Abstract
Background
PI3K/Akt/mTOR signaling is being actively pursued as a therapeutic target for breast cancer. We sought to determine if tumor heterogeneity and biospecimen variables affect the evaluation of PI3K/Akt/mTOR pathway markers.
Methods
Intraoperative image-guided core-needle biopsies (CNB), and central and peripheral surgical tumor specimens were prospectively collected in 53 patients with invasive breast cancer. Specimens were assessed with reverse phase protein arrays (RPPA) and immunohistochemistry (IHC).
Results
There was a moderate or strong correlation between the expression of 149 (97%) of the 154 different RPPA markers in the center and periphery. Correlation was higher for smaller tumors, in patients who did not undergo neoadjuvant therapy, and with shorter cold ischemia time. Of 154 markers, 132 (86%) were not statistically different between the center and periphery, and 97 (63%) were not different between the CNB and the surgical specimen (average of the central and peripheral specimen). pAkt S473 and PTEN had a significant correlation between central and peripheral specimens, and between CNB and surgical specimen. However, pAkt S473, pS6 S235/236 and pS6 240/244 levels were significantly higher in CNB than the central specimens both by RPPA and by IHC.
Conclusions
Most individual proteomic biomarkers studied do not have significant intratumoral heterogeneity. However, protein and phosphoprotein levels are affected by biospecimen type and other preanalytic variables. PI3K pathway activation is greater in CNB compared to post-excision surgical samples suggesting a potential loss of phosphorylation during surgical manipulation, or with cold ischemia of surgical specimens.
Keywords: biopsy, proteomics, tumor heterogeneity, Akt
Introduction
PI3K/Akt/mTOR signaling is being actively pursued as a therapeutic target (1). However, assessment of PI3K/Akt/mTOR pathway activity remains a genuine obstacle to personalizing therapy. Stability of phosphoproteins during sample handling is a major concern. In a multicenter trial in colorectal cancer, pAkt immunostaining was observed only in biopsies and not in surgical samples (2). Furthermore, in breast cancer, significant loss in pAkt and pErk immunostaining was found in surgical specimens that underwent routine processing compared to core needle biopsies (CNB) taken from tumors immediately after excision (3). Thus PI3K/mTOR activation status may vary based on specimen acquisition approach and processing.
FNA and CNB are especially valuable for decision-making for preoperative therapy of the primary treatment-naïve breast cancers, as well as in the metastatic setting. Previous work in our breast cancer research program demonstrated that FNAs enrich for tumor cells, retrieving 80% tumor cells compared to CNBs which contained on average of 50% tumor cells (4). However, how these tumor acquisition approaches affects protein based biomarkers of PI3K/mTOR signaling have not been determined.
There is growing concern about tumor heterogeneity (5, 6). Surgical excision allows assessment of the entire tumor, thus potentially overcoming concerns about tumor heterogeneity. However, specimen quality is dependent on a variety of post-acquisition variables, including intraoperative ischemia, specimen handling and time to preservation. Spruessel et al has shown that initial changes of gene and protein expression profiles were already observed 5–8 min after colon resection (4). The time to preservation may be especially prolonged in breast surgery, where excised specimens often undergo whole-specimen radiography followed by specimen margin inking and sectioning potentially with repeat radiography for margin assessment (7). However, in recent work, we demonstrated that mRNA expression levels of single genes and multigene signatures that are of diagnostic relevance in breast cancer were mostly unaffected by sample preservation method or prolonged cold ischemic duration (8). In our preliminary work with reverse phase protein arrays (RPPA), we found that although the functional proteomic ‘fingerprint’ was reproducible in most breast cancer samples even after a delay of 24 hours before freezing, the expression of 21/82 total and phosphoproteins was significantly affected by time to tumor freezing: including markers of PI3K/mTOR signaling (pAkt T308, PTEN, pAMPK T172, TSC2, pTSC2 T1462, S6K1, and pS6 S235/236) (9), suggesting that post-excision expression of many proteins, especially phosphoproteins may not accurately reflect the in vivo state of the proteins at the time of collection.
Immunohistochemistry (IHC) is the most commonly used approach for assessing biomarkers at the protein level but is semiquantitative. RPPA has mainly been used as a discovery tool, and has the advantage of being quantitative, allowing for assessment of hundreds of markers from a small amount of tissue in a cost-effective fashion. However, unlike IHC, where the tumor vs. stromal staining can be differentiated, in RPPA when total tumor lysate is used, the biomarker signal will be more affected by low tumor cellularity and high heterogeneity. The ideal protein assessment approach may differ from marker to marker, depending on baseline tumor cellularity, marker expression pattern and influence of sample handling.
We hypothesized that biospecimen variables and tumor heterogeneity may affect assessment of pathway activation. We therefore performed a prospective clinical study to determine whether biospecimen variables including specimen type (in vivo collection of FNA or core biopsy vs. ex vivo collection of central and peripheral surgical tumor samples) affect the functional proteomic profile by RPPA and the assessment of PI3K/Akt/mTOR pathway activation by RPPA and IHC. This work was performed to assess effect of biospecimen variables on RPPA results without microdissection. We demonstrated that most biomarkers do not have significant spatial heterogeneity in operable breast cancer. However, levels of several potentially actionable biomarkers, including markers of PI3K pathway activation, are affected by biospecimen type and biospecimen handling variables.
Materials and Methods
Human Specimen Collection
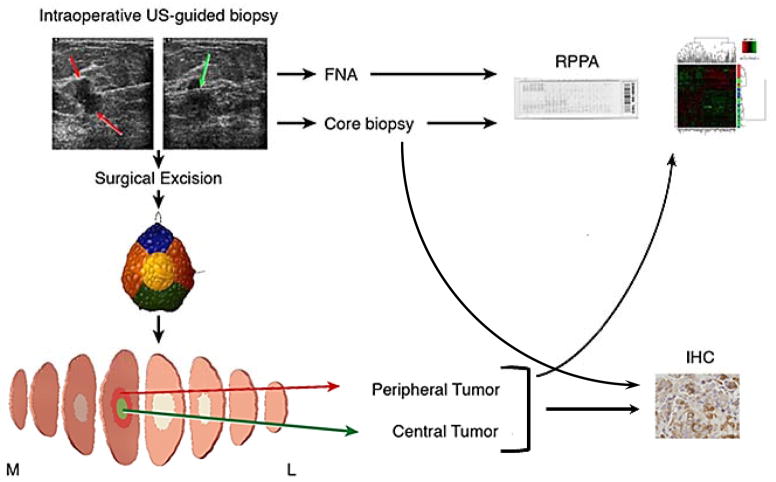
We conducted a prospective laboratory study after approval by the Institutional Review Board (Figure 1). After informed consent, 53 patients with invasive breast cancer 2 cm or greater on preoperative imaging, underwent intraoperative ultrasound-guided percutaneous FNAs (4–8 passes) and CNBs (3–4 core biopsies) after induction of anesthesia but before the beginning of the operation. The number of samples obtained per patient is listed in Supplementary Table 1. FNAs were frozen for RPPA. In addition, one CNB with the greatest tumor content upon visual inspection was macrodissected and frozen for RPPA. The other CNBs were formalin fixed and paraffin embedded (FFPE). The surgical specimen was evaluated by a dedicated breast pathologist in the frozen section suite. For breast conservation (lumpectomy) samples whole specimen radiographs were performed for confirmation of target lesion removal. For both breast conservation and mastectomy samples margins were inked and sliced specimens were radiographed as deemed necessary for margin analysis as previously described (7). A portion of central and peripheral tumor was then allocated for research; one specimen from each site was snap frozen for RPPA, another was placed in formalin. Central and peripheral research samples were acquired at the same time, and the samples were approximately the same size, and were frozen directly. Small specimens of central and peripheral tumor samples were removed from main specimen in the frozen section suite and then formalin fixed paraffin embedded directly. Surgical tumor samples were macrodissected, microdissection was not performed. The time from intraoperative specimen removal to time of freezing of the research specimen was prospectively recorded and designated as “cold ischemia time”.
