Abstract
Huntington’s Disease (HD) is a fatal neurodegenerative disorder caused by an extended polyglutamine repeat in the N-terminus of the Huntingtin protein (HTT). Reactive microglia and elevated cytokine levels are observed in the brains of HD patients, but the extent to which neuroinflammation results from extrinsic or cell-autonomous mechanisms within microglia is unknown. Using genome-wide approaches, we show that expression of mutant Huntingtin (mHTT) in microglia promotes cell-autonomous pro-inflammatory transcriptional activation by increasing the expression and transcriptional activities of the myeloid lineage-determining factors PU.1 and C/EBPs. Elevated levels of PU.1 and its target genes are observed in the brains of mouse models and HD individuals. Moreover, mutant Huntingtin-expressing microglia exhibit an increased capacity to induce neuronal death ex vivo and in vivo in the presence of sterile inflammation. These findings suggest a cell autonomous basis for enhanced microglia reactivity that may influence non-cell autonomous HD pathogenesis.
Introduction
Reactive microglia are associated with nearly all neurodegenerative diseases1, but the mechanisms underlying microglia activation and their potential contributions to disease progression remain poorly understood. In contrast to Parkinson’s and Alzheimer’s diseases that are related to a spectrum of genetic mutations and environmental factors2, 3, Huntington’s disease (HD) is a neurodegenerative disorder caused by specific expansion of a CAG repeat within the coding region of the HTT gene4. This mutation results in an elongated stretch of glutamine near the N-terminus of the Huntingtin protein (HTT) 4. Polyglutamine expansions of more than 37 residues initiate a degenerative process characterized by the loss of medium spiny neurons in the striatum5. A number of roles of this protein have been uncovered, but the precise mechanism(s) by which mutant HTT (mHTT) causes dysfunction and degeneration of neurons remains an area of intense research.
Although HD is thought to result primarily from consequences of mutant protein expression in neurons, Huntingtin is broadly expressed, including high levels of expression in microglia6, 7. Despite the fact that microglia comprise <10% of the total brain cells8, this specialized cell population rapidly responds to even minor pathological changes in the brain and may contribute directly to neuronal degeneration by producing an excess of various pro-inflammatory factors9. Several lines of evidence indicate altered microglia activation states in the context of HD. Inflammation appears early in the onset of disease10 and reactive microglia are conspicuous even in low-grade HD human brains, suggesting an early microglia response to changes in axons11. In patients’ striatum and cortex, reactive microglia occur in all grades of pathology and accumulate in relation to degree of neuronal loss11. Furthermore, an in vivo PET study showed that microglia activation correlates with severity of the pathology in HD patients12. Significant microglia activation in regions related to cognitive function in HD patients has recently been suggested to predict disease onset13. Monocytes from HD subjects express mHTT and have been reported to be hyperactive in response to stimulation14. A similar pattern was seen in macrophages and microglia derived from R6/215 and YAC14 HD mouse models. Interestingly, age-dependent changes in striatal microglial morphology and vasculature in the YAC128 mouse model of HD have been reported16. The cerebrospinal fluid and striatum of HD patients exhibit evidence of immune activation, with up-regulation of IL6, IL8 and TNFα 14, 17. These cytokines can induce a CNS inflammatory response that alters the blood brain barrier and affects neuronal function18, suggesting that they could contribute to disease progression. At a molecular level, dysfunctional kynurenine pathway19, NFκB activation20, cannabinoid receptor 2 signaling21 and P2X7 receptor involvement22 have been reported as potential mechanisms explaining HD inflammation in different mouse models.
Inflammation is normally an adaptive biological response to pathogen infection and tissue injury that serves to engage the immune system and tissue repair mechanisms. In these cases, inflammation resolves following eradication of the inciting stimulus. However, when pathological processes result in a sustained inflammatory response, persistent expression of mediators including as IL6, IL8 and TNFα can contribute to tissue damage and disease progression23. The observation of an inflammatory component to HD raises the question of whether inflammation is the response of surrounding cells to a neuron-autonomous degenerative process and/or due to microglia-autonomous immune activation resulting from expression of mutant Htt. Regardless of the mechanism(s) responsible for microglia activation, the contribution of inflammation to HD pathogenesis remains poorly understood.
Here, we investigated whether mHTT expression alters microglia function in a cell-autonomous fashion. Using genome-wide approaches, we find that mHTT expression in microglia results in increases in the expression of several inflammatory response genes even in the absence of pro-inflammatory stimuli. Unexpectedly, this phenomenon is linked to an increase in the expression and transcriptional activities of the myeloid lineage-determining factors PU.1 and C/EBPs. These transcription factors are required for the development and function of the macrophage/microglia lineage9, 23, 24 and establish the regulatory potential of these cells by selecting enhancers and promoters that are acted upon by signal-dependent transcription factors such as NFκB24. As a consequence of increased PU.1 and C/EBPs activity, mHTT expressing microglia exhibit enhanced toxic effects on wild-type neurons in comparison to wild-type microglia ex vivo and after sterile inflammation in vivo.
Results
mHTT promotes PU.1-C/EBPs-dependent pro-inflammatory gene expression
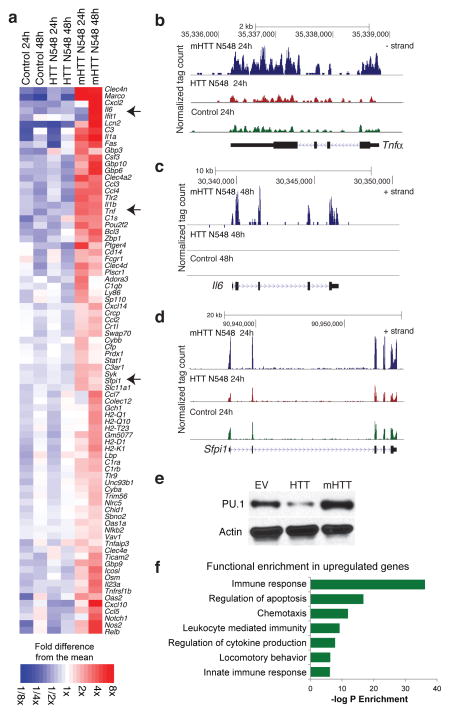
To investigate potential cell-autonomous roles of mHTT in microglia, we generated isogenic BV2 microglia cell lines stably over-expressing the first 548 aa N-terminus of human wild-type or mutant Huntingtin protein. These lines were maintained as pools of clones and cultured for 4 weeks before study to avoid acute effects of lentiviral infection on microglia activation. Deep sequencing revealed that microglia expressing mHTT N548 exhibited higher expression of mRNAs encoding pro-inflammatory factors as compared to cells expressing WT HTT N548 in the absence of pro-inflammatory stimulation (Fig. 1a, Supplemental Dataset 1). Notably, the increase in Il6 and Tnfα mRNAs (Fig. 1b,c) is similar to the pattern previously observed in HD patients14, 17. Unexpectedly, expression of Sfpi1 (encoding PU.1), a key factor in myeloid fate determination9, 24–26 was itself up-regulated in the presence of mHTT N548 at the mRNA (Fig. 1d) and protein (Fig. 1e) levels. Conversely, in presence of HTT N548, the expression of PU.1 was down-regulated (Fig. 1e). Gene Ontology analysis of the entire set of up-regulated genes, observed in presence of mHTT N548, indicated significant enrichment for terms related to innate immunity and inflammation (Fig. 1f).
Figure 1.
RNA-Seq analysis reveals that Mutant Huntingtin N-terminus expression triggers pro-inflammatory gene expression in BV2 microglia. (a) Heatmap representing RNA-Seq gene expression of up-regulated inflammatory genes in BV2 microglia cell lines transduced with lentivirus expressing mHTT N548 in comparison to empty vector (Control) and expressing HTT N548 at 24 and 48h after plating. (b–d) UCSC Browser images representing normalized RNA-Seq read density from BV2 microglia cell lines transduced with empty vector (Control), HTT N548 or mHTT N548 mapped at the (b) Tnfα, (c) Il6 and (d) Spfi1 (PU.1) genomic loci. (e) Western blots of PU.1 expression in BV2 microglia cell lines transduced with lentivirus empty vector (EV), HTT N548 or HTT N548 expression vectors. One representative experiment out of 3 biological replicates. Image shown has been cropped from the original one. Full-length blot is presented in Supplementary Figure 8. (f) Gene Ontology analysis of functional annotations associated with up-regulated genes in BV2 microglia expressing mutant Huntingtin in comparison to wild-type Huntingtin.
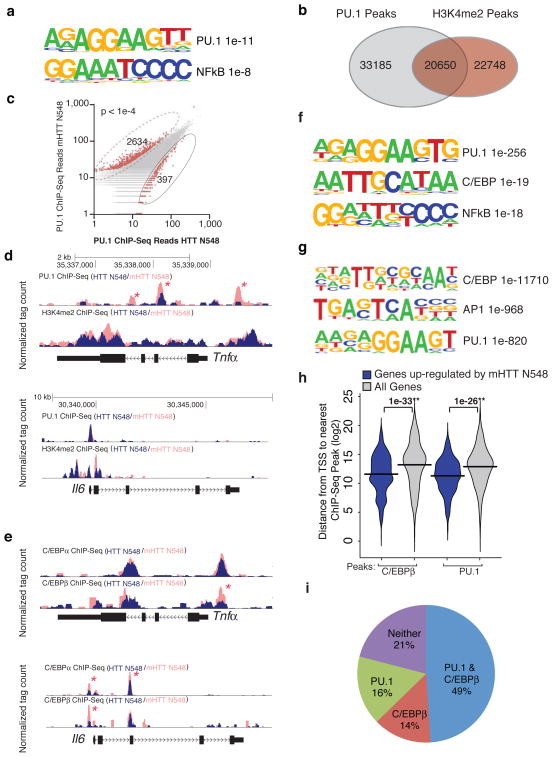
In order to investigate potential mechanisms leading to the enhanced inflammatory gene expression in microglia expressing mHTT N548, we performed de novo motif enrichment analysis of transcriptional regulatory elements associated with the up-regulated genes. Interestingly and consistent with the up-regulation of PU.1 expression, we observed a consensus PU.1 motif as the most enriched regulatory element in the enhancers/promoters of the up-regulated genes in microglia cells expressing mHTT N548 (Fig. 2a). A binding motif for NFκB was the next most enriched motif, consistent with a previously described activation of this factor in presence of mHTT20. Since PU.1 is a key factor in myeloid fate determination9, 25 and is required for selection of enhancers that are acted upon by NFκB24 these findings suggested a potential role of mHTT in priming myeloid cells toward inflammatory activation.
