Abstract
The PBAF subtype of the mammalian chromatin remodeling SWI/SNF complex has wide and diverse functions in transcription regulation and development, being both transcription activator and repressor. However, a mechanism accounting for such functional diversity remains unclear. Human PHF10/BAF45a subunit of the PBAF complex plays an important role in brain development but has not been studied sufficiently. We have shown that the PHF10 gene encodes 2 types of evolutionarily conserved, ubiquitously expressed isoforms that are incorporated into the PBAF complex in a mutually exclusive manner. One isoform contains C-terminal tandem PHD fingers, which in the other isoform are replaced by the consensus sequence for phosphorylation-dependent SUMO 1 conjugation (PDSM). PBAF complexes containing different PHF10 isoforms can bind to the promoters of the same genes but produce different effects on the recruitment of Pol II to the promoter and on the level of gene transcription. In addition, it is only the PBAF with PHD-containing isoform that activates proliferation. Our study demonstrates the existence of functionally different PBAF complexes in mammalian cell. It also provides an insight into the molecular structure and role of human PHF10/BAF45a and characterizes it as an essential PBAF subunit.
Keywords: PHF10/BAF45, SWI/SNF chromatin remodeling complex, PBAF signature subunits, PHD domains, sumoylation, PDSM motif, functionally distinct isoforms
Introduction
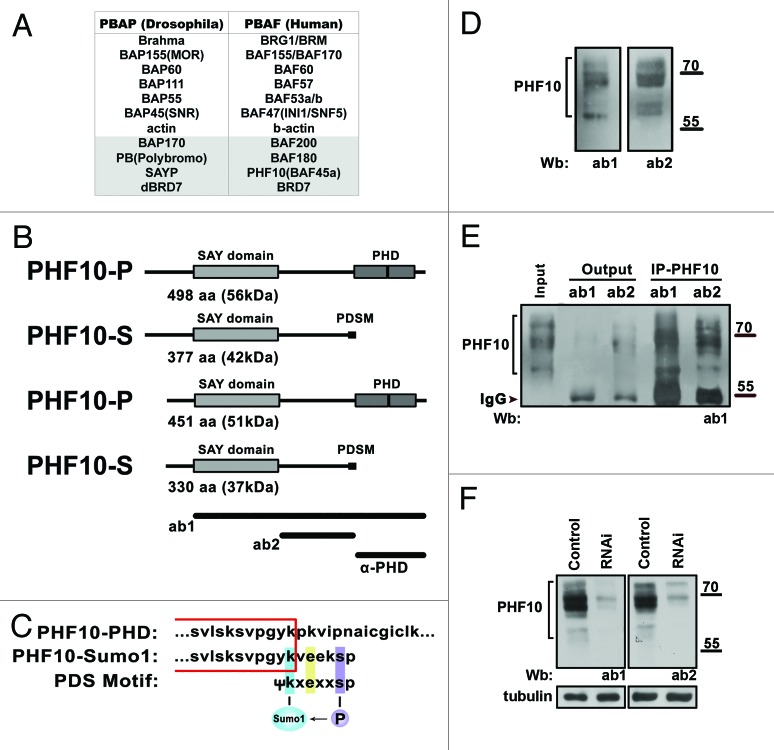
The SWI/SNF chromatin-remodeling ATP-dependent complex plays an essential role in various processes, including cell proliferation, differentiation, and development in metazoans. Such a diversity of functions is due to its variable subunit composition.1 Two types of SWI/SNF complexes, BAF/BAP and PBAF/PBAP, are distinguished in mammals and Drosophila.2 They share core proteins, including ATPase and several noncatalytic subunits, but also contain specific subunits: BAF250/Osa in the BAF/BAP type and hBAF200/dBAP170 and hBAF180/dPolybromo in the PBAF/PBAP type (Fig. 1A). It has been shown that the human BRD7 protein and its Drosophila homolog are associated with the latter type, with the corresponding complexes being designated hPBAF/dPBAP.3,4
Figure 1. The PHF10 gene encodes isoforms containing either PHD fingers or PDSM motif as the C-terminal domain. (A) The subunit composition of Drosophila and mammalian PBAF complexes. The homologous subunits are grouped horizontally. The PBAF-specific subunits are shown on a gray background. (B) The predicted structure of PHF10 isoforms. The PHF10 mRNA encodes isoforms that have an evolutionary conserved SAY domain and tandem PHD domains (PHF10-P) or PDSM motif (PHF10-S) at the C terminus. The short PHF10-P and PHF10-S lack 47 N-terminal amino acids. The full-length PHF10-P isoform corresponds to the PHF10 protein annotated earlier (NP_060758.2). The regions used to raise antibodies are shown at the bottom. (C) The consensus sequence for phosphorylation-dependent SUMO modification (PDSM) at the C terminus of PHF10-S isoforms. Sequences common to PHF10-P and PHF10-S are boxed. (D) Detection of PHF10 in HEK293 cell extract by western blotting with 2 different affinity purified anti-PHF10 antibodies (ab1 and ab2). (E) Each of anti-PHF10 antibodies precipitates PHF10 bands from the HEK293 extract. (F) The shRNAi knockdown of PHF10 leads to depletion of PHF10 isoforms from the HEK293 extract.
The composition of the BAF complex in mammals is more diverse, since some of subunits are represented by paralogous proteins alternatively incorporated in the complex. Thus, it contains either Brg1 ATPase or its close homolog Brm, both being broadly expressed but having largely different functions in transcription regulation.5,6 The BAF250 signature subunit is represented by 2 broadly expressed homologs (BAF250A and BAF250B) that are alternatively incorporated in the complex and have different effects on cell cycle activity.7,8 On the other hand, no subunit heterogeneity has as yet been demonstrated for metazoan PBAF.
Moreover, it has been shown that the Drosophila transcription co-activator SAYP is also a signature subunit of PBAP,9 and that the mammalian homolog of SAYP, PHF10/BAF45a, is associated with the neural progenitor-specific SWI/SNF complex.10 The recombinant human PHF10 has been found to interact with PBAF-specific subunits in HEK293 cells, indicating that it may be a component of PBAF.11 However, mammalian PHF10 is still regarded as a tissue-specific SWI/SNF subunit,1 and its association with SWI/SNF requires further investigation.
