Abstract
Voltage-gated sodium channels are known to be expressed in neurons and other excitable cells. Recently, voltage-gated sodium channels have been found to be expressed in human prostate cancer cells. α-Hydroxy-α-phenylamides are a new class of small molecules that have demonstrated potent inhibition of voltage-gated sodium channels. The hydroxyamide motif, an isostere of a hydantoin ring, provides an active scaffold from which several potent racemic sodium channel blockers have been derived. With little known about chiral preferences, the development of chiral syntheses to obtain each pure enantiomer for evaluation as sodium channel blockers is important. Using Seebach and Frater's chiral template, cyclocondensation of (R)-3-chloromandelic acid with pivaldehyde furnished both the cis- and trans-2,5-disubsituted dioxolanones. Using this chiral template, we synthesized both enantiomers of 2-(3-chlorophenyl)-2-hydroxynonanamide, and evaluated their ability to functionally inhibit hNav isoforms, human prostate cancer cells and xenograft. Enantiomers of lead demonstrated significant ability to reduce prostate cancer in vivo.
Keywords: Ion channel blocker, Prostate cancer, Frater, Seebach
1. Introduction
Voltage-gated sodium channels are important transmembrane proteins critical in the generation and propagation of signals in electrically excitable tissues like muscles, the heart, and nerves. They are comprised of a pore forming α-subunit, and auxiliary β-subunits.1 To date, nine α-subunit isoforms have been cloned along with four auxiliary β-subunit isoforms.2 These nine sodium channel isoforms are classified by their sensitivity to the neurotoxin tetrodotoxin (TTX). There are six TTX-sensitive isoforms: Nav1.1, Nav1.2, Nav1.3, Nav1.4, Nav1.6, and Nav1.7; and three TTX-resistant isoforms: Nav1.5, Nav1.8, and Nav1.9.3 Pharmacological modulation of voltage-gate sodium channels has proven clinically beneficial for the treatment of pain, epilepsy, depression, and cardiac arrhythmias.3
The role of ion channels in cancer is an emerging field. Recent studies have demonstrated that voltage-gated ion channels could play a role in the onset, proliferation and malignant progression of various types of cancer, such as prostate, colon, and glioma.4–7 Specifically, the voltage-gated sodium channel has been shown to play a role in cancer cell proliferation, migration, and adhesion.8 However, the signaling pathways involved in cancer progression are yet to be elucidated.9
Previously, Brown et al.10 described the design and synthesis of 2-(3-chlorophenyl)-2-hydroxynonanamide and its effect on sodium channels.11 Given the increasing focus of the effect of stereochemistry on compound efficacy and toxicity, we desired to synthesize each enantiomer of 3-chlorophenyl-α-hydroxyamide.
The chiral template described by Seebach and Frater12 utilizes cis-2,5-disubstituted 1,3-dioxolan-4-ones, as scaffolds for the stereocontrol of alkylations, aldol additions, Michael additions, nucleophilic additions, and Mannich reactions.13–18 Cyclocondensation of a mandelic acid with pivaldehyde gives the 2,5-disubstituted dioxolanone, and alkylation is directed by the tert-butyl group. Using (R)-3-chloromandelic acid we observed both cis- and trans-2,5-disubstituted dioxolanones. In this study, we employed the use of both isomers to utilize this asymmetric strategy to synthesize each enantiomer of 3-chlorophenyl-α-hydroxyamide ((±)-1) to evaluate the chiral preference for inhibiting sodium channel isoforms.
2. Chemistry
2.1. Synthesis of compounds 1 and 2
Racemic (±)-1 was synthesized using the previously described procedure shown in Scheme 1.4 Grignard addition of heptylmagnesium bromide into 3-chlorobenzonitrile followed by acidic hydrolysis gave the desired ketone 2. Treatment of the ketone with TMSCN, followed by acidic hydrolysis furnished the racemic 2-(3-chlorophenyl)-2-hydroxynonanamide ((±)-1).
Scheme 1.

Synthesis of racemic 2-(3-chloro-phenyl)-2-hydroxy-nonanoic acid amide. Reagents and conditions: (a) (i) C7H15MgBr, THF, 0°C to rt. overnight; (ii) 1 N HCl; (b) TMSCN, KCN, 18-c-6, CH2Cl2; (c) concd HCl, HCl (gas); (d) 0 °C.
2.2. Synthesis of compounds (R)-(−)-1 and (S)-(+)-1
In Scheme 2, commercially available (R)-(−)-3-chloromandelic acid was condensed with pivaldehyde to give cis isomer dioxolanones 3 with a 5:1 (cis:trans) selectivity. Interestingly, this same reaction using the unsubstituted mandelic acid gives almost exclusively the cis-isomer. The cis and trans isomers are easily separable by column chromatography and both were isolated with high purity. Alkylation of 3 was carried out under various conditions summarized in Table 1. Initially we tried to make the enolate with freshly prepared LDA at −78 °C in THF. After the complete addition of the dioxolanone the temperature was raised to 0 °C for 10 min, and then lowered back to −78 °C. Heptyl iodide was then added dropwise at −78 °C. This procedure is known as the direct addition method. Initial alkylation of the dioxolanone in THF using this addition method gave a 2:1 (cis:trans) mixture of the alkylated product. Previously, a 1:3 cyclohexane/ether solvent mixture was shown to promote diastereoselectivity in Li enolate reactions.19 Applying these conditions to our system however, resulted in no reaction. With this in mind we ran the reaction in Et2O, hoping that the less polar solvent would result in a slower reaction rate with increased selectivity. However, allowing this reaction to run overnight resulted in no conversion of the starting material. Blay et al. reported that inverse addition and the addition of 3 equiv of HMPA to these reactions resulted in increased diastereoselectivity.17 The inverse addition protocol calls for the addition of LDA to a premixed solution of the dioxolanone, heptyl iodide, and HMPA in THF at −78 °C. Indeed, when we treated the cis-dioxolanone 3 with LDA in THF–HMPA and heptyl iodide at −78 °C the alkylated product 4 was obtained as a 1:10 ratio of diastereomers. Under these conditions the major product had the heptyl chain trans to the tert-butyl group.
Scheme 2.
Synthesis of optically pure (R)- and (S)-2-(3-chloro-phenyl)-2-hydroxy-nonanoic acid amide. Reagents and conditions: (a) (1) AcCl, MeOH, 40 °C, 87%; (2) p-nitrobenzoic acid, PPh3, DEAD, THF, 86%; (3) NaN3, MeOH, 40 °C, 72%; (4) 5% NaOH, 40 °C, 96%; (b) Pivaldehyde, TfOH, pentane, Dean-Stark; (c) LDA, C7H15I, THF-HMPA, −78 °C; (d) 7 N NH3, MeOH, rt.
Table 1. Effect of solvent and addition method on diastereoselectivity of alkylation.
| Entry | Solvent | Addition method | dr (cis:trans) |
|---|---|---|---|
| 1 | THF | Direct | 2:1 |
| 2 | THF–cyclohexane | Direct | NR |
| 3 | Et2O | Direct | NR |
| 4 | THF–HMPA | Inverse | 1:10 |
| 6 | THF | Inverse | NR |
Legend. Heptyl source = C7H15I.
