Abstract
Objective
This study was designed to investigate the pathogenic contributions of fibroblast-like synoviocytes (FLS) to juvenile idiopathic arthritis (JIA) by identifying pathways with dysregulated gene expression in FLS from patients with oligoarticular JIA.
Methods
FLS were derived from synovial fluid obtained by arthrocentesis from patients with JIA undergoing intraarticular steroid injections and from orthopedic control patients. Gene expression profiles of the JIA and control FLS were obtained using the Affymetrix platform, with application of Ingenuity Pathway Analysis and Gene Set Enrichment Analysis software to define gene sets in dysregulated pathways and networks of potential pathologic relevance in this disease. Biologically relevant differentially expressed genes were confirmed by RNA and protein analysis.
Results
Exploration of global gene expression profiles of the JIA FLS revealed important dysregulated pathways, including the transforming growth factor β (TGFβ) signaling, as well as endochondral bone formation, cartilage formation, and β-catenin networks. Importantly, bone morphogenetic protein 4 (BMP-4) was significantly overexpressed in the JIA FLS. FLS from patients with oligoarticular JIA exhibit a chondrocyte phenotype, as evidenced by expression of type II collagen and aggrecan.
Conclusion
Dysregulation of the pathways involved in the pathogenesis of oligoarticular JIA were revealed through gene expression profiling. JIA FLS displayed dysregulated TGFβ signaling and exhibited a hypertrophic chondrocyte phenotype. These characteristics, along with contributions from the β-catenin network may have implications for endochondral bone formation and local growth disturbances in oligoarticular JIA. Overexpression of BMP-4 in FLS from patients with oligoarticular JIA in particular may play an important role in disease pathogenesis, with a direct effect on functional outcome and with implications for future treatment.
Juvenile idiopathic arthritis (JIA) is the most common rheumatic disease of childhood (1,2). The pathogenesis of JIA has yet to be elucidated and is likely to involve a combination of cell types in the affected joint. Although the evolution of both rheumatoid arthritis (RA) and JIA is associated with joint space narrowing, periarticular osteopenia, and erosion formation, JIA is considered to be a distinct disease (3). Unique to JIA are valgus deformity of the knee caused by local growth disturbances as well as leg length discrepancies due to condylar bony hypertrophy (4,5). Although the knee is the most frequently affected joint, there are also local growth disturbances in other areas, such as underdeveloped mandible accompanying arthritis of the temporo-mandibular joint, shortened digits, and hip abnormalities. One subtype of JIA, oligoarticular JIA, is characterized by the involvement of 4 or fewer joints within 6 months of disease onset. Localization to just a single joint or a few joints, with prominent morbidity associated with growth alterations, suggests a pivotal role of fibroblast-like synoviocytes (FLS) in the disease.
FLS are perhaps the most important indigenous cell population in the synovium. Extensive studies in adult RA have shown the existence of FLS that produce cytokines and matrix-degrading enzymes (6–10). FLS are known to play a role in cartilage destruction and inflammation in RA. Gene expression profiling of FLS from RA patients has provided insights into the mechanisms of altered proliferation of these cells in chronic arthritis (11–14) and has implicated synoviocytes in mediating joint damage. Transforming growth factor β (TGFβ) signaling is known to play a role in the pathogenesis of RA (15,16), and studies have shown constitutive up-regulation of TGFβ, its receptor, throm-bospondin 1, and the Smad-associated molecule Smad anchor for receptor activation in RA FLS as compared to FLS from patients with osteoarthritis (17). The role of the synoviocyte as the primary effector cell has not yet been examined in JIA. We propose that FLS play a central role in the pathogenesis of JIA, acting as both a gatekeeper of access to the joint space and a mediator of pathology.
Condylar bony hypertrophy is detrimental both functionally and cosmetically in children with JIA. In the present study, we investigated the phenotype of JIA FLS and their contributions to the disease, specifically with regard to condylar bony hypertrophy. Given the significant role of TGFβ and its signaling pathways in RA, we predicted that TGFβ signaling was likely to be dysregulated in JIA FLS. In FLS from both JIA patients and controls, we examined key pathways involved in chondrogenesis. Our studies revealed that bone morphogenetic protein 4 (BMP-4) and members of the TGFβ superfamily were significantly more highly expressed in FLS from JIA patients than in FLS from controls. We identified elevated levels of BMP-4 that may, in turn, play a significant role in the local growth abnormalities seen in oligoarticular JIA.
PATIENTS AND METHODS
Selection of study samples
Synovial fluid and synovial tissue samples were obtained from our Institutional Review Board–approved tissue and fluid repository. Appropriate samples were identified by chart review and JIA subtype classification. The first available sample from each patient, from steroid-naive joints, was used, except for 1 sample (J1) obtained from a patient who had received 1 steroid injection more than 6 months prior to sampling. Control samples were obtained from remnant synovial fluid and tissue samples obtained at the time of orthopedic procedures performed on nonarthritic joints. Five control samples were compared with 5 samples from patients with oligoarticular JIA. Clinical information on the samples used for microarray analysis and for additional confirmatory studies is shown in Table 1.
Table 1.
Clinical features of JIA patients and control subjects from whom synovial tissue and synovial fluid samples were obtained*
| Sample | Source of FLS |
Joint | Diagnosis | Disease duration |
Age, years | Sex | Concurrent medication |
|---|---|---|---|---|---|---|---|
| Controls | |||||||
| C1 | ST | Right knee | Skeletal dysplasia | NA | 15 | Male | None |
| C2 | SF | Knee | Torn meniscus | NA | >16 | Female | Unknown |
| C3 | ST | Left hip | Hip dysplasia | NA | 4 | Female | None |
| C4 | ST | Left knee | Patellar instability | NA | 12 | Female | None |
| C5 | ST | Right knee | Patellar instability | NA | 13 | Male | None |
| C6 | SF | Right knee | ACL tear | NA | 17 | Male | None |
| C7 | SF | Right knee | Discoid meniscus | NA | 7 | Female | None |
| C8 | SF | Right knee | Discoid meniscus | NA | 15 | Male | None |
| C9 | SF | Right knee | ACL tear | NA | 15 | Male | None |
| JIA patients | |||||||
| J1 | SF | Right knee | Oligo JIA | 18 months | 11 | Female | Naproxen |
| J2 | SF | Right knee | Oligo JIA | 9 months | 14 | Female | None |
| J3 | SF | Right knee | Oligo JIA | 24 months | 17 | Male | Naproxen |
| J4 | SF | Right knee | Oligo JIA | 7 months | 10 | Male | Naproxen |
| J5 | SF | Right knee | Oligo JIA | 10 months | 12 | Female | None |
| J6 | SF | Right knee | Oligo JIA (extended) | 10 years | 15 | Female | Methotrexate, naproxen, folic acid |
| J7 | SF | Right knee | Oligo JIA | 3 months | 6 | Male | Methotrexate, naproxen, folic acid |
| J8 | SF | Left knee | Oligo JIA (extended) | 6 weeks | 11 | Female | Naproxen, doxycycline |
| J9 | SF | Left knee | Oligo JIA | 2 months | 17 | Male | Diclofenac |
| J10 | SF | Left knee | Oligo JIA (extended) | 8 years | 12 | Male | Naproxen |
| J11 | SF | Left knee | Oligo JIA | 8 years | 13 | Male | Naproxen |
| J12 | SF | Right knee | Oligo JIA | 6 weeks | 13 | Male | Tolmetin |
| J13 | SF | Left knee | Oligo JIA | 6 months | 17 | Male | Naproxen |
Except where indicated otherwise, all patients with juvenile idiopathic arthritis (JIA) had persistent oligoarticular (oligo) disease. Patients J1 and J11 had received 1 previous injection of intraarticular steroid. FLS = fibroblast-like synoviocytes; ST = synovial tissue; NA = not applicable; SF = synovial fluid; ACL = anterior cruciate ligament.
