Abstract
Background
Bone morphogenic protein receptor 2 (BMPR2) gene mutations are the most common cause of heritable PAH (HPAH). However only 20% of mutation carriers get clinical disease. Here we explored the hypothesis that this reduced penetrance is in part due to an alteration in BMPR2 alternative splicing.
Methods and Results
Our data showed that BMPR2 has multiple alternatively spliced variants. Two of these, isoform-A (full-length) and isoform-B (missing exon 12), were expressed in all tissues analyzed. Analysis of cultured lymphocytes (CLs) of 47 BMPR2 mutation-positive HPAH-patients and 35 BMPR2 mutation-positive unaffected-carriers showed that patients had higher levels of isoform-B compared to isoform-A (B/A ratio) than carriers (P=0.002). Furthermore compared to cells with low B/A ratio, cells with high B/A ratio had lower levels of unphosphorylated cofilin following BMP stimulation. Analysis of exon 12 sequences identified an exonic splice enhancer, which binds serine arginine splicing factor 2 (SRSF2). Because SRSF2 promotes exon inclusion, reduced SRSF2 expression would mean that exon 12 would not be included in final BMPR2 mRNA (thus promoting increased isoform-B formation). Western blot analysis showed that SRSF2 expression was lower in cells from patients compared to carriers; and, siRNA-mediated knockdown of SRSF2 in pulmonary microvascular endothelial cells resulted in elevated levels of isoform-B compared to isoform-A, i.e. elevated B/A ratio.
Conclusions
Alterations in BMPR2 isoform ratios may provide an explanation of the reduced penetrance among BMPR2 mutation carriers. This ratio is controlled by an exonic splice enhancer in exon 12 and its associated splicing factor SRSF2.
Keywords: pulmonary hypertension, Alternate Splicing, BMPR2, Penetrance, SRSF2
Introduction
Mutations in the BMPR2 gene have been shown to cause heritable pulmonary arterial hypertension (HPAH), an autosomal dominant disorder with a variable age of onset.1 While ~82% of HPAH families have an identifiable mutation in the BMPR2 gene, only 20% of carriers ever develop disease.2 BMPR2 mutations can either produce stable transcripts or result in premature termination codons (PTC) resulting in the mutated transcript being rapidly degraded through the nonsense mediated decay (NMD) pathway.3, 4 NMD is an mRNA surveillance system that degrades transcripts containing PTCs to prevent translation of unnecessary or harmful transcripts. 5 Thus, individuals with PAH and NMD+ BMPR2 mutations have disease due to haploinsufficiency, whereas patients whose mutations are NMD- may have disease due to a dominant negative mechanism. Reduced penetrance is seen in both groups of patients.
The molecular mechanisms, which regulate the reduced penetrance, remain poorly defined. Several studies have explored the hypothesis that reduced penetrance is due to the effect of modifying genes; however no modifier genes have been identified which could explain reduced penetrance in majority of patients. 6-12 We recently reported that the expression of the non-mutated BMPR2 allele plays a role in HPAH penetrance.13 Patients had lower levels of expression of the normal BMPR2 allele than carriers. These data suggested for the first time that one of the important modifiers of BMPR2 related HPAH might in fact be the BMPR2 gene itself. We have delved further into which expression mechanisms of BMPR2 play a role in the reduced penetrance observed in HPAH, focusing particularly on alternative splicing
Alternative splicing is a mechanism by which a single gene can generate multiple transcripts with likely different functions through internal deletion/skipping of exons in various combinations. Alternatively splicing is a complex process that involves exonic and intronic acceptor and donor sites and intronic and exonic splice enhancers and silencers.14 A large number of proteins such as the serine-arginine rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNP) bind to these enhancers and silencers and help in splice site recognition. 15, 16 The SR proteins bind to splicing enhancers and promote exon inclusion whereas binding of hnRNPs acts as a splicing repressor promoting exon exclusion or skipping. At least 20 SR proteins have been identified from which a smaller group of 7 are termed “core” SR proteins: SRSF2 (SF2/ASF), SC35, SRp20, SRp75, SRp40, SRp55, and 9G8. 15 SR proteins bind to specific RNA sequences and assist in spliceosome recruitment through protein-protein interactions. 17, 18 This can have clinical consequences, as alternative splicing has been shown to play a role in many human diseases.19, 20
In humans as well as mice, BMPR2 has 13 exons and is alternatively spliced to produce two primary transcripts. Isoform-A is the full length BMPR2 gene product and contains all 13 exons. Isoform-B is a much rare transcript lacking exon 12.21-23 Whether this minor isoform is expressed in HPAH patients or has any role in the cellular BMPR-II function is not known. Studies have clearly shown that exon 12 is important for proper functioning of BMPR-II. Deletion of exon 12 is a common BMPR2 mutation found in HPAH patients and previous studies have shown that it can disrupt BMPR-II function in a dominant negative fashion.24-26 Furthermore studies in mice have shown that overexpression of the exon 12-truncated Bmpr2 transcript results in PH due to complex vascular lesions consisting of the narrowing at branch points of resistance-level vessels and the dropout/occlusion of very small vessels. This experimental model of PH is currently being studied.27, 28
In this manuscript we hypothesized that alternative splicing of BMPR2 plays a role in HPAH penetrance. To address this hypothesis, we BMPR2 isoform-A and isoform-B splicing in cells derived from patients and carriers. We found that patients expressed higher levels of isoform-B compared to isoform-A. Using expression analysis, sequencing, and target gene knockout we then identified a splicing enhancer and its binding splicing factor SRSF2 which appears to regulate the relative ratios of the BMPR2 isoforms in cells.
Methods
PAH patient samples
Cultured lymphocytes (CLs) from 47 BMPR2 mutation-positive HPAH patients and 35 BMPR2 mutation-positive unaffected carriers were used in this study (all the cell lines available in our cell repository at the time of study).4, 29, 30 Of the 47 patients, 27 had NMD+ and 20 had NMD-mutations while in the carrier group 20 had NMD+ mutations and 15 had NMD- mutations (Table 1). Samples were collected with informed consent under a Vanderbilt University Institutional Review Board-approved protocol. Cells were established and grown and their BMPR2 mutation and NMD status determined as previously described.3, 4, 31, 32 The full range of these mutations has been previously described in detail.3, 26
Table 1.
