Abstract
Wheat is a vital dietary component for human health and widely consumed in the world. Wheat rusts are dangerous pathogens and contribute serious threat to its production. In present study, PCR-Based DNA Markers were employed to check the rust resistance genes among 20 wheat genotypes and 22 markers were amplified. NTSYS-pc 2.2 was used to calculate genetic diversity and Nei and Li's coefficients ranged from 0.55 to 0.95. Cluster analysis was obtained using UPGMA (Unweighted Pair Group Method of Arithmetic Average) algorithm. Maximum no. of genes (23) was amplified from TW-760010 genotype whereas minimum no of genes (14) were amplified from TW-76005 genotype. The data gained from present study open up new ways to produce new varieties by breeding rust resistant germplasm to avoid the economic and food loss and varieties with improved characteristics.
Keywords: Wheat, germplasm, leaf rust, stem rust, stripe rust
Background
Wheat is the most important staple diet all over the world. It is internationally an important commodity for marketing. According to Pakistan Agriculture Research Council, its contribution to GDP is 3.0 % and it is cultivated at greater area of the country. Pakistan and India contribute 20% in annual world wheat production. Its contribution in value added in agriculture is 14.4%. In Pakistan, wheat is contained in large agro ecological area [1]. During last decade, one reason for low yield of wheat in Pakistan is due to the diseases of rust. Rusts are known to be most harmful and epidemically spread diseases of wheat. The genus of wheat rust pathogens is puccinia. There are three types of rust (1) leaf rust (2) stem rust and (3) stripe rust. The losses occurring because of each type vary from each other. The characterization of wheat rust resistance genes is necessary to control the disease so that we could incorporate these genes into the germplasm of elite varieties [2]. Crops have been improved conventionally by selecting noticeable phenotypes showing the cumulative effect of all genes and environment. Nowadays, plant breeding has been updated by molecular marker technologies. Molecular markers brought forward efficient methods of plant breeding. Methods application and automation of analysis demand the need to reduce the cost of large quantity of materials that are handled by molecular markers sufficiently [3]. Rusts are widely spread among continents by wind and act with rust resistance gene by gene-for-gene manner to induce production instability. Many new races of leaf, stripe and stem resistance have been known to evolve and showed threat for available rust resistance genes. More efficient wheat cultivars for rust resistance should be developed for production enhancement. Genetic markers and virulence characters have been utilized for rust migration patterns [4]. Various studies on wheat have been carried out using molecular marker system to characterize gene of choice, mapping of qualitative trait loci and genetic diversity analysis. This allows the marker assisted selection to occur. The present study was performed with the purpose of characterizing rust resistance genes in the wheat genotypes using different marker systems and phylogenetic analysis of the experimental data.
Methodology
Sample Collection:
A total of 20 genotypes of Wheat were obtained from different research stations of Pakistan including genotypes from Thal. Genotypes were V-4189, LASANI, V-4188, V-4022, AARI-10, AS-02, Inqlab-91, Sehar, UQAB, Pasban-90, TW-76001, TW- 76002, TW-76003, TW-76004, TW-76005, Parwaz-94, TW-76007, TW-76008, TW-76009 and TW-760010.
DNA Isolation:
For each genotype, genomic DNA was isolated by a modified Cetyl Trimethyl Ammonium Bromide (CTAB) method [5]. Then, DNA quality was determined using 0.8 % (w/v) agarose gel and DNA quantification was performed using NanoDrop- 1000 version 3.3.1 spectrophotometer.
PCR-Marker Assay:
PCR reaction mixture reagents were 10XPCR Buffer (MgCl2+ (NH4)2SO4) in 1.5 to 2.0 µl , 50mM MgCl2 in 0.75 to 1.50µl, (1.5+1.5) µl of each Primer (Forward+Reverse) in concentration of 15ng /µl, 2.0 to 5.0 µl of genomic DNA (30ng /µl), 3.0 µl of 0.2Mm of total dNTPS, 0.2-0.6 µl Taq DNA polymerase in the concentration of 5unit /µl and final volume of 16-21µl was made by adding deionized double distilled water in 5.6 to 9.5 µl quantity. Reagents were purchased from Invitrogen (USA). Polymerase chain reaction was performed using the Eppendorf Mastercycler, Germany. Mastercycler was programmed for: initial denaturation at 940C for 5-7 min, followed by 35-45 cycles of denaturing at 940C for 30 sec-1min., annealing at 360C-600C for 30 sec-1 min. and extension at 720C for 1 min. And then a final extension at 720C for 7-10 min. and then hold at 200C until the tubes were replaced. 3.0% (w/v) high resolution agarose gel was used for amplicons characterization and polyacrylamide gel electrophoresis to separate them on molecular weight basis. Gel was visualized under ultraviolet transilluminator and gel pictures were obtained using GeneSnap version 7.6.03 of Syngene gel documentation system and Gene Tool analysis system.
