Abstract
Escherichia coli RecA is the defining member of a ubiquitous class of DNA strand exchange proteins that are essential for homologous recombination, a pathway that maintains genomic integrity by repairing broken DNA1. To function, filaments of RecA must nucleate and grow on single-stranded DNA (ssDNA) in direct competition with ssDNA-binding protein (SSB), which rapidly binds and continuously sequesters ssDNA, kinetically blocking RecA assembly2,3. This dynamic self-assembly on a DNA lattice, in competition with another protein, is unique for the RecA-family relative to other filament-forming proteins such as actin and tubulin. The complexity of this process has hindered our understanding of RecA filament assembly because ensemble measurements cannot reliably distinguish between the nucleation and growth phases, despite extensive and diverse attempts2–5. Previous single-molecule assays have measured nucleation and growth of RecA—and its eukaryotic homolog RAD51—on naked dsDNA and ssDNA6–12; however, the template for RecA self-assembly in vivo is SSB-coated ssDNA3. Using single-molecule microscopy, we directly visualized RecA filament assembly on single molecules of SSB-coated ssDNA, simultaneously measuring nucleation and growth. We establish that a dimer of RecA is required for nucleation, followed by growth of the filament through monomer addition, consistent with the finding that nucleation, but not growth, is modulated by nucleotide and magnesium ion cofactors. Filament growth is bidirectional, albeit faster in the 5′→3′ direction. Both nucleation and growth are repressed at physiological conditions, highlighting the essential role of recombination mediators in potentiating assembly in vivo. We define a two-step kinetic mechanism where RecA nucleates on transiently exposed ssDNA during SSB sliding and/or partial dissociation (i.e., DNA unwrapping) and then grows. We further demonstrate that the recombination mediator protein pair, RecOR, accelerates both RecA nucleation and filament growth, and that introduction of RecF further stimulates RecA nucleation.
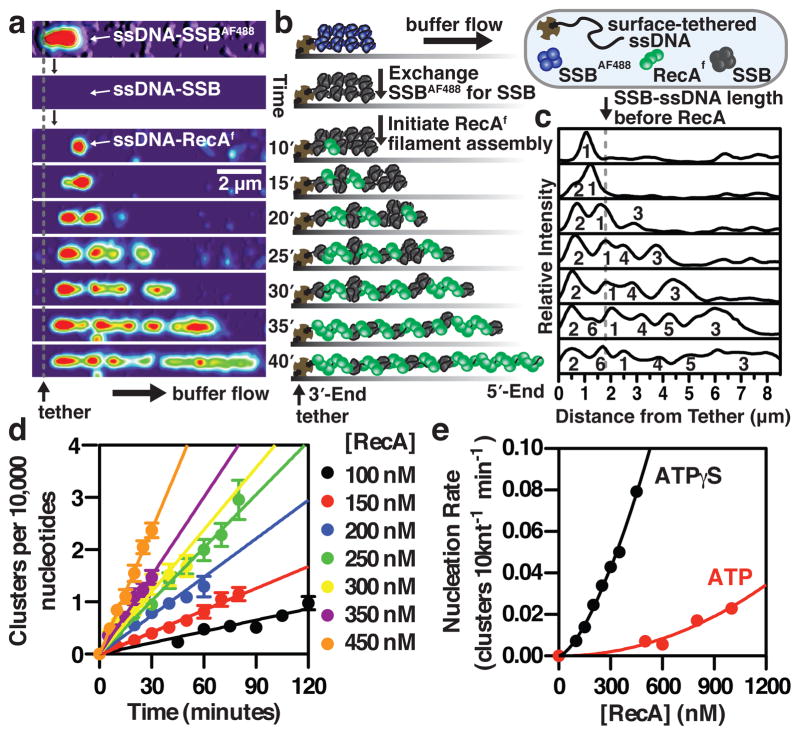
To image the assembly of RecA filaments on SSB-coated ssDNA, we first developed a procedure to generate and visualize single molecules of ssDNA. Bacteriophage λ dsDNA (48.54 kb) was biotinylated at the 3′-ends, alkali-denatured, neutralized with buffer, and saturated with a fluorescently modified SSB (SSBAF488). This complex was then attached to the surface of a streptavidin-coated glass coverslip within a microfluidic chamber, extended by buffer flow, and visualized using Total Internal Reflection Fluorescence (TIRF) microscopy (Fig. 1a, b, top panels). Because the binding affinity of SSBAF488 is attenuated13,14, we next replaced it with unlabeled wild-type SSB in situ (Fig. 1a, b, second panels and Supplementary Movie 1). The exchange of proteins in the flow cell is fast, with a half-time of approximately 2–3 seconds, resulting in a native SSB-ssDNA complex that is non-fluorescent. RecA filament assembly on the wild-type SSB-ssDNA complexes was then imaged using a fluorescent protein, RecAf, described previously7. Assembly was initiated by injecting RecAf, free SSB, and either ATPγS or ATP (plus an ATP regenerating system). RecA filament formation occurs slowly (Fig. 1a, b, third and subsequent panels), first appearing as a single spot (herein referred to as a cluster). Molecules were imaged intermittently over the course of one to two hours; when the molecules were not being imaged, both the laser excitation and flow were turned off to minimize photobleaching, reduce sample volumes, and allow filament assembly to proceed on ssDNA in its relaxed state. With time, the nascent clusters elongated, and new clusters appeared; these mixed nucleoprotein complexes comprised relatively stiff, rod-like RecA filament clusters interspersed between compacted and flexible SSB-coated ssDNA15. The composition of these intermediates was confirmed using atomic force microscopy (Supplementary Fig. 1). At the flow rate used, the SSB-ssDNA complexes are compacted to ~15% of the corresponding length of B-form dsDNA (Fig. 1a, b, top panels); in contrast, RecA-ssDNA filaments are extended to ~160% of dsDNA length14.
