Abstract
RING finger domain and RING finger-like ubiquitin ligases (E3s), such as U-box proteins, constitute the vast majority of known E3s. RING-type E3s function together with ubiquitin-conjugating enzymes (E2s) to mediate ubiquitination and are implicated in numerous cellular processes. In part because of their importance in human physiology and disease, these proteins and their cellular functions represent an intense area of study. Here we review recent advances in RING-type E3 recognition of substrates, their cellular regulation, and their varied architecture. Additionally, recent structural insights into RING-type E3 function, with a focus on important interactions with E2s and ubiquitin, are reviewed. This article is part of a Special Issue entitled: Ubiquitin-Proteasome System.
Keywords: RING finger, RING domain, U-box, ubiquitin ligase (E3), ubiquitin-conjugating enzyme (E2), ubiquitin, protein degradation, NMR spectroscopy, X-ray crystallography
1. Introduction
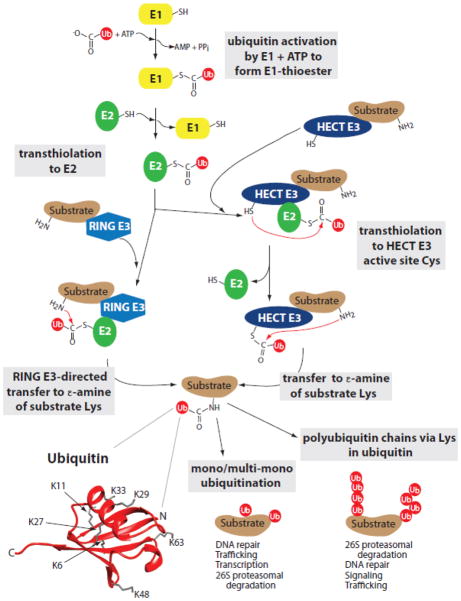
Protein ubiquitination initiates with the ATP-dependent activation of ubiquitin (Ub) by ubiquitin-activating enzyme (E1), where a thioester linkage is formed between the C-terminus of ubiquitin and the active site cysteine (Cys) of E1 (Figure 1). Ubiquitin is then transthiolated to the active site of one of ~40 different (in mammals) ubiquitin-conjugating enzymes (E2), generating an E2~Ub thioester. Specificity of ubiquitin modification is achieved largely through ubiquitin protein ligases (E3), which interact with both E2~Ub and the substrate to which ubiquitin is to be transferred. Ubiquitination generally occurs on primary amines as a consequence of nucleophillic attack on the E2~Ub linkage, resulting in stable isopeptide (or peptide) linkages with the C-terminus of ubiquitin. Ubiquitin can be transferred either to a lysine (Lys), or (less frequently) to the N-terminus, of either a substrate or another ubiquitin molecule to generate multi- or poly-ubiquitin chains. To a large extent, the nature of these ubiquitin linkages specifies the fate and function of the modified protein (see Figure 1 and legend for more detail).
Figure 1. Ubiquitination.
Protein ubiquitination requires the sequential action of three classes of enzymes. First, ubiquitin (Ub) is activated in an ATP-dependent manner by ubiquitin-activating enzyme (E1). In this step, the C-terminus of ubiquitin is linked by a thioester bond to the active site Cys of E1 (E1~Ub). Ubiquitin is then transthiolated to the active site Cys of a ubiquitin-conjugating enzyme (E2), generating an E2~Ub thioester. Ubiquitin protein ligases (E3) interact with both E2~Ub and the substrate to which ubiquitin is to be transferred, thus providing much of the specificity in the ubiquitin system. HECT-type E3s function as covalent intermediates in the ubiquitination pathway as ubiquitin is first transferred to the active site Cys of E3 via a transthiolation before conjugation to substrate. RING-type E3s instead mediate the transfer of ubiquitin directly from E2~Ub to substrate. Ubiquitination generally occurs on primary amines (Lys, and, less frequently, a free N-terminus) as a consequence of nucleophillic attack on the E2~Ub linkage, resulting in stable isopeptide (or peptide) linkages with the C-terminus of ubiquitin. Monoubiquitination plays diverse roles in processes such as DNA repair, protein trafficking, and transcription [159], and recent findings have demonstrated that multi-monoubiquitination can also target proteins for proteasomal degradation [160, 161]. Alternatively, ubiquitin can be transferred to one of the seven Lys (K) or the N-terminal Met of ubiquitin molecules [162] (PDB 1UBQ) that are already substrate-linked, generating multi- or poly-ubiquitin chains. To a large extent, the nature of the ubiquitin linkages specifies the fate and function of the modified protein. For example, chains of four or more ubiquitins linked through K48 efficiently target proteins for proteasomal degradation [163]. Similarly, K11-linked chains are integral to proteasomal targeting of anaphase-promoting complex/cyclosome (APC/C) substrates [164] and chains linked through other lysines may also play a role in proteasomal targeting [165, 166]. K63-linked and linear ubiquitin chains are implicated in non-proteasomal aspects of NF-κB signaling [167]. K63-linked chains and mono-ubiquitination are also implicated in DNA repair and the targeting of cell surface and endocytic proteins for lysosomal degradation [168, 169].
E3s function by one of two general mechanisms. They serve either as catalytic intermediates in ubiquitination, akin to E1 and E2, or they mediate the transfer of ubiquitin directly from E2~Ub to substrate. The former mechanism is used by Homologous to E6-AP Carboxy Terminus (HECT)-type E3s (reviewed in this issue by Martin et al). The latter mechanism is a characteristic of Really Interesting New Gene (RING)-type E3s. RINGs coordinate two Zn2+ ions in a cross-braced arrangement (Figure 2 and described in detail below) to create a platform for binding of E2s. The RING-like U-box family of E3s adopts a similar structure to bind E2s but without employing Zn2+ coordination. Together, RING (also known as RING finger, RING motif, or RING domain) and RING-like E3s (plant homeodomain/leukemia-associated protein (PHD/LAP) and U-box), collectively referred to as ‘RING-type’ in this review, constitute the large majority of the over 600 E3s in mammalian cells.
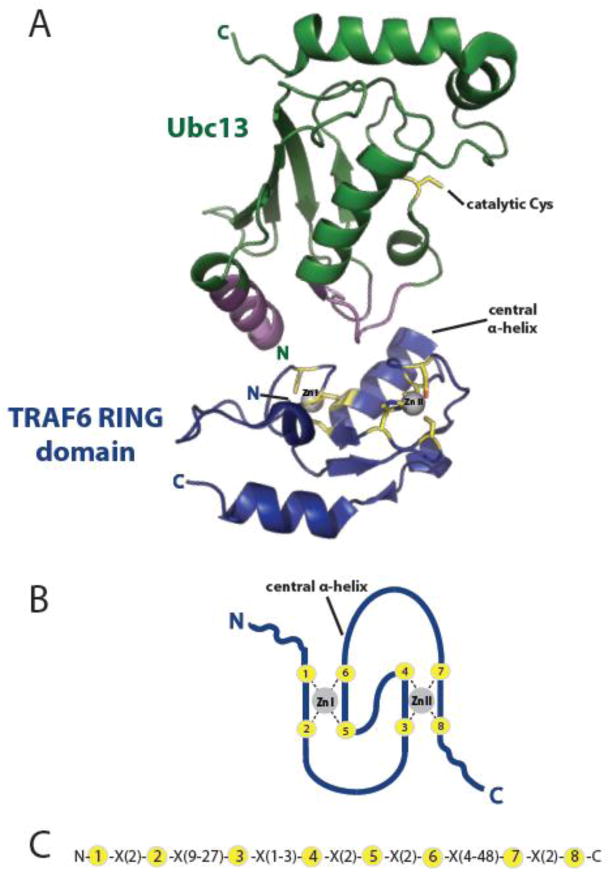
Figure 2. RING domains coordinate Zn2+ in a crossbrace arrangement that serves as a platform for E2 binding.
A) Representation of the crystal structure of the TRAF6 RING domain (blue) bound to the E2, Ubc13 (green) [89] (PDB 3HCT) highlights a stereotypical RING:E2 interaction. The catalytic Cys of Ubc13 is highlighted in yellow, while its RING domain-interacting regions are in purple. Yellow TRAF6 RING residues with sidechains shown are those that coordinate Zn2+ (C3HC3D), forming the RING crossbrace structure modeled in B). The two loops (Zn I, Zn II) and the intervening central α-helix formed by this structure together serve as a conserved platform for E2 binding. B, C) Model of the interleaved RING crossbrace structure (B) and consensus sequence (C). The eight Zn2+-coordinating residues are shown in yellow and X is any amino acid.
Any consideration of RING-type E3s must include their partners, the E2s. E2s have a core conserved ubiquitin-conjugating (UBC) domain that contains a conserved catalytic Cys (Figure 2). Some E2s have insertions within the UBC domain or N- or C-terminal extensions that confer specific functions. For HECT E3s, ubiquitin chain linkage specification lies largely with the catalytic HECT domain, but the situation is more complicated for RING-type E3s and their E2s. Some E2s are dedicated to specific ubiquitin linkages; other E2s are intrinsically more promiscuous with respect to the linkages they generate. Thus, a given RING-type E3 can generate different ubiquitin linkages depending on the E2 with which it is paired. Structural and functional properties of E2s have been reviewed in detail elsewhere [1].
Mutation of RING-type E3s, or modulation of their activity in other ways, is often associated with human disease. BRCA1, an E3 that plays critical roles in DNA repair, is mutated in familial breast and ovarian cancers [2]. Mutations in components of the FANC ubiquitin ligase, also involved in DNA repair, result in the multisystemic Fanconi Anemia syndrome [3], which includes severe developmental defects and, in children who survive, there is a marked increased risk of tumor development. Mdm2 (or Hdm2 in humans) was first characterized as a genetic amplification in mice associated with malignancy [4]. Indeed, increased activity of this E3 towards the tumor suppressor p53, either through increased Mdm2 expression or loss of a negative modulator of Mdm2 activity, is associated with human cancers, particularly those 50% that retain wild type p53 [5]. The F-box protein FBXO11, the substrate recognition component of the multi-subunit SCFFBXO11 E3 (see section 3 below), functions as a tumor suppressor by targeting BCL6, a transcription factor involved in B-cell differentiation and activation, for degradation [6]. FBXO11 is mutated or deleted in diffuse large B-cell lymphomas [6]. Mutations in VHL, the substrate recognition component of the CRL2VHL E3, lead to the malignant von Hippel-Lindau syndrome, which presumably arises because of dysregulation of HIF1-α and/or HIF2-α [5]. Mutations in the RING-IBR (in between RING)-RING E3 Parkin are associated with autosomal recessive juvenile Parkinsonism (AR-JP) [7]. Additionally, a number of viruses, for example, herpes simplex virus type 1 (HSV-1), encode RING-type E3s as virulence factors [8, 9]. In the case of HIV, the virus encodes an adaptor protein, Vpu, that redirects SCFβTrCP to downregulate CD4 [10]. The importance of RING-type E3s to human health and disease has contributed to their becoming an intensively-studied family of proteins. This review will provide an overview of their regulated function and structure and recent advances in understanding how they mediate ubiquitination by E2s.