Data Analysis:
The significant size of each fragment was determined using Gene Ruler 100 bp DNA ladder plus (Fermentas, USA) as reference. After running the gel and visualizing its picture, the data was scored. The scoring was started from lower band (small fragment size) and continued to upper band (large fragment size). Present band was scored as 1 whereas absent band was scored as 0. The number of shared amplification products were used for investigation of genetic diversity [6]. The binary data was entered in computer for further analysis by NTSYS-pc version 2.2. Dendrogram was constructed by means of Unweighted Pair Group Method of Arithmetic means (UPGMA) using similarity coefficient and similarity matrix was obtained to measure similarity range.
Results
In the present study genotypes collected from different breeding stations of Pakistan were analyzed for genomic studies of rust resistance in these genotypes using molecular marker system. A total of 30 molecular markers were employed. Out of which 22 markers Table 1 (see supplementary material) amplified the rust resistance gene.
Characterization of Rust Resistance Genes in Wheat:
There are three types of rust resistance genes in wheat; leaf rust resistance Table 2 (see supplementary material), stem rust resistance Table 3 (see supplementary material) and stripe rust resistance Table 4 (see supplementary material). Table 2 shows Leaf rust resistance markers validation for wheat genotypes. All 20 genotypes were observed with SSR marker Xgwm-234 (Figure 1) that is linked to Lr52 gene and located on the chromosome arm 5BS. This marker exhibited recombination value of 7.2 cM with Lr52 [7]. All the amplification products show the bands with the fragment size 229,250bp.
Figure 1.
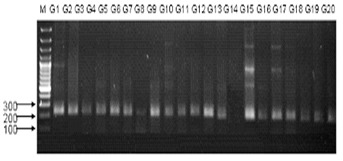
An example of SSR banding pattern obtained from primer Xgwm-234 on 20 genotypes of wheat, which are linked to the Leaf rust resistance gene Lr 52. M is DNA size markers.
Xgwm-44 marker is linked to two leaf rust resistance genes Lr21 & Lr22 genes. Lr21 & Lr22 genes are located on chromosome 7B with respect to Xgwm-44. V-4189, Lasani, V- 4188, V-4022, AARI-10, AS-02, Uqab-02, Pasban-90, TW-76001, TW-76002, TW-76004, TW-76007, TW-76008, TW-76009, TW- 76010 genotypes known to carry Lr21 & Lr22 genes at with band size of 184 and 176bp. Lr21 & Lr22 genes are also resistant to leaf rust marker Xgwm-210. Lr21 & Lr22 genes are located on chromosome 2B, 2D with respect to Xgwm-210. Its bands show the amplification in the range of 303,189bp. Xgwm-295 marker shows association with the Lr34 gene. Lr34 is located on 7D. All the amplification products show the bands with the fragment size 254,258bp. Lr1 is resistant to wheat leaf rust and shows the band with 108 bp. S30-13L is linked to leaf rust resistance gene Lr51. It is located on chromosome 1S [8]. Lr51 bands exist in the range of 790bp. Lr50 is a new leaf rust resistance gene and firstly named for leaf rust. It is known to be evolved from wild timopheevi wheat and is the only Lr gene located on the long arm of wheat homoeologous group 2 chromosomes. It shows the fragment size 110 bp. KSUD14 has the structure of a resistance gene analog (RGA) including kinase2a and kinase3 domains similar to the Cre3 gene of wheat and the rust resistance gene Rp1-D of maize [9]. KSUD-14 PCR-based assay provides an excellent marker for Lr40 and Lr21 in broadening wheat germplasm. KSUD-14 shows the band size of 885bp. Two RAPD markers S265 and S253 which flanked the locus Lr19 were converted to sequence characterized amplified region markers SCS265 and SCS253, respectively. A leaf rust resistance gene Lr19 on the chromosome 7DL of wheat derived from Agropyron elongatum was tagged with random amplified polymorphic DNA and random microsatellite markers [10]. All the amplification products were in the range of 730bp. Lr19 gene is also resistant to leaf rust marker Xgwm-37.The locus of Lr19 with respect to Xgwm-37 marker is 7D. All the amplification products show the bands with the fragment size 189 bp. All genotypes predicted Lr19 gene except the Sehar variety.
