Abstract
A fundamental question in neurobiology is how the balance between proliferation and differentiation of neuronal precursors is maintained to ensure that the proper number of brain neurons is generated. Substantial evidence implicates DYRK1A (dual specificity tyrosine-phosphorylation-regulated kinase 1A) as a candidate gene responsible for altered neuronal development and brain abnormalities in Down syndrome. Recent findings support the hypothesis that DYRK1A is involved in cell cycle control. Nonetheless, how DYRK1A contributes to neuronal cell cycle regulation and thereby affects neurogenesis remains poorly understood. In the present study we have investigated the mechanisms by which DYRK1A affects cell cycle regulation and neuronal differentiation in a human cell model, mouse neurons, and mouse brain. Dependent on its kinase activity and correlated with the dosage of overexpression, DYRK1A blocked proliferation of SH-SY5Y neuroblastoma cells within 24 h and arrested the cells in G1 phase. Sustained overexpression of DYRK1A induced G0 cell cycle exit and neuronal differentiation. Furthermore, we provide evidence that DYRK1A modulated protein stability of cell cycle-regulatory proteins. DYRK1A reduced cellular Cyclin D1 levels by phosphorylation on Thr286, which is known to induce proteasomal degradation. In addition, DYRK1A phosphorylated p27Kip1 on Ser10, resulting in protein stabilization. Inhibition of DYRK1A kinase activity reduced p27Kip1 Ser10 phosphorylation in cultured hippocampal neurons and in embryonic mouse brain. In aggregate, these results suggest a novel mechanism by which overexpression of DYRK1A may promote premature neuronal differentiation and contribute to altered brain development in Down syndrome.
Keywords: DYRK1A, phosphorylation, cell cycle, neuronal differentiation, p27Kip1, Cyclin D1, Down syndrome
Introduction
The balance between progenitor maintenance and production of differentiated neurons is crucial for normal brain development. This balance is achieved through the precise coordination of several sequential steps that include the proliferation of progenitor cells, the transition to neurogenic progenitors, the cell cycle exit of neuronal precursors, and their differentiation into neurons. Precise control of the cell cycle is essential for keeping this balance, what highlights the importance of cell cycle-regulatory proteins in neurodevelopment.1-3 For instance, a selective lengthening of G1 phase appears to be pivotal for the switch to neurogenesis.4 In particular, the antiproliferative CDK inhibitor p27Kip1 (CDKN1B) promotes cell cycle exit, whereas positive regulators of proliferation such as Cyclin D1 (CCND1) have the opposite effect.5-8
Phosphorylation is a key mechanism in the regulation of p27Kip1 and Cyclin D1 by controlling protein turnover during G1 phase.9,10 p27Kip1 is primarily modulated by phosphorylation at Ser10, which is increased during G1-G0 phase of the cell cycle and enhances p27Kip1 stability.9 Several kinases have been proposed to target this residue, including KIS (kinase interacting with stathmin), AKT, ERK2, CDK5, HIPK2 (homeodomain-interacting protein kinase 2), and DYRK1B.9,11-15 Proteasomal degradation of Cyclin D1 during G1 phase is induced by phosphorylation at Thr286 via GSK3β10, but recently DYRK1A and DYRK1B were also shown to phosphorylate this residue in non-neural cells.16 The phosphorylation of p27Kip1 and Cyclin D1 has mainly been investigated in cancer cell cycle progression, and the respective regulative kinases in neuronal progenitor cells remain uncharacterized.
The gene encoding protein kinase DYRK1A is located on human chromosome 21 and is thus 1.5-fold overexpressed in Down syndrome.17,18 Evidence from transgenic mouse models as well as studies in chicken and Drosophila (where the DYRK1A orthologs are termed Minibrain) have revealed that both overexpression and reduced expression of DYRK1A result in neurodevelopmental alterations due to deregulation of neurogenesis during brain development.19 A strong dosage effect of the human DYRK1A gene was further substantiated by the identification of patients with heterozygous loss-of-function mutations of the DYRK1A gene, who develop a severe syndrome of intellectual disability and microcephaly.20,21 Importantly, impaired balance between proliferation and cell cycle exit in neural progenitors is known to be a major cause of microcephaly.22 These findings support the hypothesis that the cellular level of DYRK1A is a critical parameter in neurogenesis, and that DYRK1A overexpression may contribute to the altered brain development in Down syndrome.23-26
Kinases of the DYRK family have been identified in various organisms as cell cycle regulators.27 In vivo and in vitro experiments indicated that DYRK1A overexpression promotes premature differentiation of neuronal progenitors as well as of PC12 cells.28-30 Moreover, we have previously reported that DYRK1A coordinates cell cycle exit and differentiation of neuronal precursors.31 Nonetheless, the mechanistic network by which DYRK1A contributes to the regulation of cell cycle withdrawal and differentiation of neuronal precursors is still poorly understood.
Here we have studied the molecular mechanisms by which DYRK1A regulates G1-phase transition, cell cycle exit, and subsequent neuronal differentiation. Using SH-SY5Y cells, a differentiable human neuronal cell model, we found that DYRK1A overexpression arrests cells in G1 phase, followed by inducing G0 cell cycle exit and neuronal differentiation. Furthermore, we provide evidence that DYRK1A stabilizes p27Kip1 by phosphorylation at Ser10 and promotes degradation of Cyclin D1 by phosphorylation at Thr286.
Results
DYRK1A reduces proliferation of SH-SY5Y cells dependent on its kinase activity and level of overexpression
To investigate the effects of DYRK1A overexpression on neuronal cells, we generated SH-SY5Y cells with a stable and tet-ON-inducible overexpression of either GFP-tagged DYRK1A or the kinase-deficient point mutant DYRK1A-K188R (DYRK1A-KR). Adequate induction of overexpression was verified by qRT-PCR (Fig. S1A) and western blot analysis (see Fig. 6). The effect of DYRK1A on cell proliferation was monitored by continuous real-time impedance measurements. Gradual induction of DYRK1A overexpression resulted in a dose-dependent suppression of cell proliferation after a lag time of 24 h (Fig. 1A). Overexpression of DYRK1A-KR did not significantly alter cell growth (Fig. 1B), indicating that the observed effect depended on DYRK1A kinase activity and was not caused by doxycycline treatment. Consistently, treatment with the DYRK1A inhibitor harmine attenuated the effect of DYRK1A overexpression (Fig. 1C), although the cells did not recover to proliferation rates of untreated control cells within the observation time.
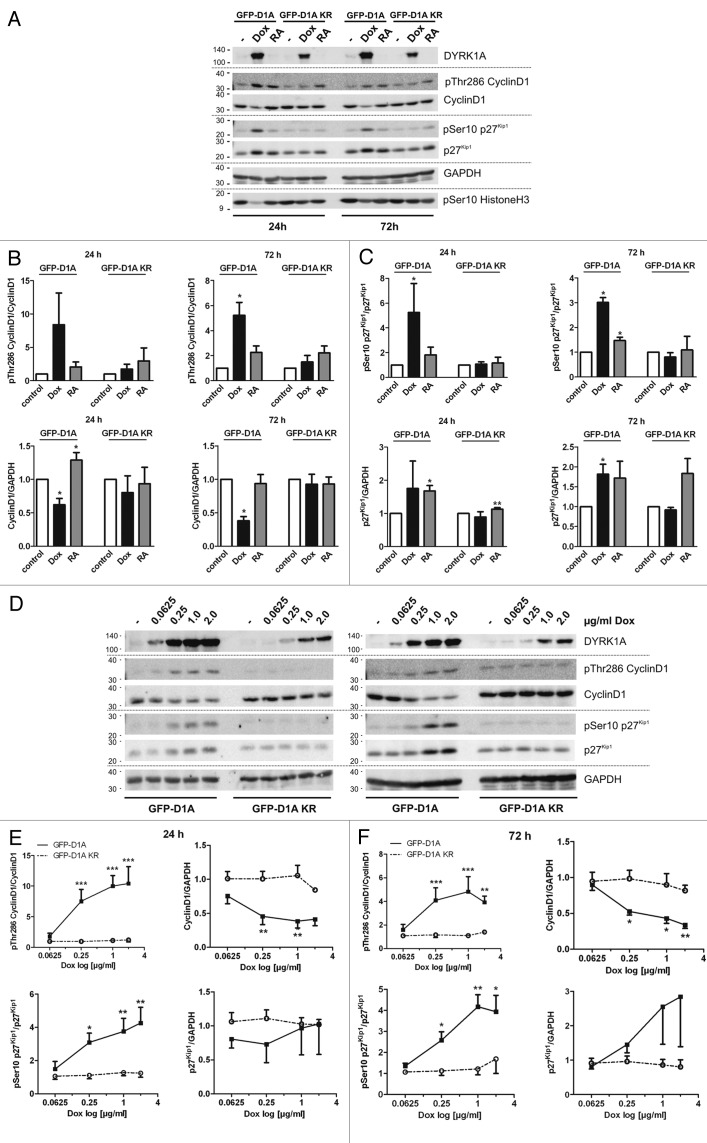
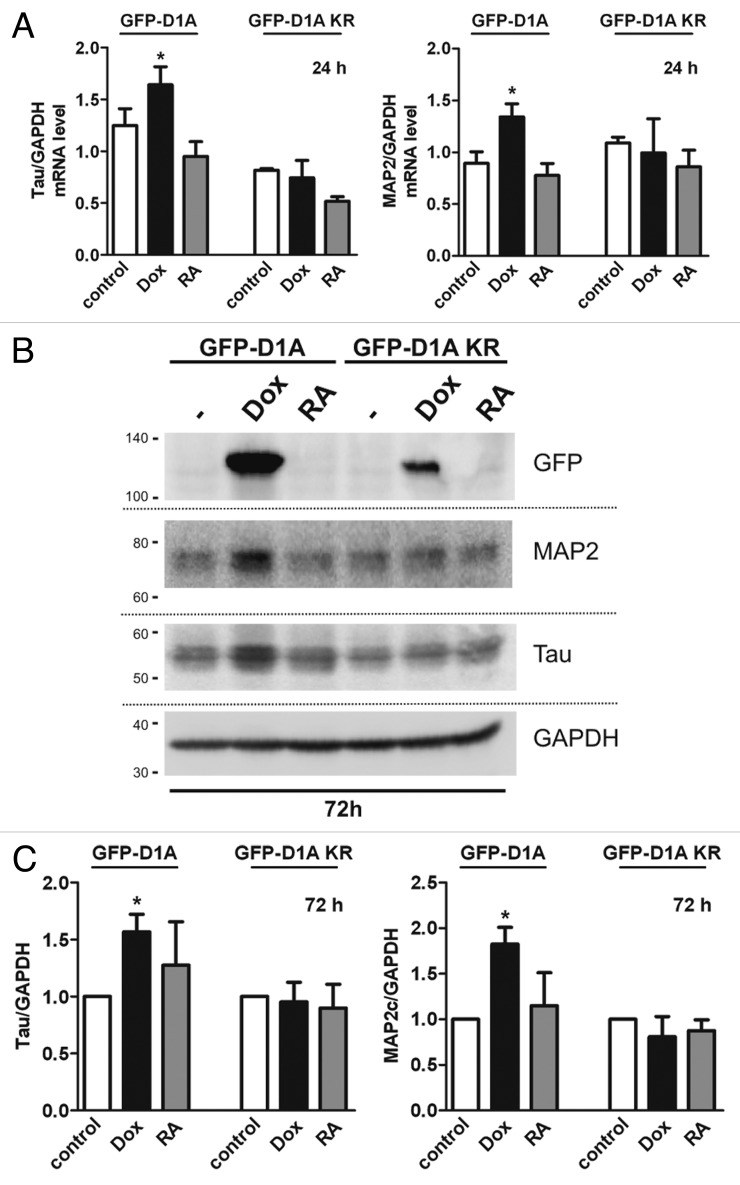
Figure 6. DYRK1A overexpression increases phosphorylation of p27Kip1 and Cyclin D1 in SH-SY5Y cells and alters their protein levels. (A) Western blot analysis of total protein extracts from SH-SY5Y cells. Cells were treated with 2 µg/ml doxycycline to induce overexpression of DYRK1A or DYRK1A-K188R or with 10 µM RA. Control cells were left untreated (−). Whole-cell lysates were prepared 24 h or 72 h after induction of DYRK1A overexpression or differentiation and analyzed by immunoblotting with the indicated antibodies. Migration of mass standards is indicated in kDa (left). Densitometric evaluation of 3 independent experiments (means + SD) is shown in panels (B; Cyclin D1) and (C; p27Kip1). (D) Overexpression of DYRK1A or DYRK1A-K188R was gradually induced using increasing concentrations of doxycyclin as indicated. Cells were lysed 24 h (left panel) or 72 h (right panel) after induction of DYRK1A overexpression, and whole-cell lysates were subjected to western blot analysis. Densitometric evaluation of 3 independent experiments (means + SEM) is summarized in panels (E) (24 h) and (F) (72 h). Ser10 and Thr286 phosphorylation were normalized to p27Kip or Cyclin D1 total protein and p27Kip or Cyclin D1 levels are shown relative to the loading control (GAPDH). One-sample t test was used to compare columns to normalized controls (B and C). One-way ANOVA + Bonferroni post-test was used to analyze effects of increasing dox concentrations. Significances are indicated for comparison of GFP-D1A with GFP-D1A-KR (F and G). p27Kip1/GAPDH data (F) was tested using Kruskal–Wallis test and Dunn multiple comparison post-test; *P ≤ 0.05.; **P ≤ 0.01***P ≤ 0.001.
