Abstract
Purpose
The goal of this study was to determine whether gene expression differences exist between inflammatory breast cancers (IBC) and stage-matched non-IBC patiens stratified by hormone receptor and HER2 status.
Experimental Design
We used Affymetrix GeneChips to analyze 82 tumor samples (25 T4d, 25 patients and 57, T4a-c patients) of newly diagnosed breast cancers. Genes that were differentially expressed between the IBC and non-IBC specimens were identified using the t-test, and differential expression of gene sets was assessed using gene set analysis. Three distinct clinical subtypes of IBC and non-IBC were compared: ER-positive/HER2-normal, HER2-amplified, and ER-negative/HER2-normal.
Results
When we compared expression data from all IBC with all non-IBC, we found no significant differences after adjusting for multiple testing. When IBC and non-IBC tumors were compared by clinical subtype, however, significant differences emerged. Complement and immune system-related pathways were overexpressed in ER-positive/HER2-normal IBC. Protein translation and mTOR signaling were overexpressed in HER2-amplified IBC. Apoptosis-, neural-, and lipid metabolism-related pathways were overexpressed in ER-negative/HER2-normal IBC compared with non-IBC of the same receptor phenotype. In this stage-matched case-control study, the survival curves of patients with IBC and non-IBC were similar for all three clinical subtypes.
Conclusions
IBC tumors can be divided into molecular and clinical subtypes similar to those of non-IBC. Clinical subtypes of IBC show molecular differences compared with similar subtypes of non-IBC.
Keywords: Inflammatory breast cancer, receptor subtypes, cDNA microarray, Gene set analysis
Introduction
Inflammatory breast cancer (IBC) is a clinical diagnosis designated as T4d disease in the TNM classification of the American Joint Committee on Cancer [1]. IBC is distinct from neglected, locally advanced breast cancer with a clinical course characterized by a rapid course of progression featuring local growth and early metastasis. IBC is a rare breast cancer that accounts for 1 to 5% of all breast cancers in the United States [2,3]. Several authors have analyzed gene expression profiles of IBC to evaluate peculiar features of this disease [4–7]. Van Laere et al. reported that the insulin-like growth factor-signaling pathway is implicated in the biology of IBC [7]. Nguyen et al. reported that IBC has higher expression of genes associated with increased metabolic rate, lipid signaling, and cell turnover [6]. All previous studies compared IBC with non-IBC simply on the bases of clinical diagnosis and without considering molecular subtypes of breast disease. However, breast cancer is no longer considered to be a single disease with variable receptor expression and histology but rather as a collection of several distinct diseases distinguished by unique clinical, biological and molecular features. These molecular and clinical subtypes can be discerned in both IBC and non-IBC.
We hypothesized that there are biological differences between IBC and non-IBC tumors according to each clinical subtype, including ER-positive/HER2-normal, HER2-amplified with any ER status, and ER−/HER2-normal cancers. We tested this hypothesis at the individual gene level and at the level of gene sets that represent biological functions by comparing IBC and non-IBC within each clinical subtype. We also studied survival for patients with IBC and non-IBC tumors matched by clinical subtype.
Materials and Methods
Patient cohorts and gene expression data
Tumor biopsies were collected prospectively from patients with locally advanced disease who were candidates for neoadjuvant therapy. Fine needle aspiration (FNA) were obtained before administration of any systemic therapy as part of an ongoing pharmacogenomic research program at The University of Texas MD Anderson Cancer Center. All patients gave written informed consent for voluntary participation. The study was approved by the Institutional Review Board. Samples were placed in RNAlater (Ambion, Austin, TX) storage reagent and stored at −80°C until gene expression analysis. This study followed a case-control design. Patients with a clinical diagnosis of IBC were selected after reviewing all medical records associated with the 440 FNA samples with complete gene expression data. Data were collected between 2000 and 2007. IBC was defined as the presence of erythema and edema (peau d’orange) involving at least one third of the skin of the breast and rapid clinical presentation and biopsy-proven invasive breast carcinoma. The presence of tumor emboli in the dermal lymphatics of the involved skin in the pathology report was not required for inclusion as IBC. Controls were selected to match the T stage, with all T4a-c tumors in the data set included as controls. We identified 25 IBCs and 57 non-IBC controls for this analysis. Clinicopathologic characteristics of the patient cohorts are presented in Table 1.
Table 1.