PCR-based microsatellite marker Xgwm-11 located in chromosome 1B were shown to be very effective for the detection of Yr15 and Yr26 genes in segregating population and can be applied in future wheat breeding strategies. Its band exits in the range of 213 bp. 20 genotypes were observed with SSR marker Xgwm-501 that is linked to the Yr5 gene resistant to wheat stripe rust (Table 4). Yr5 located on the long arm of chromosome 2B [11]. All the amplification products show the bands with the fragment size 176 bp. Leaf & Stripe resistance genes at the Lr46/Yr29 locus have 1BL [12]. Xgwm-259 is linked to both of these Leaf & Stripe resistance genes Lr46/Yr29. Lr46/Yr29 have amplification fragment size 105bp. Lr46/Yr29 are not available in the following genotypes V-4189, V-4188, Sehar, Pasban-90, TW-76005, Parwaz-94, TW-76007, TW-76008 and in TW-76009 advance line. XgWM-130 is linked to both of Lr34/Yr18 leaf & stripe rust resistance genes. It is located on chromosome 7DS [12]. Its fragment size ranges is upto 121,126bp. AS-02 and TW-76007 advance line do not show the amplification with Lr34/Yr18 leaf & stripe rust resistance genes.
Xbrac-352 is also linked with these leaf & stripe rust resistance genes Lr34/Yr18.It is present on the locus of 7DS [12]. Its band size is 150,211bp (Figure 2). Variety AS-02 and TW-76007 advance line does not show amplification. Xgwm-533 is linked to stem rust resistance gene Sr2. Sr2 is present on the locus 2-3- B. And its band size ranges upto 316bp. It does not show amplification with V-4189, TW-76001, TW-76003, TW-76004, TW-76005 and in TW-76009 advance line. XgWM-130 SSR marker is linked to Lr19/Sr25 leaf and stem rust genes. Lr19/Sr25 are present on 7D.Its fragment size ranges upto 109,111bp. Genotypes like V-4188, V-4022, Pasban-90, TW- 76003, TW-76005 and Parwaz-94 do not show the amplification with Lr19/Sr25 leaf and stem rust genes. PCR-based diagnostic marker STS-638 linked to Lr20 indicates recombination value of 7.1 cM with Lr20 [13]. It is also linked with Sr15 gene. It is resistant to both of leaf and stem rust. Its band size is 232bp and located on 7A. Lr20 & Sr15 are amplified in all genotypes except in TW-76004 advance line. CFA 2019 Sr22 is SSR marker that is used to check amplification of genotypes. Sr22 is a stem rust resistance gene and located on 7AL. Its fragment size ranges is upto 234,500bp. Lasani, V-4188, V-4022, AS-02 and TW-76004 have not Sr22 stem rust resistance gene. STM-773-2 is linked two of stem rust resistance genes Sr32 & Sr36 (Figure 3). STM-773 is present on locus of 2B. All genotypes V-4189, Lasani, V-4188, AARI-10, AS-02, Sehar, Pasban-90, V-4022, Inqlab-91, Uqab-02, TW-76001, TW-76002, TW-76003, TW- 76004, and TW-76005, Parwaz-94, TW-76007, TW-76008, TW- 76009 and TW-76010 show the amplification with the fragment size upto 192 & 162bp.
Figure 2.
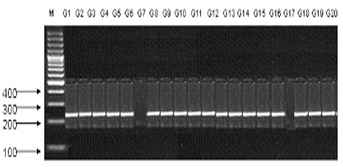
An example of SSR banding pattern obtained from primer Xbrac-352 on 20 genotypes of wheat which are linked to the Leaf rust & Stripe rust genes Lr 34/Yr18. M is DNA size markers.
Figure 3.
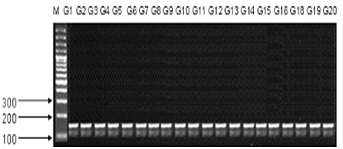
An example of SSR banding pattern obtained from primer STM-773-2 on 20 genotypes of wheat which are linked to the Stem rust genes Sr32 & Sr36. M is DNA size markers.