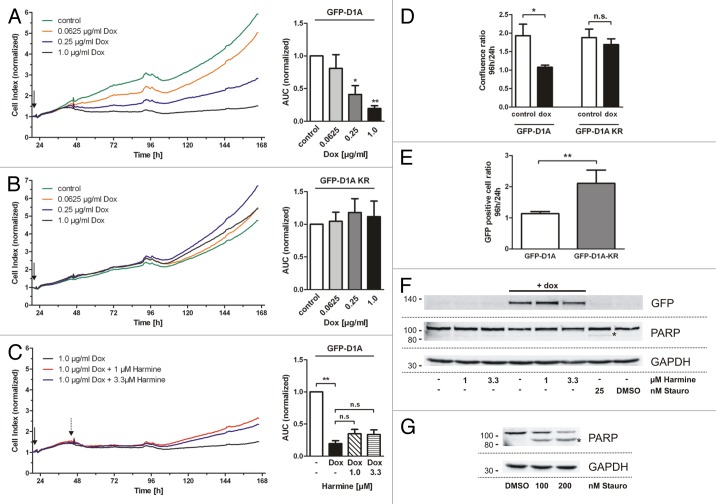
Figure 1. DYRK1A overexpression induces proliferation arrest of SH-SY5Y cells but not apoptosis. (A–C) The curves show representative real-time impedance measurements of proliferating SH-SY5Y cells after induced overexpression of wild-type DYRK1A (A; GFP-D1A) or the kinase deficient mutant DYRK1A-K188R (B; GFP-D1A KR). Panel (C) shows the proliferation of SH-SY5Y cells treated with harmine 24 h after induction of GFP-DYRK1A overexpression. DYRK1A overexpression was induced 24 h after plating with doxycycline as indicated (arrow). Column diagrams illustrate the quantitative evaluation (means + SD) of 3 independent experiments (AUC, area under the curve from 24–168 h, normalized to the untreated control cells). In panel (C), the AUCs were normalized to the untreated control cells (A). (D and E) Analysis of SH-SY5Y cell proliferation by continuous live cell imaging using the IncuCyte™ kinetic imaging system. Relative confluence of cells expressing wild-type DYRK1A or kinase-deficient DYRK1A-K188R and the number of GFP-positive cells were automatically evaluated using the IncuCyte™ software at 24 h and 96 h of doxycycline treatment. Untreated cells served as control. Confluence (D) and cell counts (E) after 96 h were standardized to the 24-h values (means + SD, n = 4 independent experiments). (F) Analysis of apoptosis in SH-SY5Y cells that were treated with doxycycline and harmine under the same conditions as in (A and C). Total protein was extracted after 72 h of treatment and analyzed for PARP activation by western blotting. Cells treated with staurosporine for 24 h served as positive control (F and G). Cleaved and activated PARP is indicated by an asterisk. GAPDH served as loading control. Statistical significance was tested with one-sample t test if comparing columns to normalized control (A–C) or one-way ANOVA and Bonferroni post-test if comparing multiple not-normalized columns (C). Kruskal–Wallis-test + Dunn multiple comparison post-test (D) and Student t test (E) were used. *P ≤ 0.05; **P ≤ 0.01; n.s, not significant.
To ensure that the observed effects of DYRK1A overexpression reflected proliferation arrest rather than cell death, cell proliferation was additionally followed with a continuous live cell imaging system. Cell confluence was determined by automated image evaluation at 24 h and 96 h after induction of DYRK1A overexpression. Consistent with the impedance measurements, confluence of SH-SY5Y cells overexpressing DYRK1A did not change within this time, whereas the confluence of control cells nearly doubled (Fig. 1D; Fig. S2). As expected, overexpression of DYRK1A-KR had no effect on confluence. Moreover, we took advantage of GFP fluorescence to monitor the number of GFP-DYRK1A-expressing cells over time. Automated analysis of the GFP-positive cell count showed that the number of wild-type DYRK1A-overexpressing cells increased by only 13% from 24 h to 96 h, while the number of DYRK1A-KR more than doubled within the same time (Fig. 1E). Furthermore, analysis of PARP (poly ADP ribose polymerase) activation excluded the possibility that the induction of apoptosis contributed to the negative effect of DYRK1A on cell growth. Taken together, these data show that DYRK1A over-activity leads to a dose-dependent reduction of SH-SY5Y cell proliferation rather than a loss of cells by induced cell death.
DYRK1A induces a G1/0-phase cell cycle arrest of SH-SY5Y cells
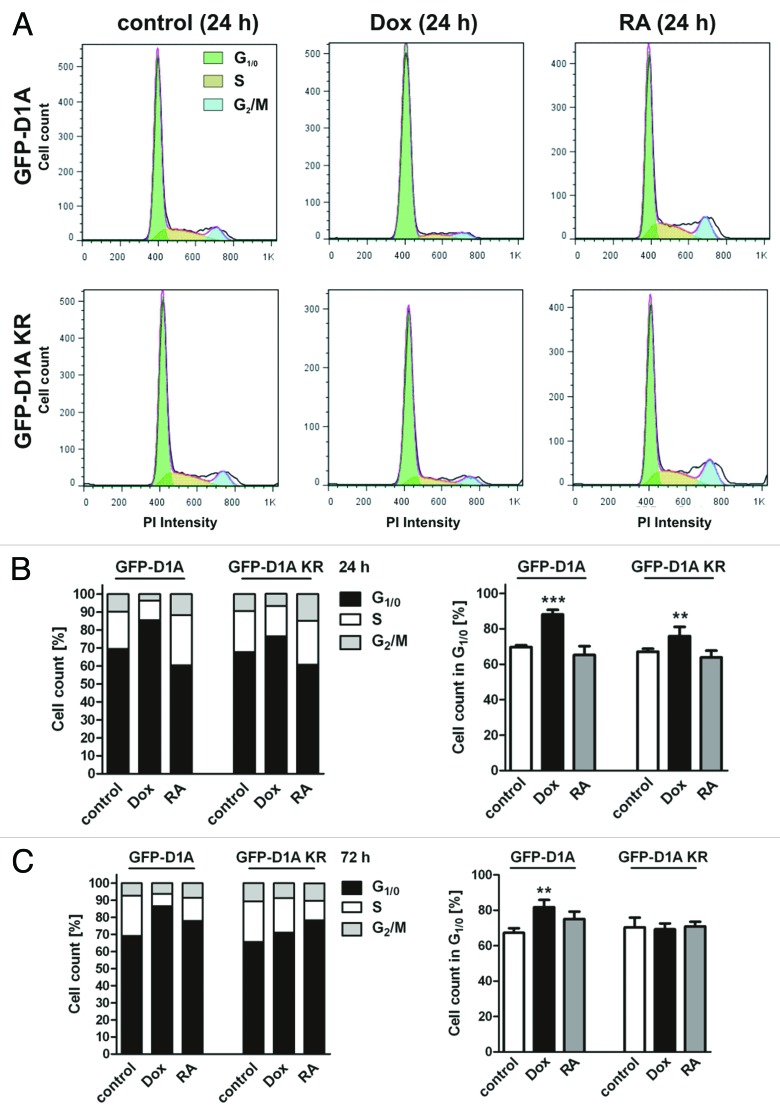
Next we analyzed SH-SY5Y cells by flow cytometry to define the effects of DYRK1A on cell cycle progression (Fig. 2). After 24 h of DYRK1A overexpression, the number of cells in S or G2/M phase was significantly reduced (12% compared with 30% in untreated cells), and more cells were found to be in G1/0 phase (88% vs. 70%) (Fig. 2B). This effect was sustained after 72 h of DYRK1A overexpression (Fig. 2C). Overexpression of DYRK1A-KR resulted in a small and transient increase of cells in G1/0 after 24 h that did not persist after 72 h. For comparison, cells were treated with retinoic acid (RA) to induce cell cycle arrest and neuronal differentiation of SH-SY5Y cells.32 RA only induced a small, non-significant increase of G1/0 cells after 72 h, possibly because its action has a slower onset or does not affect all cells.33 In summary, these results indicate that overexpression of DYRK1A arrests SH-SY5Y cells in G1/0 phase of the cell cycle.
Figure 2. DYRK1A overexpression induces G1/0 cell cycle arrest of SH-SY5Y cells. (A) Flow cytometry. Representative plots of SH-SY5Y cells after DNA staining with propidium iodide (PI). Overexpression of DYRK1A (upper panel) or DYRK1A-K188R (lower panel) was induced with 2 µg/ml doxycycline (Dox). For comparison, cell differentiation was induced with 10 µM retinoic acid (RA). Column diagrams show the cell cycle phase distribution 24 h (B) or 72 h (C) after induction of DYRK1A overexpression or RA-induced differentiation. The left panels show the quantitative evaluation of representative experiments. The right panels summarize the results of 3 independent experiments (means + SD). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; analyzed by one-way ANOVA + Bonferroni post-test comparing DYRK1A or DYRK1A-K188R data to the respective untreated controls.
Long-term overexpression of DYRK1A causes cell cycle exit of SH-SY5Y cells
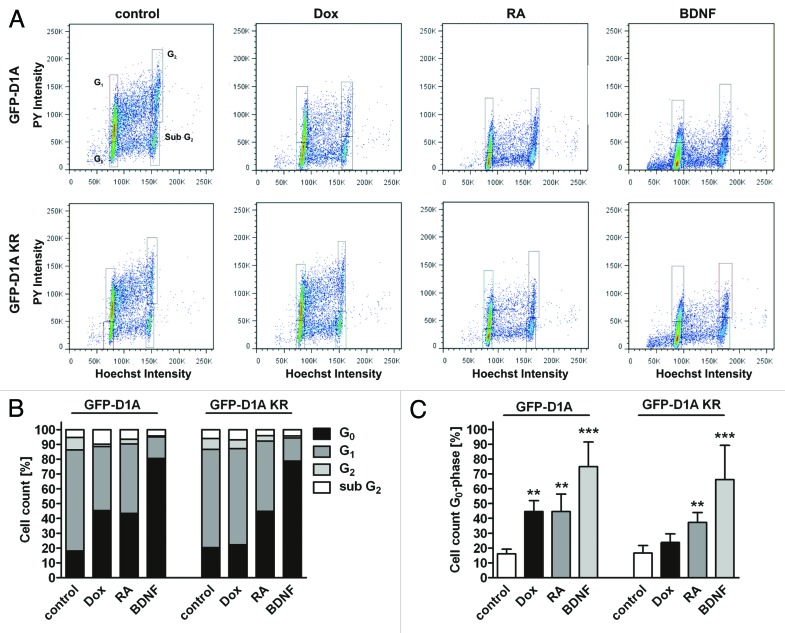
Prolonged retention of neuronal precursors in G1 phase is known to increase the probability of cell cycle exit and differentiation.34 Cells that have exited the cell cycle can be detected by their reduced RNA content.35,36 To test the hypothesis that DYRK1A overexpression induces cell cycle exit of SH-SY5Y cells, we overexpressed DYRK1A for 5 d and performed 2-dimensional FACS analyses of cellular RNA and DNA contents (Fig. 3). As positive control for differentiation and cell cycle exit, SH-SY5Y cells were treated for 5 d with RA or for 3 d with RA followed by 2 d BDNF to achieve full differentiation.33 Overexpression of DYRK1A, but not DYRK1A-KR, resulted in a large increase of cells staining low for pyronin Y (Fig. 3A and B), which identifies them as quiescent cell populations with low RNA content that have exited the cell cycle.35 The percentage of cells in G0 phase was significantly increased after DYRK1A overexpression (45% compared with 16% of control cells, Fig. 3C). Differentiation with either RA alone or RA and BDNF altered the rate of cells in G0 phase to 45% or 75%, respectively. Two-dimensional staining also revealed a subpopulation of SH-SY5Y cells in S and G2 phase with reduced RNA content. These populations have also been observed in other cell types, indicating a quiescent state (SQ- and G2Q) besides G0.35 Here, these cells may represent the intermediate and substrate type (I- and S-type) of SH-SY5Y cells that exit cell cycle but do not develop the characteristics of the neuroblastic cell type (N-type).37 These results show that long-term overexpression of DYRK1A not only prevents S-phase entry of SH-SY5Y neuroblastoma cells, but also results in subsequent cell cycle exit into G0 phase.