Cell culture
Synovial fluid or tissue was plated in 6-well plates with 15% fetal bovine serum/Dulbecco’s modified Eagle’s medium. This initial primary culture was passaged into T-75 flasks and, at confluence, was passaged 3–6 more times, and then harvested at confluence. It is well established that by the third passage, FLS are the predominant cell type in culture (18). Due to the scarcity of control samples, some control FLS were grown from synovial tissue while others were grown from synovial fluid. All JIA samples were grown from synovial fluid. To assess purity, we stained JIA FLS for Thy-1 (fibroblast marker), CD68 (macrophage marker), CD14 (monocyte marker), and CD19 (B cell marker) at passage 3 (Figure 1A). FLS cultured on Chamber Slides (Nunc) were fixed in 4% paraformaldehyde, permeabilized with Tris buffered saline– Tween (TBST), blocked with 5% donkey and goat serum, and incubated with Thy-1 plus either CD68, CD14, or CD19 antibodies for costaining. Primary antibodies were incubated on cells for 1 hour at room temperature. After incubation, secondary antibodies (1:200 dilution, Alexa Fluor 488 [green] and Alexa Fluor 594 [red]; Invitrogen) were applied for 1 hour at room temperature. Nuclei were stained with DAPI and mounted with Vectashield hard-set mounting medium (Vector).
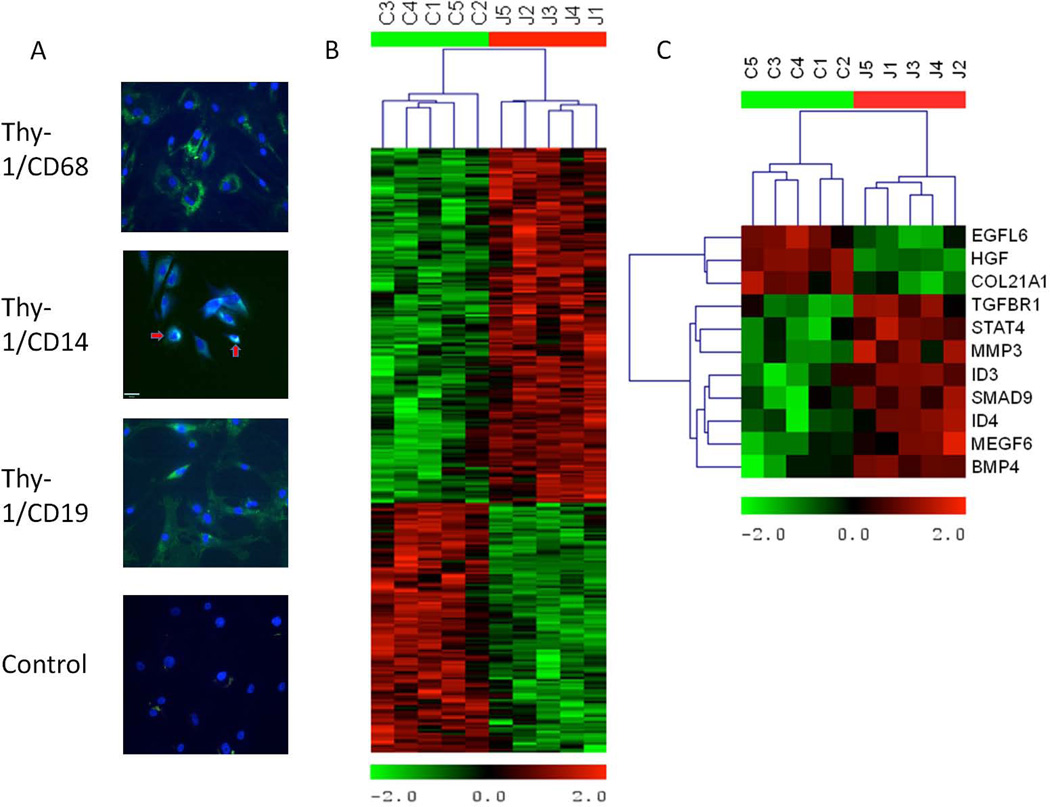
Figure 1.
Cell staining and heatmaps showing the relative expression of differentially expressed genes in fibroblast-like synoviocytes (FLS) from patients with juvenile idiopathic arthritis (JIA) and controls. A, Immunofluorescence costaining of JIA FLS from passage 3, showing that cell cultures were pure FLS. Fibroblast marker Thy-1 (green) was positive on most cell surfaces, while macrophage marker CD68 (red) and B cell marker CD19 (red) were negative. Costaining with CD14 (red) revealed 2 cells that were positive for this monocyte marker as well as for Thy-1 (arrows). This indicates that the cells are mostly FLS by passage 3 and that there are few contaminating cells. As a negative control, secondary antibodies labeled with Alexa Fluor 488 (green) and Alexa Fluor 594 (red) were applied to JIA FLS to detect any nonspecific staining. All FLS used for microarrays were from passage 6. Original magnification × 40. B, Hierarchical cluster analysis of the 305 genes that were differentially expressed between JIA patients and controls. Control genes are clustered together on the left of the heatmap, indicating similarity in gene expression pattern as compared to JIA genes, which are clustered together on the right. C, Analysis of 11 genes specifically related to transforming growth factor β (TGFβ) signaling (of the 305 differentially expressed genes). In JIA samples, genes directly involved in TGFβ signaling, including TGFβ receptor type I, bone morphogenic protein 4, and Smad9, are overexpressed. Red represents high levels of expression, and green represents low levels of expression.
Fluorescence was examined with an epifluorescence microscope. Antibodies were used at the following concentrations: Thy-1/CD90 1:40 (Novus NBP1-42067), CD68 1:50 (Novus NB600-1250), CD14 1:50 (eBioscience 14-0149-82), and CD19 1:50 (eBioscience 14-0199-82). Cultured cells were Thy-1+, CD68−, CD14+/−, and CD19−.
Gene expression profiling
RNA was extracted from cultured FLS using TRIzol and purified using a Qiagen RNeasy Mini Prep kit. Following RNA purification, samples were amplified and fluorescence labeled using a GeneChip 3′ IVT express kit (Affymetrix). Labeled RNA was then hybridized to GeneChip Human Genome U133 Plus 2.0 Arrays (Affymetrix). Arrays were stained using a GeneChip Hybridization, Wash, and Stain kit (Affymetrix), and then scanned and analyzed using Affymetrix command console software and expression console software.
