Background: New antiviral agents bind to a site on HIV integrase protein also bound by the cellular protein LEDGF/p75.
Results: Compound GSK1264 binds to this site, but it has surprising properties; it inhibits late during HIV replication, not early during integration, and it promotes abnormal multimerization.
Conclusion: GSK1264 provides new insight into HIV replication.
Significance: These observations inform the design of improved antiviral agents.
Keywords: Bioinformatics, Biophysics, Genomics, Retrovirus, X-ray Crystallography, Analytical Ultracentrifugation (AUC), Oligomerization, Size Exclusion Chromatography In-line with Multiangle Light Scattering (SEC-MALS), Small Angle X-ray Scattering (SAXS), Time-resolved FRET
Abstract
HIV-1 replication in the presence of antiviral agents results in evolution of drug-resistant variants, motivating the search for additional drug classes. Here we report studies of GSK1264, which was identified as a compound that disrupts the interaction between HIV-1 integrase (IN) and the cellular factor lens epithelium-derived growth factor (LEDGF)/p75. GSK1264 displayed potent antiviral activity and was found to bind at the site occupied by LEDGF/p75 on IN by x-ray crystallography. Assays of HIV replication in the presence of GSK1264 showed only modest inhibition of the early infection steps and little effect on integration targeting, which is guided by the LEDGF/p75·IN interaction. In contrast, inhibition of late replication steps was more potent. Particle production was normal, but particles showed reduced infectivity. GSK1264 promoted aggregation of IN and preformed LEDGF/p75·IN complexes, suggesting a mechanism of inhibition. LEDGF/p75 was not displaced from IN during aggregation, indicating trapping of LEDGF/p75 in aggregates. Aggregation assays with truncated IN variants revealed that a construct with catalytic and C-terminal domains of IN only formed an open polymer associated with efficient drug-induced aggregation. These data suggest that the allosteric inhibitors of IN are promising antiviral agents and provide new information on their mechanism of action.
Introduction
Early steps of HIV-1 replication involve reverse transcription, which produces a double-stranded cDNA copy of the viral RNA genome, and integration, which results in the incorporation of the viral DNA into host chromosomal DNA (1). Both of these steps have been targeted by clinically useful inhibitors (2, 3). The integrase strand transfer inhibitors bind to the active site of the virus-encoded integrase (IN)4 enzyme and block the initial strand transfer step that incorporates viral DNA into the host chromosome. In this report, we describe the properties of a small molecule, GSK1264, that functions as an allosteric inhibitor of IN and inhibits late during the viral replication cycle.
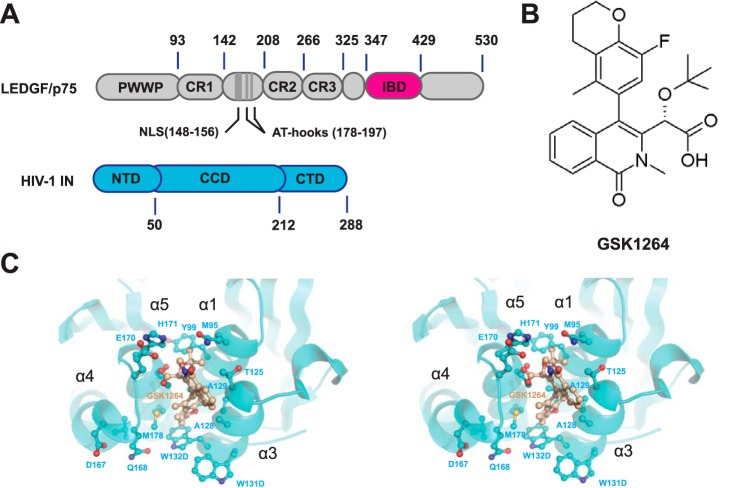
In infected cells, HIV-1 IN binds the cellular host factor, LEDGF/p75, which is a transcriptional co-activator that is the product of the PSIP1 gene (Fig. 1A). Following reverse transcription, IN binds the double-stranded viral DNA ends to form the intasome. LEDGF/p75 primarily binds to the catalytic core domain (CCD) and, to a lesser extent, the N-terminal domain (NTD) of integrase (4) via the LEDGF/p75 C-terminal integrase binding domain (IBD) (5). This interaction promotes efficient integration and the targeting of the intasome to active transcription units (6–8). LEDGF/p75 acts as a simple tether, as shown by experiments in which the LEDGF/p75 chromatin binding domain is substituted with new chromatin binding domains, resulting in retargeting of HIV-1 integration (9–11). Additionally, the LEDGF/p75·IN interaction protects IN from proteolysis and stimulates catalysis both in vitro and in vivo (4, 12–16).
FIGURE 1.
Overview of HIV-1 IN, LEDGF/p75, and GSK1264. A, HIV-1 IN and LEDGF/p75 protein domain organization. IN is a three-domain protein, composed of a zinc-binding NTD, a CCD with a DDE active site and RNase H-like fold, and a CTD with an Src homology 3-like fold. The IBD of the p75 isoform of LEDGF is shown in magenta. B, chemical structure of GSK1264. C, stereo view of the GSK1264 binding site at the IN(CCD) dimer interface, as determined by x-ray crystallography (PDB code 4OJR). Helices α3 and α1 from one monomer subunit and α5 and α4 from another are shown cradling the drug (tan).
The structure of the LEDGF/p75 IBD·IN(CCD) complex was determined by Cherepanov and colleagues (17, 18). The IBD binds at the interface of the IN(CCD) dimer, occupying a pocket at the interface. Earlier work has demonstrated that this site can be bound by small molecules (19), and subsequent screening efforts have yielded several small molecule classes that interfere with binding of the LEDGF/p75 IBD and display antiviral activity (reviewed in Refs. 20 and 21).
Here we present a detailed study of one such molecule, GSK1264 (Fig. 1B). Co-crystallization with the IN(CCD) showed that GSK1264 indeed bound the LEDGF/p75 binding site. GSK1264 inhibited most potently in the late part of the viral replication cycle, after integration. As this work was being completed, several other groups made similar findings with structurally related inhibitors (20, 22–26). Potency of GSK1264 early during infection was only modest, and there was little effect of GSK1264 on integration target site selection. In the presence of GSK1264, viral particles were produced normally late during infection, but they were reduced in infectivity. Potent inhibition correlated with increased oligomerization of IN in the presence of drug in vitro. Drug-induced oligomerization of preformed LEDGF·IN complexes was not associated with detectable displacement of LEDGF/p75. Study of truncated derivatives of IN in vitro demonstrated that drug-induced polymerization was most potent in variants containing the CCD and CTD only. Thus, compounds that bind the LEDGF/p75 site on IN are effective inhibitors whose primary effects occur at the latest steps of replication, and inhibition correlates with abnormal IN polymerization involving specific protein domains.
EXPERIMENTAL PROCEDURES
Cell Lines
The TZM-bl, 293T, and U373/CD4/CCR5 (27) cell lines were obtained through the National Institutes of Health AIDS Research and Reference Reagent Program (ARRRP) and grown as directed (28). A1953 chronic HIV producer cells were a gift from James Hoxie.
HIV-1 Infection and Integration Target Site Analysis
Infections were carried out in TZM-bl cells using standard methods and the HIV-1 strain HIV89.6 (29). Analysis of HIV-1 integration targeting was carried out as described previously (6, 30–32). All sites common among samples (including the reporter construct in the TZM-bl cells) were removed prior to analysis.
For the study of LEDGF/p75 knockdown cells, an shRNA construct (Sigma-Aldrich, TRCN0000074819) was transduced into a 293T-derived cell line, and cells were subjected to puromycin selection (1 μg/ml), yielding KD19 cells. In parallel, a matched construct encoding a GFP-targeting shRNA was introduced into the 293T cell line and compared. Knockdown was confirmed to reduce LEDGF/p75 mRNA levels by 92%, and protein was undetectable by Western blot analysis.
Protein Purification
The CCD of HIV-1 INF185K used for TR-FRET binding experiments and x-ray crystallography was expressed and purified as described in the supplemental Methods. Recombinant proteins were expressed and purified as described previously (7, 33). Complexes between LEDGF(326–530) or LEDGF(IBD) (residues 347–471) and quadramutated IN (C56S/F139D/F185H/C280S, referred to as “INQ”) or wild-type HIV-1 IN were obtained by co-expression from pETDuet (Novagen Inc., Madison, WI) in BL21 (DE3) cells (Novagen) at 37 °C. LEDGF constructs were inserted into the vector in-frame with a C-terminal Mxe intein (New England Biolabs, Ipswich, MA) containing chitin-binding domain and hexahistidine affinity tags. The domain truncations INF185H(NTD-CCD), INF185H(CCD-CTD), and INF185H(CCD) were similarly purified.
