Abstract
We analyzed the genetic diversity of Cylindrocarpon destructans isolates obtained from Korean ginseng (i.e., Panax ginseng) roots by performing virulence tests and nuclear ribosomal gene internal transcribed spacer (ITS) and mitochondrial small subunit (mt SSU) rDNA sequence analysis. The phylogenetic relationship analysis performed using ITS DNA sequences and isolates from other hosts helped confirm that all the Korean C. destructans isolates belonged to Nectria/Neonectria radicicola complex. The results of in vivo and ex vivo virulence tests showed that the C. destructans isolates could be divided into two groups according to their distinctive difference in virulence and the genetic diversity. The highly virulent Korean isolates in pathogenicity group II (PG II), together with foreign isolates from P. ginseng and P. quinquefolius, formed a single group. The weakly virulent isolates in pathogenicity group I, together with the foreign isolates from other host plants, formed another group and exhibited a greater genetic diversity than the isolates of PG II, as confirmed by the mt SSU rDNA sequence analysis. In addition, as the weakly virulent Korean isolates were genetically very similar to the foreign isolates from other hosts, they were likely to originate from hosts other than the ginseng plants.
Keywords: Cylindrocarpon destructans, Genetic diversity, Ginseng root rot, Panax ginseng, Pathogenicity
Cylindrocarpon destructans (teleomorph: Nectria/Neonectria radicicola), a soil-borne pathogenic fungus, can cause a primary root rot disease in ginseng (Panax ginseng and P. quinquefolius), reduce the yield of ginseng production, and result in great economic losses [1, 2]. In addition, C. destructans has been reported to lead to replant failure, due to its ability to survive in the soil for more than ten years after the harvest of ginseng [3]. C. destructans was originally described as Ramuraria destructans according to the root rot symptoms of American ginseng (P. quinquefolius) by Zinssmeister [4]. In South Korea, it had been identified only in ginseng, strawberries and peonies [5] since it was firstly discovered in ginseng roots by Chung [6], although it was shown to affect the roots of diverse woody plants and to cause one of the most severe fungal diseases worldwide [7, 8, 9, 10, 11].
A large number of pathogenic fungi in the genera of Cylindrocarpon, Fusarium, and Cylindrocladium, which belong to Nectriaceae (Hypocreales), show taxonomically close association with C. destructans [12]. Mantiri et al. [13] proposed the presence of approximately 125 species in the genus of Cylindrocarpon, and Booth [14] divided this genus into four groups, namely C. magnusianum, C. cylindroides, Nectria mammoidea, and C. destructans, according to the presence of microconidia and chlamydospores. Furthermore, Samuels and Brayford [15] reviewed the existing classification system and categorized Nectria radicicola, including the asexual generation of C. destructans, into three variations, known as var. radicicola (anamorph: C. destructans var. destructans), var. coprosmae (anamorph: C. destructans var. coprosmae), and var. macroconidiales (anamorph: C. macroconidiales), based on their morphological and culture characteristics.
Mantiri et al. [13] modified Booth's [14] classification and redefined the genus of Nectria into three clades (i.e., clade I: Nectria coccinea/galligena group; clade II: N. mammoidea/veuillotiana group; and clade III: N. radicicola group) through the sequence analysis of mitochondrial rDNA, and included C. destructans into clade III. Seifert et al. [16] conducted DNA sequence analysis of nuclear ribosomal internal transcribed spacer (ITS) region and partial β-tubline gene using C. destructans isolates from diverse hosts and closely related species. They reported that N. radicicola complex isolates in the clade III established by Mantiri et al. [13] could be further divided into subclades a and b, and that all the isolates from Korean and Japanese ginseng (P. ginseng) and Canadian ginseng (P. quinquefolius) are genetically close to each other and belong to subclade b. Recently, several attempts have been made to establish a new type of classification system for the Nectria/Neonectria complex and to analyze the genetic diversity using various morphological, pathogenic, and genetic analytical approaches [9, 17, 18, 19]. Although the genetic variation of Korean C. destructans population that causes root rot in P. ginseng has been evaluated by random amplified polymorphic DNA analysis [20, 21], the pathogenic and taxonomical characteristics of P. ginseng in Korea have been rarely studied. As the genetically distinct populations may show difference in host range, aggressiveness, and susceptibility to disease control treatment, a better understanding of the genetic and pathogenic variations in C. destructans will be important for developing suitable root rot disease management strategies [16].
