Abstract
Argonaute (Ago) proteins mediate post-transcriptional gene repression by binding guide microRNAs (miRNAs) to regulate targeted RNAs. To confidently assess Ago-bound small RNAs, we adapted a mouse embryonic stem cell system to express a single epitope-tagged Ago protein family member in an inducible manner. Here, we report the small RNA profile of Ago-deficient cells and show that Ago-dependent stability is a common feature of mammalian miRNAs. Using this criteria and immunopurification, we identified a new Ago-dependent class of non-canonical miRNAs derived from protein-coding gene promoters, which we name transcriptional start site miRNAs (TSS-miRNAs). A subset of promoter-proximal RNA polymerase II complexes produce hairpin RNAs that are processed in a Dgcr8/Drosha-independent, but Dicer-dependent manner. TSS-miRNA activity is detectable from endogenous levels and following over-expression of mRNA constructs. Finally, we present evidence of differential expression and conservation in humans, suggesting important roles in gene regulation.
Introduction
The transcription of protein-coding genes by RNA polymerase II (RNAPII) is regulated at several steps in the transcriptional cycle (Fuda et al., 2009). One prominent step is the regulation of elongation of promoter-proximal RNAPII (Adelman and Lis, 2012; Rougvie and Lis, 1988). This promoter-proximal RNAPII species is detected ~50 nt from transcriptional start sites (TSS) (Core et al., 2008). We have previously reported that small RNAs from promoters in both sense and antisense directions are byproducts of divergently transcribed RNAPII genes (Seila et al., 2008). In addition to these, several other promoter-proximal small RNA species have been discovered (Kanhere et al., 2010; Preker et al., 2008). Whether any of these RNA species are incorporated into the RNAi pathway has not been determined.
MicroRNAs (miRNAs) are major contributors to gene regulation (Lewis et al., 2005). The Argonaute (Ago) family of proteins function to target mRNA transcripts for post-transcriptional regulation (Wilson and Doudna, 2013). Mammals express four closely related Ago proteins, all of which can function through miRNA-directed base pairing of the seed region to a target RNA (Lewis et al., 2005; Nielsen et al., 2007). Canonical miRNAs are processed from hairpin precursors (pri-miRNAs) by the nuclear microprocessor complex, composed of DiGeorge syndrome critical region gene 8 (Dgcr8) protein and the RNAse-III like enzyme Drosha (Han et al., 2006; Lee et al., 2003). The resultant pre-miRNA hairpin is exported from the nucleus by exportin-5 (Lund et al., 2004) and cleaved by the RNAse III-enzyme Dicer, which processes the miRNA to its mature 21–24 nucleotide (nt) form (Bernstein et al., 2001; Hutvagner et al., 2001).
Mirtrons, a group of non-canonical miRNAs, are processed in a microprocessor-independent manner from hairpins within spliced introns that are substrates for Dicer (Okamura et al., 2007; Ruby et al., 2007). Additional non-canonical miRNA-like species have been reported from other classes of cellular non-coding RNAs, such as tRNA (Lee et al., 2009) and snoRNA (Ender et al., 2008) and other endogenous shRNAs (Babiarz et al., 2008). A limited number of miRNA-like small RNAs derived from within mature mRNA coding regions have also been reported in Drosophila (Berezikov et al., 2011) and the 3′ untranslated regions (UTR) of HeLa mRNA (Valen et al., 2011), but a high confidence classification of non-canonical mammalian miRNAs has not been reported. Generally, the identification of non-canonical miRNAs is determined by the detection of key features of RNAi machinery processing, such as small RNA size, predictive secondary structure and remnants of Dicer cleavage.
In order to accurately quantify the full binding potential of Ago proteins in vivo, we have generated a mouse embryonic stem cell (mESC) system expressing a single epitope-tagged Ago2 protein under control of the tetracycline-regulatory system in an otherwise Ago-family null genetic background. Using this system, we have identified a group of RNAPII protein-coding gene promoters that produce highly structured promoter-proximal RNAs that are non-canonically processed and loaded into Ago2.
Results
An inducible Ago system identifies Ago-dependent small RNAs
To accurately interrogate Ago2 function in mESCs we have adapted a previously described inducible Ago2 knockout mESC line (Su et al., 2009). E7 mESCs (Ago1−/−; Ago2−/−; Ago3−/−; Ago4−/−; hAgo2; CreERT2), lacking all mouse Ago1–4 alleles and expressing only low levels of a conditional human Ago2 transgene (hAgo2), were utilized to derive an epitope-tagged expression system (Figure 1A). E7 CreERT2-targeted hAgo2 was replaced with FLAG-HA tagged hAgo2 (FHAgo2) under the control of a TRE-Tight (TT) doxycycline(Dox)-inducible promoter (Shin et al., 2006). TT-FHAgo2 cells express FHAgo2 in a Dox-dependent manner to wildtype AB2.2 levels and allow Ago depletion upon Dox removal. As a stringent control for forthcoming Ago2 characterization, the identical procedure was used to derive cells expressing untagged hAgo2 (TT-Ago2) facilitating comparisons without confounding differences in gene expression (Figure S1A). In line with the previous report of Ago protein depletion in mESCs, long-term TT-FHAgo2 Dox starvation results in a severely delayed growth phenotype that can be reversed with Dox supplementation (Figure S1B). The cell lines derived here will allow high confidence and quantitative studies of miRNA regulation.
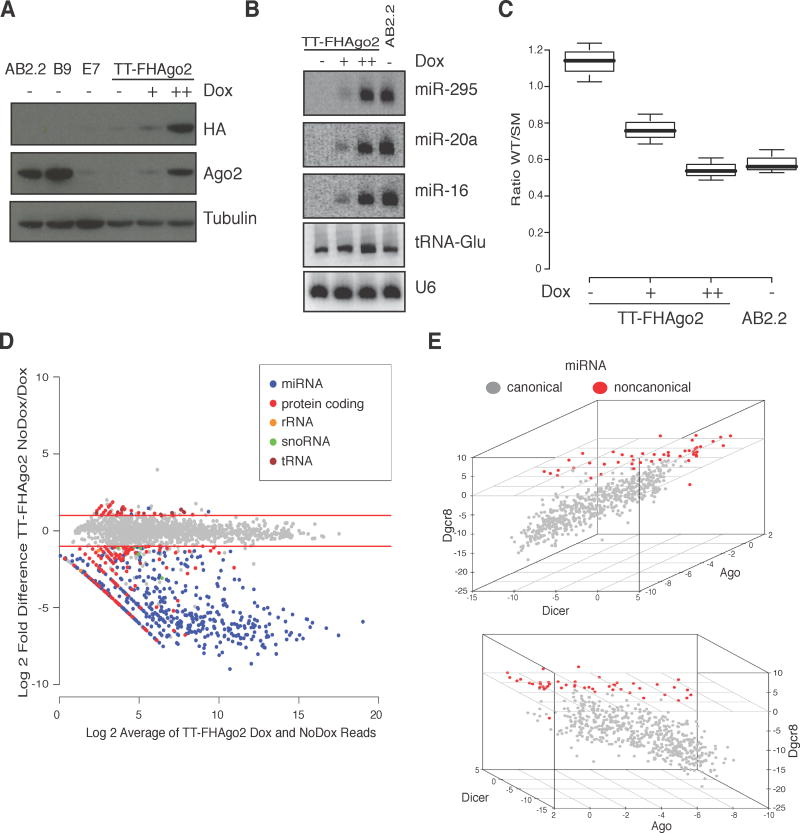
Figure 1. An inducible Ago system demonstrates miRNAs require Ago proteins for stability.