Figure 1.
Study Schema. Patients underwent intraoperative breast biopsies with fine needle aspirate (4–8 passes) and core needle biopsy (3–4 passes). The core needle biopsy with the greatest tumor cellularity was used for RPPA while the other core needle biopsies were analyzed by IHC. After surgical excision the specimen underwent routine intraoperative assessment including color-coded specimen margin inking, and sectioning and specimen mammogram as needed. Central and peripheral tumor specimen was collected for research to capture tumor heterogeneity. FNA, core needle biopsy and peripheral and central surgical resection samples underwent proteomic analysis with reverse phase protein arrays (RPPA). Core needle biopsy and both peripheral and central surgical resection samples underwent IHC for PI3K pathway markers.
The FFPE samples were assessed by hematoxylin and eosin (H&E) staining. Three patients had no tumor on H&E staining of all three samples (CNB, central and peripheral specimen); these patients were excluded from further analysis. Twelve patients had no tumor on H&E of one of the three samples; the samples without tumor were excluded from analysis.
Reverse Phase Protein Arrays
RPPA was performed in the MD Anderson Cancer Center Functional Proteomics Core Facility as described previously (10). Lysates were extracted from each tumor specimen and four replicates from each lysate were spotted on the RPPA. The cell lines were treated on separate days to obtain three biological replicates. Briefly, 200 microliters of lysis buffer was added to CNB samples. Following homogenization with ceramic beads, samples were centrifuged, and supernatant was quantitated using BCA assay kit (Thermo Scientific). The final protein concentration was adjusted to 2 μg/μl. FNA samples were lysed in 75 μl of lysis buffer that was already mixed with 25 μl of loading buffer. Following sonication, samples were quantitated with BCA assay kit and the final concentration was adjusted to 3 μg/μl. Lysates were probed with antibodies validated for RPPA that are enriched for components of PI3K/Akt/mTOR pathway (Supplementary Table 2). A total of 154 unique proteins (including phosphoproteins) and 4 replicates were evaluated. Protein levels were expressed as the mean expression values in Log2. Samples with less than 20% tumor cellularity on corresponding H&E sections were not included in statistical analysis for RPPA data.
Immunohistochemistry
IHC was performed on FFPE CNBs, and central and peripheral tumor specimens with the following antibodies: pAkt S473 (Cell Signaling #4060, 1:50, 1 hour), pS6 S235/236 (Cell Signaling # 4858, 1:50, overnight), pS6 S240/244 (Cell Signaling #2215, 1:200, overnight), pPRAS40 T246 (Cell Signaling #2997, 1:200, overnight), and p4EBP1 T70 (Cell Signaling #9455, 1:50, overnight) Ki-67 and PTEN immunostaining was performed in the MD Anderson IHC Core laboratory with the following antibodies: Ki-67 (Dako #M7240, 1:100, 20 min), and PTEN (Dako #M3627, 1:100, 15 min). Positive and negative controls were included for all assays. H score was determined by estimation of the percentage of tumor cells positively stained with weak, moderate, strong staining intensity (by AS). The cytoplasmic score was used for statistical analysis. The following formula was used: .
Statistical Analysis
The RPPA spot signal intensity data from MicroVigene was processed using the R package SuperCurve (version 1.4.3) (1), available at “http://bioinformatics.mdanderson.org/OOMPA”. RPPA superslide was normalized for FNA and surgical samples/CNB samples separately. Raw data were treated with median centering across samples; the treated data were undertaken with centering by the sample medians; and the final normalized data are obtained by applying MAD scaling to the data; the median centered values were divided by median absolute deviation to scale the data (11–13). For Spearman’s rank correlation test, p-values are computed using algorithm AS 89 (14, 15). The values of correlation were classified as follows: rs of 0.8 to 1, the correlation is very strong; between 0.6 and 0.79, correlation is strong; between 0.5 and 0.59, the correlation is moderate; between 0.3 and 0.49, the correlation is moderate to low; between 0.16 and 0.29, correlation is weak to low; and below 0.16, the correlation is too low to be meaningful. Hierarchical Ward-linkage clustering was performed based on Spearman correlation coefficients. Paired t-test was conducted to determine the difference among RPPA data of various biospecimen types, and Wilcoxon signed rank test was applied to compare IHC data. The Benjamini Hochberg procedure was employed to control the False Discovery Rate (FDR). FDR of 0.05 was considered significant.
For each of the 178 samples, we computed a PI3K pathway score, which was the sum of the normalized values (median centering and MAD scaling) of the phosphoprotein levels of Akt, mTOR, 4E-BP1, S6K, and S6, (i.e. PI3K score = pS6 240/244 + pS6 S235/236 + pS6K T389 + pmTOR S2448 + p4E-BP1 S65 + p4E-BP1 T37/46 + pPRAS40 T246 + pAkt S473 + pAkt T308). We classified the PI3K pathway scores of each sample type (FNA, CNB, central and peripheral surgical tumor) into two groups; samples were considered activated if their PI3K scores were in the top quartile within their sample type. Cohen’s kappa tests were performed to identify the agreement between PI3K pathway activity class and the biospecimen type. The Kappa agreement was characterized as values <0 as indicating no agreement, 0–0.20 as slight, 0.21–0.40 as fair, 0.41–0.60 as moderate, 0.61–0.80 as substantial, and 0.81–1 as almost perfect agreement (16).
PTEN and pAkt were defined as loss and low, respectively, if their IHC values were 0. Chi square tests were applied to check the dependency between PI3K score and PTEN loss/normal, PI3K score and pAkt high/low. The analyses were performed by using R (version 2.11.1) (R Development Core Team, 2011) (17). Supplementary Table 3 describes how each point of REMARK guidelines were addressed.
Results
Clinicopathologic Characteristics
Clinical and pathologic characteristics of the patients are listed in Supplementary Table 4. The median pathologic tumor size was 2.8 cm. Most patients (77%) had hormone-receptor positive tumors. Eleven patients had received neoadjuvant chemotherapy; these patients had their research biopsies intraoperatively (i.e. after completion of the neoadjuvant chemotherapy), followed by surgical excision, similar to the surgery first cohort. Most patients underwent a mastectomy. The specimen cold ischemia time was an average of 37 min, and ranged from 8 to 157 minutes.
A median of 6 FNA (range 3–8) and 2 core needle biopsy (range 0–3) samples were obtained from each patient. Protein yield from a single FNA sample was a mean of 1635 μg, (median yield 686 μg, range 36–8580 μg). The protein yield from a single core biopsy was a mean of 862.8 μg, (median yield 871.4 μg, range 242–1594 μg). The tumor cellularity of each sample was assessed by H&E and distribution of non-malignant components in the central specimen including contribution of epithelium, lymphocytes and endothelium and fibroblasts was detailed (Supplementary Table 5). There was no statistically significant difference in tumor cellularity between the central and peripheral samples, but the tumor cellularity was lower in the CNBs compared to the central specimens (in 47 matched samples 54.8% vs 61.2%; p=0.0689), and significantly lower in CNB compared to peripheral samples (in 43 matched samples 52.7% vs 60.7%; p=0.0476). Tumors whose corresponding FFPE samples had less that 20% tumor cellularity or that only had ductal carcinoma in situ (DCIS) on FFPE were considered inevaluable for RPPA, except for studies comparisons on effect of tumor cellularity. Tumors whose IHC sections did not contain tumor were considered inevaluable for IHC. Seven core biopsies, four central specimens and four peripheral specimens had less than 20% tumor cellularity on FFPE. Forty-four (88%) of 50 CNBs, 46 (90%) of 51 of central tumor specimens, and 43 (84%) of 51 peripheral tumor specimens were considered evaluable for RPPA and IHC.
