Summary
Virus-induced gene silencing (VIGS) was used in this study to characterize the role of thioredoxin-f and thioredoxin-m1/4 in brassinosteroid-induced changes in CO2 assimilation and cellular redox homeostasis in tomato.
Key words: Antioxidant, Benson–Calvin cycle, chloroplast, 2-Cys peroxiredoxin, glutathione, photosynthesis.
Abstract
Chloroplast thioredoxins (TRXs) and glutathione function as redox messengers in the regulation of photosynthesis. In this work, the roles of chloroplast TRXs in brassinosteroids (BRs)-induced changes in cellular redox homeostasis and CO2 assimilation were studied in the leaves of tomato plants. BRs-deficient d ^im plants showed decreased transcripts of TRX-f, TRX-m2, TRX-m1/4, and TRX-x, while exogenous BRs significantly induced CO2 assimilation and the expression of TRX-f, TRX-m2, TRX-m1/4, and TRX-x. Virus-induced gene silencing (VIGS) of the chloroplast TRX-f, TRX-m2, TRX-m1/4, and TRX-y genes individually increased membrane lipid peroxidation and accumulation of 2-Cys peroxiredoxin dimers, and decreased the activities of the ascorbate–glutathione cycle enzymes and the ratio of reduced glutathione to oxidized glutathione (GSH/GSSG) in the leaves. Furthermore, partial silencing of TRX-f, TRX-m2, TRX-m1/4, and TRX-y resulted in decreased expression of genes involved in the Benson–Calvin cycle and decreased activity of the associated enzymes. Importantly, the BRs-induced increase in CO2 assimilation and the increased expression and activities of antioxidant- and photosynthesis-related genes and enzymes were compromised in the partially TRX-f- and TRX-m1/4-silenced plants. All of these results suggest that TRX-f and TRX-m1/4 are involved in the BRs-induced changes in CO2 assimilation and cellular redox homeostasis in tomato.
Introduction
Accumulating evidence supports the hypothesis that the cellular redox signalling and hormone signalling pathways form an integrated redox–hormone network that regulates many plant growth- and defence-related pathways (Bartoli et al., 2013). Redox regulation is often mediated by thioredoxins (TRXs), which are able to (de-)activate enzymes through the reversible reduction of disulphide bonds (Buchanan and Balmer, 2005; Montrichard et al., 2009).
TRXs are ubiquitous, low molecular weight (~12kDa) proteins that catalyse thiol–disulphide exchange reactions in conjunction with a large number of enzymes and related proteins (Jacquot et al., 2002). The short peptide motif WC(G/P)PC, with two conserved cysteine residues, has been characterized as the conserved active redox site of TRXs (Jacquot et al., 1997; Buchanan and Balmer, 2005; Meyer et al., 2008). Plant cells contain numerous TRXs in the cytosol, nucleus, mitochondria, endoplasmic reticulum, and chloroplasts, which continue to be identified (Marcus et al., 1991; Baumann and Juttner, 2002; Meyer et al., 2005). Recent proteomic studies using TRX-trapping chromatography or labelled gel electrophoresis in combination with protein identification via mass spectrometry have identified >180 potential in vitro TRX target proteins (Motohashi et al., 2001; Balmer et al., 2003, 2004, 2006; Wong et al., 2004; Marchand et al., 2006; Alkhalfioui et al., 2007).
There are five types of typical TRXs present in the chloroplasts. The f- and m-type TRXs function as messengers in the Fdx (ferredoxin)/TRX system by transmitting the redox signal from Fdx:TRX reductase to the target enzymes. TRX-f has been shown to activate several chloroplast enzymes, including enzymes involved in the Benson–Calvin cycle, ATP synthesis, and fatty acid synthesis (Nishizawa and Buchanan, 1981; Wolosiuk et al., 1993; Sasaki et al., 1997; Schwarz et al., 1997; Zhang and Portis, 1999). TRX-m has been found to be involved in the inactivation of glucose-6-phosphate dehydrogenase, which is the first enzyme in the oxidative pentose phosphate cycle (Wenderoth et al., 1997). Both TRX-f and TRX-m can activate fructose-1,6-bisphosphatase (FBPase) and NADP-dependent malate dehydrogenase (NADP-MDH) in vitro, suggesting that there is some overlap in substrate specificity between the isoforms (Hodges et al., 1994; Geck et al., 1996). TRX-x, which is inactive toward FBPase and NADP-MDH, is the most efficient reductant of 2-Cys peroxiredoxin (2-CP). This property suggests that TRX-x functions specifically in resistance to oxidative stress (Collin et al., 2003). TRX-y exists in two isoforms and has been reported to act as an efficient electron donor for the chloroplast glutathione peroxidase (Navrot et al., 2006) and methionine sulphoxide reductase B2 (Vieira Dos Santos et al., 2007). Peroxiredoxin Q, which has been reported to function as an antioxidant (Rouhier et al., 2004; Lamkemeyer et al., 2006), was shown to be the best substrate of TRX-y (Collin et al., 2004). Lastly, TRX-z regulates plastid-encoded RNA polymerase-dependent transcription (Arsova et al., 2010). However, the biological functions of TRXs have been studied mostly in Arabidopsis through in vitro experiments, and detailed in vivo studies using different isoforms have not yet been conducted.
Brassinosteroids (BRs) are a group of plant steroid hormones that have been shown to play important roles in the growth, development, and stress response of plants (Clouse and Sasse, 1998; Müssig, 2005). For example, the tomato d ^im mutant with the block in C-6 oxidation in BRs biosynthesis results in a dwarf phenotype (Bishop et al., 1999). Previous studies showed that BRs can enhance stress tolerance against chilling, paraquat (PQ), and biotic stress, and this effect is largely dependent on BRs-induced hydrogen peroxide (H2O2) accumulation in the apoplast (Xia et al., 2009b ). Furthermore, BRs are involved in the regulation of CO2 assimilation in several plants. In cucumber, BRs induce transient increases in RBOH1 (respiratory burst oxidase homolog 1) expression, NADPH oxidase activity, H2O2 levels in the apoplast, and nitric oxide (NO) production (Xia et al., 2009b ; Cui et al., 2011). Additionally, BRs can induce the expression and activity of genes and enzymes involved in the Benson–Calvin cycle, and this effect is attributed to the apoplastic H2O2-induced activation of photosynthesis-related redox-sensitive enzymes (Jiang et al., 2012a ). Apoplastic H2O2 can induce an increase in the ratio of reduced glutathione (GSH) to oxidized glutathione (GSSG), resulting in increased stability of redox-sensitive enzymes (Jiang et al., 2012b ). Microarray analysis revealed that several TRX genes are targeted by BRs in Arabidopsis and rice (Müssig et al., 2002; Goda et al., 2004; Wu et al., 2008); however, their potential roles in BRs-induced cellular redox homeostasis and CO2 assimilation remain unknown.
Tomato is an important horticultural crop distributed world-wide and has been widely used to study the stress response. Virus-induced gene silencing (VIGS) has been well established and widely used for the analysis of gene functions in Solanaceous species (Liu et al., 2002a, b ; Li et al., 2006). In this study, the roles of the different chloroplastic TRXs in the BRs-induced changes in cellular redox homeostasis and CO2 assimilation were examined. Accordingly, five chloroplast TRXs were partially silenced individually using a VIGS approach, and the changes in CO2 assimilation as well as the activities of Calvin cycle and ascorbate (AsA)–GSH cycle enzymes and cellular glutathione redox homeostasis were subsequently determined in the presence or absence of exogenous BRs.
Materials and methods
Plant materials and experimental design
Experiment I
Tomato seeds from wild-type (Solanum lycopersicum L. cv. Condine Red, CR) and its partially BRs-deficient d ^im mutant were obtained from the Tomato Genetics Resource Center (University of California, Davis, CA, USA). These seeds were germinated and grown in a mixture of peat and vermiculite (1:1, v:v) under a 16h light (200 μmol m–2 s–1; at 25 °C), 8h dark (at 20 °C) cycle. To determine the role of BRs in the regulation of CO2 assimilation and the effects of BRs on the transcript levels of the chloroplastic TRX genes, plants at the four-leaf stage were sprayed with water or 0.2 μM EBR (24-epibrassinolide; Sigma, Santa Clara, CA, USA), one of the most active and stable forms of BRs. Twenty-four hours later, CO2 assimilation was measured in the third leaf from the bottom. Additionally, leaf samples were harvested from the water- or EBR-treated CR and d ^im plants, frozen immediately in liquid nitrogen, and stored at –80 °C prior to gene expression analyses.
Experiment II
Tomato (CR) seeds were germinated and grown in a mixture of peat and vermiculite (1:1, v:v) under a 16h light (200 μmol m–2 s–1; at 25 °C), 8h dark (at 20 °C) cycle. To examine the roles of the chloroplast TRXs in the regulation of CO2 assimilation and to determine whether they are involved in the BRs-induced increase in CO2 assimilation, VIGS was performed when the cotyledonary leaves were fully expanded but the true leaves had not yet appeared. Partially gene-silenced plants at the six-leaf stage were treated with 0.2 μM EBR via foliar spraying on all of the leaves; 10ml of this solution was applied per plant, and distilled water was used as a control. After ~24h, CO2 gas exchange was measured, and leaf samples were harvested for biochemical and gene expression analyses.
