Abstract
High grade serous ovarian carcinoma (HGSOC) and basal-like breast cancer (BLBC) share many features including TP53 mutations, genomic instability and poor prognosis. We recently reported that Elafin is overexpressed by HGSOC and is associated with poor overall survival. Here, we confirmed that Elafin overexpression is associated with shorter survival in 1000 HGSOC patients. Elafin confers a proliferative advantage to tumor cells through activation of the MAP kinase pathway. This mitogenic effect can be neutralized by RNA interference, specific antibodies, and a MEK inhibitor. Elafin expression in patient-derived samples was also associated with chemoresistance and strongly correlates with bcl-xL expression. We extended these findings into examination of 1100 primary breast tumors and six breast cancer cell lines. We observed that Elafin is overexpressed and secreted specifically by BLBC tumors and cell lines, leading to a similar mitogenic effect through activation of the MAP kinase pathway. Here too, Elafin overexpression is associated with poor overall survival, suggesting that it may serve as a biomarker and therapeutic target in this setting.
Keywords: Elafin, ovarian cancer, basal-like breast cancer, mitogen, MAP kinase
Introduction
Among hereditary forms of breast and ovarian cancer (OC), high grade serous ovarian carcinoma (HGSOC) 1 and basal-like breast cancers (BLBC) 1, 2 are the predominant histological subtypes that develop in BRCA mutation carriers. HGSOC and BLBC share many features, including TP53 mutations in the majority of cases 3, 4 and more genomic instability than other subtypes of breast 5 and ovarian cancers 4, leading to poor survival. Recent data from The Cancer Genome Atlas (TCGA) studies highlighted the similarities between both diseases in terms of copy number alterations, gene mutations and expression profiles 6, 7, strongly suggesting shared driving events for their carcinogenesis. Elafin is a whey acidic protein (WAP) that exhibits specific protease inhibitor activity against human neutrophil elastase and the related proteinase 3 8. Under physiologic conditions, it is produced by a subset of epithelial cells exposed to inflammatory stimuli 9, including epithelia of the female reproductive tract 10 and breast 11. Elafin has recently been validated as a biomarker predicting graft-versus host disease (GVHD) after allogeneic stem-cell transplantation 12, 13. It is related to other WAP proteins such as secretory leukocyte protease inhibitor (SLPI) and HE4 14–17; all of which are encoded on chromosome 20q11.2–13 15, 18, 19, a region commonly amplified in HGSOC 4, BLBC 6 and glioblastoma 20. High expression of Elafin correlates with chemoresistance 21 and poor survival in HGSOC 22. In this report, we examine the functional role of Elafin in HGSOC and BLBC and integrate these findings with outcome measures in these two highly related diseases.
Results
Elafin overexpression is associated with poor outcome
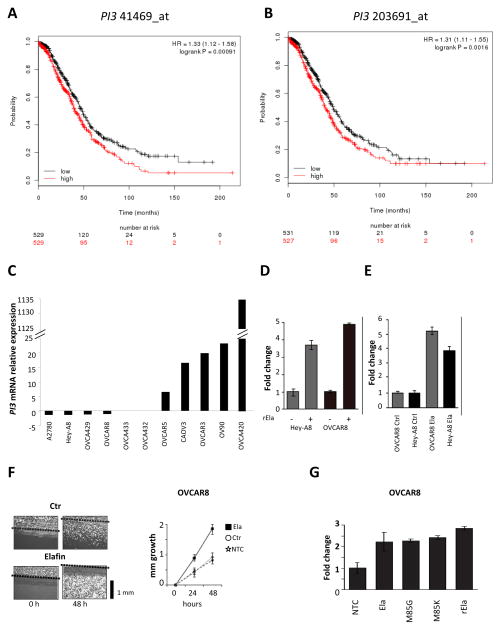
We recently showed that Elafin overexpressed at the protein level is associated with poor overall survival (OS) in HGSOC 22. To further confirm this observation, we did a meta-analysis of Elafin gene (PI3) expression among 1058 HGSOC patients using the KMplot software program 23. Using median gene expression values as the cut-off, we observed shorter OS in those patients with high PI3. This was observed with both probes, 41469_at (HR=1.33, 95%IC 1.12–1.58, p=0.00091; Figure 1A) and 203691_at (HR=1.31, 95%IC 1.11–1.55, p=0.0016; Figure 1B). These correlations remained significant in a multivariate Cox regression analysis including known clinical parameters (n=927, probe 41469_at: HR=1.36, 95%IC=1.14–1.58, p=0.006; probe 203691_at: HR=1.52, 95%IC=1.30–1.74, p=0.0002; stage: HR=1.35, 95%IC=1.13–1.47, p=0.01; grade: p=n.s.; and debulking status: p=n.s.) (Tables S1 and S2).
Figure 1. Elafin increases ovarian cancer cell proliferation.
A. Kaplan Meier analysis for Overall Survival (OS) in ovarian cancer (OC) patients according to the expression of PI3 probe 41469_at (n=1058). B. OS in OC patients according to the expression of PI3 probe 203691_at (n=1058). Log-rank test. Median value as cut-off. C. Relative PI3 mRNA expression by quantitative RT-PCR among 11 OC cell lines. D. Effect of adding 300 nM of recombinant Elafin (rElafin) on proliferation of OC cell lines Hey-A8 and OVCAR8. E. Proliferation of Hey-A8 and OVCAR8 transfected with pcDNA3.0 vector encoding Elafin (Ela) compared to cell lines transfected with empty vector (Ctr). F. Scratch test on OVCAR8 cells overexpressing Elafin (Ela), control (Ctr) and non-transfected (NTC) line. Cell growth into the scratch is plotted in the graph as mm growth over time. The black bar represents 1 mm. G. Effect of mutations in the serine proteinase domains of Elafin, M85G and M85K, on cellular proliferation rates when compared to rElafin and vector coding for non mutant Elafin. Proliferation was measured using the Celltiter Glo assay in D, E and G.
Elafin increases cellular proliferation in ovarian cancer
To further characterize the biological relationship between Elafin expression and poor survival, we analyzed a panel of 20 OC cell lines for expression of Elafin. We constructed a gene-expression profiling dataset (Figure S1A) and confirmed the results by quantitative RT-PCR (Figure 1C). We first asked whether Elafin expression has a measurable effect on cell biology by either adding recombinant Elafin (rElafin) to growth media or inducing ectopic expression of Elafin in Hey-A8 and OVCAR8. These two cell lines were selected for overexpression studies based on their relatively low endogenous Elafin expression (Figure 1C). Cell culture medium (0.5% FBS) was either mock treated or supplemented with rElafin. The addition of rElafin led to a 4–5 fold increase in proliferation in both cell lines when compared to non-stimulated controls (Figure 1D).
Next, Hey-A8 and OVCAR8 stably overexpressing a pcDNA3.0 vector encoding Elafin (Ela) or an empty vector (Ctr) were generated. Elafin was detectable in both mRNA and whole cell extracts (WCE) (Figure S1B and S1C). Ectopic expression of Elafin led to a 4–5 fold increase in proliferation in these cells compared to empty vector controls (Figure 1E).
