Abstract
The tumor suppressor protein p53 is a critical stress response transcription factor that induces the expression of genes leading to cell cycle arrest, apoptosis, and tumor suppression. We found that mammalian inositol polyphosphate multikinase (IPMK) stimulated p53-mediated transcription by binding to p53 and enhancing its acetylation by the acetyltransferase p300 independently of its inositol phosphate and lipid kinase activities. Genetic or RNA interference (RNAi)–mediated knockdown of IPMK resulted in decreased activation of p53, decreased recruitment of p53 and p300 to target gene promoters, abrogated transcription of p53 target genes, and enhanced cell viability. Additionally, blocking the IPMK-p53 interaction decreased the extent of p53-mediated transcription. These results suggest that IPMK acts as a transcriptional coactivator for p53 and that it is an integral part of the p53 transcriptional complex facilitating cell death.
INTRODUCTION
The tumor suppressor p53 is a critical transcriptional factor that senses and modulates cellular responses to injury and stress (1). Responsible for gene induction programs that affect cell cycle arrest, apoptosis, cellular senescence, and aging, p53 serves as a critical checkpoint against oncogenic insults (2, 3), with inactivating mutations demonstrated in as many as 50% of human cancers (1). In cases where the p53 gene is left intact, other mutations along the p53 pathway have been demonstrated to interfere with its function. Although some prevent the activation of p53, others ablate the functional effects of p53 downstream of its transcriptional activity (4–6). To date, the components of the p53 pathway that mediate its activation and transcriptional response remain poorly defined. Recently, Del Sal and colleagues performed a loss-of-function screen in primary human BJ fibroblasts, demonstrating that inositol poly-phosphate multikinase (IPMK) may be a critical regulator of p53 function (7).
IPMK is a broad-specificity enzyme that converts inositol 1,4,5-trisphosphate (IP3) into inositol 1,4,5,6-tetrakisphosphate (IP4) and subsequently inositol 1,3,4,5,6-pentakisphosphate (IP5). It is the rate-limiting enzyme for the generation of higher inositol polyphosphate species such as inositol pyrophosphate (IP7) (8–10), which has been shown to modulate insulin sensitivity (11), neutrophil function (12), chemotaxis and endocytosis (13, 14), and telomere maintenance (15, 16). In addition to its soluble IP3 kinase activity, mammalian IPMK also has lipid kinase activity. Thus, IPMK is a physiologic phosphatidylinositol 3-kinase (PI3K), generating phosphatidylinositol 3,4,5-trisphosphate (PIP3) from PIP2, activating Akt (also known as protein kinase B) (17), as well as PIP3-dependent transcriptional events such as those mediated by steroidogenic factor 1 (18). IPMK also manifests physiological effects that are independent of both its IP3 kinase and PI3K activities. IPMK binds to the mammalian target of rapamycin (mTOR), stabilizing and enhancing the activity of the mTOR complex 1 (mTORC1) noncatalytically (19). Because of these diverse functions, IPMK is notably pleiotropic, consistent with its designation as a “moonlighting protein” in yeast (20).
Transcriptional functions for mammalian IPMK were implied by studies in which the yeast IPMK homolog was discovered as part of a transcriptional complex regulating genes that facilitate the use of arginine—hence its designation as Arg82 (8, 21–23). In yeast, it is also critical for the efficient transcription of phosphate-responsive genes such as the phosphatase-encoding gene PHO5 through IP4- and IP5-mediated nucleosome mobilization and chromatin remodeling (24, 25). In mammals, although a substantial portion of IPMK is localized to the nucleus (26, 27), its nuclear functions have not been fully characterized. Here, we report that IPMK is a transcriptional coactivator of the tumor suppressor p53. We found that IPMK bound to p53 independently of its catalytic activity and enhanced p53 binding to the acetyltransferase p300, augmenting its acetylation. IPMK stimulated the activation and binding of p53 to its targets’ promoters with attendant transcriptional activation, facilitating p53-mediated cell death.
RESULTS
Endogenous IPMK binds to p53 during cell death
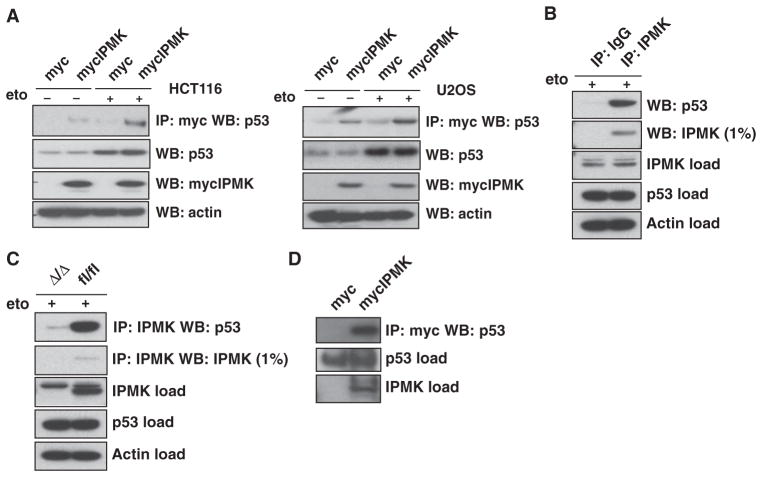
We overexpressed exogenous IPMK in the human colon cancer cell line HCT116 and the human osteosarcoma cell line U2OS. In both cell lines, exogenous IPMK bound to endogenous p53 upon treatment with etoposide, a DNA-damaging agent that canonically induces apoptosis by activating p53 (28, 29) (Fig. 1A). Endogenous IPMK also bound to p53 in etoposide-treated wild-type mouse embryonic fibroblasts (MEFs) (Fig. 1B). Such binding was lost in MEFs with tamoxifen-induced, Cre recombinase–mediated depletion of the gene encoding IPMK (Fig. 1C). The interaction between IPMK and p53 did not appear to require intervening proteins because purified IPMK bound to p53 directly in vitro (Fig. 1D).
Fig. 1.
IPMK interacts with p53. (A) HCT116 cells (left panel) and U2OS cells (right panel) transfected with plasmid encoding myc-tagged IPMK (mycIPMK) and treated overnight with 20 μM etoposide (eto) were lysed and subjected to immunoprecipitation (IP) with anti-myc agarose. IPMK and p53 were resolved from the immunoprecipitated samples by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and identified by Western blotting (WB). Inputs for p53, mycIPMK, and actin are shown in the bottom blots. (B) Wild-type primary MEFs treated with 20 μM etoposide were lysed and subjected to immunoprecipitation with normal immunoglobulin G (IgG) or antibody specific for IPMK. SDS-PAGE and Western blotting were performed as described in (A). (C) Lysates from IPMK-deficient (Δ/Δ) and floxed wild-type (fl/fl) MEFs were used as described in (B) to assess endogenous IPMK-p53 binding. (D) Immunoprecipitation of purified mycIPMK and bacterially purified p53, as assessed by immunoprecipitation with anti-myc agarose and subsequent protein resolution and identification by SDS-PAGE and Western blotting. Blots in all panels are representative of at least three experiments.
