Summary
Legionella pneumophila uses aquatic protozoa as replication niche and protection from harsh environments. Although L. pneumophila is not known to have a circadian clock, it encodes homologues of the KaiBC proteins of Cyanobacteria that regulate circadian gene expression. We show that L. pneumophila kaiB, kaiC and the downstream gene lpp1114, are transcribed as a unit under the control of the stress sigma factor RpoS. KaiC and KaiB of L. pneumophila do not interact as evidenced by yeast and bacterial two-hybrid analyses. Fusion of the C-terminal residues of cyanobacterial KaiB to Legionella KaiB restores their interaction. In contrast, KaiC of L. pneumophila conserved autophosphorylation activity, but KaiB does not trigger the dephosphorylation of KaiC like in Cyanobacteria. The crystal structure of L. pneumophila KaiB suggests that it is an oxidoreductase-like protein with a typical thioredoxin fold. Indeed, mutant analyses revealed that the kai operon-encoded proteins increase fitness of L. pneumophila in competitive environments, and confer higher resistance to oxidative and sodium stress. The phylogenetic analysis indicates that L. pneumophila KaiBC resemble Synechosystis KaiC2B2 and not circadian KaiB1C1. Thus, the L. pneumophila Kai proteins do not encode a circadian clock, but enhance stress resistance and adaption to changes in the environments.
Introduction
Legionella pneumophila is an environmental bacterium and a human pathogen. It infects and multiplies within aquatic protozoa like Acanthamoeba castellanii and also human alveolar macrophages causing a severe pneumonia known as Legionnaires’ disease (Fraser et al., 1977; McDade et al., 1977; Fields et al., 2002). Legionella is found in natural fresh waters as well as in man-made water systems, which are stressful habitats (Fields et al., 2002). Bacteria living under aerobic conditions must deal with the toxic effects of oxygen that can be generated exogenously or endogenously. In natural waters for example, solar radiation can initiate a series of photochemical reactions that produce harmful reactive oxygen species such as hydrogen peroxide and hydroxyl radicals (Scully et al., 2003). Legionella pneumophila encounters reactive oxygen species in the fresh water environment, as well as in the intracellular environment because of the oxidative burst exerted by the host cells (Newton et al., 2010). Therefore the response to oxidative stress and to toxic compounds is a key determinant of survival, spread and virulence of L. pneumophila. Indeed, L. pneumophila encodes different enzymes to defend itself against these stressors like two catalase-peroxidases: KatA being localized in the periplasm and KatB being localized in the cytoplasm (Bandyopadhyay and Steinman, 2000). These enzymes display strong peroxidase, but weak hydroperoxidase activity. In addition L. pneumophila encodes two superoxide dismutases, one Cu/Zn dependent predicted to be located in the periplasm and a Mn/Fe dependent located in the cytoplasm. These are expressed differentially depending on the growth phase (Bruggemann et al., 2006). The Cu/Zn superoxide dismutase is expressed in post exponential growth phase and is regulated by the stress sigma factor RpoS. In contrast the Mn/Fe superoxide dismutase is highly expressed during exponential growth (Bruggemann et al., 2006). Surprisingly, the typical SoxRS regulator of superoxide dismutase gene expression under conditions of oxidative stress is absent from L. pneumophila (Cazalet et al., 2004). Additionally, L. pneumophila encodes three peroxide-scavenging alkylhydroxide reductase systems that also may be important in protection from reactive oxygen species. AhpC1 (lpp3037), AhpC2 (lpp2299) and AhpD (lpp2298) protect L. pneumophila from peroxide damage as chromosomal deletion mutants were two- to eightfold more sensitive to H2O2, tert-butyl hydroperoxide, cumene hydroperoxide and paraquat than wild-type L. pneumophila (LeBlanc et al., 2008). LeBlanc and colleagues (2008) reported that ahpD is induced early in exponential phase and repressed postexponentially, concomitant with the induction of ahpC1. Our recent expression data obtained by microarray analyses showed that AhpC2D (lpp2298/lpp2299) are differently expressed in vitro and in vivo. They showed post-exponential phase up-regulation in vitro, but were expressed during replicative phase in vivo (Bruggemann et al., 2006). The gene encoding AhpC1 is predominately transcribed during replicative phase in vitro and has been shown to be up-regulated during intracellular growth in macrophages indicating a role in protection from the respiratory burst (Rankin et al., 2002). Phylogenetic analysis of the AphC1 peroxidases of Helicobacter pylori suggests that thioredoxin and thioredoxin reductase activate the enzyme in a nicotinamide adenosine dinucleotide phosphate-dependent reaction (Baker et al., 2001). In contrast, AhpC2 of L. pneumophila shares similarities to the complex AhpC/D found in Mycobacterium tuberculosis (Bryk et al., 2002). The regulation of these different oxidative stress proteins is not understood and may involve many regulators like OxyR, RpoS and perhaps other regulatory factors not yet identified. Thus, to understand how L. pneumophila adapts to conditions it faces in natural environments or within the intracellular milieu of protozoa or macrophages, its response to different stressors needs to be deciphered.
Genome sequence analyses showed that L. pneumophila encodes two genes homologous to kaiC (lpp1116) and kaiB (lpp1115) of Cyanobacteria, but that L. pneumophila lacks the kaiA gene (Cazalet et al., 2004). In Cyanobacteria, KaiABC, the circadian core genes, control rhythmic gene expression and regulate different physiological functions like chromosomal compaction and cell division to synchronize them with the Earth rotation (Smith and Williams, 2006; Dong et al., 2010). Furthermore, possession of a circadian clock has been shown to enhance the adaptive fitness of Cyanobacteria in a wide variety of environmental conditions (Woelfle et al., 2004). In prokaryotic organisms, the circadian rhythms have been described only in photosynthetic bacteria (Johnson, 2007), where they were mainly studied in Cyanobacteria and lately also in the purple bacteria Rhodobacter sphaeroides (Min et al., 2005a). Although KaiB and KaiC are found in several prokaryotes belonging to different species (Dvornyk et al., 2003; Loza-Correa et al., 2010), their implication in circadian clocks has never been shown. Thus the question arises: why are the kai genes conserved in L. pneumophila and what are their functions? Does L. pneumophila encode a circadian clock and/or are these genes involved in improving fitness in the environment like in Cyanobacteria? Here we report that the kai genes of L. pneumophila do not seem to be regulating circadian rhythms, but are implicated in stress response, in particular to oxidative and sodium stress. In line with this function, the crystal structure of KaiB resembles a thioredoxin.
Results
kaiBC genes of L. pneumophila are organized in an operon together with a regulatory protein
In the L. pneumophila genome, the kaiBC (lpp1115 and lpp1116) genes are contiguous as described in Synechococcus elongatus and other Cyanobacteria and are located next to lpp1114, a gene encoding a multidomain protein containing a Per-ARNT-Sim (PAS) sensor domain, a GGDEF and an EAL domain (Fig. 1A). GGDEF and EAL domain-containing proteins determine the cellular concentrations of c-di-GMP by mediating its synthesis and degradation, respectively (Schirmer and Jenal, 2009). The PAS domain is found in many signalling proteins that monitor changes in light, redox potential, oxygen, small ligands and overall energy of a cell (Taylor and Zhulin, 1999). Gene expression data obtained from microarray analyses and quantitative real-time polymerase chain reaction (qRT-PCR) experiments showed that in L. pneumophila kaiB and kaiC form a transcriptional unit together with lpp1114 (data not shown). We have previously undertaken a RNAseq analysis of exponentially and post exponentially grown L. pneumophila to map all transcriptional start sites (TSS) in the genome (Sahr et al., 2012). Using these data, we could map the TSS of the kai operon 67nts upstream the translational start with a predicted RpoS promoter sequence upstream the TSS (Fig. 1A). This finding is in accordance with a regulation of this operon by RpoS as indeed seen in a transcriptome analysis of a ΔrpoS mutant strain (Table S1). Sequence comparison of L. pneumophila KaiC with those of Cyanobacteria, revealed that the two Walker motifs [G or A]XXXXGK[T or S] required for the adenosine triphosphate (ATP)/guanosine 5′-triphosphate (GTP)-binding are highly conserved. Furthermore, the potential C-terminal phosphorylation sites Thr405, Ser410 and Ser411 in L. pneumophila corresponding to Thr426, Ser431 and Thr432 in S. elongatus are present (Fig. 1B).
Fig. 1.
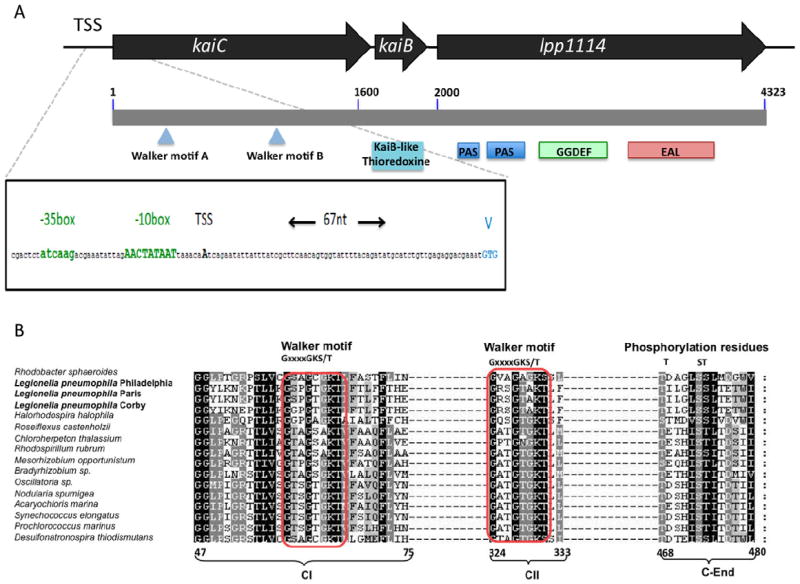
Schematic representation of the kai operon in Legionella pneumophila.
A. Black arrows represent the genes of the kai operon: kaiC (lpp1116), kaiB (lpp1115) and lpp1114. The gray bar indicates the length of the mRNA transcribed starting at the 1 + position of kaiC. Below the gray bar, the two Walker motifs found in KaiC are indicated (blue triangles), the domains encoded by each protein are represented as boxes. The transcriptional start site (TSS) is 67 nucleotides upstream the translational start (indicated by V for Valine). Upstream the TSS the −10 and −35 box of a predicted RpoS promoter sequence is shown in green.
B. Alignment of different bacterial KaiC proteins with that of L. pneumophila. Red boxes indicate the conserved walker motifs. The numbers below, indicate the amino acid position with respect to the full-length protein sequence of Synechococcus elongatus. CI stands for the first KaiC domain, CII stands for the second KaiC domain and C-End indicate the C-terminus of the protein.
Legionella pneumophila kai genes are not expressed in a circadian manner
In S. elongatus, the expression of the kaiBC mRNA shows robust circadian rhythms during continuous light (Tomita et al., 2005). Thus, our first approach was to test whether the kai genes of L. pneumophila growing in vitro show circadian activity. We examined the relative mRNA expression levels by real-time PCR of each gene of the operon (kaiC, kaiB and lpp1114) during in vitro growth in buffered yeast extract (BYE) medium. Samples were taken at different time points: after 14 h and 16 h corresponding to exponential growth phase at optical density (OD) 2.5 and 3.0, respectively, during post-exponential growth phase (OD 3.8, 20 h) and late post-exponential growth phase (OD 4.2, 24 h). As shown in Fig. 2A the kaiC, kaiB and lpp1114 mRNAs are expressed during post exponential growth phase and their expression increases until late exponential growth phase. Thus the kai operon is expressed in a typical growth phase-dependent manner as reported for many L. pneumophila genes (Bruggemann et al., 2006; Sahr et al., 2012). RNAseq analyses at exponential and post exponential growth phase confirmed this result (Sahr et al., 2012). We then further tested the expression of the kai operon during intracellular growth in A. castellanii. Amoebae were infected with wild-type L. pneumophila and its intracellular replication was monitored at 20°C, an infection temperature that mimics environmental conditions better. Samples were taken each day over 7 days and the relative mRNA expression of the kai genes was measured (Fig. 2B). Again, the expression of the kai operon genes was correlated with increased intracellular multiplication and no circadian cycles were observed. Thus, the kai operon has a growth phase dependent expression controlled by the stress sigma factor RpoS, with the highest levels during late stages of in vitro or in vivo growth.
Fig. 2.
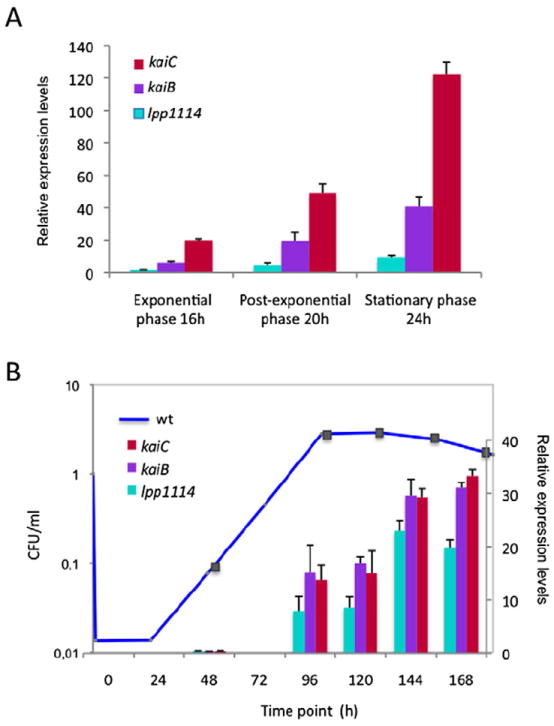
The expression of the kai operon genes is growth phase dependent in vitro and in vivo in Acanthamoeba castellanii.
A. Quantitative real-time polymerase chain reaction of the kai operon genes during 24 h in vitro growth. The bars represent the relative quantity of mRNA at the sampled time points relative to mRNA levels at exponential phase (OD 2.5). Red bars kaiC; purple bars, kaiB; cyan lpp1114.
B. Relative gene expression of the kai operon genes during intracellular growth in A. castellanii. Amoebae were infected with Legionella pneumophila wild type at MOI = 100 and the infections were followed at 20°C over 7 days (168 h). The squares on the growth curve indicate the time points (hours) of intracellular growth when the mRNA samples were taken. Transcription levels of the genes were calculated with respect to the values obtained at 72 h. The bars represent the mean of two independent experiments ± standard deviation.