Effect of Biospecimen Type on Proteomic Markers
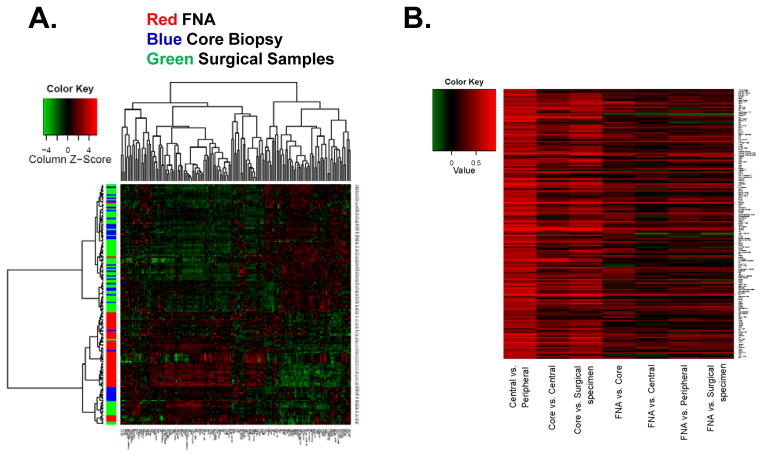
To determine the effect of biospecimen type on the functional proteomic profile, we performed unsupervised hierarchical clustering on the RPPA from FNAs, CNBs, and central and peripheral surgical samples. As demonstrated in the heatmap (Figure 2A), most of the FNA samples clustered together, while the central and peripheral samples and CNB samples co-clustered suggesting that the cellular content of the FNA is markedly different from that of the CNB and surgical samples. Notably, CNB, central and peripheral samples from the same patient often clustered together, consistent with inter-tumoral differences being greater than intratumoral differences. FNAs may not only differ because they are collected prior to any cold ischemia time, but also because there are previously reported differences in relative tumor vs. stromal cell content (18), and technical differences in RPPA sample processing and protein quantitation between FNA samples vs. CNB and surgical samples. The correlation between different biospecimens for each biomarker is listed in Supplementary Table 6 and depicted as a heatmap (Figure 2B).
Figure 2.
RPPA on different biospecimens. A. Unsupervised hierarchical clustering of RPPA results on different biospecimen types. Each column represents a different protein or phosphoprotein and each row, a different sample. Red, FNA samples; blue, core needle biopsy; green, surgical samples. B. The correlation between different biospecimens for each biomarker depicted as a heatmap. Red, high positive correlation, green negative correlation.
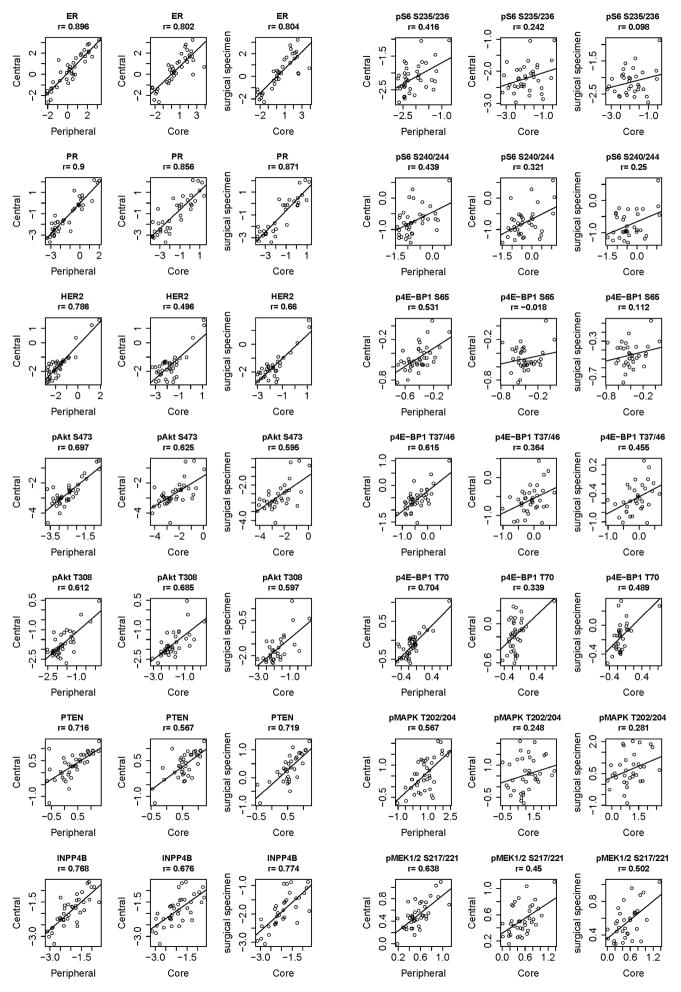
There was a moderate or strong correlation between the central and peripheral specimen results for all but 5 of the 154 proteins and phosphoproteins tested (Supplementary Table 6). There was a very strong correlation between the central and peripheral samples for ER (r=0.896), PR (r=0.90) and strong correlation for HER2 (r=0.786) (Figure 3). We also compared the central and peripheral protein expression using paired Student’s t-test (Supplementary Table 7). In 132 (86%) of 154 markers tested, at a FDR of 0.05, there was no statistical difference in the expression between central and peripheral specimens.
Figure 3.
Expression of PI3K pathway markers by RPPA in different biospecimens. Correlation between ER, PR, HER2, pAkt S473, pAkt T308, PTEN, INPP4B, pS6 S235/236, pS6 S240/244. p4E-BP1 S65, p4E-BP1 T37/46, p4E-BP1 T70, pMAPK T202/204, and pMEK1/2 S217/221 by RPPA between different biospecimens.
To determine the impact of marker assessment on in vivo biopsies compared to ex vivo excision samples, we next assessed whether RPPA results correlated between CNB and surgical specimens. There was a very strong correlation between the CNB and surgical tumor specimen (average of center and periphery) for ER (r=0.804), PR (r=0.871), and strong correlation with HER2 (r=0.66; Figure 3). Thus, our data suggest that for especially ER, PR, quantitation by RPPA does not show significant differences attributable to spatial intratumoral heterogeneity or biospecimen type. There was no statistically significant difference in expression of 97 (67%) of the 154 markers assessed in CNB, compared to the surgical samples by Student’s t-test, at a FDR of 0.05 (Supplementary Table 7).
There was a modest to poor correlation between FNA results and surgical specimens for several proteins (Supplementary Table 6). However, the FNA and surgical sample had a significant correlation for 49 proteins, and, the FNA and CNB had a correlation for 68 proteins. There was strong correlation between FNA and surgical samples for ER (r=0.606), PR (r=0.695) and fair correlation for HER2 (r=0.367). There was also a strong correlation between CNB and FNA results for ER (r=0.650), PR (r=0.642) and moderate correlation for HER2 (r=0.57).
