Abstract
Sequences of the cytochrome c oxidase subunit I (COI) mitochondrial gene from adults of 22 Culex ( Culex ) species from Argentina and Brazil were employed to assess species identification and to test the usefulness of COI for barcoding using the best close match (BCM) algorithm. A pairwise Kimura two-parameter distance matrix including the mean intra and interspecific distances for 71 COI barcode sequences was constructed. Of the 12 COI lineages recovered in the Neighbour-joining topology, five confirmed recognised morphological species ( Cx. acharistus , Cx. chidesteri , Cx. dolosus , Cx. lygrus and Cx. saltanensis ) with intraspecific divergences lower than 1.75%. Cx. bilineatus is formally resurrected from the synonymy of Cx. dolosus . Cx. maxi , Cx. surinamensis and the Coronator group species included were clustered into an unresolved lineage. The intraspecific distance of Cx. pipiens (3%) was almost twice the interspecific between it and Cx. quinquefasciatus (1.6%). Regarding the BCM criteria, the COI barcode successfully identified 69% of all species. The rest of the sequences, approximately 10%, 18% and 3%, remained as ambiguously, mis and unidentified, respectively. The COI barcode does not contain enough information to distinguish Culex ( Cux. ) species.
Keywords: Culex, identification, COI, morphology, genetic divergence
The genus Culex is one of the largest groups of the family Culicidae, containing 768 species subdivided into 26 subgenera ( Harbach 2011 ). The subgenus Culex includes 198 species, some potentially involved in the transmission of lymphatic filariasis nematodes ( Wuchereria bancrofti , Brugia malayi and Brugia timori ) ( Fontes et al. 1998 , 2005 ) and several arboviruses ( Hubálek 2000 , Komar 2003 ). Cx. quinquefasciatus is the primary vector of W. bancrofti in Northeast Brazil ( Fontes et al. 2005 ), whereas Cx. coronator and Cx . declarator were recorded as potential vectors of the Saint Louis encephalitis virus (SLEV) ( Vasconcelos et al. 1991 ). In Argentina, Cx. quinquefasciatus and Cx. interfor are primary and secondary vectors of the SLEV, respectively ( Spinsanti et al. 2009 ), while the Rio Negro virus from the Venezuelan equine encephalitis virus complex was isolated from both Cx. coronator and Cx. maxi , among other species ( Pisano et al. 2010 ). Species of the genus Culex were also implicated as vectors of the West Nile virus ( Kramer et al. 2008 ) in North America.
The current subgeneric classification of the genus Culex is based on characters of females and males ( Bram 1967 , Forattini 2002 ). Species of the subgenus Culex are subdivided into six groups, several subgroups and species complex ( Harbach 2011 ). The Neotropical species were included in the Coronator and Pipiens groups ( Harbach 2011 ). Morphological traits of the male genitalia usually allow an accurate identification of the species. Contrasting, external morphological characteristics of the females are also employed for identification. However, most of the available identification keys should be used with caution because female morphological characters may be either polymorphic or overlap among distinct species. Characters of the fourth-instar larva are also employed for identification; however, it is also possible to have either some overlap or absence of differences among species. The presence of unknown species complexes also makes species identification difficult. Complexes of morphologically similar species were demonstrated to be present in several groups of invertebrates, i.e., in anopheline mosquitoes ( Foster et al. 2013 ), wasps ( Bickford et al. 2007 ), butterflies ( Hebert et al. 2004 ), aquatic gastropod mollusks ( de Aranzamendi et al. 2009 ), among several others.
DNA sequence data are largely employed in molecular taxonomy, i.e., for species identification ( Sallum et al. 2008 , 2010 , Bourke et al. 2013 , Foster et al. 2013 ), to address phylogenetic relationships among and within groups of Culicidae ( Krzywinski et al. 2001 , Sallum et al. 2002 , Reidenbach et al. 2009 ) and to define genetic structure of vector species populations ( Mirabello & Conn 2008 ). A fragment of the cytochrome c oxidase subunit I (COI) mitochondrial gene has been largely employed for taxon barcoding ( Hebert et al. 2003a ) and as a tool to assess genetic divergence among closely related species ( Hebert et al. 2003b ). Several studies employed COI barcode sequences to identify mosquito species, i.e., mosquitoes from Canada ( Cywinska et al. 2006 ), India ( Kumar et al. 2007 ), China ( Wang et al. 2012 ) and to reveal species complexes within the subgenus Nyssorhynchus of Anopheles from the Neotropical Region ( Ruiz-Lopez et al. 2012 , Bourke et al. 2013 , Foster et al. 2013 ). Partial sequences of COI gene were also used to verify species complexes in mosquitoes ( Saeung et al. 2008 , Demari-Silva et al. 2011 ), to reveal phylogeographic patterns in Anopheles darlingi Root (Pedro & Sallum 2008 ) and to compare phylogeographic patterns between An . darlingi and Anopheles triannulatus (Neiva & Pinto) populations of several ecoregions of Brazil ( Pedro & Sallum 2009 ).
According to Hebert et al. (2004) , the mean interspecific genetic divergence should be at least 10 times higher than the average intraspecific genetic distance in order to define the presence of species complexes. Although the patterns of intra and interspecific variation in COI are similar in various animal groups ( Hebert et al. 2004 ), Ruiz-Lopez et al. (2012) suggested for mosquitoes, a mean intraspecific Kimura two-parameter (K2P) distance varying from 0.2-1.4% and a mean interspecific variation between 2-5.6%.
One of the criticisms of using the COI DNA barcode is the ambiguous identification or the absence of clusters in trees of recently diverged species ( Meyer & Paulay 2005 , Kaila & Ståhls 2006 , Lou & Golding 2010 ). In order to improve the power of the COI barcode datasets to correctly identify recently diverged species, new algorithms were developed recently by Meier et al. (2006) and van Velzen et al. (2012) . The best close match (BCM) algorithm was developed by Meier et al. (2006) to identify the best barcode matches of a query and only assigns the species name of that barcode to the query if the barcode is sufficiently similar. To determine how similar the sequences are, a threshold similarity value has to be estimated for a given dataset by obtaining a frequency distribution of all intraspecific pairwise distances and determining the distance below which 95% of all intraspecific distances are found ( Meier et al. 2006 ).
In a recent study carried out by Demari-Silva et al. (2011) , a 478 base pair fragment of the COI gene was employed to differentiate Culex species from Brazil (9 of which belong to the subgenus Culex ), to verify phylogenetic relationships of the genus Lutzia relative to Culex and the taxonomic status of the subgenus Phenacomyia within the genus Culex . Results of the analyses showed high intraspecific variation, revealing the presence of species complexes within Culex . In the present study, the COI barcode region was employed to assess both the species identification and subgroups/complexes within Culex ( Cx. ) from the Neotropics. Classification and nomenclature adopted herein were compiled by Harbach (2011) .
MATERIALS AND METHODS
Mosquito sampling - Adults of 22 species of the Pipiens and Coronator groups ( Table I ) of Culex ( Cux. ) were collected in Argentina and Brazil between 2005-2011 and employed in this study (Supplementary data 1). Females and males were obtained from individually reared fourth-instar larvae or pupae. Species identification was based on male genitalia using descriptions and redescriptions of the species and taxonomic keys ( Forattini 2002 ). Females and males from Argentina were obtained from the same larval habitats.
TABLE I. List of the species of Culex ( Culex ) from Argentina and Brazil included in the analysis.
| Group | Subgroup | Complex | Species |
|---|---|---|---|
| Coronator | - | - | Cx. camposi Dyar |
| - | - | Cx. coronator Dyar & Knab | |
| - | - | Cx. usquatus Dyar | |
| Pipiens | Apicinus | - | Cx. apicinus Philippi |
| - | - | Cx. chidesteri Dyar | |
| - | - | Cx. eduardoi Casal & Garcia | |
| - | - | Cx. mollis Dyar & Knab | |
| - | - | Cx. nigripalpus Theobald | |
| - | - | Cx. tatoi Casal & Garcia | |
| Pipiens | - | Cx. pipiens L. | |
| - | - | Cx. quinquefasciatus Say | |
| Tarsalis | - | Cx. bidens Dyar | |
| - | - | Cx. declarator Dyar & Knab | |
| - | - | Cx. interfor Dyar | |
| - | - | Cx. lygrus Root | |
| - | - | Cx. maxi Dyar | |
| - | - | Cx. saltanensis Dyar | |
| - | - | Cx. surinamensis Dyar | |
| - | Restuans | Cx. acharistus Root | |
| - | - | Cx. brethesi Dyar | |
| - | Salinarius | Cx. dolosus (Lynch Arribálzaga) | |
| - | - | Cx. spinosus Lutz |
DNA extraction, amplification and sequencing - DNA extractions were obtained either from whole adult specimens (14 individuals) or one or two legs (57 individuals) from each mosquito; most of them, preserved dry over silica gel. Genomic DNA was extracted using 50/10 µL of NaCl and 240/20 µL of Chelex-100 5% (w/v). The extract solution was vortexed and incubated at 99ºC for 10 min. After centrifugation at 13,000 rpm for 15 min, at 25ºC, the supernatant was recovered and an aliquot of 12 µL, frozen at -20ºC, was used for the polymerse chain reactions (PCRs). The remaining Chelex-DNA was stored at -80ºC in Entomological Collection Reference, School of Public Health, University of São Paulo, Brazil.
The primer pairs of Folmer et al. (1994) LCO1490 (5’-GGTCAACAAATCATAAAGATATTGG-3’) and HCO2198 (5’-TAAACTTCAGGGTGACCAAAAAATCA-3’) were used to amplify the ~658 bp fragments of COI which were trimmed between 615-654 bp. The PCR was performed in a total volume of 25 μL containing 1-6 μL of Chelex-DNA, 1 x PCR buffer (Invitrogen), 1.5 mM MgCl2 (Invitrogen), 0.1 μM each primer, 200 mM each dNTPs (Amresco), 0.625 U Taq Platinum polymerase (Invitrogen) and the remaining volume of ddH2O. The PCR thermal regime consisted of 94ºC for 3 min, 40 cycles of 94ºC for 1 min, 55ºC for 1 min and 72ºC for 1 min and a final extension at 72ºC for 7 min. For 22 samples that could not be amplified using the previously described protocol (museum samples), the reaction proceeded under the following temperature profile taken from Zapata et al. (2012) : 94ºC for 3 min, 5 cycles of 94ºC for 30 sec, 45ºC for 90 sec, 68ºC for 60 sec, 35 cycles of 94ºC for 30 sec, 51ºC for 30 sec, 68ºC for 60 sec and a final extension at 68ºC for 10 sec.
The PCR products of the COI gene were electrophoresed in 1% TAE agarose gels stained with GelRedTM Nucleic Acid Gel Stain (Biotium Inc, Hayward, USA). All sequencing reactions were carried out in both directions using an ABI Big Dye Terminator Kit v.3.1 (PE Applied Biosystems, Warrington, England) with the same set of PCR primers.
The sequencing reactions were carried in a total volume of 10 μL containing 40-50 ng of the PEG purified PCR product, 0.5 μL BigDye Terminator Ready Reaction Mix, 1 x Sequencing Buffer (Applied Biosystems), 3.6 pmol of R (HCO2198) or F (LCO1490) primers and the remaining volume of ddH2O. The sequencing reactions were purified on Sephadex G50 columns (GE Healthcare) and analysed in an Applied Biosystems 3130 DNA Analyser (PE Applied Biosystems). The sequences were edited in Sequencher v.4.9 (Genes Codes Corporation, Ann Arbor, MI, USA), primer regions were removed from sequences. Comparisons with available sequences using Basic Local Alignment Search Tool (blast.ncbi.nlm.nih.gov/Blast.cgi) were performed to check for sequence homology and species identification. DNA sequences were aligned by nucleotides using Muscle algorithm ( Edgar 2004 ) in SeaView v.4 ( Gouy et al. 2010 ) and by amino acids using TranslatorX ( Abascal et al. 2010 ).
Barcode clusters - Pairwise nucleotide sequence divergences and the mean intraspecific and interspecific distances were estimated using K2P distance ( Kimura 1980 ) implemented in MEGA v5 ( Tamura et al. 2011 ). From the 71 sequences, 63 unique sequences were recovered in DAMBE v.5.3.2 ( Xia & Xie 2001 ) and subsequently used to generate a neighbour-joining (NJ) tree, using MEGA v.5 ( Tamura et al. 2011 ) to evaluate the clustering pattern between species. The statistical support for the clusters obtained in the NJ tree was estimated using bootstrap support values (BSV) obtained with 1,000 bootstrap replicates.
Maximum parsimony (MP) was also employed to verify if MP lineages corroborate results obtained in the NJ topology. An. darlingi and Stegomyia aegypti (L.) were used as outgroups. MP analysis was carried out in MEGA v.5 ( Tamura et al. 2011 ), using the default parameters. Statistical support for the clades was estimated using BSV obtained in 1,000 bootstrap replicates.
COI and species identification - The usefulness of the COI gene for barcoding was tested using the BCM algorithm in TaxonDNA (taxondna.sf.net/). This algorithm involves matching the query sequence to the most similar barcode within a specified species threshold. TaxonDNA estimates the frequency distribution of the intraspecific distances to determine the threshold value below which 95% of all intraspecific distances are found. Queries without barcode match below the threshold value remained unidentified. Contrasting, those queries with match above the threshold value were considered a successful, ambiguous or incorrect identification. A correct identification was achieved if both names were identical, when at least two equally good best matches were found the identification was ambiguous and when the names were mismatched, the identification was a failure.
RESULTS
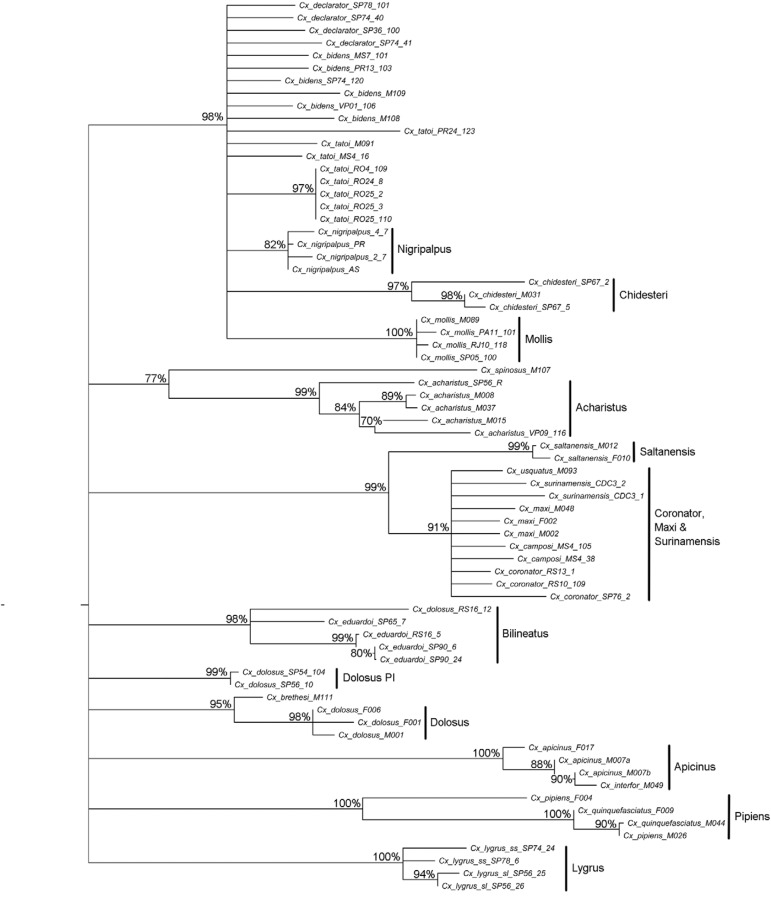
Barcode clusters - A pairwise K2P distance matrix was constructed for the 71 COI barcode sequences. K2P distances and means, between and within groups, are in Supplementary data 2, Table II , respectively. Twelve COI lineages were recovered ( Fig. 1 ), which did not entirely corroborate previously identified species and subgroups.