Figure 1. Direct visualization of RecA filament assembly on single molecules of SSB-coated ssDNA reveals that RecA nucleates as a dimer.
(a) RecAf filament assembly with ATPγS on a single molecule of SSB-coated ssDNA tethered within a microfluidic flow chamber was visualized using TIRF microscopy; montage is rendered into a topographic fluorescent intensity map. (b) Schematic and (c) fluorescent intensity profile from panel A. (d) The number of RecA clusters increases linearly with time; slope is the nucleation rate (n = 18–93 clusters for each concentration; (±s.d.)). (e) Nucleation rate increases with [RecA] according to, J=k[RecA]n, where n is 2.2 (±0.6) (ATPγS) and 1.5 (±0.1) (ATP) (error from the linear fits in (d) is smaller than the symbols).
The number of RecA clusters bound to individual SSB-ssDNA complexes was determined by comparing images of each molecule with its fluorescence intensity profile (Fig. 1a, c & Supplementary Fig. 2). The number of clusters formed increased linearly with time (Fig. 1d), allowing for direct measurement of the RecA nucleation rate, expressed as the number of clusters formed per minute per 10,000 nucleotides. When the concentration of RecA was increased, the rate of nucleation became faster, exhibiting a non-linear dependence with respect to RecA concentration (Fig. 1e). The data were fit to the relationship J = k[RecA]n, where J is the frequency of nucleation, k is a rate constant, and n is the number of protomers in a critical nucleus7,16. In the presence of ATP or ATPγS, the rate of nucleation of RecA on SSB-ssDNA exhibited a power dependence, where n is 2.2 (±0.6) or 1.5 (±0.1), respectively (Fig. 1e). Because the nucleotide-binding site of a RecA nucleoprotein filament lies in a binding pocket between monomers17, our conclusion is consistent with the expectation that a dimer would be the minimal oligomeric species that could form a stable nucleus on ssDNA.
The sizes of individual RecA filaments grew with time (Fig. 2a), and the lengths of well-resolved individual clusters were plotted as a function of time and analyzed by linear regression to determine their growth rate from the slope (Fig. 2b). Increasing RecA concentration resulted in a net increase in the growth rate as well as the distribution of rates, which varied from 10 to 100 nm min−1 (Fig. 2c); given that the RecA-ssDNA filament comprises ~6 monomers per 10 nm, these rates correspond to ~6 to 60 monomers min−1. The net growth rate increased linearly with respect to RecA concentration with a slope of 1.8 (±1.0) × 106 M−1 sec−1 in the presence of ATPγS, and 0.8 (±0.1) × 106 M−1 sec−1 in the presence of ATP (Fig. 2d). This rate measures the bimolecular rate of association of RecA for the ends of a filament, and is ~10–100 times slower than the theoretical diffusion-controlled limit18. This linear concentration dependence indicates that filaments grow primarily through monomer addition, in agreement with previous observations8. Although the variation in growth rates for different filaments was large, as is evident in their broad distribution (Fig. 2c, d), the measurement error for the growth rate of any individual filament was low (Supplementary Table 1). The nature of this broad distribution is discussed later as a salient property of RecA filament growth.
Figure 2. RecA filaments grow via rapid addition of monomers.
(a) Lengths for RecA cluster were measured and (b) analyzed (same colors as in a, plus additional data) to determine the rate (slope) and lag time (x-intercept) (100 nM RecA and ATPγS). (c) Distribution of growth rates at increasing [RecA]. (d) Mean growth rates increase linearly with [RecA]; slope is 0.16 (±0.08) nm min−1 nM−1 (ATPγS, black) (or 1.1 (±0.6) × 108 M−1min−1 at 1.5 nm/RecA17) and 0.07 (±0.01) nm min−1 nM−1 (ATP, red) (or 0.47 (±0.3) × 108 M−1min−1). (e) Lag time distribution at increasing [RecA]. (f) Lag times plotted with respect to [RecA] and fit to an inverse power law, J−1=1/k[RecA]n, where n is 1.9 (±0.3). Lines in (c, e) are Gaussian fits. See Supplementary Table 1 statistics (n = 207 filaments, total; ± s.d.).
A further measure of filament nucleation is the time at which filaments begin to grow (i.e., the lag times), which are evident in the individual growth trajectories16. The lag time for each growing cluster is an extrapolation to a real time of nucleation that is not limited by our detection system, and was determined from the x-intercept of the linear regression for each filament analyzed (Fig. 2b). Analysis of these lag times demonstrates that nucleation is slow and stochastic at low RecA concentrations and becomes dramatically shorter at high concentrations (Fig. 2e), following an inverse power dependence where n = 1.9 (±0.3) (Fig. 2f). This independent method of analysis further confirms our conclusion above that RecA filaments nucleate as a dimer (Fig. 1d, e). This is in contrast to previous observations that RecA requires a larger critical nucleus of 3–5 monomers to assemble on dsDNA7,9, possibly due to its lower affinity for dsDNA2. Our observation that neither the nucleation nor growth rate of RecA filament assembly is altered over the range of salt concentration tested (50–200 mM NaCl) is also consistent with this interpretation (Supplementary Figure 3).
