Abstract
Fat accumulation is a complex phenotype affected by factors such as neuroendocrine signaling, feeding, activity, and reproductive output. Accordingly, the most informative screens for genes and compounds affecting fat accumulation would be those carried out in whole living animals. Caenorhabditis elegans is a well-established and effective model organism, especially for biological processes that involve organ systems and multicellular interactions, such as metabolism. Every cell in the transparent body of C. elegans is visible under a light microscope. Consequently, an accessible and reliable method to visualize worm lipid-droplet fat depots would make C. elegans the only metazoan in which genes affecting not only fat mass but also body fat distribution could be assessed at a genome-wide scale. Here we present a radical improvement in oil red O worm staining together with high-throughput image-based phenotyping. The three-step sample preparation method is robust, formaldehyde-free, and inexpensive, and requires only 15 minutes of hands-on time to process a 96-well plate. Together with our free and user-friendly automated image analysis package, this method enables C. elegans sample preparation and phenotype scoring at a scale that is compatible with genome-wide screens. Thus we present a feasible approach to small-scale phenotyping and large-scale screening for genetic and/or chemical perturbations that lead to alterations in fat quantity and distribution in whole animals.
Keywords: Fat mass, body fat distribution, high-content screening, lipid, C. elegans, obesity
1. Introduction
Live staining of fats with Nile red or BODIPY-labeled fatty acids, or fixative-based staining with Sudan Black and oil red O (ORO) are common approaches to visualize fats contained in lipid droplets in mammalian specimens[1, 2]. By contrast, the use of these fat-staining techniques in C. elegans is not straightforward. Depending on the experimental conditions, feeding worms with the vital dye Nile red leads to exclusive staining of the lysosomal-related organelle compartment, or staining of this compartment in addition to the lipid droplet compartment[3–7]. Similarly, live staining with BODIPY-labeled fatty–acids, although robust, leads to staining of both the lysosomal-related organelle compartment and the lipid droplet compartment[4, 8]. Having two vesicular compartments stained with the same fluorophore complicates the use of automated scoring for lipid-droplet fats only. Feeding high concentrations of Nile red (2–10µM) improves lipid droplet staining with this dye. However, live high-concentration Nile red staining leads to heterogenic signal within and among samples[7]. Nile red exclusively stains the C. elegans lipid droplet compartment in paraformaldehyde-fixed worms[5], however paraformaldehyde fixation leads to variable staining and broken animals, which together preclude the use of automated scoring for lipid-droplet fats. Sudan Black stains lipid-vesicle fats only[9], but it is highly error-prone due to a final alcohol-based wash that introduces enormous variability. Therefore, Sudan Black requires mixing of the control and the test samples in the same tube after marking or labeling them in a way that the original populations can be distinguished after imaging (e.g., an additional fluorescent dye or an independent phenotypic distinction such as sterility or size that enables distinguishing control worms and sample worms). This requirement makes Sudan black incompatible with large-scale studies. Also, Stimulated Raman-Scattering (SRS) and Coherent Anti-Stokes Raman Scattering (CARS) have been successfully used to assess fat levels in C. elegans[6, 10, 11]. However, these techniques require manual imaging of individual worms, and experimental implementation of CARS and SRS is, in general, costly and difficult. CARS and SRS require dedicated, expensive equipment, and highly specialized expertise, which are currently not available to most researchers and certainly not to most teaching laboratory classrooms.
Our previous ORO staining protocol represented a significant advance over previous techniques[4]. First, it allowed scoring of C. elegans fat stores contained only in lipid droplets. Second, because ORO does not require alcohol-based de-staining, it limits the variability introduced by de-staining timing, which is the major caveat of Sudan black. Our ORO staining protocol is robust and correlates well with biochemically-measured lipids (total fatty-acid methyl esters by GCMS). Nevertheless, this protocol, as well as the alternative fixative-based Nile red staining protocol, includes paraformaldehyde-based fixation. Paraformaldehyde is a carcinogen, requiring the user to perform the protocol in a fume hood and increasing the cost of the procedure by generating toxic waste. Additionally, paraformaldehyde-based fixation of C. elegans generates a large proportion of broken animals, which affects staining and makes automated image-based phenotype scoring difficult.
Here, we describe a radically improved whole-animal fat screening protocol, which allows the user to phenotype a 96-well plate of RNAi- or compound-treated animals in 15 minutes of hands-on time. This method, named “quick oil red O” (qORO), does not use paraformaldehyde or other toxic fixatives. Instead, fixation is achieved with a mixture of water and isopropanol. This qORO method yields almost 100% intact worms, making it possible to quantify fat storage patterns in relation to the worm’s anatomy.
We also present a set of digital image processing and analysis tools for high- and low-throughput quantitative qORO phenotype scoring. We have previously presented image-analysis methods for scoring of ORO stained worms[4]; however, our original image analysis protocol required both expertise in code writing and purchasing the licensed software package MATLAB (The MathWorks Inc., Natick, MA)[4]. Subsequently, we presented the WormToolbox, which is part of the free and open source CellProfiler software[12–14]. Here, we describe an upgraded version of the WormToolbox and CellProfiler, including a simplified software interface and fully automated as well as semi-manual tools for the analysis of qORO images derived from high-throughput and low-sample number experiments. We have also created a set of detailed video tutorials on how to put together and optimize analysis pipelines for qORO phenotyping. Complete analysis pipelines together with sample data and video tutorials are available at http://www.cellprofiler.org/wormtoolbox.