Phylogenetic Analysis of Wheat Genotypes:
The dendrogram (Figure 4) revealed four clusters among the upland wheat genotypes. Cluster A comprised of genotypes V- 4189, TW-76009, V-4178, Uqab-02, TW-76002, TW-76010. Cluster B comprised of V-4022, TW-76003, TW-76007, Pasban- 90 and Parwaz-94. Cluster C comprised of V-4188, TW-76008, Sehar, TW-76004 and Cluster D comprised of Inqlab-91, V-3138 (Lasani), AS-02, and TW-76001. Cluster A is further divided into two sub-cluster A1 and A2. A1 has two V-4189 and TW- 76009 advance lines. A2 is consisted of four genotypes; AARI- 10, Uqab-02, TW-76002, TW-76005. Cluster B is further divided into two sub-clusters. B1 has V-4022 & TW-76003 advance lines and B2 include Pasban-90 & Parwaz-94 varieties. Cluster C is further divided into two sub-cluster C1 and C2. C1 has V-4188 & TW-76008, While C2 has Sehar & TW-76004. Cluster D consisted of D1 subgroup which has genotypes like Inqlab-91, V-3138 (Lasani), AS-02, and TW-76001. TW-76005 genotype did not make cluster and could be helpful for crop improvement programs.
Figure 4.
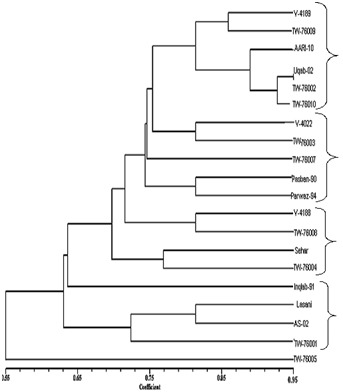
Dendrogram Constructed from Phylogenetic Software
Discussion
The objectives of the present study was to check the rust resistance genes among wheat genotypes by using PCR-Based DNA Markers and for assessing the genetic relationships among these genotypes by rust resistance markers. Statistical validation was also performed using software NTSYSPC by constructing dendrogram and similarity matrix. Molecular markers could provide useful clues about agronomical important and desirable qualitative genes by identifying or manipulating. Markers help in breeding programs by recognizing novel alleles in different species. Genetic maps of these markers have been proved useful for the introgression of novel genes into elite gemplasm [14]. Rusts pose threat to wheat having narrow genetic background due to the lack of resistance against evolving races of pathogen. It is necessary to develop new techniques for incorporating efficient resistance in wheat germplasm to avoid epidemics induced by plant pathogens. Cultivars containing better characteristics have been largely in use to stabilize wheat production. In wheat, it is known that specific gene combinations provide durable and improved resistance than using single gene because single specific gene might become susceptible to genetic shift. Genetic markers like STS (Sequence tagged site), RAPD (Randomly amplified polymorphic DNA), SSR (Simple sequence repeats) and others have been proved useful for identifying resistance among different genotypes. Various researchers have utilized these markers for the investigation of wheat cultivars for different traits [15]. 45 stem rust resistance (Sr) markers (genes), 18 yellow rust resistance (Yr) genes (markers) [16] and nearly 58 leaf rust resistance markers have been recognized for different races of the pathogen. Leaf rust resistance genes were named as Lr1 through Lr58 and mostly varieties consist of some Lr genes [17]. Microsatellite gwm 60 was the only marker that co-segregated completely linked to Lr47.Lr47 is located within an interstitial segment of Triticum speltoides Taush. 7S chromosome translocated to the short arm of chromosome 7A of bread wheat. Lr47 has been characterized using related primers and gene amplification indicated the leaf rust resistance comparing with already reported work [18]. The size of the band corresponds to Lr47 was 380-450 bp. All the genotypes possessed 380-450 bp size of band thought to be resistant against leaf rust whereas Inqlab-91 which lack 380-450 bp of band which are susceptible to leaf rust carry no Lr47 gene. Flanking microsatellite markers were identified and one tightly linked marker (gwm533) was shown to be associated with presence of Sr2. All the amplification products show the bands with the fragment size 316 bp. Genotypes like V-4189, TW-76001, TW-76003, TW-76004, TW-76005 and TW-76009 do not possess the Sr2 gene. The amplified fragments corresponded to stripe rust resistance gene Yr26 and the results were compared with the previous work which has been done by [19]. This yellow rust resistance gene was considered to be Yr26 originally thought to be also located in chromosome arm 1BS linked to marker loci Xgwm273 and Xgwm11 with genetic distances ranging from 1.075cM to 2.74cM in both combinations studied. Xgwm11 is also linked with Yr15.The size of the bands corresponds to Yr15 and Yr26 was 213 bp. It is not present in V- 4189, LASANI, V-4022, AARI-02, Inqlab-91, TW-76005 and in TW-76008 genotypes. Among 20 genotypes that used for molecular characterization of wheat genotypes for rust resistance genes with the application of genetic analysis software only TW-760010 genotype that has maximum no of genes (23). Whereas TW-76005 genotype that has minimum no of genes (14). Research was performed on improved varieties and advance varieties, known as genotypes. Varieties are very important in general cultivation in the fields. Varieties are source of resistance and used as a parent in breeding. Eight Varieties like LASANI, AARI-10, AS-02, Sehar, Pasban-90, Parwaz-94, Inqlab-91 and UQAB are involved in this research work. Pasban-90, Parwaz-94 and UQAB varieties were also used in the work which was done by [1]. LASANI possess the genes like that. The present study performed to characterize genes for rust resistance and this investigation could be helpful in predicting the occurrence of rust disease according to changing virulence of pathogen. Single gene could be enough for newly evolved races of pathogen but varieties containing efficient resistance are used economically and generally to avoid production instability [15]. It is necessary for plant breeders to choose suitable rust resistance gene or combination of these genes for avoiding rust attacks [1].