The surprising results provided by the THF–HMPA system led us to consider what effect the leaving group of the electrophile would have on this reaction. We attempted the reaction with heptyl iodide, heptyl triflate, and heptyl bromide, as summarized in Table 2. Our hypothesis was that reactivity of the electrophile would strongly affect the diastereoselectivity of the alkylation product. We predicted that using heptyl triflate would result in an increased reaction rate, allowing for better selectivity. However, addition of the triflate gave a 1:4.5 mixture of stereoisomers by 1H-NMR. Reaction with heptyl bromide was incomplete, and gave a 1:1.5 mixture of stereoisomers, thus suggesting that the heptyl iodide was the most suitable heptyl source for our synthesis resulting in a 1:10 mixture stereoisomers. Our most satisfactory result, a diastereoselectivity of 6:94, was obtained using heptyl iodide and carrying out the reaction at −78 °C for 3 h, followed by quenching the reaction at that same temperature.
Table 2. Effect of electrophile on diastereoselectivity.
| Entry | Heptyl source | dr (cis:trans) |
|---|---|---|
| 1 | C7H15OTf | 1:4.5 |
| 2 | C7H15Br | 1:1.5 + SM |
| ∼ | C7H15I | 1:10 |
| 4 | C7H15I | 6:94* |
Legend. Reaction held at −78 °C for 3–4 h and quenched immediately.
Separation of the diastereomers by column chromatography was accomplished using an ether-hexanes gradient. Treatment of alkyl dioxolanone 4, with concentrated ammonium hydroxide in ethanol furnished (R)-(−)-2-(3-chlorophenyl)-2-hydroxynonanamide ((R)-(−)-1) directly in modest yields as a 91:1 ratio of enantiomers as determined by chiral HPLC. This series of reactions proceeds with retention of stereochemistry giving the (R)-enantiomer as the main product. By utilizing the synthesized (S)-(+)-3-chloromandelic acid 5 and completing the same synthetic steps as we previously accomplished with the commercially available (R)-(−)-3-chloromandelic acid, we synthesized (S)-(+)-2-(3-chlorophenyl)-2-hydroxynonanamide ((S)-(+)-1) as a 95:5 ratio of enantiomers as determined by chiral HPLC (scheme 2). The (+)-enantiomer was confirmed by X-ray crystallography to have the S-configuration (see Supplementary data) and the ligand coordinates used as a starting geometry in our docking studies.
3. Results
3.1. Na channel activity
Using a FRET based assay to measure sodium channel dependent changes in membrane potential, (±)-1, (R)-(−)-1, (S)-(+)-1 and DPH (Fig. 1) were evaluated for their isoform specific sodium channel blocking effects and the IC50 values are shown in Table 3. Two isoforms of the channel were examined: hNav1.5, a cardiac sodium channel associated with arrhythmias, and hNav1.7, an isoform found in the peripheral nervous system.20 Using a fluorescent based assay, compound (S)-(+)-1 exhibited the greatest activity for the hNav1.5 isoform of the channel with an IC50 of 4.78 μM. One enantiomer possessed slightly greater activity than the other, and the racemic mixture was observed to have intermediate activity. Evaluation of compound (R)-(−)-1 gave an IC50 = 7.43 μM and evaluation of (±)-1, the racemic mixture, gave an IC50 = 5.78 μM. These three compounds were more potent blockers of Nav1.7 channels compared with Nav1.5 channels, but displayed a different stereoselectivity. The racemic mixture (IC50 = 1.81 μM) and (R)-(−)-1 (IC50 = 1.88 μM) were similarly active against the Nav1.7 isoform, while the (S)-(+)-1 enantiomer (IC50 = 2.62 μM) was weaker as a blocker. We observed that the (S)-(+)-1 enantiomer was preferred by hNav1.5 and (R)-(−)-1 was more active against hNav1.7. DPH was not active on either hNav1.5 or hNav1.7 at concentrations less than 100 μM.
Figure 1.
Phenytoin (DPH) and 2-(3-chloro-phenyl)-2-hydroxy-nonanoic acid amide ((±)-1).
Table 3. hNav 1.5 and hNav 1.7 IC50's for racemic, (R)-, (S)-2-(3-chloro-phenyl)-2-hydroxynonanoic acid amide, and DPH.
| Compound | hNav 1.5 IC50a (μM) | hNav 1.7 IC50a (μM) |
|---|---|---|
| (±)-1 | 5.78 ± 1.2 | 1.81 ± 0.4 |
| R-(−)-1 | 7.43 ±1.5 | 1.88 ± 0.4 |
| S-(+)-1 | 4.78 ±1.0 | 2.62 ± 0.5 |
| DPH | >100 | >100 |
Fluorescent based assay.
The effects of the racemic mixture and enantiomers of compound 1 were also examined by patch clamp electrophysiology on the human Nav channel isoform, Nav1.2, stably expressed in human embryonic kidney cells (HEK 293). Nav currents were elicited by step depolarizations from a holding potential of −60 to +10 mV for 25 ms at 15 s intervals. Sodium currents were record during a control drug free condition, after 5 min of drug perfusion and following washout. At 1 μM, (R)-(–)-1 inhibited the Nav channel current by 67.4 ± 5.3% (n = 4) and by 94.3 ± 0.6% (n = 3) at 10 μM In contrast, (S)-(+)-1 and the racemic mixture were significantly less potent than the (R)-(–)-1 enantiomer at 1 μM, (P <0.05). (S)-(+)-1 inhibited the sodium channel current by 34.9 ±2.3% (n = 3) at 1 μM and by 91.5 ±0.5% (n = 3) at 10 μM. The racemic mixture inhibited the sodium channel current by 31.9 ±2.9% (n = 3) at 1 μM and by 87.0 ± 3.1% (n = 3) at 10 μM (Fig. 2). All drug effects were fully reversible on washout.
Figure 2.
Effects of racemate and enantiomers of ICM-I-136 on hNav1.2.