Clinical characteristics of the BMPR2 mutation carriers
| AMCs (n=47) | UMCs (n=35) | P value | |
|---|---|---|---|
| Females | 35 (74.5) | 15 (48.4) | 0.029 |
| Age, yr | |||
| At diagnosis | 36.4 ± 14.1 | n/a | n/a |
| At present or at death | n/a | 60.5 ± 17.3 | n/a |
| NMD+ mutations | 27 | 20 | |
| NMD− mutations | 20 | 15 | |
| Baseline hemodynamic data at diagnosis, mean | |||
| Right atrial pressure, mmHg | 10.3 ± 6.0 | n/a | |
| Mean PAP, mmHg | 61.3 ± 12.0 | n/a | |
| Cardiac index, L•min•m−2 | 1.90 ± 0.73 | n/a | |
| Indexed PVR, U•m2 | 19.3 ± 12.7 | n/a |
Abbreviations: AMC, affected mutation carrier with HPAH; UMC, unaffected mutation carrier; NMD, nonsense mediated decay; PAP, pulmonary artery pressure; CI, cardiac index; PVR, pulmonary vascular resistance; n/a, not available
Data are presented as n (%) or mean ±SD, unless otherwise stated. A chi square test was used for P value calculation.
Determination of BMPR2 isoform mRNA levels
cDNA was synthesized from 1 μg of total cellular RNA isolated from the patient derived CL cells or pooled human tissues (Human Universal Reference Total RNA, catalogue number 636538, Clontech, Mountain View, CA 94043), including pulmonary microvascular endothelial cells (PMVEC) using the Superscript III cDNA Synthesis Kit (Life Technologies, Grand Island, NY). PMVECs were included in this analysis as they are an important cell in PAH pathogenesis. Taqman assays (4331182) were used for both isoform-A and isoform-B BMPR2 transcript detection by real-time PCR using Taqman Universal Master Mix and a 7500 Real-Time PCR system (Life Technologies, Grand Island, NY). HPRT was chosen as the housekeeping gene because it showed the least variability between CL samples.33 Relative expression levels were calculated using the comparative Ct method.34, 35
Western blot (WB) analysis
Membranes were probed with primary antibody AF811 (R&D Systems Inc., Minneapolis, MN) for 1 hour and with secondary antibody 111-035-003 (Jackson ImmunoResearch, West Grove, PA) for an additional hour. Detection was done using the Immobilon Chemiluminescent HRP substrate (Millipore, Billerica, Massachusetts). β-Actin was used as a loading control. For BMPR-II signaling analysis we used phospho-cofilin antibody #3313 (Cell Signaling Technology) and cofilin antibody #C8736 (Sigma).
Identification of exonic splice enhancer
BMPR2 sequences were scanned for exonic splice enhancers using ESE finder 3.0 as previously described (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi).36, 37
SRSF2 knockdown in PMVECs
We used Silencer Select Pre-Designed siRNA (Life Technologies, Grand Island, NY) to knock down SRSF2 (Cat. # s12730). PMVECs (Lonza, Basel, Switzerland) were transfected with 5 nM siRNA (final concentration) using Lipofectamine® RNAiMAX transfection reagent (Life Technologies, Grand Island, NY) as instructed by the manufacturer. The transfected cells were incubated for 24 hours followed by RNA and protein extraction for real-time PCR and western blot analyses.
Statistics
Categorical variables were summarized by frequencies, with dichotomous variables assessed for normality of distribution and subsequently compared using the chi square test. We compared continuous variables using Student's t-tests for normally distributed variables and Mann-Whitney U-tests for non-normally distributed variables. To adjust for potential bias related to gender and age, multivariate linear regression was used to assess the relationship of BMPR2 isoform ratios to gender and age. An α-level of 0.05 was chosen, and P-values < 0.05 were considered statistically significant. All P-values were two-tailed. Analyses were performed with Prism 5 for Mac OS X software package (GraphPad Software Inc., La Jolla, CA) and SPSS (Version 19, IBM SPSS Statistics, Armonk, NY)
Results
BMPR2 is alternatively spliced
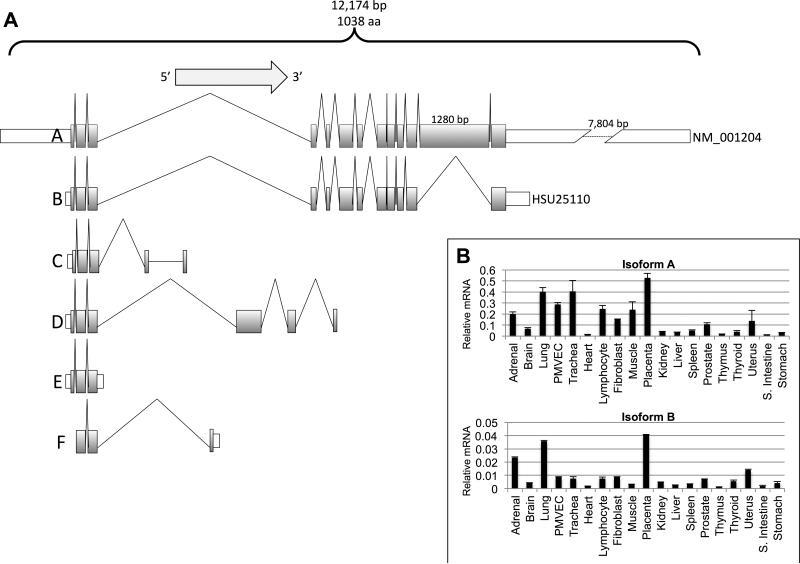
In order to determine the full extent of human BMPR2 (chromosome 2, at 2q33-q34, from 203240945 to 203432480; NCBI 37, August 2010) alternative splicing, we first analyzed 537 GenBank accessions using alignment tools in the NCBI databases. Only expressed sequence tags matching the genomic sequence with greater than 99% accuracy over their entire length were further evaluated. These analyses showed that the BMPR2 gene contains 19 distinct introns and 6 alternatively spliced potential variants (Figure 1A). Expression and sequence analysis of mRNA isolated from a variety of normal and HPAH patient tissues showed most tissues expressed isoform-A (full-length) and isoform-B (where exon 12 is spliced out), albeit isoform-A expression was significantly higher than isoform-B (Figure 1B). Interestingly lung tissues had one of the highest levels of expression of isoform A and B (Figure 1A and 1B).
Figure 1.
A) BMPR2 splice isoforms. Shaded areas indicate exons and open boxes untranslated regions. Exon 12 is the largest exon of BMPR2 and its length is indicated by the number on top. Isoform-A is the full length BMPR2 mRNA while isoform-B is lacks exon 12 and is the minor BMPR2 isoform. B) Isoform A and B mRNA levels in pooled mRNA isolated from various body tissues including PMVECs as determined by relative real-time PCR. PMVEC were included in this analysis as they are an important cell in PAH pathogenesis.