Analysis of microarray data
Expression values were calculated with the GC-RMA algorithm (19) in the gcrma Bioconductor package. Differentially expressed probe sets were determined using Significance Analysis of Microarrays program within the MultiExperiment Viewer (MeV; 10% false discovery rate [FDR]) package (20,21). Gene Set Enrichment Analysis (GSEA) (22,23) was performed on all probe sets and on selected gene sets using the signal-to-noise ratio for ranking genes and gene sets for permutation. Seven gene sets were derived from IPA (www.ingenuity.com) annotation; one was derived from KEGG (www.genome.jp/kegg) annotation. Hierarchical clustering was performed in MeV.
Western blotting
All Western blots were performed on cell culture supernatants taken from control and JIA samples. Cell medium was aspirated after 48 hours of culture and treated with complete proteinase inhibitor cocktail tablets (Roche). Protein concentrations from cell culture supernatants were measured using the Bradford assay. Protein aliquots of 30–50 µg were loaded onto Tris HCl Criterion gels. Protein concentrations vary based on antibody sensitivity and range of detection. Western blotting was performed using the Bio-Rad Criterion system. Depending on the primary antibody used, Western blots were performed under reducing or nonreducing conditions. Membranes were blocked for 30 minutes at room temperature in 5% nonfat milk and incubated overnight at 4°C with primary antibodies at the following dilutions: Aggrecan 1:500 (Millipore), BMP-2 1:500 (R&D Systems), BMP-4 1:500 (R&D Systems), cartilage oligomeric matrix protein (COMP) 1:1,000 (R&D Systems), type II collagen 1:1,000 (R&D Systems), type X collagen 1:250 (Abcam), and osteocalcin 1:500 (Santa Cruz Biotechnology). Membranes were incubated in secondary antibody for 1 hour at room temperature at dilutions ranging from 1:1,000 to 1:10,000. Membranes were then washed using TBST.
The membranes were developed using Amersham ECL Western Blotting Detection Reagent and exposed to film. Loading control proteins, such as β-actin and GAPDH, are not secreted by cell culture supernatants. To verify loadings, the entire membrane was stained with ponceau S. Once the membranes were scanned, a prominent band ~40 kd was used for each blot to represent equal protein loadings. (Full ponceau S blots are available upon request from the corresponding author.)
Reverse transcription–polymerase chain reaction (RT-PCR) analysis
RNA from the microarray analysis was used to perform RT-PCR for BMP-4, BMP-2, and aggrecan. First-strand synthesis was performed using an iScript complementary DNA synthesis kit (Bio-Rad). Following the manufacturer’s protocol, PCR was performed using Promega 2× master mixture that contains 50 µmoles/unit of Taq DNA polymerase, 3 mM MgCl2, and 400 µM dNTPs. The following primer pairs were used for PCR reactions: for BMP-4, 5′-GCT-CAA-GTC-CAC-ATA-GAG-CGA-GTG-3′ (forward) and 5′-ACT-GGT-CCA-CCA-CAA-TGT-GAC-ACG-3′ (reverse); for BMP-2, 5′-GCT-GTA-CTA-GCG-ACA-CCC-AC-3′ (forward) and 5′-TCA-TAA-AAC-CTG-CAA-CAG-CCA-ACT-CG-3′ (reverse); and for aggrecan, 5′-ATG-CTG-AGC-GCC-GGT-GTC-G-3′ (forward) and 5′-TAG-GTG-GTG-GCT-GTG-CCC-TTT-TTA-3′ (reverse). The reaction was performed as follows: 12.5 µl of 2× Master Mix containing Taq, MgCl2, and dNTPs was added to a 25-µl reaction that contained 1.0 µl of 10 µM forward and reverse primers, 100 ng of DNA template, and nuclease-free water. Complementary DNA was denatured at 94°C for 15 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 50–65°C for 30 seconds, and extension at 72°C for 1 minute. A final extension step was performed at 72°C for 7 minutes.
Enzyme-linked immunosorbent assay (ELISA)
BMP-4 ELISA was performed using a RayBiotech human BMP-4 ELISA kit. Following the manufacturer’s protocol, 100 µl of cell culture supernatant samples were loaded into the wells of a 96-well plate, along with recombinant BMP-4 protein standards, incubated overnight at 4°C, and developed using the provided horseradish peroxidase (HRP)–streptavidin system. The standard curve was plotted using SigmaPlot software. A best-fit line was drawn through the standards data points, and the unknown concentrations of the samples were calculated. COMP ELISA was performed using an R&D Systems Quantikine human COMP immunoassay. Following the manufacturer’s protocol, 50 µl of cell culture supernatants was loaded onto a 96-well plate with 100 µl of assay diluent, incubated overnight at 4°C with recombinant COMP protein standards, and developed using HRP–streptavidin after incubation with a Ready-to-Use COMP conjugate. Standards were plotted on a log graph, and a best-fit line was determined to allow calculation of the concentration in the experimental samples.
RESULTS
Significantly different gene expression patterns and signaling pathways in FLS from oligoarticular JIA patients versus controls, by microarray analysis
Five FLS samples from JIA patients and controls were analyzed by global gene expression using the Affymetrix platform. Using SAM, 305 differentially expressed probe sets were identified among oligoarticular JIA and control FLS, with an FDR of 10%. Due to redundancy inherent in the microarray chips, the 305 probe sets represent 254 unique genes. Hierarchical cluster analysis revealed that the 5 JIA samples had gene expression profiles that were very similar to each other and distinct from the 5 control group samples (Figure 1B). Of the 254 genes that met statistical significance for differential expression, 152 genes were overexpressed in JIA, while 102 genes were expressed at lower levels in JIA patients as compared to controls (data available upon request from the corresponding author).
Up-regulation of TGFβ and BMP-4 signaling, endochondral bone formation, cartilage formation, and β-catenin networks in JIA FLS
Among this group of statistically differentially expressed genes, 11 were directly regulated by or regulate TGFβ signaling (Figure 1C). Eight genes were up-regulated in JIA FLS. BMP-4 in particular was expressed at a 5.5-fold–higher level in JIA FLS than in control FLS. Smad9 (also known as Smad8) was also more highly expressed in JIA FLS. The Smad proteins associate with Smad4 and are translocated to the nucleus to allow transduction of TGFβ signaling (24). Three genes were underexpressed in the JIA FLS: growth factors epidermal growth factor–like protein 6 and hepatocyte growth factor and type XXI collagen.
We examined the gene expression data for pathways that could be involved in bony overgrowth in the inflamed joint, a key clinical feature of JIA. Gene sets for TGFβ, insulin-like growth factor (IGF), Wnt, and BMP-4 signaling pathways, and IGF-1, endochondral bone formation, cartilage formation, and β-catenin networks were generated in KEGG (TGFβ) or IPA (all others). Our data for these gene sets were examined by GSEA to determine if these pathways and networks of interest showed altered expression in JIA (Table 2). We did not detect any significant difference in the expression of the IGF, Wnt, or BMP-4 pathways or the IGF-1 network between JIA and control samples.
Table 2.
