Abstract
In the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens CHA0, the dimeric RNA-binding proteins RsmA and RsmE, which belong to the vast bacterial RsmA/CsrA family, effectively repress translation of target mRNAs containing a typical recognition sequence near the translation start site. Three small RNAs (RsmX, RsmY, RsmZ) with clustered recognition sequences can sequester RsmA and RsmE and thereby relieve translational repression. According to a previously established structural model, the RsmE protein makes optimal contacts with an RNA sequence 5′-A/UCANGGANGU/A-3′, in which the central ribonucleotides form a hexaloop. Here, we questioned the relevance of the hexaloop structure in target RNAs. We found that two predicted pentaloop structures, AGGGA (in pltA mRNA encoding a pyoluteorin biosynthetic enzyme) and AAGGA (in mutated pltA mRNA), allowed effective interaction with the RsmE protein in vivo. By contrast, ACGGA and AUGGA were poor targets. Isothermal titration calorimetry measurements confirmed the strong binding of RsmE to the AGGGA pentaloop structure in an RNA oligomer. Modeling studies highlighted the crucial role of the second ribonucleotide in the loop structure. In conclusion, a refined structural model of RsmE-RNA interaction accommodates certain pentaloop RNAs among the preferred hexaloop RNAs.
Keywords: RNA-binding protein, CsrA/RsmA, translational regulation, Shine-Dalgarno structure, Gac/Rsm pathway
Introduction
RNA-binding proteins of the RsmA/CsrA (acronyms for regulator of secondary metabolism and carbon storage regulator) family are important and widespread post-transcriptional regulators in bacteria. These proteins recognize and bind to specific motifs in target mRNAs. When binding occurs at or near the Shine-Dalgarno (SD) sequence or within the coding sequence of a target mRNA, translation is repressed and this may be accompanied by enhanced mRNA degradation.1-4 RsmA/CsrA proteins can also activate gene expression post-transcriptionally, either by binding to an upstream 5′ region or, more commonly, by indirect mechanisms.3,5 The 3D structures of the closely related Escherichia coli CsrA and Pseudomonas RsmA proteins have been determined. These proteins are homodimers consisting of two interlocking monomers, which form a β-barrel core with α-helices externalized on both sides.6-8 Mutagenesis experiments and a structural study show that protein-RNA contacts are made between the first β-strand of one subunit and the fifth β-strand of the other subunit, implying that the two RNA-binding surfaces lie on opposite sides of the protein.8-10 From current genomic data, the presence of CsrA/RsmA proteins can be inferred in 878 different bacterial strains (Pfam: PF02599).11
No low-molecular-weight effectors have been described that would inhibit or enhance the interaction of RsmA/CsrA proteins with target mRNAs. Recently, a protein (FliW) was discovered that binds to CsrA and in this way relieves CsrA-mediated translational repression of flagellin genes in Bacillus subtilis.12 In general, however, at least in γ-proteobacteria, the activity of RsmA/CsrA proteins is controlled by small RNAs (sRNAs) having a high affinity for these proteins.13 Typically, the sRNAs are expressed at high-cell population densities under the positive control of a two-component system termed GacS/GacA in Pseudomonas spp. Thus, when the GacS sensor is activated, the sRNAs (e.g., CsrB and CsrC in E. coli, RsmY and RsmZ in Pseudomonas aeruginosa or RsmX, RsmY and RsmZ in Pseudomonas fluorescens CHA0) are abundant and sequester the RsmA/CsrA proteins, resulting in translational expression of the target mRNAs.2,14-16 The sRNAs are characterized by several GGA motifs that are present in unpaired regions, especially in loops of stem-loop structures.1,2 As experimentally shown for RsmY, the GGA motifs are essential for binding RsmA and its paralogue RsmE in P. fluorescens.17,18 In pathogenic bacteria such as P. aeruginosa, Salmonella enterica or Vibrio cholerae, the Gac/Rsm signal transduction pathway is instrumental for virulence.19,20
In target mRNAs, repeated and appropriately spaced GGA motifs are also essential for effective recognition by RsmA/CsrA proteins. For several E. coli mRNAs binding of CsrA to these GGA motifs has been demonstrated and, similarly, five GGA motifs in the 5′ leader of hcnA mRNA of P. fluorescens have been shown to be involved in binding RsmA and RsmE.4,21-27 High-affinity interactions have been analyzed by two approaches. First, a SELEX (systematic evolution of ligands by exponential enrichment) experiment generated a family of RNA molecules that were recognized by CsrA. These RNAs had an 5′-RUACARGGAUGU-3′ consensus sequence (where R is A or G) and the GGA motif was most often part of a hexaloop placed on variable short stems, suggesting that both the sequence and the secondary structure of the RNA play important roles. Substitution mutations of conserved central nucleotides greatly diminished the affinity for CsrA.28 Second, NMR spectroscopy was used to investigate binding of the RsmE protein to the translation initiation region of hcnA mRNA, which encodes a subunit of hydrogen cyanide synthase and is under strong control of the Gac/Rsm system.10,14,27,29 In that work, a 12-ribonucleotide hcnA fragment containing the SD sequence (5′-UUCACGGAUGAA-3′) was artificially extended by 4 ribonucleotides at both ends such that a stable stem-loop RNA structure formed, presenting the GGA motif in the ACGGAU hexaloop. The solution structure obtained shows that the RsmE dimer binds two RNA molecules and that contacts are made at the underlined nucleotides of the hcnA RNA fragment. The structure proposes a 5′-A/UCANGGANGU/A-3′ consensus target and explains well how binding of RsmE (or RsmA) can shield the hcnA translation initiation region from the ribosome.10 The RsmE protein has been shown to act as clamp on the hcnA RNA fragment in that the nucleotides flanking the hexaloop undergo base-pairing interactions that are not stable in the absence of the protein.10 In our present work, we observed that deletion of the terminal U in the ACGGAU hexaloop resulted in loss of RsmE binding in vitro and of hcnA regulation by the Gac/Rsm system in vivo, suggesting initially that pentaloops might not interact effectively with the RsmA/CsrA binding cavity.
Intriguingly, non-canonical pentaloops incorporating a GGA motif are expected to form in some natural RsmA/CsrA target RNAs, e.g., in the sRNAs CsrB of E. coli and RsmYZ of Pseudomonas spp as well as at the SD sequence of some mRNAs, e.g., in the pgaA, csrA and nhaR mRNAs of E. coli and in pltA mRNA of P. fluorescens.1,22,25,26,30 The pltA gene is part of an operon specifying pyoluteorin biosynthetic genes.31 The expression of the secondary metabolite pyoluteorin is under strong positive control of the Gac/Rsm system in P. fluorescens.31-33 Here, we show that the pltA pentaloop (AGGGA) is an effective RsmA/RsmE target in vivo and in vitro and we provide evidence by modeling that previously unsuspected protein—RNA contacts account for this strong interaction.