(A) Western blot of doxycycline (Dox)-inducible TT-FHAgo2 mESCs after 48hr Dox starvation or treatment with low (+, 0.1 μg/mL) or high (++, 2.5 μg/mL) levels. AB2.2 (wildtype) and B9 (Ago1−/−; Ago3−/−; Ago4−/−) are included for reference of wildtype Ago2 levels. (B) Northern blot of miRNA in TT-FHAgo2 mESCs after 96 hrs Dox starvation (-) followed by 24 hrs of low (+) or high (++) Dox treatment. AB2.2 is included for wildtype reference. (C) Luciferase reporter assay for Slc31a1 3′ UTR repression relative to a seed mutant control for the samples described in panel B (completed in biological triplicate). (D) Small RNA relative fold change and mean expression level plot for TT-FHAgo2 cells after 48 hrs of high (++) Dox treatment or after 96 hrs of Dox starvation (NoDox). Small RNAs that changed less than 2-fold are shown in gray. Small RNAs that changed more than 2-fold are colored based on indicated classes. Red lines denote two-fold cutoffs. (E) Plot of log2 fold changes of canonical (gray) and non-canonical (red) miRNAs in Dicer−/− mESCs, Dgcr8−/− mESCs and TT-FHAgo2 mESCs after 96 hrs of Dox starvation. Non-canonical miRNA are defined by those that decrease by less than 2-fold with loss of either Dicer or Dgcr8. All samples were compared to their respective wildtype expression control and the three-dimensional plot is displayed at two different angles. See also Figure S1 and Table S1.
As a starting point, TT-FHAgo2 cells were used to investigate small RNA stability following loss of cellular Ago proteins. Northern blots probed for three of the most abundant mESCs miRNAs revealed a reversible Ago2 concentration dependent role in miRNA steady-state accumulation (Figure 1B). Under high Dox treatment, miRNAs increased to AB2.2 wildtype levels in agreement with Ago2 protein detection in TT-FHAgo2 cells. Using a luciferase-based reporter with a bulge seed match to the miR-290 family, Ago expression correlated with and was required for post-transcriptional miRNA targeting (Figure 1C). Since Ago2 loss is implicated in both increased miRNA turnover rates (Winter and Diederichs, 2011) and defects in miRNA biogenesis (O’Carroll et al., 2007), we tested previously characterized Ago2 mutants that are either strongly defective in small RNA binding (D529E or F2V2) (Eulalio et al., 2008; Rudel et al., 2011) or lack catalytic activity (D597A) (Liu et al., 2004) in miRNA rescue experiments in E7 cells (Figure S1C). We observed that miRNA levels were not rescued with Ago2 small RNA binding mutants and conclude that direct Ago binding to miRNA confers stability, likely bestowed by protection from nuclease degradation. As the full consequence of Ago-family protein loss on cellular small RNAs has not been determined, we globally profiled small RNA expression in Ago-deficient cells.
Cellular small RNAs were compared between TT-FHAgo2 cells expressing wildtype Ago2 levels or after Dox depletion for 96 hours, which results in undetectable Ago2 and miRNA activity (Figure 1A,B). To assess small RNA populations that are affected by Ago loss, genome-wide changes in small RNAs sequenced in a 5′ phosphate dependent cloning protocol were quantified and normalized using the procedure implemented in the DESeq package (Anders and Huber, 2010). The 20–30 nt small RNA species that decreased between the two cell states by 2-fold or more were classified and defined as Ago-dependent small RNAs (Fig. 1D). Expectedly, the predominant effect of cellular Ago loss was on the annotated miRNA population, which had a 13-fold change on average. Next, the changes across all annotated miRNAs were plotted (Figure S1D) and showed the vast majority are Ago-dependent with few miRBase-annotated miRNAs detected in mESCs unaffected by Ago loss. These exceptions are likely arms of miRNA hairpin precursors processed by Dicer but not bound by Ago, candidates for incorrect annotation or small RNA that may function outside the RNAi pathway. The second most abundant group of destabilized small RNA was derived from within protein-coding transcripts. Surprisingly, only slight changes were observed for small RNAs that have reported miRNA-like functions, such as snoRNA and tRNA fragments. These differences in steady-state accumulation likely reflect differences in direct Ago binding. The quantification of all miRBase annotated miRNAs and other small RNA regions are summarized in Table S1.
Ago-dependent small RNAs were next compared to the small RNA population of Dicer or Dgcr8 null mESCs (Babiarz et al., 2008) to access the behavior of non-canonical miRNAs (Figure 1E). As anticipated, most of the Dicer and Dgcr8-dependent miRNAs are also Ago-dependent. miRNA that are distinct in their dependence on Dgcr8 and Dicer demonstrated both Ago-dependence and independence, which likely stratifies true miRNAs from other small RNAs.
These results demonstrate that Ago-dependent stability is a fundamental characteristic of known miRNAs. Using this identified in-vivo Ago-dependence along with Ago2 immunoprecipitations (IP) followed by small RNA sequencing, we next aimed to discover new small RNA classes acting in the RNAi pathway.
Identification of Ago-bound small RNA derived from protein-coding transcripts
The generation of TT-FHAgo2 and TT-Ago2 cells permitted IP studies using the same antibody from identical cell states to generate a stringent measure for specific Ago-RNA interactions. We initiated an experimental design where RNA was collected from input samples and FLAG IPs from TT-FHAgo2 and TT-Ago2 cells expressing Ago2 at wildtype levels (Figure 2A). In all cases, samples were prepared and processed independently from biological replicates. FLAG IP resulted in near complete depletion of FHAgo2 from cell lysate (Figure S2A) demonstrating that we captured the vast majority of cellular Ago2 complexes. Immunoprecipitated small RNAs were size selected (18–75 nt) and cloned by a cDNA circularization-based procedure that will clone RNA independent of the 5′ end modification in order to capture all potential small RNA substrates. TT-FHAgo2 and TT-Ago2 input samples exhibited near identical levels of small RNA expression (Figure S2B). The processing pipeline to identify Ago-bound small RNAs is summarized in Figure S2C. An empirical false discovery rate (FDR) was determined for increasing enrichment levels based on TT-FHAgo2 and reciprocal TT-Ago2 FLAG IP comparisons. This analysis determined that an enrichment cutoff of 3-fold corresponded to a FDR < 0.02 for FHAgo2 enriched small RNAs (Figure S2D). Small RNAs enriched above this threshold or, for the subset not sequenced in TT-Ago2 cells represented by at least two reads in both TT-FHAgo2 IP biological replicates, are classified as Ago-bound (Table S2). We plotted these data comparing enrichment and abundance in TT-FHAgo2/TT-Ago2 IPs to allow comparisons between small RNA classes (Figure 2B). As previously reported, the most abundant and largest class of small RNAs bound to Ago2 was the annotated microRNA (Figure 2C). In line with our Ago-deficient small RNA profiling, most snoRNA and tRNA fragments are not bound to Ago2 apart from a few examples (Figure S2E). We focused our characterization on small RNAs derived from the genic region of protein-coding transcripts as members of this subset were selectively and reproducibly bound by Ago2 (Figure 2D).