We then performed a ‘scratch assay’ to determine whether Elafin has an impact on motility in cell wounding experiments. After 48 hours, Elafin-expressing OVCAR8 cells had fully healed the scratch (2 mm) while OVCAR8 cells with either empty vector control or non-transfected controls had only covered half of the distance (Figure 1F). Taken together, these results showed that Elafin can induce motility of OC cells. Elafin is an endogenous inhibitor of neutrophil-derived elastase and proteinase-3 24. Inhibition of protease function is mediated by a critical methionine residue (M85) located within the WAP domain and can be attenuated by mutants that replace this methionine residue with either lysine (M85K) or glycine (M85G) 25. Interestingly, both the M85K and M85G mutants exhibited a similar ability to stimulate cell proliferation as rElafin and non-mutant vector encoding for Elafin in OVCAR8 (Figure 1G), suggesting that the mitogenic properties of Elafin are independent of its anti-protease function.
SLPI, another member of the WAP domain family, has been shown to function as a serine protease inhibitor 18, 19. Compared to empty vector control and rElafin, rSLPI had no effect on cell growth of Hey-A8 (Figure S1D) suggesting a unique effect of Elafin on OC cell growth.
Elafin induces cell proliferation through the MEK-ERK pathway
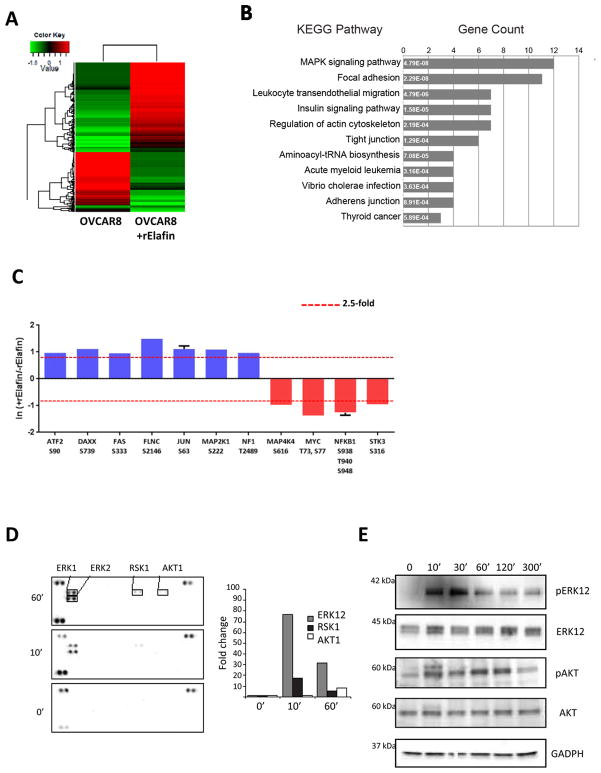
Given our evidence that Elafin induces cellular proliferation, we next sought to identify activated pathways using quantitative phosphoproteomics. We stimulated OVCAR8 cells with rElafin for 30 min and encoded the resulting tryptic peptides with isobaric stable isotope labels (Figure S2). In total we quantified 7130 distinct phosphopeptides that mapped to 2742 unique gene I.D.s. (Figure 2A). Among these data we observed changes in phosphorylation on 386 peptides, which mapped to 316 unique gene I.D.s. We queried these genes in Pathway Palette 26 and found that Elafin-mediated signaling appeared to target multiple pathways, including MAPK (p<0.001, 12 genes) (Figure 2B). Consistent with these data we observed phosphorylation on several transcription factors, including c-JUN (S63) and ATF2 (S90) (Figure 2C). We next used a reversed phase protein array technology to probe the kinetics of kinase activation resulting from Elafin stimulation. As early as 10 minutes after addition of rElafin to culture conditions, we detected an increase in the levels of phospho-ERK1/2 (pERK) and ribosomal protein S6 kinase (RSK1) (Figure 2D) confirming our mass spectrometry data. This became more pronounced at the 60 minute time point. The peak of pERK at 30 minutes was dose-dependent (Figure S1E). In addition, we observed a modest increase in the phosphorylated form of AKT1 (pAKT) at 60 minutes (Figure 2D). These observations were confirmed by western blot (WB) (Figure 2E).
Figure 2. Mitogenic effect of Elafin is mediated through the MEK/ERK pathway.
A. Multiplex iTRAQ analysis of OVCAR8 phosphoproteome at baseline and 30 minutes after adding 600 nM of rElafin. Heatmap of 404 entries, cut-off > 2.5-fold. B. Analysis of pathways altered by adding rElafin using pathway Palette software. C. Expression profile of MAP Kinase phosphopeptides activated in OVCAR8 +/− rElafin. Red lines represent global thresholds based on a ± 2.5-fold change between OVCAR8 +/− rElafin. D. Relative levels of phosphorylation of serine/threonine and receptor tyrosine kinases were determined in whole cells extracts (WCE) from OVCAR8 cells at baseline, then 10 and 60 minutes after stimulation with 300 nM of rElafin. E. Western blot analysis of pERK1/2, ERK1/2, pAKT and AKT in OVCAR8 WCE at baseline, then 10, 30, 60, 120, and 300 minutes after stimulation with 300 nM of rElafin.
Two well-characterized downstream targets of the MAPK pathway are FOS and EGR1 27. Both proteins are known to increase cellular proliferation. We detected almost a 4-fold increase in FOS mRNA levels in Elafin-expressing OVCAR8 cells, while EGR1 levels were almost 7-fold higher compared to Elafin-negative OVCAR8 cells (Figure S1F). We also confirmed higher levels of FOS and EGR1 in OVCAR8 cells treated with rElafin by WB (Figure S1G). Both FOS and EGR1 protein levels peaked 10 minutes after treatment with rElafin and then returned to baseline suggesting a possible impact on protein turnover.
The proliferative effect of Elafin can be neutralized
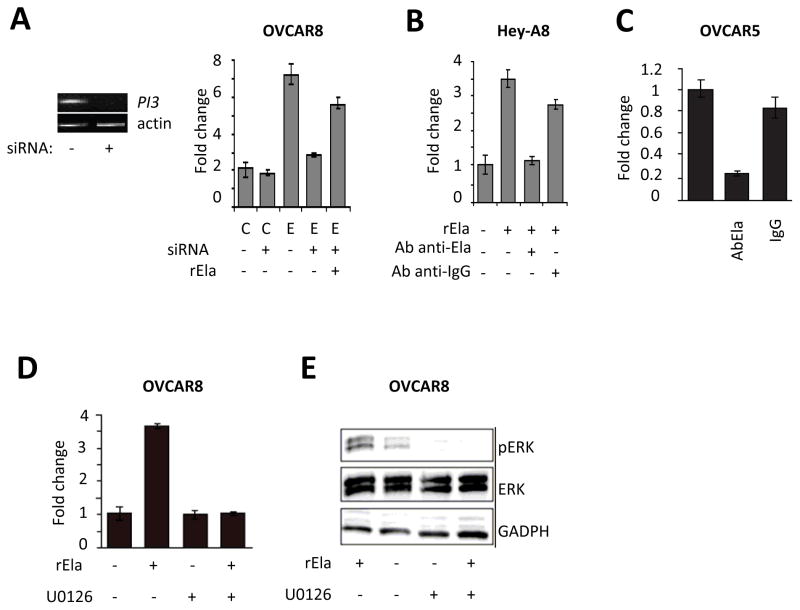
To determine whether the proliferative effects of Elafin are reversible, we used small interfering RNAs (siRNAs) to target Elafin message in OVCAR8 cells overexpressing Elafin. A pool of three siRNAs dramatically decreased Elafin mRNA levels but had no effect on proliferation of Elafin-negative cells (Figure 3A). Conversely, Elafin-positive cells were almost completely inhibited by siRNAs in their ability to proliferate compared to controls. Interestingly, proliferation was, in part, restored by adding exogenous rElafin (Figure 3A).