Results
The SD sequences of the hcnA, phlA and pltA genes markedly influence regulation by the Gac/Rsm cascade in Pseudomonas fluorescens CHA0
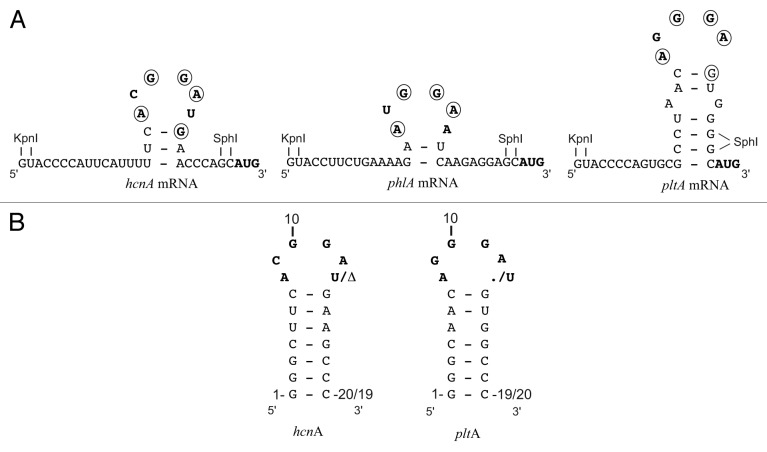
The translation initiation region of the P. fluorescens hcnA gene provides the structural basis for the present study. It was shown in previous work that, upon interaction with RsmE (a member of the RsmA/CsrA family with good solubility) the hcnA SD sequence adopts a secondary structure in which a 3-bp stem is topped by a hexaloop (Fig. 1A).10 The short stem only forms upon the addition of the RsmE protein; thus, RsmE can act as a clamp. The solution structure of the hcnA SD-RsmE complex has been determined by NMR using an hcnA oligoribonucleotide having an artificially elongated stem for improved stability of the RNA secondary structure (Fig. 1B).10 Note that in the hcnA-RsmE complex no base-pairing occurs between A8 and U13 (Fig. 1A). We verified by an RsmE cross-linking experiment (Fig. S1) and ITC dilution experiments (data not shown) that the RsmE protein forms a stable homodimer in solution, in agreement with the established dimeric structure of the RsmA and CsrA proteins.6-8,10
Figure 1. Predicted secondary structures of the hcnA, phlA and pltA mRNA leader sequences near the translation start site when complexed with RsmA/RsmE. Below are shown the hcnA and pltA RNA oligomers used for ITC measurements. The AUG start codon, the typical GGA motif and modified nucleotides are shown in boldface and nucleotides that are part of the Shine Dalgarno consensus sequence (AAGGAGGU) are encircled. The artificial restriction sites KpnI and SphI between which synthetic oligonucleotides or gene sequences were inserted are shown. The dot (.) indicates the absence of a nucleotide. The Δ indicates deletion of the nucleotide.
In the native context, i.e. downstream of four GGA motifs in the 5′ leader sequence (not shown in Fig. 1), the hcnA SD sequence ensured strong (28-fold) induction by the response regulator GacA, which activates transcription of rsmX, rsmY and rsmZ leading to titration of RsmA and RsmE. This effect can be seen with an hcnA’-‘lacZ translational fusion construct in P. fluorescens (Table 1; pME6533). Overexpression of rsmE in the wild-type mimicked the effect of a gacA mutation to some extent, albeit not completely (Table 1; pME6533). When the U13 residue of the hcnA hexaloop (Fig. 1) was deleted, regulation of hcnA’-‘lacZ expression by GacA and RsmE2+ was essentially lost (Table 1; pME9512). This result suggested that an ACGGA pentaloop may be poorly recognized by RsmE. We also constructed a single substitution mutation (A8→G) and a double mutation (A8→G plus U13→C), which encourage the formation of a G-U and a G-C base-pair, respectively, and, therefore, are predicted to favor a CGGA tetraloop. Both constructs (pME6628 and pME10102) resulted in complete loss of hcnA regulation by GacA and RsmE (Table 1). These results corroborate the structural model obtained by NMR spectroscopy10 as well as our modeling results (see below), suggesting an important role of A8 in RsmE binding.
Table 1. Post-transcriptional regulation of ‘lacZ translational fusions by GacA and RsmE: impact of mutations in SD region of mRNAs.
| β-Galactosidase activity (× 103 Miller Units)b | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHA0/ | CHA0/ | Induction | Repression | |||||||||
| Plasmida | 5′ RNA leader sequence | Genotype | CHA0 | CHA89 | pME6001 | pME6851 | factor | factor | ||||
| (wild type) | (gacA) | (vector control) | (rsmE2+) | (GacA) | (RsmE) | |||||||
| pME6533 | …GUACCCCAUUCAUUUUUCACGGAUGAACCCAGCAUG… | hcnA | 10.0 ± 1.0 | 0.35 ± 0.01 | 5.0 ± 0.4 | 1.00 ± 0.15 | 28 | 5 | ||||
| pME9512c | …GUACCCCAUUCAUUUUUCACGGA-GAACCCAGCAUG… | hcnA ΔU13 | 29.0 ± 1.5 | 22.0 ± 2.1 | 22.0 ± 1.5 | 25.0 ± 2.0 | 1.3 | 0.9 | ||||
| pME10001c | …GUACCCCAUUCAUUUUUCAGGGA-GAACCCAGCAUG… | hcnA C9G ΔU13 | 19.0 ± 1.0 | 7.0 ± 0.8 | 12.0 ± 0.5 | 9.5 ± 0.5 | 2.7 | 1.3 | ||||
| pME10002c | …GUACCCCAUUCAUUUUUCAGGAA-GAACCCAGCAUG… | hcnA C9G G11A | 28.0 ± 0.5 | 11.0 ± 1.7 | 18.0 ± 1.0 | 15.0 ± 1.0 | 2.5 | 1.2 | ||||
| ΔU13 | ||||||||||||
| pME6628 | …GUACCCCAUUCAUUUUUCGCGGAUGAACCCAGCAUG… | hcnA A8G | 4.6 ± 0.5 | 4.4 ± 0.2 | 2.40 ± 0.05 | 3.6 ± 0.2 | 1 | 0.7 | ||||
| pME10102 | …GUACCCCAUUCAUUUUUCGCGGACGAACCCAGCAUG… | hcnA A8G U13C | 6.2 ± 0.1 | 6.5 ± 0.1 | 4.5 ± 0.2 | 4.3 ± 0.3 | 1 | 1 | ||||
| pME9524 | …GUACCCCAGUGCGCCUAACAGGGAGUGGGGCAUG… | pltA | 0.36 ± 0.03 | 0.030 ± 0.005 | 0.19 ± 0.01 | 0.060 ± 0.004 | 12 | 3.2 | ||||
| pME9525 | …GUACCCCAGUGCGCCUAACAGGGAUGUGGGGCAUG… | pltA +U13 | 0.17 ± 0.01 | 0.012 ± 0.001 | 0.075 ± 0.004 | 0.012 ± 0.002 | 14 | 6.2 | ||||
| pME10101 | …GUACCCCAGUGCGCCUAACAAGGAGUGGGGCAUG… | pltA G9A | 0.40 ± 0.05 | 0.05 ± 0.01 | 0.13 ± 0.02 | 0.050 ± 0.005 | 8 | 2.6 | ||||
| pME6702 | …UCUGAAAAGAAUGGAAUCAAGAGGAAAAUG… | phlA | 1.90 ± 0.30 | 0.35 ± 0.05 | 0.90 ± 0.13 | 0.40 ± 0.05 | 5.4 | 2.2 | ||||
| pME6737 | …UCUGAAAAGACUGGAAUCAAGAGGAAAAUG… | phlA A8C | 2.90 ± 0.26 | 2.50 ± 0.15 | 0.90 ± 0.08 | 1.80 ± 0.20 | 1.2 | 0.5 | ||||
| pME9536d | …GUACCUUCUGAAAAGAAUGGAAUCAAGAGGAGCAUG… | phlA | 0.40 ± 0.08 | 0.12 ± 0.01 | 0.18 ± 0.015 | 0.14 ± 0.01 | 3.3 | 1.3 | ||||
| pME9537cd | …GUACCUUCUGAAAAGAAUGGA-UCAAGAGGAGCAUG… | phlA ΔA101 | 0.37 ± 0.01 | 0.32 ± 0.05 | 0.28 ± 0.035 | 0.32 ± 0.03 | 1.1 | 0.9 | ||||
a Bold plasmid names correspond to plasmids containing the indicated RNA sequence inserted into the KpnI and SphI restriction sites (in italics) of the hcnA leader of pME6533. bβ-Galactosidase expression of the reporter fusions was measured in P. fluorescens strains CHA0 (wild-type), CHA89 (gacA::Kmr), CHA0/pME6001 (vector control) and CHA0/pME6851 (pME6001 overexpressing rsmE) when the cells reached an OD600 of 3.0 and was determined in triplicate ± standard deviation. The GacA induction factor was calculated as the ratio of the expression in CHA0 and that in CHA89. The RsmE repression factor was calculated as the ratio of the expression in CHA0/pME6001 and that in CHA0/pME6851, respectively. cMeans no nucleotide. dIn pME9536 and pME9537, the tac promoter of pME6702 was exchanged against the hcnA promoter.