Figure 3. DYRK1A overexpression induces cell cycle exit of SH-SY5Y cells. For 2-dimensional analyses of cellular DNA and RNA and contents, SH-SY5Y cells were stained with Hoechst 33342 and pyronin Y (PY). DNA and RNA contents were analyzed by flow cytometry after 5 d of treatment with doxycycline (Dox), 10 µM RA only, or treatment with 10 µM RA followed by 50 ng/ml BDNF. Representative plots of one experiment (A), quantification of cells in G1/0- and G2-phase in this experiment (B) and the evaluation of average cell counts in G0-phase of 3 independent experiments (C). The cell population with 4n DNA content and low RNA staining is referred to as sub-G2. Means + SD; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; analyzed by 1-way ANOVA + Bonferroni post-test comparing DYRK1A or DYRK1A-K188R data to the respective untreated controls.
Overexpression of DYRK1A induces neuronal differentiation of SH-SY5Y cells
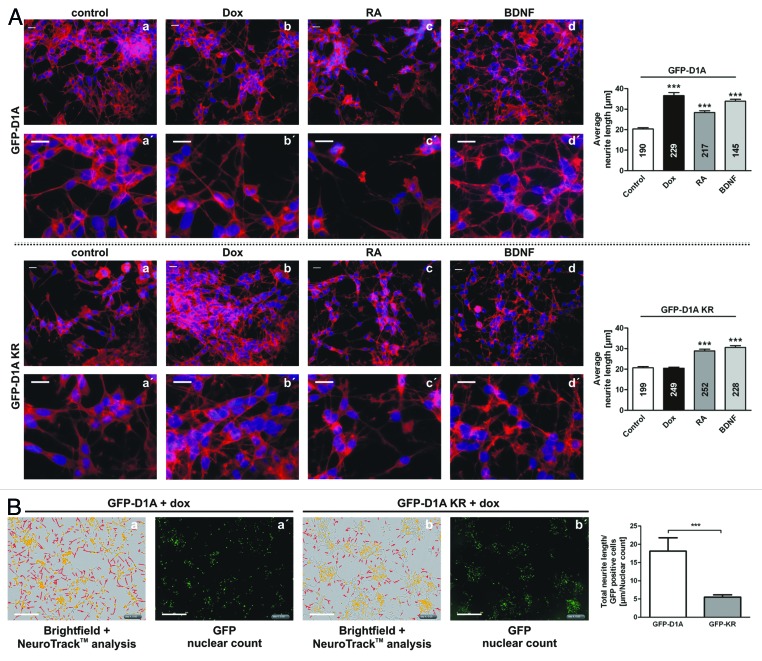
Because G0 cell cycle exit of neuronal cells is linked to differentiation, we next questioned if DYRK1A overexpression induces neuronal differentiation of SH-SY5Y cells. To this end, we visualized the differentiation-related morphological changes of SH-SY5Y cells by staining the F-actin cytoskeleton with fluorescently labeled phalloidin and manually quantified the average neurite length (Fig. 4A). As a reference, 5 d of treatment with RA/BDNF induced the expected phenotype of SH-SY5Y cells with proliferation arrest accompanied by formation of an extensive neurite network.33,38 In contrast, undifferentiated cells showed only few and shorter neurite-like extensions and a higher cell density. DYRK1A overexpression not only stopped cell proliferation, but also induced changes in the cytoskeletal organization of SH-SY5Y cells and an increased neurite length (Fig. 4A). Moreover, DYRK1A induced differentiation, as reflected by increased neurite formation, occurred earlier than neuritogenesis triggered by RA treatment (data not shown). In spite of comparable neurite lengths, the DYRK1A-overexpressing cells did not seem to attain a complete BDNF-induced morphology. Cells overexpressing DYRK1A-KR continued to proliferate and showed no evident morphological changes. To confirm the differentiation-inducing effect of DYRK1A by an observer-independent method, we employed live cell imaging with the automated NeuroTrack™ evaluation of total neurite length per cell. This analysis revealed that 96 h of DYRK1A overexpression in SH-SY5Y cells significantly increased the total neurite length per cell in comparison to cells overexpressing DYRK1A-KR (Fig. 4B).
Figure 4. DYRK1A overexpression induces neurite outgrowth of SH-SY5Y cells. (A) Fluorescence microscopic analysis of actin labeling. Cells were plated on coverslips and DYRK1A (upper panels) or DYRK1A-K188R overexpression (lower panels) was induced with 2 µg/ml doxycycline (Dox) (b). As positive control, cells were differentiated with 10 µM RA solely (c) or with 10 µM RA followed by 50 ng/ml BDNF (d). Control cells were left untreated (a). After 5 d cells were fixed and F-actin labeled with phalloidin-Alexa®-546 to visualize cellular processes. Nuclei were stained with DAPI. Scale bar = 20 µm. (a’–d’) show magnified cutouts of the respective images (a–d). Quantification of the average neurite lengths is illustrated in the column diagrams. Means + SEM; ***P ≤ 0.001; analyzed by 1-way ANOVA + Bonferroni post-test comparing DYRK1A or DYRK1A-K188R data to untreated controls. The number of measured neurites is indicated in each column. To exclude effects on neuritogenesis resulting from different cell densities, proliferation differences were compensated by plating adjusted cell numbers as described in the “Material and Methods” section. (B) Analysis of neuritogenesis using automated live cell microscopy. Overexpression of DYRK1A or DYRK1A-KR was induced with 2 µg/ml doxycycline (+dox). Total neurite length was quantified after 96 h using an IncuCyte™ kinetic imaging system with the integrated automated NeuroTrack™ image acquisition. Panels (a and b) show representative bright field images taken after 96 h with an overlay of the NeuroTrack analysis (red) and the automatically assessed count of GFP positive nuclei (yellow) as shown in (a’ and b’). Scale bar = 300 µm. The column diagram illustrates evaluation of total neurite length standardized to the total count of GFP-positive nuclei. Data are shown as means + SEM of 4 independent experiments and was analyzed using Student t test. ***, P ≤ 0.001.
Next, we analyzed the effect of DYRK1A on mRNA and protein abundance of the neuronal markers Tau (MAPT) and MAP2, which are upregulated after long-term RA treatment.32,38 Transcript levels of both markers were significantly upregulated after 24 h of DYRK1A overexpression (Fig. 5A), whereas DYRK1A-KR overexpression had no effect. After 72 h of DYRK1A overexpression, no differences in MAPT or MAP2 transcript levels were observed (data not shown). Conversely, Tau or MAP2c protein levels showed no detectable changes after 24 h (data not shown) but were significantly increased after 72 h of DYRK1A overexpression (Fig. 5B and C). These results suggest that expression of the neuronal proteins is transiently induced by DYRK1A overexpression and returns to steady-state levels in differentiated cells, although DYRK1A remains overexpressed. Unexpectedly, treatment with RA did not detectably alter MAP2 or Tau levels within 72 h. To ensure the efficacy of RA treatment in this experiment, we confirmed that the expression of the neurotrophin receptor NTRK2 (TrkB) was strongly upregulated in RA-treated SH-SY5Y cells (Fig. S1B).39 Interestingly, overexpression of DYRK1A but not DYRK1A-KR also induced a weak increase of TrkB mRNA level. Notably, endogenous DYRK1A mRNA levels were not changed during RA-induced differentiation (Fig. S1A). Taken together, our results provide evidence that overexpression of DYRK1A triggers G0 cell cycle exit and differentiation of SH-SY5Y cells.
Figure 5. DYRK1A overexpression upregulates neuronal markers in SH-SY5Y cells. (A) SH-SY5Y cells overexpressing DYRK1A or DYRK1A-K188R were treated with 2 µg/ml dox or 10 µM RA for 24 h before total RNA was extracted and analyzed by qRT-PCR. Tau and MAP2 mRNA levels were quantified relative to GAPDH mRNA. (B) Representative western blot of total protein extracts from SH-SY5Y cells 72 h after induction of DYRK1A overexpression or RA treatment. Migration of mass standards is indicated in kDa (left). (C) Densitometric evaluation of Tau and MAP2c immunoreactivity relative to GAPDH. n = 3, means + SD; *P ≤ 0.05; analyzed by one-way ANOVA + Bonferroni post-test to compare columns to none normalized controls (A) or one-sample t test to compare columns to normalized control (C).
DYRK1A alters phosphorylation and protein levels of Cyclin D1 and p27Kip1 dependent on its kinase activity and level of overexpression
Next we investigated if DYRK1A overexpression affected proteins that are known to regulate G1/0-phase transition. Western blot analysis of total protein extracts from SH-SY5Y cells overexpressing DYRK1A revealed changes in phosphorylation and protein levels of the cell cycle regulators Cyclin D1 and p27Kip1 (Fig. 6A–C). Overexpression of DYRK1A resulted in significantly decreased levels of Cyclin D1 after 24 and 72 h, concomitant with an increased phosphorylation of Thr286 (Fig. 6A and B). Furthermore, DYRK1A overexpression strongly enhanced phosphorylation of p27Kip1 at Ser10 and increased total protein levels of p27Kip1 by about 80% (Fig. 6A and C). The effects of DYRK1A on the phosphorylation and protein abundance of Cyclin D1 and p27Kip1 were already apparent after 24 h, although statistical significance of all effects was only reached after 72 h of DYRK1A overexpression. In contrast, kinase-deficient DYRK1A-KR did not alter p27Kip1 and Cyclin D1 phosphorylation or protein levels. Consistent with published results,32 treatment with RA seemed to increase p27Kip1 protein levels, although the effect on Ser10 phosphorylation was weaker than that of DYRK1A.
Because the neurodevelopmental function of DYRK1A is considered to be particularly sensitive to its expression levels, we investigated the effects on Cyclin D1 and p27Kip1 after a gradual overexpression of DYRK1A for 24 and 72 h (Fig. 6D–F). The findings confirm that in SH-SY5Y cells the effects on Cyclin D1 and p27Kip1 protein stability are well correlated with DYRK1A expression levels and expression time, and strictly depend on the catalytic activity of the kinase (Fig. 6E and F). It should be noted that wild-type DYRK1A was overexpressed at higher levels than DYRK1A-KR, although identical expression constructs were used. This effect was also observed in previous studies involving transient or stable overexpression of DYRK1A and DYRK1A-KR.40,41 Nevertheless, our results of gradual overexpression allow the comparison of DYRK1A and DYRK1A-KR at similar expression levels and support the conclusion that the effect of DYRK1A depends on its catalytic activity.
