Abstract
Oxidized 1-palmitoyl-2-arachidonyl-sn-glycero-3-phospholcholine (OxPAPC) and its component phospholipids accumulate in atherosclerotic lesions and regulate the expression of >1,000 genes, many proatherogenic, in human aortic endothelial cells (HAECs). In contrast, there is evidence in the literature that HDL protects the vasculature from inflammatory insult. We have previously shown that in HAECs, HDL attenuates the expression of several proatherogenic genes regulated by OxPAPC and 1-palmitoyl-2-(5,6-epoxyisoprostane E2)-sn-glycero-3-phosphocholine. We now demonstrate that HDL reverses >50% of the OxPAPC transcriptional response. Genes reversed by HDL are enriched for inflammatory and vascular development pathways, while genes not affected by HDL are enriched for oxidative stress response pathways. The protective effect of HDL is partially mimicked by cholesterol repletion and treatment with apoA1 but does not require signaling through scavenger receptor class B type I. Furthermore, our data demonstrate that HDL protection requires direct interaction with OxPAPC. HDL-associated platelet-activating factor acetylhydrolase (PAF-AH) hydrolyzes short-chain bioactive phospholipids in OxPAPC; however, inhibiting PAF-AH activity does not prevent HDL protection. Our results are consistent with HDL sequestering specific bioactive lipids in OxPAPC, thereby preventing their regulation of select target genes. Overall, this work implicates HDL as a major regulator of OxPAPC action in endothelial cells via multiple mechanisms.
Keywords: minimally modified low density lipoprotein, inflammatory signaling, high density lipoprotein, scavenger receptor class B type I, platelet-activating factor acetylhydrolase
Over the past 50 years, epidemiological and clinical studies have implicated the major serum lipoproteins in coronary artery disease (CAD) (1, 2). Elevated LDL is strongly associated with CAD in patient populations, and the products of the oxidative modification of LDL particles initiate an inflammatory cascade in the vessel wall that contributes to atherosclerosis (3–5). Oxidation products of 1-palmitoyl-2-arachidonyl-sn-glycero-3-phosphatidylcholine (OxPAPC) are major proatherogenic components of minimally modified LDL, which accumulate in atherosclerotic lesions and induce a strong transcriptional response of >1,000 genes in human aortic endothelial cells (HAECs) (6–9). OxPAPC treatment regulates the abundance of transcripts enriched for several gene ontology categories in HAECs, including the unfolded protein response(UPR), endocytosis, inflammation, angiogenesis, and others. Importantly, molecules that inhibit oxidized phospholipid activity have therapeutic benefits in models of atherosclerosis, implying a causal role for oxidized phospholipids in atherogenesis (10–14).
Contrary to oxidized LDL, HDL functions in many ways to reduce plaque burden and protect the vasculature (15, 16). In addition to its role in cholesterol transport, HDL possesses anti-inflammatory, antioxidant, and antithrombotic properties (17–19). These include directly inhibiting the oxidation of LDL, sequestering by-products of lipid oxidation, and promoting eNOS stability and accumulation in the vessel wall (17, 18, 20–22). In addition, members of our group have previously reported that HDL inhibits the induction by OxPAPC of chemotactic cytokines [interleukin (IL) 8, monocyte chemotactic protein 1 (MCP-1), and IL-6], UPR transcription factors [activating transcription factor 3 (ATF3), activating transcription factor 4 (ATF4), and X-box binding protein 1 (XBP1)], and sterol biosynthetic genes [LDL receptor (LDLR), Insig-1, and HMG-CoA synthase] in HAECs (23). Although research has clearly defined a significant role for HDL in vascular health, recent clinical trials and causal genetic studies have failed to corroborate the link between HDL cholesterol and CAD (24–27). However, the measurement of HDL cholesterol used in these studies may not accurately reflect important HDL functions (19, 27). Thus, further research is needed to understand the complex role of HDL in vascular biology.
The objective of the current study was to explore the ability of HDL to antagonize the effects of OxPAPC on genome-wide gene expression in HAECs. Our results demonstrate that HDL inhibits >50% of the gene expression regulated by OxPAPC but does not inhibit the activation of oxidative stress response pathways. We also examined a number of potential mechanisms of HDL protection involving both effects on endothelial cells and on OxPAPC. We conclude that HDL antagonizes OxPAPC signaling via multiple mechanisms.
MATERIALS AND METHODS
Reagents
PAPC was purchased from Avanti Polar Lipids Inc. (Alabaster, AL) and autoxidized via exposure to air for 24–48 h to produce OxPAPC as described previously (6). The 5,6-epoxyisoprostane (EI) was synthesized as described previously, and a single analog, EI4, was used in this study (28). Cholesterol-cyclodextrin complex was purchased from Sigma-Aldrich. Rabbit polyclonal antibodies targeting scavenger receptor class B type I (SR-BI) were purchased from Novus Biologicals (NB400-104 and NB400-134). HDL was isolated from human donor plasma by the Atherosclerosis Research Unit at University of California, Los Angeles (UCLA) using sequential ultracentrifugation (1.063 < d < 1.21) as described previously (29).
Cell culture and treatment conditions
HAEC cultures were isolated from aortic explants of healthy heart transplant donors and cultured in MCDB-131 complete media (Vec Technologies) as described previously (6, 8). Treatment media consisted of medium M199 (Cellgro) supplemented with 1% FBS (Hyclone, Fisher Scientific) and penicillin-streptomycin (Cellgro). All treatments with lipids were performed on primary HAECs in passages 3 through 7. After treatment, cells were lysed in Qiazol, and mRNA was isolated using the RNeasy minikit (Qiagen). For expression knockdown studies, HAECs were transfected at 50% confluence for 4 h with 40 pmol of siRNA oligos using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Transfections were performed 48 h prior to lipid treatments. siRNAs targeting SR-BI were purchased from Qiagen (cat. nos. SI03066833 and SI02777194). For experiments using platelet-activating factor acetylhydrolase (PAF-AH) inhibitors, HDL was pretreated with 1 mM Pefabloc or 100 μM methyl-γ-linolenyl fluorophosphonate (MAFP) for 30 min at 37°C. Excess inhibitor was removed by overnight dialysis before treating cells.
Microarray analysis
RNA concentrations were measured with the Nano Drop 2000 (Thermo Fisher), and quality was checked with the Agilent Bioanlalyzer 2100 (Agilent). Microarray hybridization was performed on the Illumina HT12 v4 microarray by the UCLA EpiGenetics and Genetics core. Expression profiles without background or normalization were determined using Direct Hybridization module in the GenomeStudio software. Expression profiles of multiple donors were then analyzed with the “limma” R package using ‘neqc’ function for quantile normalization and background correction, with donors used as biological replicates. Probe annotation was obtained from illuminaHumanv4.db R package, and probes that were annotated as “Bad” or “No match” were removed from downstream analysis. Contrast matrix was constructed to represent all the comparisons, and differential expression was tested using linear models and ‘ebayes’ function. All P-value cutoffs refer to adjusted P values that reflect false discovery rate correction for multiple testing.
For quantitative PCR (qPCR), cDNA was prepared using the ABI High Capacity cDNA Reverse Transcription Kit using equal amounts of input mRNA. Reactions were set up using the SYBR Green mastermix (Kapa Biosystems) and measured on the Roche LightCycler 480 (Roche Diagnostics).
Lipid extraction and quantification by HPLC-MS
OxPAPC (100 μg) was incubated with or without 100 μg of HDL in PBS lacking calcium and magnesium for 1 h at 37°C. Lipids were extracted using a modification of the method of Bligh and Dyer (30). In brief, 3 vol of chloroform-methanol (2:1) containing 0.01% butylated hydroxytoluene (BHT) was added to each sample before vortexing. Samples were then stored under argon at 4°C overnight, and 3 μg of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) was added as an internal control. An additional volume of methanol and 0.8 vol of PBS were added (final ratio chloroform-methanol-aqueous, 2:2:1.8), and the samples were vortexed and centrifuged at 2,800 rpm for 10 min (Eppendorf 5810R rotor A-4-62). The organic phase was carefully removed and dried under a stream of nitrogen. Dried samples were stored under argon at −80°C. For HPLC-MS, samples were resuspended in 1:1 chloroform-methanol and loaded onto an ACE C8 Ultra-Inert Analytical 3 micron HPLC column. Gradient elution was set at a flow rate of 0.2 ml/min. Mobile phase A consisted of 0.3% formic acid in water, and mobile phase B consisted of 0.3% formic acid in methanol. The gradient was programmed as follows: 0–11 min, linear gradient from 75.0% to 98.0% B; 11–15 min, 98.0% B; 15–15.1 min, linear gradient from 98.0% to 75.0% B; 15.1–20 min, 75.0% B. Mass spectra were measured with the LCT-Premier XE TOF Instrument controlled by MassLynx 4.1 software. The mass spectrometer was equipped with a Multi-Mode Source operated in the electrospray mode. The m/z ratios of monitored ions were as follows: 496 lysophosphatidylcholine (LPC), 594 [1-palmitoyl-2-(5-oxovaleroyl)-sn-glycero-3-phosphocholine (POVPC)], 610 [1-palmitoyl-2-glutaroyl-sn-glycero-3-phosphocholine (PGPC)], 678 (DMPC), 782 (PAPC), and 828 [1-palmitoyl-2-(5,6-epoxyisoprostane E2)-sn-glycero-3-phosphocholine (PEIPC)]. LPC, POVPC, PGPC, PAPC, and PEIPC were normalized to DMPC to quantify each species.
RESULTS
Effect of HDL on transcript levels in response to OxPAPC
We have previously reported that HDL inhibits OxPAPC-induced transcription of specific genes that were used as molecular markers of particular cellular pathways (23). Later studies demonstrated that OxPAPC induces transcriptional changes in >1,000 genes in HAECs (8). We sought to examine the effects of HDL on the HAEC response to OxPAPC in a more comprehensive manner using microarrays. To this end, we treated HAECs from four different donors with HDL, OxPAPC, and OxPAPC+HDL and compared the transcriptional response of OxPAPC-treated samples versus OxPAPC+HDL-treated samples relative to control.