Besides hydrogen cyanide, 2,4-diacetylphloroglucinol (DAPG) and pyoluteorin are two other secondary metabolites whose expression is strongly controlled by the GacS/GacA two-component system in P. fluorescens.31,32 The phlA gene is the first gene of an operon specifying DAPG biosynthesis.34 An RsmA/RsmE-binding site with a typical hexaloop (AUGGAA) located on a potential 2-bp stem lies immediately upstream of the phlA SD sequence (Fig. 1A). A translational phlA’-‘lacZ fusion (under the tac promoter, carried by plasmid pME6702) was used to demonstrate positive control by GacA (5.4-fold) and repression by overexpressed RsmE (2.2-fold). When the proximal A in the hexaloop was mutated to C, these regulatory effects were almost completely lost (Table 1; pME6737), as would be predicted from the important contacts that the proximal A residue makes with RsmE.10 The phlA 26-nt leader sequence was then transplanted into the hcnA leader context (Table 1; pME9536). Regulation by GacA and RsmE2+ was still found, but was less pronounced than in the native phlA context (Table 1; pME6702), perhaps due to perturbation of the RNA secondary structure. However, deletion of the distal A in the hexaloop abolished regulation by GacA and RsmE2+ entirely (Table 1; pME9537). This suggested that the resulting phlA pentaloop (AUGGA)—akin to the mutated hcnA pentaloop (ACGGA)—may not be able to bind RsmE.
We then turned our attention to the pltA SD sequence, which, when complexed with RsmA/RsmE, may form a pentaloop (AGGGA) topping a 6-bp stem (Fig. 1A). The pltA gene is part of the pyoluteorin biosynthetic operon.31 To facilitate comparison with the hcnA and phlA constructs, we grafted the pltA leader sequence (22 nt) into the hcnA leader. The expression of the resulting pltA’-‘lacZ fusion (in plasmid pME9524) was significantly higher in the wild-type CHA0 than in the gacA mutant CHA89 and in strain CHA0 overexpressing either rsmA or rsmE (Fig. 2). Similar results were obtained with a translational ‘lacZ reporter fusion containing the native 5′ leader of the pltA gene (data not shown). At a standardized cell concentration (OD = 3.0, corresponding to 3 × 109 CFU/ml), the GacA induction factor was 12 and the RsmE repression factor was 3.2 (Table 1; pME9524). As found previously, the empty vector plasmid pME6001 had an unexplained negative influence on lacZ expression (Fig. 2) and this effect was taken into account in the calculation of the RsmE repression factor.27 We have also previously reported that the ACGGAU hexaloop of hcnA can be changed to AGGGAU with marginal effects on the regulation by GacA, RsmA and RsmE.27 We now constructed a mutated hcnA leader having an AGGGA pentaloop. This construct (pME10001) was subject to regulation by GacA (2.7-fold) as well as RsmE2+ (1.3-fold; Table 1). We conclude from these experiments that an AGGGA pentaloop can be recognized by RsmA/CsrA-type proteins. Two further pentaloop variants, AAGGA (in plasmid pME10101) and AGGAA (in plasmid pME10002) also tested positively for regulation by the Gac/Rsm cascade (Table 1).
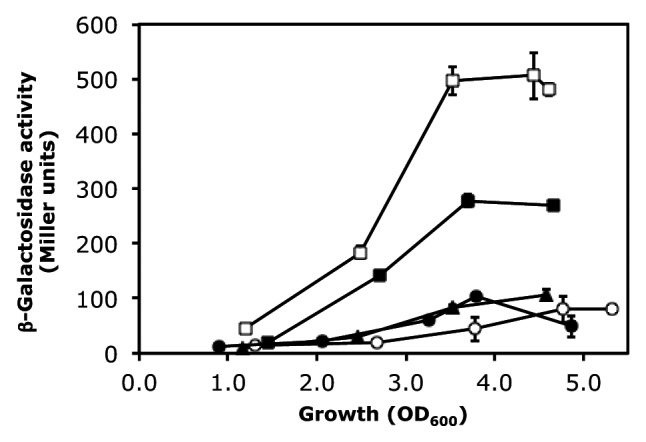
Figure 2. Cell population density-dependent β-galactosidase expression of a pltA’-‘lacZ translational fusion (pME9524) in P. fluorescens strains CHA0 (wild-type; open squares), CHA0/pME6001 (vector control; filled squares), CHA0/pME6073 (= pME6001 overexpressing rsmA; filled triangles), CHA0/pME6851 (= pME6001 overexpressing rsmE; filled circles) and CHA89 (gacA::Kmr; open circles). Each value is the average from three different cultures ± standard deviation. In some instances, the standard deviation bars are smaller than the symbols used.
We finally compared the pltA pentaloop AGGGA to the mutant hexaloop AGGGAU (in plasmid pME9525). The inserted distal U actually improved regulation by the Gac/Rsm cascade (Table 1). In conclusion, while our in vivo experiments vindicate the canonical hexaloop as a high-affinity target, we discovered that certain pentaloops can also act as effective targets.
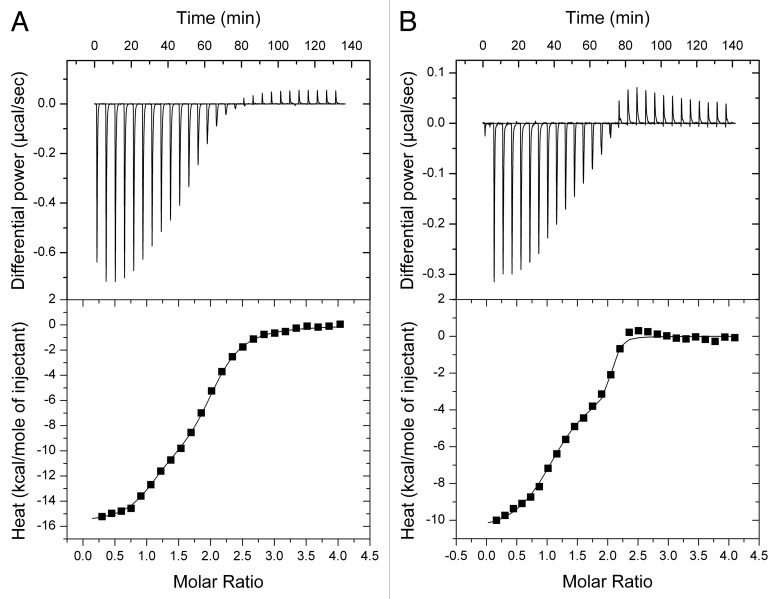
Binding affinity of RsmE for hcnA- and pltA-derived oligoribonucleotides as determined by ITC
We chose four targets derived from the hcnA and pltA SD sequences to quantitate by ITC the interaction with the RsmE protein in vitro and, in particular, to investigate the roles played by representative penta- and hexaloop sequences. As in previous work, we elongated the stem (Fig. 1B).10 The added nucleotides ensure the stability of the stem but do not interact with RsmE.10 All RNA oligomers bound specifically to RsmE and exhibited a biphasic binding isotherm (Fig. 3), which is typical of a cooperative sequential binding mechanism. The hcnA 20-mer bound tightly to the first site of the RsmE dimer with a dissociation constant KD1 of 85 nM and a favorable enthalpy change (ΔH1) (Table 2). The affinity toward the second site in the RsmE dimer was reduced 3-fold and accompanied by a less favorable enthalpy change (ΔΔH = -4.7 kcal mol−1) (Table 2). This observation indicates negative cooperative allostery, probably induced by distortion of the second binding site in the RsmE dimer as a result of binding the first oligomer. When the U residue at position 13 was deleted resulting in a pentaloop in the hcnAΔU oligomer, the binding affinity for RsmE was decreased dramatically; the first site showed a nearly 50-fold reduced affinity compared with the hcnA oligomer and the second binding site presented a 20-fold reduction of binding strength compared with the first site. In the hcnAΔU-RsmE interaction, both enthalpy changes (ΔH1 and ΔH2) were similar while the second hcnAΔU molecule interacted with a pronounced entropic penalty (ΔΔS = 9.7 cal K−1 mol−1) (Table 2), suggesting that the second site might be less distorted than the first site upon binding of the first hcnAΔU molecule and that the observed negative cooperativity is mostly entropic in nature. We will comment on the significance of the negative cooperativity in the Discussion section. The observed loss of RsmE binding upon deletion of the distal U in the target loop is in complete agreement with the in vivo data (Table 1; pME6533 and pME9512).