VIGS constructs and Agrobacterium-mediated virus infection
Five chloroplast TRX isoforms were obtained, namely TRX-f, TRX-m2, TRX-m1/4, TRX-x, and TRX-y, from the tomato genome database using the BLAST tool (http://mips.helmholtz-muenchen.de/plant/tomato/index.jsp) based on their similarity to the characterized chloroplast TRX isoforms in Arabidopsis. To generate the Tobacco rattle virus (pTRV) VIGS constructs pTRV2-TRX-f, pTRV2-TRX-m2, pTRV2-TRX-m1/4, pTRV2-TRX-x, and pTRV2-TRX-y, which were used for partial silencing of the TRX-f, TRX-m2, TRX-m1/4, TRX-x, and TRX-y genes, respectively, fragments of the corresponding genes with sizes of 210, 231, 312, 285, and 251bp were PCR amplified from tomato cDNA with primers containing XhoI (CTCGAG) and SacI (GAGCTC) restriction sites. All of the primers used are listed in Supplementary Table S1 available at JXB online. The resulting plasmids were subsequently introduced into Agrobacterium tumefaciens strain GV3101.
For virus infiltration, a mixed culture of A. tumefaciens carrying the pTRV1:pTRV2-target gene in a 1:1 ratio was infiltrated into fully expanded cotyledonary leaves of tomato plants (Ekengren et al., 2003). An Agrobacterium culture carrying the empty pTRV2 vector was also infiltrated into a set of plants, which were used as a control. The inoculated plants were maintained at 20–22 °C in a growth chamber with a 16h daylength. After ~4 weeks, quantitative real time–PCR (qRT–PCR) was performed to determine the gene silencing efficiency before the plants were used in assays (Supplementary Fig. S1 available at JXB online).
Leaf gas exchange measurements
Gas exchange analysis was conducted on the third leaf for Experiment I and on the fifth leaf for Experiment II using an open gas exchange system (LI-6400; LI-COR, Lincoln, NE, USA). The light-saturated rate of CO2 assimilation (A sat) was measured under an ambient CO2 concentration of 380 μmol mol–1 at a saturating photosynthetic photon flux density (1000 μmol m–2 s–1), at a leaf temperature of 25±1.5 °C and a relative air humidity of 80–90%. An assimilation versus intercellular CO2 concentration (A/C i) curve was determined according to von Caemmerer and Farquhar (1981). Assimilation was first measured at the ambient CO2 concentration under which the plants had grown. The atmospheric CO2 concentration was decreased to 50 μmol mol–1 in a stepwise manner and then returned to the growth concentration to check that the original rate could be regained; the rate was then finally increased stepwise to 2000 μmol mol–1 to complete the response curve. The maximum ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) carboxylation rates (V c,max) and maximum ribulose-1,5-bisphosphate (RuBP) regeneration rates (J max) were estimated from the A/C i curves using the method described by Ethier and Livingston (2004).
Determination of lipid peroxidation
Lipid peroxidation was estimated by measuring malondialdehyde (MDA) equivalents, according to the method described by Hodges et al. (1999). Leaf samples (0.3g) were ground in 3ml of ice-cold 25mM HEPES buffer (pH 7.8) containing 0.2mM EDTA and 2% polyvinylpyrrolidone (PVP). The obtained homogenates were centrifuged at 4 °C for 20min at 12000 g, and the resulting supernatants were used for analysis of MDA equivalents. The samples were mixed with 10% trichloroacetic acid (TCA) containing 0.65% 2-thiobarbituric acid (TBA) and heated at 95 °C for 25min. The content of MDA equivalents was corrected for non-MDA compounds by subtracting the absorbance at 532nm of a TBA-less solution containing the plant extract.
Non-reducing SDS–PAGE and western blot analysis of 2-Cys peroxiredoxin
Total proteins were isolated from leaf tissues in a protein extraction buffer (100mM HEPES, pH 7.5, 5mM EDTA, 5mM EGTA, 10mM Na3VO4, 10mM NaF, 50mM β-glycerophosphate, 1mM phenylmethylsulphonyl fluoride, 10% glycerol, and 7.5% polyvinylpolypyrrolidone) supplemented with 10mM N-ethylmaleimide (NEM) (thiol-blocking reagent). After centrifugation at 13 000 g for 20min, the supernatants were transferred to clean tubes, quickly frozen in liquid nitrogen, and stored at –80 °C. The protein concentration in the extracts was determined using the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA), and bovine serum albumin (BSA) was used as a standard. Reducing agents and boiling were omitted in this protocol, as it is important to maintain the remaining disulphide bonds (Muthuramalingam et al., 2010). Total protein samples (15 μg) supplemented with 5× loading buffer [225mM TRIS-HCl, pH 6.8, 5 % (w/v) SDS, 50% glycerol, 0.05 % bromophenol blue] were separated via 12% SDS–PAGE, and the redox state of 2-CP was detected through western blot analysis with a polyclonal antibody against 2-CP (Beijing Protein Innovation, Beijing, China). After incubation with a horseradish peroxidase (HRP)-linked secondary antibody (Cell Signaling Technology, Boston, MA, USA), the complexes on the blot were visualized using an enhanced chemiluminescence kit (Perkin Elmer, Wellesley, MA, USA), according to the manufacturer’s instructions. Band intensity was quantified using Quantity One software.
Determination of Rubisco, Rubisco activase (RCA), and FBPase activity
Rubisco activity was measured spectrophotometrically by coupling 3-phosphoglyceric acid formation with NADH oxidation at 25 °C according to the method described by Lilley and Walker (1974), with some modifications. Total activity was assayed after the crude extract had been activated in a 0.1ml activation mixture containing 33mM TRIS-HCl (pH 7.5), 0.67mM EDTA, 33mM MgCl2, and 10mM NaHCO3 for 15min. The initial measurements of Rubisco activity were carried out in 0.1ml of reaction medium containing 5mM HEPES-NaOH (pH 8.0), 1mM NaHCO3, 2mM MgCl2, 0.25mM dithiothreitol (DTT), 0.1mM EDTA, 1U of glyceraldehyde 3-phosphate dehydrogenase (GAPDH), 0.5mM ATP, 0.015mM NADH2, 0.5mM phosphocreatine, 0.06mM RuBP, and 10 μl of extract. The change in absorbance at 340nm was monitored for 90 s. RCA activity was determined using a Rubisco Activase Assay Kit (Genmed Scientifics, Washington, DC, USA). FBPase activity was determined by monitoring the increase in A 340 using an extinction coefficient of 6.2mM–1 cm–1 (Scheibe et al., 1986). Total activity was assayed after the crude extract had been activated in a 0.1ml activation mixture containing 100mM DTT, 2mM FBP, 10mM MgCl2, and 0.1M TRIS-HCl (pH 8.0). The initial activity was assayed immediately after homogenization. The assay mixture consisted of 0.1M HEPES-NaOH (pH 8.0), 0.5mM Na2EDTA, 10mM MgCl2, 0.3mM NADP+, 0.6mM fructose-1,6-bisphosphate, 0.6U of glucose-6-phosphate dehydrogenase from baker’s yeast (Sigma, Santa Clara, CA, USA), 1.2U of glucoe phosphate isomerase from baker’s yeast (Sigma, Santa Clara, CA, USA), and 100 μl of enzyme extract in a final volume of 1ml.
Measurements of glutathione contents and the activity of enzymes involved in the AsA–GSH cycle
For the measurement of reduced glutathione (GSH) and oxidized glutathione (GSSG), plant leaf tissue (0.3g) was homogenized in 2ml of 6% metaphosphoric acid containing 2mM EDTA and centrifuged at 4 °C for 10min at 12 000 g. After neutralization with 0.5M phosphate buffer (pH 7.5), 0.1ml of the supernatant was added to a reaction mixture containing 0.2mM NADPH, 100mM phosphate buffer (pH 7.5), 5mM EDTA, and 0.6mM 5,5’-dithio-bis (2-nitrobenzoic acid). The reaction was initiated by adding 3U of glutathione reductase (GR) and was monitored by measuring the changes in absorbance at 412nm for 1min. For the GSSG assay, GSH was masked by the addition of 40 μl of 2-vinylpyridine to the neutralized supernatant, whereas 40 μl of water was added for the total glutathione assay. The GSH concentration was obtained by subtracting the GSSG concentration from the total concentration (Rao and Ormrod, 1995).
To determine the enzymatic activities of proteins involved in the AsA–GSH cycle, leaf tissue (0.3g) was ground in 3ml of ice-cold buffer containing 25mM HEPES (pH 7.8), 0.2mM EDTA, 2mM ascorbic acid, and 2% PVP. The homogenates were centrifuged at 4 °C for 20min at 12 000 g, and the resulting supernatants were used to determine the enzymatic activity. The ascorbate peroxidase (APX) and dehydroascorbate reductase (DHAR) activities were evaluated by measuring the decrease in absorbance at 290nm and the increase in absorbance at 265nm, as described by Nakano and Asada (1981). Monodehydroascorbate reductase (MDAR) activity was measured using 1U of ascorbate oxidase, and the oxidation rate of NADH was followed at 340nm (Hossain et al., 1984). GR activity was measured according to the method reported by Halliwell and Foyer (1976), which is based on the rate at which the absorbance of NADPH decreases at 340nm. All spectrophotometric analyses were conducted in a SHIMADZU UV-2410PC spectrophotometer (Shimadzu Corporation, Kyodo, Japan).
Total RNA extraction and gene expression analysis
Total RNA was isolated from tomato leaves using the TRIZOL reagent (Sangon, Shanghai, China) according to the instructions supplied by the manufacturer. After extraction, the total RNA was dissolved in diethyl pyrocarbonate-treated water. The cDNA template for qRT-PCR was synthesized from 2 μg of total RNA using the ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan).
For qRT-PCR analysis, PCR products were amplified in triplicate using iQ SYBR Green SuperMix (Bio-Rad, Hercules, CA, USA) in 25 μl qRT-PCR assays. PCR was performed using the iCycler iQ 96-well real-time PCR Detection System (Bio-Rad, Hercules, CA, USA), and the cycling conditions consisted of denaturation at 95 °C for 3min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 30 s. The tomato actin gene was used as an internal control. Gene-specific primers were designed according to expressed sequence tag (EST) sequences and were employed for amplification as described in Supplementary Table S2 available at JXB online. Relative gene expression was calculated as described by Livak and Schmittgen (2001).