Affected BMPR2 mutation carriers have higher amounts of isoform-B mRNA relative to isoform-A
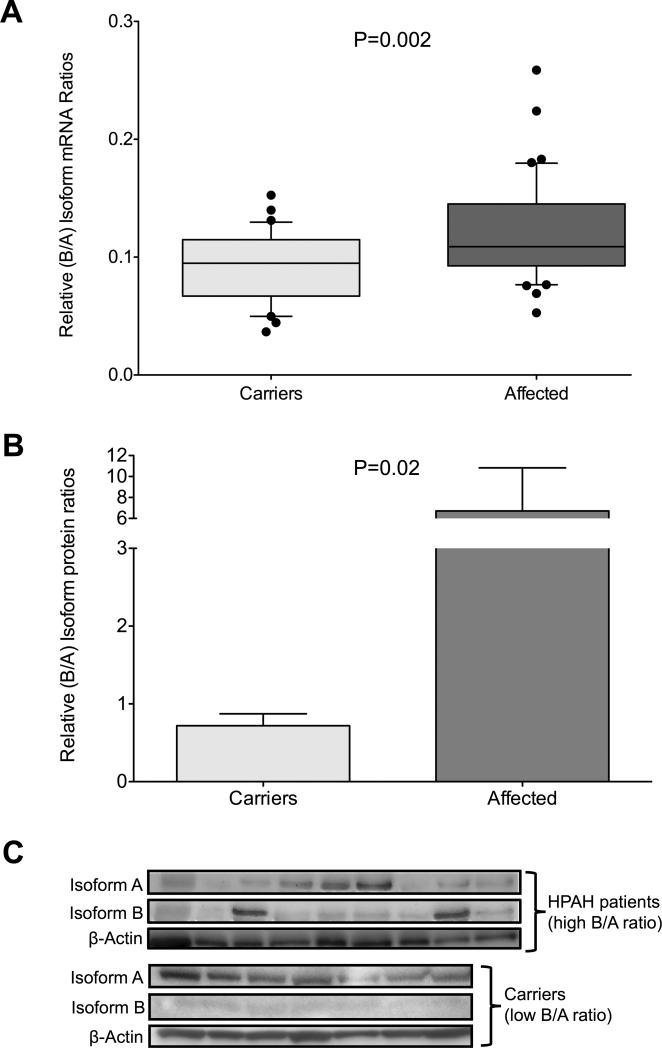
In light of the relative importance of exon 12 in BMPR-II function we hypothesized that isoform-B (lacking exon12) cellular expression levels are important for HPAH pathogenesis. To test our hypothesis we measured the relative abundance of isoform-A and -B mRNA in CLs derived from HPAH patients (47 cell lines) or carriers (35 cell lines) by isoform-specific real-time PCR and, in a smaller subset, by western blot analysis. This number comprised of all the cell lines available in our repository at that time. Characteristics of the affected mutation carrier and unaffected mutation carrier samples analyzed are shown in Table 1. Forty-seven CL cell lines carried NMD+ BMPR2 mutations while 35 carried NMD− BMPR2 mutations. The affected BMPR2 mutation carrier group contained more females and were younger at the time of sample acquisition compared to the unaffected BMPR2 mutation carrier group, as expected. RNA and protein expression data were analyzed in an aggregate fashion and showed that patients had significantly higher expression of isoform-B relative to isoform-A (B/A ratio) than unaffected mutation carriers (P=0.002) (Figure 2). This association held even when samples were analyzed based on the type of mutation, NMD+ (P=0.02) versus NMD− (P=0.03) (Supplementary Figure 1).
Figure 2. BMPR2 isoform ratios in CL derived from carriers (light gray, n=35) and affected individuals (dark gray, n=47).
A) Relative isoform-B versus isoform-A (B/A) mRNA ratio as determined by isoform specific real-time PCR. Real time PCR analysis was done three times with each sample run in triplicate. Results from one representative experiment are shown as box-and-whiskers plots with whiskers indicating Tukey whiskers and extreme data points indicated by filled circles. B and C) Western blot validation of the B/A ratios. Total cellular protein extracts from CL cell lines from 9 affected individuals with the highest B/A ratio and 7 carriers with the lowest B/A ratio were analyzed. Isoform mRNA expression data were analyzed in a nonparametric (Mann Whitney U-test) manner while WB densiometry data were analyzed using the Student's t-test. An α-level of 0.05 was chosen, and P-values < 0.05 were considered statistically significant. All P-values were two-tailed.
High relative B/A ratio adversely affects signaling from BMPR-II receptor
Since patients’ cells had a higher relative B/A ratio than carriers’, we next determined whether the higher B/A ratio had any effect on BMPR-II function on a cellular level. BMPR2 exon 12 encodes an intracytoplasmic domain, which is important for BMPR-II's interactions and regulation of actin through LIM kinase 1 (LIMK1). A normally functioning cytoplasmic domain inhibits the ability of LIMK1 to phosphorylate the actin binding/modulating protein cofilin, resulting in decreased phosphorylated-cofilin (p-cofilin) and increased unphosphorylated cofilin in cells.38 On the other hand a malfunctioning or absent cytoplasmic domain diminishes the inhibitory effects of BMPR-II on LIMK, resulting in increased p-cofilin and decreased cofilin.38, 39
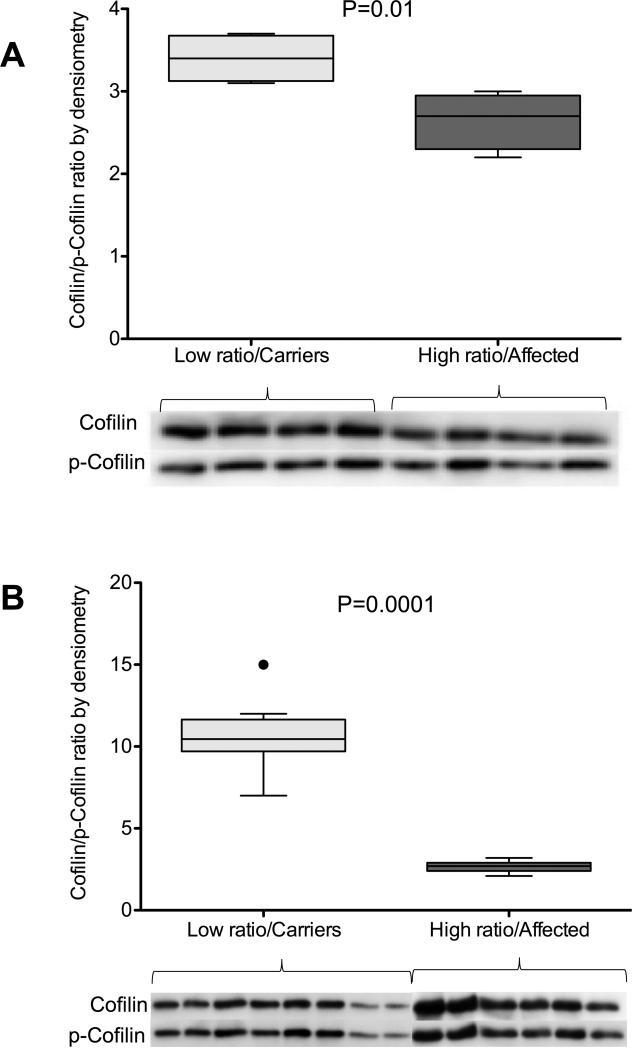
We proceeded to determine if there were differences in cofilin phosphorylation between CL cell lines with high B/A ratios and cell lines with low B/A ratios. We stimulated exponentially growing CLs with high and low B/A ratios with 10 ng BMP for 1 hour and 20 ng BMP for 2 hours. Our data show that cell lines with high relative B/A ratios had significantly lower cofilin/p-cofilin ratio than cells lines with low B/A isoform ratios (2.6 ± 0.17 vs. 3.4 ± 0.15, P=0.01, N=4) as measured by WB densiometry, in other words cells with high B/A ratios had lower cofilin and higher p-cofilin while cells with low B/A ratio had higher cofilin and lower p-cofilin, suggesting that there was defective/reduced functioning of BMPR-II receptor in these cell lines (Figure 3A).