The PHF10 and SAYP homologs in higher eukaryotes contain the SAY domain and tandem C-terminal PHD fingers.12 The PHD domains of different transcription factors have been shown to recognize the modified histone H3 tail and to be essential for the control of gene transcription.13 It is noteworthy that PHF10/SAYP is the only subunit of PBAF complex that has proved to contain the PHD fingers.11
Several pieces of evidence point out an essential function of Drosophila SAYP, a ubiquitously expressed transcription co-activator that regulates a large number of genes and participates in signaling pathways during development.14-16 SAYP forms a stable module with the dBAF170 and PB specific subunits of the PBAP complex and is necessary for the structural integrity of this module.9 SAYP is required for the recruitment of PBAP onto target genes, and its knockdown strongly decreases the level of Pol II on the promoter of SAYP-dependent gene.17 Finally, SAYP together with PB has been found to participate in Pol II pausing.18
PHF10 is responsible for the maintenance of the undifferentiated status of proliferating neural stem cells in the developing CNS.10 Its downregulation by RNAi in cell culture results in a loss of cell proliferation, suggesting a role for PHF10 in cell growth.19 However, PHF10 has not been adequately studied at the molecular level. Here we show that the PHF10 gene encodes 2 types of ubiquitously expressed, evolutionarily conserved isoforms, with the C-terminal PHD fingers in one isoform being replaced by a functionally distinct domain (PDSM motif for SUMO 1 conjugation). Both PHF10 isoforms are tightly associated with specific PBAF subunits, and the PDSM-containing isoform is sumoylated in a phosphorylation-dependent manner. The 2 isoforms are included in different PBAF complexes and have different effects on cell proliferation. Both of them can bind to promoters of the same genes, and the PHD-containing isoform (but not PDSM-containing isoform) strongly increases the presence of Pol II on the promoters. The PHD-containing isoform has proved to activate transcription of several genes studied. All this is evidence for the functional heterogeneity of PBAF complexes in mammalian cells.
Results
The PHF10 gene encodes 2 types of ubiquitously expressed, evolutionarily conserved isoforms, one of them containing C-terminal PDSM consensus sequence for phosphorylation-dependent SUMO 1 conjugation, instead of tandem PHD
The PHF10 protein contains an evolutionary conserved SAY domain of about 200 amino acids linked by a region of 120 amino acids to 2 PHD domains at the C terminus.14 Until recently, only one human transcript encoding PHF10 and one encoding a PHF10 variant lacking 2 amino acids in the SAY domain were annotated in NCBI. In addition, 4 mRNAs corresponding to the PHF10 gene were detected in human fibroblasts by northern blot analysis.19 We determined the structure of PHF10 transcripts in the HEK293 cell line (see Supplementary Material) and revealed 3 novel transcripts among them in addition to that annotated in NCBI (Fig. 1B).
The transcript annotated in NCBI encoded PHF10 (NP_060758.2) that contained the SAY domain and 2 C-terminal PHD domains. Unexpectedly, one of the novel transcripts proved to encode an isoform in which the tandem PHDs were replaced by a short stretch of 6 amino acids. As a result, it acquired a motif for SUMO conjugation (Fig. 1C) known as phosphorylation-dependent SUMOylation motif (PDSM).20 Two other novel PHF10 isoforms also had either C-terminal PHDs or PDSM but lacked 47 N-terminal amino acids (Fig. 1B). Below, we use designations PHF10-P and PHF10-S for PHD- and PDSM-containing isoforms, respectively.
An important fact is that PDSM-containing isoforms proved to be evolutionarily conserved, as well as the isoforms with PHD fingers, which is evidence for the functional significance of PDSM. A search in the NCBI database revealed an expressed sequence tag (EST) that encoded protein forms without PHD fingers in Danio rerio (BC057492.1), Xenopus laevis (NM_001093300.1), Macaca mulata (XM_001084064.2), and Otolemur garnettii (XM_003792367.1), with their C-terminal PDSM consensus sequence being remarkably conserved.
Both types of PHF10 isoforms were abundantly expressed. The PHF10-P and PHF10-S mRNA transcripts were detected at relatively high levels in total RNA samples from different human tissues (FirstChoice Human Total RNA survey panel, Applied Biosystems) and from several human cell lines (Fig. S1A and B).
For the next step of analysis, we raised and affinity purified rabbit polyclonal antibodies (ab1 and ab2) against different PHF10 fragments (Fig. 1B). Both antibodies recognized a similar set of protein bands in a molecular weight range of about 60–80 kDa in the nuclear extract from HEK293 cells (Fig. 1D). All these proteins were efficiently precipitated from the extract by anti-PHF10 antibodies and showed significant depletion after RNAi knockdown of PHF10; therefore, they indeed corresponded to PHF10 (Fig. 1E and F).
PHF10 isoforms are multiply phosphorylated
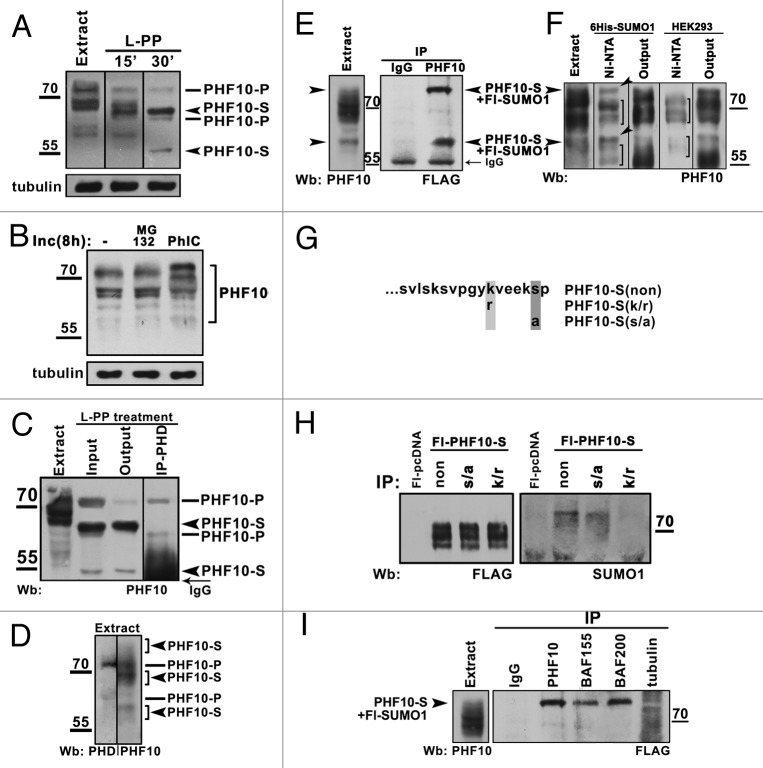
The multiplicity of PHF10 bands observed in the nuclear extract suggested that they could be a product of its posttranslational modification. Indeed, the treatment of nuclear extract with lambda protein phosphatase (L-PP) caused a significant drop in the molecular weight of observed PHF10 bands and reduced their number to 4 stable forms (Fig. 2A), indicating that PHF10 was phosphorylated at many sites. In line with this result, cell culturing in the presence of a phosphatase inhibitor (PhIC) proved to result in accumulation of high molecular weight PHF10 forms (Fig. 2B).