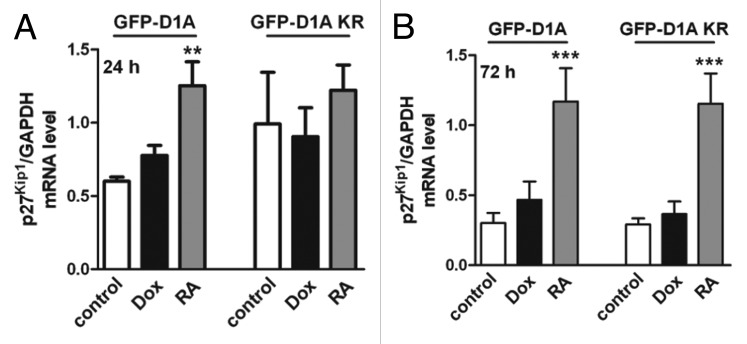
We also asked whether DYRK1A affects CDKN1B (cyclin-dependent kinase inhibitor 1B, encoding p27Kip1) mRNA levels in SH-SY5Y cells. Analysis by quantitative real-time PCR revealed no significant effect of DYRK1A overexpression on p27Kip1 mRNA levels, whereas RA upregulated p27Kip1 mRNA (Fig. 7). Thus, DYRK1A modulates p27Kip1 abundance in SH-SY5Y cells primarily by posttranslational mechanisms.
Figure 7. DYRK1A overexpression has no effect on p27Kip1 mRNA level in SH-SY5Y cells. SH-SY5Y cells were treated with doxycycline or RA for 24 h (A) or 72 h (B) before p27Kip1 mRNA levels were analyzed by qRT-PCR. p27Kip1 mRNA levels are shown as relative quantification to GAPDH mRNA levels. n = 3, means + SD; **P ≤ 0.01; ***P ≤ 0.001, analyzed by 1-way ANOVA + Bonferroni post-test to compare DYRK1A or DYRK1A-K188R data with the respective untreated controls.
DYRK1A directly phosphorylates p27Kip1 at Ser10 and Cyclin D1 at Thr286
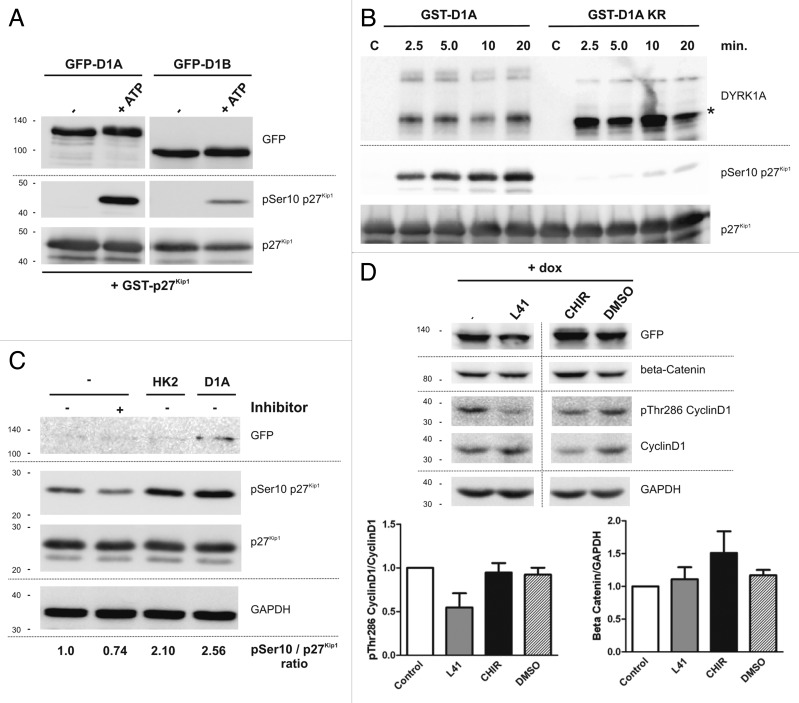
To address the question whether DYRK1A directly phosphorylated p27Kip1, we performed a kinase assay with recombinant p27Kip1 and immunoprecipitated DYRK1A overexpressed in HeLa cells. DYRK1B was used as positive control (Fig. 8A).15 To exclude the possibility that p27Kip1 was phosphorylated by other co-precipitated cellular kinases, a kinase assay was performed with bacterially expressed GST-DYRK1A (Fig. 8B). The in vitro assays prove that DYRK1A can phosphorylate Ser10 directly and not by activating another downstream kinase. To confirm that DYRK1A is able to phosphorylate p27Kip1 in a cellular environment, we used HeLa cells transiently transfected with p27Kip1. Overexpression of DYRK1A increased Ser10 phosphorylation similar to HIPK2, which is known to phosphorylate p27Kip1 at Ser10.14 Inhibition of endogenous DYRK1A with a harmine-related DYRK1A inhibitor (AnnH31, unpublished results) reduced basal Ser10 phosphorylation compared with the untreated control (Fig. 8C).
Figure 8. Phosphorylation of p27Kip1 and Cyclin D1 by DYRK1A in vitro and in cells. (A) In vitro kinase assay. Kinase assay with GFP-DYRK1A and GFP-DYRK1B immunoprecipitated from HeLa cells and subjected to a kinase assay with recombinant GST-p27Kip1. The phosphorylation reaction was started by ATP addition (+ ATP). No ATP addition served as negative control. (B) In vitro kinase assay with bacterial GST fusion proteins of DYRK1A or kinase-deficient DYRK1A-K188R and p27Kip1. Aliquots of the kinase reaction were taken at the indicated time points. Control lanes (c) were loaded only with GST-p27Kip1. The majority of GST-DYRK1A is isolated from E. coli as a catalytically active C-terminally truncated product (marked by an asterisk).79 (C) Phosphorylation of p27Kip1 by DYRK1A in HeLa cells. HeLa cells were transiently transfected with expression plasmids for p27Kip1, GFP-DYRK1A, and GFP-HIPK2 as indicated. To analyze p27Kip1 phosphorylation by endogenous DYRK1A, cells were treated with 1 µM of the DYRK1A inhibitor AnnH31 for 5 h before cell lysis. Total protein extracts then were analyzed for p27Kip1 phosphorylation by western blotting with the indicated antibodies. Relative phosphorylation of Ser10 is indicated below the bottom panel. GFP-HIPK2 was not resolved on this gel due to its large size. One representative blot from n = 3 each. (D) DYRK1A induces Cyclin D1 Thr286 phosphorylation independently of GSK3β. SH-SY5Y cells were treated with 0.5 µg/ml doxycycline (+dox) to induce DYRK1A overexpression. After 5 h, cells were additionally treated with the DYRK1A inhibitor leucettine L41 (1 µM) or the GSK3β inhibitor CHIR99021 (3 µM) for further 24 h before total cellular protein was analyzed by western blotting with the indicated antibodies. Stabilization of β catenin due to reduced GSK3β-mediated phosphorylation was used to validate GSK3β inhibition. The vertical line indicates where an irrelevant lane was deleted from the final image. Column diagrams show the densitometric quantification of Cyclin D1 Thr286 phosphorylation and total β catenin protein levels from 3 independent experiments.
Since GSK3β is known to phosphorylate Cyclin D1 at Thr286,10 we determined if the DYRK1A-mediated phosphorylation of Cyclin D1 was dependent on GSK3β. SH-SY5Y cells overexpressing DYRK1A were treated with either the GSK3β inhibitor CHIR99021 or the DYRK1A inhibitor leucettine L41.42 Inhibition of DYRK1A but not GSK3β reduced Cyclin D1 phosphorylation and increased its protein level (Fig. 8D) Thus, DYRK1A phosphorylates Cyclin D1 independently of GSK3β in SH-SY5Y cells.
To reveal possible protein–protein interactions, we immunoprecipitated GFP-DYRK1A or GFP-DYRK1A-KR from SH-SY5Y cells. Neither p27Kip1 nor Cyclin D1 were co-immunoprecipitated with DYRK1A, although binding of the known DYRK1A interaction partner WDR6843 was well detectable (Fig. S3). Taken together, these results indicate that endogenous or overexpressed DYRK1A phosphorylates p27Kip1 at Ser10, as well as Cyclin D1 at Thr286, without forming stable complexes with these substrates.
DYRK1A phosphorylates p27Kip1 at Ser10 in primary neurons and in vivo
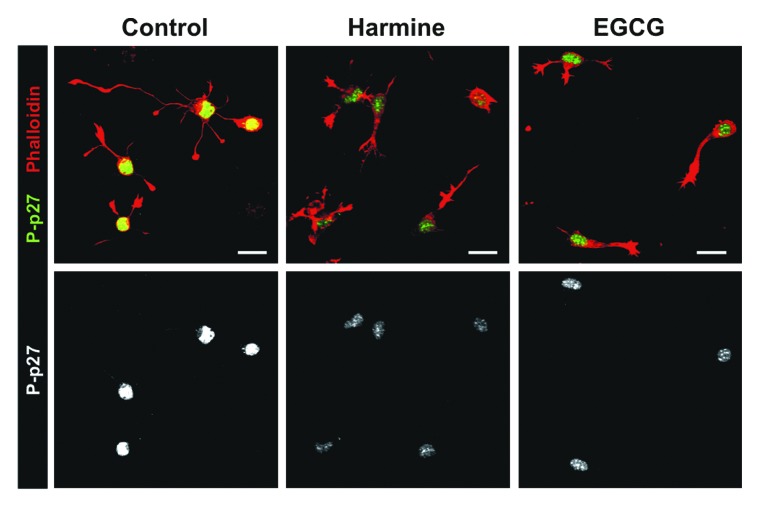
We next asked if p27Kip1 is also phosphorylated by endogenous DYRK1A in primary mouse neurons. To address this question, we treated cultured differentiating hippocampal neurons with the DYRK1A inhibitors harmine and EGCG and observed a strong reduction of pSer10 p27Kip1 immunostaining predominantly in the nucleus (Fig. 9). As we have previously reported,44 inhibition of DYRK1A also suppressed neuritogenesis.
Figure 9. Inhibition of DYRK1A decreases Ser10 phosphorylation of p27Kip1 in early differentiating mouse hippocampal neurons. Fluorescence microscopic analysis of p27Kip1 Ser10 phosphorylation in post-mitotic, differentiating mouse hippocampal neurons. Cells were cultured on coverslips in the presence of the DYRK1A inhibitors harmine (2 µM), EGCG (5 µM), or without inhibitor (control) for 20 h before cells were fixed and immunostained for pSer10 p27Kip1 (P-p27). F-actin was labeled with phalloidin-rhodamine. The upper panels show overlays of pSer10 p27Kip1 and phalloidin signals and the lower panels show pSer10 p27Kip1 only. Scale bar = 25 µm. One representative experiment is shown from n = 3.