Effect of Biospecimen Variables on Biomarker Expression
We next evaluated the effect of preanalytic biospecimen variables on biomarker expression. At FDR<0.05, in FNA samples there was no difference in protein expression between patients that were younger vs. older than median age of 51 years. In CNB, ER was higher in older patients (p<0.0001), and TIGAR and c-Kit were higher in younger patients (p=0.0008 and p=0.001, respectively). ER expression also was significantly higher in Central and Peripheral samples in patients that were older (p<0.0001 for both). There was an increased pGSK3a/b S21/9 expression in grade 3 tumors compared to grade 1 or grade 2 tumors (p=0.0343 and p=0.0001, respectively). There was no protein differentially expressed between tumors of different stages, or between small vs. large tumor size (median cut off 2.8 cm) at FDR<0.05 cut-off for any of the FNA, Core, Central and Peripheral samples.
We next determined whether preanalytic biospecimen variables impacted the correlation of expression between different biospecimen types. We expected that larger tumors would have greater tumor heterogeneity, and indeed by paired t-test, we found that correlation between central and peripheral specimens was better in tumors that were smaller than the mean (<3.6 cm), compared to tumors larger than the mean (p<0.001) (Supplementary Table 8). Interestingly larger tumors had a greater correlation between FNA vs. surgical samples (p=0.003). Tumors with shorter than the mean cold ischemia time (36.7 min) had better correlation between central vs. peripheral specimens compared to tumors with longer cold ischemia time (p<0.001) (Supplementary Table 8). There was also better correlation between FNA and central specimens in tumors with shorter cold ischemia time (p<0.001). Shorter cold ischemia time also was associated with a higher correlation between CNB and surgical specimens (p<0.001).
We also assessed the effect of surgery type on biomarker correlation. There was a stronger correlation between CNB vs. surgical samples (p<0.001) as well as FNA vs. surgical samples (p=0.005) in patients who had breast conservation compared to those who underwent mastectomy. There was also a trend towards improved correlation between central and peripheral samples in breast conservation samples (p=0.074). To better understand whether other preanalytic variables differed between breast conservation and mastectomy samples, we compared tumor size and cold ischemia time for these specimens. There was no statistically significant difference in tumor size between patients who underwent breast conservation versus mastectomy (median size 3.1 cm vs. 3.7 cm, p=0.49). However, surgery type was associated with differences in cold ischemia time. Mean cold ischemia time for breast conservation patients was 28.5 min (range 20–42 min), compared to 39.1 min (range 8–157 min) for mastectomy (p=0.021). This may be attributable to the fact that breast conservation samples routinely undergo immediate margin analysis or maybe because mastectomy specimens take longer to section.
The correlation between FNA and surgical specimens was significantly greater for non-phosphorylated proteins than phosphorylated proteins (p=0.043). There was also a trend for difference in correlation between CNB and surgical specimens (p=0.055). Thus phosphoprotein levels are more susceptible to biospecimen type/sample handling than total proteins.
We next evaluated the effect of tumor cellularity on biomarker expression (Supplementary Table 9). At FDR<0.05, there are 54 proteins in CNB data showing significant variability based on tumor cellularity. At FDR<0.05, there were 22 proteins in central tumor data showing significant variability based on tumor cellularity. There were seven common proteins in CNB and Central data that varied by tumor cellularity (Cyclin B1, Beclin, IRS, YB1, p38 MAPK, BID and eIF4G). The contribution of the tumor cell composition was then correlated with the variation in the expression of each marker between biospecimens. F-test in linear regression model was applied to check if the tumor cell composition affects the variance in the expression of each marker. The variance of most of the biomarkers could not be explained by the tumor cell composition.
The biomarker correlation in the patients who did and did not receive neoadjuvant chemotherapy was also analyzed separately (Supplementary Table 10). Correlations for samples from patients who did not receive neoadjuvant chemotherapy were significantly higher than in patients who received neoadjuvant chemotherapy when the comparing the central vs. peripheral specimen (p<0.0001), CNB vs. Central specimen (p<0.0001), CNB vs. Surgical specimen (p<0.0001), FNA vs. CNB (p<0.0001), and FNA vs. peripheral sample (p=0.0209) (Supplementary Table 11).
Effect of Biospecimen Type on PI3K/Akt/mTOR Pathway Activation
As the PI3K/Akt/mTOR pathway is a potential therapeutic target under active clinical investigation, we assessed PI3K/Akt/mTOR pathway activation by specimen type. First we assessed expression of pAkt S473 and pAkt T308, markers of activated Akt (Figure 3). pAkt S473 as measured by RPPA was correlated between all specimens and pAkt T308 was correlated between the central vs. peripheral, and core vs. surgical specimen.
PTEN and INPP4B are two tumor suppressors whose loss is associated with activation of PI3K/Akt/mTOR signaling (19). The PTEN expression of central and peripheral samples and CNB vs. surgical samples were strongly correlated, both r=0.716. The INPP4B expression of central and peripheral samples and CNB vs. surgical samples were strongly correlated, both r=0.774.
Phosphorylation of mTOR target p4E-BP1 T37/46 correlated strongly between central and peripheral samples (r=0.615) while p4E-BP1 S65 correlated moderately (r=0.531; Figure 3). p4E-BP1 T37/46 and p4E-BP1 S65 had moderate to low correlation between the CNB and surgical specimen (r=0.349 and 0.318, respectively). There was also a moderate to low correlation between central and peripheral samples for the mTOR downstream signaling markers pS6 S235/236 and pS6 240/244 (r=0.416 and r=0.439). The CNB and surgical specimens had no meaningful correlation between CNB and surgical specimen for pS6 S235/236 (r=0.098) and weak to low correlation for pS6 240/244 (r=0.25). The correlation between the CNB and FNA was somewhat stronger for pS6 S235/236 (r=0.364) and pS6 240/244 (r=0.415).
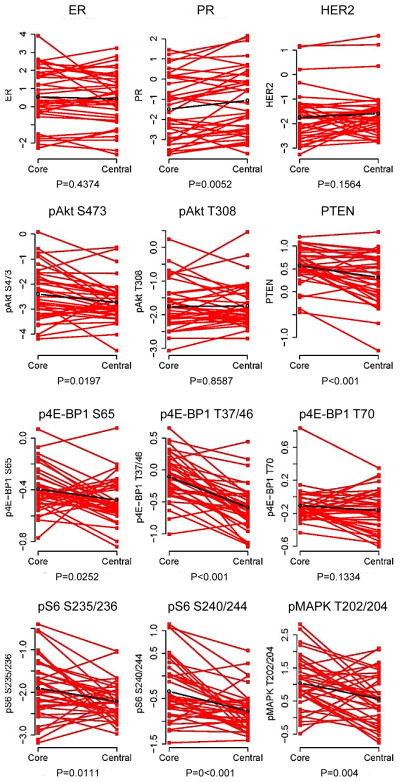
As the concordance of PI3K pathway activation in the CNB and surgical specimens was moderate to low, we next determined whether there was a significant difference in PI3K pathway activation in CNB vs. surgical specimen by paired Student’s t-test. When the CNB was compared to the central specimen, expression was higher in CNB for pAkt S473 (p=0.020), p4E-BP1 S65 (p=0.025), p4E-BP1 S37/46 (p<0.001), pS6 S235/236 (p=0.011) and pS6 S240/244 (p<0.001), as well as PTEN levels (p <0.001) and pMAPK T202/204 (p=0.004) (Figure 4). When we compared to the overall surgical specimen (average of the center and periphery), p4E-BP1 T37/46 and pS6 240/244 levels in the CNB were significantly higher (p=0.005 and p<0.001, respectively).