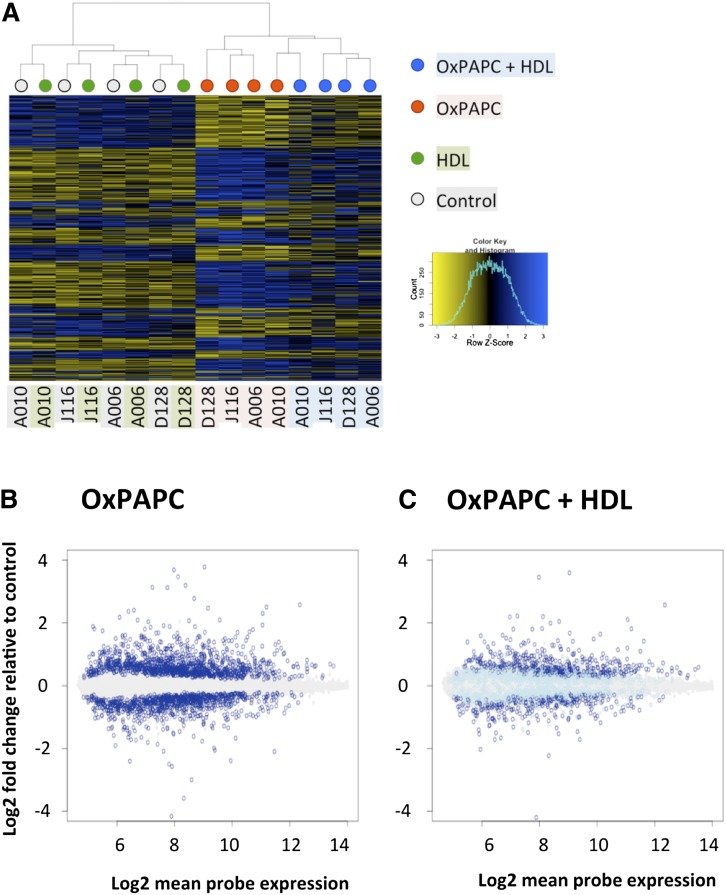
Clustering of the different donors and treatments clearly shows that HDL has a pronounced effect on expression of OxPAPC-regulated genes, while control samples treated with HDL alone cluster by donor (Fig. 1A). Markedly, HDL alone had virtually no effect on HAEC gene expression. In contrast, OxPAPC induced significant changes in the expression of 1,679 genes (FDR corrected P < 0.05). However, only 806 of these genes were significantly affected by OxPAPC in the presence of HDL (Fig. 1B, C). These results show that HDL has a broad effect on HAEC response to OxPAPC, affecting >50% of genes regulated by OxPAPC. We defined “HDL response” and “genes reversed by HDL” as the difference between HAEC transcriptional response to OxPAPC and to OxPAPC+HDL [i.e., in this experiment, HDL reversed 873 out of 1,679 genes (52%)] (Fig. 1C).
Fig. 1.
HDL affects 52% of genes regulated by OxPAPC. HAECs from four unique donors were treated with HDL (40 μg/ml), OxPAPC (40 μg/ml), or both (OxPAPC+HDL), and mRNA was isolated and subjected to microarray analysis. Differentially expressed genes as compared with control samples (false discovery rate adjusted P < 0.05) were identified in OxPAPC-treated samples and in OxPAPC+HDL-treated samples independently. OxPAPC regulated 1,679 genes out of the 14,899 genes analyzed on the array. A: Heat map showing the pattern of regulation of genes affected by OxPAPC in untreated cells, cells treated with HDL, cells treated with OxPAPC, and cells treated with OxPAPC+HDL. Dendogram on top shows clustering of the samples, where color of the nodes represents treatment group: gray for control, green for HDL, red for OxPAPC, and blue for OxPAPC+HDL. Individual donors are indicated below the heat map. Each sample was hybridized to two arrays, and expression was averaged between the technical replicates. While HDL itself has no apparent effect on gene expression (and on samples clustering), OxPAPC+HDL-treated samples clearly cluster separately from OxPAPC-treated samples, indicating a global effect of HDL on OxPAPC-related changes in gene expression. B: Log2-fold change of probe expression in OxPAPC-treated samples as a function of mean probe expression (x-axis). Genes that were not affected by OxPAPC are in gray. Genes that were significantly induced or repressed by OxPAPC are shown in dark blue. C: Fold change of expression of OxPAPC-affected genes (dark blue in B) in OxPAPC+HDL-treated samples. As in B, genes unaffected by OxPAPC are shown in gray. Genes affected by OxPAPC (dark blue in B) are shown in either dark blue or light blue. Dark blue indicates genes significantly affected by OxPAPC and not reversed by HDL, while light blue indicates genes reversed by HDL.
Many of the genes that are significantly affected by OxPAPC show a modest response in terms of fold change. For example, out of 1,679 genes affected by OxPAPC, the expression of only 240 genes was affected >2-fold (Fig. 1B and supplementary Table I). We examined whether HDL primarily affects genes that are subtly regulated by OxPAPC versus genes that show a large response to OxPAPC treatment (supplementary Table I). Surprisingly, the proportion of genes reversed by HDL was independent of the magnitude of effect of OxPAPC, and imposing fold change cutoffs only marginally increased the fraction of genes reversed by HDL to 60% (supplementary Table I).
A recent study by Romanoski et al. (8) examined the OxPAPC response in HAEC from 150 donors and demonstrated that OxPAPC responsive genes can be classified into 12 modules based on their coexpression profiles. These modules were enriched for genes from particular gene ontology (GO) categories, suggesting that they reflect activation or inhibition of specific pathways. In order to determine whether the HDL response is limited to particular pathways or modules, we compared our results with the network analysis published in that study. We found that while there is some variation in the proportion of HDL-affected genes in the different modules, there are no apparent preferences to specific modules (supplementary Table II). We note that the OxPAPC response observed in our study only partially recapitulates the OxPAPC response described previously, which is likely attributable to the different number of donors examined in each study. To identify pathways that were significantly affected by OxPAPC treatment and examine the effect of HDL on these pathways, we used the Signaling Pathway Impact Analysis (SPIA) R package (Table 1). Altogether, HDL inhibited the effect of OxPAPC on 22 out of 24 KEGG pathways. We also examined which genes were most robustly affected by HDL and found that the OxPAPC-mediated induction of several members of the Krüppel-like factor (KLF) family of transcription factors, particularly KLF2, was completely reversed by HDL cotreatment (supplementary Fig. I). Interestingly, HDL did not significantly affect the regulation of genes involved in the oxidative stress response, although several are among the most strongly induced by OxPAPC (supplementary Fig. II).
TABLE 1.
Pathways regulated by OxPAPC are reversed by HDL
| OxPAPC | OxPAPC+HDL | |||||
| KEGG ID | Name | Number of Genes in the Pathway | Number of Genes Differentially Expressed | P | Number of Genes Differentially Expressed | P |
| 4010 | MAPK signaling pathway | 208 | 53 | 9.45E-06 | 27 | 2.86E-02 |
| 5164 | Influenza Aa | 137 | 35 | 2.26E-04 | 17 | NS |
| 5134 | Legionellosisa | 44 | 17 | 1.12E-03 | 9 | NS |
| 4012 | ErbB signaling pathway | 72 | 19 | 5.64E-03 | 8 | NS |
| 4722 | Neurotrophin signaling pathway | 99 | 26 | 5.64E-03 | 12 | NS |
| 5030 | Cocaine addiction | 38 | 13 | 5.64E-03 | 7 | NS |
| 4380 | Osteoclast differentiation | 99 | 25 | 8.06E-03 | 13 | NS |
| 5162 | Measlesa | 97 | 23 | 8.06E-03 | 10 | NS |
| 5166 | HTLV-I infectiona | 213 | 41 | 8.06E-03 | 22 | NS |
| 5169 | Epstein-Barr virus infectiona | 174 | 36 | 8.06E-03 | 23 | 2.90E-02 |
| 5200 | Pathways in cancer | 253 | 48 | 8.06E-03 | 22 | NS |
| 5160 | Hepatitis Ca | 97 | 24 | 9.35E-03 | 9 | NS |
| 5168 | Herpes simplex infectiona | 145 | 30 | 9.51E-03 | 10 | NS |
| 5120 | Epithelial cell signaling in Helicobacter pylori infectiona | 61 | 15 | 2.41E-02 | 6 | NS |
| 5215 | Prostate cancer | 73 | 18 | 2.48E-02 | 6 | NS |
| 4064 | Nuclear factor κB signaling pathwaya | 70 | 15 | 2.98E-02 | 11 | NS |
| 4660 | T-cell receptor signaling pathwaya | 79 | 18 | 2.99E-02 | 8 | NS |
| 5211 | Renal cell carcinoma | 60 | 16 | 3.02E-02 | 8 | NS |
| 5203 | Viral carcinogenesisa | 162 | 31 | 3.48E-02 | 17 | NS |
| 4620 | Toll-like receptor signaling pathwaya | 74 | 18 | 4.22E-02 | 10 | NS |
| 5145 | Toxoplasmosis | 93 | 21 | 4.22E-02 | 12 | NS |
| 4360 | Axon guidance | 105 | 19 | 4.52E-02 | 10 | NS |
| 4662 | B-cell receptor signaling pathwaya | 62 | 14 | 4.52E-02 | 7 | NS |
| 5220 | Chronic myeloid leukemia | 62 | 15 | 4.80E-02 | 8 | NS |
NS, not significant. SPIA R package was used to assess enrichment of KEGG pathways for differentially expressed genes, either in OxPAPC or OxPAPC+HDL-treated samples. OxPAPC significantly affected 24 pathways, 22 of which were not significantly affected by OxPAPC+HDL treatment. Activation status provided by SPIA is an approximation and should be interpreted cautiously. ErbB, epidermal growth factor receptor (EGFR) family signaling pathway; HTLV-I, human T-lymphotropic virus 1; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Thirteen pathways that are explicitly related to immune response.