Statistical analysis
The experimental design was a completely randomized block design with four replicates. Each replicate contained 10 plants. Statistical analysis of the bioassays was performed using the SAS statistical package. The differences between the treatment means were separated using Tukey’s test at a level of P<0.05.
Results
BRs-induced changes in chloroplast TRX transcripts in tomato
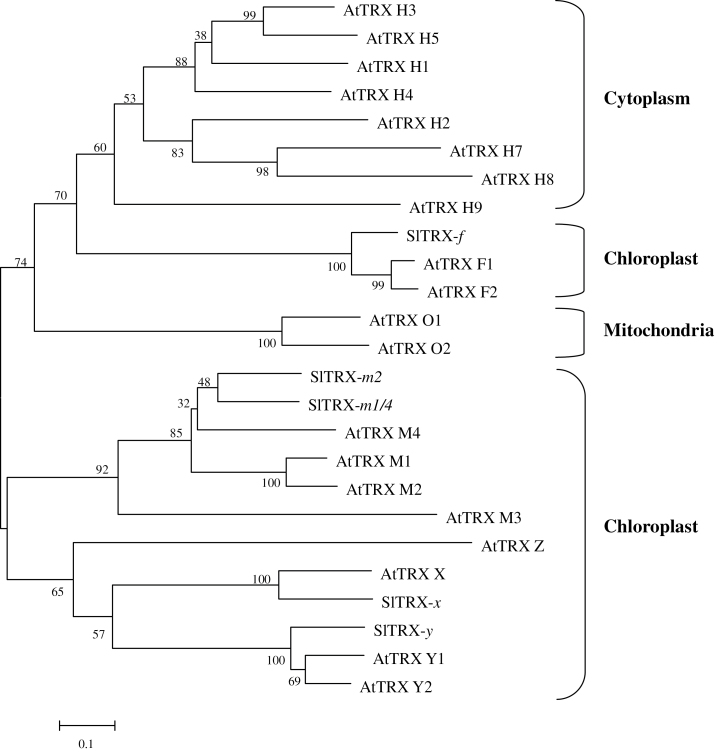
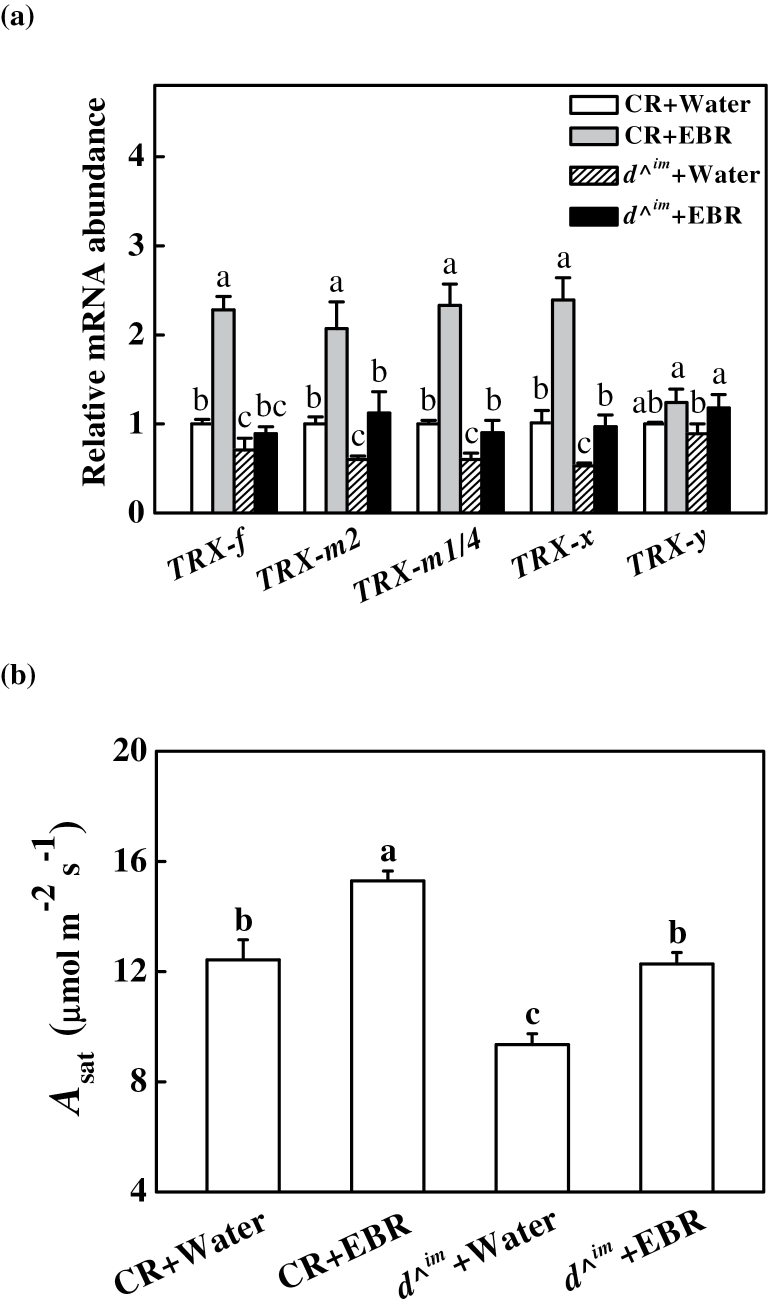
A database (Tomato Genome Sequencing Project) search based on sequence similarity with the predicted chloroplast TRXs of Arabidopsis indicated five chloroplast TRX nucleotide sequences in Solanum lycopersicum: TRX-f (Solyc05g056300), TRX-m2 (Solyc10g006970), TRX-m1/4 (Solyc12g013810), TRX-x (Solyc01g008250), and TRX-y (Solyc04g071560). A phylogenetic tree built from the alignment of these five proteins with the previously identified Arabidopsis TRXs revealed the evolutionary distances between the sequences (Fig. 1). Among these sequences, SlTRX-f, -x, and -y showed 100% similarity to the sequences of AtTRX-f, -x, and -y, while SlTRX-m2 and -m1/4 showed low similarity to their orthologues in Arabidopsis. To examine how the transcription of these TRX genes is influenced by BRs, their transcript levels were determined following EBR treatment in CR and d ^im plants. Transcript levels for TRX-f, TRX-m2, TRX-m1/4, and TRX-x were reduced by 30–40% in d ^im plants compared with CR plants, respectively, with the exception of TRX-y (Fig. 2a). In contrast, TRX-f, TRX-m2, TRX-m1/4, and TRX-x transcript levels were upregulated ~1-fold following treatment with EBR in CR plants. Similarly, no significant change was observed in TRX-y transcript levels in the EBR-treated CR plants. Interestingly, the expression of those TRX genes was restored to a level similar to that in the water-treated CR plants when d ^im plants were treated with EBR (Fig. 2a).
Fig. 1.
Phylogenetic tree of chloroplastic thioredoxins from Solanum lycopersicum (Sl) and those identified thioredoxins from Arabidopsis (At). The phylogenetic tree was constructed using MEGA 5 with the Neighbor–Joining method. Bootstrap values calculated from 1000 trials are shown at each node. The extent of divergence according to the scale (relative units) is indicated at the bottom. Predicted mature polypeptides lacking the putative transit peptide were employed for tree construction.
Fig. 2.
Effects of EBR on transcripts of chloroplastic TRX genes (a) and light-saturated rate of CO2 assimilation (A sat) in Condine Red (CR) and BRs-deficient d ^im plants (b). A sat was measured at 24h after EBR treatment in CR and d ^im plants. Leaf samples were taken at 24h after EBR treatment for gene expression analysis. Relative gene expression for each TRX gene was calculated as the water-treated CR plants (control)=1. Data are the means of four replicates with SDs. Means followed by the same letter are not significantly different according to Tukey’s test (P<0.05).
The role of chloroplast TRXs in BRs-induced CO2 assimilation
BRs levels are closely related to CO2 assimilation in cucumber plants (Yu et al., 2004; Xia et al., 2009a ). Here, it was found that the light-saturated rate of CO2 assimilation (A sat) was reduced by 24.7% in BRs-deficient d ^im mutant plants compared with wild-type CR plants. However, exogenous application of 0.2 μM EBR increased the A sat value in CR plants 24h after EBR application. Furthermore, exogenous EBR increased the A sat of the d ^im plants to the level of untreated CR plants (Fig. 2b).