Figure 4.
Comparison of expression in core needle biopsy and central specimens. ER, PR, pAkt S473, pAkt T308, pAkt T308, PTEN, pS6 S235/236, pS6 S240/244, p4E-BP1 S65, p4E-BP1 S37/46, and pMAPK T202/204 expression in the core needle biopsy and central specimen by RPPA.
PI3K/Akt/mTOR Signaling by IHC
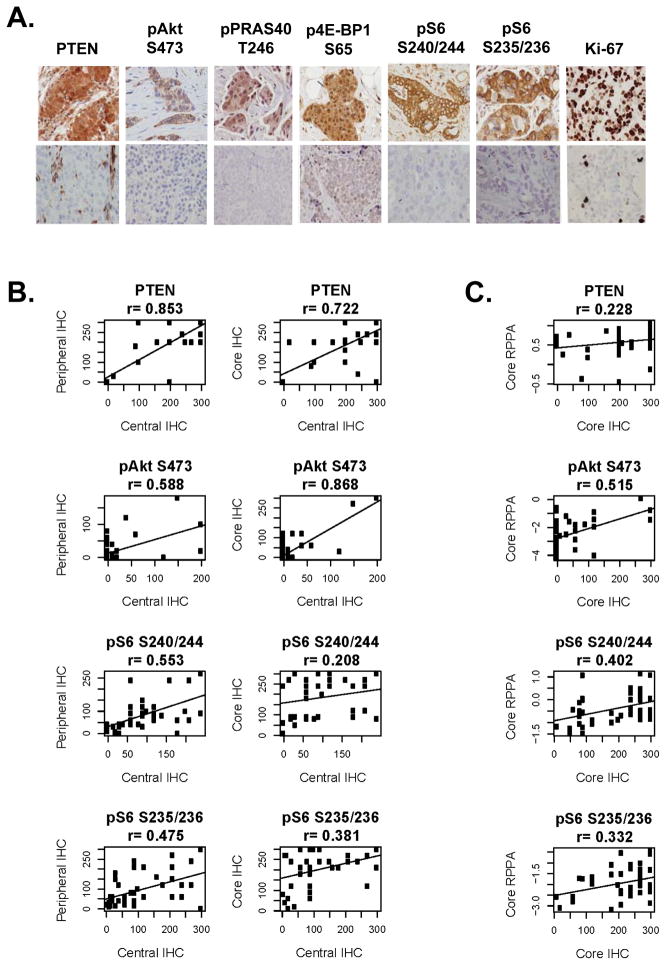
We determined PI3K/Akt/mTOR signaling using IHC. Examples of positive and negative staining for each antibody are shown in Figure 5A.
Figure 5.
IHC for PI3K markers. A. IHC for PTEN, pAkt S473, pPRAS40 T246, p4E-BP1 S65, pS6 S240/244, and Ki-67. Examples of positive (top panel) and negative (lower panel) immunostaining for each marker are included. B. Correlation of immunostaining for PTEN, pAkt S473, pS6 S240/244, and pS6 S235/236 between central and peripheral specimens (left), and central specimens and core needle biopsy (right). Immunostaining was quantitated by H-score. C. Correlation of central RPPA results with core needle biopsy specimen immunostaining for PTEN, pAkt S473, pS6 S240/244, and pS6 S235/236.
There was a very strong correlation in p4E-BP1 T70 expression in the central and peripheral tumor (r=0.81. There was a moderate correlation in central and peripheral expression of pAkt S473 (r=0.588), pPRAS40 T246 (r=0.82), pS6 S240/244 (r=0.553), and moderate to low correlation for pS6 S235/236 (r=0.475) (Figure 5B). There was a very strong correlation in PTEN expression in the central and peripheral tumor (r=0.85) (Figure 5B). When PTEN IHC was categorically classified as having PTEN loss vs. not; the central and peripheral samples had almost perfect agreement (κ = 0.916), CNB and central specimen had substantial agreement (κ = 0.682) and CNB and peripheral specimen had moderate agreement (κ = 0.579).
The CNB and central specimen immunostaining also correlated for pAkt S473 (r=0.87), PTEN (r=0.72), p4E-BP1 T70 (r=0.81), and pS6 S235/236 (r=0.38), but not for pPRAS40 T246 or pS6 S240/244. The CNB and peripheral specimen immunostaining were correlated for pAkt S473 (r=0.71), PTEN (r=0.68), p4E-BP1 S65 (r=0.63), pS6 S240/244 (r=0.47), and pS6 S235/236 (r=0.49).
We next compared PI3K pathway activation status by IHC to that of RPPA. pAkt S473 quantitation by RPPA correlated with immunostaining by IHC in central specimen (r=0.71), peripheral specimen (r=0.65), and CNB (r=0.52) (Figure 5C). RPPA quantitation of PTEN correlated with IHC results in the central specimen (r=0.41), but not in the peripheral tumor or CNB specimens. pS6 S235/236 expression by RPPA and IHC also were significantly correlated in the central specimen and in the CNB, but not tumor periphery(r= 0.24). When RPPA quantitation of FNAs was compared to IHC of central specimens, there was moderate correlation for pAkt S473 (r=0.30), PTEN (r=0.37) and pS6 S235/236 (r=0.35).
There was a very strong correlation between percentage cells positive for Ki-67 between the central and peripheral specimen (r=0.865) and between the CNB and central specimen (r=0.891). There was no significant correlation between the expression of proliferation marker PCNA (proliferating cell nuclear antigen) on RPPA and percentage cells positive for Ki-67 on IHC.
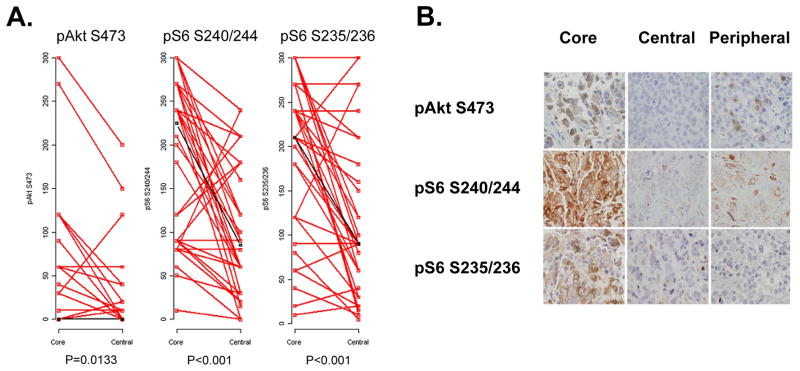
We then determined whether there were differences in immunostaining by biospecimen type. The pAkt S473 expression in the CNB was significantly higher than that in the central specimen (p=0.013, Figure 6A) but not the peripheral specimen. The pS6 S240/244 and pS6 S235/236 H scores on the CNB were significantly higher than both the central specimen and the peripheral specimen (p<0.001 for both, Figure 6A). Examples of discordant IHC results, with higher immunostaining in core biopsy than surgical specimens are shown in Figure 6B.
Figure 6.
Comparison of IHC in core biopsy and central surgical specimens. A. Immunostaining for pAkt S473, pS6 S240/244, and pS6 S235/236 in matched core biopsy and central surgical specimens. IHC immunostaining was quantitated with H score. B. Examples of discordant samples with strong immunostaining for pAkt S473, pS6 S240/244, and pS6 S235/236 in the core biopsy, but not in the matched central or peripheral specimens.