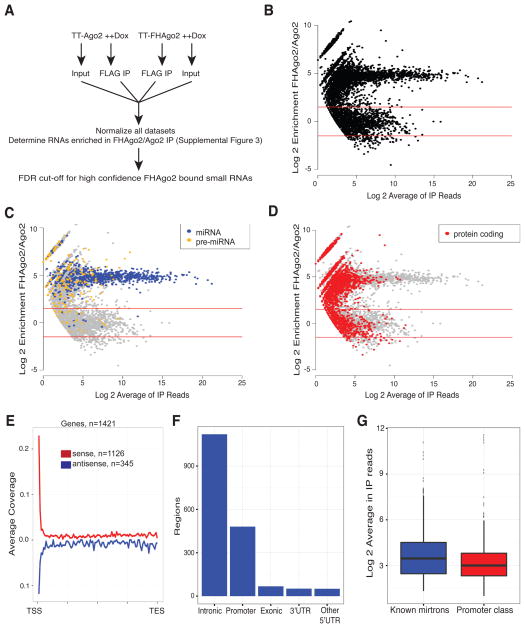
Figure 2. FHAgo2 immunoprecipitations identify small RNAs bound to Ago2 derived from protein-coding genic regions.
(A) Setup for the identification of Ago2-bound small RNAs in TT-FHAgo2 and negative control TT-Ago2 mESCs. Performed in biological duplicate. (B) Log2 mean fold-change and average reads (MA plot) of TT-FHAgo2 relative to TT-Ago2 after FLAG-IP. Red lines represent an enrichment cutoff of 3-fold empirically determined to have a false discovery rate < 0.02 for FHAgo2 enriched RNA. (C, D) MA plot from panel B color-coded to highlight various small RNA classes. Small RNAs not indicated in the figure legends are colored gray. (E) Meta profile of identified Ago2-bound regions with respect to full-length protein-coding genic regions. Red line indicate regions derived from the sense orientation and blue is antisense. The number of regions considered in each category is indicated. (F) Barplot of mRNA-derived Ago-bound regions defined by genic elements. (G) Expression comparison of TT-FHAgo2 FLAG-IP reads for promoter-proximal Ago-bound RNAs compared to candidate mirtrons. See also Figure S2, Table S2 and Table S3.
To increase the likelihood of identifying small RNAs being specifically processed for Ago binding, the Ago2-bound small RNAs derived from within mRNA transcribed regions for protein-coding genes were processed through an additional filter. We required at least five sequenced 5′ ends that vary by 2 nt or less at each region in the combined TT-FHAgo2 IP duplicates. After application of this filter, candidate Ago2-bound small RNAs from protein-coding transcripts were abundantly represented from intronic and exonic regions (Figure S3A). The intronic sequences include candidate mirtrons, some of which have been previously identified by conglomerative analysis of published mouse and human small RNA datasets (Ladewig et al., 2012). Our Ago-bound regions included 199 of these mirtron candidates, in addition to identifying 223 novel mirtron regions. These filtered candidate Ago-bound protein-coding regions are outlined in Table S3. Enriched small RNAs derived from within protein-coding RNAs were next analyzed with respect to position within the full-length gene revealing a bias toward the 5′ end of mRNA (Figure 2E). The classification based on genic structures revealed a significant group of Ago-bound small RNAs from the promoter region defined as +/− 500bp from the TSS (Figure 2F). This promoter group, which is present to a similar level as the non-canonical mirtron class (Figure 2G), was selected for further characterization.
RNAPII protein-coding gene promoters produce Ago-bound small RNAs
Meta-analysis was performed for collapsed 5′ ends of small RNAs classified as Ago-bound and within a 500-bp window flanking the TSS of protein-coding genes. To exclude potential mirtrons near the TSS, regions that overlapped with annotated introns were removed. The remaining 5′ ends displayed bimodal peaks just downstream of the TSS (Figure 3A) likely corresponding to arms of a Dicer processed product. In fact, 62 enriched promoter-proximal regions were orientated in tandem pairs downstream of the TSS suggesting processing from a hairpin precursor. Ago-bound small RNAs from antisense transcription were also observed and within a similar distance to the upstream RNAPII of divergent transcription. The promoters of protein-coding genes that overlap Ago-bound small RNAs were next separated from other protein-coding gene promoters to compare the small RNA size distribution in TT-FHAgo2 and control TT-Ago2 IP samples. The promoters overlapping Ago2-bound small RNAs showed a distinct size profile in the TT-FHAgo2 samples in the 21–24 nt range (Figure 3B, top), a size that is characteristic of known miRNAs. This difference was less pronounced in the other promoters that were not classified as producing Ago2-bound small RNA (Figure 3B, bottom). Pathway analysis using DAVID bioinformatics resources (Huang da et al., 2009a, b) of the genes that overlap Ago-bound small RNAs producing promoters shows enrichment in terms phosphoproteins and nucleus and the statistics are shown in Figure S3B. To test if the Ago-bound small RNA regions are present in the mature mRNAs produced by the shared RNAPII promoters, we cloned and sequenced full-length polyadenylated mRNA from three overlapping promoters. In each case, the region producing Ago-bound small RNA was present in the mature mRNA transcript (Figure S3C). Interestingly, the Ago-bound small RNA levels do not correlate with the cognate mESC mRNA level suggesting biogenesis may be independent of the full-length mRNA (Figure S3D, top). These results support the identification of a distinct group of RNAPII promoters of protein-coding genes that produce Ago-bound small RNAs.
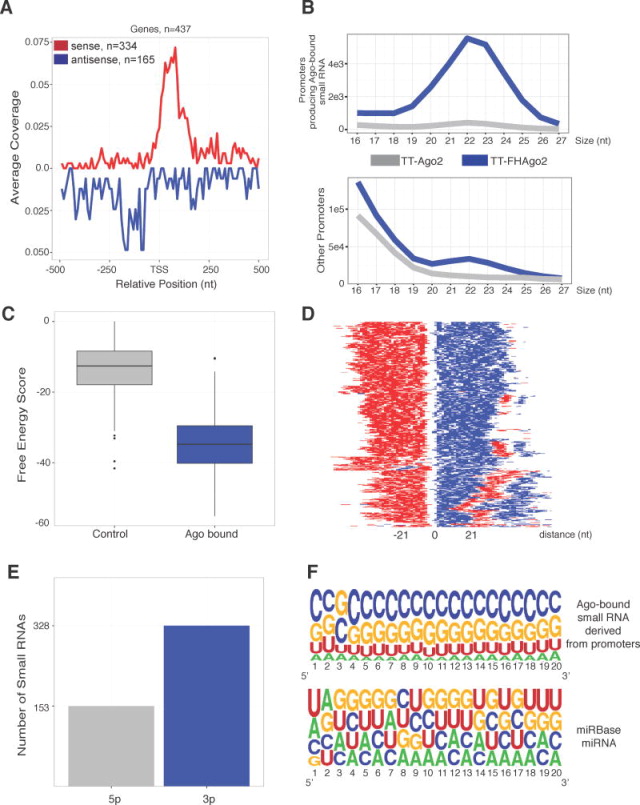
Figure 3. RNAPII protein-coding promoters can generate hairpin structures resulting in Ago-bound small RNAs.
