Abstract
Our understanding of the mechanisms underlying the development of pancreatic cancer has been greatly advanced. However, the molecular events involved in the initiation and development of pancreatic cancer remain inscrutable. None of the present medical technologies have been proven to be effective in significantly improving early detection or reducing the mortality/morbidity of this disease. Thus, a better understanding of the molecular basis of pancreatic cancer is required for the identification of more effective diagnostic markers and therapeutic targets. Non-coding RNAs (ncRNAs), generally including microRNAs and long non-coding RNAs, have recently been found to be deregulated in many human cancers, which provides new opportunities for identifying both functional drivers and specific biomarkers of pancreatic cancer. In this article, we review the existing literature in the field documenting the significance of aberrantly expressed and functional ncRNAs in human pancreatic cancer, and discuss how oncogenic ncRNAs may be involved in the genetic and epigenetic networks regulating functional pathways that are deregulated in this malignancy, particularly of the ncRNAs’ role in drug resistance and epithelial-mesenchymal transition biological phenotype, with the aim of analyzing the feasibility of clinical application of ncRNAs in the diagnosis and treatment of pancreatic cancer.
Keywords: MicroRNAs, Long non-coding RNAs, Pancreatic cancer, Diagnosis, Treatment
Core tip: The deregulation mechanisms of pancreatic cancer remain inscrutable. ncRNAs have recently been found to provide new opportunities for identifying both functional drivers and specific biomarkers of pancreatic cancer. Here, we review the expression profile of ncRNAs in human pancreatic cancer, with the aim of analyzing the feasibility of clinical application of ncRNA in pancreatic cancer’s diagnosis and treatment.
INTRODUCTION
Pancreatic cancer is a lethal malignancy with poor prognosis due to advanced stage disease at initial diagnosis, frequent recurrence and the absence of treatment strategies that specifically and effectively target these tumors[1]. Only 15% of pancreatic adenocarcinoma patients are candidates for surgical resection at the time of diagnosis[2]. Chemotherapy is considered the main treatment option for unresectable cases, while chemo-radiotherapy may improve survival and quality of life[3,4]. However, even with advancements in medicine, pancreatic cancer is still extremely resistant to the currently available regimens. The burden of pancreatic disorders is expected to increase over time[5]. This situation represents a challenge for doctors as well as scientists in seeking the best active regimen with the least side effects[6]. Therefore, there is an urgent need to understand the molecular mechanisms underlying this disease, including the genetic and epigenetic networks influencing the malignant transformation, metastasis and chemo-resistance mechanisms of pancreatic cancer.
In recent years, it has become increasingly apparent that the non-protein-coding portion of the genome is of crucial functional importance: in relation to both normal physiology and diseases[7]. The relevance of the non-coding genome to human disease has mainly been studied in the context of the widespread disruption of miRNA expression and function in human cancers. Research is also being conducted aimed at understanding the nature and extent of other ncRNAs in disease, such as PIWI-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), transcribed ultraconserved regions (T-UCRs) and large intergenic non-coding RNAs (lincRNAs)[7]. Along with miRNAs, dysregulation of these ncRNAs is being found to show key relevance to pancreatic tumorigenesis. Consequently, exploring ncRNAs as therapeutic targets and biomarkers for the diagnosis and prognosis of pancreatic cancer is of interest.
MECHANISMS OF PANCREATIC CANCER DEREGULATION
The pancreas is a glandular organ of both the digestive system and endocrine system of vertebrates and displays an astonishing capacity to carry out cellular functions. Pancreatic dysfunction can be highly deleterious. As the fourth leading cause of cancer-related death in United States and worldwide, pancreatic cancer continues to remain a devastating disease with only 5.2% of patients alive for more than 5 years[8]. Genetic analysis of pancreatic cancer indicated that multiple mutations accumulate over time with some of them being more frequent than others [such as KRAS (about 90%), p16/CDKN2A (about 75%), TP53 (about 65%), SMAD4 (about 50%)] in these tumors[9,10]. However, blocking the activity of these frequently mutated genes did not turn out to be a promising therapeutic strategy[11]. Identification of molecular mechanisms that are more directly associated with the sustained proliferation and aggressive of pancreatic cancer cells is urgently needed. In recent years it has been proposed that in addition to the accumulated mutations that favor cancerous growth, epigenetic events may also play an important role in the development and maintenance of pancreatic cancer[12]. ncRNAs are already known as master epigenetic regulators that have a role in regulating diverse cellular processes including cell proliferation, development, differentiation, apoptosis and consequently oncogenesis. In view of the differentially expressed oncogenic and tumour suppressor ncRNAs targeting multiple genes in important cancer-relevant genetic and epigenetic networks in pancreatic cancer, it is logical to suggest that distinct ncRNA expression signatures may be associated with different grades and stages and meanwhile provide therapy strategy for this malignancy.
TYPES OF NCRNAS AND THEIR FUNCTIONS
RNA is the bridge between stable DNA and versatile proteins. Traditional studies are mainly focused on protein-coding RNAs, however, the coding exons of genes account for only 1.5% of the genome[13]. With the development of new scientific technologies, especially concerning the widespread use of microarray technology in molecular biology research, significant portions of eukaryotic genomes have been found to give rise to non-protein-coding RNAs, many of which remain unannotated.
In general, ncRNAs are grouped into two major classes based on their length (Table 1). Transcripts shorter than 200 nucleotides (nt) are usually referred to as small ncRNAs, which include miRNAs, Piwi-interacting RNAs, small-interfering RNAs and some bacterial regulatory RNAs. The well-documented about 22 nt long miRNAs serve as important regulators of gene expression and as intricate components of the cellular gene expression network[14-16]. lncRNAs are mRNA-like transcripts ranging in length from 200 nt to 100 kb; they are poorly conserved and do not function as templates for protein synthesis. ncRNAs have recently been discovered to act as robust regulators of gene expression that are frequently deregulated in human cancers, providing new opportunities to unravel the aberrantly expressed cellular pathways. However, as noted above, while the biological functions and molecular mechanisms of ncRNAs have been widely investigated in cancer studies (Figure 1), their prognostic value associated with pancreatic cancer remains to be elucidated.
Table 1.