3.2. Homology and structural modeling
In an effort to rationalize the enantioselective effects of the sodium channel blockers with the sodium channel pore, and to understand the differential activity and binding event that occurs with the drug for the R and S configuration, we predicted the structure for the open and the closed Nav1.7 channel. The sodium channel pore was developed by aligning the pore-forming residues 15–101 of the X-ray structure of the open form of the KcsA potassium channel, with residues 235–410 of domain I, residues 690–799 of domain II, residues 1123–1275 of domain III, and residues 1445–1551 of domain IV of the sodium channel. KcsA potassium channel residues 22–124 were used to model the P-loop regions with the N- and C-terminal residues of these segments. The orientations of the four domains were modeled by aligning sodium channel domains I–IV with MthK channel chains A–D. The BTX binding site location, as identified by mutational studies, is on the pore-facing side of the S6 helices from domains I, III, and IV.21 Upon analysis of the homology model structure, the IS6, IIIS6 and IVS6 segments, and the residues that form the drug binding site are all conserved, and are mainly hydrophobic. We predicted both the open and the closed channel of the Nav channel based on the MthK and KcsA potassium channels as a template. In the closed channel model, F1579 and Y1586 in IVS6 were oriented toward the pore because of their possible interaction with LA drugs. In the open channel model, the bends in the S6 helices were produced at the serine sites corresponding to the glycine residues found in the MthK open channel structure. Both the R and S configuration of compound 1 were docked using AutoDock 4.022 and FlexX incorporated in Sybyl 8.0. However, the docked poses generated by both programs show different interactions with the S6 helix residues in comparison to the mutation data.23–25 To be consistent with respect to the mutation studies and previous known interactions of lidocaine analogs,26 the docked positions were remodeled using step-by-step manual docking with constrained molecular dynamics (MD) simulations followed by minimization. In the restrained MD simulations, the optimum H-bond and hydrophobic distance constraints were set between the pore forming residues and the ligand. The residues such as F1283, F1579, L1582, F1283, V1583, Y1586 in IVS6, and T1279, L1280 in IIIS6, and L788, F791, L792, in IIS6 and I433, N434, L437 in IS6, and the selectivity filter residues D400, E755, K1237 in the domains of I–IV P-loops were identified as participants in the putative binding site for our compounds.
A structural model of Nav1.7 predicted interaction with compounds (R)-(–)-1) and (S)-(+)-1 is shown in Figure 3. Our binding model suggest that compound 1 interacts with that residues F1283, F1579, L1582, V1583, Y1586 in IVS6, and T1279, L1280 in IIIS6, and L788, F791, L792, in IIS6 and F430, I433, L437 in IS6 and suggest amino acids that may contribute to potential binding interactions. In fact some of these residues are found to be important in alanine mutation experiments.22–24 As seen in Figure 3, strong hydrophobic contacts were noticed between 1 and F1283, F1579, L1582, V1583, Y1586 L1280, L788, F791, L792, I433, and L437. Enantiomeric selectivity could be rationalized for the (R)-(−)-1 isomer which may be driven by strong H-bonding between the amide functionality and T1279 (Fig. 3A). Modeling studies suggest that this interaction is only observed for the R enantiomer, and not present for the S enantiomer (Fig. 3B). This could explain the difference in sodium channel activity we observe for the R versus the S enantiomer of our compound.
Figure 3.
Binding model of compound 1 and Nav1.7. (A) Proposed binding model of R-(−)-1 with Nav1.7. (B) Proposed binding model of S-(+)-1 with Nav1.7. The sodium channel is represented as helices (cyan), ball and stick model representation for critical binding site residues (yellow), and compound 1 (atom color) with H-bond denoted by dotted lines.
3.3. Gene activity
All four compounds were also counter-screened for human ether-a-go-go-related gene (hERG) activity against the radio-ligand MK-0499. Blockade of hERG K+ channels is widely regarded as the predominant cause of drug-induced QT prolongation.27 As seen in Table 4, the racemic mixture and the enantiomers did not significantly inhibit hERG below 10 μM.
Table 4. % Inhibition for hERG assay.
| Compound | % Inhibition | ||||
|---|---|---|---|---|---|
|
|
|||||
| 0.3 μM | 1 μM | 3 μM | 10 μM | 30 μM | |
| (±)-l | 15 ± 3.0 | 17 ±3.4 | 15 ±3.0 | 51 ± 10.2 | 78 ± 15.6 |
| R-(−)-1 | 16 ± 3.2 | 26 ±5.2 | 20 ±4.0 | 57 ± 11.4 | 90 ± 18.0 |
| S-(+)-l | 10 ± 2.0 | 25 ±5.0 | 33 ± 6.6 | 65 ± 13.1 | 90 ± 18.0 |
| DPH | 9 ± 1.8 | 20 ±4.0 | 28 ±5.6 | 17 ±3.4 | 40 ± 8.0 |
3.4. Cell assays
We have an interest in evaluating the role of the Nav channels in prostate cancer. From our previous work we have demonstrated that sodium channel blockers have marked effects on prostate cancer cell proliferation.4 In this present work we have implicated several isoforms of the channel to be involved with prostate cancer cell proliferation. CWR22rv-1 whole cell lysate extracts were evaluated for expression of hNav1.5 and hNav1.7 by Western blot analysis. Both α-subunits were detected at 260 kDa with each antibody (see Supplementary data). Specific bands were also detected at lower molecular weights and are likely degradation products. Pretreating with their respective specific oligomer epitope control antigen before antibody addition eliminated the signal of both the 260 kDa band as well as the lower molecular weight bands.
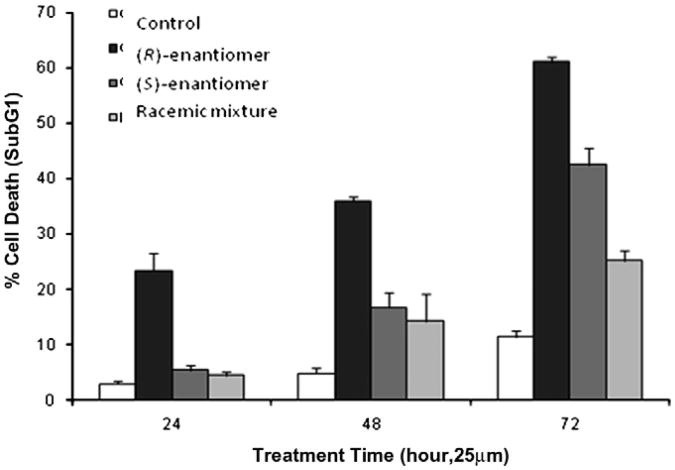
With the identification of both sodium channels in human prostate cancer cell line CWR22rv-1, we evaluated (±)-1, (R)-(−)-1, and (S)-(+)-1 for their effects on prostate cancer cell growth. We found that compound (R)-(−)-1 showed the greatest effect on CWR proliferation (Fig. 4). A concentration of 25 μM induced approximately 25% cell death after 24 h, while compounds (±)-1 and (S)-(+)-1 showed marginal effects after 24 h. After 72 h, compound (R)-(−)-1 induced cell death in 60% of the human prostate cancer cells, while compounds (±)-1 and (S)-(+)-1 induced 25% and 40% cell death, respectively.
Figure 4.
(R)-(−)-1 induces cell death of human CWR cells at 25 μM after 24 h.
3.5. Animal studies
We administered intraperitoneally (ip) 10 mg/kg of racemic (±)-1 and each enantiomer in mice bearing PC3 xenografts (Fig. 5). This dose was administered qd every other day for 24 days and effects on reducing the tumor volume were measured. We observed a statistical reduction in the prostate tumor volume after day 15. Treatment with the racemic and both enantiomers resulted in a 62% decrease in tumor volume at a 10 mg/kg dose and a qd dosing schedule. This demonstrates for the first time that Nav blockers (such as (±)-1 or its enantiomers) can significantly reduce the size of prostate tumors in vivo and tumors that are androgen insensitive.
Figure 5.