Transcriptional profiling of L. pneumophila wild type, ΔkaiC and Δlpp1114 mutant strains
KaiC has been described in Cyanobacteria as a regulatory protein. To analyse whether KaiC of L. pneumophila has regulatory functions, we constructed two mutants where either the kaiC gene (ΔkaiC : kan) or the lpp1114 gene (Δlpp1114 : apra) was replaced by an antibiotic cassette and analysed their transcriptional profiles in different conditions. Repeated efforts to construct a ΔkaiB mutant were unsuccessful. kaiC is the first gene of the operon. We thus tested the gene expression of kaiB and lpp1114 in the ΔkaiC strain to reveal possible downstream effects of the kaiC gene deletion. Indeed, qRT-PCR and microarray analyses showed that transcription of kaiB and lpp1114 was abolished in the ΔkaiC mutant strain indicating that the entire operon was not transcribed (data not shown). Thus our ΔkaiC strain is comparable with a triple mutant ΔkaiCBlpp1114.
For genome-wide transcriptional profiling of in vitro grown bacteria, RNA was sampled at OD 4.0 after growth of the wild-type and the ΔkaiC strains in BYE media at 37°C. This corresponds to the time point of the highest expression of kaiC (Fig. 2A). However, even at this time point, only very slight expression changes were observed and the kai operon genes themselves were in all experiments only weakly expressed, making the comparison and interpretation of the data difficult. Apart from the kai operon genes (lpp1114-lpp1116), four genes were identified as significantly down-regulated in the ΔkaiC mutant strain as compared with the wild type. These genes encode an esterase/lipase (lpp1856), a putative coproporphyrinogen oxidase A (lpp2849) and two hypothetical proteins (lpp1823 and lpp0954; Table 1). Our analysis did not identify any significantly up-regulated gene in the ΔkaiC strain compared with the wild type. As differences in the transcriptional profiles of L. pneumophila grown in vitro compared with in vivo exist (Bruggemann et al., 2006), we recovered RNA from L. pneumophila wild type and the two mutants after 6 days of infection of A. castellanii at 20°C, the time point where the highest level of expression of the kai genes was observed by qRT-PCR (Fig. 2B). During in vivo growth we identified five genes differentially regulated in both mutant strains [fold change (FC) > 1.5], three of them with significant changes in their expression (Table 2). Interestingly, the genes slightly down-regulated in the ΔkaiC strain, were up-regulated 1.5-fold and more in the Δlpp1114. Among the genes down-regulated in the ΔkaiC mutant are two that encode major outer membrane proteins (lpp3032, lpp3033), two hypothetical proteins (lpp1553 and lpp2209) and a small RNA (ncRNA08). This could suggest that KaiC has a negative effect on lpp1114 expression.
Table 1.
Genes down-regulated in the ΔkaiC mutant at OD 4.0 after in vitro growth in buffered yeast extract at 37°C.
| Gene ID | Product | FC |
|---|---|---|
| lpp1115 | KaiB – like circadian clock protein | 0.158* |
| lpp1116 | KaiC – like circadian clock protein | 0.167* |
| lpp1114 | Regulatory protein (GGDEF and EAL domains) | 0.237* |
| lpp1823 | Unknown | 0.506 |
| lpp0954 | Unknown | 0.57 |
| lpp1856 | Similar to esterase/lipase | 0.583 |
| lpp2849 | Similar to putative coproporphyrinogen oxidase A | 0.604 |
Significantly differentially regulated genes are marked by an asterisk.
P < 0.05.
Table 2.
Genes with differential expression in the ΔkaiC and the Δlpp1114 mutant at 144 h after in vivo growth in A. castellanii at 20°C.
| Gene ID | Product | ΔkaiC FC | Δlpp1114 FC |
|---|---|---|---|
| lpp3032 | Major outer membrane protein | 0.482* | 2.248* |
| lpp3033 | Major outer membrane protein precursor | 0.694 | 2.035* |
| lpp2209 | Unkown | 0.528* | 2.165* |
| lpp1294 | Flagelline (flaA) | 0.672 | 1.529 |
| ncRNA18 | ncRNA18 | 0.533 | 1.624 |
| lpp1114 | Regulatory protein (GGDEF/EAL domains) | 0.593 | 0.759 |
| lpp1115 | KaiB – like circadian clock protein | 0.353 | 1.093 |
| lpp1116 | KaiC – like circadian clock protein | 0.308 | 1.09 |
Significantly differentially regulated genes are marked by an asterisk.
P < 0.05.
Kai proteins provide fitness to L. pneumophila in competitive environments
In order to learn whether the Kai proteins are implicated in intracellular growth of L. pneumophila, Acanthamoeba castellanii, was infected with the wild-type strain and the ΔkaiC and Δlpp1114 mutant strains and their intracellularly replication over 72 h at 37°C and over 168 h at 20°C was recorded. No difference in intracellular replication was observed (data not shown). In order to detect more subtle differences, we conducted a competitive growth assay in A. castellanii (Kessler et al., 2013). The Δlpp1114 mutant, as compared with the wild type did not show any difference also in this assay. However, the ΔkaiC mutant, that is comparable with a knockout of the entire operon, was clearly less competitive in amoebae.Already after 2 days of infection, only half the number of mutant bacteria, as compared with wild-type bacteria, was recovered, and after 5 days, the mutant was nearly outcompeted (Fig. 3A). When complementing the ΔkaiC mutant with the entire operon on a plasmid (pMMB207_kai_operon), the phenotype was restored (Fig. 3B). Thus the kai operon mutant is likely an example of ‘soft selection’ (Wallace, 1981) where the reduced fitness of one genotype compared with another one is only seen in a competitive environment.
Fig. 3.
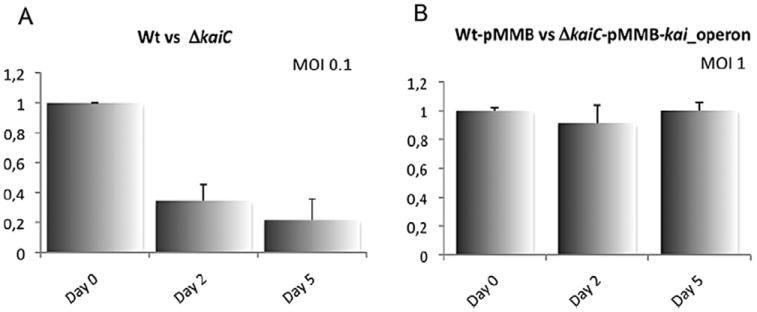
The kai operon genes confer fitness in competitive intracellular growth in Acanthamoeba castellanii.
A. Competitive assay of Legionella pneumophila wild type vs ΔkaiC after infection of A. castellanii with an MOI of 0.1. The graph represents the ratio between wild type and mutant bacteria recovered at day 0, 2 and 5. The ΔkaiC mutant is outcompeted by the wild type after two days of infection of A. castelanii.
B. Competitive assay of L. pneumophila wild type carrying the empty vector pMMB vs ΔkaiC carrying the pMMB vector encoding for the entire operon (kaiOp). Infection of A. castellanii was performed with an MOI of 1. The phenotype is restored when the ΔkaiC mutant is complemented with the entire operon in trans. The bars represent the mean of at least three independent experiments ± standard deviation.
Crystal structure of KaiB demonstrates closer resemblance to thioredoxins than to Cyanobacteria KaiB structures
In order to gain further insight into the function of the L. pneumophila KaiB and KaiC proteins, we embarked on the structural characterization of these proteins by X-ray crystallography. Diffraction quality crystals were obtained only for the L. pneumophila KaiB protein, while the KaiC crystallization was unsuccessful (data not shown). The structure of the KaiB protein was solved to 1.95 Å resolution by the single-wavelength anomalous diffraction technique using selenomethionine-enriched protein samples. The final model contained two full-length (residues 1–90) KaiB molecules in the asymmetric unit with each monomer demonstrating a slightly different conformation between residues 31 and 37 (Fig. 4A). Each KaiB molecule features a two-layer α/β sandwich composed of a four-strand mixed β-sheet surrounded by three α-helices α1, α2′ and α2″ and α5 corresponding to residues 15–27, 50–53 and 80–86 respectively. The secondary structure elements were assigned using DSSP (http://swift.cmbi.ru.nl/gv/dssp/; Kabsch and Sander, 1983) and numbered according to corresponding elements in cyanobacterial KaiB proteins (Hitomi et al., 2005).
Fig. 4.
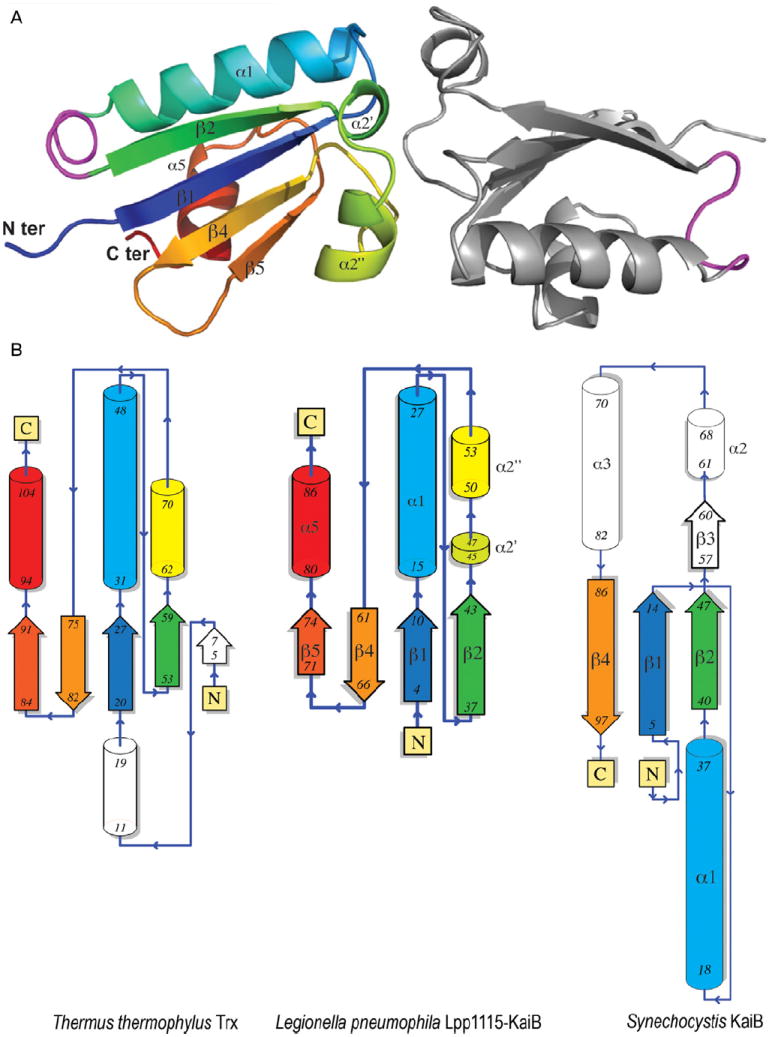
Structure of Legionella pneumophila KaiB highlights differences between Lpp1115 and other members of the KaiB protein family.
A. The structure of the L. pneumophila KaiB protein features two full-length protein (residues 1–90) molecules shown as a ribbon diagram. One of the Lpp1115-KaiB monomers is coloured in rainbow spectrum from N-terminus (blue) to C-terminus (red). The secondary structure elements in Lpp1115-KaiB are labelled (α for a helix and β for a strand) in black and numbered according to previously characterized cyanobaterial KaiB proteins. Lpp1115-KaiB N- and C-termini are also labelled. The KaiB region between residues 31 and 37 adapting different conformations in each KaiB monomer is coloured in magenta.
B. Legionella pneumophila KaiB (centre), Thermus thermophilus thioredoxin (left) and Synechocystis sp. PCC6803 (PDB 1WWJ) Thermosynechococcus elongatus BP-1 KaiB (right) secondary structure topologies illustrate the structural similarities between these protein molecules. The secondary structure elements in L. pneumophila KaiB are coloured according to rainbow spectrum of the KaiB ribbon diagram presented above. The numbers of first and last residues in each β-strand and α-helix are indicated in black. The secondary structure elements in T. thermophilus Thioredoxin (left) and T. elongatus BP-1 KaiB (right) are colored according to matching elements in the Legionella Lpp1115-KaiB structure. Pymol (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC) and PDBsum software (Laskowski, 2009) were used to generate this figure.
The comparative structural analysis using the Dali server (Holm and Rosenström, 2010; http://ekhidna.biocenter.helsinki.fi/dali_server) demonstrated that KaiB molecule bears strong resemblance to the thioredoxin family protein structures. In particular, KaiB and Thermus thermophilus thioredoxin [protein data bank (PDB) 2CVK] structures superimpose with rmsd of 2.3 Å over 84 Cα atoms while sharing only 13% of primary sequence identity (Fig. 4B). In comparison, the closest structural homologue among characterized KaiB proteins – the structure of the KaiB protein from the cyanobacterium Synechocystis sp. PCC6803 (PDB 1WWJ) superimposes with KaiB structure with rmsd of 4.3 Å over only 63 Cμ atoms. The secondary structure elements that are conserved between the L. pneumophila and Synechocystis KaiB molecules primarily include β1 (residues 4–10) and β2 (residues 37–43) strands on the main β-sheet and the α1-helix of the KaiB protein, which spans the N-terminal half (residues 1–46) of this protein (Fig. 4B). By contrast, the C-terminal half of the KaiB molecule shares very little structural similarity with the Synechocystis KaiB molecule with only the β4-strand (residues 61–66) matching to corresponding elements in the cyanobacterial protein structure. The β3-strand of the Synechocystis KaiB molecule spanning residues 57–60 contributes to the tetramerization interface critical for the circadian rhythm function of this protein (Hitomi et al., 2005). The similar region in the KaiB structure corresponds to helices α2′ (residues 45–47) and α2″ (residues 50–53). While this region is involved in the KaiB dimerization interface the orientation of the KaiB monomers in the dimer is significantly different than that of monomers in the Synechocystis KaiB tetramer. The α5-helix (residues 80–86) and the β5-strand (residues 71–74) of KaiB match to the corresponding regions in T. thermophilus thioredoxin, but not to Synechocystis KaiB structure (Fig. 4B).