2. RING dimerization
RING-type domains are found in many different structural contexts. While many exist as single-chain enzymes (Figure 3A), a notable feature of RING-type E3s is their tendency to form homodimers and heterodimers (Figure 3C–3F). Homodimeric RING-type E3s include cIAP, RNF4, BIRC7 (shown in Figure 3D), IDOL, and the U-box proteins CHIP and Prp19 [11–17]. Examples of well-characterized heterodimeric E3s include BRCA1-BARD1 (shown in Figure 3F), Mdm2-MdmX (or HdmX/Hdm4 in humans), and RING1B-Bmi1. While for homodimeric RING E3s both RINGs have the intrinsic capacity to functionally interact with E2s, this appears not to be the case for some heterodimeric RINGs. BRCA1 and RING1B each function with E2, while their partners serve to enhance activity, potentially interact with substrates, and, in the case of BRCA1-BARD1, to stabilize the complex in vivo [18–21]. For the RING of BARD1, its lack of E2 binding activity can be attributed, at least in part, to the absence of a portion of the conserved central α-helix necessary for E2 interactions [19, 22]. In contrast, there is evidence that MdmX, the ‘inactive’ partner, does physically interact with E2, in addition to having the capacity to bind the best-characterized Mdm2-MdmX substrate, p53. Importantly, while Mdm2 can homodimerize and is active, MdmX has little tendency to form a homodimer and is inactive, and the two RINGs can form an active heterodimer [23, 24]. This underscores the important role played by RING dimerization (discussed further below). Strikingly, MdmX’s lack of in vitro activity can be restored by mutating a single residue at the RING dimerization interface to that found in the analogous position of Mdm2 [25]. However, additional mutations of MdmX to mimic the nucleolar localization sequence of Mdm2, found in its RING:E2 interface, are required for in vivo activity of MdmX towards p53 [25]. Interestingly, since p53 exists largely as a homo-tetramer [26], there is the potential to assemble four Mdm2-MdmX heterodimers in close proximity. RING dimerization may also be a mode of cellular regulation of ubiquitination, as occurs for cIAP1, where its RING homodimerization interface is sequestered in a ‘closed’ inactive form until activation by IAP antagonists, such as SMAC (second mitochondrial activator of caspases) or DIABLO (direct IAP-binding protein with low isoelectric point) [27]. Binding of SMAC or DIABLO to cIAP stabilizes it in an ‘open’ conformation that allows RING dimerization and thus, presumably, E2 binding and ubiquitin transfer.
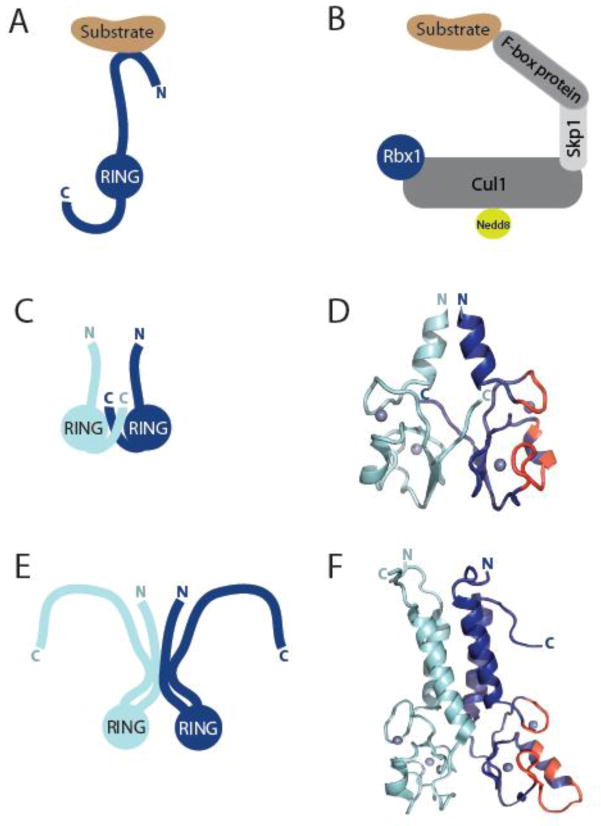
Figure 3. Architecture of RING-type E3s.
A) Model of a monomeric RING-type E3, where its RING domain would mediate binding to E2 thioester-linked to ubiquitin. Binding to substrate occurs generally through regions of the E3 other than the RING domain. B) Model of a multi-subunit RING E3 of the Cullin RING Ligase (CRL) superfamily, such as the well-studied SCF (CRL1) family, shown here. SCF consists of a cullin protein (Cul1) a small RING finger protein (Rbx1), and an adaptor protein (Skp1) that binds interchangeable substrate recognition elements (F-box proteins). The ubiquitin-like molecule, Nedd8, is reversibly conjugated to cullins and associated with activation of CRLs. C) Schematic of dimeric RING E3s, such as cIAP, RNF4, BIRC7, IDOL, Mdm2-MdmX, that dimerize through their RING domains and interleaved C-terminal tails. D) Ribbon diagram illustrating the homodimeric RING E3, BIRC7 [13] (PDB 4AUQ) as a representative of the class of dimers schematized in C. The E2-interacting residues of one RING domain are highlighted in red. E) Model of dimeric RING E3s, such as BRCA1-BARD1 and RING1B-Bmi1, where α-helices both N- and C-terminal to the RING facilitate dimerization. In the case of BRCA1-BARD1 (illustrated), this occurs through a four α-helix bundle (helices above RINGs in F). F) Ribbon diagram illustrating the heterodimeric RING dimer of BRCA1-BARD1 [19] (PDB 1JM7) modeled in E). The E2-interacting residues of the RING domain of BRCA1 are highlighted in red.
RING-type dimers are generally formed in one of two ways: 1) sequences outside the RING are primarily responsible for dimerization; or 2) the RING per se is responsible for dimerization. In both types, the two RINGs are positioned such that the E2 binding surfaces face away from each other (see Figure 3D and 3F, surface highlighted in red), indicating that a direct cooperative interaction between the two RING-bound E2s in the context of a dimer is unlikely. At present, there is more structural characterization of RING dimers of the second type (Figure 3C and 3D), which includes Mdm2-MdmX and homodimers of RNF4, IDOL, BIRC7, and cIAP. These RINGs are all found at the extreme C-terminus of the proteins containing them and the structures reveal that dimers are formed via interleaved C-termini, explaining previous reports of the importance of the C-termini in dimer stability and E3 ligase activity [11–14, 28]. In contrast, dimers of the first type are formed via interactions involving (usually) α–helical regions that flank each of the two RINGs (Figure 3E and 3F). Proteins that form RING dimers in this manner include Rad18, BRCA1-BARD1, and RING1B-Bmi1 [19, 29, 30]; these dimers have their RINGs near their N- or C-termini. These two manners of dimerization are not necessarily mutually exclusive and, in fact, the available U-box structures of CHIP and Prp19 homodimers reveal distinct dimerization interfaces involving the U-box as well as regions N- and/or C-terminal to the U-box domain [15, 17]. Some RING-type E3s have been shown to dimerize or form oligomers through domains that are structurally distinct and remote from the RING. Interestingly, proteins belonging to this group, such as Cbl family members and gp78 [31–34], contain RINGs that are neither at the N- or C-terminus of the E3. Higher-order oligomers that bring together multiple RING-type dimers have also been reported for Prp19, which is active as a tetramer [17].
3. Multi-subunit RINGs
There are RING-type E3s that exist as multi-subunit assemblies (see Figure 3B). A striking example is the Cullin RING Ligase (CRL) superfamily [35], which exhibits enormous plasticity in substrate specificity. Each CRL subfamily is characterized by a cullin protein (Cul-1, 2, 3, 4a, 4b, 5, or 7), a small RING protein (in most cases Rbx1/Roc1/Hrt1), and either an adaptor protein(s) that binds interchangeable substrate recognition elements or, in the case of CRL3, proteins that bind both to the cullin protein (Cul-3) and to substrate [36]. The CRL superfamily is exemplified by the well-studied Skp1-Cul1-F-box protein (SCF) family (Figure 3B), in which one of ~69 (in humans) interchangeable F-box proteins can potentially recognize substrates [37] (Reviewed in this issue by Bassermann et al.). Exchange of F-box proteins within the SCF scaffold takes place through a complex cycle that includes dynamic attachment and removal of the ubiquitin-like modifier, Nedd8 [38]. While the CRL superfamily overwhelmingly exhibits the greatest range of substrate recognition, other multi-subunit E3s exhibit even greater structural complexity. The anaphase promoting complex/cyclosome (APC/C) is a highly complex E3 that in humans contains 13 core subunits including a cullin-like protein and a small RING protein. It also has two interchangeable co-activator subunits, Cdc20 and Cdh1, which recognize distinct substrates and are active during different phases of the cell cycle [39] (Reviewed in this issue by Bassermann et al.).
Another multi-subunit RING-containing ligase is the Fanconi Anemia (FANC) E3. There are at least 13 complementation groups associated with this disease, and proteins corresponding to eight complementation groups are components of the FANC ubiquitin ligase, including a RING-type protein (FancL). This E3 is recruited to sites of DNA damage to effect translesional repair. Despite its complexity, the role of the FANC E3, as we currently understand it, is limited to monoubiquitination of two associated proteins that are subsequently deubiquitinated as part of the DNA repair process. Degradation of a key FANC component, FANCM, via SCFβTrCP is responsible for inactivating the function of the FANC E3 during mitosis, thereby preventing chromosomal abnormalities [3, 5, 40].
Some multi-subunit RING-type E3s contain multiple RING proteins. The yeast GID (glucose-induced degradation deficient) complex, which targets fructose-1, 6 bisphosphatase for ubiquitination in response to glucose, consists of seven subunits including two interacting RINGs [41]. The yeast PEX ubiquitin ligase, which mono-ubiquitinates the peroxisome receptor, Pex5p, and possibly other substrates, includes three distinct RING proteins as part of a multi-subunit complex [42, 43]. The specific function of each of the RINGs in such complexes is currently unknown.
Finally, there are single proteins that contain multiple RINGs. Mindbomb, involved in Notch signaling, has three RINGs in its C-terminal region, although to date only the most C-terminal of these has been studied and shown to be required for activity [44]. RING-IBR-RING (RBR) proteins are a class of ~13 proteins (in humans) that include a RING consensus sequence (RING1) followed by a Cys-rich ‘in between RING’ (IBR) region and a third domain, the RING2, that was originally characterized as a second RING-like domain. Although RBR proteins were thought to function as canonical RING E3s, recent studies have shown that they employ a RING-HECT hybrid mechanism [45–49]. The RING1 domain binds E2 (similar to the RING mechanism) but ubiquitin is transferred to a specific Cys within RING2 before being transferred to substrates (similar to the HECT mechanism). Well-known members of this family include Parkin, HHARI, HOIP, and HOIL-1L. The latter two are subunits of the Linear Ubiquitin Chain Assembly Complex (LUBAC) E3 consisting of HOIP, HOIL-1L, and Sharpin (a non-RING-containing protein). This complex plays critical roles in NF-κB activation (reviewed in this issue by Iwai et al).
