Summary
Mutations in p53 and RAS potently cooperate in oncogenic transformation and correspondingly these genetic alterations frequently coexist in pancreatic ductal adenocarcinoma (PDA) and other human cancers. Previously we identified a set of genes synergistically activated by combined RAS and p53 mutations as frequent downstream mediators of tumorigenesis. Here, we show that the synergistically activated gene Plac8 is critical for pancreatic cancer growth. Silencing of Plac8 in cell lines suppresses tumor formation by blocking autophagy, a process essential for maintaining metabolic homeostasis in PDA, and genetic inactivation in an engineered mouse model inhibits PDA progression. We show that Plac8 is a critical regulator of the autophagic machinery, localizing to the lysosomal compartment and facilitating lysosome-autophagosome fusion. Plac8 thus provides a mechanistic link between primary oncogenic mutations and the induction of autophagy, a central mechanism of metabolic reprogramming, during PDA progression.
Keywords: KRAS, p53, pancreatic ductal adenocarcinoma, mouse model, autophagosome, lysosome, Rab7, ATG12
Introduction
Pancreatic ductal adenocarcinoma (PDA) depends on a marked reprogramming of metabolic pathways, including the acquisition of autophagy dependence, for tumor cell survival and growth (Guo et al., 2011; Lock et al., 2011; Yang et al., 2011; Ying et al., 2012). Autophagy (or macro-autophagy) is the process of auto-digestion that recycles damaged organelles and enables cells to combat oxidative and nutrient stress and appears to have a context-dependent role in cancer. A present model suggests that autophagy has a dual role in cancer; suppressing cancer progression at its earliest stages, while allowing advanced cancers to meet metabolic demands (Kimmelman, 2011; White, 2012). Consistent with this, PDA and other tumor cell lines show constitutive basal autophagy in normal growth media, and pharmacologic or genetic blockade of autophagy strongly impairs tumor growth of xenograft models (Guo et al., 2011; Lock et al., 2011; O’Dell et al., 2012; Yang et al., 2011).
The spectrum of mutational events in PDA is well defined with activating RAS mutations present in most tumors (93%) and common inactivating mutational events in TP53, CDKN2A, and SMAD4 tumor suppressors and associated pathway components (Biankin et al., 2012). Analysis of human PDA pathological precursors, together with studies in genetically engineered mice, have established activating RAS mutations as a critical early events in PDA pathogenesis with cooperating loss of tumor suppressors genes occurring later (Aguirre et al., 2003; Bardeesy et al., 2006a; Hingorani et al., 2003; Hruban et al., 2001). How these mutational events in PDA cause autophagy dependence as well as the timing of autophagy activation in the course of cancer progression, have not been established. Among the commonly mutated genes in PDA, RAS has been associated with autophagy activation, though not all RAS mutant tumor demonstrate autophagy dependence (Guo et al., 2011; Yang et al., 2011). Inactivation of the p53-p19Arf tumor suppressor axis has also been implicated in autophagy control with differing effects depending on the experimental context (Balaburski et al.; Feng et al., 2005; Pimkina and Murphy, 2009; Tasdemir et al., 2008). The roles of these primary oncogenic mutations in causing autophagy activation in PDA, and a molecular basis for this phenomenon, thus remain unclear.
Previously we have shown that genes regulated synergistically in response to common cooperating oncogenic mutations found in PDA (RAS activation and functional loss of p53) are causally linked to the cancer phenotype. These genes, termed ‘cooperation response genes’ (CRG), encompass a spectrum of cellular processes down-stream of oncogenic mutations that are required for RAS/p53 mediated tumorigenesis (McMurray et al., 2008; Smith and Land, 2012). Here, we show that one such CRG, Plac8 is up-regulated by cooperating RAS and p53 mutations in PDA and critical to the growth of RAS/p53 mutant tumors by sustaining autophagy, via enabling autophagosome-lysosome fusion. Thus Plac8 provides a mechanistic link between the cooperation of two common pancreatic cancer causing gene mutations, KRAS and p53, and the elevated rates of autophagy critical to tumor growth.
Results
Plac8 expression is induced in response to Ras and p53 mutations and is critical for pancreatic tumor growth
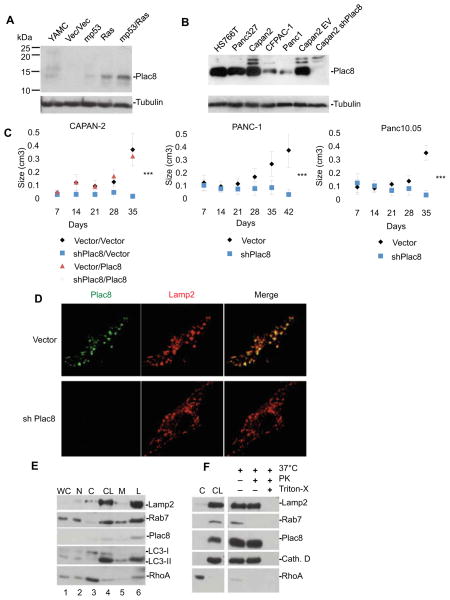
We have previously identified Plac8, a 112 amino acid cysteine-rich protein that is up-regulated at the RNA level in PDA (Buchholz et al., 2005; Lowe et al., 2007), as critical to malignant cell transformation mediated by activated RAS (RasV12) and mutant p53 (mp53) (McMurray et al., 2008). The biochemical function of Plac8 is unknown but this gene has been linked to efficient killing of phagocytosed bacteria by neutrophils, where it is enriched in a modified form of lysosomes, known as the granular fraction, critical for intracellular anti-microbial action (Ledford et al., 2007). We confirmed that Plac8 protein levels are induced synergistically in response to combined expression of mp53 and RasV12 in young adult mouse colon (YAMC) cells (Figure 1A) and can be detected in human pancreatic cancer cell lines (Figure 1B). Notably, we found that shRNA-mediated knock down of Plac8 virtually abolished tumorigenicity of the CAPAN-2, Panc-1, and Panc10.05 human PDA cell lines implanted into immuno-compromised mice (Figure 1C and Figure S1). Thus, Plac8 is cooperatively induced in response to mutations in RAS and p53, and is essential for the cancer phenotype of human PDA cell lines.
Figure 1. Plac8 expression induced by oncogenic mutations is critical for growth of human xenograft PDA cell lines and is localized to the lysosome compartment.
A) Immunoblotting of murine YAMC, vector, mp53, Ras, and and mp53/Ras transformed cells with anti-Plac8 antibody. B) Immunoblotting of human PDA cell lines including CAPAN-2 cells stably infected with Plac8shRNA expressing vector or control with anti-Plac8 antibody and β-tubulin (loading control) C) Vector control and Plac8 KD CAPAN-2 cells were injected into CD-1 nude mice, and vector control and Plac8 KD PANC-1 and Panc10.05 cell lines were injected into NOD/SCID mice. Data are presented as means +/− SD (Significance by student’s T-test, ***p<0.001) D) mp53/Ras murine cells were fixed and stained using anti-Plac8 and anti-Lamp2 antibodies, and imaged by confocal microscopy. E) Sub-cellular fractionation and immunoblotting of mp53/Ras transformed murine cells for Plac8, lysosomal proteins Rab7 and Lamp2, autophagosomal protein LC3, and a cytosolic control RhoA. Lanes are as follows; WC(1): whole cell lysate, N(2): nuclear fraction, C(3): cytosolic fraction, CL(4): crude lysosomal fraction, M(5): microsomal fraction, L(6): lysosomal fraction. F) Cytosolic fractions (C) and crude lysosomal fractions (CL) were isolated from mp53/Ras murine cells and the crude lysosomal fraction subjected to a Proteinase K protection assay. Lysosomes were, treated with Proteinase, or treated with Proteinase K and triton-X to dissolve the lysosomal membrane and immunoblotted for the external lysosomal protein Rab7, internal lysosomal proteins Lamp2 and Cathepsin D, and Plac8. RhoA served as a cytosolic fraction control.