Figure 2.
Mutant Huntingtin promotes pro-inflammatory gene expression via PU.1 and C/EBPs. (a) Motif enrichment in promoters of up-regulated genes in BV2 microglia expressing mHTT N548 in comparison to HTT N548. (b) Venn diagram representing the overlap between PU.1 peaks and H3K4me2 peaks in BV2 cells expressing HTT N548 as detected by Chip-Seq. (c) Scatter Plot of normalized tag counts for PU.1 peaks detected in microglia cell lines overexpressing wild-type vs mutant Huntingtin (data points color-coded red indicate >4 fold difference, p-value < 1e−4) (d) PU.1 and H3K4me2 Chip-Seq read density from BV2 microglia overexpressing wild-type vs mutant Huntingtin at the Tnfα and Il6 loci. Asterisks denote differential PU.1 binding. (e) Motif enrichment at PU.1 peaks specific for BV2 cells expressing mHTT N548. (f) C/EBPα,β Chip-seq read density from BV2 microglia overexpressing wild-type vs mutant Huntingtin at the Tnfα and Il6 loci. Asterisks denote differential C/EBP binding. (g) Motif enrichment at C/EBPβ peaks in BV2 cells expressing mHTT N548. (h) Distribution of distances from either mHTT N548 up-regulated genes’ promoters or all promoters to the nearest C/EBPβ and PU.1 Chip-Seq peaks. (i) Pie diagram representing the fraction of genes up-regulated by the presence of mHTT N548 with a C/EBPβ and/or PU.1 peak within 5kb of the TSS.
To establish a direct link between the increased expression of PU.1 and target genes, we performed chromatin immuno-precipitation coupled to deep sequencing (Chip-Seq) to quantify PU.1 binding in BV2 microglia cells over-expressing HTT N548 or mHTT N548. In addition, we performed Chip-Seq analysis for H3K4me2, a histone modification associated with enhancers and promoters27–29, thereby generating a microglia enhancer ‘atlas’. In BV2 microglia expressing HTT N548, PU.1 was associated with approximately half of the H3K4me2-marked regions, consistent with the binding pattern of PU.1 in macrophages24, 30 (Fig. 2b). PU.1 peaks in microglia expressing HTT N548 or mHTT N548 largely overlapped, but mHTT N548 expression was associated with a significant number of differential PU.1 peaks (defined by a normalized tag count of >4-fold comparing mHTT N548 to HTT N548), consistent with the increased expression of PU.1 (2634 differential sites in mHTT N548 cells vs 397 differential sites in HTT N548 cells, p < 1e-4) (Fig. 2c).
Genomic loci encoding mRNAs that were up-regulated in BV2 microglia expressing mHTT N548 generally exhibited higher enrichment of PU.1 binding to promoters/enhancers, exemplified by the Tnfa locus (Fig. 2d,). However, a subset of PU.1 target genes, exemplified by Il6, did not exhibit this pattern (Fig. 2d). This raised the question as to whether additional factors might cooperate with PU.1 to contribute to the observed up-regulation of PU.1 target genes. De novo motif analysis of PU.1 binding sites in the vicinity of genes up-regulated in the presence of mHTT N548 returned a motif for the C/EBP family of transcription factors (Fig. 2f). Members of the C/EBP family are required for myeloid cell development and induction of several inflammatory mediators31, 32. To investigate a potential role of C/EBPs in contributing to expression of a subset of PU.1 target genes, we performed Chip-Seq analysis for C/EBPα,β in BV2 microglia cells over-expressing HTT N548 or mHTT N548. Interestingly, we observed a stronger enrichment for C/EBPα,β on the Il6 promoter/enhancer as well as on Tnfa promoter/enhancer in mHTT N548 cells (Fig. 2e). No differences in C/ebpα,β mRNA expression levels were observed in microglia over-expressing wild-type or mHTT N548 (Supplemental Dataset 1). Motif enrichment analysis for C/EBPβ Chip-Seq confirmed the correlation between this factor and PU.1 binding (Fig. 2g). Analysis of the correlation between the distance from promoter to the nearest C/EBPβ peak as well as from promoter to the nearest PU.1 peak detected by Chip-Seq and the altered gene expression in the presence of mHTT N548 indicated a higher correlation for C/EBPβ binding and PU.1 binding (respectively 1e-33 and 1e-26) to induced genes in comparison to the total set of expressed genes (Fig. 2h). Nearly 80% of the up-regulated genes in mHTT N548-expressing cells exhibited PU.1 and/or C/EBPβ binding within 5kb of the transcriptional start site (Fig. 2i). In contrast, the corresponding frequency for all genes is ~51%. Together, these findings indicate a strong relationship of nearby PU.1 and C/EBPβ binding to up-regulated genes in mHTT N548-expressing cells.
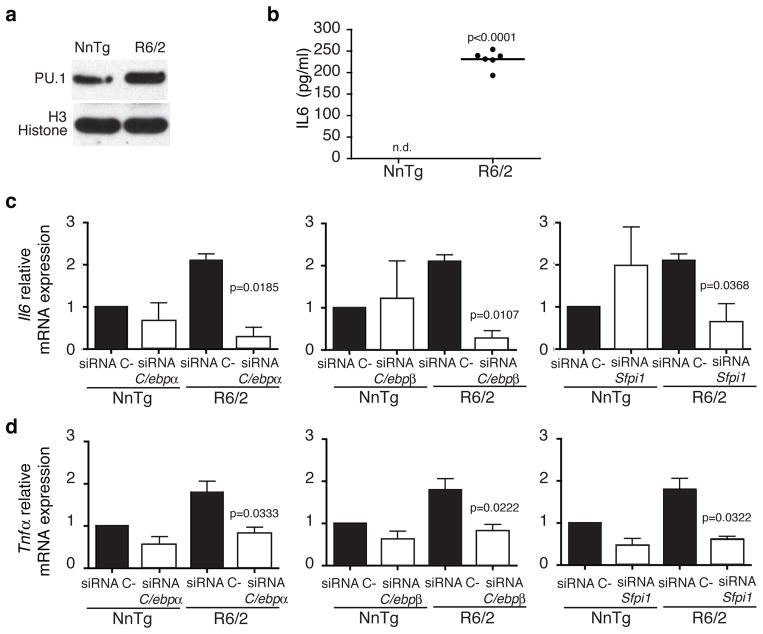
To confirm and extend the observations obtained from microglia cell lines over-expressing mHTT N548, we next measured the expression of Sfpi1 (PU.1) and representative PU.1-C/EBPs target gene mRNAs in primary microglia derived from the R6/2 mouse model33 expressing mHTT (exon 1), in the absence of pro-inflammatory stimulation. As shown in Fig. 3a Sfpi1(PU.1) expression was significantly increased in primary microglia derived from newborn R6/2 mice, both at mRNA (+50%, p<0.0009) and protein level, in comparison to primary microglia from non-transgenic littermates. Corresponding increases were observed in the expression of Il6 (3 folds increase, p=0.0035) and Tnfα mRNAs (2 folds increase, p=0.0075) in comparison to primary microglia from non-transgenic littermates. In contrast, increased expression of Sfpi1, Il6 and Tnfα was not observed in bone marrow-derived macrophages (BMDM) from R6/2 mice (Supplementary Fig. 1a–c). No difference in the level of C/ebpα,β mRNAs expression in primary microglia as well as BMDM from R6/2 and nontransgenic littermates was observed (data not shown). Thus, the observed increase of Sfpi1 expression seems to be specific for primary microglia expressing mHTT (exon 1). Supernatants collected from the same R6/2 primary microglia cultures exhibited significantly increased IL6 levels after 72h of incubation, confirming the increased Il6 mRNA expression observed in the same cells (Fig. 3b).
Figure 3.
PU.1 and PU.1-C/EBPs target genes are upregulated in primary microglia from R6/2 mice. (a) PU.1 expression in P0 primary microglia purified from nontransgenic littermates and R6/2 pups. Immunoblot represents lysates of pool of microglia cells obtained from 5 pups per group. Image shown has been cropped from the original one. Full-length blot is presented in Supplementary Figure 8. (b) IL6 protein secretion in culture supernatant from P0 primary microglia from R6/2 pups and nontransgenic littermates. Graph represents pg/ml of IL6 in pooled microglia culture supernatants from 5 pups respectively in each biological replicate, (n= 3 biological replicates, n.d.=not detected). qRT-PCR analysis for Il6 (c) and Tnαf (d) mRNAs expression levels in presence of C/ebpα,β or Spfi1 siRNAs respectively in nontransgenic littermates and R6/2 primary microglia (mean±sd, n= 3 biological replicates). In all cases p values were calculated using the two-tailed paired student t-test. Each experiment is representative of at least three independent replicates.