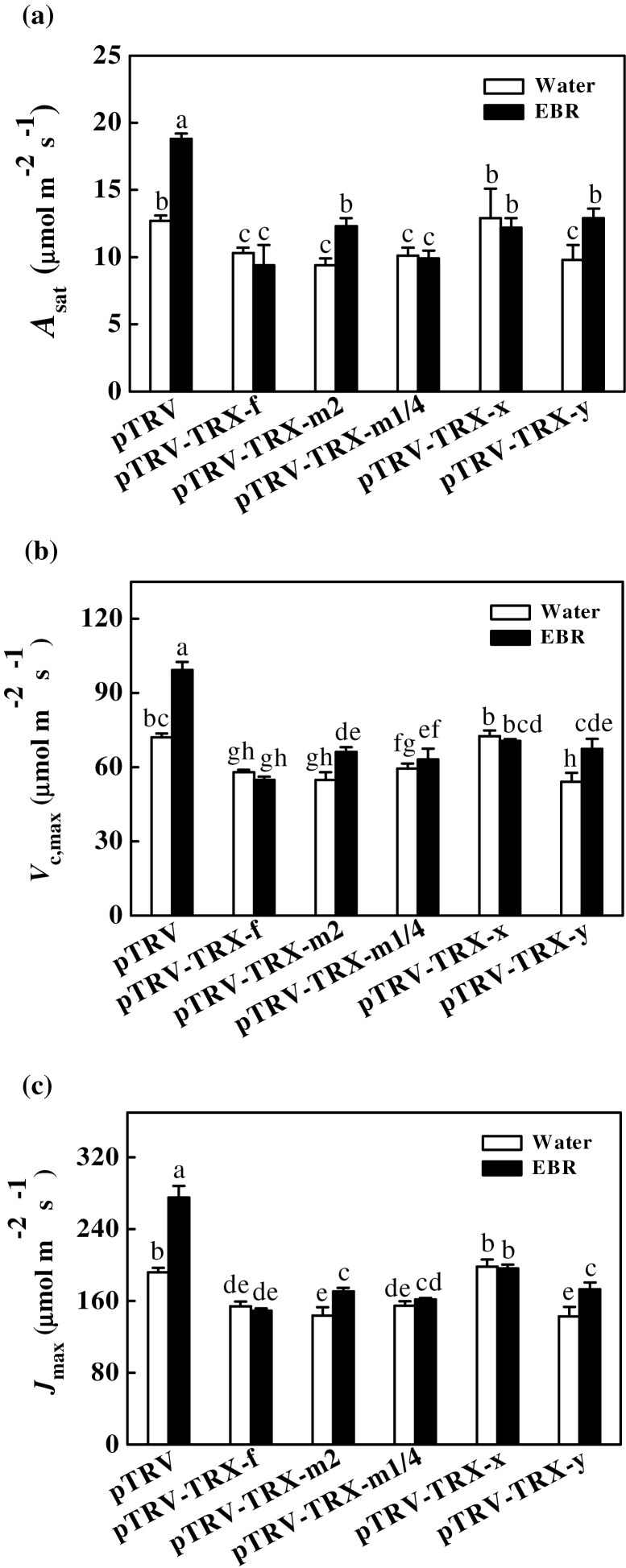
To determine the role of these TRXs in BRs-induced CO2 assimilation, the five TRX genes were partially silenced individually using a VIGS method. Transcript analysis of the leaflets in the middle of the fifth fully expanded leaves revealed that the transcripts for these genes were reduced by 65–85% in the respective silenced plants (Supplementary Fig. S1 available at JXB online). To investigate the underlying molecular mechanisms of BRs-induced CO2 assimilation, the effect of EBR on CO2 assimilation were analysed in partially TRX-f-silenced (pTRV-TRX-f,), TRX-m2-silenced (pTRV-TRX-m2), TRX-m1/4-silenced (pTRV-TRX-m1/4), TRX-x-silenced (pTRV-TRX-x), and TRX-y-silenced (pTRV-TRX-y) plants. As shown in Fig. 3a, partially silencing the TRX-f, TRX-m2, TRX-m1/4, and TRX-y genes resulted in decreases in the A sat of 18.9, 26.0, 20.5, and 22.8%, respectively, compared with the pTRV control plants. However, partially silencing TRX-x did not result in significant changes in A sat. The application of 0.2 μM EBR increased A sat in the pTRV, pTRV-TRX-m2, and pTRV-TRX-y plants by 48.0, 30.9, and 31.6%, respectively, but had little effect on the pTRV-TRX-f, pTRV-TRX-m1/4, and pTRV-TRX-x plants. Similar to the observed changes in A sat, the V c,max and the J max values were significantly decreased in the pTRV-TRX-f, pTRV-TRX-m2, pTRV-TRX-m1/4, and pTRV-TRX-y plants but were not changed in the pTRV-TRX-x plants (Fig. 3b, c). Accordingly, exogenous application of EBR increased V c,max and J max only in the pTRV, pTRV-TRX-m2, and pTRV-TRX-y plants and not in the pTRV-TRX-f, pTRV-TRX-m1/4, and pTRV-TRX-x plants. All of these results indicated that chloroplastic TRX-f, TRX-m2, TRX-m1/4, and TRX-y are involved in the regulation of CO2 assimilation, whereas only TRX-f and TRX-m1/4 play a role in the EBR-induced increase in CO2 assimilation.
Fig. 3.
Changes in light-saturated rate of CO2 assimilation (A sat) (a), maximum Rubisco carboxylation rate (V c,max) (b), and maximum RuBP regeneration rate (J max) (c) for control and EBR-treated virus-induced gene silencing (VIGS) plants. Gas exchange measurements were carried out at 24h after EBR treatment. Data are the means of four replicates with SDs. Means followed by the same letter are not significantly different according to Tukey’s test (P<0.05).
TRX-induced changes in redox homeostasis influenced by BRs
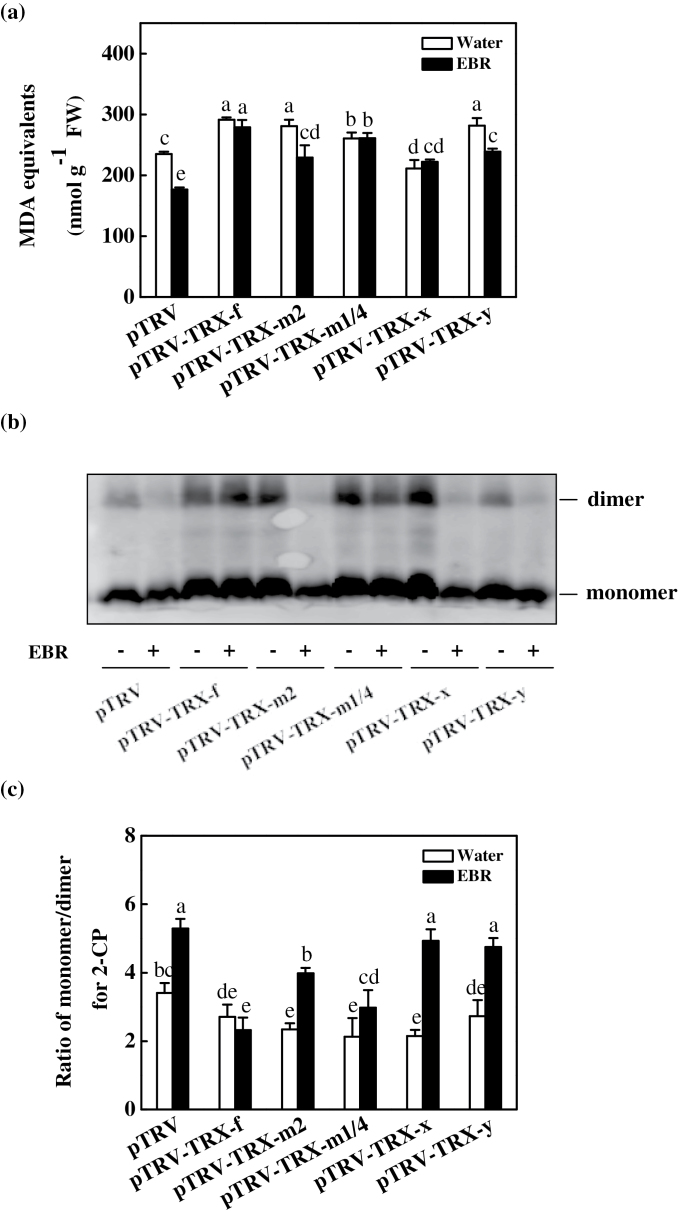
MDA is a useful indicator of reactive oxygen species (ROS)-induced lipid peroxidation in plants. Increased MDA accumulation was detected in the partially TRX-f-, TRX-m2-, TRX-m1/4-, and TRX-y-silenced plants, but not in the partially TRX-x-silenced plants. EBR treatment reduced the MDA content only in the pTRV, pTRV-TRX-m2, and pTRV-TRX-y plants and had no effect on the pTRV-TRX-f, pTRV-TRX-m1/4, and pTRV-TRX-x plants (Fig. 4a). Western blot analysis showed that 2-CP was mostly present in a reduced state (monomer), while ~22.7% was in an oxidized state (dimer) in the pTRV plants. In contrast, partial silencing of the TRXs resulted in an increase in the levels of oxidized 2-CP in the leaves, while the ratio of 2-CP monomers/2-CP dimers decreased by 20.5, 31.4, 37.5, 37.0, and 19.9% in the pTRV-TRX-f, pTRV-TRX-m2, pTRV-TRX-m1/4, pTRV-TRX-x, and pTRV-TRX-y plants, respectively, compared with the pTRV control plants. The application of 0.2 μM EBR increased the ratio of reduced 2-CP/oxidized 2-CP in the pTRV, pTRV-TRX-m2, pTRV-TRX-x, and pTRV-TRX-y plants by 55.1, 70.1, 129.3, and 74.0%, respectively, but had little effect on the pTRV-TRX-f and pTRV-TRX-m1/4 plants (Fig. 4b, c).
Fig. 4.
Changes in the lipid peroxidation and the redox state of 2-Cys peroxiredoxin (2-CP) protein in the leaves of virus-induced gene silencing (VIGS) plants as influenced by EBR application. (a) Changes in content of malondialdehyde (MDA) equivalents. (b) Changes in the redox state of 2-CP as investigated by non-reducing SDS–PAGE. The samples were separated by non-reducing SDS–PAGE and analysed in a western blot analysis with anti-2-CP. (c) The ratio of monomer/dimer for 2-CP from (b) as quantified by Quantity One. Leaf samples were taken at 24h after EBR treatment. Data are the means of four replicates with SDs. Means followed by the same letter are not significantly different according to Tukey’s test (P<0.05).