Figure 3. Blockage of Elafin abolishes its mitogenic effect on ovarian cancer.
A. Effect of a pool of 3 siRNAs against Elafin on expression of PI3 mRNA (left panel) and proliferation in OVCAR8 cells overexpressing Elafin. Effect of adding 300 nM of rElafin on proliferation (right panel). B. Effect of adding anti-Elafin antibodies on proliferation of Hey-A8 cells treated with 300 nM of rElafin. C. Effect of anti-Elafin antibodies on proliferation of OVCAR5 cells. D. MEK kinase inhibitor U0126 reverses the mitogenic effect of rElafin on OVCAR8. E. U0126 inhibits the phosphorylation of ERK induced by 300 nM of rElafin. C: control vector, E: vector overexpressing PI3, rEla: recombinant Elafin. Proliferation was measured using the Celltiter Glo assay.
Since Elafin is a secreted protein, we questioned whether blocking it with a specific antibody would neutralize its mitogenic effect. The addition of anti-Elafin antibody (Ab) to Hey-A8 cells treated with rElafin almost completely abolished its proliferative effect (Figure 3B). To determine if the same effect could be detected in cells that endogenously express and secrete Elafin, the experiment was repeated in OVCAR5 cells, a cell line with detectable levels of endogenous Elafin 22. The anti-Elafin Ab dramatically decreased cell proliferation compared to IgG control (Figure 3C).
The phosphoproteome array (Figure 2D) suggested that the proliferative effects of Elafin are mediated through the MEK-ERK signaling pathway. We therefore asked whether we could block the proliferative signals downstream using the MEK kinase inhibitor U0126. In the absence of U0126, rElafin stimulated proliferation of OVCAR8 (Figure 3D). U0126 alone had a negligible effect on proliferation in the absence of rElafin. However, when cells were treated with both rElafin and U0126 we found that U0126 completely blocked the proliferative effect of rElafin (Figure 3D) and abrogated the phosphorylation of ERK (Figure 3E).
Elafin overexpression is associated with high bcl-xL levels
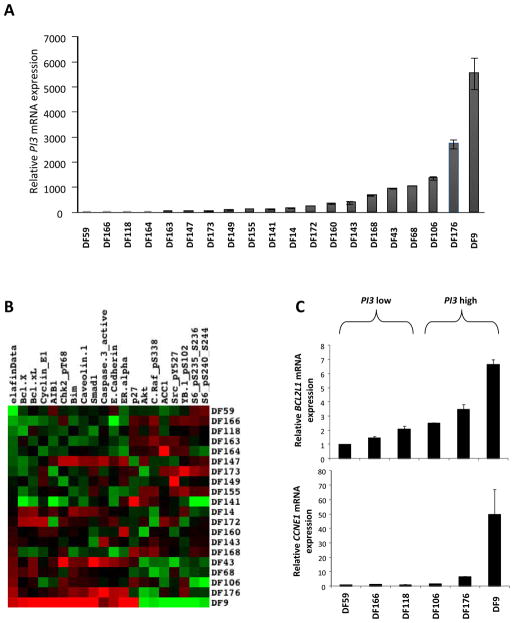
We isolated primary tumor cells from ascites of 20 patients with advanced HGSOC disease (DF lines) as previously described 22. RNA was isolated from these cells and analyzed for relative expression of PI3 by qRT-PCR (Figure 4A). We observed that the three chemo-refractory tumors (showing clinical progression during chemotherapy) were among the highest PI3 expressing tumors (DF9, DF106 and DF160), confirming our previous observation 22. Moreover, the median progression-free survival was shorter in patients with high Elafin compared to low Elafin expression (8.36 compared to 13.73 months, p=0.09), although not statistically significant due to the limited number of patients (n=20).
Figure 4. PI3 overexpression is associated with chemoresistance pathways.
A. Relative expression of PI3 mRNA in 20 HGSOC tumors by quantitative RT-PCR. B. Heatmap of RPPA of 20 HGSOC tumors clustered according to PI3 relative mRNA. C. Relative mRNA levels of BCL2L1 and CCNE1 in 3 low (DF 59, 166 and 118) and 3 high (DF106, 176 and 9) PI3 expressing tumors.
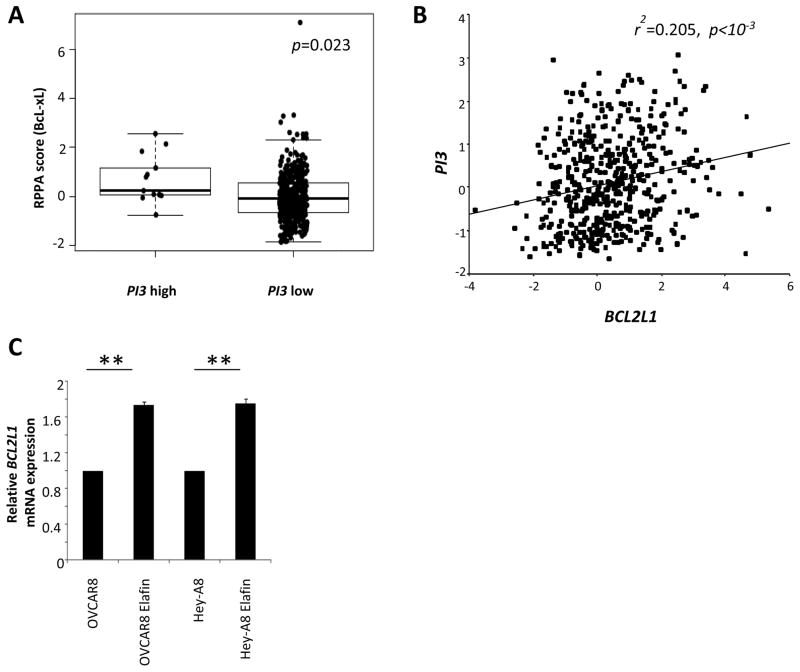
To further characterize the biology of OC cells expressing high levels of Elafin, we interrogated the intracellular proteome using a comprehensive Reverse-Phase Protein Array (RPPA) 28, 29. The 20 DF lines were clustered based on their relative PI3 mRNA levels. We observed that high-Elafin expressing tumors show high levels of proteins previously linked with chemoresistance in OC (Figure 4B and Table S3), in particular bcl-xL 30, 31 and Cyclin E1 32. We confirmed the overexpression of BCL2L1 and CCNE1 at the mRNA levels in 3 high and 3 low PI3 expressing tumors (Figure 4C). To validate these observations, we tested the correlation between PI3 relative mRNA levels and the proteins reported in the heatmap using TCGA data. We clustered the 412 TCGA HGSOC tumors into high and low PI3 sets and compared the RPPA data with all the proteins that were significant in our DF RPPA. We observed significantly increased levels of bcl-xL in high PI3 expressing tumors (p=0.023, Figure 5A). Moreover, relative mRNA levels of PI3 and BCL2L1 significantly correlated (r2=0.205, p<10−3; Figure 5B). Following this, we also checked the relative BCL2L1 mRNA levels in naive OVCAR8 and Hey-A8, and in the same cell lines overexpressing Elafin. We observed that BCL2L1 levels significantly increased in the Elafin-overexpressing cell lines (p=0.0079), reinforcing the connection between Elafin and bcl-xL.
Figure 5. Overexpression of Elafin correlates with bcl-xL in TCGA data.