Furthermore, comparative sequence analysis indicated that KaiB lacks the C-terminal sequence conserved in cyanobacterial KaiB proteins (residues 95–108 in T. elongatus KaiB protein; Fig. S1). However, this region is crucial for the function of this protein because the Glu95 to Phe102 deletion in S. elongatus PCC7942 KaiB resulted in loss of the circadian rhythm in vivo (Iwase et al., 2005). The structural comparison of KaiB and cyanobacterial KaiB molecules confirmed that the L. pneumophila protein does not contain any structural elements that would correspond to the C-terminal region of the T. elongatus KaiB protein. Taken together, the structural analysis of KaiB identifies significant differences between this protein and the structures of cyanobacterial KaiB proteins. The KaiB structure appears lacking the key functionally relevant elements conserved in cyanobacterial KaiB proteins indicating that despite remaining similarities the KaiB protein function has diversified from that of bona fide KaiB proteins involved in circadian rhythms. On the other hand the strong structural similarity between KaiB and bacterial thioredoxin proteins points to a possible function of L. pneumophila KaiB in the oxidative stress response.
Legionella pneumophila KaiB and KaiC do not interact
The crystal structure of KaiB suggested that L. pneumophila KaiB might not interact with KaiC as the C-terminal residues of KaiB are missing in L. pneumophila. To test this hypothesis, we used a yeast two-hybrid approach where KaiB was used as bait and KaiC as target. Both were expressed as fusion protein with hSos and with a myristoylation sequence, respectively, and co-expressed in the Saccharomyces cervisae mutant strain cdc25H at 37°C. If KaiB and KaiC interact, the hSos protein would activate the Ras-signalling cascade and allow the strain to grow at 37°C. However, no interaction between KaiB and KaiC of L. pneumophila was observed (data not shown). A major disadvantage of assaying protein–protein interactions in a heterologous system is that bacterial-specific post-translational modifications or protein chaperones are not present in the yeast background, which might lead to incomplete or mis-folded proteins. Thus we used a second system, a bacterial two-hybrid approach, the bacterial adenylate cyclase-based two-hybrid (BACTH) system. This method is based on the interaction-mediated reconstitution of the adenylate cyclase activity in Escherichia coli lacking endogenous adenylate cyclase (Karimova et al., 1998). It uses the catalytic domain of adenylate cyclase (CyaA) from Bordetella pertussis that consists of two fragments that are complementary to each other (T25 and T18) and that need to stay in immediate vicinity. The fragments T25 and T18 were fused to KaiC and KaiB respectively. In case the two proteins interact T25 and T18 would be closely enough to synthesize adenosine 3′5′ cyclic monophosphate that the latter binds to the catabolite activator protein and regulates the transcription of genes like the lac and mal operon. Thus, it is possible to determine if two proteins interact by plating the bacteria on selective media that allow visualizing production of β-galactosidase or maltose fermentation. Again, we did not observe any interaction of Legionella KaiB and KaiC when bacteria were plated on McConkey/maltose or LB/X-gal agar (Fig. 5A, panel 1). In order to test the hypothesis that the missing C-terminal residues are responsible for the lack of interaction, we fused the last 13 C-terminal residues (EIGDQAEDDLGLE) present in T. elongatus KaiB to the L. pneumophila KaiB protein and called it KaiB-T (Fig. S2). Then we used L. pneumophila KaiB-T and KaiC fused to T18 and T25, respectively, and repeated the bacterial BACTH two-hybrid assay. Indeed, now interaction between KaiC and KaiB-T occurred (Fig. 5A, panel 2). As positive control, we used KaiC from S. elongatus fused to T18 and L. pneumophila KaiB-T fused to T25, respectively, which indeed interacted, similarly to the L. pneumophila KaiC/KaiB-T interaction (Fig. 5A, panel 3). The quantification of the obtained β-galactosidase levels for each interaction experiment is shown in Fig. 5B. Legionella KaiC and KaiB-T showed a similar level of β-galactosidase activity like that of Cyanobacteria (Fig. 5B). We then also tested whether Lpp1114 may interact with KaiC by fusing them to T18 and T25 respectively. However, no interaction was detected (data not shown). Thus our results suggest that neither L. pneumophila KaiB and KaiC nor KaiC and Lpp1114 interact.
Fig. 5.
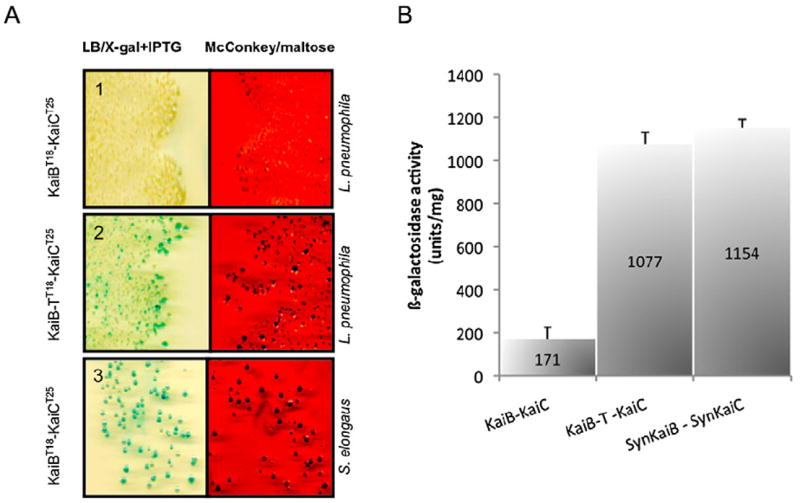
Bacterial two hybrid assay suggests that Legionella pneumophila KaiC and KaiB do not interact. The interaction test is based on the reconstitution of the adenylate cyclase activity in an Escherichia coli strain lacking endogenous adenylate cyclase.
A. KaiC and KaiB from L. pneumophila and Synechococcus elongatus were fused to T25 and T18-domain of the adenylate cyclase respectively. Interaction assay of KaiC with KaiB of L. pneumophila (Panel 1). Interaction assay KaiC with KaiB-T in which the C-terminal residues from T. elongatus are fused to L. pneumophila KaiB (Panel 2). Interaction assay with, KaiC from S. elongatus and KaiB from L. pneumophila used as a positive control showed interaction on the indicator media (Panel 3).
B. The β-galactosidase activity was measured for each combination of proteins. KaiB-T and KaiC from Legionella pneumophila display similar levels of β -galactosidase suggesting the importance of the C-terminal residues of KaiB for interaction with KaiC. The bars represent the mean values of three independent experiments ± standard deviation.
Legionella pneumophila KaiC conserves autokinase activity, but not circadian dephosporylation when KaiB is present
Bacterial proteins containing Walker motifs may display autokinase activity (Doublet et al., 1999). Cyanobacteria KaiC exhibits autokinase activity in vitro (Nishiwaki et al., 2000). Furthermore, KaiB of Cynaobacteria functions by binding the KaiC protein upon phosphorylation of the Ser431 residue (Nishiwaki et al., 2007; Rust et al., 2007; Qin et al., 2010), probably to the CII ring (Akiyama et al., 2008). Then a dephosphorylation phase follows and completes one 24 h cycle. Thus KaiB is an attenuator of phosphorylation that happens in a circadian manner (Kitayama et al., 2003). In L. pneumophila KaiC the Walker motifs and the phosphorylation residues are well conserved (Fig. 1B). To test whether L. pneumophila KaiC exhibits autokinase activity, a His6x-KaiC protein of L. pneumophila was expressed in E. coli, purified with Ni-affinity resin and incubated with [γ-32P] ATP in presence of Mg2+. As a control, we used S. elongatus His6x-KaiC. As shown in Fig. 6A, after incubation L. pneumophila and S. elongatus His6x-KaiC were indeed labelled, revealing that L. pneumophila KaiC exhibits autokinase activity. In Cyanobacteria the circadian clock can be reconstituted in vitro from the KaiA, KaiB, and KaiC proteins in the presence of ATP (Nakajima et al., 2005). We therefore tested whether KaiC of L. pneumophila can also be dephosphorylated by KaiB in vitro, and if the missing C-terminal residues in L. pneumophila KaiB play a role. We incubated L. pneumophila and S. elongatus KaiC with L. pneumophila KaiB or KaiB-T and analysed autophosphorylation, as well as dephosphorylation of KaiC. As reported previously, S. elongatus KaiC is dephosphorylated in a circadian rhythm; however, only when L. pneumophila KaiB-T was added, but not by wild-type L. pneumophila KaiB, further indicating that the C-terminal residues are essential and that KaiB of L. pneumophila does not interact with KaiC (Fig. 6B). For L. pneumophila KaiC instead, no change of the phosphorylation level over time was observed neither in presence of KaiB-T nor KaiB. Thus, our different analyses show that L. pneumophila KaiB and KaiC do not interact supporting a different function for these proteins.
Fig. 6.
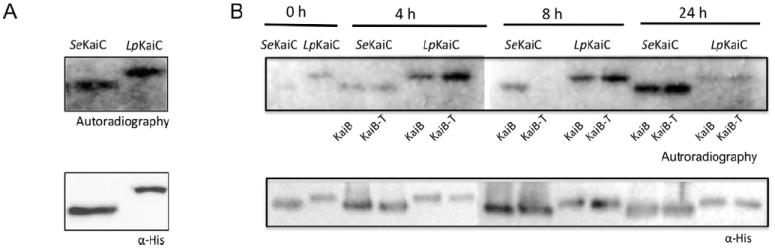
Legionella KaiC conserves autokinase activity but is not dephosphorylated by KaiB.
A. Western blot using an α-histidine antibody against the purified Legionella 6xHis-LpKaiC (65 kDa) and S. elongatus 6xHis-SeKaiC (58 kDa) proteins (lower panel). Autophosphorylation of SeKaiC and LpKaiC incubated with 5 μCi [γ-32P]-ATP for 1 h at 30°C (upper panel).
B. Kinetics of autophosphorylation and dephosphorylation of LpKaiC and SeKaiC in presence of KaiB or KaiB-T of L. pneumophila. Recombinant KaiC proteins were incubated with KaiB or KaiB-T, respectively and 5 μCi [γ -32P]-ATP at 30°C for 24 h. Samples were taken at 4 h, 8 h and 24 h of incubation and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis analyses. The phosphorylation state was detected by autoradiography (upper panel) and Western blot of the reaction mixtures with α-His as loading control (lower panel).
Legionella pneumophila ΔkaiC and Δlpp1114 are more sensitive to oxidative stress than the wild type
Legionella pneumophila lives under aerobic conditions and must deal with the toxic effect of oxygen that is generated in aquatic environments as well as when invading a eukaryotic cell. The structural analysis suggested that KaiB resembles a thioredoxin and the Lpp1114 protein, encoded in the same transcriptional unit contains a PAS domain that potentially senses intracellular redox changes. According to these findings, we hypothesized that the Kai operon may be implicated in regulating the oxidative stress response of L. pneumophila. First experiments on bacteria grown on buffered charcoal-yeast extract (BCYE) agar with different concentrations of hydrogen peroxide further substantiated this hypothesis, as the ΔkaiC mutant strain was less resistant than the wild-type strain (data not shown). In order to quantify and compare the oxidative stress response of the mutant and the wild-type strain, we first tested the sensitivity of L. pneumophila wild type to different concentrations of two oxidative reagents, H2O2 and paraquat. Legionella pneumophila wild type was highly sensitive to H2O2 in the medium as already 30 min of exposure to 1 mM H2O2 killed nearly all bacteria. In contrast, the sensitivity to paraquat was less pronounced as only 5 mM and 10 mM paraquat reduced bacterial survival significantly (Fig. 7A). We then selected two conditions for further testing the survival of the wild-type strains over longer exposure: 10 mM paraquat and 1 mM H2O2 (Fig. 7B). This showed that L. pneumophila is highly sensitive to 1 mM H2O2 as already after 10 min of exposure 80% of the bacteria were killed. However, as H2O2 does not seem to be stable in the medium, the remaining bacteria recovered and replicated. In contrast, a significant difference between the survival kinetics after exposure to 10 mM paraquat was observed with ΔkaiC and Δlpp1114 mutants showing less resistance to this oxidative stress than the wild-type strain (Fig. 7C, D). When complementing the ΔkaiC mutant with the entire operon on a plasmid (pMMB207_kaiOp) or Δlpp1114 mutant with lpp1114 (pMMB207_lppp1114) the oxidative stress resistance was similar to that of the wild-type strain (Fig. S3). Hence, our results indicate that the kai operon contributes to oxidative stress defense of L. pneumophila.
Fig. 7.
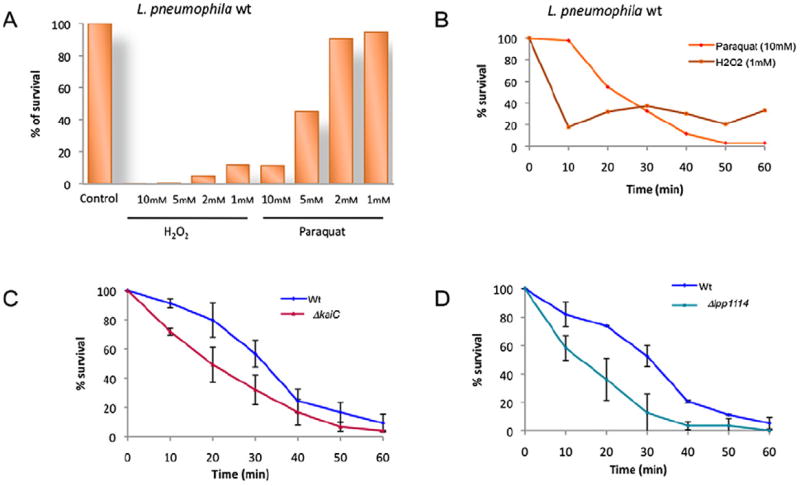
Legionella pneumophila ΔkaiC and Δlpp1114 mutants are sensitive to oxidative stress exerted by H2O2 and paraquat.
A. Percentage of wild-type L. pneumophila surviving at different concentrations of H2O2 and paraquat after 30 min of exposure.