Gene sets evaluated with GSEA software*
| Gene set | Size | ES | NES | Nominal P |
FDR q value |
Up-regulated | Significantly different† |
|---|---|---|---|---|---|---|---|
| TGFβ signaling pathway | 84 | –0.485 | –1.525 | 0.002 | 0.041 | JIA | Yes |
| Cartilage formation network | 164 | –0.370 | –1.270 | 0.033 | 0.208 | JIA | Yes |
| β-catenin network | 34 | –0.462 | –1.229 | 0.168 | 0.183 | JIA | Yes |
| Endochondral bone formation network | 45 | –0.422 | –1.203 | 0.200 | 0.168 | JIA | Yes |
| IGF signaling pathway | 96 | –0.296 | –0.930 | 0.624 | 0.619 | JIA | No |
| IGF-1 network | 63 | 0.365 | 1.054 | 0.340 | 1.000 | Control | No |
| Wnt signaling pathway | 165 | 0.319 | 1.051 | 0.314 | 0.520 | Control | No |
| BMP-4 signaling pathway | 72 | 0.291 | 0.860 | 0.761 | 0.773 | Control | No |
Gene sets were derived using IPA software to identify specific signaling pathways and molecular networks. These lists were then analyzed with Gene Set Enrichment Analysis (GSEA) software to determine if these pathways and networks were significant. It was found that the transforming growth factor β (TGFβ) signaling pathway and cartilage formation network were significantly differentially expressed, based on both the P values and the q values, while the signaling networks for β-catenin and endochondral bone formation were significantly differentially expressed based on their q values. ES = enrichment score; NES = normalized enrichment score; JIA = juvenile idiopathic arthritis; IGF = insulin-like growth factor; BMP-4 = bone morphogenetic protein 4.
P values less than 0.05 and q values less than 0.25 (a 25% false-discovery rate [FDR]) were considered significant.
The TGFβ signaling pathway and the cartilage formation network were up-regulated in JIA FLS as compared to control FLS, according to both the nominal P value and the FDR q value. For the endochondral bone formation and β-catenin networks, only the FDR q values showed significance. Although many genes in the pathways and networks were overexpressed in JIA FLS, GSEA reported the most influential genes, termed core-enrichment or leading-edge genes, in determining that an individual pathway or network was up-regulated. The interrelationships between these leading-edge genes of the TGFβ signaling, cartilage formation, endochondral bone formation, and β-catenin networks are included in data that are available upon request from the corresponding author. BMP-4 and BMP-2 were among the most influential genes determining the up-regulation of the TGFβ signaling pathway and the cartilage formation and endochondral bone formation networks.
RT-PCR confirmation of the overexpression of BMP-4 mRNA in JIA
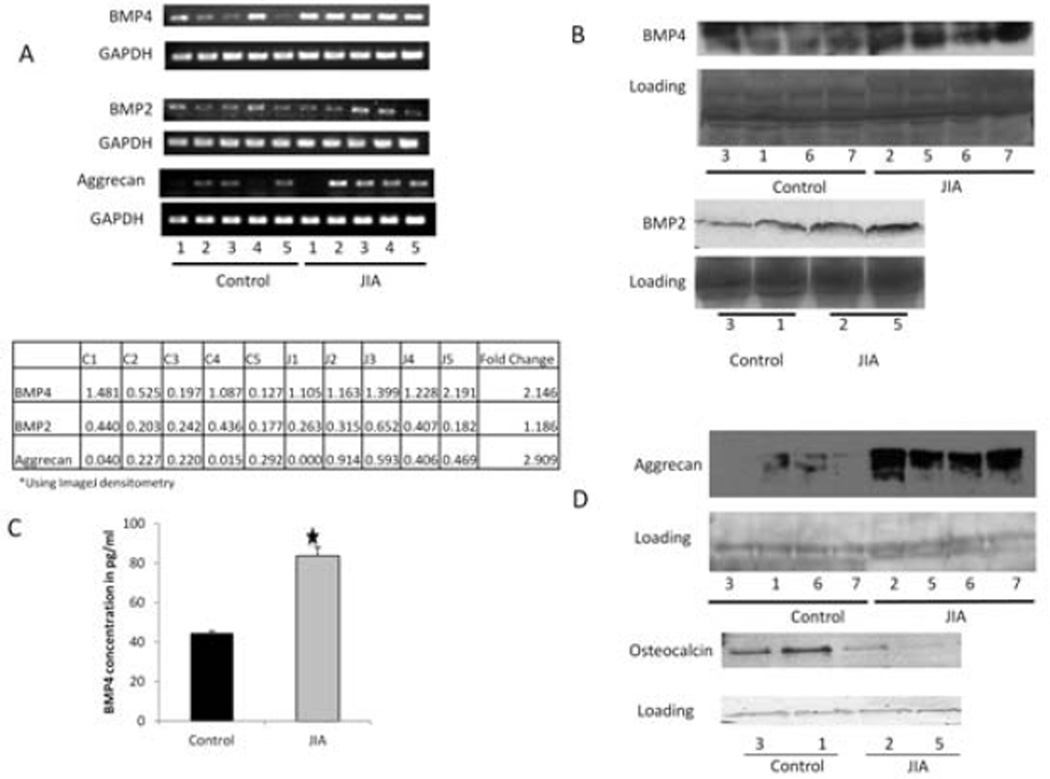
Findings of the RT-PCR analysis confirmed that BMP-4 gene expression was significantly up-regulated in JIA FLS as compared to controls, thus validating the findings of the initial microarray analysis. We compared RNA from the 5 JIA samples to RNA from the 5 control samples and, through densitometry, determined that BMP-4 had a 2.15-fold higher expression level in JIA samples (P = 0.028) (Figure 2A). Although not in our list of significantly differentially expressed genes, BMP-2 is expressed in RA FLS (25), so we examined JIA FLS for BMP-2 expression. RT-PCR analysis revealed that the level of BMP-2 in the JIA samples was not significantly different from that in the control samples (1.19-fold higher; P = 0.547) (Figure 2A).
Figure 2.
A, Reverse transcription–polymerase chain reaction (RT-PCR) analysis of bone morphogenetic protein 4 (BMP-4) and BMP-2 from total RNA extracted from synoviocytes derived from synovial fluid obtained from 5 control subjects and 5 patients with juvenile idiopathic arthritis (JIA) (top). Compared to controls, transcript levels of both BMP-4 and BMP-2 are increased in samples from JIA patients (2.15-fold [P = 0.028] and 1.9-fold [P = 0.547], respectively). Aggrecan is also overexpressed in JIA patient samples as compared to controls (2.91-fold [P = 0.008]). GAPDH was used as a loading control. Individual sample readings were determined by densitometry (bottom). B, Western blot analysis of BMP-4 and BMP-2 in cell culture supernatants from control and JIA samples, using an arbitrary band from the ponceau S-stained blot as a loading control. Total expression of BMP-4 was increased in JIA samples as compared to controls. BMP-2 expression may be slightly increased in JIA. C, BMP-4 concentrations in control and JIA samples, as determined by enzyme-linked immunosorbent assay. There is nearly twice as much BMP-4 in JIA samples than in control samples (P < 0.001). Values are the mean ± SEM. D, Western blot analysis of cartilage and bone protein marker expression in cell culture supernatants of control and JIA samples. Total aggrecan levels are increased and osteocalcin levels decreased in JIA samples as compared to controls.