TABLE II. Mean pairwise Kimura-two-parameter inter and intraspecific distances between 22 Culex ( Culex ) species.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. Cx. pipiens | 3 | ||||||||||||||||||||
| 2. Cx. quinquefasciatus | 1.6 | 0.34 | |||||||||||||||||||
| 3. Cx. spinosus | 7.5 | 7.3 | - | ||||||||||||||||||
| 4. Cx. mollis | 7.8 | 7.7 | 4.8 | 0.09 | |||||||||||||||||
| 5. Cx. declarator | 6.7 | 6.8 | 4.3 | 1.6 | 0.51 | ||||||||||||||||
| 6. Cx. dolosus | 6.4 | 6.5 | 4.1 | 4.7 | 3.6 | 1.78 | |||||||||||||||
| 7. Cx. surinamensis | 7.9 | 7.9 | 6.6 | 6.3 | 5.3 | 5.0 | 0.34 | ||||||||||||||
| 8. Cx. eduardoi | 6.4 | 6.5 | 4.7 | 5.2 | 3.9 | 2.6 | 5.4 | 0.72 | |||||||||||||
| 9. Cx. coronator | 7.4 | 7.2 | 6.1 | 6.3 | 5.1 | 4.6 | 0.9 | 5.3 | 0.57 | ||||||||||||
| 10. Cx. camposi | 7.2 | 7.4 | 6.1 | 6.3 | 5.1 | 4.7 | 0.8 | 5.2 | 0.5 | 0.51 | |||||||||||
| 11. Cx. bidens | 6.7 | 6.8 | 4.2 | 1.7 | 0.4 | 3.5 | 5.3 | 3.7 | 5.0 | 5.0 | 0.26 | ||||||||||
| 12. Cx. brethesi | 6.5 | 6.6 | 3.8 | 4.1 | 3.3 | 1.5 | 4.6 | 3.0 | 4.1 | 4.1 | 3.2 | - | |||||||||
| 13. Cx. lygrus | 7.0 | 6.9 | 6.0 | 6.7 | 5.4 | 5.4 | 6.2 | 4.8 | 6.3 | 6.2 | 5.3 | 5.4 | 0.54 | ||||||||
| 14. Cx. maxi | 7.6 | 7.7 | 6.4 | 6.3 | 5.1 | 4.7 | 0.3 | 5.2 | 0.6 | 0.4 | 5.1 | 4.2 | 6.0 | 0 | |||||||
| 15. Cx. nigripalpus | 6.7 | 6.8 | 4.6 | 1.7 | 0.9 | 3.7 | 5.4 | 4.1 | 5.1 | 5.0 | 0.9 | 3.2 | 5.8 | 5.0 | 0.17 | ||||||
| 16. Cx. saltanensis | 7.5 | 7.0 | 5.9 | 6.0 | 5.1 | 4.5 | 2.2 | 5.4 | 1.8 | 1.9 | 5.0 | 3.9 | 6.5 | 2.0 | 5.1 | 0.17 | |||||
| 17. Cx. tatoi | 6.5 | 6.5 | 4.6 | 2.0 | 1.0 | 3.8 | 5.2 | 4.1 | 5.1 | 5.1 | 1.0 | 3.5 | 5.5 | 5.1 | 1.2 | 5.2 | 0.92 | ||||
| 18. Cx. usquatus | 7.4 | 7.3 | 6.2 | 6.1 | 4.9 | 4.6 | 0.9 | 5.3 | 0.6 | 0.6 | 4.9 | 4.0 | 6.4 | 0.5 | 4.8 | 1.8 | 5.0 | - | |||
| 19. Cx. chidesteri | 8.2 | 8.4 | 5.5 | 2.9 | 2.3 | 5.3 | 6.8 | 5.4 | 6.8 | 6.8 | 2.3 | 5.0 | 7.3 | 6.8 | 2.1 | 6.4 | 2.5 | 6.6 | 0.92 | ||
| 20. Cx. apicinus | 8.3 | 8.3 | 6.9 | 6.7 | 6.2 | 6.5 | 7.6 | 6.6 | 7.8 | 7.7 | 6.1 | 6.0 | 6. 9 | 8.0 | 6.6 | 7.9 | 6.0 | 8.0 | 7.1 | 0.34 | |
| 21. Cx. acharistus | 7.5 | 7.4 | 3.6 | 5.0 | 4.6 | 4.5 | 6.1 | 5.4 | 5.8 | 5.7 | 4.6 | 4.3 | 6.6 | 5.9 | 4.6 | 5.4 | 4.8 | 5.7 | 5.7 | 7.1 | 1.2 |
| 22. Cx. interfor | 8.7 | 8.7 | 7.3 | 7.0 | 6.5 | 6.8 | 7.9 | 6.9 | 8.1 | 8.0 | 6.4 | 6.4 | 7.3 | 8.3 | 6.9 | 8.2 | 6.3 | 8.3 | 7.4 | 0.3 | 7.5 |
numbers in boldface indicate intraspecific genetic divergence.
Fig. 1. : bootstrapped neighbour-joining tree of 71 cytochrome c oxidase subunit I (COI) sequences generated from specimens belonging to 22 Culex ( Culex ) species from Argentina and Brazil based on the Kimura two-parameter distance algorithm and 1,000 replicates. Bootstrap values less than 70% are not shown. The 12 COI lineages recovered are named.
COI K2P distance between sequences generated from individuals preliminarily identified as Cx . lygrus form 1 and Cx . lygrus form 2 of the Lygrus lineage ( Fig. 1 ) ranged from 0.2-0.9% (Supplementary data 2). The split leading to both individuals was strongly supported (100% BSV) ( Fig. 1 ) and the divergence with the closest species ( Table II ), Cx. eduardoi , was 8.7 times greater than the average intraspecific distance.
The Pipiens lineage included two individuals of each Cx. pipiens and Cx. quinquefasciatus from Argentina. The average K2P distance between these species was 1.6% (0-3%) (Supplementary data 2, Table II ), approximately 0.5 the value of mean distance between specimens of Cx. pipiens (3%). The COI sequences generated from individual M044 of Cx. quinquefasciatus from Formosa province and M026 of Cx. pipiens from Córdoba province share 100% similarity. In contrast, K2P distances between these two individuals and the remaining Cx. pipiens and Cx. quinquefasciatus sequences ranged between 0.3-3% (Supplementary data 2).
Three specimens of Cx. apicinus (Apicinus subgroup) ( Table I ) and one of Cx. interfor (Tarsalis subgroup) ( Table I ) were included in the analysis. For Cx . apicinus , K2P intraspecific distance varied from 0-0.5% (0.34%) (Supplementary data 2, Table II ). Interspecific divergence relative to the closest species (except Cx. interfor ), Cx. brethesi and Cx. tatoi was 18.2 times higher than the average intraspecific divergence.
The Dolosus cluster, composed of specimens of Cx. dolosus from Argentina, formed a strongly supported grouping (98% BSV) ( Fig. 1 ) that represents the stricto sensu population. COI K2P distance within the group varied from 0-0.2% (0.13%), whereas genetic distance between it and Cx. brethesi M111 was 7.46 times greater than the intragroup K2P distance. Moreover, the Dolosus cluster did not include specimens from Brazil, which were collected in one locality from the state of Rio Grande do Sul (RS) and two localities from the state of São Paulo (SP). The specimens from RS, Cx. eduardoi RS16-5 and Cx. dolosus RS16-12, clustered with three specimens from São Paulo municipality ( Fig. 1 ), which is the type-locality of Cx. bilineatus Theobald. The K2P distance within the lineage formed by RS and SP specimens varied from 0-2.3% (1.21%) and was supported by 98% BSV ( Fig. 1 ). This cluster was herein designated as Bilineatus lineage. K2P distance between the Bilineatus and Dolosus lineages ranged from 2.8-3.3% (2.52%). Additionally, two specimens designated as Cx . dolosus SP54-104 and Cx . dolosus SP56-10 clustered together in a separate lineage from the Dolosus and Bilineatus lineages ( Fig. 1 ). Considering that Cx . dolosus SP54-104 and Cx . dolosus SP56-10 were collected at Pico do Itapeva, Serra da Mantiqueira, southeastern Brazil, the cluster composed of these specimens was herein designated as the Dolosus Pico do Itapeva (PI) Lineage ( Fig. 1 ). The COI barcode sequences generated from Cx . dolosus SP54-104 and Cx . dolosus SP56-10 shared 100% similarity. However, the COI K2P distance between Dolosus PI and the Dolosus lineage from Argentina ranged from 1.7-1.9% (1.83%), whereas that from Bilineatus lineage was from 1.9-2.8% (2.36%).
The split leading to species of the Coronator group ( Cx. coronator s.s. , Cx. camposi and Cx. usquatus ) ( Table I ), plus Cx. maxi and Cx. surinamensis from the Tarsalis subgroup ( Table I ), was supported by 91% BSV ( Fig. 1 ), whereas the K2P distance ranged from 0-1.2% (0.54%). The Saltanensis lineage ( Fig. 1 ) included two sequences generated from individuals morphologically identified as Cx . saltanensis and genetic distance within the cluster was 0.2% (Supplementary data 2). The COI K2P distance between the Saltanensis lineage and the Coronator group, Cx. maxi and Cx. surinamensis ranged from 1.6-2.3% (1.94%) and was 9.7 times greater than any intraspecific distances.
The Acharistus lineage ( Fig. 1 ) was supported by 99% BSV. It contained sequences generated from individuals identified as Cx. acharistus that were collected in both Argentina and Brazil. Intraspecific COI K2P distance varied from 0.2-1.7% (1.21%), whereas the distance between the Cx . acharistus cluster and Cx. spinosus M107 ranged from 3.3-4.2% (3.64%). These values are three-fold the intraspecific COI sequence distance within Cx . acharistus. The COI K2P distance between the Acharistus and Dolosus lineages from Argentina and the Pico do Itapeva lineage ranged from 3.8-5.1% (4.46%), which are 3.25 and 3.68 times higher than intraspecific variability estimated for Cx . acharistus , respectively, whereas between the Acharistus and Bilineatus lineages the distance is higher than 4.9%.
The Mollis lineage ( Fig. 1 ) was supported by 100% BSV and included sequences generated from representatives of Cx . mollis from Argentina and Brazil. Sequences obtained from Cx . mollis M089 and Cx . mollis SP05-100 were unique COI haplotypes. The K2P distance between the Mollis and the closest lineage formed by Cx. decla- rator , Cx . nigripalpus and Cx. bidens was lower than 2% ( Table II ). The Nigripalpus grouping was moderately supported by 82% BSV ( Fig. 1 ) and contains individuals that were morphologically identified as Cx. nigripalpus . The intraspecific K2P distance ranged from 0-0.3% (0.18%). The closest species to the Nigripalpus grouping are Cx. declarator and Cx. bidens , with a mean COI K2P distance between them of 0.9% ( Table II ). Interspecific distance values were, respectively, 4.86 and 5.14 times higher than the variability within Cx. nigripalpus .
The Chidesteri lineage (97% of BSV) ( Fig. 1 ) included sequences generated from specimens of Cx . chidesteri . The K2P intraspecific distance ranged from 0.2-1.4% (0.93%), whereas the interspecific distance between the Chidesteri and Nigripalpus lineages was 2.46 times higher than the variability within the Chidesteri group ( Table II ).
The sequences from to Cx . bidens , Cx . declarator and Cx. tatoi were not included in a cluster ( Fig. 1 ). The mean variability within Cx. declarator (0.51%) ( Table II ) was higher than the mean divergence between Cx. declarator and Cx. bidens (0.4%) (Supplementary data 2). The greatest interspecific divergence was between Cx. bidens and Cx. tatoi , which varied from 0.7-1.9% (1.02%) (Supplementary data 2, Table II ) and represents 3.92 and 1.11 times of the variability within Cx. bidens and Cx. tatoi , respectively. The COI sequences from Cx. tatoi collected in the state of Rondônia clustered together ( Fig. 1 ).
The MP tree topology ( Fig. 2 ) and the NJ topology ( Fig. 1 ) are consistent. The 12 COI lineages were recovered with both analyses with similar BSV.
Fig. 2. : phylogenetic relationships of 71 cytochrome c oxidase subunit I (COI) sequences generated from specimens belonging to 22 Culex ( Culex ) species from Argentina and Brazil based on the maximum parsimony criterion and 1,000 replicates. Anopheles darlingi and Stegomyia aegypti were used as outgroup. Bootstrap values less than 70% are not shown. The 12 COI lineages recovered are named.
COI species identification - Ninety-five percent of intraspecific K2P distances were found within the interval between 0-2.8%, the higher value was used as cut-off to define the limit for species identification. Seventy-one mosquitoes were sequenced, 69.01% of which (49 sequences) were successfully identified in accordance with the BCM, whereas 9.85% (7 sequences) of all sequences were ambiguously identified, 18.3% (13 sequences) misidentified and the remaining 2.81% ( Cx. pipiens F004 and Cx. spinosus M107) had no match below 2.95% and remained unidentified ( Table III ).
TABLE III. Identification of species based on best close match employing cytochrome c oxidase subunit I sequences.