Plac8 is predominantly located to the lysosome
The sub-cellular location of Plac8 in epithelial cells was established by immunostaining of mp53/Ras cells with Plac8-specific antibodies which showed predominantly punctate peri-nuclear staining sensitive to Plac8 knock down and partial co-localization with staining for the lysosomal protein Lamp2 (Figure 1D). Correspondingly, sub-cellular fractionation experiments indicated enrichment of Plac8 protein together with lysosomal proteins Lamp2 and Rab7 (Figure 1E) (Eskelinen et al., 2002; Gutierrez et al., 2004; Huynh et al., 2007; Jager et al., 2004). As observed for the intra-lysosomal proteins Lamp2 and Cathepsin D, Plac8 is resistant to proteinase K digestion, while the external lysosomal protein Rab7 is degraded (Figure 1F). Thus, in epithelial cells, Plac8 appears to be a predominantly intra-vesicular protein, with significant localization to the lysosomal compartment.
Plac8 facilitates autophagosome-lysosome (AL) fusion
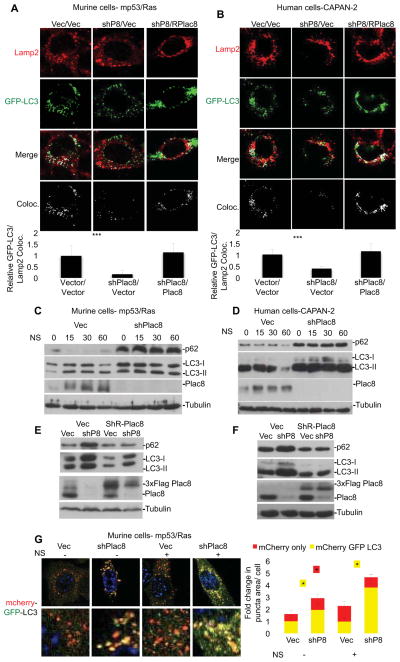
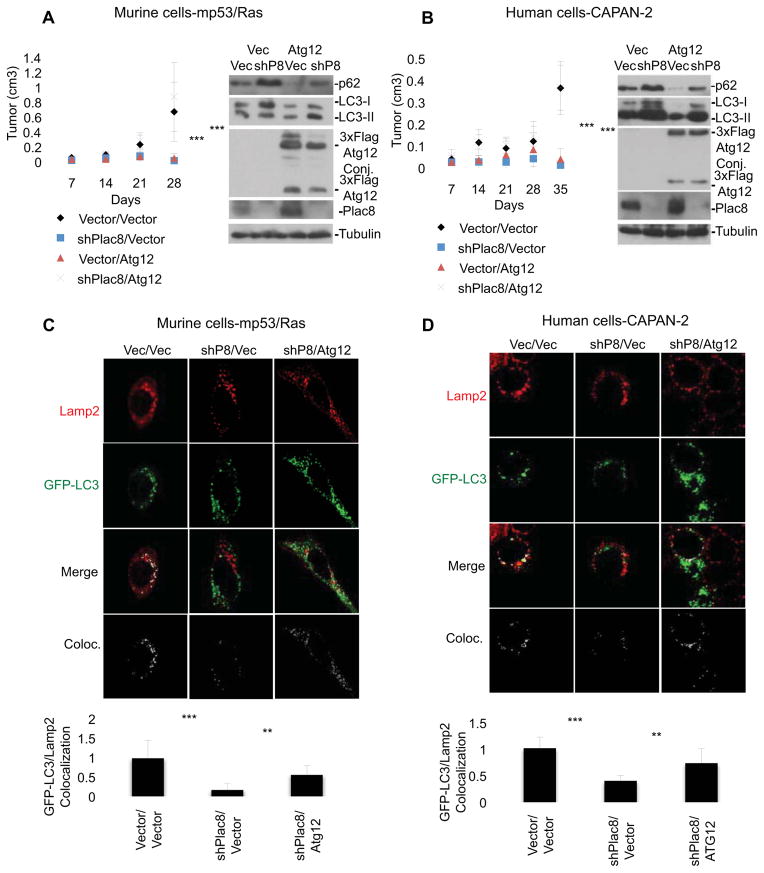
Autophagy is induced upon nutrient stress in normal cells, whereas in PDA cells autophagy is constitutively activated in the presence of nutrients (Yang et al., 2011). The mediators of this activation are unknown. As key late steps of autophagy involve autophagosome maturation and lysosomal fusion leading to autolysosome formation, we hypothesized that the localization of Plac8 to lysosomes may relate to a role in autophagy. In particular, we wished to address whether Plac8 regulates lysosomal function and/or fusion with degradative endosomes, such as phagosomes (as is the case for Plac8 function in neutrophils) or autophagosomes. To test the impact of Plac8 on AL fusion we compared co-localization of the GFP-labeled microtubule associated protein 1 light chain 3 (LC3), a marker of autophagy that associates with the autophagosome membrane after processing (Klionsky et al., 2008), and the lysosomal marker Lamp2 in control and Plac8 KD cells by confocal analysis (Morselli et al., 2010). Plac8 KD resulted in a ~80% reduction in GFP-LC3/Lamp2 co-localization in murine mp53/Ras cells and CAPAN-2 PDA cells indicating a reduction in AL fusion (Figure 2A & 2B). This effect was specific to the reduction in Plac8 levels, as fusion was rescued by the expression of an shRNA-resistant Plac8 in either cell type (Figure 2A & 2B). Moreover, consistent with our interpretation that Plac8 facilitates AL fusion Plac8 KD resulted in accumulation of autophagy markers p62 and LC3 both in normal growth medium and in nutrient- deplete HBSS (Figure 2C & 2D), with p62 and LC3 reverting to control levels in presence of a shRNA-resistant Plac8 protein (Figure 2E & 2F).
Figure 2. Plac8 KD results in the accumulation of autophagosomes and autophagy markers by inhibiting autophagosome-lysosome fusion.
A) Murine mp53/Ras transformed and B) human CAPAN-2 cells expressing GFP-LC3 and Plac8 shRNA, or Plac8 shRNA with exogenous shRNA resistant Plac8 were nutrient starved in HBSS for 15 min, fixed and stained for Lamp2. Fluorescence quantified by ImageJ. Relative ratios of autophagosomes fused with lysosomes per total autophagosomes are shown (+/− SD, ***p<0.001, determined by student’s T-test; n≥50 cells per cell line). C&D) Vector control (Vec) or Plac8 shRNA expressing mp53/Ras (C) and CAPAN-2 cells (D) were nutrient starved and immunoblotted for p62, LC3, and Plac8 (β-tubulin loading control). Lysates of mp53/Ras (E) and CAPAN-2 cells (F) expressing Vec, Plac8 shRNA, exogenous 3xFlag-tagged, shRNA-resistant Plac8, or Plac8 shRNA with exogenous 3xFlag-tagged, shRNA-resistant Plac8 were immunoblotted for p62, LC3, Rab7, and Plac8. G) mp53/Ras cells expressing mCherry-EGFP-LC3 and Vec or Plac8 shRNA were nutrient starved (NS) in HBSS or left untreated, fixed and imaged via confocal microscopy. Representative images show a merge of red and green channels (yellow represents colocalization, i.e. autophagosomes; red indicates autolysosomes). Fluorescence intensities quantified by ImageJ. Fold changes in punctae area/cell are indicated (mean +/− SD, student’s T-test: *p<0.05; n≥100 cells/cell line).
Diminished AL fusion following Plac8 KD should lead to a reduction in autophagic flux as indicated by accumulation of autophagosomes. This was confirmed using the mCherry-EGFP-LC3 reporter as a probe to estimate the relative numbers of autophagosomes and autolysosomes, as indicated by yellow (green+red) and red fluorescence, respectively (Kimura et al., 2007) in both normal growth medium and in nutrient-depleted conditions (Fig 2G). In addition, transmission electron microscopy revealed induction of vesicular structures enveloped by multiple membranes harboring cytosolic content consistent with accumulated autophagosomes in Plac8 knock-down (KD) cells that were only rarely found in vector control cells (Figure S2).