Because BMP-4 is known to play a prominent role in chondrogenesis, we also used RT-PCR to examine the expression levels of aggrecan, a highly specific chondrocyte marker (Figure 2A). Aggrecan mRNA levels were 2.91-fold higher in JIA FLS RNA samples than in control RNA samples (P = 0.008) (Figure 2A). Sample J1 contained no detectable aggrecan RNA. This sample was the only one from a patient who had received a prior intraarticular steroid injection into the same joint that was used for the microarray analysis. Aggrecan was more highly expressed in JIA FLS than in control FLS in the microarray analysis, but the difference was not statistically significant. Aggrecan is a member of the cartilage network gene set but is not one of the leading-edge genes.
Secretion of chondrocyte differentiation and extracellular matrix protein markers from JIA FLS, as characterized by proteins expressed in culture supernatants
We used Western blotting and ELISA to examine secreted proteins in culture supernatants from JIA and control FLS. BMP-4 protein expression appeared to be higher in JIA samples than in control samples, while BMP-2 protein levels appeared to be only slightly over-expressed (Figure 2B). In addition to confirming BMP-4 expression in samples used for the microarray experiments, we also detected the protein in 2 additional control FLS and 2 additional JIA FLS samples that had not been included in the original analysis (Figure 2B). ELISA showed that active BMP-4 was nearly 2-fold higher in JIA cell culture supernatants than in controls (P < 0.001) (Figure 2C).
FLS are pluripotent cells (26); thus, we considered the possibility that they could differentiate into osteoblasts or chondrocytes, which could also contribute to bony overgrowth in JIA. Consistent with the RNA levels, aggrecan protein was found to be increased in JIA samples relative to control samples by Western blotting, which was confirmed in both the original and additional samples (Figure 2D). Conversely, JIA samples expressed very low levels of osteocalcin, a typical bone cell marker (Figure 2D). Hence, JIA FLS showed preferential development of a cartilage-like phenotype.
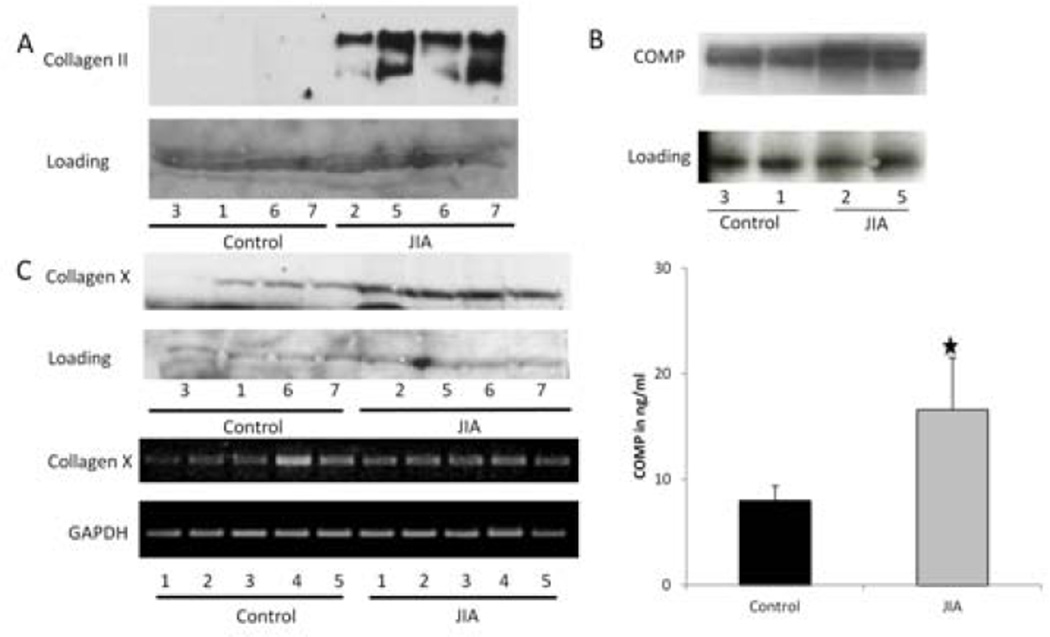
To further examine the expression of extracellular matrix protein markers, type II collagen α1 chain, COMP, and type X collagen, expression levels were measured by Western blotting. JIA FLS appeared to express type II collagen α1-chain protein at higher levels than control FLS, which was validated on both original and additional samples (Figure 3A). COMP, a noncollagenous extracellular matrix protein, was secreted at greater levels by JIA FLS than control FLS (Figure 3B). By ELISA, active COMP was nearly twice as concentrated in JIA FLS samples than in control samples (P < 0.001) (Figure 3B). Both COL2A1 and COMP were more highly expressed by JIA FLS in the microarray analysis, but the difference did not meet statistical significance. Furthermore, type II collagen α1 chain is a member of the up-regulated cartilage formation network and COMP is one of the leading-edge genes that contributed to the up-regulation of the TGFβ signaling pathway, as determined by GSEA. JIA FLS also seemed to secrete more type X collagen protein, a specific marker of chondrocyte hypertrophy, but mRNA for type X collagen was not up-regulated in JIA samples, a finding that was confirmed on both the original samples and additional samples (Figure 3C).
Figure 3.
A, Expression of type II collagen in synoviocytes from control subjects and juvenile idiopathic arthritis (JIA) patients, as determined by Western blotting, to further examine chondrocyte differentiation by JIA synoviocytes. There is increased type II collagen expression in JIA samples as compared to controls. B, Overexpression of cartilage oligomeric matrix protein (COMP), a marker of cartilage turnover, in JIA samples as compared to controls, by Western blotting (top). This increase was quantified by enzyme-linked immunosorbent assay (bottom), which revealed nearly doubled levels in JIA samples than in controls (P < 0.001). Values are the mean ± SEM. C, Expression of type X collagen in JIA and control samples, as determined by reverse transcription–polymerase chain reaction analysis. Type X collagen expression is also increased in JIA samples as compared to controls. Although type X collagen is expressed in JIA samples, it is not up-regulated at the RNA level.