SR-BI and apoA1 as potential mediators of HDL protection
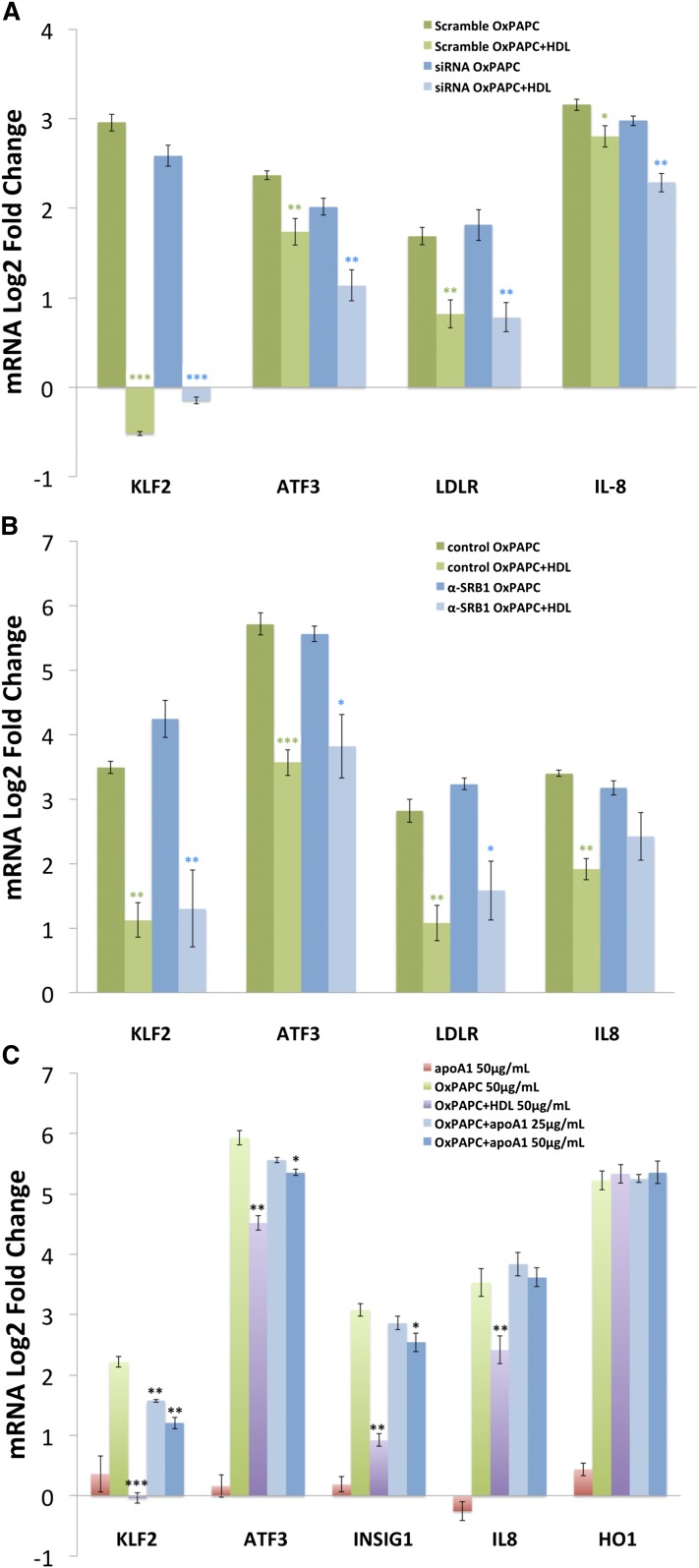
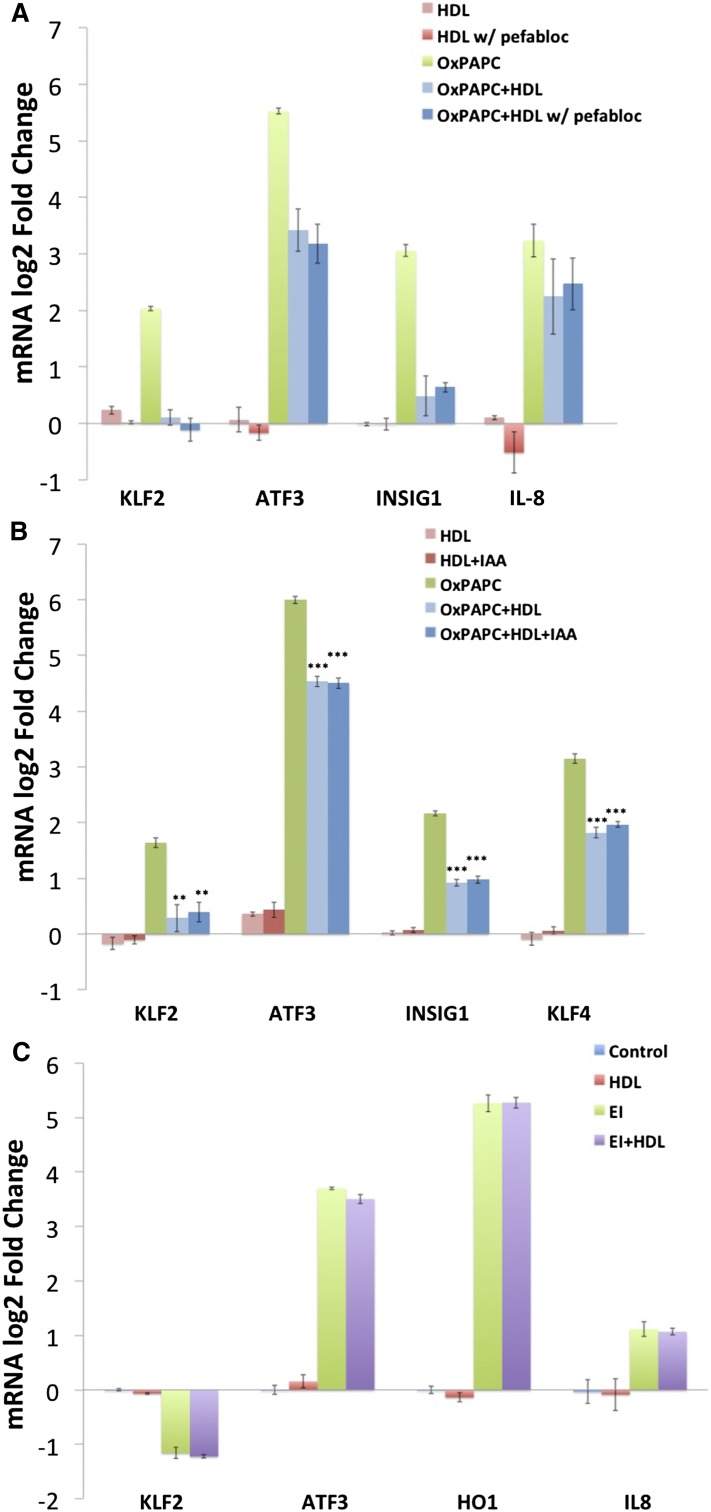
In order to gain greater insight into the mechanisms mediating the effect of HDL on OxPAPC signaling, we investigated the role of the principal HDL receptor SR-BI. In hepatocytes, SR-BI regulates the uptake of cholesterol from HDL during reverse cholesterol transport, and in endothelial cells, SR-BI has been shown to mediate eNOS activation and cholesterol repletion via interaction with apoA1 (16). Transfection of HAECs with siRNA targeting SR-BI reduced SR-BI mRNA levels by 95% with a concomitant decrease in SR-BI protein levels (supplementary Fig. III). In siRNA-transfected HAECs, HDL maintained the ability to inhibit OxPAPC-mediated induction of KLF2, ATF3, and INSIG1 expression (Fig. 2A). Similarly, cotreatment of HAECs with anti-SR-BI blocking antiserum did not attenuate the effect of HDL on OxPAPC signaling (Fig. 2B).
Fig. 2.
The effect of HDL on OxPAPC signaling is not mediated by SR-BI. HAECs were transfected with either siRNA targeting SR-BI or scramble control siRNA and treated 48 h later with media, OxPAPC (50 μg/ml), HDL (50 μg/ml), or OxPAPC and HDL (50 μg/ml) as described in the Materials and Methods. The mRNA expression levels of KLF2, ATF3, INSIG1, IL8, and HO1 were determined by qPCR and normalized to GAPDH expression levels. All data are presented as average log2-fold change of triplicates ± SEM. Asterisks indicate significance level in unpaired Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001) A: Despite effective silencing of SR-BI, the expression patterns of KLF2, ATF3, LDLR, and IL-8 after OxPAPC or OxPAPC+HDL treatment were unchanged. Results were confirmed using two separate siRNAs. B: Similarly, blocking of SR-BI using SR-BI antiserum (1:300 dilution) had no effect on the HDL response of KLF2, ATF3, LDLR, or IL-8. Rabbit IgG was used as control. C: HAECs were either untreated or treated with OxPAPC (50 μg/ml) in the presence or absence of HDL (50 μg/ml) or delipidated apoA1 (25–50 μg/ml). HDL and apoA1 were added for 1 h of pretreatment and 4 h of cotreatment. The mRNA expression levels of KLF2, ATF3, INSIG1, IL-8, and HO1 were determined by qPCR and normalized to GAPDH expression levels.
Treatment of HAECs with purified apoA1 partly mimicked HDL’s effect on OxPAPC signaling (Fig. 2C). ApoA1 at 50 μg/ml reduced the induction of KLF2, ATF3, and INSIG1 by OxPAPC. In contrast to the effect of HDL, which reduced IL-8 induction, apoA1 did not reduce IL-8 expression. Treatment with lower concentrations of apoA1 did not effect OxPAPC-mediated induction of these genes (data not shown). These results suggest that HDL, and to a lesser extent apoA1, can antagonize the effect of OxPAPC on gene expression in human endothelial cells independent of SR-BI.
Cholesterol repletion as mediator of HDL protection
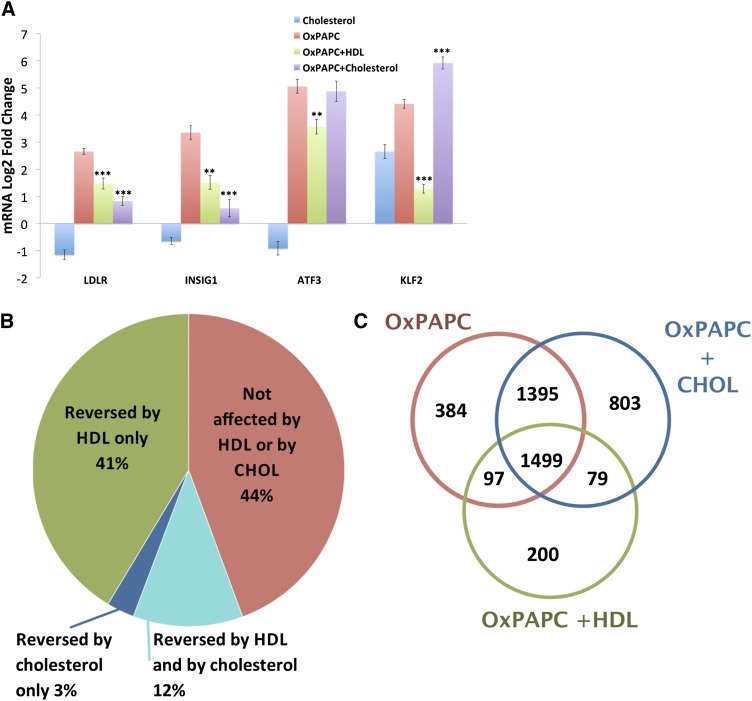
Treatment of endothelial cells with oxidized LDL or OxPAPC depletes caveolar membrane cholesterol, causing the activation of sterol-regulatory element binding protein (SREBP) and induction of SREBP target genes INSIG1 and LDLR (31). HDL can mitigate this effect by donating cholesterol to endothelial cells treated with oxidized LDL, thereby stabilizing caveolar membrane structure (32, 33). As presented in Fig. 3, cotreatment of HAECs with 20 μg/ml of cholesterol-cyclodextrin inhibited the induction of INSIG1 and LDLR by OxPAPC to a similar extent as 50 μg/ml of HDL. In contrast, cholesterol-cyclodextrin had no effect on the induction of ATF3 expression and augmented the induction of KLF2 expression in HAECs exposed to OxPAPC, a marked difference between the effects of cholesterol-cyclodextrin and HDL (Fig. 3A).
Fig. 3.