To investigate whether PU.1 and C/EBPα,β are required for controlling the enhanced pro-inflammatory gene expression observed in presence of mHTT (exon 1), we transfected primary microglia from R6/2 and non-transgenic littermates with specific siRNAs directed against C/ebpα,β or Sfpi1. The three siRNAs were effective in reducing their mRNA targets expression by ~80% in comparison to control siRNAs (Supplementary Fig. 2). Knockdown of C/ebpα,β and Sfpi1, separately, greatly reduced the levels of mRNA expression of Il6 and Tnfα in primary microglia from R6/2 in comparison to microglia transfected with control siRNA (Fig. 3c,d). These data confirm the requirement of PU.1 and C/EBPs for increased expression of these pro-inflammatory mediators in cells expressing mHTT (exon 1) in the absence of pro-inflammatory stimulation.
mHtt promotes inflammatory activation of microglia but not BMDM
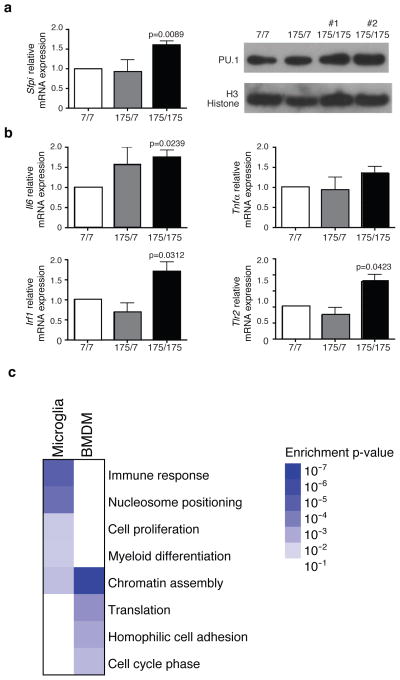
To extend these studies to a mouse model that is genetically analogous to human HD, we took advantage of a knock-in model characterized by a 175 CAG expansion in the Hdh gene34. We evaluated Sfpi1 mRNA and protein expression as well as the expression of representative PU.1-C/EBPs target genes in primary microglia derived from newborn (P0) wild-type, heterozygous and homozygous mutant Htt knock-in mice. We observed that Sfpi1 is significantly up-regulated in primary microglia from mice homozygous for mutant huntingtin (Q175/Q175), both at the mRNA and protein level (Fig. 4a), in comparison to wild-type huntingtin expressing primary microglia cells (Q7/Q7). Consistently, mRNAs from PU.1-C/EBPs target genes Il6, Tlr2 and Irf1, were up-regulated in primary microglia from homozygous mutant mice in absence of pro-inflammatory stimulation (Fig. 4b). Furthermore, we performed RNA-Seq analysis of microglia derived from wild type and knock-in Hdh175/175 adult symptomatic mice. We verified that the expression of full-length mutant Htt in vivo in microglia is able to promote a genome-wide pro-inflammatory transcription signature similar to that observed in a microglia cell line expressing the N terminus fragment of mHTT (Fig. 4c and Supplemental dataset 2). Compatible with the observation of an increased activity of the myeloid lineage determining factors PU.1-C/EBPs, we also observed an enrichment for cell proliferation and myeloid differentiation genes in the GO analysis (Fig. 4c). Although the vast majority of HD patients only carry a single allele of mutant HTT, alterations in gene expression observed in microglia from homozygous Hdh175/175 mice were less obvious in microglia from heterozygous mice. This finding suggests that genetic background, environmental factors, cellular context and age play important roles in determining penetrance of the mutant allele. Consistent with this, we performed RNA-Seq analysis of BMDM obtained from the same animals (Supplemental Dataset 2). The direct comparison of the transcriptional profiles of BMDM from wild type and knock-in Hdh175/175 mice with those determined in the corresponding microglia above revealed that expression of Q175/Q175 exerts a distinct effect on basal gene expression in BMDMs than microglia (Fig. 4c), most likely reflecting both different origins35, 36 and environmental influences. Consistent with this, global RNA-Seq analysis indicates significant differences in basal gene expression programs in WT BMDMs and microglia (Supplementary Fig 3).
Figure 4.
PU.1 and PU.1-C/EBPs target genes are up-regulated in primary microglia from Hdh175/175 Knock-in mice. (a) qRT-PCR analysis for Spfi1 (PU.1) mRNA and protein expression in primary microglia purified from Hdh7/7, Hdh175/7 and Hdh175/175 newborn (P0) pups (mean±sd, n= 3 biological replicates). Image shown has been cropped from the original one. Full-length blot is presented in Supplementary Figure 8. (b) qRT-PCR analysis for Il6, Tnfα, Irf1 and Tlr2 mRNAs expression in primary microglia purified from Hdh7/7, Hdh175/7 and Hdh175/175 pups (mean±sd, n= 3 biological replicates). All p values were determined by two-tailed paired student t-test. (c) Gene Ontology analysis reporting the biological processes enriched in genes up-regulated in Hdh175/175 versus Hdh7/7 for microglia and BMDM obtained from adult mice (18–20 months old). RNAseq results are based on mRNA extraction from pooled microglia cells or BMDM obtained from 4 mice per group.
PU.1-C/EBPs-induced inflammatory transcription is specific for mHTT in vivo
To link the observations obtained ex vivo in primary microglia from different mouse models of HD with the disease progression in vivo, we evaluated the expression of Sfpi1 and its pro-inflammatory target genes in adult R6/2 mice in comparison with non-transgenic littermates. Increased expression of Sfpi1 mRNA was already evident in pre-symptomatic mice and even greater at symptomatic stage in striatum from R6/2 mice (Supplementary Fig. 4a). Concomitantly, we detected an increase in the expression of Il6, Tnfα, Irf1 and Tlr2 mRNAs in the same region (Supplementary Fig. 4b–e). No difference in the level of C/EBPα,β mRNA expression was observed in striatum from R6/2 and nontransgenic littermates (data not shown). The levels of Sfpi1 expression as well as the pro-inflammatory gene expression correlate with the disease progression in R6/2 model of HD. In parallel, we analyzed the expression of PU.1 and specific PU.1-C/EBPα,β target pro-inflammatory genes in spinal cord, cortex and striatum of SOD1G37R mice, an animal model for Amyotrophic Lateral Sclerosis (ALS)37. ALS is also a progressive neurodegenerative disease that primarily affects motor neurons that connect the brain and spinal cord to muscles resulting in a fatal paralysis within few years from onset38. Even though microglia activation plays a key role in ALS pathogenesis39 PU.1 and specific PU.1-C/EBPα,β target pro-inflammatory genes elevated in mutant Huntingtin models were not increased in striatum, cortex or spinal cord from 1 year old symptomatic SOD1G37R mice (Supplementary Fig. 5a–e). These results are consistent with a specific, cell-autonomous role of mutant Huntingtin in promoting basal activation of pro-inflammatory genes via lineage determining factors PU.1-C/EBPα,β.
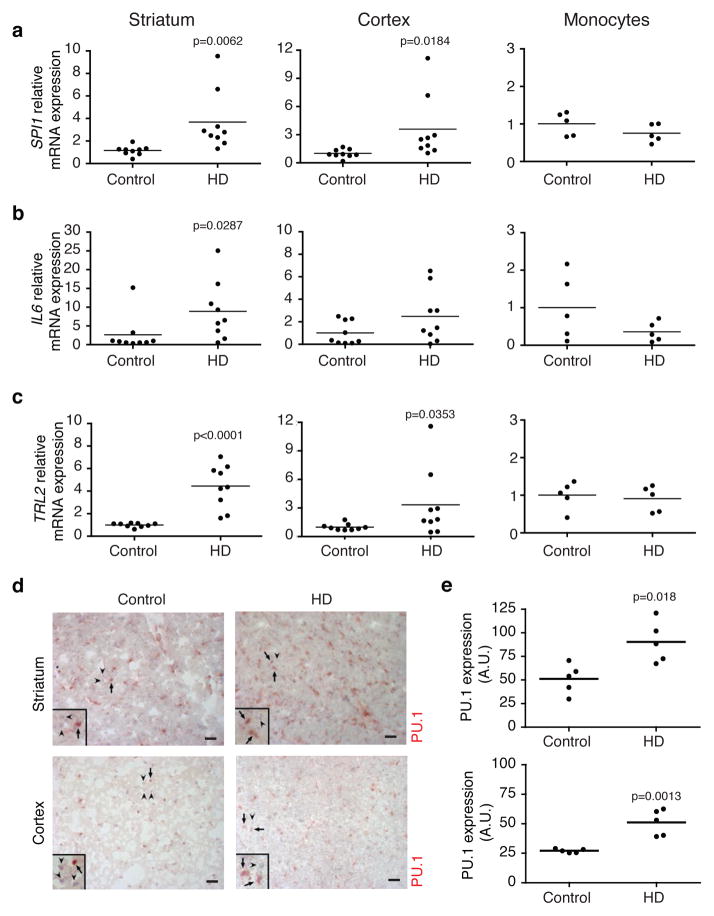
Pro-inflammatory genes expression is increased in brains of HD individuals
We next investigated the expression of SPI1(PU.1) and C/EBPα,β as well as their targets in striatum, cortex and monocytes from human HD and control individuals. SPI1 expression was increased in striatal as well as in cortical postmortem human samples from HD individuals, in concert with increased levels of IL6, IRF1 and TLR2 mRNAs in the striatum and a trend increase of IL6, TNFα and TLR2 in the cortex of the HD individuals (Fig. 5a–c and Supplementary Fig. 6a,b). No change in C/EBPα,β mRNAs levels were detected in cortical or striatal samples from HD individuals in comparison to matching controls (data not shown). The expression of SPI1 and C/EBPα,β as well as their target genes showed no significant difference in monocytes from HD patients (Fig. 5a–c and Supplementary Fig. 6a,b). These observations confirm and extend the distinct inflammatory profile observed in postmortem HD brains14, 17. We next performed immunostaining for PU.1 in frontal cortex as well as in striatum of five frozen post-mortem samples from HD individuals and matching controls (Fig. 5d). We observed an increase in the number of microglia per section (Fig. 5d), consistent with previous observations11. Quantification of the intensity and distribution of PU.1 staining further indicated a significant increase in PU.1 expression on a per cell basis (~ 2-fold, Fig. 5e). These in vivo findings in the brains of patients with HD are thus consistent with what we observed in primary microglia expressing mutant Huntingtin isolated from two different rodent models as well as in BV2 microglia expressing the mHTT N548.