Patient Characteristics at Diagnosis
| Characteristic | IBC
|
Non-IBC
|
P ¶ | ||
|---|---|---|---|---|---|
| No. of Pts | % | No. of Pts | % | ||
| Total no. of Patients | 25 | 57 | |||
| Age, years | |||||
| Mean (mini-max) | 48.4(29–78) | 52.5(24–80) | 0.121 | ||
| T at diagnosis (TNM) | |||||
| T4b-c | - | 57 | 100 | - | |
| T4d | 25 | 100 | - | ||
| Stage at diagnosis | |||||
| IIIB-IIIC | 22 | 88 | 46 | 81 | 0.624 |
| IV | 3 | 12 | 11 | 19 | |
| Supraclavicular nodes | |||||
| Yes | 10 | 40 | 12 | 27 | 0.104 |
| No | 15 | 60 | 45 | 79 | |
| Nuclear grade | |||||
| II | 3 | 12 | 13 | 23 | 0.368 |
| III | 22 | 88 | 44 | 77 | |
| Lymphatic invasion in BC | |||||
| Positive | 19 | 76 | 18 | 32 | 0.005 |
| Negative | 5 | 20 | 25 | 44 | |
| Unknown | 1 | 4 | 14 | 25 | |
| ER status* | |||||
| ER-positive | 6 | 24 | 31 | 54 | 0.016 |
| ER-negative | 19 | 76 | 26 | 46 | |
| HER2 status* | |||||
| HER2-amplified | 12 | 48 | 17 | 30 | 0.136 |
| HER2-normal | 13 | 52 | 40 | 70 | |
| Molecular subtype † | |||||
| Luminal A | 4 | 16 | 13 | 23 | <0.001 |
| Luminal B | 1 | 4 | 9 | 16 | |
| ERBB2 new positive | 10 | 40 | 15 | 26 | |
| Basal like | 7 | 28 | 17 | 30 | |
| Normal like | 3 | 12 | 3 | 5 | |
| Pathologic response ‡ | |||||
| pCR | 6 | 24 | 4 | 7 | 0.075 |
| no-pCR | 15 | 60 | 37 | 65 | |
| unknown (no surgery or lost f/u) | 4 | 16 | 16 | 28 | |
| Nodal status at surgery | |||||
| N0 | 6 | 24 | 15 | 26 | 0.795 |
| N 1–3 | 6 | 24 | 9 | 16 | |
| N ≥ 4 | 10 | 40 | 18 | 32 | |
| Unknown | 3 | 12 | 15 | 26 | |
| Neoadjuvant chemotherapy § | |||||
| FAC or FEC or AC | 2 | 8 | 6 | 11 | 0.579 |
| T/FAC or FEC or AC/ or Others | 17 | 68 | 36 | 63 | |
| T/FAC or FEC or AC/H/ or Others | 6 | 24 | 7 | 12 | |
| Others | - | 8 | 14 | ||
| Adjuvant endocrine therapy | |||||
| Yes/No | 3/22 | 17/39 | 0.098 | ||
| Event rate | |||||
| Alive/Dead | 14/11 | 37/20 | 0.468 | ||
Abbreviations: Pts, patients; IBC, Inflammatory Breast Cancer; BC, Breast Cancer; ER, estrogen receptor; pCR, pathologic complete response.
Receptor status was determined by gene expression.
Molecular subtypes were indentified by PAM50 prediction.
pCR refers to pathologic complete response both in breast and axilla.
T, paclitaxel; F, 5-fluorouracil; A, doxorubicin; E, epirubicin; C, cyclophosphamide; H, trastuzmab.
P denotes results of significant test for the comparison between IBC vs Non-IBC by Fisher exact test or t-test.
Gene expression profiling was performed using Affymetrix U133A gene chips as previously described [8]. Gene expression data were normalized with the MAS5 algorithm, mean centered to 600 and log 2 transformed before further analysis. Probe sets with the lowest 15% mean expression value were removed from all higher level analyses to reduce noise from low expressed probe sets. This left 18,940 probe sets for analysis. Cases with normalized ESR1 mRNA expression (probe set 205225_at) >10.18 were considered ER positive, and cases with HER-2 mRNA expression (probe set 216836_s_at) >12.54 were considered HER2 amplified [9]. Molecular class was assigned based on gene expression profiles using the PAM 50 centroid-based classifier [10]. All statistical analysis was performed with BRB Array Tools version 3.9.0 Alfa software (http://linus.nci.nih.gov/BRB-ArrayTools.html) and R software version 2.7.2 (http://www.r-project.org). Complete gene expression data were deposited in the Gene Expression Omnibus (GEO) data base under GSE22597.
Identification of differentially expressed genes and gene sets
We compared gene expression data between IBC and non-IBC tumors without any further stratification by clinical or molecular class using the t-test. We also performed similar class comparisons separately for each of the three clinical subtypes (ER+/HER2−, ER−/HER2−, HER2+). To adjust for the multiple comparisons, we calculated False Discovery Rates (FDR) and also assessed Global Significance using BRB Array tools. The FDR was calculated with the Significance Analysis of Microarrays (SAM) tool as the median number of false-positive genes from permutation testing divided by the number of nominally significant genes defined from the unperturbed data [11]. The Global P value is the probability of getting at least the same number of genes significant on a parametric test (e.g., t-test) at the specified P level by chance if there are no real differences between IBC and non-IBC. Significantly differentially expressed genes were also mapped to functional pathways using the Ingenuity Pathway Analysis software (IPA, http://www.ingenuity.com/).
We also examined differential expression of a priori defined gene sets using Gene Set Analysis (GSA). The goal of GSA is to determine whether members of a set of genes that correspond to a particular biological pathway tend to occur toward the top or the bottom of a rank-ordered gene list (rank ordered by differential expression between IBC versus non-IBC) [12]. In this analysis we included three gene sets that were previously reported to be discriminatory between IBC and non-IBC [13,4,14] or associated with poor prognosis in breast cancer, including a Wnt pathway gene set [15]and CD44+ related stem cell signature [16] (Supplementary Table 1). We also tested 2,113 different, functionally annotated gene sets from Gene Ontology (GO, http://www.geneontology.org) that collectively represent most known biological and metabolic pathways in human cells. Gene sets with a minimum number of 10 genes and maximum of 100 genes were selected for inclusion in this analysis. We used the Efron and Tibshirani gene set analysis method that employs “maxmean” statistics and is implemented by the BRB Array Tools [17,18]. Significance was estimated with the permutation test (n = 1000). The null hypothesis was that the average degree of differential expression of members of a given gene set between the IBC and non-IBC cohorts is the same as expected from a random set of genes of similar size.
Survival analysis
Survival was estimated by the Kaplan-Meier product limit method and compared between groups with the log-rank statistic. Relapse-free survival was defined as the time from the diagnosis to local, regional, or distant relapse. Fourteen patients with stage IV disease (three IBC and 11 non-IBC) and five additional patients who did not undergo surgery (one IBC and four non-IBC, stage III disease) were excluded from the relapse-free survival analysis. Overall survival was defined as the time from the diagnosis to death, and all patients were included in this analysis.