A. Analysis of bcl-xL protein levels in high and low PI3 tumors from the HGSOC TCGA dataset (n=412, p=0.023). Student t-test. B. Spearman test for correlation between BCL2L1 and PI3 mRNA relative levels in TCGA tumors (n=489, r2=0.205, p<10−3). C. Relative mRNA BCL2L1 in cell lines overexpressing Elafin compared to naive ones (p=0.0079). Data are median and range, Mann-Whitney test.
Elafin is highly expressed in basal-like breast carcinomas
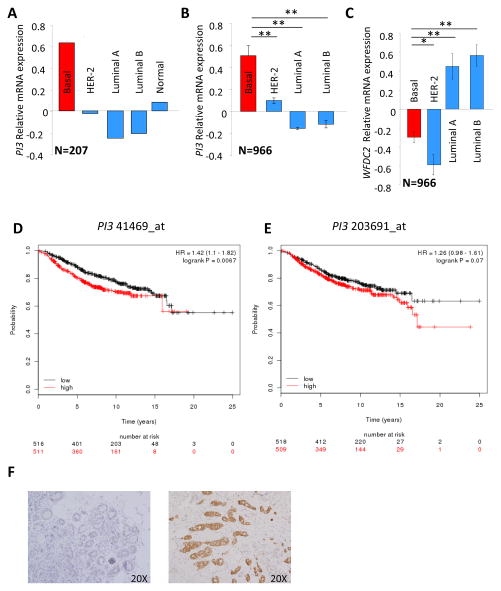
Having delineated a role for Elafin in HGSOC proliferation, we questioned whether a similar effect might hold true for breast carcinomas (BC), as these tumors arise in overlapping patient populations. A gene expression profile comparison of normal basal versus luminal epithelial cells isolated from reduction mammoplasties showed that PI3 is one of the most differentially and highly expressed genes in basal cells. In contrast, HE4 gene (WFDC2) expression was higher in luminal compared to basal epithelial cells 33. Since the normal breast basal cells share many properties with basal-like breast cancers (BLBC) 34, we analyzed Elafin expression across molecular subtypes of BC. Gene expression profiles were obtained from 7 publicly available data sets that contained 1166 primary BC. To allow for robust analysis, these data sets were further divided into a “discovery” cohort (2 data sets, 200 BC and 7 normal breast) 35, 36 and a “validation” cohort (5 data sets, n=966) 37–41. We classified breast tumors as BLBC, HER-2+, luminal A and luminal B 42. BLBC showed the highest level of PI3 expression, followed by normal breast tissues while HER-2 and luminal breast cancer samples had relatively low expression of PI3 (p<10−3, Figures 6A and B). Interestingly, WFDC2 levels were higher in luminal breast tumors than BLBC (p<10−3, Figure 6C), suggesting that whereas both genes are located in the same chromosome region (20q13), which is frequently aberrant in cancer, only PI3 overexpression is specific to BLBC.
Figure 6. Basal-like breast cancers (BLBC) overexpress Elafin.
A. Relative mRNA expression of PI3 in molecularly characterized breast tumors (n=200) and normal breast tissue (n=7). B. Relative mRNA expression of PI3 in an independent cohort molecularly characterized breast tumors (n=966, p<10−2). C. WFDC2 relative mRNA expression accordingly to BC molecular subtype (n=966, p<10−2). Student t-test. D. Kaplan Meier analysis for Overall Survival (OS) in BC patients according to the expression of PI3 probe 41469_at (n=1027) E. OS in BC patients according to the expression of PI3 probe 203691_at (n=1027). Log-rank test. Median value as cut-off. F. Elafin is detected by IHC in 12 of 16 TNBC. Left panel: negative tumor, right panel: high expressing Elafin.
We further questioned whether high Elafin levels can have an impact on BC survival, as we previously observed in OC. We used the KMplot software program to compile data from 1027 BC patients 43. Using median gene expression values as the cut-off, we observed that high Elafin expression leads to shorter OS, and this was significant with the 41469_at PI3 probe (HR=1.42, 95%IC 1.1–1.82, p=0.0067; Figure 6D) and borderline with the 203691_at probe (HR=1.26, 95%IC=0.98–1.61, p=0.07; Figure 6E). Importantly, high expression of WFDC2 was associated with significantly improved outcome in BC patients (HR=0.74, 95%IC 0.57–0.95, p=0.017; Figure S3A) highlighting the distinction between Elafin and HE4.
We then asked more specifically if overexpression of Elafin has an impact on BLBC patients’ survival. OS data were available for only 185 BLBC patients in KMplot database. When choosing the median value of PI3 expression as cut-off, we observed a trend toward worse outcome but this was not significant with both 41469_at PI3 probe (HR=1.7, 95%IC=0.95–3.05, p=0.07) and 203691_at probe (HR=1.56, 95%IC=0.87–2.79, p=0.13). To increase the statistical power of the analysis, we chose the highest quartile as cut-off and confirmed that there is a worse outcome in BLBC patients that overexpress Elafin. The difference in survival was significant with 203691_at PI3 probe (HR=1.98, 95%IC=1.06–3.68, p=0.028: Figure S3B) and borderline with 41469_at PI3 probe (HR=1.75, 95%IC=0.93–3.29, p=0.078; Figure S3C).
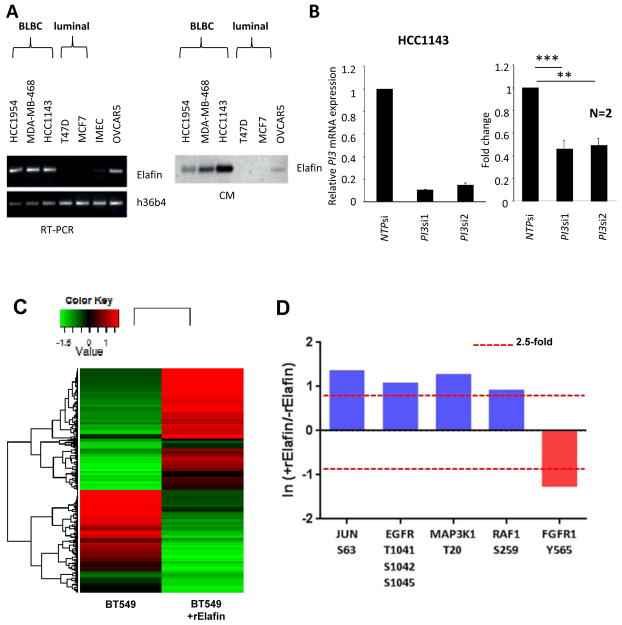
At the protein level, 12 of 16 triple negative BCs (TNBC) showed cytoplasmic Elafin staining as assessed by immunohistochemistry (Figure 6F) compared to only 3 of 12 non TNBCs. Next, we studied levels of PI3 mRNA and Elafin protein in a panel of BC cell lines that included basal-like and luminal subtypes. We observed that the BLBC cell lines, i.e. HCC1954, MDA-MB-468, and HCC1143 (all Basal A-like molecular subtypes) 44 express PI3 mRNA (Figure 7A left panel) whereas luminal BC cell lines T47D and MCF7 do not (OVCAR5 cells served as a positive control). The expression data was mirrored in secreted protein levels across the same cell lines: all three BLBC cell lines secrete Elafin in the conditioned media whereas luminal cell lines do not (Figure 7A right panel). Importantly, we did not observe expression of Elafin in WCE (Figure S4A), consistent with it being a secreted protein. Given the ability of Elafin to stimulate proliferation in OC cells, we tested whether Elafin has a mitogenic effect on BLBC. Knocking down the expression of PI3 mRNA using a pool of two different siRNAs (Figure 7B left panel) significantly reduces the proliferation of HCC1143 (p<10−2, Figure 7B right panel). Similar reduction of proliferation was observed in both MDA-MB-468 and HCC1954 cells (Figure S4C) after inhibition of PI3 expression by the same siRNA pool (Figure S4D).