Figure 5.
Inflammation in vivo in HD individuals. qRT-PCR analysis for SPI1(a), IL6 (b) and TLR2 (c), mRNAs expression in striatum (first column, n= 9 individual specimen per group), cortex (second column, n= 9 individual specimen per group) and monocytes (third column, n= 5 individual specimen per group) from controls and HD individuals (Vonsattel Grade 3 and 4). Each dot is representative of one individual. All p values were determined by unpaired student t-test. (d) PU.1 IHC staining on post-mortem frozen samples of striatum (Caudate/Putamen) and cortex (Brodman Area 4) from HD (n=5, Vonsattel Grade 3) and matching controls (n=5). One representative picture is shown for each group. (e) Densitometric analysis of PU.1 staining. Each dot is representative of PU.1 expression of one individual. (n= 5 individual specimen per group). Arbitrary Unit is defined by the number of pixels representing the area of PU.1 staining normalized by the number of PU.1+ microglia cells. PU.1+ microglia cells were distinguished morphologically from endothelial cells. All p values were determined by unpaired student t-test. Scale bar: 100μm. Insets highlight PU.1+ cells. Arrows in representative pictures and insets for each condition point to PU.1+ nuclei, arrowheads point to PU.1− nuclei.
Effect of mutant Huntingtin expressing microglia on neurons ex vivo and in vivo
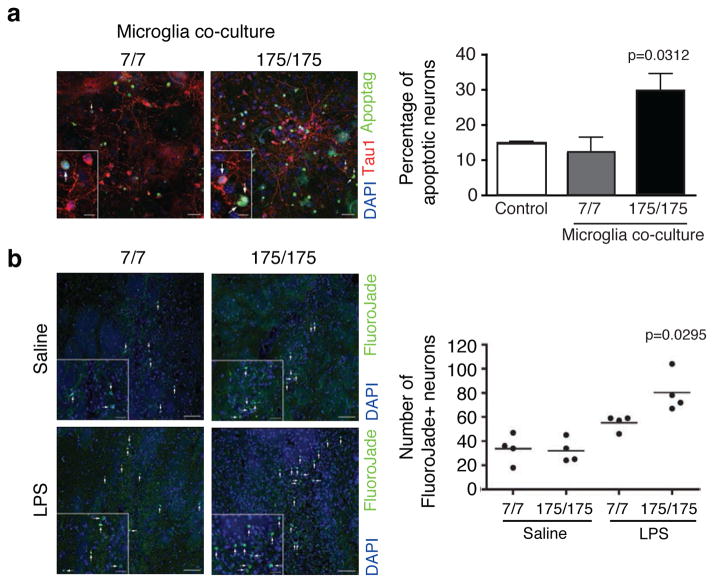
Experiments using neuron and glia co-culture in vitro suggest that activation of innate immunity in the CNS can trigger neuronal death40. We recently reported that reduced Nurr1 expression results in exaggerated inflammatory responses in microglia leading to the production of factors that cause death of tyrosine hydroxylase-expressing neurons41. Since we observed exaggerated IL6 secretion by mHTT (exon 1) expressing microglia (Fig. 3d), we investigated whether neuron-microglia co-culture would exhibit any neurotoxic effects of microglia expressing mutant Htt on wild-type neurons. This possibility was evaluated by co-culturing mouse embryonic stem cell (ESC)-derived normal neurons over a substrate of wild-type primary astrocytes. Subsequently, wild-type (Q7/Q7) or mutant Htt knock-in microglia (Q175/Q175) cells were added to the culture. As illustrated in Fig. 6a, addition of mHtt expressing microglia, but not WT microglia, increased neuronal apoptosis.
Figure 6.
Effect of mutant Huntingtin expressing microglia on primary neurons ex vivo and in vivo. (a) TUNNEL assay for neurons/astrocytes/microglia co-culture in presence of primary microglia from wild-type Hdh7/7 or homozygous mutant Huntingtin knock-in mice (Hdh175/175). Arrows in representative pictures for each condition point to apoptotic neurons. Insets highlight TUNEL positive neurons. Scale bar: 30μm in panels and 10μm in insets. Quantitative representation of the percentage of Apoptag+ neurons, obtained by the ratio between the number of triple positive cells (Apoptag+, TAU1+ and DAPI+ cells)/total number of double positive (TAU1+ and DAPI+) cells, (mean±sd, n= 3 biological replicates) with pools of microglia from 4 pups per group. Ctrl represents neurons/astrocytes co-cultured without microglia added. p value was determined by one-tailed paired student t-test. (b) FluoroJadeB staining of coronal brain sections from wild-type (Hdh7/7) or homozygous mutant Huntingtin knock-in mice (Hdh175/175) injected with saline solution or LPS (5μg). One representative image for each condition is shown (left). Arrows in representative pictures and insets for each condition point to FluoroJadeB+ neurons. Scale bar: 50μm in panels and 20μm in insets. Quantitative representation of the number of FluoroJadeB+ degenerated/dead neurons (right). Each dot represents the number of FlurojadeB+ neurons per mouse (n=4 mice per group, one-way Anova and Tukey’s post-hoc test).
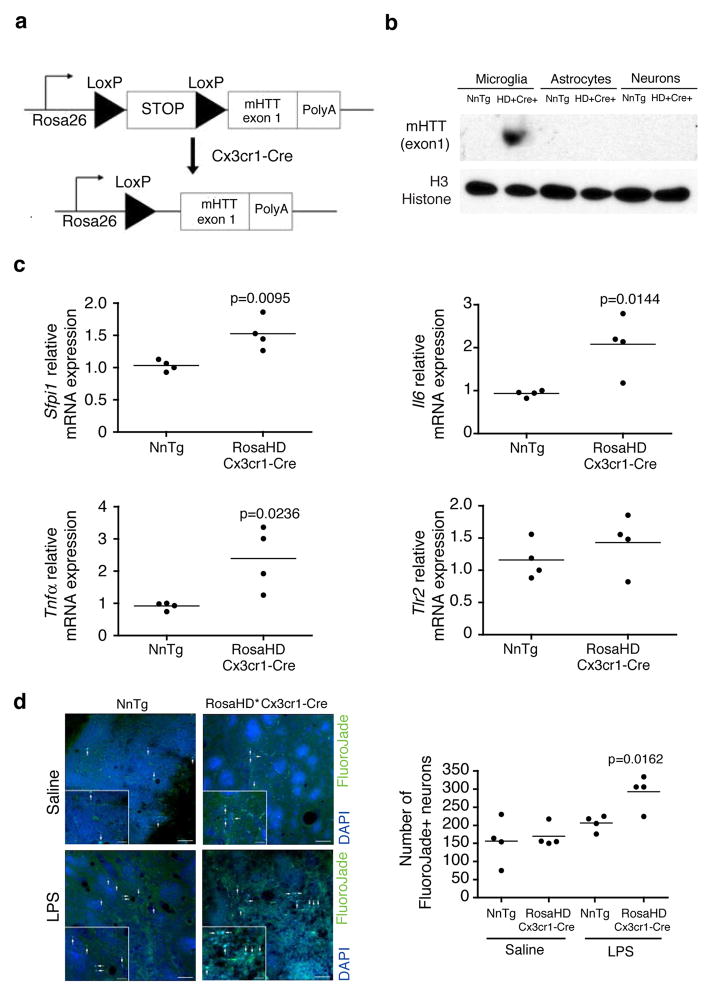
To extend these observations in vivo, we considered the possibility that sterile inflammation could be triggered in HD patients by endogenous molecules, such as DAMPs, components of dead neurons and protein aggregates. As a means of mimicking sterile inflammation, we performed stereotactic injection of LPS into the striatum of 12–15 weeks old WT or Hdh175/175 mice. Due to the fact that LPS mainly triggers microglia activation but has no direct toxic effect on neurons40, LPS-induced neurotoxicity is secondary to microglia activation. No difference was detected in the number of degenerating neurons in wild-type versus Hdh175/175 mice injected with saline solution as measured by FluorojadeB staining. This result is consistent with the lack of HD symptoms or CNS pathology in Hdh175/175 mice at this age42. In contrast, LPS induced greater neuronal death in Hdh175/175 mice in comparison to wild-type mice (Fig. 6b). As negative control, no FluorojadeB+ cells were detected in the contralateral non-injected hemispheres. This result is consistent with exaggerated inflammatory/neurotoxic responses of microglia expressing mutant Huntingtin, but does not exclude the possibility that this was also due to increased sensitivity of neurons expressing mutant Htt. To directly address this concern, we mated a mouse expressing the myeloid-specific Cre driver (Cx3cr1-Cre) to RosaHD knockin mice43 (Fig. 7a), which results in exclusive microglia-specific expression of mHTT (exon 1) in the brain (Fig. 7b). Expression of the mHTT (exon 1) in microglia is sufficient to induce an increased transcription of Sfpi1, Il6 and Tnfα mRNAs in the striatum of RosaHD x Cx3cr1-Cre mice in comparison to nontransgenic littermates (Fig. 7c). We used these and control mice to determine whether expression of mHTT (exon 1) selectively in microglia among brain cells results in exaggerated neuronal death following an inflammatory insult. We performed stereotactic injection of LPS into the striatum of 8 weeks-old RosaHD x Cx3cr1-Cre and nontransgenic littermates mice. What we observed is that mice expressing mHTT (exon 1) in microglia show an enhanced incidence of neuronal death in the presence of sterile inflammation in comparison to nontransgenic littermates (Fig. 7d). Taken together, these results demonstrate a potential contribution of microglia activation and inflammation to HD pathogenesis.
Figure 7.