Proteins were purified using nickel-nitrilotriacetic acid (Qiagen, Valencia, CA) and chitin (New England Biolabs) resins. Fusion proteins were released by intein cleavage in 50 mm DTT overnight at 4 °C. Preparations of full-length INQ alone and LEDGF(326–530) were further purified using SP-Sepharose chromatography (GE Healthcare). Proteins were concentrated at 4 °C in YM-10 Centricons (Millipore, Billerica, MA), and aliquots were flash-frozen in liquid nitrogen with 20% glycerol for storage at −80 °C. All preparations used for this study were stored in 20 mm HEPES-NaOH, pH 7.5, 450 mm NaCl, 0.1 mm EDTA, 10 μm ZnOAc2, 5 mm CHAPS, 10 mm DTT, and 20% glycerol. All biophysical analyses were performed in 0.1-μm filtered buffer composed of 20 mm HEPES-NaOH, pH 7.5, 450 mm NaCl, 0.1 mm EDTA, 10 μm ZnOAc2, 1–10 mm DTT, with or without 5 mm CHAPS.
The detergent was confirmed to be at submicellar concentrations at this ionic strength (450 mm NaCl) using both a colorimetric assay and small-angle x-ray scattering (SAXS) analysis (34, 35) (data not shown). It has been reported that detergents such as CHAPS can attenuate IN oligomerization (36). Thus, this model system provides a hypo-oligomeric background against which drug-induced multimerization is measured.
IN-LEDGF FRET Binding Assay (48)
GSK1264 was dissolved in DMSO to 10 mm and serially diluted in DMSO for assays. Reactions were performed at a 10-μl final volume in a 384-well plate format (Greiner Bio-One, San Diego, CA). The reaction buffer contained 50 mm HEPES, pH 7.5, 150 mm NaCl, 20 mm MgCl2, 0.1 mg/ml bovine serum albumin (BSA), 50 μm CHAPS, and fresh 2 mm DTT. After the addition of drug in a 0.1-μl volume, hexahistidine-tagged INF185K(CCD) was added to a final concentration of 5 nm using a Multidrop Combi reagent dispenser (Thermo Fisher Scientific). After 30 min of incubation at room temperature, another addition was made with the dispenser to deliver GST-tagged LEDGF(IBD) (residues 347–429) to a final concentration of 5 nm alongside the time-resolved FRET reagents allophycocyanin-conjugated α-hexahistidine monoclonal antibody and α-GST europium-labeled monoclonal antibody (PerkinElmer Life Sciences) at 5 nm final concentrations. After an additional 1 h of incubation at room temperature, the time-resolved FRET signal at 665 nm was recorded with a ViewLux microplate imager (PerkinElmer Life Sciences).
Size-exclusion Chromatography and Multiangle Light Scattering (SEC-MALS)
Absolute molecular weights were determined by multiangle light scattering coupled with refractive interferometric detection (Wyatt Technology Corp., Santa Barbara, CA) and a Superdex 200 10/300 GL column (GE Healthcare) at room temperature, as described previously (33).
Sedimentation Equilibrium Analysis
Sedimentation equilibrium analytical ultracentrifugation experiments were performed at 4 °C with an XL-A analytical ultracentrifuge (Beckman-Coulter, Brea, CA) and a TiAn60 rotor with two-channel charcoal-filled Epon centerpieces and quartz windows. Data were collected at 4 °C with detection at 280 nm for 5, 7.5, and 10 μm samples. Analysis was carried out using global fits to all concentrations with data acquired at 18,000, 22,000, and 24,000 rpm, with strict mass conservation using the program SEDPHAT (70).
Sedimentation Velocity Analysis
Sedimentation velocity ultracentrifugation experiments were performed at 4 °C with an XL-A analytical ultracentrifuge (Beckman-Coulter, Brea, CA) and a TiAn60 rotor with two-channel charcoal-filled Epon centerpieces and quartz windows. Samples were analyzed at an A280 of 0.5–1.2. Complete sedimentation velocity profiles were recorded every 30 s for 50–200 boundaries at 45,000 rpm. Data were fit using the c(S) distribution model of the Lamm equation as implemented in the program SEDFIT (37). After optimizing meniscus position and fitting limits, the sedimentation coefficient (S) and best fit frictional ratio (f/f0) was determined by iterative least squares analysis, and final values were corrected to 20 °C in water (s20,w). The partial specific volume (v̄) of the protein studied, including molar extinction coefficients, solvent density (ρ = 1.01 g/ml), and viscosity (η = 0.01002 poise) were derived from chemical composition by the program SEDNTERP (38).
SAXS
Data were recorded on a Rigaku S-Max3000 small-angle x-ray scattering system equipped with Osmic mirror optics (Osmic Inc., Troy, MI), a three-pinhole enclosed preflight path, an evacuated sample chamber with customized sample holder cryostated at 4 °C, and a gas-filled multiwire detector; the instrument is served by a Rigaku MicroMax-007 HF microfocus rotating anode generator (Rigaku America, Woodland, TX). Protein samples were spun at 45,000 RPM for 5 min at 4 °C in a tabletop centrifuge prior to the addition of DMSO or GSK1264 and immediate 10-min x-ray exposures. The forward scattering from the samples studied was recorded on a CCD detector and circularly averaged to yield one-dimensional intensity profiles as a function of q (q = 4πsinθ/λ, where 2θ is the scattering angle, in units of Å−1). Data were reduced using SAXSGui version 2.05.02 (JJ X-Ray Systems ApS, Lyngby, Denmark), and matching buffers were subtracted to yield the final scattering profile. The sample-to-detector distance and beam center were calibrated using silver behenate and intensity converted to absolute units (cm−1) using a known polymer standard.
SAXS Data Analysis
All of the preparations analyzed were monodisperse, as evidenced by linearity in the Guinier region of the scattering data and agreement of the I(0) and Rg values determined with inverse Fourier transform analysis by the programs GNOM (39). Guinier analyses were performed where qRg ≤ 1.4. Molecular mass was derived from I(0) measurements, using the forward scatter from the following series of protein standards of known mass at a 5 mg/ml concentration for a standard curve: cytochrome c (12.2 kDa), RNase A (13.7 kDa), myoglobin (17.7 kDa), soybean trypsin inhibitor (20.1 kDa), chymotrypsin (25 kDa), horseradish peroxidase (44 kDa), catalase (dimer of 125 kDa), γ-globulin (151 kDa), and thyroglobulin (a dimer of 670 kDa).
When fitting manually, the maximum diameter of the particle (Dmax) was adjusted in 5–10-Å increments in GNOM to maximize the goodness of fit parameter, to minimize the discrepancy between the fit and the experimental data, and to optimize the visual qualities of the distribution profile.
Turbidity Assays
Assays were performed using a Tecan 96-well plate reader (Tecan Group Ltd., Männedorf, Switzerland), which monitors absorbance at 405 nm in 1-min intervals over 10–60 min at 27 °C. Reactions were initiated by adding protein solutions to a final concentration of 9–30 μm to a buffer containing 20 mm HEPES, pH 7.5, 450 mm NaCl, 0.1 mm EDTA, 10 μm ZnOAc2, 5 mm CHAPS, 10 mm DTT, and inhibitor at 5–100 μm final concentrations or DMSO control. Experiments performed on preformed LEDGF·IN complexes were performed in the absence of CHAPS.
Structure Determination
The CCD of HIV-1 INF185K was diluted to 10 mg/ml in 10 mm HEPES, pH 7.0, 500 mm NaCl, and 3 mm DTT. Apocrystals were grown by hanging drop vapor diffusion at 4 °C. 2-μl drops were set by combining 1 μl of protein and 1 μl of 7% PEG 8000, 0.2 m ammonium sulfate, 0.1 m sodium cacodylate, pH 6.5, 5 mm manganese chloride, 5 mm magnesium chloride, and 5 mm DTT. The apocrystals were harvested and soaked with 2.5 mm GSK1264 for 3 days at 4 °C. The soaked crystals were transferred to 30% ethylene glycol in mother liquor and subsequently flash-frozen in liquid nitrogen.
X-ray diffraction data were collected at the Advanced Photon Source at Argonne National Laboratories on beam line 21-ID-G. Data collection statistics are summarized in Table 1. The structure was determined by molecular replacement using the program Phaser (40). AutoBuster (41) was used for the initial rounds of refinement and map calculations. The structure was rebuilt using the program Coot (42). Final refinements were done using the program Refmac (43).