Results
Differentially expressed genes between IBC and non-IBC
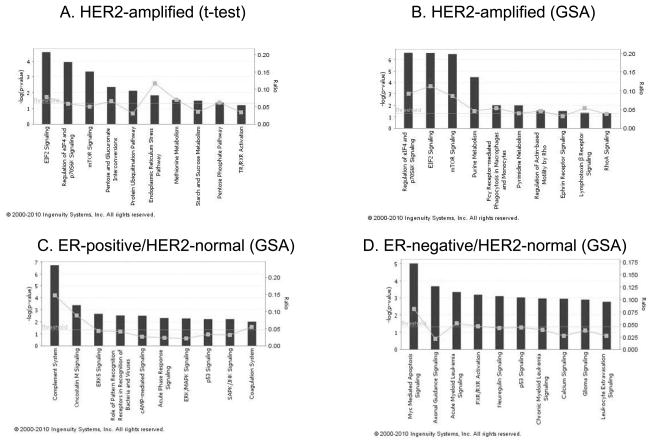
Clinical characteristics of the 82 patients included in this study are presented in Table 1. The proportions of patients with lymphatic invasion (76% vs 32%), ER-negative disease (76% versus 46%) and HER2-amplified molecular class (40% vs 26%) were greater in the IBC cohort than in the T stage-matched non-IBC group. We first compared IBC with non-IBC regardless of their clinical subtypes. At any parametric P-value threshold, the Global significance test indicated that the findings were not different from what could be expected by chance alone (Table 2). Next, we performed the same analysis by clinical subtypes. Statistically significant differences using the Global test were observed in HER2-amplified cancers (Table 2 and Supplementary Table 2). Five hundred eight probe sets were differentially expressed between HER2-amplified IBC and non-IBC at parametric P ≤ 0.01 (Global P = 0.04): 171 probe sets were overexpressed and 337 were underexpressed in IBC. However, the FDR rates associated with individual parametric P-values ranged from 1.5% to 37%, indicating a substantial potential for false discovery among these genes. We mapped the 171 probe sets overexpressed in IBC to biological pathways, using Ingenuity Pathway Analysis. The top three pathways overexpressed in HER2-amplified IBC relative to non-IBC were eIF2 signaling, regulation of eIF4 and p70S6K signaling, and mammalian target of rapamycin (mTOR) signaling (Fig. 1A). Silvera et al. have reported that eIF4GI reprograms the protein synthetic machinery for increased translation of mRNAs with internal ribosome entry sites that promote IBC tumor cell survival and formation of tumor emboli [19].
Table 2.
Class comparison test between IBC and Non-IBC in tumor subgroups
| Significantly differentially expressed probe sets (n=18,940) with t-test
| ||||||||
|---|---|---|---|---|---|---|---|---|
| No. of patients (IBC/Non-IBC) | ALL
|
ER+/HER2−
|
HER2+
|
ER−/HER2−
|
||||
| 82 (25/57)
|
27 (5/22)
|
29 (12/17)
|
26 (8/18)
|
|||||
| No. of Genes | Global P value* | No. of Genes | Global P value* | No. of Genes | Global P value* | No. of Genes | Global P value* | |
| Parametric p value† | ||||||||
| 0.05 | 1214 | 0.209 | 927 | 0.372 | 2065 | 0.051 | 989 | 0.279 |
| 0.01 | 268 | 0.167 | 195 | 0.318 | 508 | 0.043 | 144 | 0.462 |
| 0.005 | 130 | 0.177 | 90 | 0.388 | 265 | 0.039 | 68 | 0.458 |
| 0.001 | 25 | 0.188 | 23 | 0.311 | 61 | 0.030 | 16 | 0.353 |
| Efron-Tibshirani’s GSA test P-value* with prior defined gene sets
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of patients (IBC/Non-IBC) GeneList GeneSets |
No. of genes | ALL
|
ER+/HER2−
|
HER2+
|
ER−/HER2−
|
||||
| 82 (25/57)
|
27 (5/22)
|
29 (12/17)
|
26 (8/18)
|
||||||
| P -value‡ | Directions | P -value‡ | Directions | P -value‡ | Directions | P -value‡ | Directions | ||
| Wnt gene set15 | 17 | 0.545 | Non-IBC | 0.375 | IBC | 0.415 | Non-IBC | 0.040 | IBC |
| Laere genes13 | 36 | 0.395 | IBC | 0.315 | Non-IBC | 0.330 | Non-IBC | 0.195 | IBC |
| Bieche genes4 | 54 | 0.245 | IBC | 0.015 | IBC | 0.180 | Non-IBC | 0.300 | IBC |
| CD44 related signatures16 | 55 | 0.135 | Non-IBC | 0.140 | Non-IBC | 0.055 | Non-IBC | 0.415 | Non-IBC |
| Bertucci genes14 | 71 | 0.230 | Non-IBC | 0.560 | IBC | 0.045 | IBC | 0.010 | Non-IBC |
IBC: Inflammatory Breast Cancer; ER: Estrogen Receptor.
Global p value shows the probability of getting at least the same number of genes significant on a t-test (at the specified p level) by chance if there are no real differences between IBC and Non-IBC.
The parametric p-value was derived from t-test.
The Efron and Tibshirani gene set analysis method that employs “maxmean” statistics (under 1000 permutations).
Fig. 1.
Ingenuity Pathway Analysis (IPA) software results showing the top 10 canonical pathways by significant genes overexpressed in IBC. The bars indicate −log(P-value), calculated by Fisher’s exact test. The line indicates the ratio for the percentage of genes in a pathway that were found in our uploaded list. Threshold indicates the line at P = 0.05.