Further evidence indicating a role of Plac8 in facilitating autophagic flux comes from experiments in which we introduced exogenous Plac8 into YAMC cells expressing activated Ras. Exogenous Plac8 diminished the abundance of autolysosomes, as indicated by decreased red fluorescence of the mCherry-EGFP-LC3 reporter (Figure S3A). In addition, levels of endogenous p62 and LC3II proteins were diminished in reporter-free cells (Figure S3B). In contrast, all these indicators accumulated to levels above controls in presence of chloroquine (CQ), an inhibitor of lysosomal function acting via alkalinization of lysosomal pH (Klionsky et al., 2012) (Figure S3 A&B). Similarly, both the numbers of endogenous LC3 puncta and their co-localization with Lamp2 increased significantly in presence of CQ and exogenous Plac8 (Figure S3C). Together, this suggests a scenario in which Plac8 facilitates autophagic flux. Plac8 expression has minimal impact on p62 and LC3II protein levels in parental YAMC cells and in mp53 cells, i.e. YAMC cells expressing mutant p53 in absence of activated Ras (Figure S3D&E). Plac8 thus collaborates with activated Ras to elevate autophagic flux, though this is not sufficient to cause tumor growth in mice (Figure S3F).
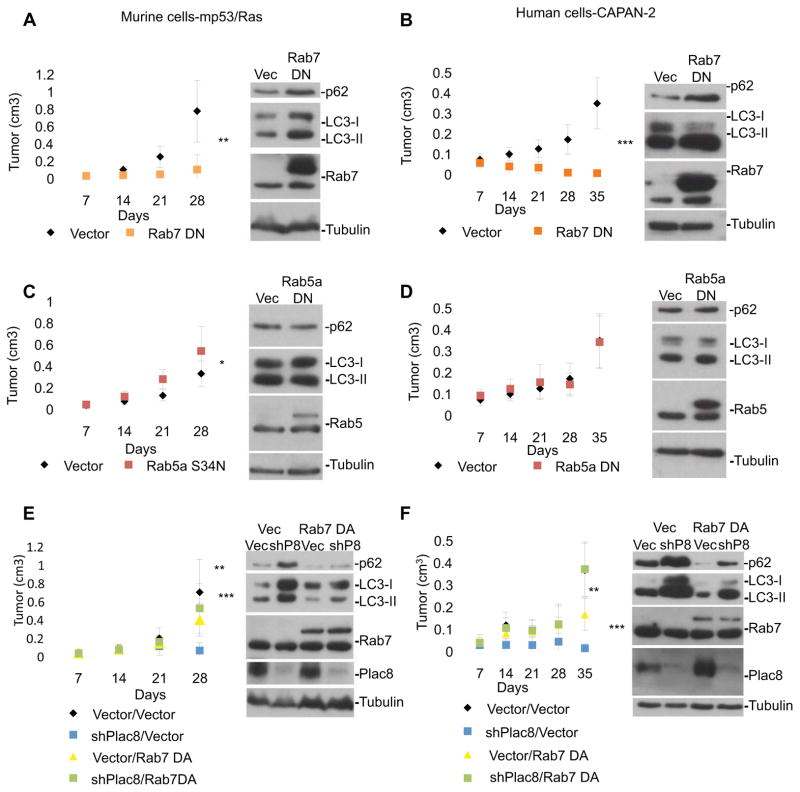
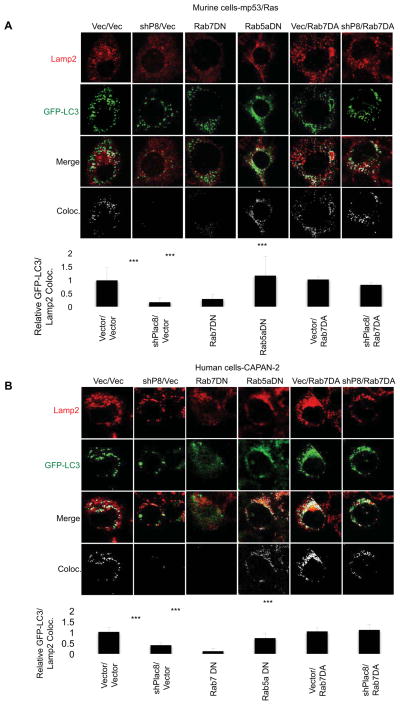
AL fusion requires the Ras-like GTPase Rab7 and is blocked by Rab7-T22N, a dominant-negative mutant (Rab7DN), thus leading to autophagosome accumulation (Ganley et al., 2011; Gutierrez et al., 2004; Jager et al., 2004). Similarly, we found that expression of Rab7DN prevented tumor growth in murine and human tumor cells, while inducing both accumulation of p62 and LC3 and decreased co-localization of Lamp2 and GFP-LC3 (Figure 3A,B and 4), consistent with a block in AL fusion. In contrast, expression of a dominant-negative mutant of Rab5a, a gene critical to early endocytosis/phagocytosis (Bucci et al., 1992; Chen and Wang, 2001; Dinneen and Ceresa, 2004), did not impact AL fusion, p62, or LC3 levels indicating that inhibition of Rab5a signaling and early endocytosis did not effect autophagy (Figure 3C,D and 4A,B). As expected, Rab5aDN inhibited endocytosis, as indicated by a decrease in uptake of fluorescently labeled dextran in the same cells, further suggesting that Rab5a-mediated endocytosis is non-essential for the tumor growth among these cells (Figure S4).
Figure 3. Activated Rab7 rescues cancer phenotype in Plac8 KD cells.
Murine mp53/Ras cells (A, C, E) and human CAPAN-2 cells (B, D, F) were grafted into CD-1 nude mice and tumor growth was monitored. Mean values of tumor volume +/− SD and student’s T-test at final time-point are indicated (*** p<0.001; ** p<0.01; *p<0.05; n≥9 tumors per condition). Cells expressing 3xFlag-tagged Rab7DN (Rab7T22N) or vector control (Vec) (A&B), 3xFlag-tagged Rab5a DN (Rab5aS34N) or Vec (C&D), and 3xFlag-tagged Rab7 DA,, Plac8 shRNA, Plac8 shRNA with 3xFlag-tagged Rab7 DA or Vec (E&F) were used. Corresponding cell lysates were immunoblotted for p62, LC3, Rab5 or Rab7 (A–F), and Plac8 in E&F (β-tubulin loading control).
Figure 4. Activated Rab7 rescues autophagosome-lysosome fusion in Plac8 KD cells.
Murine GFP-LC3-mp53/Ras cells (A) and human GFP-LC3-CAPAN-2 cells (B) expressing Vector control, Plac8 shRNA, 3xFlag-tagged Rab7DN, 3xFlag-tagged Rab5aDN, 3xFlag-tagged Rab7DA, or Plac8 shRNA and 3xFlag-tagged Rab7DA were nutrient starved in HBSS for 15 min, fixed, stained for Lamp2 and imaged via confocal microscopy. Co-localization was determined and quantified by ImageJ. The ratios of autophagosomes fused with lysosomes (co-localized signal)/total autophagosomes are indicated (mean +/− SD; student’s T-test; ***p<0.001; n≥50 cells/cell line).
Given the phenotypic similarities in the effects of Plac8 and Rab7 on autophagy and in promoting AL fusion we sought to determine if Rab7 activation could overcome the effects of Plac8 KD. Exogenous expression of dominant-active Rab7 Q67L (Rab7DA) in the presence of Plac8 KD reversed the tumor-inhibitory effect of Plac8 knockdown (Figure 3E&F), increased Lamp2 and GFP-LC3 co-localization (Figure 4A&B) and led to diminished levels of p62 and LC3 (Figure 3E&F), indicative of rescued AL fusion and autophagosomal processing. The ability of Rab7DA alone to inhibit tumor formation may thus be due to over-activation of autophagy, an effect compensated in the presence of Plac8 knockdown (Figure 3E&F).
In summary, our data indicate that knock-down of Plac8 interferes with AL fusion thus preventing downstream autophagosomal processing, while the overexpression of Plac8 can promote autophagic flux. Furthermore, activation of Rab7-dependent AL fusion bypasses the dependence on Plac8 suggesting that Plac8 functions in parallel to Rab7.