The present study offers a way of making the breeding process simpler and rust resistance more long lasting. And it provides an insight to the rust resistance genes that should facilitate efficient utilization and management of genetic stocks. It allows plant breeders to better understand the rust resistance diseases among wheat genotypes in a more systematic fashion, and to develop strategies to incorporate rust resistance genes in their breeding programs to protect crop stability. There is an urgent need to incorporate several resistance genes against major diseases into the new wheat cultivars. Marker assisted selection along with precise artificial field tests provide new and efficient possibilities for geneticists and breeders to achieve these goals.
Conclusion
The data obtained in this experiment will help in accelerating the breeding program in future, including pyramiding of different wheat resistant genes in wheat varieties and in gene stacking that is beneficial for improving the plant's immune response by having multiple lines of defence for different strains of rust.
Supplementary material
Footnotes
Citation:Haque et al, Bioinformation 10(6): 371-377 (2014)
References
- 1.Babar M, et al. Afr J Biotechnol. 2010;9:1144. [Google Scholar]
- 2.Mumtaz S, et al. Afr J Biotechnol. 2009;8:1188. [Google Scholar]
- 3.Dayteg C, Tuvesson S. Molecular Techniques in Crop Improvement, Ed. 2nd. 2009:143. [Google Scholar]
- 4.Kolmer JA, et al. Curr Opin Plant Biol. 2005;8:441. doi: 10.1016/j.pbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Iqbal MJ, et al. Theor Appl Genet. 1997;94:139. doi: 10.1007/s001220050392. [DOI] [PubMed] [Google Scholar]
- 6.Nei M, Li WH. Proc Natl Acad Sci USA. 1979;76:5269. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh B, et al. Euphytica. 2010;175:351. [Google Scholar]
- 8.Begum K, et al. Curr Res J Biol Sci. 2010;2:226. [Google Scholar]
- 9.Huang L, Gill SB. Theor Appl Genet. 2001;103:1007. [Google Scholar]
- 10.Gupta SK, et al. Theor Appl Genet. 2006;113:1027. doi: 10.1007/s00122-006-0362-7. [DOI] [PubMed] [Google Scholar]
- 11.Sui XX, et al. Phytopathology. 2009;99:1209. doi: 10.1094/PHYTO-99-10-1209. [DOI] [PubMed] [Google Scholar]
- 12.Lillemo M, et al. Theor Appl Genet. 2008;116:1155. doi: 10.1007/s00122-008-0743-1. [DOI] [PubMed] [Google Scholar]
- 13.Khan RR, et al. Theor Appl Genet. 2005;111:846. doi: 10.1007/s00122-005-0005-4. [DOI] [PubMed] [Google Scholar]
- 14.Deynze Van, et al. BMC Plant Biol. 2009;9:125. doi: 10.1186/1471-2229-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain W, et al. Afr J Biotechnol. 2001;10:8578. [Google Scholar]
- 16.Chełkowski J, Stepień L. J Appl Genet. 2001;42:117. [PubMed] [Google Scholar]
- 17.Kuraparthy V, et al. Theor Appl Genet. 2007;114:1379. doi: 10.1007/s00122-007-0524-2. [DOI] [PubMed] [Google Scholar]
- 18.Vanzetti LS, et al. Elect J Biotech. 2006;9:267. [Google Scholar]
- 19.Ali M, et al. Pak J Bot. 2010;42:383. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