2. Methods and Results
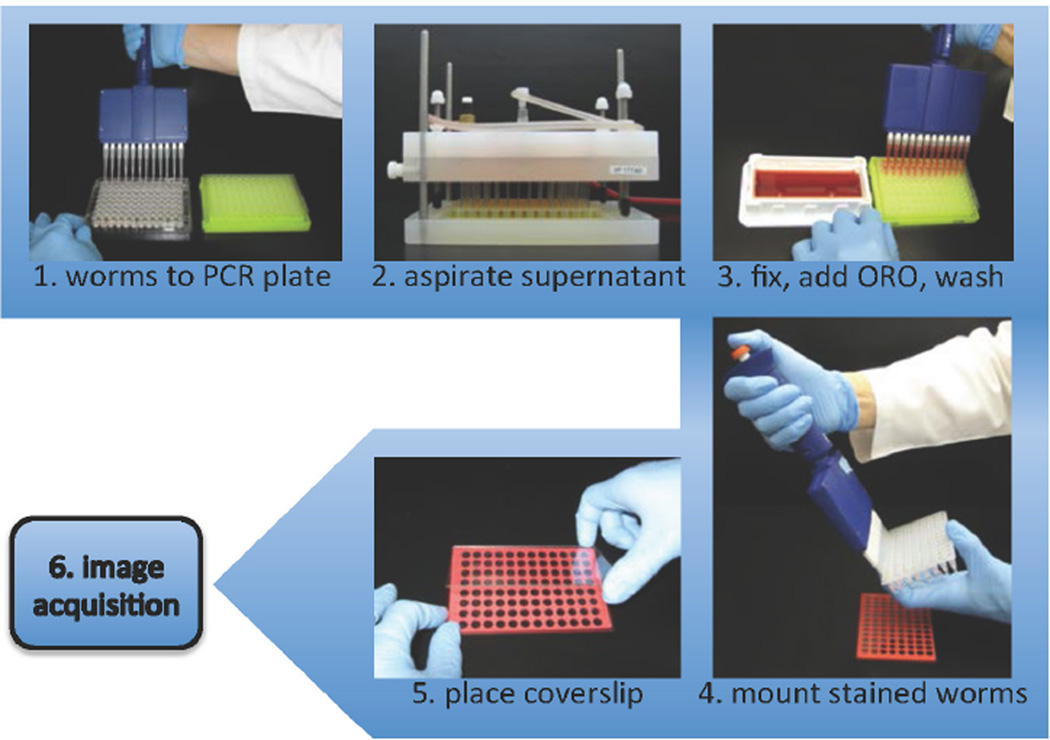
The qORO protocol is outlined in Figure 1.
Figure 1.
qORO workflow. 1. Harvest worms with S basal buffer. Transfer worms to PCR plate. 2. Let worms settle and remove supernatant. 3. Add fixative (60% isopropanol), let worms settle, stain 6–18h in 60% ORO in water (stain made one day before use, filtered, and re-filtered right before use). 4. Aspirate supernatant. Add Triton 0.01% in S basal to worm pellets. Add 2µl of mounting medium to each well and transfer 4µl of worm suspension, mix gently. 5. Place cover by minimally tilting it towards topside first. 6. Image with color camera or monochromatic camera with RGB filters.
2.1. Worm growth and harvesting
Multiple variables affect body fat levels and distribution. Consequently, it is important to carefully control the experimental conditions. Food quality and quantity, as well as fertility and age, are especially important variables. We recommend, whenever possible, to use developmental and not chronological age to compare fat levels after genetic, chemical, or environmental perturbation. General guidelines for high- and low-throughput growth and harvesting conditions are described below.
2.1.1. High-throughput screening
Worms of the desired genetic background are grown, and RNAi or compound treated in 96-well plates as previously described[15]. Fat analysis requires specific modifications to the published growth method as follows:
For F1 screening, manually pipette around 50 hatchlings into each well of a 96-well plate. If compatible with the genetic background, incubate worms at 15°C. Optimal harvesting time will depend on the genetic background and growth conditions. However, the most robust differences among true fat regulators are observed between the L4 and 1-day adult stage. Earlier time points would be informative for early onset increased fat mass phenotypes because wild-type worms show low levels of fat at the L3 stage[16]. Later time points make it difficult to assess increases in fat mass because the wild-type levels of fat reach saturation at 3-days adulthood when worms are grown at 15°C (data not shown).
Always have at least one well with the appropriate control in each 96-well plate (L4440 empty vector RNAi or compound carrier). Ideally also include a positive control. Test each RNAi clone or compound at least in duplicate, and we recommend experimental repeats to be carried out completely independent (days apart with different plates and reagent batches).
Harvest the worms growing on 96-well plates by adding 200 µL of 1× S-basal (http://www.wormbook.org/chapters/www_strainmaintain/strainmaintain.html). Worms that thereby swim out of the bacterial lawn are washed out by gently pipetting up and down 3 times using a 12-channel pipette. A second wash may help harvest more worms if they are younger than L4 or sma (small body size), but it does not increase worm recovery for wild-type L4 or later developmental stages. The S-basal suspensions of worms (up to 200µl) are transferred to the wells of meaningfully labeled 96-well PCR plates (wells should hold up to 250µL of volume per well). Let animals settle to the bottom of the wells. Aspirate up to 175µL of buffer with a VP Scientific VP177AD 96-well liquid aspirator or similar (Figure 1). Immediately add fixative (continue in Fixation and Staining below).
2.1.2. Low-throughput analyses
Prior to setting up a qORO assay, compare the developmental times (i.e. by observing gonadal development) of each new genotype or treatment to be tested so that all sample strains and the control strain are at the same developmental stage at the time of scoring. Grow strains using the same conditions (same batch of plates, same incubator, etc) to reduce variability. Seed approximately 500 worms per 6cm NGM plate containing 500µl of E. coli grown overnight to saturation (OD approx. 3–4), concentrated 2 times by centrifugation, and resuspended in S-basal. Make sure to cover as much of the surface of the lawn as possible to avoid the dietary restriction effects of leaving behavior.
Harvest worms at the desired developmental stage by washing the plates twice with 750µl of S-basal (if worms were scarce, use glass Pasteur pipettes to minimize losing animals that otherwise would stick to the surface of plastic tips). Transfer worms to meaningfully labeled 1.5mL tubes. Let worms settle to the bottom of the tubes. Aspirate as much supernatant as possible without disturbing the worms. Immediately add fixative (see Fixation and Staining below).