Concordance of PI3K Pathway Classification by RPPA
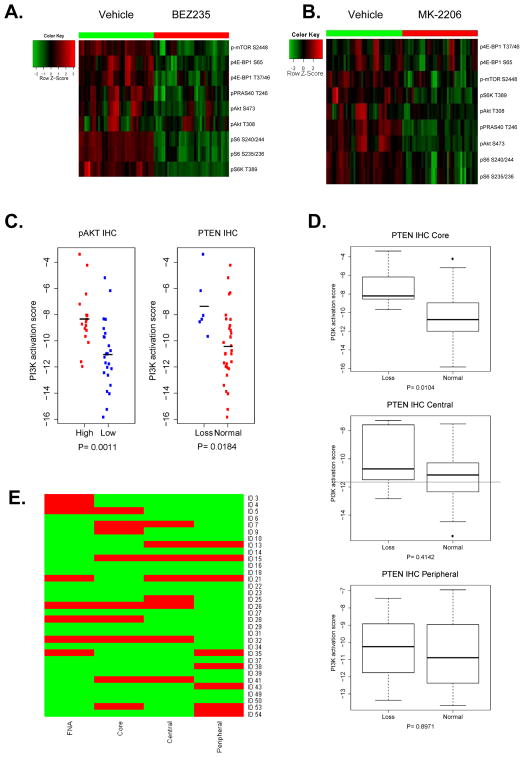
We assessed pAkt S473, pAkt T308, pPRAS40 T246, p-mTOR S2448, p4E-BP1 S65, p4E-BP T37/46, pS6K T389, pS6 240/244, pS6 S235/236 levels as indicators of high PI3K pathway activity, as they are known downstream targets (10, 20), and as their phosphorylation was found to be strongly inhibited by PI3K/mTOR inhibitor BEZ235 and Akt inhibitor MK-2206 in a panel of cancer cell lines (Figure 7A and B).
Figure 7.
Activity of PI3K pathway by RPPA sample type. Regulation of PI3K downstream signaling by PI3K/mTOR inhibitor BEZ235 (A) and Akt inhibitor MK-2206 (B). C. PI3K activation score by IHC results. PI3K score in patients with loss of PTEN on IHC (PTEN H score of 0) vs. PTEN H score>0 (left panel). PI3K score in patients with low pAkt S473 on IHC (pAkt S473 H score of 0) vs. pAkt S473 H score>0 (right panel). D. PI3K activation by biospecimen type. Activated samples by RPPA PI3K score are in red, not activated samples in green. Only patients with all sample types available are presented in this figure. E. PI3K pathway activity by RPPA for samples that had all four biospecimen types was available. Red activated, green not activated.
When all samples were analyzed together, as expected tumors with expression of pAkt S473 on IHC also had a significantly higher PI3K score than those that did not (p=0.001; Figure 7C). Further, samples with PTEN loss by IHC (H score=0) had a significantly higher PI3K pathway activation score (p=0.018; Figure 7C). The statistical difference in PI3K activity score was not preserved when the threshold for PTEN loss was elevated to H score of 50 or 100, suggesting that complete PTEN loss establishes the greatest differentiation between tumors that have PI3K activation vs. not. When PTEN IHC and PI3K activity on RPPA was analyzed separately for each sample type (core, central and peripheral), PI3K activity score was significantly higher in CNB that had PTEN loss vs. those without PTEN loss by IHC (p=0.0104), while there was no there was no difference in PI3K activity score central or peripheral samples that had PTEN loss vs. those without PTEN loss by IHC (Figure 7D).
We then classified tumors as “PI3K signaling activated” or “not activated” based on PI3K activity score calculated separately for each specimen type. F-test in linear regression model demonstrated that the variance of PI3K pathway score could not be explained by the tumor cell composition.
The samples in the top quartile of PI3K RPPA core within each specimen type were classified as “PI3K signaling activated”. In the 37 patients where RPPA results were available on both CNB and central specimens, the overall agreement between the core biopsy and central specimen was 78.4% (κ = 0.454, p=0.005). In the 39 patients for whom both central and peripheral RPPA results were available, the overall agreement was 74.4% (κ = 0.33; p=0.04). For samples that had all four biospecimen types were available, activity of PI3K pathway for each RPPA biospecimen type is shown in Figure 7E.
Discussion
To deliver on the promise of personalized therapy, it is critical to develop robust markers that can assess activation of druggable targets such activation of PI3K/Akt/mTOR signaling in patient samples. Further, in early clinical trials with targeted therapies, it is critical to incorporate pharmacodynamic studies to determine whether a new drug inhibits its target and by how much. Such studies rely on the ability to accurately assess proteomic markers in tumor samples. Marker assessment for patient selection is often performed on archival surgical tissue in FFPE; however there are significant concerns about tumor heterogeneity and marker stability, especially for phospho-proteins. Therefore, we sought to determine how tumor heterogeneity and biospecimen variables affect assessment of proteomic biomarkers and, in particular, PI3K/Akt/mTOR pathway activation. Proteomic levels were differentially affected by biospecimen type and other preanalytic variables. For example, PI3K pathway activation was greater in CNB compared to surgical samples suggesting loss of phosphorylation during tumor handling during surgery and cold ischemia after removal from the patient.
In genomic studies, intratumoral heterogeneity was observed for mutations in oncogenes and multiple tumor-suppressor genes, even with gene-expression signatures of both good and poor prognosis being detected in different regions of the same tumor (6). Thus, we were concerned that there may be significant heterogeneity in proteomic markers, and we hypothesized that there may be differences between the tumor center and periphery due to tumor hypoxia etc. Indeed the PI3K pathway classification by RPPA activation score did differ for some tumors. However, there was no statistical difference in the expression between central and peripheral specimens for most (86%) of the individual markers tested and there was a moderate to strong correlation between the central and peripheral specimen results for all but 5 markers tested. The correlation between central and peripheral samples was stronger in smaller tumors suggesting there may be greater tumor heterogeneity in larger tumors.
There is a great interest in PI3K/Akt/mTOR signaling. Our studies suggest that pAkt on RPPA and PTEN on IHC are relatively robust markers. As Akt phosphorylation is correlated with kinase activity, pAkt is one the most compelling markers to assess for pathway activation. pAkt expression on RPPA in different biopsy and surgical biospecimens showed a moderate to strong correlation. pAkt staining on IHC had a more limited range, however, pAkt expression in RPPA and IHC showed a moderate correlation, further strengthening the reliability of pAkt as a marker of pathway activity. The tumor suppressor PTEN is a critical negative regulator of PI3K signaling that is aberrantly expressed in many tumor types, and thus is already being used as a marker for patient selection in ongoing clinical trials for PI3K/Akt/mTOR inhibitors. Our results show that PTEN IHC has a very strong correlation between central and peripheral specimens and between CNB and surgical specimens. The PTEN expression of central and peripheral samples and CNB vs. surgical samples were strongly correlated, however the correlation of PTEN expression between RPPA and IHC was weak. This is likely due to the effect of stroma on RPPA results. Even if PTEN is lost in tumor cells, PTEN expression in stroma is retained, and in fact stroma serves as a positive control in IHC. On IHC, only tumor PTEN staining is scored, but on RPPA, in the absence of microdissection, quantitation of PTEN in the tumor lysate represents PTEN expression in stroma as well as tumor cells. Thus, in less cellular tumors PTEN loss may not be as apparent on RPPA, and this may contribute to weak correlation between IHC and RPPA. PTEN IHC is attractive as not only does it appear to have low interspecimen variability, but also PTEN loss and in particular complete loss (H score=0) on IHC was associated with PI3K pathway activation by RPPA. Larger studies are needed to further test the impact on “low” PTEN on pathway activation in order to set the appropriate threshold for PTEN loss that is biologically and clinically meaningful.