Gene Set Analysis (GSA) results
Individual gene (i.e., probe set) level analysis suggested relatively modest gene expression differences between IBC and non-IBC. Next, we investigated whether we could detect coordinated but relatively small scale differences in the expression of gene sets that belong to functional pathways. Such consistent but small-scale differences at the individual gene level may not be readily identified by t-statistics but gene set enrichment analysis may be able to identify them. We tested five gene sets that were previously reported to be implicated in breast cancer prognosis or in the biology of IBC [15,13,4,16,14]. None of these previously published gene sets showed significant differential expression when all IBC cases were compared with all non-IBC (Table 2). When comparisons were made within clinical subtypes, the IBC metagene by Bieche et al [4] was significantly higher in ER-positive/HER2-normal IBC than in non-IBC of the same phenotype. The gene set by Bertucci et al. [14] was significantly higher in HER2-amplified IBC and the CD44 stem cell signature had borderline significance. The Wnt gene set [15] was significantly overexpressed in ER-negative/HER2-normal IBC (Table 2). The Wnt gene set was identified in a signature that discriminates lung metastases from primary tumor in SUM1315 orthotopic xenografts. Interestingly, SUM1315 breast cancer cells were initially isolated from a breast cancer skin metastasis [20] and were reported to correlation with basal breast cancer molecular subtypes [15]. To examine possible differences between IBC and non-IBC over a broad range of biological processes, we also tested 2113 functionally annotated gene sets assembled from Gene Ontology. Many gene sets showed significant differential expression by IBC status within the three clinical subtypes (Supplementary Table 3 and Supplementary Table 4 1–4). At a P value of ≤0.01 (Efron-Tibshirani test), 28, 69, and 41 gene sets were differently expressed in ER-positive/HER2-normal, HER2-positive, and ER-negative/HER2-normal cancers, respectively. The gene sets that were significantly overexpressed in IBC in the three clinical subtypes are listed in Table 3. We mapped all the members of these overexpressed gene sets (17, 33, and 18 gene sets by each subtype) to biological pathways by IPA; the results are shown in Fig. 1B,C and D. In HER2-amplified IBC, the most significant pathways identified through GO were the same as those identified by the gene-by-gene analysis (Fig. 1A,B). In ER-positive/HER2-normal IBC, the most significant pathways overexpressed in this subtype were the complement system, oncostatin signaling, and various immune cell-related pathways (Fig. 1C). In the ER-negative/HER2-normal IBC specimens, Myc-mediated apoptosis signaling, axonal guidance signaling, and Neuregulin signaling as well as some lipid metabolism-related pathways were overexpressed. These results suggest that subtle biological differences may exist between breast cancer subtypes that show the inflammatory phenotype compared with tumors of the same subtype without inflammatory features. Interestingly, these differences seemed to vary by subtype.
Table 3.
Gene sets overexpressed in IBC compared with Non-IBC
| GO category | GO term | Number of genes | GSA test P value |
|---|---|---|---|
| Gene sets overexpressed in ER+/HER2-normal IBC compared with Non-IBC | |||
| GO:0002675 | Positive regulation of acute inflammatory response | 11 | 0.001 |
| GO:0002673 | Regulation of acute inflammatory response | 15 | 0.001 |
| GO:0050729 | Positive regulation of inflammatory response | 31 | 0.001 |
| GO:0050727 | Regulation of inflammatory response | 57 | 0.002 |
| GO:0043407 | Negative regulation of MAP kinase activity | 40 | 0.003 |
| GO:0031406 | Carboxylic acid binding | 17 | 0.005 |
| GO:0045103 | Intermediate filament-based process | 25 | 0.005 |
| GO:0010574 | Regulation of vascular endothelial growth factor production | 17 | 0.006 |
| GO:0045104 | Intermediate filament cytoskeleton organization | 23 | 0.006 |
| GO:0046928 | Regulation of neurotransmitter secretion | 12 | 0.007 |
| GO:0033549 | MAP kinase phosphatase activity | 12 | 0.008 |
| GO:0043500 | Muscle adaptation | 20 | 0.008 |
| GO:0002526 | Acute inflammatory response | 97 | 0.008 |
| GO:0014896 | Muscle hypertrophy | 15 | 0.009 |
| GO:0042531 | Positive regulation of tyrosine phosphorylation of STAT protein | 22 | 0.009 |
| GO:0051348 | Negative regulation of transferase activity | 92 | 0.009 |
| GO:0033673 | Negative regulation of kinase activity | 86 | 0.010 |
| Gene sets overexpressed in HER2-positive IBC compared with Non-IBC | |||
| GO:0051668 | Localization within membrane | 15 | 0.001 |
| GO:0051084 | ‘de novo’ posttranslational protein folding | 15 | 0.001 |
| GO:0009119 | Ribonucleoside metabolic process | 15 | 0.001 |
| GO:0043681 | Protein import into mitochondrion | 16 | 0.001 |
| GO:0009168 | Purine ribonucleoside monophosphate biosynthetic process | 22 | 0.001 |
| GO:0006458 | ‘de novo’ protein folding | 23 | 0.001 |
| GO:0019206 | Nucleoside kinase activity | 23 | 0.001 |
| GO:0042994 | Cytoplasmic sequestering of transcription factor | 11 | 0.002 |
| GO:0006626 | Protein targeting to mitochondrion | 30 | 0.002 |
| GO:0009116 | Nucleoside metabolic process | 41 | 0.002 |
| GO:0003729 | mRNA binding | 90 | 0.002 |
| GO:0043094 | Cellular metabolic compound salvage | 10 | 0.003 |
| GO:0051220 | Cytoplasmic sequestering of protein | 14 | 0.003 |
| GO:0022884 | Macromolecule transmembrane transporter activity | 15 | 0.003 |