In this study, ITS region and mitochondrial small subunit (mt SSU) rDNA which were frequently used for analyzing the genetic diversity of phytopathogenic fungi [22, 23] were sequenced from Korean ginseng isolates, and compared with those from the foreign isolates to locate the taxonomic position of the C. destructans group. The genetic diversity of C. destructans isolates was analyzed by evaluating their virulence both in vivo and ex vivo. The data obtained from this study will be useful for constructing effective treatment strategies against ginseng root rot diseases.
MATERIALS AND METHODS
Isolation of pathogenic fungus
In this study, C. destructans was isolated from diseased ginseng roots that demonstrated typical symptoms of root rot; the ginseng roots were obtained from the main ginseng-cultivating regions of South Korea (Table 1). The diseased tissues on the collected ginseng roots showing brown discoloration were washed in running water. The tissues of both healthy and rotten parts were sliced into 5-mm pieces, treated with 2% NaOCl for 1 min for surface disinfection, washed two or three times in sterilized water, and placed on filter paper for dehydration. The affected tissues, whose surface had been previously disinfected, were placed on water agar or on pentachloronitrobenzene agar medium [24] and cultured at 15℃ for seven days. Subsequently, the spores were isolated from the hypha grown on the tissues cultured in potato dextrose agar (PDA) medium at 15℃ for ten days. Individual spores were then isolated, inoculated, and cultured in PDA medium at 20℃ for 2 wk and reinoculated into PDA medium for virulence tests. Furthermore, their morphological characteristics on PDA media were observed for identification of the isolates.
Table 1.
A list of Korean ginseng (Panax ginseng) isolates of Cylindrocarpon destructans used in this study
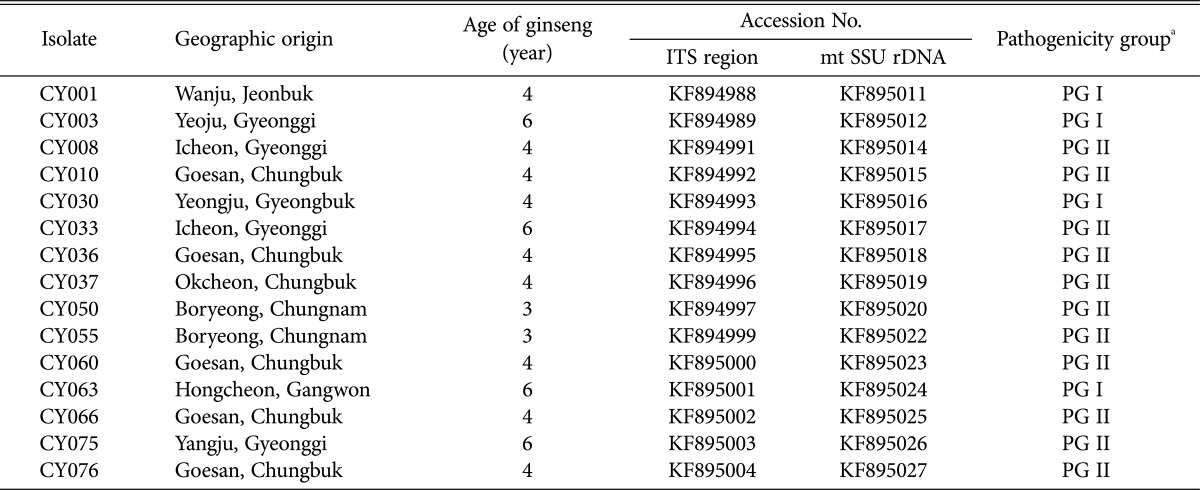
ITS region, internal transcribed spacer region; mt SSU rDNA, mitochondrial small subunit ribosomal DNA.
aPathogenicity group: PG I (lesion size < 8.1mm, diseased rate < 81%) and PG II (lesion size > 8.0mm, diseased rate > 80%).