B. Kinetics of the effect of 10 mM paraquat (purple) and 1 mM H2O2 (blue) on the survival of L. pneumophila wild type over 60 min C and D. Comparison of the survival kinetics of the wild type and ΔkaiC (C) or Δlpp1114 mutant strains (D), respectively: blue, wild-type strain; red ΔkaiC; cyan Δlpp1114. Each point represents the mean ± standard deviation value of at least two independent experiments.
To gain insight in the regulatory cascade implicated in oxidative stress response and to analyse where KaiC may act, we tested the effect of 10 mM paraquat in mutant strains that are thought to either regulate the kai operon genes like RpoS, or in genes homologous to other components in the circadian cascade. Apart from the KaiABC operon, CikA, SasA, RpaA, LabA and LdpA are implicated in this regulatory cascade in Cyanobacteria (Mackey et al., 2011). The best homologue of CikA (Gutu and O’Shea, 2013) in L. pneumophila is LetS (37% amino acid identity and e-value of 2e−43), the sensor kinase of the two component system LetA/LetS implicated in L. pneumophila virulence regulation, flagellar motility and cytotoxicity (Hammer et al., 2002; Molofsky and Swanson, 2004; Sahr et al., 2009). SasA and RpaA, the two-component system important for clock-control gene expression of S. elongatus (Taniguchi et al., 2007) have their best homologues (37% aa similarity) with StuC and CpxR of L. pneumophila respectively. stuC encodes a sensor histidine kinase protein and CpxR is a response regulator that can control the expression of different components of the Icm/Dot system (Altman and Segal, 2008). LabA and LdpA of S. elongatus are absent from L. pneumophila genome. Indeed, ΔrpoS and ΔletA are impaired for survival by the oxidative stress exerted by paraquat (Fig. S4), suggesting that LetA and RpoS are implicated in the oxidative stress response. In contrast, ΔstuC and ΔstuC/A are only very slightly affected (Fig. S4). Thus, we conclude that during oxidative stress the kai genes are regulated probably directly by RpoS and are influenced by LetA.
KaiC influences the expression of the alkyl-peroxidases ahpCD of L. pneumophila
To investigate the oxidative stress response of L. pneumophila at the transcriptional level, we first analysed the transcriptome of the L. pneumophila wild-type strain exposed to 10 mM paraquat as compared with growth in BYE medium without paraquat, using a whole genome microarray described previously (Albert-Weissenberger et al., 2010). This approach identified the significant induction of two genes, lpp2298 (31.2-fold) and lpp2299 (10.9-fold) encoding alkyl-peroxidases (ahpCD; Table 3). We confirmed this result by qRT-PCR where lpp2299 showed 74-fold up-regulation under oxidative stress conditions (data not shown). Other genes that showed up-regulation in this stress condition were feoA (lpp2712) with 2.9-fold change, feoB (lpp2711) with 1.4-fold change and frgA (lpp2846) with 2.8-fold change. The feo-operon is involved in the transport of ferrous iron into bacteria (Cartron et al., 2006) and has been described as critical for L. pneumophila growth in low-iron conditions, in host cells, and in the mammalian lung. FrgA has been described as an iron-regulated protein of L. pneumophila (Cianciotto, 2007; Hickey and Cianciotto, 1997; Robey and Cianciotto, 2002). Furthermore, two genes similar to pvcA (lpp0236) and pvcB (lpp0237) showed a fold change of 1.8 and 1.5 respectively. PvcA and PvcB are required for synthesis of pyoverdine chromophore, they serve as signalling molecules controlling gene expression inside the cell and have been reported to be induced in biofilm and regulated by iron concentrations available (Stintzi et al., 1999; Hindre et al., 2008; Table 3). Thus the general transcriptional response of L. pneumophila to oxidative stress leads to the induction of two alkyl-peroxidases and iron related genes.
Table 3.
Genes up-regulated in Legionella pneumophila after exposure to 10 mM paraquat (15 min).
| Gene ID | Product | FC |
|---|---|---|
| lpp2299 | Similar to alkyl hydroperoxide reductase AhpC | 31.177** |
| lpp2298 | Similar to alkyl hydroperoxide reductase AhpD | 10.945** |
| lpp2712 | Ferrous iron transporter A (FeoA) | 2.918 |
| lpp2846 | FrgA protein | 2.797 |
| lpp2780 | Similar to transcriptional regulator- ArsR family | 2.796 |
| lpp2711 | Ferrous iron transporter B (FeoB) | 1.365 |
| lpp0237 | Similar to pyoverdine biosynthesis protein PvcB | 1.810 |
| lpp0236 | Similar to pyoverdine biosynthesis protein PvcA | 1.525 |
Significantly differentially regulated genes are marked by an asterisk.
P < 0.001.
We then compared the transcriptional profile of the ΔkaiC and Δlpp1114 mutants to L. pneumophila wild type after exposure to 10 mM paraquat for 15 min as this was the time point where the mutants showed the highest sensitivity (Fig. 7C,D). We found that ahpC and ahpD were slightly down-regulated in both mutants (Tables 4 and 5). Furthermore, coproporphyrinogen oxidase A (lpp2849), a nicotinamide adenine dinucleotide plus hydrogen (NADH)-ubiquinone oxidoreductase (lpp0727) and fliA, coding for the sigma factor 28, were slightly down-regulated in the ΔkaiC strain (Table 4). qRT-PCR analysis of the alkyl-peroxidase encoding genes ahpC1 and ahpC2D (lpp3037 and lpp2298, lpp2299) during post-exponential growth phase under the effect of 10 mM paraquat confirmed the microarray results (data not shown). Furthermore, the genes encoding FrgA and a NADH-dependent flavin oxidoreductase (lpp2779) were down-regulated in the Δlpp1114 strain (Table 6). qRT-PCR of the Δlpp1114 strain confirmed the reduction of the alkyl-peroxidases AhpCD and also showed a down-regulation of the expression of the kai genes (data not shown). Thus, the expression of the alkyl-peroxidase operon and of other genes possibly related to the oxidative stress response of L. pneumophila is modulated by KaiC.
Table 4.
Genes down-regulated in Legionella pneumophila ΔkaiC compared with the wild type after exposure to 10 mM paraquat (15 min).
| Gene ID | Product | FC |
|---|---|---|
| lpp2299 | Similar to alkyl hydroperoxide reductase AhpC | 0.646 |
| lpp2298 | Similar to alkyl hydroperoxide reductase AhpD | 0.693 |
| lpp2141 | Global stress protein GspA | 0.71 |
| lpp1115 | KaiB – like circadian clock protein | 0.715 |
| lpp1116 | KaiC – like circadian clock protein | 0.745 |
| lpp2461 | Unkown | 0.75 |
| lpp1942 | Unkown | 0.756 |
| lpp1746 | Sigma factor 28 (fliA) | 0.758 |
| lpp0727 | Similar to nicotinamide adenine dinucleotide plus hydrogen-ubiquinone oxidoreductase | 0.764 |
| lpp1208 | Similar to protein | 0.742 |
| lpp2849 | Similar to putative coproporphyrinogen oxidase A | 0.768 |
Table 5.
Genes down-regulated in Legionella pneumophila Δlpp1114 compared with the wild type after exposure to 10 mM paraquat (15 min).
| Gene ID | Product | FC |
|---|---|---|
| lpp0623 | Unknown | 0.482* |
| lpp0988 | Unknown | 0.582* |
| ncRNA20 | Non coding small RNA | 0.582* |
| lpp1025 | Unkown | 0.622 |
| lpp2298 | Similar to alkyl hydroperoxide reductase AhpD | 0.634 |
| lpp2504 | Unknown | 0.642 |
| lpp2846 | FrgA protein | 0.647 |
| lpp1957 | Unknown | 0.667 |
| lpp0391 | Translation elongation factor G (fusA) | 0.672 |
| lpp0805 | Similar to surface antigens (17 kDa) | 0.676 |
| lpp1440 | Weak similarity to myosin | 0.68 |
| lpp2721 | RNA polymerase sigma-32 factor (rpoH) | 0.684 |
| lpp2779 | Similar to NADH-dependent flavin oxidoreductase | 0.721 |
Significantly differentially regulated genes are marked by an asterisk.
P < 0.05.
Table 6.
Genes up-regulated in Legionella pneumophila Δlpp1114 compared with the wild type after exposure to10 mM paraquat (15 min).
| Gene ID | Product | FC |
|---|---|---|
| plpp0015 | Unknown | 2.570* |
| plpp0027 | Hypothetical gene | 2.623* |
| plpp0018 | Unknown | 2.759* |
| plpp0017 | Similar to Escherichia coli TraT complement resistance protein | 2.910* |
| plpp0016 | Weakly similar to carbon storage regulator CsrA | 2.981* |
| lpp2431 | Similar to conserved hypothetical protein | 3.002* |
| lpp2430 | Unknown | 3.039* |
| plpp0022 | Similar to fimbrial protein precursor (Pilin) | 3.321* |
| lpp2884 | Similar to transposase (IS21 family) | 3.555* |
| plpp0024 | Weakly similar to conjugative transfer protein TraE | 3.566* |
| lpp2434 | Unknown | 3.583* |
| plpp0020 | Similar to transposase | 3.601* |
| plpp0023 | Weakly similar to conjugative transfer protein TraL | 3.623* |
| lpp2885 | Similar to transposase (IS21 family) | 3.630* |
| plpp0021 | Similar to plasmid DNA replication protein | 3.859* |
| plpp0023 | Unknown | 2.570* |
Significantly differentially regulated genes are marked by an asterisk.
P < 0.05.
ΔkaiC and Δlpp1114 mutants are more sensitive to sodium stress than the wild type
To further study the implication of the kai operon genes in the L. pneumophila stress response we analysed the phenotype of the mutant strains under salt stress. It is known that in broth L. pneumophila is sodium sensitive only during post exponential growth (Catrenich and Johnson, 1989). It has been shown that sodium sensitivity is also correlated with virulence of L. pneumophila as during the intracellular replication period, this bacterium is sodium resistant and lacks flagella (Bachman and Swanson, 2001; Molofsky and Swanson, 2004). Concomitant with macrophage lysis, L. pneumophila becomes sodium sensitive and flagellated (Byrne and Swanson, 1998). In order to test whether the Kai proteins are implicated in regulating this distinct phenotype, we grew the wild-type and the mutant strains ΔkaiC and Δlpp1114 to post exponential growth phase (OD 3.8) and spotted 10 μl of serial dilutions (non diluted to 10−5) of these cultures on BYCE agar plates containing 100 mM NaCl or not. Indeed, both mutants were more sensitive to sodium stress than the wild-type strain, as no growth of the mutant strains could be observed under salt stress above a dilution of 10−3. In contrast the wild-type strain grew on plates containing 100 mM NaCl up to a dilution of 10−5 (Fig. 8) indicating that the kai operon genes enhance sodium resistance of L. pneumophila in post exponential phase.
Fig. 8.
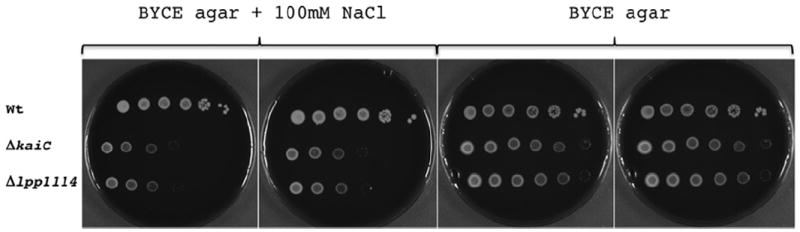
The ΔkaiC and Δlpp1114 mutant strains display a higher sensitivity to 100 mM NaCl than wild-type Legionella pneumophila. Legionella pneumophila wild-type and mutant strains were grown to OD 3.8 in buffered yeast extract (BYE) medium and 10 μl of serial dilutions (non diluted to, 10−5) were spotted onto agar containing or not 100 mM NaCl. Plates were incubated for 3 days at 37°C. Representative picture of three independent experiments.
Evolution of the KaiB and KaiC proteins in L. pneumophila
A Basic Local Alignment Search Tool (BLAST) search identified homologs of the kaiC, kaiB and lpp114 genes in all eight published L. pneumophila genome sequences. However, in L. pneumophila Lens, the kaiC gene contains a frameshift leading to a truncated gene and KaiC of L. pneumophila Alcoy has another start codon leading to a shorter protein, but the two walker motifs are conserved. Furthermore, we examined a collection of 41 L. pneumophila isolates belonging to the 15 serogroups (our unpublished data). The kai operon genes were missing in three of the 41 strains and six isolates had a frameshift in the operon sequence. Analyses of the genome sequences of five L. longbeachae strains (Cazalet et al., 2010; Kozak et al., 2010), one L. drancourtii strain (Moliner et al., 2010), two L. dumoffii strains (Qin et al., 2012) and one L. anisa strain (unpublished), and the genomes of L. hackeliae, L. micdadei and L. fallonii (our unpublished data) revealed that L. hackeliae contains the kai operon and that L. fallonii contains the kaiB and lpp1114 genes but no kaiC gene, whereas the entire operon is absent from the remaining five Legionella species. This heterogeneous distribution and the mutations present in the operons might point to the fact that this operon is on an evolutionary path to be lost from Legionella.
To get further insight into the evolution of the Kai operon proteins of L. pneumophila we undertook different phylogenetic analyses. Recently Wiegard and colleagues (2013) published the first functional analyses of paralogous KaiBC genes in Synechocystis that encodes three copies of KaiB and KaiC in its genome, showing that the paralogous KaiB2C2 genes do not seem to encode a circadian clock (Wiegard et al., 2013). To learn whether the L. pneumophila Kai proteins are related to one of the paralogous families or to the genuine Kai clock proteins of Cyanobacteria, we undertook a phylogenetic analysis including all paralogues of KaiC and KaiB that may be present in different prokaryotic genomes. We selected the representative Kai protein sequences published by Dvornyk and colleagues (2003) and included the Legionella Kai sequences. Interestingly, KaiC of L. pneumophila groups with the Synechocystis KaiC2 (sll1595) protein and not with the KaiC1 sequence that is part of the genuine circadian clock (Fig. 9).
Fig. 9.