IPMK enhances p53 transcriptional activity
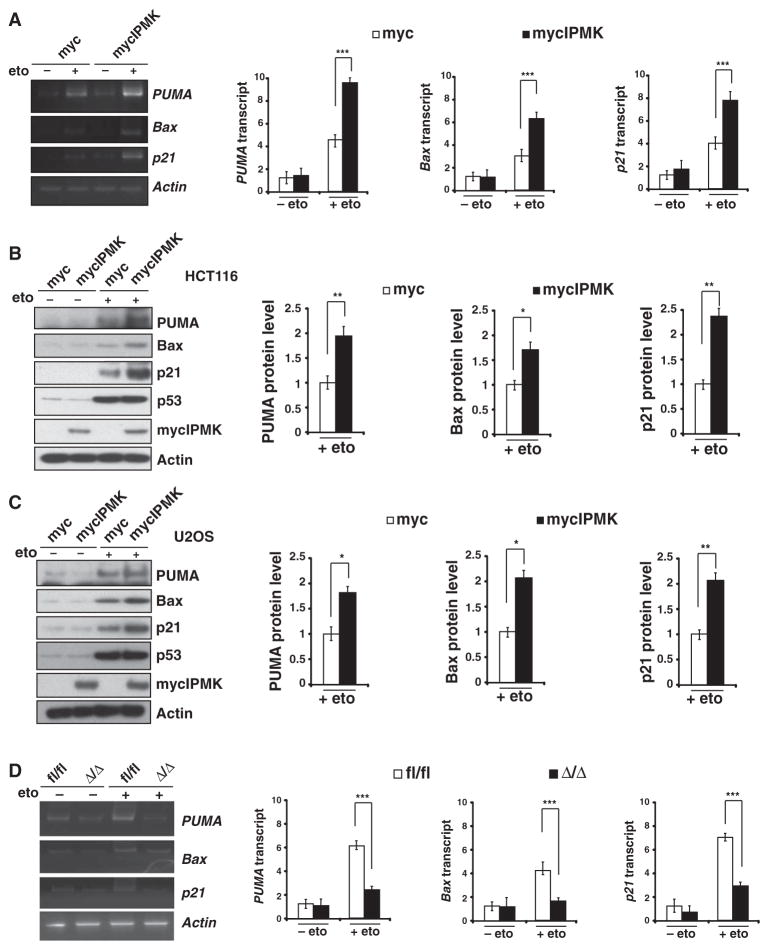
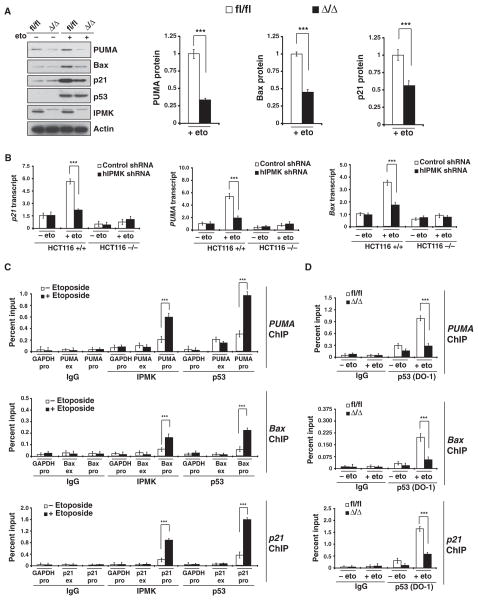
We investigated the role of IPMK-p53 association in p53 transcriptional activity by transfecting U2OS cells with exogenous IPMK and monitoring the mRNA amounts of the canonical p53 targets PUMA (p53 up-regulated modulator of apoptosis), Bax [B cell lymphoma 2 (Bcl-2)–associated X], and p21 [also known as CDKN1A (cyclin-dependent kinase inhibitor 1)] (30–34). Overexpression of IPMK increased the amounts of these mRNAs by about twofold in the presence of etoposide (Fig. 2A). Increased IPMK abundance also enhanced the production of PUMA, Bax, and p21 proteins in etoposide-treated HCT116 (Fig. 2B) and U2OS cells (Fig. 2C), as well as in U2OS cells treated with either 5-fluorouracil (fig. S1A) or doxorubicin (fig. S1B), agents that induce cell death (29, 35, 36). To explore whether endogenous IPMK regulated p53 transcriptional activity, we knocked down IPMK with short hairpin RNA (shRNA) (fig. S2) and monitored the abundance of PUMA, Bax, and p21 proteins. Depletion of IPMK resulted in decreased PUMA, Bax, and p21 abundance after treatment with etoposide (fig. S3). Commensurately, the absence of IPMK in primary MEFs resulted in a 50 to 70% decrease in the amounts of PUMA, Bax, and p21 mRNAs (Fig. 2D), as well as a 40 to 65% decrease in their corresponding protein amounts after treatment with etoposide (Fig. 3A). This regulation of PUMA, Bax, and p21 was p53-dependent, because p53-null HCT116 cells transfected with IPMK shRNA did not exhibit decreased amounts of PUMA, Bax, or p21 mRNAs (Fig. 3B). These proteins were undetectable in HCT116 p53-null cells before and after treatment with etoposide (fig. S4).
Fig. 2.
IPMK augments the transcriptional activity of p53. (A) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis (left) and quantification (right) of PUMA, Bax, and p21 mRNAs in U2OS cells transfected with plasmids encoding myc or mycIPMK and treated overnight with 10 μM etoposide. Data are means ± SEM from three experiments. ***P < 0.001, Student’s t test. (B) Western blotting analysis of PUMA, Bax, and p21 proteins in HCT116 cells transfected and treated as in (A). Data are means ± SEM from three experiments. *P < 0.05, **P < 0.01, Student’s t test. (C) Western blotting analysis of PUMA, Bax, and p21 proteins in U2OS cells transfected and treated as in (A). Data are means ± SEM from three experiments. *P < 0.05, **P < 0.01, Student’s t test. (D) qRT-PCR analysis (left) and quantification (right) of PUMA, Bax, and p21 mRNAs in fl/fl or Δ/Δ MEFs treated overnight with 20 μM etoposide. Data are means ± SEM from three experiments. ***P < 0.001, n = 3, mean ± SEM, Student’s t test.
Fig. 3.