Types of non-coding RNAs in pancreatic cancer and their functions
| Types | Name | Expression profile | Related biological function |
| Short ncRNAs | |||
| miRNAs | Let-7[117] | Up | Downregulates STAT3 phosphorylation |
| miR-15/16[17,18] | Up | Promoting tumor angiogenesis | |
| miR-34a[29] | Down | Inactivates p53 and modulates pancreatic cancer pathogenesis | |
| miR-132[88] | Down | Transcribed by RNA polymerase II and promoter methylation | |
| miR-200[36] | Down | Leading to drug resistance | |
| miR-421[30,31] | Up | Suppresses expression of DPC4/Smad | |
| piRNAs | pi-651[42] | Up? | piRNAs target transposon repression and DNA methylation |
| snoRNAs | U50, SNORD[46,118] | Up? | |
| Long ncRNAs | |||
| lincRNAs | PPP3CB/MAP3K1 4/DAPK1 loci | Up | Metastases associated with the MAPK pathway[69] |
| HOTAIR[73] | Up | Associates with the polycomb repressive complex 2 | |
| Other lncRNAs | HSATII (satellite repeat RNAs)[70] | Up | Reflect global alterations in heterochromatin silencing |
HOTAIR: Homeobox transcript antisense RNA; MAPK: Mitogen-activated protein kinase; STAT3: Signal transducer and activator of transcription 3.
Figure 1.
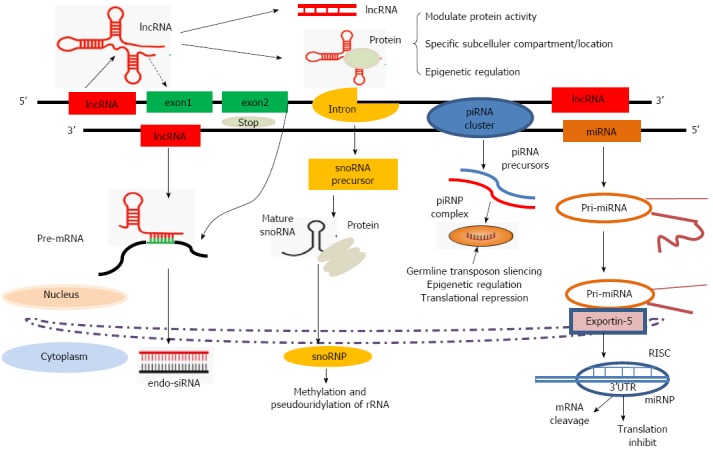
Biogenesis and functional machinery of small ncRNAs (miRNAs, piRNAs, snoRNAs) and lncRNAs. (1) miRNAs: pri-miRNA is transcribed by RNA polymerase II, then cleaved by Droshain into pre-miRNA. Pre-miRNAs are then transported to the cytoplasm via an Exportin-5-mediated mechanism and further processed by the RNase III endonuclease into mature miRNAs. Mature miRNAs are then incorporated into the RISC, where they direct the complex to specific mRNAs through complementary base pairing in their 3’UTRs; (2) piRNAs: piRNAs are mainly expressed from piRNA clusters. Subsequently, additional piRNAs influence the functions of ribonucleoprotein (piRNP) complexes in transposon repression through target degradation and epigenetic silencing; (3) snoRNAs: snoRNAs are transcribed from introns. Following splicing, debranching and trimming, mature snoRNAs are either exported, in which case they function in rRNA processing, or remain in the nucleus, where they are involved in alternative splicing; and (4) lncRNAs: lncRNAs exhibit a 5’ terminal methylguanosine cap and are often spliced and polyadenylated. lncRNAs contain structural domains that can sense or bind to other RNAs via complementary base pair interactions, in addition to interacting with proteins and possibly DNA and displaying other as yet unknown functions. endo-siRNA: Endogenous small-interfering RNA; lncRNA: Long non-coding RNA; miRNA, microRNA; piRNA: PIWI-interacting RNA; pre-miRNA: Primary miRNA; pre-miRNA: Precursor miRNA; rRNA: Ribosomal RNA; RISC: RNA-induced silencing complex; sncRNA: Small non-coding RNA; snoRNA: Small nucleolar RNA; 3’UTRs: 3’ untranslated regions.
NCRNAS IN PANCREATIC CANCER
Although all types of ncRNAs are transcribed in human cells, most of the current findings concerning ncRNAs are focused on miRNAs. Additionally, IncRNAs are also gaining prominence as emerging key elements of cellular homeostasis (Table 1), which draws our attention toward emphasizing the role of miRNAs and lncRNAs in pancreatic cancer development and progression in this review.
miRNAs in pancreatic cancer
miRNAs The most widely studied class of ncRNAs are miRNAs, which are small ncRNAs of 22 (18-25) nt in animals that mediate post-transcriptional gene silencing by controlling the translation of mRNAs into proteins[17,18]. Following the discovery that in C. elegans, the small RNAs encoded by the lin-4 gene are associated with the control of developmental timing through negatively regulating lin-14 translation[19], there was an explosion in the field of small ncRNA biology in subsequent years across different species. The biogenesis of miRNAs involves several steps (Figure 1). First, a several kilobase-long primary RNA (pri-miRNA) is transcribed by RNA polymerase II. The pri-miR is then cleaved by Droshain (an RNase III endonuclease) into a hairpin loop structure known as the precursor miRNA (pre-miRNA), after which the pre-miR is transported to the cytoplasm via a RanGTP-dependent Exportin-5-mediated mechanism, where it is further processed by the RNase III endonuclease into a 17-25 nt-long mature duplex miRNA. Mature single-stranded miRNAs are then incorporated into the RNA-induced silencing complex (RISC) and direct the complex to specific mRNAs through complementary base pairing with their 3’ untranslated regions (3’UTRs). Partial complementarity between the 3’UTR of the target gene and the seed region of the miRNA leads to mRNA decay and/or inhibition of translation[17,20]. It is predicted that on an average, each miRNA may regulate about 200 gene transcripts[21].