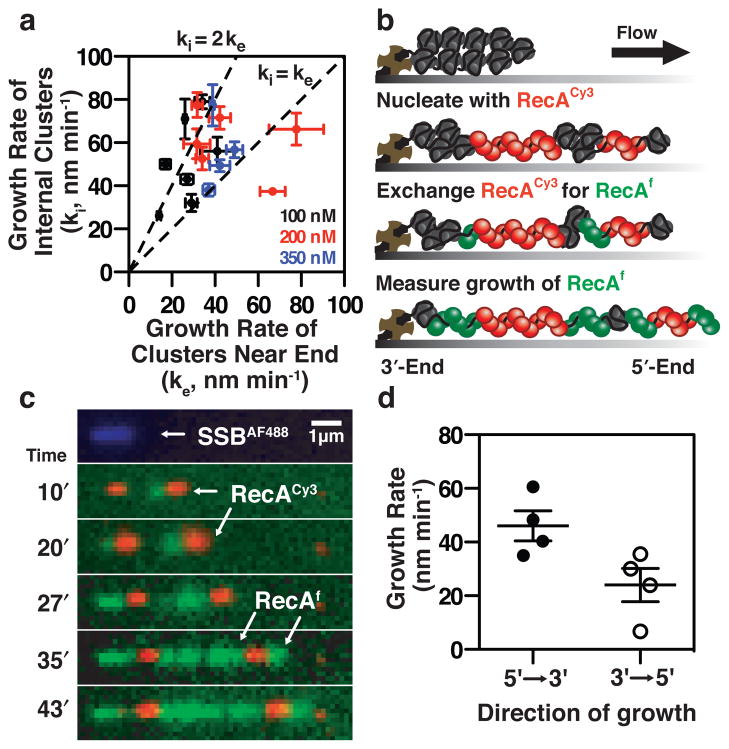
Previous single-molecule experiments monitoring RecA filament growth on both dsDNA and ssDNA demonstrated that RecA filaments could grow in both directions7,8. We sought to determine whether this would be the case in the presence of SSB, and whether growth was symmetric or asymmetric (i.e., whether there was a preferred directionality)19. We compared the growth of RecA clusters close to the point of attachment (i.e., within ~0.5 to 1 μm of the 3′-biotinylated end of the ssDNA tethered to the surface) relative to a cluster located on the same ssDNA molecule, but at an internal site. We rationalized that filaments growing from the tethered end could grow only in the 3′→5′ direction due to it being attached to the surface at the 3′-end of the ssDNA, whereas filaments forming elsewhere on the molecule would be free to grow in both directions. Therefore, if both ends were available for elongation, clusters growing at internal regions should grow faster than the clusters forming at the tethered end of the molecule. Indeed, the cluster growth rates measured at internal sites (ki) were faster than cluster growth rates measured near the ssDNA end (ke) (i.e., limited to growth in the 3′→5′ direction) (Fig. 3a). This is evident in the dot-plot in Fig. 3a where the majority of the data points fall above the dashed line representing hypothetical unidirectional growth in the 3′→5′ direction, with ki = ke; or the line representing bidirectional growth at the same rates, where ki = 2ke. The distribution of these relative rates is not affected by the concentration of RecA (Fig. 3a). While this analysis provides good evidence that RecA filament growth on SSB-coated ssDNA is bidirectional (i.e., almost all of the growth rates for internal clusters are greater than for those at an end), alone, it is insufficient to establish whether one direction grows faster than the other.
Figure 3. RecA filament growth on SSB-coated ssDNA is bidirectional.
(a) Growth rates of clusters at the tethered 3′-end of the ssDNA (ke) were plotted against growth rates of internal clusters on the same molecule (ki) (n = 18; ± s.e.m.). (b) RecACy3 (50 nM, red) clusters were pre-formed with ATPγS on SSB-coated ssDNA followed by growth with RecAf (250 nM, green). (c) Visualization of RecAf (green) growth from pre-formed RecACy3 (red) clusters, showing faster growth from the left side of the RecACy3 clusters in the 5′→3′ direction. (d) The growth rates were 44 (±11) nm min−1 in the 5′→3′ direction and 27 (±12) nm min−1 in the 3′→5′ direction; (n = 4; ± s.d.).
We therefore sought to unambiguously measure the growth rates at each end of a filament by direct single-molecule visualization. We first formed discrete clusters of RecACy3 (red) on SSB-ssDNA. The solution was then exchanged, replacing RecACy3 (red) with RecAf (green) (Fig. 3b). Growth of RecAf from the pre-formed, internal RecACy3 clusters was directly imaged and measured (Fig. 3c); growth from the left side of the RecACy3 clusters is in the 5′→3′ direction, and growth from the right side of the clusters is in the 3′→5′ direction. Filament growth from the red RecACy3, through addition of the green RecAf, is seen to commence in the leftward direction proceeding with an apparent preference for the 5′→3′ direction (i.e., towards the tethered end). The mean growth rate in the 5′→3′ direction is 44 (±11) nm min−1, whereas the growth rate in the 3′→5′ direction is 27 (±12) nm min−1 (Fig. 3d). Although our observations demonstrate that bidirectional growth on SSB-ssDNA is possible, it should not be considered obligatory during RecA filament assembly. The broad distribution of growth rates in our analysis (Fig. 2c, d), the analysis of growth rates of internal clusters relative to end clusters (Fig. 3a), and the direct visualization of bidirectional growth (Fig. 3c) suggest that filament growth proceeds through stochastic phases of unidirectional growth from either end as well as bidirectional growth from both ends.