There has been increasing concerns that specimen type (such as CNB versus surgical sample) may affect biomarkers. When standard breast cancer markers in CNB were compared to excisions, discordance between CNB and excision for HER2 and ER expression was found to be rare (1.8% and 1.2% respectively), however discordance in PR expression was common (15%) (21). A recent meta-analysis that included 21 articles involving nearly 2,500 patients compared concordance of ER and PR in CNBs and excision and found an overall agreement between CNB and surgical excisions of 92.8 % for ER (κ = 0.78) and 85.2 % for PR (κ = 0.66) (22). In contrast Baker et al reported that in colorectal cancer, pAkt S473 staining was observed only in biopsies and not in surgical samples (albeit not assessed with RPPA) (2), thus authors suggested that caution should be used when using phosphoprotein levels in surgical specimens to measure intrinsic signaling activity or drug effects because of the potential for rapid dephosphorylation. Recently, to evaluate the impact of ischemia, Mertins et al collected ovarian tumor and breast cancer xenograft and performed quantitative proteomics and phosphoproteomics with RPPA and liquid chromatography-mass spectroscopy after defined ischemic intervals (23). While the global expressed proteome and most of the >25,000 quantified phosphosites were unchanged after 60 minutes, rapid phosphorylation changes were observed in up to 24% of the phosphoproteome. Concern about stability of cell signaling markers was also raised in a recent study on breast cancer, in which loss of pAkt and pErk was found by IHC on CNB obtained ex vivo but not in surgical samples after routine processing (3). These studies suggest that loss of phospho-staining can occur during routine fixation of resection specimens. Our study design was different than the latter breast cancer study as our CNB were obtained not on surgical samples ex vivo, but in the operating room in vivo, while samples of surgical specimens were frozen or formalin fixed immediately upon specimen processing for clinical care, thus mimicking earliest potential time point for specimen collection for tumor banking or biomarker assessment. Our study findings are consistent with the previous reports in that pAkt and pS6 levels were higher in CNB, and there were several tumors in which the PI3K score was elevated in the CNB, but not in the surgical sample. Taken together, our results suggest that PI3K pathway activation is greater in vivo, with subsequent dephosphorylation/pathway inactivation ex vivo.
PI3K/Akt/mTOR pathway activity is being pursued as a potential predictor of response, marker of target inhibition and pharmacodynamic marker of response. Recently in a randomized, double-blind, placebo-controlled, phase 3 trial Jerusalem et al reported that patients with HER2 + metastatic breast cancer with high pS6 levels, derived more benefit from the addition of everolimus to trastuzumab and vinorelbine (24). However, our data and those from others suggest results of PI3K/Akt/mTOR testing such as pS6 levels may vary based on specimen acquisition approach and processing. This has several clinical implications. First, PI3K activation is likely better assessed on CNB than surgical specimens. Thus if pathway activation is to be pursued as a potential prognostic or predictive biomarker, CNBs may be preferred over surgical specimens for biomarker assessment. Expedited biospecimen processing/preservation approaches for surgical specimens may also improve accuracy. Second, this finding has study design implications for window of opportunity and neoadjuvant therapy studies planned with PI3K inhibitors. In these studies, breast tumors are treated with PI3K/Akt/mTOR targeted therapy for weeks to months after a pre-treatment biopsy to assess effects on pathway activation compared to baseline levels. As PI3K activation may be higher in CNB compared to surgical samples even without treatment, pharmacodynamic effects of targeted therapies could be best assessed either by obtaining a post-treatment CNB for comparison to pre-treatment CNB or by having a placebo controlled arm to allow for comparison of surgical specimens in both arms.
It is encouraging that there was a moderate/strong correlation between the CNB and surgical specimen for most proteins and phosphoproteins. The expression of several putative prognostic markers and druggable targets correlated significantly between the CNB and the surgical specimen, but some did not. Thus it is important to determine effect of biospecimen type for each marker used to determine its limitations. It may then be possible to identify a set of biomarkers that can be robustly measured on both CNB and surgical specimens obviating some of the concerns with sample handling. The correlation of CNB with surgical specimen was greater than that seen between FNA and surgical specimen. This is not surprising given that FNA is likely to be enriched for epithelial cells and have fewer stroma cells, and perhaps a larger contribution from blood. However it is notable that several markers were significantly correlated between FNA and surgical sample as well, including standard of care markers such as ER, PR, and HER2. Thus truly robust markers are likely to be reliable regardless of biospecimen type. Indeed, ER, PR and HER2 all tend to have a large dynamic range and to also be dichotomous markers suggesting that these variables may contribute to the robustness of the marker. This is further highlighted by the fact that several studies have successfully used FNAs to demonstrate the prognostic role of known and novel proteomic markers and to assess pharmacodynamics markers of response to targeted therapies (10, 25).
Our study has several limitations. In our study, all FNAs and core biopsies were done intraoperatively, and were immediately followed by surgical resection. However, FNA and core needle biopsies are invasive procedures that may result in wounding/bleeding/tissue damage. The local wound response itself may influence cell signaling, thus affecting proteomic profile. Further, as we collected surgical samples in the frozen section suite at the first possible time point, and small portions of tumor were formalin fixed, we are not capturing the discordance that would have emerged due to variation in fixation of larger specimens. Concordance between biospecimens has been variable, suggesting that preanalytic variables may affect markers differentially; however, in the absence of clinical outcome data, the impact of this variability cannot be assessed and thresholds for acceptable interspecimen heterogeneity has not been established for most markers. Before clinical transition of any biomarker, inter-assay reproducibility needs to be well established and clinically relevant cut-offs for each marker needs to be determined. In our analysis, correlation of biomarkers in different biospecimens varied from being strongly correlated to not being correlated. Further, discrepancies were seen between different platforms and specimens in both directions. Biomarkers that have less intraspecimen variability are likely to be more robust, more reproducible biomarkers with greater clinical utility. RPPA is a compelling research tool as it is tissue sparing, and allows for assessment of multiple markers from a small amount of protein lysate. One limitation of RPPA of macrodissected tumor lysate is that it represents the signal in the tumor cells as well as the surrounding stroma. An area of controversy has been whether laser aided microdissection is required (26). We performed RPPA after macrodissection, without laser capture microdissection, an approach also employed in The Genome Cancer Atlas for breast cancer as well as several other cancer types (26–28). As we did not do a side by side comparison of microdissected and macrodissected samples, in our study we did not test the utility of microdissection. Although IHC allows for assessment of fewer markers and may be less quantitative, it is a more readily available approach, and provides information about localization of expression, and stromal or other normal contamination is less of a concern. Other biospecimen differences such as sample type may affect results of both assay types. Optimal biospecimen type and assay approach should be considered for each biomarker and application.
In summary, overall there is limited spatial intratumoral heterogeneity for individual proteomic markers in operable breast cancer. However, some actionable markers are differentially expressed by biospecimen type and other preanalytic variables. Although expression of most markers on CNB and surgical samples correlate, some do differ. Notably, PI3K pathway activation is greater in CNB compared to surgical samples, thus CNB may be preferable to surgical samples to assess pathway activity. Careful study of preanalytic and analytic variables that influence results is needed for clinical implementation of each biomarker.
Supplementary Material
Translational Relevance.