Figure 2. Post-translational modifications of PHF10 isoforms. (A) Treatment of HEK293 cell extract with lambda protein phosphatase (L-PP) for 15 or 30 min leads to reduction of numerous PHF10 isoforms to 4 stable protein bands (arrowheads). (B) Inhibition of phosphatases stabilizes high-molecular-weight PHF10 bands. Western blotting of PHF10 in protein extracts from wild-type HEK293 cells and from HEK293 cells grown for 8 h in the presence of MG132 proteasome inhibitor or phosphatase inhibitor cocktail (PhIC). Accumulation of high-molecular-weight PHF10 bands (arrowheads) can be seen in cells grown in the presence of PhIC. (C) Immunoprecipitation of PHD-containing PHF10 forms (PHF10-P) performed with anti-PHD antibodies from the HEK293 extract treated with L-PP. The PHF10-S isoforms remain in the output material. (D) Western blotting of HEK293 extract with anti-PHD or anti-PHF10 (ab1) antibodies. The PHF10-P and PHF10-S isoforms are indicated. (E) Endogenous PHF10-S isoforms modified by SUMO 1 conjugation. FLAG-tagged SUMO 1 was transiently expressed in HEK293 cells, and the cell extract was immunoprecipitated by anti-PHF10 antibodies. Western blotting of the precipitate with anti-FLAG antibodies revealed 2 sumoylated PHF10 forms. Their electrophoretic mobility coincided with that of modified PHF-S isoforms in the HEK293 extract (left panel, indicated by arrowheads). (F) Nuclear extract from HEK293 cells transiently transfected with 10 His- SUMO 1 or from control (nontransfected) HEK293 cells was incubated with Ni-NTA, and precipitated proteins were resolved by western blotting with antibodies against PHF10. Sumoylated PHF10 bound by Ni-NTA resin (arrows) was depleted from the pool of PHF10 isoforms (see “Output”). The PHF10 forms indicated with brackets are those nonspecifically bound by Ni-NTA. (G) The amino acid sequence of wild-type and mutated PDSM. In mutants, either serine was replaced by alanine or lysine was replaced by arginine to prevent phosphorylation or sumoylation, respectively. (H) The PDSM motif of PHF10-S conjugates SUMO 1 in a phosphorylation-dependent manner. FLAG-tagged wild-type and mutated forms of PHF10-S were transiently expressed HEK293 cells and purified on anti-FLAG agarose. The recombinant PHF10 was precipitated with anti-FLAG antibodies from the cell extract, and equal amounts of precipitate were loaded onto a gel (FLAG). Staining with anti-SUMO 1 antibody revealed sumoylated recombinant PHF10 (SUMO 1). (I) The sumoylated PHF10-S is associated with PBAF. Immunoprecipitation with antibodies against BAF200 or BAF155 was performed from the extract of HEK293 cells stably transfected with FLAG-tagged SUMO 1. The western blot was developed with antibodies against FLAG to detect sumoylated PHF10 (arrowhead). Immunoprecipitation with antibodies against tubulin was used as a negative control.
To discriminate between PHD- and PDSM-containing isoforms (PHF10-P and PHF10-S), we raised an antibody against a peptide containing 2 PHD fingers (Fig. 1C; Fig. S2B). This antibody precipitated 2 out of the 4 PHF10 bands observed in the L-PP-treated extract, while the other 2 bands remained in output material, indicating that they corresponded to PDSM-containing isoforms (Fig. 2C). Even after L-PP treatment, however, the molecular weights of PHF10 bands determined by SDS-PAGE exceeded the calculated values. An abnormal electrophoretic mobility may be a common feature for PHF10 homologs from different species, as it was also shown for Drosophila SAYP.14
The PDSM motif of PHF10-S isoform conjugates SUMO 1 in a phosphorylation-dependent manner
Next we addressed the sumoylation of PHF10-S, since the motif for phosphorylation-dependent SUMO 1 conjugation (PDSM) present in this isoform has been found in numerous transcription regulators and shown to be essential for their function.20 A crude extract from HEK293 cells was resolved by SDS-PAGE and western blotted with anti-PHF10 and anti-PHD antibodies. Comparing the resultant PHF10 patterns, 3 sets of bands corresponding to PHF10-S isoforms (in a range of about 60–70 kDa) were detected (Fig. 2D). Next, we tested if any of these bands contained SUMO 1. Conjugation with SUMO 1 is a highly dynamic and transient modification, and usually only a small proportion of the total protein is sumoylated.21 To overcome this problem, the cells were transfected with FLAG-tagged SUMO 1, the cell extract was immunoprecipitated with anti-PHF10 antibodies, and the precipitate was analyzed by western blotting with anti-FLAG antibody. Two sumoylated PHF10 forms were revealed, with their electrophoretic mobility corresponding to that of PHF10-S isoforms present in the extract (Fig. 2E, indicated with arrowheads).
In a reciprocal experiment, His-tagged SUMO 1 was overexpressed in HEK293 cells. The extracts from these and control (nontransfected) cells were incubated with Ni-NTA agarose, resolved by SDS-PAGE, and western blotted with antibodies against PHF10. The SUMO 1-modified PHF10-S forms specifically bound to Ni-NTA (indicated with arrowheads) and proved to be the same as those revealed in the previous experiment. It could also be clearly seen that these forms were specifically depleted from the total PHF10 pool (Fig. 2E and F).
To confirm that the PDSM motif was essential for sumoylation, we mutated it in experiments with the PHF10-S isoform. Taking into account that phosphorylation of serine in PDSM strongly enhances lysine sumoylation,20 we replaced lysine 371 with arginine in one mutant and serine 377 with alanine in another mutant (Fig. 2G). The results showed that the mutation of serine to alanine significantly reduced the level of sumoylation, while the mutation of lysine to arginine blocked it completely, indicating that PHF10-S is sumoylated in a phosphorylation-dependent manner (Fig. 2H).
To study the association of sumoylated PHF10-S with PBAF, the extract from HEK293 cells expressing FLAG-tagged SUMO 1 was treated with antibodies against PBAF-specific subunits. Anti-BAF155 and anti-BAF200 antibodies efficiently co-precipitated SUMO 1-modified PHF10 (Fig. 2I), suggesting that sumoylated PHF10-S is associated with complex.
Both PHF10 isoforms are integral subunits of the PBAF signature module
Next, we analyzed the interaction of PHF10 isoforms with SWI/SNF. Antibodies against specific and core PBAF subunits efficiently co-precipitated differently modified PHF10 isoforms (Fig. 3A). Conversely, anti-PHF10 antibody significantly depleted specific (BAF200 and BAF180) and core (BAF155) PBAF subunits but did not co-precipitate the BAF250 subunit of the BAF complex (Fig. 3A). We also found that PHF10 isoforms were associated with PBAF in human cell lines of different origin (Fig. S2C). These data confirm that both forms of endogenous PHF10 specifically interact with the PBAF complex. They agree with results obtained by Middeljans et al.,11 who demonstrated co-precipitation of PBAF specific subunits and recombinant PHD-containing PHF10 from the HEK293 cell extract.