2.2. Stain preparation
Prepare a 0.5% Oil red O (MP, Cat. No. 155984) stock solution in high-quality 100% isopropanol. Let it rock for a day and filter through a 0.45µm filter. A stock solution kept at room temperature is good for several months.
The day before use, freshly dilute the stock to 60% with filtered water. Rock the 60% solution overnight. Filter through a 0.45µm filter. To prevent dye precipitation on the samples, filter again through a 0.45µm filter right before use.
2.3. Fixation and Staining
Sixty percent isopropanol is used as an alcohol-based fixative in several cytological techniques because of its good osmolarity, effectiveness, and safe use. We tested if resuspending harvested worms in 60% isopropanol was sufficient to fix them, and we observed perfect organ and cellular structure preservation when worms harvested from plates were resuspended in 60% isopropanol and immediately stained, indicating that resuspension in 60% isopropanol was sufficient to properly fix worms.
For large-scale qORO, add 200µl of high-quality 60% isopropanol to worms contained in the 96-well PCR plates. Let worms settle to the bottom of the wells. Aspirate up to 175µL of buffer with a 12-channel pipette or 96-well aspirator (VP-177AD, VP Scientific). Add 200 µl of freshly filtered ORO working solution and seal the plates with aluminum seals. Stain worms for 6–18h at 25°C in a wet chamber (wet paper towels in a hermetic or parafilm-wrapped plastic box); this keeps the samples humidified as the isopropanol fumes tend to unseal the plates during the incubation time.
After 6–18 h of staining aspirate the supernatant with a 12-channel pipette or a VP Scientific VP177AD 96-well aspirator or similar (shorter staining times may lead to weak signal, and longer staining times increase the changes of precipitation of the dye). Add 100µl of 0.01% Triton X-100 in S-basal. Stained worms can stay in this solution at 4°C for at least a month without altering quantities or distribution of fat. The protocol can be adapted to any standard 96-well robot that can work with 200 µl tips with at least five pads for 96-well plates (pads: sample, washing buffer, fixative, ORO dye working solution, and resuspension buffer).
For staining single samples, add 500µl of high-quality 60% isopropanol to worms contained in 1.5mL tubes. Let worms settle to the bottom of the tube. Aspirate as much supernatant as possible without disturbing the worms. Add 500 µl of freshly filtered ORO working solution. Stain worms at 25°C in a wet chamber (wet paper towels in an hermetic or parafilm-wrapped plastic box).
After 6–18h of staining, wash out the supernatant and add 250µl of 0.01% Triton X-100 in S-basal. Stained worms can stay in this solution at 4°C for at least a month without altering quantities or distribution of fat.
2.4. Sample mounting
For high-throughput image acquisition, prepare to mount worms in a 96-well slide (Trevigen) by adding 2µl of Vectashield mounting medium (Vector Labs) to all wells with a 12-well channel pipette. Without resuspending, pipette 4 µl of worms from the bottom of each sample well (a sudden upward movement accompanying the release of the plunger helps to get worms into the tip). Gently dispense the worms on the mounting medium, and pipette up and down less than 3 times to detach worms from the tip walls; avoid generating bubbles. Mix the worms with the mounting medium by gently swirling the liquids with the tip of the pipette tips. Ideally, there should be 20–30 worms per well. Add cover slip; this is the 1mm thick full plate glass used to produce the 96-well slides but without the stamped Teflon wells.
For low-throughput image acquisition, prepare 2% agar pads on as many slides as necessary. Add 3µl of mounting medium to a 24 × 24mm cover slip. Transfer 5µl (approximately 50 worms) of worm suspension to the cover slip. On the coverslip, mix the worm suspension with the mounting medium by gently swirling the liquids with the tip of the pipette tip. Flip the cover with the worms onto the agar pad and label the slide. Do not seal with nail polish; the solvent will negatively affect staining.
2.5. Image acquisition
For high-throughput image acquisition, use an automated microscope able to capture images from a 96-well slide and equipped with a high-resolution color camera (an adapter to hold slides maybe required for scopes designed to image 96-well plates). A PlanApo 4× objective enables capturing a full well in one image when using a 5.5 megapixel camera. However, higher resolution imaging can be obtained using a PlanApo 10× objective but 16 images would need to be stitched to capture more than 80% of the well with a 5.5 pixel camera. Aim to have a minimum coverage of 75% for each well in order to ensure capturing the majority of the worms, which may not be evenly distributed across the well. Minimal or no autofocusing is required when using 96-well slides. Combine multiple images of each well into a single image using software with image stitching capabilities prior to quantitative analysis. If stitching software is unavailable, simple tiling of images is possible in CellProfiler; this works best if the acquired images are precisely aligned adjacent to each other with no overlaps or gaps. It should be noted that the subsequent image analysis relies on full-length worms completely within the image field because worm detection relies on a computational model of the worm shape. Worms not completely within the image, or with segments that are not properly aligned between two adjacent individual images because of tiling errors, may be automatically discarded as debris.
2.6. Image analysis
Automated image analysis provides a quantitative, fast, and reproducible approach to phenotype scoring. The software tools presented here, as well as video tutorials and a set of test images prepared using the above qORO method, are available in the WormToolbox of the user-friendly and open source CellProfiler project (www.cellprofiler.org/wormtoolbox)[14]. We recommend the use of Google Chrome to access these resources.