Involvement of glutathione redox homeostasis in BRs-induced CO2 assimilation
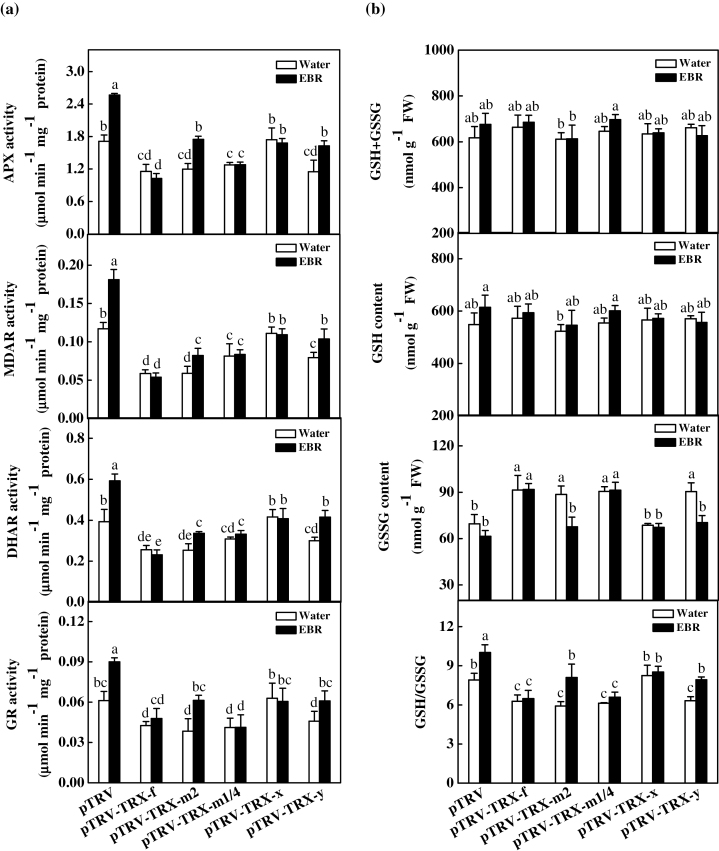
TRX can regulate the activity of the AsA–GSH cycle, which plays a critical role in maintaining the cellular redox status. In this study, the changes in the activities of APX, MDAR, DHAR, and GR, which are four important enzymes involved in the AsA–GSH cycle, were analysed. As shown in Fig. 5a, partial silencing of TRX-f, TRX-m2, TRX-m1/4, and TRX-y decreased the activity of APX, MDAR, DHAR, and GR. However, no significant effects on the activities of these enzymes were observed in the pTRV-TRX-x plants. EBR treatment significantly increased the activity of the AsA–GSH cycle in the pTRV control, pTRV-TRX-m2, and pTRV-TRX-y plants, but had no effect in the pTRV-TRX-f, pTRV-TRX-m1/4, and pTRV-TRX-x plants. To analyse the EBR-induced changes in glutathione redox homeostasis further, the effects of EBR on the contents of GSH and GSSG and their ratios in the pTRV and various pTRV-TRX plants were compared. There was generally little change in GSH content observed in the pTRV and partially TRX-silenced plants (Fig. 5b). However, partial silencing of TRX-f, TRX-m2, TRX-m1/4, and TRX-y resulted in 31.5, 27.5, 30.2, and 30.1% increases in the GSSG content, respectively, leading to a decrease in the GSH/GSSG ratio. In contrast, EBR induced a slight increase in GSH content and decrease in GSSG, resulting in a significant increase in the GSH/GSSG ratio. Similar to the observed changes in the activities of the AsA–GSH cycle enzymes, the EBR-induced increases in the glutathione pool and GSH/GSSG ratio were abolished by partially silencing TRX-f and TRX-m1/4 but not by the silencing of other TRXs (Fig. 5a, b).
Fig. 5.
Changes in the activity of key enzymes involved in the AsA–GSH cycle and glutathione redox status for control and EBR-treated virus-induced gene silencing (VIGS) plants. Leaf samples were taken at 24h after EBR treatment. Data are the means of four replicates with SDs. Means followed by the same letter are not significantly different according to Tukey’s test (P<0.05).
Gene expression and enzyme activities involved in the Benson–Calvin cycle influenced by the TRXs and BRs
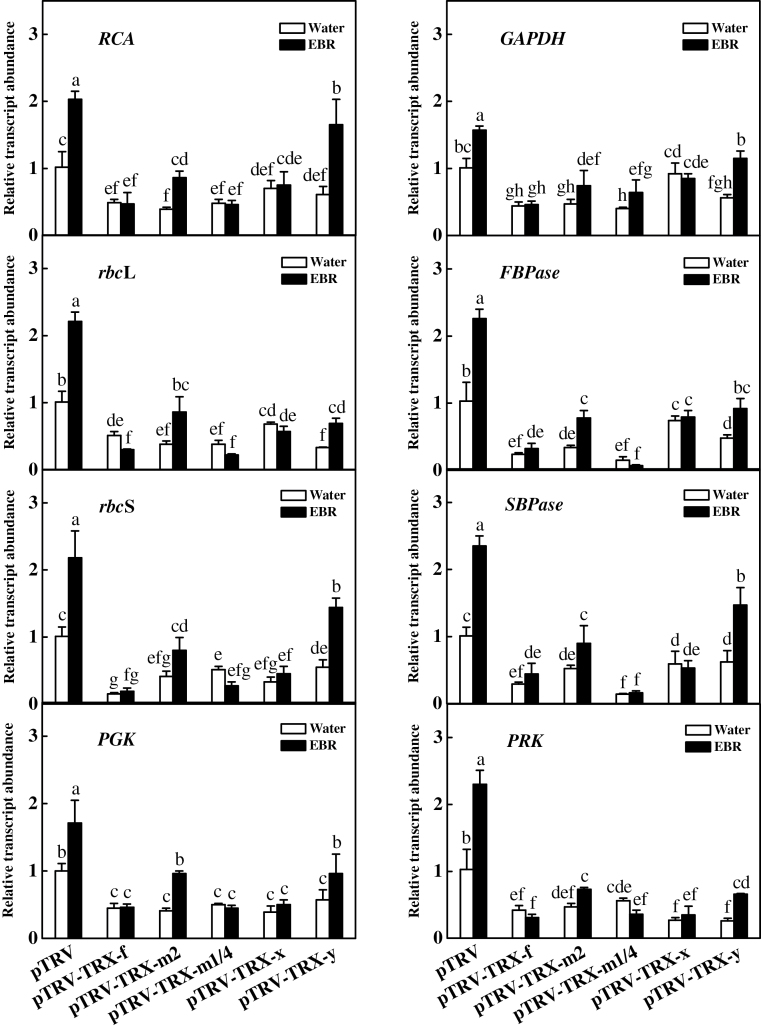
Previously, it was reported that BRs-induced CO2 assimilation is associated with increased expression of photosynthetic genes in cucumber plants (Yu et al., 2004; Xia et al., 2009a ). In this study, the transcript levels of eight Benson-Calvin cycle-related genes in the leaves of plants lacking different chloroplastic TRX genes were analysed. These tested photosynthesis-related genes included the genes encoding Rubisco activase (RCA), Rubisco large subunit (rbcL), Rubisco small subunit (rbcS), glycerate-3-phosphate kinase (PGK), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), fructose-1,6-bisphosphatase (FBPase), sedoheptulose-1,7-bisphosphatase (SBPase), and ribulose-5-phosphate kinase (PRK). As shown in Fig. 6, the transcripts of these genes were all up-regulated upon EBR treatment in the pTRV plants. In contrast, partially silencing the chloroplastic TRX genes resulted in down-regulation of these photosynthesis-related genes. Importantly, EBR-induced transcripts were detected only in the pTRV-TRX-m2 and pTRV-TRX-y plants and not in the other plants.
Fig. 6.
Changes in the expression of Benson–Calvin cycle-related genes for control and EBR-treated virus-induced gene silencing (VIGS) plants. Leaf samples were taken at 24h after EBR treatment. Data are the means of four replicates with SDs. Means followed by the same letter are not significantly different according to Tukey’s test (P<0.05).
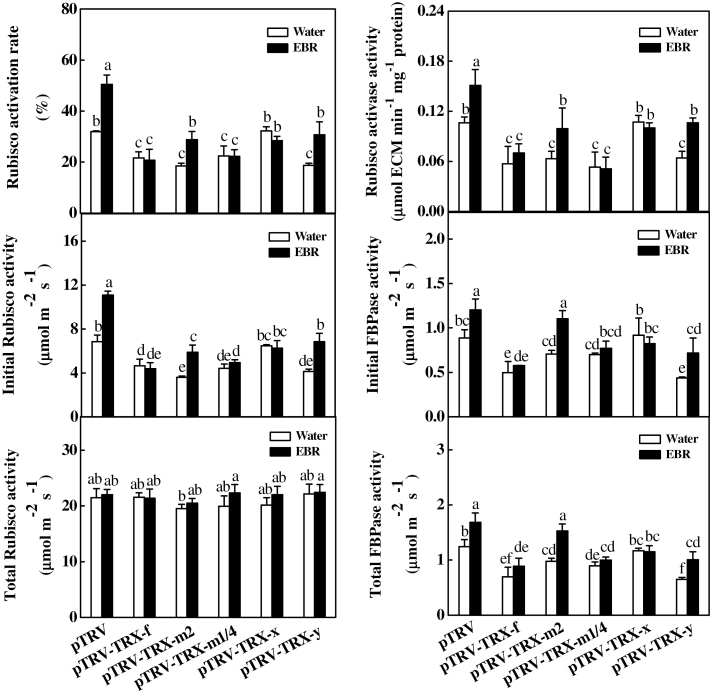
BRs levels are closely related to the activation of a subset of enzymes involved in the Benson–Calvin cycle, particularly the redox-sensitive enzymes (Yu et al., 2004; Xia et al., 2009a ). Here, the changes in the activities of RCA, Rubisco, and FBPase were analysed in VIGS plants with and without exogenous application of EBR. As shown in Fig. 7, partial silencing of these genes did not result in significant changes in total Rubisco activity but significantly decreased the initial Rubisco, initial FBPase, and Rubisco activase activities as well as the Rubisco activation rate, except in the pTRV-TRX-x plants, where these parameters were not significantly altered. For example, initial Rubisco activities decreased by 31.9, 47.4, 35.6, and 39.6%, while the Rubisco activation rate decreased by 32.1, 42.0, 29.8, and 41.3% in the pTRV-TRX-f, pTRV-TRX-m2, pTRV-TRX-m1/4, and pTRV-TRX-y plants, respectively, compared with the pTRV plants. Furthermore, exogenous EBR application increased the activity or the activation rate only in the pTRV-TRX-m2 and pTRV-TRX-y plants. The activity and the activation rates of these enzymes in the EBR-treated VIGS plants were much lower than in the pTRV control plants.