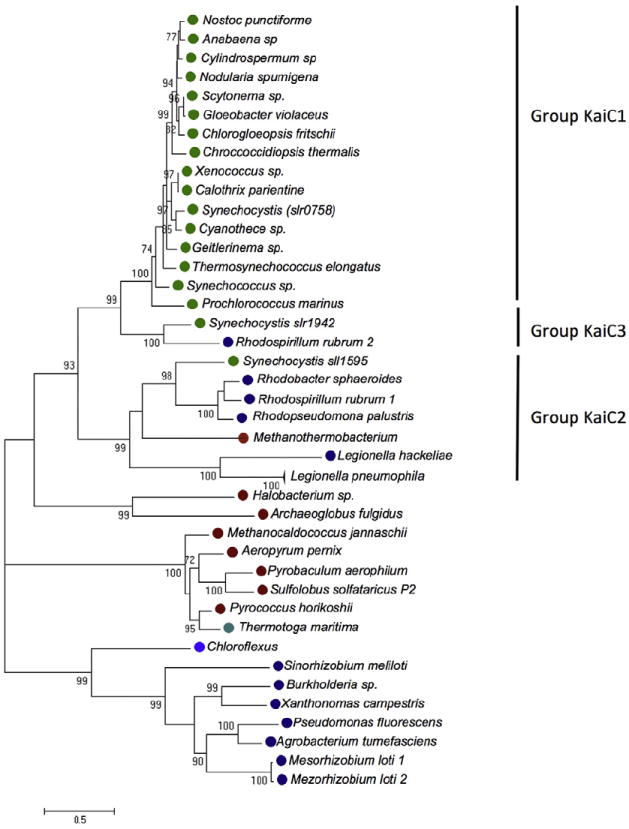
Phylogenetic tree of KaiC and its paralogues in selected organism. The tree was constructed using representative KaiC sequences as published previously (Dvornyk et al., 2003) and the KaiC sequence of Legionella pneumophila was included. KaiC of L. pneumophila groups with KaiC2 of Synechocystis. Cyanobacteria, green circles; Proteobacteria, dark blue circles; Archaea red circles; Chloroflexus, blue circles; Termotogae, cyan circles.
We then undertook phylogenetic analyses of L. pneumophila KaiB, that shares 97% amino acid similarity with KaiB of Cyanobacteria (Fig. S1). kaiB genes are present only in prokaryotic organisms that encode a kaiC gene next to it, but not all prokaryotes that contain kaiC contain kaiB as also observed previously (Dvornyk et al., 2003). We constructed the phylogenetic KaiB tree by analysing one representative KaiB protein of each prokaryote that contains it. However, given the short sequence of the KaiB protein and the high conservation of the core residues 8–94 of S. elongatus (Iwase et al., 2005), no strong evolutionary conclusion can be drawn and only few nodes of the KaiB tree have significant bootstrap support (Fig. S5). Thus we concatenated the KaiB and KaiC sequences to obtain a better resolved and supported tree. As shown in Fig. 10 the Legionella KaiCB sequences group with the KaiB2C2 genes (sll1595, sll1506) of Synechocystis and of other Proteobacteria, like KaiC alone (Fig. 9). In both phylogenetic trees (Fig. 9 and 10) the Legionella and Synechocystis KaiCB2 proteins cluster. Interestingly, when analysing the neighborhood of KaiCB of each organism it became evident that in four out of six Proteobacteria belonging to the KaiC2B2 cluster, a PAS-domain containing protein is encoded next to the kai genes like in L. pneumophila. This conserved organization suggests a link between this protein and the Kai proteins and further supports a putative role of this operon in the response to environmental signal(s).
Fig. 10.
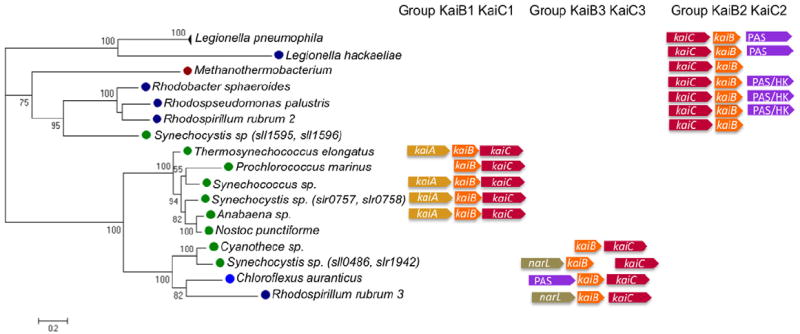
Phylogenetic tree of concatenated KaiCB sequences identifies three different groups of KaiBC systems. The tree was constructed using the same representative sequences as published by Dvornyk and colleagues (2003) with the Legionella sequences included. The Kai proteins of Legionella group with KaiC2B2 from Synechocystis (Wiegard et al., 2013), Cyanobacteria, green circles; Proteobacteria, dark blue circles; Archaea, red circles; Chloroflexus, blue circles. The right panel shows the chromosomal organization of the three different groups of kaiB and kaiC as described for the three copies of KaiC and KaiCB of Synechocystis. kaiB1C1 are the orthologous genes of the circadian genes in S. elongatus; kaiB2C2 does not seem to be involved in circadian functions; KaiB3C3 were reported to have a putative involvement in fine-tuning parameters of the circadian clock.
Taken together, our phylogenetic analyses suggest that the kai genes of L. pneumophila have been acquired by horizontal gene transfer, but probably the origin is not the operon encoding the circadian clock named KaiB1C1 in Synechocystis, but the paralogous cluster KaiC2B2 for which no clock function has been demonstrated yet.
Discussion
Bacteria are facing changing environments to which they need to adapt to survive. To respond they use signal transduction pathways that sense external or internal signals. These are processed to generate an output response that can lead to changes in gene expression, cell movement, cell growth or others. Many organisms can also generate an oscillatory response, in which the output varies over time in a repetitive manner. A striking example of such an oscillatory response is the circadian regulation of genes for oxygenic photosynthesis and nitrogen fixation in some Cyanobacteria (Mackey and Golden, 2007). This oscillatory response is regulated in Cyanobacteria by the KaiABC genes that are extensively studied in S. elongatus. Legionella pneumophila resides in aquatic reservoirs within biofilms or protozoa and encounters many different environments when it cycles between hosts. It is not known to possess circadian gene regulation or to have the capabilities of photosynthesis or nitrogen fixation. However it encodes KaiBC proteins in its genome (Gomez-Valero et al., 2011).
In this study we show that the three Kai operon genes (kaiB, kaiC and lpp1114) of L. pneumophila Paris do not seem to be related to a circadian clock. This finding is in line with our phylogenetic analysis that shows that L. pneumophila KaiBC cluster with KaiB2C2 of Synechocystis sp., a paralogous copy of KaiC1B1 that does not seem to be involved in circadian rhythms (Wiegard et al., 2013; Figs. 9 and 10). Furthermore, KaiB2 and KaiC2 do not interact (Wiegard et al., 2013). The close phylogenetic relationship between KaiC2 of Synechocystis sp. and L. pneumophila KaiC is also seen in the alignment of the three KaiC copies of Synechocystis sp. with L. pneumophila KaiC. The two main phosphorylation residues in the C-terminal region of L. pneumophila KaiC are serine/serine residues like in Synechocystis KaiC2, and not serine/threonine (S431 and T432 in Synechococcus sp.) like in KaiC1 and in other Cyanobacteria (Fig. S6). Thus, KaiBC of L. pneumophila seem to be different from the canonical circadian Kai system described for many Cyanobacteria.
Our sequence comparison and the structure of KaiB that we solved revealed that the C-terminal amino acids that are critical for the KaiB – KaiC interaction in Cyanobacteria are missing in L. pneumophila KaiB (Fig. 4). Indeed, the L. pneumophila proteins do not interact neither in a yeast-nor a bacterial two-hybrid system (Fig. 5). In contrast, a fusion protein of L. pneumophila KaiB with the 13 C-terminal residues (EIGDQAEDDLGLE) of KaiB of the Cyanobacterium Thermosynechococcus elongatus that we named KaiB-T interacts with L. pneumophila and S. elongatus KaiC. This result confirms the prediction obtained from the structure of L. pneumophila KaiB and shows that the L. pneumophila proteins do not interact and that the missing C-terminal residues are responsible. We thus reasoned that the KaiBC proteins of L. pneumophila might not encode a circadian clock, and that L. pneumophila does not show circadian gene expression regulated by the Kai proteins. Indeed, monitoring of the kai operon gene expression during 5 days of intracellular growth of L. pneumophila in A. castellanii as well as during in vitro growth in BYE medium, revealed no circadian, but growth phase-dependent gene expression with the highest expression in stationary growth phase.
The question arises what is the function of these proteins in L. pneumophila. One hint came from the KaiB structure that we solved. KaiB seems to be the most closely related to thioredoxin. Thus we hypothesized that these proteins in L. pneumophila might be involved in oxidative stress response. The knowledge concerning the genes involved in oxidative stress resistance and its regulation in Legionella is rather limited. The antioxidative defence genes of L. pneumophila described to date, are the Cu/Zn and Fe/Mn dependent superoxide dismutase, the katalase peroxidases katA and katB, or the alkyl hydroperoxide reductases ahpC1 and ahpC2D, whose expression is growth phase dependently regulated (St John and Steinman, 1996; Bandyopadhyay and Steinman, 2000; Bandyopadhyay et al., 2003; Bruggemann et al., 2006; LeBlanc et al., 2006). However, most of the regulators of the oxidative stress response pathways known in other bacteria such as SoxRS, OhrR and PerR are missing in L. pneumophila. Only a homologue of OxyR has been described (LeBlanc et al., 2008). In Pseudomonas fluorescens, a related bacterium found in aquatic environments, it is shown that the oxidative stress resistance pathway is controlled by a complex regulatory cascade, including the two-component system GacS/GacA, the mRNA binding protein RsmA, RpoS and a small RNA (rgsA; Heeb et al., 2005; González et al., 2008). A similar pathway exists in L. pneumophila, including sigma factors (RpoN, RpoS, FliA), the LetS/LetA (GacA/GacA) two-component system, the mRNA binding protein CsrA (RsmA) and two small RNAs (rsmY, rsmZ; Molofsky and Swanson, 2004; Sahr et al., 2009; Faucher et al., 2011). This pathway controls the entry of bacterial cells into the stationary or transmissive phase, which is thought to promote infection of a new host cell. It is thus tempting to speculate that this regulatory pathway in L. pneumophila may also be involved in the oxidative stress response and that KaiCB are part of this cascade.
Indeed, we show that the expression of the kai operon genes is under control of the stress sigma factor RpoS and that the ΔkaiC and Δlpp1114 strains are more sensitive to oxidative stress induced by paraquat than the wild type (Fig. 7). The effect exerted by paraquat in the kai mutants is similar or even stronger than that observed in an ΔrpoS and ΔletA mutant strain (Fig. S5). Additionally, the levels of transcription of the kai genes are reduced in the ΔletA mutant. LetA as well as RpoS respond to the alarmone ppGpp (Zusman et al., 2002; Dalebroux et al., 2010) that is required to induce the transmission phenotype in stationary phase (Bachman and Swanson, 2001; Hammer et al., 2002). In a ΔletA background, L. pneumophila is not able to express transmissive traits, including RpoS (Sahr et al., 2009). Thus, RpoS regulates directly the sensitivity to oxidative stress of ΔkaiC and Δlpp1114 and LetAinfluences the stress response indirectly. Furthermore, in response to 10 mM paraquat L. pneumophila induces the genes encoding the alkyl hydroperoxide reductases AhpC2D. To our knowledge, this is the first report of an activation of AhpC2D in response to oxidative stress. It was reported that OxyR regulates the expression of the ahpCD operon; however, we could not detect any difference in its transcription during oxidative stress due to paraquat. Interestingly, the levels of transcription of pvcA and pvcB, two proteins regulated by iron concentrations was increased during oxidative stress conditions, similar to what was seen in biofilm, where the alkyl hydroperoxidases were up-regulated together with the PvcA and PvcB (Hindre et al., 2008). In contrast, in the ΔkaiC mutant strain, the transcriptional levels of the alkyl hydroperoxides was diminished further suggesting that the kai genes are implicated in oxidative stress response. In addition to their implication in oxidative stress resistance, kai operon mutants showed also reduced growth in presence of 100 mM NaCl compared with the wild type. Interestingly, in Arabidopsis thaliana, the circadian clock associated protein LIP1, regulates light input to the clock and is required for tolerance to salt stress. However the mechanism is not understood yet (Terecskei et al., 2013). Another example of circadian clock proteins linked with salt stress is AqpZ an aquaporine regulated by daily oscillations of intracellular osmolarity in Synechocystis sp. (Akai et al., 2012).
In L. pneumophila, the kaiBC genes are in an operon with lpp1114 encoding a PAS (period circadian protein – aryl hydrocarbon receptor nuclear translocator protein – single-minded protein)-domain containing protein. PAS-domains are found from bacteria to humans, and are described to play a role in sensing changes in the environment. Furthermore, PAS proteins are related to the circadian clock of eukaryotic organisms (McIntosh et al., 2010). Thus, it seems likely that Lpp1114 senses via its PAS domain the input signal that activates the Kai proteins. The PAS domain encoded by Lpp1114 shares 70% similarity with the LOV domains of YtvA (a blue light GTP-binding receptor) from Bacillus subtilis (Jurk et al., 2011) and one from Listeria monocytogenes (lmo0799; Chan et al., 2013). LOV domains are a subset of the larger Per-ARNT-Sim (PAS) domain superfamily (Herrou and Crosson, 2011). The second class, LOV-GGDEF/EAL proteins are predicted to regulate the synthesis and hydrolysis of cyclic-di-GMP and constitute around 20% of bacterial LOV proteins. Lpp1114 might belong to this class. Interestingly, in many organisms, a PAS-domain encoding protein corresponding to this class is located adjacent to KaiC (Fig. 9). Although there is a lack of data on the role of light in the life cycle of L. pneumophila, light might function as an environmental signal that stimulates L. pneumophila in the extracellular environment to up-regulate pathways that promote infection of a new host. However, further studies are needed to determine if it plays a role in light/redox sensing. Surprisingly, the kaiC gene does not affect significantly its capacity to kill A. castellanii although the capacity to parasitize protozoa is thought to be the main force driving evolution of L. pneumophila. Thus, the acquisition of the kai operon genes may not be directly related to protozoan parasitism but may increase fitness of L. pneumophila in environments containing reactive oxygen species, e.g. in sunlight water ponds or water systems.