Detection of BMP-4, aggrecan, type II collagen, and osteocalcin in the synovial fluid
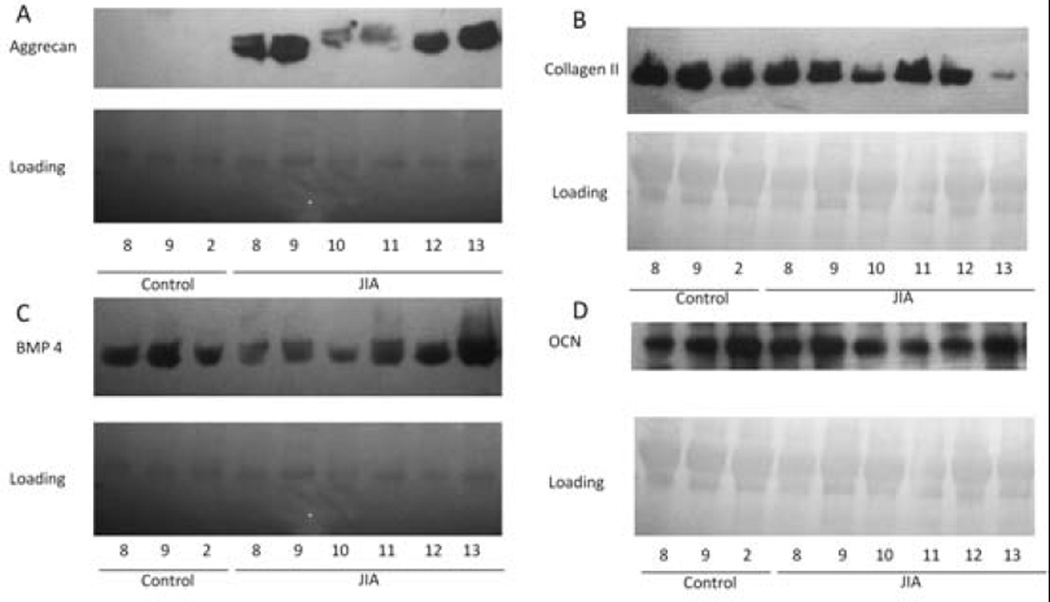
To validate important proteins in neat synovial fluid, we performed Western blotting to detect BMP-4, aggrecan, type II collagen, and osteocalcin in synovial fluid samples from 1 original control subject, 2 additional control subjects, and 6 additional JIA patients that had not previously been analyzed by microarray (Figure 4). Although the levels of BMP-4 (Figure 4C) and type II collagen (Figure 4B) were increased in cultured JIA FLS, they did not appear to be differentially expressed in synovial fluid. Protein levels of osteocalcin did appear to be lower in synovial fluid from patients with JIA, as expected (Figure 4D). Most importantly, however, protein levels of aggrecan were clearly elevated in the synovial fluid of patients with JIA, and aggrecan was only minimally present, if at all, in the synovial fluid of controls (Figure 4A).
Figure 4.
Western blots of neat synovial fluid for expression of A, aggrecan, B, type II collagen, C, bone morphogenetic protein 4 (BMP-4), and D, osteocalcin (OCN). The sample from control subject 2 was used in the original gene expression studies; the remainder of the control samples are from additional subjects. Of note, the sample from juvenile idiopathic arthritis (JIA) patient 11 was obtained from a knee joint that received 1 prior intraarticular steroid injection >1 year before the synovial fluid sample used in the analysis was obtained. The disease in JIA patients 8 and 10 ultimately evolved into extended oligoarticular JIA.
DISCUSSION
In the present study, we examined FLS from patients with oligoarticular JIA by global expression profiling to elucidate dysregulated pathways that may give insight into the pathogenesis of disease and its most conspicuous complication, local growth disturbances. Significant differences in gene expression were detected in FLS from oligoarticular JIA patients and FLS from control subjects. The TGFβ signaling pathway as well as the endochondral bone formation, cartilage formation, and β-catenin networks were up-regulated in FLS from oligoarticular JIA patients.
Important members of the TGFβ superfamily, including TGFβ receptor type I, BMP subfamily member 4, and Smad9, were more highly expressed in JIA FLS than in control FLS (Figure 1B). While up-regulation of TGFβ signaling in adult RA has also been reported, the regulation of BMPs differs from our findings for JIA. BMP-2 is increased in RA synovium, while BMPs 4 and 5 are decreased, as compared to the levels in synovium from osteoarthritis patients and normal donors (27). BMP-2 and BMP-6 have been reported in FLS from RA synovium and were thought to modulate FLS apoptosis (25). BMP-2 and BMP-6, but not BMP-4, have been shown to be up-regulated by the proinflammatory cytokines TNFα and interleukin-1β (25). Karamboulas et al (28) suggested that BMP-4 is expressed in more mature cell populations and is required for transitioning between the different phases of bone and cartilage development.
Networks of endochondral bone formation and cartilage formation were also found to be up-regulated in JIA (Table 2). Our JIA FLS samples secreted the chondrocyte markers aggrecan and type II collagen. They also secreted type X collagen, a hypertrophic chondrocyte marker. These data suggest that JIA FLS are differentiating toward the chondrocyte lineage, which could contribute to endochondral bone formation. Importantly, BMP-4 has been shown to promote endochondral bone formation through influencing the differentiation of a fibroblast cell line (NIH3T3) and a myoblast cell line (C2C12) along chondrogenic and osteogenic lineages (29). Additionally, BMP signaling in a mouse model of the spondyloarthropathies was shown to contribute to endochondral bone formation and ankylosis in these diseases (30). Chondrocytic differentiation of cultured turbinate fibroblasts has been achieved by addition of TGFβ and IGF-1 (31). It is intriguing to postulate that the combined up-regulation of endochondral bone formation and cartilage formation networks, with notable overexpression of BMP-4 and other members of the TGFβ pathway, in FLS provides a basis for stimulating aberrant endochondral bone formation, by leading to either differentiation of local fibroblasts along chondrocyte lineages or differentiation of the synoviocytes themselves. We noted the absence of aggrecan RNA in the sample from a patient who had received a previous intraarticular steroid injection. Intraarticular steroid injection has been shown to decrease the occurrence of leg length discrepancy in JIA (32), and we posit that this could be through abrogation of the chondrocyte phenotype via local steroid injection.
By GSEA, the canonical Wnt signaling pathway was shown to be down-regulated in JIA FLS as compared to controls, although the difference was not statistically significant (Table 2). When Wnt is inhibited, β-catenin is phosphorylated and prevented from translocating to the nucleus to activate the transcription factors responsible for differentiation toward osteoblasts. Gaur et al (33) suggested that inhibition of Wnt by secreted Frizzled-related protein 1 allows TGFβ to signal BMP proteins to drive chondrogenesis. This mechanism contributes to endochondral bone formation. Cells that signal through this pathway express type II and type X collagen, indicating that they are chondrogenic and hypertrophic. However, we found that the β-catenin signaling pathway was up-regulated in JIA FLS, and this did reach statistical significance (Table 2). The up-regulation of β-catenin should lead to inhibition of chondrocyte differentiation and should promote bone formation (34). The up-regulation of this pathway was based in part on the presence of β-catenin RNA transcript as one of the leading-edge genes. Further investigations are needed to determine if it is present in the FLS at the protein level and whether it is phosphorylated to be degraded or unphosphorylated for translocation to the nucleus. In addition, it may be that timing of the expression of members of the Wnt/β-catenin signaling pathway in FLS influences the outcome, initially allowing chondrocyte hypertrophy then ultimately leading to endochondral bone formation. Interestingly, Wnt-5a and Dkk-1, two of the leading-edge genes in the up-regulated cartilage formation network, activate the noncanonical Wnt/Ca2+ pathway. This pathway antagonizes the Wnt/β-catenin pathway and has been shown to degrade β-catenin (35,36). These data suggest that non-canonical Wnt signaling may play a role in the fate of the oligoarticular JIA FLS.
There are limitations to our study. The expression of chondrocyte features could arise in response to the FLS culture conditions. However, this behavior was not observed in the control FLS, which underwent the same cell culture procedures and passaging. Comparison to the control FLS should correct for genes whose expression is attributable to the culture process and do not reflect a biologic difference between groups. Furthermore, the clear difference in aggrecan protein levels in FLS from JIA synovial fluid compared to control FLS confirms that the expression of chondrocyte features is a true reflection of in vivo conditions, as opposed to an artifact from culture conditions.