Nucleation of the RecA filament on SSB-ssDNA requires formation of a RecA dimer (Fig. 1e & Fig. 2f), the interface of which is the binding site for the magnesium-bound nucleotide cofactor17. Therefore, we tested whether these cofactors might affect nucleation and growth. Increasing the concentration of Mg(OAc)2 stimulated nucleation, with a plateau occurring at concentrations equal to or greater than the ATPγS concentration (Fig. 4a), consistent with the need for this metal ion as a component of the nucleating species; in contrast, the rate of filament growth was unaffected by divalent ion (Fig. 4b). RecA binds to ssDNA with a higher affinity in the presence of ATPγS or dATP than ATP20,21. ATPγS is a non-hydrolysable analog of ATP, whereas dATP is hydrolyzed ~30% faster than ATP20; thus, a comparison of the assembly kinetics in the presence of these nucleotides experimentally uncouples hydrolysis from binding in these steps. In the presence of ATPγS and dATP, nucleation is ~10-fold and ~5-fold faster, respectively, relative to assembly with ATP (Fig. 4c), showing that stabilization is not correlated with the absence of hydrolysis; in contrast, the rate of filament growth is only modestly affected by the nucleotide cofactor and is not statistically significant (Fig. 4d). Therefore, the stabilization of nascent nuclei by ATPγS and dATP can be attributed to the nucleotide binding-linked induction of a conformation with high-affinity for ssDNA, consistent with the interpretation that nucleotide binding is important for filament assembly insomuch as it serves as a conformational modulator of RecA affinity for ssDNA that defines the stability of the critical nucleus2,21. Because the Mg++-ATP cofactor binds at adjacent RecA monomers in the filament17, our conclusions are consistent with the simplest expectations that nucleation of a filament should require minimally 2 monomers. Our observation that growth is affected by neither Mg(OAc)2 nor nucleotide cofactor—but is linearly dependent on RecA concentration—is consistent with the simplest interpretation that RecA monomers add to the ends of a filament, but require a conformational change that is slower than nucleotide binding.
Figure 4. Nucleation is modulated by ligand binding, repressed at physiological pH, and enhanced by RecOR and RecFOR; growth is enhanced by RecOR.
RecA nucleation and growth rates plotted as function of (a,b) [Mg(OAc)2], (c,d) nucleotide cofactors, and (e–f) pH (slope = −2.0 (±0.3) monomers min−1 pH−1). (a–f) Error bars are ± s.d. (g) Image montage of RecAf filament assembly on gapped DNA. (h) Nucleation enhanced by RecOR and RecFOR; (i) growth accelerated by RecOR, with no addition increase by RecFOR. (j) Kinetic model for RecA nucleation where a RecA dimer binds ssDNA transiently released from SSB through either sliding or unwrapping, which is enhanced by RecOR and RecFOR; growth via monomer addition accelerated by RecOR. [RecA] was 250 nM (a, b), 500 nM (c–f), or 1 μM (h–i).
Unexpectedly, we observed that the assembly of RecA on SSB-ssDNA at the approximately physiological pH of 7.5 was very slow; however, it was strongly stimulated by reducing the pH to 6.5, where it is predominantly a dimer22. Increasing the pH to 8.0 resulted in a dramatic monotonic inhibition of both nucleation (Fig. 4e) and growth (Fig. 4f), suggesting that the overall rates, but not the mechanism, are altered in a pH-dependent manner. The intracellular pH of E. coli is highly regulated and maintained in the range of 7.5–8.0, where the rate of uncatalyzed nucleation, seemingly paradoxically, would be very low (Fig. 4e). These findings show that, at physiological conditions, neither nucleation nor growth of RecA filaments on SSB-coated ssDNA occurs at its observed maximal rate. This finding, in turn, would suggest that cellular mediator proteins could promote RecA filament assembly on SSB-coated ssDNA by accelerating either of these steps. In E. coli, RecF, RecO, and RecR are genetically and biochemically defined mediator proteins23,24. Stimulation of assembly is via the RecOR complex, which interacts with SSB to mediate assembly onto SSB-ssDNA in a structure-independent manner, and the RecFOR complex, which acts at a junction of ssDNA with dsDNA23–25.
The addition of RecO (or RecOR) in our TIRF-based assay invariably resulted in the adsorption of SSB-coated ssDNA to the glass surface. To both circumvent this problem and to determine the mechanism by which these mediator proteins act, we switched to epifluorescent detection and optical trapping7,12,26, which moves the molecule away from the surface. To create a suitable DNA substrate with an ssDNA gap, we designed a derivative of bacteriophage λgt11 containing a φC31 attP recognition site and modified a derivative of bacteriophage M13mp7 ssDNA to contain the attB recognition site when annealed to a PCR-based segment of ssDNA. Integration of the ssDNA into the biotinylated λ DNA resulted in a dsDNA substrate containing an ssDNA gap of 8155 nucleotides that was flanked by dsDNA ‘handles’ at both ends (Supplementary Fig. 6). Using a microfluidic flow cell coupled to a fluorescence microscope and a split-beam dual optical trap26, two streptavidin-coated polystyrene beads were isolated and a single molecule of gapped DNA, biotinylated at each end, was tethered between the beads in situ (Supplementary Fig. 4 and Fig. 4g, illustration). The molecule was rotated perpendicular to the flow and successively dipped in a reaction channel containing RecAf and Mg++-ATPγS at 37°C7 (Fig. 4g, lower panel). With time, RecAf filaments nucleated on the SSB-coated ssDNA segment and grew at rates quantitatively consistent with our TIRF-based measurements, albeit faster due to the elevated temperature and RecA concentration (Fig. 4g and Supplementary Movie 2). With RecOR present, nucleation remained stochastic, but the mean rate of nucleation increased (i.e., lag times decreased), the distribution shifted to faster rates, and some nucleation events were faster than those seen for RecA alone; these characteristics were further enhanced by the addition of RecF (Fig. 4h). RecOR also accelerated RecA filament growth; however, addition of RecF had no further effect on the rate of filament growth (Fig. 4i).