(A) Meta-analysis of the 5′ ends of collapsed Ago-bound small RNAs within +/− 500 nt at the transcriptional start sites (TSS) of protein-coding genes. Red line is sense to mRNA and blue is antisense. (B) Size profile of small RNAs cloned from genes generating Ago-bound small RNAs (top) compared to other promoters in TT-FHAgo2 or TT-Ago2 FLAG-IP samples (bottom). (C) RNAfold free energy scores for predicted precursors of Ago-bound small RNA near TSS as compared to control set. (D) Heatmap representation of predicted precursors centered on the longest unstructured region. Example RNAfold output: ((((.((.(((((.(....).))))).))))))).))))). For heatmap, “(“-red, “.”-white, “)”-blue. (E) Barplot of the predicted hairpin arm (either 5p or 3p) generating Ago-bound small RNA from promoter region. (F) Nucleotide frequency plot for the first 20 nt of either Ago-bound promoter-proximal small RNAs (top) or all miRBase annotated miRNAs (bottom) generated using Weblogo (Crooks et al., 2004). See also Figure S3.
We hypothesized that the subset of promoters that produce Ago-bound small RNAs differ from other promoters due to the generation of highly structured RNA sequences near the TSS. To test this, the secondary structure was predicted for paired regions or for the most stable 50 nt extended RNA region containing the Ago2-bound small RNA with RNAfold (Hofacker, 2009) and was compared to randomly selected, size and TSS-proximity controlled sequences (Figure 3C). The free energy scores from protein-coding promoters that generate Ago-bound small RNAs predicted substantially higher structured RNA as compared to the control set. To illustrate the predicted secondary structures, output was graphically represented and aligned by centering on the largest predicted stem loop area (Figure 3D). As expected from the free energy scores hairpin structures were predicted from these sites. We find Ago-bound small RNAs are predicted from the 3p arm of hairpins at approximately a 2:1 ratio as compared to the 5p arm (Figure 3E). Finally, to explore sequence composition, the nucleotide frequency of Ago-bound RNAs derived from these promoter regions was plotted (Figure 3F, Top) and compared to all annotated mouse miRNAs (Figure 3F, Bottom). The Ago-bound small RNAs from protein-coding promoters are strikingly high in G/C content consistent with the strong secondary structure of predicted precursors and their origin from near CpG island promoters (87% of these promoters contain CpG islands). This distinct nucleotide profile potentially expands the seed pool of Ago-bound small RNAs and potential target RNAs in mESCs.
This analysis supports the presence of strong hairpin structures near the TSS of a subset of RNAPII promoters, which produce both Ago-bound small RNAs and full-length protein-coding mRNA transcripts.
TSS-miRNAs are Ago/Dicer-dependent products derived from RNAPII paused regions
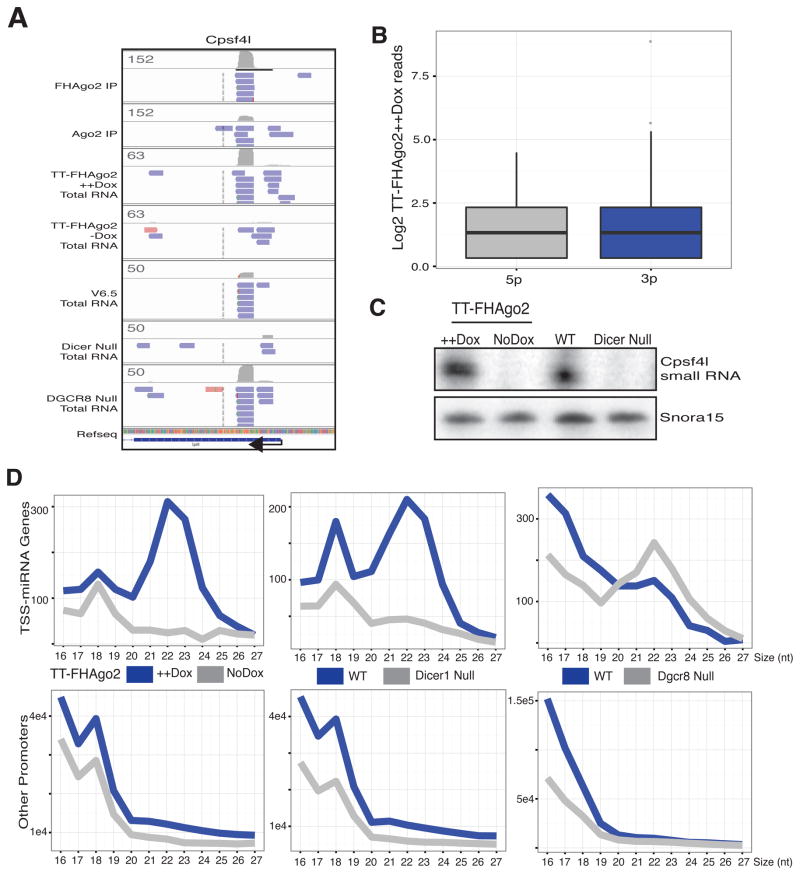
To determine if Ago-bound promoter-proximal small RNAs are dependent on RNAi proteins for steady-state accumulation in-vivo, we returned to our small RNA analysis of miRNA-deficient cells. The cleavage and polyadenylation specific factor 4-like (Cpsf4l) promoter, which produced the most abundant Ago-bound small RNA from promoter-proximal regions, was selected to display the collapsed small RNA reads from the mESC libraries considered in our analysis (Figure 4A). The genome browser shot shows the coverage and example collapsed reads for TT-FHAgo2 and TT-Ago2 FLAG IPs and three RNAi-mutant libraries with their respective wildtype control. The total small RNA libraries were sequenced in a 5′ phosphate dependent manner and therefore reveal the nature of the 5′ end of the Ago-enriched small RNA. The Ago-bound small RNA was dramatically decreased with loss of either Ago or Dicer and readily detected in Dgcr8 null cells. Previously, 17–20 nt promoter-proximal small RNAs were suggested to be RNAPII protected fragments (Valen et al., 2011) and are likely the remaining shorter reads in Ago or Dicer. In both TT-FHAgo2 ++ Dox and Dgcr8 null libraries both arms of the potential Dicer product are detected suggesting each ultimately obtain a 5′ phosphate required for sequencing. Similar results observed for two additional promoter regions are shown in Figure S4A. The abundance is these Ago-bound small RNAs are estimates at approximately 63, 12 and 3 copies per cell for Cpsf4l, Glul and Krcc1 promoters, respectively (Figure S4B). To further probe the 5′ structure, we quantified the reads corresponding to 5p and 3p arm Ago-bound products from promoter regions in TT-FHAgo2 ++ Dox samples and found each represented in this 5′ phosphate dependent library (Figure 4B), which is consistent with known requirements for Ago2 association (Chen et al., 2008). The loss of the Cpsf4l promoter-derived Ago-bound small RNA in Dicer and Ago null cells was confirmed by a splint-ligation method as compared to the Snora15 control (Figure 4C). Next, the size profile of collapsed reads from RNAPII protein-coding gene promoters that produce Ago-bound small RNAs compared to other protein-coding promoters was used to show changes in this promoter class. Ago depletion in TT-FHAgo2 mESCs resulted in loss of the 21–24 nt small RNAs generated from promoters producing Ago-bound small RNA (Figure 4D, Left). The in-vivo Ago-dependence for stability supports our Ago2 IP results and validates Ago2 association. Based on the agreement in the two experimental datasets, we name this new Ago-bound small RNA class TSS-miRNAs (Transcriptional Start Site miRNAs) based on their generation from RNAPII gene promoters.