Figure 3. Cofilin signaling in response to stimulation by BMP.
A) Cofilin to p-cofilin ratio measured by western blot analysis and presented as densiometry units in CL cell lines with low B/A ratios (light gray, N=4) and high B/A ratios (dark gray, N=4) after 1 hour of exposure to 10 ng/ml BMP. B) Cofilin to p-cofilin ratio measured by WB analysis and presented as densiometry units in CL cell lines with low B/A ratios (light gray, N=8) and high B/A ratios (dark gray, N=6) after 2 hours of exposure to 20 ng/ml BMP. Data are shown as box-and-whiskers plots with whiskers indicating Tukey whiskers and extreme data points indicated by filled circles. An α-level of 0.05 was chosen, and P-values < 0.05 were considered statistically significant. All P-values were two-tailed.
We then determined if the cofilin signaling in cell lines with high and low B/A ratio cell lines was different in response to higher or sustained stimulation of the BMPR-II receptor. We thus exposed high and low B/A ratio cell lines to higher doses of BMP (20ng) for 2 hours and then compared the levels of p-cofilin and cofilin. These data showed that while longer exposure, of the low B/A ratio cell lines, to higher dose of BMP significantly increased the cofilin/p-cofilin ratio as measured by WB densiometry, similar treatment of the high B/A ratio cell lines did not (10.6 ± 0.8, N=8 vs. 2.7 ± 0.17, N=6, P=0.0001) (Figure 3B). This suggested that cell lines with higher relative B/A ratios have significantly decreased capacity to respond to higher/longer BMP signaling compared to lower B/A ratio cell lines (Figure 3B). Thus, data presented in Figures 3A and 3B conclusively show that the relative increase in BMPR2 isoform-B levels affects the cofilin signaling function of the BMPR-II receptor.
BMPR2 exon 12 contains a splice enhancer that binds splicing factor SRSF2
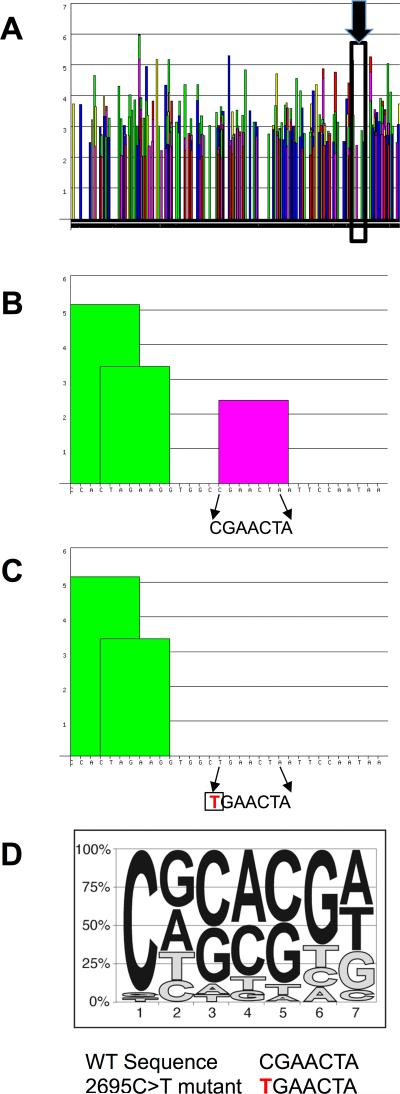
Exon inclusion and exclusion is determined by exonic and intronic splicing enhancers and silencers and the various splicing factors that bind to these. We analyzed the 1,280 bp sequence of BMPR2 exon 12 using matrices for prediction of sequences required for binding of splicing factors (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder).36, 37 This analysis revealed a large number of possible enhancer sequences that met the matrix threshold (Figure 4 A). In order to identify which, if any, of these putative enhancer sequences are involved in exon 12 inclusion versus exclusion, we queried our large HPAH BMPR2 mutation database for naturally occurring mutations in exon 12 that resulted in exclusion (deletion) of exon 12 from the BMPR2 transcript. Our query identified a 2695C>T change in exon 12 of the full-length BMPR2 transcript (NM_001204) that when present resulted in skipping/loss of exon 12.26 Further analysis of the sequence around this change using the splice enhancer prediction matrix revealed that this sequence is predicted to bind splicing factor SRSF2 (Figure 4 B) and that the C>T change alters the most important nucleotide of the consensus enhancer sequence, resulting in complete disruption of the SRSF2 binding site (Figure 4 C & D). These data suggest that exon 12 contains at least one splicing enhancer that binds splicing factor SRSF2 and can regulate BMPR2 exon 12 inclusion/exclusion.
Figure 4. Identification of a splice enhancer in exon 12 of BMPR2.
A) Bioinformatics analysis of the exon 12 sequences identified many potential splice enhancers. Columns represent significance of a particular sequence. Scores above 2 are considered significant. The splice enhancer binding SRSF2 is in the inset area marked by the black box and identified by the black arrow B) Expanded view of the inset area in A. The arrows show the 7 base pairs of an SRSF2 binding site (significant score shown by the pink column) in exon 12. C) When the first base pair of this sequence is mutated from a C to a T (shown in red), bioinformatics analysis predicts elimination of the SRSF2 binding site (graphically represented by lack of any column over the sequence) and skipping of exon 12 in the HPAH patient. D) The consensus sequence of the SRSF2 binding site is shown as a pictogram representation. The height of each letter reflects the frequency of each nucleotide at a given position after adjusting for background nucleotide composition. At each position the nucleotides are shown from top to bottom in order of decreasing frequency. The mutated base, a cytosine, is the most conserved and functionally the most important base of this enhancer.