Figure 3. Binding of RNA oligomers to RsmE measured by ITC. Typical ITC experiments of RsmE (7.6 μM of homodimer in cell) with pltA 19-mer RNA (164.1 μM in syringe) and of RsmE (7.5 μM) with pltA+U 20-mer RNA (150.3 μM) are shown in (A and B), respectively. Both oligomers bound specifically to RsmE and exhibited a biphasic binding isotherm typical of a cooperative sequential binding mechanism. The raw data for consecutive injections of RNA to RsmE (top panels) was integrated and corrected for the heat of dilution and plotted against the (oligomer)/(RsmE) ratio (lower panels). The solid line represents the best least-squares fit to the data using a sequential binding model. The corresponding results are presented in Table 2.
Table 2. Thermodynamic parameters of complex formation between RsmE and four RNA oligomers as obtained by ITCa.
| Gene | RNA sequencea | KD1 | ΔG1 | ΔH1 | ΔS1 | TΔS1 | KD2 | ΔG2 | ΔH2 | ΔS2 | TΔS2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| (nM) | (kcal/mol) | (kcal/mol) | (cal/kmol) | (kcal/mol) | (nM) | (kcal/mol) | (kcal/mol) | (cal/kmol) | (kcal/mol) | ||
| hcnA | GGGCUUCACGGAUGAAGCCC | 84.5 ± 35.4 | -9.7 ± 0.3 | -13.3 ± 0.6 | -12.1 ± 1.1 | -3.6 ± 0.3 | 271.6 ± 63.4 | -9.0 ± 0.1 | -8.6 ± 0.1 | 1.3 ± 0.2 | 0.4 ± 0.1 |
| hcnAΔU | GGGCUUCACGGA-GAAGCCC | 4140 ± 2549 | -7.4 ± 0.4 | -9.4 ± 0.6 | -6.7 ± 3.2 | -2.0 ± 1.0 | 80547 ± 51883 | -5.7 ± 0.4 | -10.5 ± 1.0 | -16.4 ± 4.7 | -4.9 ± 1.4 |
| pltA | GGGCAACAGGGA-GUGGCCC | 18.8 ± 3.3 | -10.5 ± 0.1 | -14.9 ± 0.8 | -14.6 ± 3.2 | -4.3 ± 0.9 | 588.0 ± 185.8 | -8.5 ± 0.2 | -11.5 ± 1.1 | -10.0 ± 4.2 | -3.0 ± 1.3 |
| pltA+U | GGGCAACAGGGAUGUGGCCC | 1.6 ± 1.2 | -12.1 ± 0.5 | -9.7 ± 0.7 | 8.2 ± 4.0 | 2.4 ± 1.2 | 19.8 ± 15.4 | -10.6 ± 0.5 | -3.1 ± 0.7 | 25.4 ± 4.2 | 7.6 ± 1.3 |
a The integrated heat changes were corrected for the heat of dilution and fitted with a sequential binding site model assuming a set of two identical sites (n1 = 1, n2 = 1). While ΔH and KD were experimentally determined, ΔG and TΔS were calculated from the relationships ΔG = RTlnKD and ΔG = ΔH − TΔS. The values ± standard deviations were obtained in duplicate titrations. b-, no nucleotide. Residues predicted to be in the loop are underlined.
The interaction of the pltA 19-mer RNA (Fig. 1B) was very tight, with a KD1 of 19 nM for the first site in RsmE and 31-fold lower value KD2 for the second site (Table 2). The pltA oligomer differs from the hcnAΔU oligomer mainly in the loop (AGGGA instead of ACGGA; see Fig. 1B). Additional differences occur in the stem at the second and third position from the top. However, as we will argue below (see Table 3), these base pairs make a small and a practically negligible contribution, respectively, toward the binding free enthalpy. Binding of the pltA oligomer to the first RsmE site was accompanied by a strong favorable enthalpy change, while binding to the second site was lower (ΔΔH of -3.4 kcal mol−1) (Table 2). When a uridine residue was inserted at position 13 to form a hexaloop (AGGGAU) in the pltA+U oligomer, a 10-fold increase in affinity toward the first RsmE binding site was observed, by comparison with the pltA oligomer (Table 2). As the pltA+U interaction with RsmE produced less binding heat than did pltA (ΔΔH1 of -5.2 kcal mol−1), the entropic contribution changed from strong and unfavorable to positive, thus favorable for binding (ΔΔS1 = -22.8 cal K−1 mol−1). The pltA+U oligomer bound less tightly (12-fold) to the second site than to the first site, but it is striking to observe that the interaction mechanism changed from enthalpy-driven binding (as seen for pltA) to entropy-driven binding (for pltA+U). The in vitro binding data correlate well with the regulatory patterns observed in vivo (Table 1; pME9524 and pME9525). In conclusion, the binding assays confirm that RNA hexaloops interact favorably with RsmA/CsrA-type proteins and highlight the novel insight that an AGGGA pentaloop can also provide strong interaction. In the following modeling section, we will try to rationalize these findings.
Table 3. Estimated contribution of each ribonucleotide toward the averaged binding free energy (ΔG) during formation of RNA oligomer-RsmE complexesa.
| RNA residue number |
hcnA RNA sequence |
hcnA | hcnAΔU |
pltA+U RNA sequence |
pltA | pltA+U | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| ΔG ± SD (kcal mol−1) |
ΔG ± SD (kcal mol−1) |
ΔΔG (kcal mol−1) |
ΔG ± SD (kcal mol−1) |
ΔΔG (kcal mol−1) |
ΔG ± SD (kcal mol−1) |
ΔΔG (kcal mol−1) |
||||
| 1 | G | 0.13 ± 0.26 | 0.16 ± 0.29 | 0.03 | G | 0.19 ± 0.36 | 0.06 | 0.01 ± 0.23 | -0.12 | |
| 2 | G | 1.30 ± 1.42 | 1.29 ± 1.44 | -0.01 | G | 1.13 ± 1.35 | -0.17 | 0.46 ± 1.07 | -0.84 | |
| 3 | G | 2.04 ± 1.81b | 2.14 ± 1.69b | 0.10 | G | 2.11 ± 1.66b | 0.07 | 1.25 ± 1.60b | -0.79 | |
| 4 | C | 1.45 ± 1.94 | 1.49 ± 1.87 | 0.04 | C | 0.74 ± 2.15 | -0.71 | 0.48 ± 2.13 | -0.97 | |
| 5 | U | 0.50 ± 2.22 | 0.84 ± 2.27 | 0.34 | U | 0.15 ± 2.50 | -0.35 | 0.31 ± 2.68 | -0.19 | |
| 6 | U | 1.04 ± 1.52 | 0.15 ± 1.62 | -0.89 | U | 0.76 ± 1.56 | -0.28 | 0.85 ± 1.62 | -0.19 | |
| 7 | C | 1.39 ± 1.46 | 0.15 ± 1.13 | -1.24 | C | 0.22 ± 0.83 | -1.17 | 0.98 ± 1.16 | -0.41 | |
| 8 | A | -4.49 ± 2.30 | -6.10 ± 2.32 | -1.61 | A | -7.15 ± 1.80 | -2.66 | -6.13 ± 2.28 | -1.64 | |
| 9 | C | -0.03 ± 2.20 | -1.58 ± 2.48 | -1.55 | G | -3.50 ± 2.67 | -3.47 | -1.91 ± 3.17 | -1.88 | |
| 10 | G | -4.93 ± 2.36 | -4.41 ± 2.44 | 0.52 | G | -7.54 ± 2.06 | -2.61 | -6.15 ± 2.83 | -1.22 | |
| 11 | G | -3.64 ± 2.16 | -3.78 ± 2.34 | -0.14 | G | -4.42 ± 2.30 | -0.78 | -4.71 ± 2.53 | -1.07 | |
| 12 | A | -10.44 ± 1.59 | -8.64 ± 1.39 | 1.80 | A | -8.67 ± 1.41 | 1.77 | -10.55 ± 1.53 | -0.11 | |
| 13 | U | -2.29 ± 1.65 | - | 2.29 | U | - | 2.29 | -3.45 ± 1.88 | -1.16 | |
| 14 | G | -4.69 ± 2.50 | 0.92 ± 1.70 | 5.61 | G | 1.38 ± 2.03 | 6.07 | -3.21 ± 2.60 | 1.48 | |
| 15 | A | -2.23 ± 1.37 | -1.32 ± 0.98 | 0.91 | A | -0.55 ± 1.39 | 1.68 | -0.99 ± 1.09 | 1.24 | |
| 16 | A | -0.38 ± 0.17 | -0.38 ± 0.21 | 0.00 | A | -0.35 ± 0.25 | 0.03 | -0.39 ± 0.16 | -0.01 | |
| 17 | G | -0.39 ± 0.15 | -0.39 ± 0.15 | 0.00 | G | -0.43 ± 0.18 | -0.04 | -0.30 ± 0.21 | 0.09 | |
| 18 | C | -0.34 ± 0.08 | -0.36 ± 0.09 | -0.02 | C | -0.35 ± 0.08 | -0.01 | -0.34 ± 0.08 | 0.00 | |
| 19 | C | -0.29 ± 0.06 | -0.31 ± 0.07 | -0.02 | C | -0.30 ± 0.06 | -0.01 | -0.29 ± 0.06 | 0.00 | |
| 20 | C | -0.15 ± 0.02 | -0.15 ± 0.03 | 0.00 | C | -0.15 ± 0.02 | 0.00 | -0.14 ± 0.02 | 0.01 | |
a The differences to the reference system (hcnA-RsmE complex) are represented as ΔΔG. The bold-face values indicate the contributions of the predicted loop residues. All values are given ± standard deviation. The averaged values of the contributions to the binding free energy were calculated using 2,000 RNA-dimeric RsmE interaction energies obtained from 1,000 full complex conformations. As the snapshots for each system were gathered along four independent 10 ns-long trajectories, the conformational space was sampled efficiently. The high ΔG standard deviation values result from the structural differences between the conformations and, therefore, indicate the broad distribution of the values within the experiment rather than errors between the series of measurements. Thus, they do not imply a lack of significance. bThe apparent destabilizing contribution of G3 residue, which was found in all studied complexes, might be an artifact of the MM-GBSA calculation methodology and is not considered further.