Figure 4. TSS-miRNAs are non-canonically processed, Ago and Dicer-dependent small RNAs.
(A) IGV genome browser shot for the Cpsf4l promoter small RNAs cloned from indicated samples. Collapsed reads are blue or red denoting minus strand or plus strand, respectively. Gray barplots show coverage and scale numbers reflect the raw reads. Arrow indicates the direction of transcription. (B) Quantitation of Ago-bound promoter class in TT-FHAgo2 ++ Dox total RNA sequenced in a 5′ phosphate dependent manner. These have been separated by observed or predicted arms of a potential hairpin precursor. (C) Splint ligation-mediated detection of Cpsf4l TSS-miRNA in total RNA samples from TT-FHAgo2 cells treated with high Dox (++) for 48hrs or Dox starvation for 96 hrs or wildtype and Dicer null mESCs. (D) Size profiles of small RNAs in total small RNA sequencing from 5′ phosphate dependent cloning of TT-FHAgo2 with and without Dox, Dicer or Dgcr8 null mESCs compared to their respective wildtype controls. As in Figure 3B, these are presented as RNAPII protein-coding promoters that yield Ago-bound small RNAs (TSS-miRNAs) and other annotated mouse protein-coding promoters. See also Figure S4.
Dicer protein loss also resulted in a small RNA size profile shift from TSS-miRNA promoters suggesting Dicer acts in the processing of precursor hairpins (Figure 4D, Center). No difference was observed with Dgcr8 loss implying, in contrast to canonical miRNAs, TSS-miRNAs are generated independent of this protein (Figure 4D, Right). Drosha null mouse embryonic fibroblasts (Chong et al., 2010) also showed the same TSS-miRNA size profile as wildtype indicating TSS-miRNA biogenesis is independent of the microprocessor complex (Figure S4C).
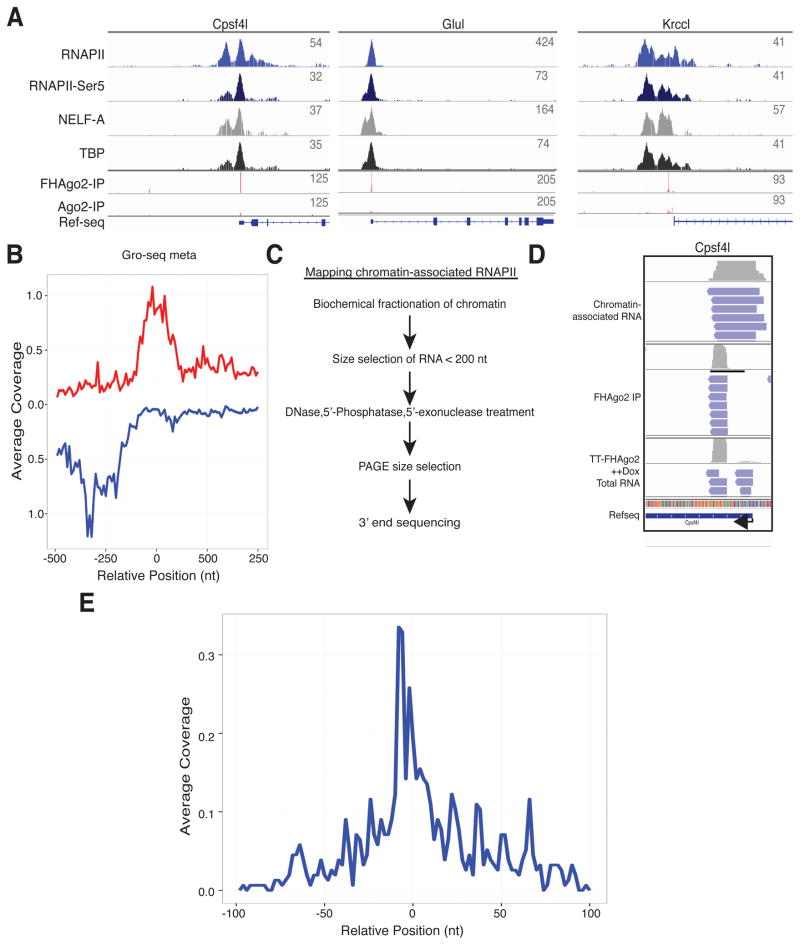
We next examined promoter-proximal RNAPII and other transcription factors from mESC ChIP-seq data (Rahl et al., 2010) near TSS-miRNA regions (Figure 5A). TSS-miRNAs showed a striking overlap with the promoter-proximal carboxyl-terminal domain serine 5 phosphorylated RNAPII associated with the transcriptional pausing complex NELF. This indicated a potential source of hairpin precursor formation. For higher resolution positioning of the RNAPII complex, global nuclear run-on (Gro-seq) mESC data (Min et al., 2011) that measures engaged RNAPII transcription at high-resolution was analyzed. Meta-analysis was performed surrounding the TSS-miRNA 3′ end of sense 3p arm products (Figure 5B). We found the sense RNAPII peak directly overlapped with the 3′ end of TSS-miRNAs. Quantitation and profiles of Gro-seq identified RNAPII complexes at these TSS-miRNA promoters compared to a matched control set showed the RNAPII levels are lower and peak slightly closer to the TSS at promoters producing TSS-miRNAs (Figure S5A). The above difference in the Gro-seq signal was not observed in ChIP-seq data suggesting a difference in transcriptional elongation at these promoters. Next, we adapted a protocol for high-resolution mapping of actively transcribing RNAPII complexes previously developed in Drosophila (Nechaev et al., 2010). In this protocol, stringent fractionation is used to isolate chromatin followed by 5′-exonuclease treatment and 3′ end sequencing allowing nucleotide level positioning of the 3′ ends of capped/structured RNAs size fractionated here to 50–100 nt for sequencing (Figure 5C). We found a striking overlap with reads from the chromatin-associated RNA and the region corresponding to the Cpsf4l TSS-miRNA that overlapped both TSS-miRNA arms and aligned to the region that included the exact 3′ end of the TSS-miRNA (Figure 5D). Meta analysis of the 3′ end of chromatin-associated capped small RNAs aligned mostly to the 3′ end of sense 3p arm TSS-miRNA supporting biogenesis from transcription and release by promoter-proximal RNAPII (Figure 5E). Biogenesis in this manner would be consistent with microprocessor-independent hairpin formation. Finally, Cpsf4l TSS-miRNA was measured in cells containing knockdown of RNAPII pause factors as previously described (Flynn et al., 2011). We found knockdown of Nelf-E and Supt5 resulted in minor, yet reproducible, increases in TSS-miRNA levels that can be decoupled from changes in mature mRNA levels (Figure S5B).
