain Clk transcription in adults. Likewise, building tools to inducibly manipulate vri expression in adult flies would enable experiments to test whether vri is required for circadian timekeeping and/or output rhythms. The traditional role for cwo as a repressor of CLK-CYC transcription is based mainly on in vitro and cell culture data, and doesn’t jibe with in vivo data suggesting that cwo contributes to activating CLK-CYC transcription (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007; Richier et al., 2008). Better tools for CWO detection and biochemistry are needed to determine how this gene contributes to the circadian timekeeping mechanism.
A major question in circadian biology is how circadian feedback loops maintain periods of ~24h. While it is clear that delays are incorporated into various steps of the feedback loop, we are now beginning to understand the mechanisms that control these delays. The delayed accumulation of PER repressor complexes in the cytoplasm (at least in the LNvs) involves regulated translation by tyf (Lim et al., 2011), but it is not as clear how much PER phosphorylation by DBT and PER stabilization by TIM and PP1 and PP2a phosphatases contribute to slowing the rate of PER accumulation in the cytoplasm (Fang et al., 2007; Kloss et al., 1998; Kloss et al., 2001; Ko et al., 2002; Price et al., 1998; Sathyanarayanan et al., 2004). Determining whether additional kinases, phosphatases, E3 ubiquitin ligases, or other factors promote PER degradation in the cytoplasm, and whether PER translation is regulated in other oscillator tissues, will be essential for understanding the extent to which these processes contribute to delays in PER cytoplasmic accumulation. SGG, CK2 and an as yet unidentified proline directed kinase phosphorylates PER and/or TIM to promote their nuclear localization (Akten et al., 2003; Ko et al., 2010; Lin et al., 2002a; Martinek et al., 2001), thus enabling these proteins to bind CLK-CYC and inhibit transcription. Movement of PER-TIM repressor complexes into the nucleus coincides with CLK phosphorylation, transcriptional repression, and the removal of CLK-CYC from E-boxes (Kim et al., 2007; Menet et al., 2010; Yu et al., 2006; Yu et al., 2009). The kinases that phosphorylate CLK have not been identified, though it is possible that NMO phosphorylates CLK to promote CLK degradation (Yu et al., 2011). To understand how PER complexes repress transcription, it is important to determine which kinases phosphorylate CLK, where CLK is phosphorylated, and how such phosphorylation represses transcription and induces CLK-CYC release from E-boxes. Kinases also contribute to light-induced and clock-dependent TIM degradation (Naidoo et al., 1999), but the identity of these kinases and their target sites on TIM haven’t been determined, but are essential to understanding how the clock maintains a ~24h period since the loss of TIM promotes PER degradation. Great strides have been made to understand how PER degradation is regulated in the nucleus. NMO phosphorylation of PER at S596 primes further phosphorylation by DBT to alter PER conformation, thus delaying phosphorylation of other DBT target sites including S47, which triggers SLIMB binding and PER degradation (Chiu et al., 2011). Determining the temporal profile of DBT phosphorylation sites on PER and the requirements for phosphorylation at specific sites would provide an unprecedented understanding of a process that is essential for setting circadian period. Once PER is degraded, CLK is returned to a dephosphorylated state that enables E-box binding and transcriptional activation by CLK-CYC (Kim and Edery, 2006; Yu et al., 2006; Yu et al., 2009). Identifying phosphatases that act on CLK during this phase of the circadian cycle will be key to understanding the extent to which CLK dephosphorylation contributes to period determination.