Modeling and analysis of RNA-RsmE interactions
To improve our understanding of the recognition specificity of the RsmE dimer for different RNA molecules and to identify the critical RNA-protein interactions, we studied in silico the structural and thermodynamic properties of the four RNA-RsmE complexes selected above (hcnA, hcnAΔU, pltA and pltA+U) using the hcnA oligomer-RsmE complex as a basis.10 We ran molecular dynamics simulations of the complexes and analyzed the RNA-protein interactions found in the resulting conformations with the Entangle program.35 We also estimated the theoretical binding free energy (ΔG) using the structures obtained by molecular dynamics simulations with the Molecular Mechanics - Generalized Born Surface Area (MM-GBSA) method. This method allows decomposing the binding free energy into residue contributions and provides insight into the origin of binding (Table 3).
We consistently observed that the free energy of binding of the RsmE dimer was more negative for one of the RNA oligomers than for the second RNA oligomer, which is in good agreement with the ITC data (Table 2). Thus, we divided the theoretical ΔG values into two groups, ΔG1 and ΔG2 (Table S1). Both sets of theoretical ΔG values correlate well with the experimental ΔG values obtained in vitro by ITC; Pearson correlation coefficients were 0.813 for the theoretical and experimental ΔG1 values and 0.811 for the theoretical and experimental ΔG2 values (Fig. S2), indicating a reasonable consistency between the theoretical and experimental results. The higher values of the theoretically estimated energies are a consequence of the approximations made in the calculations, i.e., neglecting entropy and conformational energy terms. Normally, the MM-GBSA method is used to compare the ΔG values of different conformers or mutants, but not to calculate absolute values.36
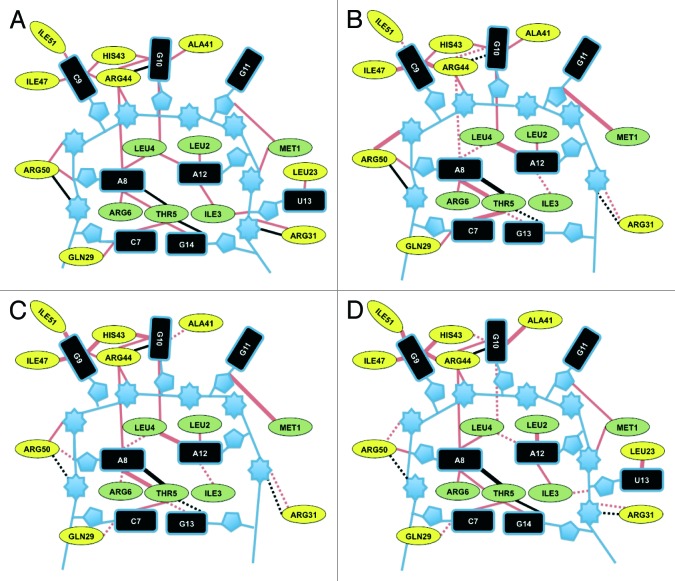
To define the role of each ribonucleotide in RNA-RsmE complex formation, we performed a detailed analysis for each of the four selected RNA-RsmE complexes (Fig. 4). We investigated the conformation and the interactions between the protein and the RNA residues in terms of contributions to the average binding free energy ΔG (Table 3). The RsmE recognition motif consists of a hexaloop in the case of the hcnA and pltA+U RNAs and of a pentaloop in the case of the hcnAΔU and pltA RNAs (Fig. 1B). Of the RNA stem-forming residues, only the conserved C7 and G14 residues made substantial contributions to the interaction energy, whereas the residues of the rest of stem did not exhibit important contributions (Table 3).
Figure 4. Schematic representation of changes in the RNA-RsmE interactions with hcnA-RsmE as a reference. The interactions of the hcnA (A), hcnAΔU (B), pltA (C) and pltA+U (D) oligomers with RsmE are shown. Red and black lines represent van der Waals interactions and hydrogen bonds, repectively. Changes relative to the RsmE-hcnA interaction (i.e., differences of > 20% in all encountered interactions) are represented as follows: thick lines, more van der Waals and hydrogen bond interactions; dashed lines, fewer van der Waals and hydrogen bond interactions. The protein residues of the RsmE N terminus are colored green and the C-terminal residues are colored yellow. The numbers of interactions were calculated with Entangle program.
Residues A12 and G10, the third and the first residue of the GGA motif, as well as G14, the first stem residue, contribute most to the binding free energy of hcnA RNA with RsmE (Table 3). Two other loop residues, A8 and G11, are less important; their interactions with RsmE have been detailed previously.10 The U13 residue contributes to the stability of the RsmE-hcnA complex through non-polar contacts with Met1 and Ile3, and a hydrogen bond between the phosphate group and the NH3 group of Met1 and C9 makes a negligible contribution to the RsmE-hcnA interactions (Table 3).
The hcnAΔU RNA containing the ACGGA pentaloop binds to RsmE with a ΔG that is less negative than the ΔG for hcnA RNA, according to ITC measurements (Table 2) and theoretical considerations (Table 3). One of the obvious reasons for the less favorable ΔG value (ΔΔG = +12.81 kcal mol−1) is the absence of the stabilizing contribution by U13, which is present in the RsmE-hexaloop RNA-complexes. However, our analysis reveals that the largest destabilizing effect comes from the lower contribution of residue G13 compared with its hcnA counterpart G14 (ΔΔG = +5.61 kcal mol−1).