We have demonstrated RecA filament nucleation on SSB-coated ssDNA is a slow and stochastic kinetic process requiring a dimer. Under physiological conditions, nascent RecA filaments grow at a rate of approximately 50 to 500 nm min−1 (~30 to 300 monomers min−1, or 0.5 to 5 monomer sec−1) (Fig. 4i), in good agreement with past ensemble work2,4. The bimolecular rate of association is rapid, but is below the theoretical diffusion-controlled limit, suggesting that filament growth proceeds through monomer addition8 followed by a required conformational change at the end of the filament. We have further demonstrated that growth of RecA clusters on SSB-ssDNA complexes is bidirectional, with preferred net growth occurring in the 5′→3′ direction. In light of these results, we define a simultaneous two-step kinetic model for nucleation of a RecA filament on SSB-coated ssDNA (Fig. 4j). In vivo, SSB binds rapidly to ssDNA, wrapping ~60–70 nucleotides of ssDNA per tetramer. Once bound, ssDNA binding proteins can slide on ssDNA27, with SSB sliding at ~260 nt2 sec−1 in ~3 nucleotide steps28. Likewise, ssDNA can locally unwrap from a subunit of the tetramer on a microsecond timescale29. This transient, limited unwrapping of ssDNA from an SSB tetramer could a create a site available for RecA binding; however, larger ssDNA segments could be rapidly rebound by another SSB tetramer29. Given that a RecA monomer requires ~3 nucleotides of ssDNA for binding28, nucleation of a RecA dimer would require the sliding of two adjacent tetramers away from each other on a nearly saturated ssDNA lattice, or the partial, transient release of ssDNA. Kinetic trapping of these transient states by RecA protomers is consistent with the slower kinetics of nucleation (requiring dimers) relative to growth (occurring with monomers). RecO binds to both SSB and RecR; however, RecO and/or RecR do not displace SSB from ssDNA30. Instead RecOR interacts with SSB to facilitate RecA filament assembly23. We propose that RecOR traps the transient states in which ssDNA is released by SSB either during sliding or unwrapping, effectively weakening the SSB-ssDNA interaction through stoichiometric interactions with SSB. Consequently, RecOR binding to SSB-ssDNA also enhances RecA filament growth. Here we establish that recombination mediators can differentially facilitate the rate-limiting step of nucleation, enhance filament growth, or both. We anticipate that the conclusions and technical approach presented here will be particularly useful, and in some cases maybe even essential, for future experiments designed to further elucidate the mechanism by which filament assembly of orthologs, such as RAD51, is regulated by the analogs, such as tumor-suppressor BRCA2 and the RAD51-paralogs, which play important but sometimes abstruse roles in the early steps of recombination-mediated DNA repair.
METHODS
Total internal reflection fluorescence microscopy & single-channel flow cells
An Eclipse TE2000-U, upright TIRF microscope (Nikon), using a CFI Plan Apo TIRF 100x, 1.45 N.A., oil-immersed objective was used as previously described26. Single channel flow cells were constructed by drilling holes into a glass microscope slide and adhering a glass coverslip using 3M Thermo-Bond Film (2.5 mil) with a channel cut out from the tape of approximate dimensions (5 mm × 35 mm × 0.1 mm). Inlet ports (PEEK tubing, 0.5 mm inner diameter) were attached to the flow cell using epoxy. The surface was functionalized by incubating the flow cell successively with the following solutions for 5 minutes each: 1 M NaOH, water, single molecule buffer (SMB: 20 mM MES (pH 6.5), 50 mM DTT, 100 mM NaCl and 20% sucrose), 0.2 mg/mL biotin-BSA, SMB, 0.2 mg/mL streptavidin (Pierce), and 0.15 mg/mL Roche Blocking Reagent. Proteins were diluted into SMB. Bacteriophage λ genomic DNA (48.5 kbps) was biotinylated by incorporating biotin-dGTP at the 3′ ends using T4 polymerase at 16°C in NEB (New England Biolabs) Buffer 2 for 10 minutes. The enzyme was inactivated with 20 mM EDTA followed by heat inactivation (70°C for 10 minutes) and removal of nucleotides with an Illustra Microspin S-400 Column (GE) equilibrated in TE (10 mM TrisOAc (pH 7.5) and 1 mM EDTA). The concentration of DNA was determined by UV absorbance, with A260 = 1 taken to be equivalent to 50 ng/μL for dsDNA. Single-stranded DNA was generated for single-molecule experiments by denaturing the biotinylated bacteriophage λ DNA (200 fmols, molecules) was denatured in 5–10 μL of 0.5 M NaOH at 37 °C for 5–10 minutes. The alkali conditions were rapidly neutralized with the addition of 0.4 mL SMB containing 200 nM SSBAF488, yielding a final concentration of ~480 nM nucleotides. Assuming a site size of 15 nucleotides per SSB monomer, SSB should be in approximately 6-fold excess over possible stoichiometric binding sites. The SSBAF488-ssDNA complexes were then injected into a flow cell and allowed to incubate for approximately 5 minutes to allow for the attachment of the biotinylated molecules to the streptavidin-coated glass surface.
RecAf filament formation on SSB-ssDNA using TIRF microscopy
SSBAF488-ssDNA complexes were attached to the functionalized surface of a flow cell as described above and imaged. The solution containing fluorescent SSBAF488 was then replaced with unlabeled wild-type SSB (500 nM) in SMB, resulting in the displacement of SSBAF488 from the ssDNA. When displacement was complete, as monitored by the rapid disappearance of fluorescence, reaction buffer containing 20 mM MES (pH 6.5), 100 mM NaCl (unless otherwise indicated), 50 mM DTT, 4 mM Mg(OAc)2, 2 mM ATPγS (unless ATP or dATP indicated), 200 nM SSB, and the indicated concentration of RecAf was injected. Experiments where Mg(OAc)2 was titrated (Fig. 4a, b) contained 250 nM RecA. Experiments comparing nucleotide cofactors and pH (Fig. 4c–f) contained 500 nM RecA. All experiments were performed at 23°C. Experiments conducted with ATP and dATP also contained 1 mM phosphoenol pyruvate (PEP) and 20 Units/mL of pyruvate kinase. Additionally, experiments performed above 500 nM total RecA contained 250 nM RecAf plus the amount of unmodified RecA necessary to achieve the final concentration indicated; this was done to reduce the extent of background fluorescence. Bulk-phase analyses of RecA filament formation on SSB-coated ssDNA, using the kinetics of ATP hydrolysis31, did not demonstrate any discernible difference for mixtures of unmodified RecA and RecAf (Supplementary Figure 5)14. Between the imaging intervals, flow was shut off in order to preserve sample and to reduce photobleaching; flow was restored approximately 4–5 seconds before laser excitation and imaging. Experiments designed to directly visualize bidirectional growth utilized RecACy3, which retains normal biochemical activity (Supplementary Fig. 6).