Effect of mutant Huntingtin expressing microglia on primary wild-type neurons in vivo. (a) The schematics of conditional HD mice in which expression of mHTT (exon 1) was dependent on the expression of Cx3cr1-driven Cre recombinase. Mutant huntingtin (exon 1) is targeted to the Rosa26 locus. The targeted locus contains the endogenous Rosa26 promoter, a transcriptional STOP sequence, two loxP sites (black triangles), mHTT exon 1 with 103 mixed CAA-CAG repeat (encoding for poly glutamine expansion) and a poly-adenylation signal (PolyA). (b) mHTT (exon 1) protein expression in primary microglia, astrocytes and neurons purified from RosaHD+ x Cx3cr1-Cre+ and nontrasngentic littermates. Immunoblot represents lysates of pool of microglia, astrocytes and neurons respectively obtained from 4 pups per group. Image shown has been cropped from the original one. Full-length blot is presented in Supplementary Figure 8. (c) qRT-PCR analysis for Sfpi1, Il6, Tnfα and Tlr2 mRNAs expression in striatum from 8 weeks old non-transgenic littermates and RosaHD x Cx3cr1-Cre mice. Each dot is representative of one mouse. All p values were determined by unpaired student t-test (n=4 mice per group). (d) FluoroJadeB staining of coronal brain sections from nontransgenic Littermates or RosaHD x Cx3cr1-Cre mice injected with saline solution or LPS (5μg). One representative image for each condition is shown (left). Arrows in representative pictures and insets for each condition point to FluoroJadeB+ neurons. Scale bar: 50μm in panels and 20μm in insets. Quantitative representation of the number of FluoroJadeB+ degenerated/dead neurons (right). For details about counting procedure see Material and Methods section. Each dot represents the number of FlurojadeB+ neurons per mouse (n=4 mice per group, one-way Anova and Tukey’s post-hoc test).
Discussion
Previous studies of the inflammatory component of HD have provided evidence for alterations in signal-dependent mechanisms in neurons, including NFκB activation20, cannabinoid receptor 2 signaling21 and P2X7 receptor involvement22. Here, we provide evidence that the expression of mutant Huntingtin in microglia is sufficient to confer a cell–autonomous increase in pro-inflammatory gene expression and exaggerated neurotoxic effects on wild-type neurons ex vivo and after pro-inflammatory stimulation in vivo. Several lines of evidence support a mechanism in which mutant Huntingtin exerts a cell-autonomous effect within microglia by increasing the expression and transcriptional activities of the myeloid lineage-determining factors PU.1 and C/EBPα,β. Binding sites for PU.1 and C/EBPs were highly enriched in enhancers and promoters associated with the genes exhibiting constitutive up-regulation in mutant Huntingtin-expressing microglia. Expression of PU.1 itself was increased at both mRNA and protein levels in mutant Huntingtin-expressing microglia and ChIP-Seq analysis demonstrated enhanced binding of PU.1 at thousands of genomic locations in these cells. Increased binding of PU.1 was associated with enhanced co-occupancy by C/EBPs, and the combination of enhanced PU.1 and C/EBP binding was highly correlated with increased expression of nearby genes, such as Il6 and Tnfα. Furthermore, since PU.1 and C/EBPα,β are lineage-determining factors, we could speculate that their increased activity in mutant Huntingtin expressing microglia and the concomitant enrichment for cell proliferation and myeloid differentiation genes in the GO analysis could be implicated in the increased density of microglia observed in HD individuals11.
Recent studies indicate that PU.1, C/EBPs and AP-1 proteins function in a collaborative manner to select a large fraction of the functional enhancers in the macrophage that are acted upon by signal-dependent transcription factors26 (Supplementary Fig. 7). Of particular relevance to the present findings, increased expression of a conditional form of PU.1 resulted in a coordinate increase in PU.1 and C/EBPαβ binding throughout the genome, without changes in C/EBPαβ expression26. Our previous studies demonstrated that enhancers selected by PU.1 and C/EBPs provide access to signal-dependent factors such as nuclear receptors and NFκB, providing the basis for cell-specific responses26. The present findings are thus consistent with a model in which mutant Huntingtin-induced PU.1 expression drives the selection PU.1-C/EBPs-dependent enhancers that promote expression of pro-inflammatory/neurotoxic mediators in microglia under basal conditions (Supplementary Fig. 7). Eventually, sterile inflammation triggered in HD patients by endogenous molecules, such as components of dead neurons and protein aggregates, could lead to a further microglia activation and result in an increased neuronal death (Supplementary Fig. 7). This mechanism would explain both increases in basal gene expression and enhanced responses to exogenous stimuli and suggests a molecular basis for the state of “priming” defined recently by Leavitt and Hayden based on morphological evidence16.
Our findings are consistent with the possibility that exaggerated microglia activation could contribute to the early inflammatory reaction observed in HD11, 16, 44. In particular, we observed that primary microglia derived from R6/2 mice constitutively express and secrete increased amounts of the pro-inflammatory cytokine IL6. Although a previous study did not find evidence for IL-6 secretion by microglia from R6/2 mice in absence of stimulation14, this apparent discrepancy with our data can easily be explained by the fact that we used a more sensitive detection system than that used in the previous publication. Our results provide a potential explanation for previous observations reporting increased IL6 in the CSF of HD patients before the clinical onset of disease45. Moreover, it has been suggested that microglia can modulate adult neurogenesis, such as when activated by inflammation46. IL6 appears to be one of the key mediators of this anti-neurogenic effect47. The abnormal IL6 secretion, reported here and by others14, 17, could therefore also help explain the impaired neurogenesis reported in several mouse models of HD48–50.
In addition to conferring a cell-autonomous basal increase in expression of pro-inflammatory and neurotoxic mediators, our findings suggest that expression of mutant Huntingtin in microglia also confers increased sensitivity to extrinsic inducers of inflammation. Because microglia are the primary cells capable of responding to LPS in the brain parenchyma, the observation that Hdh175/175 mice exhibit increased neuronal apoptosis following LPS injection is consistent with exaggerated neurotoxic responses of Hdh175/175 microglia on wild-type neurons ex vivo. The in vivo observation from Hdh175/175 mice could be explained in part by the fact that neurons expressing mHtt are more fragile than normal neurons, thus more sensitive to the toxic effect induced by mutant Huntingtin microglia. In our opinion, the in vivo experiment might reflect what could happen in HD patients, where cell-autonomous microglia hyper-activation coupled with neuronal fragility can concur to promote disease progression. Nevertheless, the experiment conducted on mice expressing mHTT (exon 1) in microglia clearly shows that microglia activation in HD is sufficient to promote neuronal degeneration and can have a potential contribution to the pathogenesis of the disease. However, further studies will be required to establish whether selective expression of mHTT in microglia is sufficient to exert neuropathological and behavioral deficits in mice.
In the context of HD, what seems to be a general transcriptional mechanism underlying macrophage and possibly microglia activation appears to be specifically related to the expression of mutant Huntingtin exclusively in microglia. In fact, the PU.1-C/EBPs-dependent enhanced basal pro-inflammatory genes expression is not observed in the CNS of an animal model for ALS carrying a mutation on SOD1 gene. This result is particularly relevant considering that ALS is a progressive neurodegenerative disease where microglia activation plays a key role in pathogenesis39
Intriguingly, effects of mutant Huntingtin on PU.1-dependent programs of gene expression appeared to be microglia-specific, as we found no differences in PU.1 levels or pro-inflammatory gene expression in bone marrow derived macrophages obtained from R6/2 and Hdh175/175 mice or monocytes from HD individuals. The basis for specific effects of mutant Huntingtin on microglia gene expression is unclear, but recent lineage tracing experiments provide evidence that microglia are derived from fetal yolk sac progenitors very early in development and represent a self-renewing population of cells that is independent of bone marrow-derived macrophages35,41. It is therefore possible that this unique origin or the specific differentiation program conferred by residence in the CNS determines the cell-autonomous functions of mutant Huntingtin in microglia. Collectively, the present findings reveal a new and unexpected role of mutant Huntingtin in disrupting the regulation of microglia identity and function and provide further impetus to better understand the contribution of microglia activation to the pathogenesis of HD.
On line Methods
Ethics statement
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of UC San Diego and every effort was made to minimize suffering. Studies of human brain samples were performed without individually identifiable information. Research undertaken on such specimens does not meet the regulatory definition of human subjects research.
Human samples
Autopsy brain samples from control and HD patients were provided to E.C. from the archives of the Harvard Brain Bank Tissue Resource Center (HBTRC, Belmont USA): Ct 6002, Ct 5919, Ct 5074, Ct 5021, AN13574, AN13112, AN08704, AN15088, AN10090, AN09667, HD 6010, HD 5570, HD 6062, HD 6062, AN06034, AN14935, AN13326, AN18743, AN10094, AN15530, AN13238, AN07121, AN04187, AN14307, AN10314, AN12699, AN13041, AN17896, AN08666, AN01077. From New York Brain Bank at Columbia University (NYBB, New York, USA): Ct-T99, Ct T-168. From Massachusetts General Hospital (MGH, Boston, USA): HD 3723, HD 3484. Control subjects were matched to HD patients for sex and age. All diagnoses were based on clinical assessment and histopathological evaluation by experienced neuropathologists according to Vonsattel classification. Fresh blood samples were obtained from Dr. Corey-Bloom at Shiley-Marcos Alzheimer’s Disease Research Center (ADRC, La Jolla, USA): 7135, 7150, 7151, 7152, 7153, 6000, 6018, 7082, 7094, 7129.
Cell lines
Murine microglial BV2 cells, primary mouse microglia, mouse astrocyte maintained with DMEM (Cellgro) supplemented with 10% FBS (low endotoxin, Hyclone) and penicillin/streptomycin (Invitrogen).