Figure 5. TSS-miRNA are derived from RNAPII promoter-proximal paused regions.
(A) Profile of chromatin-associated RNAPII, RNAPII-Ser5, NELF-A and TATA binding protein (TBP) as determined by ChIP-seq in mESC at three representative TSS-miRNA promoters. The coverage of collapsed sequenced reads is shown for TT- FHAgo2 and TT-Ago2 FLAG immunoprecipitations along with the Refseq annotation. (B) Meta-analysis of global nuclear run-on (Gro-seq) reads representing actively engaged RNAPII complex relative to the 3′ end of 3p arm TSS-miRNAs in the sense direction at position “0”. Profiles are colored red (sense) or blue (antisense) with respect to the mature mRNA. (C) Flowchart for isolation of chromatin-associated small RNA libraries. (D) IGV browser shot of TT-FHAgo2 immunoprecipitations or total RNA from TT-FHAgo2 treated with high Dox (++) for 48hrs as compared to RNA generated from chromatin-associated RNA isolation at the Cpsf4l promoter. (E) Meta plot of chromatin-associated small RNAs 3P ends aligned to the 3′ end of 3p arm TSS-miRNAs in the sense orientation at position “0”. See also Figure S5.
Functional characterization of Cpsf4l TSS-miRNA
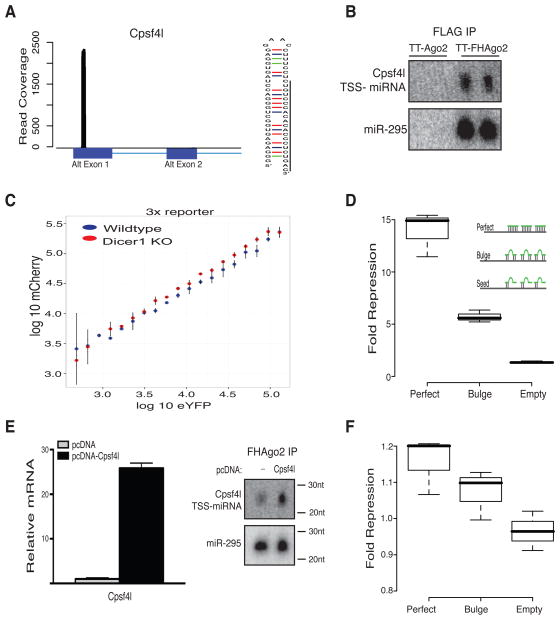
The read coverage near the Cpsf4l gene TSS showed enrichment of this small RNA compared to other exonic Cpsf4l sequence and precursor region supported a hairpin structure (Figure 6A). Northern blots were used to confirm the small RNA sequence and enrichment in FHAgo2 IPs (Figure 6B). Cpsf4l TSS-miRNA was detected specifically in TT-FHAgo2 FLAG IPs in a manner similar to a positive control miR-295.
Figure 6. Characterization of Cpsf4l TSS-miRNA post-transcriptional regulatory activity.
(A) Read coverage of TT-FHAgo2 FLAG IP small RNA near the Cpsf4l TSS with annotated exons below (left). Predicted hairpin structure for Cpsf4l TSS-miRNA precursor (right). (B) Northern blot after FLAG-IP from TT-FHAgo2 or TT-Ago2 mESCs treated 48hrs with high Dox (++). miR-295 serves as positive control. Please note: blot exposure times differed with each probe. (C) Scatterplot of single-cell bidirectional dual fluorescent reporter expression measured by flow cytometry to detect endogenous Cpsf4l TSS-miRNA activity. Points represent binned targeted mCherry log10 by control eYFP log10 expression in wildtype (blue) or Dicer−/− mESCs (red). The targeted mCherry contained three perfect matches to Cpsf4l TSS-miRNA in the 3′ UTR. Points and error bars are mean and standard deviation of independent biological replicates. (D) Ago2-dependent repression of a luciferase reporter containing a 3′ UTR with three perfect, bulged or seed mutant sites complementary to the Cpsf4l TSS-miRNA. Cpsf4l TSS-miRNAs mimic was used at 100nM with and without FHAgo2 expression (completed in biological triplicate) and normalized to the seed mutant. Inset: graphical representation of reporter constructs. (E) Relative mRNA overexpression upon transfection of pcDNA-Cpsf4l containing the full-length mRNA as compared to empty vector (left) and Northern blot following FLAG-IP in TT-FHAgo2 mESCs (right) with and without mRNA overexpression. (F) Repression of luciferase reporter constructs containing a 3′ UTR with three perfect, bulged or seed mutant sites for the Cpsf4l TSS-miRNA upon mRNA overexpression. Values for transfection with pcDNA-Cpsf4l mRNA were compared to empty vector (completed in biological triplicate). p-value < 0.05 determined for the difference between perfect sites and empty control. Please note: restricted y-axis used to show repression differences. See also Figure S6 and Table S4.
To detect endogenous Cpsf4l TSS-miRNA activity, flow cytometry measurements after transfection of a dual fluorescence bidirectional reporter construct for quantitation of miRNA activity at the single cell level was used (Mukherji et al., 2011). The dual reporter constructs contains a tet-inducible bidirectional promoter for expression of mCherry mRNA containing a 3′ UTR with miRNA binding sites and enhanced YFP (eYFP) with no miRNA sites as a transcriptional control (Figure S6A, B). Using constructs with and without three perfectly complementary Cpsf4l TSS-miRNA mCherry sites in wildtype or Dicer null cells, sequence-specific and Dicer-dependent endogenous Cpsf4l TSS-miRNA activity was measured. The cells transfected with mCherry Cpsf4l TSS-miRNA complementary sites (3x) showed decreased mCherry expression in wildtype cells compared to Dicer knockout cells (Figure 6C, p-value < 0.02). The reporter without binding sites (0x) did not show this difference suggesting sequence-specific repression (Figure S6C). This repressive activity ranged from 1–5% repression consistent with the low expression levels of Cpsf4l TSS-miRNA (Figure S6D), but supports functional Ago loading and activity in vivo. Next, Cpsf4l TSS-miRNA targeted luciferase reporter constructs were generated with 3′ UTRs harboring perfectly complementary sites, bulged complementarity sites or seed mutant sites (Figure 6D). Reporters were expressed with a Cpsf4l TSS-miRNA mimic in TT-FHAgo2 cells with or without Dox to test Ago2-dependent post-transcriptional repression. Only in the presence of Ago2 did the Cpsf4l TSS-miRNA mimic repress the perfect or bulged reporter targets in this cell population-based assay. These results confirm Cpsf4l TSS-miRNA in an Ago-dependent manner repress complementary RNA targets. Similar Ago-dependent regulation of a luciferase target was found with a second TSS-miRNA (Figure S6E).