| Query | Identification | Result |
|---|---|---|
| Cx. acharistus M008 | Cx. acharistus M037 | S |
| Cx. acharistus M015 | Cx. acharistus M037 | S |
| Cx. acharistus M037 | Cx. acharistus M008 | S |
| Cx. acharistus SP56 R | Cx. acharistus M037 | S |
| Cx. acharistus VP09 116 | Cx. acharistus M015 | S |
| Cx. apicinus F017 | Cx. apicinus M007a | S |
| Cx. apicinus M007a | Cx. apicinus M007b | S |
| Cx. apicinus M007b | Cx. apicinus M007a | S |
| Cx. bidens M108 | Cx. bidens VP01 106 and one from different species | A |
| Cx. bidens M109 | Cx. bidens SP74 120 | S |
| Cx. bidens MS7 101 | Cx. bidens PR13 103 | S |
| Cx. bidens PR13 103 | Cx. bidens MS7 101 | S |
| Cx. bidens SP74 120 | Cx. bidens VP01 106 and two from different species | A |
| Cx. bidens VP01 106 | Cx. bidens SP74 120 and two from different species | A |
| Cx. brethesi M111 | Cx. dolosus F006 | M |
| Cx. camposi MS4 105 | Cx. coronator RS13 1 | M |
| Cx. camposi MS4 38 | Cx. coronator RS13 1 | M |
| Cx. chidesteri M031 | Cx. chidesteri SP67 5 | S |
| Cx. chidesteri SP67 2 | Cx. chidesteri M031 | S |
| Cx. chidesteri SP67 5 | Cx. chidesteri M031 | S |
| Cx. coronator RS10 109 | Cx. usquatus M093 | M |
| Cx. coronator RS13 1 | Cx. camposi MS4 105 | M |
| Cx. coronator SP76 2 | Cx. camposi MS4 105 | M |
| Cx. declarator SP36 100 | Cx. declarator SP74 40 and two from different species | A |
| Cx. declarator SP74 40 | Cx. declarator SP78 101 and two from different species | A |
| Cx. declarator SP74 41 | Cx. declarator SP74 40 and one from different species | A |
| Cx. declarator SP78 101 | Cx. declarator SP74 40 and two from different species | A |
| Cx. dolosus F001 | Cx. dolosus M001 | S |
| Cx. dolosus F006 | Cx. dolosus M001 | S |
| Cx. dolosus M001 | Cx. dolosus F001 and one other | S |
| Cx. dolosus RS16 12 | Cx. eduardoi SP65 7 | M |
| Cx. dolosus SP54 104 | Cx. dolosus SP56 10 | S |
| Cx. dolosus SP56 10 | Cx. dolosus SP54 104 | S |
| Cx. eduardoi RS16 5 | Cx. eduardoi SP90 24 | S |
| Cx. eduardoi SP65 7 | Cx. eduardoi RS16 5 | S |
| Cx. eduardoi SP90 24 | Cx. eduardoi SP90 6 | S |
| Cx. eduardoi SP90 6 | Cx. eduardoi SP90 24 | S |
| Cx. interfor M049 | Cx. apicinus M007b | M |
| Cx. lygrus sl SP56 25 | Cx. lygrus s.l. SP56 26 | S |
| Cx. lygr us sl SP56 26 | Cx. lygrus s.l. SP56 25 | S |
| Cx. lygrus ss SP74 24 | Cx. lygrus s.s. SP78 6 | S |
| Cx. lygrus ss SP78 6 | Cx. lygrus s.l. SP56 26 | S |
| Cx. maxi F002 | Cx. maxi M002 | S |
| Cx. maxi M002 | Cx. maxi F002 | S |
| Cx. maxi M048 | Cx. maxi F002 and one other | S |
| Cx. mollis M089 | Cx. mollis RJ10 118 and one other | S |
| Cx. mollis PA11 101 | Cx. mollis M089 and one other | S |
| Cx. mollis RJ10 118 | Cx. mollis M089 and one other | S |
| Cx. mollis SP05 100 | Cx. mollis M089 and one other | S |
| Cx. nigripalpus 2 7 | Cx. nigripalpus PR | S |
| Cx. nigripalpus 4 7 | Cx. nigripalpus PR | S |
| Cx. nigripalpus AS | Cx. nigripalpus PR | S |
| Cx. nigripalpus PR | Cx. nigripalpus AS | S |
| Cx. pipiens F004 | Outside threshold | U |
| Cx. pipiens M026 | Cx. quinquefasciatus M044 | M |
| Cx. quinquefasciatus F009 | Cx. pipiens M026 | M |
| Cx. quinquefasciatus M044 | Cx. pipiens M026 | M |
| Cx. saltanensis F010 | Cx. saltanensis M012 | S |
| Cx. saltanensis M012 | Cx. saltanensis F010 | S |
| Cx. spinosus M107 | Outside threshold | U |
| Cx. surinamensis CDC3 1 | Cx. surinamensis CDC3 2 | S |
| Cx. surinamensis CDC3 2 | Cx. maxi F002 and two others | M |
| Cx. tatoi M091 | Cx. tatoi RO24 8 and four others | S |
| Cx. tatoi MS4 16 | Cx. tatoi PR24 123 | S |
| Cx. tatoi PR24 123 | Cx. tatoi MS4 16 | S |
| Cx. tatoi RO24 8 | Cx. tatoi RO25 110 and three others | S |
| Cx. tatoi RO25 110 | Cx. tatoi RO24 8 and three others | S |
| Cx. tatoi RO25 2 | Cx. tatoi RO24 8 and three others | S |
| Cx. tatoi RO25 3 | Cx. tatoi RO24 8 and three others | S |
| Cx. tatoi RO4 109 | Cx. tatoi RO24 8 and three others | S |
| Cx. usquatus M093 | Cx. coronator RS10 109 | M |
A: ambiguous identification; M: misidentified; S: unsuccessfully identified; U: unidentified.
DISCUSSION
Morphological identification of Culex ( Cux. ) species, particularly from females, is problematic because of both polymorphism and overlap of characters state. Characteristics of fourth-instar larva are largely employed for species identification. However, similar to females, there are overlapping and absence of morphological differences among some species. Consequently, characteristics of the male genitalia usually allow a more accurate species identification. The major objective of the current study was to determine if the COI barcode fragment contains information for the identification of the species and species complexes within the subgenus Culex of the genus Culex , employing specimens from Argentina and Brazil.
Results of all analyses recovered 12 clusters, five of which confirm currently recognised species ( Cx. acharistus , Cx. chidesteri , Cx. dolosus , Cx. lygrus and Cx. saltanensis ). The remaining specimens were not correctly recovered as individuals of the morphologically identified species. Cx. lygrus seems to be a monophyletic species because the mean intraspecific COI K2P divergence is less than 2% and the degree of divergence with the closest species is higher than 4%. According to Ruiz-Lopez et al. (2012) , intraspecific divergence within anopheline species may vary from 0-< 2%, whereas Hebert et al. (2004) proposed that the divergence with the closest species should be at least 10 times higher than the average intraspecific genetic distance. The four Cx. lygrus sequences were successfully identified according to the BCM criterion. Based on morphological traits of the male genitalia, specimens of Cx . lygrus can be subdivided into two subgroups: Cx. lygrus form 1 ( Cx. lygrus SP74-24 and Cx. lygrus SP78-6) and Cx. lygrus form 2 ( Cx. lygrus SP56-25 and Cx. lygrus SP56-26). COI barcode sequences generated from Cx. lygrus form 1 clustered together with Cx. lygrus form 2 because the intergroup distance varied between 0.3-0.9% (0.6%) and the variability between both subgroups was only 0.86 and three times greater than the variation within Cx. lygrus form 1 and Cx. lygrus form 2, respectively (Supplementary data 2). Cx. lygrus was described by Root (1927) . The type-locality is Magé, state of Rio de Janeiro, Brazil. Comparing both ventral and dorsal divisions of the lateral plate of the male genitalia of Cx . lygrus forms 1 and 2 with that illustrated by Root (1927) , it seems that form 1 is morphologically more similar to the Magé specimen than form 2. However, in considering COI barcode sequence similarity, we can infer that specimens may belong to a single species and thus differences observed represent polymorphisms or COI barcode does not contain enough information to indicate Cx . lygrus forms 1 and 2 are distinct species.
The Pipiens subgroup belongs to the Pipiens group compiled by Harbach (2011) . The nominal subgroup is formed by Cx. australicus Dobrotworsky & Drummond, Cx. globocoxitus Dobrotworsky, Cx. pipiens pallens Coquillett, Cx. pipiens pipiens and Cx. quinquefasciatus . Cx. p. pipiens and Cx. quinquefasciatus occur in the Neotropics ( Sirivanakarn & White 1978 ). COI sequences generated from four individuals clustered together with 100% BSV. The K2P divergence within the Pipiens lineage varied from 0-3%. A similar K2P range (0.1-2.8%) was estimated for Cx. quinquefasciatus intraspecific divergence using a COI fragment of 556 bp generated by Quintero and Navarro (2012) . The most divergent individual was Cx. pipiens F004 from Argentina. Based on the BCM criterion ( Table III ), both Cx. quinquefasciatus sequences were misidentified as Cx. p. pipiens and Cx. pipiens M026 sequence was misidentified as Cx. quinquefasciatus M044. Cx. p. pipiens F004 could not be identified as either of the two species. The high intracluster variation due to the Cx. p. pipiens F004 sequence could be explained because Cx. p. pipiens F004 and Cx. p. pipiens M026 belong to different species complexes although no morphological differences were found between the specimens, which were collected at the same locality in Achiras, Córdoba province. On the other hand, it seems that the low COI K2P genetic distances observed between the two sequences of Cx. quinquefasciatus (F009 and M044) and Cx. p. pipiens M026 do not reflect the conspicuous morphological differences observed in the male genitalia. The absence of genetic divergence between two sequences geographically distant (M044 and M026) identified as separate species (and confirmed to rule out misidentification) and the great genetic distance between sequences from the same locality (F004 and M026) identified as Cx. pipiens support the idea of Reddy et al. (2012) about how controversial genomes of the Cx. pipiens complex are despite having been entirely sequenced ( Arensburger et al. 2010 ). Aditionally, intermediate forms between Cx. pipiens and Cx. quinquefasciatus were reported in Córdoba province ( Brewer et al. 1987 ), an established intergradation area which also include Santa Fe province ( Almirón et al. 1995 ). Due to our questionable results about the Pipiens lineage, we agree with Reddy et al. (2012) that comparative genomic studies will help to understand the incipient speciation in this species complex.
The cluster leading to Cx . apicinus and Cx . interfor is supported by 100% BSV, with the latter species forming a lineage with Cx . apicinus M007b. Cx. interfor is a species of the Tarsalis subgroup ( Table I ), whereas Cx . apicinus belongs to the Apicinus subgroup ( Table I ), both subgroups of the Pipiens group. Cx. interfor is morphologically similar to Cx. bidens . Identification of these species is problematic when employing only characteristics of the fourth-instar larva and female. However, Cx. apicinus is easily distinguished from other members of the subgenus based on features of the male genitalia ( Harbach et al. 1986 , Sallum et al. 1996 ), including Cx. inerfor based also on features of females and larvae ( Rossi et al. 2008 ). Consequently, having both species clustered together was a totally unexpected result that might be caused by both the limited information of the COI barcode fragment and poor resolution of the K2P distance analysis. Results of the BCM analyses show that Cx . apicinus is a good species.
The grouping herein designated as the Dolosus lineage ( Fig. 1 ) includes specimens from Argentina and supports the specific status of Cx. dolosus s.s. , a species included in the Salinarius complex of the Pipiens group ( Harbach 2011 ). It is noteworthy that Cx. dolosus s.s. clustered with Cx. brethesi from the Restuans complex ( Table I ). This was an unexpected result, mainly because these two species are easily recognised by traits of the male genitalia and fourth-instar larvae. Interestingly, individuals identified as Cx . dolosus from Pico do Itapeva clustered in a lineage that is separate from Cx . dolosus s.s. Moreover, individuals preliminarily identified as Cx . eduardoi from SP and RS and Cx. dolosus also from RS formed a strongly supported lineage, separate from individuals from Pico do Itapeva and Argentina. This lineage is herein designated as the Bilineatus cluster because it includes individuals from the type-locality of Cx . bilineatus in São Paulo municipality. The species was described by Theobald (1903) and synonymised with Cx . dolosus by Lane (1951) . However, our results suggest that Cx . bilineatus is a valid species that is morphologically similar to both Cx . dolosus s.s. and Cx . eduardoi . We therefore formally resurrect Cx . bilineatus from the synonymy of Cx . dolosus and retain Cx . eduardoi as a valid species until COI data can be obtained from individuals from its type locality in Perú. Regarding Cx . dolosus , it is noteworthy that Senise and Sallum (2008) demonstrated that the population from Pico do Itapeva is an undescribed species which has been misidentified as Cx . dolosus s.s. COI barcode data support Senise and Sallum’s (2008) hypothesis and the population from Pico do Itapeva will be formally named as a new species in another publication.
The COI lineage composed of Cx . camposi , Cx . coronator , Cx . maxi , Cx . usquatus , Cx . s altanensis and Cx . surinamensis is supported by 99% BSV. Cx. maxi , Cx . saltanensis and Cx . surinamensis are from the Tarsalis subgroup of the Pipiens group ( Table I ); whereas Cx . camposi , Cx . coronator and Cx . usquatus belong to the Coronator group ( Table I ). Two individuals of Cx . saltanensis formed a strongly supported lineage (99% BSV). Intraspecific COI K2P distance within Cx. saltanensis is on average lower than 2%. Furthermore, the interspecific distance ratio between Cx. saltanensis and the cluster composed of the Coronator lineage, Cx. maxi and Cx. surinamensis is approximately the value proposed by Hebert et al. (2004) . The BCM criterion supports Cx. saltanensis as a valid species that can be identified based on COI barcode sequence ( Table III ). Females of the three species share some morphological similarities, which make it difficult to distinguish these species. However, morphological characteristics of male genitalia and fourth-instar larvae, as well as COI barcode sequence, can be useful for an accurate identification. The remaining species are clustered into a moderately supported, unresolved lineage (91% BSV). Results of the analyses revealed that the COI barcode fragment does not contain enough information to identify these species. Moreover, it seems that the low COI K2P genetic distances observed among species from Argentina and Brazil do not reflect the conspicuous morphological differences observed in male genitalia and fourth-instar larvae. Despite that, the grouping consisting of Cx. coronator , Cx. maxi , Cx. surinamensis and Cx. saltanensis was not unexpected due to the fact that Laurito and Almirón (2013) found a close relationship between them and an unresolved polytomy comprised species of the Coronator group ( Cx. camposi , Cx. coronator , Cx. ousqua , Cx. usquatissimus and Cx. usquatus ) in their phylogenetic study of the subgenus Culex from Argentina based on morphological characters. In that work, the Pipiens group was recovered as polyphyletic relative to the Sitiens and the Coronator groups in contrast to the study of Harbach et al. (2012) in which the Pipiens group was recovered as monophyletic in the absence, however, of species of the Coronator group.
According to the criterion of Ruiz-Lopez et al. (2012) for the species status, Cx. acharistus seems to be a single species because the average intraspecific COI K2P divergence is less than 2% and the divergence from the closest species is between 2-5.6%. The five Cx. acharistus sequences were successfully identified according to the BCM criterion ( Table III ). Morphological features of the male genitalia, pupa and fourth-instar larva specimens were compared following Laurito et al. (2009) . This analysis showed that specimens from remote localities (Supplementary data 1) have the apical third of the gonostylus with minute annulations, the absence of dorsal and lateral arms of the phallosome, the typical shapes of dorsomentum and siphon and the seta 1-A near the middle of the antenna in larvae.
The mean interspecific divergence between Cx. mollis and the closest species is lower than 2%, but at least 25 times the divergence within the species. All the sequences of Cx. mollis were successfully identified based on the BCM criterion ( Table III ). Demari-Silva et al. (2011) distinguished and established relationships between 17 Culex species, using a shorter fragment than the one assessed in the present study: 478 bp of the COI gene, which includes part of the barcode region. The authors found a high intraspecific variation (0%-2.3%) between Cx. mollis sequences, indicating they comprise a complex. This idea was also supported by a deeper morphological analysis with striking differences in the male genitalia. The low intraspecific divergence (0.08%) and the correct identification of the Cx. mollis sequences included here leads us to conclude that the samples belong to one of the two complexes found by Demari-Silva et al. (2011) . The specimens are also similar morphologically and were collected in remote localities from Argentina and Brazil (Supplementary data 1).
Even though mean divergence between the Chidesteri and Nigripalpus lineages is 2.10%, the upper limit of the criterion of intraspecific distance of Ruiz-Lopez et al. (2012) , the intercluster divergence is 11 times higher than the intracluster divergence of the Nigripualpus lineage. The morphological differences of both male genitalia and fourth-instar larvae allow an unequivocal identification, as well as the COI barcode region included in the BCM analyses, which show that Cx . chidesteri and Cx. nigripalpus are separate species ( Table III ).
The remainder of the included sequences, which correspond to morphological specimens identified as Cx . bidens , Cx . declarator and Cx. tatoi , were not included in a cluster. The criteria of neither Ruiz-Lopez et al. (2012) nor Hebert et al. (2004) could be used to differentiate these morphological groups. On the other hand, all Cx. tatoi and most of the Cx. bidens sequences were successfully identified based on the BCM criterion, but not the Cx. declarator sequences which were ambiguously identified. As has been mentioned for other lineages in this study, the COI K2P genetic distances observed between the sequences of Cx. bidens , Cx. declarator and Cx. tatoi do not reflect the conspicuous morphological differences observed in male genitalia, revealing that the COI barcode fragment does not contain enough information to identify these species.
In accordance with Dai et al. (2012) and Bourke et al. (2013) , a single molecular marker can not successfully resolve and identify all the species included in a group or subgroup. However, in the latter study, a multilocus barcode composed of COI and ITS2 resolved all species in a NJ tree and successfully identified all specimens to species using the BCM approach. The use of combined datasets, including both multilocus and morphological strategies for phylogenetic analyses is proposed in order to identify those species which remain unclear, as was suggested for other dipteran families, such as Muscidae ( Renaud et al. 2012 ) and Sarcophagidae ( Meiklejohn et al. 2013 ).
In our study, only 42% of the samples were clustered with their conspecifics in the NJ tree and 69.01% of the sequences were successfully identified in accordance with the BCM criterion. Because the COI barcode fragment does not contain enough information to distinguish between morphologically well-defined species of the subgenus Culex from Argentina and Brazil, no interspecific boundary values could be established as in other Diptera ( Smith et al. 2006 , Rivera & Currie 2009 , Renaud et al. 2012 ), including Culicidae ( Cywinska et al. 2006 , Ruiz-Lopez et al. 2012 , Wang et al. 2012 ). Frey et al. (2013) revealed that COI fails to distinguish some tephritid sibling species, which may reflect a very recent ancestry. Regarding mosquito identification in the study of Kumar et al. (2007) , two closely related species of the genus Ochlerotatus could not be identified by the COI barcode region, which showed a negligible genetic divergence.