Upstream activation of autophagy can compensate for Plac8 knock down
In order to understand the relationship between Plac8 and autophagy activation we tested the capacity of upstream autophagy activation to compensate for Plac8 loss and diminished rates of AL fusion. We found that ectopic expression of Atg12, a ubiquitin-like protein covalently linked to ATG5 that forms a complex essential to autophagosome formation (Mizushima et al., 1998; Mizushima et al., 2001; Ohsumi, 2001), restored tumor formation capacity to Plac8 KD cells and increased AL fusion in these cells (Figure 5AD). Notably, in cells expressing ectopic Atg12 in the presence of Plac8 KD, p62 and LC3-II abundance is restored to levels found in unperturbed cells (Figure 5A&B), reflective of an autophagy rate restored to levels observed in the native cancer cells. In contrast, exogenous Atg12 expression in the absence of Plac8 KD was tumor inhibitory (Figure 5A&B), possibly due to pro-apoptotic functions (Rubinstein et al., 2011) or through hyper-activation of autophagy to levels inconsistent with survival. The reduction in steady-state levels of p62 and the increased ratio of LC3-II/LC3-I protein in cells expressing ectopic Atg12 support this latter hypothesis. Thus, while impaired AL fusion caused by Plac8 KD impedes PDA growth, upstream activation of the pathway can compensate for this. Moreover, consistent with an essential role of autophagy for the cancer phenotype, Atg12 KD inhibited tumor formation together with a decreased ratio of LC3-II/LC3-I and p62 accumulation (Tanida et al., 2002) in both mp53/Ras and CAPAN-2 cells (Figure S5).
Figure 5. Over-expression of Atg12 rescues Plac8 KD inhibition of cancer phenotype and autophagosome-lysosome fusion.
Murine mp53/Ras cells (A) and human CAPAN-2 cells (B) were grafted into CD-1 nude mice and tumor growth was monitored. Mean values of tumor volume +/− SD and student’s T-test at final time-point are indicated (***p<0.001; n≥9 tumors per condition). Cells expressing vector control (Vec), 3xFlag-tagged Atg12, Plac8 shRNA, or Plac8 shRNA and 3xFlag-tagged Atg12 were used. Corresponding cell lysates were immunoblotted for p62, LC3, Atg12 and Plac8. GFP-LC3-mp53/Ras cells (C) and GFP-LC3-CAPAN-2 cells (D) expressing Vec, Plac8 shRNA, or Plac8 shRNA and 3xFlag-tagged Atg12 were nutrient starved in HBSS for 15 min fixed, stained for Lamp2 and imaged via confocal microscopy. Co-localization was determined and quantified by ImageJ. The ratios of autophagosomes fused with lysosomes (co-localized signal)/total autophagosomes are indicated (mean +/− SD; student’s T-test ***p<0.001; **p<0.01; n≥50 cells/line).
Increased expression of Plac8 was noted in mp53/Ras cells in response to ectopic Rab7DA or ATG12 expression (Figures 3E, 3F, 5A and 5B). This may occur as a response to stress, or as part of the program stimulating autophagy, considering Plac8 is able to stimulate autophagic flux in the presence of activated Ras (Figure S3).
Synergistic induction of autophagy by activated Ras and mutant p53
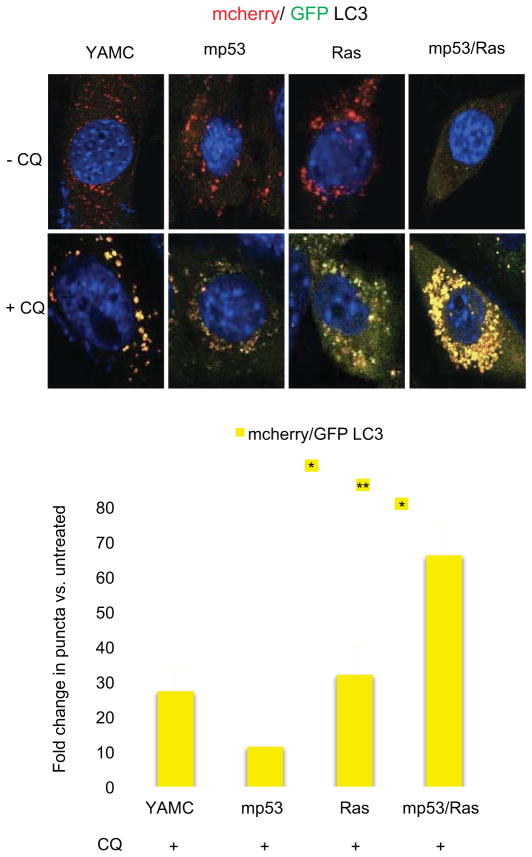
Mutations in both KRAS and p53 have been linked with alterations in autophagy, however, their combined impact as well as the associated mechanisms are unknown (Guo et al., 2011; Livesey et al., 2012; Lock et al., 2011; Morselli et al., 2011; Tasdemir et al., 2008; Yang et al., 2011). Given the identification of Plac8 as a gene cooperatively up-regulated by these mutations and its role in promoting AL fusion we asked if autophagy rates would be regulated similarly by concurrent oncogenic mutations. To test this possibility, we compared accumulation of LC3 punctae following chloroquine treatment in YAMC cells, mp53, Ras, and mp53/Ras cells using the mCherry-EGFP-LC3 reporter. Notably, a significant increase in punctae was detected only in mp53/Ras cells, indicating that autophagic flux is induced synergistically by mutant p53 and activated Ras (Figure 6).
Figure 6. Synergistic induction of autophagic flux by mutant p53 and activated Ras.
YAMC, mp53, Ras and mp53/Ras cells expressing mCherry-EGFP-LC3 were exposed to 25μM chloroquine (CQ) for 24 hours or left untreated, fixed, and imaged using confocal microscopy. Representative images show the merge of red and green channels, with yellow representing signal colocalization. Autophagosomes (yellow) and autolysosomes (red) quantified using ImageJ. Fold-changes in punctae areas/cell are indicated (mean +/− SD; student’s T-test; **p<0.01; *p<0.05; n≥50 cells/line).
Plac8 depletion does not alter lysosomal pH
Inhibitors of AL fusion such as bafilomycin A1 or CQ act via disrupting lysosomal pH (Klionsky et al., 2012; Yamamoto et al., 1998). Plac8 KD however had no effect on lysosomal pH (Figure S6A). Consistent with this finding lysosomal pH-dependent endocytic degradation of epidermal growth factor receptors (EGFR) (Ganley et al., 2011) was unaffected by Plac8 KD (Figure S6B). Similarly, Plac8 KD has no detectable effect on endocytosis, as visualized by cellular dextran uptake (Figure S6C). Taken together, our data suggest that Plac8 functions via mechanisms that modulate rates of AL fusion without affecting lysosomal pH or endocytic degradation.
Inhibition of autophagosome-lysosome fusion by CQ has also been associated with an increase in cellular reactive oxygen species (ROS) suggesting a role for autophagy in coping with oxidative stress (Yang S, 2011). While we found CQ-mediated increases of ROS levels in mp53/Ras cells, Plac8 KD had no impact on cellular ROS, as detectable by DCF staining (Figure S6 D&E), suggesting that inhibition of autophagosome-lysosome fusion is not necessarily associated with increases in cellular ROS. Conversely, induction of cellular ROS following CQ exposure thus may be unrelated to modulation of autophagic flux.