| GO:0042273 | Ribosomal large subunit biogenesis | 18 | 0.003 |
| GO:0010466 | Negative regulation of peptidase activity | 21 | 0.003 |
| GO:0005758 | Mitochondrial intermembrane space | 34 | 0.003 |
| GO:0046128 | Purine ribonucleoside metabolic process | 13 | 0.004 |
| GO:0019201 | Nucleotide kinase activity | 30 | 0.004 |
| GO:0008344 | Adult locomotory behavior | 35 | 0.004 |
| GO:0019059 | Initiation of viral infection | 42 | 0.004 |
| GO:0006446 | Regulation of translational initiation | 72 | 0.004 |
| GO:0015935 | Small ribosomal subunit | 89 | 0.004 |
| GO:0030159 | Receptor signaling complex scaffold activity | 24 | 0.005 |
| GO:0031970 | Organelle envelope lumen | 41 | 0.005 |
| GO:0046148 | Pigment biosynthetic process | 54 | 0.007 |
| GO:0015934 | Large ribosomal subunit | 99 | 0.007 |
| GO:0006144 | Purine base metabolic process | 24 | 0.008 |
| GO:0009146 | Purine nucleoside triphosphate catabolic process | 13 | 0.009 |
| GO:0042274 | Ribosomal small subunit biogenesis | 22 | 0.009 |
| GO:0009161 | Ribonucleoside monophosphate metabolic process | 33 | 0.009 |
| GO:0006195 | Purine nucleotide catabolic process | 20 | 0.010 |
| GO:0030041 | Actin filament polymerization | 81 | 0.010 |
| Gene sets overexpressed in ER-negative/HER2-normal IBC compared with Non-IBC | |||
| GO:0035094 | Response to nicotine | 19 | 0.001 |
| GO:0043279 | Response to alkaloid | 27 | 0.001 |
| GO:0005548 | Phospholipid transporter activity | 35 | 0.001 |
| GO:0048545 | Response to steroid hormone stimulus | 97 | 0.001 |
| GO:0006800 | Oxygen and reactive oxygen species metabolic process | 78 | 0.002 |
| GO:0002437 | Inflammatory response to antigenic stimulus | 14 | 0.003 |
| GO:0005100 | Rho GTPase activator activity | 26 | 0.003 |
| GO:0014902 | Myotube differentiation | 16 | 0.004 |
| GO:0016528 | Sarcoplasm | 34 | 0.004 |
| GO:0008367 | Bacterial binding | 12 | 0.005 |
| GO:0014070 | Response to organic cyclic substance | 37 | 0.006 |
| GO:0043627 | Response to estrogen stimulus | 62 | 0.006 |
| GO:0002861 | Regulation of inflammatory response to antigenic stimulus | 13 | 0.007 |
| GO:0048019 | Receptor antagonist activity | 14 | 0.008 |
| GO:0016529 | Sarcoplasmic reticulum | 33 | 0.008 |
| GO:0007631 | Feeding behavior | 41 | 0.008 |
| GO:0002683 | Negative regulation of immune system process | 71 | 0.009 |
| GO:0006656 | Phosphatidylcholine biosynthetic process | 13 | 0.010 |
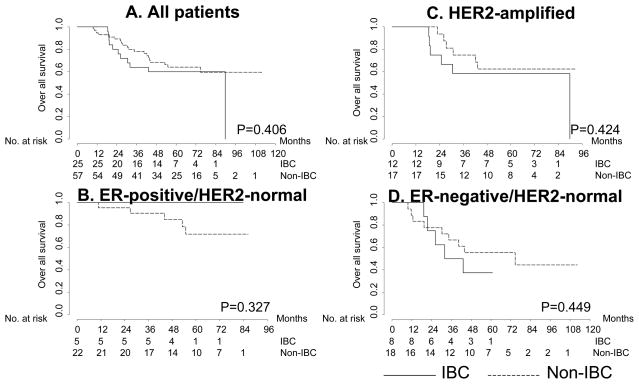
Subtype-specific survival of IBC and non-IBC
We examined whether the survival of patients with the three differs between IBC and non-IBC, with or without stratifying by clinical subtype. The median follow-ups were 53 months (range: 18.5–89.8) and 57.2 months (range: 9.6–112.5) for the IBC and non-IBC cohorts, respectively. In this case-control study, the overall survival results did not significantly differ for IBC and non-IBC, overall or within each clinical subset (Fig. 2). Among the IBC cases under consideration, 0 out of 5 patients with ER-positive/HER2-normal tumors, 6 (50%) of 12 with HER2-positive, and 5 (62.5%) of 8 with ER-negative/HER2-normal had died. Among the non-IBC cases, 5 (22.7%) out of 22 patients with ER-positive/HER2-normal tumors, 6 (35.3%) of 17 with HER2-positive, and 9 (50%) of 18 with ER-negative/HER2-normal had died. Relapse-free survival was worse in ER-negative/HER2-normal IBC patients than for the non-IBC of the same phenotype (Supplementary Fig. 1). There was also no significant difference in overall or relapse-free survival by clinical subtype between the IBC and non-IBC cohorts (Fig. 3). A trend was observed in both IBC and non-IBC for better survival in ER-positive/HER-normal cases, but the small sample limited the power of survival analysis by subtype. Also, survival analysis might have been affected by an imbalance for targeted therapy (trastuzumab: HER2 antibody). Six (50%) of 12 patients with HER2-amplified IBC and 11 (65%) of 17 patients with HER2-amplified non-IBC had never received either neoadjuvant or adjuvant therapy.
Fig. 2.
Kaplan-Meier curves showing overall survival by breast cancer phenotype for each receptor status.
Fig. 3.
Kaplan-Meier curves showing overall survival by receptor subtype for each breast cancer phenotype.
Discussion
This is the first study to systematically examine gene expression differences between IBC and non-IBC patients according to clinically relevant molecular subtypes defined based on ER and HER2 expression. Moreover, we also carefully matched the IBC and non-IBC cohorts for T stage. All patients in this analysis had clinical T4 and represented locally advanced breast cancer with and without inflammatory features. In a global comparison of IBC versus non-IBC, we detected no significant gene expression differences at the individual gene level between these two cohorts of cases after adjusting for multiple comparisons. This is in contrast to several published reports that found significant gene expression differences between IBC and non-IBC [4–7]. This could probably be related to differences in the clinical characteristics of the comparison cohort. A closer analysis of the published literature reveals that many of the previous reports have not included stage II cancers in the comparison cohort [4,5]. Some of the previous reports have not adjusted for multiple comparisons or used reverse transcription polymerase chain reaction (RT-PCR) to quantify gene expression differences, and RT-PCR has a different sensitivity and dynamic range of mRNA detection compared with gene expression arrays [4].