4. RING-type E3s and their substrates
There is enormous diversity in substrate ubiquitination and its regulation, as the targets of RING-type E3s are incredibly varied. RING-type E3s are implicated as tumor suppressors, oncogenes, and mediators of endocytosis, and play critical roles in complex multi-step processes such as DNA repair and activation of NF-κB signaling. A RING-type E3 may have multiple substrates and several E3s can target the same substrate. Not surprisingly, the mechanisms of substrate recognition by RING-type E3s are highly varied, and occur in the context of networks of interactions that often also include HECT E3s and deubiquitinating enzymes (DUBs). Substrates may bind directly to a RING-type E3 or may associate indirectly. The capacity of RING-type E3s for self-ubiquitination, first utilized as a means of assessing their potential to function with E2s [50], frequently occurs in vivo, as does ubiquitination of RING E3s by heterologous RING or HECT-type E3s as part of regulatory networks [51].
E3-substrate interactions may be constitutive, and, in such cases, regulation can occur at the level of E3 transcription or degradation. The complexity of such trans-regulation is illustrated by the interplay of SCF and APC/C E3s during the cell cycle where, for example, APC/CCdh1 targets the F-box protein Skp2 for degradation in early G1, thereby stabilizing p27 and preventing premature G1-S transition, and SCFβTrCP targets the ‘pseudo-substrate’ and suppressor of APCCdc20, EMI1, for degradation in late G2 [5] (reviewed in this issue by Bassermann et al.). Similarly, SCFFBXO11 ubiquitinates and targets Cdt2, the conserved substrate recognition subunit of CRL4Cdt2, for degradation, stabilizing its substrates, such as p21 and Set8, and allowing for cells to properly exit the cell cycle [52, 53].
An emerging theme is a role for metabolites in substrate recognition and E3 activity. As above, the effect may be via a direct interaction between a metabolite and a RING-type E3, or may be indirect, for example via interaction with the substrate. In the latter category, sterols serve as feedback regulators of their own synthesis by regulating the association of Insig-1 with the ER-resident RING E3 gp78 and hence the stability of the former, which is critical to regulation of cholesterol biosynthesis [54]. The plant hormones auxin and jasmonic acid are examples of regulation by a direct metabolite:E3 interaction [55, 56]. These hormones bind directly to SCF complexes and target transcriptional repressors for ubiquitination and degradation. This strategy provides a way to de-repress gene expression and alter the transcriptional profile in response to environmental factors in plants. Another intriguing example of regulation by a metabolite is a report that the RING E3 TRAF-2 is inactive due to its RING structure being unsuitable for E2 interactions, but is activated by its association with sphingosine-1-phosphate [57]. A structural understanding of how this occurs awaits further studies. Nevertheless, as nature rarely uses a good idea just once, it seems likely that additional examples of small molecule or metabolite activation (or inhibition) of E3s will be uncovered in the future.
The most common means of regulating substrate ubiquitination is by post-translational modifications that alter either ligase activity or substrate recognition by RING-type E3s. Examples of regulation via protein phosphorylation are widespread. Regulated substrate phosphorylation on either Ser or Thr residues allows for the recognition of numerous substrates by SCFβTrCP and SCFFbw7 [37]. Tyrosine phosphorylation of activated receptor tyrosine kinases (RTKs) facilitates their recognition by the Cbl family of RING E3s (Cbl, Cbl-b and Cbl-c) [58]. Cbl-c phosphorylation also modulates the dynamic interaction of its RING with E2~Ub (see below). The net result of activation of Cbl family members includes the ubiquitination of RTKs, leading to their lysosomal degradation and an attenuation of signaling. Both phosphorylation of the core RING subunit and dephosphorylation of Cdc20 play roles in the activation of the APC/CCdc20 [59]. The level of complexity that phosphorylation offers is exemplified by p53 and Mdm2. Regulated phosphorylation of specific residues on either p53 or Mdm2 can either inhibit or enhance their interaction. Also, Mdm2 degradation as a consequence of self-ubiquitination is enhanced by phosphorylation, which prevents interaction of Mdm2 with the DUB HAUSP/USP7. The failure of Mdm2 to be deubiquitinated by USP7 leads to degradation of Mdm2 and increased p53 activity under conditions of genotoxic stress [5] (reviewed in this issue by Vousden et al).
A growing number of other post-translational modifications are implicated in regulation of the ubiquitin system, including substrate modification by hydroxylation, glycosylation, acetylation, methylation, modification by poly(ADP-ribose) (PAR), and attachment of ubiquitin-like modifiers. The HIF1-α and HIF2-α transcription factors undergo proline hydroxylation in response to increased oxygen levels and become substrates for the multi-subunit RING E3, CRL2VHL. The hydroxylation reaction is mediated by proline-hydroxylase domain (PHD) proteins, which are themselves targets for SIAH RING E3s. SIAH expression is positively regulated by HIF transcriptionally, creating a positive feedback loop to increase HIF1-α levels [5]. A subfamily of F-box proteins, denoted as Fbs1-5, has the potential to recognize glycosylated proteins in the cytosol, which have presumably been transported out of the endoplasmic reticulum (ER). This provides a means to target these displaced glycoproteins for ubiquitination and proteasomal degradation [60].
Modification of proteins with the small ubiquitin-like modifier (SUMO) family of proteins can inhibit substrate degradation by competing for specific sites of ubiquitination or by altering the localization of modified proteins. Perhaps a more general role for sumoylation is to target proteins for ubiquitination, particularly those modified with chains of SUMO (in metazoans SUMO 2/3). This occurs by SUMO-targeted ubiquitin ligases (STUbLs), which contain multiple SUMO-interacting motifs (SIMs) in addition to their E3 ligase domains [61].
The RING E3 RNF146 uses a WWE domain to recognize substrates modified with poly(ADP-ribose) (PAR). In response to Wnt signaling, axin is PARsylated by tankyrase and subsequently ubiquitinated by RNF146, ultimately resulting in its proteasomal degradation. In vitro, PAR as been shown to stimulate the ligase activity of RNF146, though the mechanism is presently unknown [62–64]. There are other RING-type E3s that contain WWE domains, including the Deltex family, whose members play an important role in Notch signaling. Thus, it is likely that additional examples of regulation by PARsylation will emerge in the future.
There is a complex dynamic between ubiquitination, acetylation, and methylation of lysines as part of the histone code [65]. Additionally, there is data to suggest that specific lysines in p53 that are targets of Mdm2-MdmX are also acetylated, thereby preventing their ubiquitination [66]. The acetylation of Mdm2 itself may decrease its activity towards p53 [67]. There is also evidence for interplay between acetylation and ubiquitination of transcription factors such as estrogen receptor-α[68].
Finally, for E3s to target substrates they must exist in the same cellular compartment. Some RING-type E3s have nuclear localization signals and many RING-type E3s are transmembrane proteins targeted to sites such as the ER, plasma membrane, endosomes, peroxisomes, and mitochondria [42, 69–73].
5. RING-type domain structure
RING structure is conformed as a consequence of a cross-braced arrangement of eight Zn2+ coordinating residues, generally Cys and His, with conserved spacing between these residues (Figure 2B and 2C). Canonical RINGs have either one or two His in the linear arrangement of coordinating residues, denoted C3H2C3 or C3HC4, however other variations exist. The PHD/LAP finger found in the transcription factor NF-X1 and the MARCH family of membrane-bound E3s is defined by its C4HC3 consensus. RINGs having a C8 configuration (CNOT4) or an Asp residue in the final position (e.g. Rbx1 and TRAF6) have been shown to have ligase activity [74–76]. Thus, it has become apparent that categorizing RINGs by the linear arrangement of coordinating residues has little to do with functional properties of the domain. Nevertheless, context does matter, as swapping Zn2+ liganding residues in a C3H2C3 RING to create a C3HC4 configuration resulted in loss of activity for AO7 (RNF25), one of the first RING E3s studied [50]. NF-X1 contains a sequence in which both a RING and a PHD/LAP motif are recognizable, but only the PHD/LAP consensus is functional [77]. Unlike RING domain E3s, U-box proteins do not coordinate Zn2+ but adopt a RING-like tertiary structure for binding E2, stabilized by non-covalent interactions among core amino acids [16]. Additionally, some pathogenic bacteria have evolved proteins that show no sequence homology to eukaryotic RING or U-box domains, yet fold into highly similar structures and display robust ubiquitin ligase activity [78, 79].
Crystallographic and NMR-based analyses have revealed that RINGs and U-boxes have a common mode of interaction with E2s (Figure 2A). The key structural elements are two loop-like regions, which, in the case of RINGs, coordinate Zn2+. The loops surround a shallow groove formed by the central α-helix. Together these elements serve as a platform for interactions with the UBC domain of E2s (Figure 2A). The E2 surface that interacts with the RING domain overlaps with the region that interacts with E1, leading to the notion that dissociation of E2s from RINGs is required for an E2 to be ‘reloaded’ with ubiquitin by E1 [80–82]. A characteristic of RING:E2 interactions is that they are generally of low affinity, typically with Kd values in the high micromolar range. Thus, even though a RING domain may function robustly with an E2, assessing physical interactions between these proteins using standard ‘pulldown’ approaches is often not fruitful. Exceptions to this feature include E3s such as gp78 [83], Rad18 [84], and AO7 [50] (S. Li, Y. Liang, X. Ji, & A.M.W., unpublished observations), which contain regions outside the RING motif that bind E2s through distinct interfaces, resulting in high affinity interactions (see below).
RING:E2 interactions typically involve conserved, bulky hydrophobic side chains. Mutation of these side chains mitigates E2 binding and causes decreased levels of ubiquitination activity in vitro, as demonstrated for c-Cbl Trp408Ala [85], CNOT4 Leu16Ala [74], and BRCA1 Ile26Ala [86]. However, because a given RING can function with a cohort of E2s with varying binding affinities [86, 87], mutation of such RING domain:E2 residues may yield unexpected results. For example, residual activity is observed in vitro for the BRCA1 Ile26Ala mutant with select E2 pairings (J.N.P., D.M. Wenzel, & R.E.K., unpublished observations), and the analogous mutation in other RING E3s does not consistently eliminate activity (J. Callis, UC Davis, personal communications). Thus, the relationship between E2 binding and activity remains to be fully characterized and will require mutants where, in the context of a correctly folded RING (i.e. retaining its Zn2+-coordinating residues), E2 binding is abolished. Conversely, in vivo analysis of RING function demands a truly ‘ligase-dead’ mutant that retains E2~Ub binding. Identification of such mutants awaits a more thorough definition of RING catalytic function.
6. RINGs as Activators of E2~Ub
In contrast to HECT-type E3 ligases, RING-type E3s lack a bona fide catalytic center. A lingering question in the field has been whether RINGs serve solely to position E2~Ub relative to the substrate, or if they also serve as activators of E2~Ub. Clearly, unwanted ubiquitination events in the cell are detrimental, so it follows that the reactive E2~Ub species must be activated at the opportune moment for transfer. Specialized examples have indeed provided evidence in support of an activating role for RING E3s. One involves the E2 Ubc13 (Ube2N in humans), which catalyzes free, K63-linked polyubiquitin chains in the presence of its accessory protein, Mms2. A crystal structure of the Mms2:Ubc13~Ub complex revealed that Mms2 binds an incoming substrate ubiquitin in a way that orients the ubiquitin K63 directly towards the Ubc13~Ub thioester [88]. Thus, this heterodimeric E2 carries its own substrate-binding domain and does not require an E3 to coordinate substrate. Nevertheless, ubiquitin chain formation by Mms2:Ubc13 is dramatically enhanced in the presence of a minimal RING domain [89]. A second example, inspired by work of the late Cecile Pickart, showing E2-catalyzed Ub transfer onto free Lys, demonstrates an increased rate of ubiquitin discharge to small molecule nucleophiles in the presence of minimal RING domains [45, 90]. Use of these substrate-independent assays has allowed the observation of a catalytic role for RING-type E3s in ubiquitin transfer reactions uncoupled from proximity effects afforded by E3:substrate interactions.