FLS grown from adherent synovial fluid cells display the same protein markers of differentiation and functional behavior as FLS cultured from synovial membranes and are therefore a good substitute, since synovial tissue is not always available, especially when studying early stages of disease (18,37). Importantly, all of the control FLS, regardless of the source, clustered together in their pattern of differentially expressed genes (Figure 1B).
In addition to cell staining (Figure 1), to ensure that the cells we characterized were a pure sample of FLS, we looked at the microarray data for expression levels of markers of various cell types found in the synovial fluid. Epithelial marker CD90 (Thy-1) was highly expressed, whereas expression levels of CD14 (monocyte marker) and CD68 (macrophage marker) were variable and in the moderate-to-low range. No expression was detected for the α-subunit of CR3 (neutrophils, macrophages), CD3 (T cells), and CD19 (B cells).
JIA FLS display a chondrocyte phenotype. To ensure that we grew fibroblasts, as opposed to chondrocytes, from the synovial fluid, we interrogated the microarray data for specific fibroblast markers. Both control and JIA samples expressed RNA transcripts for CD90 (Thy-1), vimentin, and prolyl 4-hydroxylase α-peptide 1 (P4HA1), P4HA2, and P4HA3, confirming their fibroblast-like characteristics. It is possible that there is a mixed population of cells, with some FLS differentiating along the chondrocyte lineage and others maintaining the fibroblast phenotype. Fibrochondrocytes derived from rabbit knee meniscus displayed more fibroblast-like or more chondrocyte-like features, depending on the basal nutrient medium (38).
Oligoarticular JIA is more likely to go into remission than other subtypes of JIA (39) or adult RA (40). Perhaps there is a question of balance in the joints of these patients. Sekiya et al (41) suggest that a large amount of cartilage can form from treating mesenchymal stem cells with BMP proteins. It is further suggested that these mesenchymal stem cells migrate to the injured cartilage and, when treated with BMPs, become chondrocytes. JIA FLS may be playing a reparative role in these joints by renewing cartilage to prevent further joint destruction. If this is the case, the significant difference in BMP-4 expression in our JIA FLS samples could potentially be a new therapeutic target. Also, differences in BMP-4 gene expression could provide a way to differentiate between JIA subtypes and eventually predict disease course.
The importance of these characterizations may have relevance for endochondral bone formation that leads to the bony overgrowth seen in JIA. These data suggest that JIA FLS are differentiating toward the chondrocyte lineage, which would place them in a pivotal position to contribute to the local growth disturbances seen in oligoarticular JIA. Characterization of JIA FLS has an impact on our understanding of the pathogenesis of JIA and on future treatment considerations.
ACKNOWLEDGMENTS
We would like to thank N. Carolyn Schanen, MD, PhD, for scientific guidance and mentorship, Robert W. Mason, PhD, for critical review of the manuscript, Rose O’Connor, PhD, for technical guidance, James N. Jarvis, MD, for helpful discussions, Nemours/AI DuPont Hospital for Children, Division of Orthopedics, for control samples, Erin Boyle for technical assistance, and Timothy Stetson for cell culture work. We are grateful for the technical support of the COBRE-funded Biomolecular Core Laboratory at Nemours Biomedical Research.
Supported by the NIH (Centers of Biomedical Research Excellence [COBRE] grant 8P20-GM-103464 and an American Recovery and Reinvestment Act grant supplement to P20-RR-020173), the Arthritis Foundation (Innovative Research grant), the Nancy Taylor Foundation for Chronic Diseases, Nemours Biomedical Research, the Nemours Fund for Children’s Health, the Delaware Community Foundation, and the Open Net Foundation. The Biomolecular Core Laboratory at Nemours Biomedical Research is funded by the NIH COBRE program.
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Brescia had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Brescia, Simonds, Fawcett, Rose.
Acquisition of data. Brescia, Simonds.
Analysis and interpretation of data. Brescia, Simonds, McCahan, Rose.
REFERENCES
- 1.Lawrence RC, Helmick CG, Arnett FC, Deyo RA, Felson DT, Giannini EH, et al. Estimates of the prevalence of arthritis and selected musculoskeletal disorders in the United States. Arthritis Rheum. 1998;41:778–799. doi: 10.1002/1529-0131(199805)41:5<778::AID-ART4>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 2.Manners PJ, Bower C. Worldwide prevalence of juvenile arthritis why does it vary so much? J Rheumatol. 2002;29:1520–1530. [PubMed] [Google Scholar]
- 3.Prahalad S, Glass DN. Is juvenile rheumatoid arthritis/juvenile idiopathic arthritis different from rheumatoid arthritis? Arthritis Res Ther. 2002;4:303–310. [Google Scholar]
- 4.Simon S, Whiffen J, Shapiro F. Leg-length discrepancies in monoarticular and pauciarticular juvenile rheumatoid arthritis. J Bone Joint Surg Am. 1981;63:209–215. [PubMed] [Google Scholar]
- 5.MacRae VE, Farquharson C, Ahmed SF. The pathophysiology of the growth plate in juvenile idiopathic arthritis. Rheumatology (Oxford) 2006;45:11–19. doi: 10.1093/rheumatology/kei091. [DOI] [PubMed] [Google Scholar]
- 6.Yamanishi Y, Firestein GS. Pathogenesis of rheumatoid arthritis: the role of synoviocytes. Rheum Dis Clin North Am. 2001;27:355–371. doi: 10.1016/s0889-857x(05)70206-4. [DOI] [PubMed] [Google Scholar]
- 7.Firestein GS. Invasive fibroblast-like synoviocytes in rheumatoid arthritis: passive responders or transformed aggressors? [review] Arthritis Rheum. 1996;39:1781–1790. doi: 10.1002/art.1780391103. [DOI] [PubMed] [Google Scholar]
- 8.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 9.Kinne RW, Boehm S, Iftner T, Aigner T, Vornehm S, Weseloh G, et al. Synovial fibroblast-like cells strongly express jun-B and C-fos proto-oncogenes in rheumatoid- and osteoarthritis. Scand J Rheumatol Suppl. 1995;101:121–125. doi: 10.3109/03009749509100913. [DOI] [PubMed] [Google Scholar]
- 10.Neumann E, Gay RE, Gay S, Muller-Ladner U. Functional genomics of fibroblasts. Curr Opin Rheumatol. 2004;16:238–245. doi: 10.1097/00002281-200405000-00012. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe N, Ando K, Yoshida S, Inuzuka S, Kobayashi M, Matsui N, et al. Gene expression profile analysis of rheumatoid synovial fibroblast cultures revealing the overexpression of genes responsible for tumor-like growth of rheumatoid synovium. Biochem Biophys Res Commun. 2002;294:1121–1129. doi: 10.1016/S0006-291X(02)00608-3. [DOI] [PubMed] [Google Scholar]