Image processing and analysis
For each time interval, ~ 25–35 frames (~2–4 seconds at 100 ms exposure) were averaged using ImageJ v.1.41o. A theoretical Point Spread Function (PSF) was generated using the ImageJ plugin, Deconvolution Lab, and subsequently deconvolved using the Tikhonov-Miller algorithm32. Images collected during a time-lapse experiment were then compiled into a stack, aligned to correct for stage drift and the intensity was normalized. Those stacks were then converted to 3D surface plots using the ImageJ 3D Surface Plot function, which applies a nearest neighbor smoothing function. Plots were directly compared to fluorescent intensity profiles in ImageJ to more accurately visually analyze clusters of RecAf forming at longer time intervals. Only molecules where SSBAF448-ssDNA complexes initially appeared as distinct single molecules were analyzed. Nucleation data were normalized by taking the number of clusters per molecule and dividing by the relative length of the SSBAF448-ssDNA of that molecule to the longest molecules from that particular experiment. As the length of the SSBAF448-ssDNA complexes changes with flow cell variation (which affects flow rate and extension) as well as buffer conditions14, this serves as an internal normalization factor for each experiment.
Atomic force microscopy imaging of SSB-coated and RecA-coated ssDNA complexes
SSB-coated ssDNA complexes were formed by incubating 35 μM (nucleotides) M13 ssDNA with 1.75 μM SSB for 5–10 minutes at 37°C in 20 mM TrisOAc (pH 7.5), 20 mM NaCl, 1 mM Mg(OAc)2, 0.5 mM ATPγS, 0.1 μM spermidine-HCl and 7 μM RecA. The nucleoprotein complexes were purified away from free protein using a C-1000 spin column washed with binding buffer, adsorbed onto freshly cleaved mica for 1 minute, rinsed with water, stained with 0.02% uranyl acetate and visualized using a Nanoscope IV in tapping mode with an Al-T300 tip (Budget Sensors). Uranyl acetate stain facilitates AFM imaging of the SSB-ssDNA complex33.
ATPase assays for monitoring the kinetics of RecA binding to SSB-coated ssDNA
ATPase assays comparing relative amounts of RecAf and unmodified RecA were performed in 20 mM MES (pH 6.5), 20% sucrose, 50 mM DTT, 100 mM NaCl, 0.2 mg/mL NADH, 20 U/mL pyruvate kinase and lactate dehydrogenase, 4 mM Mg(OAc)2, 2 mM ATP, 1 mM phosphoenolpyruvate, 3 μM M13 ssDNA, 1.5μM SSB, and 1 μM total RecA at the ratio of RecAf:RecAindicated. All assays were performed at 37°C with a 5-minute pre-incubation of all components except for RecA, which was used to initiate the reaction. The change in absorbance (λ = 340 nm) was converted to μmols ATP using the extinction coefficient for NADH = 6.22 × 10−3μM cm−1. The data were then fit to a second order polynomial (Y = A+Bx+Cx2, where the intercept, A, was constrained to the time at mixing, 0 seconds). The derivative of the fit was taken and the slope determined to be the change in hydrolysis rate as a function of time, expressed as Δv/Δt (i.e., the instantaneous change in the reaction velocity with time inμmols/min2), which defines the acceleration of the reaction velocity, and is a measure of RecA filament formation on SSB-ssDNA complexes31.
Fluorescent modification and activity of RecACy3
RecACy3 was made as described previously for RecAf by covalently modifying the N-terminus of the protein with Cy3-succidimyl ester (GE Healthcare) instead of 5,6-carboxyfluorescein-succidimyl ester7. The ssDNA binding activity of RecACy3 was determined by monitoring the ATP hydrolysis rate of RecA at increasing concentrations of polydeoxythymidylate ssDNA. The ATP hydrolysis rate was calculated from the linear decrease in the absorbance of NADH at 340 nm. The reaction buffer contained 25 mM TrisOAc (pH 7.5), 8 mM Mg(OAc)2, 1 mM ATP, 1.5 mM PEP, 1 mM DTT, 30 U/mL pyruvate kinase, 20 U/mL lactate dehydrogenase and 2 mg/mL NADH at 37°C.