Isolation of genomic DNA from C. destructans
After the pure culture, the isolates were inoculated into a potato dextrose broth medium and subject to stationary culture at 20℃ for ten days for isolating the genomic DNA. The mycelia were then harvested, split into 1.5-mL Effendorf tubes, lyophilized, and ground to extract the genomic DNA, as per the method of Doyle and Doyle [25].
Virulence test
Ex vivo and in vivo tests were performed to determine the virulence of C. destructans isolates obtained from the ginseng roots (Table 1, Fig. 1). In the ex vivo test, the surface of a 2-year-old ginseng plant was disinfected with 2% NaOCl. And the isolates cultured in PDA medium for 14 days were used as an inoculum. We placed a piece of paper towel wetted with sterilized water in a sealed container disinfected with 70% ethanol, fixed it onto the ginseng root pierced using plastic pipette tips (5 mm in diameter) containing the cultured mycelia, and stored it in an incubator for 4 wk, before measuring the size of the lesions. An average value of the lesion size was obtained to provide a mean value for the six replicate roots for each treatment. In the in vivo test, ginseng seedlings were dipped in the mycelia and spores suspension, which was prepared by suspending the ground isolates cultured at 20℃ for 14 days after inoculation into a PDA medium in 40 mL of sterilized water, and transplanted into the preplanting field for ginseng cultivation. Seven months later, an average value of diseased rate (%) was assessed to provide a mean value for the ten roots for each treatment. The ginseng isolates were divided into two groups according to the results of the virulence tests. The isolates showing an average lesion size less than 8.1 mm and diseased rate lower than 81% were categorized as pathogenicity group I (PG I); and those with an average lesion size more than 8.0 mm and diseased rate higher than 80% were classified as pathogenicity group II (PG II).
Fig. 1.
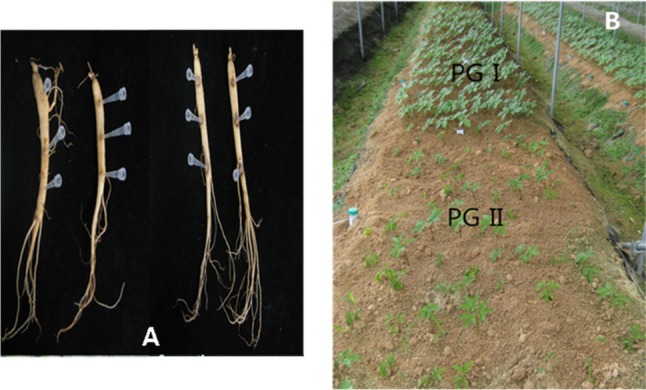
Optical images showing symptoms of root rot due to Cylindrocarpon destructans on wounded 2-year-old ginseng plants ex vivo (A), and field-grown ginseng plants affected by weakly (PG I) and highly (PG II) aggressive C. destructans isolates in vivo (B).
Genetic relatedness analysis
PCR amplification of approximately 600-bp region of ITS and mt SSU rDNA region was performed using ITS1 and ITS4 primers [26] and NMS1 and NMS2 primers [27], respectively. For each amplification, a 0.5 pmol primer, 2 ng of genomic DNA, 0.2mM deoxynucleotide triphosphate (dNTP), 10mM Tris-HCl, 50 mM KCl, 1.5mM MgCl2, and 2.5 units of Taq DNA polymerase were added to sterilized water to obtain a final volume of 50 µL. The PCR conditions used in analyzing the ITS region were as follows: an initial denaturation step at 95℃ for 3 min; followed by 35 cycles of 95℃ for 35 sec, 55℃ for 1 min, and 72℃ for 1 min; and a final elongation step at 72℃ for 8 min. The PCR conditions for mt SSU rDNA region analysis were as follows: an initial denaturation step at 95℃ for 3 min; followed by 34 cycles of 95℃ for 1min, 56℃ for 1 min, and 72℃ for 1 min; and a final elongation step at 72℃ for 4min. Each of the PCR products were analyzed by electrophoresis on a 1.5% agarose gel and stained with ethidium bromide for visualizing its amplification and size.