Taken together, our findings indicate that the Kai proteins of L. pneumophila do not encode a circadian clock but resemble the KaiB2C2 proteins of Synechocystis sp. and are involved in stress response. Phylogenetic analyses suggest that they have been acquired by horizontal gene transfer from Cyanobacteria, but it seems likely that they have diverged during evolution.
Material and methods
Bacterial strains and growth conditions
Legionella pneumophila strain Paris and derivates were grown on BCYE agar or in ACES (N-(2-acetamido)-2-aminoethanesulfonic acid)-BYE broth. Legionella pneumophila was routinely grown on BCYE agar for 2–3 days at 37°C. For growth in liquid medium, L. pneumophila was inoculated at an OD 0.5–0.8 at 37°C under continuous shaking. Bacterial growth was monitored by measuring the optical density (OD) of the culture at a wavelength of 660 nm using a spectrophotometer (Eppendorf France S.A.S., Le Pecq, France). E. coli strains were grown in Luria-Bertani (LB) broth or LB-agar. When necessary antibiotics were added at the following final concentrations: ampicillin at 100 μg ml−1 for E. coli; kanamycin at 12.5 μg ml−1 for L. pneumophila and 50 μg ml−1 for E. coli; chloramphenicol at 10 μg ml−1 for L. pneumophila, at 20 μg ml−1 for E. coli; apramycin at 15 μg ml−1 for L. pneumophila. Recombinant expression of the KaiC and KaiB proteins was done in E. coli BL21 (DE3).
DNA manipulation and construction of mutants
The ΔkaiC mutant was obtained by replacing the kaiC gene with a kanamycin cassette and the Δlpp1114 mutant by replacing lpp1114 with an apramycin cassette. Legionella pneumophila ΔkaiC mutant strain was constructed as described previously (Bruggemann et al., 2006). To construct the ΔkaiC mutant strain, the chromosomal region containing the kaiC gene was PCR-amplified using the primers KaiC-F ATCGCATCAACCCCTCCTCATT and KaiC-R TGCCAACAAGCTCTGTATGGGG. The product was cloned in the pGEM-T easy vector (Promega Corporation, Madison, WI, USA) and on the resulting plasmid an inverse PCR was performed using the primers KaiC_inv_BamHI-F CGGGATCCCGCATATC CCTTAATTCC and KaiC_inv_BamHI_R CGGGATCCCG CTCTTGAATCTTGAGTTGAGCACG. The PCR product was digested using BamHI enzyme and ligated with a BamHI-digested kanamycin resistance cassette. The Δlpp1114 was constructed by a three-step PCR as previously described (Rolando et al., 2013). In the first step, we amplified the apramycin-resistant cassette and the upstream and downstream sequence of the lpp1114, flanked by short overlapping regions of the apramycin cassette next to the insertion site. For the upstream sequence we used the primers Lpp1114_mut-for TCGT AAGCTGTCTATCCCAGAG and Lpp1114_inv_rev GCT GATGGAGCTG CACATGAATGCTCCAGAATGGGGAG AAAACC, for the downstream sequence Lpp1114_inv_for GAGCGGATCGGGGATTGTCTTTCCGGATCGATGATG ATAA TGC and Lpp1114_mut-rev AACAAGGGGATA GCAGAACAAG. In a second step, the full-length recombinant DNA was synthesized by combining the three PCR fragments obtained in step one. For complementation experiments, the entire operon, genes kaiC, kaiB and lpp1114, was cloned in the vector pMMB207c. As control L. pneumophila Paris wild type and the kaiC mutant containing the empty plasmid pMMB207 were used respectively. Mutants and plasmids used in this study are listed in Table 7. E. coli DH5α and BL21 (DE3) were used for amplification of recombinant plasmid DNA with backbones: pMMB207, pGEM-T easy vector (Promega), pGEX vector (GE Healthcare en France, Aulnay sous bois, France), pQE30 (QIAGEN Sciences, Germantown, MD, USA), TOPOXL (Invitrogen, Life Technologies SAS, Saint Aubin, France). Genomic and plasmid DNA were prepared, amplified and sequenced according to standard procedures. Nucleotide and protein sequence were analysed using NCBI website (http://www.ncbi.nlm.nih.gov/), ExPAsy (http://www.expasy.ch), T-Coffee (http://www.tcofee.org) and DNA sequence assembly (Gap4).
Table 7.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Properties | Reference |
|---|---|---|
| Strain | ||
| Escherichia coli | ||
| DH5a | F-φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rk-. mk+) phoA supE44 thi-1 gyrA96 relA1 λ- | Invitrogen |
| BL21 | F-ompT hsdSB (rB-mB-) gal dcm rne131 (DE3) | Invitrogen |
| Legionella pneumophila | ||
| CIP 107629 | L. pneumophila strain Paris | Cazalet etal., (2004) |
| ΔkaiC | Paris lpp1116:Km | This study |
| Δlpp1114 | Paris lpp1114:Apm | This study |
| ΔkaiC/pkaiOp | Paris lpp1116:Km carrying plpp1116/lpp1115/lpp1114 | This study |
| Δlpp1114/plpp1114 | Paris lpp1114:Ap carrying plpp1114 | This study |
| Plasmids | ||
| pGEM-T Easy | Cloning of PCR products (Ampr) | Promega |
| TOPO XL | Cloning of long PCR products (> 3 kb; Kmr) | Invitrogen |
| pMMB207C | Legionella expression vector (Cmr) | Chen et al. (2004) |
| pkaiOp | pMMB207C containing the kai operon | This study |
| plpp1114 | pMMB207C containing gene lpp1114 | This study |
| pQE30 | Expression vector N-terminal His6x tag | Qiagen |
| pKNT25 | Expression vector N-terminal T25 (Kmr) | Euromedex |
| pUT18 | Expression vector N-terminal T18 (Apr) | Euromedex |
| pGEX | Expression vector N-terminal GST-tag (Amr) | GE healthcare |
Amp, ampicillin; Ap, apramycin resistance; Cm, chloramphenicol; Km, kanamycin resistance; PCR, polymerase chain reaction.
Intracellular growth in A. castellanii
Intracellular growth of L. pneumophila in A. castellanii was done as previously described (Lomma et al., 2010). Briefly, 3-day-old cultures of A. castellanii incubated at 20°C in PYG 712 medium (2% bactotryptone, 0.1% yeast extract, 0.1 M glucose, 4 mM MgSO4, 0.4 M CaCl2 0.1% sodium citrate dihydrate, 0.05 mM Fe(NH4)2(SO4)2 × 6H2O, 2.5 mM NaH2PO3, 2.5 mM K2HPO3) were washed using infection buffer (PYG 712 medium without tryptone, glucose and yeast extract) seeded in 25 cm2 flasks and adjusted to 105 cells per ml. Legionella pneumophila grown on BYCE agar and diluted in PBS and added to the flask containing A. castellanii at multiplicity of infection (MOI) of 0.1 or 100. After an hour of incubation at 37°C or 20°C to allow the invasion of L. pneumophila, attached A. castellanii were washed carefully twice with infection buffer. Intracellular replication was monitored by taking 300 μl of sample at each time point. To lyse the amoeba, 300 μl the cells were centrifuged (3 min, 14 000 rpm) and vortexed and 100 μl were plated on BYCE agar in serial dilutions.
Competitive assay during infection of A. castellanii
For competitive infection assays, L. pneumophila strains Paris wild type and the ΔkaiC or Δlpp1114 mutants or the wild-type strain carrying the empty plasmid and the complemented strains ΔkaiC, respectively, were compared. The competition assay was done as previously described (Kessler et al., 2013). Briefly, L. pneumophila and the respective competitor were both grown to stationary phase on BYCE agar and mixed in equal ratios adjusted to OD 1 in PBS. This mix of bacteria was added to the flask containing A. castellanii prepared as described earlier at an MOI of 0.1. After 1 h invasion at 37°C the layer of A. castellanii was washed twice with infection buffer. To monitor the total number of bacteria, a 300 μl sample was taken, centrifuged to 14 000 rpm, vortexed and plated on BYCE agar with and without corresponding antibiotic for bacterial selection. After 2 days of incubation at 37°C, a second sample was taken and seeded in BCYE agar plates and BCYE agar plates containing selective antibiotic. Additionally, new 3-day-old cultures of A. castellanii in PYG 712 medium were washed twice and adjusted to 105 cells per ml and 100 μl of 1 × 10−3 dilution of the bacteria sample was added to the flask starting a new round of infection. Again after 2 days of incubation the same procedure described earlier was performed.
RNA isolation and labelling for array hybridization
For in vitro-grown L. pneumophila, total RNA for array hybridization and real-time PCR was extracted as described previously (Sahr et al., 2012). For total RNA extraction of L. pneumophila growing intracellularly in A. castellanii, an additional FastPrep step was added to the protocol prior to RNA extraction as described previously (Bruggemann et al., 2006). RNA was reverse-transcribed and indirectly labelled with Cy5 and Cy3 (GE Healthcare) by using the cDNA Superscript Indirect Labeling Kit (Invitrogen).
L. pneumophila genome microarray and hybridization experiments
For transcriptional experiments, microarrays for L. pneumophila were used as previously described (Bruggemann et al., 2006). Array hybridization was performed using 250 pmol of labelled cDNA. At least, two biological replicates were performed for each condition including dye swap. Microarray slides were scanned using GENEPix 4200 AL scanner (Axon Instruments, Molecular Devices, LLC, Sunnyvale, CA, USA) and the laser power was adjusted to balance the two channels. The images were analysed using the GENEPIX PRO 6.0 software. For statistical analysis the R software was used (http://www.r-project.org/) and the microarray data was normalized using a loess based method using the BioConductor package marray (http://www.bioconductor.org/packages/bioc/stable/src/contrib/html/marray.html). The gene names in the tables (lpp) refer to the strain Paris, available at the LegioList web server (http://genolist.pasteur.fr/LegioList).
Real-time PCR
qRT-PCR was performed to confirm transcriptome results using the total RNA in the same conditions. After DNase treatment, cDNA synthesis was performed with 5 μg RNA and reverse transcriptase (400 U μl−1, Invitrogen) according to the manufacturer’s instructions for 2–3 h at 42°C. The experiment was done in 20 μl reaction volume containing 1.2 ng of cDNA, 10 μl SYBR PCR master mix (Applied Biosystems, Life Technologies SAS) and gene-specific primers (300 nM; Table S2). Each sample was done in triplicate and with different cDNA dilutions (non diluted, 1:10, 1:100, 1:1000). The quantity of cDNA for each tested gene was normalized to the constitutively expressed genes, letA and tldD. The relative gene expression was calculated as the ratio normalized target concentrations (ΔΔCt) using the REST software (http://www.gene-quantification.de/rest.html). Primers were designed using the PRIME3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) leading to amplification products in a size range of 150–250 bp. The amplification was performed using an ABI Prism 7900 HT sequence detection system (PE Applied Biosystems).
Oxidative stress induced by 10 mM paraquat
Legionella pneumophila and its derivates were grown in 50 ml BYE medium until exponential growth (OD 2.5). The cultures were adjusted to OD 1.0 and two aliquots of 5 ml of this culture were taken, centrifuged at 5000 g for 5 min at room temperature and the pellet was resuspend in 5 ml of fresh BYE medium. A final concentration of 10 mM of Paraquat (Sigma-Aldrich, Lyon, France) was added and incubated for 1 h at 37°C under shaking at 250 rpm. Every 10 min a sample of 300 μl was taken and serial dilutions were plated on BYCE to estimate the number of bacterial survival. In parallel, the second aliquot was recovered after 15 min of incubation with 10 mM paraquat, and the RNA was extracted as described earlier, for further transcriptional analysis.
Sodium sensitivity
Sodium sensitivity was tested with L. pneumophila grown to post-exponential growth (OD 3.8) in liquid broth. Ten microliters of the culture was pipette in serial dilutions on BCYE agar plates containing or lacking 100 mM NaCl.
Two-hybrid experiments to test protein–protein interaction
We used a yeast two-hybrid approach based on the Cytotrap vector kit (Stratagene, Agilent Technologies, Inc., Life Sciences and Chemical Analysis Group, Santa Clara, CA, USA) according to the manufactures instructions. In this method KaiB was used as bait and KaiC. Briefly, kaiB and kaiC were cloned in frame into the pSos and pMyr vectors, respectively, and expressed as fusion protein. hSos-kaiB was obtained by amplifying kaiB using the primer pSos-KaiB-FAGGATCCCCATGGCCATGACTAAG CTAAAGCTG and pSos-KaiB-R GTTATTTCTGCTTTGG AAATGGATTAGACGTCGACGCGCGCA. The product was cloned in pSos; pMyr-kaiC was obtained by amplifying kaiC using the primers pMyr-KaiC-F GCCCGGGC CTCGAGGGTGAATACCAATAAAGCAAAAACGGGA and pMyr-KaiC-R GAGTCCAACAAATTGTTAAGAGAGTAA TCGAGGTCGACTAAT and the product was cloned in pMyr fusing kaiC to a myristoylation sequence at the N-terminus. Both fusions were co-expressed in the Saccharomyces cervisae mutant strain cdc25H at 37°C.
Secondly we used a bacterial two-hybrid approach based on the BACTH kit (Euromedex, Souffelweyersheim, France). In this method, the T25 and T18 domains were separately fused to the N-termini of KaiC and KaiB/KaiB-T respectively. pKT25-kaiC was obtained by amplifying kaiC using the primer T25-KaiC-F TCTAGAGGTGAAT ACCAATAAAGCAAAAACG and T25-KaiC-R GAGCT CGGCTCTCTTAACAATT TGTTGGACTC; pUT18-kaiB was obtained by amplifying kaiB using the primers T18-KaiB-F CTGCAGGATGACTCGAACTAAGCTAAAGC and T18-KaiB-R GAGCTCGGAT CCATTTCCAAAGCAGAAA TAAC. To construct pUT18C-kaiB-T, the kaiB sequence was used as a template and amplified using the primers T18-KaiB-F and T18-KaiB-T-R GAGCTCGGTTCCAA GCCTAAGTCATCCTC. As control S. elongatus kaiC and kaiB were cloned in pKT25 and pUT18C respectively. The primers used to amplify kaiC were T25-CyKaiC-XbaI-F CTGCAGGATGAGCCCTCGTAAAACCTAC and T25-CyKaiC-KpnI-R GGTACCCGG CTCTCCGGCCCTTT TTCTTG; and for kaiB T18-CyKaiB-PstI-F GAGCTCGG GAAGTCGTCGGAATCTTGAAG and T18-CyKaiB-SacI-R TCTAGAGATGACTTCC GCTGAGATGACTAG. All constructs were confirmed by sequencing.