Regarding the current classification of the subgenus Culex , Harbach (2011) pointed out that the two groups which comprise the subgenus are both complex assemblages of species that do not fit readily into either group. Furthermore, Harbach et al. (2012) noted that evidence from their study and previous studies indicates that the subgenus is polyphyletic and has been retained as a taxon of convenience. The last classification review, on a worldwide sense, of the subgenus was done more than 80 years ago ( Edwards 1932 ). During this period, a lot of taxonomic studies carried out that diminish the number of unknown life stages of mosquito species, new molecular markers proved to be informative tools ( Miller et al. 1996 , Mukabayire et al. 1999 , Hebert et al. 2003a , Chen et al. 2004 , Yao et al. 2010 , Dai et al. 2012 , Morais et al. 2012 ) and stronger statistical methodologies have been developed that engender trust in morphologically based classifications (NJ, parsimony, maximum likelihood, bayesian inference). A natural classification will be achieved when further phylogenetic studies include integrated morphological and molecular character treatments, as many group members as possible and broader sampling of each species is undertaken to evaluate inherent polymorphism.
Supplementaty data 1
Information for species included in the study, including specimen numbers, localities, geographical coordinates and GenBank accessions.
| Species | Sample ID | Sex | Locality | State | Country | Latitude | Longitude | Collector | Identifier | Voucher location | GenBank |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cx. acharistus | M008 | ♂ | Chepes | LR | Argentina | 31º16’S | 66º35’60’’W | ML-AMV | WRA-ML | CIEC-UNC | KF919245 |
| Cx. acharistus | M037 | ♂ | Vaquerías | C | Argentina | 31º07’S | 64º33’’W | ML-AMV | WRA-ML | CIEC-UNC | KF919247 |
| Cx. acharistus | M015 | ♂ | San Carlos de Bariloche | RN | Argentina | 41º07’43.60’’S | 71º25’12.27’’W | MB | WRA | CIEC-UNC | KF919246 |
| Cx. acharistus | SP56_R | ♂ | Mairiporã | SP | Brazil | 23º19′08″S | 46º35’13”W | MAMS | MAMS | FSP-USP | KF919248 |
| Cx. acharistus | VP09_116 | ♂ | Pindamonhangaba | SP | Brazil | 22º45′31.7″S | 45º30’55.8’’W | MAMS | MAMS | FSP-USP | KF919249 |
| Cx. apicinus | F017 | ♀ | P.N. Lihué Calel | LP | Argentina | 38º00’13.5’’S | 65º35’47.4’’W | ML-AMV | WRA | CIEC-UNC | KF919250 |
| Cx. apicinus | M007a | ♂ | Buena Esperanza | SL | Argentina | 34º45’S | 65º15’W | IR | WRA | CIEC-UNC | KF919244 |
| Cx. apicinus | M007b | ♂ | Buena Esperanza | SL | Argentina | 34º45’S | 65º15’W | IR | WRA | CIEC-UNC | KF919251 |
| Cx. bidens | MS07_101 | ♂ | Aquidauana | MS | Brazil | 19º30′29.9″S | 55º37’42.4’’W | MAMS | MAMS | FSP-USP | KF919203 |
| Cx. bidens | PR13_103 | ♂ | Querência do Norte | PR | Brazil | 23º05′26.3″S | 53º30’15.5’’W | MAMS | MAMS | FSP-USP | KF919204 |
| Cx. bidens | SP74_120 | ♂ | Pariquera-Açu | SP | Brazil | 24º42′54″S | 47º52’52”W | MAMS | MAMS | FSP-USP | KF919205 |
| Cx. bidens | M109 | ♂ | Bernardo do Irigoyen | Mi | Argentina | 26º14’47.69’’S | 53º38’56.49’’W | JMD | JMD-GCR | CIEC-UNC | KF919202 |
| Cx. bidens | VP01_106 | ♂ | Aparecida do Norte | SP | Brazil | 22º50′34.4″S | 45º14’45.6’’W | MAMS | MAMS | FSP-USP | KF919206 |
| Cx. bidens | M108 | ♂ | La Plata | BA | Argentina | 34º55’3.24’’S | 58º1’16.46’’W | VM | GCR | CIEC-UNC | KF919201 |
| Cx. brethesi | M111 | ♂ | La Para | C | Argentina | 30º61’S | 62º55’W | ML-AMV | WRA-ML | CIEC-UNC | KF919207 |
| Cx. camposi | MS4_105 | ♀ | Aquidauana | MS | Brazil | 19º29′59.4″S | 55º36’33.8’’W | MAMS | MAMS | FSP-USP | KF919208 |
| Cx. camposi | MS4_38 | ♂ | Aquidauana | MS | Brazil | 19º29′59.4″S | 55º36’33.8’’W | MAMS | MAMS | FSP-USP | KF919200 |
| Cx. chidesteri | SP67_2 | ♀ | Dourado | SP | Brazil | 22º04′32.5″S | 48º26’14.5”W | MAMS | MAMS | FSP-USP | KF919252 |
| Cx. chidesteri | M031 | ♂ | San Pedro | Mi | Argentina | 26º39’21.2’’S | 54º10’46’’W | EAL | ML-WRA | FSP-USP | KF919243 |
| Cx. chidesteri | SP67_5 | ♂ | Dourado | SP | Brazil | 22º04′32.5″S | 48º26’14.5”W | MAMS | MAMS | FSP-USP | KF919253 |
| Cx. coronator s.s. | RS13_1 | ♂ | Trevo da Praia Grande | RS | Brazil | 29º13′06.2″S | 49º53’56.2”W | MAMS | MAMS | FSP-USP | KF919199 |
| Cx. coronator s.s. | RS10_109 | ♂ | Maquiné | RS | Brazil | 29º39′35.4″S | 50º13’03”W | MAMS | MAMS | FSP-USP | KF919209 |
| Cx. coronator s.s. | SP76_2 | ♂ | Pariquera-Açu | SP | Brazil | 24º42′54″S | 47º52’52”W | MAMS | MAMS | FSP-USP | KF919210 |
| Cx. declarator | SP78_101 | ♂ | Pariquera-Açu | SP | Brazil | 24º42′54″S | 47º52’52”W | MAMS | MAMS | FSP-USP | KF919194 |
| Cx. declarator | SP74_40 | ♂ | Pariquera-Açu | SP | Brazil | 24º42′54″S | 47º52’52”W | MAMS | MAMS | FSP-USP | KF919212 |
| Cx. declarator | SP36_100 | ♂ | Inúbia Paulista | SP | Brazil | 21º40′21.3″S | 50º57’40.2’’W | MAMS | MAMS | FSP-USP | KF919211 |
| Cx. declarator | SP74_41 | ♂ | Pariquera-Açu | SP | Brazil | 24º42′54″S | 47º52’52”W | MAMS | MAMS | FSP-USP | KF919193 |
| Cx. dolosus | SP54_104 | ♂ | Pindamonhangaba | SP | Brazil | 22º45′50″S | 45º30’87’’W | MAMS | MAMS | FSP-USP | KF919196 |
| Cx. dolosus | SP56_10 | ♂ | Mairiporã | SP | Brazil | 23º19′08″S | 46º35’13”W | MAMS | MAMS | FSP-USP | KF919216 |
| Cx. dolosus | F006 | ♀ | Achiras | C | Argentina | 33º09’59.5’’S | 64º59’22.4’’W | ML-AMV | WRA | CIEC-UNC | KF919214 |
| Cx. dolosus | F001 | ♀ | Tanti | C | Argentina | 31º21’17’’S | 64º35’32.8’’W | WRA-ML | WRA | CIEC-UNC | KF919213 |
| Cx. dolosus | M001 | ♂ | Tanti | C | Argentina | 31º21’17’’S | 64º35’32.8’’W | WRA-ML | WRA | CIEC-UNC | KF919215 |
| Cx. dolosus | RS16_12 | ♂ | São Franscisco de Paula | RS | Brazil | 29º29′50.3″S | 50º21’04.1”W | MAMS | MAMS | FSP-USP | KF919195 |
| Cx. eduardoi | SP65_7 | ♀ | Dourado | SP | Brazil | 22º08′04.9″S | 48º23’30.2”W | MAMS | MAMS | FSP-USP | KF919217 |
| Cx. eduardoi | RS16_5 | ♂ | São Franscisco de Paula | RS | Brazil | 29º29′50.3″S | 50º21’04.1”W | MAMS | MAMS | FSP-USP | KF919198 |
| Cx. eduardoi | SP90_6 | ♂ | São Paulo | SP | Brazil | 23º32′51″S | 46º38’10”W | MAMS | MAMS | FSP-USP | KF919218 |
| Cx. eduardoi | SP90_24 | ♂ | São Paulo | SP | Brazil | 23º32′51″S | 46º38’10”W | MAMS | MAMS | FSP-USP | KF919219 |
| Cx. interfor | M049 | ♂ | Altos de Chipión | C | Argentina | 30º54’S | 62º18’W | ML-AMV | WRA-ML | CIEC-UNC | KF919254 |
| Cx. lygrus s.l. | SP56_26 | ♀ | Mairiporã | SP | Brazil | 23º19′08″ S | 46º35’13” W | MAMS | MAMS | FSP-USP | KF919223 |
| Cx. lygrus s.l. | SP56_25 | ♀ | Mairiporã | SP | Brazil | 23º19′08″ S | 46º35’13” W | MAMS | MAMS | FSP-USP | KF919222 |
| Cx. lygrus s.s. | SP74_24 | ♂ | Pariquera-Açu | SP | Brazil | 24º42′54″S | 47º52’52”W | MAMS | MAMS | FSP-USP | KF919220 |
| Cx. lygrus s.s. | SP78_6 | ♂ | Pariquera-Açu | SP | Brazil | 24º42′54″S | 47º52’52”W | MAMS | MAMS | FSP-USP | KF919221 |
| Cx. maxi | M048 | ♂ | La Para | C | Argentina | 30º60’S | 62º56’W | ML-AMV | WRA | CIEC-UNC | KF919226 |
| Cx. maxi | F002 | ♀ | Chamical | LR | Argentina | 30º21’S | 52º17’W | ML-AMV | WRA | CIEC-UNC | KF919224 |
| Cx. maxi | M002 | ♂ | Chamical | LR | Argentina | 30º22’S | 66º19’W | ML-AMV | WRA | CIEC-UNC | KF919225 |
| Cx. mollis | M089 | ♂ | P.N. Iguazú | Mi | Argentina | 25º39’17.77’’S | 54º27’26.29’’W | EAL | GCR | CIEC-UNC | KF919255 |
| Cx. mollis | PA11_101 | ♂ | Belterra | PA | Brazil | 2º37′58.5″S | 54º58’32.9’’W | MAMS | MAMS | FSP-USP | KF919256 |
| Cx. mollis | RJ10_118 | ♂ | Nova Iguaçu | RJ | Brazil | 22º45′37.16″S | 43º26′51.82″W | MAMS | MAMS | FSP-USP | KF919192 |
| Cx. mollis | SP05_100 | ♂ | Pariquera-Açu | SP | Brazil | 24º44′48″S | 47º56’55’’W | MAMS | MAMS | FSP-USP | KF919257 |
| Cx. nigripalpus | 4_7 | ♀ | Pariquera-Açu | SP | Brazil | 24º42′54″S | 47º52’52”W | MAMS | MAMS | FSP-USP | KF919228 |
| Cx. nigripalpus | PR | ♂ | Parelheiros | SP | Brazil | 23º51’00.00’’S | 46º43’58.70’’W | MAMS | MAMS | FSP-USP | KF919231 |
| Cx. nigripalpus | 2_7 | ♀ | Pariquera-Açu | SP | Brazil | 24º42′54″S | 47º52’52”W | MAMS | MAMS | FSP-USP | KF919227 |
| Cx. nigripalpus | AS | ♂ | São Paulo | SP | Brazil | 23º32’40.32’’S | 46º43’58.70’’W | MAMS | MAMS | FSP-USP | KF919229 |
| Cx. pipiens | F004 | ♀ | Achiras | C | Argentina | 33º10’26.6’’S | 64º57’39.4’’W | ML-AMV | WRA | CIEC-UNC | KF919187 |
| Cx. pipiens | M026 | ♂ | Achiras | C | Argentina | 33º10’26.6’’S | 64º57’39.4’’W | ML-AMV | WRA | CIEC-UNC | KF919189 |
| Cx. quinquefasciatus | F009 | ♀ | Córdoba | C | Argentina | 31º21’S | 64º05’W | WRA | ML-WRA | CIEC-UNC | KF919190 |
| Cx. quinquefasciatus | M044 | ♂ | Clorinda | Fo | Argentina | 25º17’11’’S | 57º43’02’’W | GA | WRA | CIEC-UNC | KF919188 |
| Cx. saltanensis | M012 | ♂ | Chamical | LR | Argentina | 30º14’58.8’’S | 66º25’44.5’’W | ML-AMV | WRA-ML | CIEC-UNC | KF919230 |
| Cx. saltanensis | F010 | ♀ | Altos de Chipión | C | Argentina | 31º29’18’’S | 64º17’23’’W | ML-AMV | WRA | CIEC-UNC | KF919232 |
| Cx. spinosus | M107 | ♂ | Posadas | Mi | Argentina | 27º25’59.85’’S | 55º53’40.16’’W | GCR | GCR | CIEC-UNC | KF919191 |
| Cx. surinamensis | CDC3_2 | ♂ | Cacaulândia | RO | Brazil | 10º20′21″ S | 62º53’43” W | MAMS | MAMS | FSP-USP | KF919197 |
| Cx. surinamensis | CDC3_1 | ♂ | Cacaulândia | RO | Brazil | 10º20′21″ S | 62º53’43” W | MAMS | MAMS | FSP-USP | KF919233 |
| Cx. tatoi | PR24_123 | ♂ | Doutor Camargo | PR | Brazil | 23º35′51.4″ S | 52º17’31.4’’W | MAMS | MAMS | FSP-USP | KF919236 |
| Cx. tatoi | M091 | ♂ | Corpus | Mi | Argentina | 27º06’22.90’’S | 55º31’19.73’’W | RSt | GCR | CIEC-UNC | KF919234 |
| Cx. tatoi | MS4_16 | ♂ | Aquidauana | MS | Brazil | 19º29′59.4″S | 55º36’33.8’’W | MAMS | MAMS | FSP-USP | KF919235 |
| Cx. tatoi | RO4_109 | ♂ | Monte Negro | RO | Brazil | 10º17′56.1″S | 63º14’22.5’’W | MAMS | MAMS | FSP-USP | KF919237 |
| Cx. tatoi | RO24_8 | ♂ | Monte Negro | RO | Brazil | 10º18′03.5″S | 63º14’09.1’’W | MAMS | MAMS | FSP-USP | KF919238 |
| Cx. tatoi | RO25_2 | ♂ | Monte Negro | RO | Brazil | 10º18′03.5″S | 63º14’09.1’’W | MAMS | MAMS | FSP-USP | KF919239 |
| Cx. tatoi | RO25_3 | ♂ | Monte Negro | RO | Brazil | 10º18′03.5″S | 63º14’09.1’’W | MAMS | MAMS | FSP-USP | KF919240 |
| Cx. tatoi | RO25_110 | ♂ | Monte Negro | RO | Brazil | 10º18′03.5″S | 63º14’09.1’’W | MAMS | MAMS | FSP-USP | KF919241 |
| Cx. usquatus | M093 | ♂ | Puerto Iguazú | Mi | Argentina | 25º36’36.42’’S | 54º33’37’’W | EAL | GCR | CIEC-UNC | KF919242 |
AMV: A.M. Visintin; BA: Buenos Aires; C: Córdoba; CIEC: Entomological Research Center of Cordoba; EAL: E.A. Lestani; Fo: Formosa; FSP: School of Public Health; GA: G. Albrieu; GCR: G.C. Rossi; IR: I. Roccia; JMD: J.M D’oria; LP: La Pampa; LR: La Rioja; MAMS: M.A.M. Sallum; MB: M. Bonino; Mi: Misiones; ML: M. Laurito; MS: Mato Grosso do Sul; PA: Pará; P.N.: Parque Nacional; PR: Paraná; RJ: Rio de Janeiro; RN: Río Negro; RO: Rondônia; RS: Rio Grande do Sul; RSt: R. Stetson; SL: San Luis; SP: São Paulo; UNC: National University of Córdoba; USP: São Paulo University; VM: V. Micielli; WRA: W.R. Almirón.