miRNAs in pancreatic cancer development and progression: miRNAs have emerged as critical components of networks of complex functional pathways controlling important cellular processes, such as proliferation, differentiation, apoptosis, stress response and drug resistance[22]. Recent analyses of tumor miRNA expression profiles have demonstrated that the miRNAs that are abnormally expressed in tumors usually exhibit reduced expression levels compared to their expression levels in normal tissues[23]. The role of miRNAs in tumorigenesis and tumor development has recently been reviewed in detail elsewhere[24], and it is therefore only briefly recapped here. First, downregulated expression of a tumor suppressor miRNA can lead to the expression of miRNA target genes that promote tumorigenesis, causing excessive cell proliferation and abnormal differentiation and resulting in tumorigenesis. On the other hand, the overexpression of oncogenic miRNAs can lead to decreased expression of target genes with tumor suppressor functions, thereby promoting tumor initiation and development. Another role of oncogenic miRNAs is the regulation of tumor angiogenesis. The cellular levels of miR-15b and miR-16 are downregulated under hypoxic conditions, leading to diminished inhibitory effects of miR-15b and miR-16 on VEGF and thereby promoting tumor angiogenesis[25]. miRNAs such as miR-10b and miR-373 are also important drivers of tumor metastasis[26,27]. A number of expression profiling studies have demonstrated that deregulation of miRNA expression occurs in pancreatic cancer tissues and cell lines. An extensive analysis of miRNA expression profiles in tissue samples from normal pancreases and patients with chronic pancreatitis and pancreatic ductal adenocarcinoma (PDAC) using microarray technology revealed that some miRNAs, including miR-29c and miR-96, were differentially expressed in both the chronic pancreatitis and PDAC samples, whereas the expression of miR-196s, miR-203 and miR-210 was altered only in PDAC tissues[28]. Moreover, p53 inactivation contributes to the reduction of miR-34a levels, which raises the possibility that loss of miR-34a function modulates the pathogenesis of pancreatic cancer[29]. Our previous findings have identified miR-421 and miR-483-3p as potent regulators of DPC4/Smad4, which may provide a novel therapeutic strategy for the treatment of DPC4/Smad4-driven pancreatic cancer[30,31]. Taken together, these findings indicate that miRNAs play an important role in the development and progression of pancreatic cancer.
miRNAs in pancreatic drug resistance: Chemotherapy represents an important therapeutic strategy for most patients with pancreatic cancer. Prior to the 1990s, 5-fluorouracil (5-FU) was the accepted monotherapy. However, soon after Burris and colleagues reported the results of a phase III clinical trial directly comparing gemcitabine and 5-FU[3], showing that gemcitabine confers significantly increased median survival, gemcitabine became the first-line treatment and gold standard for pancreatic cancer chemotherapy. Nevertheless, drug resistance can cause failure of this treatment[6]. Despite investigations into the mechanisms underlying drug resistance over the past 50 years, much is still unknown about exactly how this phenomenon occurs. The three most common reasons for the acquisition of drug resistance are the expression of energy-requiring transporters, insensitivity to drug-induced apoptosis and the induction of drug-detoxification mechanisms[32]. The tumor cell microenvironment (e.g., interactions between cell surface integrins and extracellular matrix components) is responsible for innate drug resistance[33]. miRNAs appear to be critical regulators of drug resistance in pancreatic cancer cells[34]. For example, in one study, the levels of the oncogenic miR-155 were shown to increase after pancreatic cancer cells were treated with gemcitabine[35]. miR-200a, miR-200b and miR-200c are all downregulated in pancreatic cancer cells that are resistant to gemcitabine[36]. A recent report suggests that miR-34 is involved in the self-renewal of pancreatic cancer stem cells, while the loss of miR-34 in pancreatic cancer is associated with an enrichment of cancer stem cells that are insensitive to chemotherapy[37] (Figure 2). Evidence also indicates that miRNAs might regulate the epithelial-mesenchymal transition (EMT) through the regulation of cadherin1 and other molecules[38], which mediate various types of cellular drug-resistance mechanisms (Figure 2). Many members of the Let-7 family are downregulated in EMT-type cells that are resistant to gemcitabine. In an investigation of the expression levels of miR-200 and Let-7 in EMT-phenotype pancreatic cancer cells that are resistant to gemcitabine, re-expression of the downregulated miR-200 family upregulates cadherin1 and downregulates ZeB1 and vimentin (EMT inducers). Knockdown of ZeB1 in mesenchymal Panc1 cells results in the upregulation of miR-200c[36]. Therefore, re-expression of specific miRNAs could serve as a new strategy for the treatment of pancreatic cancer via the targeted elimination of either cancer stem cells or EMT-phenotype cells that contribute to tumor recurrence and metastasis.
Figure 2.
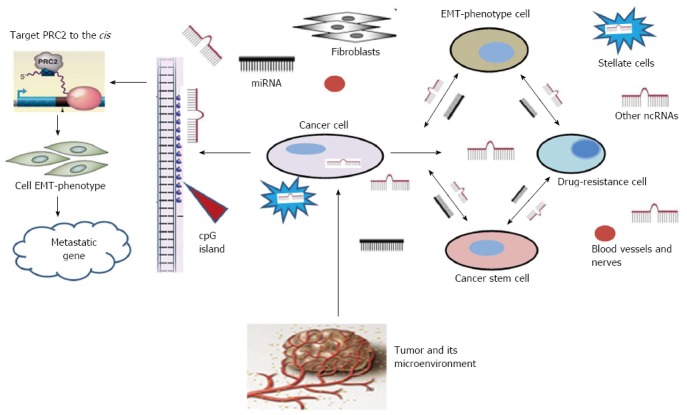
Pancreatic cancer cell microenvironment (the cellular elements are predominately mesenchymal cells such as pancreatic stellate cells, fibroblasts, blood vessels and nerves) and the links between cancer cells and drug-resistant cells determined through the epithelial-mesenchymal transition-phenotype as well as between cancer stem cells and the ncRNAs in their surroundings. PRC2: Polycomb repressive complex 2; EMT: Epithelial-mesenchymal transition.
Other small ncRNAs in pancreatic cancer
Other ncRNAs, such as piRNAs and snoRNAs, are also reported to be associated with cancer. Thousands of different piRNA species have been found in mammals[39]. piRNAs are generated from single-stranded RNA precursors through a Dicer-independent mechanism[40], providing essential protection for germ-cell genomes against the activity of transposable elements[41] (Figure 1). Regarding the role of piRNAs in cancer, they were first described to be over-expressed in seminomas but not in non-seminomas of the adult testis. Recent studies have highlighted the role of piRNAs in the regulation of tumorigenesis. For example, the expression of piR-651 in gastric, colon, lung, and breast cancer tissues is higher compared to normal adjacent tissues[42]. The levels of piR-651 have also been associated with the stage of tumor-node-metastasis (TNM)[43].