Figure 7. Overexpression of Elafin in BLBC leads to mitogen effect.
A. PI3 expression by RT-PCR in BLBC cell lines (HCC1954, MDA-MB-468 and HCC1143) and luminal cell lines (T47D and MCF7) (left panel) and secretion of Elafin in the conditioned media (right panel). OVCAR5 served as positive control. IMEC expresses low levels of PI3. B. Effect of siRNA anti-Elafin (PI3si1 and PI3si2) on relative expression of PI3 mRNA assessed by qRT-PCR in HCC1143 compared to non-target pool (NTPsi). Effect of PI3si1 and PI3si2 on HCC1143 proliferation when compared to NTPsi (p=0.0008 for PI3si1 and p=0.004 for PI3si2; n=2). **: p<10−2, ***: p<10−3. Proliferation was measured using the Celltiter Glo assay. Mann-Whitney test. C. Multiplex iTRAQ analysis of BT549 phosphoproteome at baseline and 30 minutes after adding 600 nM of rElafin. Heatmap of 352 entries, cut-off > 2.5-fold. D. Regulated MAP Kinase phosphopeptides in BT549 by rElafin. Red lines represent global thresholds based on a ± 2.5-fold change between BT549 +/− rElafin.
Elafin induces activation of MEK-ERK pathway in BLBC
Having observed a mitogenic effect of Elafin in BLBC cell lines, we questioned if it also acts through the MAP kinase pathway. Like HGSOC, not all BLBC cell lines overexpressed Elafin. Indeed, we found that BT549 did not express 45 or secrete Elafin (Figure S4B). When we added rElafin to the culture media of BT549 cells, we observed a significant modification (cut-off of 2.5 fold change) in 352 peptides (Figure 7C). Specific analysis of the MAP Kinase pathway showed an increase in c-JUN, MAP3K1 and EGFR (Figure 7D), consistent with our ovarian cancer cell line results.
Discussion
We recently reported that approximately 40% of HGSOCs express high levels of Elafin, and that this correlates with poor OS 22. Here, we confirm this relationship in a larger cohort of HGSOC patients and replicate this finding in a related malignancy, BLBC. We show that Elafin, when present either as an exogenous or an endogenous source, increases cellular proliferation. Elafin activates the MEK-ERK signaling pathway 46 and induces mediators of proliferation, in particular the JNK subgroup with phosphorylation of ATF2 and c-JUN 47, 48. To our knowledge, this is the first report demonstrating a mitogenic role for Elafin in HGSOC and BLBC cells and suggests that Elafin may be a therapeutic target in this setting.
We 22 and others 21 have shown that overexpression of Elafin in HGSOC is associated with platinum resistance. Here we propose that Elafin contributes to chemoresistance in OC through enhanced expression of bcl-xL, a potent anti-apoptotic protein involved in resistance to both platinum agents and paclitaxel. We also show an association between high Elafin expression and other chemoresistance markers, including Cyclin E.
Among the different molecular subtypes of breast cancer, only BLBC express Elafin at the mRNA level. Overexpression of Elafin was associated with shorter OS in BC patients, and more specifically in BLBC patients. BLBC shares many features with HGSOC. First, among hereditary forms of breast and ovarian cancer, HGSOC 1 and BLBC 1, 2 are the predominant histological subtypes that develop in BRCA mutation carriers. Second, the majority of HGSOCs 4 and BLBCs 3 harbor TP53 mutations, and shows increased genomic instability compared to other breast 5 and ovarian 4 cancer subtypes. Third, both tumors are associated with poor outcome 49, 50. In this regard they resemble another aggressive malignant tumor, glioblastoma multiforme (GBM). GBMs exhibit high genomic instability, frequent TP53 mutations 20, and expresses high levels of Elafin that lead to poorer survival 51.
Previous studies have suggested that Elafin is transcriptionally downregulated in breast tumor-derived cell lines compared to normal breast cells 45, 52, 53. This is true for cell lines of luminal origin, like MCF7 45, 53 and T47D 45. However, normal basal (myoepithelial) breast cells overexpress Elafin gene 33 and consequently BLBC cell lines such as MDA-MB-157 and MDA-MB-436 overexpress it too 53. Our data are consistent with these results since we did not observe secretion of Elafin by luminal cell lines but only by the three BLBC lines 44. Thus, Elafin may have different functional roles in luminal breast cancers versus basal-like breast cancer.
Recent studies from our laboratory and others point to the fallopian tube (FT) secretory epithelial cell as a possible cell-of-origin for the majority of HGSOC 54–56. Normal FT epithelial cells secrete high levels of Elafin when compared to other epithelia of the female reproductive tract 10, 57. In addition, PI3, the gene encoding Elafin, is one of the most differentially expressed genes in basal compared to luminal cells in the normal breast 33. Thus, high expression of Elafin seems to be a feature that is preserved in tumors possibly originating from these epithelia. While it is not clear what function Elafin serves in normal epithelial cells, its role in wound repair and innate immunity 58 suggests that Elafin expression in these cells enables them to quickly respond to tissue stresses associated with injury, monthly ovulation, or pregnancy through autocrine and paracrine loops. It is conceivable that carcinomas that express Elafin may effectively be co-opting the normal role of Elafin in tissue repair and homeostasis to promote tumor growth. Finally, Elafin is an extracellular protein and is biologically active as both a cell-associated and secreted protein. Indeed, when we measure this protein in both conditioned media and whole cell extract (WCE), we do not find Elafin in the WCE. As Elafin is a protein secreted into different body fluids including blood 11, it would be interesting to evaluate Elafin as a potential serum biomarker for early diagnosis of BLBC and HGSOC in high risk patient populations such as women with BRCA mutations.
Material and Methods
Cell lines
Twenty established ovarian cancer cell lines (A2780, Hey-A8, OVCAR3, OVCAR5, OVCAR8, OV90, OVCA420, OVCA429, OVCA432, OVCA433, ES-2, TOV112D, TOV21G, IGROV1, SW626, MCAS, SKOV3, PEO4, PA1, and CaOV3,), six breast cancer cell lines (T47D, MCF7, HCC1954, MDA-MB-468, BT549 and HCC1143), and immortalized mammary epithelial cells (IMEC) were used to evaluate the expression of Elafin. MDA-MB-468, BT549 and HCC1143 were purchased from ATCC. Cell lines were propagated in RPMI 1640 (Invitrogen, Carlsbad, CA) or DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Invitrogen) at 37°C in a 5% CO2-containing atmosphere.
Stable cell lines
The coding sequence of Elafin was cloned into the mammalian vector pcDNA3.0 (Invitrogen, Carlsbad, CA). pcDNA-Elafin vector (Addgene plasmid 18102) was transfected into Hey-A8 and OVCAR8 using FuGENE6 (Roche, Indianapolis, IN). After transfection, Elafin expression was confirmed by both RT-PCR and Western blot. Pools of transfected lines were cultured in media supplemented with 500 μg/ml of Genticin (Invitrogen, Carlsbad, CA). Empty pcDNA3.0 vector was used as a negative control. To knock down Elafin expression, we used small interfering RNAs (siRNA) that target Elafin message (Qiagen, Germantown, MD). In the Elafin-overexpressing OVCAR8 cell line, we used a pool of three siRNAs (Hs_PI3_8, HsPI3_5 and Hs_PI3_7). In HCC1143, we used two siRNAs targeting Elafin separately (Hs_PI3_8 and HsPI3_5) and a non-target pool (NTP) (allstars negative control siRNA, QIAGEN) as a control. RT-PCR confirmed successful transfection.