A Solgent PCR Purification Kit (Solgent Co., Ltd., Seoul, Korea) was used to purify the amplified PCR products, according to the manufacture's protocol and sequenced by Genotech Co., Ltd. (Daejeon, Korea). Sequences generated from the present study were deposited in Genbank (Table 1). The PHYDIT program ver. 3.2 [28] was used to align the sequences, and the unclearly aligned fragments were excluded from the analysis. Furthermore, PHYLIP 3.57c Package [29] was used to produce a neighbor-joining tree based on Kimura's two-parameter model [30], and an analysis of 1,000 bootstrap samples was carried out to assess its reliability.
RESULTS AND DISCUSSION
Virulence test
Virulence tests were performed on all the Korean ginseng isolates of C. destructans that formed white or brown colonies with microconidia, macroconidia, and chlamydospores on PDA media (data not shown). C. destructans isolates obtained from the diseased ginseng tissues can be divided into two groups based on their virulence (Table 1, Fig. 1) determined by the in vivo and ex vivo tests. In the in vivo test, an entire rotten root and weakened ginseng tissues were observed in the ginseng plants with withered leaves in the aerial part or in case of leaves failing to grow; and in such a case, normal ginseng growth was not observed, and the isolates were found to be highly virulent (PG II) after inoculation. Whereas, in the case where the root rot was found only in some parts of the plants, normal ginseng growth was observed, and weakly virulent isolates (PG I) were identified postinoculation. Kim [31] observed that the symptoms of root rot in ginseng growth varied with the soil types, plant ages, and cultivating seasons, and he assumed that they also varied with the differences in the virulence among the C. destructans isolates rather than with the environmental factors. Matuo and Miyazawa [7] reported that the C. destructans isolates obtained from tea plants and cedar trees were weakly virulent against ginseng seedlings, although the isolates from ginseng plants were highly virulent. In this study, remarkable difference in virulence was identified among the C. destructans isolates grown on P. ginseng, and the isolates could be clearly divided into two groups-PG I and PG II-on the basis of their virulence.
Analysis of genetic diversity
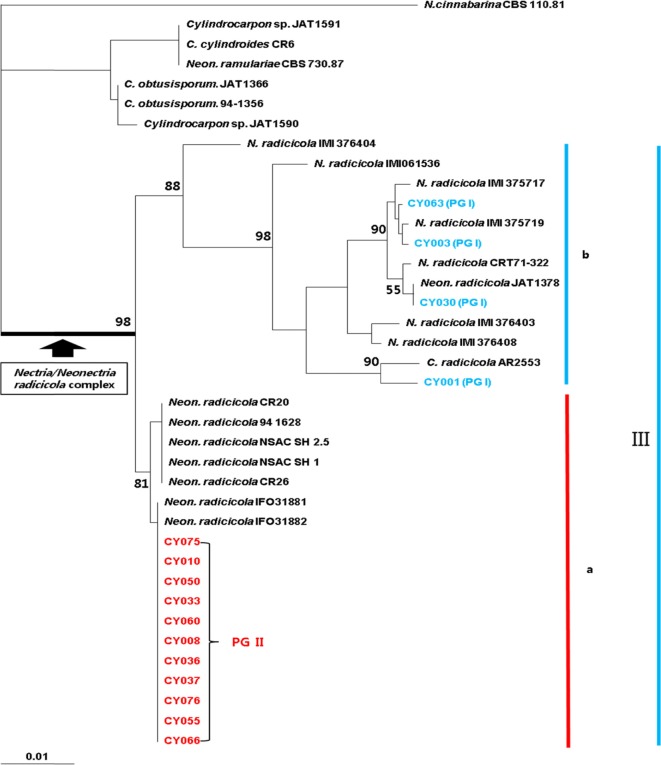
DNA sequence analysis of ITS region is often used to examine the genetic diversity of phytopathogenic fungi [22]. Seifert et al. [16] reported the presence of two groups, subclades a and b, in clade III, including the Nectria/Neonectria radicicola complex. In this study, the DNA sequences of ITS region from 15 C. destructans isolates of rotten ginseng roots collected from several different regions in Korea were used to analyze the genetic relatedness of C. destructans