Mutation of Plac8 extends survival in a Kras-p53 PDA engineered mouse model
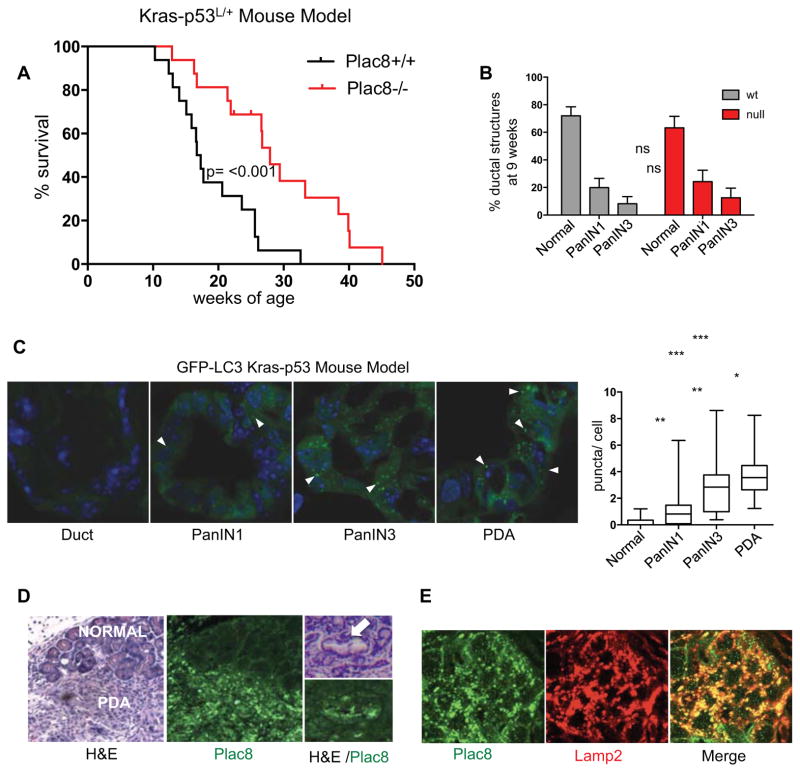
To extend our findings in xenograft models, we evaluated the importance of Plac8 in PDA progression and biology using a Pdx1-Cre; LSL-KrasG12D; p53L/+ genetically engineered mouse PDA model. This model exhibits successive stages of cancer progression (referred to as pancreatic intraepithelial neoplasia or PanIN I-III) leading to PDA, that typifies the human disease. Tumor progression is associated with loss of the wild-type p53 allele, and thus all ensuing tumors are p53 null (Bardeesy et al., 2006a). Cohorts of Pdx 1-Cre; LSL-KrasG12D; p53L/+ mice with concurrent germ-line Plac8+/+, or Plac8−/− mutations, hence-forth referred to as PDA-Plac8wt and PDA-Plac8null were created using established strains (Bardeesy et al., 2006a; Ledford et al., 2007). Mice were born at expected mendelian ratios and were indistinguishable from each other with regards to outward appearance and activity through the first 8 weeks of life as expected (Ledford et al., 2007). Individuals were followed longitudinally until signs of illness necessitated necropsy (see Methods). Overall survival of the PDA-Plac8null cohort (OS 27.9 weeks) was significantly longer than the PDA-Plac8wt cohort (OS 17.0 weeks, p=0.0006) (Figure 7A) demonstrating in vivo that genetic inactivation of Plac8 impedes cancer progression and resulting death. Thus Plac8 has a central role in PDA pathogenesis.
Figure 7. Plac8 mutation impedes advanced tumor progression in the Kras-p53 mouse PDA model.
A) Kaplan-Meier analysis of cohorts of Pdx1-Cre;LSL-KrasG12D;p53L/+ mice with concurrent germ-line Plac8+/+ or Plac8−/− mutations. Overall survival is increased in Plac8null cohort (27.9 weeks) compared with Plac8wt (OS 17. weeks, p=0.0006). B) Whole pancreatic histological analysis of Pdx1-Cre;LSL-KrasG12D;p53L/+ mice with concurrent germ-line Plac8−/− mutations (n=6) vs. Plac8 wt (n=10) at 9 weeks of age demonstrated similar burdens of PanIN. Data are presented as a fraction of the total with confidence intervals. 3/10 wild-type compared to 0/6 Plac8−/− harbored advanced PDA. C) Epithelial cell GFP punctae (white arrows) harbored within normal ducts, pancreatic intraepithelial neoplasia I and III (PanIN I&III), and PDA (n=50 cells/lesion) were quantified from cohort of Pdx1-Cre; LSL-KrasG12D;p53L/+;GFP-LC3 mice (n=6 mice; significance determined by the Mann Whitney test ***p<0.001; **p<0.01; *p<0.05). D) Immunoflourescent-staining of Plac8 in normal pancreatic duct, PanIN (white arrow) and PDA from Pdx1-Cre; LSL-KrasG12D;p53L/+ mice. Adjacent H&E stained sections were evaluated to determine histologic classification. E) PDA tissue from Pdx1-Cre;LSL-KrasG12D;p53L/+ stained with Plac8 and Lamp2 antibodies, visualized with Alexa 488 (green) and Alexa 555 (red), respectively.
The increased survival of the PDA-Plac8null cohort could be due to an impact on tumor progression in the early stages of tumor initiation, namely the formation of PanIN precursor lesions, or in the later stages of fully advanced PDA growth. To differentiate these possibilities we conducted a histological survey of the pancreas of PDA-Plac8null and PDA-Plac8wt at 9 weeks of age just prior to the onset of lethal advanced tumors. Quantification of normal ducts, as well as, early and late PanIN revealed no significant differences in the prevalence of premalignant lesions between the groups (Figure 7B and Figure S7). Therefore, Plac8 loss has minimal effect on the early stages of tumor initiation, indicating that Plac8 rather contributes to the progression of advanced PanINs and PDA.
To gain further insight into the relationship between Plac8, autophagy control, and PDA pathogenesis, we crossed a GFP-LC3 transgenic strain with our Kras-p53 model, enabling us to measure autophagy in vivo (Kuma et al., 2004). In this model (GFP-LC3; Pdx1-Cre; LSL-KrasG12D; p53L/+), in which the wild-type p53 allele is lost during the course of PanIN progression towards cancer (Bardeesy et al., 2006a), we found an increase in LC3 punctae, indicating increased numbers of epithelial autophagosomes, with each successive histological stage of cancer progression, with highest levels in PDA, and virtually no punctae in normal ducts or low grade PanIN-1 (Figure 7C). Consistent with this finding, immunofluorescent-staining of histological sections indicated Plac8 expression in PanINs and PDA, whereas Plac8 staining is not detectable in normal pancreatic ducts. In addition, Plac8 staining co-localizes with Lamp2 staining, confirming lysosomal localization of Plac8 in PDA (Figure 7D&E) Taken together, the marked and specific activation of autophagy in advanced PDA matches well with the specific impact of Plac8 inactivation on the later stages of disease.
Discussion
Synergistic regulation downstream of cooperating oncogenic mutations has emerged as a reliable indicator of genes critical to malignant cell transformation in a variety of contexts including cancer-initiating cells (McMurray et al., 2008; Ashton et al., 2012). Here we identify such a gene, Plac8, as a novel regulator of autophagosome-lysosome fusion required for PDA growth, thus providing a mechanistic link between oncogenic mutations and the activation of autophagy in cancer. Activated by oncogenic KRAS and p53 loss-of-function, two of the most commonly occurring mutations in cancer, Plac8 expression is required for growth of human pancreatic cancer cells as xenografts in mice, as well as activation of autophagy in vitro and in vivo where we observe that both RAS and p53 mutations are critical for elevated autophagy rates. Our data also suggest that the role of Plac8 in facilitating autophagy is critical to the cancer phenotype, as the requirement of Plac8 for both tumorigenicity and autophagy can be compensated by over-expression of Atg12, a gene critical for autophagosome formation (Mizushima et al., 1998), or by constitutively activated Rab7, a gene encoding a GTP-binding protein stimulating autophagosome-lysosome fusion (Gutierrez et al., 2004; Jager et al., 2004). This is consistent with an interpretation that tumor growth of Plac8-deficient cells is compromised due to suboptimal autophagic flux. Notably, we also show that Plac8 may offer a potential therapeutic window and point of intervention, as Plac8 germ-line mutation in an engineered PDA model inhibits cancer progression and significantly improves survival while having a minimal impact on the overall fitness of the animals. This suggests that Plac8, and regulation of autophagosome-lysosome fusion, has specific relevance to regulation of autophagy during malignant cell transformation. Independent support for the oncogenic potential of the Plac8 gene comes from several in vivo genetic screens in which Plac8 has been identified repeatedly as a target for retroviral insertion mutagenesis, implicating its de-regulation as a driving force in carcinogenesis (Kool et al., 2010; Williams et al., 2006).