Supplementaty data 2
Pairwise Kimura two-parameter distances between 71 cytochrome c oxidase subunit I sequences of Culex ( Culex ) specimens from Argentina and Brazil.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 . Cx. pipiens F004 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| 2. Cx. quinquefasciatus M044 | 2.96 | |||||||||||||||||||||||||||||||||||||||||||||||||
| 3. Cx. pipiens M026 | 2.96 | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4. Cx. quinquefasciatus F009 | 2.96 | 0.34 | 0.34 | |||||||||||||||||||||||||||||||||||||||||||||||
| 5. Cx. spinosus M107 | 7.51 | 7.51 | 7.51 | 7.13 | ||||||||||||||||||||||||||||||||||||||||||||||
| 6. Cx. mollis RJ10 118 | 7.70 | 7.89 | 7.89 | 7.51 | 4.74 | |||||||||||||||||||||||||||||||||||||||||||||
| 7. Cx. declarator SP74 41 | 6.57 | 6.76 | 6.76 | 6.39 | 4.74 | 1.72 | ||||||||||||||||||||||||||||||||||||||||||||
| 8. Cx. declarator SP78 101 | 6.39 | 6.94 | 6.94 | 6.57 | 4.20 | 1.55 | 0.86 | |||||||||||||||||||||||||||||||||||||||||||
| 9. Cx. dolosus RS16 12 | 6.21 | 6.96 | 6.96 | 6.58 | 5.29 | 5.67 | 4.93 | 4.20 | ||||||||||||||||||||||||||||||||||||||||||
| 10 . Cx. dolosus SP54 104 | 5.65 | 5.83 | 5.83 | 5.47 | 3.666 | 4.02 | 3.30 | 2.59 | 2.80 | |||||||||||||||||||||||||||||||||||||||||
| 11. Cx. surinamensis CDC3 2 | 7.51 | 8.08 | 8.08 | 7.70 | 6.57 | 6.39 | 5.65 | 5.11 | 5.84 | 4.20 | ||||||||||||||||||||||||||||||||||||||||
| 12. Cx. eduardoi RS16 5 | 6.40 | 6.76 | 6.76 | 6.39 | 4.74 | 5.11 | 4.389 | 3.66 | 2.26 | 2.26 | 5.47 | |||||||||||||||||||||||||||||||||||||||
| 13. Cx. coronator RS13 1 | 6.94 | 7.51 | 7.51 | 7.13 | 6.02 | 6.20 | 5.47 | 4.92 | 5.66 | 3.66 | 0.51 | 5.29 | ||||||||||||||||||||||||||||||||||||||
| 14. Cx. camposi MS4 38 | 6.76 | 7.70 | 7.70 | 7.32 | 6.20 | 6.20 | 5.47 | 4.92 | 5.84 | 4.20 | 0.68 | 5.47 | 0.51 | |||||||||||||||||||||||||||||||||||||
| 15. Cx. bidens M108 | 6.76 | 7.32 | 7.32 | 6.94 | 4.56 | 1.90 | 1.20 | 0.34 | 4.56 | 2.95 | 5.45 | 4.02 | 5.28 | 5.28 | ||||||||||||||||||||||||||||||||||||
| 16. Cx. bidens M109 | 6.57 | 7.13 | 7.13 | 6.76 | 4.38 | 1.72 | 1.03 | 0.17 | 4.38 | 2.77 | 5.28 | 3.84 | 5.10 | 5.10 | 0.51 | |||||||||||||||||||||||||||||||||||
| 17. Cx. bidens MS7 101 | 6.20 | 6.76 | 6.76 | 6.39 | 4.02 | 1.72 | 1.03 | 0.17 | 4.02 | 2.42 | 5.28 | 3.48 | 4.78 | 5.10 | 0.51 | 0.34 | ||||||||||||||||||||||||||||||||||
| 18. Cx. bidens PR13 103 | 6.20 | 6.76 | 6.76 | 6.39 | 4.02 | 1.72 | 1.03 | 0.17 | 4.02 | 2.42 | 5.28 | 3.48 | 4.74 | 5.10 | 0.514 | 0.34 | 0 | |||||||||||||||||||||||||||||||||
| 19. Cx. bidens SP74 120 | 6.39 | 6.94 | 6.94 | 6.57 | 4.20 | 1.55 | 0.86 | 0 | 4.02 | 2.59 | 5.10 | 3.66 | 4.992 | 4.92 | 0.34 | 0.17 | 0.17 | 0.17 | ||||||||||||||||||||||||||||||||
| 20. Cx. bidens VP01 106 | 6.39 | 6.94 | 6.94 | 6.57 | 4.20 | 1.55 | 0.86 | 0 | 4.02 | 2.59 | 5.10 | 3.66 | 4.992 | 4.92 | 0.34 | 0.17 | 0.172 | 0.17 | 0 | |||||||||||||||||||||||||||||||
| 21. Cx. brethesi M111 | 6.20 | 6.76 | 6.76 | 6.39 | 3.89 | 4.02 | 3.66 | 3.12 | 2.96 | 1.55 | 4.38 | 3.14 | 3.89 | 4.38 | 3.48 | 3.30 | 2.95 | 2.95 | 3.12 | 3.12 | ||||||||||||||||||||||||||||||
| 22. Cx. camposi MS4 105 | 6.94 | 7.51 | 7.51 | 7.13 | 6.02 | 6.201 | 5.47 | 4.92 | 5.66 | 3.66 | 0.51 | 5.29 | 0 | 0.51 | 5.28 | 5.10 | 4.74 | 4.74 | 4.92 | 4.92 | 3.84 | |||||||||||||||||||||||||||||
| 23. Cx. coronator RS10 109 | 7.70 | 7.13 | 7.13 | 6.76 | 6.02 | 5.83 | 5.10 | 4.56 | 6.03 | 3.66 | 0.86 | 5.29 | 0.69 | 0.86 | 4.92 | 4.74 | 4.74 | 4.74 | 4.56 | 4.56 | 4.20 | 0.68 | ||||||||||||||||||||||||||||
| 24. Cx. coronator SP76 2 | 7.32 | 7.51 | 7.51 | 7.13 | 6.39 | 6.58 | 5.88 | 5.28 | 6.039 | 4.02 | 0.86 | 5.66 | 0.34 | 0.86 | 5.65 | 5.47 | 5.10 | 5.10 | 5.28 | 5.285 | 4.20 | 0.34 | 0.68 | |||||||||||||||||||||||||||
| 25. Cx. declarator SP36 100 | 6.57 | 7.13 | 7.13 | 6.76 | 4.20 | 1.55 | 1.03 | 0.17 | 4.38 | 2.77 | 5.28 | 3.84 | 5.10 | 5.10 | 0.51 | 0.34 | 0.34 | 0.34 | 0.17 | 0.17 | 3.12 | 5.10 | 4.74 | 5.47 | ||||||||||||||||||||||||||
| 26. Cx. declarator SP74 40 | 6.39 | 6.94 | 6.94 | 6.57 | 4.20 | 1.55 | 0.86 | 0 | 4.20 | 2.59 | 5.10 | 3.67 | 4.92 | 4.92 | 0.34 | 0.17 | 0.17 | 0.17 | 0 | 0 | 3.12 | 4.92 | 4.56 | 5.23 | 0.17 | |||||||||||||||||||||||||
| 27. Cx. dolosus F001 | 6.58 | 7.14 | 7.14 | 6.76 | 4.02 | 4.75 | 4.38 | 3.84 | 3.32 | 1. 09 | 5.10 | 3.14 | 4.56 | 5.10 | 4.20 | 4.02 | 3.66 | 3.66 | 3.84 | 3.89 | 1.03 | 4.56 | 4.92 | 4.92 | 3.84 | |||||||||||||||||||||||||
| 28. Cx. dolosus F006 | 6.39 | 6.95 | 6.95 | 6.58 | 3.84 | 4.56 | 4.20 | 3.66 | 3.14 | 1.72 | 4.92 | 2.96 | 4.38 | 4.92 | 4.02 | 3.84 | 3.48 | 3.48 | 3.66 | 3.66 | 0.85 | 4.38 | 4.74 | 4.74 | 3.66 | |||||||||||||||||||||||||
| 19. Cx. dolosus M001 | 6.58 | 7.14 | 7.14 | 6.765 | 4.02 | 4.75 | 4.38 | 3.84 | 3.32 | 1. 09 | 5.10 | 3.14 | 4.56 | 5.10 | 4.20 | 4.02 | 3.66 | 3.66 | 3.84 | 3.84 | 1.03 | 4.56 | 4.92 | 4.92 | 3.84 | |||||||||||||||||||||||||
| 30. Cx. dolosus SP56 10 | 5.65 | 5.83 | 5.83 | 5.47 | 3.66 | 4.018 | 3.30 | 2.59 | 2.80 | 0 | 4.20 | 2.26 | 3.66 | 4.20 | 2.95 | 2.77 | 2.42 | 2.42 | 2.59 | 2.56 | 1.55 | 3.66 | 3.66 | 4.02 | 2.77 | |||||||||||||||||||||||||
| 31. Cx. eduardoi SP65 7 | 6.02 | 6.02 | 6.02 | 5.65 | 4.38 | 4.74 | 4.02 | 3.30 | 1.20 | 1.90 | 5.29 | 1.20 | 5.10 | 5.29 | 3.66 | 3.49 | 3.12 | 3.12 | 3.30 | 3.30 | 2.78 | 5.10 | 5.10 | 5.476 | 3.48 | |||||||||||||||||||||||||
| 32. Cx. eduardoi SP90 6 | 6.21 | 6.95 | 6.95 | 6.58 | 4.92 | 5.29 | 4.56 | 3.84 | 2.08 | 2.43 | 5.29 | 0.17 | 5.10 | 5.29 | 4.20 | 4.02 | 3.66 | 3.66 | 3.84 | 3.84 | 2.96 | 5.10 | 5.47 | 5.47 | 4.02 | |||||||||||||||||||||||||
| 33. Cx. eduardoi SP90 24 | 6.21 | 6.95 | 6.95 | 6.58 | 4.92 | 5.29 | 4.56 | 3.84 | 2.08 | 2.43 | 5.29 | 0.17 | 5.10 | 5.29 | 4.20 | 4.02 | 3.66 | 3.66 | 3.84 | 3.84 | 2.96 | 5.10 | 5.47 | 5.47 | 4.02 | |||||||||||||||||||||||||
| 34. Cx. lygrus ss SP74 24 | 7.32 | 7.51 | 7.51 | 7.13 | 6.39 | 6.76 | 5.65 | 5.29 | 5.65 | 5.10 | 6.02 | 5.10 | 6.20 | 6.02 | 5.65 | 5.47 | 5.47 | 5.47 | 5.29 | 5.29 | 5.65 | 6.20 | 6.02 | 6.57 | 5.47 | |||||||||||||||||||||||||
| 35. Cx. lygrus ss SP78 6 | 6.76 | 6.94 | 6.94 | 6.57 | 5.83 | 6.57 | 5.47 | 5.11 | 5.10 | 4.56 | 6.20 | 4.74 | 6.02 | 6.20 | 5.47 | 5.29 | 4.93 | 4.93 | 5.11 | 5.11 | 5.47 | 6.02 | 6.20 | 6.3 | 5.29 | |||||||||||||||||||||||||
| 36. Cx. lygrus sl SP56 25 | 6.94 | 7.13 | 7.13 | 6.76 | 6.02 | 6.76 | 6.02 | 5.29 | 5.29 | 4.74 | 6.39 | 4.92 | 6.20 | 6.39 | 5.65 | 5.47 | 5.11 | 5.11 | 5.23 | 5.29 | 5.28 | 6.20 | 6.39 | 6.57 | 5.47 | |||||||||||||||||||||||||
| 37. Cx. lygrus sl SP56 26 | 6.76 | 6.94 | 6.94 | 6.57 | 5.83 | 6.57 | 5.83 | 5.11 | 5.10 | 4.56 | 6.20 | 4.74 | 6.02 | 6.20 | 5.47 | 5.29 | 4.93 | 4.93 | 5.11 | 5.11 | 5.10 | 6.02 | 6.20 | 6.39 | 5.29 | |||||||||||||||||||||||||
| 38. Cx. maxi F002 | 7.32 | 7.89 | 7.89 | 7.51 | 6.39 | 6.20 | 5.47 | 4.92 | 5.66 | 4.02 | 0.17 | 5.29 | 0.345 | 0.51 | 5.28 | 5.10 | 5.10 | 5.10 | 4.92 | 4.92 | 4.20 | 0.34 | 0.68 | 0.68 | 5.10 | |||||||||||||||||||||||||
| 39. Cx. maxi M002 | 7.32 | 7.89 | 7.89 | 7.51 | 6.39 | 6.20 | 5.47 | 4.92 | 5.66 | 4.02 | 0.17 | 5.29 | 0.34 | 0.51 | 5.28 | 5.10 | 5.10 | 5.10 | 4.92 | 4.92 | 4.20 | 0.34 | 0.68 | 0.68 | 5.10 | |||||||||||||||||||||||||
| 40. Cx. maxi M048 | 7.32 | 7.89 | 7.89 | 7.51 | 6.39 | 6.20 | 5.47 | 4.92 | 5.66 | 4.02 | 0.17 | 5.29 | 0.34 | 0.51 | 5.28 | 5.10 | 5.10 | 5.10 | 4.92 | 4.92 | 4.20 | 0.34 | 0.68 | 0.68 | 5.10 | |||||||||||||||||||||||||
| 41. Cx. nigripalpus 2 7 | 6.39 | 6.94 | 6.94 | 6.57 | 4.56 | 1.72 | 1.03 | 0.86 | 4.38 | 3.12 | 5.28 | 4.20 | 5.109 | 5.10 | 1.20 | 1.03 | 1.03 | 1.03 | 0.86 | 0.86 | 3.30 | 5.10 | 5.10 | 5.47 | 1.03 | |||||||||||||||||||||||||
| 42. Cx. nigripalpus 4 7 | 6.57 | 7.13 | 7.13 | 6.76 | 4.74 | 1.722 | 1.03 | 0.86 | 4.56 | 3.30 | 5.28 | 4.38 | 5.10 | 5.10 | 1.20 | 1.03 | 1.03 | 1.03 | 0.86 | 0.86 | 3.30 | 5.10 | 5.10 | 5.47 | 1.03 | |||||||||||||||||||||||||
| 43. Cx. nigripalpus AS | 6.39 | 6.94 | 6.94 | 6.57 | 4.56 | 1.55 | 0.86 | 0.68 | 4.38 | 3.12 | 5.10 | 4.20 | 4.92 | 4.92 | 1.03 | 0.86 | 0.86 | 0.86 | 0.68 | 0.68 | 3.12 | 4.92 | 4.92 | 5.28 | 0.86 | |||||||||||||||||||||||||