IPMK enhances p53 localization to target promoters and augments expression of p53 downstream targets. (A) Western blotting analysis of p53 and its targets PUMA, Bax, and p21 in fl/fl or Δ/Δ MEFs treated overnight with 20 μM etoposide. **P < 0.01, n = 3, mean ± SEM, Student’s t test. (B) Amounts of p21, PUMA, and Bax mRNAs in wild-type (+/+) and p53-null (−/−) HCT116 cells. ***P < 0.001, n = 3, mean ± SEM, Student’s t test. (C) Detection of IPMK at the promoters or exon regions of PUMA, Bax, and p21 by ChIP analysis of lysates from etoposide-treated wild-type MEFs. Data are means ± SEM from three experiments. ***P < 0.001, Student’s t test. (D) ChIP analysis of p53 binding to the promoter regions of PUMA, Bax, and p21 in etoposide-treated fl/fl or Δ/Δ MEFs. Data are means ± SEM from three experiments. ***P < 0.001, Student’s t test.
We sought to determine whether IPMK stimulated the transcriptional activity of p53 by serving as a component of the p53 transcriptional complex. We performed chromatin immunoprecipitation (ChIP) assays with antibodies against IPMK to monitor its association with the PUMA, Bax, and p21 promoters and with p53. We found that IPMK and p53 were indeed localized to PUMA, Bax, and p21 promoters in etoposide-treated primary MEFs (Fig. 3C). IPMK was also recruited to the PUMA promoter in etoposide-treated U2OS cells transfected with plasmid encoding IPMK (fig. S5). Additionally, IPMK augmented the formation of the p53 transcriptional complex because depletion of IPMK in primary MEFs decreased the extent of recruitment of p53 to the PUMA, Bax, and p21 promoters by 60 to 80% (Fig. 3D and fig. S6).
IPMK enhances p53 acetylation and histone acetylation via p300
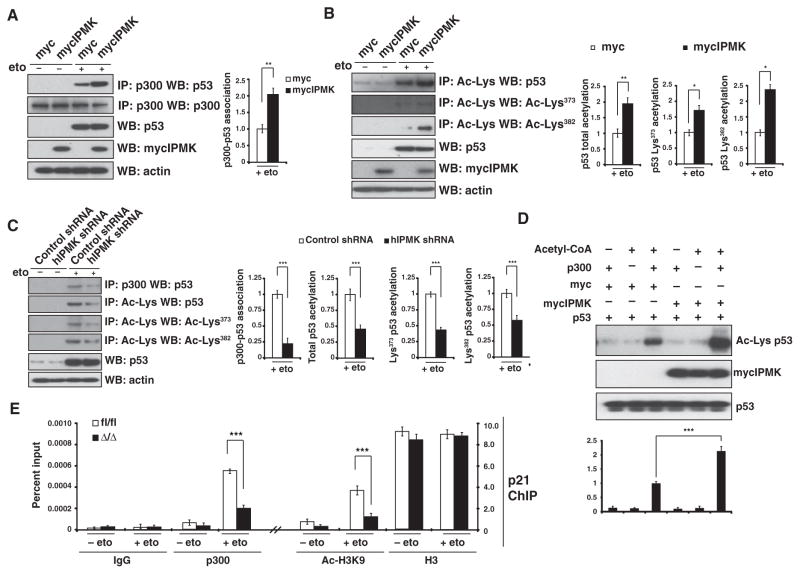
We next investigated molecular mechanisms responsible for the stimulation of p53 transcriptional activity by IPMK. The histone acetyltransferase p300 acetylates p53 and is a coactivator for p53 transcriptional activity (37–40). Overexpression of IPMK enhanced the binding of p53 to p300 in etoposide-treated U2OS cells (Fig. 4A), which correlated with an increase in the acetylation of p53 at Lys373 and Lys382 (Fig. 4B). In etoposide-treated HCT116 cells, shRNA-mediated knockdown of IPMK reduced the binding of p300 to p53 by about 75% (Fig. 4C) and reduced both the total and lysine-specific (Lys373 and Lys382) acetylation of p53 by about 50% (Fig. 4C). To further explore whether IPMK directly mediated the acetylation of p53, we examined the interaction of these proteins in vitro. Purified mycIPMK doubled the acetylation of purified p53 mediated by p300 (Fig. 4D), indicating that IPMK induces the acetylation of p53 directly through p300.
Fig. 4.
IPMK stimulates p53 acetylation and histone H3 acetylation by recruiting p300. (A) Immunoprecipitation and Western blotting were used to detect binding between p300 and p53 in lysates from HCT116 cells transfected with plasmids encoding myc or mycIPMK and treated with 10 μM etoposide overnight. Data are means ± SEM from three experiments. **P< 0.01, Student’s t test. (B) Immunoprecipitation for acetylated lysine (Ac-Lys) and Western blotting for acetylated p53 at Lys373 (Ac-Lys373) and Lys382 (Ac-Lys382) in lysates from etoposide-treated HCT116 cells transfected with plasmids encoding myc or mycIPMK. Data are means ± SEM from three experiments. **P < 0.01, *P < 0.05, Student’s t test. (C) Immunoprecipitation and Western blotting were used to detect acetylated p53 and p300-p53 binding in lysates from etoposide-treated HCT116 cells transfected with either control shRNA or IPMK-specific shRNA. Data are means ± SEM from four experiments. ***P < 0.001, Student’s t test. (D) Western blotting and quantification of the acetylation of purified p53 (Ac-Lys) when exposed to combinations of purified IPMK and p300 in the presence of acetyl-CoA (coenzyme A). Data are means ± SEM from three experiments. ***P < 0.001, Student’s t test. (E) ChIP analysis of acetylation (Ac-H3K9 compared with H3) and p300 binding (p300 compared with IgG) at the promoter region of p21 in etoposide-treated fl/fl and Δ/Δ MEFs. Data are means ± SEM from three experiments. ***P < 0.001, Student’s t test.
Because p53 transcriptional activity is also dependent on chromatin assembly and accessibility, we investigated whether IPMK also regulated the p300-mediated acetylation of histones at p53 target genes. Genetic deletion of IPMK in primary MEFs resulted in about 60% reduction in the binding of p300 to the promoter of p21 after treatment with etoposide and a significant decrease in the acetylation of Lys9 on histone H3 at the p21 promoter (Fig. 4E). Thus, the findings indicated that IPMK’s stimulation of p53 transcriptional activity is mediated through the p300-mediated acetylation of p53 and histones at promoters of p53 targets.