snoRNAs are small non-coding RNAs with lengths ranging from 60 to 300 nt. snoRNAs are normally located within the introns of protein-coding genes and are transcribed by RNA polymerase II (Figure 1), although in some cases, they can be found within the introns of lncRNAs. snoRNAs and snoRNPs are likely to contribute to tumorigenesis through an effect on ribosomes and protein translation, and snoRNAs might also be involved in the regulation of gene expression by giving rise to other regulatory RNA species, such as miRNAs[44]. Initial insights into the potential roles of snoRNAs in cancer were provided by a study in which substantial downregulation of snoRNAs was observed in meningiomas compared with normal brains[45]. Other studies have shown that a germline homozygous 2 bp (TT) deletion in the snoRNA U50 is associated with the development of prostate cancer[46]. However, the mechanisms through which both piRNAs and snoRNAs function to target pancreatic cancer remain unknown. The currently available data suggest important roles of piRNAs and snoRNAs that go beyond the regulation of the genome in germline tissues, and further studies are needed to reveal their specific roles in pancreatic tumorigenesis.
lncRNAs in pancreatic cancer
An updated view of lncRNAs: lncRNAs are a heterogeneous group of non-coding transcripts more than 200 nt long. This class of ncRNAs makes up the largest portion of the mammalian non-coding transcriptome[7]. Although the proverbial “dark matter” of the genome was initially argued to be spurious transcriptional noise, recent evidence suggests that it may play a major biological role in cellular development, differentiation and metabolism[47]. lncRNAs often overlap with, or are interspersed between, multiple coding and non-coding transcripts[48,49]. Many transcripts resist classification into any particular category and instead exhibit a combination of these qualities. Based on their genomic proximity to protein-coding genes, lncRNAs are generally classified into five types: (1) sense; (2) antisense; (3) bidirectional; (4) intronic; and (5) intergenic[7,50]. In many cases, the secondary structure of lncRNAs dictates their function. For example, conservation of the secondary structure of the lncRNA maternally expressed gene 3 (MEG3) and its propensity to fold into thermodynamically stable secondary and higher-order structures maintains the tumor suppressor function of this lncRNA, rather than its primary sequence[51]. By virtue of their ability to base pair with other RNAs, lncRNAs can act as highly specific sensors of the expression of mRNAs, miRNAs and other lncRNAs (Figure 1). The conversion of modified nucleotides for detection via high throughput sequencing is currently revealing widespread nucleotide modifications throughout the transcriptome[52]. There is a growing catalog of bifunctional mRNAs: coding transcripts can lose their ability to encode a protein, and noncoding transcripts can acquire a coding function[53-55]. Many of these posttranscriptional modifications are reversible, and given the range of modifications and targets observed, they may constitute an additional layer of posttranscriptional regulation, analogous to the epigenetic landscape regulated by lncRNAs[56].
There are three types of well-known lncRNAs: homeobox transcript antisense RNA (HOTAIR), lincRNAs and T-UCRs. lncRNAs are essential in many physiological processes, such as X-chromosome inactivation in mammals, in which the X-inactivation-specific transcript (XIST) lncRNA (17 kb) recruits the polycomb complex to silence the X chromosome from which it is transcribed[57]. lincRNAs, which are transcribed from intergenic regions, are associated with active transcription in the regions across which transcriptional elongation takes place[58]. Thus far, the aberrant regulation of T-UCR expression in cancers has been found to occur in two main ways: through altered interactions with miRNAs[59] and via hypermethylation of CpG islands in their promoters[60]. Among various examples of the involvement of lncRNAs in cancer, the role of HOTAIR in human neoplasia is the best understood[61]. HOTAIR might play an active role in modulating the cancer epigenome and mediating cell transformation[61].
lncRNAs and pancreatic cancer: Although the functional annotation of lncRNAs on a genomic scale has remained elusive for quite some time, lncRNAs may play critical roles in the regulation of multiple biological processes[62], including epigenetic regulation, transcriptional regulation, the processing of small RNAs, and other regulatory functions[63-65]. Due to advancements in cancer transcriptome profiling and the accumulating evidence indicating the functions of lncRNAs, these ncRNAs are emerging as new factors in the cancer paradigm, showing potential roles in both oncogenic and tumor-suppressive pathways[66]. Recent studies have demonstrated that certain lncRNAs are specifically associated with certain types of cancer, and their expression level may function as an indicator of metastasis or prognosis[61,67,68]. Despite the many lncRNAs that have been identified or validated in human cancer tissues, there is a paucity of information regarding the expression of lncRNAs in pancreatic cancer. Tahira et al[69] were the first to perform a microarray interrogation of protein-coding genes and putative lncRNAs and indicated that subsets of intronic/intergenic lncRNAs are expressed in both pancreatic tumor and non-tumor tissue samples. They identified loci harboring intronic lncRNAs (PPP3CB, MAP3K1 4 and DAP K1 loci) that were differentially expressed in PDAC metastases and were enriched in genes associated with the MAPK pathway. In an interesting study performed by Ting et al[70], aberrant overexpression of satellite repeat RNAs (HSATII) ranging from 100 to 5000 nt was observed in patients with PDAC. The overexpression of satellite transcripts in cancer may reflect global alterations in heterochromatin silencing and could potentially be useful biomarkers for pancreatic cancer detection[70].
HOTAIR is a long intervening non-coding RNA (lincRNA) that associates with the polycomb repressive complex 2 (PRC2), and its overexpression is correlated with poor survival in breast, nasopharyngeal and liver cancer patients[61,71,72]. Kim et al[73] showed that knockdown of HOTAIR in PANC1 and L3.6pL pancreatic cancer cells decreases cell proliferation, alters cell cycle progression and induces apoptosis, demonstrating an expanded function of HOTAIR in pancreatic cancer cells. HOTAIR is a negative prognostic factor for pancreatic cancer patients and exhibits pro-oncogenic activity in both in vitro and in vivo bioassays.