| 44. Cx. saltanensis M012 | 7.70 | 7.13 | 7.13 | 6.79 | 5.83 | 5.83 | 5.10 | 4.92 | 5.66 | 3.66 | 2.25 | 5.29 | 1.73 | 2.25 | 5.28 | 5.10 | 4.74 | 4.74 | 4.92 | 4.92 | 3.84 | 1.72 | 1.72 | 2.08 | 5.10 | |||||||||||||||||||||||||
| 45. Cx. nigripalpus PR | 6.39 | 6.94 | 6.94 | 6.57 | 4.56 | 1.55 | 0.86 | 0.68 | 4.38 | 3.12 | 5.10 | 4.20 | 4.92 | 4.92 | 1.03 | 0.86 | 0.86 | 0.86 | 0.68 | 0.68 | 3.12 | 4.92 | 4.92 | 5.28 | 0.86 | |||||||||||||||||||||||||
| 46. Cx. saltanensis F010 | 7.89 | 7.32 | 7.32 | 6.94 | 6.02 | 6.02 | 5.28 | 5.10 | 5.84 | 3.84 | 2.08 | 5.47 | 1.55 | 2.08 | 5.47 | 5.28 | 4.92 | 4.92 | 5.10 | 5.10 | 4.02 | 1.55 | 1.55 | 1.90 | 5.28 | |||||||||||||||||||||||||
| 47. Cx. surinamensis CDC3 1 | 7.89 | 8.08 | 8.08 | 7.70 | 6.57 | 6.02 | 5.65 | 5.10 | 6.22 | 4.20 | 0.34 | 5.47 | 0.86 | 1.03 | 5.47 | 5.28 | 5.28 | 5.28 | 5.10 | 5.10 | 4.74 | 0.86 | 0.86 | 1.20 | 5.28 | |||||||||||||||||||||||||
| 48. Cx. tatoi M091 | 5.83 | 6.76 | 6.76 | 6.39 | 4.38 | 2.43 | 1.38 | 1.20 | 4.20 | 2.95 | 5.65 | 3.66 | 5.10 | 5.47 | 1.55 | 1.38 | 1.03 | 1.03 | 1.20 | 1.20 | 3.30 | 5.10 | 5.47 | 5.47 | 1.38 | |||||||||||||||||||||||||
| 49. Cx. tatoi MS4 16 | 6.20 | 6.39 | 6.39 | 6.02 | 4.38 | 1.72 | 1.03 | 0.86 | 4.57 | 2.59 | 4.92 | 4.02 | 4.74 | 4.74 | 1.20 | 1.03 | 1.03 | 1.03 | 0.86 | 0.86 | 3.30 | 4.74 | 4.38 | 5.10 | 1.03 | |||||||||||||||||||||||||
| 50. Cx. tatoi PR24 123 | 6.95 | 6.76 | 6.76 | 6.39 | 5.10 | 2.07 | 1.73 | 1.55 | 4.94 | 3.304 | 4.92 | 4.38 | 5.10 | 5.10 | 1.90 | 1.73 | 1.73 | 1.73 | 1.55 | 1.55 | 4.02 | 5.10 | 4.74 | 5.47 | 1.73 | |||||||||||||||||||||||||
| 51. Cx. tatoi RO4 109 | 6.20 | 6.76 | 6.76 | 6.39 | 4.56 | 1.90 | 0.86 | 0.68 | 4.75 | 3.12 | 5.28 | 4.20 | 5.10 | 5.10 | 1.03 | 0.86 | 0.86 | 0.86 | 0.68 | 0.68 | 3.48 | 5.10 | 4.92 | 5.47 | 0.86 | |||||||||||||||||||||||||
| 52. Cx. tatoi RO24 8 | 6.20 | 6.76 | 6.76 | 6.39 | 4.56 | 1.90 | 0.86 | 0.68 | 4.75 | 3.12 | 5.28 | 4.20 | 5.10 | 5.10 | 1.03 | 0.86 | 0.86 | 0.86 | 0.68 | 0.68 | 3.48 | 5.10 | 4.92 | 5.47 | 0.86 | |||||||||||||||||||||||||
| 53. Cx. tatoi RO25 2 | 6.20 | 6.76 | 6.76 | 6.39 | 4.56 | 1.90 | 0.86 | 0.68 | 4.75 | 3.12 | 5.28 | 4.20 | 5.10 | 5.10 | 1.3 | 0.86 | 0.86 | 0.86 | 0.68 | 0.68 | 3.48 | 5.10 | 4.92 | 5.47 | 0.86 | |||||||||||||||||||||||||
| 54. Cx. tatoi RO25 3 | 6.20 | 6.76 | 6.76 | 6.39 | 4.56 | 1.90 | 0.86 | 0.68 | 4.75 | 3.12 | 5.28 | 4.20 | 5.10 | 5.10 | 1.03 | 0.86 | 0.86 | 0.86 | 0.68 | 0.68 | 3.48 | 5.10 | 4.92 | 5.47 | 0.86 | |||||||||||||||||||||||||
| 55. Cx. tatoi RO25 110 | 6.20 | 6.76 | 6.76 | 6.39 | 4.56 | 1.90 | 0.86 | 0.68 | 4.75 | 3.12 | 5.28 | 4.20 | 5.10 | 5.10 | 1.03 | 0.86 | 0.86 | 0.86 | 0.68 | 0.68 | 3.48 | 5.10 | 4.92 | 5.47 | 0.86 | |||||||||||||||||||||||||
| 56. Cx. usquatus M093 | 7.32 | 7.51 | 7.51 | 7.13 | 6.20 | 6.02 | 5.28 | 4.74 | 5.84 | 3.89 | 0.68 | 5.47 | 0.51 | 0.69 | 5.10 | 4.92 | 4.92 | 4.92 | 4.74 | 4.74 | 4.02 | 0.51 | 0.51 | 0.86 | 4.92 | |||||||||||||||||||||||||
| 57. Cx. chidesteri M031 | 7.51 | 8.46 | 8.46 | 8.08 | 5.28 | 2.78 | 2.43 | 1.91 | 5.11 | 4.38 | 6.58 | 5.11 | 6.58 | 6.58 | 2.25 | 2.08 | 2.08 | 2.08 | 1.90 | 1.90 | 4.74 | 6.58 | 6.201 | 6.95 | 2.08 | |||||||||||||||||||||||||
| 58. Cx. apicinus M007a | 8.27 | 8.65 | 8.65 | 8.27 | 7.14 | 6.76 | 6.40 | 6.21 | 6.97 | 6.22 | 7.91 | 6.78 | 7.71 | 7.91 | 6.58 | 6.40 | 6.02 | 6.02 | 6.21 | 6.21 | 6.23 | 7.71 | 7.91 | 8.10 | 6.40 | |||||||||||||||||||||||||
| 59. Cx. acharistus M008 | 7.70 | 7.51 | 7.51 | 7.13 | 3.49 | 4.74 | 4.74 | 4.56 | 5.85 | 3.66 | 5.83 | 5.48 | 5.28 | 5.83 | 4.92 | 4.74 | 4.38 | 4.38 | 4.56 | 4.56 | 3.84 | 5.29 | 5.287 | 5.65 | 4.56 | |||||||||||||||||||||||||
| 60. Cx. acharistus M015 | 7.51 | 7.70 | 7.70 | 7.32 | 3.66 | 5.10 | 4.74 | 4.56 | 5.67 | 3.84 | 6.21 | 5.30 | 5.65 | 5.83 | 4.92 | 4.74 | 4.38 | 4.38 | 4.56 | 4.56 | 4.38 | 5.65 | 5.65 | 6.02 | 4.74 | |||||||||||||||||||||||||
| 61. Cx. acharistus M037 | 7.51 | 7.32 | 7.32 | 6.94 | 3.31 | 4.74 | 4.74 | 4.56 | 5.67 | 3.89 | 6.02 | 5.29 | 5.47 | 5.65 | 4.92 | 4.74 | 4.38 | 4.38 | 4.56 | 4.56 | 4.02 | 5.47 | 5.47 | 5.83 | 4.56 | |||||||||||||||||||||||||
| 62. Cx. acharistus SP56 R | 7.32 | 7.69 | 7.69 | 7.32 | 3.48 | 4.92 | 4.56 | 4.38 | 5.85 | 4.02 | 5.83 | 5.11 | 5.65 | 5.47 | 4.38 | 4.56 | 4.56 | 4.56 | 4.38 | 4.38 | 4.56 | 5.65 | 5.28 | 6.02 | 4.565 | |||||||||||||||||||||||||
| 63. Cx. acharistus VP09 116 | 7.32 | 7.51 | 7.51 | 7.13 | 4.20 | 5.28 | 4.92 | 4.74 | 5.85 | 4.38 | 6.39 | 5.84 | 6.39 | 6.20 | 5.10 | 4.92 | 4.92 | 4.92 | 4.74 | 4.74 | 4.92 | 6.39 | 6.02 | 6.76 | 4.92 | |||||||||||||||||||||||||
| 64. Cx. apicinus F017 | 7.89 | 8.27 | 8.27 | 7.89 | 6.57 | 6.39 | 6.02 | 5.84 | 6.41 | 5.66 | 7.52 | 6.59 | 7.33 | 7.52 | 6.21 | 6.03 | 5.65 | 5.65 | 5.84 | 5.8 | 5.67 | 7.33 | 7.52 | 7.71 | 6.02 | |||||||||||||||||||||||||
| 65. Cx. apicinus M007b | 8.27 | 8.65 | 8.65 | 8.27 | 7.14 | 6.76 | 6.40 | 6.21 | 6.98 | 6.22 | 7.91 | 6.78 | 7.71 | 7.91 | 6.59 | 6.40 | 6.02 | 6.02 | 6.21 | 6.21 | 6.23 | 7.71 | 7.91 | 8.10 | 6.40 | |||||||||||||||||||||||||
| 66. Cx. chidesteri SP67 2 | 8.27 | 8.65 | 8.65 | 8.27 | 5.83 | 2.96 | 2.61 | 2.43 | 6.04 | 4.93 | 7.14 | 6.03 | 7.14 | 7.14 | 2.78 | 2.61 | 2.61 | 2.61 | 2.43 | 2.43 | 5.30 | 7.14 | 6.76 | 7.52 | 2.61 | |||||||||||||||||||||||||
| 67. Cx. chidesteri SP67 5 | 7.70 | 8.65 | 8.65 | 8.27 | 5.47 | 2.96 | 2.61 | 2.08 | 5.30 | 4.56 | 6.76 | 5.29 | 6.76 | 6.76 | 2.43 | 2.25 | 2.25 | 2.25 | 2.08 | 2.08 | 4.93 | 6.76 | 6.39 | 7.14 | 2.25 | |||||||||||||||||||||||||
| 68. Cx. interfor M049 | 8.46 | 8.84 | 8.84 | 8.46 | 7.32 | 6.95 | 6.58 | 6.40 | 6.78 | 6.41 | 8.10 | 6.97 | 7.91 | 8.10 | 6.77 | 6.587 | 6.21 | 6.20 | 6.40 | 6.40 | 6.42 | 7.91 | 8.10 | 8.30 | 6.58 | |||||||||||||||||||||||||
| 69. Cx. mollis M089 | 7.70 | 7.89 | 7.89 | 7.51 | 4.74 | 0 | 1.72 | 1.55 | 5.67 | 4.02 | 6.39 | 5.11 | 6.20 | 6.20 | 1.90 | 1.72 | 1.72 | 1.72 | 1.55 | 1.55 | 4.02 | 6.20 | 5.83 | 6.58 | 1.55 | |||||||||||||||||||||||||
| 70. Cx. mollis PA11 101 | 7.89 | 8.08 | 8.08 | 7.70 | 4.92 | 0.17 | 1.90 | 1.72 | 5.85 | 4.20 | 6.58 | 5.29 | 6.39 | 6.39 | 2.07 | 1.90 | 1.90 | 1.90 | 1.72 | 1.72 | 4.20 | 6.40 | 6.02 | 6.77 | 1.72 | |||||||||||||||||||||||||
| 71. Cx. mollis SP05 100 | 7.70 | 7.89 | 7.89 | 7.51 | 4.74 | 0 | 1.72 | 1.55 | 5.67 | 4.02 | 6.39 | 5.11 | 6.21 | 6.20 | 1.90 | 1.72 | 1.72 | 1.72 | 1.55 | 1.55 | 4.02 | 6.20 | 5.83 | 6.58 | 1.55 | |||||||||||||||||||||||||
| 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49 | 50 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27. Cx. dolosus F001 | 3.84 | ||||||||||||||||||||||||
| 28. Cx. dolosus F006 | 3.66 | 0.17 | |||||||||||||||||||||||
| 19. Cx. dolosus M001 | 3.8 | 0 | 0.17 | ||||||||||||||||||||||
| 30. Cx. dolosus SP56 10 | 2.59 | 1.90 | 1.72 | 1.90 | |||||||||||||||||||||
| 31. Cx. eduardoi SP65 7 | 3.30 | 3.14 | 2.96 | 3.14 | 1.90 | ||||||||||||||||||||
| 32. Cx. eduardoi SP90 6 | 3.84 | 2.96 | 2.78 | 2.96 | 2.43 | 1.37 | |||||||||||||||||||
| 33. Cx. eduardoi SP90 24 | 3.84 | 2.96 | 2.78 | 2.96 | 2.43 | 1.37 | 0 | ||||||||||||||||||
| 34. Cx. lygrus ss SP74 24 | 5.29 | 6.39 | 6.20 | 6.39 | 5.10 | 4.74 | 5.29 | 5.29 | |||||||||||||||||
| 35. Cx. lygrus ss SP78 6 | 5.11 | 5.83 | 5.65 | 5.83 | 4.56 | 4.20 | 4.92 | 4.92 | 0.682 | ||||||||||||||||
| 36. Cx. lygrus sl SP56 25 | 5.29 | 6.02 | 5.83 | 6.02 | 4.74 | 4.38 | 5.10 | 5.10 | 0.86 | 0.51 | |||||||||||||||
| 37. Cx. lygrus sl SP56 26 | 5.11 | 5.83 | 5.65 | 5.83 | 4.56 | 4.20 | 4.92 | 4.92 | 0.68 | 0.34 | 0.17 | ||||||||||||||
| 38. Cx. maxi F002 | 4.92 | 4.92 | 4.74 | 4.92 | 4.02 | 5.10 | 5.10 | 5.10 | 5.83 | 6.02 | 6.20 | 6.02 | |||||||||||||
| 39. Cx. maxi M002 | 4.92 | 4.92 | 4.74 | 4.92 | 4.02 | 5.10 | 5.10 | 5.10 | 5.83 | 6.02 | 6.20 | 6.02 | 0 | ||||||||||||
| 40. Cx. maxi M048 | 4.92 | 4.92 | 4.74 | 4.92 | 4.02 | 5.10 | 5.10 | 5.10 | 5.83 | 6.02 | 6.20 | 6.02 | 0 | 0 | |||||||||||
| 41. Cx. nigripalpus 2 7 | 0.86 | 3.84 | 3.66 | 3.84 | 3.12 | 3.84 | 4.02 | 4.02 | 5.83 | 5.65 | 5.83 | 5.65 | 5.10 | 5.10 | 5.10 | ||||||||||
| 42. Cx. nigripalpus 4 7 | 0.86 | 4.02 | 3.84 | 4.02 | 3.30 | 4.02 | 4.207 | 4.02 | 6.02 | 5.83 | 6.02 | 5.83 | 5.10 | 5.10 | 5.10 | 0.34 | |||||||||