TABLE 1.
Crystallographic data and results
| HIV-1 IN(CCD)·GSK1264 | |
|---|---|
| Data collection | |
| Source | APS 21IDG |
| Wavelength (Å) | 0.97856 |
| Resolution range (Å) | 28.4-1.82 (1.89-1.82)a |
| Space group | P 31 21 |
| Unit cell parameters | a = b = 72.664 Å, c = 65.453 Å, α = β = 90º, γ = 120º |
| No. of measured reflections | 149,182 (14,898) |
| No. of unique reflections | 17,968 (1,750) |
| Multiplicity | 8.3 (8.5) |
| Completeness (%) | 98.3 (98.1) |
| Mean I/σ(I) | 21.6 (6.2) |
| Wilson B-factor (Å2) | 32.5 |
| Rmerge (%)a | 4.4 (41) |
| Refinement | |
| Rwork (%)b | 18.9 (23.0) |
| Rfree (%)c | 20.1 (24.4) |
| No. of atoms | 1,146 |
| Protein | 1,001 |
| Ligands | 44 |
| Water | 101 |
| Root mean square deviation | |
| Bond lengths (Å) | 0.008 |
| Bond angles (degrees) | 1.20 |
| Ramachandran plot (%) | |
| Most favored region | 99 |
| Additionally allowed region | 1 |
| Average B-factor (Å2) | |
| Overall | 40.9 |
| Protein | 40.2 |
| Ligands | 34.7 |
| Water | 49.9 |
a Numbers in parentheses represent values in the highest resolution shell.
b Rmerge = Σ|Ih − 〈Ih〉|/ΣIh, where 〈Ih〉 is the average intensity over symmetry equivalent measurements.
c R-factor = Σ|Fobs − Fcalc|/ΣFobs, where summation is data used in refinement.
d The summation for Rfree was calculated with 5% of the data.
RESULTS
GSK1264 Inhibits Binding of LEDGF(IBD) to INF185K(CCD)
GSK1264 (Fig. 1B) was identified as a disruptor of the LEDGF/p75 IBD interactions with IN(CCD) in vitro.5 GSK1264 inhibited IN 3′-end processing in vitro (data not shown), comparable with other compounds that interfere with LEDGF/IN interactions in vitro (23, 44–46).
GSK1264 was tested in an INF185K(CCD) versus LEDGF(IBD) time-resolved FRET binding assay (47). The pIC50 was determined to be 8.2 ± 0.14 nm (n = 7, where pIC50 is the negative log of the IC50 in moles/liter; IC50 is the concentration required to achieve 50% inhibition). In multicycle viral replication assays (MT4 cell assay), GSK1264 inhibited HIV-1 replication with an IC50 of ∼38 nm.
X-ray Crystallography Reveals That GSK1264 Binds to the IN(CCD) Dimer Interface
Crystals of INF185K(CCD) were soaked with GSK1264, and the structure was determined by x-ray crystallography (Fig. 1C). In this structure, electron density consistent with bound GSK1264 could be identified and unambiguously modeled. As is observed in other crystal structures of IN(CCD) and inhibitors of this class, the drug is bound within a pocket defined by helices α1 and α3 from one monomer subunit and α4 and α5 from another. The carboxylate moiety of the compound interacts with the backbone amides of IN residues Glu-170 and His-171, with additional hydrogen bonds to the carboxylate moiety provided by the hydroxyl oxygen of Thr-174. As was observed with tert-butoxy-(4-phenyl-quinolin-3-yl) acid inhibitor co-crystallized with the IN(CCD) (44), the tert-butoxy moiety binds to a complementary pocket lined by residues Gln-95, Tyr-99, Thr-125, and Thr-174. The substituent at R4 of the isoquinoline core is pointed toward Trp-132 and adjacent to Ala-128, almost at a 45º angle with respect to the plane of the dimer interface and parallel to α3. The positioning of the allosteric inhibitor GSK1264 spatially coincides with that observed for other multimodal inhibitors studied by x-ray crystallography (22–24, 44, 48).
Inhibition of Early Steps of HIV-1 Infection by GSK1264
Fig. 2A shows the effects of GSK1264 on the early steps of HIV-1 infection. TZM-bl cells (28) were infected with HIV-1, and the efficiency of infection was read out as luciferase production from a long terminal repeat (LTR)-luciferase reporter. Thus, the assay measures early infection steps through gene expression from the viral LTR. Efficient infection was seen at concentrations of 250 nm GSK1264 and lower, but inhibition was seen at 1,250 nm. Thus, inhibition as measured in the TZM-bl assay, which requires only infection steps through transcription, was much less potent than that measured in the multicycle viral replication assay (which was ∼38 nm).
FIGURE 2.
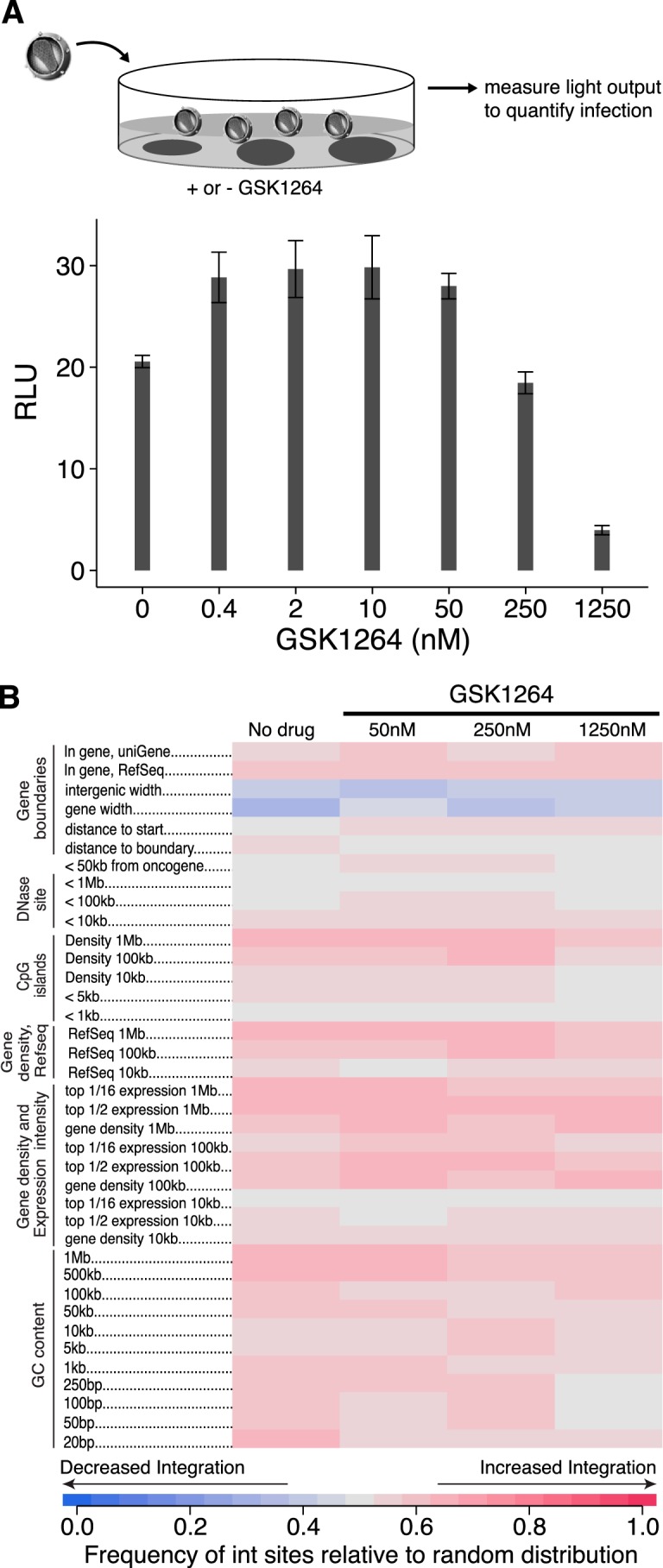
Effects of GSK1264 on early infection steps. A, inhibition of early steps of viral replication requires high concentrations of GSK1264. TZM-bl indicator cells were infected with the indicated amounts of HIV89.6 (in ng p24 antigen), and luciferase expression from an LTR-luc reporter was quantified. B, integration target site selection. The infected cells studied in A were analyzed by ligation-mediated PCR to quantify integration site distributions. Results are summarized in the indicated heat map using the ROC area method for comparing integration site distributions on the human genome with random distributions. Each column shows an integration site data set, with the concentration of GSK1264 added to cell cultures indicated at the top. Each row shows a form of genomic annotation. The key at the bottom shows the frequency of association for integration sites compared with the random distributions. Red, positive association; blue, negative association. The numbers on the left indicate the interval size over which the densities of integration sites were compared with the indicated form of genomic annotation. None of the slight differences seen between samples achieved statistical significance. For the experiment in B, we used cellular DNA from infections in A to maintain linkage between experiments; however, low multiplicity of infection was used in A to minimize cells with multiple proviruses, so as a consequence the number of integration sites recovered was modest (645 total) (supplemental Table S1). RLU, relative light units. Error bars, S.D.