Fig. 7.
Changes in the activity of Rubisco activase, Rubisco, and FBPase for control and EBR-treated virus-induced gene silencing (VIGS) plants. Leaf samples were taken at 24h after EBR treatment. Data are the means of four replicates with SDs. Means followed by the same letter are not significantly different according to Tukey’s test (P<0.05).
Discussion
TRX-f, TRX-m2, TRX-m1/4, and TRX-y are involved in the regulation of cellular redox homeostasis via the AsA–GSH cycle and 2-CP
TRXs are involved in the regulation of ROS metabolism and CO2 assimilation. The results of the present study demonstrated that TRXs can differentially regulate oxidative stress in tomato plants. In rice, Ostrxm RNAi plants exhibit increased H2O2 accumulation in the leaves (Chi et al., 2008). Histochemical staining demonstrated that more O2 ·− and H2O2 accumulates in the leaves of the VIGS-TRX-f/TRX-m pea plants compared with control plants, confirming that TRX-f and TRX-m are important for the metabolic balance of ROS in plant cells (Luo et al., 2012). Here, it was found that partially silencing TRX-f, TRX-m2, TRX-m1/4, or TRX-y resulted in increased oxidative stress, as indicated by the increased MDA content in the leaves, while partially silencing TRX-x did not induce significant changes in the MDA content of the leaves (Fig. 4a). These findings revealed that chloroplastic TRX-f, TRX-m2, and TRX-m1/4 as well as TRX-y are involved in ROS metabolism. It seems likely that TRX-x plays an insignificant role in ROS metabolism, as partially silencing TRX-x did not induce oxidative stress in the leaves.
ROS accumulation is closely related to the ROS scavenging capacity of cells, which is largely dependent on antioxidant enzymes, such as APX, MDAR, DHAR, and GR, and non-enzymatic antioxidants, such as AsA and GSH. Proteomic studies in Arabidopsis have determined that several key enzymes involved in the AsA–GSH cycle, such as GR, are redox sensitive (Ströher and Dietz, 2008; Wang et al., 2012), and biochemical and genetic assays have identified the NADPH-dependent thioredoxin system as a backup system for GR1 (Marty et al., 2009). Until now, few studies have investigated the role of TRXs in the regulation of antioxidant activity in plants. Here, it is demonstrated that partially silencing the TRX-f, TRX-m2, TRX-m1/4, and TRX-y genes resulted in decreased activity of the enzymes APX, MDAR, DHAR, and GR, while partially silencing the TRX-x gene did not induce significant changes in the activity of these enzymes (Fig. 5a). Additionally, partially silencing TRX-f, TRX-m2, TRX-m1/4, and TRX-y did not alter the total glutathione (GSH+GSSG) or GSH content but did induce an increase in the GSSG content, leading to a decrease in the GSH/GSSG ratio (Fig. 5b). These results indicated that TRX-f, TRX-m2, TRX-m1/4, and TRX-y do not influence GSH biosynthesis but affect glutathione homeostasis by regulating GR activity, as observed in both the glutathione-deficient cad2 and GR-deficient gr1 Arabidopsis mutants (Meyer et al., 2007; Marty et al., 2009). However, such an effect was not observed in the pTRV-TRX-x plants. All of these results suggested that the enzymes involved in the AsA–GSH cycle are largely modified by TRX-f, TRX-m2, TRX-m1/4, and TRX-y, and that the oxidative stress induced in partially TRX-f, TRX-m2, TRX-m1/4, and TRX-y-silenced plants is at least in part attributable to the reduced activity of the antioxidant enzymes. The finding that the cellular glutathione redox status could be regulated by chloroplastic TRXs further revealed the existence of cross-talk between the TRX system and the glutathione system.
In addition to the AsA–GSH cycle, peroxiredoxin, which is activated by TRX, acts as an alternative pathway to remove H2O2 from chloroplasts (Foyer and Shigeoka, 2011). The 2-CP enzyme exhibits broad substrate specificity, showing activity toward both hydrogen peroxides and complex alkyl hydroperoxides. During the peroxide reduction reaction, 2-CP is alternatively oxidized and reduced as it catalyses the electron flow from an electron donor to peroxide. In vitro tests have demonstrated that TRX-x is by far the most efficient in Arabidopsis (Collin et al., 2003). However, only the oxidized dimeric form of 2-CP was detected in the Ostrxm RNAi plants, suggesting that Ostrxm could also regulate the catalytic activity of 2-CP by reducing redox-active cysteine residues in plants (Chi et al., 2008). The present study revealed increased accumulation of oxidized 2-CP in all of the partially TRX-silenced plants, with the partial silencing of TRX-x, TRX-m1/4, and TRX-m2 being more significant, suggesting that these TRXs could differentially regulate 2-CP (Fig. 4b, c).
TRX-f and TRX-m1/4 play important roles in BRs-induced changes in cellular redox homeostasis
As observed in previous research, foliar application of EBR relieved oxidative stress (Xia et al., 2009b ). In addition to increasing the activity of the AsA–GSH cycle, EBR also induced an increase in the ratio of the reduced monomers to the oxidized dimers of 2-CP (Fig. 4c). Importantly, partially silencing TRX-f and TRX-m1/4 compromised the EBR-induced decrease in MDA content and increase in the activity of antioxidant enzymes, as well as the associated changes in glutathione metabolism (Figs 4a, 5a, b), suggesting that EBR partially alleviates oxidative stress by regulating TRX-f and TRX-m1/4.
It was found that the TRXs are actively involved in the regulation of photosynthesis-related gene transcripts, enzyme activity, and, ultimately, the CO2 assimilation capacity. Ostrxm plants exhibit abnormal chloroplast development and growth inhibition in rice plants, displaying decreased levels of several chloroplast proteins that are critical for photosynthesis and biogenesis (Chi et al., 2008). Silencing TRX-f and TRX-m (VIGS-TRX-f/TRX-m) in pea plants results in a significant reduction in Mg chelatase activity and the 5-aminolaevulinic acid synthesizing capacity as well as decreased transcript levels of RBCS and chlorophyll biosynthesis-related genes and a decreased photosynthetic capacity (Luo et al., 2012). In the present study, it was found that partially silencing the chloroplastic TRXs, except for TRX-x, resulted in a reduced photosynthetic capacity in tomato leaves (Fig. 3), supporting the hypothesis that TRX-f, TRX-m2, TRX-m1/4, and TRX-y are actively involved in the regulation of CO2 assimilation. In Arabidopsis, the maximal (F v/F m) and effective quantum yields of photosystem II (PSII; ФPSII) in the trx f1.1 and trx f1.2 mutant plants are not different from those in wild-type plants (Thormählen et al., 2013). In agreement with this finding, it was observed that only the pTRV-TRX-m1/4 plants showed a decrease in F v/F m, by 15.4% (Supplementary Fig. S2 available at JXB online). All of these results suggested that the TRXs do not affect CO2 assimilation by modulating PSII electron transport.
TRX-f and TRX-m1/4 play important roles in BRs-induced changes in CO2 assimilation
Previous studies have shown that chloroplastic TRXs participate in the regulation of the Calvin cycle and associated processes (Balmer et al., 2003). In addition to the down-regulation of genes involved in the Benson–Calvin cycle, partially silencing TRX-f, TRX-m2, TRX-m1/4, and TRX-y also resulted in a significant decrease in the initial Rubisco and FBPase activities as well as RCA activity (Fig. 7). An in vitro assay revealed that AtTRX-f is able to activate FBPase and NADP-MDH most efficiently, followed by AtTRX-m1, AtTRX-m2, and AtTRX-m4, whereas AtTRX-m3, AtTRX-x, and AtTRX-y cannot efficiently activate NADP-MDH from sorghum or from Chlamydomonas (Collin et al., 2003, 2004; Lemaire et al., 2003, 2005). The simultaneous decrease in V c,max and initial Rubisco activity, with little change in total Rubisco activity, indicates that chloroplastic TRX-f, TRX-m2, TRX-m1/4, and TRX-y mainly regulate the activation state of Rubisco via the action of Rubisco activase (Figs 3b, 7). The regeneration of RuBP is dependent on both the photosynthetic electron transport chain and the enzymes downstream of Rubisco in the Calvin cycle (Long et al., 2006). In addition to the decrease in RCA activity, initial FBPase activity was also decreased in the plants in which TRX-f, TRX-m2, TRX-m1/4, and TRX-y were partially silenced (Fig. 7). Accordingly, the lack of direct activation of RCA or FBPase was involved in the decrease in J max. Interestingly, the observed changes in the activities of these enzymes were in agreement with cellular glutathione homoeostasis (Fig. 5b). In a previous study, it was found that several redox-sensitive Benson–Calvin cycle enzymes, such as RCA, can undergo disulphide bond interchange, leading to changes in the activities of these enzymes (Jiang et al., 2012b ). It is likely that the oxidized cellular environment induced by the partial silencing of TRX-f, TRX-m2, TRX-m1/4, and TRX-y can directly modify the structure of these redox-sensitive enzymes.