IPMK is required for p53-mediated cell death
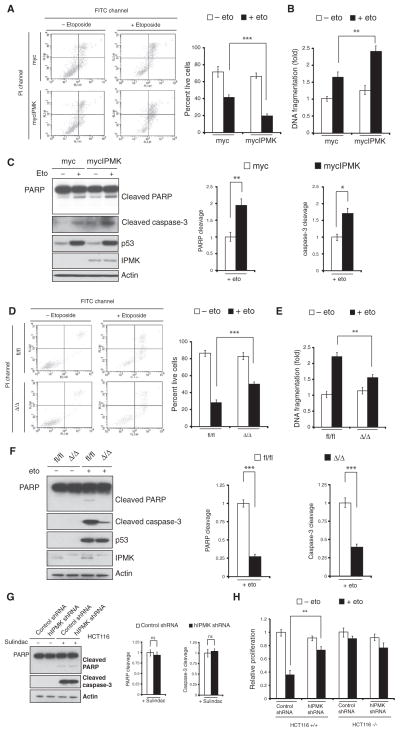
As a tumor suppressor, p53 activates proapoptotic genes to induce cell death (41). We explored whether IPMK, through its interaction with p53, played a role in this mechanism. U2OS cells transfected with plasmid encoding mycIPMK showed increased apoptosis after treatment with etoposide compared with cells transfected with control plasmid encoding myc (Fig. 5A). Reduced cell proliferation was also detected in these cells treated with either etoposide (fig. S7A) or 5-fluorouracil (fig. S7B). In U2OS cells treated with etoposide, IPMK overexpression stimulated DNA fragmentation by more than 30% (Fig. 5B) and increased the abundance of cleaved PARP [poly(ADP-ribose) polymerase] and caspase-3 (Fig. 5C), indicating augmented apoptosis. By contrast, genetic deletion of IPMK in etoposide-treated MEFs significantly increased cell viability (Fig. 5D) and proliferation (fig. S7C), decreased DNA fragmentation (Fig. 5E), and reduced the abundance of cleaved PARP and caspase-3 (Fig. 5F) compared to treated MEFs that were sufficient in IPMK, indicating that IPMK induces apoptosis. However, IPMK shRNA did not affect PARP cleavage or caspase-3 cleavage in HCT116 cells treated with sulindac, a cyclooxygenase inhibitor that induces p53-independent apoptosis (Fig. 5G). Moreover, depletion of IPMK in p53-null HCT116 cells did not significantly affect cell proliferation after etoposide treatment (Fig. 5H), suggesting that the ability of IPMK to induce cell death is p53-dependent.
Fig. 5.
IPMK mediates cell death. (A) Flow cytometry analysis with annexin V–FITC (fluorescein isothiocyanate) and propidium iodide (PI) of live (lower left quadrant), dead (upper right), and apoptosing (lower right) U2OS cells transfected with plasmids encoding myc or mycIPMK and treated with 20 μM etoposide. (B and C) DNA fragmentation (B) and Western blot analysis (C) of PARP and caspase-3 cleavage in transfected, etoposide-treated U2OS cells. Data are means ± SEM from three experiments. **P < 0.01, *P < 0.05, Student’s t test. (D to F) Analysis of cell viability (D), DNA fragmentation (E), and PARP and caspase-3 cleavage (F) in fl/fl and Δ/Δ MEFs treated with 20 μM etoposide. Data are means ± SEM from three experiments. ***P < 0.001, **P < 0.01, Student’s t test. (G) Western blot analysis of PARP and caspase-3 cleavage in HCT116 cells transfected with control or hIPMK shRNA and treated with 120 μM sulindac. Data are means ± SEM from three experiments (ns, no significance; Student’s t test). (H) Cell proliferation was measured by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay in wild-type (+/+) and p53-null (−/−) HCT116 cells transfected with control or hIPMK-specific shRNA and treated with 10 μM etoposide. Data are means ± SEM from three experiments. **P < 0.01, Student’s t test.
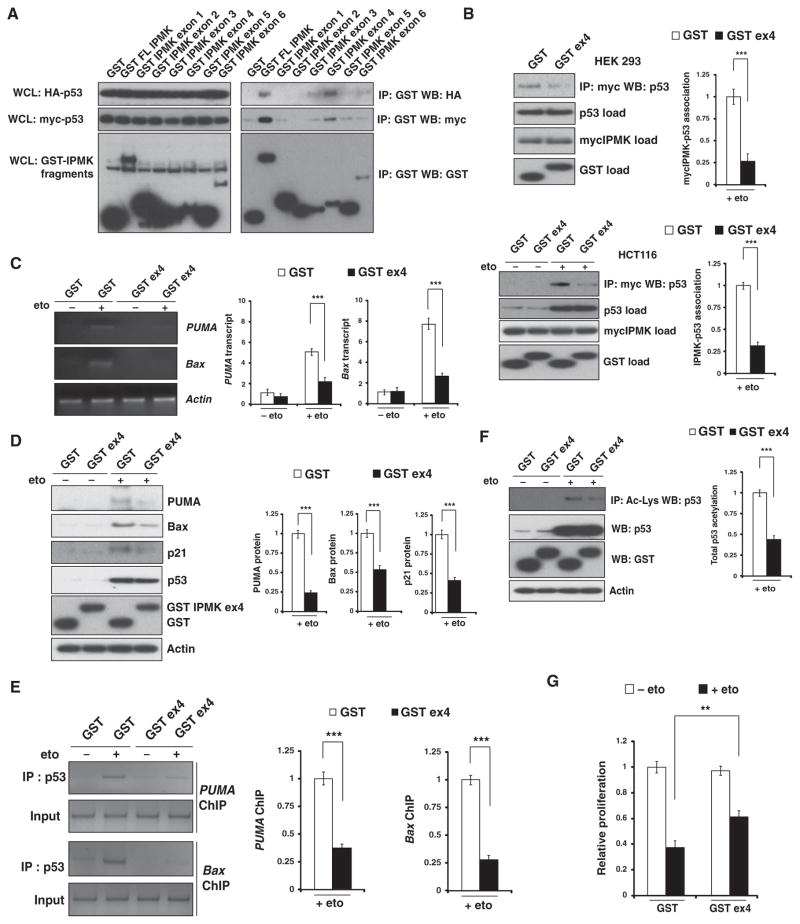
The association between IPMK and p53 is required for coactivation of p53 transcriptional activity
In experiments with glutathione S-transferase (GST)–tagged full-length and fragments of IPMK, we mapped sites on IPMK that bound to p53. Binding was primarily mediated by a region of the IPMK protein encoded by exon 4 (Fig. 6A), a fragment that contains amino acids 125 to 184 and both the conserved inositol phosphate–binding P-C-x-x-D-x-K-x-G motif and the linker region leading up to but excluding the catalytic domain necessary for adenosine 5′-triphosphate binding and catalysis (42–44). This 60–amino acid residue fragment could be used as a dominant-negative construct because its over-expression prevented full-length IPMK from binding to p53 in etoposide-treated human embryonic kidney (HEK) 293 and HCT116 cells (Fig. 6B). Additionally, overexpression of the fragment decreased the amount of PUMA and Bax mRNA transcripts (Fig. 6C), the abundance of PUMA, Bax, and p21 proteins (Fig. 6D), and the recruitment of p53 to PUMA and Bax promoters (Fig. 6E) in etoposide-treated U2OS cells, indicating that binding between IPMK and p53 stimulated the transcriptional activity of p53. Consistent with our findings that IPMK stimulated the acetylation of p53 by p300 (Fig. 3), overexpression of the exon 4–encoded fragment markedly diminished the acetylation of p53 (Fig. 6F). Disruption of the binding of IPMK to p53 also enhanced the proliferation of etoposide-treated U2OS cells by almost 50% (Fig. 6G). Together, these data suggest that a direct association between IPMK and p53 is necessary for IPMK to stimulate the transcriptional activity of p53.