Effects of (±)-1 and enantiomers of 1 on human prostate xenograft PC3. Legend. Compounds were dosed ip once a day every other day for 26 days. Studies with N = 6.
4. Conclusions
The relationship between voltage-gated ion channels and cancer represents an exciting new area of investigation, and the synthesis of active chiral inhibitors for selected isoforms may provide potential therapeutic opportunities. We have successfully demonstrated that by using the Seebach and Frater chiral template, both enantiomers of 2-(3-chlorophenyl)-2-hydroxynonanamide can be synthesized. Our biological evaluation of compounds (±)-1, (R)-(−)-1, and (S)-(+)-1, shows a preference for the R enantiomer over the S enantiomer in CWR22rv-1 cells. Our model of (R)-(−)-1 docked in the sodium channel shows a critical hydrogen bond interaction of the amide group with the T1279 residue in the surrounding protein. This interaction is not present with (S)-(+)-1, and is a potential reason for its reduced sodium channel activity.
Finally, our xenograft studies provide evidence that treatment with Nav blockers (such as (±)-1 or its enantiomers) can significantly reduce the size of prostate tumors in vivo including tumors that are androgen insensitive. Since 80–90% of prostate cancer patients develop androgen-independent tumors 12–33 months after androgen ablation, these findings may have significant clinical potential for this phenotype.28
5. Experimental section
5.1. Chemistry
5.1.1. Computational chemistry
Multiple sequence alignment of the S6 transmembrane residues from domains I, III, and IV was carried out using PSI-BLAST and CLUSTALW.29 Homology modeling of the S5, the P-loops, and S6 from all four domains was accomplished using open MthK channel X-ray structure (PDB: 1lnq) as a template. Non-homologous regions in the longer P-loops of domains I and III, which correspond to putative glycosylation sites, were deleted. The P-loops and the N and C termini were modeled based on homologous segments of the KcsA channel structure (PDB: 1bl8). Sodium channel sequences were aligned versus the MthK channel using ClustalW, and the structure was modeled employing the program Modeler 8.1. To avoid close contacts of the side chain atoms with each other, different rotamer states of the residues were considered to find the state with minimal clashes but favorable interactions. Local minimization of side chain atoms was also performed. Docking studies between the ligand and the sodium channel were carried out using the program autodock 4.022 with all the parameters set to default. Molecular dynamics simulations were carried out using AMBER 8.030 with default parameters.
5.1.2. General remarks
Chemicals were purchased from Aldrich Chemical Company, and were used without any further purification. Dry solvents were dried over 4 Å molecular sieves prior to use. Air-sensitive reactions were carried out in oven-dried glassware under an N2 atmosphere. Flash column chromatography separations were done on a Biotage SP1 system monitoring at 254 nm. All NMR spectra were recorded on a Varian 400 spectrometer, operating at 400 MHz for 1H and 100 MHz for 13C NMR. Optical rotations were taken on a Bellingham & Stanley ADP220 polarimeter using a 25 mm cell. Chiral HPLC analysis was carried out on a Shimadzu LCMS-2010EV using a ChiralPak AS column monitoring at 254 nm.
5.1.3. (2R,5R)-2-(tert-Butyl)-5-(3-chlorophenyl)-1,3-dioxolan-4-one (3)
To a suspension of R-(−)-3-chloromandelic acid (6.0 g, 32.15 mmol) in dry pentane was added pivaldehyde (4.2 mL, 38.59 mmol), followed by the addition of triflic acid (0.11 mL, 1.29 mmol) at room temperature. A Dean–Stark trap was attached added to the flask, and the reaction mixture was warmed to 36 °C and allowed to reflux for 6 h. The mixture was cooled to room temperature and 8% aqueous NaHCO3 solution was added and the reaction was concentrated to remove pentane. The slurry was filtered and concentrated to give the product as a 5:1 mixture of diastereomers. The diastereomers were separated by flash column chromatography eluting with 1:5 EtOAc/hexanes to give 4.26 g of pure product as a fluffy white solid. mp 84–86 °C; 1H NMR (400 MHz, CDCl3, δ): 1.06 (s, 9H), 5.19 (s, 1H), 5.30 (d, J=1 Hz, 1H), 7.33– 7.34 (m, 3H), 7.44 (m, 1H). 13C NMR (100 MHz, CDCl3, δ): 20.1, 34.2, 91.1, 121.6, 127.3, 128.1, 134.2, 136.1, 173.5. −72.0 (c 1.00, CHCl3).
5.1.4. S-(+)-3-Chloromandelic acid (5)
To a round bottom flask was added 50 mL of MeOH and three drops of AcCl, 1.61 g (8.60 mmol) of (R)-3-chloromandelic acid was then added in one portion. The mixture was allowed to stir at rt. overnight. The reaction was then poured in to 150 mL of H2O containing 10 mL of saturated NaHCO3 and extracted with CH2Cl2 (3 × 30 mL). The combined organic fractions were washed with brine, dried (Na2SO4), filtered and concentrated to give 1.53 g (87%) of the product as a thick viscous oil which was sufficiently pure for the next step. 1H NMR (400 MHz, CDCl3, δ): 3.73 (s, 3H), 3.78 (d, J = 6 Hz, 1H), 5.13 (d, J = 5 Hz, 1H), 7.26–7.28 (m,3H), 7.40–7.41 (m, 1H). 13C NMR (100 MHz, CDCl3, δ): 53.4, 72.4, 125.0, 127.0, 128.8, 130.1, 134.7,140.3,173.7. −168 (c 1.00, CHCl3).
To a stirred solution of 0.350 g (1.74 mmol) of (R)-3-chloromandelic acid methyl ester in dry THF was added 0.913 g (3.48 mmol) of PPh3 and 0.582 g (3.48 mmol) of p-nitrobenzoic acid. The solution was cooled to 0 °C and 1.31 mL (3.48 mmol) of a 40% wt. solution of DEAD was slowly added. The reaction mixture was allowed to stir at rt. for 6 h under N2. THF was then removed in vacuo and the crude product partitioned between H2O and EtOAc. The combined organic layers were washed with brine, dried (Na2SO4), filtered and concentrated. The residue was purified by flash column chromatography eluting with 1:19 EtOAc/hexanes then 1:9 EtOAc/hexanes to give 0.523 g (86%) of the product as a sticky yellow oil. 1H NMR (400 MHz, CDCl3, δ): 3.74 (s, 3H), 6.14 (s, 1H), 7.34–7.36 (m, 2H), 7.42–7.45 (m, 1H), 7.53–7.54 (m, 1H), 8.25 (s, 4H). 13C NMR (100 MHz, CDCl3, δ): 53.2, 74.8, 123.9, 126.1, 127.9, 130.0, 130.5, 131.3, 134.5, 135.1, 135.4, 151.1, 164.0, 168.5.