- 12.Pufe T, Bartscher M, Petersen W, Tillmann B, Mentlein R. Expression of pleiotrophin, an embryonic growth and differentiation factor, in rheumatoid arthritis. Arthritis Rheum. 2003;48:660–667. doi: 10.1002/art.10839. [DOI] [PubMed] [Google Scholar]
- 13.Lacey D, Sampey A, Mitchell R, Bucala R, Santos L, Leech M, et al. Control of fibroblast-like synoviocyte proliferation by macrophage migration inhibitory factor. Arthritis Rheum. 2003;48:103–109. doi: 10.1002/art.10733. [DOI] [PubMed] [Google Scholar]
- 14.Kurowska M, Rudnicka W, Kontny E, Janicka I, Chorazy M, Kowalczewski J, et al. Fibroblast-like synoviocytes from rheumatoid arthritis patients express functional IL-15 receptor complex: endogenous IL-15 in autocrine fashion enhances cell proliferation and expression of Bcl-xL and Bcl-2. J Immunol. 2002;169:1760–1767. doi: 10.4049/jimmunol.169.4.1760. [DOI] [PubMed] [Google Scholar]
- 15.Brennan FM, Chantry D, Turner M, Foxwell B, Maini R, Feldmann M. Detection of transforming growth factor-β in rheumatoid arthritis synovial tissue: lack of effect on spontaneous cytokine production in joint cell cultures. Clin Exp Immunol. 1990;81:278–285. doi: 10.1111/j.1365-2249.1990.tb03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bira Y, Tani K, Nishioka Y, Miyata J, Sato K, Hayashi A, et al. Transforming growth factor β stimulates rheumatoid synovial fibroblasts via the type II receptor. Mod Rheumatol. 2005;15:108–113. doi: 10.1007/s10165-004-0378-2. [DOI] [PubMed] [Google Scholar]
- 17.Pohlers D, Beyer A, Koczan D, Wilhelm T, Thiesan HJ, Kinne RW. Constitutive upregulation of the transforming growth factor-β pathway in rheumatoid arthritis synovial fibroblasts. Arthritis Res Ther. 2007;9:R59. doi: 10.1186/ar2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stebulis JA, Rossetti RG, Atez FJ, Zurier RB. Fibroblast-like synovial cells derived from synovial fluid. J Rheumatol. 2005;32:301–306. [PubMed] [Google Scholar]
- 19.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. JASA. 2004;99:909–917. [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 22.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heldin CH, Miyazono K, ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 25.Lories RJ, Derese I, Ceuppens JL, Luyten FP. Bone morphogenetic proteins 2 and 6, expressed in arthritic synovium, are regulated by proinflammatory cytokines and differentially modulate fibroblast-like synoviocyte apoptosis. Arthritis Rheum. 2003;48:2807–2818. doi: 10.1002/art.11389. [DOI] [PubMed] [Google Scholar]
- 26.De Bari C, Dell’Accio F, Tylzanowski P, Luyten FP. Multipotent mesenchymal stem cells from adult human synovial membrane. Arthritis Rheum. 2001;44:1928–1942. doi: 10.1002/1529-0131(200108)44:8<1928::AID-ART331>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 27.Bramlage CP, Haupl T, Kaps C, Ungethum U, Krenn V, Pruss A, et al. Decrease in expression of bone morphogenetic proteins 4 and 5 in synovial tissue of patients with osteoarthritis and rheumatoid arthritis. Arthritis Res Ther. 2006;8:R58. doi: 10.1186/ar1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karamboulas K, Dranse HJ, Underhill TM. Regulation of BMP-dependent chondrogenesis in early limb mesenchyme by TGFβ signals. J Cell Sci. 2010;123:2068–2076. doi: 10.1242/jcs.062901. [DOI] [PubMed] [Google Scholar]
- 29.Li G, Peng H, Corsi K, Usas A, Olshanski A, Huard J. Differential effect of BMP4 on NIHβT3 and C2C12 cells: implications for endochondral bone formation. J Bone Miner Res. 2005;20:1611–1623. doi: 10.1359/JBMR.050513. [DOI] [PubMed] [Google Scholar]
- 30.Lories RJ, Derese I, Luyten FP. Modulation of bone morphogenetic protein signaling inhibits the onset and progression of ankylosing enthesitis. J Clin Invest. 2005;115:1571–1579. doi: 10.1172/JCI23738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SW, Cho JH, Hong MW, Rhie JW, Yoon HR. Induction of chondrogenic differentiation in cultured fibroblasts isolated from the inferior turbinate. Otolaryngol Head Neck Surg. 2008;139:143–148. doi: 10.1016/j.otohns.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Sherry DD, Stein LD, Reed AM, Schanberg LE, Kredich DW. Prevention of leg length discrepancy in young children with pauciarticular juvenile rheumatoid arthritis by treatment with intraarticular steroids. Arthritis Rheum. 1999;42:2330–2334. doi: 10.1002/1529-0131(199911)42:11<2330::AID-ANR11>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 33.Gaur T, Rich L, Lengner CJ, Hussain S, Trevant B, Ayers D, et al. Secreted frizzled related protein 1 regulates Wnt signaling for BMP2 induced chondrocyte differentiation. J Cell Physiol. 2006;208:87–96. doi: 10.1002/jcp.20637. [DOI] [PubMed] [Google Scholar]
- 34.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/β-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 35.Ishitani T, Kishida S, Hyodo-Miura J, Ueno N, Yasuda J, Waterman M, et al. The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca2+ pathway to antagonize Wnt/β-catenin signaling. Mol Cell Biol. 2003;23:131–139. doi: 10.1128/MCB.23.1.131-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ, Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent β-catenin degradation. J Cell Biol. 2003;162:899–908. doi: 10.1083/jcb.200303158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scanu A, Oliviero F, Braghetto L, Ramonda R, Luisetto R, Calabrese F, et al. Synoviocyte cultures from synovial fluid. Reumatismo. 2007;59:66–70. doi: 10.4081/reumatismo.2007.66. In Italian. [DOI] [PubMed] [Google Scholar]
- 38.Webber RJ, Harris MG, Hough AJ., Jr Cell culture of rabbit meniscal fibrochondrocytes: proliferative and synthetic response to growth factors and ascorbate. J Orthop Res. 1985;3:36–42. doi: 10.1002/jor.1100030104. [DOI] [PubMed] [Google Scholar]
- 39.Wallace CA, Huang B, Bandeira M, Ravelli A, Giannini EH. Patterns of clinical remission in select categories of juvenile idiopathic arthritis. Arthritis Rheum. 2005;52:3554–3562. doi: 10.1002/art.21389. [DOI] [PubMed] [Google Scholar]
- 40.Eberhardt K, Fex E. Clinical course and remission rate in patients with early rheumatoid arthritis: relationship to outcome after 5 years. Br J Rheumatol. 1998;37:1324–1329. doi: 10.1093/rheumatology/37.12.1324. [DOI] [PubMed] [Google Scholar]
- 41.Sekiya I, Larson BL, Vuoristo JT, Reger RL, Prockop DJ. Comparison of effect of BMP-2, −4, and −6 on in vitro cartilage formation of human adult stem cells from bone marrow stroma. Cell Tissue Res. 2005;320:269–276. doi: 10.1007/s00441-004-1075-3. [DOI] [PubMed] [Google Scholar]