Nevertheless, only a small number of lncRNAs have been well characterized in pancreatic cancer, and little of the underlying molecular mechanism has been elucidated. The potential importance of lncRNAs in pancreatic cancer demands further study.
ncRNAs are linked to epigenetic mechanism of pancreatic cancer in the microenvironment
Pancreatic cancer and its microworld: Several lines of evidence indicate that tumorigenesis is a complex and multistep process. Cancers acquire the same set of functional capabilities during development and progression through various mechanistic strategies. The importance of the local tumor microenvironment to tumor progression has been recognized for many years and was highlighted in several reviews[74-76]. The cellular elements of pancreatic tumor environment are predominately mesenchymal cells such as pancreatic stellate cells, fibroblasts, blood vessels and nerves (Figure 2). Using in vitro and in vivo models containing tumor cells and stromal cells, investigators have shown that stromal signals and cell-to-cell interactions including proteases, growth factors, and mediators of invasion are critical determinants of pancreatic cancer behavior[77,78]. As pancreatic cancer has an especially abundant stroma originating from abundant pancreatic stellate cells, lacking of suitable models limited the exploration of interactions between pancreatic cancer cells and the adjacent stromal cells[79]. It was found that conditioned medium from human pancreatic stellate cells (HPSCs) stimulated pancreatic tumor cell proliferation and metastasis in vitro and in vivo via mitogen-activated protein kinase (MAPK) and Akt pathways[80]. However, simple models composed of cancer cells and their adjacent stromal cells in the tumor progress have several important limitations. Fortunately, with the development of ncRNA detection techniques, measurable biologically ncRNAs secreted from variational stroma complex provided novel research models in the exploration of role of tumor microenvironment in pancreatic carcinogenesis.
Epigenetics and pancreatic cancer: Epigenetics is a term used to describe mitotically and meiotically heritable states of gene expression that are not due to changes in DNA sequences[81], such as DNA methylation and histone tail modifications. Epigenetic regulations are important in all aspects of biology[82], and studies conducted during the past decade have shown that they play a key role in carcinogenesis and tumor progression[83]. It is evident that multiple epigenetic mechanisms are crucial in the development and progression of PDAC. In addition to genetic changes, epigenetic alterations add another layer of complexity and contribute to the heterogeneity of PDAC. Ongoing studies on chromatin dynamics are revealing the existence of robust machinery that can mediate epigenetic changes in pancreatic cells[12]. Epigenetic biomarkers, such as miRNAs, DNA methylation and satellite repeats, can be utilized to assess pancreatic cancer risk, progression and therapeutic responses; the use of these markers is regarded as the road to the early detection of pancreatic cancer. Numerous drugs that target specific enzymes involved in the epigenetic regulation of gene expression are emerging as an effective and valuable approach to chemotherapy[84]. A noteworthy characteristic of epigenetic-based inheritance is its reversibility, which contrasts with the stable nature of DNA sequence-based alterations.
ncRNAs are linked to epigenetic mechanisms associated with the biological phenotype of pancreatic cancer: Epigenetics influence genomic inheritance via miRNA-dependent mechanisms as well as DNA and histone modifications[12]. Members of the miR-200 family have been identified as modulators of the EMT due to negative activity against zinc finger E-box-binding homeobox1 and 2 (ZEB1 and ZEB2), which function as repressors of EMT-opposing genes[38,85,86]. The miR-34 family has been shown to be primarily inactivated by aberrant CpG methylation in PDAC, revealing an interesting example of an epigenetic mechanism influencing the functioning of other mechanisms[87]. We have reported that the downregulation of miR-132 in pancreatic cancer tissues is due to promoter methylation, which consequently impairs the binding of essential transcription factors[88]. miR-107 and miR-148a have also been observed to be hypermethylated in the promoter region and, thus, down-regulated in pancreatic cancer[89,90]. Using a pyrosequencing technique, Wang et al[91] showed that the promoters of miR-124 family members (miR-124-1, miR-124-2 and miR-124-3) are highly methylated in pancreatic cancer tissues compared with non-cancerous tissues, and in vitro studies have shown that miR-124 inhibits cell proliferation, invasion and metastasis.
Important roles of lncRNAs have been described in epigenetic processes, and this broad topic has been reviewed elsewhere[92]. Lee noted that lncRNAs are implicated in almost every epigenetic regulation event[93]. The intriguing story of lncRNAs was initially related to the phenomena of genomic imprinting and X-chromosome inactivation[94,95]. The lncRNA AS1DHRS4 is transcribed from the locus of the dehydrogenase/reductase SDR family member 4 (DHRS4) gene and recruits DNA methyl transferases and other factors to the DHRS4 gene cluster, inducing DNA methylation in the DHRS4L2 promoter region[96]. Two other lncRNAs have been reported to induce either DNA methylation in specific regions of the Kcnq1 locus or demethylation at the Sphk1 CpG island[97,98]. Another potential mechanism is that lncRNAs recruit demethylases and/or acetylases to the promoter regions of oncogenes, and thus, the lncRNAs might direct the transcriptional activation of such protein-coding genes[99]. Some potential epigenetic mechanisms involving lncRNAs related to pancreatic cancer are as follows[99]: (1) lncRNAs affect DNA methylation; (2) lncRNAs alter nucleosome positioning; and (3) lncRNAs display an in-cis function or carry out trans-regulation.
It is evident that multiple epigenetic mechanisms are indeed crucial in the development and progression of pancreatic cancer. As one category of epigenetic alterations, our understanding of ncRNA-epigenetic-based events provides a new research model for studying PDAC.
NCRNA-BASED DIAGNOSIS AND TREATMENT OF PANCREATIC CANCER
Regarding the identification of molecular markers for pancreatic cancer diagnosis/prognosis, some promising candidate genes have been proposed[100]. However, none of these candidates have been proven to be effective in significantly improving early detection or reducing the mortality/morbidity of this disease. Thus, a better understanding of the molecular basis of pancreatic cancer is required for the identification of more effective diagnostic markers and therapeutic targets.
Not only are numerous miRNAs preferentially expressed between pancreatic cancer tissues and normal tissues, they are also extremely stable in both blood plasma and serum[101]. Studies have demonstrated that miRNAs that are differentially expressed in PDAC can be profiled in blood as a minimally invasive biomarker assay for pancreatic cancer[102]. Consequently, miRNA levels serve as suitable tumor markers and biomarkers for testing conducted for clinical diagnosis. It has been suggested that endocrine tumors of the pancreas can be distinguished from acinar-type tumors based on a set of 10 miRNAs. These miRNAs are potentially associated with normal endocrine differentiation or endocrine tumorigenesis[103], indicating the diagnostic utility of miRNA expression signatures in pancreatic cancer. Another report addressing serum samples from pancreatic cancer patients and matched cancer-free controls indicates that numerous miRNAs display significantly different expression levels and present high sensitivity and specificity for distinguishing the various stages of pancreatic cancer compared to cancer-free controls[104]. miR-210 and miR-1290 were subsequently found to show ideal diagnostic performance in a qRT-PCR miRNA array analysis of sera from pancreatic cancer patients and controls[105,106]. A recent study analyzed the expression of several miRNAs in different types of pancreatic disease to determine if miRNA expression could aid in the diagnosis of PDAC and its precursor, pancreatic intraepithelial neoplasm (PanIN)[107]. Results showed that compared to the non-neoplastic parenchyma, miR-148a was significantly underexpressed, whereas miR-196a and miR-10b were highly overexpressed in PanIN. miR-217 expression was shown to decrease in PanIN compared to that in the non-neoplastic tissue[107], suggested that these miRNA markers may be involved in an early event in pancreatic carcinogenesis. These findings provide compelling reasons to pursue the targeting of miRNAs as novel molecular diagnostic markers and for use in therapeutic approaches in the early stage of pancreatic carcinogenesis.