Cholesterol loading mimics some aspects of the HDL response. To determine the extent to which HDL’s effect on OxPAPC signaling in HAECs is mediated by cholesterol repletion, we compared the effect of cholesterol loading using cholesterol-cyclodextrin with that of HDL. HAECs were pretreated for 1 h with media containing 1% FBS with or without HDL (50 μg/ml) or cholesterol-cyclodextrin (20 μg/ml). Media or OxPAPC was then added to a final concentration of 50 μg/ml. A: After a 4 h treatment, mRNA was isolated and quantified by qPCR. The expression of SREBP target genes (LDLR and INSIG1) was induced by OxPAPC and similarly affected by HDL and cholesterol-cyclodextrin cotreatment. In contrast, HDL inhibited the OxPAPC-mediated induction of ATF3 and KLF2, whereas cholesterol-cyclodextrin had no effect and enhanced the upregulation of ATF3 and KLF2, respectively. Data are presented as average log2-fold change of triplicates ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001 for comparison with OxPAPC log2-fold changes. P values defined by unpaired Student’s t-test. B, C: To further characterize the similarity between the effects of HDL and cholesterol-cyclodextrin on OxPAPC signaling in HAECs, we repeated the treatment described previously on three additional HAEC donors and quantified global mRNA expression by microarray. B: Pie chart summarizing the overlap between the effects of HDL and cholesterol-cyclodextrin on genes affected by OxPAPC. Consistent with our previous experiment, HDL reversed a majority (53%) of genes affected by OxPAPC. In contrast, cholesterol-cyclodextrin reversed only 23% of genes reversed by HDL. Only 3% of genes affected by OxPAPC were reversed by cholesterol but not by HDL. C: Venn diagram, comparing cholesterol treatment versus HDL treatment. Cholesterol-cyclodextrin, when combined with OxPAPC, had synergistic effects on gene expression as reflected by the number of genes (i.e., 803) affected only in presence of both. OxPAPC and HDL, however, had a limited synergistic effect on gene expression.
To assess more accurately which aspects of the HDL response can be mimicked by cholesterol loading, we used microarrays to compare genome-wide expression of HAECs treated with OxPAPC (50 μg/ml), OxPAPC (50 μg/ml) + HDL (50 μg/ml), and cholesterol-cyclodextrin (20 μg/ml) + OxPAPC (50 μg/ml). We observed dramatic differences between the effects of cholesterol loading and HDL on HAEC response to OxPAPC. HDL reversed 1,779 (53%) of 3,375 genes affected by OxPAPC, while cholesterol loading reversed 481 genes (15%) (Fig. 3B). A total of 384 genes were reversed by both HDL and cholesterol loading, representing 21.5% of all genes reversed by HDL. Moreover, while cholesterol loading alone had minimal effects on gene expression, cotreatment with OxPAPC synergistically effected the expression of 882 genes, while HDL+OxPAPC synergistically regulated 279 genes (Fig. 3C). We note that in this experiment, OxPAPC regulated the expression of 3,375 genes, which is about twice as many as we observed in our previous experiment. This difference can be ascribed both to a higher concentration of OxPAPC used in this experiment as well as to donor-specific variation in HAEC response, which has been previously observed (8, 34).
We next examined whether the effects of cholesterol loading and HDL are comparable at the pathway rather than individual gene level. Comparison of gene ontology categories affected by OxPAPC versus OxPAPC+HDL and OxPAPC + cholesterol loading (supplementary Table III) showed that cholesterol loading inhibited 43% of the GO biological processes reversed by HDL. Among the top 10 GO processes affected by OxPAPC treatment and reversed by HDL, only “positive regulation of defense response” was similarly reversed by cholesterol loading (Table 2). Other gene ontology categories induced by OxPAPC and inhibited both by HDL and cholesterol loading include “toll signaling pathway” and “regulation of innate immune response,” suggesting that cholesterol repletion may mediate anti-inflammatory properties of HDL. However, we also observe GO categories regulated by HDL for which cholesterol loading did not have an observable effect. These include categories related to blood vessel formation such as blood vessel morphogenesis, vasculature development, and smooth muscle cell differentiation. These effects are consistent with HDL not only reducing the inflammatory effects of oxidized phospholipids, but also promoting vascular integrity (35, 36). In addition, preliminary results suggest that HDL protects against endothelial barrier disruption induced by OxPAPC as measured by transendothelial electrical resistance (data not shown). In total, the effects of HDL on endothelial cells mediated by SR-BI, apoA1, and cholesterol repletion are unable to explain a majority of the inhibition we observe on OxPAPC signaling, suggesting that HDL may directly affect OxPAPC composition and activity.
TABLE 2.
GO biological processes inhibited by HDL but not by cholesterol-cyclodextrin
| GO ID | Term | N Genes in Pathway | Expected N Significant Genes | N Significant OxPAPC | POxPAPC | POxPAPC+HDL | POxPAPC+CHOL |
| GO:0051145 | Smooth muscle cell differentiation | 22 | 5.82 | 17 | 1.60E-06 | NS | 0.00015 |
| GO:0007049 | Cell cycle | 1,137 | 300.9 | 373 | 2.50E-06 | NS | 4.20E-06 |
| GO:0001568 | Blood vessel development | 341 | 90.24 | 139 | 6.90E-06 | NS | 1.40E-05 |
| GO:0042326 | Negative regulation of phosphorylation | 155 | 41 | 68 | 1.80E-06 | NS | 5.00E-06 |
| GO:0001944 | Vasculature development | 358 | 94.74 | 146 | 1.00E-05 | NS | 9.20E-06 |
| GO:2000112 | Regulation of cellular macromolecule biosynthesis | 2,227 | 589.37 | 701 | 4.10E-05 | NS | 0.00017 |
| GO:0031349 | Positive regulation of defense response | 153 | 40.49 | 69 | 4.40E-05 | NS | NS |
| GO:0006809 | Nitric oxide biosynthetic process | 35 | 9.26 | 22 | 6.70E-05 | NS | 0.00046 |
| GO:0001816 | Cytokine production | 263 | 69.6 | 105 | 6.90E-05 | NS | 0.0008 |
| GO:2000113 | Negative regulation of cellular macromolecule biosynthetic process | 633 | 167.52 | 223 | 0.00014 | NS | 0.00033 |
Enrichment of GO terms was assessed using topGO R package. Biological processes that showed significant enrichment in OxPAPC-treated sample but not in OxPAPC+HDL-treated sample were ranked by enrichment P value in OxPAPC-treated sample. Top 10 biological processes are shown (see supplementary Table III for the full list). Notably, 9 of the 10 most significant process reversed by HDL were not reversed by cholesterol loading. CHOL, cholesterol.
Cell-independent effects of HDL on OxPAPC
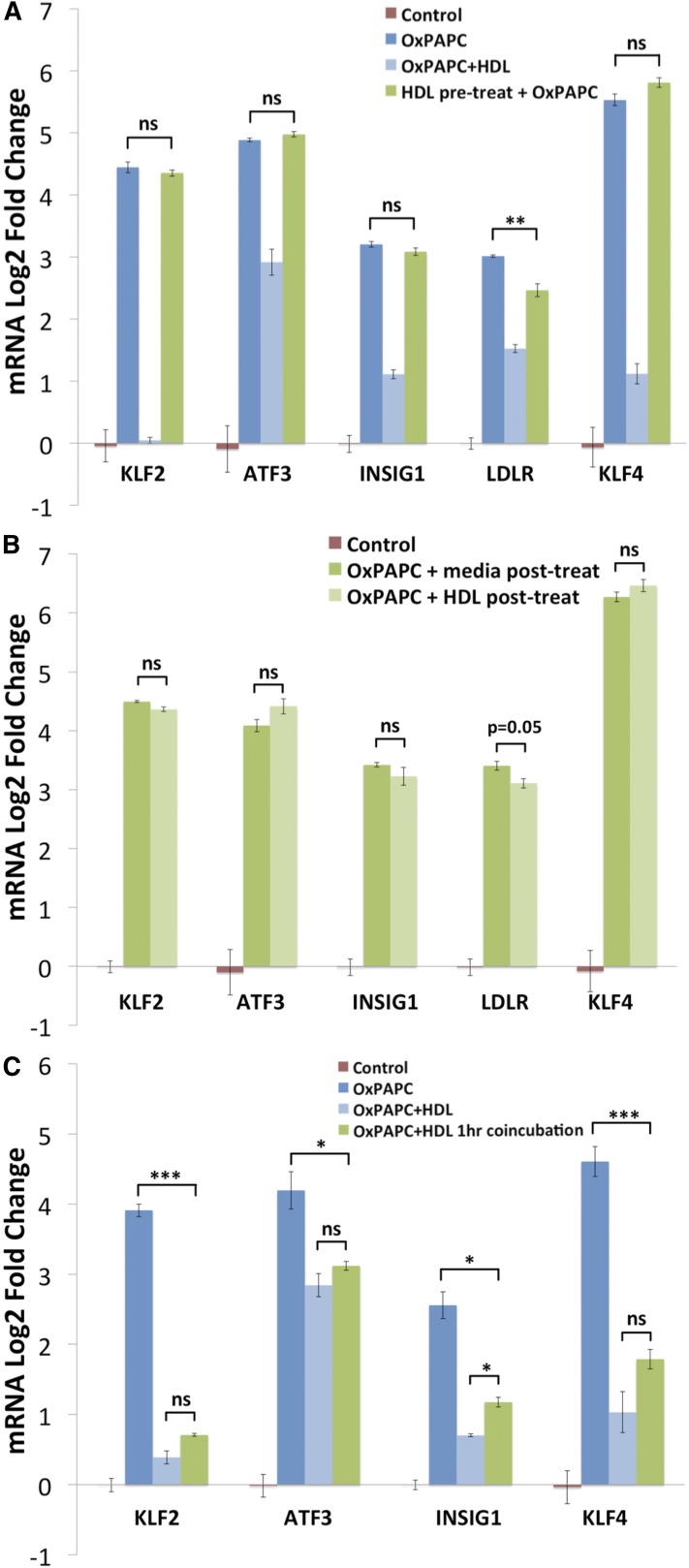
We assessed whether cotreatment with OxPAPC is necessary and sufficient for HDL protection of HAECs by testing several different treatment conditions: pretreatment alone with HDL preceding OxPAPC treatment, posttreatment with HDL following OxPAPC treatment, and coincubation of OxPAPC and HDL preceding cotreatment. The transcriptional responses of HAECs following each of these conditions were compared in parallel with our standard treatment condition shown in Fig. 1 (1 h HDL pretreatment followed by 4 h treatment with OxPAPC in the presence of HDL).
As presented in Fig. 4, neither pretreatment alone with HDL nor posttreatment with HDL were able to inhibit the induction of KLF2, ATF3, INSIG1, LDLR, and KLF4 by OxPAPC to the same extent as our standard condition. Furthermore, coincubation of OxPAPC with HDL, without pretreatment of HAECs, was as effective as our standard condition at attenuating the OxPAPC response in HAECs (Fig. 4C). These data indicate the effect of HDL on the OxPAPC response in HAECs requires cotreatment and suggest that HDL inhibits OxPAPC activity via direct interactions.
Fig. 4.