Lack of Effect of GSK1264 on Integration Targeting
The LEDGF/p75 protein binds both HIV-1 IN and chromatin at active transcription units, thereby directing HIV-1 DNA integration to these sites (6, 8, 49–51). We thus investigated whether integration targeting was altered in the presence of GSK1264. For this study, we used the infected cells generated in the study in Fig. 2A. Integration site distributions were mapped in genomic DNA using ligation-mediated PCR and 454/Roche deep sequencing. Results are summarized as a heat map in Fig. 2B.
Each integration site data set was matched with random sequences for statistical analysis, and departures from random by the experimental data relative to genomic features were scored using the ROC (receiver operating characteristic) area method (30). Samples were then compared in the presence and absence of GSK1264. The samples tested were from the infections in Fig. 2A, so at 1,250 nm, efficient inhibition was observed.
As can be seen by comparing rows in the heat map in Fig. 2B, only slight differences were seen between samples. No comparisons achieved statistical significance. The study was limited by the relatively modest number of integration sites available due to the low multiplicity of infection used in Fig. 2A but is still sufficient to conclude that concentrations of GSK1264 that are sufficient to achieve significant inhibition in the TZM-bl assay (1,250 nm) did not have major effects on integration site targeting at this depth of analysis.
We next investigated whether competition between LEDGF and GSK1264 was having any detectable influence early during infection. We compared the IC50 of GSK1264 in test infections on 293T-derived cells knocked down for LEDGF using shRNA versus control cells. We found that there was a drop in IC50 from 55.2 ± 10.2 nm in the wild-type cells to 11.0 ± 1.4 nm in the LEDGF knockdown cells. As controls, an integrase strand transfer inhibitor and a nonnucleoside reverse transcriptase inhibitor were tested and found to show no difference between wild-type and LEDGF knockdown cells. This 5-fold differential seen with GSK1264 was less than for BI-D, another compound of this class, which showed a 30-fold differential (23). Thus, competition between LEDGF and GSK1264 early during infection is less pronounced for GSK1264 than for another compound in this class.
Potent Inhibition of Late Replication Steps by GSK1264 in a Virus Prebinding Format
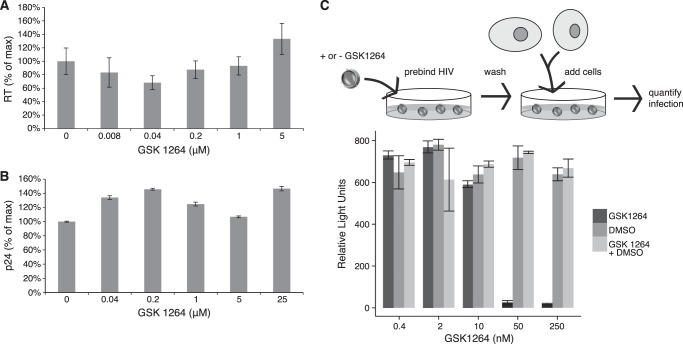
In contrast to the modest potency observed for inhibition of early steps in the single round assay, inhibition of late steps was potent. Two assay formats were used to make this point.
In the first, 293T cells producing HIVNL4–3 virus were treated with GSK1264 or DMSO carrier, and infectivity was analyzed. Virus production quantified by RT assay (Fig. 3A) or p24 antigen yield (Fig. 3B) showed no significant difference in the presence of GSK1264 up to 5 μm. To test the infectivity of these particles, virus produced in the presence of GSK1264 or DMSO control was applied to U373/CD4/CCR5 target cells. Potent inhibition was detected, with an estimated IC50 of 12 nm. In contrast, infection of the U373/CD4/CCR5 cells in the presence of different concentrations of GSK12654 with HIVNL4–3 (where virus was not treated with drug) yielded an IC50 of 557 nm. Thus, there was a 46-fold difference between the efficiency of infection late versus early.
FIGURE 3.
Inhibition of the late steps of HIV-1 replication by GSK1264. A, lack of inhibition of particle production measured using an RT assay of particle-associated RT activity. Infected cells were treated with the indicated concentration of GSK1264, and cell-free particles were assayed for RT activity. Drug concentrations are as indicated on the x axis. B, lack of inhibition of particle production measured using a p24 assay of particle production. Infected cells were treated with the indicated concentration of GSK1264, and cell-free particles were assayed for p24 content. C, potent inhibition late during infection. After chronic virus producer cells were treated with GSK1264, viral supernatants were harvested and applied to retronectin-coated plates. Plates were washed, and then TZM-bl indicator cells were added, and luciferase activity was assayed. As a control, a mixture of drug-treated and control viruses was worked up similarly, and infection was shown to be normal (GSK1264 + DMSO), indicating that drug was not carried over in quantities sufficient to be inhibitory; nor was drugged virus able to inhibit untreated virus. Error bars, S.D.
A complication arises in analyzing virus stocks produced in the presence of drug, which is that compound carried over with the viral stock may influence the course of the infection in the newly infected target cells (although the above study was designed to dilute drug in viral stocks extensively before application to target cells). To eliminate this concern, in a second study, HIV-1 particles were prebound to retronectin-coated plates and washed, and then TZM-bl target cells were added. In this format, inhibition by GSK1264 remained effective (Fig. 3C), with complete inhibition of viral replication seen between 10 and 50 nm. As a control, non-drugged and drugged viruses were mixed, bound to the plate, and washed, and then cells were added (Fig. 3C, GSK1264 + DMSO). Infection was efficient, indicating that the drug was not carried over and that the virus produced in the presence of drug did not inhibit infection by non-drug-treated virus.
We thus conclude that GSK1264 exerts its effects primarily in the late steps of the viral replication cycle, resulting in the formation of replication-deficient particles, a conclusion that has also been reached by others in studies of structurally related molecules (20, 22, 23, 25, 26).
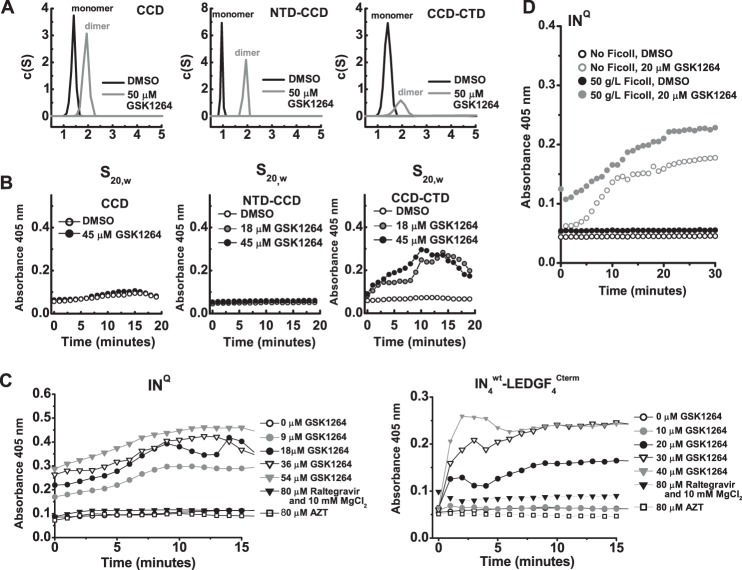
Effects of GSK1264 on the Solution Properties of Full-length IN in Vitro
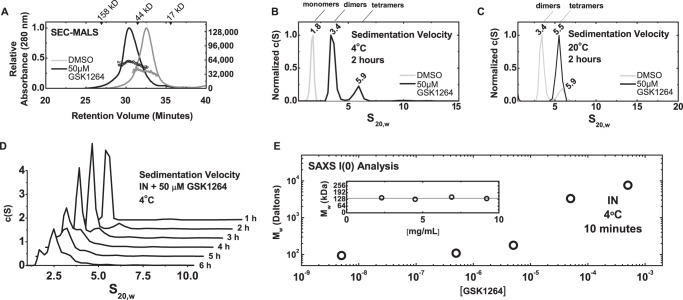
We analyzed IN by several biophysical methods to assess the effects of GSK1264 on its solution properties in vitro (Fig. 4, A–E). For this, we examined a mutant version of INQ (C56S, F139D, F185H, and C280S) (33, 52–54) that has more favorable solubility properties than wild-type IN alone. Global fitting of sedimentation equilibrium data with strict mass conservation obtained from recombinant INQ at 4 °C was best described by a monomer-dimer-tetramer association, with dissociation constants of ∼5 μm for Kd1–2 and ∼86 μm for Kd2–4, respectively (data not shown). Attempts to analyze INQ in the presence of 50 μm GSK1264 were stymied by loss of soluble mass. However, the data that were obtained through two speeds (∼40 h at 4 °C) and across three concentrations could be well described by a dimer-tetramer-octamer association with dissociation constants of ∼4 μm for Kd2–4 and ∼86 μm for Kd4–8, respectively. However, such an analysis cannot reliably discriminate a 2-4-8 association from other models, such as 2-4-16 or 2-5-7.