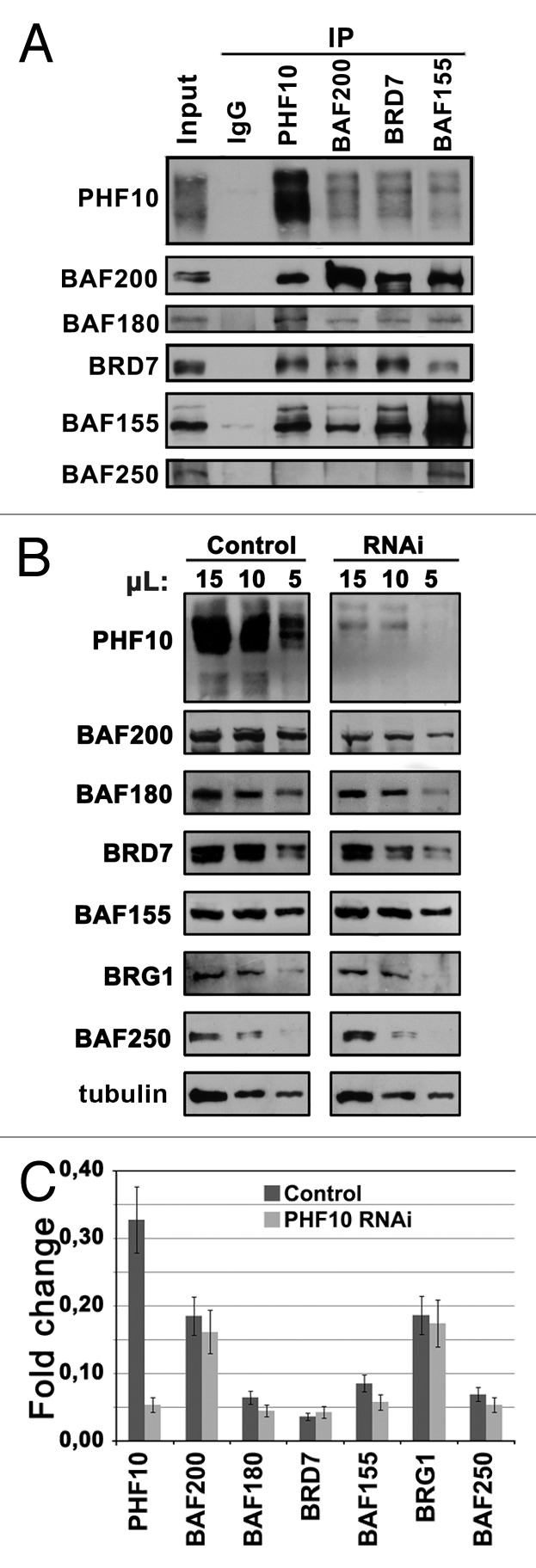
Figure 3. PHF10 isoforms are associated with PBAF signature subunits. (A) Immunoprecipitation (IP) of PBAF from HEK293 extract with antibodies against PHF10 (ab1), BAF200, and BRD7 specific PBAF subunits and a BAF155 core SWI/SNF subunit. (B) The RNAi knockdown of PHF10 leads to a decrease in the level of specific PBAF subunits in the cells. HEK293 cells were stably transfected with the construct expressing shRNA for PHF10 RNAi or with an empty vector (control). The equal amounts of protein extracts from PHF10 RNAi or control cells were loaded onto the gel, and the western blot was developed with antibodies against PBAF subunits. Tubulin was used as a loading control. (C) Transcription levels of different PBAF subunits against the background of PHF10 RNAi and in control cells as measured by qPCR.
As shown previously, the signature subunits of Drosophila PBAP are tightly associated with each other.9,22 SAYP depletion interferes with association of other signature subunits with the core complex, leading to their degradation and reduction of their total level in cells.17 We tested whether PHF10 depletion would have the same influence on PBAF specific subunits (Fig. 3B and C). RNAi of total PHF10 proved to significantly reduce the levels of BAF200, BAF180, and BRD7 (Fig. 3B), with the levels of BAF155 and BRG1 core subunits and the BAF250 specific subunit of BAF remaining unchanged. The observed effect was not caused by the drop of transcription against the background of PHF10 RNAi (Fig. 3C). Therefore, RNAi-mediated depletion of PHF10 destabilizes the signature subunits but does not affect the core complex, revealing that, similar to its Drosophila counterpart, PHF10 is stably associated with the rest of the PBAF signature module.
PHD fingers- and PDSM motif-containing isoforms are associated with different PBAF complexes
The next question was as to whether the PHF10-P and PHF10-S isoforms were subunits of the same or different PBAF complexes. In the experiments described below, we focused on PHF10-P and PHF10-S that had a N-terminal, 47-amino acids extension (Fig. 1B), since their amount in HEK293 cell extract was significantly greater than that of short forms.
PHF10-P was depleted from the extract with anti-PHD antibodies (Fig. 4A, lanes 1–5). A portion of PBAF co-precipitated with PHF10-P, while the rest remained in the extract (Fig. 4A, lower panel). The PBAF complex from the output material was precipitated with antibody against BAF200 (Fig. 4A, lanes 6, 7). The PHF10-S isoform present in the extract was completely depleted with anti-BAF200 antibodies, indicating that the PBAF complex contained either PHF10-P or PHF10-S isoform. The same result was obtained when analyzing FLAG-tagged PHF10 isoforms in stably transfected cells (Fig. 4B). In each case, anti-FLAG antibody treatment resulted in complete depletion of the recombinant isoform, while the alternative isoform remained in the output material. These results demonstrate that PHD fingers- and PDSM motif-containing PHF10 isoforms are alternative subunits of PBAF.
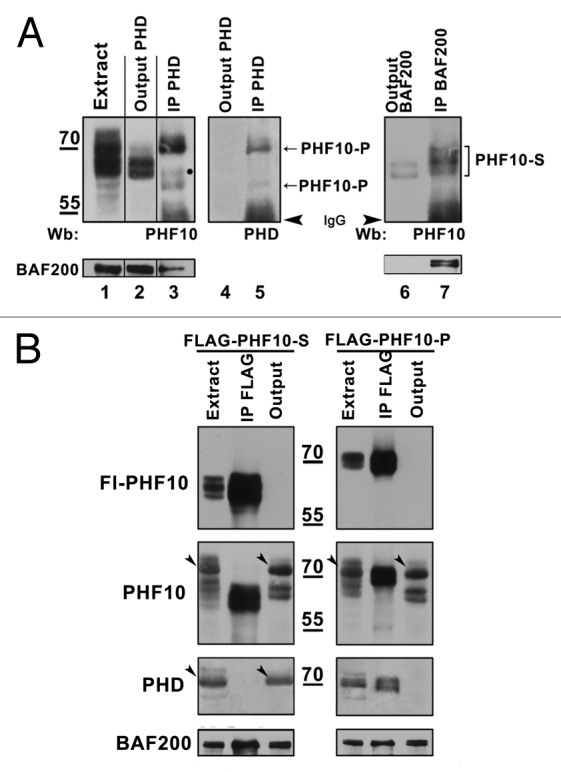
Figure 4. PHF10 isoforms containing PHD fingers or PDSM motif are associated with distinct PBAF complexes. (A) PBAF complexes containing distinct PHF10 isoforms are present in the HEK293 cell extract. The PHF10-P associated PBAF complexes were depleted from the extract with anti-PHD antibodies (lanes 1–5), while the complexes that contained PHF10-S remained in the output material and were further precipitated with anti-BAF200 antibodies (lanes 6–7). The western blot was developed with antibodies against PHF10 (lanes 1–3, 6, 7) or anti-PHD (lanes 4, 5). A significant amount of BAF200 co-precipitated with each of the PHF10 isoforms (lower panel), suggesting that PBAF complexes with PHF10-P or PHF10-S were both well represented in the cells. (B) Recombinant FLAG-tagged PHF10-S (left panel) or PHF10-P (right panel) in stably transfected HEK293 cells do not co-precipitate with endogenous PHF10-P and PHF10-Sl isoforms. The western blot was developed with anti-FLAG, anti-PHF10, and anti-PHD antibodies. Each recombinant isoform (panel FLAG-PHF10) was completely depleted from the extract, while the endogenous isoform with a different C-terminal domain remained in the output material (panels PHF10, PHD). Endogenous isoforms PHF10-P (left panel) and PHF10-Sl (right panel) are indicated by arrowheads.