In all available E2:E3 structures, the RING-type domain binds the E2 on a surface that is remote from the active site Cys (and therefore from the ubiquitin thioester) (Figure 2). The non-contiguous E3-binding and active sites on the E2 imply that the role played by a RING to facilitate ubiquitin transfer may be indirect and, therefore, allosteric. However, apo- and E3-bound E2 structures are largely indistinguishable and fail to suggest a mechanism for the allostery. Recent structural studies characterizing the interactions of the more relevant E2~Ub conjugated species with RING-type domains have provided much needed insight. Notably, a solution-based study of E2~Ub conjugates established their dynamic nature and that E2 and the thioester-linked ubiquitin adopt an array of ‘open’ and ‘closed’ conformations [91] (Figure 4A). Three structures of E2~Ub conjugates of the UbcH5 family (Ube2D1-3) in complex with RING-type E3s (RNF4:UbcH5A~Ub, E4B:UbcH5C~Ub, and BIRC7:UbcH5B~Ub) provide the first glimpses at an E3:E2 complex poised to transfer ubiquitin [13, 92, 93]. A striking common feature is a ‘closed’ conformation of the E2~Ub conjugate in which the Ile44 hydrophobic surface of ubiquitin is positioned against the 310 helix, active site, and helix 2 of E2 (Figure 4A–C). In solution, where multiple species can exist simultaneously, E3 binding promotes a population shift in the highly flexible E2~Ub towards closed conformations which, based on activity data, primes the active site for transfer (Figure 4A) [13, 93, 94]. The closed E2~Ub states are readily disrupted and even conservative mutations of hydrophobic residues in the interface between E2 and ubiquitin can destabilize the closed state and greatly decrease E3-stimulated ubiquitin transfer [93]. It is interesting to note that E2~Ub conjugates that populate closed conformations to a significant extent in the absence of an E3, such as Ubc13 [91], Ubc1 [95], Ube2S [96], and Cdc34 [97], also demonstrate E3-independent ubiquitin transfer. In contrast, the UbcH5 family of E2s, which shows robust activity with a large number of RING-type E3s, has almost undetectable E3-independent activity, consistent with its populating mainly open states in the absence of an E3 [91].
Figure 4. Activation of E2~Ub conjugates by RING-type domains.
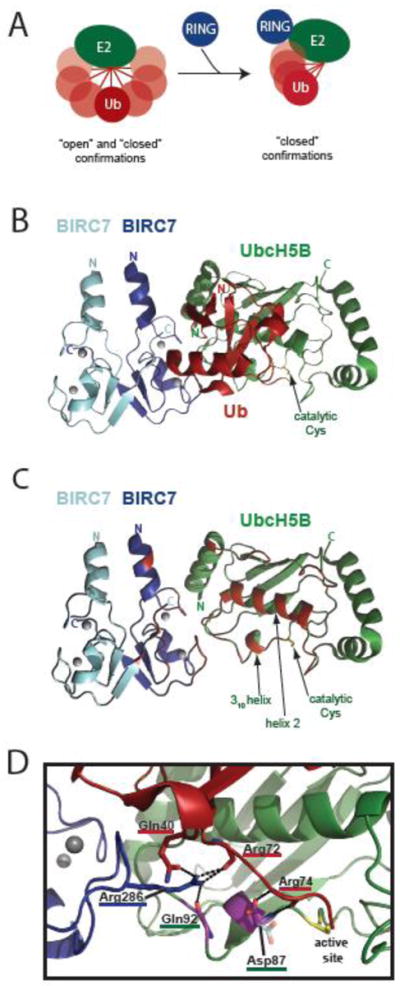
A) Schematic of ubiquitin thioester linked to E2 sampling ‘open’ and ‘closed’ confirmations in the absence of a RING domain (left). RING binding to E2~Ub promotes increased occupancy of closed confirmations needed for ubiquitin transfer (right). B) Ribbon diagram of BIRC7-BIRC7:UbcH5B~Ub (PDB 4AUQ) as an example of a RING-type E3:E2~Ub ternary complex. E3, E2, and ubiquitin are blue, green, and red, respectively. The catalytic Cys of UbcH5B is labeled. Gray circles are the zincs coordinated by the BIRC7 RINGs. C) The ribbon diagram shown in B, with the ubiquitin molecule removed and the residues on E2 or the RING domains that would contact ubiquitin in red. The 310 helix, helix 2, and catalytic Cys of UbcH5B are labeled. D) A closer view of the E2 active site and ubiquitin C-terminus from the BIRC7-BIRC7:UbcH5B~Ub structure shown in B and C. The UbcH5B helix 2 (green) is positioned directly above Arg72. Residues from BIRC7, UbcH5B, and ubiquitin are underlined in blue, green, and red respectively. The hydrogen bonding network created by BIRC7 Arg286 (blue) and the E2 backbone of Gln92, the ubiquitin backbone of Arg72, and the side chain of ubiquitin Gln40 is shown. Residues colored in purple correspond to those that display NMR spectral effects specifically arising from the E3:E2 hydrogen bond. The ‘up’ conformation of the Asp87 side chain seen in this structure is shown in purple and red, the ‘down’ conformation, frequently seen in structures in the absence of covalently-bound ubiquitin is shown coming off the backbone directly below in semi-transparent cyan and orange (taken from PDB 3UGB [170]). Contacts made by the ‘up’ Asp87 conformation to Arg74 (red) of ubiquitin are shown. The side chains of UbcH5B Gln92, Ub Arg72, and Ub Arg74 are not shown for clarity.
Several non-mutually exclusive possibilities for how RING binding promotes closed E2~Ub conformations are suggested by the recent RING-type E3:E2~Ub structures. One possibility is through direct RING-ubiquitin interactions. Available structures, however, show that the extent to which the RING directly interacts with ubiquitin varies among E3s (Figure 4B and 4C). The interleaved homodimeric RING structures of BIRC7 and RNF4 show additional interactions required for activity between the opposing (i.e. non-E2-binding) RING and the conjugated ubiquitin [13, 92, 98] (Figure 4C, see the C-terminus of BIRC7 highlighted in red). As discussed above, E3s including Mdm2-MdmX, XIAP, and IDOL also adopt an interleaved RING dimer structure and therefore may adopt a similar strategy to enhance closed E2~Ub states. However, monomeric RING-type E3s such as the U-box E3 E4B, or dimeric E3s such as BRCA1-BARD1, which dimerize through regions outside of their RINGs, either lack the additional ubiquitin-interacting surface used by the interleaved dimers or this surface is not available to interact with ubiquitin. The presence or absence of additional RING contacts with ubiquitin is consistent with reports that some E3s exhibit a higher affinity toward E2~Ub than the isolated E2, while others bind them with affinities that are indistinguishable.
A second non-conflicting possibility for how RING binding promotes closed E2~Ub conformations is through allosteric activation in the context of the E2~Ub. All three of the recent RING-type E3:E2~Ub structures contain an Arg in loop 2 of the RING-type domain that is critical for E3-enhanced activity (e.g. Figure 4D, Arg286 of BIRC7, shown in blue). Crystal structures of the dimeric RNF4 and BIRC7 complexes show the side chain of this Arg hydrogen bonding with the backbone of the E2 (Gln92), the backbone of ubiquitin (Arg72), and the side chain of ubiquitin residue Gln40 (Figure 4D). While mutation of the analogous Arg (Arg1143Ala) in the monomeric E4B only modestly decreases its binding affinity for E2, it leads to a substantial loss of closed E2~Ub conformations. Coincident with this is a disappearance of select NMR spectral perturbations near the E2 310 helix associated with functional E2 binding and the formation of closed E2~Ub conformations. The Arg-induced alterations near the E2 310 helix span much of the distance between the RING-binding surface and active site of E2, prompting Pruneda et al. to propose that the interaction between this Arg and the backbone of E2 is the allosteric ‘link’ underlying the observed E2~Ub closed conformation and rearrangements near the active site (Fig. 4D, magenta). Notably, a similar allosteric path had been proposed previously for UbcH5 on the basis of a statistical coupling analysis of E2 primary sequences [99]. Interestingly, as noted above, the analogous RING loop 2 Arg also contacts ubiquitin in the crystal structures of RNF4 and BIRC7, and mutation of ubiquitin Gln40 (Gln40Ala or Gln40Arg) affects E3-enhanced activity. However, the significance of the Arg interaction with Gln40 in promoting E2~Ub closed confirmations in the context of RING binding awaits further analysis. Notably, only an Arg at this position can provide multiple hydrogen-bond donors; even a Lys, the next the most prevalent amino acid found at this position in RING-type domains, cannot do so.
Intriguingly, one E2 residue may serve as a molecular ‘gate’ to allow the C-terminus of ubiquitin to access the closed E2~Ub conformations favorable for ubiquitin transfer. This residue, Asp87 in UbcH5 (Ube2D) family members, resides on one side of the opening that leads to the active site Cys (Figure 4D). In the structures of the RNF4 and BIRC7 RINGs bound to UbcH5~Ub, the Asp side chain is positioned to form hydrogen bonds to the backbone of the ubiquitin C-terminal tail. How RING binding promotes the positioning of this Asp is unclear, but this may occur through the allosteric link between the critical Arg of the RING with Gln92, which neighbors Asp87. Given the lack of observable changes in structure in this region when RING-type E3:E2 and RING-type E3:E2~Ub structures are compared, the effect is likely to be a subtle one, probably involving small but important changes in electrostatics. The steric and chemical nature of this molecular gate is critical for activity, as even a Glu substitution severely impacts ubiquitination [45]. The residue corresponding to Asp87 is conserved as Asp, Asn, or Ser in most E2s, with the exceptions of UbcH7 and UbcH8, neither of which has been shown to function with RING-type E3s. Notably, the SUMO-specific E2, Ubc9, which encodes a Ser at the position analogous to Asp87 in UbcH5C, makes a similar contact with the C-terminus of SUMO in the SUMO-RanGAP1-Ubc9-Nup358 structure, consistent with a general mechanistic feature of ubiquitin and ubiquitin-like transfer [100].
In sum, it is now clear that RING-type E3s are more than mere molecular scaffolds. By binding E2~Ub conjugates and promoting closed conformations, RING-type E3s activate their cognate E2s to stimulate ubiquitin transfer. Several non-mutually exclusive mechanisms for how RING domains promote the structural arrangements associated with increased ubiquitin transfer activity have been suggested by recent studies, but all require further vetting with other RING-type domains and E2s. Nevertheless, these studies suggest that it will be possible to create ‘catalytic’ mutations within RING-type E3s that can be used in place of, or together with, E2-binding mutations (such as the widely used BRCA1-Ile26Ala) to create more profoundly ligase-dead versions of RING-type E3s. Such strategies will pave the way for the generation of new tools to be used in investigations of E3 cellular functions and protein substrates.