FIGURE 4.
Multimerization of HIV-IN induced by GSK1264. A, aggregation of INQ as a function of drug concentration. Shown is SAXS I(0) analysis of the weight-averaged mass of INQ solutions at a 5-mg/ml concentration as a function of drug concentration. These measurements were recorded over 10-min time courses at 4 °C immediately after the addition of drug or DMSO. I(0) calculations were relative to a to a standard curve of nine proteins of known mass and concentration (supplemental Fig. 2). Shown in the inset are mass determinations for INQ solutions at four different concentrations. B and C, sedimentation velocity analysis. c(S) analysis of sedimentation velocity data is shown for full-length 30 μm INQ at 4 °C (B) and 20 °C (C) in the presence of DMSO (gray) or 50 μm drug (black) is shown. Species assigned as monomers (∼1.8 S), dimers (∼3.4 S), and tetramers (∼5.5 S) are denoted. Distributions were derived from the fitting of the Lamm equation to the experimental data collected in the first 2 h of the experiment, as implemented in the program SEDFIT (70). D, time-dependent loss of soluble mass evidenced by sedimentation velocity analysis. Distributions derived using different time ranges of data recorded. Seen is a diminution of the ∼3.4 S species (dimers) coinciding with the loss of overall soluble mass (as evidenced by a decrease in the area under the curve). E, representative SEC-MALS analysis. Shown as lines are the absorbance profiles (left axis) of INQ samples injected at 100 μm after a 30-min incubation at room temperature with either DMSO (gray) or 200 μm drug (black), as a function of elution time from a Superdex-200 10/300 column at room temperature. The peak eluent is at ∼0.1 mg/ml concentrations, as determined by refractive index. Corresponding circles denote molecular masses determined by in-line light scattering (right axis) in Da. An IN monomer has a predicted molecular mass of ∼32 kDa.
Simulations using the monomer-dimer-tetramer association model over the 1–100 μm range predicted that IN alone forms a mixture of monomer, dimer, and tetramer species (supplemental Fig. 1). Such heterogeneity precludes structural analysis by small-angle x-ray scattering. However, weight-averaged mass determination is possible using I(0) analysis, where mass is derived by comparison of the incident intensities of known concentration with mass standards of known mass and concentration. We found that the weight-average mass (Mw) of IN was consistent with ∼128 kDa from 1–5 mg/ml concentrations (Fig. 4A and Table 2), in line with our simulations. Titration of the GSK1264 compound with IN at 4 °C over a 10-min period revealed a concentration-dependent aggregation of IN, where the determined mass did not terminate with tetramer or octamer but instead arrived in the megadalton range. These observations were corroborated by sedimentation velocity analyses (Fig. 4, B–D), which allowed for the discrimination of discrete oligomers stimulated by the compound.
TABLE 2.
Parameters derived from SAXS analysis of IN preparations
| Sample | Concentrationa | Guinier |
GNOMc |
Molecular mass by I(0) | ||||
|---|---|---|---|---|---|---|---|---|
| qRgb | Rg | I(0) | Rg | I(0) | Dmax | |||
| mg/ml | Å | a.u.d | Å | a.u. | Å | Da | ||
| INQ | 9.2 | 0.70–1.4 | 38.7 ± 2.5 | 0.032 ± 0.002 | 43.3 | 0.03 | 139.5 | 131,520 |
| 6.9 | 0.68–1.4 | 41.7 ± 1.6 | 0.070 ± 0.002 | 41.9 | 0.07 | 125.0 | 125,253 | |
| 4.5 | 0.70–1.4 | 38.8 ± 1.6 | 0.017 ± 0.001 | 39.3 | 0.02 | 121.5 | 115,233 | |
| 2.3 | 0.79–1.5 | 43.3 ± 2.2 | 0.057 ± 0.003 | 42.4 | 0.05 | 131.5 | 150,304 | |
| INQ + 5 μm GSK1264 | 2.0 | 0.40–1.4 | 38.4 ± 3.6 | 0.015 ± 0.001 | 37.2 | 0.01 | 114.0 | 94,200 |
| INQ + 500 μm GSK1264 | 2.0 | 0.40–1.4 | 31.6 ± 2.3 | 0.013 ± 0.001 | 35.6 | 0.01 | 110.0 | 108,500 |
| INQ + 5 mm GSK1264 | 2.0 | 0.41–1.4 | 33.3 ± 2.5 | 0.015 ± 0.001 | 33.3 | 0.01 | 110.0 | 178,100 |
| INQ + 50 mm GSK1264 | 2.0 | 0.53–1.4 | 79.5 ± 8.1 | 0.050 ± 0.005 | 70.4 | 0.04 | 222.7 | 3,316,000 |
| INQ + 500 mm GSK1264 | 2.0 | 1.05–2.0 | 99.6 ± 6.3 | 0.046 ± 0.005 | 147.3 | 0.08 | 400.0 | 7,614,000 |
a As determined by absorbance using a theoretical extinction coefficient.
b Where q = 4πsinθ/λ, where 2θ is the scattering angle, in units of Å−1.
c Ref. 39.
d a.u., arbitrary units.
To study the drug-induced oligomerization of IN further, we employed SEC-MALS coupled with dynamic light scattering (Fig. 4E). In this approach, absolute concentration determination is coupled with 18-angle light scattering to determine the weight-averaged mass of the eluent. At room temperature and at eluted concentrations of ∼0.1 mg/ml, IN was an apparent monomer. However, the slope of the mass profile and the breadth of the eluent peak were indicative of a self-association (Fig. 4E). The retention properties and mass profiles observed were concentration-dependent, consistent with the modest micromolar dissociation constants determined by centrifugation. After preincubation with saturating amounts of GSK1264 (200 μm) for 15 min at room temperature, an increase in apparent mass was observed, consistent with a population of dimers and higher order species (Fig. 4A), similar to previous studies with related molecules (23, 55, 56). Similar tests of raltegravir and azidothymidine did not show aggregation (data not shown). The Rh profile across the monomer and dimer peaks provided by in-line dynamic light scattering were nearly identical, suggestive of the transition from an extended monomer to more compact dimer (data not shown).
The Effect of GSK1264 on Preformed LEDGF·IN Complexes in Vitro
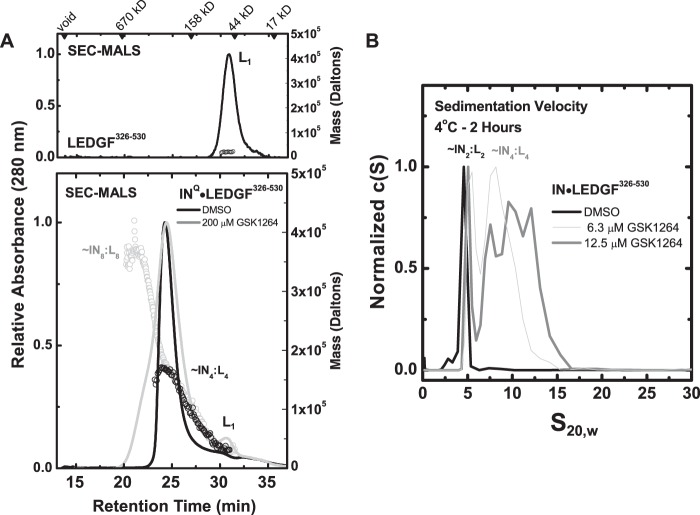
We next examined the effect of GSK1264 on preformed LEDGF·IN complexes. By co-expression, monodisperse complexes of LEDGF·IN can be obtained in 4:4 or 4:2 stoichiometries, depending on the mutational background and length of the C-terminal domain of LEDGF/p75 used (33).