Quantitative phosphoproteomic analysis of Elafin induced signaling pathways
OVCAR8 and BT549 cell lines cultured as described above were harvested at baseline and 30 minutes after treatment with 600 nM of rElafin. Cells were washed once in ice-cold PBS, snap frozen in liquid nitrogen, and lysed using 7.2 M guanidine hydrochloride with 100 mM ammonium bicarbonate. Proteins were reduced (DTT), alkylated (IAA), and digested overnight with trypsin (1:10) at 37°C. Resulting peptides were desalted and labeled with iTRAQ-4plex reagents according to the manufacturer’s protocol (OVCAR8-elafin 114, OVCAR8+elafin 115, BT549-elafin 116, BT549+elafin 117). Phosphopeptides were enriched by Fe-NTA IMAC, and analyzed at a depth of 40 fractions using RP-SAX-RP interfaced to an LTQ-Orbitrap Velos mass spectrometer (ThermoFisher Scientific, San Jose, CA). Hierarchical clustering was performed using z-scores derived from iTRAQ reporter ion intensities (R version 2.15.1, heatmap.2 algorithm with default arguments). Details have been published in 59.
Kaplan Meier Curves
Kaplan–Meier curves were generated using the KMplot software, from a database of public microarray datasets (http://kmplot.com/analysis). Results were collected from 1058 HGSOC patients 23 and 1027 BC patients 43. Kaplan–Meier plots were generated for each probe of PI3 (203691_at and 41469_at) and WFDC2 (203892_at). To analyze the prognostic value of each probe, the samples were split into two groups according to the median expression of the probe. Multivariate Cox regression was computed using WinSTAT 2007 from Microsoft Excel (Robert K. Fitch Software, Germany).
Data analysis from TCGA database
Gene-expression (GE) profiling data were downloaded from the HGSOC TCGA database 4 using the interface website http://www.cbioportal.org/public-portal/. Relative expression of PI3 in 489 tumors was correlated with BCL2L1 relative expression using the Pearson correlation test. Bcl-xL relative expression (from RPPA data) was compared in PI3 high and PI3 low tumors using a Student’s t-test (n=412).
Breast tumor array
Data from 7 published studies 35–41 evaluating GE arrays from early stage BC and normal breast were compiled (n=1173) and divided into “discovery” (2 data sets, 200 BC and 7 normal breast) 35, 36 and “validation” cohorts (5 data sets, n=966) 37–41. Bioinformatics methods were previously detailed 60.
RT-PCR and Quantitative PCR
RNA was purified using TriZol (Invitrogen, Carlsbad, CA), then RNeasy MinElute Cleanup kit (Qiagen, Valencia, CA). cDNA was synthesized from each cell line with 1 ug of RNA using SuperscriptIII™cDNA synthesis Kit (Invitrogen, Carlsbad, CA). PCR primers are listed in Supplemental Table 4. PCR products were identified on a 2.5% TAE agarose gel. Quantitative amplification data was generated from 10 ng of cDNA on a Mastercycler ep realplex2 (Eppendorf, Hauppauge, NY) in conjunction with Kapa SYBR® (Kapa Biosystems, Woburn, MA).
Protein isolation and western blot analysis
Whole-cell protein lysates were prepared using RIPA lysis buffer supplemented with protease and phosphatase inhibitors (Roche, Basel, Switzerland). Cells were lysed for 40 minutes at 4°C and whole cell extracts (WCE) were frozen at −80°C until further use. Conditioned medium (CM) was prepared as follows: approximately 30 × 106 cells were seeded individually into 15 cm cell culture dishes per cell line. When cells became confluent and covered up to 80–90% of the dish’s surface, RPMI 1640 or DMEM medium was discarded, and the cells were rinsed three times with serum-free medium. Twenty ml of serum-free medium was added, and the plates were incubated for an additional 72 hours. The CM were collected and spun down to remove cellular debris, then concentrated 75× using Vivaspin 20 mL tubes (SartoriusStedim, Goettingen, Germany). Western blots (WB) were done as previously described 22. Elafin antibody was generated by the investigators as previously described 22. All other antibodies were purchased from Cell Signaling Technologies (Danvers, MA).
Proteome arrays
Relative levels of phosphorylation of serine/threonine and receptor tyrosine kinases were determined using the Proteome Profiler™ Array ARY-002 (R&D systems, Minneapolis, MN). OVCAR8 cells were stimulated with 300 nM recombinant Elafin (rElafin), and WCE was harvested at baseline, 10 and 60 minutes after stimulation with rElafin. Intensity of the dots was measured using the Spot densitometer algorithm bundled with FluorChem HD2 imaging system (Alpha Innotech, San Leandro, CA).
Proliferation assay
Proliferation was measured using the Celltiter Glo assay (Promega, Madison, WI). Briefly, a total of 1000 cells per well were seeded in sextets on a 96 well plate, and luminescence was measured in a Modulus plate reader (Turner Biosystems, Sunnyvale, CA) at 2 and 72 hours. Fold change was calculated by comparing luminescence reading between the two time points. Cells treated with 300 nM rElafin or rSLPI (R&D systems, Minneapolis, MN) were grown in medium containing only 0.5% FBS, otherwise cells were cultured as described above. In the antibody blocking experiment, 200 ng/ml of affinity-purified Elafin antibody or IgG purified from pre-immune sera of the same rabbit was used.
MEK inhibitor
OVCAR8 cells were pretreated with 10uM of U0126 (Cell Signaling Technologies), a selective MEK kinase inhibitor, 2 hours before their stimulation with 300 nM rElafin. WCE was collected 10 minutes after stimulation and analyzed by WB.
Scratch-wound assay
OVCAR8 cells were grown to full confluence in 10 cm cell culture dishes before a wound was generated using a sterile cell scraper. Fresh media was added to the plate, and wound-healing ability of the cells was monitored over time. Growth of the cells into the wound area was measured using QCapture imaging software Version 3.1.1 (Qiamging, Surrey, BC, Canada). All scratch-wound assays were repeated at least three times.
Elafin Mutants
Two point mutations that change amino acid 85 from a Methionine to either a Glycine (M85G) or Lysine (M85K) were introduced into the pcDNA-Elafin vector sequence using GeneTailor® (Invitrogen, Carlsbad, CA) and transfected into OVCAR8. Mutations were confirmed by DNA sequencing. Stable cell lines were constructed as described above.
Gene expression-profiling of ovarian cancer cell lines
Gene expression analyses used GeneChip© Human Genome U133 Plus 2.0 (Affymetrix, CA) oligonucleotide microarrays. Preparation of cDNA, hybridizations, washes, and detection were done as recommended by the supplier. Normalized gene expression values for PI3 in the 20 OC cell lines were calculated using RMA (Robust Multichip Average) method available in Bioconductor software package 61. The relative PI3 expression values were plotted from the median centered data using the ggplot2 module available in the R programming environment 62. Expression data will be publicly available (GEO) after publication.