The effect of HDL on the OxPAPC response is mediated predominantly by direct interactions between HDL and OxPAPC. A: HDL pretreatment alone has minimal effect on the OxPAPC response. HAECs were pretreated for 2 h with HDL (50 μg/ml), washed three times with PBS to remove HDL, and then cells treated for 4 h with OxPAPC (50 μg/ml). In contrast to the standard cotreatment (shown as OxPAPC+HDL), pretreatment alone with HDL did not inhibit the effect of OxPAPC on the expression of KLF2, ATF3, INSIG1, and KLF4 and modestly reduced the induction of LDLR. For the standard treatment, cells were incubated without (dark blue bar) or with HDL (light blue bar) for 1 h, and then OxPAPC was added to the media for 4 h. B: Posttreatment with HDL does not inhibit the effect of OxPAPC on HAEC gene expression. HAECs were treated for 3 h with OxPAPC (50 μg/ml), washed three times with PBS to remove OxPAPC, and posttreated with either control media or media containing HDL (50 μg/ml) for an additional 3 h. Posttreatment with HDL did not reduce the induction of KLF2, ATF3, INSIG1, and KLF4 and only marginally affected the induction of LDLR compared with posttreatment with media alone. C: In contrast, coincubation of OxPAPC with HDL reduces the OxPAPC response to a similar extent as our standard HAEC pretreatment with HDL followed by cotreatment. OxPAPC (50 μg/ml) was incubated for 1 h at 37°C in medium M199 + 1% FBS with or without HDL (50 μg/ml). The solution was then used to treat HAECs for 4 h in parallel with standard control, OxPAPC (50 μg/ml), and OxPAPC+HDL (50 μg/ml) treatments. In both cases, OxPAPC was cotreated with HDL for 4 h. Data are presented as average log2-fold change of triplicate samples ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001 for indicated comparisons. P values defi ned by unpaired Student’s t-test.
The specificity of HDL protection suggests that HDL affects OxPAPC activity in a lipid-specific manner
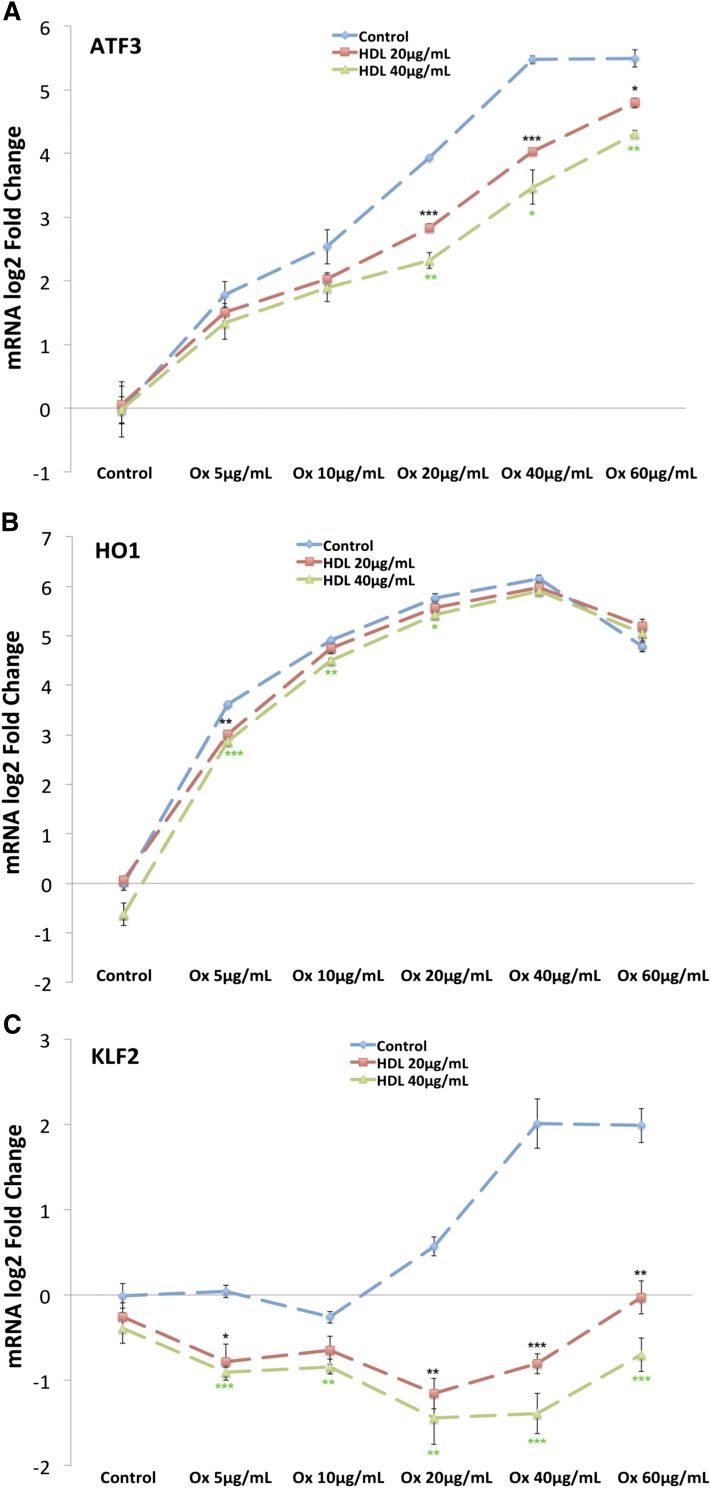
The results of the microarray studies suggest that the effects of HDL on OxPAPC signaling in HAECs are gene specific, with 48% of genes regulated by OxPAPC unaffected by cotreatment with HDL (Fig. 1). One possible explanation for this finding is that HDL nonselectively sequesters all components of OxPAPC, thereby effectively reducing the exposure of HAECs to bioactive oxidized phospholipids. In this scenario, genes that are sensitive to low concentrations of OxPAPC might be relatively unaffected by HDL at low concentrations and affected at higher concentrations. To address the role of nonspecific sequestration, we examined OxPAPC dose-response curves for several transcripts in the absence or presence of HDL at several concentrations. For the UPR transcription factor ATF3, HDL shifted the OxPAPC dose-response curve to the right in a concentration-dependent manner that is consistent with the sequestration hypothesis (Fig. 5A). In the dose response, HDL reversed the induction of KLF2 at all concentrations of OxPAPC and HDL tested (Fig. 5C). While not affected by HDL treatment alone, KLF2 expression was repressed in the presence of both OxPAPC and HDL, indicating a synergistic effect of cotreatment. In contrast, the OxPAPC dose-response curve of HO1, a marker of the oxidative stress response in HAECs, was marginally affected by HDL cotreatment at either 20 μg/ml or 40 μg/ml of HDL, consistent with our microarray results and previous findings (23) (Fig. 5B). We also examined the effect of HDL on the abundance of 12 oxidative stress response-related transcripts (supplementary Fig. II). OxPAPC significantly affected the expression of 6 of these transcripts, and HDL did not reverse these changes. Together, these results confirm that the effects of HDL on OxPAPC signaling are gene specific and argue against a model in which HDL nonselectively sequesters all the bioactive lipid components of OxPAPC.
Fig. 5.
HDL has a gene-specific effect on OxPAPC signaling in HAECs. HAECs were pretreated in the absence or presence of HDL at 20 μg/ml or 40 μg/ml for 1 h as described in the Materials and Methods. Subsequently, OxPAPC was added to a final concentration of 5 μg/ml, 10 μg/ml, 20 μg/ml, 40 μg/ml, or 60 μg/ml. After a 4 h cotreatment, mRNA was isolated from cells and quantified by qPCR. The expression of ATF3 (A) was induced in the presence of OxPAPC, and HDL cotreatment caused a dose-dependent shift in the dose-response curve. B: In contrast, HDL had a minimal effect on HO1 induction at all concentrations of OxPAPC tested. C: Unlike ATF3 and HO1, the expression of KLF2 was induced by OxPAPC and repressed in the presence of both OxPAPC and HDL, indicating a syngergistic effect of cotreatment. Points along the dose-response curve were calculated as average log2-fold change versus media control. All treatments were run in triplicate, and error bars correspond to ± SEM. Asterisks indicate significance level (*P < 0.05, **P < 0.01, ***P < 0.001).
Role of phospholipid hydrolysis by HDL-associated enzymes in HDL protection
Various enzymes on HDL hydrolyze oxidized phospholipids (29, 37, 38). PAF-AH is a calcium-independent type VII phospholipase associated with HDL that hydrolyzes biologically active oxidized phospholipids in minimally modified LDL (mmLDL), thereby limiting their inflammatory effects on endothelial cells, including the ability to promote monocyte-endothelial interactions (29, 39). We assessed the enzymatic effects of HDL on OxPAPC by incubating OxPAPC with HDL and reisolating phospholipids for analysis by HPLC-MS. Under these conditions, HDL minimally reduced the abundance of PEIPC (m/z 828.3), while strongly augmenting the hydrolysis of POVPC (m/z 594.2) and reducing the levels of PGPC (m/z 610.2) by ∼50% (Table 3). Native PAPC (m/z 782.3) was minimally hydrolyzed in the presence of HDL. Consistent with the degradation of these phospholipids, the levels of LPC increased nearly 20-fold in the samples of OxPAPC coincubated with HDL relative to the net abundance in OxPAPC and HDL incubated separately. Addition of Pefabloc or MAFP, nonreversible inhibitors of PAF-AH, markedly reduced the degradation of POVPC and PGPC as well as the accumulation of LPC (Table 3). Surprisingly, the loss of PEIPC was enhanced by the inactivation of PAF-AH. Combined with the fact that this experiment was done using buffer lacking calcium, which precludes activity of calcium-dependent phospholipases, these results demonstrate that HDL effectively catalyzes the degradation of several components of OxPAPC in a PAF-AH-dependent manner.
TABLE 3.
PAF-AH-dependent effects of HDL on OxPAPC degradation
| OxPAPC+HDL | OxPAPC+HDL+Pefabloc | OxPAPC+HDL+MAFP | |
| m/z 782.3 (PAPC) | 0.83 | 0.98 | 1.07 |
| m/z 594.2 (POVPC) | 0.14 | 0.64 | 0.50 |
| m/z 610.2 (PGPC) | 0.61 | 1.53 | 1.00 |
| m/z 828.3 (PEIPC) | 0.74 | 0.34 | 0.79 |
| m/z 496.2 (LPC) | 19.5 | 2.53 | 1.30 |
OxPAPC was incubated with PBS alone, HDL, HDL treated with 1 mM Pefabloc, or HDL treated with 100 μM MAFP for 1 h at 37°C. Total lipids were extracted and quantified by HPLC-MS as described in the Materials and Methods. Ion abundances were normalized to a DMPC internal standard and are presented as fold changes relative to the net abundance in OxPAPC alone (POVPC, PGPC, and PEIPC) or OxPAPC alone + HDL alone (PAPC and LPC).
To test whether PAF-AH activity is important in mediating the effects of HDL on HAECs exposed to OxPAPC, we incubated HDL with Pefabloc and assessed its ability to inhibit OxPAPC signaling. HDL treated with Pefabloc was as effective as untreated HDL at attenuating the induction of KLF2, ATF3, and INSIG1 by OxPAPC (Fig. 6A). IL-8 and HO1 induction was unaffected by inhibitor-treated and untreated HDL in this experiment. These results suggest that the hydrolysis of PGPC and POVPC is not responsible for HDL protection against OxPAPC signaling. However, we were unable to inhibit the hydrolysis of PEIPC by HDL using Pefabloc and cannot rule out the possibility that hydrolysis of PEIPC by other HDL enzymes contributes to the protective effects of HDL (Table 3).