SRSF2 regulates BMPR2 exon 12 inclusion
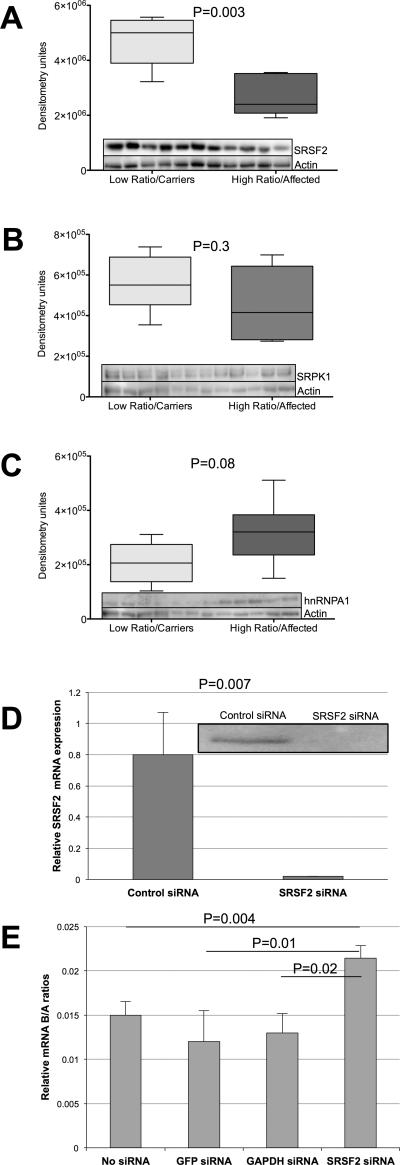
Since naturally occurring mutations in the SRSF2 binding sequence lead to exon skipping, we wondered whether SRSF2 plays a direct role in BMPR2 alternative splicing and thus cellular levels of isoform-A and -B. Generally, it is well known that SRSF2 promotes exon inclusion. Thus, we first determined whether there is any difference in cellular SRSF2 levels between the high (patients) and low B/A ratio cell lines (carriers). Six cell lines with a high B/A ratio and five with a low B/A ratio were analyzed by western blot analysis. We also analyzed levels of two other splicing factors, hnRNPA1 and SRPK, with binding sites in exon 12. This analysis showed that CL cell lines with a high B/A ratio had low levels of SRSF2 compared to cell lines with a low B/A ratio, as measured by WB densiometry, and that this difference was significant (2.72 × 106 ± 3.36 × 105, N=5 vs. 4.722 × 106 ± 3.6 × 105, N=6, P=0.003) (Figure 5A). There were no statistical differences in levels of SRPK (4.52 × 105 ± 7.25 × 104, N=5 vs. 5.59 × 105 ± 5.56 × 104, N=6 P=0.3) (Figure 5B) or hnRNPA1 (3.18 × 105 ± 4.81 × 104, N=5 vs. 2.06 × 105 ± 3.07 × 104, N=6, P=0.08), (Figure 5C) between cells with high and low B/A ratios, as measured by WB densiometry, though hnRNPA1 levels were suggestive of an association. These data indicate that SRSF2 might be a more important regulator of BMPR2 isoform ratios than SRPK and hnRNPA1 in the cell lines analyzed. They are also consistent with the splicing patterns, since (as discussed above) SRSF2 promotes exon inclusion, and so we would expect that cell lines with low levels of isoform-B would have higher amounts of SRSF2 and vice versa. In order to more directly identify a role for SRSF2 in BMPR2 alternative splicing, we used siRNA to knock down SRSF2 (Figure 5D) in PMVECs. We then tested the relative quantities of BMPR2 isoform-A and-B in these cells. While control siRNA did not alter the B/A ratio, SRSF2 knockdown resulted in a higher B/A ratio (Figure 5E). These data are consistent with the notion that SRSF2 regulates BMPR2 isoform levels.
Figure 5. Splicing factor levels in high and low B/A ratio cell lines.
A) SRSF2 levels as detected by western blot analysis and shown as densitometry units in low (light gray, n=5) and high (dark gray, n=6) B/A ratio cell lines. B) SRPK1 levels as detected by western blot analysis and shown as densitometry units in low (light gray, n=5) and high (dark gray, n=6) B/A ratio cell lines. C) hnRNP1 levels as detected by western blot analysis and shown as densitometry units in low (light gray, n=5) and high (dark gray, n=6) B/A ratio cell lines. D) Real time PCR (graph) and western blot (inset) confirmation of siRNA-mediated SRSF2 knockdown in PMVECs. E) Determination of B/A ratio by real-time PCR in PMVECs after siRNA-mediated knockdown of SRSF2. An α-level of 0.05 was chosen, and P-values < 0.05 were considered statistically significant. All P-values were two-tailed.
Discussion
One of the most perplexing features of BMPR2-associated HPAH is the observed reduced penetrance. Nearly 80% of mutation carriers never manifest disease, but they can produce offspring that do.13 Here, we present data that suggest that alternative splicing of BMPR2 is one factor that contributes to this reduced penetrance.
Our data show that cells from patients are more likely to have higher levels of isoform-B relative to levels of isoform-A (B/A ratio) compared to carriers. Thus if the wild-type BMPR2 allele in a BMPR2 mutation carrier is spliced in such a manner that there is more of isoform-B relative to isoform-A, then that mutation carrier is more likely to develop PAH.
We further show that cells with high B/A isoform ratios have higher p-cofilin and lower cofilin (a reduced capacity to unphosphorylate cofilin) under both short-term and sustained BMP stimulation. Thus increased levels of isoform-B relative to isoform-A reduce the cells’ ability to inhibit cofilin phosphorylation in response to BMP. This increase in B/A ratio is in part due to an exonic splice enhancer that binds a serine arginine-rich splicing factor, SRSF2. Carriers had higher cellular levels of SRSF2 than patients, and SRSF2 knockdown in PMVECs resulted in increased B/A isoform ratios, implicating SRSF2 directly in BMPR2 alternative splicing in a lung cell line thought to be important in HPAH pathogenesis. Thus the data presented here add HPAH to the list of pulmonary diseases in which alternative splicing plays a role.
Our data showing that cellular isoform-B levels may have a role in BMPR-II function and HPAH penetrance are not surprising given that isoform-B lacks exon 12 of the BMRP2 gene. At ~1,280 base pairs, exon 12 is the largest exon of the gene and forms part of the intracytoplasmic tail domain that is important for interactions of BMPR-II with β-actin. BMPR2 mutations that result in skipping of exon 12 cause HPAH through what is thought to be a dominant negative effect, since overexpression of exon12-truncated Bmpr2 in mice produces PH.27, 28 One of the more surprising observations presented here was that CL cell lines with increased B/A isoform ratios have significantly decreased capacity to respond to overstimulation of the BMPR-II receptor as compared to cell lines with low B/A ratios. Thus, in the setting where one BMPR2 allele is abnormal because of a mutation, additional stress on BMPR2-II signaling in the form of increased expression of a naturally occurring dominant negative isoform (isoform-B) likely further impairs receptor function and is thus more likely to contribute to disease.