Co-expression of INQ·LEDGF(326–530), which contains the C-terminal domain of LEDGF/p75, provides a 4:4 complex (33) (Fig. 5A). SEC-MALS analysis of the complex preincubated with 200 μm GSK1264 at room temperature for 15 min showed stimulated oligomerization, with a heterogeneous mass profile up to at least an LEDGF-bound IN octamer. LEDGF appeared to be mostly retained within the aggregate because relatively little LEDGF monomer was detected in the absorbance, refractive index, and light scattering data. A similar result was obtained after GSK1264 treatment at lower concentrations over a 2-h time course by sedimentation velocity analysis at 4 °C, where heterogeneous aggregates of the LEDGF·IN complex were observed (Fig. 5B). Similar aggregation was seen in the presence of GSK1264 for INQ·LEDGF(IBD), which is purified as a 4:2 complex (33) (data not shown).
FIGURE 5.
GSK1264 stimulates the oligomerization of preformed INQ·LEDGF(326–530) complexes. A, shown are SEC-MALS analyses for recombinant LEDGF(326–530) alone (top), injected at 10 mg/ml, and co-expressed preparations of INQ·LEDGF(326–530) (bottom), injected at 15.8 mg/ml and incubated with DMSO (black) or 200 μm compound (gray) for 30 min at room temperature before injection onto a Superdex 200 10/300 column at room temperature in the presence of 5 mm CHAPS. Shown as lines are the absorbance profiles of the eluent, and the corresponding open circles denote molecular masses determined by in-line light scattering (right axis). By refractive index, the eluted concentrations of protein were less than 0.1 mg/ml. In these buffer conditions and at these concentrations, little liberated LEDGF(326–530) was observed. B, sedimentation velocity analysis of the drug-induced oligomerization of preformed 5 μm INQ·LEDGF(326–530) complexes, performed at 4 °C. Although higher order soluble species were observed to form in a drug-dependent fashion during the time course of this experiment, no evidence of liberated LEDGF(326–530) alone (s20,w of 1.9 (33)) was obtained. In both panels, the stoichiometry inferred for each species is denoted.
Drug-induced Aggregation of IN Is Promoted by the CTD
We next investigated the IN domains required for GSK1264-induced oligomerization. Oligomerization of IN involves both the NTD (57) and CTD (54, 58), and the crystallographic lattices seen in a number of retroviral integrase structures provide evidence for domain swap interactions between IN dimers mediated by both. Sedimentation velocity analysis (Fig. 6A) performed at low micromolar concentrations of IN at 4 °C over 2 h demonstrated clear stimulation of dimerization by GSK1264 of INF185H(CCD) alone, INF185H(NTD-CCD), and INF185H(CCD-CTD). However, in contrast to other constructs, the INF185H(CCD-CTD) showed clear evidence of loss of soluble mass over the time course of this experiment, as evidenced by a decrease in sample absorbance and the diminution of the area under the curve of the c(S) distribution. Similarly, in samples incubated on ice, a clear effect was observed for INF185H(CCD-CTD) at ∼10–30 μm protein concentrations; visible heavy precipitation was evident after 15 min, whereas less precipitation was seen with full-length IN and the other truncated variants (data not shown).
FIGURE 6.
The concentration-dependent formation of insoluble IN aggregates by GSK1264 requires IN(CTD) and is stimulated by crowding agents. A, sedimentation velocity analysis of drug-induced oligomerization of IN domain truncations. In each panel, c(S) analysis is shown for INF185H(CCD) (30 μm; top), INF185H(NTD-CCD) (24 μm; middle), or INF185H(CCD-CTD) (19 μm; bottom) in the presence of DMSO (black) or 50 μm drug (gray). Species assigned as monomers and dimers are denoted. By quantitating the area under the c(S) distribution, it was observed that drug-induced oligomerization of INF185H(CCD-CTD) (bottom) coincided with loss of soluble mass. B, turbidity analysis of IN domain truncation aggregation. INF185H(CCD) (36.6 μm), INF185H(NTD-CCD) (11.2 μm), or INF185H(CCD-CTD) (13.1 μm) was mixed with DMSO or GSK1264 to a final concentration of 18–45 μm, and aggregation was monitored over a 20-min time course (x axis) at 27 °C, using absorbance readings at 405 nm (y axis). C, concentration-dependent aggregation of 9 μm INQ alone (left) and co-expressed 12.5 μm INwt·LEDGF(326–530) (4:4 stoichiometry; right). GSK1264 was compared with Raltegravir (IN active site-targeted inhibitor) and azidothymidine (AZT) (nucleotide reverse transcriptase inhibitor). D, crowding agents stimulate drug-induced aggregation of IN. Full-length INQ (9 μm) was mixed with DMSO or 20 μm GSK1264 in the presence or absence of 50 g/liter Ficoll, and aggregation was monitored over a 60-min time course at 27 °C by absorbance readings at 405 nm. The presence of the crowding agent amplified the drug-induced aggregation of IN.
To quantitate the aggregation reaction, we devised a turbidity assay by monitoring absorbance at 405 nm. At this wavelength, there is no contributing absorbance from protein or GSK1264 alone. Over a 60-min time course at room temperature, only the full-length IN and the INF185H(CCD-CTD) construct showed evidence of drug-dependent aggregation (Fig. 6B). Thus, we conclude that GSK1264-induced aggregation of IN in this assay minimally requires interactions mediated by the CCD and CTD. As controls, aggregation was tested in the presence of raltegravir and azidothymidine (Fig. 6C), and none was seen for INQ, wild-type IN bound to LEDGF(326–530) (4:4; Fig. 6C), and INQ bound with LEDGF(IBD) (4:2; data not shown).
It has been suggested that IN is oligomeric in virions (59) and that oligomerization is stimulated by LEDGINs (a group of allosteric IN inhibitors) (23, 25, 26). We found that the drug-induced aggregation of INQ was increased by the addition of the crowding agent Ficoll, potentially mimicking the crowded environment during assembly and in particles. Ficoll added to INQ in the absence of drug had no effects on its aggregation rate under the conditions tested (Fig. 6D).
DISCUSSION
Here we report a study of GSK1264, an allosteric inhibitor of HIV-1 IN that blocks viral replication. GSK1264 is a compound of the group variously named ALLINIs (20), NCINIs (26), LEDGINs (48, 60, 61), and the tert-butoxy-(4-phenyl-quinolin-3-yl) acid molecules (44, 46). GSK1264 was isolated in an assay requiring displacement of LEDGF from IN, but surprisingly, these compounds seem to act mainly independently of LEDGF in vivo. Instead, these inhibitors have been linked to post-integration effects associated with the self-association of IN (20, 22–26). We show that the GSK1264 compound elicits concentration and time-dependent polymerization of IN, ultimately leading to the formation of polydisperse, insoluble aggregates. Inhibition did not block particle release but resulted in production of particles impaired for subsequent steps of replication, consistent with a defect in reverse transcription or other steps in the next round of replication (20, 22–26). However, purified particles were fully competent for reverse transcription after disruption and assay in vitro (Fig. 3A), indicating that the enzyme is present and functional, implicating defects in other properties, such as higher order assembly.
We did not observe displacement of LEDGF(IBD) from preformed IN·LEDGF complexes in the presence of GSK1264 in vitro, despite induction of efficient aggregation. It is unclear whether IN protein is saturated with LEDGF in vivo, but here we show that both 4:2 and 4:4 complexes can show stimulated oligomerization in the presence of GSK1264 without measureable LEDGF displacement. A simple model is that LEDGF remains trapped in the aggregate after compound binding. It is unknown whether this would be the case in the context of preintegration complexes in vivo.
A recent study suggested that competition between an ALLINI (BI-D) and LEDGF played out differently at early and late steps of viral replication. Relatively potent late inhibition by BI-D was not affected by the presence of LEDGF, but weaker inhibition of early replication was diminished (23). This suggests that there was competition between LEDGF and BI-D binding to early replication complexes in vivo. For BI-D, the difference in inhibition early versus late in the presence of LEDGF was 26-fold. For GSK1264, the differential early versus late was greater (46-fold), and the effects of LEDGF knockdown on early steps were more modest (only 5-fold versus 30-fold). Biophysical data showed no detectable displacement of LEDGF by GSK1264, and GSK1264 did not strongly disrupt integration target site selection, all consistent with the idea that GSK1264 may be less effective at displacing LEDGF from IN than is BI-D.