Reverse phase protein array of DF cell lines
Primary HGSOC cells (DF) were collected from ascites of 20 patients treated at Dana Farber Cancer Institute with institutional review board approval. A vial from each of the DF samples was thawed and plated onto 10 cm cell culture dishes. After 48 hours, the attached cells were trypsinized and split into 3 wells of a 6-well cell culture dish. Cells were allowed to re-attach overnight to form confluent monolayers. Reverse phase protein array (RPPA) analysis on tumor cells was performed as previously described 28, 29 (Supplemental Table 5). Relative protein levels were then determined for each sample using software specifically developed for RPPA analyses (http://www.vigenetech.com). Log transformed intensity data were subjected to Student’s t-tests in bioConductor R. Relative expression of PI3 was determined by quantitative RT-PCR (QRT-PCR) in the 20 DF lines. Significant antibody probes were defined as p<0.05. Log transformed and centered heatmaps were generated using Cluster 3.0 and Java TreeView 1.1.1 63.
Tissue samples and immunohistochemistry
Sections of formalin-fixed, paraffin-embedded (FFPE) TNBC (n=16) and non-TNBC (n=12) were evaluated for the expression of Elafin by immunohistochemistry (IHC) as previously described 22.
Supplementary Material
Acknowledgments
The authors thank Drs. Michelle Hirsch for critical review of the manuscript, Laura Selfors of biocomputing analyzes of the RPPA data, and members of the Drapkin laboratory for fruitful discussions. This work was supported by the NIH/NCI SPORE in Ovarian Cancer P50-CA105009 (RD), U01 CA-152990 (RD), R21 CA-156021 (RD); NIH/NINDS P01 NS047572 (JM), the Honorable Tina Brozman ‘Tina’s Wish’ Foundation (JM, RD), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (GBM and RD), The Robert and Debra First Fund (RD), The Gamel Family Fund (RD), the Ovarian Cancer Research Fund (RD), The Madeline Franchi Ovations for the Cure Fund (AC), the Mary Kay Foundation (RD), the Sandy Rollman Ovarian Cancer Foundation (RD), the Dana-Farber Cancer Institute (DFCI) Strategic Initiative (JM), the Executive Council of the Susan Smith Center for Women’s Cancers at the DFCI, the Triple Negative Breast Cancer Foundation (SG), and the New Jersey Commission on Cancer Research (SG). SILG is a recipient of grants from Arthur Sachs/Fulbright/Harvard, La Fondation Philippe and La Fondation de France - “Recherche clinique en cancérologie – Aide à la mobilité des chercheurs”.
Footnotes
Conflict of interest
All the authors declare no conflict of interest.
References
- 1.Mavaddat N, Barrowdale D, Andrulis IL, Domchek SM, Eccles D, Nevanlinna H, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21(1):134–47. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26(15):2568–81. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 3.Manie E, Vincent-Salomon A, Lehmann-Che J, Pierron G, Turpin E, Warcoin M, et al. High frequency of TP53 mutation in BRCA1 and sporadic basal-like carcinomas but not in BRCA1 luminal breast tumors. Cancer research. 2009;69(2):663–71. doi: 10.1158/0008-5472.CAN-08-1560. [DOI] [PubMed] [Google Scholar]
- 4.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwei KA, Kung Y, Salari K, Holcomb IN, Pollack JR. Genomic instability in breast cancer: pathogenesis and clinical implications. Molecular oncology. 2010;4(3):255–66. doi: 10.1016/j.molonc.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang ZC, Birkbak NJ, Culhane AC, Drapkin R, Fatima A, Tian R, et al. Profiles of genomic instability in high-grade serous ovarian cancer predict treatment outcome. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(20):5806–15. doi: 10.1158/1078-0432.CCR-12-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams SE, Brown TI, Roghanian A, Sallenave JM. SLPI and elafin: one glove, many fingers. Clin Sci (Lond) 2006;110(1):21–35. doi: 10.1042/CS20050115. [DOI] [PubMed] [Google Scholar]
- 9.Pfundt R, van Ruissen F, van Vlijmen-Willems IM, Alkemade HA, Zeeuwen PL, Jap PH, et al. Constitutive and inducible expression of SKALP/elafin provides anti-elastase defense in human epithelia. The Journal of clinical investigation. 1996;98(6):1389–99. doi: 10.1172/JCI118926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh M, Shen Z, Fahey JV, Cu-Uvin S, Mayer K, Wira CR. Trappin-2/Elafin: a novel innate anti-human immunodeficiency virus-1 molecule of the human female reproductive tract. Immunology. 2010;129(2):207–19. doi: 10.1111/j.1365-2567.2009.03165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulasingam V, Diamandis EP. Proteomics analysis of conditioned media from three breast cancer cell lines: a mine for biomarkers and therapeutic targets. Molecular & cellular proteomics : MCP. 2007;6(11):1997–2011. doi: 10.1074/mcp.M600465-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Paczesny S, Braun TM, Levine JE, Hogan J, Crawford J, Coffing B, et al. Elafin is a biomarker of graft-versus-host disease of the skin. Science translational medicine. 2010;2(13):13ra2. doi: 10.1126/scitranslmed.3000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine JE, Logan BR, Wu J, Alousi AM, Bolanos-Meade J, Ferrara JL, et al. Acute graft-versus-host disease biomarkers measured during therapy can predict treatment outcomes: a Blood and Marrow Transplant Clinical Trials Network study. Blood. 2012;119(16):3854–60. doi: 10.1182/blood-2012-01-403063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drapkin R, von Horsten HH, Lin Y, Mok SC, Crum CP, Welch WR, et al. Human epididymis protein 4 (HE4) is a secreted glycoprotein that is overexpressed by serous and endometrioid ovarian carcinomas. Cancer research. 2005;65(6):2162–9. doi: 10.1158/0008-5472.CAN-04-3924. [DOI] [PubMed] [Google Scholar]
- 15.Bouchard D, Morisset D, Bourbonnais Y, Tremblay GM. Proteins with whey-acidic-protein motifs and cancer. The lancet oncology. 2006;7(2):167–74. doi: 10.1016/S1470-2045(06)70579-4. [DOI] [PubMed] [Google Scholar]
- 16.Hellstrom I, Raycraft J, Hayden-Ledbetter M, Ledbetter JA, Schummer M, McIntosh M, et al. The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer research. 2003;63(13):3695–700. [PubMed] [Google Scholar]
- 17.Moore RG, McMeekin DS, Brown AK, DiSilvestro P, Miller MC, Allard WJ, et al. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecologic oncology. 2009;112(1):40–6. doi: 10.1016/j.ygyno.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clauss A, Lilja H, Lundwall A. A locus on human chromosome 20 contains several genes expressing protease inhibitor domains with homology to whey acidic protein. The Biochemical journal. 2002;368(Pt 1):233–42. doi: 10.1042/BJ20020869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundwall A, Clauss A. Identification of a novel protease inhibitor gene that is highly expressed in the prostate. Biochemical and biophysical research communications. 2002;290(1):452–6. doi: 10.1006/bbrc.2001.6224. [DOI] [PubMed] [Google Scholar]