For β-galactosidase activities for the BACTH measurements, cells were grown to OD 4. The culture was diluted 1:10 and the OD at 600 nm was measured. An aliquot of 20 μl of the culture was added to 80 μl of permeabilization solution (Solution A: 0.8 mg ml−1 hexadecyltrimethylammonium bromide, 0.4 mg ml−1 sodium deoxycholate, 70 mM NaH2PO4 (pH 7), 30 mM NaH2PO4 20 mM KCl 2 mM MgSO4, and 5.4 β-mercaptoethanol). The permeabilization solution mixture was kept for 30 min at 30°C. Then 600 μl of substrate solution (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mMKCl, 1 mg ml−1 o-nitrophenyl-B-D-galactopyranoside and 2.7 μl ml−1 β-mercaptoethanol) was added to start the reaction. After 30 min to 1 h at 30°C, the reactions were stopped by adding 1 ml of 1 M Na2CO3. Enzyme activity was expressed as the rate of change A420 per h per A600 unit of culture; the enzymatic activity was calculated according to the following formula: Enzymatic activity = 200 × {[(OD420 – OD420 in control tube)/min of incubation] × dilution factor}. The enzymatic activity was given in units per mg considering 1 ml of culture at OD600 = 1 corresponds to 300 μg dry weight bacteria.
Purification of His6-KaiC protein
The kaiC genes of L. pneumophila and S. elongatus were cloned into the expression vector pQE30 using the restriction sites of the primers SalI_KaiC_F TTTGTCGAC TAGTGAATACCAATAAAGCAAAAACG and PstI_KaiC_R TTCTGCAG TTACTCTCTTAACAATTTGTTGG for L. pneumophila and Sal_KaiC_F TTTGTCGACTAATGAC TTCCGCTGAGATGACTAGCC HindIII_KaiC_R TTTA AGC TTCTAGCTCTCCGGCCCTTTTTC for S. elongatus. Both kaiC genes were fused to the His6x tag at the N-terminus. For expression of His-KaiC, BL21 (DE3) cells were transformed with pQE30_kaiC and cultured at 25°C for 50 h without IPTG (Iwasaki et al., 1999). Total protein was extracted and affinity purification of the His6-KaiC protein from cell extracts of each transformant using a nickel-charged resin (Qiagen) was performed. To wash the column buffers with increasing concentration of imidazole were used: W-Buffer (25 mM Tris pH 8, 0.5 M NaCl, 10 mM/30 mM/50 mM/70 mM Imidazole, 5 mM MgCl2). Proteins were eluted with 500 mM Imidazole and separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) (10%), blotted onto polyvinylidene difluoride membranes and incubated with anti-histidine-antibody (AbCam #ab14923). Primary antibodies were detected with anti-rabbit HRP conjugates IgG (DakoCytomation, Dako France S.A.S., Les Ulis Cedex, France ) and immunoblots were revealed with a G : Box system (Syngene Europe office, Cambridge, UK).
KaiB crystallization and structure determination
For structural analysis, L. pneumophila KaiB and KaiC genes were cloned into a modified pET15b vector (Novagen, EMD Millipore Corporation, Billerica, MA, USA) in which the tobacco etch virus protease cleavage site replaced the thrombin cleavage site and a double stop codon was introduced downstream the cloning site (Zhang et al., 2001). Selenomethionine enriched protein samples was obtained by culturing the E. coli BL21(DE3)-RIPL (Agilent Technologies Canada, Inc., Mississauga, Canada) cells containing KaiB or KaiC expressing constructs in M9 SeMET High-Yield growth media (Shanghai Medicilon Inc., Shanghai, China) at 37°C to an optical density (at 600 nm) of approximately 1.2. At this point 0.4 mM IPTG was added to induce KaiC protein expression. After induction, the cells were incubated overnight with shaking at 20°C.
The harvested cells were resuspended in 250 mM NaCl 5% glycerol, 100 mM NH4 sulphate, 50 mM phosphate–citrate buffer pH 7.5, 0.5 mM tris(2-carboxyethyl) phosphine (TCEP), 0.08% n-Dodecyl-β-D-maltopyranoside (DDM) and 5 mM imidazole and lysed by sonication. The lysate was clarified by centrifugation (30 min at 17 000 × g) and the clarified soluble fraction was applied to a metal chelate affinity column charged with Ni2+ pre-equilibrated with 300 mM NaCl, 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) Na pH 7.5, 5% glycerol, 5 mM imidazole. The bound protein was washed with 300 mM NaCl, 50 mM HEPES Na pH 7.5, 5% glycerol, 30 mM imidazole buffer and eluted with 300 mM NaCl, 5% glycerol, 50 mM HEPES pH 7.5, and 250 mM imidazole buffer, Next the polyhistidine tag was cleaved from the fusion protein by treatment with recombinant His-tagged Tobacco Etch Virus (TEV) protease (Tropea et al., 2009) followed by dialyses at 4°C in 50 mM HEPES (pH 7.5), 300 mM NaCl, 5% glycerol and 0.5 mM TCEP. The cleaved protein was then resolved from the cleaved His-tag and the His-tagged protease by flowing the mixture through a second Ni2+-chelated affinity column.
Before submission to crystallization trial protein was dialysed in 300 mM NaCl, 10 mM HEPES Na pH 7.5 and 0.5 mM TCEP. Particulate matter was removed from protein samples by passing it through a 0.2 μm Ultrafree-MC centrifugal filter (EMD Millipore Corporation). No crystallization was observed for KaiC protein samples. The KaiB protein was crystallized by hanging-drop vapour diffusion by mixing 2 μL of the protein solution (20 mg ml−1) with 2 μL of a precipitant solution, 0.2 M Na Acetate, 0.1 M Bis-Tris pH 6.0, 35% w/v PEG 8 K and 1 mM Zn acetate. Prior to data collection, crystals were transferred into paratone-N and cryo-cooled in a steam of liquid nitrogen.
The crystal structure of L. pneumophila KaiB was determined by the single anomalous dispersion method using selenomethionine enriched protein crystals. Initial phases were calculated using Solve (Terwilliger and Berendzen, 1999) from Phenix software suite (Adams et al., 2010) and improved by solvent flattening and histogram matching using Resolve (Terwilliger and Berendzen, 1999) prior to model building. The KaiB native crystal diffracted to a 1.95 Å resolution and its structure was solved by molecular replacement using Phaser from the CCP4 suite. The obtained structure model was manually refined using Coot (Emsley and Cowtan, 2004) and Refine from the Phenix suite. The KaiB final model contains 181 residues per asymmetric unit and 150 water molecules; 98.8% and 1.2% residues belonged to the most and additionally favoured regions of the Ramachandran plot respectively. The complete data reduction and the refinement statistics are summarized in Table S3.
Protein phosphorylation assay
The autokinase assay was performed at 30°C as described previously, with some modifications (Nishiwaki et al., 2000). In brief, 1 μg of KaiC was incubated for 1 h at 30°C in 10 μl of incubation buffer (20 mM Tris pH 8, 150 mM NaCl, 5 mM MgCl2) containing 5 μCi of [γ-32P]-ATP. Reaction mixtures were subjected to SDS-PAGE (10%) followed by autoradiography.
Phylogenetic analysis of the KaiB and KaiC sequences
Homologues amino acid sequences of the KaiC protein were identified by BlastP search against 1408 completely sequenced genomes of bacteria and archea available at the NCBI database, using the protein sequence of KaiC of Synechococcus elongatus. Only hits covering at least 80% of the length of the protein of S. elongatus with a minimum e-value of e−5 were taking into account. We then selected the representative protein of each species of bacteria and archea that was giving the best hit. Finally, a total of 53 sequences containing KaiC were retrieved belonging each to a different species. In each of those the presence of the KaiB protein sequence was searched by BlastP using the S. elongatus KaiB sequence as reference. KaiB was identified in 23 of the 53 organisms. Then the KaiC and KaiB sequence of these 23 organisms were aligned separately and concatenated using the programme Expresso (Taly et al., 2011). For the alignments, the best evolutionary model was selected using the program Prottest (Abascal et al., 2005) with the Akaike information criterion to select the best-fit model for amino acid substitutions. Then phylogenetic trees for KaiB and Kaic, respectively, were constructed using both a distance method (neighbour-joining) and maximum likelihood with 300 bootstrap replicates. Distance methods were run in MEGA4 (Tamura et al., 2007) and the KaiBC tree was obtained by using the program Phyml (Guindon et al., 2005). To construct the KaiB tree, we used the 23 sequences identified, aligned them using Expresso, the best evolutionary model was selected using Prottest and the phylogenetic tree was constructed using a distance and maximum likelihood using 300 bootstrap replicates.
Supplementary Material
Fig. S1. Alignment of KaiB sequences of different prokaryotes. In blue, the residues that are well conserved among different species. In bold letter are marked the acidic residues found in the C-terminal described as important for interaction with KaiC for Cyanobacteria [red box (Iwase et al. 2005)]. Legionella pneumophila KaiB misses the acidic residues, although it shares 97% similarity to the KaiB of Cyanobacteria.
Fig. S2. Sequence of the Legionella pneumophila fusion protein KaiB-T. Red, the C-terminal residues from T. elongatus that were fused to KaiB of L. pneumophila.
Fig. S3. Complementation of ΔkaiC and Δlpp1114 mutants under oxidative stress conditions.
A. The sensitivity to 10 mM paraquat of the ΔkaiC strain is complemented by the complete operon (ΔkaiC/pMMB207_kaioperon) in trans.
B. The sensitivity to 10 mM paraquat of the Δlpp1114 strain is complemented by lpp1114 (Δlpp1114_pMMB207lpp1114) in trans. Blue, wild-type strain; red ΔkaiC strain; orange ΔkaiC/pMMB207_kaioperon; cyan Δlpp1114; green Δlpp1114_pMMB207lpp1114. Each point represents the mean ± SD of at least two independent experiments.
Fig. S4. Comparison of the survival kinetics of wt Legionella pneumophila and letA, rpoS, stuC and stuA/C mutan strains. Legionella pneumophila ΔrpoS and ΔletA mutants are sensitive to oxidative stress exerted 10 mM paraquat. In agreement with our findings RpoS seems to be regulating the kai operon genes. The ΔrpoS mutant (red curve) has a strong sensitivity to the effect of paraquat in comparison to the wt strain. Interestingly, also the ΔletA mutant shows strong sensitivity to the effects of paraquat. StuC does not show any higher sensitivity to paraquat than the wt, but the ΔstuA/C double mutant is slightly more sensitive as compared with the wt.
Fig. S5. Phylogenetic tree of the aminoacid sequences of KaiB. The tree was obtained by likelihood method. Bootstrap values > 50 are shown in the corresponding nodes.
Fig. S6. Alignment of the KaiC sequence of Legionella and Cyanobacteria. The alignment shows that the Walker motifs in the KaiCI and KaiCII (CI and CII respectively) domains are well conserved in Legionella. The two main phosphorylation residues in cyanobacterial KaiC are serine/threonine (S431 and T432 in Synechococcus sp.) and there is a third phosphorylation site (T426 in Synechococcus). All of them are well conserved in all species (red boxes). Interestingly the Synechocystis KaiC2 shows serine/serine phosphorylation residues like Legionella.
Table. S1. Expression of the kai operon genes in an ΔrpoS mutant.
Table. S1. Kai operon genes are down-regulated in the ΔrpoS mutant.
Table. S2. Primers used for quantitative real-time polymerase chain reaction.
Table. S3. Primers used for quantitative real-time polymerase chain reaction.
Acknowledgments
We would like to thank Frank Kunst for initiating this project, Philippe Glaser for helpful discussions and Peter Stogios (University of Toronto) for help in structure deposition. This work received financial support from the Institut Pasteur, the Centre National de la Recherche (CNRS), the Institut Carnot-Pasteur MI, the Fondation pour la Recherche Médicale (FRM) grant N° DEQ20120323697, the French region Ile-de-France (DIM Malinf), grant n°ANR-10-LABX-62-IBEID to C. Buchrieser and the U.S. Government National Institutes of Health (grants GM074942 and GM094585 to A. Savchenko through the Midwest Center for Structural Genomics). M. Loza Correa is holder of a contract from the Ministère délégué à l’enseignement supérieur et à la recherche” (France) and CONACYT. M. Rolando is holder of a Roux contract financed by the Institut Pasteur.