The pltA RNA—containing an AGGGA pentaloop—forms a stable complex with RsmE as it compensates the loss of U13 and the weakening of the G14-RsmE interactions by stronger G9, G10 and A8 interactions with the protein. The calculated binding free energy is more favorable for pltA-RsmE than for hcnAΔU-RsmE (-74.15 vs. -64.41 kcal mol−1; Table 3). In particular, the contribution of G9 in pltA RNA is predicted to be stronger than that C9 in hcnA by -3.47 kcal mol−1. Unlike C, G has atoms with hydrogen-bonding potential positioned at the Hoogsteen edge and is involved in two hydrogen bonds with the neighboring Arg50 residue (Fig. 4). This additional stabilization from G9-Arg50 hydrogen bonds also leads to an increased number of hydrophobic contacts between G9 and the Ile51, Ile47 and His43 side chains (Fig. 4). In addition, the better anchoring of G9 to RsmE leads to more stable interactions of A8 and G10 compared with the corresponding residues in hcnA and hcnAΔU RNAs (Fig. 4, Table 3).
The more favorable binding free energy of the artificial pltA+U RNA, compared with hcnA RNA, originates from stronger interactions of all loop residues (Table 3). The hydrogen bonds formed by G9 with Arg50 causes better anchoring of the AGGGAU loop to RsmE and a higher contribution of G9 and of neighboring residues to the binding free energy, as in the case of pltA RNA (Fig. 4).
Discussion
Proteins of the RsmA/CsrA family are global post-transcriptional regulators in a large variety of bacteria. The rsmA regulon in P. aeruginosa and the csrA regulon in E. coli have been estimated to comprise 10–15% of all transcripts.24,37 Similarly, in P. fluorescens strain Pf-5, a gacA-null mutation affects the steady-state concentration of about 10% of all transcripts.31 In strain CHA0, which is very closely related to strain Pf-5, the RsmA and RsmE proteins regulate the expression of genes involved in exoproducts (including the hcnA, pltA and phlA genes encoding hydrogen cyanide, pyoluteorin and DAPG biosynthetic enzymes, respectively) and the oprF gene for a major porin.14,18,38,39 The expression of both RsmA and RsmE increases with increasing cell density; however, the amount of RsmE being subject to control by the GacS/GacA system shows stronger variation.18
Here, we have experimentally determined the affinity of RsmE for the main target in the pltA mRNA (Table 2), which consists of a stem topped with a pentaloop (Fig. 1A). Because of the remarkable sequence conservation of RsmA/CsrA proteins, we expect that most of the interactions that the RsmE protein undergoes with target RNAs, according to an NMR study and to the present modeling calculations, will also apply to the interactions of other proteins in the same family.10 This view is supported by the fact that the RNA sequences recognized by these proteins share common features, notably the common GGA motif.1-3 In particular, RsmA and RsmE appear to have similar target recognition specificities.18,27
An important initial result was our observation that the RsmE dimer binds to its two RNA targets with differential affinity (Table 2) and this behavior was confirmed by modeling studies (Table S1). The affinity of the RsmE protein was consistently higher for first RNA oligomer than for the second, separate RNA molecule (Table 2). We cannot predict from these experiments whether two (or more) appropriately spaced recognition sites in an mRNA would result also in negative cooperativity or, on the contrary, in positive cooperativity. However, the observed negative cooperativity with a single-site target may well have important consequences in vivo. If an RsmE dimer complexed with one target RNA has a reduced affinity for a second, separate target RNA, this may prevent the formation of multimeric RsmE/RNA lattices, which would be detrimental to the cells.
Most mRNA targets of RsmA/CsrA proteins contain repeated GGA motifs. For instance, the hcnA 5′ leader mRNA has five GGA motifs, all of which contribute to regulation by GacA and RsmA/E and allow the formation of at least three distinct complexes between RsmE and this leader RNA in vitro.27 In the phlA and pltA mRNA leader sequences, the typical GGA motif present in the SD sequence is flanked by three and two upstream GGA motifs, respectively. A decisive interaction between the hcnA leader and RsmA/E takes place at the GGA motif belonging to the SD sequence; without this interaction, regulation by the GacS/GacA system is lost.27 We suspect that in general, the strongest and presumably primary interaction of RsmA/CsrA proteins with target mRNAs takes place at or near the translation start site, while further appropriately spaced binding sites may have a modulating role.4,26,27
As can be seen from the data of Table 1, mutations in the SD region not only affected post-transcriptional regulation by the Gac/Rsm system, but also influenced the translation efficiency in the wild-type over a 100-fold range. This variation mostly reflects differential translation initiation efficiencies but may also be due to differential mRNA stabilities.
In both the consensus SELEX RNA and in the hcnA RNA structural model, the target RNA interacting with CsrA or RsmE, respectively, adopts a hexaloop structure placed on a short stem.10,28 Our present study confirms the effectiveness of such a conformation, in particular by demonstrating the extremely high affinity of the artificial pltA+U RNA for RsmE (Tables 1–3). By contrast, the hcnAΔU variant, which exhibits a stem-pentaloop structure, is a very poor target for RsmE both in vivo and in vitro (Tables 1–3). Interestingly, the principal reason for this poor interaction appears to be a weakened contact between one uppermost stem ribonucleotide (G13) and RsmE rather than a poor affinity of the pentaloop for RsmE (Table 3). As a major caveat then, the hexaloop model should not be used as a generalized model to predict targets of RsmA/CsrA proteins. Indeed, as we have shown here, the stem-pentaloop structure in the pltA SD sequence serves as an efficient target for RsmE in P. fluorescens. This view is supported by the fact that the pgaA, csrA and nhaR mRNAs, which are efficient targets for the CsrA protein of E. coli, are also predicted to adopt stem-pentaloop structures.4,22,26 Furthermore, the titrating sRNAs CsrB of E. coli and RsmY and RsmZ of Pseudomonas spp contain several predicted pentaloop targets.1,30
In hcnA and hcnAΔU RNAs, the second loop residue (C9) is not able to form any hydrogen bond on its Hoogsteen edge end and, as a result, no interactions with Arg50 of RsmE are possible. We can hypothesize that a U residue in position 9 (as in phlA mRNA; Table 1) would also fail to interact with Arg50 of RsmE. By contrast, a G in this position (as in pltA and pltA+U RNAs) has the capacity to form two hydrogen bonds with Arg50 (Fig. 4).35 In pltA RNA, the G9 interaction with RsmE additionally strengthens the contacts of the neighboring residues with RsmE (Table 3). Indeed, the SELEX consensus sequence RUACARGGAUGU (28) reflects a preference for G or A (= R) over U or C at position 9, albeit in a hexaloop configuration.
The entropy effects in the ITC measurements (Table 2) suggest that a hexaloop structure (of the ARGGAU type) may be more dynamic and flexible than a pentaloop structure, enabling strong interactions with RsmE. Nevertheless, as we have demonstrated in the present study, an AGGGA pentaloop placed on a short stem can be a very effective target of RsmE. In P. fluorescens Pf-5, 4.4% of all SD sequences have an AGGGA motif and 1.1% present this pentaloop motif placed on a putative short stem. These motifs are predicted candidates for regulation by the Gac/Rsm system and include the recognized target genes prnA, pchR, pchP and hcp as well as around 40 previously unrecognized, putative candidates.
Materials and Methods
Strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table 4. The oligonucleotides used are listed in Table S2. P. fluorescens and E. coli strains were grown in nutrient yeast broth (NYB) medium with shaking (180 rpm) at 30°C and 37°C, respectively.41 Triton X-100 was added at 0.05% (w/v) to liquid cultures to avoid cell aggregation. Antibiotics, when required, were added at the following final concentrations: kanamycin, 50 μg/ml; tetracycline, 125 μg/ml; and gentamicin, 10 μg/ml for P. fluorescens and kanamycin, 50 μg/ml or 25 μg/ml for E. coli.