Fig. 6.
The effect of HDL on OxPAPC signaling in HAECs is not dependent on the activity of PAF-AH or direct binding to cysteine residues but is specific to phospholipid components of OxPAPC. HDL (5 mg/ml) was incubated with 1 mM Pefabloc at 37°C for 1 h to inactivate lipoprotein-associated PAF-AH. The HDL was then dialyzed overnight and used in cotreatments with OxPAPC. After the inactivation of PAF-AH, HDL (50 μg/ml) reduced the effects of OxPAPC on markers of inflammatory response (IL-8 and KLF2), ER stress (ATF3), and cholesterol depletion (INSIG1) (A). Shown are the results from one of three representative experiments. Data are presented as average log2-fold change of duplicate samples ± (Max − Min)/2. B: HDL (5 mg/ml) was incubated with 100 mM iodoacetamide at 37°C for 1 h to irreversibly block free thiol residues on HDL. The HDL was then dialyzed overnight and used in HAEC treatments as described in the Materials and Methods. Treatment with iodoacetamide did not affect the ability of HDL to inhibit OxPAPC signaling in HAECs indicating that OxPAPC binding to cysteine residues on HDL is not mediating HDL protection. Data are presented as average log2-fold change of triplicates ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001 for comparison with OxPAPC log2-fold changes. P values defined by unpaired Student’s t-test. C: HAECs were treated with 1 μg/ml EI in the absence or presence of 50 μg/ml HDL for 4 h as described in the Materials and Methods. Unlike for OxPAPC, HDL cotreatment did not mitigate the effects of EI on the expression of IL-8, KLF2, ATF3, or HO1 as determined by qPCR. Data are presented as average log2-fold change versus control treated samples. All treatments were run in triplicate, and error bars correspond to SEM.
HDL protection does not involve OxPAPC binding to cysteine residues on HDL
Previous studies have shown that circulating HDL can accumulate oxidized phospholipids and that apoA1 can bind oxidized phospholipids in OxPAPC (40, 41). In addition, we previously demonstrated that bioactive phospholipid components of OxPAPC can form Michael adducts with cysteine residues on proteins in HAECs (42). We therefore hypothesized that a protective mechanism of HDL might involve covalent binding of OxPAPC to protein constituents of HDL via nucleophilic cysteine residues. However, irreversibly blocking cysteine residues on HDL using 100 mM iodoacetamide did not affect the ability of HDL to inhibit OxPAPC signaling in HAECs, implying that either noncysteine nucleophiles or noncovalent interactions may predominate in HDL regulation of OxPAPC activity (Fig. 6B).
Oxidized phospholipids are the target of the HDL protection
Previous work from our group demonstrated that the effect of HDL on one major component of OxPAPC, PEIPC, was similar to that seen for total OxPAPC. HDL reduced the induction of ATF3 and LDLR by PEIPC while HO1 induction was unaffected (23). We therefore addressed the question of whether HDL can protect against the effects of the epoxyisoprostane (EI) formed from the sn-2 hydrolysis of PEIPC. EI has recently been characterized and shown to recapitulate ∼50% of the effects of PEIPC, including the activation of oxidative stress response and ER stress response pathways (28). However, in contrast to OxPAPC and PEIPC, EI induces an anti-inflammatory phenotype in HAECs. Cotreatment of HAECs with EI and HDL demonstrated that HDL is unable to inhibit the effects of EI on the expression of several genes regulated by both PEIPC and EI (Fig. 6C). This suggests that oxidized phospholipids and not their fatty acids are targets of HDL protection.
DISCUSSION
HDL effects on transcription in the presence and absence of OxPAPC
The aim of this current study was to determine the effects of HDL on the genome-wide transcriptional response of HAECs exposed to OxPAPC and identify the proximate mechanisms mediating these effects. For this study, we used OxPAPC rather than individual oxidized phospholipids because circulating HDL interacts with multiple oxidized phospholipids that accumulate in both blood and the vessel wall (43–45). These combined phospholipids would likely interact differently with HDL than would individual phospholipids. Our microarray data consistently demonstrated that HDL attenuates 50–60% of the transcriptional changes induced by OxPAPC, and this effect is observed in multiple donors. Pathway analysis using SPIA also indicated that HDL acts on the majority of OxPAPC affected pathways, and comparison with OxPAPC network analysis suggested that the HDL effect is not limited to particular modules of genes coregulated by OxPAPC (8). Furthermore, we demonstrate that there is no relationship between the extent of OxPAPC gene regulation and the ability of HDL to inhibit OxPAPC signaling.
Previously published studies explored the role of HDL alone as a transcriptional regulator using microarrays of human endothelial cells treated with fetal or reconstituted HDL (46, 47). Both studies reported extensive regulation of transcription by reconstituted HDL (rHDL), although for the vast majority of genes, the fold change was small. In addition, McGrath et al. (46) showed that for some of the most regulated genes, the effect of rHDL was time and concentration dependent, and these authors observed no effect on gene expression after a 4 h treatment. More recently, Tabet et al. (48) found that human coronary artery endothelial cells treated with 1 mg/ml of plasma-isolated HDL for 16 h differentially expressed 120 genes, an effect partly mediated by transfer of microRNA-223 from HDL. In contrast, our study found that treatment with HDL alone at 50 μg/ml does not alter gene expression in HAECs after 5 h. These results were robust across several concentrations of HDL, donors, and experiments suggesting that concentration, exposure time, endothelial subtype, and use of rHDL versus serum isolated are important variables in assessing the effects of HDL on human endothelial cells.
Mechanisms of HDL protection
We examined several cell-dependent mechanisms by which HDL might attenuate the OxPAPC response in HAECs. SR-BI mediates several of the vasoprotective effects of HDL including the inhibition of adhesion molecule expression in endothelial cells following proinflammatory stimuli, the induction of cyclooxygenase 2 and prostacyclin production, and the stimulation of endothelial cell migration and reendothelialization following vascular injury (49–52). A majority of these responses are dependent on NO production following SR-BI mediated eNOS activation (17, 53). In contrast to these studies, we found that knockdown of SR-BI and cotreatment with blocking antibodies toward SR-BI had no effect on the ability of HDL to inhibit OxPAPC signaling (Fig. 2). Signaling via SR-BI by HDL is primarily mediated by apoA1, and in this study, we found that apoA1 had a small but significant effect on the activity of OxPAPC. We hypothesize that this modest effect of apoA1 may be due to binding of specific bioactive oxidized phospholipids rather than signaling through SR-BI (16, 40).
We also examined the possibility that despite the results with apoA1 and SR-BI, cholesterol transport was involved in the HDL protection against OxPAPC-dependent gene regulation. HDL participates in bidirectional cholesterol flux with the endothelium, thereby supporting cellular cholesterol homeostasis (16, 54). In contrast, it has been shown that OxPAPC depletes caveolar membrane cholesterol from HAECs leading to the activation of SREBP and induction of IL-8 and LDLR expression (31). To determine the extent to which cholesterol repletion could account for the observed effects of HDL on OxPAPC signaling, we collected microarray expression data from HAECs treated with cholesterol-cyclodextrin, HDL, and OxPAPC. Cotreatment of HAECs with cholesterol-cyclodextrin and OxPAPC recapitulated HDL’s effect on key SREBP target genes, INSIG1 and LDLR, as well as key inflammatory pathways (Fig. 3A and supplementary Table III). However, only 23% of the genes affected by OxPAPC treatment and reversed by HDL cotreatment were similarly reversed by cholesterol-cyclodextrin. The remaining 77% of transcripts unaffected by cholesterol-cyclodextrin were enriched among GO categories related to blood vessel development, consistent with reports of HDL activating endothelial repair mechanisms (16). These results indicate that cholesterol repletion is insufficient to explain a majority of the HDL response, though it likely plays a role in regulating genes affected by OxPAPC and HDL related to cholesterol homeostasis.
Because the major known protective effects of HDL on endothelial cells did not account for the ability of HDL to robustly inhibit OxPAPC signaling, we tested the hypothesis that HDL affects OxPAPC composition and activity directly. Our evidence that HDL cotreatment with OxPAPC is required for HDL protection supports this hypothesis. Coincubation of OxPAPC with HDL, without pretreatment of HAECs with HDL, was sufficient to reduce the induction of KLF2, ATF3, INSIG1, and KLF4 by OxPAPC; whereas pretreatment or posttreatment alone with HDL, without cotreatment with OxPAPC, only modestly reduced the induction of LDLR and did not affect the upregulation of KLF2, ATF3, INSIG1, and KLF4 (Fig. 4). These data strongly suggest that the effect of HDL on the OxPAPC response in HAECs is mediated predominantly by direct interactions between HDL and OxPAPC.
We next addressed the question of how the HDL OxPAPC interaction mediates protection. The large number of OxPAPC-regulated genes affected by HDL suggested that nonspecific sequestration of OxPAPC lipids by HDL might account for the changes in gene expression. However, our data provide evidence against this mechanism. Transcriptional changes of multiple genes in dose-response experiments, using varying concentrations of OxPAPC and HDL, are inconsistent with nonspecific sequestration of OxPAPC by HDL. This is further supported by the fact that OxPAPC retains the ability to activate oxidative stress response pathways during cotreatment with HDL (supplementary Fig. II).
As possible direct and specific effects of HDL on OxPAPC activity, we examined the role of HDL-mediated degradation or binding to select oxidized lipids. Our group and others have shown that several HDL-associated enzymes catalyze the degradation of oxidized phospholipids including those contained in OxPAPC (29, 38, 39). Because our HDL preparations are isolated in the presence of EDTA and lack calcium-dependent phospholipase activity, we investigated the role of calcium-independent PAF-AH in mediating the HDL response. Coincubation of HDL with OxPAPC induced a large accumulation of LPC in cell-free conditions, an effect inhibited by the addition of PAF-AH inhibitors (Table 3). The increase in LPC was accompanied by a marked 86% decrease in the abundance of POVPC, a 39% reduction of PGPC, and a 26% reduction of PEIPC. Inhibition of PAF-AH activity using Pefabloc or MAFP attenuated the degradation of POVPC and PGPC. However, incubation of HDL with Pefabloc did not affect HDL’s ability to inhibit OxPAPC signaling, indicating that the degradation of POVPC and PGPC via PAF-AH does not play a role in the HDL response (Fig. 6A). PEIPC was only slightly reduced by incubation of OxPAPC with HDL, suggesting that degradation of PEIPC plays a minimal role in HDL protection. Five isomers of PEIPC are produced from the oxidation of PAPC, and it is possible that enzymes on HDL are active toward only some of these molecules (55). In addition, other phospholipases carried by HDL may play a more predominate role in attenuating OxPAPC activity. Still, these data suggest that HDL catalyzes the degradation of OxPAPC by PAF-AH in a phospholipid-specific manner, but this degradation is not responsible for HDL protection.