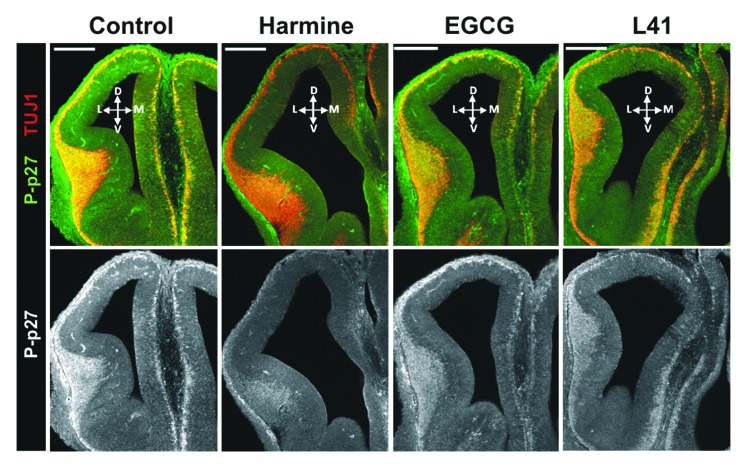
Because DYRK1A has been implicated in different processes of brain development,19 we assessed whether DYRK1A phosphorylates p27Kip1 in vivo. We used mouse whole-embryo cultures (E12), where all relevant neurodevelopmental stages are coexisting and DYRK1A is highly expressed in the telencephalon.45 As shown in Figure 10, most of pSer10 p27Kip1 immunostaining in E12 embryonic mouse brain was overlapping with that of β-III-tubulin (TUJ1), a well-known marker of early differentiating neurons.46 At the same time, the ventricular zone, the area where neuronal progenitors and newborn precursors localize, was mostly devoid of pSer10 p27Kip1 immunostaining. This strongly suggests that pSer10 p27Kip1 is mainly present in differentiating neurons. In order to assess whether DYRK1A contributes to p27Kip1 Ser10 phosphorylation in differentiating brain neurons, we cultured E12 embryos for 6 h in the presence of EGCG, harmine, and leucettine L41, 3 different and chemically unrelated DYRK1A inhibitors.47 This time period is substantially shorter than the duration of a cell cycle, which at this stage has been estimated to last for 20–24 h.4,48 Harmine, EGCG, and leucettine L41 have been profiled against large panels of protein kinases47 and do not inhibit the other kinases that are known to phosphorylate p27Kip1 at Ser10 (KIS, AKT, CDK5, HIPK2)11-14,49 except for DYRK1B, which is a regulator of p27Kip1 in cancer cells.15,50 All 3 DYRK1A inhibitors substantially decreased pSer10 p27Kip1 immunostaining in the mouse telencephalon (Fig. 10). This reduction was not an indirect consequence of the inhibitors affecting cell cycle, since we did not observe substantial alterations of p27Kip1 total protein levels, as well as no increase of proliferation or cell death as determined by EdU labeling and detection of activated caspase 3 (Fig. S4). These data strongly indicate that DYRK1A accounts for a large part of pSer10 p27Kip1 in differentiating mouse brain neurons.
Figure 10. Inhibition of DYRK1A decreases Ser10 phosphorylation of p27Kip1 in the embryonic mouse telencephalon. Confocal images of one hemisphere showing the telencephalon of E12 mouse embryos cultured for 6 h in the presence of the DYRK1A inhibitors harmine (4 µM), EGCG (10 µM), leucettine L41 (8 µM), or without inhibitor (control). Brain slices were stained for pSer10 p27Kip1 (P-p27) and β-III-tubulin (TUJ1) as indicated. The dorso–ventral (D-V) and medial–lateral (M-L) orientation is indicated. Scale bar = 200 µm. One representative experiment is shown from n = 3.
Discussion
There is a growing body of evidence that overexpression of DYRK1A in Down syndrome affects brain development and neuronal differentiation.19,29,51-56 DYRK1A was recently identified as a negative regulator of G1–S transition, but the underlying mechanisms in neurons remain largely unknown.40,41,54 We therefore addressed the question how DYRK1A overexpression influences neuronal cell cycle progression and differentiation.
DYRK1A promotes neuronal differentiation by inducing G1-phase arrest and G0 cell cycle exit
We employed the human SH-SY5Y neuroblastoma cell line as a model system to investigate the effect of controlled DYRK1A overexpression on cell cycle progression and neuronal differentiation. After differentiation with RA and BDNF, SH-SY5Y cells exit the cell cycle and achieve neuronal characteristics. Therefore they are well established as a cell model for neurobiological questions.32,33,38,57
Here we show that DYRK1A suppressed proliferation of SH-SY5Y cells in a dose- and kinase activity-dependent manner. This finding supports the validity of our cell model, since DYRK1A overexpression has previously been reported to attenuate the proliferation of neural progenitors in the developing mouse cerebral cortex and neural precursor cells derived from human embryonic stem cells.29,54 The majority of SH-SY5Y cells ceased proliferation and arrested in G1 phase after 24 h of DYRK1A overexpression, whereas RA arrests the cell cycle of SH-SY5Y cells in a delayed manner after at least 2–3 d.32 Therefore, our results indicate a direct effect of DYRK1A on the current G1-phase once expression levels and activity reach a certain threshold. Enhanced DYRK1A activity was required to maintain cell cycle arrest, since pharmacological inhibition of DYRK1A after 24 h partially restored cell proliferation. Consistent with previous reports that DYRK1A acts as a negative regulator of apoptosis and a positive regulator of cell survival, DYRK1A overexpression did not result in enhanced cell death of SH-SY5Y cells.58,59
Long-term overexpression of DYRK1A increased a population of SH-SY5Y cells with reduced cellular DNA and RNA contents, which characterize cells that have entered a permanently non-proliferative G0 state (quiescence/differentiation).36 Accordingly, the largest fraction of quiescent cells was observed after induction of full differentiation with RA and BDNF.33 DYRK1A overexpressing SH-SY5Y cells not only exited cell cycle into G0 but also developed morphological and molecular hallmarks of neuronal differentiation, including enhanced neurite outgrowth and expression of the neuronal markers Tau and MAP2. Compared with RA,39 DYRK1A overexpression resulted in a much weaker increase of TrkB mRNA (Fig. S1B). Taking also into account the faster effect on G1 arrest, we conclude that DYRK1A overexpression and RA treatment promote SH-SY5Y differentiation in different ways. DYRK1A primarily induces cell cycle arrest of SH-SY5Y by phosphorylation of cell cycle proteins, which then favors neuronal differentiation, whereas RA induces changes of differentiation-related genes (such as TrkB) in the first place before cells exit the cell cycle.60,61 Nonetheless, elevated DYRK1A activity eventually induced the expression of neuronal markers.
We and others have previously shown that DYRK1A overexpression enhances neurite outgrowth in rat PC12 cells and that DYRK1A inhibition interferes with neurite formation of mouse hippocampal neurons.28,30,44 Consistent with our previous findings in the developing vertebrate central nervous system,31 our present results provide evidence that DYRK1A regulates G1–S transition of neuronal cells and that an elevated activity of DYRK1A promotes proliferation arrest, cell cycle exit, and premature differentiation.
DYRK1A phosphorylates p27Kip1 at Ser10 and Cyclin D1 at Thr286
Cell cycle exit and differentiation of neural precursors are regulated by p27Kip1 and Cyclin D/CDK complexes.1,3,62 Here we show that DYRK1A increased p27Kip1 protein levels in SH-SY5Y cells by phosphorylation at Ser10, a phosphorylation site known to induce p27Kip1 stabilization.9,63 Interestingly, phosphorylation of p27Kip1 on Ser10 in neural stem cells has been shown to be correlated with cell cycle exit and progression to neuronal differentiation and neurite outgrowth.49 In addition, phosphorylation of Ser10 by an unidentified protein kinase has previously been reported to account for the increased half-life and accumulation of p27Kip1 during RA-induced differentiation of neuroblastoma cells.64 In SH-SY5Y cells, phosphorylation and stabilization of p27Kip1 were well correlated with the time course and level of DYRK1A overexpression, and DYRK1A was capable of directly phosphorylating Ser10 in recombinant p27Kip1. In contrast to our previous observations in neuronal precursors of developing chicken spinal chord and mouse telencephalon,31 p27Kip1 mRNA levels were not significantly changed by DYRK1A overexpression in SH-SY5Y cells. Here we show that inhibition of DYRK1A activity with different inhibitors reduced p27Kip1 Ser10 phosphorylation in differentiating hippocampal neurons and developing mouse telencephalon. Overexpression of p27Kip1 has been shown to lengthen G1 phase of neural progenitor cells in the mouse embryo.65 However, we detected p27Kip1 Ser10 phosphorylation primarily in early differentiating neurons but not in the ventricular zone of mouse telencephalon, where neural progenitors and newborn neuronal precursors are located. Thus, our results identify DYRK1A as the predominant kinase that phosphorylates and stabilizes p27Kip1 during neuronal differentiation. Consistently, beyond its function as an inhibitor of CDK activity in the nucleus, p27Kip1 is also thought to be a cytosolic mediator of neuronal differentiation, microtubule polymerization, and migration.7,34,66 Here we propose that DYRK1A overexpression promotes neuronal differentiation by increasing p27Kip1 levels through phosphorylation at Ser10.
Additionally, we provide evidence that DYRK1A reduces Cyclin D1 protein levels in SH-SY5Y cells by phosphorylation on Thr286. Phosphorylation of Cyclin D1 on Thr286 induces the ubiquitinylation by the Skp1-Cul1-F box (SCF) ubiquitin ligase and subsequent proteasomal degradation.67 Although GSK3β was initially reported to phosphorylate Cyclin D1 on Thr286,68 DYRK1B was later proposed to promote Cyclin D1 degradation by phosphorylation at Thr288.69 Two recent reports have now made clear that either DYRK1A or DYRK1B can promote Cyclin D1 degradation via direct phosphorylation of Thr286 but do not target Thr288.16,41 Consistent with our results for the effect of DYRK1A in SH-SY5Y cells, Ashford et al.16 have found in the pancreatic carcinoma cell line PANC-1 that the DYRK1B-driven phosphorylation at Thr286 and turnover of Cyclin D1 were not reduced by GSK3β inhibition. Thus, DYRK1A and DYRK1B regulate Cyclin D1 stability independent of GSK3β activity at least in some cell types.
DYRK1A overexpression has been shown to induce nuclear export of ectopically expressed Cyclin D1 in neuronal cells,29 which is considered an S phase-specific regulatory mechanism.68,70 Here we demonstrate that DYRK1A induces degradation of Cyclin D1 in SH-SY5Y cells concurrent with enhanced phosphorylation on Thr286. This effect occurred within 24 h after induction of DYRK1A overexpression and thus could account for G1-phase arrest and subsequent neuronal differentiation of SH-SY5Y cells. In fact, a previous study has shown that silencing of Cyclin D1 expression sufficed to induce cell cycle exit and subsequent differentiation of neuroblastoma cells.68
In contrast to Chen et al.,41 who showed a stable interaction of overexpressed Cyclin D1 with DYRK1A in cotransfected HEK293T cells, we did not detect endogenous Cyclin D1 or p27Kip1 in co-immunoprecipitates of GFP-DYRK1A in SH-SY5Y cells (Fig. S3). It remains to be determined whether this discrepancy reflects different experimental conditions or differences in protein interactions between the HEK293 cell line and neuronal cells.
DYRK1A in neuronal cell cycle regulation
The observation that DYRK1A regulates protein turnover of p27Kip1 and Cyclin D1 in neuronal cells is in accordance with numerous previous reports that kinases of the DYRK family function as regulators of protein stability in cell cycle control.27 However, our results do not exclude the possibility that other downstream targets of DYRK1A contribute to its differentiation-inducing effect. DYRK1A has been proposed to promote cell cycle exit and quiescence by phosphorylation of LIN52 on Ser28, which is required for the assembly of the DREAM complex.40 However, Chen et al.41 have recently provided evidence that the stabilization of the DREAM complex may be a long-term effect of DYRK1A in the maintenance of quiescence rather than a direct regulatory mechanism of G1 cell cycle exit. For example, it is conceivable that DYRK1A induced depletion of nuclear Cyclin D1 promotes DREAM complex formation by reducing CyclinD/CDK4-dependent phosphorylation of p130/RBL2.71,72 Another substrate of DYRK1A that may contribute to neuronal cell cycle regulation and differentiation is p53.54,73 Phosphorylation at Ser15 by DYRK1A enhances expression of p53 target genes such as the CDK inhibitor p21CIP1 and thereby attenuates G1–S phase transition.
Conclusion
In conclusion, we have shown that overexpression of DYRK1A arrests proliferating neuronal cells in G1 phase within about 24 h, which can be best explained by the rapid degradation of Cyclin D1.41 Thereafter, DYRK1A-dependent stabilization of p27Kip1, combined with further Cyclin D1 depletion, may promote cell cycle exit into G0 and subsequent neuronal differentiation.34,72 Because of its well-documented overexpression in trisomy 21,17,18 DYRK1A may affect brain development in Down syndrome by deregulating G1 phase of the proliferating neuronal precursor and inducing premature differentiation, which would result in a depletion of the neural progenitor pool in brain development and eventually in an inappropriate number of neurons.19,25
Materials and Methods
Generation of tetON-DYRK1A SH-SY5Y neuroblastoma cells
A lentiviral vector system was used to generate SH-SY5Y sublines for controllable overexpression of GFP-DYRK1A or GFP-DYRK1A-K188R. In brief, rat DYRK1A and DYRK1A-K188R cDNA were amplified out of pEGFP-DYRK1A and integrated into the lentiviral FUW-tetON backbone (after removing the hMYC insert from Addgene plasmid 20723 with EcoRI) by homologous recombination in Escherichia coli,74 resulting in FUW-tetON-GFP-DYRK1A and FUW-tetON-GFP-DYRK1A-K188R. Co-transduction of the FUW-tetON vector with FUW-M2rtTA (Addgene 20342), which encodes the tetracycline-dependent transactivator, allows for tetracycline-regulated expression of the transgene.75 Lentivirus production was performed as described.30 Co-transduction of SH-SY5Y cells then was titrated with different amounts of FUW-tetON-GFP-DYRK1A/KR and FUW-M2rtTA containing lentivirus until achieving maximal transduction efficiency as controlled by doxycycline-induced GFP fluorescence. These cell populations were considered as stably transduced, amplified, and frozen. All experiments were performed with cells derived from the same transduction experiment.