The knowledge that miRNAs regulate their targets through base pairing has led to the use of antisense oligonucleotides (ASOs) to inhibit miRNA function therapeutically. ASOs inhibit miRNAs based on base pair complementarity. Three main classes of ASOs have been developed: locked nucleic acids (LNAs), anti-miRNA oligonucleotides (AMOs) and antagomirs, which incorporate different chemical modifications to increase their stability and efficacy[108-110]. However, as cancer progression is influenced by multiple miRNAs, recent research in this field suggests that several miRNAs can be simultaneously inhibited using mixed ASOs targeting multiple miRNAs[111]. One multiple-target anti-miRNA AMO was designed to suppress miR-21, miR-155 and miR-17-5p, which are oncogenic miRNAs that are frequently overexpressed in many types of tumors, showing promising results related to inhibiting cancer growth.
Dozens of lncRNAs have been found to be dysregulated in pancreatic cancer, raising the possibility that lncRNAs may represent promising biomarkers for the diagnosis and prognosis of this disease. The strategy of inhibiting the function of deregulated miRNAs could also be useful for lncRNAs. The progress in the use of RNAi-mediated gene silencing for the treatment of different diseases is encouraging, as this strategy provides a straightforward approach for selectively silencing oncogenic lncRNAs. Thus far, few lncRNAs have been characterized as potential biomarkers in human body fluids. For example, the lncRNA PCA3 has been demonstrated as a more specific and sensitive marker of prostate cancer in urine samples from patients compared to the widely used serum prostate-specific antigen (PSA)[112,113], highlighting the advantages of PCA3 over PSA and enabling noninvasive diagnosis. Targeting human H19 for the treatment of bladder cancer with a plasmid-based system was found to be successful[114]. Decreased expression of the lncRNA HOTAIR has been shown to alter cell cycle progression and induce apoptosis, displaying a unique association with the treatment of pancreatic cancer patients, and overexpression of HOTAIR may serve as a potential biomarker of pancreatic cancer prognosis[73]. Circulating lncRNAs may be promising biomarkers for cancer diagnosis, and knockdown of lincRNAs can be achieved using siRNAs[61,115]. However, successful inhibition of long ncRNAs appears to be more difficult than inhibiting miRNAs, presumably due to the extensive secondary structure of lncRNAs. It is therefore urgent to ask important questions such as the following[66]: (1) How stable are circulating lncRNAs, and is their stability altered in various disease states? (2) although the RNA contents of microvesicles and exosomes reported thus primarily consist of small miRNAs, is it possible that long protein-coding mRNAs and lncRNAs are also packaged into microparticles in a manner similar to miRNAs? (3) as considerable numbers of circulating lncRNAs may be dysregulated in various human diseases, might these lncRNAs be causing the disease, or do they become altered as a consequence of the disease itself? These ncRNAs could represent an unexpected source of potential diagnostic and prognostic biomarkers that might aid in addressing the great challenges related to the battle against pancreatic cancer[66].
However, the overall treatment strategy is to correct imbalances of ncRNAs (Figure 3), including drugs or adenovirus-associated vectors enhancing ncRNA processing or synthetic-anti ncRNAs, epigenetic ncRNA silencing or other strategies. ncRNAs may represent a gold mine for all diagnostic and therapeutic strategies. More studies need to be conducted to confirm this phenomenon and how these ncRNAs are involved in the early stage of pancreatic carcinogenesis.
Figure 3.
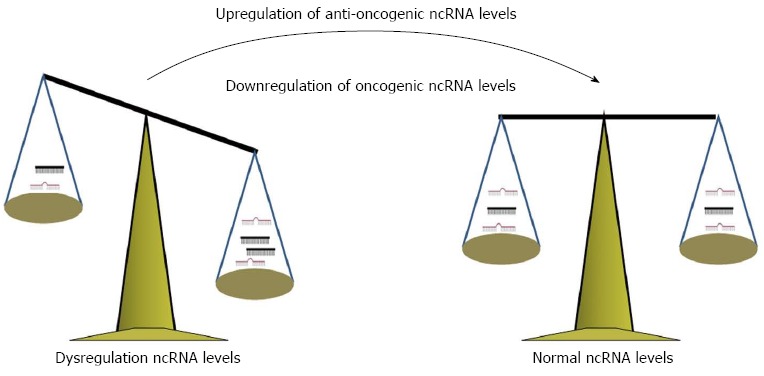
Overall therapeutic strategy is to correct imbalances of ncRNAs, including inhibiting/restoring their functions via biological and/or biochemical techniques. ncRNA: Non-coding RNA.
CONCLUSION
The outlook for patients with PDAC is bleak, primarily because these tumors are detected late and are often too advanced for surgical resection[116]. The number of patients who die annually from pancreatic cancer continues to increase, despite an overall decrease in cancer deaths worldwide. It is our goal to be able to reduce the local invasiveness of pancreatic cancer, allow more patients to undergo curative surgical resection and prevent local recurrence, which is observed in 80% of pancreatic cancer patients. Research on ncRNAs has not only increased greatly but has changed in character during the past quarter century, providing new research methodology to improve our understanding of the biological functions, molecular mechanisms and prognostic value associated with pancreatic cancer in the curable stage.
A major challenge remains regarding the elucidation of the biological pathways or signaling networks underlying cancer development and how specific ncRNAs interact or contribute to malignant transformation. There is increasing evidence suggesting an association of ncRNAs with disease, and knowledge of disease-related ncRNAs is essential in relation to the pathogenesis of pancreatic cancer at the molecular level. We must also continue to expand our research into the associated genetic and epigenetic mechanisms to obtain complete picture regarding the alterations of gene expression that occur in pancreatic cancer.