The prepared nitroester (4.58 g, 22.85 mmol) was added to 7.43 g (114.23 mmol) of NaN3 and the mixture was heated for 1 h at 40 °C. The solvent was then removed and the residue purified by flash column chromatography eluting with 1:5 EtOAc/hexanes to give 3.291 g (72%) of a white solid. 1H NMR (400 MHz, CDCl3, δ): 3.53 (d, J = 6 Hz, 1H), 3.76 (s, 3H), 5.13 (d, J = 5 Hz, 1H), 7.28–7.30 (m, 3H), 7.40–7.41 (m, 1H). 13C NMR (100 MHz, CDCl3, δ): 53.4, 72.3, 125.0, 126.9, 128.9, 130.1, 134.7, 140.3, 173.8.
To a 3.091 g (15.41 mmol) of (S)-3-chloromandelic acid methyl ester was added 100 mL of 5% NaOH. The reaction mixture was heated to 40 °C for one and then allowed to cool to rt. The reaction was then acidified with 1 N and extracted with EtOAc (3 × 20 mL). The combined organic fractions were dried (Na2SO4), filtered and concentrated in vacuo. The solid residue was recrystallized from hot toluene to give 2.768 g (96%) of a white crystalline solid 5. 1H NMR (400 MHz, DMSO-d6, δ): 5.05 (s, 1H), 7.32–7.36 (m, 3H). 13C NMR (100 MHz, DMSO-d6, δ): 72.4, 126.0, 127.0, 128.2, 130.7, 133.5, 143.3, 174.2. +123 (c 3.00, H2O).
5.1.5. (2S,5S)-2-(tert-Butyl)-5-(3-chlorophenyl)-1,3-dioxolan-4-one (6)
Prepared in the same manner as (2R,5R)-2-(tert-butyl)-5-(3-chlorophenyl)-5-heptyl-1,3-dioxolan-4-one (3) using (S)-(+)-3-chloromandelic acid 5. Mp 40–42 °C; 1H NMR (400 MHz, CDCl3, δ): 1.06 (s, 9H), 5.24 (s, 1H), 5.30 (d, J = 1 Hz, 1H), 7.33–7.34 (m, 3H), 7.44 (m, 1H). 13C NMR (100 MHz, CDCl3, δ): 21.1, 35.2, 91.6, 122.9, 127.3, 128.1, 133.5, 137.1, 173.5. −4.0 (c 1.00, CHCl3).
5.1.6. (2R,5R)-2-(tert-Butyl)-5-(3-chlorophenyl)-5-heptyl-1,3-dioxolan-4-one (4)
To a flame-dried round bottom flask equipped with a magnetic stir bar was added diisopropylamine in THF under N2. The flask was cooled to -78 °C and BuLi was added in one portion. The cooling bath was removed and replaced with an ice-water bath. Meanwhile, (2R,5R)-2-(tert-butyl)-5-(3-chlorophenyl)-1,3-dioxolan-4-one, heptyl iodide, and HMPA were dissolved in THF and place in a separate flame dried round bottom under an N2 atmosphere. The flask was cooled to −78 °C and the previously prepared LDA was added dropwise over 15 min. The reaction mixture was maintained at a constant temperature of −78 °C for 3 h at which time it was quenched with saturated NH4Cl solution. The product was then extracted with Et2O, the organic layers combined, dried (Na2SO4), filtered and concentrated in vacuo. The crude 96:4 mixture of diastereomers was separated by flash column chromatography carefully eluting with 1% Et2O in hexanes. 1H NMR (400 MHz, CDCl3, δ): 0.84 (t, J = 7 Hz, 3H), 0.96 (s, 9H), 1.16–1.35 (m, 10H), 1.88–2.06 (m, 2H), 5.35 (s, 1H), 7.26–7.30 (m, 2H), 7.50–7.54 (m, 1H), 7.62–7.63 (m, 1H). 13C NMR (100 MHz, CDCl3, δ): 14.3, 22.8, 23.7, 23.8, 29.2, 29.5, 31.9, 35.2, 38.9, 82.3, 109.1, 123.4, 125.4, 128.3, 129.7, 134.5, 140.5, 173.5. −40 (c 1.00, CH2Cl2).
5.1.7. R-(−)-2-(3-Chlorophenyl)-2-hydroxy-nonanoic acidamide ((R)-(−)-1)
To a flame dried round bottom flask was added the dioxolanone 4 (0.948 g, 2.69 mmol) in 5 mL or dry MeOH. To this was added 20 mL of 7 N NH3 in MeOH dropwise. This was allowed to stir over 16 h while monitoring by TLC (1:1 hexanes/EtOAc). The reaction mixture was then poured into water and extracted with CH2Cl2 (4 × 10 mL). The combined organic layers were then washed with brine, dried (Na2SO4), filtered and concentrated. The residue was purified by flash column chromatography eluting with 1:1 hexanes/EtOAc to yield 0.659 g (86%). mp 78–79 °C; 1H NMR (400 MHz, CDCl3, δ): 0.85 (t, J = 7 Hz, 3H), 1.23–1.29 (m, 10H), 1.94–2.01 (m, 1H), 2.16–2.22 (m, 1H), 3.19 (s, 1H), 5.65 (br s, 1H), 6.47 (br s, 1H), 7.23–7.28 (m, 2H), 7.44–7.47 (m, 1H), 7.58– 7.59 (m, 1H). 13C NMR (100 MHz, CDCl3, δ): 14.3, 22.8, 23.5, 29.3, 29.8, 32.0, 39.6, 78.8, 123.9, 125.9, 128.1, 129.9, 134.6, 144.6, 176.3. −24 (c 0.50, CHCl3).
5.1.8. (2S,5S)-2-(tert-Butyl)-5-(3-chlorophenyl)-5-heptyl-1,3-dioxolan-4-one (7)
Prepared in the same manner as (2R,5R)-2-(tert-butyl)-5-(3-chlorophenyl)-5-heptyl-1,3-dioxolan-4-one using (2S,5S)-2-(tert-butyl)-5-(3-chlorophenyl)-1,3-dioxolan-4-one. 1.414 g yield (71%). 1H NMR (400 MHz, CDCl3, δ): 0.84 (t, J = 7 Hz, 3H), 0.96 (s, 9H), 1.16–1.35 (m, 10H), 1.88–2.06 (m, 2H), 5.35 (s, 1H), 7.26– 7.30 (m, 2H), 7.50–7.54 (m, 1H), 7.62–7.63 (m, 1H). 13C NMR (100 MHz, CDCl3, δ): 14.3, 22.8, 23.7, 23.8, 29.2, 29.5, 31.9, 35.2, 38.9, 82.3, 109.1, 123.4, 125.4, 128.3, 129.7, 134.5, 140.5, 173.5.
5.1.9. S-(+)-2-(3-Chlorophenyl)-2-hydroxy-nonanoic acid amide ((S)-(+)-1)
Prepared in the same manner as (R)-(−)-2-(3-chlorophenyl)-2-hydroxy-nonanoic acid amide using (2S,5S)-2-(tert-butyl)-5-(3-chlorophenyl)-5-heptyl-1,3-dioxolan-4-one. 0.683 g yield (60%) mp 78–79 °C; 1H NMR (400 MHz, CDCl3, δ): 0.85 (t, J = 7 Hz, 3H), 1.23–1.29 (m, 10H), 1.94–2.01 (m, 1H), 2.16–2.22 (m, 1H), 3.19 (s, 1H), 5.65 (br s, 1H), 6.47 (br s, 1H), 7.23–7.28 (m, 2H), 7.44– 7.47 (m, 1H), 7.58–7.59 (m, 1H). 13C NMR (100 MHz, CDCl3, δ): 14.3, 22.8, 23.5, 29.3, 29.8, 32.0, 39.6, 78.8, 123.9, 125.9, 128.1, 129.9, 134.6, 144.6, 176.3. +24 (c 0.50, CHCl3).