Another interesting question was whether the difference in PHF10 subunit could lead to change in the composition of the PBAF complex. To find the answer, the nuclear extract from HEK293 cells stably transfected with FLAG-tagged PHF10-P or PHF10-S was fractionated on a Superose-6 gel filtration column, and PHF10-containing complexes were purified from the corresponding fractions using an antibody against FLAG (Fig. S3A). The precipitated proteins were resolved by SDS-PAGE and identified by MALDI-TOF analysis (Fig. S3B). The results demonstrated that both complexes had the same subunit composition. However, this result does not exclude the possibility that corresponding PHF10 isoforms may facilitate PBAF interaction with some other transcriptional complexes or proteins. Therefore, our next purpose was to determine if PBAF complexes containing the PHF10-P or PHF10-S isoform have the same functions in the cell.
PHF10-P and PHF10-S have different effects on cell proliferation capacity
Two types of PHF10 isoforms were expected to have distinct activities in the cells, since they contained functionally unrelated C-terminal domains. As shown previously, PHF10 RNAi resulted in a loss of cell proliferation.19 Therefore, we studied the effect of PHF10-P and PHF10-S on the proliferation capacity of stably transfected HEK293 cells. The mRNA and protein levels of PHF10-P and PHF10-S were approximately the same in both lines (data not shown), and the ratio of endogenous and FLAG-tagged PHF10 isoforms was about 1:3 in each cell line. Similar to the endogenous protein, FLAG-tagged PHF10 isoforms had nuclear localization (Fig. S3C) and were incorporated in the PBAF complex (Fig. 5A), which was indicative of their functionality.
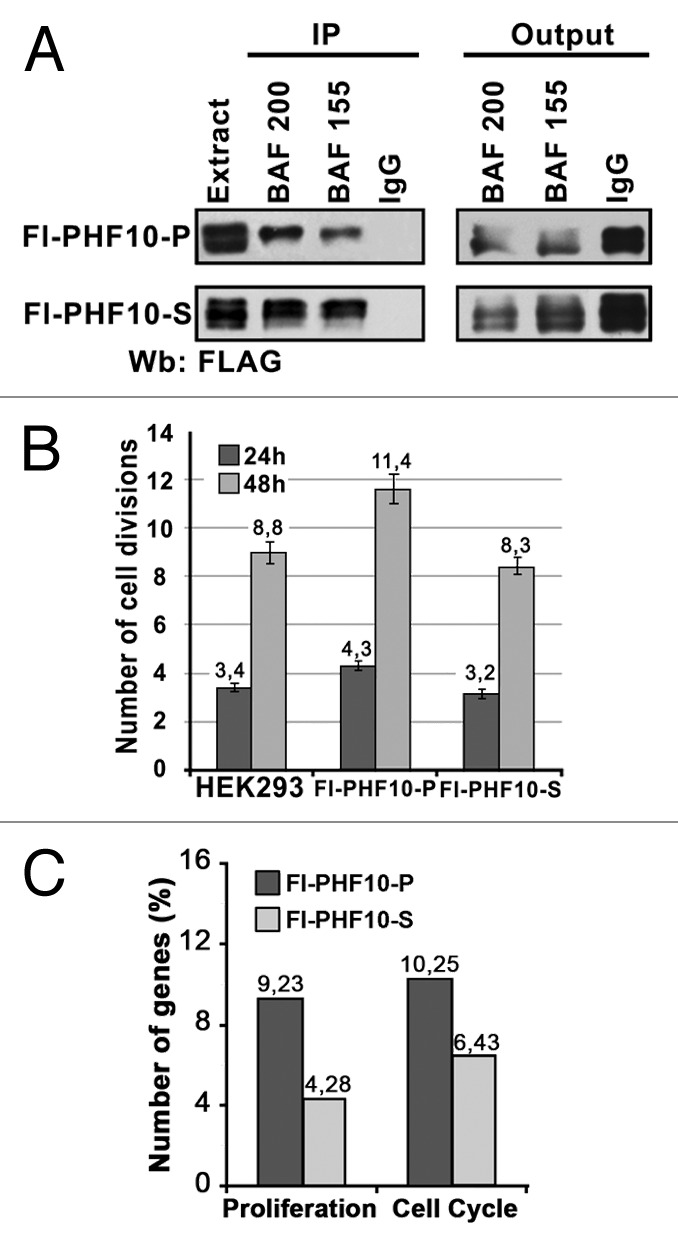
Figure 5. PHF10 isoforms containing PHD fingers or PDSM motif have different effects on cell proliferation. (A) Recombinant PHF10-P and PHF10-S isoforms are associated with PBAF. Immunoprecipitation with antibodies against BAF200 or BAF155 was performed from the extract of HEK293 cells stably transfected with FLAG-tagged PHF10-P or PHF10-S. The western blot was developed with antibodies against FLAG to detect recombinant PHF10. (B) Numbers of cell divisions recorded 24 and 48 h after plating in a wild-type HEK293 culture and in cells stably transfected with FLAG-PHF10-P or FLAG- PHF10-S. Cells were stained with CellTrace Violet Kit and counted in a flow cytometer. The proportion of necrotic cells (propidium iodide staining) in any sample did not exceed 5%. (C) The results of gene expression analysis with Illumuna HumanHT-12 microarray. The percentages of genes involved in proliferation and cell cycle (DAVID Functional Annotation Tool) relative to the total number of differentially expressed genes are shown for FLAG-PHF10-P (black bars) or FLAG- PHF10-S (gray bars) transfected cells.
The numbers of cell divisions in Fl-PHF10-P and Fl-PHF10-S stable cell lines and in nontranfected (control) HEK293 cells were measured 24 and 48 h after plating. The cells were stained with a CellTrace Violet Cell Proliferation Kit (Life Technologies) and scored by flow cytometry. The expression of FLAG-PHF10-P provided for an increase in the number of cell divisions compared with the control, which was detected after 24 h and became more significant after 48 h (Fig. 5B). In contrast, the number of divisions of FLAG-PHF10-S transfected cells after 48 h was slightly lower than in the control. Thus, the PHF10-P isoform significantly activates cell proliferation, while the PHF10-S isoform has different effect, causing a slight but statistically significant decrease in the proliferation capacity of the cells.