Fig. 6.
Overexpression of a fragment of IPMK inhibits endogenous IPMK function. (A) Immunoprecipitation and Western blotting analysis were used to detect binding between p53 and fragments of IPMK in p53-null (−/−) HCT116 cells cotransfected with plasmids encoding GST, GST-tagged full-length (FL) IPMK, or GST-tagged exon-encoded IPMK fragments, he-magglutinin (HA)–tagged p53, or myc-tagged p53. Left, whole-cell lysates (WCLs); right, immunoprecipitate fractions. (B) Binding between mycIPMK and endogenous p53 in HEK 293 cells (top) and HCT116 cells (bottom) cotransfected with plasmids encoding mycIPMK and either GST or a GST-tagged IPMK fragment encoded by exon 4 (GST ex4) and treated with 20 μM etoposide. Data are means ± SEM from three experiments. ***P < 0.001, Student’s t test. (C and D) Abundance of PUMA and Bax mRNAs (C) and proteins (D) in U2OS cells transfected with plasmids encoding GST or GST ex4 and treated with 10 μM etoposide overnight. Data are means ± SEM from three experiments. ***P < 0.001, Student’s t test. (E) ChIP analysis of p53 binding to the PUMA and Bax promoters in U2OS cells trans-fected with plasmids encoding GST or GST ex4 and treated with etoposide. Data are means ± SEM from three experiments. ***P < 0.001, Student’s t test. (F and G) Abundance of acetylated p53 (F) and relative proliferation (G) in etoposide-treated U2OS cells transfected with plasmids encoding either GST or GST ex4. Data are means ± SEM from three experiments. ***P < 0.001, **P < 0.01, Student’s t test.
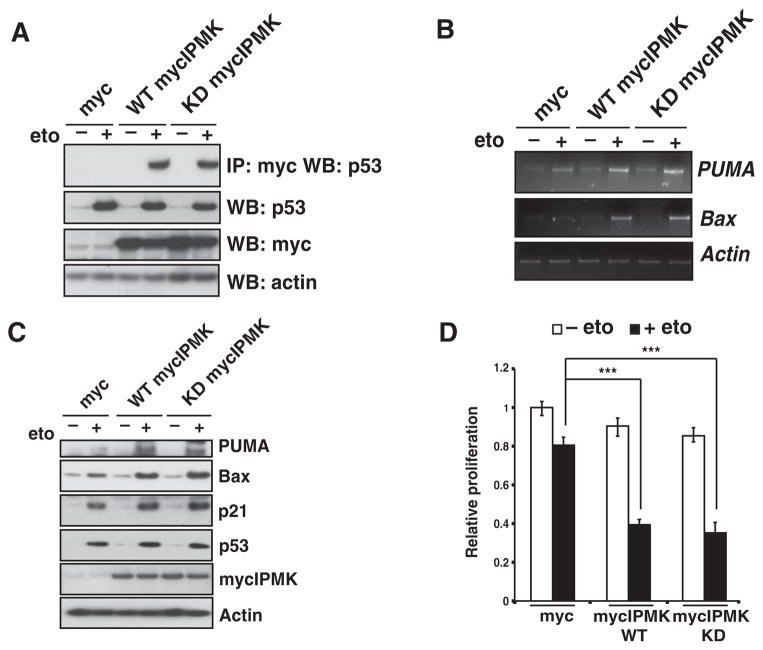
IPMK does not require catalytic activity to enhance p53-mediated cell death
We next investigated whether the catalytic activity of IPMK was required to stimulate p53 transcriptional activity. Because the single K129A mutant of IPMK has trace catalytic activity (17), we used a myc-tagged K129A-S235A IPMK double mutant that is completely devoid of kinase activity (19). In transfected U2OS cells, kinase-deficient K129A-S235A IPMK bound to endogenous p53 to the same extent as did wild-type IPMK (Fig. 7A). Overexpression of kinase-deficient IPMK in U2OS cells increased the abundance of mRNAs (Fig. 7B) and proteins (Fig. 7C) of p53 targets to a similar extent as that seen in cells transfected with plasmid encoding wild-type IPMK. Furthermore, overexpression of kinase-deficient IPMK inhibited cell proliferation in etoposide-treated U2OS cells to a similar extent as did wild-type IPMK (Fig. 7D). Thus, IPMK’s stimulation of p53 transcriptional activity is independent of its catalytic activity.
Fig. 7.
IPMK coactivation of p53 transcriptional activity is kinase-independent. (A) Immunoprecipitation and Western blotting were used to detect binding between p53 and IPMK in lysates from U2OS cells transfected with plasmids encoding wild-type (WT) mycIPMK and kinase-deficient (KD) mycIPMK constructs and treated with 20 μM etoposide. Blots are representative of three experiments. (B and C) Detection of PUMA and Bax mRNAs (B) and proteins (C) in transfected, etoposide-treated U2OS cells. Blots are representative of three experiments. (D) Cell proliferation of transfected, etoposide-treated U2OS cells. Data are means ± SEM from three experiments. ***P < 0.001, one-way analysis of variance (ANOVA).
DISCUSSION
We showed that mammalian IPMK is a physiologic transcriptional coactivator of p53. IPMK bound to p53 and participated in a transcriptional complex at the promoters of p53 target genes. IPMK stimulated the binding of p53 to the acetyltransferase p300, increasing the acetylation of p53 and its binding to cognate promoters. Moreover, IPMK stimulated the recruitment of p300 to the promoters of p53 target genes, enhancing histone acetylation and promoter activity. IPMK enhanced p53-mediated cell death, indicating a physiological role for the IPMK-p53 interaction.
Studies by Lindner and associates (45, 46) and our laboratory (47) show that another inositol phosphokinase, IP6K2 (inositol hexakisphosphate kinase-2), generates IP7 and physiologically induces p53-mediated cell death (48). However, in contrast to IPMK, which coactivated the transcription of both cell cycle arrest and apoptotic p53 targets, namely, p21 and PUMA, respectively, IP6K2 biases p53-dependent gene transcription toward apoptotic rather than cycle arrest genes (47).