isolates with Nectria/Neonectria radicicola complex, as suggested by Seifert et al. [16] (Table 2, Fig. 2). All the Korean P. ginseng isolates of C. destructans, together with the isolates in clade III, formed a single group, with 98% bootstrap support. Among them, the highly virulent C. destructans (PG II) isolates from diverse ginseng-cultivation regions in Korea showed 100% similarity, and together with Japanese (IFO31881, IFO31882) and Canadian (NSAC SH 1, NSAC SH 2.5) ginseng isolates in clade III, formed the subclade a. Moreover, they showed 100% DNA sequence homology to the Japanese P. ginseng isolates, but not to the Canadian P. quinquefolius isolates. Though Seifert et al. [16] reported that all Korean, Japanese, and Canadian ginseng isolates belonged to subclade a in clade III, we observed 97.0~100% similarity among the weakly virulent P. ginseng isolates in PG I, which formed subclade b, along with the foreign isolates from diverse hosts. This finding indicates a great genetic diversity among the weakly virulent isolates, which is contrary to the results observed for the highly virulent isolates that form a single group with a high genetic homology.
Table 2.
Identification of reference isolates used in the molecular analysis
Fig. 2.
Phylogenetic analysis of Korean ginseng Cylindrocarpon destructans isolates with respect to the Nectria/Neonectria radicicola complex of Seifert et al. [16] by using internal transcribed spacer sequence region. The bootstrap analysis was performed with 1,000 replications.
Mantiri et al. [13] elucidated phylogenetic relationships among Neonectria species with Cylindrocarpon anamorphs using mt SSU rDNA region, and identified four well-supported clades or groups of the Neonectria complex. Based on this result, we analyzed the genetic relatedness between the DNA sequences of the PCR products obtained from Korean ginseng isolates of C. destructans and the DNA sequences of foreign Neonectria/Cylindrocarpon isolates and closely related species, which belonged to the Neonectria complex, as identified by GeneBank (Table 2, Fig. 3). In the analysis of the ITS region, both weakly (PG I) and highly (PG II) virulent isolates formed a monophyletic clade or group, and yet they formed different clusters in group III of Neonectria. Apart from the isolates in PG II, which showed 100% DNA sequence homology to each other and formed a specific group, the isolates in PG I showed 99.5~100% homology, and together with the foreign isolates from various hosts, formed an individual cluster.
Fig. 3.
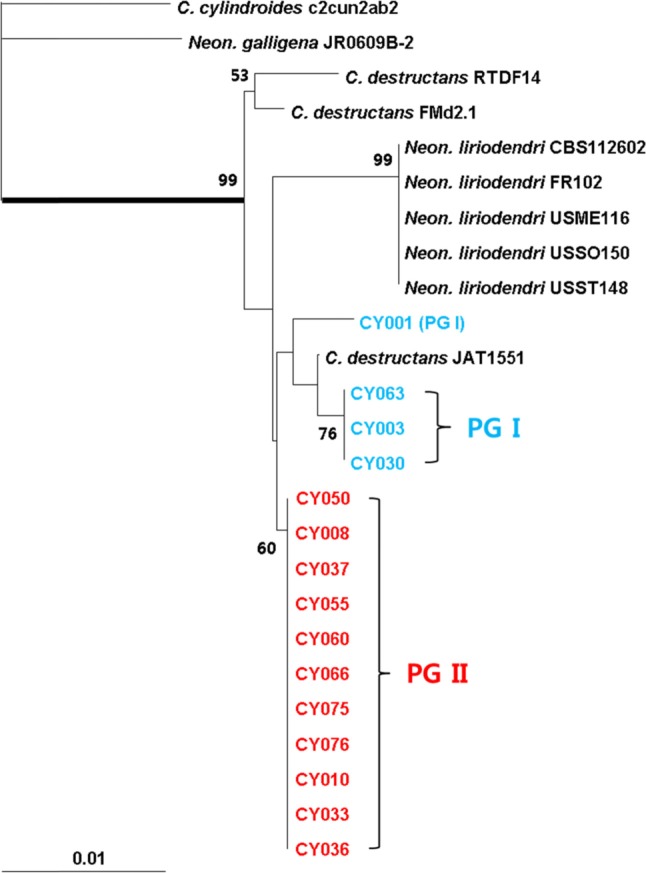
Phylogenetic analysis of other host plants and closely related isolates with respect to Korean ginseng Cylindrocarpon destructans isolates by using mitochondrial small subunit rDNA. The bootstrap analysis was performed with 1,000 replications.