Previously, it was found that inhibition of the biosynthesis of BRs using an inhibitor decreased CO2 assimilation, while exogenously applied BRs increased CO2 assimilation in cucumber plants (Yu et al., 2004; Xia et al., 2009a ). Here, it was demonstrated that exogenous EBR application resulted in enhanced CO2 assimilation in tomato plants, while BRs-deficient d ^im mutant plants exhibited decreased CO2 assimilation compared with wild-type CR plants (Fig. 2b), providing further evidence that BRs levels are closely related to the CO2 assimilation capacity of plants.
BRs can differentially induce TRX transcripts in Arabidopsis and rice (Müssig et al., 2002; Goda et al., 2004; Wu et al., 2008). In the present study, it was found that BRs were able to up-regulate TRX-f, TRX-m2, TRX-m1/4, and TRX-x but did not affect the transcription of TRX-y (Fig. 2a). In contrast, partially silencing TRX-x did not induce any changes in CO2 assimilation, the activities of antioxidant enzymes, redox homeostasis, or the expression and activities of photosynthesis-related genes and enzymes (Figs 3a, 5a, 5b, 6, 7), suggesting that TRX-f, TRX-m2, and TRX-m1/4 are potentially involved in the BRs-induced increase in CO2 assimilation. However, the partial silencing of TRX-f, TRX-m2, and TRX-m1/4 resulted in decreases in CO2 assimilation, the expression of Benson–Calvin cycle-related genes, and the activities of associated enzymes, while EBR induced these changes only in pTRV-TRX-m2 plants and not in pTRV-TRX-f or pTRV-TRX-m1/4 plants (Figs 3a, 6, 7). These results suggest that TRX-f and TRX-m1/4 are the TRXs responsible for BRs-induced CO2 assimilation.
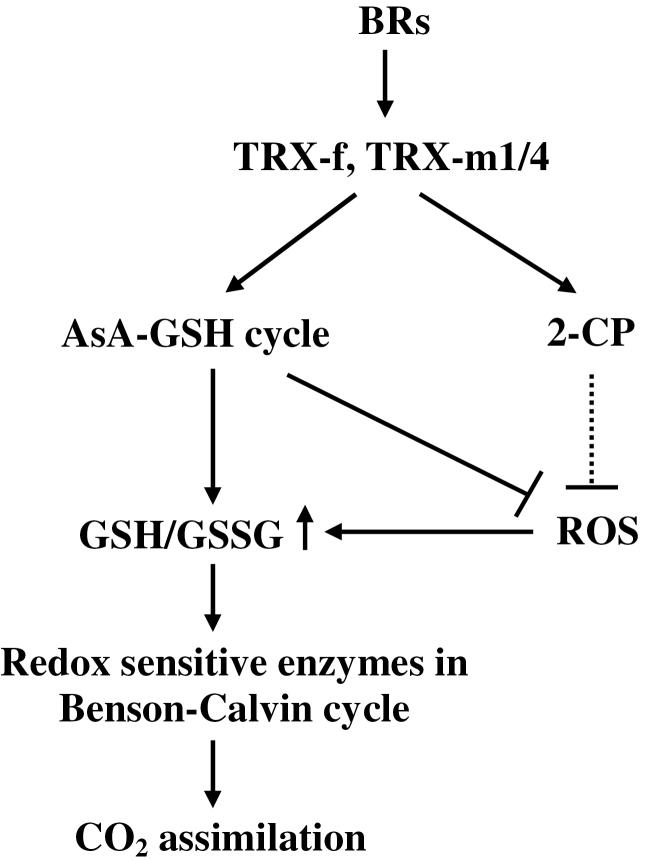
Many processes involved in plant growth, development, and metabolism are under the tight control of the TRXs (Schürmann and Buchanan, 2008). The results of this study demonstrated that the chloroplastic TRXs of tomato play different roles in cellular redox homeostasis and CO2 assimilation, and BRs regulate cellular redox homeostasis and CO2 assimilation in a TRX-f- and TRX-m1/4-dependent manner (Fig. 8). Among the examined TRXs, TRX-f, TRX-m2, TRX-m1/4, and TRX-y were all able to activate the antioxidant system by up-regulating the AsA–GSH cycle and the Benson–Calvin cycle, while TRX-x is not involved in the regulation of these processes. In addition to their direct effects on gene transcription, BRs also increased the antioxidant capacity, leading to a reduced cellular environment, which could directly reduce specific disulphide bridges in photosynthesis-related redox-sensitive enzymes, leading to the activation of these enzymes and therefore playing an important role in the regulation of normal photosynthesis.
Fig. 8.
A proposed model for BRs-induced increase in CO2 assimilation in tomato plants.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Relative mRNA abundance of TRX-f, TRX-m2, TRX-m1/4, TRX-x, and TRX-y in respective virus-induced gene silencing (VIGS) plants.
Figure S2. Changes in maximum quantum yield of PSII (F v/F m) in the leaves of pTRV and various partially TRX-silenced plants.
Table S1. PCR primers designed for vector construction.
Table S2. Gene-specific primers designed for qRT-PCR.
Acknowledgements
We are grateful to Professor C.H. Foyer for her critical suggestions regarding this study. This work was supported by the National Basic Research Program of China (2009CB119000).
References
- Alkhalfioui F, Renard M, Vensel WH, Wong J, Tanaka CK, Hurkman WJ, Buchanan BB, Montrichard F. 2007. Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds. Plant Physiology 144, 1559–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustün S, Melzer M, Petersen K, Lein W, Börnke F. 2010. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana . The Plant Cell 22, 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schürmann P, Buchanan BB. 2003. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proceedings of the National Academy of Sciences, USA 100, 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schürmann P, Hurkman WJ, Buchanan BB. 2006. A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proceedings of the National Academy of Sciences, USA 103, 2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Tanaka CK, et al. 2004. Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proceedings of the National Academy of Sciences, USA 101, 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli CG, Casalongué CA, Simontacchi M, Marquez-Garcia B, Foyer CH. 2013. Interactions between hormone and redox signalling pathways in the control of growth and cross tolerance to stress. Environmental and Experimental Botany 94, 73–88 [Google Scholar]
- Baumann U, Juttner J. 2002. Plant thioredoxins: the multiplicity conundrum. Cellular and Molecular Life Sciences 59, 1042–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JDG, Kamiya Y. 1999. The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proceedings of the National Academy of Sciences, USA 96, 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. 2005. Redox regulation: a broadening horizon. Annual Review of Plant Biology 56, 187–220 [DOI] [PubMed] [Google Scholar]
- Chi YH, Moon JC, Park JH, Kim H-S, Zulfugarov IS, Fanata WI, Jang HH, Lee JR, Lee YM, Kim ST. 2008. Abnormal chloroplast development and growth inhibition in rice thioredoxin m knock-down plants. Plant Physiology 148, 808–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. 1998. Brassinosteroids: essential regulators of plant growth and development. Annual Review of Plant Physiology and Plant Molecular Biology 49, 427–451 [DOI] [PubMed] [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M. 2003. The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. Journal of Biological Chemistry 278, 23747–23752 [DOI] [PubMed] [Google Scholar]
- Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, Knaff DB, Dietz KJ, Issakidis-Bourguet E. 2004. Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiology 136, 4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JX, Zhou YH, Ding JG, Xia XJ, Shi K, Chen SC, Asami T, Chen Z, Yu JQ. 2011. Role of nitric oxide in hydrogen peroxide-dependent induction of abiotic stress tolerance by brassinosteroids in cucumber. Plant, Cell and Environment 34, 347–358 [DOI] [PubMed] [Google Scholar]
- Ekengren SK, Liu YL, Schiff M, Dinesh-Kumar SP, Martin GB. 2003. Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. The Plant Journal 36, 905–917 [DOI] [PubMed] [Google Scholar]
- Ethier GJ, Livingston NJ. 2004. On the need to incorporate sensitivity to CO2 transfer conductance into the Farquhar–von Caemmerer–Berry leaf photosynthesis model. Plant, Cell and Environment 27, 137–153 [Google Scholar]
- Foyer CH, Shigeoka S. 2011. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiology 155, 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geck MK, Larimer FW, Hartman FC. 1996. Identification of residues of spinach thioredoxin f that influence interactions with target enzymes. Journal of Biological Chemistry 271, 24736–24740 [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. 2004. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis . Plant Physiology 134, 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Foyer CH. 1976. Ascorbic acid, metal-ions and the superoxide radical. Biochemistry Journal 155, 697–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges DM, DeLong JM, Forney CF, Prange RK. 1999. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611 [DOI] [PubMed] [Google Scholar]
- Hodges M, Miginiac-Maslow M, Decottignies P, Jacquot JP, Stein M, Lepiniec L, Cretin C, Gadal P. 1994. Purification and characterization of pea thioredoxin f expressed in Escherichia coli . Plant Molecular Biology 26, 225–234 [DOI] [PubMed] [Google Scholar]
- Hossain M, Nakano Y, Asada K. 1984. Monodehydroascorbate reductase in spinach-chloroplast and its participation in regeneration of ascorbate for scavenging hydrogen-peroxide. Plant and Cell Physiology 25, 385–395 [Google Scholar]
- Jacquot JP, Gelhaye E, Rouhier N, Corbier C, Didierjean C, Aubry A. 2002. Thioredoxins and related proteins in photosynthetic organisms: molecular basis for thiol dependent regulation. Biochemical Pharmacology 64, 1065–1069 [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Lancelin JM, Meyer Y. 1997. Thioredoxins: structure and function in plant cells. New Phytologist 136, 543–570 [DOI] [PubMed] [Google Scholar]
- Jiang YP, Cheng F, Zhou YH, Xia XJ, Mao WH, Shi K, Chen ZX, Yu JQ. 2012. a Cellular glutathione redox homeostasis plays an important role in the brassinosteroid-induced increase in CO2 assimilation in Cucumis sativus . New Phytologist 194, 932–943 [DOI] [PubMed] [Google Scholar]
- Jiang YP, Cheng F, Zhou YH, Xia XJ, Mao WH, Shi K, Chen ZX, Yu JQ. 2012. b Brassinosteroid-induced CO2 assimilation is associated with increased stability of redox-sensitive photosynthetic enzymes in the chloroplasts in cucumber plants. Biochemical and Biophysical Research Communications 426, 390–394 [DOI] [PubMed] [Google Scholar]