Culture and treatment of cell lines, differentiation, and transfection
The following substances were used for treatment of cultured cells: Doxycycline (Sigma), all-trans retinoic acid (RA) (Sigma), brain-derived neurotrophic factor (BDNF) (Peprotech), harmine (Fluka), CHIR99021 (Cayman Chemical), staurosporine (Enzo Life Sciences). Leucettine L41 was a kind gift from Laurent Meijer (ManRos Therapeutics).
SH-SY5Y-tetON-GFP-DYRK1A/DYRK1A-K188R cells were cultured in Dulbecco modified Eagle medium (DMEM) high glucose (PAA) supplemented with 10% FCS (PAA) at 37 °C, 5% CO2, and constant humidity. At 75% confluence, cells were washed twice with PBS, trypsinized, and resuspended in fresh medium. Cells were centrifuged for 5 min at 500 g, resuspended in fresh medium, and then plated onto multiwell plates or dishes according to the experimental requirements. DYRK1A overexpression was induced by treatment with 2 µg/ml doxycycline or as indicated. Neuronal differentiation was induced with 10 µM retinoic acid (RA). For terminal differentiation, the RA-containing medium was removed after 3 d and replaced by serum-free DMEM high glucose containing 50 ng/ml brain derived neurotrophic factor (BDNF) for 2 more days. For long-term differentiation with BDNF, culture dishes were coated with 0.05 mg/ml collagen from rat tail.
HeLa cells were cultured in Quantum medium for HeLa cells (PAA) at 37 °C, 5% CO2, and constant humidity. For transfection, 100 000 cells were seeded into 6-well plates and 0.5 µg of pCS2-p27Kip1 (kindly provided by J Vervoorts-Weber, Institute of Biochemistry, RWTH Aachen University) were either transfected alone or together with 0.5 µg pEGFP-DYRK1A or pEGFP-HIPK276 (kindly provided by ML Schmitz, Justus-Liebig-University) using FuGene HD transfection reagent (Promega).
Real-time cell analysis (RTCA)
The xCELLigence RTCA system with a 96-well E-Plate (Roche) was used for continuous impedance-based monitoring of cell proliferation. Before plating cells, the baseline impedance of 200 µl medium per well was measured. After removing 100 µl of medium, 7500 SH-SY5Y-tetON-GFP-DYRK1A or tetON-GFP-DYRK1A-K188R cells/well were added in a volume of 100 µl. Impedance measurements were taken at intervals of 15 min for a total experimental time of 7 d. After cell adhesion overnight, 100 µl medium/well were carefully replaced by 100 µl fresh medium containing doxycycline to reach the intended final concentrations. Harmine was added 24 h after doxycycline induction. All handling outside the incubator was performed using a 37 °C pre-warmed hot plate to minimize impedance measurement artifacts. Impedance values were averaged over triplicate wells and automatically converted to a dimensionless value called cell index (CI) by the xCELLigence software (RTCA 1.2.1, Roche), representing the density of cells per well. Data was exported to GraphPad Prism 5.0 (GaphPad software) for determination of the respective area under the curve.
Cell cycle analysis and flow cytometry
One-dimensional DNA content measurements were performed after staining with propidium iodide (PI). In brief, 750 000 SH-SY5Y-tetON-GFP-DYRK1A or tetON-GFP-DYRK1A-K188R cells/well were seeded into 6-well plates for 24 h measurements. For analysis after 72 h of cultivation, reduced proliferation of DYRK1A-overexpressing and differentiating cells was compensated by plating cells as follows: 350 000 control cells or DYRK1A-KR-overexpressing cells and 500 000 cells overexpressing DYRK1A or undergoing RA or RA/BDNF differentiation. The day after plating, cells were treated with 2 µg/ml doxycycline to induce DYRK1A overexpression or with 10 µM RA to induce differentiation. After 24 or 72 h of cultivation, cells were harvested in cold PBS and fixed at −20 °C overnight at a final concentration of 75% EtOH. Cells then were transferred to FACS tubes and spun down for 5 min at 500 g, resuspended in 1 ml cold PBS, and rehydrated for 10 min. After additional centrifugation, the pellet was resuspended in 400 µl of PI staining solution (50 µg/ml PI [Sigma], 20 µg/ml RNase A in PBS) and stained for 30 min on ice.
Two-dimensional (multiparameter) DNA and RNA analysis36 was performed using 300 000 plated control or DYRK1A-KR overexpressing cells and 500 000 plated cells overexpressing wild-type DYRK1A or undergoing RA/BDNF-induced differentiation. After fixation, cell pellets were resuspended in 400 µl DNA staining solution containing 2 µg/ml Hoechst 33342 (Sigma) and incubated for 30 min at room temperature. Afterwards 4 µg/ml Pyronin Y (Sigma) was added to stain RNA, and tubes were kept on ice for 20 min. Dyes were washed out by centrifuging the samples at 500 g for 5 min, washing the pellet with 1 ml cold PBS, centrifuging again, and finally resuspending in 400 µl PBS.
Nucleic acid contents were analyzed using an LSR Fortessa flow cytometer (BD Bioscience), and data were evaluated with FlowJo 7.6.5 software (Tree Star). For one-dimensional DNA measurements the Dean–Jett–Fox algorithm was used to evaluate cell cycle phase distribution (Constrains: G2 width = G1). In the case of 2-dimensional DNA and RNA measurements, gates were applied and cell cycle phases determined according to Darzynkiewicz and Shapiro.35,36
Extraction of total protein, SDS-PAGE, western blotting, and immunodetection
To compensate for the treatment-dependent differences in cell proliferation, the numbers of SH-SY5Y-tetON-GFP-DYRK1A/DYRK1A-K188R cells seeded into 6-well plates were adjusted as described for FACS analysis. After 24 or 72 h, cells were carefully washed once and then collected in cold PBS, centrifuged for 5 min at 500 g, and the cell pellet lysed in 75 µl of hot SDS-lysis buffer (20 mM Tris-HCL pH 7.4, 1% SDS in ddH20). After boiling (5 min at 98 °C), samples were sonicated on ice for 3 min and subsequently centrifuged for 5 min at 20 000 g. The supernatant was collected, and sample amounts were adjusted according to the bicinchoninic acid protein assay. Protein extraction of transfected HeLa cells was performed using 150 µl SDS-lysis buffer and the same lysis protocol. Transfer to nitrocellulose membranes was performed by tank blotting for 2 h at 200 mA. Afterwards, membranes were blocked in 5% BSA/TBST, and incubation of primary antibodies followed overnight. After incubation with HRP-coupled secondary antibodies, chemiluminescence detection was performed with a LAS 3000 CCD imaging system (Fujifilm), and densitometric signal quantification followed by using the AIDA imager software (Raytest). For immunodetection the following antibodies were used: GAPDH (mouse, CST), Cyclin D1 (mouse, CST), pThr286 Cyclin D1 (rabbit, CST), p27Kip1 (mouse, BD Biosciences), pSer10 p27Kip1 (rabbit, Abcam), PARP (rabbit, CST), Beta Catenin (mouse, BD Biosciences), pSer10 HistoneH3 (rabbit, CST), (MAP2 (mouse, Sigma), Tau-13 (mouse, SantaCruz), DYRK1A (mouse, Abnova), WDR68 (rabbit, Abcam), GFP (goat, Rockland). Secondary HRP-coupled antibodies (donkey) were from Rockland.
RNA isolation and quantitative real-time PCR
Total RNA was isolated from 2.0 × 106 plated SH-SY5Y cells at the indicated time points using the NucleoSpin II kit (Macherey-Nagel) with the QIAcube workstation (Qiagen) according to the manufacturer’s instructions. The sequences of PCR primers and the PCR conditions are given in the supplemental material.
In vitro kinase assays
GFP-DYRK1A and GFP-DYRK1B were immunoprecipitated from transiently transfected HeLa cells with the help of the GFP trap M reagent (ChromoTek). A fusion protein of human p27Kip1 and glutathione S transferase (GST-p27Kip1) was produced in Escherichia coli from the plasmid pGEX-5x-3-p27Kip1 (kindly provided by J Vervoorts-Weber, Institute of Biochemistry, RWTH Aachen University) and purified by affinity adsorption to glutathione Sepharose. The immobilized kinases were incubated with 400 ng GST-p27 for 1 h at 30 °C in 30 µl kinase buffer (25 mM Hepes pH 7.4, 0.5 mM dithiothreitol, 5 mM MgCl2) in the presence or absence of 500 µM ATP, and phosphorylation of Ser10 was detected by western blot analysis. Kinase assays with bacterial GST fusion proteins of DYRK1A or DYRK1A-K188R and GST-p27Kip1 were performed in 100 µl kinase buffer with 100 mM ATP at room temperature.
Culture of primary hippocampal neurons and whole-mouse embryos
Mice (ICR strain) were housed, bred, treated, and used in experiments according to the corresponding Spanish laws (particularly, 32/2007 [Nov 7] and 1201/2005 [Oct 10]). Specific experimental procedures were approved by the Commission of Ethics in Animal Experimentation of the Instituto de Neurociencias and the Bioethics Commission of the CSIC (Spanish Upper Research Council). Hippocampal neurons were isolated from E17.5 mouse embryos and cultured according to Kaech and Banker77 for 18–24 h as previously described.44 The DYRK1A inhibitors harmine (2 µM) and EGCG (5 µM) were added when changing the plating medium to neuronal medium. This time point is considered as t0, since no signs of neuronal differentiation are observed before. After 20 h of inhibitor treatment, neurons were fixed with 4% paraformaldehyde.
Dissection and culture of mouse embryos was performed essentially as described by Takahashi et al.78 E12 embryos were cultured in the presence or absence of the DYRK1A inhibitors harmine (4 µM), EGCG (10 µM), leucettine L41 (8 µM) or without inhibitor for 6 h. EdU (5-ethynyl-2’-deoxyuridine) (Invitrogen/Life Technologies) was added to the culture medium at a final concentration of 20 µM 1 h prior to stopping the culture. The brains of the embryos were immediately dissected in cold PBS and fixed in 4% paraformaldehyde. After washing out the fixative, brains were embedded in 4% low gelling temperature agarose and sectioned in frontal orientation with a vibratome.
Fluorescence imaging and continuous live cell imaging
For fluorescence imaging, SH-SY5Y cells were seeded on collagen-coated 12-mm glass coverslips (Marienfeld) in 24-well plates as follows: 30 000 control or DYRK1A-KR-overexpressing cells and 60 000 cells overexpressing DYRK1A or undergoing RA or RA/BDNF differentiation. After 5 d of treatment, cells were fixed in 4% paraformaldehyde (Sigma) for 20 min at 4 °C. Cells were then permeabilized for 30 min at room temperature (5% BSA, 0.1% Triton X-100, in PBS) and stained with Alexa®-546-coupled phalloidin (Invitrogen; 1:40 in 5% BSA, 0.1% Triton X-100, in PBS). Cells were washed twice with PBS and mounted on object slides using DAPI containing Vectashield (Vectorlabs). Fluorescence imaging was performed using an Axiovert 200 M inverted microscope (Zeiss) and images were edited using the AxioVision 4.7 software (Zeiss). Neurite lengths were measured using the AxioVision software, and only neurites that were clearly definable over their entire length were selected for evaluation.