Cabral et al. [17] recently renamed the Nectria/Neonectria radicicola complex as Ilyonectria radicicola complex after analyzing its morphological characteristics and multi-gene relatedness, and reclassified the fungi in this complex into 15 species under the genus of Ilyonectria based on their morphological characteristics and genetic diversity. They also reported the presence of genetically diverse fungi within the group of P. quinquefolius isolates, which were originally divided into four species: I. mors-panacis, I. robusta, I. panacis, and I. crassa. In particular, both Japanese P. ginseng isolates and those highly virulent to P. quinquefolius were included in I. mors-panacis. The Japanese isolates (IFO31881) in this study showed 100% hereditary homology to the highly virulent Korean isolates in PG II group. Thus, they were expected to belong to I. mors-panacis, as per the classification scheme of Cabral et al. [17].
In this study, we found that the P. ginseng isolates collected from several regions in Korea could be divided into two distinct groups on the basis of their virulence, although they all belonged to the same complex of Nectria/Neonectria radicicola. All the highly virulent isolates were genetically homologous and formed a specific single group, whereas the weakly virulent isolates were genetically similar to the isolates from other hosts and showed a significant genetic diversity. These results suggest that the domestic group of weakly virulent isolates may possibly originate from hosts other than the ginseng plants.
ACKNOWLEDGEMENTS
This study was supported by the research grant funded by Chungnam National University, Korea.
References
- 1.Chung HS. Studies on Cylindrocarpon destructans (Zins.) Scholten causing root rot of ginseng. Rep Tottori Mycol Inst Jpn. 1975;12:127–138. [Google Scholar]
- 2.Rahman M, Punja ZK. Factors influencing development of root rot on ginseng caused by Cylindrocarpon destructans. Phytopathology. 2005;95:1381–1390. doi: 10.1094/PHYTO-95-1381. [DOI] [PubMed] [Google Scholar]
- 3.Kang SW, Yeon BY, Hyeon GS, Bae YS, Lee SW, Seong NS. Changes of soil chemical properties and root injury ratio by progress years of post-harvest in continuous cropping soils of ginseng. Korean J Med Crop Sci. 2007;15:157–161. [Google Scholar]
- 4.Zinssmeister CL. Ramularia root rots of ginseng. Phytopathology. 1918;8:557–571. [Google Scholar]
- 5.Korean Society of Plant Pathology. List of plant diseases in Korea. 5th ed. Suwon: Korean Society of Plant Pathology; 2009. p. 91.p. 284.p. 288. [Google Scholar]
- 6.Chung HS. Ginseng disease. Research reports of the Korean Society of Plant Protection. Seoul: Korean Society of Plant Protection; 1979. pp. 107–144. [Google Scholar]
- 7.Matuo T, Miyazawa Y. Scientific name of Cylindrocarpon sp. causing root rot of ginseng. Ann Phytopathol Soc Jpn. 1984;50:649–652. [Google Scholar]
- 8.Domsch KH, Gans W, Andreson TH. Compendium of soil fungi. Vol. 1. Nectria radicicola Gerlach & L. Nilsson. New York: Academic Press; 1980. pp. 503–508. [Google Scholar]
- 9.Halleen F, Fourie PH, Crous PW. A review of black foot disease of grapevine. Phytopathol Mediterr. 2006;45:S55–S67. [Google Scholar]
- 10.Tewoldemedhin YT, Mazzola M, Mostert L, McLeod A. Cylindrocarpon species associated with apple tree roots in South Africa and their quantification using real-time PCR. Eur J Plant Pathol. 2011;129:637–651. [Google Scholar]