We have previously shown that expression of the WT-BMPR2 allele also plays a role in disease penetrance in that patients are more likely to have lower levels of WT-BMPR2 expression than unaffected mutation carriers.13 This, together with the data presented here, suggests that the BMPR2 gene itself may be the most important modifier of HPAH penetrance. Because of limitations of our earlier study, we are unable at this time to determine the relative contributions of WT BMPR2 expression and BMPR2 alternative splicing to the penetrance of HPAH. It is likely, however, that there is some overlap, at a molecular level, between these mechanisms.
In this study we identified one splice enhancer and one splicing factor, SRSF2 that regulate BMPR2 alternative splicing. SPRK1 and hnRNPA1 levels were not different between the high and low B/A isoform ratio cell lines. This may suggest that these splicing factors are not involved in BMPR2 alternative splicing or that our western blot analyses were not adequately powered to detect a difference of expression of these splicing factors. As shown in Figure 5A exon 12 contains a large number of possible enhancer and silencer sequences thus increasing the likelihood that additional enhancers and their associated factors play a role in BMPR2 alternative splicing. Our hnRNPA1 expression data hint at this possibility. hnRNPA1 is an exon exclusion factor and its increased function and levels can lead to an exon being excluded. So, one would predict that hnRNPA1 would be higher in cells lines with high B/A ratios and low in low cell lines with low B/A ratios. Our data are suggestive of this (Figure 5C). Thus, the molecular mechanisms that lead to a high B/A ratio in patients might include both low SRSF2 and high hnRNPA1. It is likely that alterations of SRSF2 levels also affect alternative splicing of many other genes in addition to BMPR2, so that the eventual effect on penetrance may be due to a global affect on alternative splicing. Elucidation of the global alternative splicing pattern will be important and may provide further insights into the pathways and cellular processes that are involved. In addition to cellular levels, processes like phosphorylation, poly(ADP-ribosyl)ation, sumoylation and/or arginine methylation can also affect splicing factor function; however, whether any of these are at play here is not known and will require significant additional work.40
We at this time do not know why SRSF2 expression is higher in some mutation carriers. It is known that SRSF2 can auto regulate it's expression by activating the NMD pathway, however expression levels of SRSF2 have not been previously associated with any disease.41 Similarly the role of any polymorphisms in SRSF2 expression or function remains undefined. Interestingly mutations in SRSF2 were recently shown to be associated with worse outcomes in myelodysplastic syndromes.42 SRSF2 can also auto regulate it's function by post translational modifications.43 Thus the higher expression of SRSF2 in some mutation carriers may be secondary to one of the reasons discussed above or simply baseline expression differences due to cis or trans regulatory effects on the SRSF2 gene.
Obviously in a patient there might be other mechanisms at play. Several studies, including our own, have shown that environment including exposure to drugs and chemicals can alter splicing factor levels.44 Thus, dynamic changes to alternative splicing, in response to medication, drugs and environmental signals, could lead to added stress on BMPR-II signaling and push a mutation carrier towards disease. This may be particularly important when HPAH patients are under treatment where the pharmacological milieu may in fact act to dynamically alter alternative splicing, leading to worsening of disease. However, since our analyses were done on CL cells, the differences in SRSF2 expression reported here are more likely to be due to genetic/inherited factors. Nevertheless, our studies do suggest that further investigations are needed to determine the effects of environmental signals (e.g., hypoxia and oxygen) as well the common pharmacological therapies typically used in HPAH on BMPR2 as well as global alternative splicing. We also do not know if other splicing isoforms (identified through our EST analysis; Figure 1) might also play a role in HPAH pathogenesis. Certainly we did not detect any evidence of their expression in patient-derived CLs or normal PMVECs. However, we do not know whether the same is the case in patient-derived PMVECs as well. Those studies will require availability of additional resources such as IPS-derived PMVECs from HPAH patients, which are not currently available.
Use of CLs in this study is a relative weakness of this study. However the use of patient CLs is an unavoidable limitation due to the non-availability of patient PMVECs in sufficient numbers. Furthermore since splicing could be impacted by the overall disease milieu of a patient, primary cells from patients may not be an ideal model to study baseline genetic variation in splicing patterns even if they were available. Thus one relative advantage of CLs is that they are removed from the immediate environment and are thus consequently not affected by any potential drug or paracrine disease effects. Additionally CLs can be easily obtained not only from HPAH patients, but also from asymptomatic mutation carriers and from control subjects. A second concern about the use of CLs is whether the process of establishing these cell lines can in itself lead to the changes in gene expression. However we feel that this is unlikely to be the case for two reasons, A) even if the derivation influenced gene expression and /or splicing, we would expect that it would influence affected and unaffected CL's equally, B) we have shown previously that the derivation of CL cell lines did not affect BMPR2 gene expression.13, 45 Thus it is likely that the differences observed here reflect underlying genetic differences. Therefore even though CLs have several relative weaknesses when used as a discovery tool they can help formulate hypotheses that can then be tested in a cell line of interest. This study provides a good example of this concept.
In conclusion, we have shown that alternative splicing of BMPR2 has a role in HPAH penetrance. If a mutation carrier splices his BMPR2 gene in a manner that results in more isoform-B transcript relative to isoform-A, then that mutation carrier is more likely to develop PAH. In light of what is known about how drugs and environmental factors can affect splicing, our findings have implications for HPAH pathogenesis, treatment and follow-up.
Supplementary Material
Clinical Summary.
One of the most perplexing features of HPAH is its reduced penetrance. Nearly 80% of mutation carriers have no clinical symptoms, but they can produce offspring that are affected by HPAH. Thus, disease development cannot be predicted in a mutation carrier, creating anxiety in the patient and uncertainty about treatment in the physician. The data presented in this manuscript point to a novel explanation for the reduced penetrance seen in HPAH. Our data show that BMPR2 alternative splicing plays a role in this reduced penetrance. BMPR2 mutation carriers were more likely to have PAH if they had higher levels of an alternatively spliced BMPR2 transcript, isoform-B relative to the full length BMPR2 transcript. Thus our data suggest that while a BMPR2 mutation creates baseline susceptibility, an important determinant of disease penetrance appears to be the higher relative expression of the alternative spliced BMPR2 isoform-B. These data emphasize the importance of BMPR2 alternative splicing in HPAH and raise some intriguing question. 1) Can a predictive model of disease based on expression levels of the BMPR2 alternative splicing be developed? This predictive model will have obvious clinical utility in predicting which mutation carriers may eventually develop disease and thus will require appropriate follow-up. 2) Since splicing can be manipulated in vivo by drugs our findings suggest that manipulation of BMPR2 alternative splicing should be explored as a potential new intervention in HPAH.