Mice and Isolation of Primary Cells
C57BL/6, R6/2 and RosaHD43 mice were purchased from Jackson Lab. Tg(Cx3cr1-cre)MW126Gsat mice have been generated by Nathaniel Heintz, Ph.D., The Rockefeller University, GENSAT and purchased from MMRRC (UC Davis) and together with HdhQ175/Q175, and SOD1G37R housed according to UCSD protocols. mHTT (exon 1) expression has been verified by Western Blot on microglia, astrocytes and neurons cells lysates obtained from RosaHD+ x Cx3cr1-Cre+ and nontransgenic littermates. Primary microglia cells and astrocytes have been purified from the cerebrum of P0 pups. After 10–14 days of culture, microglia cells were isolated from astrocytes as described previously41. Primary neurons were purified from mice cerebrum using Papain Dissociation System (LKL003153, Worthington) according to manufacturer’s instructions. BMDM have been obtained from adult mice as previously reported24. Human monocytes have been purified from freshly collected blood samples by Ficoll-Plaque and selected using Pan Monocytes Isolation Kit (Miltenyi, #130-096-537) according to manufacturer’s instructions.
Reagents
N-terminus human wild-type and N-terminus mutant huntingtin have been cloned from pCAG-Htt19550-15Q and pCAG-Htt1955-128Q respectively into MCS of pCDH-CMV-MCS-EF1-Puro (System Bioscience) using EcoRI and NotI. Lentiviral production and BV2 cells transduction were performed according to the manufacturer’s protocol. Control cell line was generated by transducing BV2 cells with Lentivirus obtained from pCDH-CMV-MCS-EF1-Puro (empty vector). Validation of plasmids used in this study was performed by Western blotting.
SiRNA for C/EBPα sequence: 5′-ACAACAUCGCGGUGCGCAAUU-3′. SiRNAs for C/EBPβ, PU.1 and siGENOME Non-targeting have been purchased fro Thermo Scientific (ON-TARGETplus SMART pool 043110, 041420, 001210 respectively) and has been transfected in primary mouse microglia by Lipofectamin 2000 (Invitrogen) according with manufacturer’s instructions.
Adult microglia Purification
Mice were deeply anaesthetized and then perfused intracardially with ice-cold DPBS (Mediatech 21-031CV). Cortex, striatum, and hippocampus were extracted and gently homogenized in staining buffer (HBSS (Life Technologies, 14175-095), 1% BSA, 1mM EDTA) on ice using a 2 ml polytetrafluoroethylene pestle (Wheaton), first in a 14 ml round-bottom tube (BD Falcon, 352059) and then in a 2 ml grinder chamber (Wheaton, 358029). Homogenates were filtered onto a 70 μm cell strainer (BD Falcon, 352350) and centrifuged for 10 min at 400g. Cell pellets were resuspended in 6 ml of 37% isotonic Percoll (Sigma, P4937) and then underlayed with 5 ml of 70% isotonic Percoll in a 15ml centrifuge tube (Corning, 430790). Tubes were then centrifuged at 600g for 40 min at 18°C, with no acceleration or decelaration. Cells at the 37–70% Percoll interface were recovered and washed once in 15 ml HBSS. Cell were then incubated in staining buffer on ice with CD16/CD32 (eBioscience, clone 93) antibody for 25 min, and then with CD11b-PE (BioLegend 101208, clone M1/70) and CD45-Alexa488 (BioLegend 103122, clone 3-F11) antibodies for 30 min. Cells were then washed once and filtered onto a 40 μm cell strainer (BD Falcon, 352340). Sorting was performed on a BD Influx cell sorter. Microglia were defined as singlets, CD11b+CD45Low events, and emcompassed 90–95% of all CD11b+ events.
Microglia and ESC-derived neuron co-culture
Mouse E14 ESC were maintained on gelatin coated dishes in ES medium (Glasgow Minimum Essential Medium, Sigma G5154) supplemented with 15% FBS (Atlanta Biologicals), 2mM L-Glutamine (Gibco), 1X non-essential amino acids (Gibco), 1X sodium pyruvate (Gibco), and 55μM of 2-mercaptoethanol (Gibco), 1000U/ml LIF (Millipore)) similar to described previously51. Neural progenitor cells (NPCs) were generated and differentiated as described previously52. ESCs were grown in suspension in ES medium without LIF for the first day and in N2/B27 medium (DMEM/F12-Glutamax Medium (Gibco) supplemented with 1X B27 (Gibco) and 1X N2 (Gibco) supplements) supplemented with 500ng/ml Noggin (PeproTech) for four more days. Next, embryoid bodies were dissociated, plated on and maintained on laminin coated dishes in N2/B27 medium supplemented with 20ng/ml EGF (PeproTech), 20ng/ml FGF2 (Stemgent), and 10μg/ml heparin (Sigma). NPCs were differentiated into neurons in N2/B27 medium supplemented with 500μg/ml cAMP (Sigma), 0.2μM ascorbic acid (Sigma), 20ng/ml BDNF (R&D), and 20ng/ml GDNF (R&D) at least for five days before starting a co-culture with primary mouse astrocytes and medium was supplemented with 2% FBS (Hyclone). Primary microglias were plated on neuron-astrocyte co-cultures at 30,000 cells. Microglias triple co-cultures were maintained for another 24h before proceeding with immunohistochemistry.
TUNEL Staining and quantification
Cells were fixed with 4% paraformaldehyde-PBS for 15 min, blocked and permeabilized with donkey serum (10%)+Triton X-100 (0.1%) in PBS and were incubated overnight with mouse monoclonal TAU1 (1:250; Covance). Next day, cells were incubated with secondary antibodies against mouse and 1μg/ml DAPI. TUNEL staining was performed using ApopTag Fluorescein Direct In Situ Apoptosis Detection Kit (Millipore) as described by the manufacturer. Briefly, cells were post-fixed in cooled Ethanol: Acetic Acid (2:1) and incubated with TdT enzyme at 37°C for 1h. For quantification, epifluorescence images for 20 random fields were taken using Stereo Investigator Software (MBF Biosciences) and number of DAPI+ nuclei, TAU1+ neurons and TUNEL+ apoptotic neurons were counted using ImageJ (NIH). The number of apoptotic neurons has been obtained by the ratio between the number of triple positive cells (Apoptag+, TAU1+ and DAPI+ cells)/total number of double positive TAU1+ and DAPI+ cells. At both steps researchers were blind to the experimental conditions.
Stereotaxic injection of LPS in the mouse SN in vivo
Mice (12–15 weeks of age for HdhQ7/Q7 and HdhQ175/Q175 or 8 weeks of age for RosaHD x Cx3cr1-Cre and nontransgenic littermates) were anesthetized using a mixture of ketamine/xylazine (100mg/kg, 10mg/kg) and immobilized in a stereotaxic apparatus. The stereotaxic injection site into the right SN was AP +0.9mm, ML +2.25mm, DV −3mm from bregma. A stainless steel cannula (5μl Hamilton syringe) was inserted and a single 1μl injection of 5μg of LPS (Sigma) or 1μl of PBS was delivered over a 2 min period into the same coordinates.
Immunofluorescence (IF) and quantification
One week after injection, experimental animals were anesthetized and perfused transcardially with 0.9% saline followed by 4% paraformaldehyde. The brain samples were post-fixed with 4% paraformaldehyde overnight and equilibrated in 30% sucrose. Coronal sections of 40μm were prepared with a sliding microtome and stored in cryoprotectant (ethyleneglycol, glycerol, 0.1M phosphate buffer pH 7.4, 1:1:2 by volume) at −20°C. IHC and co-labeling IF for free-floating sections were performed with FluorojadeB (Chemicon, #AG310) as follows. Tissue sections were mounted onto gelatinized slides and allowed to dry at RT. Slides were immersed in EtOH solution at different descending concentration and H2O all for 1 min each. Then, slides have been incubated in 0.06% KmNO4 for 5 min and washed in H2O for 1 min. Fluorojade solution at 0.001% had been added to the slides and left in incubation for 30 min. Finally, after washing and drying overnight slides have been immersed in Neoclear and mounted on coverslip. 4,6–Diamidino-2-phenylindole (DAPI, 1:1000, Roche) was used to reveal nuclei. In the case of WT and Hdh175/175 mice experiment, to determine the number of FluoroJadeB+ degenerated/dead neurons, the number of double positive cells (FluoroJadeB+ and DAPI+) were counted in 6 contiguous sections per mouse. In the case of RosaHD x Cx3cr1-Cre and nontransgenic littermates experiment, massive cell death was observed after LPS injection for both genotypes. Thus, to determine the number of FluoroJadeB+ degenerated/dead neurons, the number of double positive cells (FluoroJadeB+ and DAPI+) as well as the green puncta clusters within/surrounding DAPI nuclei were counted as positive in 9 contiguous sections per each mouse. Scoring was performed by an investigator who was blinded to the experimental conditions.
Immunohistochemistry (IHC) and quantification
Post-mortem frozen brain samples (Brodman Area 4, Caudate-Putamen) were cryosectioned at 5 microns and used in the immunoshitochemsitry assays. Controls included sections of human spleen (control for macrophages), rabbit IgG (negative control), diluting buffer (BSA, 1% bovine serum albumin in PBS phosphate buffered saline) and rabbit anti-Von Willebrand factor (blood vessel marker, positive control). Each step of the assay was followed by buffer rinses. Endogenous Peroxidase was blocked with 0.03% H2O2 (Fisher # H325-100) for 30 min. Endogenous collagen was blocked with 1% BSA/PBS (Sigma #A4503G) for 30 min. Sections were fixed with 10 % neutral buffered Formalin (Fisher SF93-4) for 30 min, washed with PBS and overlaid with, either Rabbit IgG (Dako #N1699) or Rabbit anti PU.1 1:100 (Cell Signalling #2266S) overnight in a humid chamber at 4°C. Binding was detected using HRP labeled anti-rabbit (Jackson #111-035-144) at 1:500 for 30 min at room temperature. Substrate color development was performed using the AEC kit (Vectorlab#H 5501). Nuclei were counterstained using Mayer’s hematoxylin and slides were aqueous mounted for viewing and digital photomicrography using an Olympus BH2 microscope, equipped with a digital Magnafire camera. For quantification, 5 images for each sample were captured. To determine the intensity of signal, the number of pixels representing the area of PU.1staining was calculated automatically using Magic Wand Tool (Adobe Photoshop). For each image, the number of PU.1+ microglia cells is represented by the number of noncontiguous area where PU.1 staining merging nuclear staining automatically revealed by the software. PU.1+ microglia cells were distinguished morphologically from endothelial cells.