Although the basic molecular events that control light-induced phase shifting by CRY have been characterized, it is apparent that CRY mediates phase delays in locomotor activity independent of TIM degradation. Where light is detected to induce phase delays and how CRY acts to shift the LNv clock represent major gaps in our understanding. A subset of DN1s send projections to LNvs (Helfrich-Forster et al., 2007; Kaneko and Hall, 2000), and certain DN1s also express CRY (Benito et al., 2008; Yoshii et al., 2008). If the CRY positive DN1s also projection to LNvs, these cells could receive light and signal the LNvs just as the H-B eyelet receives light and signals the LNvs (Helfrich-Forster et al., 2001). CRY is required for light shifting in LNvs in the absence of TIM degradation (Tang et al., 2010), which suggests that CRY induces phase delays in LNvs by altering the level or activity of some other core feedback loop component. Determining how LNvs receive light signals during the early evening and shift the oscillator in a CRY-dependent manner is critical for a comprehensive understanding of behavioral phase resetting to light. However, CRY is not the only photoreceptor capable of entraining behavioral rhythms to LD cycles. H-B eyelet neurons may effect light-induced entrainment of behavior by signaling lLNvs, which then relay this information to the sLNvs (Shang et al., 2008; Sheeba et al., 2010). Additional studies are needed to solidify this pathway, define the cellular pathways through which photoreceptors in the compound eyes and ocelli effect light entrainment, and determine how light information from other photoreceptor cells effect shifts in the sLNv molecular oscillator. Lastly, recent work shows that light and temperature information is integrated in DN1s to modulate the pattern of locomotor activity rhythms during LD conditions (Zhang et al., 2010a; Zhang et al., 2010b). How temperature information is received by DN1s, integrated with CRY-dependent light signals, and sent to LNvs to modify behavioral activity are important challenges for future studies, as is the integration of olfactory-based social cues that also modulate behavioral activity.
Although interlocked transcriptional feedback loops function to keep circadian time in eukaryotes, this is not the case in cyanobacteria. The core timekeeping mechanism in cyanobacteria does not require transcription, but instead consists of a ~24h phosphorylation/dephosphorylation cycle mediated by the KaiA, KaiB and KaiC proteins. When KaiA, KaiB, KaiC are incubated with ATP, KaiA stimulates KaiC autophosphorylation and KaiB blocks KaiA to stimulate KaiC dephosphorylation, which results in a KaiC phosphorylation cycle with a ~24h period (Nakajima et al., 2005). Although this cycle necessarily occurs independent of transcription, the Kai oscillator drives rhythms in virtually all transcripts by periodically altering chromosome compaction (Smith and Williams, 2006; Woelfle and Johnson, 2006; Woelfle et al., 2007). Although the core KaiC phosphorylation oscillator functions under most conditions in vitro, transcriptional rhythms contribute to the stability and entrainment of the Kai oscillator in vivo (see review by (Dong and Golden, 2008)). Recent studies show that circadian clocks in some eukaryotic organisms and cell types can operate in the absence of rhythmic transcription. Red blood cells lack nuclei and mitochondria, and thus no transcription can occur. However, the dimerization of peroxyredoxin enzymes, which function to reduce reactive oxygen species, cycles with a circadian rhythm that is temperature entrainable (O’Neill and Reddy, 2011). In fact, binding of reactive oxygen species is what catalyzes the formation of peroxyredoxin dimers, which are then converted back to monomers via reduction by thioredoxin (O’Neill and Reddy, 2011). A similar phenomenon has been observed in the unicellular eukaryotic algae Ostreococcus tauri, which cease transcriptional activity in the dark, yet circadian oscillations in peroxyredoxin dimerization persist (O’Neill et al., 2011). Rhythms in peroxyredoxin dimerization also occur under conditions when the transcriptional feedback loops are operating in Ostreococcus and mouse embryonic fibroblasts, thus both circadian oscillators function simultaneously in the same cell (O’Neill and Reddy, 2011; O’Neill et al., 2011). These oscillators apparently interact at some level as mutant mouse embryonic fibroblasts that abolish transcriptional feedback loops lengthen the period of peroxyredoxin dimerization rhythms (O’Neill and Reddy, 2011). Given that peroxyredoxin dimerization rhythms are seen in mouse embryonic fibroblasts, rhythms in peroxyredoxin dimerization likely occur in Drosophila. Once the oscillator that drives peroxyredoxin dimerization rhythms is identified, it will be fascinating to determine the extent to which this oscillator influences transcription-based oscillators.