To further support TSS-miRNA biogenesis during mRNA transcription, we overexpressed full-length spliced Cpsf4l mRNA and measured the changes in TSS-miRNA production and repressive activity. Cpsf4l mRNA overexpression resulted in increased mature TSS-miRNA loaded into Ago2 (Figure 6E). To determine if the small RNA derived from Cpsf4l mRNA overexpression was functional, miRNA luciferase reporter constructs were co-transfected with the Cpsf4l mRNA overexpression construct (Figure 5F, fold repression scale 0.8–1.2). We observed statistically significant repression of the perfectly complementary reporter and increased repression of the bulged targeting construct indicating overexpressed TSS-miRNA mediated Ago2-dependent repression. These data also indicate that increased transcription results in increased production of both mRNA and functional mature TSS-miRNA, which occurs in a splicing-independent manner. TargetScan (Lewis et al., 2005) predictions for Cpsf4l TSS-miRNA are included in Table S4.
As small RNAs targeting promoter regions have previously been suggested to regulate gene transcription (Janowski et al., 2005; Kim et al., 2008) we also tested if TSS-miRNAs could act to regulate its cognate full-length mRNA or RNA transcribed in the antisense direction (Figure S6F,G). At both regions examined, RNA changes across the TSS-miRNA region in Dicer nulls were not rescued with a TSS-miRNA mimic. These data indicate exogenously introduced TSS-miRNA have limited potential for feedback regulation for the tested RNA targets.
Together, these experiments confirm that TSS-miRNAs derived from RNAPII transcription are processed into small RNAs, loaded into Ago2 and bind complementary RNA for repression.
TSS-miRNAs are differentially expressed in mouse and human tissue and show evidence of conservation
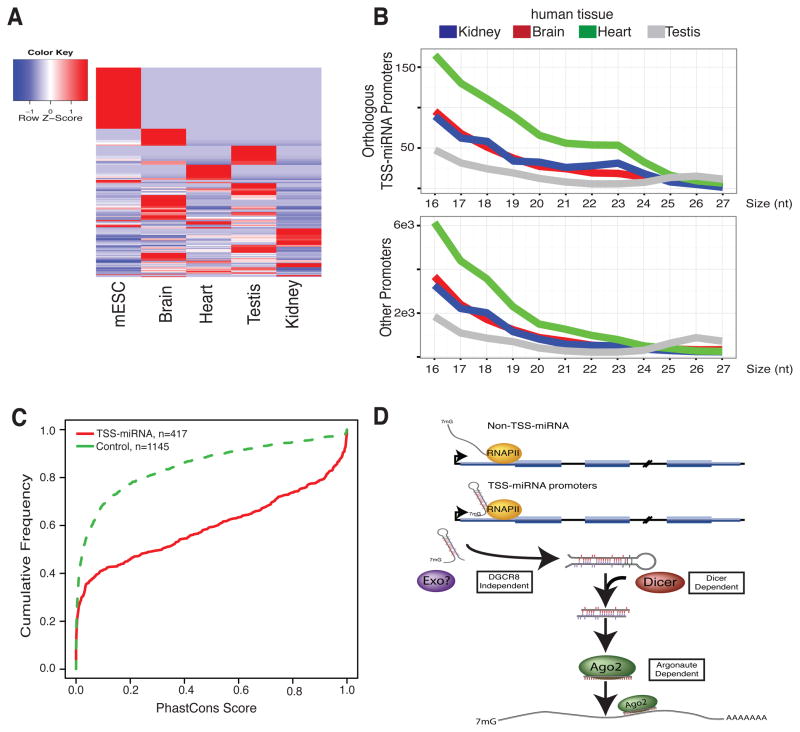
To address if TSS-miRNAs are detected in other mouse or human samples, high-sequencing depth small RNA data from mouse and human tissues (Meunier et al., 2013) were analyzed. We exclusively examined TSS-miRNA promoters identified in mESCs for quantitation. However, this does not exclude the possibility of other tissue-specific TSS-miRNAs. First, the detection of TSS-miRNAs in different tissue types supported widespread and tissue specific expression in mice. The variation in TSS-miRNA levels observed in the mouse tissues was quantified and indicated differential expression dependent upon the biological context (Figure 7A, Figure S7A). These changes again did not correlate with mature mRNA levels in tissue-matched samples (Figure S7C). Next, examination of the orthologous mESC TSS-miRNA RNAPII promoters in human tissue datasets showed similar distinctions in the size profiles of collapsed small RNA reads indicating conservation of some TSS-miRNA expression in humans (Figure 7B, Figure S7B). To further examine the sequence conservation, the phastCons (Siepel et al., 2005) score from the UCSC 30-way placental mammal annotation was quantified over TSS-miRNA regions and compared to expression level, CpG island and nucleotide composition matched regions with similar size and distance distribution from the TSS (Figure 7C) showing association with conserved sequence elements. Together these data support that TSS-miRNAs are expressed to different levels during cellular differentiation and are likely a class of non-canonical miRNA also acting in humans.
Figure 7. TSS-miRNAs are differentially expressed in mouse tissue samples and conserved in humans.
(A) Heatmap of mESC TSS-miRNAs expression from four mouse tissue types colored by z-score of expression as indicated in the color legend. (B) Small RNA size profile from collapsed reads of human tissues for orthologous mESC TSS-miRNAs RNAPII promoters and other annotated protein-coding RNAPII promoters. Colors reflect human tissue sample as indicated. (C) Cumulative distribution function plot comparing the average phastCons score from UCSC placental mammal alignments between TSS-miRNAs and a matched control sequence set. (D) Model figure of biogenesis of TSS-miRNAs. See also Figure S7.
Discussion
As the primary mediator of miRNA function, describing the complete contingent of Ago-bound small RNAs is key to understanding the full role of RNAi in mammalian cells. Here, we have introduced an adapted mESC system that allows for Dox-inducible expression of a single Ago protein. The advantages of this system include control of FHAgo2 expression levels from wildtype to undetectable and near complete depletion from cell lysates by FLAG-IP. We describe the global changes of small RNAs upon complete loss of all Ago proteins. We find canonical and non-canonical miRNAs are destabilized upon Ago loss and can be rescued by expressing Ago, confirming that Ago proteins are required for stability of small RNAs acting in the RNAi pathway.