5.2. X-ray crystallography
Both (ICM-1-136-1) (S)-2-(3-chloro-phenyl)-2-hydroxy-nonanoic acid amide and (ICM-1-136-2) (R)-2-(3-chloro-phenyl)-2-hydroxy-nonanoic acid amide were crystallized from 70% ethanol using slow evaporation method at room temperature. The diffraction data were collected at 100 K using a Rigaku R-axis Rapid diffractometer, equipped with a Mo Kα radiation source (60 kV, 40 mA). The radiation was monochromatized with graphite monochromator. HKL-200031,32 was used for control of the data collection as well as for data reduction. The structure was solved and refined by the HKL-3000SM system33 which is integrated with SHELXS, SHELXL34 and O.35 Absolute configurations of both compounds were determined using anomalous dispersion. Details of data collection, processing and refinement (see Supplementary Table 1). Interestingly both compounds crystallized with two molecules in asymmetric unit (see Supplementary Fig. 1). The molecules of the (+)-2-(3-chloro-phenyl)-2-hydroxy-nonanoic acid amide forming crystals have two different conformations of the aliphatic chains.
5.3. Biology
5.3.1. Cell culture for western blots
The CWR22rv-1 cell line was obtained from the American Tissue Culture Collection (Manassas, VA). All cell lines were maintained in RPMI-1640 with L-glutamine (CellGro, Lawrence, KS) supplemented with 5% heat-inactivated fetal bovine serum (Sigma, St. Louis, MO). LNCaP media was additionally supplemented with 0.1 nM DHT (Sigma, St. Louis, MO). Cells were seeded into Corning T-75 flasks (Fisher, Pittsburg, PA) and incubated at 37 °C, 5% CO2, and 100% relative humidity. Cultures were subcultured once per week via trypsinization.
Western protocols were adapted from Collins et al.36 Cells were trypsin-harvested, washed and flash frozen prior to lysis. Lysates were prepared in modified radioimmunoprecipitation (RIPA) buffer (Sigma, St. Louis, MO) plus 50 mM Tris-HCl, 5 mM EDTA, 150 mM NaCl, 30 mM NaPPi, 50 mM NaF, 1 mM Na orthovanadate, 1% Triton-X 100, 0.1% SDS, 0.5% Na deoxycholate, 1 mM phenylm-ethylsulphonyl fluoride (SIGMA, St. Louis, MO) and 1% protease inhibitor cocktail (SIGMA, St. Louis, MO) for 2 h. Insoluble debris was removed by centrifugation at 15,000g for 45 min. Total protein was determined using the Bradford method (Bio-Rad, Hercules, CA). Equivalent amounts of protein from different lysate samples (30 Lig/well) were denatured by boiling for 5 min and were resolved against Seeblue2 (Invitrogen, Carlsbad, CA) by SDS-PAGE using 4% tris-glycine gels (Invitrogen, Carlsbad, CA) in Tris-Gly SDS buffer (Biorad) at 85 V for 30 min and then 125 V for 2 h. Protein was transferred to methanol-pretreated PVDF membranes at 4 °C in Tris-glycine transfer buffer (Bio-Rad, Hercules, CA) at 30 V for 16 h. Membranes were washed in 0.1% PBS-Tween, blocked for 1 h in blocking buffer (50 mM Tris-Cl, 150 mM NaCl, and 10g/L BSA in diH20), and subsequently incubated with either 625 ng/mL (1:2000) human Nav1.5 antibody pretreated with 1:10 blocking peptide in blocking buffer with 500 mg/L NaN3 at 4 °C for 16 h or 7 μg/mL (1:1000) rat Nav1.7 (13/15 sequence homology to human) pretreated with 1:1 blocking peptide under similar conditions. Membranes were washed four times for 15 min in 0.1% PBS-Tween, blocked for 30 min in blocking buffer, and incubated in 75 ng/mL (3:40,000) horse radish peroxidase-conjugated goat anti-rabbit secondary antibody in blocking buffer for 1 h. Membranes were washed in dH2O. After the last of four washes for min with 0.1% PBS-Tween, the blots were developed using the ECL chemiluminescence system (Amersham, Piscataway, NJ) and visualized by exposure to Biomax MR Film (Kodak, Rochester, NY). Membranes were washed in dH2O and then washed twice for 5 min with 0.1% PBS-Tween. Blots were then stripped with stripping buffer (0.375 M Tris-HCl, 12% SDS, 60 mM HCl, pH 6.8) for 1 h at 56 °C and again washed in dH2O and then washed twice for 5 min with 0.1% PBS-Tween. Membranes were blocked for 1 h in blocking buffer and subsequently incubated with either 625 ng/mL (1:2000) human Nav1.5 antibody without blocking peptide pretreatment in blocking buffer with 500 mg/L NaN3 at 4 °C for h or 7 Lig/mL (1:1000) rat Nav1.7 without blocking peptide pretreatment in blocking buffer with 500 mg/L NaN3 at 4 °C for 16 h. The western procedure was repeated as described above.
5.3.2. Cell culture and treatment
CWR22Rv1 cells (ATCC, Manassas, VA) were seeded in 6 well tissue culture plate at density of 300,000 cells/well and maintained in RPMI 1640 (Mediatech, Herndon, VA) containing 10% fetal bovine serum, 2.5 mM L-glutamine, and penicillin-streptomycin (100 IU/mL and 100 μg/mL, respectively) at 37 °C with 5% CO2. Cells were then switched to serum free RPMI media overnight prior to drug treatment. Compounds were dissolved in 100% dimethylsufoxide (DMSO) and diluted to the desired concentrations in serum free RPMI. Cells were treated with indicated drug concentrations (in triplicate per treatment point) for 24, 48, 72 h. After treatment, cells were harvested by trypsinization and fixed in 70% ethanol. The fixed cells were then stained with propidium iodide (50 μg/mL) after treatment with RNase (5 μg/mL). The stained cells were analyzed for DNA content using FACSsort (Becton Dickinson). Cell cycle fractions were quantified with Cell Quest (Becton Dickinson) or ModFit LT (Verity Software House).