Acknowledgements
I want to thank Dr. Wangjie Yu for comments on the manuscript and Isaac Edery for communicating unpublished results. This work was supported by NIH grant NS052854.
References
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Bae K, Lee C, Hardin PE, Edery I. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex. J Neurosci. 2000;20:1746–1753. doi: 10.1523/JNEUROSCI.20-05-01746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Lee C, Sidote D, Chuang KY, Edery I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol Cell Biol. 1998;18:6142–6151. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargiello TA, Jackson FR, Young MW. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312:752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- Bargiello TA, Young MW. Molecular genetics of a biological clock in Drosophila. Proc Natl Acad Sci U S A. 1984;81:2142–2146. doi: 10.1073/pnas.81.7.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylies MK, Bargiello TA, Jackson FR, Young MW. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;326:390–392. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Vosshall LB, Sehgal A, Young MW. New short period mutations of the Drosophila clock gene per. Neuron. 1992;9:575–581. doi: 10.1016/0896-6273(92)90194-i. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J, Houl JH, Roman GW, Hardin PE. The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS. J Biol Rhythms. 2008;23:296–307. doi: 10.1177/0748730408318588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J, Hoxha V, Lama C, Lazareva AA, Ferveur JF, Hardin PE, Dauwalder B. The circadian output gene takeout is regulated by Pdp1epsilon. Proc Natl Acad Sci U S A. 2010;107:2544–2549. doi: 10.1073/pnas.0906422107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J, Zheng H, Hardin PE. PDP1epsilon functions downstream of the circadian oscillator to mediate behavioral rhythms. J Neurosci. 2007;27:2539–2547. doi: 10.1523/JNEUROSCI.4870-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke I, Lang P, Hofbauer A, Engelmann W, Helfrich-Forster C. Preliminary action spectra suggest that the clock cells of Drosophila are synchronized to the external LD-cycle by the compound eyes plus extraretinal photoreceptors. In: Schnitzler NEaH-U., editor. 24th Gottingen Neurobiology Conference. Vol. I. Gottingen, Germany: Thieme; 1996. [Google Scholar]
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Boothroyd CE, Wijnen H, Naef F, Saez L, Young MW. Integration of light and temperature in the regulation of circadian gene expression in Drosophila. PLoS Genet. 2007;3:e54. doi: 10.1371/journal.pgen.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Kay SA. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WF, Low KH, Lim C, Edery I. Thermosensitive splicing of a clock gene and seasonal adaptation. Cold Spring Harb Symp Quant Biol. 2007;72:599–606. doi: 10.1101/sqb.2007.72.021. [DOI] [PubMed] [Google Scholar]
- Chen WF, Majercak J, Edery I. Clock-gated photic stimulation of timeless expression at cold temperatures and seasonal adaptation in Drosophila. J Biol Rhythms. 2006;21:256–271. doi: 10.1177/0748730406289306. [DOI] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Liu Y. Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc Natl Acad Sci U S A. 2001;98:7408–7413. doi: 10.1073/pnas.121170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J, Ko HW, Edery I. NEMO/NLK primes phosphorylation of a time-delay phospho-cluster on PERIOD revealing a novel mechanism for how circadian clock speed is set. 2011 [Google Scholar]
- Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 2008;22:1758–1772. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Nitabach MN. Circadian biology: environmental regulation of a multi-oscillator network. Curr Biol. 2010;20:R322–R324. doi: 10.1016/j.cub.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Collins B, Mazzoni EO, Stanewsky R, Blau J. Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr Biol. 2006;16:441–449. doi: 10.1016/j.cub.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Collins BH, Rosato E, Kyriacou CP. Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc Natl Acad Sci U S A. 2004;101:1945–1950. doi: 10.1073/pnas.0308240100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin KD, Huang ZJ, Rosbash M. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron. 1995;14:365–3672. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- Cusumano P, Klarsfeld A, Chelot E, Picot M, Richier B, Rouyer F. PDF-modulated visual inputs and cryptochrome define diurnal behavior in Drosophila. Nat Neurosci. 2009;12:1431–1437. doi: 10.1038/nn.2429. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Dauwalder B, Tsujimoto S, Moss J, Mattox W. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 2002;16:2879–2892. doi: 10.1101/gad.1010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dissel S, Codd V, Fedic R, Garner KJ, Costa R, Kyriacou CP, Rosato E. A constitutively active cryptochrome in Drosophila melanogaster. Nat Neurosci. 2004;7:834–840. doi: 10.1038/nn1285. [DOI] [PubMed] [Google Scholar]