Our conclusion that IPMK is an indispensable coactivator of p53-mediated transcription and cell death is consistent with a study in human primary BJ fibroblasts that identified IPMK as a potential target in p53-dependent oncogene-induced senescence (7). Furthermore, our findings that IPMK stimulated p53-mediated transcription independently of its catalytic function is consistent with evidence that its homolog in yeast does not require its catalytic metabolites to enhance transcription mediated by Mcm1 in response to arginine (49–51). This noncatalytic action of IPMK is also supported by observations that IPMK stabilizes the amino acid–induced mTORC1 independently of any kinase activity (19).
The direct role of IPMK in the activation of p53 may have therapeutic implications, particularly because direct interaction between IPMK and p53 appeared to be critical for the functional effects. Overexpression of a fragment of IPMK, encoded by exon 4 of IPMK, acted like a dominant-negative construct by abrogating the binding of full-length IPMK to p53. In turn, blocking this interaction decreased the acetylation of p53 and its binding to cognate promoters, reduced the transcription and translation of PUMA, Bax, and p21, and decreased cell death after treatment with etoposide. The ability of a dominant-negative construct to prevent the functional interaction between IPMK and p53 suggests that low–molecular weight drugs acting in a similar manner might prevent the acetylation and activation of p53. Cell death initiated by p53 is implicated in disorders such as Huntington’s disease (52) and stroke (53). Therefore, drugs that selectively block IPMK-p53 binding might provide therapeutic benefit.
Recently, Ingraham and associates have delineated a transcriptional activation role for IPMK that requires its PI3K activity (18). In contrast to its protranscriptional activity, IPMK may also have a role in transcriptional repression. In yeast, deletion of the IPMK homolog activates a subset of genes that are transcriptionally inactive in high-phosphate conditions (51). Watson et al. (54) reported the crystal structure of histone deacetylase 3 and the activation domain of nuclear co-repressor 2, which requires the presence of the IPMK metabolite IP4 at the protein-protein interface. How IPMK alternates between its transcriptional coactivation and presumed co-repressive functions remains to be elucidated. Conceivably, inter- and intracellular signaling networks might control a switch between the PI3K, IP3 kinase, and noncatalytic states of IPMK, altering its function to elicit optimal cellular response to changes in the environment. It is tempting to speculate about potential signaling mechanisms, such as differential phosphorylations, that could cause conformational changes in IPMK to induce these switches.
MATERIALS AND METHODS
Reagents and antibodies
Anti-rabbit and anti-mouse IgG agarose were purchased from eBioscience. Anti-myc agarose was obtained from Sigma. Antibodies against actin horseradish peroxidase (HRP) (C4), p300 (N-15), p53 (human DO-1 and FL-393), Bax (N-20), p21 (C-19), and normal IgG were obtained from Santa Cruz Biotechnology. Antibodies against PUMA (human), acetyl-lysine, PARP, and cleaved caspase-3 were purchased from Cell Signaling Technology. Antibodies against acetyl-K373 p53, acetyl-K382, acetyl-H3K9, or his-tone H3 were purchased from Millipore. Antibody against PUMA (mouse) was purchased from ProSci, HA from Covance, and p53 (mouse) from Novocastra. Antibody against IPMK raised in rabbit was produced in-house.
Generation of inducible floxed IPMK mice
Floxed IPMK mice were generated at Ozgene. A loxP site was inserted between exons 5 and 6. Floxed IPMK mice were mated with knock-in mice (Jackson Laboratory) carrying the tamoxifen-inducible Cre-ERT2 (Cre recombinase–estrogen receptor T2) driven by the ubiquitin C promoter (stock number 008085).
Generation of fl/fl or Δ/Δ primary MEFs
Primary Cre-ERT2/floxed/floxed IPMK MEFs (fl/fl) were generated as described previously (19). Depletion of IPMK in Cre-ERT2/floxed/floxed MEFs (Δ/Δ) was achieved by adding 4-hydroxytamoxifen (1 μM) for 48 hours. Ethanol-treated MEFs were used as a control.
Protein purification and interaction assays
Purified p300 protein was purchased from ProteinOne. GST-tagged p53 was prepared according to the manufacturer’s recommendations (Pharmacia Biotech) and purified through the affinity matrix glutathione-Sepharose (Amersham Biosciences). PreScission Protease (GE Healthcare Life Sciences) cleaved p53 from the GST tag. mycIPMK and recombinant myc were purified from HEK 293 cells 48 hours after transfection with pCMV mycIPMK or pCMV myc plasmid, respectively. Immunoprecipitation of the myc tag was performed with 2 mg of protein lysates in radioimmuno-precipitation assay (RIPA) buffer incubated overnight with anti-myc agarose beads at 4°C. Beads were pelleted and washed with RIPA buffer containing 600 mM NaCl six times before bacterially purified p53 was added. Binding was allowed to occur for 2 hours at 4°C. Immunoprecipitates were washed four more times before SDS sample buffer loading dye was added. Immunoprecipitated samples were resolved by PAGE, and proteins were detected by Western blotting.
GST-tagged IPMK exon fragments were coexpressed in p53-null HCT116 cells with either myc- or HA-tagged p53. Immunoprecipitation of the GST-tag was performed with 500 μg of protein lysate with glutathione-Sepharose beads and washed four times in RIPA buffer. Beads were pelleted and washed three times before adding SDS sample buffer and loading dye. Immunoprecipitated samples were resolved by PAGE, and tagged p53 was detected by either anti-myc HRP antibody or anti-HA HRP antibody.
Cell culture and transfection
HEK 293, HCT116, and U2OS cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and 2 mM L-glutamine at 37°C with a 5% CO2 atmosphere in a humidified incubator. For transient transfection of cells with expression constructs, we used PolyFect Reagent (Qiagen) for HEK 293 cells and Lipofectamine 2000 (Invitrogen) for HCT116 and U2OS cells according to the manufacturers’ protocols. Thirty hours after transfection, cells were treated overnight with etoposide (10 to 20 μM), 5-fluorouracil (200 to 400 μM), or doxorubicin (1 to 2 μM) (Sigma). For shRNA experiments, HCT116 cells were transfected with either nontargeted shRNA or shRNA targeting IPMK (OriGene) with Lipofectamine 2000, and 72 hours after transfection, cells were treated overnight with etoposide or sulindac (Sigma).