To further evaluate the role of PHF10 isoforms in cell proliferation, we performed global gene expression profiling using the Illumina HumanHT-12 microarray. The mRNA was extracted from control cells and cell lines stably transfected with FLAG-PHF10-P or FLAG-PHF10-S. Two samples of each cell line were used for analysis. In general, PHF10-P and PHF10-S overexpression had distinct but overlapping effects on the gene expression profile (the Pearson correlation coefficient was 0.87). We identified 195 genes that changed their expression level by a factor of 2 or greater in FLAG-PHF10-P cells and 280 such genes in FLAG-PHF10-S cells (Table S1). Then DAVID Functional Annotation Tool was used to functionally annotate differentially expressed genes. In line with a wide role of PBAF in transcription regulation, genes involved in different processes of biosynthesis, transport, and metabolism were identified with a high score (data not shown). We focused on genes involved in cell division and proliferation and found that about 9% of all genes identified in FLAG-PHF10-P cells were involved in proliferation and about 10% in cell cycle control (compared with 4 and 6%, respectively, in FLAG-PHF10-S cells) (Fig. 5C).
To validate these results, the transcription level of several of these genes was measured by quantitative RT-PCR. In agreement with the results of the microarray test, their transcription level proved to be increased against the background of PHF10-P overexpression and decreased against the background of PHF10-S overexpression (data not shown).
PHF10-P facilitates Pol II recruitment on promoter
The next question we addressed was that of probable difference in transcription regulation between PHF10-P and PHF10-S. First we assayed the presence of these isoforms on the promoters of NOV and ZMIZ1 genes, which are involved in the regulation of proliferation and cell growth and are upregulated in PHF10-P transfected cells. According to the epigenomics data (http://www.ncbi.nlm.nih.gov/epigenomics), several subunits of the SWI/SNF complex (BRG1, BAF155, Ini1, BAF170) have been found on the promoters of these genes in the HeLa cell line.
In ChIP experiments with anti-FLAG antibody, FLAG-tagged PHF10-P, and PHF10-S isoforms in the corresponding transfected cell lines showed approximately the same level of binding to the promoters of NOV and ZMIZ1 (Fig. 6A). Therefore, the presence of PHD or PDSM in PBAF does not determine the specificity of the complex to particular genes but appears to be relevant mainly for transcription regulation. The level of other tested PBAF subunits was also approximately the same on the promoters bound by either PHF10-P or PHF10-S, i.e., the affinity of the complex to the promoter did not change depending on PHF10 isoform (Fig. 6A). The level of PBAF subunits was increased in transfected cells, compared with the initial (non-transfected) HEK293 cell culture. This is evidence that PHF10 is essential for PBAF recruitment, as previously shown for its Drosophila homolog.17
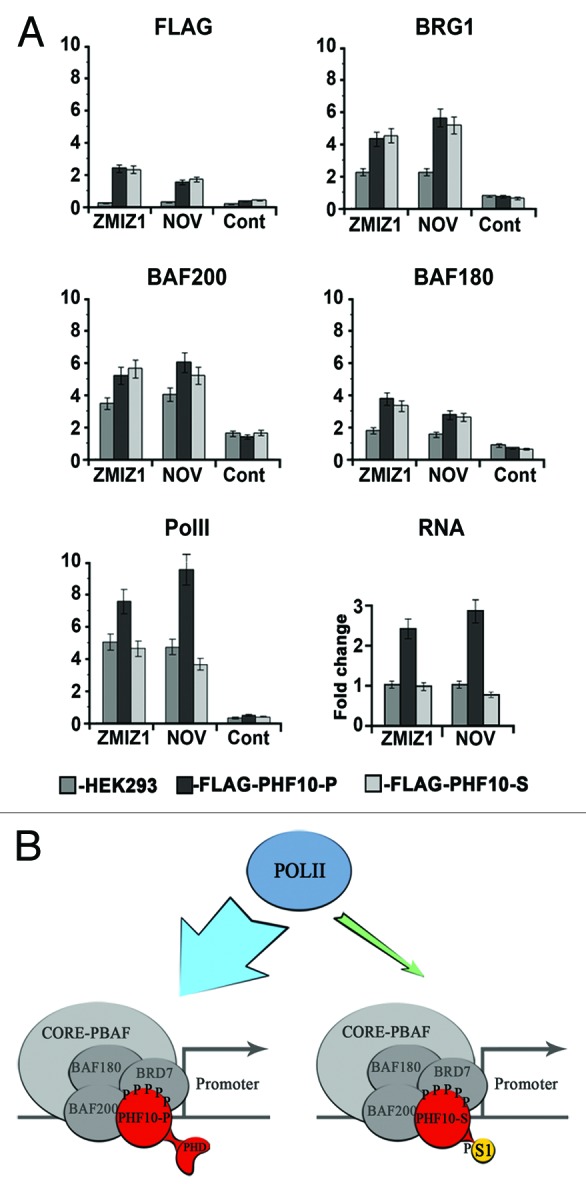
Figure 6. PHF10-P and PHF10-S have different effects on Pol II recruitment to promoter and on transcription level. (A) The presence of FLAG-PHF10-P, FLAG-PHF10-S, PBAF subunits, and Pol II on the promoters of ZMIZ1 and NOV genes and their transcription level (RNA) in HEK293 and stably transfected cells as determined by ChIP. The intergenic region was used as a control. (C) Two types of PBAF complexes that contain PHF10 with either PHD fingers or a PDSM motif at the C terminus are present in human cells. PHF10 is an intrinsic subunit of the PBAF signature module and is stably associated with BAF200 and BAF155. Both types of PHF10 isoforms are phosphorylated, and the PDSM isoform is also sumoylated. The isoform containing the PHD fingers, but not that with the PDSM motif, facilitates the recruitment of Pol II to the promoter and enhances the transcription level of the gene.
The level of Pol II on the promoter in PHF10-P transfected cells was much higher than in the initial cell culture, while that in PHF10-S transfected cells remained almost unchanged and even decreased slightly (Fig. 6A, Pol II). In line with this fact, PHF10-P and PHF10-S proved to have opposite effects on NOV and ZMIZ1 transcription (Fig. 6A, RNA).
It should be noted that the same situation was observed on the promoter regions of several other genes: PHF10-P overexpression resulted in an increased level of Pol II, while PHF10-S overexpression had no such effect, and this was not due to changes in the overall Pol II expression level in the transfected cells (data not shown). Thus, it is PHF10-P, but not PHF10-S, that facilitates Pol II recruitment on the promoter.