- Dong G, Golden SS. How a cyanobacterium tells time. Curr Opin Microbiol. 2008;11:541–546. doi: 10.1016/j.mib.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruille R, Emery P. A plastic clock: how circadian rhythms respond to environmental cues in Drosophila. Mol Neurobiol. 2008;38:129–145. doi: 10.1007/s12035-008-8035-y. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci U S A. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Hall JC, Rosbash M. A unique circadian-rhythm photoreceptor. Nature. 2000a;404:456–457. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Helfrich-Forster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000b;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Fang Y, Sathyanarayanan S, Sehgal A. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21:1506–1518. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- Fujii S, Krishnan P, Hardin P, Amrein H. Nocturnal male sex drive in Drosophila. Curr Biol. 2007;17:244–251. doi: 10.1016/j.cub.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N, Saez L, Delahaye-Brown AM, Myers MP, Sehgal A, Young MW, Weitz CJ. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- George H, Terracol R. The vrille gene of Drosophila is a maternal enhancer of decapentaplegic and encodes a new member of the bZIP family of transcription factors. Genetics. 1997;146:1345–1363. doi: 10.1093/genetics/146.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser FT, Stanewsky R. Synchronization of the Drosophila circadian clock by temperature cycles. Cold Spring Harb Symp Quant Biol. 2007;72:233–242. doi: 10.1101/sqb.2007.72.046. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Hardin PE. Central and peripheral circadian oscillator mechanisms in flies and mammals. J Cell Sci. 2002;115:3369–3377. doi: 10.1242/jcs.115.17.3369. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- Golombek DA, Rosenstein RE. Physiology of circadian entrainment. Physiol Rev. 2010;90:1063–1102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- Griffin EA, Jr., Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Chelot E, Papin C, Limbourg-Bouchon B, Rouyer F. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- Hao H, Allen DL, Hardin PE. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE. Transcription regulation within the circadian clock: the E-box and beyond. J Biol Rhythms. 2004;19:348–360. doi: 10.1177/0748730404268052. [DOI] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Hardin PE. Essential and expendable features of the circadian timekeeping mechanism. Curr Opin Neurobiol. 2006;16:686–692. doi: 10.1016/j.conb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci U S A. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C, Shafer OT, Wulbeck C, Grieshaber E, Rieger D, Taghert P. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol. 2007;500:47–70. doi: 10.1002/cne.21146. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30:249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- Houl JH, Ng F, Taylor P, Hardin PE. CLOCK expression identifies developing circadian oscillator neurons in the brains of Drosophila embryos. BMC Neurosci. 2008;9:119. doi: 10.1186/1471-2202-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu KG, Reichart H, Stark WS. Electrophysiological characterization of Drosophila ocelli. J Comp Physiol. 1978;126:15–24. [Google Scholar]
- Huang ZJ, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- Ivanchenko M, Stanewsky R, Giebultowicz JM. Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks. J Biol Rhythms. 2001;16:205–215. doi: 10.1177/074873040101600303. [DOI] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Schoer R, Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6:e119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT, 2nd, Nelson S, Rosbash M. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009 doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21:1675–1686. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Keegan KP, Pradhan S, Wang JP, Allada R. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput Biol. 2007;3:e208. doi: 10.1371/journal.pcbi.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Bae K, Ng FS, Glossop NR, Hardin PE, Edery I. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron. 2002;34:69–81. doi: 10.1016/s0896-6273(02)00639-6. [DOI] [PubMed] [Google Scholar]