We also present evidence that direct interactions between OxPAPC and HDL involving binding to cysteine residues are not involved in mediating the HDL response. The major bioactive component of OxPAPC in regulating transcription, PEIPC, has been shown to bind cysteine residues of proteins in HAECs, and this interaction is important in mediating the effects of OxPAPC on gene expression (42). Furthermore, human apoA1 as well as the apoA1 mimetic 4-F peptide bind OxPAPC with high affinity (40). However, irreversibly blocking cysteine residues on HDL with iodoacetamide did not attenuate HDL’s effect on OxPAPC signaling in HAECs (Fig. 6B). OxPAPC may form adducts with proteins on HDL at nucleophilic residues other than cysteine as Szapacs and colleagues (56) demonstrated using a biotin-sulfoxide analogue of 1-palmitoyl-2-linoleoylglycerylphosphatidylchole (PLPBSO). More recently, a novel enrichment method was used to demonstrate the covalent modification of oxidized HDL by a variety of oxidized phospholipids primarily at lysine and histidine residues (57). Assessing the role of these interactions in mediating the HDL response warrants further investigation. Furthermore, while we have not delineated all the mechanisms by which HDL inhibits the effects of OxPAPC on HAECs, we present evidence that this protection is specific for oxidized phospholipids by showing that the effects of EI, which regulates many of the same genes as OxPAPC and PEIPC (28), are undiminished by cotreatment with HDL. Additional research may identify other bioactive constituents of OxPAPC, or products of OxPAPC degradation, which remain active toward HAECs in the presence of HDL.
In summary, we have shown that HDL can block the regulation of >50% of the 1,000 or more genes regulated by OxPAPC in primary HAECs, a cell type particularly relevant to atherosclerosis development. In addition, we have preliminary evidence that HDL also blocks the rapid effect of OxPAPC on endothelial monolayer permeability. While recent genetic studies and clinical trials cast doubt on whether HDL cholesterol is causally associated with CAD (24–26), our results support the view that in vitro a relatively short exposure to HDL has atheroprotective effects, which are likely mediated by multiple mechanisms acting in concert. Cholesterol loading and apoA1 are partially involved in the protective effects of HDL; however, neither is sufficient to explain it entirely. Our evidence suggests that HDL interacts directly with OxPAPC to affect composition and activity. We hypothesize that this interaction involves oxidized phospholipids being degraded by phospholipases carried by HDL, binding to nucleophilic residues on HDL, and integrating into the phospholipid monolayer of HDL, altogether reducing the activation of some but not other OxPAPC signaling pathways. Taken together, our study provides important insight into the complexity of the effects of HDL on endothelial cells and highlights the importance of more systematic and global approaches to understanding HDL function.
Supplementary Material
Acknowledgments
The authors would like to thank the Atherosclerosis Research Unit at UCLA and in particular Alan Wagner, Elizabeth Tarling, and Thomas de Aguiar Vallim for technical advice and valuable discussion; and Sangderk Lee at the University of Kentucky for important insight.
Footnotes
Abbreviations:
- ATF
- activating transcription factor
- CAD
- coronary artery disease
- DMPC
- 1,2-dimyristoyl-sn-glycero-3-phosphocholine
- EI
- 5,6-epoxyisoprostane
- GO
- gene ontology
- HAEC
- human aortic endothelial cell
- IL
- interleukin
- INSIG1
- insulin-induced gene 1
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- KLF
- Krüppel-like factor
- LPC
- lyso-phosphatidylcholine
- MAFP
- methyl-γ-linolenyl fluorophosphonate
- OxPAPC
- oxidized 1-palmitoyl-2-arachidonyl-sn-glycero-3-phospholcholine
- PAF-AH
- platelet-activating factor acetylhydrolase
- PEIPC
- 1-palmitoyl-2-(5,6-epoxyisoprostane E2)-sn-glycero-3-phosphocholine
- PGPC
- 1-palmitoyl-2-glutaroyl-sn-glycero-3-phosphocholine
- POVPC
- 1-palmitoyl-2-(5-oxovaleroyl)-sn-glycero-3-phosphocholine
- qPCR
- quantitative PCR
- rHDL
- reconstituted high-density lipoprotein
- SR-BI
- scavenger receptor class B type I
- SREBP
- sterol-regulatory element binding protein
- SPIA
- Signaling Pathway Impact Analysis
- UPR
- unfolded protein response
This work was supported by National Institutes of Health Grants HL094322 (A.J.L.), HL30568 (J.A.B., A.J.L.), HL064731 (J.A.B.), and HL69766 (J.R.S.). MS instrumentation was made available through the support of Greg Khitrov (University of California, Los Angeles Molecular Instrumentation Center – Mass Spectrometry Facility in the Department of Chemistry). The MS data were collected through a project described and supported by Grant Number S10-RR025631 from the National Center for Research Resources.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables and three figures.
REFERENCES
- 1.Kannel W. B., Dawber T. R., Frideman G. D., Glennon W. E., McNamara P. M. 1964. Risk factors in coronary heart disease. An evaluation of several serum lipids as predictors of coronary heart disease: the Framingham Study. Ann. Intern. Med. 61: 888–899. [DOI] [PubMed] [Google Scholar]
- 2.Kannel W. B., Dawber T. R., Kagan A., Revotskie N., Stokes J., III 1961. Factors of risk in the development of coronary heart disease—six-year follow-up experience: the Framingham Study. Ann. Intern. Med. 55: 33–50. [DOI] [PubMed] [Google Scholar]
- 3.Lusis A. J. 2000. Atherosclerosis. Nature. 407: 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaya J. 2013. The association between biomarkers in the blood and carotid plaque composition-focusing on oxidized lipids, oxysterols and plaque status. Biochem Pharmacol. 86: 15–18. [DOI] [PubMed] [Google Scholar]
- 5.Berliner J. A., Watson A. D. 2005. A role for oxidized phospholipids in atherosclerosis. N. Engl. J. Med. 353: 9–11. [DOI] [PubMed] [Google Scholar]
- 6.Watson A. D., Leitinger N., Navab M., Faull K. F., Hörkkö S., Witztum J. L., Palinski W., Schwenke D., Salomon R. G., Sha W., et al. 1997. Structural identification by mass spectrometry of oxidized phospholipids in minimally oxidized low density lipoprotein that induce monocyte/endothelial interactions and evidence for their presence in vivo. J. Biol. Chem. 272: 13597–13607. [DOI] [PubMed] [Google Scholar]
- 7.Berliner J. A., Leitinger N., Tsimikas S. 2009. The role of oxidized phospholipids in atherosclerosis. J. Lipid Res. 50 (Suppl.): S207–S212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romanoski C. E., Che N., Yin F., Mai N., Pouldar D., Civelek M., Pan C., Lee S., Vakili L., Yang W-P., et al. 2011. Network for activation of human endothelial cells by oxidized phospholipids: a critical role of heme oxygenase 1. Circ. Res. 109: e27–e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S., Birukov K. G., Romanoski C. E., Springstead J. R., Lusis A. J., Berliner J. A. 2012. Role of phospholipid oxidation products in atherosclerosis. Circ. Res. 111: 778–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faria-Neto J. R., Chyu K-Y., Li X., Dimayuga P. C., Ferreira C., Yano J., Cercek B., Shah P. K. 2006. Passive immunization with monoclonal IgM antibodies against phosphorylcholine reduces accelerated vein graft atherosclerosis in apolipoprotein E-null mice. Atherosclerosis. 189: 83–90. [DOI] [PubMed] [Google Scholar]
- 11.Caligiuri G., Khallou-Laschet J., Vandaele M., Gaston A-T., Delignat S., Mandet C., Kohler H. V., Kaveri S. V., Nicoletti A. 2007. Phosphorylcholine-targeting immunization reduces atherosclerosis. J. Am. Coll. Cardiol. 50: 540–546. [DOI] [PubMed] [Google Scholar]
- 12.Binder C. J., Hörkkö S., Dewan A., Chang M-K., Kieu E. P., Goodyear C. S., Shaw P. X., Palinski W., Witztum J. L., Silverman G. J. 2003. Pneumococcal vaccination decreases atherosclerotic lesion formation: molecular mimicry between Streptococcus pneumoniae and oxidized LDL. Nat. Med. 9: 736–743. [DOI] [PubMed] [Google Scholar]
- 13.Tsimikas S., Miyanohara A., Hartvigsen K., Merki E., Shaw P. X., Chou M-Y., Pattison J., Torzewski M., Sollors J., Friedmann T., et al. 2011. Human oxidation-specific antibodies reduce foam cell formation and atherosclerosis progression. J. Am. Coll. Cardiol. 58: 1715–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lichtman A. H., Binder C. J., Tsimikas S., Witztum J. L. 2013. Adaptive immunity in atherogenesis: new insights and therapeutic approaches. J. Clin. Invest. 123: 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon D. J., Rifkind B. M. 1989. High-density lipoprotein—the clinical implications of recent studies. N. Engl. J. Med. 321: 1311–1316. [DOI] [PubMed] [Google Scholar]
- 16.Riwanto M., Landmesser U. 2013. High density lipoproteins and endothelial functions: mechanistic insights and alterations in cardiovascular disease. J. Lipid Res. 54: 3227–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mineo C., Shaul P. W. 2013. Regulation of signal transduction by HDL. J. Lipid Res. 54: 2315–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mineo C., Deguchi H., Griffin J. H., Shaul P. W. 2006. Endothelial and antithrombotic actions of HDL. Circ. Res. 98: 1352–1364. [DOI] [PubMed] [Google Scholar]
- 19.Rye K-A., Barter P. J. 2014. Cardioprotective functions of HDLs. J. Lipid Res. 55: 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terasaka N., Westerterp M., Koetsveld J., Fernández-Hernando C., Yvan-Charvet L., Wang N., Sessa W. C., Tall A. R. 2010. ATP-binding cassette transporter G1 and high-density lipoprotein promote endothelial NO synthesis through a decrease in the interaction of caveolin-1 and endothelial NO synthase. Arterioscler. Thromb. Vasc. Biol. 30: 2219–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rämet M. E., Rämet M., Lu Q., Nickerson M., Savolainen M. J., Malzone A., Karas R. H. 2003. High-density lipoprotein increases the abundance of eNOS protein in human vascular endothelial cells by increasing its half-life. J. Am. Coll. Cardiol. 41: 2288–2297. [DOI] [PubMed] [Google Scholar]
- 22.Mineo C., Shaul P. W. 2003. HDL stimulation of endothelial nitric oxide synthase: a novel mechanism of HDL action. Trends Cardiovasc. Med. 13: 226–231. [DOI] [PubMed] [Google Scholar]