7. E2-binding domains distinct from the RING domain
7.1 E2-binding domains found in RING proteins
In addition to canonical RING:E2 interactions, RING-type E3s can also modulate E2 function using UBC-interacting domains separate from their RINGs. Of the non-RING domains characterized structurally, most contact the ‘backside’ of the E2 UBC, a region centered on its β-sheet, opposite its active site, and distinct from both RING- and E1-interacting regions. The backside of some E2s interacts non-covalently with ubiquitin via a hydrophobic patch formed by Leu8, Ile44, and Val70 within the ubiquitin β-sheet [84, 101, 102] (Figure 5A). This surface of ubiquitin is commonly recognized by ubiquitin-binding domains (UBDs) [103], and similar to the interaction of ubiquitin with UBDs, the ubiquitin:E2 backside interaction is of relatively low affinity (Kd of ~300 μM) [102]. At least in the case of BRCA1-BARD1, the ubiquitin:E2 backside interaction enhances processive polyubiquitination [102], potentially by facilitating increased E2~Ub self-assembly. Several ubiquitin E2-variant (UEV) proteins also bind ubiquitin via their backside site and the yeast ubiquitin E2 Ubc4 interacts with the Ubl-protein, Nedd8, using the same interface [104–106]. Arabidopsis thaliana membrane-anchored Ub-fold (MUB) proteins bind to the backside of some E2s and target them to the plasma membrane [107]. MUBs structurally resemble ubiquitin, but their C-terminal CAAX motif, which specifies lipidation [108], precludes potential activation by E1. There are also several examples of backside binding to the SUMO E2, Ubc9 (see below).
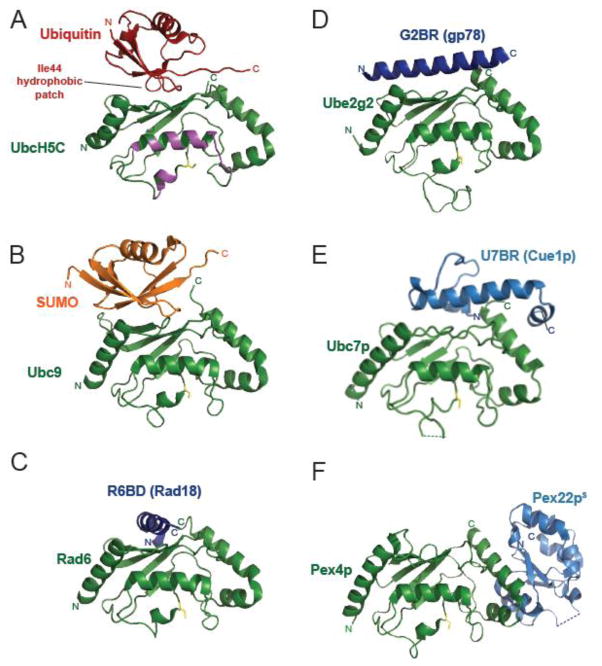
Figure 5. E2 binding domains other than the RING modulate ubiquitination by binding to the ‘backside’ of E2s.
A–F) Structures of A) ubiquitin (red, PDB 2FUH [102]); B) SUMO (orange, PDB 2UYZ [151]); or the non-RING E2 binding domains (blue) found in C) Rad18 (R6BD, PDB 2YBF [84]); D) gp78 (G2BR, PDB 3H8K [83]); E) Cue1p (U7BR, PDB 4JQU [141]); or F) Pex22p (Pex22pS, PDB 2Y9M [148]) binding to the ‘backside’ of their respective E2s (labeled and shown in green). The catalytic cysteine of each E2 is highlighted in yellow. The thioester-linked ubiquitin interaction surface is colored in magenta in panel A.
The RING E3 Rad18 contains a C-terminal E2 backside-binding domain. Rad18 and its cognate E2, Rad6 (Rad6a/Ube2A and Rad6b/Ube2B in mammals), are conserved and essential members of the DNA damage response. Together, Rad18 and Rad6 specifically monoubiquitinate proliferating cell nuclear antigen (PCNA), a modification that recruits translesion DNA polymerases to stalled replication forks [109]. Rad18 interacts with Rad6 both canonically through its RING domain and via the C-terminal domain, called the Rad6-binding domain (R6BD) [110–113] (Figure 5C). A crystal structure of the R6BD in complex with human Rad6b revealed that the R6BD forms a ‘kinked α-helix’ that binds the backside of the E2 [84]. The backside-binding site of the R6BD on Rad6 significantly overlaps with the low affinity site of non-covalent ubiquitin binding and with a Kd of ~60 μM, the R6BD can effectively compete with ubiquitin for binding to Rad6 [84]. R6BD binding does not alter the rate of formation of the Rad6~Ub thioester. Instead, the R6BD either in trans or in the context of intact Rad18 limits the inherent ubiquitin chain-forming activity of Rad6 [84]. Such modulation of activity likely serves to direct Rad6 towards monoubiquitination, rather than polyubiquitination of PCNA, but this hypothesis has been difficult to assess directly as mutation of the R6BD also reduces PCNA monoubiquitination.
Bre1 and Ubr1, two other RING E3s that can partner with Rad6 in yeast, also contain secondary binding sites for Rad6 [114–116]. The human RAD6/BRE1 pair monoubiquitinates histone H2B in mammalian cells [115], raising the interesting possibility that Bre1 also restricts the activity of Rad6 to monoubiquitination through its secondary E2 binding site. The non-canonical Rad6-binding region of Ubr1, called the basic residues-rich (BRR) domain, mediates a low affinity (millimolar level) interaction with the backside of Rad6 [84, 116]. Unlike the R6BD, though, the BRR domain does not outcompete ubiquitin for binding to the backside of Rad6 [84]. The Ubr1/Rad6 pair polyubiquitinates N-end rule substrates [116–118], making it unlikely that binding through the BRR domain negatively regulates Rad6 function in a manner similar to the R6BD.
Another E2 backside-binding domain is found in the human pro-metastatic [119] RING E3 gp78, which functions in ER-associated degradation (ERAD) with the E2 Ube2g2 (originally known as MmUbc7 [120]). The Ube2g2-binding region (G2BR) of gp78 is a discrete C-terminal domain that is necessary and sufficient for interaction with Ube2g2 [121]. The G2BR forms a high-affinity (Kd of ~20 nM) complex with the backside of Ube2g2 that would preclude ubiquitin from binding to this same surface [83]. In contrast to the R6BD, G2BR binding to Ube2g2 stimulates polyubiquitin chain formation, even when added in trans to reactions containing gp78 lacking the G2BR. Intriguingly, polyubiquitin chain formation by two other Ube2g2-interacting RING E3s, hsHRD1 and Trc8, can also be stimulated by the G2BR when provided in trans.
Crystal structures of the G2BR bound to Ube2g2 reveal it to be a single α-helix (Figure 5D). Relative to their respective E2s, the G2BR is oriented approximately perpendicular to the R6BD [32, 83, 84]. The effect of G2BR binding on ubiquitination by Ube2g2 is mediated by two distinct allosteric mechanisms [83]. First, G2BR binding ‘locks’ loops surrounding Ube2g2’s catalytic cysteine into more ‘closed’ orientations and reorients the catalytic cysteine away from the accessible active site region defined by the orientation of the loops. These changes are associated with a decreased rate of E1-dependent ubiquitin loading of Ube2g2. Second, the G2BR increases the affinity of Ube2g2 for the gp78 RING domain almost 50-fold. The latter effect is most significant in the overall stimulation of ubiquitin transfer, as the increased affinity of Ube2g2 for the gp78 RING domain accounts for the observed stimulation of ubiquitination.
The high affinity interaction of gp78 with Ube2g2 afforded by binding of two discrete domains raises the question of how Ube2g2 can be reloaded with ubiquitin by E1, whose binding site on E2 overlaps with the site of RING binding [80–82]. Reloading of an E2 with ubiquitin by E1 (or at least, dissociation of a discharged E2 to allow association of a charged E2~Ub) must occur to achieve substrate polyubiquitination, although the G2BR-dependent transfer of active-site cysteine-linked ubiquitin chains from Ube2g2 to substrate has been reported [32, 122]. In this system, a full description of the dynamics of the G2BR and RING domain interactions with E2, in the context of the putative oligomerization of gp78 through a distinct region [32], will be needed to provide further insight into mechanisms of ubiquitination. Interestingly, the non-RING-containing protein, ancient ubiquitous protein 1 (AUP1), also contains a G2BR-like region that binds and recruits Ube2g2 to lipid droplets [123, 124]. Whether the AUP1 G2BR-like region has an effect on ubiquitination by Ube2g2 is not yet known and a precise role for Ube2g2 in lipid droplets has not yet been described.
The N-termini of Cbl family RING E3s are capable of modulating affinity for the Cbl E2, UbcH5B. As mentioned above, Cbl proteins down-regulate signaling pathways by mediating ubiquitination, endocytosis, and ultimately lysosomal degradation of RTKs such as epidermal growth factor receptor (EGFR) [85, 125–127]. The three Cbl proteins have a conserved N-terminal tyrosine kinase-binding (TKB) domain, connected by a short linker to their RING domain [128]. The N-termini of Cbl proteins inhibit their RING-dependent autoubiquitination, and Cbl activation requires phosphorylation of a conserved tyrosine in the linker region immediately N-terminal to the RING [129, 130]. Intriguingly, recent structural studies with Cbl and Cbl-b revealed that linker tyrosine phosphorylation and substrate binding combine to stimulate release of the E2 binding face of the RING from auto-inhibition by the TKB region [131, 132]. Together, phosphorylation and substrate binding to the E3 serve to enhance binding of both Cbl and Cbl-b to E2. On the other hand, for Cbl-c, tyrosine phosphorylation decreases the affinity of the E3 for E2 by >3-fold (to a Kd of ~1 μM) [129]. The decrease in affinity may allow more rapid cycling of E2 on and off of Cbl-c, thereby potentially explaining the inhibition of autoubiquitination by the N-terminus for this E3. However, how these two disparate findings fit together will require further investigation.