Table 4. Strains and plasmids used in this study.
| Strain, plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E.coli | ||
| BL21(DE3) | F-ompT hsdSB (rB-,mB-) gal dcm rne-131 (λDE3) | Invitrogen |
| P. fluorescens | ||
| CHA0 | wild type | 51 |
| CHA89 | gacA::Kmr | 32 |
| Plasmid | ||
| pME6001 | Cloning vector, pBBR1MCS derivative, Gmr | 14 |
| pME6073 | pME6001 derivative carrying rsmA under Plac control, Gmr | 14 |
| pME6359 | pME6032 derivative carrying rsmZ under Ptac control, Tcr | 41 |
| pME6533 | pME3219 with artificial restriction sites for KpnI and SphI in hcnA leader | 14 |
| pME6628 | pME6533 derivative with A8 → G mutation | This study |
| pME6702 | Ptac-phlA'-'lacZ containing 92 nucleotides of phlA, Tcr | 18 |
| pME6737 | pME6702 derivative with A8 → C mutation | This study |
| pME7609 | pET28b derivative carrying rsmE, Kmr | 10 |
| pME6851 | pME6001 derivative carrying rsmE under Plac control, Gmr | 18 |
| pME9512 | pME6533 derivative with T13 deleted | This study |
| pME9524 | pME6533 derivative with 22 nucleotides of pltA SD (+128 to ATG), inserted between KpnI and SphI in hcnA leader | This study |
| pME9525 | pME6533 derivative with 22 nucleotides of pltA SD (+128 to ATG) containing an additional T at position 13, inserted between KpnI and SphI in hcnA leader | This study |
| pME9536 | pME6533 derivative with 26 nucleotides of phlA SD (+ 376 to ATG), inserted between KpnI and SphI in hcnA leader | This study |
| pME9537 | pME6533 derivative with 26 nucleotides of phlA SD 376 to ATG) with deletion in A101 between KpnI and SphI in hcnA leader | This study |
| pME10001 | pME6533 derivative with C9 → G mutation and T13 deleted | This study |
| pME10002 | pME6533 derivative with C9 → G and G11 → A mutations and T13 deleted | This study |
| pME10101 | pME6533 derivative with 22 nucleotides of pltA SD (+128 to ATG) with G9 → A mutation, inserted between KpnI and SphI in hcnA leader | This study |
| pME10102 | pME6533 derivative with A8 → G and T13 → C mutations | This study |
a Oligonucleotides used for plasmid constructions are given in Table S1. The same numbers are used for oligonucleotides and corresponding plasmids. SD, Shine-Dalgarno.
DNA manipulations and plasmid preparations
These were performed according to standard protocols.42 The plasmids containing mutated hcnA leader sequences were constructed by inserting the synthetic double-stranded oligonucleotides 6628-6628rev, 9512-9512rev, 10001-10001rev, 10002-10002rev or 10102-10102rev (Table S2) into the unique KpnI and SphI restriction sites of pME6533 resulting in pME6628, pME9512, pME10001, pME10002 and pME10102, respectively. The derivatives of pME6533 containing the wild-type and mutant pltA SD or the phlA SD were constructed by inserting into pME6533, digested with KpnI and SphI, the synthetic double-stranded oligonucleotides 9524-9524rev, 9525-9525rev, 9536-9536rev or 9537-9537rev (Table S2), resulting in pME9524, pME9525, pME9536 and pME9537, respectively. Sequences were confirmed by commercial nucleotide sequencing (Microsynth AG).
Protein purification
Histidine-tagged RsmE (RsmE6H) was overproduced in E. coli BL21(DE3)/pME7609 and purified by affinity chromatography on nickel-nitrilotriacetic acid agarose beads (Qiagen) according to the manufacturer’s recommendations with modifications.10 The washing procedure was performed in three steps using, respectively, 50 mM K phosphate buffer pH 8.0, 300 mM NaCl, 20 mM imidazole; 50 mM K phosphate buffer pH 8.0, 1 M NaCl, 20 mM imidazole; and 50 mM K phosphate buffer pH 8.0, 300 mM NaCl, 100 mM imidazole. The elution fraction containing the protein was dialyzed against 50 mM K phosphate buffer pH 8.0, 300 mM NaCl at 4°C and stored at -20°C. The concentration of RsmE was estimated with the Bradford Bio-RAD protein assay method (Bio-RAD) using bovine serum albumin as the standard.
Isothermal titration calorimetry (ITC)
ITC experiments were performed at 25°C and 307 rpm stirring speed on a VP-ITC instrument (MicroCal). The reference cell was filled with a solution of 0.1% sodium azide, and the calorimeter was calibrated as recommended by the manufacturer. The RNA oligomers (HPLC purified) were synthesized by Microsynth AG and used without further purification. In order to induce stem-loop formation, oligomers were dissolved in water, heated to 95°C (10 min) and shock-frozen in liquid nitrogen (3 min). Samples of protein and RNA oligomers were simultaneously dialyzed against the same batch of titration buffer (50 mM K phosphate buffer pH 7.9, 300 mM NaCl), and concentrations were determined after dialysis. The sample cell (1.45 ml) was loaded with 4–36 μM protein (values referring to the homodimer) while the RNA oligomer concentration in the syringe was 140–800 μM. A typical titration experiment consisted of a first control injection of 2 μl followed by 27 injections, each of 10 μl and 10 sec duration, with a 5 min interval in between. Each titration experiment was followed by a control experiment performed under identical conditions but replacing the protein solution with titration buffer. Raw data were collected, integrated, corrected for ligand heats of dilution and fitted to a sequential binding sites model assuming a set of two identical sites (n1 = 1, n2 = 1) using the Origin software supplied with the instrument. The measurements were performed in duplicate.
β-Galactosidase assays
P. fluorescens strains were grown at 30°C in 50-ml flasks containing 20 ml of NYB supplemented with Triton X-100. Specific activities were determined by the Miller method.43
Modeling
The NMR structure of the hcnA-RsmE complex (PDB code 2JPP) was used as a starting point to model the structure of some of the experimentally studied RNA oligomers, i.e., hcnA, hcnAΔU, pltA and pltA+U bound to the RsmE protein dimer.10 The first of the 10 models available in the NMR structure was chosen to set up the hcnA-RsmE complex and to model the other systems. The stem of the hcnA RNA was kept, while the mutations and deletions in the RNA loop sequence were performed with the UCSF Chimera program for visualization and analysis of molecular structures.44 After deletion of the looped-out U13 residue in hcnA RNA, the system was minimized with the Molecular Modeling Tools for Structural Biology (MMTSB) toolset using the molecular simulation package CHARMM22 (version c34b1) with 500 steps of steepest descent minimization to close the backbone break.45,46
All systems were set up for Molecular Dynamics (MD) simulation with another molecular simulation package, Gromacs (4.0), in a dodecahedron box filled with water molecules described by the TIP3P model of water model and equilibrated at 300 K and 1 bar.47,48 The necessary numbers of Na+ and Cl- ions were added to achieve the 0.18 M salt concentration reported for the experimental conditions of the NMR experiment.10 Four state-of-the-art molecular dynamics simulations, 10 ns in length, were performed for each system with the CHARMM27 force field using the Gromacs engine (version 4.0), at 300 K and 1 bar.46,49 The systems had previously been minimized with 300, 400, 500 and 600 steps of steepest descent to create different starting points for the four MD runs. The structures extracted from the MD simulations were analyzed with the program to explore the RNA-protein interactions, Entangle; van der Waals contacts and hydrogen bond were analyzed.35
The free energy of binding, ∆G, between each RNA molecule and the RsmE dimer was calculated with the MM-GBSA method.50 The ∆G values were obtained for each system and, subsequently, decomposed into per-residue contributions.36 In this approach, ∆G is estimated as the sum of the gas phase energies, solvation free energies and entropic contributions, averaged over a series of conformations obtained during MD simulations. The gas phase contribution to the binding free energy is equal to the sum of the van der Waals and electrostatic interaction energies between the binding partners. These terms are obtained by molecular mechanics (MM) calculations. The electrostatic contribution to the solvation energy is calculated by using a generalized Born model (an implicit solvation model approximating the water as a continuum with water-like electrostatic properties) as a difference between the solvation energy of the complex and that of the separated binding partners. The non-polar solvation free energy is assumed to be proportional to the solvent accessible surface area buried upon complexation. The entropy term was omitted in the free energy of binding calculations.
Supplementary Material
Acknowledgments
This work was supported by the Sandoz Family Foundation (Programme for academic promotion) to K.L. D.H. is grateful to the Swiss National Science Foundation for support. We thank Nicolas Pradervand for performing RsmE cross-linking experiments, Dr Michel Cuendet for his help in developing the RNA parameters for Gromacs, Prof. Shamoo from the Rice University, for kindly providing the Linux version of the Entangle program and the VITAL-IT group of the Swiss Institute of Bioinformatics for providing computational resources. The bioinformatics aspects of this work were performed at the Protein Modeling Facility of the Lausanne University.