- 20.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei H, Hellstrom KE, Hellstrom I. Elafin selectively regulates the sensitivity of ovarian cancer cells to genotoxic drug-induced apoptosis. Gynecologic oncology. 2012;125(3):727–33. doi: 10.1016/j.ygyno.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clauss A, Ng V, Liu J, Piao H, Russo M, Vena N, et al. Overexpression of elafin in ovarian carcinoma is driven by genomic gains and activation of the nuclear factor kappaB pathway and is associated with poor overall survival. Neoplasia. 2010;12(2):161–72. doi: 10.1593/neo.91542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocrine-related cancer. 2012;19(2):197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 24.Verrier T, Solhonne B, Sallenave JM, Garcia-Verdugo I. The WAP protein Trappin-2/Elafin: a handyman in the regulation of inflammatory and immune responses. The international journal of biochemistry & cell biology. 2012;44(8):1377–80. doi: 10.1016/j.biocel.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Doucet A, Bouchard D, Janelle MF, Bellemare A, Gagne S, Tremblay GM, et al. Characterization of human pre-elafin mutants: full antipeptidase activity is essential to preserve lung tissue integrity in experimental emphysema. The Biochemical journal. 2007;405(3):455–63. doi: 10.1042/BJ20070020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Askenazi M, Li S, Singh S, Marto JA. Pathway Palette: a rich internet application for peptide-, protein- and network-oriented analysis of MS data. Proteomics. 2010;10(9):1880–5. doi: 10.1002/pmic.200900723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodge C, Liao J, Stofega M, Guan K, Carter-Su C, Schwartz J. Growth hormone stimulates phosphorylation and activation of elk-1 and expression of c-fos, egr-1, and junB through activation of extracellular signal-regulated kinases 1 and 2. J Biol Chem. 1998;273(47):31327–36. doi: 10.1074/jbc.273.47.31327. [DOI] [PubMed] [Google Scholar]
- 28.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular cancer therapeutics. 2006;5(10):2512–21. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 29.Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, et al. A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-microdissected Human Breast Cancers. Clinical proteomics. 2010;6(4):129–51. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams J, Lucas PC, Griffith KA, Choi M, Fogoros S, Hu YY, et al. Expression of Bcl-xL in ovarian carcinoma is associated with chemoresistance and recurrent disease. Gynecologic oncology. 2005;96(2):287–95. doi: 10.1016/j.ygyno.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 31.Jinawath N, Vasoontara C, Jinawath A, Fang X, Zhao K, Yap KL, et al. Oncoproteomic analysis reveals co-upregulation of RELA and STAT5 in carboplatin resistant ovarian carcinoma. PloS one. 2010;5(6):e11198. doi: 10.1371/journal.pone.0011198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Etemadmoghadam D, deFazio A, Beroukhim R, Mermel C, George J, Getz G, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15(4):1417–27. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huper G, Marks JR. Isogenic normal basal and luminal mammary epithelial isolated by a novel method show a differential response to ionizing radiation. Cancer research. 2007;67(7):2990–3001. doi: 10.1158/0008-5472.CAN-06-4065. [DOI] [PubMed] [Google Scholar]
- 34.Pazaiti A, Fentiman IS. Basal phenotype breast cancer: implications for treatment and prognosis. Womens Health (Lond Engl) 2011;7(2):181–202. doi: 10.2217/whe.11.5. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 36.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer cell. 2006;9(2):121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Li Q, Eklund AC, Juul N, Haibe-Kains B, Workman CT, Richardson AL, et al. Minimising immunohistochemical false negative ER classification using a complementary 23 gene expression signature of ER status. PloS one. 2010;5(12):e15031. doi: 10.1371/journal.pone.0015031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–64. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13 (11):3207–14. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer research. 2008;68(13):5405–13. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 41.Bittner M. Expression project for Oncology (expO) [cited; Available from: www.intgen.org/expo/
- 42.Haibe-Kains B, Desmedt C, Loi S, Culhane AC, Bontempi G, Quackenbush J, et al. A three-gene model to robustly identify breast cancer molecular subtypes. J Natl Cancer Inst. 2012;104(4):311–25. doi: 10.1093/jnci/djr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123(3):725–31. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 44.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer cell. 2006;10(6):515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M, Zou Z, Maass N, Sager R. Differential expression of elafin in human normal mammary epithelial cells and carcinomas is regulated at the transcriptional level. Cancer research. 1995;55(12):2537–41. [PubMed] [Google Scholar]
- 46.Aung G, Niyonsaba F, Ushio H, Ikeda S, Okumura K, Ogawa H. Elafin and secretory leukocyte protease inhibitor stimulate the production of cytokines and chemokines by human keratinocytes via MAPK/ERK and NF-kappaB activation. Journal of dermatological science. 2011;63(2):128–31. doi: 10.1016/j.jdermsci.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 47.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103(2):239–52. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 48.Morton S, Davis RJ, Cohen P. Signalling pathways involved in multisite phosphorylation of the transcription factor ATF-2. FEBS letters. 2004;572(1–3):177–83. doi: 10.1016/j.febslet.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 49.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13 (15 Pt 1):4429–34. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 50.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA: a cancer journal for clinicians. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 51.Saidi A, Javerzat S, Bellahcene A, De Vos J, Bello L, Castronovo V, et al. Experimental anti-angiogenesis causes upregulation of genes associated with poor survival in glioblastoma. International journal of cancer Journal international du cancer. 2008;122(10):2187–98. doi: 10.1002/ijc.23313. [DOI] [PubMed] [Google Scholar]
- 52.Caruso JA, Hunt KK, Keyomarsi K. The neutrophil elastase inhibitor elafin triggers rb-mediated growth arrest and caspase-dependent apoptosis in breast cancer. Cancer research. 2010;70(18):7125–36. doi: 10.1158/0008-5472.CAN-10-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yokota T, Bui T, Liu Y, Yi M, Hunt KK, Keyomarsi K. Differential regulation of elafin in normal and tumor-derived mammary epithelial cells is mediated by CCAAT/enhancer binding protein beta. Cancer research. 2007;67(23):11272–83. doi: 10.1158/0008-5472.CAN-07-2322. [DOI] [PubMed] [Google Scholar]
- 54.Levanon K, Crum C, Drapkin R. New insights into the pathogenesis of serous ovarian cancer and its clinical impact. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26(32):5284–93. doi: 10.1200/JCO.2008.18.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karst AM, Drapkin R. Ovarian cancer pathogenesis: a model in evolution. J Oncol. 2010;2010:932371. doi: 10.1155/2010/932371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karst AM, Levanon K, Drapkin R. Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(18):7547–52. doi: 10.1073/pnas.1017300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levanon K, Ng V, Piao HY, Zhang Y, Chang MC, Roh MH, et al. Primary ex vivo cultures of human fallopian tube epithelium as a model for serous ovarian carcinogenesis. Oncogene. 2010;29(8):1103–13. doi: 10.1038/onc.2009.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw L, Wiedow O. Therapeutic potential of human elafin. Biochem Soc Trans. 2011;39(5):1450–4. doi: 10.1042/BST0391450. [DOI] [PubMed] [Google Scholar]
- 59.Ficarro SB, Zhang Y, Carrasco-Alfonso MJ, Garg B, Adelmant G, Webber JT, et al. Online nanoflow multidimensional fractionation for high efficiency phosphopeptide analysis. Molecular & cellular proteomics : MCP. 2011;10(11):O111 011064. doi: 10.1074/mcp.O111.011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vermeer PD, Bell M, Lee K, Vermeer DW, Wieking BG, Bilal E, et al. ErbB2, EphrinB1, Src kinase and PTPN13 signaling complex regulates MAP kinase signaling in human cancers. PloS one. 2012;7(1):e30447. doi: 10.1371/journal.pone.0030447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 62.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2009. [Google Scholar]
- 63.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.