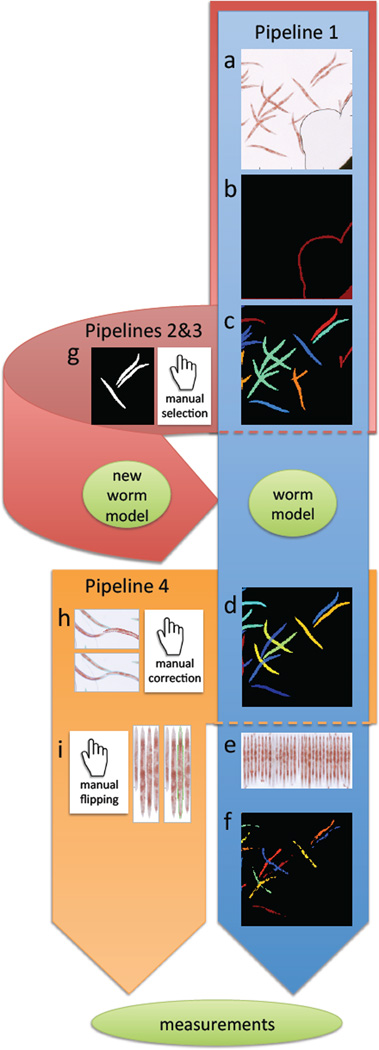
The procedures are described as pipelines consisting of a number of steps, or modules, each contributing to the final result of the analysis. Here we describe the general steps of four different pipelines: Pipeline 1 for high-throughput qORO phenotyping, Pipelines 2 and 3 for creating a customized worm model (used for untangling of clustered worms or to score an experiment in which worms significantly differ in size or illumination from the prebuilt pipelines available at cellprofiler.org/wormtoolbox), and Pipeline 4 for low-throughput phenotyping with manual interaction. An overview of the pipelines is outlined in Figure 2.
Figure 2.
Overview of image analysis pipelines. Pipeline 1 in the tutorial (blue), is fully automated, and consists of the following steps: a) Load input images from qORO assay (here showing only part of full well for better visualization of worms). b) Pre-process to remove artifacts such as bubbles and well edges. c) Detect worms and clusters of worms. Individual worms/worm clusters are randomly colored. d) Delineate individual worms by model-based worm untangling (this step is called “segmentation”). Here, each individual worm has a unique (and arbitrary) color. e) Digitally straighten worms and align them to a simple worm atlas for measurement of stain distribution. f) Detect fatty regions within worms. Once worms and fatty regions are identified, a large number of measurements are extracted from individual worms, such as the intensity of stain, fat stain distribution, fat region size, worm width, etc. All measurements are exported to a spreadsheet or database for further exploration and analysis. Pipeline 2 and 3: If the provided default worm model does not fit the input data (e.g. due to a different image resolution, worm age range, or different worm strain), a new worm model can be built using the pipelines 2 an 3 in the tutorial (pink) by g) manual selection of non-touching worms that build up a training set for a new model. Pipeline 4: For low-throughput experiments, it is possible to manually curate segmentation results using pipeline 4 in the tutorial (orange) by h) manual editing of the output from step (d) of the blue pipeline to correct errors, and i) manual flipping of digitally straightened worms so that they all align with their heads or tails up (in cases where the orientation of fat within the worm is important for the study). The same set of measurements as described in part (f) above can be extracted from such manually-edited segmentation results.
The example images presented here come from a control 96-well qORO experiment. The animals have been treated with RNAi against either the insulin receptor daf-2 (which makes the animals fat), peb-1 (DNA-binding protein required for normal development of the pharynx[17], which defect likely affects feeding making animals slim), or with an empty RNAi vector (L4440) as negative control.
The user starts the analysis by opening CellProfiler (available for download from www.cellprofiler.org) and simply dragging and dropping the image folders into the ‘File list’ panel. In order to keep track of the different treatments and positions in the 96-well plate, metadata (information about each image) can be extracted from file- and folder names, as detailed in the tutorials and example pipelines. Note that it is also possible to load metadata, such as treatment doses, drug- or RNAi names etc. from a text file.
The qORO input data is color images with a bright background. CellProfiler was originally designed for fluorescence microscopy images with a dark background, so images are first inverted so that the worms appear bright on a dark background, and then separated into their red, green and blue color channels. Before identifying the worms, the red image channel is pre-processed to identify and remove artifacts such as non-uniform illumination, well edges and bubbles that may influence automated analysis. Several different options for compensating for uneven illumination are available through the ‘CorrectIllumination’ modules. For the presented dataset, however, illumination is comparably uniform, so this step is omitted and the first step of the analysis is removal of well edges and bubbles. These regions of the image are identified based on being significantly darker (brighter in the inverted image) than the worms, as illustrated in Figure 2b. Once artifacts are removed, worms and worm clusters are separated from the image background by foreground/background image segmentation, Figure 2c. This step relies on a sufficient difference in contrast between the worm bodies and the image background. Adjusting image exposure time during image acquisition may improve the result of this analysis step, but these settings should thereafter be kept constant throughout the entire experiment.
The steps above serve to identify worms or worm clusters, rather than individual worms. As the goal of the quantitative analysis is to extract measurements relevant to worm phenotype from individual worms, the next step in the analysis pipeline is to identify individual worms by worm untangling. The ‘Worm untangling’ module makes use of a worm model, which is a mathematical description of the acceptable variation in worm shape and posture[14, 18, 19]. A worm model called ‘DefaultWormModel.xml’ is provided together with the example pipelines at www.cellprofiler.org/wormtoolbox, but the procedure for creating a new worm module is also described in Pipelines 2 and 3. To use or test the provided worm model, click on the .xml file and save the file to your hard drive in the same folder as where you have your input data. This model has been created to fit the provided sample data set, and may be used in Pipelines 1 and 4.
The ‘Worm untangling’ step results in two different outlines of the worms: one that includes, and one that excludes regions where the worms overlap. This means that overlapping worm regions can be included when measuring size but excluded when measuring, for example, the mean intensity of a worm. Once individual worms are identified, the user can select which measurements to extract. In CellProfiler, measurements are grouped by category. For example, the ‘MeasureObjectIntensity’ module includes total intensity, mean intensity, intensity mass displacement, minimum intensity, etc., on a per-object basis.