Preparation of the gapped ssDNA substrate for the single-molecule studies
An engineered derivative of bacteriophage λgt11 containing a φC31 attP recognition site, was created as follows. An EcoRI fragment derived from pDHJS AN+34 containing a kanamycin resistance cassette and the φC31 attP recognition site was ligated into EcoRI-digested bacteriophage λgt11 DNA. The attP-containing derivative of bacteriophage λgt11 is hereafter called bacteriophage λKytos. A modified a derivative of bacteriophage M13mp7 ssDNA contained the attB recognition site, from which a 500 bp dsDNA containing the φC31 attB at its center was generated by PCR using Phusion High Fidelity PCR Master Mix from NEB. After heat denaturation, it was annealed to the M13mp7 ssDNA derivative. Integration of the ssDNA plasmid into λKytos dsDNA resulted in a gapped DNA substrate (Supplementary Figure 4c). φC31 integrase was used to recombine λKytos dsDNA and the annealed 13mp7 ssDNA containing the attB recognition site. The construct pHS62, containing the full coding sequence of φC31 integrase, was kindly provided by Margaret Smith35. Biotin was incorporated into the cos sites of λKytos in a reaction consisting of 10 mM Tris-HCl (pH 7.9), 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 33 μM dATP, 33 μM dTTP, 3 3μM dCTP, 33 μM biotin-dGTP, 17 ng/μlλKytos, and 0.17 U/μl Klenow exo− DNA polymerase. After 15 minutes at 22°C, the polymerase was heat inactivated at 70°C for 20 minutes.
Epifluorescence microscopy, optical trapping, and multi-channel flow cells
Infrared optical trapping capability was achieved on the same TE-2000-U microscope (Nikon) used for the TIRF-based assay with the addition of a polarizer (Newport) to split the beam, generating two traps, and a steering mirror (Newport) to control the x-y position of one of the beams26. Excitation of the sample in epifluorescence mode was achieved using a Cyan 488 nm laser (Picarro) by adjusting the angle of the laser to pass completely through the sample chamber (i.e., not in TIRF-mode). The fluorescence emission was directed through a polychroic mirror (515/30 nm and 600/40 nm, Chroma). Images were captured on a DU- 897E iXon CCD camera (Andor). Channels were laser etched into glass slides (Fisher Scientific 25×75×1 mm) covered with an adhesive abrasive blasting mask (Rayzist Photomask, Inc.) using a 30 Watt Mini-24 Laser Engraver (Epilog Lasers). The slides were sandblasted using 220 grit silicon carbide (Electro Abrasives) to remove residual laser-ablated glass from the channels, resulting in channels ~100–150 μm deep. Holes were drilled using a diamond-bit Dremel tool, washed with 2% Hellmanex and rinsed with water and methanol. Cover glass (Corning No. 1, 24×60 mm) was cleaned in 1 M KOH/MeOH with sonication for one hour, rinsed with water and methanol and dried. The cleaned cover glass was attached to the etched microscope slide with UV Optical adhesive #74 (Norland Products) applied through capillary action on a 45°C heat block. The adhesive was cured by placing the flow cell 30 cm from a 100 Watt HBO lamp (Zeiss, Inc.) for 60 minutes followed by curing at 50 °C overnight. PEEK tubing with 0.5 mm inner diameter (Upchurch Scientific) was inserted into each of the etched holes to create inlet and outlet connection ports using epoxy (Devcon, “5 minute”). The flow cell was mounted to the microscope and attached to a syringe pump (KD Scientific). The temperature of the objective lens was held at 37°C by circulating water through a brass and copper collar machined to fit around the objective lens. The flow cell had six laminar flow channels containing the following components in addition to 20% sucrose and 50 mM DTT: (Ch 1) 100 mM NaHCO3 (pH 8.3), 100 mM NaCl, 1 mM Mg(OAc)2, 5 nM YOYO-1, and 0.2% streptavidin-coated polystyrene beads (1 μm, Bangs); (Ch 2) 100 mM NaHCO3 (pH 8.3), 100 mM NaCl, 1 mM Mg(OAc)2, 50 nM YOYO-1, 4 pM gapped DNA, and 300 nM SSB; (Ch 3) 100 mM NaHCO3 (pH 8.3), 100 mM NaCl, 1 mM Mg(OAc)2, 50 nM YOYO-1, and 100 nM SSB; (Ch 4) 100 mM NaHCO3 (pH 8.3), and 100 nM SSB; (Ch 5) 20 mM Tris(OAc) (pH 7.5), 200 mM NaCl, 4 mM Mg(OAc)2, 2 mM ATPγS and 200 nM SSB; and (Ch 6) 20 mM Tris(OAc) (pH 7.5), 200 mM NaCl, 4 mM Mg(OAc)2, 2 mM ATPγS, 200 nM SSB, and 1 μM RecA. When indicated, RecO and RecR were present at 100 nM each in Ch 6, while RecF was present at 50 nM. Sterile 0.2 μm filtered sucrose solutions were degassed for at least one hour in a vacuum chamber before the addition of buffer and DTT, and degassed for an additional 15 minutes; reactions were then assembled at room temperature protected from light. Optically trapped molecules were moved between flow channels by movement of the sample stage, which was automated and synchronized with both laser excitation and camera acquisition during dipping experiments using custom software coded in LabView. RecAf filaments were successively transferred from Ch 6 to Ch 5, imaged for 1 second, and immediately transferred back to Ch 6. The time at which a RecAf cluster first appeared was determined to be the apparent lag time. Images were processed in ImageJ by frame averaging and contrast enhancement.
Supplementary Material
Web summary.
Single-molecule analysis of RecA filament assembly on its in vivo substrate, SSB-coated ssDNA, reveals that a dimer of RecA is required for nucleation, and is followed by bidirectional growth of the filament through monomer addition; the recombination mediator RecOR accelerates nucleation and growth, and addition of RecF further stimulates nucleation.
Acknowledgments
We are grateful to members of the laboratory for their comments on this work. J.C.B. was funded by an NIH Pre-doctoral Training Program in Molecular & Cellular Biology (T32 GM007377), C.C.D and J.L.P were funded by the NIH T32 Training Program in Oncogenic Signals and Chromosome Biology (CA10052159), J.L.P was also funded by an NIH Postdoctoral Fellowship (CA136103), and S.C.K. was supported by the National Institutes of Health (GM62653 and GM64745).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions J.C.B., J.L.P., and C.C.D., and S.C.K. conceived the general ideas, designed the experiments and interpreted the data. J.C.B., J.L.P. and C.C.D. performed experiments. J.C.B. and S.C.K. wrote the manuscript.