After fixation, mouse hippocampal neurons were permeabilized as described for SH-SY5Y cells and incubated with a pSer10 p27Kip1 antibody (rabbit, Santa Cruz) overnight as recommended by the manufacturer. Fluorescence labeling then was performed using a secondary anti-rabbit biotin labeled antibody (Jackson ImmunoResearch) and Cy2-coupled Streptavidin (GE Healthcare). To visualize cell morphology, the actin cytoskeleton was stained with phalloidin-Rhodamine (1:100, Sigma). Images were collected by confocal microscopy.
After sectioning the telencephalon of cultured mouse embryos, EdU incorporation and immunostaining (rabbit anti pSer10 p27Kip1, Santa Cruz; mouse anti-p27Kip1, BD Biosciences; rabbit anti-Caspase 3, CST and mouse anti-TUJ1, Covance) of floating sections was performed essentially as previously described.31 Images were collected by confocal microscopy.
Continuous live cell imaging was performed using an IncuCyte™ Zoom kinetic imaging system (Essen Biosciences). For analysis of proliferation and differentiation, 90 000 SH-SY5Y-tetON-GFP-DYRK1A or tet-ON-GFP-DYRK1A-K188R cells were seeded into 24-well plates. DYRK1A or DYRK1A-K188R overexpression was induced the next day with 2 µg/ml doxycycline. Subsequently, cell proliferation was followed by automated continuous live cell imaging. The whole well was imaged by recording 9 phase-contrast and GFP images per well in intervals of 1 h at the same positions (10× objective lens) for 96 h in total. For analysis of proliferation, well confluence (%) was automatically calculated at 24 h and 96 h by using the Basic Analyzer segmentation mask of the IncuCyte Zoom™ software (v2013B, Essen Biosciences). The number of GFP-positive cells was assessed using the Basic Analyzer segmentation mask (nuclear count/mm2 image area).
To analyze changes in neurite outgrowth by DYRK1A or DYRK1A-K188R overexpression, images recorded after 96 h of cultivation were analyzed using the automated NeuroTrack™ image acquisition module of the IncuCyte Zoom™ software. The NeuroTrack algorithm automatically defines cell bodies and neurite extensions (as illustrated in Fig. 4B) to calculate and quantify the collective neurite length per image area (mm/mm2 image area). The neurite length was standardized to the total count of GFP-positive nuclei (nuclear count/mm2) yielding the neurite length per cell (mm/nuclear count).
Statistics
Statistical analyses were performed with JMP 10.0.0 (SAS) and GraphPad Prism 5.0 (GaphPad software). All data were tested for homoscedasticity. Equality of variances was analyzed by Bartlett test and normality of residues by Shapiro–Wilk test. In case of heteroscedasticity data were transformed by Box–Cox transformation to achieve homoscedasticity. Parametric data were compared either by one-sample t test (comparison to normalized control values), Student t test (comparison of 2 groups) or one-way ANOVA followed by Βonferroni multiple comparison post-test (multiple comparisons of selected pairs of columns). If homoscedasticity was not achieved after Box–Cox transformation, non-parametric Kruskal–Wallis test followed by Dunn post-test (multiple comparisons of selected pairs of columns) was performed. The used statistical tests are also indicated in the respective figure legends.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We are grateful to Simone Bamberg-Lemper, Birgit Feulner, Nadine Ruske, and Sofia Jimenez Garcia for excellent technical assistance. We wish to thank Florian Glenewinkel for carrying out the p27Kip1 in vitro kinase assay, Alexandra Alves-Sampaio for establishing the mouse embryo culture, Marco Schlepütz for profound statistical support. We also thank Jörg Vervoorts-Weber (Institute of Biochemistry, RWTH Aachen University, Germany), Lienhard Schmitz (Justus-Liebig-University, Giessen, Germany), and Laurent Meijer (ManRos Therapeutics, Roscoff, France) for providing reagents. This work was funded by grants from the German research foundation (DFG grant Be 1967/2–1) to W.B., from the Fondation Jérôme Lejeune (France) to W.B. and F.J.T., as well as by grants of the MINECO (BFU2012–38892) and Generalitat Valenciana (FPA/2012/067) to F.J.T. U.S. was supported by a doctoral fellowship from the Friedrich-Naumann-Foundation, subsidized by the federal government of Germany.
Glossary
Abbreviations:
- BDNF
brain-derived neurotrophic factor
- CDK
cyclin-dependent kinase
- ERK
extracellular signal-regulated kinase
- EGCG
epigallocatechin gallate
- GFP
green fluorescent protein
- GSK3β
glycogen synthase kinase 3β
- MAP2
microtubule-associated protein 2
- qRT-PCR
quantitative real-time PCR
- RA
retinoic acid
References
- 1.Frank CL, Tsai L-H. Alternative functions of core cell cycle regulators in neuronal migration, neuronal maturation, and synaptic plasticity. Neuron. 2009;62:312–26. doi: 10.1016/j.neuron.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salomoni P, Calegari F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 2010;20:233–43. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Hindley C, Philpott A. Co-ordination of cell cycle and differentiation in the developing nervous system. Biochem J. 2012;444:375–82. doi: 10.1042/BJ20112040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calegari F, Haubensak W, Haffner C, Huttner WB. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci. 2005;25:6533–8. doi: 10.1523/JNEUROSCI.0778-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doetsch F, Verdugo JM-G, Caille I, Alvarez-Buylla A, Chao MV, Casaccia-Bonnefil P. Lack of the cell-cycle inhibitor p27Kip1 results in selective increase of transit-amplifying cells for adult neurogenesis. J Neurosci. 2002;22:2255–64. doi: 10.1523/JNEUROSCI.22-06-02255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohnuma S, Harris WA. Neurogenesis and the cell cycle. Neuron. 2003;40:199–208. doi: 10.1016/S0896-6273(03)00632-9. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen L, Besson A, Roberts JM, Guillemot F. Coupling cell cycle exit, neuronal differentiation and migration in cortical neurogenesis. Cell Cycle. 2006;5:2314–8. doi: 10.4161/cc.5.20.3381. [DOI] [PubMed] [Google Scholar]
- 8.Lange C, Huttner WB, Calegari F. Cdk4/Cyclin D1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–31. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Ishida N, Kitagawa M, Hatakeyama S, Nakayama K. Phosphorylation at serine 10, a major phosphorylation site of p27(Kip1), increases its protein stability. J Biol Chem. 2000;275:25146–54. doi: 10.1074/jbc.M001144200. [DOI] [PubMed] [Google Scholar]
- 10.Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–72. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 11.Boehm M, Yoshimoto T, Crook MF, Nallamshetty S, True A, Nabel GJ, Nabel EG. A growth factor-dependent nuclear kinase phosphorylates p27(Kip1) and regulates cell cycle progression. EMBO J. 2002;21:3390–401. doi: 10.1093/emboj/cdf343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita N, Sato S, Katayama K, Tsuruo T. Akt-dependent phosphorylation of p27Kip1 promotes binding to 14-3-3 and cytoplasmic localization. J Biol Chem. 2002;277:28706–13. doi: 10.1074/jbc.M203668200. [DOI] [PubMed] [Google Scholar]
- 13.Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. Cdk5 phosphorylates and stabilizes p27kip1 contributing to actin organization and cortical neuronal migration. Nat Cell Biol. 2006;8:17–26. doi: 10.1038/ncb1338. [DOI] [PubMed] [Google Scholar]
- 14.Pierantoni GM, Esposito F, Tornincasa M, Rinaldo C, Viglietto G, Soddu S, Fusco A. Homeodomain-interacting protein kinase-2 stabilizes p27(kip1) by its phosphorylation at serine 10 and contributes to cell motility. J Biol Chem. 2011;286:29005–13. doi: 10.1074/jbc.M111.230854. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Deng X, Mercer SE, Shah S, Ewton DZ, Friedman E. The cyclin-dependent kinase inhibitor p27Kip1 is stabilized in G(0) by Mirk/dyrk1B kinase. J Biol Chem. 2004;279:22498–504. doi: 10.1074/jbc.M400479200. [DOI] [PubMed] [Google Scholar]
- 16.Ashford AL, Oxley D, Kettle J, Hudson K, Guichard S, Cook SJ, Lochhead PA. A novel DYRK1B inhibitor AZ191 demonstrates that DYRK1B acts independently of GSK3β to phosphorylate cyclin D1 at Thr(286), not Thr(288) Biochem J. 2014;457:43–56. doi: 10.1042/BJ20130461. [DOI] [PubMed] [Google Scholar]
- 17.Guimera J, Casas C, Estivill X, Pritchard M. Human minibrain homologue (MNBH/DYRK1): characterization, alternative splicing, differential tissue expression, and overexpression in Down syndrome. Genomics. 1999;57:407–18. doi: 10.1006/geno.1999.5775. [DOI] [PubMed] [Google Scholar]
- 18.Dowjat WK, Adayev T, Kuchna I, Nowicki K, Palminiello S, Hwang YW, Wegiel J. Trisomy-driven overexpression of DYRK1A kinase in the brain of subjects with Down syndrome. Neurosci Lett. 2007;413:77–81. doi: 10.1016/j.neulet.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tejedor FJ, Hämmerle B. MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J. 2011;278:223–35. doi: 10.1111/j.1742-4658.2010.07954.x. [DOI] [PubMed] [Google Scholar]
- 20.Møller RS, Kübart S, Hoeltzenbein M, Heye B, Vogel I, Hansen CP, Menzel C, Ullmann R, Tommerup N, Ropers H-H, et al. Truncation of the Down syndrome candidate gene DYRK1A in two unrelated patients with microcephaly. Am J Hum Genet. 2008;82:1165–70. doi: 10.1016/j.ajhg.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Courcet J-B, Faivre L, Malzac P, Masurel-Paulet A, Lopez E, Callier P, Lambert L, Lemesle M, Thevenon J, Gigot N, et al. The DYRK1A gene is a cause of syndromic intellectual disability with severe microcephaly and epilepsy. J Med Genet. 2012;49:731–6. doi: 10.1136/jmedgenet-2012-101251. [DOI] [PubMed] [Google Scholar]
- 22.Kawauchi T, Shikanai M, Kosodo Y. Extra-cell cycle regulatory functions of cyclin-dependent kinases (CDK) and CDK inhibitor proteins contribute to brain development and neurological disorders. Genes Cells. 2013;18:176–94. doi: 10.1111/gtc.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hämmerle B, Elizalde C, Galceran J, Becker W, Tejedor FJ. The MNB/DYRK1A protein kinase: neurobiological functions and Down syndrome implications. J Neural Transm Suppl. 2003;67:129–37. doi: 10.1007/978-3-7091-6721-2_11. [DOI] [PubMed] [Google Scholar]
- 24.Park J, Song W-J, Chung KC. Function and regulation of Dyrk1A: towards understanding Down syndrome. Cell Mol Life Sci. 2009;66:3235–40. doi: 10.1007/s00018-009-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dierssen M. Down syndrome: the brain in trisomic mode. Nat Rev Neurosci. 2012;13:844–58. doi: 10.1038/nrn3314. [DOI] [PubMed] [Google Scholar]
- 26.Haydar TF, Reeves RH. Trisomy 21 and early brain development. Trends Neurosci. 2012;35:81–91. doi: 10.1016/j.tins.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker W. Emerging role of DYRK family protein kinases as regulators of protein stability in cell cycle control. Cell Cycle. 2012;11:3389–94. doi: 10.4161/cc.21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly PA, Rahmani Z. DYRK1A enhances the mitogen-activated protein kinase cascade in PC12 cells by forming a complex with Ras, B-Raf, and MEK1. Mol Biol Cell. 2005;16:3562–73. doi: 10.1091/mbc.E04-12-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yabut O, Domogauer J, D’Arcangelo G. Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells. J Neurosci. 2010;30:4004–14. doi: 10.1523/JNEUROSCI.4711-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stefos GC, Soppa U, Dierssen M, Becker W. NGF upregulates the plasminogen activation inhibitor-1 in neurons via the calcineurin/NFAT pathway and the Down syndrome-related proteins DYRK1A and RCAN1 attenuate this effect. PLoS One. 2013;8:e67470. doi: 10.1371/journal.pone.0067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hämmerle B, Ulin E, Guimera J, Becker W, Guillemot F, Tejedor FJ. Transient expression of Mnb/Dyrk1a couples cell cycle exit and differentiation of neuronal precursors by inducing p27KIP1 expression and suppressing NOTCH signaling. Development. 2011;138:2543–54. doi: 10.1242/dev.066167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuende J, Moreno S, Bolaños JP, Almeida A. Retinoic acid downregulates Rae1 leading to APC(Cdh1) activation and neuroblastoma SH-SY5Y differentiation. Oncogene. 2008;27:3339–44. doi: 10.1038/sj.onc.1210987. [DOI] [PubMed] [Google Scholar]