Discussion
Here we describe the alternative protein isoform encoded by the PHF10 gene as a novel subunit of the signature module of a mammalian PBAF chromatin-remodeling complex. Instead of C-terminal PHD fingers characteristic of the PHF10 isoform described previously,12,19 the novel isoform contains the PDSM consensus sequence that conjugates SUMO 1 in a phosphorylation-dependent manner. Similar to the isoform with PHD fingers (PHF10-P), the PDSM-containing isoform (PHF10-S) is evolutionarily conserved, which implies its significant role in transcription regulation. The 2 types of PHF10 isoforms are ubiquitously expressed in human cells and are subunits of different PBAF complexes (Fig. 6B). Their C-terminal domains are functionally unrelated, suggesting that the roles of these isoforms in control of gene expression differ significantly. Thus, it is PHF10-P, but not PHF10-S, that stimulates cell cycle progression and increases the level of promoter-bound Pol II, functioning as a transcription co-activator. The structural and functional differences between the PHF10 isoforms appear to account for the functional diversity of corresponding PBAF complexes in mammalian cells.
Several pieces of evidence indicate that PHF10 is an important subunit of the PBAF. Both types of PHF10 isoforms are ubiquitously expressed and subject to post-translational modifications, suggesting that they are actively regulated. The PHF10 isoforms associate with other specific subunits of the PBAF complex (BAF180, BAF200, and BRD7), forming a stable module (Fig. 6B). The conserved SAY domain present in both isoforms is apparently responsible for this association, similar to what has been shown for the Drosophila SAYP protein.17 The knockdown of PHF10 leads to a drop in the level of specific PBAF subunits, which means that such treatment affects the integrity of the architecture of the signature module. These data are in line with the results of previous studies,9,17 showing that Drosophila SAYP is critical for stable assembly of the PBAP signature module.
The PHD family is one of the largest families of epigenetic effectors capable of recognizing post-translational modifications of the H3 histone tail. Our data show that the PHD fingers of PHF10 facilitate transcription, since the PHF10-P isoform acts as a transcription co-activator: it promotes Pol II recruitment to gene promoters and enhances transcription of corresponding genes. This is in line with our previous data that deletion of PHDs from Drosophila SAYP reduces the transcription level of SAYP-dependent genes.17
The tandem PHD fingers of PHF10 are evolutionarily conserved, being present in PHF10 homologs from different species. Importantly, PHF10 is the only PBAF subunit that has been found to contain such domains. The BAF45b, BAF45c, and BAF45d subunits of the neuron-specific BAF chromatin remodeling complex also have tandem PHD fingers, which are highly similar to those of PHF10.10 This is evidence for the significance of this double PHD type for SWI/SNF function.
Our data show that a significant proportion of PBAF complexes contain the PHF10-S isoform and that both types of complexes (containing either PHD fingers or PDSM motif) are more or less equally represented in cells. The substitution of PHF10-S for PHF10-P does not lead to change in the subunit composition of the complex and does not determine its promoter specificity. However, the 2 isoforms have different roles in gene regulation, since PHF10-S, unlike PHF10-P, does not enhance transcription and the level of Pol II recruitment to promoters.
Sumoylation of PHF10-S occurs within the complex, since both sumoylated and not sumoylated PHF10-S are associated with PBAF. Its functional role is as yet unclear. The PDSM motif has been found in mutually unrelated regulatory proteins, with sumoylation either potentiating or repressing its transcription activation function.20 Sumoylation of PHF10-S may attract certain repressive complexes. Thus, it has been shown that SUMO promotes HDAC-mediated transcriptional repression.21,23 Further studies are underway in our laboratory to elucidate the role of PHF10-S sumoylation.
No subunit heterogeneity has as yet been demonstrated for metazoan PBAF. The composition of the BAF complex in mammals is more diverse, since some of subunits are represented by paralogous proteins alternatively incorporated in the complex. Thus, it contains either Brg1 ATPase or its close homolog Brm, both being broadly expressed but having largely different functions in transcription regulation.5,6 The BAF250 signature subunit is represented by 2 broadly expressed homologs (BAF250A and BAF250B) that are alternatively incorporated in the complex and have different effects on cell cycle activity.7,8 Our data demonstrate that PHF10 isoforms with different functions in transcription regulation are mutually exclusive components of PBAF. This fact may explain the dual function of PBAP as a transcription activator or repressor.3,24
Materials and Methods
RNAi knockdown
To downregulate PHF10 expression, the pSuper RNAi System (OligoEngine) was used. Two 19-nt fragments of the PHF10 sequence19 were cloned as hairpins in pSuper and used for simultaneous transfection. Control cells were transfected by empty pSuper. Clones were selected with puromycin (1 μg/mL) and individually tested for PHF10 mRNA downregulation. Six clones with the highest rate of PHF10 downregulation were pooled for further experiments.
Sequence accession numbers
The accession numbers of the PHF10 transcripts and corresponding proteins are as follows: KC839988 (PHF10-P short), KC839989 (PHF10-S long), and KC839990 (PHF10-S short).
Nuclear extract preparation
HEK293 nuclei were lysed in 10 mM HEPES (pH 7.9) containing 5 mM MgCl2, 0.5% Nonidet P-40, 0.45 M NaCl, 1 mM DTT, and a protease inhibitor cocktail (PIC) (Roche). The lysate was centrifuged at 10 000 rpm, 4 °C, for 10 min, and the supernatant was diluted 4-fold with the same buffer but without NaCl. The extract was treated with DNase I (USB, 0.6 units/mL) and RNase (Stratagene, 10 units/mL) in the presence of additional inhibitors (final concentrations): 20 μM MG132 (Calbiochem, Millipore), 1% PhIC (Sigma-Aldrich), and 10 mM N-ethylmaleimide (NEM, Sigma). This was followed by treatment with lambda protein phosphatase (New England Biolabs) according to the manufacturer’s protocol. Immunoprecipitation was performed by adding 15 μL of antibody-saturated protein A Sepharose beads (Sigma-Aldrich) to the cell extract (2 × 106 cells per round) and incubating the mixture overnight at 4 °C on a shaker. The beads were washed 3 times with 1× PBS containing 0.5 M NaCl and 0.1% Triton X-100, and the bound proteins were eluted with 2 bead volumes of 1× PBS with 0.5% N-lauroylsarcosine. Their purification on Ni-NTA Sepharose (GE Healthcare, 15 μL) was performed under the same conditions.
Antibodies and immunostaining
Polyclonal antibodies were generated by immunizing rabbits with the following His-tagged polypeptides: amino acids 89–498 (ab1), 238–261 (ab2), and 379–498 (ab3, or α-PHD) of PHF10 (NP_060758.2); BAF200, amino acids 1058–1468; BAF155, amino acids 752–1105; BAF180, amino acids 1–431; and BRD7, amino acids 1–284 and 284–562. All antibodies were affinity purified. Antibody to α-FLAG (M2 monoclone) was from Sigma-Aldrich; to α-tubulin-b (E7 monoclone) from DSHB; to α-SUMO-1 (#4930) from Cell Signaling Technology. Antibodies to α-BAF250 and α-Brg1 were kindly provided by Peter Verrijzer. Immunostaining of endogenous or FLAG-tagged recombinant PHF10 isoforms in HEK293 cells was performed as described,25 using affinity purified primary antibodies ab1 (1:50), ab2 (1:50), or α-PHD (1:250) and secondary anti-rabbit Alexa-488 Fluor-conjugated antibodies (Invitrogen). Stained preparations on glass slides were mounted in mounting medium (Vector Laboratories) and examined under a TCS SP2 confocal microscope or a DMR/HC5 fluorescence microscope (Leica) with an HCX PZ Fluotar × 100/1.3 objective lens. Microscopic images were made with a Leica DC350 F digital camera.