Acknowledgments
Funding Sources: Research presented here was supported by NHLBI 1R01HL102020 to RH
Footnotes
Conflict of Interest Disclosures: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lane KB, Machado RD, Pauciulo MW, Thomson JR, Phillips JA, 3rd, Loyd JE, Nichols WC, Trembath RC. Heterozygous germline mutations in bmpr2, encoding a tgf-beta receptor, cause familial primary pulmonary hypertension. The international pph consortium. Nat Genet. 2000;26:81–84. doi: 10.1038/79226. [DOI] [PubMed] [Google Scholar]
- 2.Newman JH, Wheeler L, Lane KB, Loyd E, Gaddipati R, Phillips JA, 3rd, Loyd JE. Mutation in the gene for bone morphogenetic protein receptor ii as a cause of primary pulmonary hypertension in a large kindred. N Engl J Med. 2001;345:319–324. doi: 10.1056/NEJM200108023450502. [DOI] [PubMed] [Google Scholar]
- 3.Cogan JD, Pauciulo MW, Batchman AP, Prince MA, Robbins IM, Hedges LK, Stanton KC, Wheeler LA, Phillips JA, 3rd, Loyd JE, Nichols WC. High frequency of bmpr2 exonic deletions/duplications in familial pulmonary arterial hypertension. Am J Respir Crit Care Med. 2006;174:590–598. doi: 10.1164/rccm.200602-165OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cogan JD, Vnencak-Jones CL, Phillips JA, 3rd, Lane KB, Wheeler LA, Robbins IM, Garrison G, Hedges LK, Loyd JE. Gross bmpr2 gene rearrangements constitute a new cause for primary pulmonary hypertension. Genet Med. 2005;7:169–174. doi: 10.1097/01.gim.0000156525.09595.e9. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez CI, Bhattacharya A, Wang W, Peltz SW. Nonsense-mediated mrna decay in saccharomyces cerevisiae. Gene. 2001;274:15–25. doi: 10.1016/s0378-1119(01)00552-2. [DOI] [PubMed] [Google Scholar]
- 6.Phillips JA, 3rd, Poling JS, Phillips CA, Stanton KC, Austin ED, Cogan JD, Wheeler L, Yu C, Newman JH, Dietz HC, Loyd JE. Synergistic heterozygosity for tgfbeta1 snps and bmpr2 mutations modulates the age at diagnosis and penetrance of familial pulmonary arterial hypertension. Genet Med. 2008;10:359–365. doi: 10.1097/GIM.0b013e318172dcdf. [DOI] [PubMed] [Google Scholar]
- 7.Abraham WT, Raynolds MV, Badesch DB, Wynne KM, Groves BM, Roden RL, Robertson AD, Lowes BD, Zisman LS, Voelkel NF, Bristow MR, Perryman MB. Angiotensin-converting enzyme dd genotype in patients with primary pulmonary hypertension: Increased frequency and association with preserved haemodynamics. J Renin Angiotensin Aldosterone Syst. 2003;4:27–30. doi: 10.3317/jraas.2003.003. [DOI] [PubMed] [Google Scholar]
- 8.Hoeper MM, Tacacs A, Stellmacher U, Lichtinghagen R. Lack of association between angiotensin converting enzyme (ace) genotype, serum ace activity, and haemodynamics in patients with primary pulmonary hypertension. Heart. 2003;89:445–446. doi: 10.1136/heart.89.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koehler R, Olschewski H, Hoeper M, Janssen B, Grunig E. Serotonin transporter gene polymorphism in a cohort of german patients with idiopathic pulmonary arterial hypertension or chronic thromboembolic pulmonary hypertension. Chest. 2005;128:619S. doi: 10.1378/chest.128.6_suppl.619S. [DOI] [PubMed] [Google Scholar]
- 10.Machado RD, Koehler R, Glissmeyer E, Veal C, Suntharalingam J, Kim M, Carlquist J, Town M, Elliott CG, Hoeper M, Fijalkowska A, Kurzyna M, Thomson JR, Gibbs SR, Wilkins MR, Seeger W, Morrell NW, Gruenig E, Trembath RC, Janssen B. Genetic association of the serotonin transporter in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2006;173:793–797. doi: 10.1164/rccm.200509-1365OC. [DOI] [PubMed] [Google Scholar]
- 11.Willers ED, Newman JH, Loyd JE, Robbins IM, Wheeler LA, Prince MA, Stanton KC, Cogan JA, Runo JR, Byrne D, Humbert M, Simonneau G, Sztrymf B, Morse JA, Knowles JA, Roberts KE, McElroy JJ, Barst RJ, Phillips JA., III Serotonin transporter polymorphisms in familial and idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med. 2006;173:798–802. doi: 10.1164/rccm.200509-1361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Remillard CV, Tigno DD, Platoshyn O, Burg ED, Brevnova EE, Conger D, Nicholson A, Rana BK, Channick RN, Rubin LJ, O'Connor D T, Yuan JX. Function of kv1.5 channels and genetic variations of kcna5 in patients with idiopathic pulmonary arterial hypertension. Am J Physiol Cell Physiol. 2007;292:C1837–1853. doi: 10.1152/ajpcell.00405.2006. [DOI] [PubMed] [Google Scholar]
- 13.Hamid R, Cogan JD, Hedges LK, Austin E, Phillips JA, 3rd, Newman JH, Loyd JE. Penetrance of pulmonary arterial hypertension is modulated by the expression of normal bmpr2 allele. Hum Mutat. 2009;30:649–654. doi: 10.1002/humu.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: Diversification, exon definition and function. Nat Rev Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 15.Long JC, Caceres JF. The sr protein family of splicing factors: Master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 16.Han SP, Tang YH, Smith R. Functional diversity of the hnrnps: Past, present and perspectives. Biochem J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 17.Shen H, Green MR. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 18.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 19.Orengo JP, Cooper TA. Alternative splicing in disease. Adv Exp Med Biol. 2007;623:212–223. doi: 10.1007/978-0-387-77374-2_13. [DOI] [PubMed] [Google Scholar]
- 20.Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim Biophys Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu F, Ventura F, Doody J, Massague J. Human type ii receptor for bone morphogenic proteins (bmps): Extension of the two-kinase receptor model to the bmps. Mol Cell Biol. 1995;15:3479–3486. doi: 10.1128/mcb.15.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenzweig BL, Imamura T, Okadome T, Cox GN, Yamashita H, ten Dijke P, Heldin CH, Miyazono K. Cloning and characterization of a human type ii receptor for bone morphogenetic proteins. Proc Natl Acad Sci U S A. 1995;92:7632–7636. doi: 10.1073/pnas.92.17.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beppu H, Minowa O, Miyazono K, Kawabata M. Cdna cloning and genomic organization of the mouse bmp type ii receptor. Biochem Biophys Res Commun. 1997;235:499–504. doi: 10.1006/bbrc.1997.6816. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Dunmore BJ, Morrell NW. Bone morphogenetic protein type ii receptor mutations causing protein misfolding in heritable pulmonary arterial hypertension. Proc Am Thorac Soc. 2010;7:395–398. doi: 10.1513/pats.201002-024AW. [DOI] [PubMed] [Google Scholar]