- Kim EY, Edery I. Balance between DBT/CKIepsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci U S A. 2006;103:6178–6183. doi: 10.1073/pnas.0511215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu W, Hardin PE, Edery I. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol. 2007;27:5014–5028. doi: 10.1128/MCB.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Vitaterna MH, Chang AM, Dove WF, Pinto LH, Turek FW, Takahashi JS. The mouse Clock mutation behaves as an antimorph and maps within the W19H deletion, distal of Kit. Genetics. 1997a;146:1049–1060. doi: 10.1093/genetics/146.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian clock gene. Cell. 1997b;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivimae S, Saez L, Young MW. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol. 2008;6:e183. doi: 10.1371/journal.pbio.0060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Kloss B, Rothenfluh A, Young MW, Saez L. Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron. 2001;30:699–706. doi: 10.1016/s0896-6273(01)00320-8. [DOI] [PubMed] [Google Scholar]
- Knowles A, Koh K, Wu JT, Chien CT, Chamovitz DA, Blau J. The COP9 signalosome is required for light-dependent timeless degradation and Drosophila clock resetting. J Neurosci. 2009;29:1152–1162. doi: 10.1523/JNEUROSCI.0429-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3beta/SGG in circadian clocks. J Neurosci. 2010;30:12664–12675. doi: 10.1523/JNEUROSCI.1586-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Hamblen-Coyle MJ, Jamison CF, Hall JC. An ultrashort clock mutation at the period locus of Drosophila melanogaster that reveals some new features of the fly’s circadian system. J Biol Rhythms. 1994;9:189–216. doi: 10.1177/074873049400900303. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Pittendrigh CS, Orr D. Reciprocal behaviour associated with altered homeostasis and photosensitivity of Drosophila clock mutants. Journal of Neurogenetics. 1989;6:1–10. doi: 10.3109/01677068909107096. [DOI] [PubMed] [Google Scholar]
- Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400:375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- Krishnan B, Levine JD, Lynch MK, Dowse HB, Funes P, Hall JC, Hardin PE, Dryer SE. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- Krishnan P, Chatterjee A, Tanoue S, Hardin PE. Spike amplitude of single-unit responses in antennal sensillae is controlled by the Drosophila circadian clock. Curr Biol. 2008;18:803–807. doi: 10.1016/j.cub.2008.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp JJ, Kent C, Billeter JC, Azanchi R, So AK, Schonfeld JA, Smith BP, Lucas C, Levine JD. Social experience modifies pheromone expression and mating behavior in male Drosophila melanogaster. Curr Biol. 2008;18:1373–1383. doi: 10.1016/j.cub.2008.07.089. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol Cell Biol. 1999;19:5316–8325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Advanced analysis of a cryptochrome mutation’s effects on the robustness and phase of molecular cycles in isolated peripheral tissues of Drosophila. BMC Neurosci. 2002a;3:5. doi: 10.1186/1471-2202-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Resetting the circadian clock by social experience in Drosophila melanogaster. Science. 2002b;298:2010–2012. doi: 10.1126/science.1076008. [DOI] [PubMed] [Google Scholar]
- Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol. 2007;17:1082–1089. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Lee J, Choi C, Kilman VL, Kim J, Park SM, Jang SK, Allada R, Choe J. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature. 2011;470:399–403. doi: 10.1038/nature09728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci U S A. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FJ, Song W, Meyer-Bernstein E, Naidoo N, Sehgal A. Photic signaling by cryptochrome in the Drosophila circadian system. Mol Cell Biol. 2001;21:7287–7294. doi: 10.1128/MCB.21.21.7287-7294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002a;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- Lin JM, Schroeder A, Allada R. In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J Neurosci. 2005;25:11175–11183. doi: 10.1523/JNEUROSCI.2159-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Han M, Shimada B, Wang L, Gibler TM, Amarakone A, Awad TA, Stormo GD, Van Gelder RN, Taghert PH. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2002b;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low KH, Lim C, Ko HW, Edery I. Natural variation in the splice site strength of a clock gene and species-specific thermal adaptation. Neuron. 