Plac8 is a lysosomal protein that facilitates autophagosome-lysosome fusion without affecting endocytic protein degradation and lysosomal pH. This is in contrast to Rab7 and SNARE proteins such as VAMP8, that impact both autophagosome-lysosome fusion and endocytic protein degradation (Furuta N, 2010; Gutierrez et al., 2004; Klionsky et al., 2012; Wong et al., 1998). The apparently minimal impact of Plac8 deficiency on normal physiologic processes (Ledford et al., 2007) fits well with the mutant phenotypes of several other genes involved in the autophagosome maturation including Lamp2 and Tecpr1 (Chen et al., 2012; Eskelinen et al., 2002; Ogawa et al., 2011; Tanaka et al., 2000). Unlike many Atg-family mutant mice which are not viable (Komatsu et al., 2005; Kuma et al., 2004), Plac8, Lamp2, and Tecpr1 mutant mice can all survive through adulthood, though all suffer from a diminished capacity to clear infections, due to defects in lysosomal function (Beertsen et al., 2008; Binker et al., 2007; Ledford et al., 2007; Ogawa et al., 2011). Thus, later stages of autophagy and autophagosome maturation may provide a variety of attractive therapeutic opportunities where inhibition of steps in this process are likely better tolerated by normal tissues but not by tumors that depend on a high autophagic rate.
Plac8 was identified originally as a leukocyte inhibitory factor regulated gene in the mouse uterus and also in genome-wide expression analysis of the placenta, from where its name is derived (Galaviz-Hernandez et al., 2003). It has been functionally ascribed to the regulation of a number of distinct cellular processes (Rogulski et al., 2005; Wu et al., 2010). Plac8 knockout mice have been found to suffer increased adiposity at advanced ages leading to the discovery of a role in brown fat differentiation through induction of C/EBPbeta expression (Jimenez-Preitner et al., 2011). Of note we did not find significant differences in levels of C/EBPbeta expression among PDA cell lines with or without Plac8. As our data points towards an extra-nuclear location in epithelial cells, within the lysosomal compartment where it facilitates autophagosome-lysosome fusion, it seems plausible that Plac8 may have differing roles among different cell types.
Here we find that concurrent mutation of KRAS and p53 is critical for maximal induction of autophagy, and correspondingly, we see a step-wise incremental increase in LC3 puncta in vivo with each histological stage through the course of PDA progression. Thus, we find that the cooperative effects of KRAS and p53 drive activation of autophagy rather than either mutation alone. This is consistent with established associations between autophagy dependence and RAS mutant tumor cell lines, many of which harbor p53 mutations (Guo et al., 2011; Lock et al., 2011; O’Dell et al., 2012; Yang et al., 2011). Others have reported conflicting roles of p53 in regulating autophagy (Morselli et al., 2011; Tasdemir et al., 2008), with a recent report showing that in a setting of homozygous p53 deletion during embryonal development loss of autophagy accelerates pancreatic tumor progression (Rosenfeldt et al., 2013). We elected to study autophagy in vivo using a model in which loss of p53 function occurs in a step-wise progressive manner relying on the spontaneous loss of a heterozygous WT p53 allele (Bardeesy et al., 2006a) as a late mutational event mimicking human PDA progression (Hruban et al., 2001; Luttges et al., 2001). In this context, we observe full autophagy activation at later stages of tumor formation. Correspondingly, it is at this point in advanced PanIN and PDA, when autophagy is most active, that we observe a delay in tumor progression in the absence of Plac8. In contrast, we do not see any effect of loss of Plac8 in a PDA model in which both copies of the conditional p53 allele are somatically mutated during embryogenesis (Figure S8) suggesting that the timing of complete p53 loss in the natural history and progression of PDA impacts the resulting tumor’s dependencies.
Pancreatic ductal adenocarcinoma (PDA) remains a significant challenge clinically despite iteration of many of the pathways and processes downstream of the signature genetic lesions (Hezel et al., 2006; Hidalgo, 2010). Further highlighting this is the fact that the most significant treatment advance in the past decade has come from a new combination of traditional chemotherapies rather than any particular novel genetic or biologic revelation (Conroy et al., 2011). While insights into improving drug delivery, modulation of the tumor micro-environment, and additional downstream effectors of KRAS have recently pointed to alternative therapeutic strategies, new processes and targets critical to PDA growth are needed (DeNicola et al., 2011; Frese et al., 2012; Olive et al., 2009; Provenzano et al., 2012; Rhim et al., 2012). While pharmacologic inhibition of autophagy is under ongoing evaluation clinically there are limited agents available, moreover autophagy plays a critical role in normal tissue homeostasis and energy balance making the identification of a therapeutic window potentially difficult. Thus, regulators and effectors of autophagy specific to tumors, such as Plac8, could potentially offer valuable targets for intervention.
Experimental Procedures
Cells
YAMC, mp53, Ras, mp53/Ras cell derivation and maintenance were described previously (McMurray et al. 2008; Xia and Land, 2007). CAPAN-2, PANC-1 and Panc10.05 cell lines were obtained from ATCC.
Antibodies and compounds
The following antibodies were used: rabbit polyclonal antibody raised against the c-terminal 16 amino acids of murine Plac8 (Ledford et al., 2007) (Pocono Rabbit Farm & Laboratories, Inc.), rabbit polyclonal anti-β-Tubulin antibody (H-235, Santa Cruz), rat monoclonal anti-Lamp2 antibody (GL2A7, Abcam), mouse monoclonal anti-Rab7 antibody (R8779, Sigma), rabbit polyclonal anti-LC3B antibody (L7543, Sigma), mouse monoclonal anti-RhoA antibody (sc-418, Santa Cruz), goat polyclonal anti-Cathepsin D antibody (sc-6486, Santa Cruz), guinea pig polyclonal anti-p62 antibody (GP62-C, PROGEN), mouse monoclonal anti-3xFlag antibody and HRP-conjugated anti-3xFlag antibody (F2426 and A8592, Sigma), rabbit polyclonal anti-Rab5 antibody (ab18211, Abcam), rabbit polyclonal anti-Atg12 antibody (A8731, Sigma), EGFR antibody (sc 03).
The following compounds were used: bafilomycin A1 (Sigma B1793), chloroquine (Sigma 6628), cycloheximide (Sigma C7698, Lysosensor Yellow Blue DND-160 and DCF-DA (Molecular Probes, L-7545 and C6827, respectively).
Plasmids
Plasmids for retro-viral Plac8 KD, and shRNA-resistant Plac8 expression were described previously (McMurray et al., 2008). cDNAs for Rab7, Rab5a, and Atg12 were PCR-cloned in pBabe-hygro containing an in-frame, N-terminal 3xFlag-tag and sequence verified. Dominant-negative and activated Rab7 and Rab5a mutants were generated via site-directed mutagenesis and sequence-verified. N-terminal 3xFlag tagged DN Rab7, DA Rab7, DN Rab5a and Atg12 were PCR-cloned in pLenti/Ubc/V5 (Invitrogen) and sequence-verified. cDNA for LC3 was PCR-cloned in pEGFP (Clonetech) and subsequently PCR-cloned into lenti-viral vector FG12. The mCherry-EGFP-LC3 puro construct (Addgene 22418) (N’Diaye et al., 2009) was cloned into pBabe hygro. Atg12 shRNA molecules were designed (http://jura.wi.mit.edu/bioc/siRNAext/home.php) and cloned into the pSuper-retro system (Oligoengine). Atg12 lenti-viral KD constructs were obtained from Openbiosystems.