When it comes to qORO quantitation, and this would be true for broadly distributed fluorescent markers also, the pattern of distribution of the stain in the worm provides important information on the worm phenotype. In order to compare stain distribution between worms, individual worms can be digitally straightened and aligned to a very simple atlas or grid, enabling measurement of stain intensity in a number of longitudinal and/or transversal sub-regions. This task is performed by the ‘StraightenWorms’ module (Figure 2e). Additionally, qORO-phenotypic classification benefits from identifying the fatty sub-regions within each worm. This is done using a fixed intensity threshold in the module called IdentifyPrimaryObjects (Figure 2f). It is important to optimize this threshold for each new experiment, and also to keep it constant throughout the analyses of the experiment to ensure that measurements are comparable. The identified fatty regions are then coupled with a given worm using the ‘RelateObjects’ module. Any fatty region measurement, such as size and shape, is averaged and linked to a specific worm or worm sub-region using this module.
Once a pipeline has been created and optimized, it can be run on a large number of images by batch processing or using a computer cluster. Nevertheless, it is important to keep in mind that an automated approach optimized for one data set may not work optimally on a different data set. Parameter settings may have to be re-optimized if, for example, the imaging system, staining protocol, or worm strain is changed. When analyzing your own data, the provided worm model may not provide the desired results due to experimental differences such as a different image resolution or worm strain. To create a new worm model, 40–60 worms representing the variability within an experiment must be manually selected from your own experiment. These sample worms are referred to as a training set and this training set is used to create a new worm model. The procedure for manually selecting and saving single worms for a training set is described in Pipeline 2. After visual confirmation of the selected worms, a model is created using Pipeline 3.
The fully automated worm detection sometimes fails to create a perfect delineation of every worm in the image. Some errors may be acceptable in favor of a fully automated and unbiased analysis. However, for some treatments leading to high phenotypic variation, it may be necessary to manually inspect and correct errors. We have therefore included Pipeline 4. This pipeline loads the segmentation masks created by Pipeline 1, and allows manual correction of the worm outlines. Pipeline 4 also enables digital straightening of the worms and it allows the user to orient worms (heads- or tails-up), therefore making measurements of fat distribution possible.
2.7. Data analysis
CellProfiler extracts a large number of different measurements per worm. Individual measurements can be exported to a comma-delimited spreadsheet for further data processing using an appropriate program such as Microsoft Excel. However, it may be difficult to decide which particular measurement(s) best discriminate between different phenotypes. To facilitate phenotypic classification we use Classifier tool available in CellProfiler Analyst (CPA) to interactively train a machine-learning classifier to automatically find the best combination of measurements for discriminating between different phenotypes[20]. CPA 2.0 can be downloaded from (http://www.cellprofiler.org/downloadCPA.shtml) and a detailed manual and a users forum are accessible online. We will here provide basic guidelines in the use of CPA to score qORO but we encourage users to take advantage of the extensive tutorials available online.
In order to explore data in CellProfiler Analyst (CPA), the CellProfiler user has to create a database. The provided CellProfiler example pipeline exports all measurements to an SQLite database that enables the measurements in the database to be further explored using CPA[21]. When creating a CellProfiler pipeline, select the option to create a “properties file” output in the ‘ExportToDatabase’ module. The properties file is a simple text file that describes where raw data and extracted measurements are located, and when launching CPA, the user will be prompted to select this properties file. It is important to note that the properties file contains information of file location and CPA will fail to locate files if they are moved from their original location unless the path in the properties file is appropriately updated to reflect the actual location of the files. For training CPA to classify ORO phenotypes, prior to launching CPA, the user would need to edit the CellProfiler properties output file in a regular text editor. CellProfiler Analyst was originally designed for fluorescent images of cells, so a few modifications are needed to optimize viewing of brightfield images of worms, which are also often larger than cells. The following additions/changes to the properties file are needed prior to launching CPA (each on separate lines):
image_channel_colors = red, green, blue, red, green, blue
channels_per_image = 3,3
image_channel_blend_modes = subtract, subtract, subtract, add, add, add
image_tile_size = 250
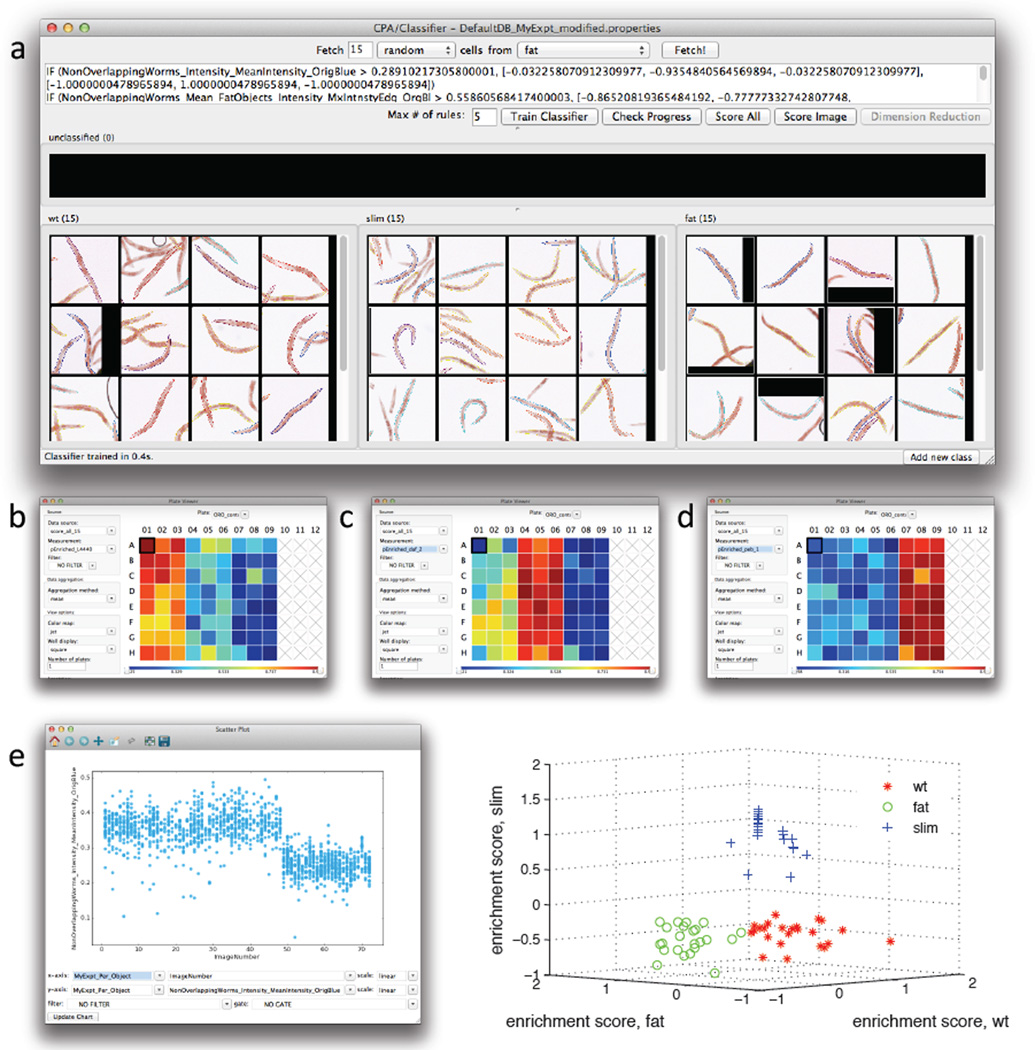