RNA Isolation and Quantitative PCR
Total RNA was isolated by RNAeasy kit (Qiagen) from BV2 cells, microglia, monocytes and BMDM. Cortex and striatum from R6/2, SOD1G37R, RosaHD x Cx3cr1-Cre mice as well as HD post-mortem brain samples have been processed with Trizol (Invitrogen) to extract total RNA. One microgram of total RNA was used for cDNA synthesis using Superscript III (Invitrogen), and quantitative PCR was performed with SYBR-GreenER (Invitrogen) detected by 7300 Real Time PCR System (ABI). The sequences of qPCR primers used for mRNA quantification in this study were obtained from PrimerBank53. For human samples the results were normalized over expressed Alu repeats54
Western blotting
BV2 cells, primary microglia, astrocyets and neurons were lysed with buffer C (NaCl 150mM, TrisHCl pH 7.5 250mM, Triton X-100 1%, glycerol 10%, EDTA 5mM, EGTA 1mM) plus PMSF 1mM, DTT 1mM, Complete Protease Inhibitor Cocktail (Roche). Lysates have been incubated in ice for 1h. Finally, lysates have been cleared by spinning 4°C 13000rpm for 10 min. The protein concentrations in the supernatants were then determined using the BCA protein assay (Biorad) as described by the manufacturers. A total of 100μg of protein was separated by SDS-polyacrylamide electrophoresis using NuPAGE 4–12% Bis-Tris Gel (Invitrogen), then transferred electrophoretically to a PVDF membrane. The membranes were blocked by incubation with 5% (w/v) non-fat milk in Tris-buffered saline containing 0.1% (v/v) Tween-20 (TBST) for 60 min at room temperature. The membranes were then incubated overnight at 4°C with primary antibody anti-PU.1 1:1,000 (sc-352, Santa Cruz), anti-Actin 1:10,000 (CP01, Calbiochem), anti-Histone H3 1:1,000 (Cell Signaling, #4620) and anti-Polyglutamine 1:500 (MAB1574, Millipore) respectively. The membranes were washed in TBST (3×5 min) before 1h incubation with an anti-rabbit or anti-mouse secondary peroxidase linked antibody accordingly at room temperature. The immunoblots were then visualized using the enhanced chemiluminescence method (Amersham).
ELISA for IL6
IL6 quantification in cell culture supernatant have been performed with Mouse Interleukin-6 (IL-6) ELISA kit from Thermo Scientific, according to manufacturer’s instructions.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously24. Briefly, 20×106 cells were crosslinked in Formaldehyde/PBS 1% for 10 min at RT. After quenching the reaction by adding 125mM glycine, cells were washed 2X with PBS and were centrifuged (8 min, 800g, 4°C). Cells were resuspended in swelling buffer (10mM HEPES/KOH pH 7.9, 85mM KCl, 1mM EDTA, 0.5% IGEPAL CA-630, 1X protease inhibitor cocktail (Roche), 1mM PMSF) for 5 min. Cells were spun down and resuspended in 500μl lysis buffer (50mM Tris-HCl pH 7.4, 1% SDS, 0.5% EmpigenBB, 10mM EDTA, 1X protease inhibitor cocktail (Roche), 1mM PMSF) and chromatin was sheared to an average DNA size of 300–400bp by administering 5 pulses of 10 sec duration at 10 W power output with 30 sec pause on ice using a Misonix 3000 sonicator. The lysate was cleared by centrifugation (5 min, 16000g, 4°C), and supernatant was diluted 2.5-fold with 750μl dilution buffer (20mM Tris-HCl pH 7.4, 100mM NaCl, 0.5% Triton X-100, 2mM EDTA, 1X protease inhibitor cocktail (Roche)). The diluted lysate was pre-cleared by rotating for 2h at 4°C with 120μl 50% rProtein A sepharose Fast Flow (GE Healthcare). The beads were discarded, and 1% of the supernatant were kept as ChIP input. The protein of interest was immunoprecipitated by rotating the supernatant with 2.5 μg antibody overnight at 4°C, then adding 50μl blocked rProtein A sepharose and rotating the sample for an additional 1h at 4°C. The beads were pelleted (2 min, 1000g, 4°C), the supernatant discarded, and the beads were transferred in 400μl wash buffer I (WBI) (20mM Tris-HCl pH 7.4, 150mM NaCl, 0.1% SDS, 1% Triton X-100, 2mM EDTA) into 0.45 μm filter cartridges (Ultrafree MC, Millipore), spun dry (1 minute, 2200g, 4°C), washed one more time with WBI, and twice each with WBII (20mM Tris-HCl pH 7.4, 500mM NaCl, 1% Triton X-100, 2mM EDTA), WBIII (10mM Tris-HCl pH 7.4, 250mM LiCl, 1% IGEPAL CA-630, 1% Na-deoxycholate, 1mM EDTA), and TE. Immunoprecipitated chromatin was eluted twice with 100 μl elution buffer each (100mM NaHCO3, 1% SDS) into fresh tubes for 20 min. Eluates were pooled, the Na+ concentration was adjusted to 300mM with 5M NaCl and crosslinks were reversed overnight at 65°C in a hybridization oven. The samples were sequentially incubated at 37°C for 2h each with 0.33mg/ml RNase A and 0.5mg/ml proteinase K. The DNA was isolated using the QiaQuick PCR purification kit (Qiagen) according to the manufacturer’s instructions. Antibodies used: anti-PU.1 (sc-352), anti-C/EBPα (sc-61), anti-C/EBPβ (sc-150), control rabbit IgG (sc-2027) (Santa Cruz Biotech), anti-H3K4me2 (cat#07-030, Millipore).
RNA sequencing
RNA was purified using RNeasy Mini Kit (Qiagen) and enriched for Poly(A)-RNA with MicroPoly(A) Purist Kit (Ambion). Subsequently, RNA was treated with TURBO DNase (Ambion), fragmented with RNA Fragmentation Reagents (Ambion) and purified by a P-30 column (Biorad). Fragmented RNA was dephosphorylated with Antarctic phosphatase (New England Biolabs) heat inactivated and precipitated over-night. Poly(A)-tailing and cDNA synthesis was performed as previously described55. For reverse transcription, oligos with custom barcodes (underlined) were used: 5′-Phos-CA/TG/AC/GT GATCGTCGGACTGTAGAACTCT/idSp/CAAGCAGAAGACGGCATACGATTTT TTTTTTTTTTTTTTTTVN-3′. Subsequently, exonuclease was used to remove the excess oligo. After heat-inactivation, RNA was hydrolyzed by alkaline treatment (100mM NaOH) and heat at 95°C for 25 min. The cDNA fragments of 50–150 nucleotides were purified on a denaturing Novex 10% polyacrylamide TBE-urea gel (Invitrogen). The recovered cDNA was circularized, linearized, amplified for 12 cycles, and gel purified as previously described55. The library was sequenced on the Illumina HiSeq 2000 according to the manufacturer’s instructions. Reads were aligned to the mouse mm9 genome (NCBI Build 37) using Tophat. RNA-Seq experiments were normalized and visualized by using HOMER (http://biowhat.ucsd.edu/homer/) to generate custom tracks for the UCSC Genome Browser (http://genome.ucsc.edu/). Gene expression values were generating for RefSeq annotated transcripts using HOMER and differential expression calculations were performed using edgeR27. Gene Ontology analysis was performed using DAVID (http://david.abcc.ncifcrf.gov/). Gene expression clustering was performed using Cluster 3.0 and visualized using Java TreeView. Promoters of regulated genes were analyzed for enriched motifs using HOMER24
ChIP-sequencing and data analysis
DNA from chromatin immunoprecipitation (10–50ng) was adapter-ligated and PCR amplified according to the manufacturer’s protocol (Illumina). ChIP fragments were sequenced for 36 or 50 cycles on an Illumina HiSeq 2000 according to the manufacturer’s instructions. Reads were aligned to the mouse mm9 genome assembly (NCBI Build 37) using Bowtie allowing up to 2 mismatches. Only tags that mapped uniquely to the genome were considered for further analysis. ChIP-Seq experiments were normalized and visualized by using HOMER (http://biowhat.ucsd.edu/homer/) to generate custom tracks for the UCSC Genome Browser (http://genome.ucsc.edu/). Peak finding, motif finding, and peak annotation were performed using HOMER. Peaks were assigned to gene targets based on the closest RefSeq defined TSS. Randomizations were performed during the HOMER motif finding algorithm. These involved selecting random fragments of genomic DNA to be used as control regions for motif discovery. Randomly selected fragments of DNA were selected to normalize GC% content.
Statistical Analyses
Standard deviation and Student’s t-test and one way-Anova were performed with the Prism 4 program. p < 0.05 was considered significant. Unpaired t-test was used for comparisons between control and transgenic mice within one group and between control and HD samples. Data are presented as mean ±sd. For each experiment, a minimum sample size of 3 biological replicates was analyzed in each experimental condition. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those generally employed in the field. The estimate of the variance within each group was calculated and found to be similar between groups that are being statistically compared.
Supplementary Material
Acknowledgments
We thank E. Mejia for FluoroJadeB staining, Dr. H. Kordasiewicz for providing R6/2 mice, M. McAlonis, J. Artates and J. Boubaker for stereotaxic injection, UCSD Histology Core for PU.1 staining, UCSD Human Embryonic Stem Cell Core Facility at Sanford Consortium for Regenerative Medicine for assistance with cell sorting, Dr. J. Corey-Bloom for providing blood samples from HD patients, Dr. M. Hayden for providing HTT and mHTT N548 aa original cDNA, and the Harvard Brain Tissue Resource Center (HBTRC, Belmont USA) which is supported in part by PHS R24 MH068855, New York Brain Bank at Columbia University (NYBB, New York, USA) and Massachusetts General Hospital (MGH, Boston, USA) for HD post mortem samples. These studies were supported by NIH grants DK091183, DK063491, GM 069338 and CA17390 to CKG.