Measuring cell proliferation capacity
The cells were vitally stained with CellTraceViolet Cell Proliferation Kit (Invitrogene) according to the manufacturer’s protocol and plated in the wells of 6-well Corning culture plates at 30% confluence. Their fluorescence was measured in a Gallios flow cytometer (Beckman Coulter). Three wells per experimental point were used, and the mean level of fluorescence was calculated. The experiment was performed in 5 replications.
Microarray analysis
The mRNA was extracted from control cells and cell lines stably transfected with FLAG-PHF10-P or FLAG-PHF10-S. Two samples from each cell line were used for analysis with Illumina HumanHT-12 microarray. The resulting data were filtered with a threshold detection value of P = 0.05 and then normalized by the quantile normalization method. The sample-to-reference ratio was used to identify genes induced or reduced more than 2-fold (at P ≤ 0.05), which were classified as significantly changed genes.
ChIP and qPCR analysis
ChIP was performed as described26 with DNA sheared to about 500 bp. Approximately 3 × 106 cells and 10 mg of an antibody were taken per experiment. The primers used for analysis are given in the Supplemental Materials. After ChIP, the recovered DNA was analyzed by qPCR with Chromo-4 (Bio-Rad). Each point was measured in at least 3 experiments, and the mean value was calculated. A fragment of intergenic region was used as a negative control in each experiment.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to NE Vorobyeva and NA Gorgolyuk for critical reading of the manuscript and to P Verrijzer and Yu Shidlovskii for a kind gift of antibodies. This study was supported by the program “Molecular and Cellular Biology” of the Russian Academy of Sciences, RFBR [13-04-40378-H].
References
- 1.Wu JI. Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta Biochim Biophys Sin (Shanghai) 2012;44:54–69. doi: 10.1093/abbs/gmr099. [DOI] [PubMed] [Google Scholar]
- 2.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Kaeser MD, Aslanian A, Dong MQ, Yates JR, 3rd, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem. 2008;283:32254–63. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moshkin YM, Chalkley GE, Kan TW, Reddy BA, Ozgur Z, van Ijcken WF, Dekkers DH, Demmers JA, Travers AA, Verrijzer CP. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol Cell Biol. 2012;32:675–88. doi: 10.1128/MCB.06365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–95. doi: 10.1016/S1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 6.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–6. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagl NG, Jr., Zweitzig DR, Thimmapaya B, Beck GR, Jr., Moran E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66:1289–93. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- 8.Nagl NG, Jr., Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 2007;26:752–63. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalkley GE, Moshkin YM, Langenberg K, Bezstarosti K, Blastyak A, Gyurkovics H, Demmers JA, Verrijzer CP. The transcriptional coactivator SAYP is a trithorax group signature subunit of the PBAP chromatin remodeling complex. Mol Cell Biol. 2008;28:2920–9. doi: 10.1128/MCB.02217-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–15. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Middeljans E, Wan X, Jansen PW, Sharma V, Stunnenberg HG, Logie C. SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS One. 2012;7:e33834. doi: 10.1371/journal.pone.0033834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vorobyeva NE, Soshnikova NV, Kuzmina JL, Kopantseva MR, Nikolenko JV, Nabirochkina EN, Georgieva SG, Shidlovskii YV. The novel regulator of metazoan development SAYP organizes a nuclear coactivator supercomplex. Cell Cycle. 2009;8:2152–6. doi: 10.4161/cc.8.14.9115. [DOI] [PubMed] [Google Scholar]
- 13.Musselman CA, Kutateladze TG. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res. 2011;39:9061–71. doi: 10.1093/nar/gkr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shidlovskii YV, Krasnov AN, Nikolenko JV, Lebedeva LA, Kopantseva M, Ermolaeva MA, Ilyin YV, Nabirochkina EN, Georgiev PG, Georgieva SG. A novel multidomain transcription coactivator SAYP can also repress transcription in heterochromatin. EMBO J. 2005;24:97–107. doi: 10.1038/sj.emboj.7600508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panov VV, Kuzmina JL, Doronin SA, Kopantseva MR, Nabirochkina EN, Georgieva SG, Vorobyeva NE, Shidlovskii YV. Transcription co-activator SAYP mediates the action of STAT activator. Nucleic Acids Res. 2012;40:2445–53. doi: 10.1093/nar/gkr1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vorobyeva NE, Nikolenko JV, Krasnov AN, Kuzmina JL, Panov VV, Nabirochkina EN, Georgieva SG, Shidlovskii YV. SAYP interacts with DHR3 nuclear receptor and participates in ecdysone-dependent transcription regulation. Cell Cycle. 2011;10:1821–7. doi: 10.4161/cc.10.11.15727. [DOI] [PubMed] [Google Scholar]
- 17.Vorobyeva NE, Soshnikova NV, Nikolenko JV, Kuzmina JL, Nabirochkina EN, Georgieva SG, Shidlovskii YV. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc Natl Acad Sci U S A. 2009;106:11049–54. doi: 10.1073/pnas.0901801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vorobyeva NE, Nikolenko JV, Nabirochkina EN, Krasnov AN, Shidlovskii YV, Georgieva SG. SAYP and Brahma are important for ‘repressive’ and ‘transient’ Pol II pausing. Nucleic Acids Res. 2012;40:7319–31. doi: 10.1093/nar/gks472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banga SS, Peng L, Dasgupta T, Palejwala V, Ozer HL. PHF10 is required for cell proliferation in normal and SV40-immortalized human fibroblast cells. Cytogenet Genome Res. 2009;126:227–42. doi: 10.1159/000251960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA. 2006;1034:5–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Dominguez M. SUMO Tasks in Chromatin Remodeling. Chromatin Remodeling. InTech, 2013. DOI: 10.5772/55395 [DOI] [Google Scholar]
- 22.Moshkin YM, Mohrmann L, van Ijcken WF, Verrijzer CP. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol Cell Biol. 2007;27:651–61. doi: 10.1128/MCB.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol Cell. 2004;13:611–7. doi: 10.1016/S1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 24.Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddison PJ. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–91. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurskiy D, Orlova A, Vorobyeva N, Nabirochkina E, Krasnov A, Shidlovskii Y, Georgieva S, Kopytova D. The DUBm subunit Sgf11 is required for mRNA export and interacts with Cbp80 in Drosophila. Nucleic Acids Res. 2012;40:10689–700. doi: 10.1093/nar/gks857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vorobyeva NE, Mazina MU, Golovnin AK, Kopytova DV, Gurskiy DY, Nabirochkina EN, Georgieva SG, Georgiev PG, Krasnov AN. Insulator protein Su(Hw) recruits SAGA and Brahma complexes and constitutes part of Origin Recognition Complex-binding sites in the Drosophila genome. Nucleic Acids Res. 2013;41:5717–30. doi: 10.1093/nar/gkt297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.