The Classifier tool of CPA needs to be “trained” to recognize worm phenotypes such that CPA can determine the likelihood that a given well shows worms belonging to a given class (i.e. wild type, slim, or fat); the user does so by hand-sorting examples of worms into phenotypic class bins. To change the default Classifier class bin names (“positive”, “negative”) to more meaningful class names (“fat”, “wild-type”) right-click in the corresponding bin. Click the ‘Add new class’ button located at the bottom-right of the interface (Figure 3A) if more than two phenotypic variants are expected (i.e. to add “slim” worms). Click the ‘Fetch’ button to retrieve image tiles of random worms and drag the tiles to their corresponding phenotypic bin. If multiple worms appear in the image, the worm that CPA is presenting to the user for classification is the one labeled with a little square when the user scrolls over the image with the mouse. Once a reasonable number of worm samples are dragged to their corresponding phenotypic bins (in the example of Figure 3a, fifteen example worms were used for each phenotype class or bin), press the ‘Train Classifier’ button. Test the classification accuracy by fetching animals of a particular class, i.e. change ‘random’ class drop down to ‘fat’, and fetch. In this example, verify that only or mostly fat worms are fetched after training. If satisfied with the training, press the ‘Score All’ button to automatically classify every worm in every well of the experiment: the result is a table displaying an “enrichment score” for each well. The enrichment score is a measure of the likelihood that a well is “enriched” for the phenotype of interest (i.e. a well with an enrichment score of 0.94 for High ORO phenotype is likely to contain mostly fat worms, and a well with an enrichment score for Low ORO of 0.94 is likely to contain mostly slim worms); note that the sum of enrichment scores for all classes does not add to 1. CPA enrichment scores can be visualized as a table, heat map, histogram, scatter plot, etc. To use these alternative views, the enrichment score table needs to be saved as a database and images have to be linked to positions in the plate. To do this, select 'File' from the main menu in the table and then select 'Save table to database'. Chose a short name without spaces or special characters, e.g. “results”. At the next prompt, choose 'Store permanently' and press ‘OK’. As an example of an alternative view of the scoring data, open the ‘PlateViewer’ in CPA. There, under the ‘Data source’ drop-down (located to the left of the plate layout display), choose *OTHER TABLE* and at the prompt, select the name of the enrichment score table just saved. At the next prompt, select 'Per-well' and press OK. At the next prompt, you have to select ‘Plate_ID’ (or image number) to correspond to ‘Image_Metadata_Plate’ (or image number), then press ‘OK’.
Figure 3.
Use of the machine learning capabilities of CellProfiler Analyst to classify worms as having a wild-type, fat, or slim phenotype. Twenty-four wells (columns 1–3) contained vector-control treated worms (wild type), 24 wells (columns 4–6) contained daf-2 RNAi treated worms (fat), and 24 wells (columns 7–9) contained peb-1 RNAi treated worms. a) Fifteen training individual worms were randomly selected among the wells from each of the three RNAi treatments, and used for training the classifier. After training, the classifier was applied to the full dataset (consisting of 1527 worms). Enrichment scores were then calculated and displayed as a plate layout heatmap in CellProfiler Analyst for each of the phenotypes b) wild-type worm enrichment, c) fat worm enrichment, and d) slim worm enrichment. As can be seen, the three well columns containing each of these controls scores highest (yellow/red) for the expected phenotype whereas the remaining columns show lower scores (blue). e) The most significant classification rule, or feature measurement, identified by CellProfiler Analyst (shown in the top window of a), was the mean intensity of all pixels within a worm in the blue image channel. If this per-worm measure is scatter-plotted versus image number (the sequential order of images in the experiment, left to right across the plate), it is clear that this measurement is significantly lower for peb-1 (the right-hand third of the data points) as compared to daf-2 and L4440 (the left-hand two-thirds of the data points) but it is not sufficient to separate all three phenotypes. This indicates that more than a single measurement is necessary to separate all three phenotypes from each other and thus that the machine-learning Classifier step is necessary. Indeed, a 3D scatterplot of per-well enrichment scores from the machine-learning Classifier step (f) clearly shows that the three phenotypes can be separated if a combination of differentiating parameters is used instead of a single measurement.
In the ‘Plate Viewer’, the default measurement displayed is the ‘ImageNumber’ index, which is uninformative. To view a heat map of phenotypes, select the desired enrichment score from the ‘Measurement’ drop-down to the left of the plate layout display. As an example, a heat map plate layout is shown in Figure 3b–d, where the the phenotypes are High ORO (Figure 3b), wild-type (Figure 3c), or Low ORO (Figure 3d): the redder wells likely have an overrepresentation of fat, wild type, or slim animals, whereas the bluer ones are likely underrepresented for that phenotype. Individual measurements may also be exported to a comma-delimited spreadsheet for downsstream data processing using other analysis software. Additional video tutorials for how to use CPA are provided at www.cellprofiler.org/wormtoolbox.