In vitro acetylation assay
Acetyl-CoA (20 μM), p300 (100 ng), and recombinant myc (100 ng) or mycIPMK were added to 30 μl of reaction buffer [20 mM tris-HCl (pH 8.0), 20% glycerol, 100 mM KCl, 1 mM dithiothreitol, and 0.2 mM EDTA]. After incubation at 30°C for 1 hour, the reaction was stopped by the addition of 10 μl of SDS sample buffer loading dye. The samples were subjected to SDS-PAGE and Western blotting analysis.
ChIP assay
ChIP assays were performed as described previously (55). In brief, intact cells were treated with 2 mM disuccinimidyl glutarate (Pierce) to cross-link protein complexes, followed by formaldehyde to link protein to DNA covalently. Cells were lysed, the nucleoprotein complexes were sonicated, and the cross-linked DNA-protein complexes were enriched by immunoprecipitation. The retrieved complexes were analyzed by PCR amplification to detect and quantify specific DNA targets. For real-time PCR, we used Brilliant SYBR Green Master Mix (Stratagene) according to the manufacturer’s protocol. The following were the ChIP primers: human p21 promoter, 5-GTGGCTCTGATTGGCTTTCTG-3 and 5-CTGAAAA-CAGGCAGCCCAAG-3; human PUMA promoter, 5-GCGAGACTGTG-GCCTTGTGT-3 and 5-CGTTCCAGGGTCCACAAAGT-3; human Bax promoter, 5-TAATCCCAGCGCTTTGGAA-3 and 5-TGCAGAGAC-CTGGATCTAGCAA-3; mouse p21 promoter, 5-GTGGCTCTGATTG-GCTTTCTG-3 and 5-CTGAAAACAGGCAGCCCAAG-3; mouse PUMA promoter, 5-CTGTGGCCTTGTGTCTGTGAG-3 and 5-CGCGGACAA-GTCAGGACTTG-3; mouse Bax promoter, 5-AGCGTTCCCCTAGCCT-CTTT-3 and 5-GCTGGGCCTGTATCCTACATTCT-3.
Quantitative RT-PCR analysis
Total RNA was extracted from cells with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. qRT-PCR for the genes encoding p21, Bax, PUMA, and actin used SuperScript One Step RT-PCR with TaqMan Probes (Invitrogen).
Apoptosis and cell proliferation
U2OS cells were transfected with the indicated plasmids separately and treated with 10 to 20 μM etoposide for 16 hours in the presence of 10% FBS. IPMK floxed (IPMKfl/fl) or deleted (IPMKΔ/Δ) MEFs were treated with 20 μM etoposide for 16 hours in the presence of 10% FBS. Apoptosis was determined by detection of the externalization of phosphatidylserine from the inner to the outer leaflet of the plasma membrane with the annexin V–FITC detection kit (Roche Biosciences) and flow cytometry (FACSCalibur, Becton Dickinson) (56). Apoptosis was also quantified in U2OS cells and fl/fl or Δ/Δ MEFs with an enzyme immunoassay (Roche Applied Science) using a combination of antibodies that recognize histones and DNA, as described previously (57). Apoptosis was also determined by detecting the amounts of cleaved PARP and cleaved caspase-3 in cell lysates by Western blotting. Cell proliferation was measured with the MTT assay as described previously (47).
Image quantification and statistical analysis
Images were quantified with ImageJ software. Data are presented as means ± SEM from at least three independent experiments. P values were calculated by two-tailed Student’s t test or one-way ANOVA with Tukey’s post hoc test using Minitab 13 software (Minitab).
Supplementary Material
Fig. S1. IPMK regulates expression of p53 downstream targets.
Fig. S2. IPMK is depleted with shRNA.
Fig. S3. Protein abundance of p53 targets is decreased by IPMK depletion.
Fig. S4. Abundance of p53 target proteins increased by IPMK is p53-dependent.
Fig. S5. IPMK binds the PUMA promoter.
Fig. S6. Genetic deletion of IPMK decreases p53 binding to its target promoters.
Fig. S7. IPMK inhibits cell proliferation.
Acknowledgments
We thank A. Resnick, M. Saleh, R. Tyagi, M. Ma, J. Sbodio, R. Tokhunts, R. Mealer, P. Scherer, M. Harraz, and other members of the Solomon H. Snyder laboratory for helpful discussions. We thank B. Ziegler for organizing the manuscript. Funding: This work was funded by the U.S. Public Health Service grant DA-000266 and the Medical Scientist Training Program T32 grant (to R.X.).
Footnotes
Author contributions: R.X., N.S., B.D.P., and S.H.S. designed the research; R.X., N.S., B.D.P., A.M.S., F.R., M.S.V., and J.X. performed the experiments; R.X., N.S., and B.D.P. analyzed the data; and R.X. and S.H.S. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Vousden KH, Lu X. Live or let die: The cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguría A, Zaballos A, Flores JM, Barbacid M, Beach D, Serrano M. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 4.Bartkova J, Horejsí Z, Sehested M, Nesland JM, Rajpert-De Meyts E, Skakkebaek NE, Stucki M, Jackson S, Lukas J, Bartek J. DNA damage response mediators MDC1 and 53BP1: Constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene. 2007;26:7414–7422. doi: 10.1038/sj.onc.1210553. [DOI] [PubMed] [Google Scholar]
- 5.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 6.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drost J, Mantovani F, Tocco F, Elkon R, Comel A, Holstege H, Kerkhoven R, Jonkers J, Voorhoeve PM, Agami R, Del Sal G. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol. 2010;12:380–389. doi: 10.1038/ncb2038. [DOI] [PubMed] [Google Scholar]
- 8.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 9.Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 10.Leyman A, Pouillon V, Bostan A, Schurmans S, Erneux C, Pesesse X. The absence of expression of the three isoenzymes of the inositol 1,4,5-trisphosphate 3-kinase does not prevent the formation of inositol pentakisphosphate and hexakisphosphate in mouse embryonic fibroblasts. Cell Signal. 2007;19:1497–1504. doi: 10.1016/j.cellsig.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 11.Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, Juluri KR, Maag D, Kim S, Huang AS, Dailey MJ, Saleh M, Snowman AM, Moran TH, Mezey E, Snyder SH. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasad A, Jia Y, Chakraborty A, Li Y, Jain SK, Zhong J, Roy SG, Loison F, Mondal S, Sakai J, Blanchard C, Snyder SH, Luo HR. Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat Immunol. 2011;12:752–760. doi: 10.1038/ni.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc Natl Acad Sci USA. 2002;99:14206–14211. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, Huang Y, Nagata E, Devreotes P, Snyder SH. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 15.Ponnusamy S, Alderson NL, Hama H, Bielawski J, Jiang JC, Bhandari R, Snyder SH, Jazwinski SM, Ogretmen B. Regulation of telomere length by fatty acid elongase 3 in yeast. Involvement of inositol phosphate metabolism and Ku70/80 function. J Biol Chem. 2008;283:27514–27524. doi: 10.1074/jbc.M802980200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc Natl Acad Sci USA. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maag D, Maxwell MJ, Hardesty DA, Boucher KL, Choudhari N, Hanno AG, Ma JF, Snowman AS, Pietropaoli JW, Xu R, Storm PB, Saiardi A, Snyder SH, Resnick AC. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc Natl Acad Sci USA. 2011;108:1391–1396. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blind RD, Suzawa M, Ingraham HA. Direct modification and activation of a nuclear receptor–PIP2 complex by the inositol lipid kinase IPMK. Sci Signal. 2012;5:ra44. doi: 10.1126/scisignal.2003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Kim SF, Maag D, Maxwell MJ, Resnick AC, Juluri KR, Chakraborty A, Koldobskiy MA, Cha SH, Barrow R, Snowman AM, Snyder SH. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell Metab. 2011;13:215–221. doi: 10.1016/j.cmet.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gancedo C, Flores CL. Moonlighting proteins in yeasts. Microbiol Mol Biol Rev. 2008;72:197–210. doi: 10.1128/MMBR.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bechet J, Greenson M, Wiame JM. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 22.Dubois E, Messenguy F. Pleiotropic function of ArgRIIIp (Arg82p), one of the regulators of arginine metabolism in Saccharomyces cerevisiae. Role in expression of cell-type-specific genes. Mol Gen Genet. 1994;243:315–324. doi: 10.1007/BF00301067. [DOI] [PubMed] [Google Scholar]
- 23.Messenguy F, Dubois E. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2586–2592. doi: 10.1128/mcb.13.4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- 25.Steger DJ, Haswell ES, Miller AL, Wente SR, O’Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nalaskowski MM, Deschermeier C, Fanick W, Mayr GW. The human homologue of yeast ArgRIII protein is an inositol phosphate multikinase with predominantly nuclear localization. Biochem J. 2002;366:549–556. doi: 10.1042/BJ20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resnick AC, Snowman AM, Kang BN, Hurt KJ, Snyder SH, Saiardi A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc Natl Acad Sci USA. 2005;102:12783–12788. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordaliza M, García PA, del Corral JM, Castro MA, Gómez-Zurita MA. Podophyllotoxin: Distribution, sources, applications and new cytotoxic derivatives. Toxicon. 2004;44:441–459. doi: 10.1016/j.toxicon.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Hande KR. Etoposide: Four decades of development of a topoisomerase II inhibitor. Eur J Cancer. 1998;34:1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 30.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 31.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 32.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 33.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci USA. 2003;100:1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longley DB, Harkin DP, Johnston PG. 5-Fluorouracil: Mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 36.Tan C, Tasaka H, Yu KP, Murphy ML, Karnofsky DA. Daunomycin, an anti-tumor antibiotic, in the treatment of neoplastic disease. Clinical evaluation with special reference to childhood leukemia. Cancer. 1967;20:333–353. doi: 10.1002/1097-0142(1967)20:3<333::aid-cncr2820200302>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 37.Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 38.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 42.Endo-Streeter S, Tsui MK, Odom AR, Block J, York JD. Structural studies and protein engineering of inositol phosphate multikinase. J Biol Chem. 2012;287:35360–35369. doi: 10.1074/jbc.M112.365031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmes W, Jogl G. Crystal structure of inositol phosphate multikinase 2 and implications for substrate specificity. J Biol Chem. 2006;281:38109–38116. doi: 10.1074/jbc.M606883200. [DOI] [PubMed] [Google Scholar]
- 44.Saiardi A, Nagata E, Luo HR, Sawa A, Luo X, Snowman AM, Snyder SH. Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc Natl Acad Sci USA. 2001;98:2306–2311. doi: 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrison BH, Bauer JA, Hu J, Grane RW, Ozdemir AM, Chawla-Sarkar M, Gong B, Almasan A, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 sensitizes ovarian carcinoma cells to multiple cancer therapeutics. Oncogene. 2002;21:1882–1889. doi: 10.1038/sj/onc/1205265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morrison BH, Haney R, Lamarre E, Drazba J, Prestwich GD, Lindner DJ. Gene deletion of inositol hexakisphosphate kinase 2 predisposes to aerodigestive tract carcinoma. Oncogene. 2009;28:2383–2392. doi: 10.1038/onc.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koldobskiy MA, Chakraborty A, Werner JK, Jr, Snowman AM, Juluri KR, Vandiver MS, Kim S, Heletz S, Snyder SH. p53-mediated apoptosis requires inositol hexakisphosphate kinase-2. Proc Natl Acad Sci USA. 2010;107:20947–20951. doi: 10.1073/pnas.1015671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-β in ovarian carcinoma cells. J Biol Chem. 2001;276:24965–24970. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bosch D, Saiardi A. Arginine transcriptional response does not require inositol phosphate synthesis. J Biol Chem. 2012;287:38347–38355. doi: 10.1074/jbc.M112.384255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dubois E, Dewaste V, Erneux C, Messenguy F. Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS Lett. 2000;486:300–304. doi: 10.1016/s0014-5793(00)02318-8. [DOI] [PubMed] [Google Scholar]
- 51.El Alami M, Messenguy F, Scherens B, Dubois E. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol Microbiol. 2003;49:457–468. doi: 10.1046/j.1365-2958.2003.03562.x. [DOI] [PubMed] [Google Scholar]
- 52.Bae BI, Xu H, Igarashi S, Fujimuro M, Agrawal N, Taya Y, Hayward SD, Moran TH, Montell C, Ross CA, Snyder SH, Sawa A. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Cheng T, Liu D, Griffin JH, Fernández JA, Castellino F, Rosen ED, Fukudome K, Zlokovic BV. Activated protein C blocks p53-mediated apoptosis in ischemic human brain endothelium and is neuroprotective. Nat Med. 2003;9:338–342. doi: 10.1038/nm826. [DOI] [PubMed] [Google Scholar]
- 54.Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-κB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 56.Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L, Fitzgerald P, Guio-Carrion A, Waterhouse NJ, Li CW, Mari B, Barbry P, Newmeyer DD, Beere HM, Green DR. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 57.Frade JM, Rodríguez-Tébar A, Barde YA. Induction of cell death by endogenous nerve growth factor through its p75 receptor. Nature. 1996;383:166–168. doi: 10.1038/383166a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. IPMK regulates expression of p53 downstream targets.
Fig. S2. IPMK is depleted with shRNA.
Fig. S3. Protein abundance of p53 targets is decreased by IPMK depletion.
Fig. S4. Abundance of p53 target proteins increased by IPMK is p53-dependent.
Fig. S5. IPMK binds the PUMA promoter.
Fig. S6. Genetic deletion of IPMK decreases p53 binding to its target promoters.
Fig. S7. IPMK inhibits cell proliferation.