5.3.3. Fluorescent based sodium channel assay
A functional fluorescent assay for Nav1.5 and Nav1.7 channel activity was performed as previously described.37 Tissue culture media and CC2-DMPE and DiSBAC2 were purchased from Invitrogen Corporation, Carlsbad CA, and pluronic acid from Molecular Probes, Eugene, OR. HEK293 cells stably transfected with either hNaV1.5 or hNaV1.7 were plated at approximately 11,000-20,000 cells/well in flat bottom, poly-D-lysine coated black-wall, clear-bottom, 384-well plates (Becton Dickinson, Bedford, MA), and incubated overnight at 37 °C in a 10% CO2 atmosphere in growth medium. Cells were washed with 0.03 mL of Dulbecco's phosphate buffered saline (D-PBS) containing calcium and magnesium. Cells were then incubated with 0.025 mL of a solution containing 10 μM CC2-DMPE and 0.02% pluronic acid in D-PBS with calcium and magnesium, supplemented with 10 mM glucose, and 10 mM Hepes-NaOH, pH 7.4. After incubation in the dark for 45 min at 25 °C, cells were washed twice with 0.03 mL of (in mM): 165 NaCl, 4.5 KCl, 2 CaCl2, 1 MgCl2, 10 glucose, 10 Hepes-NaOH, pH 7.4 (VIPR-S). Afterwards, cells were incubated in the dark at 25 °C for 45 min with 0.025 mL of a solution containing 5 μM DiSBAC2(3) in VIPR-S, in the absence or presence of test compound. At the end of the incubation period, the plate was placed in a FLIPRTETRA instrument (MDS Analytical Technologies, Sunnyvale CA), illuminated at 390-420 nm, and fluorescence emissions were recorded at approximately 1 Hz at 460 and 580 nm. Following a 7 s baseline reading, 0.025 mL of VIPR-S containing 10 μM veratridine for Nav1.5 or 20 μM veratridine for Nav1.7 was added to each well, and the emissions of both dyes were recorded for an additional 33 s. The change in fluorescence resonance energy transfer (FRET) ratio (F/F0) was calculated as:
where A and I represent the readings after or before addition of veratridine, respectively, at the indicated wavelengths. For I, the readings from 2 to 5 s were averaged, and for A, readings from 3 s after the signal had reached a plateau level (usually within 30–40 s) were also averaged. Triplicate measurements were performed for each experimental condition and the data were averaged.
5.3.4. HERG binding assay
MK-499, a potent hERG blocker, was used in a ligand binding assay to evaluate the binding of test compounds to hERG potassium channels as previously described.38 Test compounds were incubated with 0.05 nM of radiolabeled MK-499 ([35S]MK-499; specific Activity = 1,279,010 μCi/μmol) in an assay buffer solution (70 mM NaCl, 60 mM KCl, 10 mM HEPES/NaOH (pH 7.4), 2 mM MgCl2, 1 mM CaCl2) also containing membranes isolated from HEK-293 cells stably expressing hERG channels in 96-well polypropylene deep well assay plates at room temperature (25 °C) for >90 min. Assay plates were rinsed three time with buffer (130 mM NaCl, 10 mM HEPES/NaOH (pH 7.4), 2 mM MgCl2, 1 mM CaCl2) and the solution transferred to a Packard FilterMate Universal Harvester apparatus and filtered using PerkinElmer UniFilter-96 GF/C 96-well white microplates pre-soaked with 0.3% BSA (10 mL/L of 30% BSA). Plates were dried overnight at 37 °C, or for 2 h at 56 °C. The bottom of the plates were sealed and 0.025 mL of Microscint 0 was added to each well. The plates were top sealed and counted for 2 min/well in a Packard TopCount Scintillation counter. Test compounds were evaluated in a five point titration format using half-log steps from 0.3 to 30 μM. Percent inhibition of [35S]MK-499 was calculated relative to high control values (no unlabeled MK-499 added) and values obtained in the presence of 1 μM unlabeled MK-499. Less than 5% of the added radioactivity was retained on the filters. Assays were performed in triplicate.
5.3.5. Nav electrophysiology studies
Sodium currents were recorded using the whole-cell configuration of the patch clamp recording technique with an Axopatch 200 amplifier (Axon Instruments, Foster City, CA). All voltage protocols were applied using pCLAMP 9 software (Axon, USA) and a Digidata 1322A (Axon, USA). Currents were amplified and low pass filtered (2 kHz) and sampled at 33 kHz. Borosilicate glass pipettes were pulled using a Brown-Flaming puller (model P87, Sutter Instruments Co, Novato, CA) and heat polished to produce electrode resistances of 1.5–3.0 MΩ when filled with the following electrode solution (in mM); CsCl 130, MgCl2 1, MgATP 5, BAPTA 10, HEPES 5 (pH adjusted to 7.4 with CsOH). Cells were plated on glass coverslips and superfused with solution containing the following composition; (in mM) NaCl 130, KCl 4, CaCl2 1, MgCl2 5, HEPES 5, and glucose 5 (pH adjusted to 7.4 with NaOH). Compounds were prepared as 100 mM stock solutions in dimethyl sulfoxide (DMSO) and diluted to desired concentration (1 or 10 μM) in perfusion solution. All experiments were performed at room temperature (20–22 °C). After establishing whole-cell, a minimum series resistance compensation of 75% was applied and cells were held at −100 mV for 5 min to account for equilibrium gating shifts. Sodium currents were evoked by stepping to +10 mV from a holding potential of −60 mV for 25 ms at 15 s intervals. After control recordings, test compounds were applied for 5 min to allow for bath equilibration. Tonic block was assessed by comparing peak sodium current in drug free conditions to peak current when drug was present. Data analysis was performed using Clampfit software (v9, Axon Instruments, CA, USA) and Origin (v6, Microcal Software, MA, USA).
5.3.6. Animals
Balb/c mice and Athymic Balb/c Nude mice were purchased from the National Cancer Institute (NCI). Animals were housed 4–6 per cage with microisolater tops and provided food (Furina mice chow) and water ad libitum. The light cycle was regulated automatically (12 hours light/dark cycle) and temperature was maintained at 23 ± 1 °C. All animals were allowed to acclimate to this environment fro one week prior to experimental manipulations. The Georgetown University Animal Care and Use Committee approved all animal studies in accordance with the guideline adopted by the National Institute of Health.
5.3.7. Cell culture for xenograft
PC-3 cell line (ATCC, Manassas, VA) was cultured in RPMI-1640 with L-glutamine (Mediatech Inc., Herdon, VA) containing 5% fetal bovine serum (FBS), 2.5 mM L-glutamine at 37 °C with 5% CO2.
5.3.8. Xenograft study
Male athymic balbc/nude mice (18–22 g) were injected with 3 × 106 (0.3 mL) of the human prostate cancer cells (PC3). The human prostate cancer cells were injected in the subcutaneous tissue of the right axillary region of the mice. One week after the injection, the mice were randomly sorted into four groups with 6 mice per group. Stock solutions of compounds (±)-1, (R)-(−)-1 and (S)-(+)-1 were obtained by dissolving 1 mg of compound in 1 μl DMSO. The stock of each compound was added to polyethylene glycol 400 (PEG) (Hampton) and PBS in a 1:1 ratio. The test concentrations were obtained by diluting with PEG/PBS. The tumor-bearing mice received an intraperitoneal injection (IP) with either 10 mg/kg of (±)-1, R-(−)-1 or S-(+)-1 or vehicle control respectively once every other day for 4 weeks. At the same time, the tumor size of each mouse was measured by caliper and calculated by the formula: Length × width × height/2.