Importantly, all of these previous reports that suggested gene expression differences between IBC and non-IBC reported small-scale differences. One report assessed 538 genes and identified 22 that were differentially expressed [4]. Another report evaluated over 8,000 genes and found 105 that were differentially expressed [5]. A third report, similar to our analysis, found no significant differences at the individual gene level but observed some differences at the gene set level. [6] A fourth group of investigators tested close to 6,000 genes and found 50 that could discriminate between IBC and non-IBC [7]. When we tested these previously identified gene sets, we could only partially confirm the results from these publications in the current data set. Several of the gene sets were significantly overexpressed in IBC but only in particular clinical subtypes and not during a global comparison of all IBC with all non-IBC cohorts (as previously reported). This discrepancy reflects the unstable nature of these observations due to small and variable sample sizes of all current studies and the potential confounding effect of a variable clinical subtype distribution from study to study. The numbers of IBC cases included in the comparisons in published studies ranged from 14 to 37, and all previous analyses compared IBC with non-IBC without adjusting for molecular class or clinical receptor phenotype. Subtle imbalances in receptor phenotype between the IBC and non-IBC cohorts could lead to the identification of significantly differentially expressed genes that reflect the large-scale gene expression differences between molecular classes that were unevenly distributed across the two comparison cohorts, rather than the IBC-specific gene expression differences.
In the current study, we compared IBC and non-IBC after stratifying both groups into three clinically relevant subtypes of breast cancers. We found that individual gene level analysis showed significant differences only within the HER2-amplified group. Indeed, the natural history of HER2-amplified breast cancer has been dramatically changed by treatment with trastuzumab, an anti-HER2 antibody [21,22]. This observation has also been noted in IBC [23–27]. Several studies have reported the efficacy of trastuzumab in the treatment of HER2-amplified IBC [23–26]. Kaufman et al. reported that treatment with an EGFR/HER2 tyrosine kinase inhibitor, lapatinib, provided clinical efficacy in HER2-amplified IBC during phase II trials [27]. For treatment of the other subtypes of IBC, no similar targeted drugs are available. We speculate the unique activity could stem from the different HER2 pathway involvement between IBC and non-IBC. In the other clinical subtypes, individual gene level analysis showed no significant differences.
When gene sets were tested for differential expression, different gene sets distinguished IBC from non-IBC in each clinical subtype (Fig. 1). This suggests the possibility that different biological pathways are involved with the pathogenesis of the inflammatory phenotype in different molecular subsets of breast cancers. We found that complement and immune systems were highly associated with the ER-positive/HER2-normal IBC phenotype. Activation and regulation of complement pathways have been reported to play important roles in inflammatory mediators [28]. In addition, we found that protein translation and mTOR signaling were overexpressed in HER2-amplified tumors. mTOR, which functions downstream of the PI3K/Akt signaling pathway, is also being explored in therapies for HER2-amplified breast cancer [29]. mTOR inhibitors being tested in clinical trials for patients with breast cancer and other solid tumors include CCI-779 [30], RAD001 [31], and AP23573 [32]. We speculate that the importance of the mTOR pathway could be tested as potential target for HER2-amplified IBC by using one of the mTOR inhibitors. On the other hand, GSA showed that apoptosis, neural, and lipid metabolism pathways were overexpressed in ER-negative/HER2-normal IBC. There is biological plausibility to implicate these processes in IBC features. It is known that the neurotransmitter pathway and lipid metabolism play important roles in the regulation of cell migration and apoptosis [33–35]. These observations suggest the possibility of potential novel strategies to modulate ER-negative/HER2-normal IBC. However, the lack of overlap between the discriminating gene sets is also partly due to false-negative findings owing to the small sample sizes for each cohort, leading to limited power for detecting differentially expressed gene sets.
Our data suggest that future studies involving IBC should consider the various clinical subtypes separately. These subtypes may have important clinical and molecular differences that justify their separate analysis. The different clinical prognoses of patients with the different clinical subtypes of breast cancer are well recognized, and similar differences in outcome may also exist among patients with IBC. This current study did not find any difference in overall survival related to different clinical subtypes of IBC, but we observed a worse relapse-free survival for patients with ER-negative/HER2-normal IBC compared with non-IBC of the same phenotype. However, this study was limited in its power to detect any but the most dramatic differences in survival because of the small sample size, which is due to the rarity of this disease entity (particularly when IBC is further subdivided by clinical phenotype). It may be necessary to create national and international cooperative networks to ensure sufficient sample sizes and homogeneous and mutually compatible analysis platforms for future studies of IBC.
Supplementary Material
Kaplan-Meier curves showing relapse-free survival by breast cancer phenotype in each receptor subtype.
Acknowledgments
Grant support:
This work was supported by the followings: the Breast Cancer Research Foundation (LP and WFS), MD Anderson Cancer Center Faculty Incentive Funds (WFS), the Commonwealth Cancer Foundation (LP and WFS) and MD Anderson’s Cancer Center Support Grant (CA016672)(NTU), the National Institute of Health R01CA138239-01(WAW), The State of Texas Grant for Rare and Aggressive Cancers(MC and WAW), the American Airlines Komen Foundation Promise Grant KGO81287(WAW). Morgan Welch Inflammatory Breast Cancer Research Program(MC).