2008;60:1054–1067. doi: 10.1016/j.neuron.2008.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Chen WF, Edery I. Splicing of the period gene 3’-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol. 2004;24:3359–3372. doi: 10.1128/MCB.24.8.3359-3372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Sidote D, Hardin PE, Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron. 1999;24:219–230. doi: 10.1016/s0896-6273(00)80834-x. [DOI] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, Kasukawa T, Dauwalder B, Itoh TQ, Takahashi K, Ueda R, Hardin PE, Tanimura T, Ueda HR. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007;21:1687–1700. doi: 10.1101/gad.1552207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 2010;24:358–367. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A, Emery-Le M, Emery P. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron. 2007;53:689–701. doi: 10.1016/j.neuron.2007.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Song W, Hunter-Ensor M, Sehgal A. A role for the proteasome in the light response of the timeless clock protein. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nambu JR, Lewis JO, Wharton KA, Jr., Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Rosbash M. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell. 2004;13:213–223. doi: 10.1016/s1097-2765(03)00503-3. [DOI] [PubMed] [Google Scholar]
- O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohata K, Nishiyama H, Tsukahara Y. Action spectrum of the circadian clock photoreceptor in Drosophila melanogaster. In: Touitou Y, editor. Biological Clocks: Mechanisms and Applications. Amsterdam: Elesevier; 1998. pp. 167–171. [Google Scholar]
- Pearn MT, Randall LL, Shortridge RD, Burg MG, Pak WL. Molecular, biochemical, and electrophysiological characterization of Drosophila norpA mutants. J Biol Chem. 1996;271:4937–4945. doi: 10.1074/jbc.271.9.4937. [DOI] [PubMed] [Google Scholar]
- Peschel N, Chen KF, Szabo G, Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol. 2009;19:241–247. doi: 10.1016/j.cub.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Peschel N, Veleri S, Stanewsky R. Veela defines a molecular link between Cryptochrome and Timeless in the light-input pathway to Drosophila’s circadian clock. Proc Natl Acad Sci U S A. 2006;103:17313–17318. doi: 10.1073/pnas.0606675103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS, Minis DH. The Entrainment of Circadian Oscillations by Light and Their Role as Photoperiodic Clocks. American Naturalist. 1964;98:261–294. [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Price JL, Dembinska ME, Young MW, Rosbash M. Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 1995;14:4044–4049. doi: 10.1002/j.1460-2075.1995.tb00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, Hall JC, Rosbash M. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell. 1984;38:701–710. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Richier B, Michard-Vanhee C, Lamouroux A, Papin C, Rouyer F. The clockwork orange Drosophila protein functions as both an activator and a repressor of clock gene expression. J Biol Rhythms. 2008;23:103–116. doi: 10.1177/0748730407313817. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Edery I, Hall JC, Rosbash M. The analysis of new short-period circadian rhythm mutants suggests features of D. melanogaster period gene function. J Neurogenet. 1992;8:101–113. doi: 10.3109/01677069209084155. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Kumar S, Chen CH, Chen D, Hay B, Sehgal A. Identification of novel genes involved in light-dependent CRY degradation through a genome-wide RNAi screen. Genes Dev. 2008;22:1522–1533. doi: 10.1101/gad.1652308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Posttranslational Regulation of Drosophila PERIOD Protein by Protein Phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Saunders DS, Gillanders L, Lewis RD. Light-pulse Locomotor Phase Response Curves for the Activity Rhythm in Period Mutants of Drosophila melanogaster. Journal of Insect Physiology. 1994;40:957–968. [Google Scholar]