7.2. Enhancement of RING E3 activity by non-RING accessory proteins
E2-binding domains that enhance RING-dependent ubiquitination have been identified in several non-RING-containing accessory proteins. A paradigm example is Cue1p, a transmembrane protein that recruits the yeast ERAD E2, Ubc7p, to the ER-localized HRD1 (Der3p) and DOA10 RING E3 complexes [133–135]. Cue1p is also required for the stability of Ubc7p [136–138]. Cue1p’s function expands beyond these roles, as a stable, membrane-anchored form of Ubc7p still requires Cue1p for ERAD [139, 140]. Ubc7p is the yeast ortholog of mammalian Ube2g2, and like the G2BR, Cue1p stimulates RING-dependent ubiquitination with Ubc7p [139, 140]. E2 binding by Cue1p occurs through a C-terminal domain called the Ubc7p binding region (U7BR) [136, 140], which shows only modest overall (~15%) sequence similarity to the significantly shorter G2BR. The U7BR folds into a multi-helical structure whose central helix is ~40% identical to the G2BR and orients on the backside of Ubc7p similarly to the G2BR on Ube2g2 (Figure 5E [141]). Also, like the G2BR, the U7BR increases the affinity of Ubc7p for RING domains [141]. However, U7BR binding also imparts several distinct allosteric effects on Ubc7p. The E2’s catalytic Cys is more accessible when Ubc7p is bound to the U7BR, an effect that correlates with an enhancement in the rate of ubiquitin loading of Ubc7p by E1 and an increase in the rate of RING-independent ubiquitin transfer when U7BR is bound. Uniquely, Cue1p’s ability to enhance ubiquitin loading indicates that Ubc7p could remain associated with the HRD1 or DOA10 E3 complexes via Cue1p while being reloaded with ubiquitin by E1, something that RING binding precludes. In support of this, modeling of E1 onto the Ubc7p:U7BR structure based on the recent crystal structure of Schizosaccharomyces pombe ubiquitin E1 (Uba1) in complex with S. pombe Ubc4 [142] (PDB 4II2) indicates that E1 and U7BR could be mutually bound to Ubc7p. Interestingly, similar analysis predicts that simultaneous binding of E1 and G2BR to Ube2G2 would be precluded; however, this is not the case for the R6BD or Pex22pS (see below). It is interesting to consider how the U7BR, being part of a protein distinct from the RING itself, affords flexibility to the system and could allow Ubc7p to simultaneously bind to E1 or pair with various RING E3s in vivo.
Another example of activation of an E2 by a non-RING E3 protein is the yeast E2 Pex4p, which is recruited to the peroxisome by the transmembrane protein, Pex22p [143–145]. Pex4p functions with a complex of RING E3s (Pex2p, Pex10p, Pex12p) [146] to monoubiquitinate Pex5p on a specific Cys residue, required for efficient peroxisomal protein import [147]. Pex22p binds to Pex4p with high affinity (Kd of ~2 nM) and its binding stimulates the formation of lysine-linked ubiquitin chains on Pex4p in the absence of a RING domain. Pex22p:Pex4p binding is also required for RING-dependent substrate ubiquitination in vivo [148]. The enhancement of ubiquitin transfer activity does not appear to be mediated by an enhancement in the rate of E1 loading of Pex4p [148]. A crystal structure of the soluble portion of Pex22p (Pex22pS) with Pex4p reveals that Pex22pS adopts a novel mixed β-sheet and α-helix fold that contacts the C-terminal α3 and α4 helices of the E2, adjacent to the backside region [148] (Figure 5F). The molecular mechanism of Pex4p stimulation by Pex22p remains unknown, and the binding to this distinct region of the E2 by Pex22p may reflect a novel mode of regulation as well. Further investigation will reveal whether Pex22p and AUP1 (described above) have similar or distinct mechanistic effects on their respective E2s as the other E2 binding domains whose functions have now been described.
7.3 Parallels in the SUMO system
Few E3 ligases have been identified and characterized for SUMO and, to date, no HECT-type SUMO E3s have been identified. Although the known SUMO E3 ligases are not structurally similar to RINGs, like RING-type E3s, they facilitate direct transfer of SUMO from the sole SUMO E2, Ubc9, to a substrate Lys. Thus, it is informative to compare and contrast features and strategies used by the SUMO system to those of the ubiquitin system. Ubc9 interacts non-covalently with SUMO via the analogous E2 backside site and SUMO β-sheet [149–151] (Figure 5B). Like ubiquitin binding, non-covalent SUMO binding to Ubc9 promotes SUMO chain formation on target proteins [149, 151]. Furthermore, the SUMO E3 Nup358/Ran-binding protein 2 (RanBP2), which is involved in nucleocytoplasmic trafficking [152–154], despite being structurally distinct from SUMO, contacts the Ubc9 backside using the same residues on Ubc9, making their binding mutually exclusive [149]. How SUMO and Nup358/RanBP2 interact in vivo to modulate function remains to be determined.
SUMO accessory proteins also contain domains that, despite very little sequence identity, structurally mimic SUMO and bind the backside of Ubc9. An example is Rad60/Esc2 and its human ortholog, nuclear factor of activated T-cells (NFAT)-interacting protein of 45kDa (Nip45), which contains a C-terminal SUMO-like domain (SLD) that is a structural mimic for SUMO with respect to the Ubc9-interacting region [155, 156]. Notably, binding of SLD2 to the backside of Ubc9 inhibits SUMO chain elongation in vitro and is important for survival of genotoxic stress in yeast [155, 156].
Another putative SUMO accessory protein, the RWD domain-containing protein RSUME, enhances SUMO conjugation in several ways including: stimulating the loading of Ubc9 with SUMO, mediating E3-independent SUMO transfer to substrates, such as IκB and HIF-1α, and stimulating sumoylation by the SUMO E3, PIAS [157]. RSUME binds to both SUMO and Ubc9 and enhances the non-covalent SUMO binding to Ubc9 [157]. Interestingly, like other RWD domains [158], the structure of RSUME closely resembles that of E2 Ub-conjugating enzymes, despite limited sequence homology (PDB 2EBK).
8. Perspective and Future Directions
In the 14 years since RING finger function was discovered, our knowledge of RING-type ubiquitin ligases has increased dramatically. Through their targeting of a diverse array of substrates, we are beginning to appreciate the range of roles played by this family of E3s in development, in maintaining homeostasis, and in response to cellular signals. Many challenges remain, however, as exemplified by the fact that substrates for most RING-type E3:E2 pairs are not yet known,
For some E3s, insights have emerged as to how their activity and substrate interactions can be regulated. We now know that a wide variety of protein-protein interactions are employed in substrate recognition and that post-translational protein modifications are, in many cases, critical to substrate binding. It is also evident that multiple substrates can be targeted by one RING-type E3, and that multiple E3s can target the same substrate. However, at both the cellular and organismal levels, the overall significance of E3 redundancy in substrate ubiquitination is, in general, poorly understood.
For substrates that are ubiquitinated on specific sites, with few exceptions, the factors that specify these sites are unknown. Progress will require a marked expansion of our understanding of the positioning of both E2~Ub and the substrate in the context of the entire RING-type ligase. Related to this, an emerging concept is that some E2s function with RING-type ligases as ‘chain initiators,’ which put the first ubiquitin on a substrate, while others are ‘chain builders’ that add to an existing ubiquitin chain. Progress in parsing this issue has been slowed by the fact that the most extensively studied E2s, the UbcH5 (Ube2D) family, can perform both functions. In considering chain building, an important outstanding issue is whether different E2s provide specific local environments around their active sites favorable to particular lysines on acceptor ubiquitins, and thereby favor certain ubiquitin chain linkages. If this is the case, understanding the nature of these local environmental factors, and how they are influenced by RING binding, will be crucial to appreciating how the myriad of ubiquitin signals is generated.
Until recently, the molecular basis by which RING-type domains stimulate the transfer of ubiquitin from E2 was enigmatic. As reviewed herein, the pieces are falling into place for some E2:E3 pairs, as both crystal and solution structures of RING-type domains complexed with E2~Ub rather than E2 alone have recently been described and corroborated with functional data. These findings are all consistent with critical roles for both a RING-induced closed E2~Ub conformation and the RING’s Zn II loop (or the equivalent in U-box proteins) in facilitating activation. While these paradigm-shifting observations provide insights into some aspects of the RING:E2 interface, they do not fully account for defects in ubiquitination seen with mutations in other regions of this interface. It will be necessary to expand upon the small number of productive E3:E2 pairs that have been studied in depth to fully understand this critical interface. Similarly, it is important to determine the details and significance of the RING:E2 binding interface for RING-IBR-RING E3s, as we now know these function as classic catalysts, akin to HECT-type E3s.
A common feature of RING-type domains is a tendency to form active homo- and/or heterodimers. For C-terminal interleaved dimers, the distal RING-type domain provides additional contacts to ubiquitin (E2~Ub) to facilitate the closed E2~Ub conformation and promote catalysis. E3 dimerization/oligomerization, in instances where multiple RING-type domains can bind E2~Ub, could also enhance the probability of successful ubiquitin chain formation by increasing the local concentration of RING-type domains accessible to substrate. In the cases of BRCA1-BARD1 and RING1B-Bmi1, which dimerize through non-RING interactions, positioning of the second inactive (i.e., non-E2-binding) RING finger is not predicted to play a direct role in the binding of E2~Ub. In these cases, the function of dimerization in ubiquitination remains enigmatic.
A central remaining question in RING-type E3-mediated ubiquitination is what regulates the processivity of ubiquitination and thus the fate of the substrate. The answer is likely complicated and includes E3 dimerization/oligomerization, the affinity for substrate, the relative affinities for E2 versus E2~Ub, and ubiquitin-binding domains intrinsic to E3s or E3 complexes. All of these potentially positive factors are, of course, countered by DUBs that are associated with E3s or substrates. Another factor that, in some cases, facilitates ubiquitination is the non-covalent binding of ubiquitin to the backside of a subclass of E2s. An emerging factor, reviewed herein, is the contribution of non-RING regions of E3s binding to E2s using surfaces distinct from the shared RING- and E1-interacting interface. In some cases, these interactions compete with non-covalent ubiquitin backside binding and limit ubiquitination. In other cases, binding to a similar region of the E2 increases the affinity of the E2:E3 interaction and thereby enhances processivity of ubiquitination. These secondary sites of E2 binding may also provide a means to tether the E2 to the E3 complex, without continuous RING finger binding, and thereby provide a potential means to ‘reload’ E2 with ubiquitin (E2~Ub) without dissociation from the E3 complex. Whether such E2-specific binding is of general importance in vivo in determining combinatorial specificity in RING-type domain:E2 interactions and in the processivity of ubiquitination now become important questions.
Finally, RING-type E3s and their substrates are implicated in a wide variety of human diseases ranging from viral infections to neurodegenerative disorders to cancer. Thus, they are attractive targets for therapeutic development. However, the lack of a catalytic center in RING-type domains makes targeting strategies more difficult. As biochemical and biophysical/structural approaches converge on developing an understanding of specific aspects of RING-type E3s and their interactions with E2s, the potential for generating therapeutics by incorporation of structure-based design becomes increasingly promising.
Highlights.