- 25.Rudarakanchana N, Flanagan JA, Chen H, Upton PD, Machado R, Patel D, Trembath RC, Morrell NW. Functional analysis of bone morphogenetic protein type ii receptor mutations underlying primary pulmonary hypertension. Hum Mol Genet. 2002;11:1517–1525. doi: 10.1093/hmg/11.13.1517. [DOI] [PubMed] [Google Scholar]
- 26.Machado RD, Aldred MA, James V, Harrison RE, Patel B, Schwalbe EC, Gruenig E, Janssen B, Koehler R, Seeger W, Eickelberg O, Olschewski H, Elliott CG, Glissmeyer E, Carlquist J, Kim M, Torbicki A, Fijalkowska A, Szewczyk G, Parma J, Abramowicz MJ, Galie N, Morisaki H, Kyotani S, Nakanishi N, Morisaki T, Humbert M, Simonneau G, Sitbon O, Soubrier F, Coulet F, Morrell NW, Trembath RC. Mutations of the tgf-beta type ii receptor bmpr2 in pulmonary arterial hypertension. Hum Mutat. 2006;27:121–132. doi: 10.1002/humu.20285. [DOI] [PubMed] [Google Scholar]
- 27.West J, Harral J, Lane K, Deng Y, Ickes B, Crona D, Albu S, Stewart D, Fagan K. Mice expressing bmpr2r899x transgene in smooth muscle develop pulmonary vascular lesions. Am J Physiol Lung Cell Mol Physiol. 2008;295:L744–755. doi: 10.1152/ajplung.90255.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson JA, Hemnes AR, Perrien DS, Schuster M, Robinson LJ, Gladson S, Loibner H, Bai S, Blackwell TR, Tada Y, Harral JW, Talati M, Lane KB, Fagan KA, West J. Cytoskeletal defects in bmpr2-associated pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol. 2012;302:L474–484. doi: 10.1152/ajplung.00202.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cogan JD, Pauciulo MW, Batchman AP, Prince MA, Robbins IM, Hedges LK, Stanton KC, Wheeler LA, Phillips JA, 3rd, Loyd JE, Nichols WC. High frequency of bmpr2 exonic deletions/duplications in familial pulmonary arterial hypertension. Am J Respir Crit Care Med. 2006;174:590–598. doi: 10.1164/rccm.200602-165OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machado RD, Pauciulo MW, Thomson JR, Lane KB, Morgan NV, Wheeler L, Phillips JA, 3rd, Newman J, Williams D, Galie N, Manes Ak, McNeil K, Yacoub M, Mikhail G, Rogers P, Corris P, Humbert M, Donnai D, Martensson G, Tranebjaerg L, Loyd JE, Trembath RC, Nichols WC. Bmpr2 haploinsufficiency as the inherited molecular mechanism for primary pulmonary hypertension. Am J Hum Genet. 2001;68:92–102. doi: 10.1086/316947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bird AG, McLachlan SM, Britton S. Cyclosporin a promotes spontaneous outgrowth in vitro of epstein-barr virus-induced b-cell lines. Nature. 1981;289:300–301. doi: 10.1038/289300a0. [DOI] [PubMed] [Google Scholar]
- 32.Oh HM, Oh JM, Choi SC, Kim SW, Han WC, Kim TH, Park DS, Jun CD. An efficient method for the rapid establishment of epstein-barr virus immortalization of human b lymphocytes. Cell Prolif. 2003;36:191–197. doi: 10.1046/j.1365-2184.2003.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamid R, Patterson J, Brandt SJ. Genomic structure, alternative splicing and expression of tg-interacting factor, in human myeloid leukemia blasts and cell lines. Biochim Biophys Acta. 2008;1779:347–355. doi: 10.1016/j.bbagrm.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Bustin SA. Quantification of mrna using real-time reverse transcription pcr (rt-pcr): Trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 36.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of sf2/asf-specific exonic splicing enhancers. Hum Mol Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 37.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. Esefinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foletta VC, Lim MA, Soosairajah J, Kelly AP, Stanley EG, Shannon M, He W, Das S, Massague J, Bernard O. Direct signaling by the bmp type ii receptor via the cytoskeletal regulator limk1. J Cell Biol. 2003;162:1089–1098. doi: 10.1083/jcb.200212060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eaton BA, Davis GW. Lim kinase1 controls synaptic stability downstream of the type ii bmp receptor. Neuron. 2005;47:695–708. doi: 10.1016/j.neuron.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh G, Adams JA. Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 2011;278:587–597. doi: 10.1111/j.1742-4658.2010.07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. Sc35 autoregulates its expression by promoting splicing events that destabilize its mrnas. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thol F, Kade S, Schlarmann C, Loffeld P, Morgan M, Krauter J, Wlodarski MW, Kolking B, Wichmann M, Gorlich K, Gohring G, Bug G, Ottmann O, Niemeyer CM, Hofmann WK, Schlegelberger B, Ganser A, Heuser M. Frequency and prognostic impact of mutations in srsf2, u2af1, and zrsr2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 43.Edmond V, Moysan E, Khochbin S, Matthias P, Brambilla C, Brambilla E, Gazzeri S, Eymin B. Acetylation and phosphorylation of srsf2 control cell fate decision in response to cisplatin. EMBO J. 2011;30:510–523. doi: 10.1038/emboj.2010.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poling JS, Phillips JA, 3rd, Cogan JD, Hamid R. Pharmacologic correction of dominant-negative gh1 deficiency causing mutations. Clin Transl Sci. 2011;4:175–179. doi: 10.1111/j.1752-8062.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West J, Cogan J, Geraci M, Robinson L, Newman J, Phillips JA, Lane K, Meyrick B, Loyd J. Gene expression in bmpr2 mutation carriers with and without evidence of pulmonary arterial hypertension suggests pathways relevant to disease penetrance. BMC Med Genomics. 2008;1:45. doi: 10.1186/1755-8794-1-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.