References
- 1.Singletary SE, Allred C, Ashley P, Bassett LW, Berry D, Bland KI, Borgen PI, Clark G, Edge SB, Hayes DF, Hughes LL, Hutter RV, Morrow M, Page DL, Recht A, Theriault RL, Thor A, Weaver DL, Wieand HS, Greene FL. Revision of the american joint committee on cancer staging system for breast cancer. J Clin Oncol. 2002;20 (17):3628–3636. doi: 10.1200/JCO.2002.02.026. [DOI] [PubMed] [Google Scholar]
- 2.Levine PH, Steinhorn SC, Ries LG, Aron JL. Inflammatory breast cancer: The experience of the surveillance, epidemiology, and end results (seer) program. J Natl Cancer Inst. 1985;74 (2):291–297. [PubMed] [Google Scholar]
- 3.Cristofanilli M, Buzdar AU, Hortobagyi GN. Update on the management of inflammatory breast cancer. Oncologist. 2003;8 (2):141–148. doi: 10.1634/theoncologist.8-2-141. [DOI] [PubMed] [Google Scholar]
- 4.Bieche I, Lerebours F, Tozlu S, Espie M, Marty M, Lidereau R. Molecular profiling of inflammatory breast cancer: Identification of a poor-prognosis gene expression signature. Clin Cancer Res. 2004;10 (20):6789–6795. doi: 10.1158/1078-0432.CCR-04-0306. 10/20/6789 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Bertucci F, Finetti P, Rougemont J, Charafe-Jauffret E, Cervera N, Tarpin C, Nguyen C, Xerri L, Houlgatte R, Jacquemier J, Viens P, Birnbaum D. Gene expression profiling identifies molecular subtypes of inflammatory breast cancer. Cancer Res. 2005;65 (6):2170–2178. doi: 10.1158/0008-5472.CAN-04-4115. 65/6/2170 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Nguyen DM, Sam K, Tsimelzon A, Li X, Wong H, Mohsin S, Clark GM, Hilsenbeck SG, Elledge RM, Allred DC, O’Connell P, Chang JC. Molecular heterogeneity of inflammatory breast cancer: A hyperproliferative phenotype. Clin Cancer Res. 2006;12 (17):5047–5054. doi: 10.1158/1078-0432.CCR-05-2248. 12/17/5047 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Van Laere S, Van der Auwera I, Van den Eynden G, Van Hummelen P, van Dam P, Van Marck E, Vermeulen PB, Dirix L. Distinct molecular phenotype of inflammatory breast cancer compared to non-inflammatory breast cancer using affymetrix-based genome-wide gene-expression analysis. Br J Cancer. 2007;97 (8):1165–1174. doi: 10.1038/sj.bjc.6603967. 6603967 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ, Rouzier R, Sneige N, Ross JS, Vidaurre T, Gomez HL, Hortobagyi GN, Pusztai L. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24(26):4236–4244. doi: 10.1200/JCO.2006.05.6861. JCO.2006.05.6861 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Gong Y, Yan K, Lin F, Anderson K, Sotiriou C, Andre F, Holmes FA, Valero V, Booser D, Pippen JE, Jr, Vukelja S, Gomez H, Mejia J, Barajas LJ, Hess KR, Sneige N, Hortobagyi GN, Pusztai L, Symmans WF. Determination of oestrogen-receptor status and erbb2 status of breast carcinoma: A gene-expression profiling study. Lancet Oncol. 2007;8(3):203–211. doi: 10.1016/S1470-2045(07)70042-6. S1470-2045(07)70042-6 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. JCO.2008.18.1370 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. 0506580102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Laere S, Van der Auwera I, Van den Eynden GG, Fox SB, Bianchi F, Harris AL, van Dam P, Van Marck EA, Vermeulen PB, Dirix LY. Distinct molecular signature of inflammatory breast cancer by cdna microarray analysis. Breast Cancer Res Treat. 2005;93 (3):237–246. doi: 10.1007/s10549-005-5157-z. [DOI] [PubMed] [Google Scholar]
- 14.Bertucci F, Finetti P, Rougemont J, Charafe-Jauffret E, Nasser V, Loriod B, Camerlo J, Tagett R, Tarpin C, Houvenaeghel G, Nguyen C, Maraninchi D, Jacquemier J, Houlgatte R, Birnbaum D, Viens P. Gene expression profiling for molecular characterization of inflammatory breast cancer and prediction of response to chemotherapy. Cancer Res. 2004;64 (23):8558–8565. doi: 10.1158/0008-5472.CAN-04-2696. 64/23/8558 [pii] [DOI] [PubMed] [Google Scholar]
- 15.DiMeo TA, Anderson K, Phadke P, Feng C, Perou CM, Naber S, Kuperwasser C. A novel lung metastasis signature links wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69 (13):5364–5373. doi: 10.1158/0008-5472.CAN-08-4135. 0008-5472.CAN-08-4135 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, Halushka MK, Sukumar S, Parker LM, Anderson KS, Harris LN, Garber JE, Richardson AL, Schnitt SJ, Nikolsky Y, Gelman RS, Polyak K. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11 (3):259–273. doi: 10.1016/j.ccr.2007.01.013. S1535-6108(07)00029-3 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Jiang Z, Gentleman R. Extensions to gene set enrichment. Bioinformatics. 2007;23 (3):306–313. doi: 10.1093/bioinformatics/btl599. btl599 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Efron B, Tishirani R. On testing the significance of sets of genes. Ann Appl Stat. 2007;1 (1):107–129. [Google Scholar]