RING-type E3s constitute a large and diverse class of ubiquitin ligases
RING-type E3s exist as monomers, dimers, and in complex multi-subunit assemblies
RING-type E3s recognize substrates by a variety of mechanisms
Recent structural studies have provided insight into how RINGs function with E2s
Recent studies provide a role for non-RING E2-binding domains in altering function
Acknowledgments
The study of RING-type E3s continues to grow extremely rapidly. We regret that it was possible to only cite a fraction of the outstanding primary publications in this field. This work was supported by the National Institute of General Medical Sciences grants R01 GM088055 and R01 GM098503 (R.E.K.) and by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (A.M.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J. 2011;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 3.Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2011;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 8.Boutell C, Everett RD. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol. 2012;94:465–481. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- 9.Nathan JA, Lehner PJ. The trafficking and regulation of membrane receptors by the RING-CH ubiquitin E3 ligases. Exp Cell Res. 2009;315:1593–1600. doi: 10.1016/j.yexcr.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 11.Mace PD, Linke K, Feltham R, Schumacher FR, Smith CA, Vaux DL, Silke J, Day CL. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 12.Liew CW, Sun H, Hunter T, Day CL. RING domain dimerization is essential for RNF4 function. Biochem J. 2010;431:23–29. doi: 10.1042/BJ20100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Fairall L, Goult BT, Calkin AC, Hong C, Millard CJ, Tontonoz P, Schwabe JW. The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes Dev. 2011;25:1262–1274. doi: 10.1101/gad.2056211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Z, Kohli E, Devlin KI, Bold M, Nix JC, Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vander Kooi CW, Ohi MD, Rosenberg JA, Oldham ML, Newcomer ME, Gould KL, Chazin WJ. The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry. 2006;45:121–130. doi: 10.1021/bi051787e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joukov V, Chen J, Fox EA, Green JB, Livingston DM. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc Natl Acad Sci U S A. 2001;98:12078–12083. doi: 10.1073/pnas.211427098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 20.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 22.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 23.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyappan S, Wollscheid HP, Rojas-Fernandez A, Marquardt A, Tang HC, Singh RK, Scheffner M. Turning the RING domain protein MdmX into an active ubiquitin-protein ligase. J Biol Chem. 2010;285:33065–33072. doi: 10.1074/jbc.M110.115113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman PN, Chen X, Bargonetti J, Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci U S A. 1993;90:3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dueber EC, Schoeffler AJ, Lingel A, Elliott JM, Fedorova AV, Giannetti AM, Zobel K, Maurer B, Varfolomeev E, Wu P, Wallweber HJ, Hymowitz SG, Deshayes K, Vucic D, Fairbrother WJ. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science. 2011;334:376–380. doi: 10.1126/science.1207862. [DOI] [PubMed] [Google Scholar]
- 28.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem. 2006;281:20643–20649. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- 30.Huang A, Hibbert RG, de Jong RN, Das D, Sixma TK, Boelens R. Symmetry and asymmetry of the RING-RING dimer of Rad18. J Mol Biol. 2011;410:424–435. doi: 10.1016/j.jmb.2011.04.051. [DOI] [PubMed] [Google Scholar]
- 31.Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci U S A. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozlov G, Peschard P, Zimmerman B, Lin T, Moldoveanu T, Mansur-Azzam N, Gehring K, Park M. Structural basis for UBA-mediated dimerization of c-Cbl ubiquitin ligase. J Biol Chem. 2007;282:27547–27555. doi: 10.1074/jbc.M703333200. [DOI] [PubMed] [Google Scholar]
- 34.Bartkiewicz M, Houghton A, Baron R. Leucine zipper-mediated homodimerization of the adaptor protein c-Cbl. A role in c-Cbl’s tyrosine phosphorylation and its association with epidermal growth factor receptor. J Biol Chem. 1999;274:30887–30895. doi: 10.1074/jbc.274.43.30887. [DOI] [PubMed] [Google Scholar]
- 35.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 36.Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duda DM, Scott DC, Calabrese MF, Zimmerman ES, Zheng N, Schulman BA. Structural regulation of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2011;21:257–264. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, Robinson CV, da Fonseca PC, Barford D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 40.Kee Y, Kim JM, D’Andrea AD. Regulated degradation of FANCM in the Fanconi anemia pathway during mitosis. Genes Dev. 2009;23:555–560. doi: 10.1101/gad.1761309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menssen R, Schweiggert J, Schreiner J, Kusevic D, Reuther J, Braun B, Wolf DH. Exploring the topology of the Gid complex, the E3 ubiquitin ligase involved in catabolite-induced degradation of gluconeogenic enzymes. J Biol Chem. 2012;287:25602–25614. doi: 10.1074/jbc.M112.363762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Platta HW, El Magraoui F, Baumer BE, Schlee D, Girzalsky W, Erdmann R. Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol. 2009;29:5505–5516. doi: 10.1128/MCB.00388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams C, van den Berg M, Geers E, Distel B. Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem Biophys Res Commun. 2008;374:620–624. doi: 10.1016/j.bbrc.2008.07.054. [DOI] [PubMed] [Google Scholar]
- 44.Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 45.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wenzel DM, Klevit RE. Following Ariadne’s thread: a new perspective on RBR ubiquitin ligases. BMC Biol. 2012;10:24. doi: 10.1186/1741-7007-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smit JJ, Monteferrario D, Noordermeer SM, van Dijk WJ, van der Reijden BA, Sixma TK. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 2012;31:3833–3844. doi: 10.1038/emboj.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 2012;13:840–846. doi: 10.1038/embor.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, Youle RJ. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol. 2013;200:163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weissman A, Shabek N, Ciechanover A. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nature Reviews Molecular Cell Biology. 2011;12:605–620. doi: 10.1038/nrm3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abbas T, Mueller AC, Shibata E, Keaton M, Rossi M, Dutta A. CRL1-FBXO11 Promotes Cdt2 Ubiquitylation and Degradation and Regulates Pr-Set7/Set8-Mediated Cellular Migration. Mol Cell. 2013;49:1147–1158. doi: 10.1016/j.molcel.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rossi M, Duan S, Jeong YT, Horn M, Saraf A, Florens L, Washburn MP, Antebi A, Pagano M. Regulation of the CRL4(Cdt2) Ubiquitin Ligase and Cell-Cycle Exit by the SCF(Fbxo11) Ubiquitin Ligase. Mol Cell. 2013;49:1159–1166. doi: 10.1016/j.molcel.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JN, Song B, DeBose-Boyd RA, Ye J. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J Biol Chem. 2006;281:39308–39315. doi: 10.1074/jbc.M608999200. [DOI] [PubMed] [Google Scholar]
- 55.Stone SL, Callis J. Ubiquitin ligases mediate growth and development by promoting protein death. Curr Opin Plant Biol. 2007;10:624–632. doi: 10.1016/j.pbi.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 56.Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- 57.Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, Milstien S, Spiegel S. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryan PE, Davies GC, Nau MM, Lipkowitz S. Regulating the regulator: negative regulation of Cbl ubiquitin ligases. Trends Biochem Sci. 2006;31:79–88. doi: 10.1016/j.tibs.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 59.Labit H, Fujimitsu K, Bayin NS, Takaki T, Gannon J, Yamano H. Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. EMBO J. 2012;31:3351–3362. doi: 10.1038/emboj.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida Y, Tanaka K. Lectin-like ERAD players in ER and cytosol. Biochim Biophys Acta. 2009;1800:172–180. doi: 10.1016/j.bbagen.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 61.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, Schirle M, Shi X, Hild M, Bauer A, Myer VE, Finan PM, Porter JA, Huang SM, Cong F. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 63.Kang HC, Lee YI, Shin JH, Andrabi SA, Chi Z, Gagne JP, Lee Y, Ko HS, Lee BD, Poirier GG, Dawson VL, Dawson TM. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci U S A. 2011;108:14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Callow MG, Tran H, Phu L, Lau T, Lee J, Sandoval WN, Liu PS, Bheddah S, Tao J, Lill JR, Hongo JA, Davis D, Kirkpatrick DS, Polakis P, Costa M. Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS One. 2011;6:e22595. doi: 10.1371/journal.pone.0022595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26. doi: 10.3389/fonc.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X, Taplick J, Geva N, Oren M. Inhibition of p53 degradation by Mdm2 acetylation. FEBS Lett. 2004;561:195–201. doi: 10.1016/S0014-5793(04)00168-1. [DOI] [PubMed] [Google Scholar]
- 68.Ma Y, Fan S, Hu C, Meng Q, Fuqua SA, Pestell RG, Tomita YA, Rosen EM. BRCA1 regulates acetylation and ubiquitination of estrogen receptor-alpha. Mol Endocrinol. 2009;24:76–90. doi: 10.1210/me.2009-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Fruh K. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J Virol. 2004;78:1109–1120. doi: 10.1128/JVI.78.3.1109-1120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.El Magraoui F, Baumer BE, Platta HW, Baumann JS, Girzalsky W, Erdmann R. The RING-type ubiquitin ligases Pex2p, Pex10p and Pex12p form a heteromeric complex that displays enhanced activity in an ubiquitin conjugating enzyme-selective manner. FEBS J. 2012;279:2060–2070. doi: 10.1111/j.1742-4658.2012.08591.x. [DOI] [PubMed] [Google Scholar]
- 71.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin Cell Dev Biol. 2007;18:770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, Collart MA, Boelens R, Timmers HT. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 76.Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, Harper JW, Conaway JW. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 77.Fang S, Lorick KL, Jensen JP, Weissman AM. RING finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin Cancer Biol. 2003;13:5–14. doi: 10.1016/s1044-579x(02)00095-0. [DOI] [PubMed] [Google Scholar]
- 78.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 79.Wu B, Skarina T, Yee A, Jobin MC, Dileo R, Semesi A, Fares C, Lemak A, Coombes BK, Arrowsmith CH, Singer AU, Savchenko A. NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases. PLoS Pathog. 2010;6:e1000960. doi: 10.1371/journal.ppat.1000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 81.Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 82.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 83.Das R, Mariano J, Tsai YC, Kalathur RC, Kostova Z, Li J, Tarasov SG, McFeeters RL, Altieri AS, Ji X, Byrd RA, Weissman AM. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hibbert RG, Huang A, Boelens R, Sixma TK. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc Natl Acad Sci U S A. 2011;108:5590–5595. doi: 10.1073/pnas.1017516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 86.Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D, 3rd, Fukuda M, Ohta T, Klevit R. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci U S A. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Christensen DE, Klevit RE. Dynamic interactions of proteins in complex networks: identifying the complete set of interacting E2s for functional investigation of E3-dependent protein ubiquitination. FEBS J. 2009;276:5381–5389. doi: 10.1111/j.1742-4658.2009.07249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 89.Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, Zheng L, Rich RL, Campos AD, Myszka DG, Lenardo MJ, Darnay BG, Wu H. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pickart CM, Rose IA. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985;260:1573–1581. [PubMed] [Google Scholar]