- Sawyer LA, Hennessy JM, Peixoto AA, Rosato E, Parkinson H, Costa R, Kyriacou CP. Natural variation in a Drosophila clock gene and temperature compensation. Science. 1997;278:2117–2120. doi: 10.1126/science.278.5346.2117. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–166. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers MP, Young MW. Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci U S A. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Holmes TC. Persistence of morning anticipation behavior and high amplitude morning startle response following functional loss of small ventral lateral neurons in Drosophila. PLoS ONE. 2010;5:e11628. doi: 10.1371/journal.pone.0011628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18:1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci U S A. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler ML, Kuropatwinski KK, Schumer M, Antoch MP. A serine cluster mediates BMAL1-dependent CLOCK phosphorylation and degradation. Cell Cycle. 2009;8:4138–4146. doi: 10.4161/cc.8.24.10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Nawathean P, Fernandez Mde L, Menet JS, Ceriani MF, Rosbash M. The Drosophila circadian network is a seasonal timer. Cell. 2007;129:207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Tang CH, Hinteregger E, Shang Y, Rosbash M. Light-mediated TIM degradation within Drosophila pacemaker neurons (s-LNvs) is neither necessary nor sufficient for delay zone phase shifts. Neuron. 2010;66:378–385. doi: 10.1016/j.neuron.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue S, Krishnan P, Chatterjee A, Hardin PE. G protein-coupled receptor kinase 2 is required for rhythmic olfactory responses in Drosophila. Curr Biol. 2008;18:787–794. doi: 10.1016/j.cub.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. Control of Daily Transcript Oscillations in Drosophila by Light and the Circadian Clock. PLoS Genet. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfle MA, Johnson CH. No promoter left behind: global circadian gene expression in cyanobacteria. J Biol Rhythms. 2006;21:419–431. doi: 10.1177/0748730406294418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfle MA, Xu Y, Qin X, Johnson CH. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc Natl Acad Sci U S A. 2007;104:18819–18824. doi: 10.1073/pnas.0706069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Lee JE, Padgett RW, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Todo T, Wulbeck C, Stanewsky R, Helfrich-Forster C. Cryptochrome operates in the compound eyes and a subset of Drosophila’s clock neurons. J. Comp. Neurol. 2008 doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- Yoshitane H, Takao T, Satomi Y, Du NH, Okano T, Fukada Y. Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol Cell Biol. 2009;29:3675–3686. doi: 10.1128/MCB.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- Yu Q, Jacquier AC, Citri Y, Hamblen M, Hall JC, Rosbash M. Molecular mapping of point mutations in the period gene that stop or speed up biological clocks in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1987;84:784–788. doi: 10.1073/pnas.84.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119:4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- Yu W, Houl JH, Hardin PE. NEMO kinase contributes to core period determination by slowing the pace of the Drosophila circadian oscillator. Curr Biol. 2011 doi: 10.1016/j.cub.2011.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–7233. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Price JL, Hardin PE. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol. 2009;29:1452–1458. doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Lin F, Zheng X, Sehgal A. Serotonin modulates circadian entrainment in Drosophila. Neuron. 2005;47:115–127. doi: 10.1016/j.neuron.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol. 2007;24:948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- Zehring WA, Wheeler DA, Reddy P, Konopka RJ, Kyriacou CP, Rosbash M, Hall JC. P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell. 1984;39:369–376. doi: 10.1016/0092-8674(84)90015-1. [DOI] [PubMed] [Google Scholar]
- Zeng H, Hardin PE, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. Embo J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chung BY, Lear BC, Kilman VL, Liu Y, Mahesh G, Meissner RA, Hardin PE, Allada R. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol. 2010a;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lear BC, Seluzicki A, Allada R. The CRYPTOCHROME photoreceptor gates PDF neuropeptide signaling to set circadian network hierarchy in Drosophila. Curr Biol. 2009;19:2050–2055. doi: 10.1016/j.cub.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol. 2010b;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Koh K, Sowcik M, Smith CJ, Chen D, Wu MN, Sehgal A. An isoform-specific mutant reveals a role of PDP1 epsilon in the circadian oscillator. J Neurosci. 2009;29:10920–10927. doi: 10.1523/JNEUROSCI.2133-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Sauman I, Yuan Q, Casselman A, Emery-Le M, Emery P, Reppert SM. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 2008;6:e4. doi: 10.1371/journal.pbio.0060004. [DOI] [PMC free article] [PubMed] [Google Scholar]