- 19.Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, Hochman T, Formenti SC, Schneider RJ. Essential role for eif4gi overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol. 2009;11 (7):903–908. doi: 10.1038/ncb1900. ncb1900 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi OP, Ethier SP. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer. 1999;81 (8):1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, Cameron D, Dowsett M, Barrios CH, Steger G, Huang CS, Andersson M, Inbar M, Lichinitser M, Lang I, Nitz U, Iwata H, Thomssen C, Lohrisch C, Suter TM, Ruschoff J, Suto T, Greatorex V, Ward C, Straehle C, McFadden E, Dolci MS, Gelber RD. Trastuzumab after adjuvant chemotherapy in her2-positive breast cancer. N Engl J Med. 2005;353 (16):1659–1672. doi: 10.1056/NEJMoa052306. 353/16/1659 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, Swain SM, Pisansky TM, Fehrenbacher L, Kutteh LA, Vogel VG, Visscher DW, Yothers G, Jenkins RB, Brown AM, Dakhil SR, Mamounas EP, Lingle WL, Klein PM, Ingle JN, Wolmark N. Trastuzumab plus adjuvant chemotherapy for operable her2-positive breast cancer. N Engl J Med. 2005;353 (16):1673–1684. doi: 10.1056/NEJMoa052122. 353/16/1673 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Van Pelt AE, Mohsin S, Elledge RM, Hilsenbeck SG, Gutierrez MC, Lucci A, Jr, Kalidas M, Granchi T, Scott BG, Allred DC, Chang JC. Neoadjuvant trastuzumab and docetaxel in breast cancer: Preliminary results. Clin Breast Cancer. 2003;4 (5):348–353. doi: 10.3816/cbc.2003.n.040. [DOI] [PubMed] [Google Scholar]
- 24.Hurley J, Doliny P, Reis I, Silva O, Gomez-Fernandez C, Velez P, Pauletti G, Powell JE, Pegram MD, Slamon DJ. Docetaxel, cisplatin, and trastuzumab as primary systemic therapy for human epidermal growth factor receptor 2-positive locally advanced breast cancer. J Clin Oncol. 2006;24(12):1831–1838. doi: 10.1200/JCO.2005.02.8886. JCO.2005.02.8886 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Limentani SA, Brufsky AM, Erban JK, Jahanzeb M, Lewis D. Phase ii study of neoadjuvant docetaxel, vinorelbine, and trastuzumab followed by surgery and adjuvant doxorubicin plus cyclophosphamide in women with human epidermal growth factor receptor 2-overexpressing locally advanced breast cancer. J Clin Oncol. 2007;25 (10):1232–1238. doi: 10.1200/JCO.2005.05.3306. JCO.2005.05.3306 [pii] [DOI] [PubMed] [Google Scholar]
- 26.Yamauchi H, Cristofanilli M, Nakamura S, Hortobagyi GN, Ueno NT. Molecular targets for treatment of inflammatory breast cancer. Nat Rev Clin Oncol. 2009;6 (7):387–394. doi: 10.1038/nrclinonc.2009.73. nrclinonc.2009.73 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Kaufman B, Trudeau M, Awada A, Blackwell K, Bachelot T, Salazar V, DeSilvio M, Westlund R, Zaks T, Spector N, Johnston S. Lapatinib monotherapy in patients with her2-overexpressing relapsed or refractory inflammatory breast cancer: Final results and survival of the expanded her2+ cohort in egf103009, a phase ii study. Lancet Oncol. 2009;10 (6):581–588. doi: 10.1016/S1470-2045(09)70087-7. S1470-2045(09)70087-7 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Gelderman KA, Tomlinson S, Ross GD, Gorter A. Complement function in mab-mediated cancer immunotherapy. Trends Immunol. 2004;25(3):158–164. doi: 10.1016/j.it.2004.01.008. S1471490604000262 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Nahta R, Yu D, Hung MC, Hortobagyi GN, Esteva FJ. Mechanisms of disease: Understanding resistance to her2-targeted therapy in human breast cancer. Nat Clin Pract Oncol. 2006;3 (5):269–280. doi: 10.1038/ncponc0509. ncponc0509 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Atkins MB, Hidalgo M, Stadler WM, Logan TF, Dutcher JP, Hudes GR, Park Y, Liou SH, Marshall B, Boni JP, Dukart G, Sherman ML. Randomized phase ii study of multiple dose levels of cci-779, a novel mammalian target of rapamycin kinase inhibitor, in patients with advanced refractory renal cell carcinoma. J Clin Oncol. 2004;22(5):909–918. doi: 10.1200/JCO.2004.08.185. JCO.2004.08.185 [pii] [DOI] [PubMed] [Google Scholar]
- 31.Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O’Reilly T, Natt F, Hall J, Lane HA, Thomas G. The mtor inhibitor rad001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120 (6):747–759. doi: 10.1016/j.cell.2004.12.040. S0092-8674(05)00087-5 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Mita MM, Mita AC, Chu QS, Rowinsky EK, Fetterly GJ, Goldston M, Patnaik A, Mathews L, Ricart AD, Mays T, Knowles H, Rivera VM, Kreisberg J, Bedrosian CL, Tolcher AW. Phase i trial of the novel mammalian target of rapamycin inhibitor deforolimus (ap23573; mk-8669) administered intravenously daily for 5 days every 2 weeks to patients with advanced malignancies. J Clin Oncol. 2008;26 (3):361–367. doi: 10.1200/JCO.2007.12.0345. 26/3/361 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Drell TLt, Joseph J, Lang K, Niggemann B, Zaenker KS, Entschladen F. Effects of neurotransmitters on the chemokinesis and chemotaxis of mda-mb-468 human breast carcinoma cells. Breast Cancer Res Treat. 2003;80 (1):63–70. doi: 10.1023/A:1024491219366. [DOI] [PubMed] [Google Scholar]
- 34.Crawford KW, Bowen WD. Sigma-2 receptor agonists activate a novel apoptotic pathway and potentiate antineoplastic drugs in breast tumor cell lines. Cancer Res. 2002;62 (1):313–322. [PubMed] [Google Scholar]
- 35.Elstner E, Muller C, Koshizuka K, Williamson EA, Park D, Asou H, Shintaku P, Said JW, Heber D, Koeffler HP. Ligands for peroxisome proliferator-activated receptorgamma and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in bnx mice. Proc Natl Acad Sci U S A. 1998;95 (15):8806–8811. doi: 10.1073/pnas.95.15.8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kaplan-Meier curves showing relapse-free survival by breast cancer phenotype in each receptor subtype.