Footnotes
Supported by National Natural Science Foundation of China, No. 81172597, No. 81102078, No. 81301933 and No. 81301764; and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD)
P- Reviewer: Chen RF, Shi CJ S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Zhang DN
References
- 1.Kaur S, Kumar S, Momi N, Sasson AR, Batra SK. Mucins in pancreatic cancer and its microenvironment. Nat Rev Gastroenterol Hepatol. 2013;10:607–620. doi: 10.1038/nrgastro.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camacho D, Reichenbach D, Duerr GD, Venema TL, Sweeney JF, Fisher WE. Value of laparoscopy in the staging of pancreatic cancer. JOP. 2005;6:552–561. [PubMed] [Google Scholar]
- 3.Burris HA, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, Cripps MC, Portenoy RK, Storniolo AM, Tarassoff P, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 4.Cardenes HR, Moore AM, Johnson CS, Yu M, Helft P, Chiorean EG, Vinson J, Howard TJ, Stephens AW, Tai DF, et al. A phase II study of gemcitabine in combination with radiation therapy in patients with localized, unresectable, pancreatic cancer: a Hoosier Oncology Group study. Am J Clin Oncol. 2011;34:460–465. doi: 10.1097/COC.0b013e3181e9c103. [DOI] [PubMed] [Google Scholar]
- 5.Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1252–1261. doi: 10.1053/j.gastro.2013.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao H, LE D, Yang LX. Current status in chemotherapy for advanced pancreatic adenocarcinoma. Anticancer Res. 2013;33:1785–1791. [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 9.Schneider G, Schmid RM. Genetic alterations in pancreatic carcinoma. Mol Cancer. 2003;2:15. doi: 10.1186/1476-4598-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 11.Chiu J, Yau T. Metastatic pancreatic cancer: are we making progress in treatment? Gastroenterol Res Pract. 2012;2012:898931. doi: 10.1155/2012/898931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCleary-Wheeler AL, Lomberk GA, Weiss FU, Schneider G, Fabbri M, Poshusta TL, Dusetti NJ, Baumgart S, Iovanna JL, Ellenrieder V, et al. Insights into the epigenetic mechanisms controlling pancreatic carcinogenesis. Cancer Lett. 2013;328:212–221. doi: 10.1016/j.canlet.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet. 2010;11:559–571. doi: 10.1038/nrg2814. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 16.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 17.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 18.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 19.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Sen S. MicroRNA functional network in pancreatic cancer: from biology to biomarkers of disease. J Biosci. 2011;36:481–491. doi: 10.1007/s12038-011-9083-4. [DOI] [PubMed] [Google Scholar]
- 23.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 24.Tie J, Fan D. Big roles of microRNAs in tumorigenesis and tumor development. Histol Histopathol. 2011;26:1353–1361. doi: 10.14670/HH-26.1353. [DOI] [PubMed] [Google Scholar]
- 25.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 27.Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 29.Seux M, Iovanna J, Dagorn JC, Dusetti NJ. MicroRNAs in pancreatic ductal adenocarcinoma: new diagnostic and therapeutic clues. Pancreatology. 2009;9:66–72. doi: 10.1159/000178876. [DOI] [PubMed] [Google Scholar]
- 30.Hao J, Zhang S, Zhou Y, Liu C, Hu X, Shao C. MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer. Biochem Biophys Res Commun. 2011;406:552–557. doi: 10.1016/j.bbrc.2011.02.086. [DOI] [PubMed] [Google Scholar]
- 31.Hao J, Zhang S, Zhou Y, Hu X, Shao C. MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer. FEBS Lett. 2011;585:207–213. doi: 10.1016/j.febslet.2010.11.039. [DOI] [PubMed] [Google Scholar]
- 32.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 33.Damiano JS. Integrins as novel drug targets for overcoming innate drug resistance. Curr Cancer Drug Targets. 2002;2:37–43. doi: 10.2174/1568009023334033. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Li Y, Ahmad A, Banerjee S, Azmi AS, Kong D, Sarkar FH. Pancreatic cancer: understanding and overcoming chemoresistance. Nat Rev Gastroenterol Hepatol. 2011;8:27–33. doi: 10.1038/nrgastro.2010.188. [DOI] [PubMed] [Google Scholar]
- 35.Xia QS, Ishigaki Y, Sun L, Chen R, Motoo Y. [Effect of anti-cancer drugs on the expression of BIC/miR-155 in human pancreatic cancer PANC-1 cells] Zhonghua Yi Xue Zazhi. 2010;90:123–127. [PubMed] [Google Scholar]
- 36.Li Y, VandenBoom TG, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 39.Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 41.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 42.Qiao D, Zeeman AM, Deng W, Looijenga LH, Lin H. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21:3988–3999. doi: 10.1038/sj.onc.1205505. [DOI] [PubMed] [Google Scholar]
- 43.Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 44.Scott MS, Ono M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie. 2011;93:1987–1992. doi: 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang LS, Lin SY, Lieu AS, Wu TL. Differential expression of human 5S snoRNA genes. Biochem Biophys Res Commun. 2002;299:196–200. doi: 10.1016/s0006-291x(02)02623-2. [DOI] [PubMed] [Google Scholar]
- 46.Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou W, Dong JT. Implication of snoRNA U50 in human breast cancer. J Genet Genomics. 2009;36:447–454. doi: 10.1016/S1673-8527(08)60134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 49.Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, Gingeras TR. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, Zhou Y, Klibanski A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151:939–947. doi: 10.1210/en.2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kellner S, Burhenne J, Helm M. Detection of RNA modifications. RNA Biol. 2010;7:237–247. doi: 10.4161/rna.7.2.11468. [DOI] [PubMed] [Google Scholar]
- 53.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. Proto-genes and de novo gene birth. Nature. 2012;487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng D, Frankish A, Baertsch R, Kapranov P, Reymond A, Choo SW, Lu Y, Denoeud F, Antonarakis SE, Snyder M, et al. Pseudogenes in the ENCODE regions: consensus annotation, analysis of transcription, and evolution. Genome Res. 2007;17:839–851. doi: 10.1101/gr.5586307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 57.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 58.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 60.Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, Croce CM, Calin GA, Esteller M. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glazko GV, Zybailov BL, Rogozin IB. Computational prediction of polycomb-associated long non-coding RNAs. PLoS One. 2012;7:e44878. doi: 10.1371/journal.pone.0044878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N, Isaacs WB. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 64.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S, Brown CJ, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–165. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 67.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 68.Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol. 2009;21:688–692. doi: 10.1097/meg.0b013e328306a3a2. [DOI] [PubMed] [Google Scholar]