Competing Interests Statement The Authors have no competing financial interests.
References
- 1.Kowalczykowski SC, Eggleston AK. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski SC. Biochemistry of genetic recombination: Energetics and mechanism of DNA strand exchange. Annu Rev Biophys Biophys Chem. 1991;20:539–575. doi: 10.1146/annurev.bb.20.060191.002543. [DOI] [PubMed] [Google Scholar]
- 3.Kowalczykowski SC, Clow J, Somani R, Varghese A. Effects of the Escherichia coli SSB protein on the binding of Escherichia coli RecA protein to single-stranded DNA: Demonstration of competitive binding and the lack of a specific protein-protein interaction. J Mol Biol. 1987;193:81–95. doi: 10.1016/0022-2836(87)90629-2. [DOI] [PubMed] [Google Scholar]
- 4.Thresher RJ, Christiansen G, Griffith JD. Assembly of presynaptic filaments. Factors affecting the assembly of RecA protein onto single-stranded DNA. J Mol Biol. 1988;201:101–113. doi: 10.1016/0022-2836(88)90442-1. [DOI] [PubMed] [Google Scholar]
- 5.Cazenave C, Toulme JJ, Helene C. Binding of RecA protein to single-stranded nucleic acids: spectroscopic studies using fluorescent polynucleotides. EMBO J. 1983;2:2247–2251. doi: 10.1002/j.1460-2075.1983.tb01730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shivashankar GV, Feingold M, Krichevsky O, Libchaber A. RecA polymerization on double-stranded DNA by using single-molecule manipulation: the role of ATP hydrolysis. Proc Natl Acad Sci U S A. 1999;96:7916–7921. doi: 10.1073/pnas.96.14.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 8.Joo C, et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 9.Handa N, Amitani I, Gumlaw N, Sandler SJ, Kowalczykowski SC. Single molecule analysis of a red fluorescent RecA protein reveals a defect in nucleoprotein filament nucleation that relates to its reduced biological functions. J Biol Chem. 2009;284:18664–18673. doi: 10.1074/jbc.M109.004895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modesti M, et al. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 11.van der Heijden T, et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Natl Acad Sci U S A. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillingham MS, et al. Fluorescent single-stranded DNA binding protein as a probe for sensitive, real-time assays of helicase activity. Biophys J. 2008;95:3330–3339. doi: 10.1529/biophysj.108.133512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell JC. Ph D thesis. University of California; Davis: 2011. Dynamic Assembly of RecA Filaments on Single Molecules of SSB-coated ssDNA. [Google Scholar]
- 15.Griffith JD, Harris LD, Register J., III Visualization of SSB-ssDNA complexes active in the assembly of stable recA-DNA filaments. Cold Spring Harb Symp Quant Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 16.Kowalczykowski S, Steinhardt J. Kinetics of hemoglobin S gelation followed by continuously sensitive low-shear viscosity. J Mol Biol. 1977;115:201–213. doi: 10.1016/0022-2836(77)90097-3. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 18.Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biophys Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 19.Register JC, III, Griffith J. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J Biol Chem. 1985;260:12308–12312. [PubMed] [Google Scholar]
- 20.Menetski JP, Kowalczykowski SC. Enhancement of Escherichia coli recA protein enzymatic function by dATP. Biochemistry. 1989;28:5871–5881. doi: 10.1021/bi00440a025. [DOI] [PubMed] [Google Scholar]
- 21.Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J Mol Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 22.McEntee K, Weinstock GM, Lehman IR. Binding of the recA protein of Escherichia coli to single- and double-stranded DNA. J Biol Chem. 1981;256:8835–8844. [PubMed] [Google Scholar]
- 23.Umezu K, Chi NW, Kolodner RD. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc Natl Acad Sci U S A. 1993;90:3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 25.Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev. 2009;23:1234–1245. doi: 10.1101/gad.1780709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forget AL, Kowalczykowski SC. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature. 2012;482:423–427. doi: 10.1038/nature10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohman TM, Kowalczykowski SC. Kinetics and mechanism of the association of the bacteriophage T4 gene 32 (helix destabilizing) protein with single-stranded nucleic acids. Evidence for protein translocation. J Mol Biol. 1981;152:67–109. doi: 10.1016/0022-2836(81)90096-6. [DOI] [PubMed] [Google Scholar]
- 28.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuznetsov SV, Kozlov AG, Lohman TM, Ansari A. Microsecond dynamics of protein-DNA interactions: direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. J Mol Biol. 2006;359:55–65. doi: 10.1016/j.jmb.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 30.Umezu K, Kolodner RD. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J Biol Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
- 31.Kowalczykowski SC, Krupp RA. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein: Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J Mol Biol. 1987;193:97–113. doi: 10.1016/0022-2836(87)90630-9. [DOI] [PubMed] [Google Scholar]
- 32.Napolitano MA, et al. Brg1 chromatin remodeling factor is involved in cell growth arrest, apoptosis and senescence of rat mesenchymal stem cells. J Cell Sci. 2007;120:2904–2911. doi: 10.1242/jcs.004002. [DOI] [PubMed] [Google Scholar]
- 33.Hamon L, et al. High-resolution AFM imaging of single-stranded DNA-binding (SSB) protein--DNA complexes. Nucleic Acids Res. 2007;35:e58. doi: 10.1093/nar/gkm147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plank JL, Hsieh TS. A novel, topologically constrained DNA molecule containing a double Holliday junction: design, synthesis, and initial biochemical characterization. J Biol Chem. 2006;281:17510–17516. doi: 10.1074/jbc.M602933200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci U S A. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.