| 43. Cx. nigripalpus AS | 0.68 | 3.84 | 3.66 | 3.84 | 3.12 | 3.84 | 4.02 | 4.02 | 5.83 | 5.65 | 5.83 | 5.65 | 4.92 | 4.902 | 4.92 | 0.17 | 0.17 | ||||||||
| 44. Cx. saltanensis M012 | 4.92 | 4.56 | 4.38 | 4.56 | 3.66 | 5.10 | 5.47 | 5.47 | 6.77 | 6.20 | 6.39 | 6.20 | 2.08 | 2.08 | 2.08 | 5.10 | 5.10 | 4.92 | |||||||
| 45. Cx. nigripalpus PR | 0.68 | 3.84 | 3.66 | 3.84 | 3.12 | 3.84 | 4.02 | 4.02 | 5.83 | 5.65 | 5.83 | 5.65 | 4.92 | 4.92 | 4.92 | 0.17 | 0.17 | 0 | 4.92 | ||||||
| 46. Cx. saltanensis F010 | 5.10 | 4.74 | 4.56 | 4.74 | 3.84 | 5.29 | 5.65 | 5.65 | 6.95 | 6.39 | 6.57 | 6.39 | 1.90 | 1.90 | 1.90 | 5.28 | 5.28 | 5.10 | 0.17 | 5.10 | |||||
| 47. Cx. surinamensis CDC3 1 | 5.10 | 5.47 | 5.29 | 5.47 | 4.20 | 5.29 | 5.66 | 5.66 | 6.02 | 6.20 | 6.39 | 6.20 | 0.51 | 0.51 | 0.51 | 5.65 | 5.65 | 5.47 | 2.25 | 5.47 | 2.08 | ||||
| 48. Cx. tatoi M091 | 1.20 | 3.30 | 3.12 | 3.30 | 2.95 | 3.667 | 3.48 | 3.48 | 6.0 | 5.47 | 5.65 | 5.47 | 5.47 | 5.47 | 5.47 | 1.20 | 1.37 | 1.20 | 5.47 | 1.20 | 5.65 | 6.02 | |||
| 49. Cx. tatoi MS4 16 | 0.86 | 4.02 | 3.84 | 4.02 | 2.59 | 3.66 | 4.20 | 4.20 | 5.65 | 5.47 | 5.65 | 5.47 | 4.74 | 4.74 | 4.74 | 1.03 | 1.03 | 0.86 | 4.38 | 0.85 | 4.56 | 4.92 | 1.37 | ||
| 50. Cx. tatoi PR24 123 | 1.55 | 4.75 | 4.57 | 4.75 | 3.30 | 4.02 | 4.57 | 4.57 | 6.02 | 5.84 | 6.02 | 5.83 | 4.74 | 4.74 | 4.74 | 1.73 | 1.72 | 1.55 | 4.74 | 1.55 | 4.92 | 4.56 | 2.08 | 0.68 | |
| 51. Cx. tatoi RO4 109 | 0.69 | 4.20 | 4.02 | 4.20 | 3.12 | 3.84 | 4.38 | 4.38 | 5.47 | 5.29 | 5.47 | 5.29 | 5.10 | 5.10 | 5.10 | 1.20 | 1.20 | 1.03 | 5.28 | 1.03 | 5.47 | 5.28 | 1.20 | 1.20 | 1.90 |
| 52. Cx. tatoi RO24 8 | 0.69 | 4.20 | 4.02 | 4.20 | 3.12 | 3.84 | 4.38 | 4.38 | 5.47 | 5.29 | 5.47 | 5.29 | 5.10 | 5.10 | 5.10 | 1.20 | 1.20 | 1.03 | 5.28 | 1.03 | 5.47 | 5.28 | 1.20 | 1.20 | 1.90 |
| 53. Cx. tatoi RO25 2 | 0.69 | 4.20 | 4.02 | 4.20 | 3.12 | 3.84 | 4.38 | 4.38 | 5.47 | 5.29 | 5.47 | 5.296 | 5.10 | 5.10 | 5.10 | 1.20 | 1.20 | 1.03 | 5.28 | 1.003 | 5.47 | 5.28 | 1.20 | 1.20 | 1.90 |
| 54. Cx. tatoi RO25 3 | 0.69 | 4.20 | 4.02 | 4.20 | 3.12 | 3.84 | 4.38 | 4.38 | 5.47 | 5.29 | 5.47 | 5.296 | 5.10 | 5.10 | 5.10 | 1.20 | 1.20 | 1.03 | 5.28 | 1.03 | 5.47 | 5.28 | 1.20 | 1.20 | 1.90 |
| 55. Cx. tatoi RO25 110 | 0.69 | 4.20 | 4.02 | 4.20 | 3.12 | 3.84 | 4.38 | 4.38 | 5.47 | 5.29 | 5.47 | 5.29 | 5.10 | 5.10 | 5.10 | 1.20 | 1.20 | 1.03 | 5.28 | 1.03 | 5.47 | 5.28 | 1.20 | 1.20 | 1.90 |
| 56. Cx. usquatus M093 | 4.74 | 4.74 | 4.56 | 4.74 | 3.84 | 5.23 | 5.29 | 5.29 | 6.20 | 6.39 | 6.57 | 6.39 | 0.51 | 0.51 | 0.51 | 4.92 | 4.92 | 4.74 | 1.90 | 4.74 | 1.73 | 1.03 | 5.28 | 4.56 | 4.92 |
| 57. Cx. chidesteri M031 | 1.90 | 5.48 | 5.29 | 5.48 | 4.38 | 4.56 | 5.29 | 5.29 | 7.13 | 6.94 | 7.13 | 6.941 | 6.58 | 6.58 | 6.58 | 2.08 | 2.07 | 1.90 | 6.20 | 1.90 | 6.39 | 6.58 | 2.43 | 2.07 | 2.78 |
| 58. Cx. apicinus M007a | 6.21 | 6.98 | 6.79 | 6.98 | 6.22 | 6.21 | 6.97 | 6.97 | 7.51 | 6.95 | 7.14 | 6.95 | 8.10 | 8.10 | 8.10 | 6.77 | 6.77 | 6.58 | 7.91 | 6.58 | 8.09 | 7.52 | 6.39 | 6.59 | 6.97 |
| 59. Cx. acharistus M008 | 4.56 | 4.02 | 3.84 | 4.02 | 3.66 | 5.11 | 5.66 | 5.66 | 7.13 | 6.57 | 6.76 | 6.57 | 5.65 | 5.65 | 5.65 | 4.56 | 4.74 | 4.56 | 4.92 | 4.56 | 5.10 | 5.83 | 4.74 | 4.02 | 4.38 |
| 60. Cx. acharistus M015 | 4.56 | 4.57 | 4.38 | 4.57 | 3.84 | 4.93 | 5.48 | 5.48 | 6.95 | 6.39 | 6.57 | 6.39 | 6.02 | 6.02 | 6.02 | 4.56 | 4.74 | 4.56 | 5.28 | 4.56 | 5.47 | 6.21 | 4.74 | 4.02 | 4.74 |
| 61. Cx. acharistus M037 | 4.56 | 4.20 | 4.02 | 4.20 | 3.84 | 4.92 | 5.48 | 5.48 | 6.94 | 6.39 | 6.57 | 6.39 | 5.83 | 5.83 | 5.83 | 4.56 | 4.74 | 4.56 | 5.10 | 4.56 | 5.28 | 6.02 | 4.74 | 4.02 | 4.38 |
| 62. Cx. acharistus SP56 R | 4.38 | 4.74 | 4.56 | 4.74 | 4.02 | 4.92 | 5.29 | 5.29 | 6.39 | 6.20 | 6.39 | 6.20 | 5.65 | 5.65 | 5.65 | 4.38 | 4.56 | 4.38 | 5.28 | 4.38 | 5.47 | 5.83 | 4.92 | 3.84 | 4.56 |
| 63. Cx. acharistus VP09 116 | 4.74 | 5.11 | 4.93 | 5.11 | 4.38 | 5.47 | 6.03 | 6.03 | 7.13 | 6.94 | 7.13 | 6.94 | 6.39 | 6.39 | 6.39 | 4.74 | 4.92 | 4.74 | 6.02 | 4.74 | 6.20 | 6.39 | 5.29 | 4.20 | 4.92 |
| 64. Cx. apicinus F017 | 5.84 | 6.41 | 6.22 | 6.41 | 5.66 | 5.66 | 6.78 | 6.78 | 6.95 | 6.39 | 6.57 | 6.39 | 7.71 | 7.71 | 7.71 | 6.40 | 6.39 | 6.21 | 7.52 | 6.21 | 7.71 | 7.14 | 6.02 | 6.02 | 6.40 |
| 65. Cx. apicinus M007b | 6.21 | 6.98 | 6.79 | 6.98 | 6.22 | 6.21 | 6.97 | 6.97 | 7.51 | 6.95 | 7.14 | 6.95 | 8.10 | 8.10 | 8.10 | 6.77 | 6.77 | 6.58 | 7.91 | 6.58 | 8.09 | 7.52 | 6.39 | 6.59 | 6.96 |
| 66. Cx. chidesteri SP67 2 | 2.43 | 6.04 | 5.85 | 6.04 | 4.93 | 5.66 | 6.22 | 6.22 | 7.70 | 7.51 | 7.70 | 7.51 | 7.14 | 7.14 | 7.14 | 2.25 | 2.25 | 2.08 | 6.39 | 2.08 | 6.57 | 7.14 | 2.96 | 2.25 | 2.61 |
| 67. Cx. chidesteri SP67 5 | 2.08 | 5.66 | 5.48 | 5.66 | 4.56 | 4.74 | 5.48 | 5.48 | 7.32 | 7.13 | 7.32 | 7.13 | 6.76 | 6.76 | 6.76 | 2.25 | 2.25 | 2.08 | 6.39 | 2.08 | 6.57 | 6.76 | 2.61 | 2.25 | 2.96 |
| 68. Cx. interfor M049 | 6.40 | 7.17 | 6.98 | 7.17 | 6.41 | 6.40 | 7.16 | 7.16 | 7.71 | 7.14 | 7.32 | 7.14 | 8.29 | 8.29 | 8.29 | 6.96 | 6.96 | 6.78 | 8.10 | 6.77 | 8.29 | 7.71 | 6.58 | 6.77 | 7.15 |
| 69. Cx. mollis M089 | 1.55 | 4.75 | 4.56 | 4.75 | 4.02 | 4.74 | 5.29 | 5.29 | 6.76 | 6.57 | 6.76 | 6.57 | 6.20 | 6.20 | 6.20 | 1.72 | 1.72 | 1.55 | 5.83 | 1.55 | 6.02 | 6.02 | 2.43 | 1.72 | 2.07 |
| 70. Cx. mollis PA11 101 | 1.72 | 4.93 | 4.75 | 4.93 | 4.20 | 4.92 | 5.48 | 5.48 | 6.706 | 6.57 | 6.76 | 6.57 | 6.39 | 6.39 | 6.39 | 1.90 | 1.90 | 1.72 | 6.02 | 1.72 | 6.20 | 6.20 | 2.61 | 1.90 | 2.25 |
| 71. Cx. mollis SP05 100 | 1.55 | 4.75 | 4.56 | 4.75 | 4.02 | 4.74 | 5.29 | 5.29 | 6.76 | 6.57 | 6.76 | 6.57 | 6.20 | 6.20 | 6.20 | 1.72 | 1.72 | 1.55 | 5.83 | 1.55 | 6.01 | 6.02 | 2.43 | 1.72 | 2.07 |
| 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 52. Cx. tatoi RO24 8 | 0 | |||||||||||||||||||
| 53. Cx. tatoi RO25 2 | 0 | 0 | ||||||||||||||||||
| 54. Cx. tatoi RO25 3 | 0 | 0 | 0 | |||||||||||||||||
| 55. Cx. tatoi RO25 110 | 0 | 0 | 0 | 0 | ||||||||||||||||
| 56. Cx. usquatus M093 | 5.10 | 5.10 | 5.10 | 5.10 | 5.10 | |||||||||||||||
| 57. Cx. chidesteri M031 | 2.25 | 2.25 | 2.25 | 2.25 | 2.25 | 6.39 | ||||||||||||||
| 58. Cx. apicinus M007a | 5.84 | 5.84 | 5.84 | 5.84 | 5.84 | 8.10 | 6.95 | |||||||||||||
| 59. Cx. acharistus M008 | 4.92 | 4.92 | 4.92 | 4.92 | 4.92 | 5.47 | 5.65 | 7.34 | ||||||||||||
| 60. Cx. acharistus M015 | 4.92 | 4.92 | 4.92 | 4.92 | 4.92 | 5.83 | 5.65 | 6.96 | 0.86 | |||||||||||
| 61. Cx. acharistus M037 | 4.93 | 4.93 | 4.93 | 4.93 | 4.93 | 5.65 | 5.65 | 7.15 | 0.17 | 0.68 | ||||||||||
| 62. Cx. acharistus SP56 R | 4.74 | 4.74 | 4.74 | 4.74 | 4.74 | 5.47 | 5.10 | 7.34 | 1.72 | 1.55 | 1.55 | |||||||||
| 63. Cx. acharistus VP09 116 | 5.10 | 5.10 | 5.10 | 5.10 | 5.10 | 6.20 | 5.47 | 7.53 | 1.37 | 1.20 | 1.20 | 1.72 | ||||||||
| 64. Cx. apicinus F017 | 5.47 | 5.47 | 5.47 | 5.47 | 5.47 | 7.71 | 6.58 | 0.51 | 6.77 | 6.40 | 6.59 | 6.77 | 6.96 | |||||||
| 65. Cx. apicinus M007b | 5.84 | 5.84 | 5.84 | 5.84 | 5.84 | 8.01 | 6.95 | 0 | 7.34 | 6.96 | 7.15 | 7.34 | 7.53 | 0.51 | ||||||
| 66. Cx. chidesteri SP67 2 | 2.78 | 2.78 | 2.78 | 2.78 | 2.78 | 6.9 | 1.20 | 7.52 | 5.84 | 5.84 | 5.84 | 5.65 | 5.65 | 7.14 | 7.52 | |||||
| 67. Cx. chidesteri SP67 5 | 2.43 | 2.43 | 2.43 | 2.43 | 2.43 | 6.58 | 0.17 | 7.14 | 5.84 | 5.84 | 5.84 | 5.28 | 5.65 | 6.76 | 7.14 | 1.38 | ||||
| 68. Cx. interfor M049 | 6.02 | 6.02 | 6.02 | 6.02 | 6.02 | 8.29 | 7.14 | 0.17 | 7.53 | 7.15 | 7.34 | 7.53 | 7.72 | 0.68 | 0.17 | 7.71 | 7.33 | |||
| 69. Cx. mollis M089 | 1.90 | 1.90 | 1.90 | 1.90 | 1.90 | 6.02 | 2.78 | 6.76 | 4.74 | 5.10 | 4.74 | 4.92 | 5.28 | 6.39 | 6.76 | 2.96 | 2.96 | 6.95 | ||
| 70. Cx. mollis PA11 101 | 2.07 | 2.07 | 2.07 | 2.07 | 2.07 | 6.20 | 2.96 | 6.95 | 4.92 | 5.219 | 4.92 | 5.10 | 5.47 | 6.587 | 6.95 | 3.14 | 3.14 | 7.14 | 0.17 | |
| 71. Cx. mollis SP05 100 | 1.90 | 1.90 | 1.90 | 1.90 | 1.90 | 6.02 | 2.78 | 6.76 | 4.74 | 5.10 | 4.74 | 4.92 | 5.28 | 6.39 | 6.76 | 2.96 | 2.96 | 6.95 | 0 | 0.17 |
Acknowledgments
To B Demari-Silva, who helped MAMS with field collections, and to GC Rossi and AM Visintin, who assisted M Laurito.