Immunofluorescent staining of cells
Cells were plated onto collagen-1-coated 22mm glass coverslips (BD Biosciences). Cells were fixed and incubated with primary antibody overnight at 4°C while shaking followed by appropriate secondary antibodies. Immunostained cells were 1) washed 3x for 10 minutes with PBS, 2) mounted in VectaShield Mounting Media (Vector Labs), and 3) analyzed and imaged using a SP5 Leica inverted confocal microscope.
Subcellular fractionation and lysosome isolation
mp53/Ras cells were grown for two days at 39°C in RPMI with 10% (v/v) FBS, before pelleting at 1,200rpm for 5 minutes at 4°C. A fraction was retained and lysed in RIPA buffer for the whole cell lysate (WC) sample. Lysosomes were then isolated using a Lysosomal Isolation Kit (Sigma). In short, the cell pellet was resuspended, then homogenized in a Dounce homogenizer, and centrifuged at 1000 x g for 10 minutes. The pellet was saved as the nuclear fraction (N) sample. The supernatant was then centrifuged at 20,000xg for 20 minutes and supernatant was removed and saved as the cystosolic fraction (C). The pellet was resuspended in a small aliquot as the crude lysosomal fraction (CL). To separate lysosomes from other organelles 505μl of Optiprep and 275μl of Optiprep dilution buffer were added per 800ul of resuspended crude lysosomal fraction. CaCl2 was added to final concentration of 8mM, followed by 15 min incubation on ice and centrifugation at 5000xg for 10 minutes at 4°C. Purified lysosomal fraction (L) (supernatant) and microsomal pellet (M) were retained. Pellets were resuspended in RIPA, protein was determined by Bradford assay. SDS sample buffer was added to all samples.
Proteinase-K (PK) treatment of lysosomes
The crude lysosomal fraction was isolated as described above and resuspended in 50mM Tris buffer, pH 7.4. Crude lysosomal fractions were divided and treated for 30 min at 37°C with: 1) buffer 2) 0.5μg/ml PK 3) 0.5μg.ml PK + 1.0% Triton-X. Samples were then placed on, ice PK activity was quenched with 1mM PMSF and SDS sample buffer was added.
Xenograft assays
Matrigel (BD biosciences) was mixed with PANC-1 and Panc10.05 cell/medium mixture in a 1:1 ratio. Cell mixtures were then injected bilaterally into the flanks of CD-1 nude mice (Crl:CD-1-Foxnnu, Charles River Laboratories) in the following cell numbers: mp53/Ras – 5×105 cells, CAPAN-2 - 5×105 cells. Cell mixtures were injected into the flanks of NOD/SCID mice (Charles River Laboratories) in the following cell numbers per injection: PANC-1 – 2.5×106 cells and Panc10.05 – 1×106 cells. Tumor size was measured every 7 days by caliper and volume calculated by the formula volume=(4/3)Πr3, using the average of two orthogonal radius measurements; significance of difference in tumor size was calculated by T-test.
Quantification of GFP-LC3 and mCherry-EGFP-LC3 punctae, and of Lamp2/GFP-LC3 co-localization
Cells were fixed, stained, mounted, and imaged by the methods described above. GFP-LC3 punctae were determined using the ImageJ plug-in Watershed Segmentation. The image produced by selecting Object/Background binary was inverted and overlayed on top of the GFP-LC3 image. The resulting image was quantified by measuring the mean green and blue signals per image and dividing the blue signal by the total green signal to get the amount of punctae per total GFP-LC3 expressed in the cell and then normalized to the mean YAMC ratio of punctae formation. To quantify GFP-LC3 co-localization the ImageJ plug-in Co-localization Finder was used to determine co-localization (in white) of red (Lamp2) and green (GFP-LC3) signal. The mean co-localized signal is then divided by the GFP-LC3 signal to derive a ratio of co-localization per GFP-LC3 signal and these ratios were then normalized to the mean values derived from vector control or YAMC cells. The mCherry-EGFP-LC3 reporter was used to determine ratios of autophagosomes (red and green co-localized signal) and autolysosomes (red signal only). The Red and Green Punctae Co-localization plugin in ImageJ was used to derive the puncta areas per cell. The area value for autolysosomes was determined by subtracting the red and green co-localized signal from the red puncta area and normalized to the mean of the mCherry-EGFP-LC3 puncta area of vector control or untreated control samples.
Mice
Mutant mouse strains: All animal studies were conducted in accordance with the AAALAC accredited University Committee on Animal Resources (UCAR). All mouse strains used in these studies have been previously described and characterized (Bardeesy et al., 2006b; Kuma et al., 2004; Ledford et al., 2007). Specifically KrasG12D, p53L/L, Plac8−/−, GFP-LC3 and Pdx-Cre mutant strains were intercrossed to achieve the desired cohorts as outlined above. The genetic background was mixed. Individual mice within experimental cohorts were followed until signs of illness including poor grooming, abdominal bloating, diminished activity or weight loss at which point a full necropsy was performed followed by histological analysis.
Histology and in vivo GFP-LC3- analysis
A board-certified pathologist with a specialization in pancreatic histopathology independently reviewed and classified tumors. Lymph nodes, lungs and spleen were included in a survey for metastasis in all individuals with tumors. To determine the extent of PanIN formation at 9 weeks of age the whole pancreas from wild-type and Plac8 null Kras-p53 PDA mice (n=6–10/group) was sectioned lengthwise. Ductal structures were analyzed using H&E stained sections and classified as normal, PanIN1 or PanIN3. To determine formation of LC3 punctae in vivo frozen sections from compound GFP-LC3 Kras-p53 mutant pancreas were analyzed as previously described by confocal microscopy (Kuma et al., 2004). Adjacent H&E stained sections were evaluated by Dr. Whitney-Miller to determine histologic classification (normal ducts, PanIn1 or 3, or PDA). In summary n=3 mice encompassing each histologic type of lesion were evaluated. Punctae and nuclei from ductal structures (Normal and PanIN 1–3) and well-differentiated PDA (in which a distinction could be made histologically between tumor and stroma) were manually counted as well as determined by ImageJ. As results between manual counts and those determined by ImageJ analysis were consistent, ImageJ analysis is presented.
Survival analysis
Survival was determined using the Kaplan-Meier method and comparisons between groups were determined using the Log-rank test. All statistical analyses were performed using Prism statistical software version 4.0a May 11, 2003. Animals that displayed signs of illness and were found to have advanced cancers on necropsy were included as events as were all animals that died prior to developing signs of illness that had a pancreatic mass and histological diagnosis of adenocarcinoma identified at autopsy. Animals that died for reasons other than advanced cancer, as determined by the absences of pancreatic pathology on autopsy were censored.
Immunofluorescent staining of Tumor sections
Whole pancreas from wild type and Plac8 null Kras-p53 PDA mice was paraffin embedded and sectioned lengthwise. Slides were baked in a 60°C oven for one hour followed by routine deparafinization. Antigen retrieval was then performed in a pressure cooker with 1x Rodent decloaker (Biocare Medical). Slides were blocked with 3% hydrogen peroxide, rinsed well in phosphate buffered saline and incubated with Plac8 antibody at 1:500 dilution at room temperature, and then anti-rabbit Alexa 488 secondary antibody for 30 minutes at room temperature. The tissue sections were next incubated with the Lamp2 and anti-rat Alexa 555 secondary antibody sequentially as described above. The sections were rinsed again, dehydrated in 100% ethanol, mounted and imaged using confocal microscopy.
Supplementary Material
Autophagy and Plac8 are critical for progression to advanced stage pancreatic cancer
Plac8 enhances autophagic flux by facilitating autophagosome-lysosome fusion
Plac8 links cooperating oncogenic mutations to autophagy activation in cancer
Plac8 deficiency impedes tumor growth but is well tolerated by normal tissues
Acknowledgments
We would like to thank Dr. Beverly H. Koller for Plac8−/− mice, Karen Bentley for electron microscopy, Dr. Shanshan Pei for FACS analysis, Loralee McMahon for histology, Nicole Scott for advice and Drs. Dirk Bohman and Mark Noble for critical reading of the manuscript. This work has been supported by NIH grants CA90663, CA120317 and CA138249 to HL, and CA122835 and CA172302 to AFH. AFH has also been supported by a HHMI early career award, the Edelman-Gardner Cancer Research Foundation award, a University of Rochester Buswell Fellowship, and the generous support of the Pancreatic Cancer Association of Western New York and the Michael F. Contestabile Memorial Golf Tournament.