We investigated the domain requirements for aggregation with GSK1264, which suggested that polymerization of CTDs mediates self-association and formation of an uncapped polymer. In support of this, the HIV-1 IN(CCD-CTD) crystal structure (Protein Data Bank entry 1EX4 (53)) reveals a tetramer packing interface that is mediated solely by CTD-CTD interactions and bears no resemblance to the tetramers observed in the prototype foamy virus integrase in complex with DNA (62–66). Thus, we hypothesize that the open polymer formed in the presence of allosteric inhibitors proceeds through polymerization of the CTD as in the 1EX4 crystal lattice.
Inhibition by GSK1264 and related molecules may be promoted by IN protein crowding late during viral replication, as emphasized here by increased precipitation in the presence of Ficoll. A typical HIV-1 virion has an internal volume of ∼980,000 nm3 and a total macromolecular concentration exceeding 500 g/liter (67–69). Allosteric inhibitors have been shown to impair maturation of particle cores (23, 25, 26). As the nascent virion assembles, the crowded environment that arises may promote aggregation induced by GSK1264 and related compounds. Thus, potency may be amplified by the environment encountered by IN during assembly late during the viral replication cycle.
Acknowledgments
We are grateful to members of the Bushman, Van Duyne, and Jeffrey groups for help and suggestions. We thank Meredith Jackrel and Elizabeth Sweeney (University of Pennsylvania) for technical assistance with turbidity assays. We also acknowledge Yingnian Shen, Tim Broderick, and Kurt Weaver for the cloning, expression, and purification of the IN-CCD protein used in the crystallographic studies. The following reagent was obtained through the National Institutes of Health (NIH) AIDS Reagent Program, Division of AIDS, NIAID, NIH: U373-MAGI-CCR5E from Dr. Michael Emerman.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI 052845 (to F. D. B.). GlaxoSmithKline owns the patent on the compound used in this study.

This article contains supplemental Table S1 and Figs. 1 and 2.
The atomic coordinates and structure factors (code 4OJR) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Patent filing: M. DeLa Rosa, S. N. Haydar, B. A. Johns, and E. J. Velthuisen (January 23, 2012) Isoquinline Compounds and Methods for Treating HIV. WO 2012/102985. The compound referred to here as GSK1264 is compound 159 in the filing.
- IN
- integrase
- INQ
- quadramutated IN (C56S/F139D/F185H/C280S)
- NTD
- N-terminal domain
- CCD
- catalytic core domain
- CTD
- C-terminal domain
- LEDGF
- lens epithelium-derived growth factor
- SEC-MALS
- size-exclusion chromatography in-line with multiangle light scattering
- SAXS
- small-angle X-ray scattering
- IBD
- integrase binding domain.
REFERENCES
- 1. Craigie R., Bushman F. D. (2012) HIV DNA integration. Cold Spring Harb. Perspect. Med. 2, a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Summa V., Petrocchi A., Bonelli F., Crescenzi B., Donghi M., Ferrara M., Fiore F., Gardelli C., Gonzalez Paz O., Hazuda D. J., Jones P., Kinzel O., Laufer R., Monteagudo E., Muraglia E., Nizi E., Orvieto F., Pace P., Pescatore G., Scarpelli R., Stillmock K., Witmer M. V., Rowley M. (2008) Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 51, 5843–5855 [DOI] [PubMed] [Google Scholar]
- 3. Arts E. J., Hazuda D. J. (2012) HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2, a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y. (2003) LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278, 33528–33539 [DOI] [PubMed] [Google Scholar]
- 5. Cherepanov P., Devroe E., Silver P. A., Engelman A. (2004) Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 279, 48883–48892 [DOI] [PubMed] [Google Scholar]
- 6. Schröder A. R., Shinn P., Chen H., Berry C., Ecker J. R., Bushman F. (2002) HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, 521–529 [DOI] [PubMed] [Google Scholar]
- 7. Ciuffi A., Diamond T. L., Hwang Y., Marshall H. M., Bushman F. D. (2006) Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum. Gene Ther. 17, 960–967 [DOI] [PubMed] [Google Scholar]
- 8. Shun M. C., Raghavendra N. K., Vandegraaff N., Daigle J. E., Hughes S., Kellam P., Cherepanov P., Engelman A. (2007) LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Silvers R. M., Smith J. A., Schowalter M., Litwin S., Liang Z., Geary K., Daniel R. (2010) Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum. Gene Ther. 21, 337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferris A. L., Wu X., Hughes C. M., Stewart C., Smith S. J., Milne T. A., Wang G. G., Shun M. C., Allis C. D., Engelman A., Hughes S. H. (2010) Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc. Natl. Acad. Sci. U.S.A. 107, 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gijsbers R., Ronen K., Vets S., Malani N., De Rijck J., McNeely M., Bushman F. D., Debyser Z. (2010) LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol. Ther. 18, 552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. (2003) HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, 372–381 [DOI] [PubMed] [Google Scholar]
- 13. Hendrix J., Gijsbers R., De Rijck J., Voet A., Hotta J., McNeely M., Hofkens J., Debyser Z., Engelborghs Y. (2011) The transcriptional co-activator LEDGF/p75 displays a dynamic scan-and-lock mechanism for chromatin tethering. Nucleic Acids Res. 39, 1310–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Llano M., Vanegas M., Fregoso O., Saenz D., Chung S., Peretz M., Poeschla E. M. (2004) LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 78, 9524–9537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turlure F., Maertens G., Rahman S., Cherepanov P., Engelman A. (2006) A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 34, 1653–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pandey K. K., Sinha S., Grandgenett D. P. (2007) Transcriptional coactivator LEDGF/p75 modulates human immunodeficiency virus type 1 integrase-mediated concerted integration. J. Virol. 81, 3969–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hare S., Shun M. C., Gupta S. S., Valkov E., Engelman A., Cherepanov P. (2009) A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 5, e1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cherepanov P., Sun Z. Y., Rahman S., Maertens G., Wagner G., Engelman A. (2005) Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 12, 526–532 [DOI] [PubMed] [Google Scholar]
- 19. Molteni V., Greenwald J., Rhodes D., Hwang Y., Kwiatkowski W., Bushman F. D., Siegel J. S., Choe S. (2001) Identification of a small molecule binding site at the dimer interface of the HIV integrase catalytic domain. Acta Crystallogr. D Biol. Crystallogr. 57, 536–544 [DOI] [PubMed] [Google Scholar]
- 20. Engelman A., Kessl J. J., Kvaratskhelia M. (2013) Allosteric inhibition of HIV-1 integrase activity. Curr. Opin. Chem. Biol. 17, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Christ F., Debyser Z. (2013) The LEDGF/p75 integrase interaction, a novel target for anti-HIV therapy. Virology 435, 102–109 [DOI] [PubMed] [Google Scholar]
- 22. Le Rouzic E., Bonnard D., Chasset S., Bruneau J. M., Chevreuil F., Le Strat F., Nguyen J., Beauvoir R., Amadori C., Brias J., Vomscheid S., Eiler S., Lévy N., Delelis O., Deprez E., Saïb A., Zamborlini A., Emiliani S., Ruff M., Ledoussal B., Moreau F., Benarous R. (2013) Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage. Retrovirology 10, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jurado K. A., Wang H., Slaughter A., Feng L., Kessl J. J., Koh Y., Wang W., Ballandras-Colas A., Patel P. A., Fuchs J. R., Kvaratskhelia M., Engelman A. (2013) Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc. Natl. Acad. Sci. U.S.A. 110, 8690–8695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feng L., Sharma A., Slaughter A., Jena N., Koh Y., Shkriabai N., Larue R. C., Patel P. A., Mitsuya H., Kessl J. J., Engelman A., Fuchs J. R., Kvaratskhelia M. (2013) The A128T resistance mutation reveals aberrant protein multimerization as the primary mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 288, 15813–15820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Desimmie B. A., Schrijvers R., Demeulemeester J., Borrenberghs D., Weydert C., Thys W., Vets S., Van Remoortel B., Hofkens J., De Rijck J., Hendrix J., Bannert N., Gijsbers R., Christ F., Debyser Z. (2013) LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology 10, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Balakrishnan M., Yant S. R., Tsai L., O'Sullivan C., Bam R. A., Tsai A., Niedziela-Majka A., Stray K. M., Sakowicz R., Cihlar T. (2013) Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PloS One 8, e74163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vodicka M. A., Goh W. C., Wu L. I., Rogel M. E., Bartz S. R., Schweickart V. L., Raport C. J., Emerman M. (1997) Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology 233, 193–198 [DOI] [PubMed] [Google Scholar]