- Lamkemeyer P, Laxa M, Collin V, et al. 2006. Peroxiredoxin Q of Arabidopsis thaliana is attached to the thylakoids and functions in context of photosynthesis. The Plant Journal 45, 968–981 [DOI] [PubMed] [Google Scholar]
- Lemaire SD, Collin V, Keryer E, Quesada A, Miginiac-Maslow M. 2003. Characterization of thioredoxin y, a new type of thioredoxin identified in the genome of Chlamydomonas reinhardtii . FEBS Letters 543, 87–92 [DOI] [PubMed] [Google Scholar]
- Lemaire SD, Quesada A, Merchan F, Corral JM, Igeno MI, Keryer E, Issakidis-Bourguet E, Hirasawa M, Knaff DB, Miginiac-Maslow M. 2005. NADP-malate dehydrogenase from unicellular green alga Chlamydomonas reinhardtii. A first step toward redox regulation? Plant Physiology 137, 514–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Xie QG, Smith-Becker J, Navarre DA, Kaloshian I. 2006. Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling cascades. Molecular Plant-Microbe Interactions 19, 655–664 [DOI] [PubMed] [Google Scholar]
- Lilley RM, Walker DA. 1974. An improved spectrophotometric assay for ribulose-bisphosphate carboxylase. Biochimica et Biophysica Acta 358, 226–229 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. 2002. a Virus-induced gene silencing in tomato. The Plant Journal 31, 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. 2002. b Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. The Plant Journal 30, 415–429 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Long SP, Zhu X, Naidu SL, Ort DR. 2006. Can improvement in photosynthesis increase crop yields? Plant, Cell and Environment 29, 315–330 [DOI] [PubMed] [Google Scholar]
- Luo T, Fan TT, Liu YN, Rothbart M, Yu J, Zhou SX, Grimm B, Luo MZ. 2012. Thioredoxin redox regulates ATPase activity of magnesium chelatase CHLI subunit and modulates redox-mediated signaling in tetrapyrrole biosynthesis and homeostasis of reactive oxygen species in pea plants. Plant Physiology 159, 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand C, Le Marechal P, Meyer Y, Decottignies P. 2006. Comparative proteomic approaches for the isolation of proteins interacting with thioredoxin. Proteomics 6, 6528–6537 [DOI] [PubMed] [Google Scholar]
- Marcus F, Chamberlain SH, Chu C, Masiarz FR, Shin S, Yee BC, Buchanan BB. 1991. Plant thioredoxin h: an animal-like thioredoxin occurring in multiple cell compartments. Archives of Biochemistry and Biophysics 287, 195–198 [DOI] [PubMed] [Google Scholar]
- Marty L, Siala W, Schwarzländer M, Fricker MD, Wirtz M, Sweetlove LJ, Meyer Y, Meyer AJ, Reichheld JP, Hell R. 2009. The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis . Proceedings of the National Academy of Sciences, USA 106, 9109–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R. 2007. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. The Plant Journal 52, 973–986 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Reichheld JP, Vignols F. 2005. Thioredoxins in Arabidopsis and other plants. Photosynthesis Research 86, 419–433 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Siala W, Bashandy T, Riondet C, Vignols F, Reichheld JP. 2008. Glutaredoxins and thioredoxins in plants. Biochimica et Biophysica Acta 1783, 589–600 [DOI] [PubMed] [Google Scholar]
- Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB. 2009. Thioredoxin targets in plants: the first 30 years. Journal of Proteomics 72, 452–474 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T. 2001. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proceedings of the National Academy of Sciences, USA 98, 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig C. 2005. Brassinosteroid-promoted growth. Plant Biology 7, 110–117 [DOI] [PubMed] [Google Scholar]
- Müssig C, Fischer S, Altmann T. 2002. Brassinosteroid-regulated gene expression. Plant Physiology 129, 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuramalingam M, Dietz K-J, Ströher E. 2010. Thiol–disulfide redox proteomics in plant research. Methods in Molecular Biology 639, 219–238 [DOI] [PubMed] [Google Scholar]
- Nakano Y, Asada K. 1981. Hydrogen peroxide is scavenged by ascorbate specific peroxidase in spinach chloroplasts. Plant and Cell Physiology 22, 867–880 [Google Scholar]
- Navrot N, Collin V, Gualberto J, Gelhaye E, Hirasawa M, Rey P, Knaff DB, Issakidis E, Jacquot JP, Rouhier N. 2006. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiology 142, 1364–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa AN, Buchanan BB. 1981. Enzyme regulation in C4 photosynthesis. Purification and properties of thioredoxin-linked fructose bisphosphatase and sedoheptulose bisphosphatase from corn leaves. Journal of Biological Chemistry 256, 6119–6126 [PubMed] [Google Scholar]
- Rao MV, Ormrod DP. 1995. Ozone exposure decreases UVB sensitivity in a UVB-sensitive flavonoid mutant of Arabidopsis . Photochemistry and Photobiology 61, 71–78 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Gualberto JM, et al. 2004. Poplar peroxiredoxin Q. A thioredoxin-linked chloroplast antioxidant functional in pathogen defense. Plant Physiology 134, 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Hatano M. 1997. Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proceedings of the National Academy of Sciences, USA 94, 11096–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R, Fickenscher K, Ashton AR. 1986. Studies on the mechanism of the reductive activation of NADP-malate dehydrogenase by thioredoxin m and low molecular weight thiols. Biochimica et Biophysica Acta 870, 191–197 [Google Scholar]
- Schürmann P, Buchanan BB. 2008. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxidants and Redox Signaling 10, 1235–1273 [DOI] [PubMed] [Google Scholar]
- Schwarz O, Schurmann P, Strotmann H. 1997. Kinetics and thioredoxin specificity of thiol modulation of the chloroplast H+-ATPase. Journal of Biological Chemistry 272, 16924–16927 [DOI] [PubMed] [Google Scholar]
- Ströher E, Dietz KJ. 2008. The dynamic thiol–disulphide redox proteome of the Arabidopsis thaliana chloroplast as revealed by differential electrophoretic mobility. Physiologia Plantarum 133, 566–583 [DOI] [PubMed] [Google Scholar]
- Thormählen I, Ruber J, Roepenack-Lahaye EV, Ehrlich S-M, Massot V, Hümmer C, Tezycka J, Issakidis-Bourguet E, Geigenberger P. 2013. Inactivation of thioredoxin f1 leads to decreased light activation of ADP-glucose pyrophosphorylase and altered diurnal starch turnover in leaves of Arabidopsis plants. Plant, Cell and Environment p36, 16–29 [DOI] [PubMed] [Google Scholar]
- Vieira Dos Santos C, Laugier E, Tarrago L, Massot V, Issakidis–Bourguet E, Rouhier N, Rey P. 2007. Specificity of thioredoxins and glutaredoxins as electron donors to two distinct classes of Arabidopsis plastidial methionine sulfoxide reductases B. FEBS Letters 581, 4371–4376 [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Farquhar GD. 1981. Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153, 376–387 [DOI] [PubMed] [Google Scholar]
- Wang H, Wang SB, Lu YQ, Alvarez S, Hicks LM, Ge XC, Xia YJ. 2012. Proteomic analysis of early-responsive redox-sensitive proteins in Arabidopsis . Journal of Proteome Research 11, 412–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenderoth I, Scheibe R, von Schaewen A. 1997. Identification of the cysteine residues involved in redox modification of plant plastidic glucose-6-phosphate dehydrogenase. Journal of Biological Chemistry 272, 26985–26990 [DOI] [PubMed] [Google Scholar]
- Wolosiuk RA, Ballicora MA, Hagelin K. 1993. The reductive pentose phosphate cycle for photosynthetic CO2 assimilation: enzyme modulation. FASEB Journal 7, 622–637 [DOI] [PubMed] [Google Scholar]
- Wong JH, Cai N, Balmer Y, Tanaka CK, Vensel WH, Hurkman WJ, Buchanan BB. 2004. Thioredoxin targets of developing wheat seeds identified by complementary proteomic approaches. Phytochemistry 65, 1629–1640 [DOI] [PubMed] [Google Scholar]
- Wu CY, Trieu A, Radhakrishnan P, et al. 2008. Brassinosteroids regulate grain filling in rice. The Plant Cell 20, 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XJ, Huang LF, Zhou YH, Mao WH, Shi K, Wu JX, Asami T, Chen ZX, Yu JQ. 2009. a Brassinosteroids promote photosynthesis and growth by enhancing activation of Rubisco and expression of photosynthetic genes in Cucumis sativus. Planta 230, 1185–1196 [DOI] [PubMed] [Google Scholar]
- Xia XJ, Wang YJ, Zhou YH, Tao Y, Mao WH, Shi K, Asami T, Chen ZX, Yu JQ. 2009. b Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiology 150, 801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JQ, Huang LF, Hu WH, Zhou YH, Mao WH, Ye SF, Nogues S. 2004. A role for brassinosteroids in the regulation of photosynthesis in Cucumis sativus . Journal of Experimental Botany 55, 1135–1143 [DOI] [PubMed] [Google Scholar]
- Zhang N, Portis AR., Jr 1999. Mechanism of light regulation of Rubisco: a specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proceedings of the National Academy of Sciences, USA 96, 9438–9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.