- 69.Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 72.Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 75.Fidler IJ. The organ microenvironment and cancer metastasis. Differentiation. 2002;70:498–505. doi: 10.1046/j.1432-0436.2002.700904.x. [DOI] [PubMed] [Google Scholar]
- 76.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 77.Cheng N, Bhowmick NA, Chytil A, Gorksa AE, Brown KA, Muraoka R, Arteaga CL, Neilson EG, Hayward SW, Moses HL. Loss of TGF-beta type II receptor in fibroblasts promotes mammary carcinoma growth and invasion through upregulation of TGF-alpha-, MSP- and HGF-mediated signaling networks. Oncogene. 2005;24:5053–5068. doi: 10.1038/sj.onc.1208685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tuxhorn JA, Ayala GE, Smith MJ, Smith VC, Dang TD, Rowley DR. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin Cancer Res. 2002;8:2912–2923. [PubMed] [Google Scholar]
- 79.Farrow B, Albo D, Berger DH. The role of the tumor microenvironment in the progression of pancreatic cancer. J Surg Res. 2008;149:319–328. doi: 10.1016/j.jss.2007.12.757. [DOI] [PubMed] [Google Scholar]
- 80.Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, Ji B, Evans DB, Logsdon CD. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68:918–926. doi: 10.1158/0008-5472.CAN-07-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 82.Waddington CH, Pantelouris EM. Transplantation of nuclei in newt’s eggs. Nature. 1953;172:1050–1051. doi: 10.1038/1721050a0. [DOI] [PubMed] [Google Scholar]
- 83.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 85.Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, Wellner U, Dimmler A, Faller G, Schubert J, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30:770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6:e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J, Zhou J, Wu J, Shao C. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32:1183–1189. doi: 10.1093/carcin/bgr105. [DOI] [PubMed] [Google Scholar]
- 89.Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, Goggins MG, Mendell JT, Maitra A. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9:293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hanoun N, Delpu Y, Suriawinata AA, Bournet B, Bureau C, Selves J, Tsongalis GJ, Dufresne M, Buscail L, Cordelier P, Torrisani J. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin Chem. 2010;56:1107–1118. doi: 10.1373/clinchem.2010.144709. [DOI] [PubMed] [Google Scholar]
- 91.Wang P, Chen L, Zhang J, Chen H, Fan J, Wang K, Luo J, Chen Z, Meng Z, Liu L. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene. 2014;33:514–524. doi: 10.1038/onc.2012.598. [DOI] [PubMed] [Google Scholar]
- 92.Saxena A, Carninci P. Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. Bioessays. 2011;33:830–839. doi: 10.1002/bies.201100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 94.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 95.Wan LB, Bartolomei MS. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- 96.Li Q, Su Z, Xu X, Liu G, Song X, Wang R, Sui X, Liu T, Chang X, Huang D. AS1DHRS4, a head-to-head natural antisense transcript, silences the DHRS4 gene cluster in cis and trans. Proc Natl Acad Sci USA. 2012;109:14110–14115. doi: 10.1073/pnas.1116597109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mohammad F, Pandey GK, Mondal T, Enroth S, Redrup L, Gyllensten U, Kanduri C. Long noncoding RNA-mediated maintenance of DNA methylation and transcriptional gene silencing. Development. 2012;139:2792–2803. doi: 10.1242/dev.079566. [DOI] [PubMed] [Google Scholar]
- 98.Imamura T, Yamamoto S, Ohgane J, Hattori N, Tanaka S, Shiota K. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem Biophys Res Commun. 2004;322:593–600. doi: 10.1016/j.bbrc.2004.07.159. [DOI] [PubMed] [Google Scholar]
- 99.Beckedorff FC, Amaral MS, Deocesano-Pereira C, Verjovski-Almeida S. Long non-coding RNAs and their implications in cancer epigenetics. Biosci Rep. 2013;33 doi: 10.1042/BSR20130054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.López-Casas PP, López-Fernández LA. Gene-expression profiling in pancreatic cancer. Expert Rev Mol Diagn. 2010;10:591–601. doi: 10.1586/erm.10.43. [DOI] [PubMed] [Google Scholar]
- 101.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 104.Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610–618. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 105.Li A, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH, Goggins M. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19:3600–3610. doi: 10.1158/1078-0432.CCR-12-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, Koong AC. Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer. Transl Oncol. 2010;3:109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xue Y, Abou Tayoun AN, Abo KM, Pipas JM, Gordon SR, Gardner TB, Barth RJ, Suriawinata AA, Tsongalis GJ. MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm. Cancer Genet. 2013;206:217–221. doi: 10.1016/j.cancergen.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 108.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 109.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 110.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44:55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 111.Lu Y, Xiao J, Lin H, Bai Y, Luo X, Wang Z, Yang B. A single anti-microRNA antisense oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers an improved approach for microRNA interference. Nucleic Acids Res. 2009;37:e24. doi: 10.1093/nar/gkn1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tinzl M, Marberger M, Horvath S, Chypre C. DD3PCA3 RNA analysis in urine--a new perspective for detecting prostate cancer. Eur Urol. 2004;46:182–16; discussion 187. doi: 10.1016/j.eururo.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 113.Hessels D, Klein Gunnewiek JM, van Oort I, Karthaus HF, van Leenders GJ, van Balken B, Kiemeney LA, Witjes JA, Schalken JA. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44:8–15; discussion 15-6. doi: 10.1016/s0302-2838(03)00201-x. [DOI] [PubMed] [Google Scholar]
- 114.Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. 2010;12:607–616. [PubMed] [Google Scholar]
- 115.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Frampton AE, Fletcher CE, Gall TM, Castellano L, Bevan CL, Stebbing J, Krell J. Circulating peripheral blood mononuclear cells exhibit altered miRNA expression patterns in pancreatic cancer. Expert Rev Mol Diagn. 2013;13:425–430. doi: 10.1586/erm.13.31. [DOI] [PubMed] [Google Scholar]
- 117.Patel K, Kollory A, Takashima A, Sarkar S, Faller DV, Ghosh SK. MicroRNA let-7 downregulates STAT3 phosphorylation in pancreatic cancer cells by increasing SOCS3 expression. Cancer Lett. 2014;347:54–64. doi: 10.1016/j.canlet.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kiss-László Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]