- 33.Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Ceña V, Gallego C, Comella JX. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem. 2000;75:991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen L, Besson A, Heng JI-T, Schuurmans C, Teboul L, Parras C, Philpott A, Roberts JM, Guillemot F. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006;20:1511–24. doi: 10.1101/gad.377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darzynkiewicz Z, Traganos F, Melamed MR. New cell cycle compartments identified by multiparameter flow cytometry. Cytometry. 1980;1:98–108. doi: 10.1002/cyto.990010203. [DOI] [PubMed] [Google Scholar]
- 36.Shapiro HM. Flow cytometric estimation of DNA and RNA content in intact cells stained with Hoechst 33342 and pyronin Y. Cytometry. 1981;2:143–50. doi: 10.1002/cyto.990020302. [DOI] [PubMed] [Google Scholar]
- 37.Melino G, Thiele CJ, Knight RA, Piacentini M. Retinoids and the control of growth/death decisions in human neuroblastoma cell lines. J Neurooncol. 1997;31:65–83. doi: 10.1023/A:1005733430435. [DOI] [PubMed] [Google Scholar]
- 38.Constantinescu R, Constantinescu AT, Reichmann H, Janetzky B. Neuronal differentiation and long-term culture of the human neuroblastoma line SH-SY5Y. J Neural Transm Suppl. 2007;72:17–28. doi: 10.1007/978-3-211-73574-9_3. [DOI] [PubMed] [Google Scholar]
- 39.Kaplan DR, Matsumoto K, Lucarelli E, Thiele CJ, Eukaryotic Signal Transduction Group Induction of TrkB by retinoic acid mediates biologic responsiveness to BDNF and differentiation of human neuroblastoma cells. Neuron. 1993;11:321–31. doi: 10.1016/0896-6273(93)90187-V. [DOI] [PubMed] [Google Scholar]
- 40.Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011;25:801–13. doi: 10.1101/gad.2034211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J-Y, Lin J-R, Tsai F-C, Meyer T. Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle. Mol Cell. 2013;52:87–100. doi: 10.1016/j.molcel.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tahtouh T, Elkins JM, Filippakopoulos P, Soundararajan M, Burgy G, Durieu E, Cochet C, Schmid RS, Lo DC, Delhommel F, et al. Selectivity, cocrystal structures, and neuroprotective properties of leucettines, a family of protein kinase inhibitors derived from the marine sponge alkaloid leucettamine B. J Med Chem. 2012;55:9312–30. doi: 10.1021/jm301034u. [DOI] [PubMed] [Google Scholar]
- 43.Miyata Y, Nishida E. DYRK1A binds to an evolutionarily conserved WD40-repeat protein WDR68 and induces its nuclear translocation. Biochim Biophys Acta. 2011;1813:1728–39. doi: 10.1016/j.bbamcr.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 44.Göckler N, Jofre G, Papadopoulos C, Soppa U, Tejedor FJ, Becker W. Harmine specifically inhibits protein kinase DYRK1A and interferes with neurite formation. FEBS J. 2009;276:6324–37. doi: 10.1111/j.1742-4658.2009.07346.x. [DOI] [PubMed] [Google Scholar]
- 45.Hämmerle B, Elizalde C, Tejedor FJ. The spatio-temporal and subcellular expression of the candidate Down syndrome gene Mnb/Dyrk1A in the developing mouse brain suggests distinct sequential roles in neuronal development. Eur J Neurosci. 2008;27:1061–74. doi: 10.1111/j.1460-9568.2008.06092.x. [DOI] [PubMed] [Google Scholar]
- 46.Lee MK, Tuttle JB, Rebhun LI, Cleveland DW, Frankfurter A. The expression and posttranslational modification of a neuron-specific beta-tubulin isotype during chick embryogenesis. Cell Motil Cytoskeleton. 1990;17:118–32. doi: 10.1002/cm.970170207. [DOI] [PubMed] [Google Scholar]
- 47.Becker W, Soppa U, Tejedor FJ. DYRK1A: a potential drug target for multiple Down syndrome neuropathologies. CNS Neurol Disord Drug Targets. 2014;13:26–33. doi: 10.2174/18715273113126660186. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi T, Nowakowski RS, Caviness VS., Jr. The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci. 1995;15:6046–57. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng Y-L, Li B-S, Rudrabhatla P, Shukla V, Amin ND, Maric D, Kesavapany S, Kanungo J, Pareek TK, Takahashi S, et al. Phosphorylation of p27Kip1 at Thr187 by cyclin-dependent kinase 5 modulates neural stem cell differentiation. Mol Biol Cell. 2010;21:3601–14. doi: 10.1091/mbc.E10-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin K, Ewton DZ, Park S, Hu J, Friedman E. Mirk regulates the exit of colon cancer cells from quiescence. J Biol Chem. 2009;284:22916–25. doi: 10.1074/jbc.M109.035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altafaj X, Dierssen M, Baamonde C, Martí E, Visa J, Guimerà J, Oset M, González JR, Flórez J, Fillat C, et al. Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down’s syndrome. Hum Mol Genet. 2001;10:1915–23. doi: 10.1093/hmg/10.18.1915. [DOI] [PubMed] [Google Scholar]
- 52.Fotaki V, Dierssen M, Alcántara S, Martínez S, Martí E, Casas C, Visa J, Soriano E, Estivill X, Arbonés ML. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol Cell Biol. 2002;22:6636–47. doi: 10.1128/MCB.22.18.6636-6647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahn K-J, Jeong HK, Choi H-S, Ryoo S-R, Kim YJ, Goo J-S, Choi S-Y, Han J-S, Ha I, Song W-J. DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiol Dis. 2006;22:463–72. doi: 10.1016/j.nbd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 54.Park J, Oh Y, Yoo L, Jung M-S, Song W-J, Lee S-H, Seo H, Chung KC. Dyrk1A phosphorylates p53 and inhibits proliferation of embryonic neuronal cells. J Biol Chem. 2010;285:31895–906. doi: 10.1074/jbc.M110.147520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurabayashi N, Sanada K. Increased dosage of DYRK1A and DSCR1 delays neuronal differentiation in neocortical progenitor cells. Genes Dev. 2013;27:2708–21. doi: 10.1101/gad.226381.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hibaoui Y, Grad I, Letourneau A, Sailani MR, Dahoun S, Santoni FA, Gimelli S, Guipponi M, Pelte MF, Béna F, et al. Modelling and rescuing neurodevelopmental defect of Down syndrome using induced pluripotent stem cells from monozygotic twins discordant for trisomy 21. EMBO Mol Med. 2014;6:259–77. doi: 10.1002/emmm.201302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agholme L, Lindström T, Kågedal K, Marcusson J, Hallbeck M. An in vitro model for neuroscience: differentiation of SH-SY5Y cells into cells with morphological and biochemical characteristics of mature neurons. J Alzheimers Dis. 2010;20:1069–82. doi: 10.3233/JAD-2010-091363. [DOI] [PubMed] [Google Scholar]
- 58.Seifert A, Allan LA, Clarke PR. DYRK1A phosphorylates caspase 9 at an inhibitory site and is potently inhibited in human cells by harmine. FEBS J. 2008;275:6268–80. doi: 10.1111/j.1742-4658.2008.06751.x. [DOI] [PubMed] [Google Scholar]
- 59.Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem. 2010;285:13223–32. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maden M, Hind M. Retinoic acid, a regeneration-inducing molecule. Dev Dyn. 2003;226:237–44. doi: 10.1002/dvdy.10222. [DOI] [PubMed] [Google Scholar]
- 61.Korecka JA, van Kesteren RE, Blaas E, Spitzer SO, Kamstra JH, Smit AB, Swaab DF, Verhaagen J, Bossers K. Phenotypic characterization of retinoic acid differentiated SH-SY5Y cells by transcriptional profiling. PLoS One. 2013;8:e63862. doi: 10.1371/journal.pone.0063862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Molenaar JJ, Ebus ME, Koster J, van Sluis P, van Noesel CJM, Versteeg R, Caron HN. Cyclin D1 and CDK4 activity contribute to the undifferentiated phenotype in neuroblastoma. Cancer Res. 2008;68:2599–609. doi: 10.1158/0008-5472.CAN-07-5032. [DOI] [PubMed] [Google Scholar]
- 63.Vervoorts J, Lüscher B. Post-translational regulation of the tumor suppressor p27(KIP1) Cell Mol Life Sci. 2008;65:3255–64. doi: 10.1007/s00018-008-8296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borriello A, Cucciolla V, Criscuolo M, Indaco S, Oliva A, Giovane A, Bencivenga D, Iolascon A, Zappia V, Della Ragione F. Retinoic acid induces p27Kip1 nuclear accumulation by modulating its phosphorylation. Cancer Res. 2006;66:4240–8. doi: 10.1158/0008-5472.CAN-05-2759. [DOI] [PubMed] [Google Scholar]
- 65.Mitsuhashi T, Aoki Y, Eksioglu YZ, Takahashi T, Bhide PG, Reeves SA, Caviness VS., Jr. Overexpression of p27Kip1 lengthens the G1 phase in a mouse model that targets inducible gene expression to central nervous system progenitor cells. Proc Natl Acad Sci U S A. 2001;98:6435–40. doi: 10.1073/pnas.111051398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Godin JD, Thomas N, Laguesse S, Malinouskaya L, Close P, Malaise O, Purnelle A, Raineteau O, Campbell K, Fero M, et al. p27(Kip1) is a microtubule-associated protein that promotes microtubule polymerization during neuron migration. Dev Cell. 2012;23:729–44. doi: 10.1016/j.devcel.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 67.Lin DI, Barbash O, Kumar KGS, Weber JD, Harper JW, Klein-Szanto AJP, Rustgi A, Fuchs SY, Diehl JA. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell. 2006;24:355–66. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zou Y, Ewton DZ, Deng X, Mercer SE, Friedman E. Mirk/dyrk1B kinase destabilizes cyclin D1 by phosphorylation at threonine 288. J Biol Chem. 2004;279:27790–8. doi: 10.1074/jbc.M403042200. [DOI] [PubMed] [Google Scholar]
- 70.Alt JR, Cleveland JL, Hannink M, Diehl JA. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14:3102–14. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sandoval R, Pilkinton M, Colamonici OR. Deletion of the p107/p130-binding domain of Mip130/LIN-9 bypasses the requirement for CDK4 activity for the dissociation of Mip130/LIN-9 from p107/p130-E2F4 complex. Exp Cell Res. 2009;315:2914–20. doi: 10.1016/j.yexcr.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13:585–95. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tedeschi A, Di Giovanni S. The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep. 2009;10:576–83. doi: 10.1038/embor.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, Buchholz F, Muyrers JPP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–8. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 75.Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–53. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hofmann TG, Möller A, Sirma H, Zentgraf H, Taya Y, Dröge W, Will H, Schmitz ML. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- 77.Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–15. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 78.Takahashi M, Nomura T, Osumi N. Transferring genes into cultured mammalian embryos by electroporation. Dev Growth Differ. 2008;50:485–97. doi: 10.1111/j.1440-169X.2008.01046.x. [DOI] [PubMed] [Google Scholar]
- 79.Himpel S, Panzer P, Eirmbter K, Czajkowska H, Sayed M, Packman LC, Blundell T, Kentrup H, Grötzinger J, Joost HG, et al. Identification of the autophosphorylation sites and characterization of their effects in the protein kinase DYRK1A. Biochem J. 2001;359:497–505. doi: 10.1042/0264-6021:3590497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.