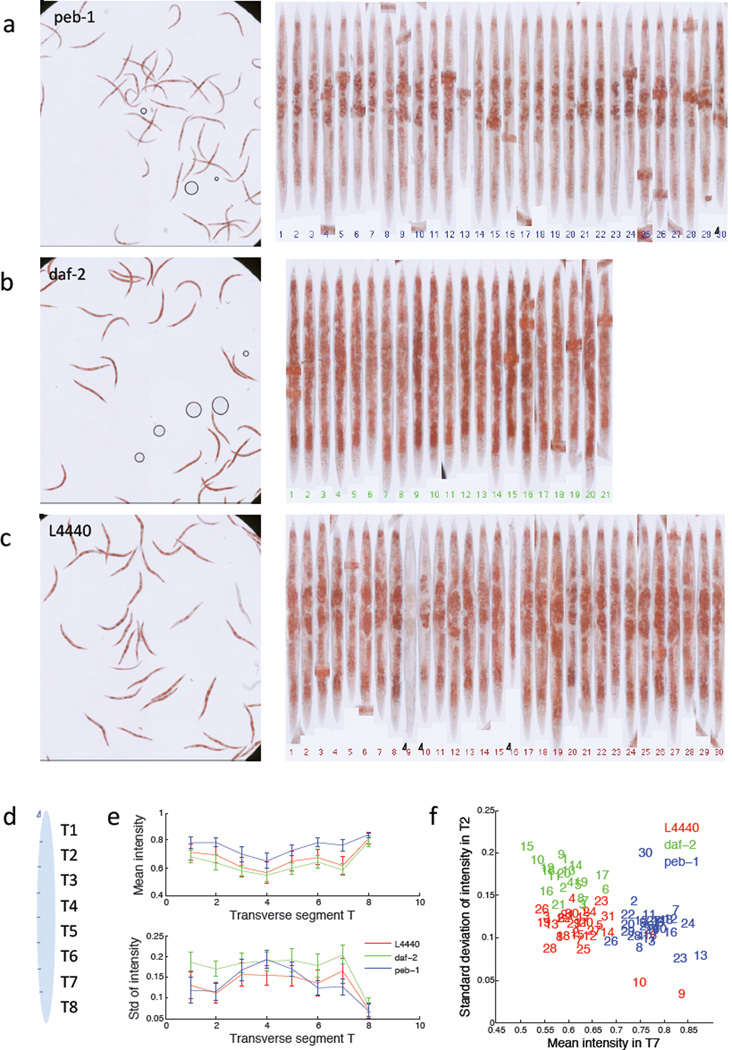
To illustrate the rich phenotypic descriptions that are made possible using the worm straightening and atlas fitting of Pipeline 4, we selected one well from each of the three RNAi treatments (Figure 4a–c), manually edited any segmentation errors, and then manually flipped the straightened worms to align them tail up. In this case we only used the measurements of mean and standard deviation of stain intensity in each of eight transverse segments across the worm body (Figure 4d). As illustrated in Figure 4e–f, two measurements are sufficient to separate the three phenotypes. It is interesting to note that the outliers represent worms that deviate also in visual phenotype (circled worm numbers in Figure 4a and c), illustrating that this approach also provides valuable information on within-population heterogeneity.
Figure 4.
Result of the low-throughput worm pipeline 4. Segmentation errors from the automated worm detection were manually corrected (Pipeline 4). Worms were then automatically digitally straightened, and manually ‘flipped’ to ensure they all have the same orientation. a–c) show the input data and aligned worms from a single sample well from peb-1, daf-2 and L4440 respectively. d) A very simple anatomical atlas (in this case 8 transverse segments) is fitted to each worm, and the mean and standard deviation of ORO staining intensity is measured for each segment. e) Segmentation allows scoring of differences in body fat distribution. Mean and standard deviation of the ORO signal intensity in transverse segments T1–T8 is plotted for all analyzed worms. It is observed that, in agreement with established manual scoring of fat staining, the intensity measurement of T7 (anterior intestinal cells) has the best discriminative power to classify fat, slim, and wild-type worms. f) Scatter plot of measurements. Note that the outliers 9, 10 and 16 for L4440 (red) and 30 for peb-1 (blue) also represent measurements from worms that visually have a deviating phenotype (circled in a and c).
3. Discussion
In this paper, we describe a radically improved whole-animal fat scoring protocol, quick Oil red O (qORO). qORO allows users to process a 96-well plate of RNAi or compound treated animals in 15 minutes of hands-on time. qORO also enables accurate fat assessment in a small number of samples for routine or follow up analyses. With qORO, intact-animal recoveries are close to 100% by comparison to <40% using the original ORO and post-fixation Nile red methods[5, 22], making this protocol amenable to high-quality automated scoring. The method is based on an isopropanol/water mixture for preservation and staining, which has the advantages of lower cost, negligible exposure to toxic compounds, and reduced usage of dye reagents. Furthermore, the method does not require alcohol-based de-staining, avoiding the variability introduced by time of de-staining, which is one of the major caveats of the alternative lipid staining method, Sudan black. This protocol allows the user to perform the whole procedure on the bench top without the need of a chemical hood. Additionally, unlike fluorescent dyes, qORO is not light sensitive. qORO stained worm samples can be stored for weeks without affecting the quality of the signal, enabling processing of hundreds of worm populations in the same day and imaging at a later time. The qORO method requires about 20 times less labor per sample. Because the qORO protocol saves time, researchers can increase the number of samples processed at once and thus improve data quality, while yielding quantitative results.