5.3.9. Data analysis
Statistical analyses were performed using the standard one-way ANOVA or ANOVA on ranks followed by a Tukey or Dunn's post hoc test. Data is reported as mean ± S.E.M.
Supplementary Material
Acknowledgments
This work was supported by NIH grant CA105435-04, Merck Academic Development Award, and the Georgetown University Drug Discovery Program. The authors would also like to thank Dr. H.A. Hartmann for the HEK 293 cells stably expressing hNav1.2 and the shared resources at the Lombardi Comprehensive Cancer Center.
Footnotes
Supplementary data: Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmc.2011.08.061.
References and notes
- 1.Catterall WA. Neuron. 2000;26:13. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 2.Goldin AL. Annu Rev Physiol. 2001;63:871. doi: 10.1146/annurev.physiol.63.1.871. [DOI] [PubMed] [Google Scholar]
- 3.Baker MD, Wood JN. Trends Pharmacol Sci. 2001;22:27. doi: 10.1016/s0165-6147(00)01585-6. [DOI] [PubMed] [Google Scholar]
- 4.Anderson JD, Hansen TP, Lenkowski PW, Walls AM, Choudhury IM, Schenck HA, Friehling M, Hoell GM, Patel MK, Sikes RA, Brown ML. Mol Cancer Ther. 2003;2:1149. [PubMed] [Google Scholar]
- 5.Laniado ME, Fraser SP, Djamgoz MB. A Prostate (NY, NY, U S) 2001;46:262. doi: 10.1002/1097-0045(20010301)46:4<262::aid-pros1032>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 6.Preussat K, Beetz C, Schrey M, Kraft R, Wolfl S, Kalff R, Patt S. Neurosci Lett. 2003;346:33. doi: 10.1016/s0304-3940(03)00562-7. [DOI] [PubMed] [Google Scholar]
- 7.Wang XT, Nagaba Y, Cross HS, Wrba F, Zhang L, Guggino SE. Am J Pathol. 2000;157:1549. doi: 10.1016/S0002-9440(10)64792-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith P, Rhodes NP, Shortland AP, Fraser SP, Djamgoz MBA, Ke Y, Foster CS. FEBS Lett. 1998;423:19. doi: 10.1016/s0014-5793(98)00050-7. [DOI] [PubMed] [Google Scholar]
- 9.Fiske JL, Fomin VP, Brown ML, Duncan RL, Sikes RA. Cancer Metastasis Rev. 2006;25:493. doi: 10.1007/s10555-006-9017-z. [DOI] [PubMed] [Google Scholar]
- 10.Brown ML, Zha CC, Van Dyke CC, Brown GB, Brouillette WJ. J Med Chem. 1999;42:1537. doi: 10.1021/jm980556l. [DOI] [PubMed] [Google Scholar]
- 11.Ko SH, Jochnowitz N, Lenkowski PW, Batts TW, Davis GC, Martin WJ, Brown ML, Patel MK. Neuropharmacology. 2006;50:865. doi: 10.1016/j.neuropharm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Seebach D, Sting AR, Hoffmann M. Angew Chem, Int Ed Engl. 1996;35:2708. [Google Scholar]
- 13.Nagase R, Oguni Y, Misaki T, Tanabe Y. Synthesis. 2006;22:3915. [Google Scholar]
- 14.Misaki T, Ureshino S, Nagase R, Oguni Y, Tanabe Y. Org Process Res Dev. 2006;10:500. [Google Scholar]
- 15.Liu YM, Liu H, Zhong BH, Liu KL. Synth Commun. 2006;36:1815. [Google Scholar]
- 16.Grover PT, Bhongle NN, Wald SA, Senanayake CH. J Org Chem. 2000;65:6283. doi: 10.1021/jo005562f. [DOI] [PubMed] [Google Scholar]
- 17.Blay G, Fernandez I, Monje B, Munoz MC, Pedro JR, Vila C. Tetrahedron. 2006;62:9174. [Google Scholar]
- 18.Blay G, Cardona L, Torres L, Pedro JR. Synthesis. 2007;1:108. [Google Scholar]
- 19.Caine DS, Paige MA. Synlett. 1999;9:1391. [Google Scholar]
- 20.George AL., Jr J Clin Invest. 2005;115:1990. doi: 10.1172/JCI25505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linford NJ, Cantrell AR, Qu Y, Scheuer T, Catterall WA. Proc Natl Acad Sci USA. 1998;95:13947. doi: 10.1073/pnas.95.23.13947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. J Comput Chem. 1998;19:1639. [Google Scholar]
- 23.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Science. 1994;265:1724. doi: 10.1126/science.8085162. [DOI] [PubMed] [Google Scholar]
- 24.Yarov-Yarovoy V, Brown J, Sharp EM, Clare JJ, Scheuer T, Catterall WJ. Biol Chem. 2001;276:20. doi: 10.1074/jbc.M006992200. [DOI] [PubMed] [Google Scholar]
- 25.Yarov-Yarovoy V, McPhee JC, Idsvoog D, Pate C, Scheuer T, Catterall WA. J Biol Chem. 2002;277:35393. doi: 10.1074/jbc.M206126200. [DOI] [PubMed] [Google Scholar]
- 26.Lipkind GM, Fozzard HA. Mol Pharmacol. 2005;68:1611. doi: 10.1124/mol.105.014803. [DOI] [PubMed] [Google Scholar]
- 27.Aronov AM. J Med Chem. 2006;49:6917. doi: 10.1021/jm060500o. [DOI] [PubMed] [Google Scholar]
- 28.Hellerstedt BA, Pienta KJ. CA CancerJ Clin. 2002;52:154. doi: 10.3322/canjclin.52.3.154. [DOI] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER 8. San Francisco: University of California; 2004. [Google Scholar]
- 31.Otwinowski Z, Minor W. In: Macromolecular Crystallography, Part A. Carter CW Jr, Sweet RM, editors. Academic Press; New York: 1997. pp. 307–326. [Google Scholar]
- 32.Otwinowski Z, Borek D, Majewski W. Acta Crystallogr, Sect A. 2003;59:228. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 33.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. Acta Crystallogr, Sect D. 2006;62:859. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 34.Sheldrick GMA. Acta Crystallogr, Sect A. 2008;64:112. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 35.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr, Sect A. 1991;47:110. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 36.Collins SP, Reoma JL, Gamm DM, Uhler MD. Biochem J. 2000;345:673. [PMC free article] [PubMed] [Google Scholar]
- 37.Felix JP, Williams BS, Priest BT, Brochu RM, Dick IE, Warren VA, Yan L, Slaughter RS, Kaczorowski GJ, Smith MM, Garcia ML. Assay Drug Dev Technol. 2004;2:260. doi: 10.1089/1540658041410696. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Penna KD, Wang H, Karczewski J, Connolly TM, Koblan KS, Bennett PB, Salata JJ. Am J Physiol Heart Circ Physiol. 2003;284:H256. doi: 10.1152/ajpheart.00220.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.