There is great interest in using protein based biomarkers, especially phosphoproteins for assessment of pathway activation, as potential predictors of response to targeted therapies. Our results demonstrate that most individual proteomic biomarkers did not have significant intratumoral heterogeneity. However, protein and phosphoprotein levels were affected by biospecimen type and other preanalytic variables. PI3K pathway activation was greater in CNB compared to surgical samples. Use of archival samples, especially surgical samples may introduce additional variability to biomarker assessment. These findings have implications for study design for biomarker-driven targeted therapy trials as well as window of opportunity studies. These studies also highlight the need for biospecimen science to reduce postexcision degradation of phosphorylation as well as the need for identifying the most robust markers least prone to intratumoral heterogeneity and biospecimen and assay-related variation.
Acknowledgments
Funding: This work was supported by a Society of Surgical Oncology Clinical Investigator Award in Breast Cancer Research sponsored by Susan G. Komen for the Cure (FMB), Susan G. Komen for the Cure SAC10006 (FMB, KAD), Stand Up to Cancer Dream Team Translational Research Grant, a Program of the Entertainment Industry Foundation administered by the American Association for Cancer Research (SUC2-AACR-DT0209) (AMG, GBM, AA, FMB), the Kleberg Foundation (FMB, AMG, GM), and NCRR Grant 3UL1RR024148 (FMB, KAD) and UL1TR000371 (AA, FMB, KAD), and the National Cancer Institute through R21CA159270 (FMB, AMG, ET) and The University of Texas MD Anderson Cancer Center Support Grant (P30 CA016672).
References
- 1.McAuliffe PF, Meric-Bernstam F, Mills GB, Gonzalez-Angulo AM. Deciphering the role of PI3K/Akt/mTOR pathway in breast cancer biology and pathogenesis. Clin Breast Cancer. 2010;10 (Suppl 3):S59–65. doi: 10.3816/CBC.2010.s.013. [DOI] [PubMed] [Google Scholar]
- 2.Baker AF, Dragovich T, Ihle NT, Williams R, Fenoglio-Preiser C, Powis G. Stability of phosphoprotein as a biological marker of tumor signaling. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:4338–40. doi: 10.1158/1078-0432.CCR-05-0422. [DOI] [PubMed] [Google Scholar]
- 3.Pinhel IF, Macneill FA, Hills MJ, Salter J, Detre S, A’Hern R, et al. Extreme loss of immunoreactive p-Akt and p-Erk1/2 during routine fixation of primary breast cancer. Breast cancer research : BCR. 2010;12:R76. doi: 10.1186/bcr2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spruessel A, Steimann G, Jung M, Lee SA, Carr T, Fentz AK, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. BioTechniques. 2004;36:1030–7. doi: 10.2144/04366RR04. [DOI] [PubMed] [Google Scholar]
- 5.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabioglu N, Hunt KK, Sahin AA, Kuerer HM, Babiera GV, Singletary SE, et al. Role for intraoperative margin assessment in patients undergoing breast-conserving surgery. Annals of surgical oncology. 2007;14:1458–71. doi: 10.1245/s10434-006-9236-0. [DOI] [PubMed] [Google Scholar]
- 8.Hatzis C, Sun H, Yao H, Hubbard RE, Meric-Bernstam F, Babiera GV, et al. Effects of tissue handling on RNA integrity and microarray measurements from resected breast cancers. Journal of the National Cancer Institute. 2011;103:1871–83. doi: 10.1093/jnci/djr438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, et al. A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-microdissected Human Breast Cancers. Clin Proteomics. 2010;6:129–51. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meric-Bernstam F, Akcakanat A, Chen H, Do KA, Sangai T, Adkins F, et al. PIK3CA/PTEN mutations and Akt activation as markers of sensitivity to allosteric mTOR inhibitors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:1777–89. doi: 10.1158/1078-0432.CCR-11-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudoit S, Yang YH. Bioconductor R packages for exploratory analysis and normalization of cDNA microarray data. In: Parmigiani G, Garrett ES, Irizarry RA, Zeger SL, editors. The Analysis of Gene Expression Data: Methods and Software. New York: Springer; 2002. [Google Scholar]
- 12.Yang YH, Dudoit S, Luu P, Speed TP. Normalization for cDNA microarray data. In: Bittner ML, Chen Y, Dorsel AN, Dougherty ER, editors. Microarrays: Optical Technologies and Informatics. SPIE-International Society for Optical Engine; 2001. [Google Scholar]
- 13.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic acids research. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Best DJ, Roberts DE. Algorithm AS 89. The upper tail probabilities of Spearman’s rho. Applied Statistics. 1975:24. [Google Scholar]
- 15.Hollander M, Wolfe DA. Nonparametric Statistical Methods. New York: John Wiley & Sons; 1973. [Google Scholar]
- 16.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–74. [PubMed] [Google Scholar]
- 17.Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics (Oxford, England) 2007;23:1986–94. doi: 10.1093/bioinformatics/btm283. [DOI] [PubMed] [Google Scholar]
- 18.Symmans WF, Ayers M, Clark EA, Stec J, Hess KR, Sneige N, et al. Total RNA yield and microarray gene expression profiles from fine-needle aspiration biopsy and core-needle biopsy samples of breast carcinoma. Cancer. 2003;97:2960–71. doi: 10.1002/cncr.11435. [DOI] [PubMed] [Google Scholar]
- 19.Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–25. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol. 2009;27:2278–87. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnedos M, Nerurkar A, Osin P, A’Hern R, Smith IE, Dowsett M. Discordance between core needle biopsy (CNB) and excisional biopsy (EB) for estrogen receptor (ER), progesterone receptor (PgR) and HER2 status in early breast cancer (EBC) Annals of oncology : official journal of the European Society for Medical Oncology/ESMO. 2009;20:1948–52. doi: 10.1093/annonc/mdp234. [DOI] [PubMed] [Google Scholar]
- 22.Li S, Yang X, Zhang Y, Fan L, Zhang F, Chen L, et al. Assessment accuracy of core needle biopsy for hormone receptors in breast cancer: a meta-analysis. Breast cancer research and treatment. 2012;135:325–34. doi: 10.1007/s10549-012-2063-z. [DOI] [PubMed] [Google Scholar]
- 23.Mertins P, Yang F, Liu T, Mani DR, Petyuk VA, Gillette MA, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteomics. 2014 doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerusalem G, Andre F, Chen D. Evaluation of everolimus (EVE) in HER2+ advanced breast cancer (BC) with activated PI3K/mTOR pathway: Exploratory biomarker observations from the BOLERO-3 trial. European Cancer Congress; Amsterdam, Netherlands. 2013; p. Abstract LBA16. [Google Scholar]
- 25.Gonzalez-Angulo AM, Hennessy BT, Meric-Bernstam F, Sahin A, Liu W, Ju Z, et al. Functional proteomics can define prognosis and predict pathologic complete response in patients with breast cancer. Clin Proteomics. 2011;8:11. doi: 10.1186/1559-0275-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller C, Decarvalho AC, Mikkelsen T, Lehman NL, Calvert V, Espina V, et al. Glioblastoma cell enrichment is critical for analysis of phosphorylated drug targets and proteomic-genomic correlations. Cancer Res. 2014;74:818–28. doi: 10.1158/0008-5472.CAN-13-2172. [DOI] [PubMed] [Google Scholar]
- 27.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akbani R, Ng PKS, Werner H, Shahmoradgoli M, Zhang F, Ju Z, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nature Comms. 2014 doi: 10.1038/ncomms4887. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.