One large-scale, biochemical lipid screen has been reported using Drosophila[23], and while highly informative, biochemically-based screens do not enable analyses of body fat distribution or parallel observation of pleiotropies. A screen for membrane and nuclear hormone receptors regulating organismal fat has been carried out using Stimulated Raman Scattering (SRS) in C. elegans[10]. Although dye-free imaging of lipids in whole-animals constitutes a great advance towards unperturbed in vivo lipid measurements, SRS quantification is currently based on the analysis of fat content in 2 pairs of cells of the intestine of 5 randomly but manually selected worms per treatment. Limited sampling dramatically reduces the power of these screens, especially in whole animal RNAi screens where there is significant within-population heterogeneity[14]. Moreover, two pairs of intestinal cells do not inform about body fat distribution at the organismal level. By contrast, automated whole-population high-content image-based screens enable unbiased scoring of not only fat amounts, but also fat distribution. In addition, high-content screening of worm populations permits to evaluate the impact of each genetic modification or chemical treatment on growth and reproduction, among other relevant parameters scorable in full-well images containing dozens of worms.
In addition, we present an upgraded set of image analysis tools for automated quantification of the qORO signal. The analysis tools can either be used in a fully automated mode, or allow the user to interact with the data. The image analysis tools are provided to the research and teaching communities through www.cellprofiler.org/wormtoolbox. This resource is free, user-friendly, and requires no more than a standard computer. The analysis pipelines and video tutorials allow any inexperienced user without programming background to analyze their images. Furthermore, analysis pipelines can easily be included as supplementary information upon publication of research results, facilitating reproducible science.
Functional genomics is an emerging field with the ambitious task of comprehensively determining the function of a large number of genes. We present here an improved fat staining method for C. elegans that is compatible with high-content screening and automated scoring. We have highlighted the benefits of using the presented protocol, including increases in throughput and robustness, and making large screens for fat regulators possible in whole animals. Additionally, automated image capture and analysis enables objective scoring and reanalysis of primary data. Finally, the simplicity and affordability of the qORO method makes it compatible with the teaching laboratory, increasing the interest of future scientists in the biological and medical relevant area of fat metabolism, and in computational analysis, an increasingly important component of modern biological research.
Acknowledgements
Funding for this work was provided by the National Institutes of Health to C.W. (R01 GM095672), to A.E.C. (R01 GM089652), and to E.O’R (R00 DK087928).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bonilla E, Prelle A. J Histochem Cytochem. 1987;35:619–621. doi: 10.1177/35.5.3559182. [DOI] [PubMed] [Google Scholar]
- 2.Fowler SD, Greenspan P. J Histochem Cytochem. 1985;33:833–836. doi: 10.1177/33.8.4020099. [DOI] [PubMed] [Google Scholar]
- 3.Schroeder LK, Kremer S, Kramer MJ, Currie E, Kwan E, Watts JL, Lawrenson AL, Hermann GJ. Mol Biol Cell. 2007;18:995–1008. doi: 10.1091/mbc.E06-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks KK, Liang B, Watts JL. PLoS One. 2009;4:e7545. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yen K, Le TT, Bansal A, Narasimhan SD, Cheng JX, Tissenbaum HA. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barros AG, Liu J, Lemieux GA, Mullaney BC, Ashrafi K. Methods Cell Biol. 2012;107:383–407. doi: 10.1016/B978-0-12-394620-1.00013-8. [DOI] [PubMed] [Google Scholar]
- 8.Mak HY. J Lipid Res. 2012;53:28–33. doi: 10.1194/jlr.R021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKay RM, McKay JP, Avery L, Graff JM. Dev Cell. 2003;4:131–142. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang MC, Min W, Freudiger CW, Ruvkun G, Xie XS. Nature methods. 2011;8:135–138. doi: 10.1038/nmeth.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le TT, Duren HM, Slipchenko MN, Hu CD, Cheng JX. J Lipid Res. 2010;51:672–677. doi: 10.1194/jlr.D000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wählby C, Kamentsky L, Liu ZH, Riklin-Raviv T, Conery AL, O'Rourke EJ, Sokolnicki KL, Visvikis O, Ljosa V, Irazoqui JE, Golland P, Ruvkun G, Ausubel FM, Carpenter AE. Nature methods. 2012;9:714–716. doi: 10.1038/nmeth.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Rourke EJ, Conery AL, Moy TI. Methods Mol Biol. 2009;486:57–75. doi: 10.1007/978-1-60327-545-3_5. [DOI] [PubMed] [Google Scholar]
- 16.O'Rourke EJ, Ruvkun G. Nat Cell Biol. 2013;15:668–676. doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thatcher JD, Fernandez AP, Beaster-Jones L, Haun C, Okkema PG. Dev Biol. 2001;229:480–493. doi: 10.1006/dbio.2000.9978. [DOI] [PubMed] [Google Scholar]
- 18.Riklin-Raviv TR, Ljosa V, Conery AL, Ausubel FM, Carpenter AE, Golland P, Wählby C. Med Image Comput Comput Assist Interv. 2010;13:634–641. doi: 10.1007/978-3-642-15711-0_79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wählby C, Riklin-Raviv T, Ljosa V, Conery AL, Golland P, Ausubel FM, Carpenter AE. Proceedings / IEEE International Symposium on Biomedical Imaging: from nano to macro. IEEE International Symposium on Biomedical Imaging. 2010;2010:552–555. doi: 10.1109/ISBI.2010.5490286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, Sabatini DM. Proc Natl Acad Sci U S A. 2009;106:1826–1831. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P, Carpenter AE. BMC Bioinformatics. 2008;9:482. doi: 10.1186/1471-2105-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pospisilik JA, Schramek D, Schnidar H, Cronin SJ, Nehme NT, Zhang X, Knauf C, Cani PD, Aumayr K, Todoric J, Bayer M, Haschemi A, Puviindran V, Tar K, Orthofer M, Neely GG, Dietzl G, Manoukian A, Funovics M, Prager G, Wagner O, Ferrandon D, Aberger F, Hui CC, Esterbauer H, Penninger JM. Cell. 2010;140:148–160. doi: 10.1016/j.cell.2009.12.027. [DOI] [PubMed] [Google Scholar]