Footnotes
Accession numbers: All ChIP-Seq and RNA-Seq, data sets will be deposited in the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/).
COMPETING FINANCIAL INTERESTS:
The authors declare no competing financial interests.
AUTHORS’ CONTRIBUTIONS:
C.K.G and A.C. developed the study, conceived the experimental plans and analyzed the data. C.B. analyzed the genome-wide data. A.C. performed most of the biological, biochemical and molecular experiments. B.K. conceived and performed the TUNNEL assay experiment. G.D performed microglia purification from adult mice brains. C.L.T. provided Hdh175/175 SOD1G37R tissues, mice and performed the in vivo experiment. E.C. and C.Z. participated in the elaboration of the project, provided original constructs and mRNA from post-mortem human samples. D.W.C. and F.G. participated in experimental design and provided essential resources and reagents. C.K.G and A.C. interpreted the data and wrote the manuscript. All of the authors read and edited the manuscript. C.K.G supervised the entire work, directed the strategies, provided financial support and gave final approval of the version to be published.
Bibliography
- 1.Streit WJ, Walter SA, Pennell NA. Reactive microgliosis. Prog Neurobiol. 1999;57:563–581. doi: 10.1016/s0301-0082(98)00069-0. [DOI] [PubMed] [Google Scholar]
- 2.Samii A, Nutt JG, Ransom BR. Parkinson’s disease. Lancet. 2004;363:1783–1793. doi: 10.1016/S0140-6736(04)16305-8. [DOI] [PubMed] [Google Scholar]
- 3.Schellenberg GD, Montine TJ. The genetics and neuropathology of Alzheimer’s disease. Acta Neuropathol. 2012;124:305–323. doi: 10.1007/s00401-012-0996-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.HDRCG A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 5.Reiner A, et al. Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci U S A. 1988;85:5733–5737. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrante RJ, et al. Heterogeneous topographic and cellular distribution of huntingtin expression in the normal human neostriatum. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1997;17:3052–3063. doi: 10.1523/JNEUROSCI.17-09-03052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trottier Y, et al. Cellular localization of the Huntington’s disease protein and discrimination of the normal and mutated form. Nature genetics. 1995;10:104–110. doi: 10.1038/ng0595-104. [DOI] [PubMed] [Google Scholar]
- 8.Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 1990;39:151–170. doi: 10.1016/0306-4522(90)90229-w. [DOI] [PubMed] [Google Scholar]
- 9.Kierdorf K, et al. Microglia emerge from erythromyeloid precursors via Pu.1-and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 10.Tai YF, et al. Microglial activation in presymptomatic Huntington’s disease gene carriers. Brain. 2007;130:1759–1766. doi: 10.1093/brain/awm044. [DOI] [PubMed] [Google Scholar]
- 11.Sapp E, et al. Early and progressive accumulation of reactive microglia in the Huntington disease brain. J Neuropathol Exp Neurol. 2001;60:161–172. doi: 10.1093/jnen/60.2.161. [DOI] [PubMed] [Google Scholar]
- 12.Pavese N, et al. Microglial activation correlates with severity in Huntington disease: a clinical and PET study. Neurology. 2006;66:1638–1643. doi: 10.1212/01.wnl.0000222734.56412.17. [DOI] [PubMed] [Google Scholar]
- 13.Politis M, et al. Microglial activation in regions related to cognitive function predicts disease onset in Huntington’s disease: a multimodal imaging study. Hum Brain Mapp. 2011;32:258–270. doi: 10.1002/hbm.21008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjorkqvist M, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J Exp Med. 2008;205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crocker SF, Costain WJ, Robertson HA. DNA microarray analysis of striatal gene expression in symptomatic transgenic Huntington’s mice (R6/2) reveals neuroinflammation and insulin associations. Brain Res. 2006;1088:176–186. doi: 10.1016/j.brainres.2006.02.102. [DOI] [PubMed] [Google Scholar]
- 16.Franciosi S, et al. Age-dependent neurovascular abnormalities and altered microglial morphology in the YAC128 mouse model of Huntington disease. Neurobiol Dis. 2011;45:438–449. doi: 10.1016/j.nbd.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Silvestroni A, Faull RL, Strand AD, Moller T. Distinct neuroinflammatory profile in post-mortem human Huntington’s disease. Neuroreport. 2009;20:1098–1103. doi: 10.1097/WNR.0b013e32832e34ee. [DOI] [PubMed] [Google Scholar]
- 18.van Gool WA, van de Beek D, Eikelenboom P. Systemic infection and delirium: when cytokines and acetylcholine collide. Lancet. 2010;375:773–775. doi: 10.1016/S0140-6736(09)61158-2. [DOI] [PubMed] [Google Scholar]
- 19.Giorgini F, Guidetti P, Nguyen Q, Bennett SC, Muchowski PJ. A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nature genetics. 2005;37:526–531. doi: 10.1038/ng1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khoshnan A, et al. Activation of the IkappaB kinase complex and nuclear factor-kappaB contributes to mutant huntingtin neurotoxicity. J Neurosci. 2004;24:7999–8008. doi: 10.1523/JNEUROSCI.2675-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palazuelos J, et al. Microglial CB2 cannabinoid receptors are neuroprotective in Huntington’s disease excitotoxicity. Brain: a journal of neurology. 2009;132:3152–3164. doi: 10.1093/brain/awp239. [DOI] [PubMed] [Google Scholar]
- 22.Diaz-Hernandez M, et al. Altered P2X7-receptor level and function in mouse models of Huntington’s disease and therapeutic efficacy of antagonist administration. Faseb J. 2009;23:1893–1906. doi: 10.1096/fj.08-122275. [DOI] [PubMed] [Google Scholar]
- 23.Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010;140:771–776. doi: 10.1016/j.cell.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonifer C, Hoogenkamp M, Krysinska H, Tagoh H. How transcription factors program chromatin--lessons from studies of the regulation of myeloid-specific genes. Semin Immunol. 2008;20:257–263. doi: 10.1016/j.smim.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Walsh JC, et al. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 27.Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22:490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brykczynska U, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 29.Regha K, et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell. 2007;27:353–366. doi: 10.1016/j.molcel.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghisletti S, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Tsukada J, Yoshida Y, Kominato Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54:6–19. doi: 10.1016/j.cyto.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 32.Huber R, Pietsch D, Panterodt T, Brand K. Regulation of C/EBPbeta and resulting functions in cells of the monocytic lineage. Cell Signal. 2012;24:1287–1296. doi: 10.1016/j.cellsig.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 34.Heikkinen T, et al. Characterization of neurophysiological and behavioral changes, MRI brain volumetry and 1H MRS in zQ175 knock-in mouse model of Huntington’s disease. PLoS One. 2012;7:e50717. doi: 10.1371/journal.pone.0050717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 37.Wong PC, et al. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 38.Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 39.Boillee S, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 40.Lehnardt S, et al. Activation of innate immunity in the CNS triggers neurodegeneration through a Toll-like receptor 4-dependent pathway. Proc Natl Acad Sci U S A. 2003;100:8514–8519. doi: 10.1073/pnas.1432609100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saijo K, et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menalled LB, et al. Comprehensive Behavioral and Molecular Characterization of a New Knock-In Mouse Model of Huntington’s Disease: zQ 175. PLoS One. 2013;7:e49838. doi: 10.1371/journal.pone.0049838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu X, et al. Pathological cell-cell interactions elicited by a neuropathogenic form of mutant Huntingtin contribute to cortical pathogenesis in HD mice. Neuron. 2005;46:433–444. doi: 10.1016/j.neuron.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 44.Kraft AD, Kaltenbach LS, Lo DC, Harry GJ. Activated microglia proliferate at neurites of mutant huntingtin-expressing neurons. Neurobiol Aging. 2011 doi: 10.1016/j.neurobiolaging.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bjorkqvist M, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J Exp Med. 2008;205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Battista D, Ferrari CC, Gage FH, Pitossi FJ. Neurogenic niche modulation by activated microglia: transforming growth factor beta increases neurogenesis in the adult dentate gyrus. Eur J Neurosci. 2006;23:83–93. doi: 10.1111/j.1460-9568.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- 47.Monje ML, Toda H, Palmer TD. Inflammatory blockade restores adult hippocampal neurogenesis. Science. 2003;302:1760–1765. doi: 10.1126/science.1088417. [DOI] [PubMed] [Google Scholar]
- 48.Ransome MI, Hannan AJ. Impaired basal and running-induced hippocampal neurogenesis coincides with reduced Akt signaling in adult R6/1 HD mice. Mol Cell Neurosci. 2013;54:93–107. doi: 10.1016/j.mcn.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 49.Simpson JM, et al. Altered adult hippocampal neurogenesis in the YAC128 transgenic mouse model of Huntington disease. Neurobiology of disease. 2011;41:249–260. doi: 10.1016/j.nbd.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 50.Kohl Z, et al. Impaired adult olfactory bulb neurogenesis in the R6/2 mouse model of Huntington’s disease. BMC Neurosci. 2010;11:114. doi: 10.1186/1471-2202-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaspard N, et al. Generation of cortical neurons from mouse embryonic stem cells. Nat Protoc. 2009;4:1454–1463. doi: 10.1038/nprot.2009.157. [DOI] [PubMed] [Google Scholar]
- 52.Marchetto MC, et al. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marullo M, et al. Expressed Alu repeats as a novel, reliable tool for normalization of real-time quantitative RT-PCR data. Genome Biol. 2010;11:R9. doi: 10.1186/gb-2010-11-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.