- 23.Gharavi N. M., Gargalovic P. S., Chang I., Araujo J. A., Clark M. J., Szeto W. L., Watson A. D., Lusis A. J., Berliner J. A. 2007. High-density lipoprotein modulates oxidized phospholipid signaling in human endothelial cells from proinflammatory to anti-inflammatory. Arterioscler. Thromb. Vasc. Biol. 27: 1346–1353. [DOI] [PubMed] [Google Scholar]
- 24.Rader D. J., Tall A. R. 2012. The not-so-simple HDL story: is it time to revise the HDL cholesterol hypothesis? Nat. Med. 18: 1344–1346. [DOI] [PubMed] [Google Scholar]
- 25.Voight B. F., Peloso G. M., Orho-Melander M., Frikke-Schmidt R., Barbalic M., Jensen M. K., Hindy G., Hólm H., Ding E. L., Johnson T., et al. 2012. Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet. 380: 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.AIM-HIGH Investigators, Boden W. E., Probstfield J. L., Anderson T., Chaitman B. R., Desvignes-Nickens P., Koprowicz K., McBride R., Teo K., Weintraub W. 2011. Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. N. Engl. J. Med. 365: 2255–2267. [DOI] [PubMed] [Google Scholar]
- 27.Rosenson R. S., Brewer H. B., Ansell B., Barter P., Chapman M. J., Heinecke J. W., Kontush A., Tall A. R., Webb N. R. 2013. Translation of high-density lipoprotein function into clinical practice: current prospects and future challenges. Circulation. 128: 1256–1267. [DOI] [PubMed] [Google Scholar]
- 28.Zhong W., Springstead J. R., Al-Mubarak R., Lee S., Li R., Emert B., Berliner J. A., Jung M. E. 2013. An epoxyisoprostane is a major regulator of endothelial cell function. J. Med. Chem. 56: 8521–8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watson A. D., Navab M., Hama S. Y., Sevanian A., Prescott S. M., Stafforini D. M., McIntyre T. M., Du B. N., Fogelman A. M., Berliner J. A. 1995. Effect of platelet activating factor-acetylhydrolase on the formation and action of minimally oxidized low density lipoprotein. J. Clin. Invest. 95: 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bligh E. G., Dyer W. J. 1959. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 31.Yeh M., Cole A. L., Choi J., Liu Y., Tulchinsky D., Qiao J-H., Fishbein M. C., Dooley A. N., Hovnanian T., Mouilleseaux K., et al. 2004. Role for sterol regulatory element-binding protein in activation of endothelial cells by phospholipid oxidation products. Circ. Res. 95: 780–788. [DOI] [PubMed] [Google Scholar]
- 32.Everson W. V., Smart E. J. 2001. Influence of caveolin, cholesterol, and lipoproteins on nitric oxide synthase: implications for vascular disease. Trends Cardiovasc. Med. 11: 246–250. [DOI] [PubMed] [Google Scholar]
- 33.Uittenbogaard A., Shaul P. W., Yuhanna I. S., Blair A., Smart E. J. 2000. High density lipoprotein prevents oxidized low density lipoprotein-induced inhibition of endothelial nitric-oxide synthase localization and activation in caveolae. J. Biol. Chem. 275: 11278–11283. [DOI] [PubMed] [Google Scholar]
- 34.Romanoski C. E., Lee S., Kim M. J., Ingram-Drake L., Plaisier C. L., Yordanova R., Tilford C., Guan B., He A., Gargalovic P. S., et al. 2010. Systems genetics analysis of gene-by-environment interactions in human cells. Am. J. Hum. Genet. 86: 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Argraves K. M., Gazzolo P. J., Groh E. M., Wilkerson B. A., Matsuura B. S., Twal W. O., Hammad S. M., Argraves W. S. 2008. High density lipoprotein-associated sphingosine 1-phosphate promotes endothelial barrier function. J. Biol. Chem. 283: 25074–25081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkerson B. A., Grass G. D., Wing S. B., Argraves W. S., Argraves K. M. 2012. Sphingosine 1-phosphate (S1P) carrier-dependent regulation of endothelial barrier: high density lipoprotein (HDL)-S1P prolongs endothelial barrier enhancement as compared with albumin-S1P via effects on levels, trafficking, and signaling of S1P1. J. Biol. Chem. 287: 44645–44653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leitinger N., Watson A. D., Hama S. Y., Ivandic B., Qiao J-H., Huber J., Faull K. F., Grass D. S., Navab M., Fogelman A. M., et al. 1999. Role of group II secretory phospholipase A2 in atherosclerosis: 2. Potential involvement of biologically active oxidized phospholipids. Arterioscler. Thromb. Vasc. Biol. 19: 1291–1298. [DOI] [PubMed] [Google Scholar]
- 38.Subramanian V. S., Goyal J., Miwa M., Sugatami J., Akiyama M., Liu M., Subbaiah P. V. 1999. Role of lecithin-cholesterol acyltransferase in the metabolism of oxidized phospholipids in plasma: studies with platelet-activating factor-acetyl hydrolase-deficient plasma. Biochim. Biophys. Acta. 1439: 95–109. [DOI] [PubMed] [Google Scholar]
- 39.McIntyre T. M., Prescott S. M., Stafforini D. M. 2009. The emerging roles of PAF acetylhydrolase. J. Lipid Res. 50 (Suppl.): S255–S259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Lenten B. J., Wagner A. C., Jung C-L., Ruchala P., Waring A. J., Lehrer R. I., Watson A. D., Hama S., Navab M., Anantharamaiah G. M., et al. 2008. Anti-inflammatory apoA-I-mimetic peptides bind oxidized lipids with much higher affinity than human apoA-I. J. Lipid Res. 49: 2302–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowry V. W., Stanley K. K., Stocker R. 1992. High density lipoprotein is the major carrier of lipid hydroperoxides in human blood plasma from fasting donors. Proc. Natl. Acad. Sci. USA. 89: 10316–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Springstead J. R., Gugiu B. G., Lee S., Cha S., Watson A. D., Berliner J. A. 2012. Evidence for the importance of OxPAPC interaction with cysteines in regulating endothelial cell function. J. Lipid Res. 53: 1304–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oskolkova O. V., Afonyushkin T., Preinerstorfer B., Bicker W., von Schlieffen E., Hainzl E., Demyanets S., Schabbauer G., Lindner W., Tselepis A. D., et al. 2010. Oxidized phospholipids are more potent antagonists of lipopolysaccharide than inducers of inflammation. J. Immunol. 185: 7706–7712. [DOI] [PubMed] [Google Scholar]
- 44.Podrez E. A., Poliakov E., Shen Z., Zhang R., Deng Y., Sun M., Finton P. J., Shan L., Febbraio M., Hajjar D. P., et al. 2002. A novel family of atherogenic oxidized phospholipids promotes macrophage foam cell formation via the scavenger receptor CD36 and is enriched in atherosclerotic lesions. J. Biol. Chem. 277: 38517–38523. [DOI] [PubMed] [Google Scholar]
- 45.Podrez E. A. 2010. Anti-oxidant properties of high-density lipoprotein and atherosclerosis. Clin. Exp. Pharmacol. Physiol. 37: 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGrath K. C. Y., Li X. H., Puranik R., Liong E. C., Tan J. T. M., Dy V. M., DiBartolo B. A., Barter P. J., Rye K-A., Heather A. K. 2009. Role of 3beta-hydroxysteroid-delta 24 reductase in mediating antiinflammatory effects of high-density lipoproteins in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 29: 877–882. [DOI] [PubMed] [Google Scholar]
- 47.Augsten M., Hackl H., Ebner B., Chemelli A., Glatter O., Marsche G., Lang U., Desoye G., Wadsack C. 2011. Fetal HDL/apoE: a novel regulator of gene expression in human placental endothelial cells. Physiol. Genomics. 43: 1255–1262. [DOI] [PubMed] [Google Scholar]
- 48.Tabet F., Vickers K. C., Cuesta Torres L. F., Wiese C. B., Shoucri B. M., Lambert G., Catherinet C., Prado-Lourenco L., Levin M. G., Thacker S., et al. 2014. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat. Commun. 5: 3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura T., Tomura H., Mogi C., Kuwabara A., Damirin A., Ishizuka T., Sekiguchi A., Ishiwara M., Im D-S., Sato K., et al. 2006. Role of scavenger receptor class B type I and sphingosine 1-phosphate receptors in high density lipoprotein-induced inhibition of adhesion molecule expression in endothelial cells. J. Biol. Chem. 281: 37457–37467. [DOI] [PubMed] [Google Scholar]
- 50.Seetharam D., Mineo C., Gormley A. K., Gibson L. L., Vongpatanasin W., Chambliss K. L., Hahner L. D., Cummings M. L., Kitchens R. L., Marcel Y. L., et al. 2006. High-density lipoprotein promotes endothelial cell migration and reendothelialization via scavenger receptor-B type I. Circ. Res. 98: 63–72. [DOI] [PubMed] [Google Scholar]
- 51.Zhu W., Saddar S., Seetharam D., Chambliss K. L., Longoria C., Silver D. L., Yuhanna I. S., Shaul P. W., Mineo C. 2008. The scavenger receptor class B type I adaptor protein PDZK1 maintains endothelial monolayer integrity. Circ. Res. 102: 480–487. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Q-H., Zu X-Y., Cao R-X., Liu J-H., Mo Z-C., Zeng Y., Li Y-B., Xiong S-L., Liu X., Liao D-F., et al. 2012. An involvement of SR-B1 mediated PI3K-Akt-eNOS signaling in HDL-induced cyclooxygenase 2 expression and prostacyclin production in endothelial cells. Biochem. Biophys. Res. Commun. 420: 17–23. [DOI] [PubMed] [Google Scholar]
- 53.Yuhanna I. S., Zhu Y., Cox B. E., Hahner L. D., Osborne-Lawrence S., Lu P., Marcel Y. L., Anderson R. G., Mendelsohn M. E., Hobbs H. H., et al. 2001. High-density lipoprotein binding to scavenger receptor-BI activates endothelial nitric oxide synthase. Nat. Med. 7: 853–857. [DOI] [PubMed] [Google Scholar]
- 54.Yancey P. G., Bortnick A. E., Kellner-Weibel G., de la Llera-Moya M., Phillips M. C., Rothblat G. H. 2003. Importance of different pathways of cellular cholesterol efflux. Arterioscler. Thromb. Vasc. Biol. 23: 712–719. [DOI] [PubMed] [Google Scholar]
- 55.Watson A. D., Subbanagounder G., Welsbie D. S., Faull K. F., Navab M., Jung M. E., Fogelman A. M., Berliner J. A. 1999. Structural identification of a novel pro-inflammatory epoxyisoprostane phospholipid in mildly oxidized low density lipoprotein. J Biol Chem. 274: 24787–98. [DOI] [PubMed] [Google Scholar]
- 56.Szapacs M. E., Kim H. Y., Porter N. A., Liebler D. C. 2008. Identification of proteins adducted by lipid peroxidation products in plasma and modifications of apolipoprotein A1 with a novel biotinylated phospholipid probe. J. Proteome Res. 7: 4237–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao D., Willard B., Podrez E. A. 2014. Analysis of covalent modifications of proteins by oxidized phospholipids using a novel method of peptide enrichment. Anal. Chem. 86: 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.