REFERENCES
- Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignments of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:7–13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almirón WR, Humeres SG, Gardenal CN. Distribution and hybridization between Culex pipiens and Culex quinquefasciatus (Diptera: Culicidae) in Argentina. Mem Inst Oswaldo Cruz. 1995;90:469–473. doi: 10.1590/s0074-02761998000100011. [DOI] [PubMed] [Google Scholar]
- Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, Campbell CL, Campbell KS, Casola C, Castro MT, Chandramouliswaran I, Chapman SB, Christley S, Costas J, Eisenstadt E, Feschotte C, Fraser-Liggett C, Guigo R, Haas B, Hammond M, Hansson BS, Hemingway J, Hill SR, Howarth C, Ignell R, Kennedy RC, Kodira CD, Lobo NF, Mao C, Mayhew G, Michel K, Mori A, Liu N, Naveira H, Nene V, Nguyen N, Pearson MD, Pritham EJ, Puiu D, Qi Y, Ranson H, Ribeiro JM, Roberston HM, Severson DW, Shumway M, Stanke M, Strausberg RL, Sun C, Sutton G, Tu ZJ, Tubio JM, Unger MF, Vanlandingham DL, Vilella AJ, White O, White JR, Wondji CS, Wortman J, Zdobnov EM, Birren B, Christensen BM, Collins FH, Cornel A, Dimopoulos G, Hannick LI, Higgs S, Lanzaro GC, Lawson D, Lee NH, Muskavitch MA, Raikhel AS, Atkinson PW. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickford D, Lohman DJ, Sodhi NS, Ng PK, Meier R, Winker K, Ingram K, Das I. Cryptic species as a window on diversity and conservation. Trends Ecol Evol. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Bourke BP, Oliveira TP, Suesdek L, Bergo ES, Sallum MA. A multi-locus approach to barcoding in the Anopheles strodei subgroup (Diptera: Culicidae) 111Parasit Vectors. 2013;6 doi: 10.1186/1756-3305-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram RA. Classification of Culex subgenus Culex in the New World (Diptera, Culicidae) Proc US Natl Mus. 1967;120:1–122. [Google Scholar]
- Brewer M, Buffa L, Almirón W. Culex pipiens quinquefasciatus y Culex pipiens pipiens (Diptera: Culicidae) en Córdoba, Argentina. Rev Per Entomol. 1987;29:69–72. [Google Scholar]
- Chen B, Harbach RE, Butlin RK. Genetic variation and population structure of the mosquito Anopheles jeyporiensis in southern China. Mol Ecol. 2004;10:3051–3056. doi: 10.1111/j.1365-294X.2004.02309.x. [DOI] [PubMed] [Google Scholar]
- Cywinska A, Hunter FF, Hebert PDN. Identifying Canadian mosquito species through DNA barcodes. Med Vet Entomol. 2006;20:413–424. doi: 10.1111/j.1365-2915.2006.00653.x. [DOI] [PubMed] [Google Scholar]
- Dai QY, Gao Q, Wu CS, Chesters D, Zhu CD, Zhang AB. Phylogenetic reconstruction and DNA barcoding for closely related pine moth species (Dendrolimus) in China with multiple gene markers. 3254PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0032544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aranzamendi MC, Gardenal CN, Martin JP, Bastida R. Limpets of the genus Nacella (Patellogastropoda) from the southwestern Atlantic: species identification based on molecular data. J Molluscan Stud. 2009;75:241–251. [Google Scholar]
- Demari-Silva B, Vesgueiro FT, Sallum MA, Marrelli MT. Taxonomic and phylogenetic relationships between species of the genus Culex (Diptera: Culicidae) from Brazil inferred from the cytochrome c oxidase I mitochondrial gene. J Med Entomol. 2011;48:272–279. doi: 10.1603/me09293. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards FW. Genera Insectorum. Diptera. Family Culicidae, fascicle 194. Louis Desmet-Verteneuil; Belgium: 1932. 258 [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Fontes G, Braun RF, Fraiha H, Neto, Vieira JB, Padilha SS, Rocha RC, da Rocha EM. Filariose linfática em Belém, estado do Pará, norte do Brasil e a perspectiva de eliminação. Rev Soc Bras Med Trop. 2005;38:131–136. doi: 10.1590/s0037-86822005000200001. [DOI] [PubMed] [Google Scholar]
- Fontes G, Rocha EMM, Brito AC, Antunes CMF. Lymphatic filariasis in Brazilian urban area (Maceió, Alagoas) Mem Inst Oswaldo Cruz. 1998;93:705–710. doi: 10.1590/s0074-02761998000600001. [DOI] [PubMed] [Google Scholar]
- Forattini OP. Culicidologia médica: identificação, biologia e epidemiologia. Vol. 2. EDUSP; São Paulo: 2002. 864 [Google Scholar]
- Foster PG, Bergo ES, Bourke BP, Oliveira TP, Nagaki SS, Sant’Ana DC, Sallum MAM. Phylogenetic analysis and DNA-based species confirmation in Anopheles (Nyssorhynchus) 54063PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0054063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey JE, Guillén L, Frey B, Samietz J, Rull J, Aluja M. Developing diagnostic SNP panels for the identification of true fruit flies (Diptera: Tephritidae) within the limits of COI-based species delimitation. 106BMC Evol Biol. 2013;13 doi: 10.1186/1471-2148-13-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. Seaview version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree bulding. 221-224Mol Biol Evol. 2010;27 doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Harbach RE. Classification within the cosmopolitan genus Culex (Diptera: Culicidae): the foundation for molecular systematics and phylogenetic research. 1-14Acta Trop. 2011;120 doi: 10.1016/j.actatropica.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Harbach RE, Jackob WL, Peyton EL. Recognition of Culex bidens Dyar and Culex interfor Dyar (Diptera: Culicidae) as separate species. Mosq Syst. 1986;18:139–144. [Google Scholar]
- Harbach RE, Kitching IK, Culverwell CL, Dubois J, Linton Y-M. Phylogeny of mosquitoes of tribe Culicini (Diptera: Culicidae) based on morphological diversity. Zool Scr. 2012;5:499–514. [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, de Waard JR. Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci. 2003a;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the Neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Ratnasingham S, de Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B Biol Sci. 2003b;270:96–99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubálek Z. European experience with the West Nile virus ecology and epidemiology: could it be relevant for the New World? Viral Immunol. 2000;13:415–426. doi: 10.1089/vim.2000.13.415. [DOI] [PubMed] [Google Scholar]
- Kaila L, Ståhls G. DNA barcodes: evaluating the potential of COI to differentiate closely related species of Elachista (Lepidoptera: Gelechioidea: Elachistidae) from Australia. Zootaxa. 2006;1170:1–26. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Komar N. West Nile virus: epidemiology and ecology in North America. Adv Virus Res. 2003;61:185–234. doi: 10.1016/s0065-3527(03)61005-5. [DOI] [PubMed] [Google Scholar]
- Kramer LD, Styer LM, Ebel GD. A global perspective on the epidemiology of West Nile virus. Annu Rev Entomol. 2008;53:61–81. doi: 10.1146/annurev.ento.53.103106.093258. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Wilkerson RC, Besansky NJ. Toward understanding Anophelinae (Diptera, Culicidae) phylogeny: insights from nuclear single-copy genes and weight of evidence. Syst Biol. 2001;50:540–546. [PubMed] [Google Scholar]
- Kumar NP, Rajavel AR, Natarajan R, Jambulingam P. DNA Barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae) J Med Entomol. 2007;44:1–7. doi: 10.1603/0022-2585(2007)44[1:dbcdso]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Lane J. Synonymy of Neotropical Culicidae (Diptera) Proc Entomol Soc Wash. 1951;53:333–336. [Google Scholar]
- Laurito M, Almirón WR. Phylogenetic relationships of Culex (Culex) species (Diptera, Culicidae) from Argentina based on morphological characters. Zootaxa. 2013;3652:117–155. doi: 10.11646/zootaxa.3652.1.4. [DOI] [PubMed] [Google Scholar]
- Laurito M, Almirón WR, Rossi GC. Morphological redescription of immature and adult stages of Culex (Culex) acharistus Root (Diptera: Culicidae) Zootaxa. 2009;2263:21–30. [Google Scholar]
- Lou M, Golding GB. Assigning sequences to species in the absence of large interspecific differences. Mol Phylogenet Evol. 2010;56:187–194. doi: 10.1016/j.ympev.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Meier R, Shiyang K, Vaidya G, Ng PK. DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol. 2006;55:715–728. doi: 10.1080/10635150600969864. [DOI] [PubMed] [Google Scholar]
- Meiklejohn KA, Wallman JF, Pape T, Cameron SL, Dowton M. Utility of COI, CAD and morphological data for resolving relationships within the genus Sarcophaga (sensu lato) (Diptera: Sarcophagidae): a preliminary study. Mol Phylogenet Evol. 2013;69:133–141. doi: 10.1016/j.ympev.2013.04.034. [DOI] [PubMed] [Google Scholar]
- Meyer CP, Paulay G. DNA barcoding: error rates based on comprehensive sampling. 422PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Crabtree MB, Savage HM. Phylogeny of fourteen Culex mosquito species, including the Culex pipiens complex, inferred from the internal transcribed spacers of ribosomal DNA. Insect Mol Biol. 1996;5:93–107. doi: 10.1111/j.1365-2583.1996.tb00044.x. [DOI] [PubMed] [Google Scholar]
- Mirabello L, Conn JE. Population analysis using the nuclear white gene detects Pliocene/Pleistocene lineage divergence within Anopheles nuneztovari in South America. Med Vet Entomol. 2008;22:109–119. doi: 10.1111/j.1365-2915.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Morais SA, Almeida FD, Suesdek L, Marrelli MT. Low genetic diversity in Wolbachia infected Culex quinquefasciatus (Diptera: Culicidae) from Brazil and Argentina. Rev Inst Med Trop Sao Paulo. 2012;54:325–329. doi: 10.1590/s0036-46652012000600007. [DOI] [PubMed] [Google Scholar]
- Mukabayire O, Boccolini D, Lochouarn L, Fontenille D, Besansky NJ. Mitochondrial and ribosomal internal transcribed spacer (ITS2) diversity of the African malaria vector Anopheles funestus. Mol Ecol. 1999;8:289–297. doi: 10.1046/j.1365-294x.1999.00567.x. [DOI] [PubMed] [Google Scholar]
- Pedro M, Sallum MA. Spatial expansion and population structure of the Neotropical malaria vector, Anopheles darlingi (Diptera: Culicidae) Biol J Linn Soc Lond. 2008;97:854–866. [Google Scholar]
- Pedro P, Sallum MAM. Spatial expansion and population structure of the Neotropical malaria vector, Anopheles darlingi. Biol J Linn Soc Lond. 2009;97:854–866. [Google Scholar]
- Pisano MB, Ré VE, Díaz LA, Farías A, Stein M, Sánchez-Seco MP, Tenorio A, Almirón WR, Contigiani MS. Enzootic activity of Pixuna and Rio Negro viruses (Venezuelan Equine Encephalitis complex) in a Neotropical region of Argentina. Vector Borne Zoonotic Dis. 2010;10:199–201. doi: 10.1089/vbz.2008.0156. [DOI] [PubMed] [Google Scholar]
- Quintero L, Navarro JC. Intraspecific phylogeny and genetic variability of Culex quinquefasciatus say (Diptera: Culicidae) with mitochondrial genes ND5 and COI. Bol Mal Salud Amb. 2012;52:45–65. [Google Scholar]
- Reddy NBP, Labbe P, Corbel V. Culex genome is not just another genome for comparative genomics. 63Parasit Vectors. 2012;5 doi: 10.1186/1756-3305-5-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidenbach KR, Cook S, Bertone MA, Harbach RE, Wiegmann BM, Besansky NJ. Phylogentic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. 298BMC Evol Biol. 2009;9 doi: 10.1186/1471-2148-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud AK, Savage J, Adamowicz SJ. DNA barcoding of northern Nearctic Muscidae (Diptera) reveals high correspondence between morphological and molecular species limits. 24BMC Evol Biol. 2012;23 doi: 10.1186/1472-6785-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera J, Currie D. Identification of Nearctic black flies using DNA barcodes (Diptera: Simuliidae) Mol Ecol Resour. 2009;9:224–236. doi: 10.1111/j.1755-0998.2009.02648.x. [DOI] [PubMed] [Google Scholar]
- Root FM. Studies on Brazilian mosquitoes III. The genus Culex. Am J Hyg. 1927;7:574–598. [Google Scholar]
- Rossi GC, Laurito M, Almirón WR. Morphological description of the pupa and redescription of the adults and larva of Culex (Culex) apicinus Philippi (Diptera: Culicidae) Zootaxa. 2008;1941:31–42. [Google Scholar]
- Ruiz-Lopez F, Wilkerson RC, Conn JE, McKeon SN, Levin DM, Quiñones ML, Póvoa MM, Linton Y-M. DNA barcoding reveals both known and novel taxa in the Albitarsis group (Anopheles: Nyssorhynchus) of Neotropical malaria vectors. 44Parasit Vectors. 2012;5 doi: 10.1186/1756-3305-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeung A, Baimai V, Otsuka Y, Rattanarithikul R, Somboon P, Junkum A, Tuetun B, Takaoka H, Choochote W. Molecular and cytogenetic evidence of three sibling species of the Anopheles barbirostris form A (Diptera: Culicidae) in Thailand. Parasitol Res. 2008;102:499–507. doi: 10.1007/s00436-007-0788-0. [DOI] [PubMed] [Google Scholar]
- Sallum MAM, Almirón WR, Forattini OP. Culex (Culex) interfor Dyar (Diptera: Culicidae), morphological description including previously unknown life stages. Mem Inst Oswaldo Cruz. 1996;91:563–570. [Google Scholar]
- Sallum MAM, Foster PG, dos Santos CL, Flores DC, Motoki MT, Bergo ES. Resurrection of two species from synonymy of Anopheles (Nyssorhynchus) strodei Root and characterization of a distinct morphological form from the Strodei complex (Diptera: Culicidae) J Med Entomol. 2010;47:504–526. doi: 10.1603/me09229. [DOI] [PubMed] [Google Scholar]
- Sallum MAM, Marrelli MT, Nagaki SS, Laporta GZ, dos Santos CL. Insight into Anopheles (Nyssorhynchus) (Diptera: Culicidae) species from Brazil. J Med Entomol. 2008;45:970–981. doi: 10.1603/0022-2585(2008)45[970:iiandc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Sallum MAM, Schultz TR, Foster PG, Aronstein K, Wirtz RA, Wilkerson RC. Phylogeny of Anophelinae (Diptera: Culicidae) based on nuclear ribosomal and mitochondrial DNA sequences. Syst Entomol. 2002;27:361–382. [Google Scholar]
- Senise LV, Sallum MAM. Redescription of Culex (Culex) dolosus (Lynch Arribálzaga) (Diptera: Culicidae), based on specimens from Pico do Itapeva, Serra da Mantiqueira, São Paulo, Brazil. Zootaxa. 2008;1683:51–62. [Google Scholar]
- Sirivanakarn S, White GB. Neotype designation of Culex quinquefasciatus say (Diptera: Culicidae) Proc Entomol Soc Wash. 1978;80:360–372. [Google Scholar]
- Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PDN. DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae) Proc Natl Acad Sci USA. 2006;103:3657–3662. doi: 10.1073/pnas.0511318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinsanti LI, Diaz LA, Contigiani MS. Eco-epidemiología del virus encefalitis St. Louis en Córdoba, Argentina. Rev Fac Cienc Med. 2009;66:52–59. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald FV. A monograph of the Culicidae or mosquitoes. III. British Museum; London: 1903. 359 [Google Scholar]
- van Velzen R, Weitschek E, Felici G, Bakker FT. DNA barcoding of recently diverged species: relative performance of matching methods. 30490PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0030490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos PFC, Travassos-da-Rosa JFS, Travassos-da-Rosa APA, Dégallier N, Pinheiro FP, Sá GC., Filho Epidemiologia das encefalites por arbovírus na Amazônia brasileira. Rev Inst Med Trop Sao Paulo. 1991;33:465–476. [PubMed] [Google Scholar]
- Wang G, Li C, Guo X, Xing D, Dong Y, Wang Z, Zhang Y, Liu M, Zheng Z, Zhang H, Zhu X, Wu Z, Zhao T. Identifying the main mosquito species in China based on DNA barcoding. 47051PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0047051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Xie Z. DAMBE: data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Yao H, Song J, Liu C, Luo K, Han J, Li Y, Pang X, Xu H, Zhu Y, Xiao P, Chen S. Use of ITS2 region as the universal DNA barcode for plants and animals. 13102PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata S, Mejía L, Le Pont F, León R, Pesson B, Ravel C, Bichaud L, Charrel R, Cruaud C, Trueba G, Depaquit J. A study of a population of Nyssomyia trapidoi (Diptera: Psychodidae) caught on the Pacific coast of Ecuador. 144Parasit Vectors. 2012;5 doi: 10.1186/1756-3305-5-144. [DOI] [PMC free article] [PubMed] [Google Scholar]