- 28. Platt E. J., Wehrly K., Kuhmann S. E., Chesebro B., Kabat D. (1998) Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72, 2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doranz B. J., Rucker J., Yi Y., Smyth R. J., Samson M., Peiper S. C., Parmentier M., Collman R. G., Doms R. W. (1996) A dual-trophic primary HIV-1 isolate that uses Fusin and the β-chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell 85, 1149–1158 [DOI] [PubMed] [Google Scholar]
- 30. Berry C., Hannenhalli S., Leipzig J., Bushman F. D. (2006) Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2, e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang G. P., Ciuffi A., Leipzig J., Berry C. C., Bushman F. D. (2007) HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 17, 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ciuffi A., Ronen K., Brady T., Malani N., Wang G., Berry C. C., Bushman F. D. (2009) Methods for integration site distribution analyses in animal cell genomes. Methods 47, 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gupta K., Diamond T., Hwang Y., Bushman F., Van Duyne G. D. (2010) Structural properties of HIV integrase. Lens epithelium-derived growth factor oligomers. J. Biol. Chem. 285, 20303–20315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Urbani A., Warne T. (2005) A colorimetric determination for glycosidic and bile salt-based detergents: applications in membrane protein research. Anal. Biochem. 336, 117–124 [DOI] [PubMed] [Google Scholar]
- 35. Lipfert J., Columbus L., Chu V. B., Lesley S. A., Doniach S. (2007) Size and shape of detergent micelles determined by small-angle x-ray scattering. J. Phys. Chem. B 111, 12427–12438 [DOI] [PubMed] [Google Scholar]
- 36. Deprez E., Tauc P., Leh H., Mouscadet J. F., Auclair C., Brochon J. C. (2000) Oligomeric states of the HIV-1 integrase as measured by time-resolved fluorescence anisotropy. Biochemistry 39, 9275–9284 [DOI] [PubMed] [Google Scholar]
- 37. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) Computer-aided interpretation of analytical sedimentation data for proteins in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., Horton J. C. eds), pp. 90–125 The Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 39. Semenyuk A. V., Svergun D. I. (1991) GNOM: a program package for small-angle scattering data-processing. J. Appl. Crystallogr. 24, 537–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smart O. S., Womack T. O., Flensburg C., Keller P., Paciorek W., Sharff A., Vonrhein C., Bricogne G. (2012) Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D 68, 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 44. Tsiang M., Jones G. S., Niedziela-Majka A., Kan E., Lansdon E. B., Huang W., Hung M., Samuel D., Novikov N., Xu Y., Mitchell M., Guo H., Babaoglu K., Liu X., Geleziunas R., Sakowicz R. (2012) New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J. Biol. Chem. 287, 21189–21203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kessl J. J., Jena N., Koh Y., Taskent-Sezgin H., Slaughter A., Feng L., de Silva S., Wu L., Le Grice S. F., Engelman A., Fuchs J. R., Kvaratskhelia M. (2012) Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 287, 16801–16811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fenwick C. W., Tremblay S., Wardrop E., Bethell R., Coulomb R., Elston R., Faucher A. M., Mason S., Simoneau B., Tsantrizos Y., Yoakim C. (2011) Resistance studies with HIV-1 non-catalytic site integrase inhibitors. Antivir. Ther. 16, Suppl. 1, A9 [Google Scholar]
- 47. Hou Y., McGuinness D. E., Prongay A. J., Feld B., Ingravallo P., Ogert R. A., Lunn C. A., Howe J. A. (2008) Screening for antiviral inhibitors of the HIV integrase-LEDGF/p75 interaction using the AlphaScreen luminescent proximity assay. J. Biomol. Screen. 13, 406–414 [DOI] [PubMed] [Google Scholar]
- 48. Christ F., Voet A., Marchand A., Nicolet S., Desimmie B. A., Marchand D., Bardiot D., Van der Veken N. J., Van Remoortel B., Strelkov S. V., De Maeyer M., Chaltin P., Debyser Z. (2010) Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat. Chem. Biol. 6, 442–448 [DOI] [PubMed] [Google Scholar]
- 49. Mitchell R. S., Beitzel B. F., Schroder A. R. W., Shinn P., Chen H. M., Berry C. C., Ecker J. R., Bushman F. D. (2004) Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2, 1127–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J. R., Bushman F. (2005) A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11, 1287–1289 [DOI] [PubMed] [Google Scholar]
- 51. Marshall H. M., Ronen K., Berry C., Llano M., Sutherland H., Saenz D., Bickmore W., Poeschla E., Bushman F. D. (2007) Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PloS One 2, e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Alian A., Griner S. L., Chiang V., Tsiang M., Jones G., Birkus G., Geleziunas R., Leavitt A. D., Stroud R. M. (2009) Catalytically-active complex of HIV-1 integrase with a viral DNA substrate binds anti-integrase drugs. Proc. Natl. Acad. Sci. U.S.A. 106, 8192–8197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen J. C., Krucinski J., Miercke L. J., Finer-Moore J. S., Tang A. H., Leavitt A. D., Stroud R. M. (2000) Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. U.S.A. 97, 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bischerour J., Leh H., Deprez E., Brochon J. C., Mouscadet J. F. (2003) Disulfide-linked integrase oligomers involving C280 residues are formed in vitro and in vivo but are not essential for human immunodeficiency virus replication. J. Virol. 77, 135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Demeulemeester J., Tintori C., Botta M., Debyser Z., Christ F. (2012) Development of an AlphaScreen-based HIV-1 integrase dimerization assay for discovery of novel allosteric inhibitors. J. Biomol. Screen. 17, 618–628 [DOI] [PubMed] [Google Scholar]
- 56. Tintori C., Demeulemeester J., Franchi L., Massa S., Debyser Z., Christ F., Botta M. (2012) Discovery of small molecule HIV-1 integrase dimerization inhibitors. Bioorg. Med. Chem. Lett. 22, 3109–3114 [DOI] [PubMed] [Google Scholar]
- 57. Zheng R., Jenkins T. M., Craigie R. (1996) Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. U.S.A. 93, 13659–13664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jenkins T. M., Engelman A., Ghirlando R., Craigie R. (1996) A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 271, 7712–7718 [DOI] [PubMed] [Google Scholar]
- 59. Petit C., Schwartz O., Mammano F. (1999) Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J. Virol. 73, 5079–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Christ F., Shaw S., Demeulemeester J., Desimmie B. A., Marchand A., Butler S., Smets W., Chaltin P., Westby M., Debyser Z., Pickford C. (2012) Small-molecule inhibitors of the LEDGF/p75 binding site of integrase block HIV replication and modulate integrase multimerization. Antimicrob. Agents Chemother. 56, 4365–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Peat T. S., Rhodes D. I., Vandegraaff N., Le G., Smith J. A., Clark L. J., Jones E. D., Coates J. A., Thienthong N., Newman J., Dolezal O., Mulder R., Ryan J. H., Savage G. P., Francis C. L., Deadman J. J. (2012) Small molecule inhibitors of the LEDGF site of human immunodeficiency virus integrase identified by fragment screening and structure based design. PloS One 7, e40147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hare S., Gupta S. S., Valkov E., Engelman A., Cherepanov P. (2010) Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hare S., Maertens G. N., Cherepanov P. (2012) 3′-processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 31, 3020–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hare S., Smith S. J., Métifiot M., Jaxa-Chamiec A., Pommier Y., Hughes S. H., Cherepanov P. (2011) Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572). Mol. Pharmacol. 80, 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hare S., Vos A. M., Clayton R. F., Thuring J. W., Cummings M. D., Cherepanov P. (2010) Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc. Natl. Acad. Sci. U.S.A. 107, 20057–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yin Z., Lapkouski M., Yang W., Craigie R. (2012) Assembly of prototype foamy virus strand transfer complexes on product DNA bypassing catalysis of integration. Protein Sci. 21, 1849–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. del Alamo M., Rivas G., Mateu M. G. (2005) Effect of macromolecular crowding agents on human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 79, 14271–14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Benjamin J., Ganser-Pornillos B. K., Tivol W. F., Sundquist W. I., Jensen G. J. (2005) Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J. Mol. Biol. 346, 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Briggs J. A., Simon M. N., Gross I., Kräusslich H. G., Fuller S. D., Vogt V. M., Johnson M. C. (2004) The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11, 672–675 [DOI] [PubMed] [Google Scholar]
- 70. Vistica J., Dam J., Balbo A., Yikilmaz E., Mariuzza R. A., Rouault T. A., Schuck P. (2004) Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 326, 234–256 [DOI] [PubMed] [Google Scholar]