Glossary
Abbreviations:
- SD
Shine-Dalgarno
- SELEX
systematic evolution of ligands by exponential enrichment
- ITC
isothermal titration calorimetry
- MMTSB
molecular modelling tools for structural biology
- MD
molecular dynamics
- MM-GBSA
molecular mechanics - generalized born surface
- MM
molecular mechanics
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–63. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–53. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 3.Timmermans J, Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci. 2010;67:2897–908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yakhnin H, Baker CS, Berezin I, Evangelista MA, Rassin A, Romeo T, et al. CsrA represses translation of sdiA, which encodes the N-acylhomoserine-L-lactone receptor of Escherichia coli, by binding exclusively within the coding region of sdiA mRNA. J Bacteriol. 2011;193:6162–70. doi: 10.1128/JB.05975-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei BL, Brun-Zinkernagel A-M, Simecka JW, Prüss BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–56. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 6.Gutiérrez P, Li Y, Osborne MJ, Pomerantseva E, Liu Q, Gehring K. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J Bacteriol. 2005;187:3496–501. doi: 10.1128/JB.187.10.3496-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rife C, Schwarzenbacher R, McMullan D, Abdubek P, Ambing E, Axelrod H, et al. Crystal structure of the global regulatory protein CsrA from Pseudomonas putida at 2.05 A resolution reveals a new fold. Proteins. 2005;61:449–53. doi: 10.1002/prot.20502. [DOI] [PubMed] [Google Scholar]
- 8.Heeb S, Kuehne SA, Bycroft M, Crivii S, Allen MD, Haas D, et al. Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J Mol Biol. 2006;355:1026–36. doi: 10.1016/j.jmb.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 9.Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J Biol Chem. 2006;281:31832–42. doi: 10.1074/jbc.M606057200. [DOI] [PubMed] [Google Scholar]
- 10.Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, et al. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol. 2007;14:807–13. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 11.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–22. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukherjee S, Yakhnin H, Kysela D, Sokoloski J, Babitzke P, Kearns DB. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol Microbiol. 2011;82:447–61. doi: 10.1111/j.1365-2958.2011.07822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romeo T, Vakulskas CA, Babitzke P. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol. 2013;15:313–24. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blumer C, Heeb S, Pessi G, Haas D. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc Natl Acad Sci USA. 1999;96:14073–8. doi: 10.1073/pnas.96.24.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci USA. 2005;102:17136–41. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, et al. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol. 2009;73:434–45. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valverde C, Lindell M, Wagner EGH, Haas D. A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J Biol Chem. 2004;279:25066–74. doi: 10.1074/jbc.M401870200. [DOI] [PubMed] [Google Scholar]
- 18.Reimmann C, Valverde C, Kay E, Haas D. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J Bacteriol. 2005;187:276–85. doi: 10.1128/JB.187.1.276-285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol. 2007;10:182–8. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Valverde C, Haas D. Small RNAs controlled by two-component systems. In: Bacterial Signal Transduction: Network and Drug targets. R. Utsumi, ed., Landes Bioscience, Austin, Texas, USA. 2008:54-79. [Google Scholar]
- 21.Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol. 2002;44:1599–610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–63. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 23.Mercante J, Edwards AN, Dubey AK, Babitzke P, Romeo T. Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J Mol Biol. 2009;392:511–28. doi: 10.1016/j.jmb.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Vinella D, et al. Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol. 2011;80:1561–80. doi: 10.1111/j.1365-2958.2011.07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yakhnin H, Yakhnin AV, Baker CS, Sineva E, Berezin I, Romeo T, et al. Complex regulation of the global regulatory gene csrA: CsrA-mediated translational repression, transcription from five promoters by Eσ⁷⁰ and Eσ(S), and indirect transcriptional activation by CsrA. Mol Microbiol. 2011;81:689–704. doi: 10.1111/j.1365-2958.2011.07723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pannuri A, Yakhnin H, Vakulskas CA, Edwards AN, Babitzke P, Romeo T. Translational repression of NhaR, a novel pathway for multi-tier regulation of biofilm circuitry by CsrA. J Bacteriol. 2012;194:79–89. doi: 10.1128/JB.06209-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lapouge K, Sineva E, Lindell M, Starke K, Baker CS, Babitzke P, et al. Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol Microbiol. 2007;66:341–56. doi: 10.1111/j.1365-2958.2007.05909.x. [DOI] [PubMed] [Google Scholar]
- 28.Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005;11:1579–87. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laville J, Blumer C, Von Schroetter C, Gaia V, Défago G, Keel C, et al. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J Bacteriol. 1998;180:3187–96. doi: 10.1128/jb.180.12.3187-3196.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heeb S, Heurlier K, Valverde C, Cámara M, Haas D, Williams P. Post-transcriptional regulation in Pseudomonas spp. via the Gac/Rsm regulatory network. In Pseudomonas. J.L. Ramos, ed., Kluwer Academic/Plenum Publishers, New York, 2004; 2:239-255. [Google Scholar]
- 31.Hassan KA, Johnson A, Shaffer BT, Ren Q, Kidarsa TA, Elbourne LD, et al. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ Microbiol. 2010;12:899–915. doi: 10.1111/j.1462-2920.2009.02134.x. [DOI] [PubMed] [Google Scholar]
- 32.Laville J, Voisard C, Keel C, Maurhofer M, Défago G, Haas D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc Natl Acad Sci USA. 1992;89:1562–6. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corbell N, Loper JE. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J Bacteriol. 1995;177:6230–6. doi: 10.1128/jb.177.21.6230-6236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kidarsa TA, Goebel NC, Zabriskie TM, Loper JE. Phloroglucinol mediates cross-talk between the pyoluteorin and 2,4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol Microbiol. 2011;81:395–414. doi: 10.1111/j.1365-2958.2011.07697.x. [DOI] [PubMed] [Google Scholar]
- 35.Morozova N, Allers J, Myers J, Shamoo Y. Protein-RNA interactions: exploring binding patterns with a three-dimensional superposition analysis of high resolution structures. Bioinformatics. 2006;22:2746–52. doi: 10.1093/bioinformatics/btl470. [DOI] [PubMed] [Google Scholar]
- 36.Zoete V, Irving MB, Michielin O. MM-GBSA binding free energy decomposition and T cell receptor engineering. J Mol Recognit. 2010;23:142–52. doi: 10.1002/jmr.1005. [DOI] [PubMed] [Google Scholar]
- 37.Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72:612–32. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frapolli M, Pothier JF, Défago G, Moënne-Loccoz Y. Evolutionary history of synthesis pathway genes for phloroglucinol and cyanide antimicrobials in plant-associated fluorescent pseudomonads. Mol Phylogenet Evol. 2012;63:877–90. doi: 10.1016/j.ympev.2012.02.030. [DOI] [PubMed] [Google Scholar]
- 39.Crespo MC, Valverde C. A single mutation in the oprF mRNA leader confers strict translational control by the Gac/Rsm system in Pseudomonas fluorescens CHA0. Curr Microbiol. 2009;58:182–8. doi: 10.1007/s00284-008-9306-6. [DOI] [PubMed] [Google Scholar]
- 40.Burrowes E, Baysse C, Adams C, O’Gara F. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology. 2006;152:405–18. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- 41.Heeb S, Blumer C, Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol. 2002;184:1046–56. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual 3rd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2001. [Google Scholar]
- 43.Miller V. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1972. [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Feig M, Karanicolas J, Brooks CL., 3rd MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graph Model. 2004;22:377–95. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr., Nilsson L, Petrella RJ, Roux B, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;3:435–47. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 48.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–35. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 49.Bjelkmar P, Larsson P, Cuendet MA, Hess B, Lindahl E. Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J Chem Theory Comput. 2010;6:459–66. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- 50.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–97. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 51.Haas D, Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307–19. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 52.Dubey AK, Baker CS, Suzuki K, Jones AD, Pandit P, Romeo T, et al. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J Bacteriol. 2003;185:4450–60. doi: 10.1128/JB.185.15.4450-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.