We have utilized this system to discover a novel class of Ago-bound small RNAs, TSS-miRNAs, which are derived from RNAPII protein-coding gene promoters (Figure 7D). We show that a subset of RNAPII promoters have a predicted proclivity to generate stable hairpin structures that produce Ago-bound 21–24mer TSS-miRNAs. Similar to other miRNAs, Ago protein is required for TSS-miRNA steady-state accumulation, supporting their in-vivo Ago association and reducing concerns of post-lysis binding. Maturation of TSS-miRNAs is Dicer-dependent and microprocessor independent. Dgcr8/Drosha independence, lack of correlation to mature mRNA levels, and a striking alignment to promoter-proximal RNAPII all suggest early termination of RNAPII transcription generates TSS-miRNA hairpin precursors. In fact, the 3′ end of TSS-miRNAs corresponds to the position of paused, engaged RNAPII. Based on the position of 5p arm TSS-miRNA downstream of the TSS and their 5′ phosphates, we propose the cap structure is removed from products released by promoter-proximal RNAPII and processed at the 5′ end. Since decapping and 5′-3′ exonuclease activities have recently been suggested to occupy promoter regions (Brannan et al., 2012), this likely occurs before action of Dicer. What is distinct about TSS-miRNA promoters? Based on the link between the formation of RNA hairpins and RNAPII pausing in the bacterial intrinsic transcriptional termination, we propose the hairpin structure destabilizes the RNAPII complex and releases a precursor for incorporation into the RNAi pathway. In bacteria, RNA hairpins have been shown to weaken RNA:DNA interactions (Farnham and Platt, 1981; Nudler et al., 1997) by inducing allosteric conformational changes in the polymerase within conserved polymerase subunits (Liphardt et al., 2001). The release of short, promoter-derived hairpins has also been previously reported from Polycomb-targeted promoters in mESCs (Kanhere et al., 2010); however, we see no correlation between TSS-miRNAs promoters and Polycomb-regulated genes.
TSS-miRNAs are able to function in an Ago-dependent manner to regulate reporter constructs in a siRNA or miRNA-like fashion. Probably because of the proximity to CpG islands within promoter regions, TSS-miRNAs are significantly GC-rich. TSS-miRNAs can be detected in small RNA-seq data across multiple mouse tissues and show differential expression. Furthermore, some TSS-miRNAs were found at orthologous human promoters potentially indicating a conserved biological function. The application of a single-cell based reporter assay allowed detection of activity from endogenous lowly expressed TSS-miRNA. Consistent with the low regulatory activity on reporters and likely low ratio of TSS-miRNA to target mRNA pool, we do not detect global shifts in predicted targets with Dicer loss (data not shown). Thus, the relative contribution of TSS-miRNAs to the total silencing potential of mammalian Ago proteins is unclear and will be aided by the development of sensitive methods to identify in vivo targets of lowly expressed miRNA. Similar to miRNAs located within introns, the linked expression by shared promoter activity of TSS-miRNAs and mRNA may result in co-regulation of molecular pathways.
The possibility of single-stranded RNA loading into Ago proteins (Lima et al., 2012; Okamura et al., 2013) expands the potential for miRNAs processed from various cellular RNAs and underscores the importance of stringent criterion for miRNA classification. As demonstrated with the identification of TSS-miRNAs, the rigorous measurement of Ago association in IP experiments and in-vivo Ago-dependence for accumulation could be used in concert to validate true small RNAs acting in the RNAi pathway. Generally, RNA fragments such as those derived from snoRNA and tRNA, are not Ago-bound suggesting an underlying specificity of association: a specificity likely mediated by Dicer processing or hairpin formation. Future characterization of novel small RNAs with RNAi activities will be aided by classification based on both Ago-association and Ago-dependent stability.
Experimental Procedures
Detailed procedures are described in the Extended Experimental Procedures.
Generation of doxycycline inducible FHAgo2 clonal cell lines
E7 cells were infected with pSLIK lentivirus (Shin et al., 2006) with TRE-Tight (TT) Ago2 or FHAgo2. For clonal selection, TT-FHAgo2 or TT-Ago2 cells were plated with 1μM 4-hydroxytamoxifen (4-OHT) (Sigma) and doxycycline (Sigma) and grown until single colonies were isolated by trypsinization in sterile cloning cylinders (Bel-Art Products). Loss of the 4-OHT regulated hAgo2 transgene was verified by selection in Blasticidin S (Invitrogen) and by PCR amplification of the BsdS gene from 50 μg genomic DNA isolated with the GenElute Mammalian Genomic DNA Miniprep kit (Sigma).
Small RNA cloning
RNA between 18–75 nt was size selected via denaturing polyacrylamide gels. 5′ phosphate independent cloning was performed with a protocol adapted from (Churchman and Weissman, 2011). A detailed protocol is available on request. 5′ phosphate dependent cloning was performed using the NEBNext Small RNA Library Prep Set for Illumina (New England Biolabs).
Supplementary Material
Acknowledgments
We thank R. Blelloch and X. Wang for cells and the Swanson Biotechnology Center at the Koch Institute and the MIT BioMicro Center for sequencing support. This work was supported by United States Public Health Service grants RO1 GM34277 from the NIH, PO1-CA42063 from the NCI to PAS and partially by Koch Institute Support (core) grant P30-CA14051 from the NCI. JRZ and TJK were supported by NIH NSRA F32CA139902 and F32GM101872, respectively. The content of the project described is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The authors wish to dedicate this paper to the memory of MIT Police Officer Sean Collier, for his caring service to the MIT community and for his sacrifice.
Footnotes
Accession Number
The NCBI Gene Expression Omnibus number for sequencing data: GSE50595.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes & development. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Molecular cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, Meister G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes & development. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Molecular cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Farnham PJ, Platt T. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic acids research. 1981;9:563–577. doi: 10.1093/nar/9.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci U S A. 2011;108:10460–10465. doi: 10.1073/pnas.1106630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-Dgcr8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hofacker IL. RNA secondary structure analysis using the Vienna RNA package. In: Baxevanis Andreas D, et al., editors. Current protocols in bioinformatics. Unit 12. Chapter 12. 2009. p. 12. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat Chem Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Molecular cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Res. 2012;22:1634–1645. doi: 10.1101/gr.133553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lima WF, Prakash TP, Murray HM, Kinberger GA, Li W, Chappell AE, Li CS, Murray SF, Gaus H, Seth PP, et al. Single-stranded siRNAs activate RNAi in animals. Cell. 2012;150:883–894. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute 2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu H, Khaitovich P, Kaessmann H. Birth and expression evolution of mammalian microRNA genes. Genome Res. 2013;23:34–45. doi: 10.1101/gr.140269.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes & development. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nature genetics. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes & development. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ladewig E, Zhou L, Lai EC. Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes & development. 2013;27:778–792. doi: 10.1101/gad.211698.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel S, Wang Y, Lenobel R, Korner R, Hsiao HH, Urlaub H, Patel D, Meister G. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic acids research. 2011;39:2330–2343. doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci U S A. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes & development. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valen E, Preker P, Andersen PR, Zhao X, Chen Y, Ender C, Dueck A, Meister G, Sandelin A, Jensen TH. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat Struct Mol Biol. 2011;18:1075–1082. doi: 10.1038/nsmb.2091. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA biology. 2011;8:1149–1157. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.