- 91.Pruneda JN, Stoll KE, Bolton LJ, Brzovic PS, Klevit RE. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme~ubiquitin conjugate. Biochemistry. 2011;50:1624–1633. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Soss SE, Klevit RE, Chazin WJ. Activation of UbcH5c〜Ub Is the Result of a Shift in Interdomain Motions of the Conjugate Bound to U-Box E3 Ligase E4B. Biochemistry. 2013;52:2991–2999. doi: 10.1021/bi3015949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 96.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nakatani Y, Kleffmann T, Linke K, Condon SM, Hinds MG, Day CL. Regulation of ubiquitin transfer by XIAP, a dimeric RING E3 ligase. Biochem J. 2012;450:629–638. doi: 10.1042/BJ20121702. [DOI] [PubMed] [Google Scholar]
- 99.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci U S A. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miura T, Klaus W, Gsell B, Miyamoto C, Senn H. Characterization of the binding interface between ubiquitin and class I human ubiquitin-conjugating enzyme 2b by multidimensional heteronuclear NMR spectroscopy in solution. J Mol Biol. 1999;290:213–228. doi: 10.1006/jmbi.1999.2859. [DOI] [PubMed] [Google Scholar]
- 102.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 103.Randles L, Walters KJ. Ubiquitin and its binding domains. Front Biosci. 2012;17:2140–2157. doi: 10.2741/4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP. Ubiquitin recognition by the human TSG101 protein. Mol Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- 105.Teo H, Veprintsev DB, Williams RL. Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J Biol Chem. 2004;279:28689–28696. doi: 10.1074/jbc.M400023200. [DOI] [PubMed] [Google Scholar]
- 106.Lewis MJ, Saltibus LF, Hau DD, Xiao W, Spyracopoulos L. Structural basis for non-covalent interaction between ubiquitin and the ubiquitin conjugating enzyme variant human MMS2. J Biomol NMR. 2006;34:89–100. doi: 10.1007/s10858-005-5583-6. [DOI] [PubMed] [Google Scholar]
- 107.Dowil RT, Lu X, Saracco SA, Vierstra RD, Downes BP. Arabidopsis membrane-anchored ubiquitin-fold (MUB) proteins localize a specific subset of ubiquitin-conjugating (E2) enzymes to the plasma membrane. J Biol Chem. 2011;286:14913–14921. doi: 10.1074/jbc.M110.158808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Downes BP, Saracco SA, Lee SS, Crowell DN, Vierstra RD. MUBs, a family of ubiquitin-fold proteins that are plasma membrane-anchored by prenylation. J Biol Chem. 2006;281:27145–27157. doi: 10.1074/jbc.M602283200. [DOI] [PubMed] [Google Scholar]
- 109.Lee KY, Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol Cells. 2008;26:5–11. [PMC free article] [PubMed] [Google Scholar]
- 110.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bailly V, Prakash S, Prakash L. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol Cell Biol. 1997;17:4536–4543. doi: 10.1128/mcb.17.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Back JW, Notenboom V, de Koning LJ, Muijsers AO, Sixma TK, de Koster CG, de Jong L. Identification of cross-linked peptides for protein interaction studies using mass spectrometry and 18O labeling. Anal Chem. 2002;74:4417–4422. doi: 10.1021/ac0257492. [DOI] [PubMed] [Google Scholar]
- 113.Notenboom V, Hibbert RG, van Rossum-Fikkert SE, Olsen JV, Mann M, Sixma TK. Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res. 2007;35:5819–5830. doi: 10.1093/nar/gkm615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem. 2009;284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xie Y, Varshavsky A. The E2-E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Madura K, Dohmen RJ, Varshavsky A. N-recognin/Ubc2 interactions in the N-end rule pathway. J Biol Chem. 1993;268:12046–12054. [PubMed] [Google Scholar]
- 118.Watkins JF, Sung P, Prakash S, Prakash L. The extremely conserved amino terminus of RAD6 ubiquitin-conjugating enzyme is essential for amino-end rule-dependent protein degradation. Genes Dev. 1993;7:250–261. doi: 10.1101/gad.7.2.250. [DOI] [PubMed] [Google Scholar]
- 119.Tsai YC, Mendoza A, Mariano JM, Zhou M, Kostova Z, Chen B, Veenstra T, Hewitt SM, Helman LJ, Khanna C, Weissman AM. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat Med. 2007;13:1504–1509. doi: 10.1038/nm1686. [DOI] [PubMed] [Google Scholar]
- 120.Tiwari S, Weissman AM. Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s) J Biol Chem. 2001;276:16193–16200. doi: 10.1074/jbc.M007640200. [DOI] [PubMed] [Google Scholar]
- 121.Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci U S A. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 123.Spandl J, Lohmann D, Kuerschner L, Moessinger C, Thiele C. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem. 2011;286:5599–5606. doi: 10.1074/jbc.M110.190785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Klemm EJ, Spooner E, Ploegh HL. Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J Biol Chem. 2011;286:37602–37614. doi: 10.1074/jbc.M111.284794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 127.Yokouchi M, Kondo T, Houghton A, Bartkiewicz M, Horne WC, Zhang H, Yoshimura A, Baron R. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J Biol Chem. 1999;274:31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- 128.Meng W, Sawasdikosol S, Burakoff SJ, Eck MJ. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature. 1999;398:84–90. doi: 10.1038/18050. [DOI] [PubMed] [Google Scholar]
- 129.Ryan PE, Sivadasan-Nair N, Nau MM, Nicholas S, Lipkowitz S. The N terminus of Cbl-c regulates ubiquitin ligase activity by modulating affinity for the ubiquitin-conjugating enzyme. J Biol Chem. 2010;285:23687–23698. doi: 10.1074/jbc.M109.091157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kassenbrock CK, Anderson SM. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J Biol Chem. 2004;279:28017–28027. doi: 10.1074/jbc.M404114200. [DOI] [PubMed] [Google Scholar]
- 131.Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat Struct Mol Biol. 2012;19:184–192. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kobashigawa Y, Tomitaka A, Kumeta H, Noda NN, Yamaguchi M, Inagaki F. Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc Natl Acad Sci U S A. 2011;108:20579–20584. doi: 10.1073/pnas.1110712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 134.Deak PM, Wolf DH. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J Biol Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- 135.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 137.Gardner RG, Shearer AG, Hampton RY. In vivo action of the HRD ubiquitin ligase complex: mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol Cell Biol. 2001;21:4276–4291. doi: 10.1128/MCB.21.13.4276-4291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ravid T, Hochstrasser M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol. 2007;9:422–427. doi: 10.1038/ncb1558. [DOI] [PubMed] [Google Scholar]
- 139.Bazirgan OA, Hampton RY. Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem. 2008;283:12797–12810. doi: 10.1074/jbc.M801122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kostova Z, Mariano J, Scholz S, Koenig C, Weissman AM. A Ubc7p-binding domain in Cue1p activates ER-associated protein degradation. J Cell Sci. 2009;122:1374–1381. doi: 10.1242/jcs.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Metzger MB, Liang YH, Das R, Mariano J, Li S, Li J, Kostova Z, Byrd RA, Ji X, Weissman AM. A Structurally Unique E2-Binding Domain Activates Ubiquitination by the ERAD E2, Ubc7p, Through Multiple Mechanisms. Mol Cell. 2013;50:516–527. doi: 10.1016/j.molcel.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Olsen SK, Lima CD. Structure of a Ubiquitin E1-E2 Complex: Insights to E1-E2 Thioester Transfer. Mol Cell. 2013;49:884–896. doi: 10.1016/j.molcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Collins CS, Kalish JE, Morrell JC, McCaffery JM, Gould SJ. The peroxisome biogenesis factors pex4p, pex22p, pex1p, and pex6p act in the terminal steps of peroxisomal matrix protein import. Mol Cell Biol. 2000;20:7516–7526. doi: 10.1128/mcb.20.20.7516-7526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Koller A, Snyder WB, Faber KN, Wenzel TJ, Rangell L, Keller GA, Subramani S. Pex22p of Pichia pastoris, essential for peroxisomal matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisomal membrane. J Cell Biol. 1999;146:99–112. doi: 10.1083/jcb.146.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell. 2005;17:3422–3435. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kragt A, Voorn-Brouwer T, van den Berg M, Distel B. The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J Biol Chem. 2005;280:7867–7874. doi: 10.1074/jbc.M413553200. [DOI] [PubMed] [Google Scholar]
- 147.Rucktaschel R, Girzalsky W, Erdmann R. Protein import machineries of peroxisomes. Biochim Biophys Acta. 2011;1808:892–900. doi: 10.1016/j.bbamem.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 148.Williams C, van den Berg M, Panjikar S, Stanley WA, Distel B, Wilmanns M. Insights into ubiquitin-conjugating enzyme/co-activator interactions from the structure of the Pex4p:Pex22p complex. EMBO J. 2011;31:391–402. doi: 10.1038/emboj.2011.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Capili AD, Lima CD. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J Mol Biol. 2007;369:608–618. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Duda DM, van Waardenburg RC, Borg LA, McGarity S, Nourse A, Waddell MB, Bjornsti MA, Schulman BA. Structure of a SUMO-binding-motif mimic bound to Smt3p-Ubc9p: conservation of a non-covalent ubiquitin-like protein-E2 complex as a platform for selective interactions within a SUMO pathway. J Mol Biol. 2007;369:619–630. doi: 10.1016/j.jmb.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Saitoh H, Pu R, Cavenagh M, Dasso M. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc Natl Acad Sci U S A. 1997;94:3736–3741. doi: 10.1073/pnas.94.8.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 155.Prudden J, Perry JJ, Arvai AS, Tainer JA, Boddy MN. Molecular mimicry of SUMO promotes DNA repair. Nat Struct Mol Biol. 2009;16:509–516. doi: 10.1038/nsmb.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sekiyama N, Arita K, Ikeda Y, Hashiguchi K, Ariyoshi M, Tochio H, Saitoh H, Shirakawa M. Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45. Proteins. 2010;78:1491–1502. doi: 10.1002/prot.22667. [DOI] [PubMed] [Google Scholar]
- 157.Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, Stalla GK, Holsboer F, Arzt E. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell. 2007;131:309–323. doi: 10.1016/j.cell.2007.07.044. [DOI] [PubMed] [Google Scholar]
- 158.Nameki N, Yoneyama M, Koshiba S, Tochio N, Inoue M, Seki E, Matsuda T, Tomo Y, Harada T, Saito K, Kobayashi N, Yabuki T, Aoki M, Nunokawa E, Matsuda N, Sakagami N, Terada T, Shirouzu M, Yoshida M, Hirota H, Osanai T, Tanaka A, Arakawa T, Carninci P, Kawai J, Hayashizaki Y, Kinoshita K, Guntert P, Kigawa T, Yokoyama S. Solution structure of the RWD domain of the mouse GCN2 protein. Protein Sci. 2004;13:2089–2100. doi: 10.1110/ps.04751804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ramanathan HN, Ye Y. Cellular strategies for making monoubiquitin signals. Crit Rev Biochem Mol Biol. 2012;47:17–28. doi: 10.3109/10409238.2011.620943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dimova NV, Hathaway NA, Lee BH, Kirkpatrick DS, Berkowitz ML, Gygi SP, Finley D, King RW. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012;14:168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shabek N, Herman-Bachinsky Y, Buchsbaum S, Lewinson O, Haj-Yahya M, Hejjaoui M, Lashuel HA, Sommer T, Brik A, Ciechanover A. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 162.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 163.Pickart CM. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 164.Wickliffe KE, Williamson A, Meyer HJ, Kelly A, Rape M. K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol. 2011;21:656–663. doi: 10.1016/j.tcb.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schmukle AC, Walczak H. No one can whistle a symphony alone - how different ubiquitin linkages cooperate to orchestrate NF-kappaB activity. J Cell Sci. 2012;125:549–559. doi: 10.1242/jcs.091793. [DOI] [PubMed] [Google Scholar]
- 168.Ramaekers CH, Wouters BG. Regulatory functions of ubiquitin in diverse DNA damage responses. Curr Mol Med. 2011;11:152–169. doi: 10.2174/156652411794859269. [DOI] [PubMed] [Google Scholar]
- 169.Clague MJ, Liu H, Urbe S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev Cell. 2012;23:457–467. doi: 10.1016/j.devcel.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 170.Page RC, Pruneda JN, Amick J, Klevit RE, Misra S. Structural insights into the conformation and oligomerization of E2~ubiquitin conjugates. Biochemistry. 2012;51:4175–4187. doi: 10.1021/bi300058m. [DOI] [PMC free article] [PubMed] [Google Scholar]