Footnotes
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2014–2015. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akai M, Onai K, Morishita M, Mino H, Shijuku T, Maruyama H, et al. Aquaporin AqpZ is involved in cell volume regulation and sensitivity to osmotic stress in Synechocystis sp. strain PCC 6803. J Bacteriol. 2012;194:6828–6836. doi: 10.1128/JB.01665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama S, Nohara A, Ito K, Maéda Y. Assembly and disassembly dynamics of the cyanobacterial periodosome. Mol Cell. 2008;29:703–716. doi: 10.1016/j.molcel.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Albert-Weissenberger C, Sahr T, Sismeiro O, Hacker J, Heuner K, Buchrieser C. Control of flagellar gene regulation in Legionella pneumophila and its relation to growth phase. J Bacteriol. 2010;192:446–455. doi: 10.1128/JB.00610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman E, Segal G. The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J Bacteriol. 2008;90:1985–1996. doi: 10.1128/JB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman MA, Swanson MS. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol Microbiol. 2001;40:1201–1214. doi: 10.1046/j.1365-2958.2001.02465.x. [DOI] [PubMed] [Google Scholar]
- Baker LM, Raudonikiene A, Hoffman PS, Poole LB. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J Bacteriol. 2001;183:1961–1973. doi: 10.1128/JB.183.6.1961-1973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay P, Steinman HM. Catalase-peroxidases of Legionella pneumophila: cloning of the katA gene and studies of KatA function. J Bacteriol. 2000;182:6679–6686. doi: 10.1128/jb.182.23.6679-6686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay P, Byrne B, Chan Y, Swanson MS, Steinman HM. Legionella pneumophila catalase-peroxidases are required for proper trafficking and growth in primary macrophages. Infect Immun. 2003;71:4526–4535. doi: 10.1128/IAI.71.8.4526-4535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, et al. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 2006;8:1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- Byrne B, Swanson MS. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. Feo – transport of ferrous iron into bacteria. Biometals. 2006;19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- Catrenich CE, Johnson W. Characterization of selective inhibition of growth of virulent Legionella pneumophila by supplemented Mueller–Hinton medium. Infect Immun. 1989;57:1862–1864. doi: 10.1128/iai.57.6.1862-1864.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- Cazalet C, Gomez-Valero L, Rusniok C, Lomma M, Dervins-Ravault D, Newton HJ, et al. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 2010;6:e1000851. doi: 10.1371/journal.pgen.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RH, Lewis JW, Bogomolni RA. Photocycle of the LOV-STAS protein from the pathogen Listeria monocytogenes. Photochem Photobiol. 2013;89:361–369. doi: 10.1111/php.12004. [DOI] [PubMed] [Google Scholar]
- Chen J, de Felipe KS, Clarke M, Lu H, Anderson OR, Segal G, Shuman HA. Legionella effectors that promote nonlytic release from protozoa. Science. 2004;303:1358–1361. doi: 10.1126/science.1094226. [DOI] [PubMed] [Google Scholar]
- Cianciotto N. Iron acquisition by Legionella pneumophila. Biometals. 2007;20:323–331. doi: 10.1007/s10534-006-9057-4. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, Swanson MS. Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol Microbiol. 2010;76:200–219. doi: 10.1111/j.1365-2958.2010.07094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Yang Q, Wang Q, Kim YI, Wood TL, Osteryoung KW, et al. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140:529–539. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublet P, Vincent C, Grangeasse C, Cozzone AJ, Duclos B. On the binding of ATP to the autophosphorylating protein, Ptk, of the bacterium Acinetobacter johnsonii. FEBS Lett. 1999;445:137–143. doi: 10.1016/s0014-5793(99)00111-8. [DOI] [PubMed] [Google Scholar]
- Dvornyk V, Vinogradova O, Nevo E. Origin and evolution of circadian clock genes in prokaryotes. Proc Natl Acad Sci USA. 2003;100:2495–2500. doi: 10.1073/pnas.0130099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Faucher SP, Mueller CA, Shuman HA. Legionella pneumophila transcriptome during intracellular multiplication in human macrophages. Front Microbiol. 2011;2:60. doi: 10.3389/fmicb.2011.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, Sharrar RG, et al. Legionnaires’ disease: description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- Gomez-Valero L, Rusniok C, Jarraud S, Vacherie B, Rouy Z, Barbe V, et al. Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics. 2011;12:536. doi: 10.1186/1471-2164-12-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González N, Heeb S, Valverde C, Kay E, Reimmann C, Junier T, Haas HD. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics. 2008;9:167. doi: 10.1186/1471-2164-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online – a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutu A, O’Shea EK. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol Cell. 2013;50:288–294. doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Tateda ES, Swanson MS. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol Microbiol. 2002;44:107–118. doi: 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- Heeb S, Valverde C, Gigot-Bonnefoy C, Haas D. Role of the stress sigma factor RpoS in GacA/RsmA-controlled secondary metabolism and resistance to oxidative stress in Pseudomonas fluorescens CHA0. FEMS Microbiol Lett. 2005;243:251–258. doi: 10.1016/j.femsle.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Herrou J, Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol. 2011;9:713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey EK, Cianciotto N. An iron- and fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect Immun. 1997;65:133–143. doi: 10.1128/iai.65.1.133-143.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindre T, Bruggemann H, Buchrieser C, Hechard Y. Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology. 2008;154:30–41. doi: 10.1099/mic.0.2007/008698-0. [DOI] [PubMed] [Google Scholar]
- Hitomi K, Oyama T, Han S, G E, Arvai AS. Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm. J Biol Chem. 2005;280:19127–19135. doi: 10.1074/jbc.M411284200. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server Issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Taniguchi Y, Ishiura M, Kondo T. Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J. 1999;18:1137–1145. doi: 10.1093/emboj/18.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase R, Imada K, Hayashi F, Uzumaki T, Morishita M, Onai K, et al. Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex. J Biol Chem. 2005;280:43141–43149. doi: 10.1074/jbc.M503360200. [DOI] [PubMed] [Google Scholar]
- Johnson CH. Bacterial circadian programs. Cold Spring Harb Symp Quant Biol. 2007;72:395–404. doi: 10.1101/sqb.2007.72.027. [DOI] [PubMed] [Google Scholar]
- Jurk M, Dorn M, Schmieder P. Blue flickers of hope: secondary structure, dynamics, and putative dimerization interface of the blue-light receptor YtvA from Bacillus subtilis. Biochemistry. 2011;50:8163–8171. doi: 10.1021/bi200782j. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A, Schell U, Sahr T, Tiaden A, Harrison C, Buchrieser C, Hilbi H. The Legionella pneumophila orphan sensor kinase LqsT regulates competence and pathogen-host interactions as a component of the LAI-1 circuit. Environ Microbiol. 2013;15:646–662. doi: 10.1111/j.1462-2920.2012.02889.x. [DOI] [PubMed] [Google Scholar]
- Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak NA, Buss M, Lucas CE, Frace M, Govil D, Travis T, et al. Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J Bacteriol. 2010;192:1030–1044. doi: 10.1128/JB.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc JJ, Davidson RJ, Hoffman PS. Compensatory functions of two alkyl hydroperoxide reductases in the oxidative defense system of Legionella pneumophila. J Bacteriol. 2006;188:6235–6244. doi: 10.1128/JB.00635-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc JJ, Brassinga AK, Ewann F, Davidson RJ, Hoffman PS. An ortholog of OxyR in Legionella pneumophila is expressed postexponentially and negatively regulates the alkyl hydroperoxide reductase (ahpC2D) operon. J Bacteriol. 2008;190:3444–3455. doi: 10.1128/JB.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomma M, Dervins-Ravault D, Rolando M, Nora T, Newton HJ, Sansom FM, et al. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 2010;12:1272–1291. doi: 10.1111/j.1462-5822.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- Loza-Correa M, Gomez-Valero L, Buchrieser C. Circadian clock proteins in prokaryotes: hidden rhythms? Front Microbiol. 2010;1:130. doi: 10.3389/fmicb.2010.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, Dowdle WR. Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med. 1977;297:1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu Rev Physiol. 2010;72:625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- Mackey SR, Golden SS. Winding up the cyanobacterial circadian clock. Trends Microbiol. 2007;1:381–388. doi: 10.1016/j.tim.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Mackey SR, Golden SS, Ditty JL. The itty-bitty time machine genetics of the cyanobacterial circadian clock. Adv Genet. 2011;74:13–53. doi: 10.1016/B978-0-12-387690-4.00002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Guo H, Xiong J. Rhythmic gene expression in a purple photosynthetic bacterium, Rhodobacter sphaeroide. FEBS Lett. 2005a;579:808–812. doi: 10.1016/j.febslet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Moliner C, Fournier PE, Raoult D. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol Rev. 2010;34:281–294. doi: 10.1111/j.1574-6976.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Swanson MS. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol. 2004;53:29–40. doi: 10.1111/j.1365-2958.2004.04129.x. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Newton HJ, Ang DK, van Driel IR, Hartland EL. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev. 2010;23:274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc Natl Acad Sci USA. 2000;97:495–499. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, Takao T, Kondo T. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin T, Cui Y, Cen Z, Liang T, Ren H, Yang X, et al. Draft genome sequences of two Legionella dumoffii strains, TEX-KL and NY-23. J Bacteriol. 2012;194:1251–1252. doi: 10.1128/JB.06352-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Byrne M, Mori T, Zou P, Williams DR, McHaourab H, Johnson CH. Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc Natl Acad Sci USA. 2010;107:14805–14810. doi: 10.1073/pnas.1002119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin S, Li Z, Isberg RR. Macrophage-induced genes of Legionella pneumophila: protection from reactive intermediates and solute imbalance during intracellular growth. Infect Immun. 2002;70:3637–3648. doi: 10.1128/IAI.70.7.3637-3648.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey M, Cianciotto N. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect Immun. 2002;70:5659–5669. doi: 10.1128/IAI.70.10.5659-5669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando M, Sanulli S, Rusniok C, Gomez-Valzero L, Bertholet C, Sahr T, et al. Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe. 2013;13:395–405. doi: 10.1016/j.chom.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahr T, Bruggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol. 2009;72:741–762. doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahr T, Rusniok C, Dervins-Ravault D, Sismeiro O, Coppee JY, Buchrieser C. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol. 2012;9:503–519. doi: 10.4161/rna.20270. [DOI] [PubMed] [Google Scholar]
- St John G, Steinman HM. Periplasmic copperzinc superoxide dismutase of Legionella pneumophila: role in stationary-phase survival. J Bacteriol. 1996;178:1578–1584. doi: 10.1128/jb.178.6.1578-1584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- Scully NM, Cooper WJ, Tranvik LJ. Photo-chemical effects on microbial activity in natural waters: the interaction of reactive oxygen species and dissolved organic matter. FEMS Microbiol Ecol. 2003;46:353–357. doi: 10.1016/S0168-6496(03)00198-3. [DOI] [PubMed] [Google Scholar]
- Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Johnson Z, Stonehouse M, Ochsner U, Meyer JM, Vasil ML, Poole K. The pvc gene cluster of Pseudomonas aeruginosa: role in synthesis of the pyoverdine chromophore and regulation by PtxR and PvdS. J Bacteriol. 1999;181:4118–4124. doi: 10.1128/jb.181.13.4118-4124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taly JF, Magis C, Bussotti G, Chang JM, Tommaso PD, Erb I, et al. Using the T-coffee package to build multiple sequence alignments of protein, RNA, DNA sequences and 3D structures. Nat Protoc. 2011;6:1669–1682. doi: 10.1038/nprot.2011.393. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Katayama M, Ito R, Takai N, Kondo T, Oyama T. labA: a novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes Dev. 2007;21:60–70. doi: 10.1101/gad.1488107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terecskei K, Tóth R, Gyula P, Kevei E, Bindics J, Coupland G, et al. The circadian clock-associated small GTPase LIGHT INSENSITIVE PERIOD1 suppresses light-controlled endoreplication and affects tolerance to salt stress in Arabidopsis. Plant Physiol. 2013;161:278–290. doi: 10.1104/pp.112.203356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- Tropea JE, Cherry S, Waugh DS. Expression and purification of soluble His(6)-tagged TEV protease. Methods Mol Biol. 2009;498:297–307. doi: 10.1007/978-1-59745-196-3_19. [DOI] [PubMed] [Google Scholar]
- Wallace B. Basic Population Genetics. New York: Columbia University; 1981. [Google Scholar]
- Wiegard A, Dorrich AK, Deinzer HT, Beck C, Wilde A, Holtzendorff J, Axmann IM. Biochemical analysis of three putative KaiC clock proteins from Synechocystis sp. PCC 6803 suggests their functional divergence. Microbiology. 2013;159:948–958. doi: 10.1099/mic.0.065425-0. [DOI] [PubMed] [Google Scholar]
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Zhang RG, Skarina T, Katz JE, Beasley S, Khachatryan A, Vyas S, et al. Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase. Structure. 2001;9:1095–1106. doi: 10.1016/s0969-2126(01)00675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman T, Gal-Mor O, Segal G. Characterization of a Legionella pneumophila relA insertion mutant and toles of RelA and RpoS in virulence gene expression. J Bacteriol. 2002;184:67–75. doi: 10.1128/JB.184.1.67-75.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Alignment of KaiB sequences of different prokaryotes. In blue, the residues that are well conserved among different species. In bold letter are marked the acidic residues found in the C-terminal described as important for interaction with KaiC for Cyanobacteria [red box (Iwase et al. 2005)]. Legionella pneumophila KaiB misses the acidic residues, although it shares 97% similarity to the KaiB of Cyanobacteria.
Fig. S2. Sequence of the Legionella pneumophila fusion protein KaiB-T. Red, the C-terminal residues from T. elongatus that were fused to KaiB of L. pneumophila.
Fig. S3. Complementation of ΔkaiC and Δlpp1114 mutants under oxidative stress conditions.
A. The sensitivity to 10 mM paraquat of the ΔkaiC strain is complemented by the complete operon (ΔkaiC/pMMB207_kaioperon) in trans.
B. The sensitivity to 10 mM paraquat of the Δlpp1114 strain is complemented by lpp1114 (Δlpp1114_pMMB207lpp1114) in trans. Blue, wild-type strain; red ΔkaiC strain; orange ΔkaiC/pMMB207_kaioperon; cyan Δlpp1114; green Δlpp1114_pMMB207lpp1114. Each point represents the mean ± SD of at least two independent experiments.
Fig. S4. Comparison of the survival kinetics of wt Legionella pneumophila and letA, rpoS, stuC and stuA/C mutan strains. Legionella pneumophila ΔrpoS and ΔletA mutants are sensitive to oxidative stress exerted 10 mM paraquat. In agreement with our findings RpoS seems to be regulating the kai operon genes. The ΔrpoS mutant (red curve) has a strong sensitivity to the effect of paraquat in comparison to the wt strain. Interestingly, also the ΔletA mutant shows strong sensitivity to the effects of paraquat. StuC does not show any higher sensitivity to paraquat than the wt, but the ΔstuA/C double mutant is slightly more sensitive as compared with the wt.
Fig. S5. Phylogenetic tree of the aminoacid sequences of KaiB. The tree was obtained by likelihood method. Bootstrap values > 50 are shown in the corresponding nodes.
Fig. S6. Alignment of the KaiC sequence of Legionella and Cyanobacteria. The alignment shows that the Walker motifs in the KaiCI and KaiCII (CI and CII respectively) domains are well conserved in Legionella. The two main phosphorylation residues in cyanobacterial KaiC are serine/threonine (S431 and T432 in Synechococcus sp.) and there is a third phosphorylation site (T426 in Synechococcus). All of them are well conserved in all species (red boxes). Interestingly the Synechocystis KaiC2 shows serine/serine phosphorylation residues like Legionella.
Table. S1. Expression of the kai operon genes in an ΔrpoS mutant.
Table. S1. Kai operon genes are down-regulated in the ΔrpoS mutant.
Table. S2. Primers used for quantitative real-time polymerase chain reaction.
Table. S3. Primers used for quantitative real-time polymerase chain reaction.