- 11.Abreo E, Martinez S, Bettucci L, Lupo S. Morphological and molecular characterisation of Campylocarpon and Cylindrocarpon spp. associated with black foot disease of grapevines in Uruguay. Aust Plant Pathol. 2010;39:446–452. [Google Scholar]
- 12.Rossman AY, Samuels GJ, Rogerson CT, Lowen R. Genera of Bionectriaceae, Hypocreaseae, and Nectriaceae (Hypocreales, Ascomycetes) Stud Mycol. 1999;42:1–248. [Google Scholar]
- 13.Mantiri FR, Samuels GJ, Rahe JE, Honda BM. Phylogenetic relationships in Neonectria species having Cylindrocarpon anamorphs inferred from mitochondrial ribosomal DNA sequences. Can J Bot. 2001;79:334–340. [Google Scholar]
- 14.Booth C. The genus Cylindrocarpon. CAB Int Mycol Inst Mycol Pap. 1966;104:1–56. [Google Scholar]
- 15.Samuels GJ, Brayford D. Variation in Nectria radicicola and its anamorph, Cylindrocarpon destructans. Mycol Res. 1990;94:433–442. [Google Scholar]
- 16.Seifert KA, McMullen CR, Yee D, Reeleder RD, Dobinson KF. Molecular differentiation and detection of ginseng-adapted isolates of the root rot fungus Cylindrocarpon destructans. Phytopathology. 2003;93:1533–1542. doi: 10.1094/PHYTO.2003.93.12.1533. [DOI] [PubMed] [Google Scholar]
- 17.Cabral A, Groenewald JZ, Rego C, Oliveira H, Crous PW. Cylindrocarpon root rot: multi-gene analysis reveals novel species within the Ilyonectria radicicola species complex. Mycol Prog. 2012;11:655–688. [Google Scholar]
- 18.Chaverri P, Salgado C, Hirooka Y, Rossman AY, Samuels GJ. Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs. Stud Mycol. 2011;68:57–78. doi: 10.3114/sim.2011.68.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo J, Zhuang WY. Three new species of Neonectria (Nectriaceae, Hypocreales) with notes on their phylogenetic positions. Mycologia. 2010;102:142–152. doi: 10.3852/08-224. [DOI] [PubMed] [Google Scholar]
- 20.Jun NJ. Study on new host range and genetic diversity for ginseng root rot pathogen, Cylindrocarpon destructans in Korea [dissertation] Daejeon: Chungnam National University; 2008. [Google Scholar]
- 21.Seo MW, Kim SI, Song JY, Kim HG. Genetic diversity of Korean Cylindrocarpon destructans based on virulence assay and RAPD analysis. Korean J Mycol. 2011;39:16–21. [Google Scholar]
- 22.O'Donnell K. Ribosomal DNA internal transcribed spacers are highly divergent in the phytopathogenic ascomycete Fusarium sambucinum (Gibberella pulicaris) Curr Genet. 1992;22:213–220. doi: 10.1007/BF00351728. [DOI] [PubMed] [Google Scholar]
- 23.O'Donnell K, Nirenberg HI, Aoki T, Cigelnik E. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience. 2000;41:61–78. [Google Scholar]
- 24.Punja ZK. Fungal pathogens of American ginseng (Panax quinquefolium) in British Columbia. Can J Plant Pathol. 1997;19:301–306. [Google Scholar]
- 25.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 26.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 27.Li KN, Rouse DI, German TL. PCR primers that allow intergeneric differentiation of ascomycetes and their application in Verticillium spp. Appl Environ Microbiol. 1994;60:4324–4331. doi: 10.1128/aem.60.12.4324-4331.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chun J. Computer-assisted classification and identification of actinomycetes [dissertation] Newcastle upon Tyne: University of Newcastle upon Tyne; 1995. [Google Scholar]
- 29.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 30.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 31.Kim SI. Characteristics of occurrence and cultural, chemical control of ginseng root rot [dissertation] Daejeon: Chungnam National University; 2006. [Google Scholar]