Footnotes
The authors have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaburski GM, Hontz RD, Murphy ME. p53 and ARF: unexpected players in autophagy. Trends Cell Biol. 20:363–369. doi: 10.1016/j.tcb.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006a;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, et al. From the Cover: Both p16Ink4a and the p19Arf-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006b;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beertsen W, Willenborg M, Everts V, Zirogianni A, Podschun R, Schroder B, Eskelinen EL, Saftig P. Impaired phagosomal maturation in neutrophils leads to periodontitis in lysosomal-associated membrane protein-2 knockout mice. J Immunol. 2008;180:475–482. doi: 10.4049/jimmunol.180.1.475. [DOI] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binker MG, Cosen-Binker LI, Terebiznik MR, Mallo GV, McCaw SE, Eskelinen EL, Willenborg M, Brumell JH, Saftig P, Grinstein S, et al. Arrested maturation of Neisseria-containing phagosomes in the absence of the lysosome-associated membrane proteins, LAMP-1 and LAMP-2. Cell Microbiol. 2007;9:2153–2166. doi: 10.1111/j.1462-5822.2007.00946.x. [DOI] [PubMed] [Google Scholar]
- Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- Buchholz M, Braun M, Heidenblut A, Kestler HA, Kloppel G, Schmiegel W, Hahn SA, Luttges J, Gress TM. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene. 2005;24:6626–6636. doi: 10.1038/sj.onc.1208804. [DOI] [PubMed] [Google Scholar]
- Chen D, Fan W, Lu Y, Ding X, Chen S, Zhong Q. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell. 2012;45:629–641. doi: 10.1016/j.molcel.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang Z. Regulation of intracellular trafficking of the EGF receptor by Rab5 in the absence of phosphatidylinositol 3-kinase activity. EMBO Rep. 2001;2:68–74. doi: 10.1093/embo-reports/kve005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardiere C, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneen JL, Ceresa BP. Expression of dominant negative rab5 in HeLa cells regulates endocytic trafficking distal from the plasma membrane. Exp Cell Res. 2004;294:509–522. doi: 10.1016/j.yexcr.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL, Illert AL, Tanaka Y, Schwarzmann G, Blanz J, Von Figura K, Saftig P. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell. 2002;13:3355–3368. doi: 10.1091/mbc.E02-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese KK, Neesse A, Cook N, Bapiro TE, Lolkema MP, Jodrell DI, Tuveson DA. nab-Paclitaxel Potentiates Gemcitabine Activity by Reducing Cytidine Deaminase Levels in a Mouse Model of Pancreatic Cancer. Cancer Discov. 2012;2:260–269. doi: 10.1158/2159-8290.CD-11-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta NFN, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21:1001–1010. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaviz-Hernandez C, Stagg C, de Ridder G, Tanaka TS, Ko MS, Schlessinger D, Nagaraja R. Plac8 and Plac9, novel placental-enriched genes identified through microarray analysis. Gene. 2003;309:81–89. doi: 10.1016/s0378-1119(03)00508-0. [DOI] [PubMed] [Google Scholar]
- Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Hruban RH, Iacobuzio-Donahue C, Wilentz RE, Goggins M, Kern SE. Molecular pathology of pancreatic cancer. Cancer J. 2001;7:251–258. [PubMed] [Google Scholar]
- Huynh KK, Eskelinen EL, Scott CC, Malevanets A, Saftig P, Grinstein S. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 2007;26:313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Jimenez-Preitner M, Berney X, Uldry M, Vitali A, Cinti S, Ledford JG, Thorens B. Plac8 is an inducer of C/EBPbeta required for brown fat differentiation, thermoregulation, and control of body weight. Cell Metab. 2011;14:658–670. doi: 10.1016/j.cmet.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool J, Uren AG, Martins CP, Sie D, de Ridder J, Turner G, van Uitert M, Matentzoglu K, Lagcher W, Krimpenfort P, et al. Insertional mutagenesis in mice deficient for p15Ink4b, p16Ink4a, p21Cip1, and p27Kip1 reveals cancer gene interactions and correlations with tumor phenotypes. Cancer Res. 2010;70:520–531. doi: 10.1158/0008-5472.CAN-09-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Ledford JG, Kovarova M, Koller BH. Impaired host defense in mice lacking ONZIN. J Immunol. 2007;178:5132–5143. doi: 10.4049/jimmunol.178.8.5132. [DOI] [PubMed] [Google Scholar]
- Livesey KM, Kang R, Vernon P, Buchser W, Loughran P, Watkins SC, Zhang L, Manfredi JJ, Zeh HJ, 3rd, Li L, et al. p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Res. 2012;72:1996–2005. doi: 10.1158/0008-5472.CAN-11-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe AW, Olsen M, Hao Y, Lee SP, Taek Lee K, Chen X, van de Rijn M, Brown PO. Gene expression patterns in pancreatic tumors, cells and tissues. PloS one. 2007;2:e323. doi: 10.1371/journal.pone.0000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttges J, Galehdari H, Brocker V, Schwarte-Waldhoff I, Henne-Bruns D, Kloppel G, Schmiegel W, Hahn SA. Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis. The American journal of pathology. 2001;158:1677–1683. doi: 10.1016/S0002-9440(10)64123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray HR, Sampson ER, Compitello G, Kinsey C, Newman L, Smith B, Chen SR, Klebanov L, Salzman P, Yakovlev A, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453:1112–1116. doi: 10.1038/nature06973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Shen S, Ruckenstuhl C, Bauer MA, Marino G, Galluzzi L, Criollo A, Michaud M, Maiuri MC, Chano T, et al. p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle. 2011;10:2763–2769. doi: 10.4161/cc.10.16.16868. [DOI] [PubMed] [Google Scholar]
- N’Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10:173–179. doi: 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Grose V, Rossi RM, Zhu AX, Land H, Bardeesy N, et al. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 2012;72:1557–1567. doi: 10.1158/0008-5472.CAN-11-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshikawa Y, Kobayashi T, Mimuro H, Fukumatsu M, Kiga K, Piao Z, Ashida H, Yoshida M, Kakuta S, et al. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe. 2011;9:376–389. doi: 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimkina J, Murphy ME. ARF, autophagy and tumor suppression. Autophagy. 2009;5:397–399. doi: 10.4161/auto.5.3.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulski K, Li Y, Rothermund K, Pu L, Watkins S, Yi F, Prochownik EV. Onzin, a c-Myc-repressed target, promotes survival and transformation by modulating the Akt-Mdm2-p53 pathway. Oncogene. 2005;24:7524–7541. doi: 10.1038/sj.onc.1208897. [DOI] [PubMed] [Google Scholar]
- Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol Cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Smith B, Land H. Anticancer activity of the cholesterol exporter ABCA1 gene. Cell Rep. 2012;2:580–590. doi: 10.1016/j.celrep.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GT, Hughes JP, Stoneman V, Anderson CL, McCarthy NJ, Mourtada-Maarabouni M, Pickard M, Hedge VL, Trayner I, Farzaneh F. Isolation of genes controlling apoptosis through their effects on cell survival. Gene Ther Mol Biol. 2006;10:255–262. [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Zhang T, Xu Y, Subramaniam VN, Griffiths G, Hong W. Endobrevin, a novel synaptobrevin/VAMP-like protein preferentially associated with the early endosome. Mol Biol Cell. 1998;9:1549–1563. doi: 10.1091/mbc.9.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SF, Huang Y, Hou JK, Yuan TT, Zhou CX, Zhang J, Chen GQ. The downregulation of onzin expression by PKCepsilon-ERK2 signaling and its potential role in AML cell differentiation. Leukemia. 2010;24:544–551. doi: 10.1038/leu.2009.280. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell structure and function. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SWX, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras Maintains Pancreatic Tumors through Regulation of Anabolic Glucose Metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.